Abstract
A novel genomic island (LGI1) was discovered in Listeria monocytogenes isolates responsible for the deadliest listeriosis outbreak in Canada, in 2008. To investigate the functional role of LGI1, the outbreak strain 08-5578 was exposed to food chain-relevant stresses, and the expression of 16 LGI1 genes was measured. LGI1 genes with putative efflux (L. monocytogenes emrE [emrELm]), regulatory (lmo1851), and adhesion (sel1) functions were deleted, and the mutants were exposed to acid (HCl), cold (4°C), salt (10 to 20% NaCl), and quaternary ammonium-based sanitizers (QACs). Deletion of lmo1851 had no effect on the L. monocytogenes stress response, and deletion of sel1 did not influence Caco-2 and HeLa cell adherence/invasion, whereas deletion of emrE resulted in increased susceptibility to QACs (P < 0.05) but had no effect on the MICs of gentamicin, chloramphenicol, ciprofloxacin, erythromycin, tetracycline, acriflavine, and triclosan. In the presence of the QAC benzalkonium chloride (BAC; 5 μg/ml), 14/16 LGI1 genes were induced, and lmo1861 (putative repressor gene) was constitutively expressed at 4°C, 37°C, and 52°C and in the presence of UV exposure (0 to 30 min). Following 1 h of exposure to BAC (10 μg/ml), upregulation of emrE (49.6-fold), lmo1851 (2.3-fold), lmo1861 (82.4-fold), and sigB (4.1-fold) occurred. Reserpine visibly suppressed the growth of the ΔemrELm strain, indicating that QAC tolerance is due at least partially to efflux activity. These data suggest that a minimal function of LGI1 is to increase the tolerance of L. monocytogenes to QACs via emrELm. Since QACs are commonly used in the food industry, there is a concern that L. monocytogenes strains possessing emrE will have an increased ability to survive this stress and thus to persist in food processing environments.
INTRODUCTION
Listeria monocytogenes is a foodborne pathogen capable of causing severe human disease in at-risk populations. It is especially known for causing complications during pregnancy and severe infections in unborn fetuses, neonates, and elderly and immunocompromised individuals, for whom 20 to 40% of infections can lead to death (1). In addition to being widespread in the natural environment, L. monocytogenes is frequently associated with food processing environments and is most problematic in ready-to-eat (RTE) foods. Challenges in controlling these ubiquitous bacteria in RTE products and their processing environments are associated with the ability of L. monocytogenes to form biofilms, to grow at temperatures below refrigeration temperatures, to resist antimicrobial compounds, and to tolerate acidic and high-salt conditions (2–7). However, there is a large variation among strains when it comes to stress survival and the ability to adapt, grow, and persist in the food chain, as well as the potential to cause human illness (8, 9).
Studies have shown that point mutations and premature stop codons in inlA (10–15) and prfA (9, 15, 16) that lead to formation of truncated InlA and PrfA proteins and attenuated virulence occur in 35 to 45% of L. monocytogenes strains found in food processing environments. Similarly, L. monocytogenes isolates originating from human clinical isolates, foods, and processing environments differ in their relative capacity to adapt to cold temperatures, though precise mechanisms that result in different adaptation rates remain elusive (12, 17–19). Various degrees of tolerance and resistance of L. monocytogenes to antimicrobials have also been noted, due to modifications of the cell membrane that reduce permeability, the acquisition of genes conferring resistance, and/or the function of several efflux pumps (20–24).
In Canada, a specific clone has predominated in outbreaks and sporadic cases of listeriosis across the country for more than 2 decades (25). This clone belongs to serotype 1/2a, clonal complex 8 (CC8; includes sequence type 8 [ST8], ST120, ST232, ST289, ST292, ST387, and ST551), and more specifically, it includes single-locus variants of multilocus sequence type 120 and pulsed-field gel electrophoresis (PFGE) profile LMACI.0001/LMAAI.0001 (25). Examination of 1,061 L. monocytogenes isolates collected from 1995 to 2010 revealed the presence of this clone in 22.3% of isolates, with nationwide distribution believed to have occurred by the mid-1990s (25). This particular PFGE clone was linked to RTE deli meats implicated in the nationwide outbreak of listeriosis in 2008 (26). Isolates associated with this outbreak also possessed a previously unreported genomic island, LGI1 (26). Subsequent testing of the 71 human clinical L. monocytogenes isolates collected in Canada between 1988 and 2010 revealed the presence of LGI1 in 61% of isolates (25). Notably, all of the isolates possessing LGI1 belonged to CC8 and, with the exception of one serotype 3a isolate, were exclusively of serotype 1/2a (25). A small survey of retail RTE foods in the Vancouver metropolitan area (British Columbia, Canada) also found LGI1 in a serotype 1/2a and ST120 isolate recovered from an RTE fish product (27); however, it was absent from 58 L. monocytogenes serotype 1/2a isolates, including CC8 isolates, recovered from clinical samples in Switzerland (28). Presently, the worldwide prevalence of the island among L. monocytogenes isolates recovered from the food chain and from clinical samples and its role are not known.
