Abstract
Human alkyladenine DNA glycosylase (AAG) initiates the base excision repair pathway by excising alkylated and deaminated purine lesions. In vitro biochemical experiments demonstrate that AAG uses facilitated diffusion to efficiently search DNA to find rare sites of damage, and suggest that electrostatic interactions are critical to the searching process. However, it remains an open question whether DNA searching limits the rate of DNA repair in vivo. We constructed AAG mutants with altered searching ability and measured their ability to protect yeast from alkylation damage in order to address this question. Each of the conserved arginine and lysine residues that are near the DNA binding interface were mutated and the functional impacts were evaluated using kinetic and thermodynamic analysis. These mutations do not perturb catalysis of N-glycosidic bond cleavage, but they decrease the ability to capture rare lesion sites. Nonspecific and specific DNA binding properties are closely correlated, suggesting that the electrostatic interactions observed in the specific recognition complex are similarly important for DNA searching complexes. The ability of the mutant proteins to complement repair-deficient yeast cells is positively correlated with the ability of the proteins to search DNA in vitro, suggesting that cellular resistance to DNA alkylation is governed by the ability to find and efficiently capture cytotoxic lesions. It appears that chromosomal access is not restricted and toxic sites of alkylation damage are readily accessible to a searching protein.
INTRODUCTION
DNA repair proteins must search the genome to locate sites of damage. Biophysical and biochemical studies have provided insight into the mechanisms by which this search process is achieved on the biological timescale. Collectively, these pathways are known as facilitated diffusion to emphasize that the searching process is more efficient than would be possible by conventional three-dimensional diffusion.1,2 Although the in vitro evidence for facilitated diffusion is compelling, there is no direct evidence that these pathways contribute to biological function in the context of chromatin. On the one hand, nucleosomal structure is predicted to limit accessibility and introduce a rate-limiting remodeling step that is not dependent on the searching protein.3,4 On the other hand, chromatin dynamics could be sufficiently rapid that sites are available to the searching protein. In this case, the search for specific sites would constitute the rate-limiting step and defects in searching would decrease biological fitness. We have addressed whether the searching ability of a DNA repair enzyme is important for function in a eukaryotic cell.
Human alkyladenine DNA glycosylase (AAG; also known as methylpurine DNA glycosylase, MPG) initiates the base excision repair pathway, and it is the only DNA glycosylase in human cells to remove alkylated purine bases, such as 1,N6-ethenoadenine (εA), 3-methyladenine and 7-methylguanine.5–8 In addition to alkylated bases, AAG efficiently recognizes and removes deaminated purines hypoxanthine (Hx), oxanine and xanthine.9–11 Previous studies show that AAG searches for its target lesion sites on DNA via facilitated diffusion.12,13 The crystal structure of AAG in complex with specific DNA reveals a positively charged binding interface and a backbone bending angle of 22° across the central eight base pairs.14,15 It is not known whether AAG uses the same binding interface for nonspecific DNA binding, as specific binding of repair proteins often involves conformational changes in both protein and DNA.16–18
Baker’s yeast (Saccharomyces cerevisiae) is a well-characterized model for studying both eukaryotic chromatin and repair of DNA alkylation. Yeast lack a homolog of AAG, but have an analogous DNA glycosylase called Mag1 that uses an alternative protein fold to initiate repair of alkylated DNA bases. Deletion of Mag1 sensitizes yeast to DNA damaging agents, largely due to the cytotoxicity of 3-methylpurines.19 The mag1 strains can be complemented by human AAG, providing a heterologous system to study the DNA repair function of AAG.15,20 An advantage of this system is that it isolates the repair protein from potential protein-protein interactions. Previous studies examined the effect of chromatin on DNA alkylation and repair,21 and the depletion of the RSC chromatin remodeling complex results in increased sensitivity to DNA alkylating agents.22 Taken together, these observations validate yeast as a model to understand the importance of DNA searching in repair of damaged chromatin.
We systematically dissected the contributions of conserved electrostatic interactions in the DNA binding groove of AAG and found that individual residues did not affect the transition state for N-glycosidic bond hydrolysis (kchem), but contributed as much as 200-fold to the efficient capture of specific sites of DNA damage. The ability to capture sites of damage is reflected in several kinetic parameters, including kcat/KM, fraction processive, and efficiency of excision. We introduced these mutant proteins into yeast cells to address the question of whether the ability to locate and capture sites of damage is rate-limiting for DNA repair in a eukaryotic nucleus. The positive correlation between biological function and substrate capture provides experimental evidence for the importance of DNA searching in vivo and reveals that AAG searches the genome very efficiently.
