ABSTRACT
Uptake and trafficking of metals and their delivery to their respective metalloproteins are important processes. Cells need precise control of each step to avoid exposure to excessive metal concentrations and their harmful consequences. Copper (Cu) is a required micronutrient used as a cofactor in proteins. However, in large amounts, it can induce oxidative damage; hence, Cu homeostasis is indispensable for cell survival. Biogenesis of respiratory heme-Cu oxygen (HCO) reductases includes insertion of Cu into their catalytic subunits to form heme-Cu binuclear centers. Previously, we had shown that CcoA is a major facilitator superfamily (MFS)-type bacterial Cu importer required for biogenesis of cbb3-type cytochrome c oxidase (cbb3-Cox). Here, using Rhodobacter capsulatus, we focused on the import and delivery of Cu to cbb3-Cox. By comparing the CcoA amino acid sequence with its homologues from other bacterial species, we located several well-conserved Met, His, and Tyr residues that might be important for Cu transport. We determined the topology of the transmembrane helices that carry these residues to establish that they are membrane embedded, and substituted for them amino acids that do not ligand metal atoms. Characterization of these mutants for their uptake of radioactive 64Cu and cbb3-Cox activities demonstrated that Met233 and His261 of CcoA are essential and Met237 and Met265 are important, whereas Tyr230 has no role for Cu uptake or cbb3-Cox biogenesis. These findings show for the first time that CcoA-mediated Cu import relies on conserved Met and His residues that could act as metal ligands at the membrane-embedded Cu binding domain of this transporter.
IMPORTANCE
Cu is a micronutrient that is both essential and toxic; hence, its cellular homeostasis is crucial. Respiratory cbb3-type cytochrome c oxidases (cbb3-Cox) are Cu-containing energy-transducing enzymes that are important for many microaerophilic processes, including photosynthesis, respiration, and bacterial pathogenesis. How Cu is incorporated into cbb3-Cox enzymes is not well known. So far, CcoA is the only known major facilitator superfamily (MFS)-type transporter required for Cu import into the bacterial cytoplasm and for cbb3-Cox biogenesis. This study shows that the membrane-embedded, universally conserved Met and His residues of CcoA are essential for its Cu import function and also for its role in cbb3-Cox biogenesis, shedding light on the mechanism of function of this bacterial prototypical Cu importer.
INTRODUCTION
Copper (Cu) is a transition metal that is required for many important cellular processes exploiting its redox properties, such as aerobic respiration (e.g., cytochrome c oxidases [Cox]) (1), oxidation of chemicals (e.g., multicopper oxidases) (2), and elimination of free oxygen radicals (e.g., Cu-Zn superoxide dismutases) (3). However, like many essential metals, excessive concentrations of Cu are toxic to cells. Consequently, cells tightly control Cu homeostasis so that required amounts of Cu are incorporated into the cuproproteins while its excess is eliminated from the cells.
Remarkable progress has been achieved in our understanding of Cu homeostasis and delivery to cuproproteins (4, 5), motivated by pronounced toxicity of Cu and severe diseases associated with defective Cu chaperones and Cu enzymes (6). A major cellular Cu efflux mechanism is provided by the P-type ATPases (P1B subgroup), which are crucial for Cu homeostasis and detoxification in many prokaryotes and eukaryotes (7, 8). In humans, for example, the proteins named ATP7A and ATP7B are vital for health, as their absence or malfunction cause the Menkes and Wilson diseases, which are manifested by growth defects and deterioration of the nervous system (9). Similarly, increased production of these proteins is associated with resistance to chemotherapy (10) and with Alzheimer’s disease (11).
Bacteria contain multiple P1B-type transporters that export Cu from the cytoplasm to confer cellular metal tolerance. In addition, these Cu exporters play different physiological roles depending on their efflux rates. Pseudomonas aeruginosa (12), Rhodobacter capsulatus (13, 14), Rubrivivax gelatinosus (15), and many other species produce at least two highly homologous P1B-type ATPases that export Cu out of the cytoplasm. The high-flux Cu exporter CopA is dedicated for detoxification, whereas the low-flux Cu exporter CcoI (homologue of P. aeruginosas CopA2 and R. gelatinosus CtpA) is required for biogenesis of HCO reductases. Specifically, CcoI and its homologues are proposed to supply Cu to the heme-Cu binuclear centers of the cbb3-type cytochrome c oxidases (cbb3-Cox) (13–15).
Molecular mechanisms underlying Cu transport across the cytoplasmic membrane are not well understood due to the different types of transporters that mediate Cu import and export in various organisms. The three-dimensional (3D) structure of the P1B-type transporter of Legionella pneumophila (LpCopA) revealed the precise locations of its characteristic motifs, such as the ATP binding, CPC phosphorylation and phosphatase domains (16), and also indicated a possible Cu entry and transport path for this family of transporters. LpCopA has two Cu+ binding domains: the N-terminally located metal-binding domain (MBD) containing a CXXC motif, and the transmembrane (TM) MBD with a conserved CPC motif. Cu+ is conveyed from the cytosol exposed MBD, or from a soluble cytoplasmic Cu chaperone (e.g., CopZ) (5, 17, 18), to the “entry site” of a P1B-type ATPase via a docking platform, and then to the high-affinity site, which includes the CPC motif. Cu is released to the extracytoplasmic side of the membrane via a mechanism involving conformational changes driven by ATP hydrolysis (16).
Another Cu-exporting system, called the CusCFBA complex, spans the inner and outer membranes of some bacteria. This complex forms a different kind of Cu transporter that eliminates Cu from both the cytoplasm and the periplasm (19). CusA is located in the inner membrane as the central component of CusCFBA and contains at its TM MBD three pairs of Met residues as well as other periplasm-exposed Met residues involved in Cu+ transport (20, 21). CusB is a membrane fusion protein bridging the inner and outer membranes and interacts with CusA (22). Like CusA, CusB also binds Cu+ via three conserved Met residues in a similar coordination (23). CusBA might form a Met-rich wire where the Met residues of CusB help to transfer Cu+ into a periplasmic cleft of CusA (22, 24). CusC is an outer membrane porin, whereas CusF is a periplasmic chaperone that uses trigonal coordination via Met, Met, and His residues to deliver Cu to CusCBA complex (24).
In eukaryotes, the Ctr (Cu transporter) proteins are major cytoplasmic Cu importers located in the plasma membrane (25, 26). They convey extracellular Cu to cytoplasmic chaperones like Atx1/Atox1 (for P-type ATPase) and Ccs1 (for cytoplasmic Zn-Cu superoxide dismutase) or to mitochondrial Cox17 (for Cox) (27, 28). Ctr proteins and their physiological partners have been well studied due to their importance for human diseases and related therapies (29–31). Differently from CopA, Ctr is a homotrimer that forms a cone-shaped pore at its center, and each monomer has three TM helices (30, 32). Although the numbers of extracellular N-terminal Met motifs of Ctr proteins vary among members of the Ctr family, they were shown to bind Cu and enhance the transport efficiency (29, 33). Indispensable residues for the function of Ctr1-mediated Cu transport include the last methionine of the Met motifs, located approximately 20 amino acids before the first TM helix, as well as the Met-X-X-X-Met motif in the second TM helix. The TM MBD within the Ctr pore is composed of three Met residues (one per monomer), forming a trigonal planar coordination similar to that seen in CusCBA complexes (23). It is still uncertain how Cu is transported through the pore and which residues are involved in this process. Additional Tyr, Cys, and His residues also seem important for Cu import, possibly coordinating conformational changes that occur during transport (29). In yeasts, apart from the Ctr-like proteins, Saccharomyces cerevisiae Pic2 imports Cu into the mitochondrial matrix, whereas Schizosaccharomyces pombe Mfc1 is required for Cu accumulation into the yeast forespore during meiosis (34, 35).
So far, homologues of Ctr proteins have not been found in bacteria. Thus, how Cu enters the bacterial cytoplasm remained hitherto unknown (36). Some studies suggested that specific P1B-type ATPases might be involved in Cu uptake in Enterococcus hirae (CopB) and cyanobacteria (CtaA), respectively (37, 38). However, all known bacterial P1B-type ATPases export metals from the cytoplasm (39, 40); hence, whether they play a role in Cu import is unclear. Currently, the only well-documented bacterial Cu importer is R. capsulatus CcoA, which is a major facilitator superfamily (MFS)-type transporter (41, 42). In this species, a CcoA-deficient strain is defective in cbb3-Cox biogenesis, and compared with a wild-type strain, contains smaller amounts of cellular Cu (like a mutant lacking cbb3-Cox) (41). The inability of a mutant lacking CcoA to take up radioactive 64Cu (14) and the heterologous complementation of a Ctr-less S. pompe mutant with R. capsulatus CcoA (43) showed unequivocally that CcoA is a cytoplasmic Cu importer required for cbb3-Cox production in R. capsulatus (41).
