ABSTRACT
Bacterial type IV secretion systems (T4SSs) can function to export or import DNA, and can deliver effector proteins into a wide range of target cells. Relatively little is known about the structural organization of T4SSs that secrete effector proteins. In this report, we describe the isolation and analysis of a membrane-spanning core complex from the Helicobacter pylori cag T4SS, which has an important role in the pathogenesis of gastric cancer. We show that this complex contains five H. pylori proteins, CagM, CagT, Cag3, CagX, and CagY, each of which is required for cag T4SS activity. CagX and CagY are orthologous to the VirB9 and VirB10 components of T4SSs in other bacterial species, and the other three Cag proteins are unique to H. pylori. Negative stain single-particle electron microscopy revealed complexes 41 nm in diameter, characterized by a 19-nm-diameter central ring linked to an outer ring by spoke-like linkers. Incomplete complexes formed by Δcag3 or ΔcagT mutants retain the 19-nm-diameter ring but lack an organized outer ring. Immunogold labeling studies confirm that Cag3 is a peripheral component of the complex. The cag T4SS core complex has an overall diameter and structural organization that differ considerably from the corresponding features of conjugative T4SSs. These results highlight specialized features of the H. pylori cag T4SS that are optimized for function in the human gastric mucosal environment.
IMPORTANCE
Type IV secretion systems (T4SSs) are versatile macromolecular machines that are present in many bacterial species. In this study, we investigated a T4SS found in the bacterium Helicobacter pylori. H. pylori is an important cause of stomach cancer, and the H. pylori T4SS contributes to cancer pathogenesis by mediating entry of CagA (an effector protein regarded as a “bacterial oncoprotein”) into gastric epithelial cells. We isolated and analyzed the membrane-spanning core complex of the H. pylori T4SS and showed that it contains unique proteins unrelated to components of T4SSs in other bacterial species. These results constitute the first structural analysis of the core complex from this important secretion system.
INTRODUCTION
Helicobacter pylori is a Gram-negative bacterium that colonizes the stomach in about half of the world’s population (1, 2). Persistent H. pylori infection is a risk factor for the development of gastric cancer or peptic ulcer disease (3–5). Adverse disease outcomes associated with H. pylori infection occur predominantly in persons infected with strains that produce an effector protein known as CagA (6, 7), which is translocated into host cells through a type IV secretion system (T4SS) (8, 9). The cagA gene and genes required for T4SS-dependent secretion of CagA are located within a 40-kb region of the H. pylori chromosome known as the cag pathogenicity island (cag PAI) (10–14). CagA is the only protein known to be secreted by the H. pylori cag T4SS. Upon entry into host cells, CagA causes alterations in cell signaling that are linked to gastric carcinogenesis (15–18). H. pylori-induced gastric inflammation also contributes to carcinogenesis. H. pylori can stimulate gastric epithelial cells to produce and secrete interleukin-8 (IL-8 [a proinflammatory cytokine]) through a T4SS-dependent, CagA-independent process associated with peptidoglycan entry into host cells (9, 19).
T4SSs are a versatile family of secretion systems found in a wide variety of bacterial species (20–22). Three main functional categories of T4SSs are recognized: conjugation systems, effector translocator systems, and DNA release/uptake systems (20). Effector proteins or DNA-protein complexes can be delivered into many types of recipient cells, including mammalian cells, plant cells, fungi, or other bacteria. Examples of DNA-translocating T4SSs include the VirB/VirD4 system found in Agrobacterium tumefaciens (which translocates T-DNA into plant cells, causing crown-gall disease) and plasmid conjugation systems (such as the Escherichia coli pKM101-encoded Tra system). Effector protein-translocating T4SSs are exemplified by the Dot/Icm systems in Legionella and Coxiella and the H. pylori cag T4SS. Studies of T4SSs from A. tumefaciens and plasmid conjugation systems have revealed the existence of a transmembrane channel known as the core complex (23–26). The T4SS core complex encoded by the E. coli pKM101 conjugative plasmid is composed of three proteins, VirB7/TraN, VirB9/TraO, and VirB10/TraF, which are organized into a ring-like structure, 18.5 nm in diameter, with 14-fold symmetry (24, 25). In comparison to the structural organization of T4SSs that mediate conjugative transfer of DNA, relatively little is known about the structural organization of T4SSs that translocate effector proteins.
Among 18 genes within the H. pylori cag pathogenicity island (PAI) that are required for cag T4SS-dependent phenotypes (9), several encode proteins that share some sequence relatedness to components of T4SSs in other bacterial species. Notably, the level of sequence conservation is low, and nine of the cag PAI genes required for CagA translocation are unique to H. pylori, with no obvious homologs in other bacteria (13, 27). This suggests that there are important differences between the H. pylori cag T4SS and T4SSs in other bacterial species. Models for the structural organization of the H. pylori cag T4SS have been proposed (13, 28–30), but there are numerous limitations and controversial features of the current models, due to a lack of detailed biochemical and structural data. In this report, we describe isolation of the H. pylori cag T4SS core complex, biochemical analyses of its composition, and single-particle electron microscopy (EM) analysis of its structure. We show that the H. pylori cag T4SS core complex contains proteins that are unrelated to components of T4SSs in other bacterial species and that the architecture of the H. pylori core complex differs considerably from that of previously studied T4SS core complexes.
RESULTS
Purification of CagF and copurification of additional Cag proteins.
Previous studies have shown that the H. pylori cagA effector protein interacts with another cag PAI-encoded protein, CagF (31–33). CagF is required for CagA translocation into eukaryotic host cells, and it has been proposed that CagF is a chaperone for CagA (31–33). We hypothesized that the CagF-CagA complex physically interacts with components of the cag T4SS machinery and that the CagF-CagA complex could be used as bait for purifying subassemblies of the cag T4SS. To test this hypothesis, we generated three H. pylori strains that produced hemagglutinin (HA) epitope-tagged forms of CagF; two (HA-CagF and ΔcagF/HA-CagF) produced CagF with an N-terminal tag, and one (CagF-HA) produced CagF with a C-terminal tag (see Fig. S1a and b in the supplemental material). The HA-CagF and CagF-HA strains retained the endogenous cagF gene (encoding an untagged form of CagF), whereas the ΔcagF/HA-CagF strain encoded only the tagged form of CagF. All three strains retained the ability to induce IL-8 secretion in human gastric epithelial (AGS) cells, indicating that the cag T4SS is functional in each of these strains (see Fig. S1c).
We immunopurified HA-tagged CagF from lysates of the three H. pylori strains using a monoclonal HA antibody, and an untagged wild-type (WT) strain was processed in parallel as a negative control. SDS-PAGE and silver stain analyses showed distinct proteins in preparations derived from the strains producing HA-tagged CagF that were absent in preparations from the untagged WT control strain (see Fig. S1d in the supplemental material). Immunoblotting showed that CagF and CagA were immunopurified from the strains producing HA-tagged CagF, whereas neither of these proteins was immunopurified from the untagged WT control strain (Fig. 1). We then immunoblotted the samples with a panel of antibodies against several components of the cag T4SS and found that Cag3, CagT, CagM, CagX, and CagY copurified with CagF and CagA (Fig. 1). Since similar results were obtained with all three strains producing HA-tagged CagF (Fig. 1), we conducted subsequent experiments using strains producing CagF with an N-terminal HA tag (either HA-CagF or ΔcagF/HA-CagF).
FIG 1 .