RESULTS AND DISCUSSION
Choice of Mutants to Investigate the Electrostatic Contributions of DNA Binding Residues
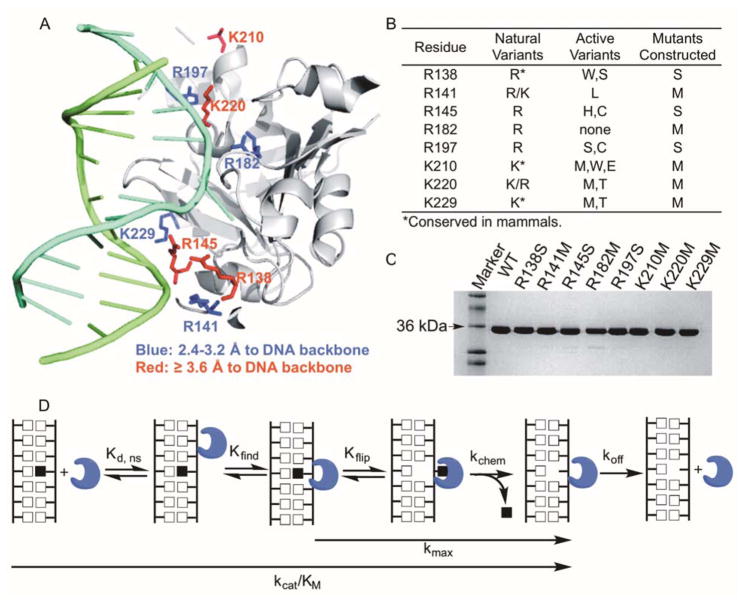
AAG binds to DNA via a positively charged DNA binding cleft containing highly conserved Arg and Lys residues.19,20 These individual residues are highlighted in the structure of AAG bound to εA-DNA (Figure 1A),15 and are distributed across the binding interface, with four residues upstream and four residues downstream of the active site. Half of the residues are close enough to the phosphate backbone to make direct interactions (Figure 1A, blue residues), and the others are more distant with space for intervening water molecules. Whereas most of these side chains are well ordered, the side chain of K210 appears to be mobile with poor electron density.15 These residues are conserved in mammals, and most of them are universally conserved among AAG homologs (Figure 1B). Additional insight into the functional importance of these individual conserved residues was obtained by Guo and colleagues, who introduced random mutations throughout the AAG gene and selected for the ability of the mutant proteins to complement repair-deficient bacteria.23 Active variants were found at all of these positions except for R182, three out of the five Arg residues could be replaced by either Ser or Cys, and all three Lys residues could be replaced by Met (Figure 1B). Therefore, we used site directed mutagenesis to create individual Arg→Ser, Arg→Met, or Lys→Met mutations. Subsequent biochemical characterization was used to evaluate the choice of mutants for study.
Figure 1.
AAG mutants tested in this study. (A) Structure of AAG bound to εA-DNA (PDB ID 1F4R).15 The εA-containing strand of the DNA molecule is colored in cyan and the opposing strand is in green. (B) Known AAG variants and mutations chosen for removal of positively charged residues at the DNA binding interface. Active variants were previously identified in a bacterial complementation assay.23 (C) SDS-PAGE of the purified AAG variants. (D) Minimal reaction scheme for AAG-catalyzed glycosylase activity.
Wild-type and mutant AAG proteins were expressed in E. coli and purified to homogeneity (Figure 1C). Most of the mutants were obtained in similar yields as the wild-type enzyme, but the R182M mutant partitioned mostly to the insoluble fraction and was obtained in much lower yield. Steady-state glycosylase assays confirmed that all AAG variants were active and exhibited burst kinetics for the excision of Hx. This burst amplitude was used to precisely determine the concentration of active enzyme (Supporting Figure S2).12 The R182M mutant protein showed much lower percentage of active enzyme than the other mutant proteins, suggesting that this mutation is destabilizing. This position is invariant in nature and was intolerant to change in the previous study.23 All other AAG mutants exhibited active concentrations similar to the wild-type enzyme.
Catalysis of N-Glycosidic Bond Hydrolysis
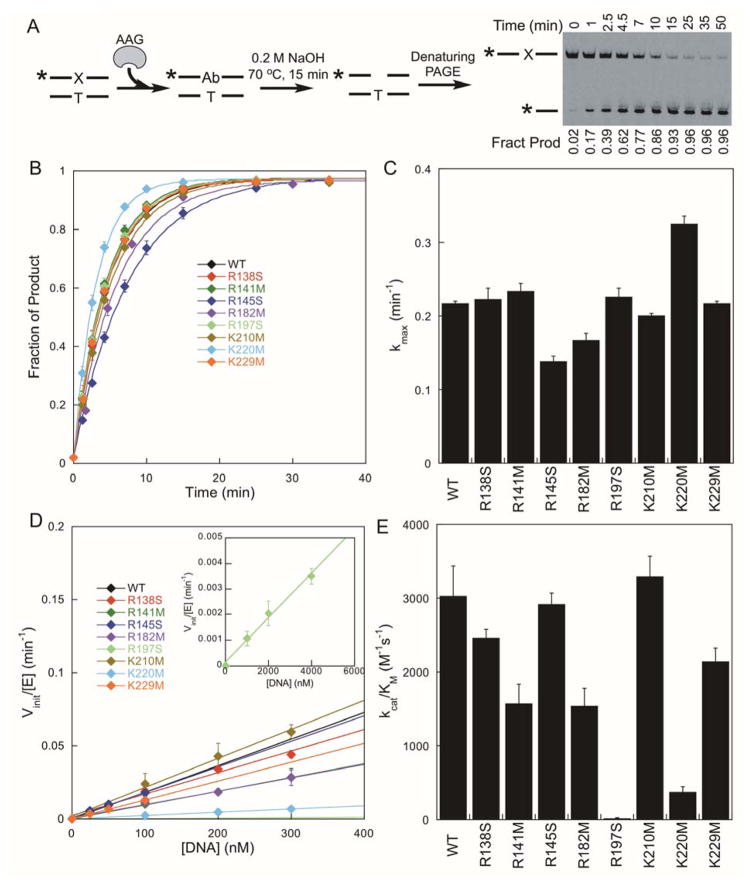
To evaluate the catalytic activity of the AAG mutant proteins, gel-based single-turnover glycosylase assays were performed with enzyme in excess over εA-DNA substrate (Figure 2A). At high concentration of enzyme the single-turnover reaction includes all of the steps from the formation of the initial AAG•DNA complex up to and including N-glycosidic bond cleavage (Figure 1D). For the wild-type enzyme the N-glycosidic bond hydrolysis step is rate limiting for the excision of εA, because the searching step is very fast and the base flipping reaction is highly favorable (kmax = kchem).24,25 All of the mutant enzymes exhibited characteristic exponential reaction kinetics that reached the same reaction endpoint and were independent of the concentration of enzyme (Figure 2B). The wild-type AAG showed a kmax of 0.22 min−1 in this assay (Figure 2C), consistent with the previously reported results.12,26 The rate constants for hydrolysis by the AAG mutants were within 50% of the value measured for the wild-type enzyme (Figure 2C; Supporting Table S1). This strongly suggests that none of these mutations perturb the overall structure of AAG, because the stabilization of the transition state is not affected by the mutation. We conclude that these mutations, that replace positively charged amino acids without altering the rate of N-glycosidic bond cleavage, are appropriate for examining individual electrostatic contributions to DNA searching.