The large group of MFS-type transporters convey across the cytoplasmic membranes a wide spectrum of substrates (http://www.tcdb.org), including sugars (44, 45), drugs (46), and siderophores (47, 48), and among them, CcoA is the first MFS member shown to import Cu (14, 41). Its predicted topology exhibits all characteristics of the MFS transporters, and in addition, it contains multiple Met motifs that could bind Cu. In this work, the amino acid sequence of R. capsulatus CcoA was aligned with its homologues from other bacterial species to locate its conserved metal binding motifs and probe their role in Cu import. Among the universally conserved residues of CcoA homologues, two salient motifs (Met233XXXMet237 and His261XXXMet265 in R. capsulatus) were found, and their Met and His residues were replaced with amino acids that do not coordinate metal atoms. These CcoA mutants were characterized for their ability to import 64Cu and to produce cbb3-Cox to gain an understanding of the mode of Cu recognition and transport by CcoA as a prototypical bacterial Cu importer.
RESULTS
Predicting the Cu binding residues of CcoA.
R. capsulatus CcoA is the bacterial prototype of cytoplasmic Cu importers; thus, it is important to define its MBD and identify its liganding residues. The CcoA homologues are common among the alpha-, beta-, and gamma-proteobacteria but not in the Cytophaga-Flexibacter-Bacteroides group (41). The amino acid sequences of CcoA homologues from representative species (e.g., Paraccocus denitrificans, Agrobacterium fabrum and Bradyrhizobium japonicum from alphaproteobacteria; Cupriavidus metallidurans and Bordetella parapertussis from betaproteobacteria; Pseudomonas stutzeri, Pseudomonas aeruginosa, Pseudomonas putida, Photobacterium profondicum, Shewanella pealeana, and Vibrio species MED 222 from gammaproteobacteria) were aligned with that from R. capsulatus (Fig. 1A) (for a complete alignment, see Fig. S1 in the supplemental material). This alignment shows a number of well-conserved residues, many of which are hallmarks of MFS-type transporters, like the Arg-rich motifs located in the cytoplasmic loops between its TM helices 2 and 3 and helices 8 and 9 (41). Remarkably, though, additional conserved sequences rich in Met and His residues (often implicated in Cu binding) are seen in a region encompassing TM7, TM8, and the loop joining them. In particular, two highly conserved motifs (M233XXXM237 and H261XXXM265 in R. capsulatus) seem specific to CcoA homologues (Fig. 1A). Less conserved MXC or MXXC sequences are also present either before (e.g., R. capsulatus M223XC225M227) or after (e.g., P. aeruginosa Met234XXC237) the fully conserved M233XXXM237 motif (see Fig. S1 in the supplemental material).
FIG 1 .
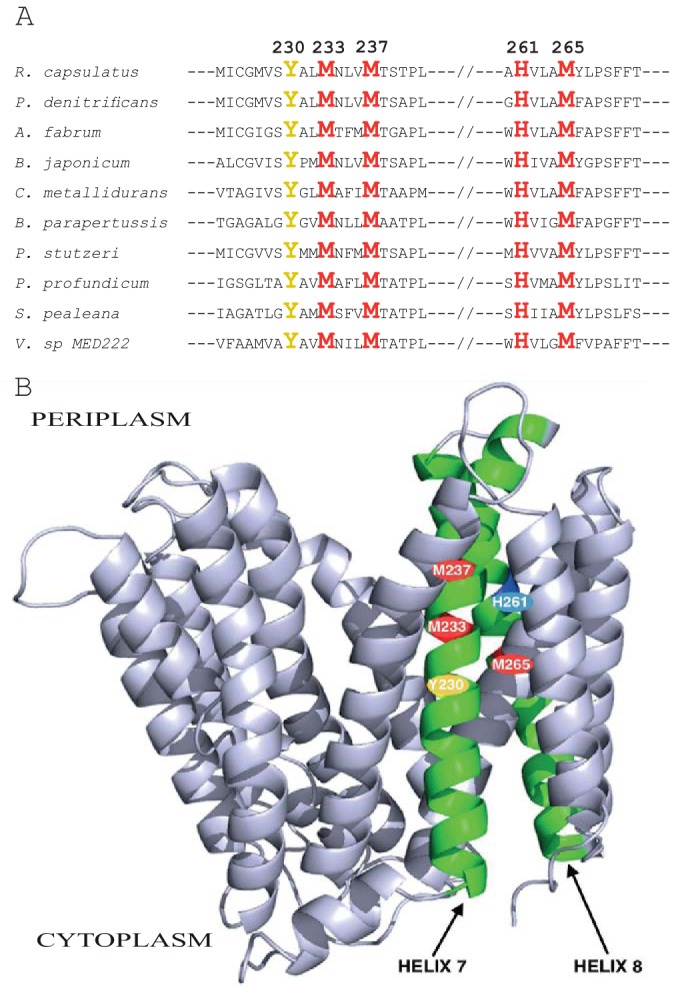
Sequence alignment of R. capsulatus CcoA homologues and its homology-based 3D structure. (A) Partial alignment of the R. capsulatus CcoA amino acid sequence with its homologues from selected proteobacteria. The conserved Met, His, and Tyr residues are highlighted, and their amino acid numbers in R. capsulatus CcoA are indicated (see Table S1 for the corresponding amino acid positions in the other species). (B) Homology model of R. capsulatus CcoA constructed with Swiss-Model (http://swissmodel.expasy.org) using the E. coli YaJR protein (PDB code 3WDO), which is the closest homologue of R. capsulatus CcoA (16% sequence identity), as the template.
Currently, there is no three-dimensional (3D) structure for CcoA or its homologues, although several MFS-type transporters with different substrates have been crystallized (49). Close examination of a CcoA structural homology model, constructed using the 3D structure (PDB code 3WDO) of its most homologous (16% amino acid sequence identity) MFS member (i.e., Escherichia coli YajR, which has no Met motifs and no known substrate [50]) showed that many of the positions corresponding to the conserved Met residues of CcoA were aligned across the membrane, starting at the centers of TM7 and TM8 and extending toward their cytoplasmic and periplasmic ends, respectively (Fig. 1B). Although the individual residues involved in substrate coordination and transport, and their positions within the proteins, are variable among the MFS transporters (49), the conserved Met and His residues within the TM7 and TM8 helices of CcoA pointed out a possible TM MBD for Cu. In addition, the predicted kinked TM8 of CcoA (Fig. 1B) with its conserved Pro268 (see Fig. S1 in the supplemental material) is reminiscent of the kinked TM7 of the MFS member XylE, which is involved in coordinating its substrate d-xylose (45).
Membrane-embedded location of the conserved Met and His residues of CcoA.