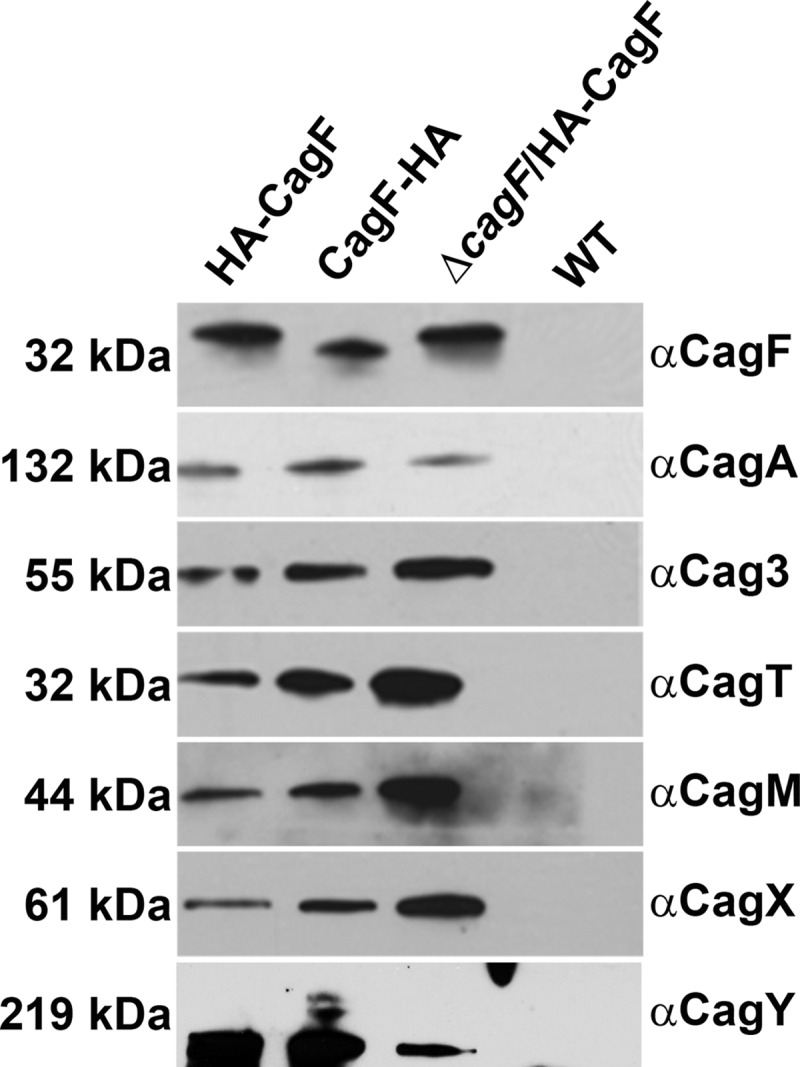
Analysis of proteins that copurify with CagF. Immunoblot analysis of preparations immunopurified from three strains producing HA-tagged CagF using a monoclonal HA antibody and of a control preparation from an untagged WT strain that was processed in parallel. Samples were immunoblotted with the indicated antisera. Results are representative of at least two independent experiments.
Analysis by mass spectrometry indicated that the seven Cag proteins described above (CagF, CagA, Cag3, CagT, CagM, CagX, and CagY) were the most abundant proteins in preparations derived from the HA-CagF-producing strain and that each of these proteins was enriched in preparations from the HA-CagF-producing strain compared to a control preparation from the untagged WT strain (Table 1; see also Table S1 in the supplemental material for complete results). The copurification of several H. pylori proteins encoded by genes outside the cag PAI and produced at high levels by the bacteria (Table 1) most likely reflects nonspecific interactions (34).
TABLE 1 .
Cag proteins that copurify with HA-CagF
| Identified protein | Gene numberb | Assigned spectral countsa |
|
|---|---|---|---|
| HA-CagF | WT | ||
| CagA | HP0547 | 178 | 0 |
| CagF | HP0543 | 56 | 1 |
| CagX | HP0528 | 40 | 0 |
| CagM | HP0537 | 38 | 0 |
| Cag3 | HP0522 | 44 | 0 |
| CagT | HP0532 | 26 | 0 |
| CagY | HP0527 | 18 | 0 |
| No. of Cag spectra | 400 | 1 | |
| No. of non-Cag spectra | 69 | 23 | |
| Total no. of spectra | 469 | 24 | |
An H. pylori strain producing HA-tagged CagF (HA-CagF) was grown in liquid culture, and HA-CagF was immunoaffinity purified using a monoclonal anti-HA antibody. An untagged wild-type strain (WT) was processed in parallel as a control. The protein content of these preparations was analyzed by the use of Multidimensional Protein Identification Technology (MudPIT). The list of H. pylori proteins identified was filtered via IDPicker 3.0 using a <5% peptide false-discovery rate (FDR) and a minimum of 2 distinct peptides per protein. The table shows numbers of peptides matched to individual H. pylori proteins and lists cag PAI-encoded proteins. The complete results are shown in Table S1 in the supplemental material.
Genbank accession number NC_000915.
The immunopurifications described above were performed using lysates of H. pylori grown in liquid culture. CagA is not actively secreted by the bacteria under such conditions, but H. pylori contact with gastric epithelial cells triggers translocation of CagA into the epithelial cells (8, 35). We hypothesized that H. pylori contact with gastric epithelial cells leads to activation of the cag T4SS, possibly resulting in altered interactions of the CagF-CagA complex with T4SS components. Therefore, we cocultured AGS gastric epithelial cells with HA-CagF-producing H. pylori (ΔcagF/HA-CagF) and purified HA-CagF from lysates of the coculture. AGS cells cocultured with untagged WT bacteria were processed in parallel as a negative control. Mass spectrometry analysis revealed that preparations isolated from the coculture contained the same seven Cag proteins detected in the earlier experiments (see Table S2 in the supplemental material). Low levels of several other Cag proteins were also detected (see Table S2). Importantly, Cag3, CagT, CagM, CagX, and CagY reproducibly copurified with CagF and CagA in experiments conducted with either bacterial cell-epithelial cell cocultures or bacterial liquid cultures.
Isolation of a Cag protein complex.
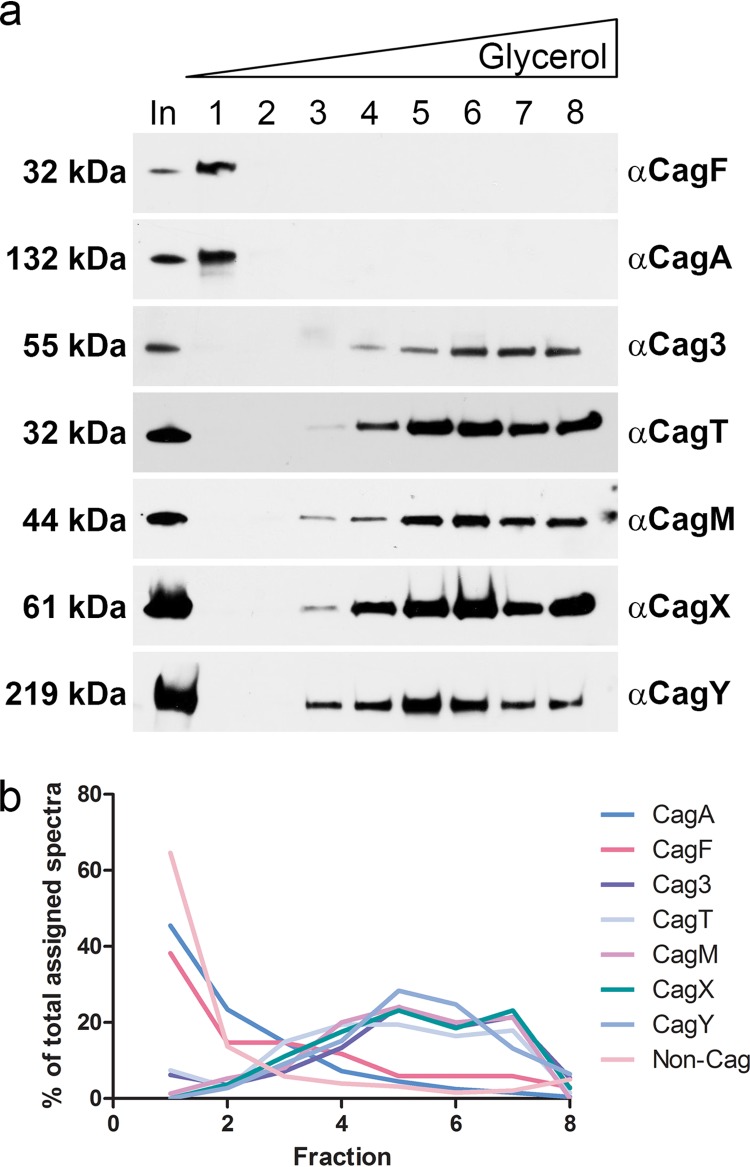
To investigate whether the proteins copurifying with CagF were constituents of one or more protein complexes, we analyzed immunopurified samples prepared as described above by velocity sedimentation in glycerol density gradients. Immunoblot analysis indicated that Cag3, CagT, CagM, CagX, and CagY cosedimented in the bottom fractions of the gradient (i.e., in the high-molecular-mass fractions) (Fig. 2a). This result provides evidence that Cag3, CagT, CagM, CagX, and CagY are components of a large protein complex. CagA and CagF were detected in the first fraction (taken from the top of the gradient), separately from the other Cag proteins, suggesting that CagF and CagA dissociate from the larger complex during gradient centrifugation. Mass spectrometry analysis of the gradient fractions (Fig. 2b; see also Table S3 in the supplemental material) confirmed that Cag3, CagT, CagM, CagX, and CagY were all distributed in the same gradient fractions (supporting the conclusion that these are components of a protein complex), whereas CagF and CagA were found predominantly in fractions from the top of the gradient (Fig. 2b). Each of the non-Cag proteins present in the sample was detected predominantly in the first fraction (taken from the top of the gradient) instead of the fractions containing Cag3, CagT, CagM, CagX, and CagY (Fig. 2b). Thus, the stringent conditions of the glycerol density gradient allowed purification to homogeneity of a large stable complex of Cag proteins distinct from CagA, CagF, and non-Cag proteins (Fig. 2b; see also Table S3 in the supplemental material).