Figure 2.
Excision of εA by AAG variants. (A) Gel-based glycosylase assay and an example gel for the single-turnover excision of εA by wild-type AAG. (B) Representative time courses for the formation of abasic DNA product from εA-DNA under single-turnover conditions, fit with single exponentials. (C) Maximal single-turnover rate constants are the mean ± SD (n = 3). (D) Determination of kcat/KM values from multiple-turnover glycosylase assays. At low concentration of substrate, the dependence on [DNA] is linear and the slope is equal to kcat/KM. The R197S mutant required higher concentrations of substrate (see inset). (E) Values of kcat/KM are the mean ± SD (n≥3).
Determination of kcat/KM Values
We next measured the catalytic specificity (kcat/KM) of the AAG mutant proteins toward the εA-DNA substrate. At a low concentration of salt the binding of AAG to DNA is always productive, and kcat/KM reflects the association rate constant. At a high concentration of salt the binding of AAG to DNA is reversible, and kcat/KM includes all the steps from the binding of substrate up to and including the first irreversible step, which corresponds to N-glycosidic bond cleavage. Thus, it is most informative to measure kcat/KM under high salt conditions.27 As the rates of N-glycosidic bond hydrolysis were very similar for all of the mutant proteins, differences in the kcat/KM value mainly reflect differences in specific DNA binding.
The initial rates for multiple-turnover glycosylase activity were measured as a function of substrate concentration ([substrate] ≪ KM). As expected, the initial rates were linearly dependent upon the concentration of DNA substrate (Figure 2D). The wild-type enzyme exhibits a kcat/KM value of 3000 ± 400 M−1s−1 under these conditions, and the mutants showed a wide range of kcat/KM values (Figure 2E). The R197S and K220M had the most dramatic decreases in kcat/KM at 15 ± 2 M−1s−1 and 360 ± 80 M−1s−1, which correspond to 200-fold and 8-fold decreases compared to the wild-type enzyme. Both R197 and K220 interact with the lesion-containing strand (Figure 1A), raising the possibility that electrostatic interactions with this strand are more important than electrostatic interactions with the opposing strand (R138, R141, R145, and K229). However, R182 also contacts the lesion-containing strand and the R182M mutation caused only a 2-fold decrease in kcat/KM compared to the wild-type enzyme. These observations provide a hierarchy of the functional importance of DNA binding residues and underscore the difficulty in predicting energetic contributions of interactions identified in crystal structures.
We note that the R145S and K210M mutants exhibit kcat/KM values that are identical within error to that of the wild-type enzyme (Figure 2E; Supporting Table S1). This suggests that these residues do not contribute to specific DNA binding, but as they are conserved, they could contribute to DNA searching. Therefore, we measured the searching ability of these and the other mutant proteins.
Searching Ability
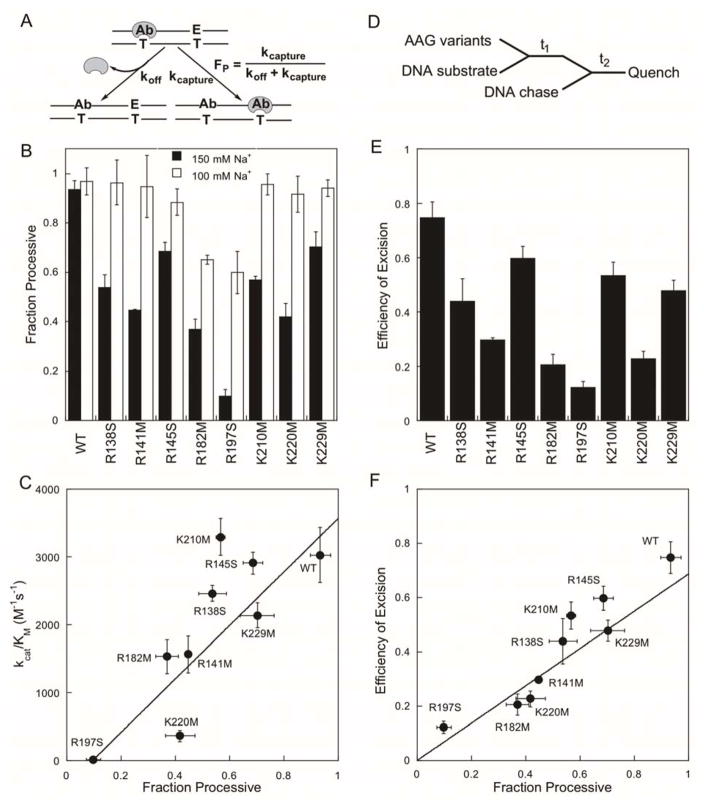
Multiple-turnover processivity assays were performed to quantify the searching ability of each enzyme. This approach utilizes an oligonucleotide containing two εA-sites, and measures the ability of the enzyme to successfully diffuse along the DNA to locate and excise both sites of damage in a single binding encounter (Figure 3A).12 The fraction processive (FP) provides a quantitative measure of the partitioning between the successful capture of a second site and dissociation (FP = kcapture/(koff + kcapture)) and this is an appropriate measurement to describe the process by which a searching protein locates and processes a specific site within a DNA polymer.
Figure 3.
Processivity and efficiency of excision for AAG-catalyzed excision of εA. (A) Multiple-turnover processivity assay to measure partitioning between dissociation (koff) and excision of a second lesion site (kcapture). (B) The fraction processive (mean ± SD, n≥3) was measured at 150 mM Na+ (black bars) or 100 mM Na+ (open bars). (C) Searching efficiency (FP) at 150 mM Na+ and catalytic specificity (kcat/KM; Figure 2E) follow a linear dependence (R2 = 0.82). (D) Pulse-chase assay to measure partitioning between N-glycosidic bond cleavage and dissociation. (E) Efficiency of excision values are the mean ± SD (n=3). (F) Searching efficiency (FP) and efficiency of excision follow a linear dependence (slope = 0.69; R2 = 0.85). Fits were weighted by errors in y-values.