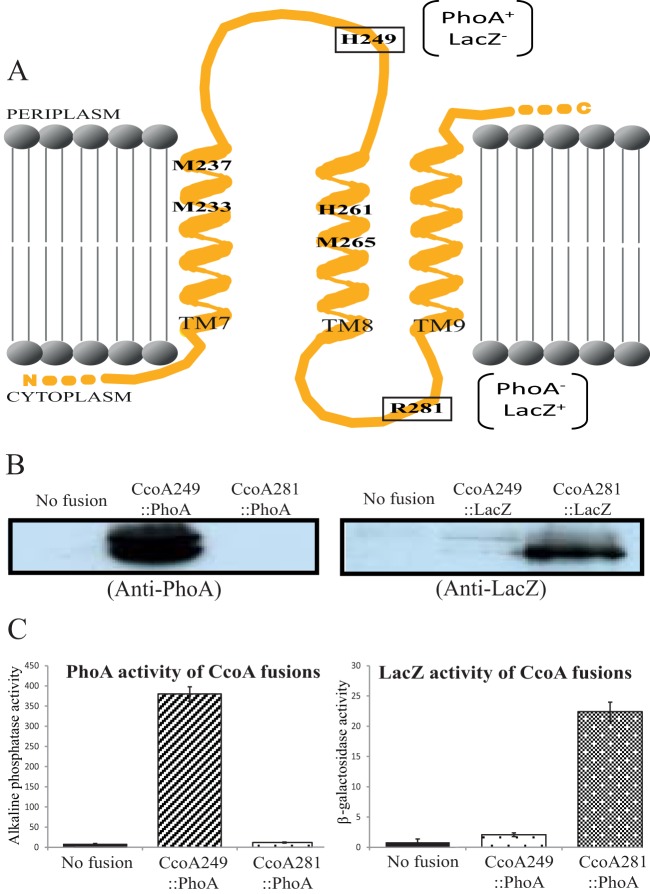
Despite the wide spectrum of substrates transported by the MFS-type transporters, their 3D structures (e.g., LacY [PDB code 1PV7], GlpT [PDB code 1PW4], and FucP [PDB code 3O7Q]) show a common canonical MFS fold with 12 TM segments, which is consistent with the topological model derived from the primary sequence of CcoA (Fig. 1). In order to experimentally establish the predicted membrane-embedded locations of the M233XXXM237 and H261XXXM265 motifs on TM7 and TM8, respectively, we used ccoA translational fusions with E. coli periplasmic phoA and cytoplasmic lacZ (Fig. 2). These reporter proteins were fused in frame after truncation of CcoA at His249 and Arg281 residues, predicted to be at the loops between TM7 and TM8 and between TM8 and TM9 on the periplasmic and cytoplasmic faces of the membrane, respectively (Fig. 2A). Plasmids carrying these fusions (see Table S1 in the supplemental material) were introduced into R. capsulatus ΔccoA (strain SE8) (and also wild-type MT1131, with similar outcomes), and their ability to produce active alkaline phosphatase or β-galactosidase was determined. A strain carrying the ccoA249::phoA fusion at position 249 (predicted to be periplasmic) formed blue colonies on 5-bromo-4-chloro-3-indolyl phosphate (XP)-supplemented MPYE plates, whereas that carrying the ccoA281::phoA fusion at position 281 (predicted to be cytoplasmic) did not. Chromatophore membranes isolated from these strains were tested for the presence of the fusion proteins using E. coli PhoA polyclonal antibodies, and assayed for their alkaline phosphatase activities. A CcoA-PhoA fusion protein was seen in chromatophore fractions of a strain carrying ccoA249::phoA, whereas no fusion protein was detected under the conditions used (presumably being rapidly degraded) in a strain carrying ccoA281::phoA (Fig. 2B). As expected, chromatophore membranes of control strains lacking the fusion constructs showed no CcoA-PhoA protein. Next, the alkaline phosphatase activities of the fusion strains were determined using p-nitrophenyl phosphate as a substrate (see Materials and Methods). Quantitative data indicated that, in agreement with the colony colors on XP plates, significant PhoA activity was detected in the strain carrying the ccoA249::phoA fusion construct, whereas no such activity was seen in the case of the ccoA281::phoA fusion construct (Fig. 2C). Considering that the PhoA moiety has to be exposed to the periplasm for full activity (51, 52), overall data indicated that the loop containing His249 between TM7 and TM8 is periplasmic.
FIG 2 .
Membrane topology of the TM7, -8, and -9 of R. capsulatus CcoA. (A) The TMRPres2D program predicted topology of the TM7, TM8, and TM9 of CcoA was used to fuse LacZ and PhoA to His249, located between TM7 and TM8, and to Arg281, located between TM8 and TM9. The conserved residues Met233, Met237, His261, and Met265, located in TM7 and TM8, and the PhoA and LacZ phenotypes of the fusions are indicated. (B) Immunoblot analysis of CcoA-PhoA and CcoA-LacZ fusions. Chromatophore membranes from cells carrying no fusion (ΔccoA) or the CcoA-LacZ (CcoA249::LacZ and CcoA281::LacZ) and CcoA-Pho (CcoA249::PhoA and CcoA281::PhoA) protein fusions were probed with polyclonal antibodies against E. coli alkaline phosphatase (anti-PhoA) or β-galactosidase (anti-LacZ), as indicated. The double band seen with the CcoA249::PhoA fusion protein was attributed to proteolytic degradation (38). (C) β-Galactosidase (for the CcoA-LacZ fusions) and alkaline phosphatase (for the CcoA-PhoA fusions) activities of chromatophore membranes of R. capsulatus cells harboring the appropriate fusions.
Complementary analyses were also conducted using the ccoA249::lacZ and ccoA281::lacZ fusions (see Table S1 in the supplemental material). The ability of strains carrying these lacZ fusions to produce an active β-galactosidase was the converse of that of strains carrying the corresponding phoA fusions, as indicated qualitatively by the colony color on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)-supplemented MPYE plates. A strain carrying the ccoA281::lacZ fusion at position 281 formed blue colonies, whereas that carrying ccoA249::lacZ did not. Immunoblot analysis using E. coli LacZ polyclonal antibodies was performed, and β-galactosidase activities were determined using chromatophore membranes (Fig. 2B). While chromatophore membranes of a strain lacking the fusion constructs showed no CcoA-LacZ fusion protein, only those carrying ccoA281::lacZ produced a fusion protein of the expected size. Finally, β-galactosidase activities of chromatophore membranes assayed using o-nitrophenyl-galactose (ONPG) as a substrate in a strain carrying ccoA281::lacZ were significantly higher than those detected in the presence of the ccoA249::lacZ fusion (Fig. 2C). Considering that the LacZ moiety is active only in the cytoplasm (51, 52), the data indicated that the loop between TM8 and TM9, where Arg281 is located, is cytoplasmic. Therefore, the experimentally deduced TM locations of the M233XXXM237 and H261XXXM265 motifs agreed with the homology model of CcoA (Fig. 1B). The data furthermore indicated that CcoA fusions carrying either a cytoplasmic PhoA or a periplasmic LacZ moiety were rapidly degraded.
A genetic system for structure-function studies of R. capsulatus CcoA.
A genetic system was developed to produce and purify large amounts of CcoA, to mutate its desired residues to probe their roles, and to score the associated cbb3-Cox phenotypes. R. capsulatus ccoA was cloned into the arabinose (l-Ara) inducible plasmid pBAD/Myc-His A (Invitrogen, Inc.) with its C-terminus fused to a tandem Myc/His tag, to yield pBK68 (Fig. 3A). Immunoblot analysis using anti-Myc antibodies of the E. coli ΔaraC mutant (LMG194) (see Table S1 in the supplemental material) harboring pBK68 showed that R. capsulatus CcoA was readily produced and membrane localized upon l-Ara induction (Fig. 3B). Next, the plasmid pBK68 was cloned into the broad-host-range plasmid pRK415, yielding pBK69 (Fig. 3A). Upon conjugation into R. capsulatus ΔccoA mutant (41) and induction by 0.5% l-Ara, transconjugants carrying pBK69 produced CcoA and also complemented fully its cbb3-Cox-deficient phenotype in the presence of 0.5% l-Ara. These colonies were positive for α-naphthol + dimethyl-p-phenylene diamine staining (Nadi+), reflecting the presence of cbb3-Cox activity (see Materials and Methods). Immunoblot data showed that both E. coli and R. capsulatus strains grown in the presence of l-Ara produced copious amounts of Myc-tagged CcoA with an apparent molecular mass of 37 kDa (Fig. 3B).
FIG 3 .
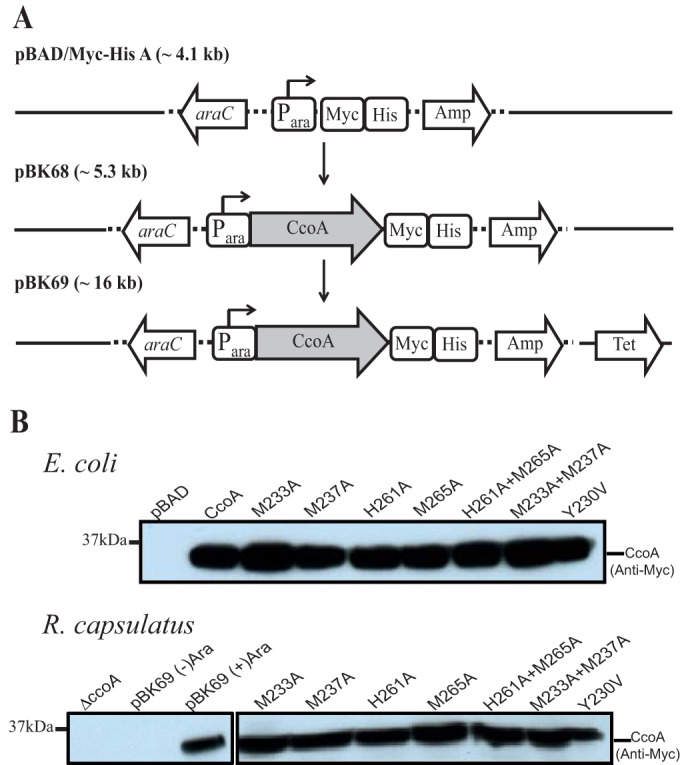
l-Arabinose inducible production of R. capsulatus CcoA in E. coli and R. capsulatus. (A) R. capsulatus ccoA was first cloned into the expression plasmid pBAD/Myc-His (pBK68) to provide an inducible and C-terminally Myc-His epitope-tagged ccoA construct. The plasmid pBK68 was then integrated into the broad-host-range plasmid pRK415 to yield pBK69, which is conjugally transferable into R. capsulatus. (B) Immunoblot analysis of R. capsulatus CcoA expressed in E. coli and in R. capsulatus. Membranes prepared from l-Ara induced E. coli (LMG194) and R. capsulatus ΔccoA (SE8) strains harboring appropriate plasmids expressing wild-type CcoA or mutant derivatives (as indicated), grown with or without 0.5% l-Ara, and probed with anti-Myc monoclonal antibodies.