The 50-kb LGI1 element carries a combination of putative antimicrobial resistance, stress response, and virulence genes, thereby possibly enhancing the capacity of L. monocytogenes to survive in the food chain and to cause human listeriosis (26). The presence of genes that are typically involved in stress responses, such as those encoding a two-component signal transduction system possessing a response regulator (locus 1851) and a sensor histidine kinase (locus 1852), a putative small multidrug-resistant (SMR) efflux pump (encoded by L. monocytogenes emrE[emrELm]; locus 1862), and a putative small RNA polymerase sigma-24 subunit (locus 1859), indicates that strains possessing LGI1 may be better equipped to survive environmental and/or food processing stresses (26). It is also tempting to speculate that the island contributes to virulence, considering that it was found in a number of clinical isolates examined over more than 2 decades. The presence of genes homologous to type IV secretion system genes (e.g., vriB4, virD4, cpa, and tad), as well as a putative adhesin gene (i.e., sel1), further supports this notion, albeit evidence of increased virulence due to LGI1 activity is currently lacking. In fact, the functions of genes located on LGI1 and their contributions to fitness and/or virulence of L. monocytogenes have not yet been reported. The purpose of the present study was to explore whether LGI1 contributes to increased tolerance of L. monocytogenes to food-related stresses. The L. monocytogenes 08-5578 strain, responsible for a nationwide listeriosis outbreak in Canada in 2008, was exposed to a variety of stresses (e.g., hot and cold temperatures, UV light, and sanitizers), and the expression of selected LGI1 genes was measured. The roles of specific LGI1 genes were investigated further by use of gene deletions (e.g., deletions of emrELm, encoding a putative efflux pump; lmo1851, encoding a response regulator [locus 1851]; and sel1, encoding a putative adhesin [locus 1861]) and the exposure of L. monocytogenes 08-5578 and its isogenic mutants to diverse stresses encountered in the food chain (e.g., acid, cold, and saline conditions). The contribution of sel1 to adherence to human cell lines (Caco-2 and HeLa) was also examined in further defining the mechanism of virulence.
MATERIALS AND METHODS
Bacterial isolates and media.
Listeria monocytogenes strains and plasmids used in this study are listed in Table 1. All LGI1 deletion mutants were generated in L. monocytogenes 08-5578, a clinical strain responsible for the Canadian deli meat listeriosis outbreak in 2008 (26). This strain does not possess genes encoding previously described SMR efflux pump systems in L. monocytogenes (e.g., bcrABC and qacH) (26). Additionally, when phenotypic changes in mutants were observed, other strains possessing LGI1 (CC8+ LGI1+; n = 8) (25), strains from CC8 that do not possess LGI1 (n = 4), and strains belonging to serotype 1/2a but not to CC8 and not possessing LGI1 (n = 2) were exposed to identical conditions to investigate whether the same growth behavior was seen across unrelated strains possessing LGI1 (Table 1). Listeria monocytogenes EGD-SmR (29) and 81-0861 (25) were used as LGI1 serotype 1/2a and 4b negative controls, respectively.
TABLE 1.
Bacterial strains and plasmids used in the present study
| Strain or plasmid | Descriptionc | Presence of LGI1a | Reference or source |
|---|---|---|---|
| Strains used for mutant constructs | |||
| L. monocytogenes strains | |||
| 08-5578 | Wild-type clinical strain (Ontario); serotype 1/2a, ST120, CC8 | + | 26 |
| 08-5578 Δlmo1851 | 08-5578 with 408-bp in-frame deletion within the lmo1851 gene | Δlmo1851 | This study |
| 08-5578 ΔemrE | 08-5578 with 240-bp in-frame deletion within the emrE gene | ΔemrE | This study |
| 08-5578 Δsel1 | 08-5578 with 2,598-bp in-frame deletion within the sel1 gene | Δsel1 | This study |
| E. coli strains | |||
| DH5α | DH5α with pKSV7 | − | This study |
| One Shot TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | − | Invitrogen |
| TOP10 Δlmo1851 | TOP10 carrying pKSV7 with 08-5578 lmo1851 insert with deletion | Δlmo1851 | This study |
| TOP10 ΔemrE | TOP10 carrying pKSV7 with 08-5578 emrE insert with deletion | ΔemrE | This study |
| TOP10 Δsel1 | TOP10 carrying pKSV7 with 08-5578 sel1 insert with deletion | Δsel1 | This study |
| Plasmids used for mutant constructs | |||
| pKSV7 | Gram-positive and Gram-negative temperature-sensitive shuttle vector; CHLr (L. monocytogenes) AMPr (E. coli); multiple-cloning sites; lacZ β-laccat pE194tsb | 37 | |
| pKSV7:ΔemrE | pKSV7 with 08-5578 emrE insert with deletion | This study | |
| pKSV7:Δlmo1851 | pKSV7 with 08-5578 lmo1851 insert with deletion | This study | |