Control experiments with wild-type AAG at 150 mM Na+ yielded an average FP value of close to unity, consistent with previous studies (Figure 3B, black bars; Supporting Figure S3).12,13 Under these conditions, all of the AAG mutant proteins showed decreased processivity compared to the wild-type enzyme. The R197S mutant had the most dramatic decrease in processivity (FP = 0.10; Figure 3B). Processive searching by AAG is mediated by electrostatic interactions and the FP value is strongly dependent on the salt concentration.12 Therefore, we performed the processivity assay at decreased concentration of salt (100 mM) to test if the mutant proteins are still capable of efficient searching (Figure 3B, open bars). Each of the mutants showed increased FP values at the lower salt concentration. Two of the more deleterious mutants, R182M and R197S, remained less processive than the wild-type protein even at this lower salt concentration (Figure 3B; FP ~ 0.6). We conclude that all of the mutant proteins are capable of facilitated diffusion, but the individual mutations have different effects on the processivity of AAG.
Decreased processivity could be due to decreased ability to diffuse along DNA and/or to decreased efficiency of removing the lesion after it is encountered. To distinguish these possibilities, we used pulse-chase assays that measure partitioning of a lesion-bound glycosylase between base excision and dissociation (Figure 3D).13,28 The efficiency of excision is plotted in Figure 3E for each AAG variant (see Supporting Figure S4 for experimental data and controls). The magnitude of the effects on processivity and efficiency of excision are very similar for the mutant proteins and there is a linear relationship between these two parameters (Figure 3F). This observation suggests that diffusion remains rapid for all of the mutant proteins and is not rate-limiting. The main effect of these charge neutralization mutants is decreased efficiency with which the damaged base is recognized and excised once it is encountered.
We considered the hypothesis that some of the DNA binding residues might be more important for nonspecific binding than for specific binding. Although the K210M and R145S mutants exhibit decreased processivity without a decrease in kcat/KM, the defect in processivity is relatively small. A comprehensive plot comparing kcat/KM and FP values for all of the mutant proteins shows significant scatter, making it hard to evaluate whether these mutants truly separate specific and nonspecific DNA binding (Figure 3C; R2 = 0.82). Overall, the roughly linear correlation between processivity and catalytic specificity suggests that a dominant effect of the charge neutralization mutants is decreased specific DNA binding affinity. This can also be inferred from the positive correlation between catalytic specificity and efficiency of excision (Supporting Figure S4).
Kinetic Characterization of Hx Excision
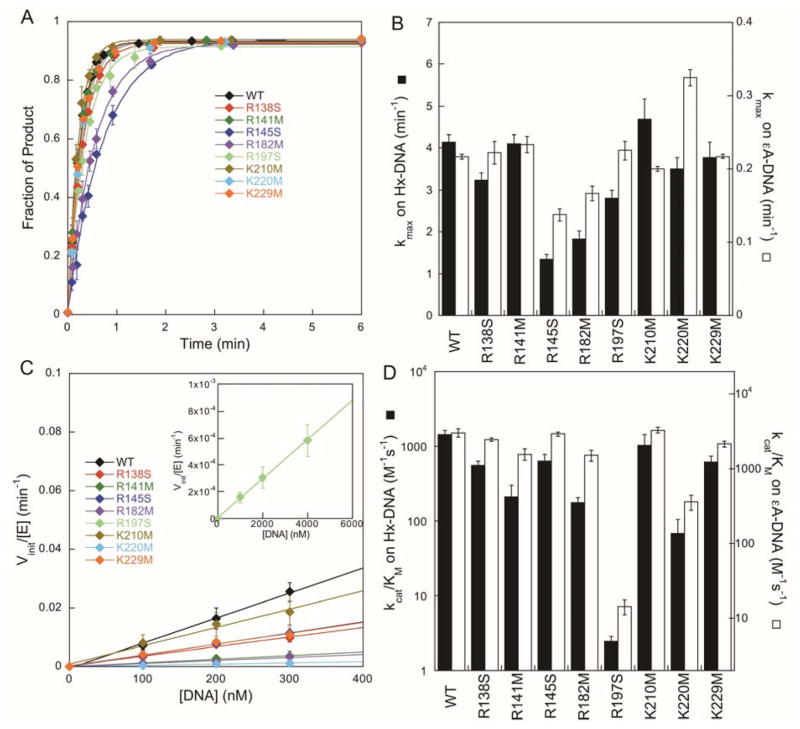
As AAG can recognize a broad range of substrates, it is important to consider the possibility that the mutant proteins could have differential effects on recognition of different substrates. To test the specificity of the mutants, we performed a parallel analysis for the excision of Hx, a deaminated purine, under single- and multiple-turnover conditions. Although Hx binds much more weakly than εA to AAG, it is excised more quickly.26 Therefore, we performed these experiments under slightly different experimental conditions (as noted in the Supporting Methods). The maximal single-turnover rates for excision of Hx, independent of the concentration of AAG, were obtained from single exponential time courses (Figure 4A). The kcat/KM values were determined from the dependence of the initial rates on Hx-DNA concentration (Figure 4C). Despite the different reaction conditions and the very different kinetic parameters for excision of Hx, as compared to excision of εA, all of the mutants showed very similar defects with the two substrates (Figure 4B and D). This indicates that mutation of the conserved DNA binding residues affects DNA binding affinity with minimal change in lesion specificity.
Figure 4.
Excision of Hx by AAG variants. (A) Representative time courses for the single-turnover excision of Hx, fit with single exponentials. (B) Values of kmax for excision of Hx (mean ± SD; n=3) are depicted as solid bars and the open bars show the rate constants for excision of εA from Figure 2C. (C) Determination of kcat/KM values from multiple-turnover glycosylase assays as in Figure 2D. The R197S mutant required higher concentrations of substrate (see inset). (D) Values of kcat/KM are the mean ± SD (n≥3) and are depicted as solid bars. The kcat/KM values for excision of εA are replotted from Figure 2E for comparison (open bars).