By using this genetic system, structure-function studies were initiated starting with the conserved Met and His motifs of CcoA. Site-directed single (M233A, M237A, H261A, and M265A) and double (M233A+M237A and H261A+M265A) mutations in CcoA were obtained using pBK68. In addition, considering that in human Ctr1 (hCtr1), the Tyr156 located near the Met ligands plays a structural role in bringing together the residues that are at the entrance to its Cu translocation path (29), the conserved Tyr230 located near the Met and His residues of CcoA was mutated to Val to probe its role in Cu transport. Upon l-Ara induction, mutant CcoA variants were produced in E. coli (LMG194) and also in R. capsulatus ΔccoA (SE8) upon their transfer into the broad-host-range plasmid pRK415. Immunoblotting data obtained by using anti-Myc antibodies to membrane fractions from appropriate cells showed no difference in l-Ara-induced expression between wild-type CcoA and mutant variants with both single and double mutations (Fig. 3B).
Met233 and His261 of CcoA are essential for Cu uptake and active cbb3-Cox production.
Previously, we showed that mutants lacking CcoA were unable to accumulate radioactive 64Cu in a temperature-dependent manner (14). In order to assess the direct effect(s) of mutations on CcoA function, the kinetics of 64Cu uptake was determined using whole cells incubated at 35°C versus 0°C. Upon l-Ara induction, E. coli strain LMG194 producing wild-type R. capsulatus CcoA (Fig. 3B) exhibited significantly higher levels of Cu uptake than the same strain harboring pBAD/Myc-His lacking ccoA (Fig. 4A, inset). This showed that R. capsulatus CcoA was functional for Cu uptake in E. coli. We therefore chose this species for uptake studies to avoid the frequently observed bypass suppressors (the Nadi+ phenotype and Cu sensitivity [CuS]) of CcoA that interfere with Cu homeostasis in R. capsulatus (14). Temperature-dependent 64Cu uptake kinetics using cells expressing the M233A and H261A single CcoA mutants displayed only background (i.e., similar to the control strain lacking ccoA) levels of Cu uptake. Cells expressing the M233A+M237A or H261A+M265A double mutants also behaved like those expressing the M233A or H261A single mutants. In contrast, cells expressing the M237A and M265A single mutants exhibited a Cu uptake that was slightly lower than that of cells expressing either a wild-type CcoA (Fig. 4A and B) or its Y230V variant (not shown). The data thus established that the M233 and H261 residues of CcoA are essential for its ability to import 64Cu into the cells.
FIG 4 .
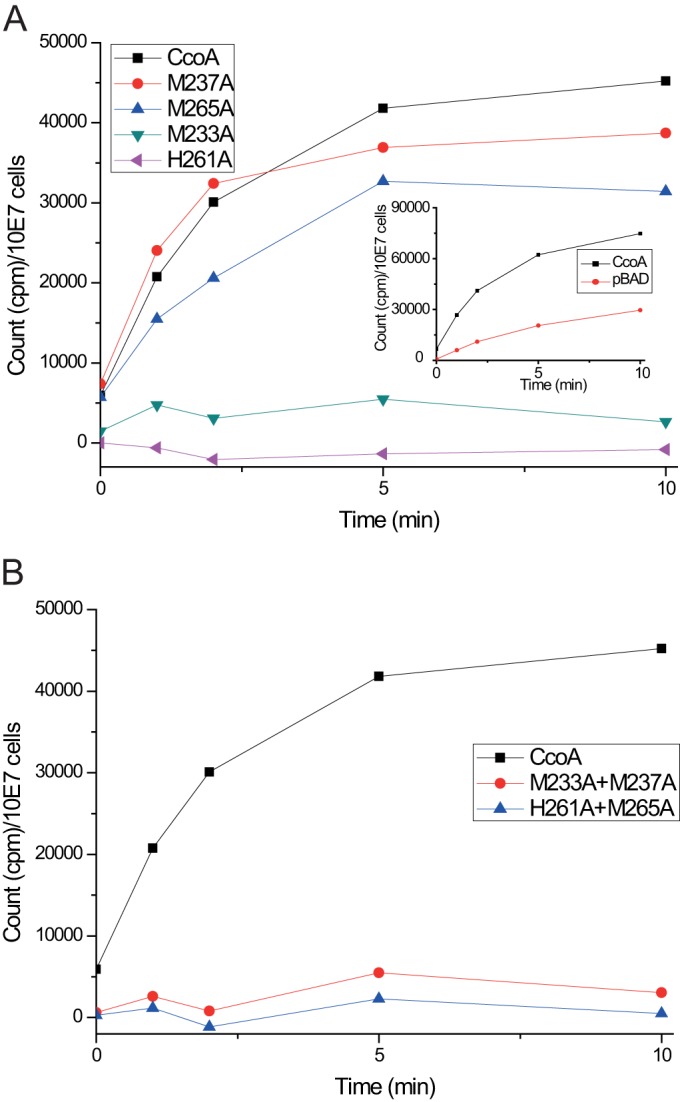
Whole-cell uptake of radioactive 64Cu by native and mutant variants of CcoA. Uptake kinetics was determined using E. coli LMG194 expressing native, single-mutant (A) and double-mutant (B) derivatives of CcoA, as indicated. The strain LMG194 carrying pBAD/Myc-His without ccoA was used to monitor the CcoA-independent background 64Cu uptake in E. coli (A, inset). This CcoA-independent 64Cu uptake was subtracted from the data obtained using native and mutant CcoA variants, and corrected values were plotted as a function of time. Each assay was repeated at least three times using independently grown cultures, and for each time point, the measured values were within ±10% of each other.
Next, the CcoA mutants were tested for their effects on cbb3-Cox production, which relies on CcoA activity (41), because CcoA provides cytoplasmic Cu for heme-Cu center synthesis. The ability to complement the cbb3-Cox deficient phenotype of a ΔccoA mutant (SE8) was qualitatively tested using Nadi staining of colonies grown on MPYE supplemented with 0.5% l-Ara (Table 1). The wild-type cbb3+ Nadi+ (MT1131), Δcbb3 Nadi− (GK32), and ΔccoA Nadi− (SE8) strains were used as positive and negative controls together with the l-Ara-inducible merodiploid ΔccoA/ccoA (pBK69/SE8) strain, which is fully complemented for the cbb3-Cox defect (cbb3+ Nadi+). The CcoA M233A and H261A single mutants, as well as the M233A+M237A and H261A+M265A mutants, were Nadi− (i.e., unable to complement the ΔccoA [SE8] strain). On the other hand, the M265A single mutant was Nadi+/−, suggesting that it partially complemented the absence of CcoA, whereas the M237A and Y230V mutants were Nadi+, indicating that they complemented fully the absence of CcoA and contained active cbb3-Cox (Table 1). As expected, all mutants were phenotypically complemented by the addition of 5 µM CuSO4 to MPYE medium like a ΔccoA mutant (41) via the CcoA-independent low-affinity Cu uptake pathway (data not shown). Moreover, they were all able to grow in the presence of 100 µM CuSO4, indicating that the Nadi+ and Nadi+/− phenotypes of the Y230V and M237A mutants and the M265A mutants, respectively, were not due to mutations in the P1B-type Cu exporter CopA, whose inactivation suppresses the ΔccoA-dependent cbb3-Cox defect at the expense of rendering cells Cu sensitive (14).
TABLE 1 .
Nadi phenotype and KCN-sensitive TMPD oxidase activities of wild-type and CcoA mutant R. capsulatus strains

Nadi phenotype tested on enriched medium as described in Materials and Methods.
One hundred percent for a wild-type strain (MT1131) corresponds to 2.6 µmol of TMPD oxidized/min/mg of protein.
One hundred percent for a ΔccoA strain complemented with a wild-type copy of ccoA (ΔccoA/ccoA) corresponds to 3.9 µmol (i.e., 150% of wild type) of TMPD oxidized/min/mg of protein.
The cbb3-Cox activity of the CcoA mutants was also determined quantitatively using tetramethyl-p-phenylenediamine (TMPD) as an artificial electron donor (Table 1). Wild-type R. capsulatus (MT1131) exhibited ~2.6 μmol of TMPD oxidized/min/mg of total membrane proteins (referred to as 100%), which was abolished by 200 µM KCN, indicating that this activity was specific to cbb3-Cox. As expected, the Δcbb3 (GK32) and ΔccoA (SE8) strains showed basically no activity, whereas the l-Ara-inducible ccoA+/ΔccoA strain exhibited increased (~150%) cbb3-Cox activity compared to a wild-type strain, probably due to overproduction of CcoA in this strain. The M233A and H261A mutants as well as M233A+M237A and H261A+M265A mutants showed ~2 to 9% cbb3-Cox activity compared to their parental (i.e., l-Ara-induced ccoA+/ΔccoA) strain, confirming that the M233A and H261A mutations abolished not only the CcoA import activity but also the production of active cbb3-Cox enzyme. On the other hand, the M265A, M237A, and Y230V mutants showed ~36, 73, and 85% of wild-type cbb3-Cox activity, in line with their Nadi phenotypes (Table 1).