| pKSV7:Δsel1 | pKSV7 with 08-5578 sel1 insert with deletion | This study | |
| Control strains | |||
| EGD-SmR | EGD derivative resistant to streptomycin; serotype 1/2a, non-CC8 | − | 29 |
| 81-0861 | Isolated from coleslaw (Nova Scotia); serotype 4b, ST1, CC1 | − | 25 |
| Other L. monocytogenes strains used in stress experiments | |||
| 01-1465 | Clinical human blood isolate (Ontario); serotype 1/2a, ST120, CC8 | + | 25 |
| 01-2417 | Clinical human blood isolate (British Columbia); serotype 1/2a, ST120, CC8 | + | 25 |
| 01-7210 | Liverwurst sausage isolate (British Columbia); serotype 1/2a, ST120, CC8 | + | 25 |
| 02-4056 | Clinical human blood isolate (Ontario); serotype 1/2a, ST311, CC155 | − | 25 |
| 03-0402 | Clinical human blood isolate (Alberta); serotype 1/2a, ST120, CC8 | + | 25 |
| 06-6956 | Clinical human blood isolate (Quebec); serotype 1/2a, ST37, CC37 | − | 25 |
| 08-6040 | RTE meat isolate (Ontario); serotype 1/2a, ST120, CC8 | + | 25 |
| 95-0093 | Clinical human blood isolate (Alberta); serotype 1/2a, ST120, CC8 | + | 25 |
| 95-0151 | Clinical human blood isolate (Ontario); serotype 1/2a, ST120, CC8 | + | 25 |
| 99-3046 | Clinical human blood isolate (Ontario); serotype 1/2a, ST120, CC8 | − | 25 |
| 01-5373 | Clinical human blood isolate (Ontario); serotype 1/2a, ST120, CC8 | − | 25 |
| 03-5833 | Clinical human blood isolate (Alberta); serotype 1/2a, ST120, CC8 | − | This study |
| 08-5375 | Clinical human blood isolate (Ontario); serotype 1/2a, ST120, CC8 | − | 25 |
| LR39-1 | RTE fish isolate (British Columbia); serotype 1/2a, ST120, CC8 | + | 27 |
Presence (+) or absence (−) of Listeria genomic island 1 (LGI1).
Thermosensitive replication origin of plasmid pE194.
CC8, clonal complex 8; based on multilocus sequence typing.
Strains and transformants were stored long-term at −80°C in tryptic soy broth (TSB; Acumedia, Neogen, Lansing, MI) supplemented with 20% (wt/vol) glycerol (L. monocytogenes) or in Luria-Bertani (LB; Difco, Becton Dickinson, Sparks, MD) broth with 20% glycerol (Escherichia coli). Prior to use, the strains were streaked from frozen stocks onto tryptic soy agar (TSA; Acumedia) supplemented with 0.6% yeast extract (YE; Thermo Fisher Scientific, Ottawa, ON, Canada) or onto LB agar, followed by 24 h of incubation at 37°C. With the exception of specific sanitizer stress survival studies, which were performed in TSB, brain heart infusion broth (BHI; Difco) was used to grow strains prior to stress experiments. Specific conditions are described below for each stress treatment. Recovery of survivors following exposure to stress conditions was performed on TSA-YE, with incubation at 37°C for 24 to 48 h.
Antimicrobial agents and MIC determinations.
Antimicrobial agents used in the study included the following: (i) the antibiotics chloramphenicol (CHL; 2.5, 5, 10, 15, and 20 μg/ml), ciprofloxacin (CIP; 2.5, 5, 10, 15, and 20 μg/ml), erythromycin (ERY; 1, 2.5, 5, 10, and 15 μg/ml), gentamicin (GEN; 1, 2.5, 5, and 10 μg/ml), and tetracycline (TET; 2.5, 5, 10, 15, and 30 μg/ml); (ii) quaternary ammonium compounds (QACs), such as 10% E-San (2.5, 5, 10, 15, and 20 μl/ml), a sanitizer containing 5% N-alkyl dimethyl benzyl ammonium chloride (Epsilon Chemicals Ltd., Edmonton, AB, Canada), and a benzalkonium chloride (BAC; 5, 10, 20, 25, and 30 μg/ml) with alkyl distribution from C8H17 to C16H33 (Acros Organics, NJ); (iii) acriflavine (12, 16, 20, and 24 μg/ml; Sigma-Aldrich Canada Co., Oakville, ON, Canada), a cationic dye used in enrichment media during isolation of Listeria spp.; and (iv) triclosan (Irgasan; 5-chloro-2-[2,4-dichlorophenoxy] phenol) (1, 2, 4, and 8 μg/ml; Sigma-Aldrich), a broad-spectrum antimicrobial agent that inhibits enoyl acyl carrier protein reductase in fatty acid synthesis (30). Antibiotic stock solutions were prepared according to the manufacturer's instructions and were stored at −20°C for up to 2 months. Other antimicrobial agents were stored according to the manufacturer's recommendations (i.e., at 4°C or at room temperature). Working solutions of water-soluble agents (1,000 μg/ml) were prepared by diluting the concentrated sanitizers in sterile deionized water on the day of the experiment, and these solutions were stored at 4°C and used within 3 h of preparation. CIP was dissolved in dimethyl sulfoxide (DMSO) and sterile deionized water (1:9 [vol/vol]), whereas triclosan was dissolved in 70% ethanol. A working solution of reserpine (10,000 μg/ml; Sigma-Aldrich), an efflux inhibitor (31), was prepared in DMSO and added to bacterial cultures to a final concentration of 20 μg/ml. The highest volumes of DMSO used for dissolving CIP and reserpine and of 70% ethanol used for dissolving triclosan were applied as controls to check for a diluent effect.