FIG 2 .
Sedimentation of H. pylori cag T4SS core complex in density gradients. (a) Preparations resulting from immunoaffinity purification of HA-CagF were analyzed by velocity sedimentation in glycerol density gradients, and gradient fractions (collected from the top of the gradient) were immunoblotted with the indicated antisera. In, input sample before it was applied to the gradient. Results are representative of two independent experiments. (b) The protein content of gradient fractions was analyzed by 1D mass spectrometry (see Table S3 in the supplemental material for complete results). The distribution of CagF, CagA, Cag3, CagT, CagM, CagX, CagY, and total non-Cag proteins in individual gradient fractions is shown.
Cag3, CagT, CagM, CagX, and CagY are all known to be required for T4SS function, including translocation of CagA into host cells (9, 36). Cag3, CagM, and CagT are unique to H. pylori and do not exhibit sequence relatedness to constituents of T4SSs in other bacteria, whereas CagX and CagY are orthologs of the VirB9/TraO and VirB10/TraF components of core complexes in DNA-translocating T4SSs (13). Therefore, we reasoned that the Cag protein complex isolated in these experiments corresponds to the cag T4SS core complex.
Analysis of complexes formed by mutant strains.
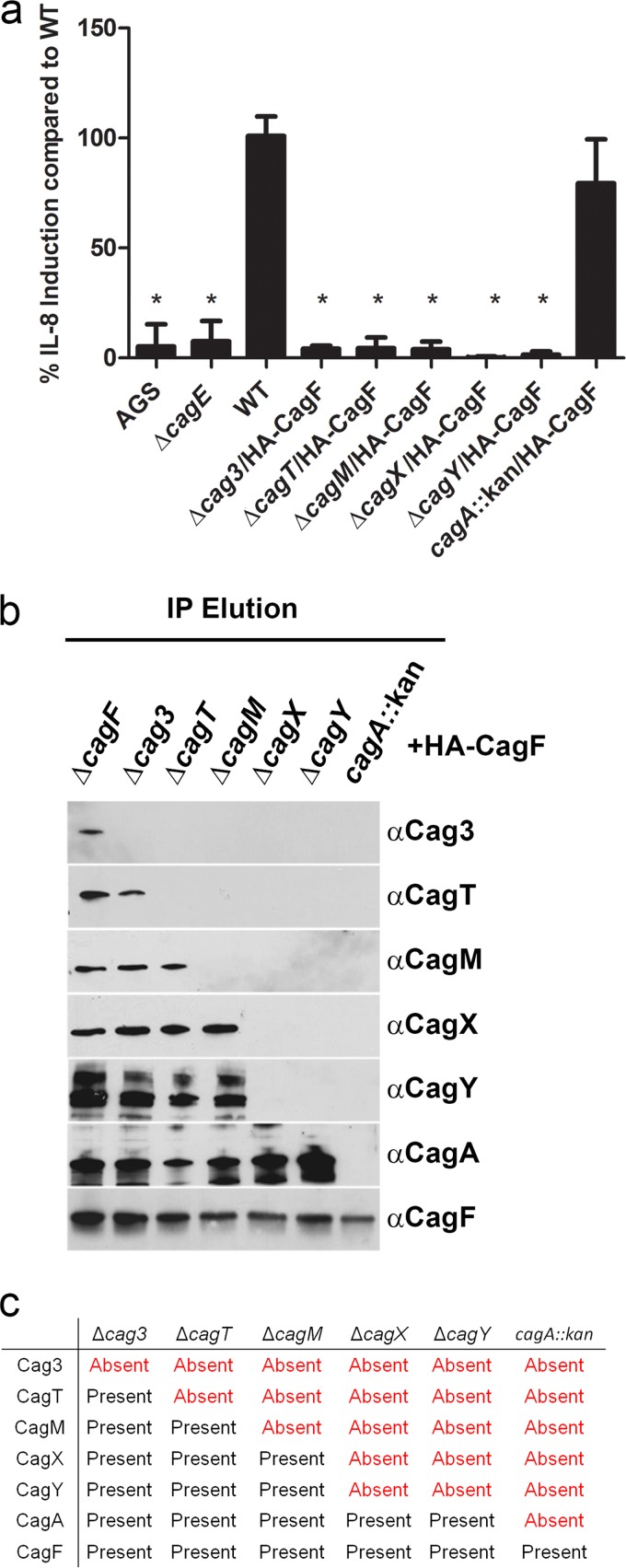
We hypothesized that several of the Cag proteins identified in the immunopurification experiments would be required for assembly or stability of the core complex, whereas others would be nonessential for core complex assembly (despite being required for T4SS function). For example, the Cag proteins that have orthologs in T4SSs of multiple bacterial species are likely to be required for core complex formation, whereas proteins unique to H. pylori (Cag3, CagT, and CagM) might be dispensable. To test this hypothesis, we analyzed mutant strains in which cagY, cagX, cagT, cag3, and cagM were individually deleted, and each mutant was engineered to produce HA-CagF, thereby permitting isolation of the core complex. Consistent with the results of previous studies (9), these five mutant strains were unable to induce IL-8 secretion in AGS cells (indicating the absence of a functional cag T4SS) (Fig. 3a). We also generated a cagA mutant strain that expressed HA-CagF, and, consistent with previous results (9), this strain retained the capacity to induce IL-8 secretion (Fig. 3a). We then sought to isolate Cag protein complexes from the mutant strains by immunopurifying HA-tagged CagF (using the method shown in Fig. 1) and analyzed the resulting preparations by immunoblotting. In experiments performed with ΔcagX or ΔcagY mutants, we successfully purified CagF and CagA but were unable to copurify core complex components (Fig. 3b and c), despite evidence that these proteins were present in lysates from these mutants (see Fig. S2 in the supplemental material). Similarly, in an experiment performed with the cagA mutant strain, we successfully purified CagF but were unable to copurify any components of the core complex (Fig. 3b and c). In contrast, we were able to immunopurify multiple Cag proteins as well as CagF and CagA from the Δcag3, ΔcagT, and ΔcagM mutant strains. In experiments performed with the Δcag3 strain, all components of the core complex (except Cag3) copurified with CagF and CagA. Preparations immunopurified from the ΔcagT strain lacked Cag3 and CagT, and preparations immunopurified from the ΔcagM strain lacked Cag3, CagT, and CagM (Fig. 3b and c). These experiments provided evidence that incomplete protein complexes can assemble in the absence of Cag3, CagT, or CagM (Fig. 3b and c).
FIG 3 .
Immunopurification of HA-CagF from a panel of cag mutant strains. (a) AGS cells were cocultured with the WT strain or the indicated mutants, and IL-8 secretion was quantified by enzyme-linked immunosorbent assay (ELISA). “AGS” indicates cells without added H. pylori. Values represent means ± standard deviations (SD), based on analysis of at least six replicate samples. Levels of IL-8 production induced by mutant strains were compared to levels induced by the WT strain (analysis of variance [ANOVA] followed by Dunn’s multiple-comparison test). *, P < 0.001. (b) HA-CagF was immunoaffinity purified from the strains analyzed in panel A. The affinity-purified samples (immunopurification [IP] elution) were then immunoblotted with the indicated antisera. Strains are designated at the top of the panel, and antisera are designated at the right side of the panel. (c) Summary of results from the experiments presented in panel B, indicating the presence or absence of Cag proteins in the preparations immunopurified from mutant strains. Results are representative of three independent experiments.
Ultrastructure analysis of the core complex.