Overall, the data on 64Cu uptake and cbb3-Cox production established that the M233 and H261 residues of CcoA are essential for cytoplasmic Cu import and cbb3-Cox production, whereas M237 and M265 are less critical and Y230 is dispensable.
DISCUSSION
Currently, the R. capsulatus CcoA, which is the prototypical MFS-type Cu importer, is the only well-established bacterial Cu importer (41, 42). Thus, understanding how CcoA mediates Cu entry across the cytoplasmic membrane is important. In the present study, we initiated mutational and functional analyses of R. capsulatus CcoA as a step toward this goal. By comparing the amino acid sequences of CcoA homologues with those of MFS-type transporters, we identified two universally conserved, and functionally important, Met-rich motifs (M233XXXM237 and H261XXXM265) located on the TM7 and TM8 of CcoA. The widespread distribution of CcoA homologues among the bacterial species allowed extensive comparisons to define these motifs as possible landmarks of a metal-importing subgroup of MFS-type transporters (see Fig. S1 in the supplemental material). Subsequent mutagenesis studies established that of the conserved residues of CcoA Met233 and His261 are absolutely required for its role in respect to both Cu import and cbb3-Cox production.
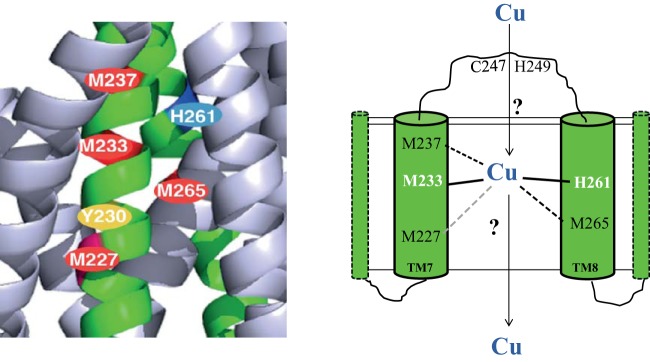
In cuproproteins, Cu atoms are often coordinated with His, Cys, or Met residues. The diversity, spatial arrangement, and solvent accessibility of these ligands define the specific role of Cu atoms acting as structural elements, redox cofactors of specific enzymes, or transporters of substrates. Proteins that employ Cu as a redox cofactor bind it with a strong coordination geometry via a large number of ligands to prevent its loss during its different redox states (53). For instance, the CuB center of the HCO reductases (e.g., cbb3-Cox) is coordinated with three His residues (54). On the other hand, Cu trafficking chaperones and transporters seem to use weak coordination geometry and a small number of ligands to facilitate sequential transfer from the donor to the acceptor proteins. Although the exact roles of Met233 and His261 are not yet completely established, our findings indicate that they may serve as obligate ligands during Cu transport. Met237 and Met265 are also important for CcoA function, but their roles appear to be less critical than those of Met233 and His261. A possibility is that Cu might be coordinated with CcoA via a trigonal planar geometry using two Met (e.g., Met233 and either Met237 or Met265) and one His (His261) residues, similar to E. coli CusF (55) (Fig. 5). The milder CcoA defect seen in the absence of either Met237 or Met265 might reflect the ability of these residues to substitute for each other, provided that no other residue (e.g., the nonconserved Met227 located on TM7) acts as a surrogate in their absence. In support of this hypothesis, ongoing work indicates that cbb3-Cox production is severely affected (~7% of the wild-type level) in the Met237A+Met265A double mutant of CcoA. Another possibility is that Cu might be held via a tetrahedral geometry involving, in addition to Met233 and H261 both Met237 and Met265, similar to Pseudomonas syringae CopC, which can bind either Cu1+ or Cu2+ (56) (Fig. 5). Future studies that are beyond the scope of this work will further address this issue.
FIG 5 .
Proposed transmembrane Cu binding sites of CcoA. A hypothetical model of R. capsulatus CcoA illustrating its putative trigonal (Met233, His261, and Met265) or tetrahedral (with additional Met237 or Met227) Cu-ligating amino acid residues is shown. For the sake of simplicity, only TM7 and TM8 of CcoA are shown. The Cys247 and His249 residues that are located on the periplasmic loop between TM7 and TM8 and might be involved in early steps of Cu recognition are also shown.
Whether the molecular mechanism of CcoA Cu binding and transport is similar to or different from that of other Cu transporters is not yet known. In the case of the P1B-type transporter CopA, the TM MBD has high affinity for Cu, involving six invariant residues located in TM4, TM5, and TM6, contributing to two metal-binding sites, of which one contains the two Cys residues of the CPC motif (16). In the case of hCTR1, the Met residues of the MXXXM motif located at the extracytoplasmic portion of TM2 are thought to provide Cu ligands to facilitate its entry to the central pore of the trimeric hCTR1 (29, 30). The passage of Cu through the pore is thought to be associated with conformational changes, delivering Cu to the putative HCH binding motif located at the C-terminal site, on the opposite end of the pore (30). Finally, the CusA subunit of the CusBCA efflux pump provides a different Cu binding mode, with three distinct pairs of Met residues located in the TM MBD of CusA, potentially forming its metal binding site (21). However, none of these functionally important structural elements is readily recognized in R. capsulatus CcoA and its homologues. Furthermore, unlike the Tyr156 located in the short loop between the TM2 and TM3 hCTR1 that affects Cu import (29), no similar Tyr residue is found in the short loop between the TM7 and TM8 in R. capsulatus CcoA. Although sequence alignments highlighted a well-conserved Tyr residue at TM7 (Y230) near the M233XXXM237 motif of CcoA (Fig. 1), mutagenesis data showed that this residue has no effect on Cu import or cbb3-Cox production, further suggesting that the molecular mechanism of Cu import by the MFS-type Cu importer CcoA is different from that of the Ctr-type importers.
Physiological partners of CcoA?
How Cu accesses the TM MBD of CcoA from the periplasm and how Cu is released to the cytoplasm are intriguing questions. Most P1B-type Cu exporters contain one or more cytoplasmic MBDs with CXXC motifs. These MBDs might be involved in transferring Cu to the TM MBDs and also exert regulatory roles (16, 18). In the case of the Ctr proteins, their extracellular N-terminal MBDs, which are rich in Met and His residues, also provide similar physiological roles in accepting Cu from the donor proteins, facilitating metal entry into their membrane pores (29). In CcoA, additional nonconserved Met motifs with currently undefined roles (e.g., Met30XMet32 in TM1 and Met69XXXMet73, located in the cytoplasmic loop between TM2 and TM3) (see Fig. S1 in the supplemental material) as well as the Cys247 and His249 residues located in the periplasmic loop separating the TM7 and TM8, are also present (Fig. 5). Although currently unknown, these residues could also be involved in conducting Cu to the TM MBD or to the cytoplasm of R. capsulatus CcoA.
Membrane-integral Cu transporters usually work together with soluble Cu chaperones as their immediate metal donors or acceptors. For instance, the cytoplasmic CopZ (5) and the periplasmic CusF (20, 24) chaperones are Cu donors to CopA and CusCAB, respectively. In R. capsulatus, the periplasm exposed membrane-anchored SenC (homologue of Sco1) (57, 58) and the periplasmic soluble Pcc1 (homologue of PCuAC) (P.-I. Trasnea, M. Utz, B. Khalfaoui-Hassani, S. Lagies, F. Daldal, and H.-G. Koch, personal communication) are also involved in cbb3-Cox biogenesis (59). Like CcoA, their absences can be bypassed phenotypically upon exogenous Cu supplementation, suggesting that they are required under low Cu availability. However, whether these or other Cu chaperones interact directly with CcoA and whether CcoA is an Cu importer exclusively dedicated to cbb3-Cox or is a general cytoplasmic Cu supplier are unknown. Ongoing studies are addressing these issues to further elucidate the physiological role of CcoA as a prototypical Cu importer in bacteria.
MATERIALS AND METHODS
Growth conditions, strains, and plasmids used.