MICs were determined using a slightly modified agar dilution method (e.g., for antibiotics and QACs) described by Elhanafi et al. (23) and a broth microdilution protocol (e.g., for acriflavine and triclosan) described by Kovacevic et al. (32). Briefly, for the agar dilution method, strains were grown on Mueller-Hinton agar (MHA-B) (1.2% agar; Difco) supplemented with 5% defibrinated sheep blood (Alere Inc., Ottawa, ON, Canada) and incubated at 37°C overnight. Two colonies were transferred to 200 μl of Mueller-Hinton broth (MHB; Difco), and 5 μl of suspension was spotted in duplicate onto MHA-B plates containing appropriate concentrations of antimicrobials/other compounds. Following 48 h of incubation at 30°C, MICs were determined as the lowest assessed concentrations that prevented confluent growth. For the broth microdilution method, plates were incubated at 30°C in a SpectraMax M2 plate reader (SoftMax Pro 6.3 software; Molecular Devices, Sunnyvale, CA), with the optical density at 600 nm (OD600) measured at 30-min intervals for 24 h. All MIC experiments were performed at least three times.
Growth in the presence of sublethal concentrations of antimicrobials.
Growth of L. monocytogenes in the presence of sublethal concentrations of E-San (0.8 and 1.6 μl/ml) and BAC (1 and 2 μg/ml), with or without reserpine (20 μg/ml), was assessed using a SpectraMax plate reader (Molecular Devices, Sunnyvale, CA) at 30°C. Briefly, single colonies were inoculated into 5 ml of TSB and incubated at 30°C with shaking (200 rpm). Following 16 h of incubation, cultures were diluted 1:100 in TSB containing appropriate concentrations of test compounds. Aliquots (200 μl) for each strain and treatment were transferred in duplicate into a 96-well plate. The OD600 levels were monitored at 30-min intervals for 24 h. The OD600 data were fitted to growth curves to obtain the lag-phase duration (LPD), maximum growth rate (MGR), and maximum density, using the DMFit 3.0 Excel add-in program (ComBase; Computational Microbiology Research Group, Institute of Food Research, Colney, Norwich, United Kingdom), based on the models of Baranyi and Roberts (33). Experiments were performed at least three times. With each run, blank controls containing TSB only or TSB with appropriate concentrations of the tested antimicrobial were included, and their values were subtracted from those for the strains containing the respective treatments. The correspondence between the OD600 values and viable cell counts was examined by plating onto TSA-YE at seven time points (i.e., 0, 1, 3, 5.5, 8, 10, and 24 h) representing the early logarithmic, late logarithmic, and late stationary growth phases at 30°C.
Acid stress survival.
Survival of L. monocytogenes 08-5578 and its LGI1 mutant derivatives under acidic conditions was assessed according to protocols described by Ells and Truelstrup Hansen (34) and Oliver et al. (35). Briefly, BHI (50 ml) was adjusted to pHs of 4.5, 3.5, and 2.5 (Pinnacle pH meter; Nova Analytics Corporation) with 6 N HCl, and 9.9-ml samples were distributed into sterile tubes (i.e., five for each pH). Overnight cultures (10 μl) grown in BHI at 30°C (with shaking at 200 rpm) were inoculated into 5 ml of fresh BHI. Following incubation for 16 h at 30°C, 2 ml of culture was collected, centrifuged at 6,000 × g for 10 min at room temperature, washed twice in 0.1% peptone water (2 ml), resuspended in 2 ml of 0.1% peptone water, and added to pH-adjusted BHI to obtain 107 to 108 CFU/ml (counts were confirmed by enumeration on TSA-YE plates). Cultures were vortexed and incubated at 30°C with shaking (200 rpm). A 100-μl aliquot was removed immediately (t = 0) and at 1, 2, 4, 6, 8, 10, and 24 h, diluted in buffered peptone water (BPW; Acumedia), and spread onto TSA-YE plates, in duplicate. Plates were incubated at 37°C, and counts were recorded after 24 h. Experiments were repeated three times.
Cold growth evaluation.
Cold growth adaptation of L. monocytogenes and its LGI1 mutant derivatives was evaluated according to the protocol described by Kovacevic et al. (12).
Salt stress survival.