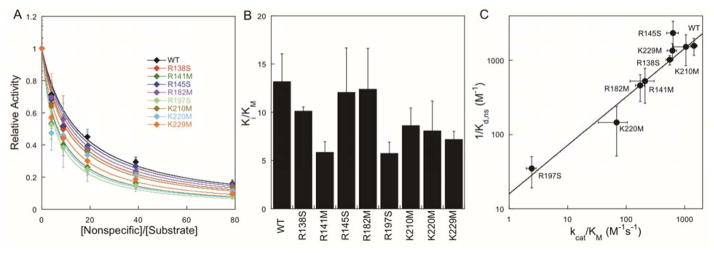
Affinity for Nonspecific DNA
To test if these mutations have a differential effect on nonspecific DNA binding, we measured the relative affinity for nonspecific (undamaged) DNA. Using a competitive inhibition model, the relative Ki/KM values were determined for each of the AAG mutants by varying the ratio of nonspecific inhibitor to substrate DNA and measuring the relative glycosylase activity (Figure 5).29 The apparent Kd for the undamaged DNA was calculated from these data and the independently determined kinetic constants (Supporting Table S1). The nonspecific DNA binding affinity (1/Kd,ns) is strongly correlated with the catalytic specificity (kcat/KM), suggesting that the strengths of the DNA binding interactions are similar for nonspecific and specific binding modes (Figure 5C).
Figure 5.
Binding affinity of AAG variants for nonspecific DNA measured by competitive inhibition. (A) Multiple-turnover competition assays between Hx-DNA and undamaged (25A•T) DNA were fit by the model for competitive inhibition. (B) Relative Ki/KM values from the nonlinear regression. (C) There is a linear dependence between the affinity for nonspecific DNA (1/Kd,ns) which was calculated in Supporting Table S1 and the catalytic specificity (kcat/KM) for excision of Hx (R2 = 0.95; weighted by errors in y-values).
Although the magnitude of the mutational effects could not be predicted from the structure, the general trends are fully consistent with the crystal structure. Electrostatic interactions between the protein and the DNA contribute to DNA binding without significantly changing the rate of N-glycosidic bond hydrolysis. These mutants exhibit defects in DNA searching that vary by more than an order of magnitude, affording the opportunity to investigate to what extent searching ability contributes to DNA repair in the cell.
Function of AAG Mutants in vivo
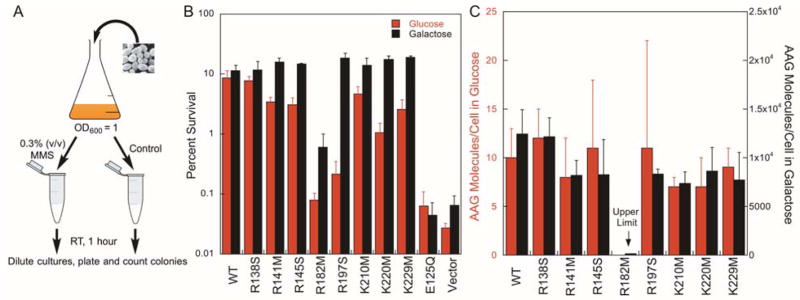
We performed complementation assays in yeast to quantify the in vivo activity of the AAG variants. The sensitivity of Mag1 deletion strains to methylmethane sulfonate (MMS) can be complemented by expression of human AAG from a vector that is under control of the inducible Gal promoter, making this an ideal system to measure the function of AAG in a eukaryotic cell.15,20 Galactose induces a high level of AAG expression, but in the absence of galactose there is still a low level of expression that is sufficient to complement the Mag1 deletion. Yeast expression plasmids were constructed with each of the individual point mutations. As additional controls, we also transfected empty vector and a catalytically inactive mutant, E125Q.15,30 The yeast cells expressing different AAG variants were incubated in the presence or absence of 0.3% (v/v) MMS and the number of viable cells was counted by plating (Figure 6A). This treatment generates approximately 3 alkylated base lesions per kb (Supporting Figure S5).
Figure 6.
Functional assay for the ability of AAG variants to complement Mag1 deficient yeast. (A) Schematic for measuring the MMS-sensitivity of yeast in liquid culture. (B) Yeast containing either empty vector or AAG variant were grown in the presence of either glucose (constitutive) or galactose (inducing) and exposed to MMS to determine the survival frequency (mean ± SD, n≥3). The catalytically inactive mutant (E125Q) and empty vector served as negative controls. (C) The number of AAG molecules per cell was determined using glycosylase assays in cell extracts (Supporting Table S2). Different Y-axes were used for glucose and galactose, because the AAG concentration was increased by ~1000-fold by galactose-induction. Values are the mean ± SD (n=4).
Under the conditions of this assay, less than 0.1% of the Mag1 deletion yeast survived MMS treatment if they did not express an active form of AAG (empty vector and E125Q inactive mutant; Figure 6 & Supporting Table S1). In contrast, ~10% of the yeast survived if they expressed even a low amount of wild-type AAG. This provides a range of two orders of magnitude to quantify the biological function of the AAG variants. We first compared uninduced expression of AAG, because this is expected to mimic the physiological ratio of enzyme to DNA that might be found in a human cell. The different AAG mutants provided varying degrees of protection to yeast cells (Figure 6B, red bars), and cells expressing the R182M and R197S mutants had more than 10-fold decreased survival compared to those expressing the wild-type enzyme.