We next analyzed the ultrastructure of the cag T4SS core complex. H. pylori strain HA-CagF (containing an intact cag PAI) was grown in liquid culture, complexes were isolated using the CagF-based immunopurification procedure, and the resulting complexes were visualized using negative-stain single-particle EM. EM analysis revealed homogeneous ring-shaped complexes, characterized by well-defined central and outer rings (Fig. 4a). We also used negative-stain single-particle EM to analyze fractions of a glycerol gradient to which the immunopurified sample had been applied, and detected the ring-shaped complexes in the gradient fractions that contained Cag3, CagT, CagM, CagX, and CagY but not in the fractions collected from the top of the gradient (see Fig. S3 in the supplemental material). Since the gradient fractions containing these five Cag proteins were free of other non-Cag proteins (Fig. 2b; see also Table S3 in the supplemental material), this result provides evidence that the complexes visible by EM were composed of Cag proteins.
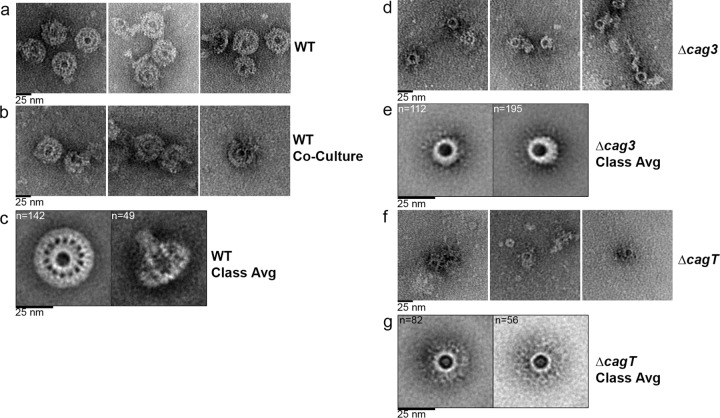
FIG 4 .
Negative-stain EM analysis of the H. pylori cag T4SS core complex. (a) Representative negative-stain images of WT cag T4SS complexes purified from a strain with an intact cag PAI that was engineered to produce HA-CagF. (b) Representative negative-stain images of WT cag T4SS complexes purified from an HA-CagF-producing strain (ΔcagF/HA-CagF) cocultured with AGS cells. (c) Class averages (Class Avg) showing an en face view and a side view of WT complexes. (d) Representative negative-stain images of cag T4SS complexes purified from a Δcag3 strain engineered to produce HA-CagF. (e) Class averages of the cag T4SS complex purified from a Δcag3 strain. (f) Representative negative-stain images of cag T4SS complexes purified from a ΔcagT strain engineered to produce HA-CagF. (g) Class averages of the cag T4SS complex purified from a ΔcagT strain. Numbers of particles used for generating each average are shown in upper left corner of panels. All scale bars represent 25 nm.
CagA is not actively secreted during H. pylori growth in liquid culture, but coculture of H. pylori with gastric epithelial cells is a trigger for translocation of CagA into host cells (8, 35). Therefore, although we did not detect substantial changes in the protein composition of the immunopurified samples in response to epithelial cell contact (see Table S2 in the supplemental material), we hypothesized that the structural organization of the complexes might change when H. pylori contacts gastric epithelial cells. To investigate this possibility, we cocultured AGS gastric epithelial cells with HA-CagF-producing H. pylori, purified HA-CagF from lysates of the coculture, and then analyzed the immunopurified core complexes by negative-stain EM. The protein complexes isolated from H. pylori-AGS cocultures were similar in appearance to the complexes isolated from H. pylori liquid cultures, but the complexes isolated from cocultures were present in relatively low abundance and had a less homogeneous appearance (suggestive of degradation) than complexes isolated from liquid cultures (Fig. 4b). Similarly, the complexes in fractions collected from glycerol gradients had suboptimal morphology and exhibited increased clumping compared to complexes in preparations that were not passed through a gradient (see Fig. S3 in the supplemental material). Consequently, all of the subsequent experiments focused on complexes isolated from H. pylori grown in liquid culture without use of gradient procedures.
To further characterize the structural features of the complexes, we collected a data set containing a larger number of particles, classified them into 10 class averages using reference-free alignment (see Fig. S4a in the supplemental material), and then selected five of these as references for an additional round of reference-based alignment (see Fig. S4a and b). This analysis revealed well-defined classes, corresponding to en face and side views of the T4SS complex (Fig. 4c). The en face class shows a well-defined complex, 41 nm in diameter, composed of a central and outer ring connected by short linkers. The central ring is 19 nm in diameter. Fourteen linkers or “spokes” connect the central and outer ring. The side view reveals a stalk-like structure about 40 nm in length, most likely corresponding to the central ring seen in the en face views, that extends away from the ring-like portions of the complex.
Ultrastructure analysis of complexes formed by mutant strains.
The immunoprecipitation analyses of mutant strains shown in Fig. 3 suggested that incomplete complexes could be formed in the absence of Cag3, CagT, or CagM (Fig. 3b and c). To investigate these incomplete complexes further, we applied the CagF-based immunopurification procedure to these mutant strains and then analyzed the resulting complexes by negative-stain EM. We first imaged preparations derived from the ΔcagX mutant (containing CagA and CagF but not any components of the core complex, based on immunoblotting results) (Fig. 3b and c). Consistent with the absence of core complex components detectable by immunoblotting, we did not visualize any ring-like structures in these preparations.
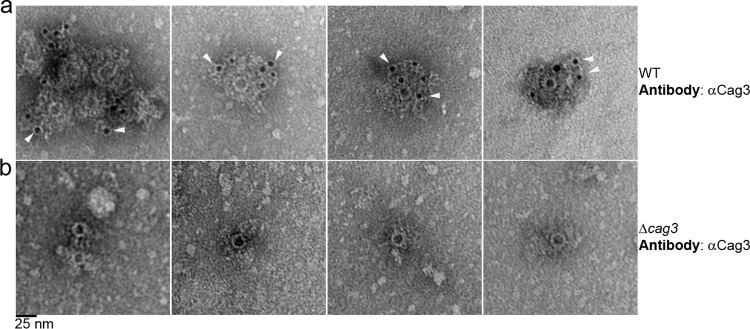
We then imaged complexes isolated from the Δcag3 mutant (lacking Cag3 but containing CagX, CagY, CagT, and CagM, based on immunoblot analysis results). The complexes detected in these preparations contained a well-defined ring, but there was a marked reduction in peripheral content and no evidence of a well-organized outer ring (Fig. 4d). To further characterize the structural features of complexes purified from the Δcag3 mutant, we collected a data set of particles and generated class averages (see Fig. S4c and d in the supplemental material). These averages revealed en face views of a 19-nm-diameter ring (Fig. 4e). Unlike images and classes of wild-type T4SS core complexes, the 2D class averages of complexes isolated from the Δcag3 mutant contained no outer ring or connecting spokes (Fig. 4e). The absence of peripheral components in complexes from the Δcag3 mutant without any obvious perturbation of the 19-nm-diameter ring provides evidence that Cag3 is localized to the periphery of the complex. To further test this hypothesis, we immunolabeled purified WT core complexes as well as complexes from the Δcag3 mutant with primary polyclonal antibodies reactive to Cag3, followed by a secondary antibody conjugated to 5-nm-diameter gold particles. Imaging of the labeled wild-type complexes by negative-stain EM revealed gold particles in the periphery of the complexes, but no gold particles were associated with the central ring (Fig. 5a). Mutant complexes lacking Cag3 were not labeled with gold particles (Fig. 5b), demonstrating the specificity of the immunolabeling. These immunogold EM results corroborated the conclusion that Cag3 is a peripheral component of the cag T4SS core complex.
FIG 5 .
Localization of Cag3 by immunogold labeling and negative-stain EM. Negative-stain EM images of WT core complexes, isolated from a strain with an intact cag PAI (a) or a Δcag3 mutant (b) and subjected to immunogold labeling with primary anti-Cag3 antiserum followed by 5-nm-diameter-gold-particle-conjugated secondary antibodies, are shown. Arrowheads indicate representative gold particles. Images are representative of results from three independent experiments. Scale bar, 25 nm.