The bacterial strains used in this work are described in Table S1 in the supplemental material. Escherichia coli strains were grown at 37°C in Luria Bertani (LB) liquid or solid medium, supplemented with antibiotics (100, 50, and 12.5 µg/ml ampicillin [Amp], kanamycin [Kan] and tetracycline [Tet], respectively), and l-Ara (0.5%) as appropriate. The purple nonsulfur facultative photosynthetic R. capsulatus strains were grown at 35°C under respiratory (aerobic dark) conditions in liquid or solid enriched MPYE medium, as described earlier (60). Growth media were supplemented with antibiotics (10, 2.5, and 10 µg/ml kanamycin, tetracycline, and spectinomycin, respectively) and l-Ara (0.5%) as needed.
Molecular biology techniques.
Standard molecular genetic techniques were performed as described by Sambrook and Russell (61). The plasmids and primers used in this work are listed in Tables S1 and S2 in the supplemental material, respectively. The 1.2-kb R. capsulatus ccoA gene was amplified using CcoA-For and CcoA-Rev primers (see Table S2 in the supplemental material) into which NcoI and HindIII restriction sites, respectively, had been introduced. The PCR fragment digested with these enzymes was inserted into a similarly digested plasmid, pBAD/Myc-His, yielding plasmid pBK68, which carried an l-Ara-inducible form of R. capsulatus ccoA. Next, the plasmid pBK68 was cut with NsiI and ligated to a PstI-cut pRK415 derivative, yielding the plasmid pBK69 carrying the l-Ara inducible form of ccoA, which can be conjugated into R. capsulatus, as needed.
Construction of CcoA mutants.
Plasmid pBK68 was used as a template for site-directed mutagenesis to construct ccoA derivatives carrying the desired point mutations. Appropriate forward and reverse mutagenic primers (see Table S2 in the supplemental material) were used to generate pBK78 (M233A), pBK79 (M237A), pBK80 (H261A), pBK81 (M265A), pBK72 (M233AM237A), pBK71 (H261AM265A), and pBK73 (Y230V). Similar to pBK69, these plasmids were cut with the restriction enzyme NsiI and ligated to a PstI-digested pRK451 derivative, yielding pBK82 (M233A), pBK83 (M237A), pBK84 (H261A), pBK85 (M265A), pBK75 (M233A+M237A), pBK74 (H261A+M265A), and pBK76 (Y230V).
Construction of ccoA::phoA and ccoA::lacZ fusions.
For ccoA::phoA fusions, the CcoA-XhoIFor primer located 400 bp upstream of the ATG of CcoA was used as a forward primer. This forward primer was used in conjunction with the reverse primers BamHI249Rev and BamHI281Rev to amplify the DNA fragments of ccoA encoding its N-termini up to the His249 and Arg281 residues of CcoA, respectively. The PCR products obtained were digested with XhoI and BamHI and cloned into the similarly digested pTFD342 to yield pBK86 (ccoA249::phoA) and pBK88 (ccoA281::phoA). Similarly, the primer CcoA-PstIFor, also located 400 bp upstream of the ATG of ccoA, was used as a forward primer for the ccoA::lacZ fusions, and BamHI249Rev and BamHI281Rev were used as reverse primers to amplify the corresponding DNA fragments of ccoA. The PCR products obtained were digested with PstI and BamHI restriction enzymes and cloned into similarly digested pXCA601 to yield pBK87 (ccoA249::lacZ) and pBK89 (ccoA281::lacZ) fusion constructs.
Chromatophore membrane preparation, SDS-PAGE, and immunodetection.
Intracytoplasmic membrane vesicles (chromatophore membranes) were prepared in 20 mM Tris-HCl (pH 7.0) containing 150 mM NaCl and 1 mM phenylmethylsulfonyl fluoride (PMSF), as described earlier (62). Total protein concentrations were determined using the bicinchoninic acid assay according to the supplier’s recommendations (Sigma Inc.; procedure TPRO-562), and SDS-PAGE (12%) analyses were conducted as described in reference 63. Prior to loading, 40 µg of samples were solubilized in a loading buffer to yield a final concentration of 62.5 mM Tris-HCl (pH 6.8), 2% SDS, 2% β-mercaptoethanol, and 0.01% bromophenol blue by incubation at room temperature for 10 min. The gels were electroblotted onto Immobilon-P polyvinylidene difluoride (PVDF) membranes (Millipore Inc., Billerica, MA), and probed with either anti-His tag (clone HIS.H8 from Millipore Inc.) or anti-c-Myc tag (clone 9E10 from University of Pennsylvania Cell Center) monoclonal antibodies or with anti-E. coli alkaline phosphatase (Bio-Design, Inc.) or anti-E. coli β-galactosidase (Bio-Design, Inc.) polyclonal antibodies. Antigen-antibody complexes were detected using horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG secondary antibodies (GE, Health care Inc.), as appropriate. Signal detection used the Supersignal West Pico chemiluminescence substrate from Thermo Fisher Inc., as recommended by the supplier.
In vivo and in vitro enzyme assays.
The in vivo cbb3-Cox activity of R. capsulatus colonies was visualized qualitatively using Nadi staining solution, made by mixing a 1:1 (vol/vol) ratio of 35 mM α-naphthol and 30 mM N,N,N′,N′-dimethyl-p-phenylene diamine (DMPD) dissolved in ethanol and water, respectively (64). The in vitro cbb3-Cox activity was measured quantitatively using R. capsulatus chromatophore membranes and tetramethyl-p-phenylenediamine (TMPD) by monitoring spectrophotometrically its oxidized form at 562 nm (ε562 = 11.7), at room temperature, in a stirred cuvette. Ten micrograms of R. capsulatus chromatophore membrane proteins was added to 1 ml of assay buffer (25 mM Tris-HCl [pH 7.0], 150 mM NaCl), and the enzymatic reaction was started by the addition of a final concentration of 1 mM TMPD. When tested for cyanide sensitivity, chromatophore membranes were incubated with 200 µM of KCN for 2 min prior to TMPD addition. The cbb3-Cox activity was calculated by subtracting from the TMPD oxidase activity the portion that is KCN insensitive and corresponded to 2.6 μmol of TMPD oxidized/min/mg of total membrane proteins for a wild-type strain.
The alkaline phosphatase or β-galactosidase activities of strains carrying the ccoA::phoA or ccoA::lacZ fusions were visualized qualitatively in vivo using MPYE plates containing 40 µg/ml of the chromogenic indicator 5-bromo-4-chloro-3-indolyl phosphate (XP) or 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), respectively. Alkaline phosphatase activities were measured quantitatively in vitro using 100 µg of chromatophore membrane proteins and p-nitrophenyl phosphate as a substrate, as described in reference 65. The absorbance at 420 nm resulting from the pigments of R. capsulatus chromatophore membranes was recorded before addition of the substrate and subtracted from the final absorbance obtained at 420 nm after incubation with XP. Alkaline phosphatase activity units were calculated using the following formula: [A420(after addition of substrate) − A420(before addition of substrate)] × 1,000/min/mg of total protein. The β-galactosidase activities were assayed at 420 nm using 100 µg of chromatophore membrane proteins and o-nitrophenyl-β-galactoside (ONPG) as a substrate. The absorbance at 420 nm resulting from the pigments of R. capsulatus chromatophore membranes was also recorded before the addition of 2 mM ONPG and subtracted from the final absorbance obtained at 420 nm at the end of incubation. The hydrolysis of ONPG, expressed as moles of ONPG hydrolyzed per minute per milligram of proteins, was calculated using the following formula: ΔOD420 × 106/4,860/min/mg of protein (66).
64Cu uptake assays.
Assays measuring uptake of radioactive 64Cu were performed as described in reference 14. The radioactive 64Cu (specific activity, 1.84 × 104 mCi/µmol) was obtained from the Mallinckrodt Institute of Radiology, Washington University Medical School. E. coli strains carrying appropriate l-Ara-inducible CcoA variants on pBAD/Myc-His derivatives were grown overnight in 10 ml of LB supplemented with 0.5% l-Ara, centrifuged, washed with the assay buffer (50 mM sodium citrate [pH 6.5] and 5% glucose), and resuspended in 1 ml of the same buffer. All cultures were normalized to the same number of cells (7.5 × 108 cells) per 500 µl of assay based on their OD600 values, and cells were preincubated at 35°C or 0°C for 10 min before the assay. The uptake activity was initiated by addition of 107 cpm of 64Cu, determined immediately before use (the half-life of 64Cu isotope is ~12 h). At each time point (0, 1, 2, 5, and 10 min), 50 µl of 1 mM CuCl2 and 50 µl of 50 mM EDTA (pH 6.5) were added to stop the reaction, and the samples were placed on ice. At the end of the assay, cells were pelleted, and the pellets were washed twice with 100 µl of ice-cold 50 mM EDTA solution, resuspended in 1 ml of scintillation liquid, and counted by using a scintillation counter (Coulter-Beckman Inc.) with a wide open window. The uptake activities obtained were corrected for endogenous activities observed in the absence of CcoA (in pBAD/LMG194) (see Table S1 in the supplemental material) and plotted as a function of time to compare the Cu uptake abilities of wild-type CcoA and its mutant derivatives. All strains of both R. capsulatus and E. coli expressed similar amounts of l-Ara-inducible CcoA proteins, as indicated by immunodetection using appropriate anti-tag antibodies, as described above.