To assess the growth and survival of the isolates at different salt concentrations, L. monocytogenes 08-5578 and its LGI1 mutant derivative strains were exposed to NaCl solutions (5 to 20% [wt/vol]; Fisher Scientific) according to the protocol described by Ells and Truelstrup Hansen (34), with slight modifications. Briefly, 10-μl aliquots of overnight cultures grown in BHI at 30°C with shaking (200 rpm) were inoculated into 5 ml of fresh BHI. Following incubation for 16 h at 30°C (with shaking at 200 rpm), cultures were centrifuged (3,000 × g, 10 min) at room temperature, washed twice in 0.1% peptone water, and diluted to 107 to 108 CFU/ml in BHI containing different concentrations of NaCl. Aliquots (200 μl) for each strain and treatment were transferred in duplicate into a 96-well plate. OD600 levels were monitored in a SpectraMax plate reader (Molecular Devices) at 30°C at 30-min intervals for 24 h. In parallel, cultures exposed to 10 and 20% NaCl were vortexed and incubated at 30°C with shaking (200 rpm). A 100-μl aliquot was removed immediately (t = 0) and after 1, 2, 4, 6, 8, 10, and 24 h and then serially diluted in BPW and spread onto TSA-YE plates, in duplicate. Plates were incubated at 37°C, and counts were recorded after 24 h. Experiments were repeated three times.
Construction and selection of deletion mutants.
Nonpolar deletion mutants of putative efflux pump (emrE homolog; locus tag LM5578_RS09350 replaced the old locus tag LM5578_1862), two-component regulator (lmo1851; LM5578_1851), and adhesin (sel1; LM5578_1866) genes were generated in L. monocytogenes 08-5578 (GenBank accession number CP001602) by using the allelic exchange protocol described by Camilli et al. (36). A list of oligonucleotide primers, thermocycling conditions, and restriction endonucleases used is provided in Table 2. Pfu Turbo CX DNA polymerase (2.5 U) (Agilent Technologies Inc., Mississauga, ON, Canada) was used according to the manufacturer's instructions for all PCRs, with 0.4 μM (each) oligonucleotide primers and with L. monocytogenes 08-5578 genomic DNA, isolated by use of a DNeasy blood and tissue kit (Qiagen), as the template. PCR fragments (e.g., SOE-AB and SOE-CD) were purified using a QIAquick PCR purification kit (Qiagen, Toronto, ON, Canada) and subsequently used as templates in a PCR with SOE-A and SOE-D primers. The resulting SOE-AD PCR product was electrophoresed (Bio-Rad horizontal electrophoresis system) in a 1% agarose gel (Fisher Scientific), and ethidium bromide-stained bands were visualized using Image Master VSD (Amersham Pharmacia Biotech, Uppsala, Sweden) to confirm the presence of a single band of the appropriate size. When more than one band was present, the band of the appropriate size was cut out from the agarose gel and further purified using a QIAquick gel extraction kit (Qiagen). The SOE-AD PCR product and the suicide shuttle vector pKSV7 (Cornell University, Ithaca, NY) (37) were purified by use of a QIAquick PCR purification kit (Qiagen), digested with appropriate endonucleases (FastDigest; Fisher Scientific) (Table 2), and confirmed by running in a 1% agarose gel stained with ethidium bromide (Image Master VSD). Once confirmed, products were purified once more, and the SOE-AD PCR product was ligated (T4 ligase; Thermo Scientific) into pKSV7. The vector containing the gene of interest was first inserted into One Shot TOP10 chemically competent E. coli cells (Invitrogen, Carlsbad, CA) via electroporation (11 kV/cm; 5-ms time constant) (Bio-Rad Gene Pulser; Bio-Rad, Hercules, CA) and subsequently electroporated into L. monocytogenes (11 kV/cm; 5-ms time constant). Escherichia coli transformants were selected on LB agar plates containing 100 μg/ml of ampicillin (AMP100; Sigma-Aldrich). Plasmids were obtained from E. coli by using a GeneJET plasmid miniprep kit (Thermo Scientific), sequenced at the Nucleic Acid Protein Service Unit (NAPS) of the University of British Columbia with NAPS-prepared primers (−21M13 and M13R) to confirm the absence of nucleotide deletions and polymorphisms, and subsequently electroporated into 100 μl of competent L. monocytogenes 08-5578 cells. Following passaging and screening for vector excision, allelic exchange was confirmed by PCR amplification with SOE-A and SOE-D primers. Mutants were sequenced at NAPS (University of British Columbia) by use of SOE-A and SOE-D primers (lmo1851) or XF and XR primers (emrELm and sel1).
TABLE 2.