We also imaged complexes isolated from the ΔcagT mutant (lacking both Cag3 and CagT). Similarly to complexes isolated from the Δcag3 mutant, the complexes isolated from the ΔcagT mutant contained a single well-defined ring but no well-organized outer ring (Fig. 4f). The ΔcagT mutant complexes were less abundant than the Δcag3 mutant complexes. In order to characterize the structural features of complexes purified from the ΔcagT mutant, we collected a data set of particles and generated class averages (see Fig. S4e and f in the supplemental material). The en face views indicate that the diameter of the ring in complexes from the ΔcagT mutant is similar to the diameters of the rings in complexes from the Δcag3 mutant and WT strains (Fig. 4g), but the ring in complexes from the ΔcagT mutant appears to have a reduced thickness. Unlike images and classes of wild-type T4SS core complexes, the 2D class averages of complexes isolated from the ΔcagT mutant contained no outer ring or connecting spokes (Fig. 4g). Immunogold labeling of CagT was attempted using complexes purified from WT and ΔcagT strains; however, the results were inconclusive due to the nonspecific binding of the anti-CagT polyclonal antibody under these conditions. We also analyzed preparations derived from the ΔcagM mutants (which lack Cag3, CagT, and CagM) but were unable to visualize any ring-shaped complexes in these preparations. The absence of detectable ring-like complexes in preparations from the ΔcagM mutant could be due to a reduced stability of these complexes, or, alternatively, ring-shaped complexes may fail to assemble in the absence of CagM. Collectively, the structural differences between mutant and WT complexes, combined with specific antibody labeling, confirm that the ring-like structures seen by negative-stain EM correspond to the T4SS core complex.
DISCUSSION
In this report, we describe the isolation and analysis of a T4SS core complex composed of five different proteins encoded by the H. pylori cag pathogenicity island (CagX, CagY, CagM, CagT, and Cag3). Previous studies have detected interactions among several of these Cag proteins (30, 37–39), but the results reported here represent the first successful efforts to isolate and analyze the structure of a complex containing these proteins. Multiple lines of evidence indicate that the complex visualized by EM is composed of Cag proteins. Specifically, glycerol gradient fractions free of contamination from non-Cag proteins contained complexes detectable by EM (see Fig. S3 and Table S3 in the supplemental material), mutant strains lacking Cag3 or CagT formed complexes with a morphology different from that of wild-type complexes (Fig. 4), and Cag3 was detected as a component of the complexes by immunogold staining (Fig. 5).
Each of the 5 protein components of the core complex (CagX, CagY, CagM, CagT, and Cag3) is required for cag T4SS-dependent phenotypes, including CagA translocation into host cells and induction of IL-8 secretion by gastric epithelial cells (9). Most of these proteins are also required for production of T4SS-associated pilus structures at the bacterium-host cell interface (36). Previous studies have localized all 5 of these proteins at the outer membrane of H. pylori (10, 34). CagX and CagY exhibit low-level sequence relatedness to VirB9 and VirB10 components of T4SSs in other bacterial species, but CagM, CagT, and Cag3 are not closely related to any other bacterial proteins. Sequence analysis of CagT suggests that it may be a lipoprotein, leading to the suggestion that it may be a VirB7 homolog (14). CagA exhibits a high level of sequence variation in comparisons of H. pylori strains from different geographic areas, but, in contrast, there is relatively little phylogeographic variation in sequences of the five core complex components (10).
The methodology used in this study for isolating the H. pylori cag T4SS core complex relied on immunopurification of CagF, a cag PAI-encoded protein that binds the CagA effector protein (31–33). Importantly, velocity sedimentation experiments performed with glycerol gradients indicated that CagF and CagA are not integral components of the core complex. The CagF-based purification strategy conducted with a cagA mutant strain failed to yield core complex components, which suggests that CagA directly interacts with the core complex. The interaction of CagA (and possibly CagF) with the core complex is presumably relatively weak, as expected for an interaction of an effector protein with secretion machinery.
The CagF-based immunopurification strategy led to successful isolation of core complexes when applied to bacteria grown in broth culture (a condition under which CagA is not actively secreted), as well as when applied to bacteria cocultured with gastric epithelial cells (a condition that promotes CagA translocation into the epithelial cells). These results indicate that the core complex is assembled prior to H. pylori contact with host cells and CagA translocation. We speculate that CagA is bound to assembled core complexes, ready to be translocated as soon as the bacteria receive the appropriate signal. The molecular mechanisms that regulate CagA secretion (i.e., inhibition of secretion during bacterial growth in broth culture and activation of secretion when H. pylori contacts gastric epithelial cells) remain poorly understood.
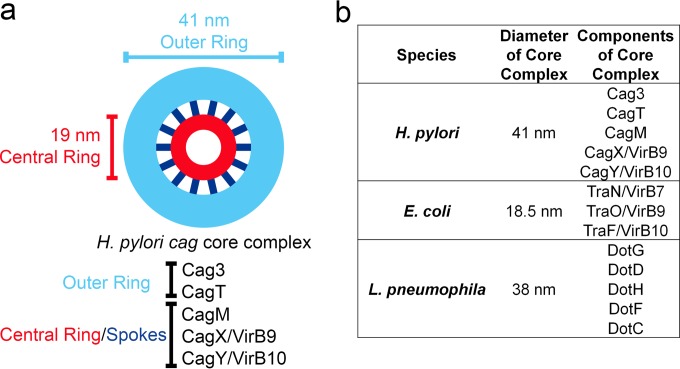
EM analysis of the isolated core complexes revealed homogeneous ring-shaped structures, 41 nm in diameter, characterized by a central ring connected to an outer ring by thin spoke-like linkers (Fig. 6a). The presence of 14 visible spoke-like linkers in the class averages suggests that the complex may have 14-fold rotational symmetry, but three-dimensional (3D) structural analysis will be required for an accurate assessment of the symmetry. A side view of the complex reveals a long stalk-like domain 40 nm in length that likely corresponds to the central ring seen in the en face view. Since all of the core complex components identified in the current study (CagX, CagY, CagT, CagM, and Cag3) have been detected previously on the external surface of H. pylori or as components of the outer membrane (30, 34), we propose that the complex is associated primarily with the outer membrane.
FIG 6 .
Schematic model of the H. pylori cag T4SS core complex. (a) Prominent features of the complex include an outer ring (light blue) and central ring (red) connected by 14 spokes (dark blue). Analysis of complexes from mutant strains suggests that Cag3 and CagT are localized to the periphery of the intact wild-type complex. (b) Comparison of the diameters and components of T4SS core complexes from H. pylori, E. coli, and L. pneumophila.
Analyses of complexes isolated from a Δcag3 mutant strain, along with immunogold labeling experiments, indicate that Cag3 is localized to the periphery of the core complex. Specifically, the complexes formed by the Δcag3 mutant strain (containing CagX, CagY, CagM, and CagT, based on immunoblot analysis results) have an intact 19-nm-diameter ring but exhibit a significantly reduced overall diameter compared to wild-type complexes, attributable to loss of the outer ring and spoke-like linkers. We speculate that the substantial loss of peripheral components in complexes from the Δcag3 mutant reflects not only the absence of Cag3 but also an alteration in the structure of residual peripheral components so that they are structurally disordered and no longer visible by EM. Complexes isolated from a ΔcagT mutant strain (containing CagX, CagY, and CagM, based on immunoblot analysis results) also contained an intact 19-nm-diameter ring and exhibited a loss of visible peripheral components. The complexes from the ΔcagT mutant appeared similar to the Δcag3 complexes except that the ring appeared to have a reduced thickness. The incomplete complexes derived from Δcag3 or ΔcagT mutant strains might correspond to intermediate or premature stages in the assembly process that leads to formation of the complete core complex.
Previous studies have analyzed the 3D structure of the T4SS core complex encoded by the E. coli pKM101 conjugative plasmid, which is composed of three proteins, VirB7/TraN, VirB9/TraO, and VirB10/TraF (24). The pKM101 T4SS core complex is organized into a ring-like structure, 18.5 nm in diameter, with 14-fold symmetry (24) and is localized primarily to the outer membrane. Core complexes isolated from the A. tumefaciens VirB/D4 T4SS are similar in size (about 20 nm in diameter) (40). Therefore, the H. pylori cag T4SS core complex (41 nm in diameter) is substantially larger in size than either the pKM101-encoded core complex (25) or A. tumefaciens core complex (40) (Fig. 6b). Moreover, the H. pylori core complex is composed of five Cag proteins, whereas core complexes from the pKM101 and A. tumefaciens T4SSs contain only three (VirB7/TraN, VirB9/TraO, and VirB10/TraF) (24, 25) (Fig. 6b). Despite these differences in size and overall architecture, it is notable that 14 linkers or spokes connecting the central ring to the outer ring could be visualized in the class average of H. pylori cag T4SS complexes, which suggests that the cag T4SS core complex may have 14-fold rotational symmetry similar to that of the pKM101 core complex. It is also notable that the central ring of the H. pylori cag T4SS core complex has a diameter (19 nm) similar to those seen with the T4SS core complexes of E. coli and A. tumefaciens (24, 40), suggesting that the overall structural organization may be conserved.