SUPPLEMENTAL MATERIAL
Alignment of CcoA homologues from various bacterial species. Residues that are conserved at 80% or higher are indicated in red (identical amino acid) and green (similar amino acids). Download
All strains and plasmids used in this work and related references.
All oligonucleotide primers used in this work.
ACKNOWLEDGMENTS
We thank Alexander Kelly for his help with the homology modeling of CcoA and alignment of its homologues from other bacterial species.
This work was supported primarily by a grant from the National Institutes of Health [NIH GM38237 to F.D.] and partially (for protein purification and characterization) by a grant from the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of Department of Energy [DOE DE-FG02-91ER20052 to F.D.]; HGK was supported by the Deutsche Forschungsgemeinschaft (DFG ITRG 1478) and the German French PhD College on Membranes and Membrane Proteins.
This article is a direct contribution from a Fellow of the American Academy of Microbiology.
Footnotes
Citation Khalfaoui-Hassani B, Verissimo AF, Koch H, Daldal F. 2016. Uncovering the transmembrane metal binding site of the novel bacterial major facilitator superfamily-type copper importer CcoA. mBio 7(1):e01981-15. doi:10.1128/mBio.01981-15.
REFERENCES
- 1.Yoshikawa S, Shimada A, Shinzawa-Itoh K. 2015. Respiratory conservation of energy with dioxygen: cytochrome c oxidase. Met Ions Life Sci 15:89–130. [DOI] [PubMed] [Google Scholar]
- 2.Martins LO, Durão P, Brissos V, Lindley PF. 2015. Laccases of prokaryotic origin: enzymes at the interface of protein science and protein technology. Cell Mol Life Sci 72:911–922. doi: 10.1007/s00018-014-1822-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desideri A, Falconi M. 2003. Prokaryotic Cu,Zn superoxide dismutases. Biochem Soc Trans 31:1322–1325. doi: 10.1042/bst0311322. [DOI] [PubMed] [Google Scholar]
- 4.Osman D, Cavet JS. 2008. Copper homeostasis in bacteria. Adv Appl Microbiol 65:217–247. doi: 10.1016/S0065-2164(08)00608-4. [DOI] [PubMed] [Google Scholar]
- 5.Robinson NJ, Winge DR. 2010. Copper metallochaperones. Annu Rev Biochem 79:537–562. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lutsenko S. 2010. Human copper homeostasis: a network of interconnected pathways. Curr Opin Chem Biol 14:211–217. doi: 10.1016/j.cbpa.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Argüello JM, Eren E, González-Guerrero M. 2007. The structure and function of heavy metal transport P1B-ATPases. Biometals 20:233–248. doi: 10.1007/s10534-006-9055-6. [DOI] [PubMed] [Google Scholar]
- 8.Chan H, Babayan V, Blyumin E, Gandhi C, Hak K, Harake D, Kumar K, Lee P, Li TT, Liu HY, Lo TC, Meyer CJ, Stanford S, Zamora KS, Saier MH Jr.. 2010. The p-type ATPase superfamily. J Mol Microbiol Biotechnol 19:5–104. doi: 10.1159/000319588. [DOI] [PubMed] [Google Scholar]
- 9.Cox DW, Moore SD. 2002. Copper transporting P-type ATPases and human disease. J Bioenerg Biomembr 34:333–338. doi: 10.1023/A:1021293818125. [DOI] [PubMed] [Google Scholar]
- 10.Leonhardt K, Gebhardt R, Mössner J, Lutsenko S, Huster D. 2009. Functional interactions of Cu-ATPase ATP7B with cisplatin and the role of ATP7B in the resistance of cells to the drug. J Biol Chem 284:7793–7802. doi: 10.1074/jbc.M805145200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Z, White C, Lee J, Peterson TS, Bush AI, Sun GY, Weisman GA, Petris MJ. 2010. Altered microglial copper homeostasis in a mouse model of Alzheimer’s disease. J Neurochem 114:1630–1638. doi: 10.1111/j.1471-4159.2010.06888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.González-Guerrero M, Raimunda D, Cheng X, Argüello JM. 2010. Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Mol Microbiol 78:1246–1258. doi: 10.1111/j.1365-2958.2010.07402.x. [DOI] [PubMed] [Google Scholar]
- 13.Koch HG, Winterstein C, Saribas AS, Alben JO, Daldal F. 2000. Roles of the ccoGHIS gene products in the biogenesis of the cbb(3)-type cytochrome c oxidase. J Mol Biol 297:49–65. doi: 10.1006/jmbi.2000.3555. [DOI] [PubMed] [Google Scholar]
- 14.Ekici S, Turkarslan S, Pawlik G, Dancis A, Baliga NS, Koch HG, Daldal F. 2014. Intracytoplasmic copper homeostasis controls cytochrome c oxidase production. mBio 5:e01055-13. doi: 10.1128/mBio.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassani BK, Astier C, Nitschke W, Ouchane S. 2010. CtpA, a copper-translocating P-type ATPase involved in the biogenesis of multiple copper-requiring enzymes. J Biol Chem 285:19330–19337. doi: 10.1074/jbc.M110.116020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gourdon P, Liu XY, Skjørringe T, Moller LB, Pedersen BP, Nissen P. 2011. Crystal structure of a copper-transporting PIB-type ATPase. Nature 475:59–64. doi: 10.1038/nature10191. [DOI] [PubMed] [Google Scholar]
- 17.Banci L, Bertini I, Ciofi-Baffoni S, Del Conte R, Gonnelli L. 2003. Understanding copper trafficking in bacteria: interaction between the copper transport protein CopZ and the N-terminal domain of the copper ATPase CopA from Bacillus subtilis. Biochemistry 42:1939–1949. doi: 10.1021/bi027096p. [DOI] [PubMed] [Google Scholar]
- 18.González-Guerrero M, Argüello JM. 2008. Mechanism of Cu+-transporting ATPases: soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proc Natl Acad Sci U S A 105:5992–5997. doi: 10.1073/pnas.0711446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mealman TD, Blackburn NJ, McEvoy MM. 2012. Metal export by CusCFBA, the periplasmic Cu(I)/Ag(I) transport system of Escherichia coli. Curr Top Membr 69:163–196. doi: 10.1016/B978-0-12-394390-3.00007-0. [DOI] [PubMed] [Google Scholar]
- 20.Franke S, Grass G, Rensing C, Nies DH. 2003. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bacteriol 185:3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long F, Su CC, Zimmermann MT, Boyken SE, Rajashankar KR, Jernigan RL, Yu EW. 2010. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature 467:484–488. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su CC, Long F, Zimmermann MT, Rajashankar KR, Jernigan RL, Yu EW. 2011. Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature 470:558–562. doi: 10.1038/nature09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bagai I, Liu W, Rensing C, Blackburn NJ, McEvoy MM. 2007. Substrate-linked conformational change in the periplasmic component of a Cu(I)/Ag(I) efflux system. J Biol Chem 282:35695–35702. doi: 10.1074/jbc.M703937200. [DOI] [PubMed] [Google Scholar]
- 24.Mealman TD, Bagai I, Singh P, Goodlett DR, Rensing C, Zhou H, Wysocki VH, McEvoy MM. 2011. Interactions between CusF and CusB identified by NMR spectroscopy and chemical cross-linking coupled to mass spectrometry. Biochemistry 50:2559–2566. doi: 10.1021/bi102012j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dancis A, Yuan DS, Haile D, Askwith C, Eide D, Moehle C, Kaplan J, Klausner RD. 1994. Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport. Cell 76:393–402. doi: 10.1016/0092-8674(94)90345-X. [DOI] [PubMed] [Google Scholar]
- 26.Ohrvik H, Thiele DJ. 2014. How copper traverses cellular membranes through the mammalian copper transporter 1, Ctr1. Ann N Y Acad Sci 1314:32–41. doi: 10.1111/nyas.12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim BE, Nevitt T, Thiele DJ. 2008. Mechanisms for copper acquisition, distribution and regulation. Nat Chem Biol 4:176–185. doi: 10.1038/nchembio.72. [DOI] [PubMed] [Google Scholar]
- 28.Cobine PA, Pierrel F, Winge DR. 2006. Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim Biophys Acta 1763:759–772. doi: 10.1016/j.bbamcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Eisses JF, Kaplan JH. 2005. The mechanism of copper uptake mediated by human CTR1: a mutational analysis. J Biol Chem 280:37159–37168. doi: 10.1074/jbc.M508822200. [DOI] [PubMed] [Google Scholar]