Overexpression of the AAG mutants is expected to decrease the average search size required and compensate for diminished searching ability. Therefore, we cultured the cells in galactose media to induce the overexpression of the AAG variants, and again measured the susceptibility to MMS. Under these conditions most of the AAG variants gave the same level of protection as wild-type AAG, except for R182M, which conferred approximately ten-fold less protection compared to other AAG constructs (Figure 6B, black bars). As expected, the galactose condition did not significantly affect MMS sensitivity of yeast, because the strains that harbored empty vector or the inactive E125Q mutant showed similar survival in either glucose or galactose media. Overall, these experiments demonstrate that the DNA binding mutants are functional in the yeast nucleus.
Determination of the Abundance of AAG
Changes in protection could be due to differences in either their activity or their abundance, and thus it is critical to evaluate the expression levels for each of the AAG variants. We performed activity-based assays using the log-phase cultures that were used in the MMS sensitivity assays (Figure 6C; see Supporting Figure S6 and Table S2 for the calculation of the amounts of protein). As an internal control, the endogenous uracil DNA glycosylase activity was also measured to confirm that the quality of the extracts was uniform (Supporting Figure S7). Most of the AAG variants, including the wild-type protein and the majority of the active mutants, have the same level of expression within error (Figure 6C and Supporting Table S2). In contrast, the R182M mutant is present at less than 1% the level of the wild-type enzyme. Due to its extremely low level of expression, the R182M mutant was omitted from the correlation of in vitro and in vivo parameters.
The activity assay in cell extracts provides a convenient method to estimate the absolute concentration of AAG present on a per cell basis, and to compare this to the situation in a human cell. There is a 500-fold difference in genome size between a haploid yeast (12 Mbp) and a diploid human cell (6000 Mbp), and therefore about 500-fold less protein would be required in yeast to maintain the same ratio of protein to DNA. Estimates of AAG abundance in human cells vary from 500 to 10,000 molecules per cell in a variety of human cells.31,32 The average corresponds to 11 molecules per cell in yeast, which is very similar to the ~10 protein molecules that were obtained in yeast under the uninduced (glucose) conditions (Figure 6C & Supporting Table S2). In contrast, induction by galactose results in approximately 1000-fold higher levels of protein. Therefore, the basal expression in glucose is an appropriate surrogate for DNA repair activity in a human cell and defective mutants in the yeast system are expected to have a similar effect in human cells.
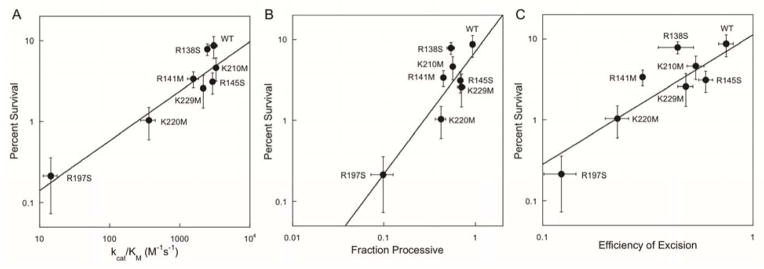
Correlation of In Vivo and In Vitro Parameters for AAG Function
To determine which of the in vitro kinetic parameters of the AAG mutants best predict the functional complementation results in vivo, we plotted the kinetic data relative to the yeast cell survival data. The plot of kmax versus survival rate did not show any clear correlation (Supporting Figure S8). However, roughly linear correlations of kcat/KM, FP, and efficiency of excision were observed (Figure 7). As all three of these kinetic parameters are measurements of productive capture of a site of damage, we conclude that this property is important for cell survival under conditions of exogenous DNA damage.
Figure 7.
Survival of yeast cells is correlated with in vitro parameters describing the efficiency of capturing a site of damage. Survival from exposure to MMS is linearly correlated with catalytic specificity (A; R2 = 0.82), fraction processive (B; R2 = 0.67), and efficiency of excision (C; R2 = 0.67) for AAG-catalyzed excision of εA (fits were weighted by errors in y-values). See the Supporting Information for other correlative plots (Supporting Figure S8).
These observations suggest that AAG efficiently searches chromatin in yeast, and that the overall efficiency of repair depends on this searching ability because mutants with decreased searching ability perform more poorly. These conclusions are very similar to those reached in some earlier studies that examined the importance of DNA searching in bacteria. Mutants of T4 endonuclease V (also known as T4-pdg, pyrimidine dimer glycosylase) with diminished processivity have significantly reduced survival following UV challenge as measured by colony-forming ability of E. coli cells.33,34 Similarly, the ability of EcoRV mutants to search DNA in vitro is positively correlated with the in vivo protection of E. coli from phage.35 It has also been shown that the ability of MutY glycosylase mutants to conduct cellular repair is affected by changes that alter searching and lesion capture.36 Although the physical state of genomic DNA is quite different in prokaryotic and eukaryotic cells, these studies suggest that searching proteins are not limited by accessibility.
It is important to note that our observations regarding AAG were made using exogenous DNA damaging agents and relatively high levels of DNA damage. Chromosomal accessibility may be an important factor when considering other forms of DNA damage, particularly oxidative or hydrolytic damage that may occur even in chromatin. There are several examples using reconstituted nucleosomes in vitro that illustrate that damaged sites on the interior of the nucleosome cannot be accessed without disrupting the nucleosome.3,4 Furthermore, the timescale for repair of UV-induced lesions in yeast appears to be sensitive to chromatin state.37
Insights into Structure/Function of AAG
We explored the functional importance of conserved Arg and Lys residues along the DNA binding cleft of AAG and find that their effects on specific and nonspecific DNA binding are similar. Catalysis is not perturbed by the individual mutations, demonstrating that the network of interactions with DNA is sufficient to optimally position the extrahelical base when any single interaction is disrupted. We cannot precisely define the energetic contribution of each individual residue, but have identified several substitutions that are well-tolerated in vitro and in vivo (R138S, R141M, R145S, K210M, K229M). Two of these substitutions (R145S and K210M) have no significant effect on kcat/KM, but exhibit defects in searching efficiency. This suggests that these contacts are more important for DNA searching than for specific DNA binding, which is consistent with their location far from the base binding pocket. It will be interesting to test whether combinations of these and other mutants might allow a more complete separation of searching from specific site recognition, as this would map out the structural determinants of the poorly characterized searching complexes.