Only two components of the H. pylori cag T4SS core complex exhibit sequence similarity to components of previously studied T4SS core complexes. Specifically, CagX and CagY exhibit sequence relatedness to VirB9 and VirB10 (Fig. 6b). We propose that CagX and CagY are localized to the central ring of the cag T4SS core complex. Consistent with this view, the complexes isolated from Δcag3 and ΔcagT mutants contained CagX and CagY and had intact rings (with diameters similar to that of the central ring of WT complexes) but lacked the structurally organized outer ring and connecting spokes visualized in WT T4SS complexes.
The exact composition of core complexes in effector protein-translocating T4SSs has not previously been determined for most bacterial species, though the Legionella Dot/Icm core complex is reported to contain at least five proteins (DotC, DotD, DotF, DotG, and DotH) (41). Recently published EM images of the Legionella Dot/Icm T4SS core complex indicate that it is organized as a circular structure 38 nm in diameter (41). Thus, the size of the H. pylori core complex analyzed in the current study is more similar to that of the Legionella Dot/Icm core complex than to those of Agrobacterium or pKM101 core complexes (Fig. 6b). Another notable similarity between the H. pylori cag and Legionella Dot/Icm T4SSs is that the core complexes of the two systems have constituents not widely conserved in T4SSs of other bacterial species. Three H. pylori core complex constituents (Cag3, CagM, and CagT) seem to be unique species-specific adaptations, and three Legionella core complex constituents (DotC, DotF, and DotD) are absent from T4SSs in most other bacterial species. The H. pylori cag T4SS translocates CagA across the plasma membrane of gastric epithelial cells, whereas the Legionella Dot T4SS translocates several hundred effector proteins across endosomal membranes in eukaryotic cells. The existence of specialized adaptations in H. pylori and Legionella T4SS core complexes is consistent with the considerably different functions of the T4SSs in these two species.
In future studies, it will be important to investigate the molecular mechanisms by which the H. pylori cag T4SS mediates CagA translocation into host cells. One possibility is that CagA may pass directly through the central pore visible within the core complex. Analysis of the crystal structure of CagA (amino-terminal residues 1 to 876) indicates that it has dimensions of 8 by 11 by 5.5 nm (17), so either a folded CagA protein or an unfolded CagA protein could conceivably fit through the ~10-nm pore of the core complex. The mechanisms by which CagA enters host cells after translocation across the bacterial envelope remain a topic of inquiry. T4SS-associated pili assembled at the bacterium-host cell interface presumably contribute to the entry of CagA into host cells (42–44), and it has been suggested that binding of CagA to phosphatidylserine or integrins on the surface of host cells also may be important for entry into host cells (18, 45).
In summary, this report provides the first in-depth analysis of the H. pylori cag T4SS core complex, which has a key role in the pathogenesis of H. pylori-associated gastric diseases, including gastric cancer. The results show that the cag T4SS core complex is considerably larger and more complex than core complexes from conjugative T4SSs. Finally, the results highlight the existence of unique species-specific components of the H. pylori cag core complex, which presumably represent specialized adaptations optimized for type IV secretion in the human gastric mucosal environment.
MATERIALS AND METHODS
H. pylori culture methods and construction of mutants.
For descriptions of the H. pylori culture methods and construction of mutants, please refer to Text S1 in the supplemental material.
Isolation of the H. pylori cag T4SS core complex.
H. pylori strains were grown in liquid culture for 16 h (resulting in values of optical density at 600 nm [OD600] of 0.4 to 0.8). Bacterial cells were pelleted at 3,300 × g for 15 min at 4°C and resuspended in radioimmunoprecipitation assay (RIPA) buffer (1% NP-40, 0.25% deoxycholate, 10 mM HEPES, and 100 mM NaCl [pH 7.0] supplemented with 1 mM phenylmethylsulfonyl fluoride [PMSF] and protease inhibitors [Complete Mini Protease Inhibitor tablet; Roche]) for initial experiments or in RIPA buffer containing 300 mM NaCl for experiments involving electron microscopy and density gradients. The bacterial suspensions were sonicated on ice and incubated for 1 h at 4°C, and the bacterial lysates were then centrifuged (21,000 × g) to pellet insoluble material. Monoclonal anti-HA antibodies were noncovalently linked to protein G Dynabeads (Invitrogen) and incubated with the clarified bacterial lysates at room temperature for 30 min. Beads were washed three times at room temperature with the same buffers used for bacterial lysis, and proteins were selectively eluted with wash buffer containing 200 µg/ml HA peptide (YPYDVPDYA; GenScript). To isolate the core complex from H. pylori cocultured with gastric epithelial cells, subconfluent AGS cells were cocultured with H. pylori (multiplicity of infection [MOI] of 100) for 4 h. After rinsing with phosphate-buffered saline (PBS) to remove nonadherent bacteria, adherent bacteria and epithelial cells were resuspended in RIPA buffer, and the protocol described above was used.
Glycerol gradients, Western blotting, and mass spectrometric analysis.
For details on glycerol gradients, Western blotting, and mass spectrometric analysis, please refer to Text S1 in the supplemental material.
Negative-stain electron microscopy.
Samples were prepared for EM as previously described (46). In brief, 5-µl aliquots of samples were absorbed to a glow-discharged 400-mesh copper grid covered with carbon-coated collodion film. Grids were washed in four drops of water and then stained with two drops of uranyl formate (0.75%). Samples were imaged on a Morgagni electron microscope (FEI, Hillsboro, OR) operated at an acceleration voltage of 100 kV and equipped with a 1kx1k charge-coupled-device (CCD) camera (ATM). Images were recorded at a magnification of ×28,000. Images used to generate class averages were collected using a TF20 electron microscope (FEI, Hillsboro, OR) equipped with a field emission gun with an acceleration voltage of 200 kV under low-dose conditions at ×62,000 magnification and a defocus value of ~1.5 µm on a 4kx4k Gatan Ultrascan CCD camera.
Class average generation.
Images were converted to mrc (mixed raster content) and binned using a value of 2, resulting in images of 3.5 Å/pixel. For alignment and averaging, 445, 416, and 149 images of wild-type complexes, Δcag3 complexes, and ΔcagT complexes, respectively, were selected with Boxer and windowed with a 180-pixel side length (63 nm). Image analysis was conducted with Spider (47). The images were rotationally and translationally aligned and subjected to 10 cycles of multireference alignment and K-means classification. For analysis of wild-type complexes, particles were classified into 10 class averages and then 5 representative classes were chosen as references for multireference alignment. For analysis of Δcag3 complexes, particles were classified into 20 class averages and then 4 representative projections were chosen as references for another cycle of multireference alignment. For analysis of ΔcagT complexes, particles were first classified into 10 class averages and then 3 representative projections were chosen as references for another cycle of multireference alignment.
Immunogold labeling of protein complexes.
For details on immunogold labeling of protein complexes, please refer to Text S1 in the supplemental material.