- 30.De Feo CJ, Aller SG, Siluvai GS, Blackburn NJ, Unger VM. 2009. Three-dimensional structure of the human copper transporter hCTR1. Proc Natl Acad Sci U S A 106:4237–4242. doi: 10.1073/pnas.0810286106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graper ML, Huster D, Kaler SG, Lutsenko S, Schilsky ML, Thiele DJ. 2014. Introduction to human disorders of copper metabolism. Ann NY Acad Sci 1314:v–vi. doi: 10.1111/nyas.12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aller SG, Unger VM. 2006. Projection structure of the human copper transporter CTR1 at 6-A resolution reveals a compact trimer with a novel channel-like architecture. Proc Natl Acad Sci U S A 103:3627–3632. doi: 10.1073/pnas.0509929103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puig S, Lee J, Lau M, Thiele DJ. 2002. Biochemical and genetic analyses of yeast and human high affinity copper transporters suggest a conserved mechanism for copper uptake. J Biol Chem 277:26021–26030. doi: 10.1074/jbc.M202547200. [DOI] [PubMed] [Google Scholar]
- 34.Vest KE, Leary SC, Winge DR, Cobine PA. 2013. Copper import into the mitochondrial matrix in Saccharomyces cerevisiae is mediated by Pic2, a mitochondrial carrier family protein. J Biol Chem 288:23884–23892. doi: 10.1074/jbc.M113.470674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beaudoin J, Ioannoni R, López-Maury L, Bähler J, Ait-Mohand S, Guérin B, Dodani SC, Chang CJ, Labbé S. 2011. Mfc1 is a novel forespore membrane copper transporter in meiotic and sporulating cells. J Biol Chem 286:34356–34372. doi: 10.1074/jbc.M111.280396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol 10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Odermatt A, Suter H, Krapf R, Solioz M. 1993. Primary structure of two P-type ATPases involved in copper homeostasis in Enterococcus hirae. J Biol Chem 268:12775–12779. [PubMed] [Google Scholar]
- 38.Tottey S, Rich PR, Rondet SA, Robinson NJ. 2001. Two Menkes-type ATPases supply copper for photosynthesis in Synechocystis PCC 6803. J Biol Chem 276:19999–20004. doi: 10.1074/jbc.M011243200. [DOI] [PubMed] [Google Scholar]
- 39.Palmgren MG, Nissen P. 2011. P-type ATPases. Annu Rev Biophys 40:243–266. doi: 10.1146/annurev.biophys.093008.131331. [DOI] [PubMed] [Google Scholar]
- 40.Rosenzweig AC, Argüello JM. 2012. Toward a molecular understanding of metal transport by P(1B)-type ATPases. Curr Top Membr 69:113–136. doi: 10.1016/B978-0-12-394390-3.00005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ekici S, Yang H, Koch HG, Daldal F. 2012. Novel transporter required for biogenesis of cbb3-type cytochrome c oxidase in Rhodobacter capsulatus. mBio 3:e00293-11. doi: 10.1128/mBio.00293-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Argüello JM, Raimunda D, Padilla-Benavides T. 2013. Mechanisms of copper homeostasis in bacteria. Front Cell Infect Microbiol 3:73. doi: 10.3389/fcimb.2013.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beaudoin J, Ekici S, Daldal F, Ait-Mohand S, Guérin B, Labbé S. 2013. Copper transport and regulation in Schizosaccharomyces pombe. Biochem Soc Trans 41:1679–1686. doi: 10.1042/BST2013089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. 2003. Structure and mechanism of the lactose permease of Escherichia coli. Science 301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 45.Sun L, Zeng X, Yan C, Sun X, Gong X, Rao Y, Yan N. 2012. Crystal structure of a bacterial homologue of glucose transporters GLUT1-4. Nature 490:361–366. doi: 10.1038/nature11524. [DOI] [PubMed] [Google Scholar]
- 46.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. 2006. Structure of the multidrug transporter EmrD from Escherichia coli. Science 312:741–744. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furrer JL, Sanders DN, Hook-Barnard IG, McIntosh MA. 2002. Export of the siderophore enterobactin in Escherichia coli: involvement of a 43-kDa membrane exporter. Mol Microbiol 44:1225–1234. doi: 10.1046/j.1365-2958.2002.02885.x. [DOI] [PubMed] [Google Scholar]
- 48.Miethke M, Schmidt S, Marahiel MA. 2008. The major facilitator superfamily-type transporter YmfE and the multidrug-efflux activator Mta mediate bacillibactin secretion in Bacillus subtilis. J Bacteriol 190:5143–5152. doi: 10.1128/JB.00464-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan N. 2013. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem Sci 38:151–159. doi: 10.1016/j.tibs.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 50.Jiang D, Zhao Y, Wang X, Fan J, Heng J, Liu X, Feng W, Kang X, Huang B, Liu J, Zhang XC. 2013. Structure of the YajR transporter suggests a transport mechanism based on the conserved motif A. Proc Natl Acad Sci U S A 110:14664–14669. doi: 10.1073/pnas.1308127110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boyd D, Manoil C, Beckwith J. 1987. Determinants of membrane protein topology. Proc Natl Acad Sci U S A 84:8525–8529. doi: 10.1073/pnas.84.23.8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silhavy TJ, Beckwith JR. 1985. Uses of lac fusions for the study of biological problems. Microbiol Res 49:398–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rubino JT, Franz KJ. 2012. Coordination chemistry of copper proteins: how nature handles a toxic cargo for essential function. J Inorg Biochem 107:129–143. doi: 10.1016/j.jinorgbio.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 54.Buschmann S, Warkentin E, Xie H, Langer JD, Ermler U, Michel H. 2010. The structure of cbb3 cytochrome oxidase provides insights into proton pumping. Science 329:327–330. doi: 10.1126/science.1187303. [DOI] [PubMed] [Google Scholar]
- 55.Loftin IR, Franke S, Roberts SA, Weichsel A, Héroux A, Montfort WR, Rensing C, McEvoy MM. 2005. A novel copper-binding fold for the periplasmic copper resistance protein CusF. Biochemistry 44:10533–10540. doi: 10.1021/bi050827b. [DOI] [PubMed] [Google Scholar]
- 56.Arnesano F, Banci L, Bertini I, Mangani S, Thompsett AR. 2003. A redox switch in CopC: an intriguing copper trafficking protein that binds copper(I) and copper(II) at different sites. Proc Natl Acad Sci U S A 100:3814–3819. doi: 10.1073/pnas.0636904100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swem DL, Swem LR, Setterdahl A, Bauer CE. 2005. Involvement of SenC in assembly of cytochrome c oxidase in Rhodobacter capsulatus. J Bacteriol 187:8081–8087. doi: 10.1128/JB.187.23.8081-8087.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lohmeyer E, Schröder S, Pawlik G, Trasnea PI, Peters A, Daldal F, Koch HG. 2012. The ScoI homologue SenC is a copper binding protein that interacts directly with the cbb(3)-type cytochrome oxidase in Rhodobacter capsulatus. Biochim Biophys Acta 1817:2005–2015. doi: 10.1016/j.bbabio.2012.06.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ekici S, Pawlik G, Lohmeyer E, Koch HG, Daldal F. 2012. Biogenesis of cbb(3)-type cytochrome c oxidase in Rhodobacter capsulatus. Biochim Biophys Acta 1817:898–910. doi: 10.1016/j.bbabio.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darrouzet E, Daldal F. 2002. Movement of the iron-sulfur subunit beyond the ef loop of cytochrome b is required for multiple turnovers of the BC 1 complex but not for single turnover Qo site catalysis. J Biol Chem 277:3471–3476. doi: 10.1074/jbc.M107974200. [DOI] [PubMed] [Google Scholar]
- 61.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 62.Darrouzet E, Valkova-Valchanova M, Daldal F. 2002. The [2Fe-2S] cluster E(m) as an indicator of the iron-sulfur subunit position in the ubihydroquinone oxidation site of the cytochrome bc1 complex. J Biol Chem 277:3464–3470. doi: 10.1074/jbc.M107973200. [DOI] [PubMed] [Google Scholar]
- 63.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 64.Koch HG, Hwang O, Daldal F. 1998. Isolation and characterization of Rhodobacter capsulatus mutants affected in cytochrome cbb3 oxidase activity. J Bacteriol 180:969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brickman E, Beckwith J. 1975. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J Mol Biol 96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 66.Deshmukh M, Brasseur G, Daldal F. 2000. Novel Rhodobacter capsulatus genes required for the biogenesis of various c-type cytochromes. Mol Microbiol 35:123–138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of CcoA homologues from various bacterial species. Residues that are conserved at 80% or higher are indicated in red (identical amino acid) and green (similar amino acids). Download
All strains and plasmids used in this work and related references.
All oligonucleotide primers used in this work.