METHODS
Proteins
Mutations in full-length human AAG were generated using site-directed mutagenesis and confirmed by DNA sequencing. Expression and purification was modified slightly from the previously described procedure (see Supporting Information).38 Protein concentrations were corrected for the fraction active (see Supporting Figure S2).
Oligonucleotides
DNA substrates were from Integrated DNA Technologies or the Keck Center at Yale University and purified by denaturing PAGE as previously described.12 Lesion-containing oligonucleotides were labeled with a terminal fluorescein as indicated (Supporting Figure S1). Concentrations were determined from the absorbance at 260 nm using the calculated extinction coefficients.
Glycosylase Assays
Reactions were carried out in 50 mM NaMES pH 6.1, 10% (v/v) glycerol, 0.1 mg/mL BSA, 1 mM DTT, 1 mM EDTA and varying concentrations of NaCl to obtain the desired concentration of Na+. Temperature was 37 °C, except for single-turnover Hx excision which was 25 °C. Reactions were initiated by the addition of enzyme to a final reaction volume of 20–60 μL. Aliquots were withdrawn at various times and quenched with NaOH (0.2 M final concentration). Samples were heated at 70 °C for 15 min, loading buffer consisting of 10 mM EDTA and 98% formamide was added, and the DNA species were resolved on 14% (w/v) denaturing polyacrylamide gels with 8 M urea. Gels were scanned with a Typhoon Trio+ fluorescence imager (GE Healthcare), and fluorescein signal was detected using an excitation wavelength of 532 nm and a 526SP emission filter. Fluorescence signal was quantified with ImageQuant TL and corrected for background signal. The intensity of each DNA band was converted into a fraction of the total DNA by dividing its intensity by the sum of the intensities for all of the DNA species in the sample. Fits were to theoretical equations (KaleidaGraph; Synergy Software). Experimental details for single-turnover and multiple-turnover kinetics, processivity assays, pulse-chase assays, and competitive inhibition assays are described in the Supporting Information.
Yeast Survival Assay
The pYES2-N169S AAG plasmid was generously provided by M. Wyatt (University of South Carolina).39 Wild-type and mutant forms of AAG were constructed by site-directed mutagenesis and the sequences were confirmed by sequencing both strands. The BY4741-Mag1Δ yeast strain (MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 mag1::KanMX) was a gift from D. Engelke (University of Michigan, Ann Arbor).40 Cells grown in glucose or galactose were treated with 0.3% (v/v) MMS for one hour and the percent survival was determined by dividing the number of viable colonies by the number of colonies in controls that were not treated with MMS (Supporting Table S1).
Determination of AAG Expression Level in Yeast Cells
Whole cell extracts were made from the same cultures used for yeast survival and assayed for AAG and uracil glycosylase (UNG) activity. UNG activity did not vary between the different samples, suggesting that the extracts were of uniform quality. Specific activity of each AAG variant was determined using the recombinant proteins. Concentration of AAG in the extracts was converted into the number of molecules per cell (Supporting Table S2).
Supplementary Material
Acknowledgments
We thank members of the O’Brien laboratory for critical comments. This work was supported by a grant from the National Institutes of Health to P.O. (GM108022) and by a Rackham International Fellowship and a Barbour Fellowship from the University of Michigan to Y.Z. The University of Michigan Sequencing Core received support from the National Institutes of Health (P30CA046592).
Footnotes
Notes
The authors declare no competing financial interest.
Supporting information includes detailed experimental methods, eight figures, and two tables (18 pages).
This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1 Models and theory. Biochemistry (Mosc) 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 2.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beard BC, Wilson SH, Smerdon MJ. Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes. Proc Natl Acad Sci U S A. 2003;100:7465–7470. doi: 10.1073/pnas.1330328100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hara R, Mo J, Sancar A. DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol Cell Biol. 2000;20:9173–9181. doi: 10.1128/mcb.20.24.9173-9181.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engelward BP, Weeda G, Wyatt MD, Broekhof JL, de Wit J, Donker I, Allan JM, Gold B, Hoeijmakers JH, Samson LD. Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94:13087–13092. doi: 10.1073/pnas.94.24.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hang B, Singer B, Margison GP, Elder RH. Targeted deletion of alkylpurine-DNA-N-glycosylase in mice eliminates repair of 1,N6-ethenoadenine and hypoxanthine but not of 3,N4-ethenocytosine or 8-oxoguanine. Proc Natl Acad Sci U S A. 1997;94:12869–12874. doi: 10.1073/pnas.94.24.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Connor TR. Purification and characterization of human 3-methyladenine-DNA glycosylase. Nucleic Acids Res. 1993;21:5561–5569. doi: 10.1093/nar/21.24.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singer B, Antoccia A, Basu AK, Dosanjh MK, Fraenkel-Conrat H, Gallagher PE, Kusmierek JT, Qiu ZH, Rydberg B. Both purified human 1,N6-ethenoadenine-binding protein and purified human 3-methyladenine-DNA glycosylase act on 1,N6-ethenoadenine and 3-methyladenine. Proc Natl Acad Sci U S A. 1992;89:9386–9390. doi: 10.1073/pnas.89.20.9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hitchcock TM, Dong L, Connor EE, Meira LB, Samson LD, Wyatt MD, Cao W. Oxanine DNA glycosylase activity from mammalian alkyladenine glycosylase. J Biol Chem. 2004;279:38177–38183. doi: 10.1074/jbc.M405882200. [DOI] [PubMed] [Google Scholar]