Oligonucleotide primers used for mutant constructiona
| Gene target and primer name | PCR conditions | Oligonucleotide sequence (5′-3′)b | Product size (bp) (primers used) | Digestion enzyme |
|---|---|---|---|---|
| emrE | 95°C for 2 min; 30 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 1.7 min; 72°C for 10 min | 1,588 (AD) | ||
| SOE-A | CCC CTG CAG AGA CCC TCG GCT TTG CGT CC | 881 (AB) | PstI | |
| SOE-B | GCA GGG GTT GTA GGC CTG AAC | |||
| SOE-C | GTT CAG GCC TAC AAC CCC TGC AAG TTC AAG TAC GAT AGC GAC | 707 (CD) | ||
| SOE-D | CCC GGT ACC GAT GGC GTG AAA ACG GCG GC | KpnI | ||
| emrE-XF | GCC ACA AAA GGG CAG GTT | |||
| emrE-XR | TAA AGC TCT CCC GCA GTA CC | |||
| lmo1851 | 95°C for 2 min; 30 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 1.3 min; 72°C for 2 min | 1,083 (AD) | ||
| SOE-A | CCC CTG CAG ATC CAT TAG AGC ATC AAT TTG | 537 (AB) | PstI | |
| SOE-B | TTA CTA AAA GAA ATC AGT TCT | |||
| SOE-C | AGA ACT GAT TTC TTT TAG TAA ATT AGC CAC TTC ATC TTC TAT | 546 (CD) | ||
| SOE-D | CCC GGT ACC CAT TAT AGC AAC TTG ATT GTG | KpnI | ||
| sel1 | 95°C for 2 min; 35 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 1.5 min; 72°C for 10 min | 3,603 (AD) | ||
| SOE-A | GG TCT AGA GCT GCT TGA TGA GGT ATG C | 501 (AB) | XbaI | |
| SOE-B | GCA TTC CAC ATT GAC CGC | |||
| SOE-C | GCG GTC AAT GTG GAA TGC CGG TAA CAG TAG CTT GCT ATC ATC | 504 (CD) | ||
| SOE-D | GG GGT ACC ACA TGA GCC TAT CAG AAT TAA CCC | KpnI | ||
| sel1-XF | CAT CTA CAC CGA CAA ATA CCG CA | |||
| sel1-XR | GCA ATC TTG TGC GAG TCT TTC |
All primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA).
Endonuclease restriction sites are underlined, and regions complementary to SOE-B primers are italicized.
Preparation of L. monocytogenes competent cells.
Listeria monocytogenes 08-5578 cells were first grown overnight in BHI at 37°C with shaking (200 rpm). Fresh BHI (50 ml) was inoculated with the overnight culture (500 μl) and subsequently incubated at 37°C with shaking until the culture reached an OD600 of 0.2 (iMark! microplate absorbance reader; Bio-Rad, Hercules, CA). Penicillin G (50 μl of a 100-mg/ml stock; Sigma-Aldrich, Oakville, ON, Canada) was added to the culture, and cells were incubated for an additional 2 h at 37°C with shaking. Following incubation, the culture was chilled on ice for 15 min, transferred to four centrifuge tubes, and centrifuged (5,939 × g; Eppendorf 5415 R centrifuge) for 1 min. The supernatant was discarded, and 1.2 ml of HEPES (1 mM; Sigma-Aldrich) with sucrose (0.5 M; Sigma-Aldrich) and glycerol (10% [wt/vol]; Fisher Scientific) was added to each tube. The contents of the tubes were mixed by gentle pipetting up and down five times. Tubes were centrifuged (5,939 × g) and supernatants discarded. Pellets were rehydrated with 100 μl of HEPES-sucrose-glycerol solution and either used immediately following preparation or stored at −80°C until use.
RNA isolation and cDNA preparation.
Total RNAs were recovered from cultures under the various stress conditions tested, including cold, heat, UV, and BAC stress treatments and normal laboratory conditions as a control treatment, for TaqMan-based semiquantitative real-time PCR (sqRT-PCR) assay. RNA was isolated from each sample by using an RNeasy minikit with RNAprotect reagent (Qiagen) according to the manufacturer's instructions. Recovered RNA was treated with Ambion Turbo DNA-free DNase (Ambion Inc., Austin, TX) according to the manufacturer's double-digestion protocol to eliminate any genomic DNA contamination. Samples were immediately placed at −80°C or converted to cDNA by using the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen), following the manufacturer's instructions. The same amount of total RNA of each sample was also subjected to the cDNA synthesis reaction without the reverse transcriptase enzyme. This provided control samples to assess the potential residual DNA contamination of each sample.
For quantitative real-time PCR (qRT-PCR) experiments to examine the expression levels of selected genes during BAC exposure, L. monocytogenes 08-5578 was grown in 25 ml of TSB at 30°C with shaking (200 rpm). Following 14 h of incubation, 5 ml was used as a control (no treatment), whereas BAC was added to another 5 ml of culture, to a final concentration of 10 μg/ml. Following 1 h of incubation at 30°C with shaking, cultures were used directly for RNA extraction, using an RNA PowerSoil total RNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA). Recovered RNA samples were treated with RTS DNase (Mo Bio) and immediately placed at −80°C or converted to cDNA by using a QuantiTect reverse transcription kit (Qiagen, Toronto, ON, Canada). RNAs were quantified and checked for quality by spectrophotometry (NanoDrop ND1000; Thermo Scientific, Toronto, ON, Canada) and gel electrophoresis.
Exposure to food chain-relevant stresses to assess LGI1 gene expression.