SUPPLEMENTAL MATERIAL
Generation of H. pylori strains that produce HA-tagged CagF. (a) Schematic of plasmids encoding epitope-tagged forms of CagF (with either an N-terminal or C-terminal epitope HA tag). The sequences were introduced into the H. pylori ureAB chromosomal locus, yielding strains designated HA-CagF or CagF-HA, respectively. The HA-CagF and Cag-HA strains retained the endogenous cagF gene (encoding an untagged form of CagF). We also generated a ΔcagF/HA-CagF strain, which encodes only the tagged form of CagF. In this strain, the endogenous copy of cagF was deleted, and a gene encoding HA-CagF was introduced into the urease locus. (b) Immunoblotting of the indicated strains with an anti-HA antibody showed the production of HA-tagged forms of CagF. (c) AGS cells were infected with the wild-type (WT) strain or the indicated strains producing HA-tagged CagF, and IL-8 production was quantified by ELISA. AGS indicates results for cells without added H. pylori. Values represent means ± SD, based on analysis of at least six replicate samples. Levels of IL-8 production induced by mutant strains were compared to levels induced by the WT strain (ANOVA followed by Dunn’s multiple-comparison test). *, P < 0.001. (d) SDS-PAGE and silver stain analysis of preparations immunopurified from strains producing HA-tagged CagF (using a monoclonal HA antibody) and analysis of a control preparation from an untagged WT strain that was processed in parallel. Download
Analysis of Cag protein production in a panel of cag mutant strains. Immunoblotting of whole-cell lysates from mutant strains, each expressing HA-CagF from a gene in the urease locus, with the indicated antisera. HspB (a GroEL heat shock protein) was analyzed as a loading control. Strains are designated at the top of the figure, and antisera are designated to the right of the figure. Arrows indicate immunoreactive bands corresponding to Cag proteins, and other bands correspond to cross-reactive proteins. Two CagF bands correspond to the tagged and the untagged forms of the protein. Download
Negative-stain EM analysis of glycerol gradient fractions containing the cag T4SS core complex. Preparations resulting from immunoaffinity purification of HA-CagF were analyzed by velocity sedimentation in glycerol density gradients as shown in Fig. 2, and gradient fractions were analyzed by negative-stain EM. (a) Representative negative-stain EM images of fraction 1 (top of the gradient), which contains CagA and CagF but no core complex components. (b) Representative negative-stain EM images of fraction 6 (near bottom of the gradient), which contains cag core complex components. Fraction 1 contained no complexes as visualized by EM (a), while cag T4SS complexes were observed in images taken from fraction 6 (b). Scale bar, 25 nm. Download
Negative-stain 2D class averages of WT cag T4SS complexes and complexes purified from Δcag3 or ΔcagT mutant strains. (a) Reference-free alignment of 445 particles isolated from an HA-CagF-producing strain with an intact cag PAI into 10 classes. Five classes (marked with %, @, $, #, and & symbols) were chosen as references for an additional round of multireference alignment. (b) Result of reference-based alignment. (c) Reference-free alignment of 416 particles isolated from a Δcag3/HA-CagF-producing strain into 20 classes. Four classes (marked with %, @, $, and # symbols) were chosen as references for an additional round of multireference alignment. (d) Result of reference-based alignment. (e) Reference-free alignment of 149 particles isolated from a ΔcagT/HA-CagF-producing strain into 10 classes. Three classes (marked with %, @, and $ symbols) were chosen as references for an additional round of multireference alignment. (f) Result of reference-based alignment. Numbers of particles used for generating each average are shown in upper left-hand corner of the panels. Side length of panels, 63 nm. Download
Proteins that copurify with HA-CagF.
Proteins that copurify with HA-CagF when H. pylori ΔcagF/HA-CagF is cocultured with AGS cells.
Protein composition of fractions resulting from the sedimentation of an HA-CagF-immunopurified sample in a density gradient.
Supplemental Materials and Methods. Download
ACKNOWLEDGMENTS
The work described in this paper was supported by NIH AI039657, CA116087, T32 AI007281, and F31 AI112324 and by the Department of Veterans Affairs (Merit Review grant 2I01BX000627). Proteomics experiments were supported by the Vanderbilt Digestive Diseases Research Center (P30DK058404) and Vanderbilt-Ingram Cancer Center (P30 CA068485).
We thank Charles Sanders for providing helpful advice and the Vanderbilt Center for Structural Biology for support of the microscope facility.
A.E.F.-C., T.L.C., and M.D.O. conceived and designed the experiments. A.E.F.-C. and W.H.M. performed the experiments. A.E.F.-C., T.M.P., B.J.V., T.L.C., and M.D.O. analyzed the data. All authors wrote and approved the manuscript.
Footnotes
Citation Frick-Cheng AE, Pyburn TM, Voss BJ, McDonald WH, Ohi MD, Cover TL. 2016. Molecular and structural analysis of the Helicobacter pylori cag type IV secretion system core complex. mBio 7(1):e02001-15. doi:10.1128/mBio.02001-15.
REFERENCES
- 1.Robin Warren J, Marshall B. 1983. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 321:1273–1275. doi: 10.1016/S0140-6736(83)92719-8. [DOI] [PubMed] [Google Scholar]
- 2.Cover TL, Blaser MJ. 2009. Helicobacter pylori in health and disease. Gastroenterology 136:1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peek RM Jr, Blaser MJ. 2002. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer 2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 4.Atherton JC. 2006. The pathogenesis of Helicobacter pylori-induced gastroduodenal diseases. Annu Rev Pathol 1:63–96. doi: 10.1146/annurev.pathol.1.110304.100125. [DOI] [PubMed] [Google Scholar]
- 5.Suerbaum S, Michetti P. 2002. Helicobacter pylori infection. N Engl J Med 347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 6.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci U S A 90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaser MJ, Perez-Perez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH, Stemmermann GN, Nomura A. 1995. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res 55:2111–2115. [PubMed] [Google Scholar]
- 8.Odenbreit S, Püls J, Sedlmaier B, Gerland E, Fischer W, Haas R. 2000. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287:1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 9.Fischer W, Püls J, Buhrdorf R, Gebert B, Odenbreit S, Haas R. 2001. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol 42:1337–1348. doi: 10.1046/j.1365-2958.2001.02714.x. [DOI] [PubMed] [Google Scholar]
- 10.Olbermann P, Josenhans C, Moodley Y, Uhr M, Stamer C, Vauterin M, Suerbaum S, Achtman M, Linz B. 2010. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet 6:e1001069. doi: 10.1371/journal.pgen.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tegtmeyer N, Wessler S, Backert S. 2011. Role of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J 278:1190–1202. doi: 10.1111/j.1742-4658.2011.08035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci U S A 93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer W. 2011. Assembly and molecular mode of action of the Helicobacter pylori cag type IV secretion apparatus. FEBS J 278:1203–1212. doi: 10.1111/j.1742-4658.2011.08036.x. [DOI] [PubMed] [Google Scholar]
- 14.Akopyants NS, Clifton SW, Kersulyte D, Crabtree JE, Youree BE, Reece CA, Bukanov NO, Drazek ES, Roe BA, Berg DE. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol 28:37–53. doi: 10.1046/j.1365-2958.1998.00770.x. [DOI] [PubMed] [Google Scholar]
- 15.Nešić D, Buti L, Lu X, Stebbins CE. 2014. Structure of the Helicobacter pylori CagA oncoprotein bound to the human tumor suppressor ASPP2. Proc Natl Acad Sci U S A 111:1562–1567. doi: 10.1073/pnas.1320631111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatakeyama M. 2014. Helicobacter pylori CagA and gastric cancer: a paradigm for hit-and-run carcinogenesis. Cell Host Microbe 15:306–316. doi: 10.1016/j.chom.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi T, Senda M, Morohashi H, Higashi H, Horio M, Kashiba Y, Nagase L, Sasaya D, Shimizu T, Venugopalan N, Kumeta H, Noda N, Inagaki F, Senda T, Hatakeyama M. 2012. Tertiary structure-function analysis reveals the pathogenic signaling potentiation mechanism of Helicobacter pylori oncogenic effector CagA. Cell Host Microbe 12:20–33. doi: 10.1016/j.chom.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan-Türköz B, Jiménez-Soto LF, Dian C, Ertl C, Remaut H, Louche A, Tosi T, Haas R, Terradot L. 2012. Structural insights into Helicobacter pylori oncoprotein CagA interaction with β1 integrin. Proc Natl Acad Sci U S A 109:14640–14645. doi: 10.1073/pnas.1206098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, Athman R, Mémet S, Huerre MR, Coyle AJ, DiStefano PS, Sansonetti PJ, Labigne A, Bertin J, Philpott DJ, Ferrero RL. 2004. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol 5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 20.Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fronzes R, Christie PJ, Waksman G. 2009. The structural biology of type IV secretion systems. Nat Rev Microbiol 7:703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallden K, Rivera-Calzada A, Waksman G. 2010. Type IV secretion systems: versatility and diversity in function. Cell Microbiol 12:1203–1212. doi: 10.1111/j.1462-5822.2010.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christie PJ, Cascales E. 2005. Structural and dynamic properties of bacterial type IV secretion systems. Mol Membr Biol 22:51–61. doi: 10.1080/09687860500063316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fronzes R, Schäfer E, Wang L, Saibil HR, Orlova EV, Waksman G. 2009. Structure of a type IV secretion system core complex. Science 323:266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. 2009. Structure of the outer membrane complex of a type IV secretion system. Nature 462:1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, Lu F, Redzej A, Fronzes R, Orlova EV, Waksman G. 2014. Structure of a type IV secretion system. Nature 508:550–553. doi: 10.1038/nature13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cendron L, Zanotti G. 2011. Structural and functional aspects of unique type IV secretory components in the Helicobacter pylori cag-pathogenicity island. FEBS J 278:1223–1231. doi: 10.1111/j.1742-4658.2011.08038.x. [DOI] [PubMed] [Google Scholar]