- 10.Saparbaev M, Laval J. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc Natl Acad Sci U S A. 1994;91:5873–5877. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dianov G, Lindahl T. Preferential recognition of I. T base-pairs in the initiation of excision-repair by hypoxanthine-DNA glycosylase. Nucleic Acids Res. 1991;19:3829–3833. doi: 10.1093/nar/19.14.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedglin M, O’Brien PJ. Human alkyladenine DNA glycosylase employs a processive search for DNA damage. Biochemistry (Mosc) 2008;47:11434–11445. doi: 10.1021/bi801046y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hedglin M, O’Brien PJ. Hopping enables a DNA repair glycosylase to search both strands and bypass a bound protein. ACS Chem Biol. 2010;5:427–436. doi: 10.1021/cb1000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau AY, Scharer OD, Samson L, Verdine GL, Ellenberger T. Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell. 1998;95:249–258. doi: 10.1016/s0092-8674(00)81755-9. [DOI] [PubMed] [Google Scholar]
- 15.Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc Natl Acad Sci U S A. 2000;97:13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalodimos CG, Biris N, Bonvin AM, Levandoski MM, Guennuegues M, Boelens R, Kaptein R. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 17.Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 18.Warshel A. Electrostatic origin of the catalytic power of enzymes and the role of preorganized active sites. J Biol Chem. 1998;273:27035–27038. doi: 10.1074/jbc.273.42.27035. [DOI] [PubMed] [Google Scholar]
- 19.Veatch W, Okada S. Radiation-induced breaks of DNA in cultured mammalian cells. Biophys J. 1969;9:330–346. doi: 10.1016/S0006-3495(69)86390-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glassner BJ, Rasmussen LJ, Najarian MT, Posnick LM, Samson LD. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc Natl Acad Sci U S A. 1998;95:9997–10002. doi: 10.1073/pnas.95.17.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Czaja W, Bespalov VA, Hinz JM, Smerdon MJ. Proficient repair in chromatin remodeling defective ino80 mutants of Saccharomyces cerevisiae highlights replication defects as the main contributor to DNA damage sensitivity. DNA repair. 2010;9:976–984. doi: 10.1016/j.dnarep.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bespalov VA, Conconi A, Zhang X, Fahy D, Smerdon MJ. Improved method for measuring the ensemble average of strand breaks in genomic DNA. Environ Mol Mutagen. 2001;38:166–174. doi: 10.1002/em.1068. [DOI] [PubMed] [Google Scholar]
- 23.Guo HH, Choe J, Loeb LA. Protein tolerance to random amino acid change. Proc Natl Acad Sci U S A. 2004;101:9205–9210. doi: 10.1073/pnas.0403255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolfe AE, O’Brien PJ. Kinetic mechanism for the flipping and excision of 1,N(6)-ethenoadenine by human alkyladenine DNA glycosylase. Biochemistry (Mosc) 2009;48:11357–11369. doi: 10.1021/bi9015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendershot JM, O’Brien PJ. Critical role of DNA intercalation in enzyme-catalyzed nucleotide flipping. Nucleic Acids Res. 2014;42:12681–12690. doi: 10.1093/nar/gku919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Brien PJ, Ellenberger T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J Biol Chem. 2004;279:9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
- 27.Baldwin MR, O’Brien PJ. Defining the functional footprint for recognition and repair of deaminated DNA. Nucleic Acids Res. 2012;40:11638–11647. doi: 10.1093/nar/gks952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hedglin M, Zhang Y, O’Brien PJ. Isolating contributions from intersegmental transfer to DNA searching by alkyladenine DNA glycosylase. J Biol Chem. 2013;288:24550–24559. doi: 10.1074/jbc.M113.477018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baldwin MR, O’Brien PJ. Human AP endonuclease 1 stimulates multiple-turnover base excision by alkyladenine DNA glycosylase. Biochemistry (Mosc) 2009;48:6022–6033. doi: 10.1021/bi900517y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Brien PJ, Ellenberger T. Human alkyladenine DNA glycosylase uses acid-base catalysis for selective excision of damaged purines. Biochemistry (Mosc) 2003;42:12418–12429. doi: 10.1021/bi035177v. [DOI] [PubMed] [Google Scholar]
- 31.Madhukar NS, Warmoes MO, Locasale JW. Organization of enzyme concentration across the metabolic network in cancer cells. PLoS One. 2015;10:e0117131. doi: 10.1371/journal.pone.0117131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. The quantitative proteome of a human cell line. Mol Syst Biol. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dowd DR, Lloyd RS. Biological consequences of a reduction in the non-target DNA scanning capacity of a DNA repair enzyme. J Mol Biol. 1989;208:701–707. doi: 10.1016/0022-2836(89)90160-5. [DOI] [PubMed] [Google Scholar]
- 34.Dowd DR, Lloyd RS. Biological significance of facilitated diffusion in protein-DNA interactions. Applications to T4 endonuclease V-initiated DNA repair. J Biol Chem. 1990;265:3424–3431. [PubMed] [Google Scholar]
- 35.Jeltsch A, Wenz C, Stahl F, Pingoud A. Linear diffusion of the restriction endonuclease EcoRV on DNA is essential for the in vivo function of the enzyme. EMBO J. 1996;15:5104–5111. [PMC free article] [PubMed] [Google Scholar]
- 36.Brinkmeyer MK, Pope MA, David SS. Catalytic contributions of key residues in the adenine glycosylase MutY revealed by pH-dependent kinetics and cellular repair assays. Chem Biol. 2012;19:276–286. doi: 10.1016/j.chembiol.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bucceri A, Kapitza K, Thoma F. Rapid accessibility of nucleosomal DNA in yeast on a second time scale. EMBO J. 2006;25:3123–3132. doi: 10.1038/sj.emboj.7601196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baldwin MR, O’Brien PJ. Nonspecific DNA binding and coordination of the first two steps of base excision repair. Biochemistry (Mosc) 2010;49:7879–7891. doi: 10.1021/bi100889r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Connor EE, Wilson JJ, Wyatt MD. Effects of substrate specificity on initiating the base excision repair of N-methylpurines by variant human 3-methyladenine DNA glycosylases. Chem Res Toxicol. 2005;18:87–94. doi: 10.1021/tx049822q. [DOI] [PubMed] [Google Scholar]
- 40.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.