Listeria monocytogenes 08-5578 was exposed to various stress conditions representative of those encountered in the food chain. Briefly, single colonies grown overnight at 37°C on TSA-YE plates were inoculated into BHI. Cultures were incubated at 37°C with shaking (250 rpm) for 18 h to reach the stationary growth phase, and total RNAs were isolated to represent the normal laboratory growth conditions. In the cold stress model, the stationary-phase cultures were pelleted by centrifugation (5,939 × g) and resuspended in fresh BHI. Each culture was further subdivided into two aliquots, which were incubated at 4°C and 35°C for 4 h, and total RNAs were then extracted. To assess expression in the heat stress model, the stationary-phase cultures were pelleted by centrifugation (5,939 × g), resuspended in fresh BHI, and subdivided into two aliquots, which were incubated for 4 h at 52°C and 35°C. Following individual treatments, total RNAs were isolated from the cultures. To assess gene expression following DNA damage with UV light, the stationary-phase cultures were exposed to UV light for 0, 0.17, 1, 5, 10, 15, and 30 min, and RNA was extracted at each time point. Cultures were also exposed to QACs by growing cells in BHI containing 0, 1, 5, and 10 μg/ml BAC at 37°C for 18 h, followed by RNA extraction. All experiments were performed in duplicate. Following each stress treatment, the expression of 16 genes (Table 3) within LGI1, representing various functional units and operons within LGI1, was measured using TaqMan sqRT-PCR. Reactions were performed using ABI TaqMan universal PCR master mix (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions, with 0.4 μM (each) oligonucleotide primers (Table 3), 0.2 μM TaqMan probe, and 5 μl cDNA template per 25-μl reaction mixture. Thermocycling conditions included denaturation at 95°C for 8 min and 35 cycles of 95°C for 10 s and 60°C for 30 s, using a Cepheid SmartCycler (Cepheid, Sunnyvale, CA). Two housekeeping genes, an rRNA gene and bglA, were used to ensure standardization of the cDNA samples based upon the crossing point (CP) values as computed by the SmartCycler software (Cepheid) (38). The cDNA was diluted if necessary to obtain standardized concentrations. LGI1 genes were determined to be expressed based on the CP values computed by the SmartCycler software (Cepheid).
TABLE 3.
Oligonucleotides used for TaqMan sqRT-PCR LGI1 gene expression experiments
| 08-5578 LGI1 target gene | Sequence (5′–3′) |
Length of PCR product (bp) | ||
|---|---|---|---|---|
| Forward primer | TaqMan probe | Reverse primer | ||
| bglAa | AGCTGCTGCTGCAAACCAAT | CGAAGGCGCTTACAACGTCGATGG | AGTAACATCTTGAACGGAAAGTCCTT | 73 |
| rRNA genea | CTTCCGCAATGGACGAAAGT | TGACGGAGCAACGCCGCGT | TTACGATCCGAAAACCTTCTTCA | 66 |
| lmo1901 | TGATCCGCCGTATTACGAAAC | AGGGTCACTACGCTGTGGTGTTCCGA | AAGCCGTGCATGATCTTCCT | 70 |
| lmo1891 | ACGGCAGGCAGTGGTTATG | TGCCAACACAGGTGAGCAGGAAGA | AAGGCAGTTCCTTTTCCTCAGA | 67 |
| lmo1889 | GGCGGGAAAACCGAGAAG | AACACTTTGACAGCCCAGAACACGCC | TGCGGCGTACCACATTGA | 70 |
| lmo1885 | CGGCGATGTGGTGGAATAC | TGTCGGGCTTGGAGGTAACAGTT | GAGCCAGCGGAGCATTTC | 65 |
| lmo1883 | TTCACCTCACTATGCTCCACTACTG | CTATGCGGTTGGTGACGGGAATGTAGA | CATTCCACCACCGCCATT | 72 |
| lmo1882 | CCGCTTTTGCAGCAGGAA | AGGCGATGTTGCCGGAGCGATT | GGGATGCAGTCGTCCATGTC | 66 |
| lmo1876 | CACTTACCGCTGCGTTTGG | CGCTCCGAGAATACCCGACTGCTACA | TGTCGAAGAATTGCCTGCTCTT | 69 |
| lmo1873 | ACAGCATTCGTATGAAAGCCATT | TTTACGCTTTGTTTTTCGGCACGGAA | TCGTCAGCGTTCATTTTGAGA | 73 |
| lmo1866 | CACGTCATTAGTCTTCGCAGAGA | ATGCTGCCCGCCTTGGATATGACA | ATTAAATCCTGCCATTGCTTTCC | 74 |
| lmo1865 | AGTTCCTATCCCTGCGGAAGA | CATCCACAGCTTATGCTTCGGGATGAA | TTGGGATACTTGCCCGCATA | 70 |
| lmo1864 | ACCGGGAGATGCTGACAATT | CTGATGAATACGGTGGCAAGCTGCC | CTGCGAAAATCATCACACGATT | 69 |
| lmo1863 | TCTCCCGCAGTACCGTGAA | CGTGCCATTGCTGACCTTGAA | CCTTGGAGAGATGGCCAGTCT | 63 |
| lmo1862 | AAATGTCTGATGGACTTACAAAGCTATT | CCAAGTGCAGGGATGTTCATAGCATTTATCCTA | GCACTAACCGGTATTTTTTTTAATGC | 109 |
| lmo1861 | GGAAATAGAAGCTTTCGCCAGTAT | CGCAGGAACAACGTAGGGCTATATCCG | GGAACTCCCGAGAAAAGAAAATG | 79 |
| lmo1859 | AAGAGCGCGAAGCTGAAAGATA | AAGGAAGTGCATTCATCAATTTGAGCTTTCC | CCTCATCTTGGAATCGTTCCA | 78 |
| lmo1857 | CACTGCCGCCAAGAAAAAAA | CCAGCTTCTTGACAGTAATGGTTTTGACCGA | GCAACTCTGTCCGAGCAATTT | 77 |
Housekeeping gene not carried on LGI1. The relevant primers were described by Tasara and Stephan (38).