- 28.Terradot L, Waksman G. 2011. Architecture of the Helicobacter pylori cag-type IV secretion system. FEBS J 278:1213–1222. doi: 10.1111/j.1742-4658.2011.08037.x. [DOI] [PubMed] [Google Scholar]
- 29.Bourzac KM, Guillemin K. 2005. Helicobacter pylori-host cell interactions mediated by type IV secretion. Cell Microbiol 7:911–919. doi: 10.1111/j.1462-5822.2005.00541.x. [DOI] [PubMed] [Google Scholar]
- 30.Kutter S, Buhrdorf R, Haas J, Schneider-Brachert W, Haas R, Fischer W. 2008. Protein subassemblies of the Helicobacter pylori Cag type IV secretion system revealed by localization and interaction studies. J Bacteriol 190:2161–2171. doi: 10.1128/JB.01341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pattis I, Weiss E, Laugks R, Haas R, Fischer W. 2007. The Helicobacter pylori CagF protein is a type IV secretion chaperone-like molecule that binds close to the C-terminal secretion signal of the CagA effector protein. Microbiology 153:2896–2909. doi: 10.1099/mic.0.2007/007385-0. [DOI] [PubMed] [Google Scholar]
- 32.Bonsor DA, Weiss E, Iosub-Amir A, Reingewertz TH, Chen TW, Haas R, Friedler A, Fischer W, Sundberg EJ. 2013. Characterization of the translocation-competent complex between the Helicobacter pylori oncogenic protein CagA and the accessory protein CagF. J Biol Chem 288:32897–32909. doi: 10.1074/jbc.M113.507657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Couturier MR, Tasca E, Montecucco C, Stein M. 2006. Interaction with CagF is required for translocation of CagA into the host via the Helicobacter pylori type IV secretion system. Infect Immun 74:273–281. doi: 10.1128/IAI.74.1.273-281.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voss BJ, Gaddy JA, McDonald WH, Cover TL. 2014. Analysis of surface-exposed outer membrane proteins in Helicobacter pylori. J Bacteriol 196:2455–2471. doi: 10.1128/JB.01768-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein M, Rappuoli R, Covacci A. 2000. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc Natl Acad Sci U S A 97:1263–1268. doi: 10.1073/pnas.97.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson EM, Gaddy JA, Voss BJ, Hennig EE, Cover TL. 2014. Genes required for assembly of pili associated with the Helicobacter pylori cag type IV secretion system. Infect Immun 82:3457–3470. doi: 10.1128/IAI.01640-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Busler VJ, Torres VJ, McClain MS, Tirado O, Friedman DB, Cover TL. 2006. Protein-protein interactions among Helicobacter pylori cag proteins. J Bacteriol 188:4787–4800. doi: 10.1128/JB.00066-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinto-Santini DM, Salama NR. 2009. Cag3 is a novel essential component of the Helicobacter pylori Cag type IV secretion system outer membrane subcomplex. J Bacteriol 191:7343–7352. doi: 10.1128/JB.00946-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jurik A, Hausser E, Kutter S, Pattis I, Prassl S, Weiss E, Fischer W. 2010. The coupling protein Cagbeta and its interaction partner CagZ are required for type IV secretion of the Helicobacter pylori CagA protein. Infect Immun 78:5244–5251. doi: 10.1128/IAI.00796-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarkar MK, Husnain SI, Jakubowski SJ, Christie PJ. 2013. Isolation of bacterial type IV machine subassemblies. Methods Mol Biol 966:187–204. doi: 10.1007/978-1-62703-245-2_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kubori T, Koike M, Bui XT, Higaki S, Aizawa S-, Nagai H. 2014. Native structure of a type IV secretion system core complex essential for Legionella pathogenesis. Proc Natl Acad Sci U S A 111:11804–11809. doi: 10.1073/pnas.1404506111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohde M, Püls J, Buhrdorf R, Fischer W, Haas R. 2003. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol 49:219–234. doi: 10.1046/j.1365-2958.2003.03549.x. [DOI] [PubMed] [Google Scholar]
- 43.Shaffer CL, Gaddy JA, Loh JT, Johnson EM, Hill S, Hennig EE, McClain MS, McDonald WH, Cover TL. 2011. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. PLoS Pathog 7:e1002237. doi: 10.1371/journal.ppat.1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, Misselwitz R, Berger J, Sewald N, König W, Backert S. 2007. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature 449:862–866. doi: 10.1038/nature06187. [DOI] [PubMed] [Google Scholar]
- 45.Murata-Kamiya N, Kikuchi K, Hayashi T, Higashi H, Hatakeyama M. 2010. Helicobacter pylori exploits host membrane phosphatidylserine for delivery, localization, and pathophysiological action of the CagA oncoprotein. Cell Host Microbe 7:399–411. doi: 10.1016/j.chom.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Ohi M, Li Y, Cheng Y, Walz T. 2004. Negative staining and image classification—powerful tools in modern electron microscopy. Biol Proced Online 6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J. 2008. Spider image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc 3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Generation of H. pylori strains that produce HA-tagged CagF. (a) Schematic of plasmids encoding epitope-tagged forms of CagF (with either an N-terminal or C-terminal epitope HA tag). The sequences were introduced into the H. pylori ureAB chromosomal locus, yielding strains designated HA-CagF or CagF-HA, respectively. The HA-CagF and Cag-HA strains retained the endogenous cagF gene (encoding an untagged form of CagF). We also generated a ΔcagF/HA-CagF strain, which encodes only the tagged form of CagF. In this strain, the endogenous copy of cagF was deleted, and a gene encoding HA-CagF was introduced into the urease locus. (b) Immunoblotting of the indicated strains with an anti-HA antibody showed the production of HA-tagged forms of CagF. (c) AGS cells were infected with the wild-type (WT) strain or the indicated strains producing HA-tagged CagF, and IL-8 production was quantified by ELISA. AGS indicates results for cells without added H. pylori. Values represent means ± SD, based on analysis of at least six replicate samples. Levels of IL-8 production induced by mutant strains were compared to levels induced by the WT strain (ANOVA followed by Dunn’s multiple-comparison test). *, P < 0.001. (d) SDS-PAGE and silver stain analysis of preparations immunopurified from strains producing HA-tagged CagF (using a monoclonal HA antibody) and analysis of a control preparation from an untagged WT strain that was processed in parallel. Download
Analysis of Cag protein production in a panel of cag mutant strains. Immunoblotting of whole-cell lysates from mutant strains, each expressing HA-CagF from a gene in the urease locus, with the indicated antisera. HspB (a GroEL heat shock protein) was analyzed as a loading control. Strains are designated at the top of the figure, and antisera are designated to the right of the figure. Arrows indicate immunoreactive bands corresponding to Cag proteins, and other bands correspond to cross-reactive proteins. Two CagF bands correspond to the tagged and the untagged forms of the protein. Download
Negative-stain EM analysis of glycerol gradient fractions containing the cag T4SS core complex. Preparations resulting from immunoaffinity purification of HA-CagF were analyzed by velocity sedimentation in glycerol density gradients as shown in Fig. 2, and gradient fractions were analyzed by negative-stain EM. (a) Representative negative-stain EM images of fraction 1 (top of the gradient), which contains CagA and CagF but no core complex components. (b) Representative negative-stain EM images of fraction 6 (near bottom of the gradient), which contains cag core complex components. Fraction 1 contained no complexes as visualized by EM (a), while cag T4SS complexes were observed in images taken from fraction 6 (b). Scale bar, 25 nm. Download
Negative-stain 2D class averages of WT cag T4SS complexes and complexes purified from Δcag3 or ΔcagT mutant strains. (a) Reference-free alignment of 445 particles isolated from an HA-CagF-producing strain with an intact cag PAI into 10 classes. Five classes (marked with %, @, $, #, and & symbols) were chosen as references for an additional round of multireference alignment. (b) Result of reference-based alignment. (c) Reference-free alignment of 416 particles isolated from a Δcag3/HA-CagF-producing strain into 20 classes. Four classes (marked with %, @, $, and # symbols) were chosen as references for an additional round of multireference alignment. (d) Result of reference-based alignment. (e) Reference-free alignment of 149 particles isolated from a ΔcagT/HA-CagF-producing strain into 10 classes. Three classes (marked with %, @, and $ symbols) were chosen as references for an additional round of multireference alignment. (f) Result of reference-based alignment. Numbers of particles used for generating each average are shown in upper left-hand corner of the panels. Side length of panels, 63 nm. Download
Proteins that copurify with HA-CagF.
Proteins that copurify with HA-CagF when H. pylori ΔcagF/HA-CagF is cocultured with AGS cells.
Protein composition of fractions resulting from the sedimentation of an HA-CagF-immunopurified sample in a density gradient.
Supplemental Materials and Methods. Download