Exposure to benzalkonium chloride to assess LGI1 gene expression.
qRT-PCR assays to examine transcript levels of two known efflux pump genes in Listeria (lde and mdrL), sigB, and LGI1-carried emrE, lmo1851, and lmo1861 were performed following 1 h of exposure of L. monocytogenes 08-5578 to BAC (10 μg/ml) at 30°C. A list of primers used and their relative efficiencies are provided in Table 4. Primers were designed using Geneious 5.4 software (Biomatters Ltd., Auckland, New Zealand) and were optimized to achieve specific target gene amplification (a product with a single melting peak) and PCR efficiencies between 97 and 105% (Table 4). cDNA templates derived from L. monocytogenes 08-5578 treated with BAC (10 μg/ml) for 30 min at 30°C were used for PCR optimization and amplification efficiency evaluation. Reactions were carried out in a final reaction volume of 20 μl containing 1 μl cDNA template, 10 μl SsoAdvanced SYBR Green supermix (Bio-Rad), and 0.25 μM (each) forward and reverse oligonucleotides (Table 4). Thermocycling conditions included initial denaturation at 95°C for 3 min followed by 39 cycles of 95°C for 10 s, 56°C for 5 s, and 72°C for 12 s, melting curve measurement (65 to 95°C in 0.5°C increments for 5 s), and cooling (4°C), using a CFX96 Touch real-time PCR detection system (Bio-Rad). Target gene transcript levels were quantified using CFX Manager 3.1 software (Bio-Rad). Relative changes in expression levels for genes of interest relative to those of the control grown in TSB without sanitizer treatment were normalized against the housekeeping 16S rRNA gene, encoding the RNA component of the smaller subunit of the bacterial ribosome (38). Cycle threshold standard deviations (SD) for all genes were ≤0.3. Gene expression fold changes reported here represent the means and SD for three independent assays with each sample run in duplicate.
TABLE 4.
Oligonucleotides used for qRT-PCR experimentsa
| Primer | Oligonucleotide sequence (5′-3′) | Product size (bp) | Primer efficiency (%) |
|---|---|---|---|
| 16S rRNA-Fb | TTA GCT AGT TGG TAG GGT | 318 | 91.3 |
| 16S rRNA-Rb | AAT CCG GAC AAC GCT TGC | ||
| emrE-JKq-F | GTT GCT ATA GCG GTG ATT GGA GT | 102 | 104.3 |
| emrE-JKq-R | GTT CAG GCC TAC AAC CCC TG | ||
| lde-JKq-F | TCC CAA TGG CTT TCG CAC AA | 136 | 99.4 |
| lde-JKq-R | ATT CGA CCT GCA ACC TCA CG | ||
| lmo1861-JKq-F | GCT TAC AGA AGA AGG AGC GCA | 101 | 99.6 |
| lmo1861-JKq-R | CCC TAC GTT GTT CCT GCG G | ||
| mdrL-JK2q-F | TCG AGC TGG TTG GGG TTT TG | 96 | 97.1 |
| mdrL-JK2q-R | ATC CCA ATT GCA TGG CCT GG | ||
| sigBq-Fc | TGT TGG TGG TAC GGA TG | 221 | 100 |
| sigBq-Rc | CAT TCT GCA ACG CCT C |
Adhesion and invasion assays.
The adhesion and invasion efficiencies of L. monocytogenes 08-5578 (wild type [WT]) and its Δsel1 mutant were assessed according to the protocol described by Kovacevic et al. (12), using the TC-7 subclone of Caco-2 cells (39) and HeLa (ATCC CCL-2) cells (1 × 105 cells per well; passages 5 to 20). Bacterial cultures grown statically overnight in BHI (5 ml) at 30°C were pelleted by centrifugation (5,939 × g at 22°C; Eppendorf 5415 R centrifuge), washed once, resuspended in 1× Dulbecco's phosphate-buffered saline (DPBS; HyClone) with magnesium and calcium, and adjusted to an OD600 of 0.5 (Genesys 10UV system; Thermo Spectronic, Rochester, NY). Prior to infection, bacterial cultures were diluted in Dulbecco's modified Eagle's medium (DMEM) to approximately 5 × 106 CFU/ml as assessed by plating on TSA-YE plates. Bacterial suspensions (0.5 ml) were added to Caco-2 and HeLa cells and incubated at 37°C for 30 min and 1 h to allow bacterial adherence and entry, respectively. Infected cells were washed three times with DPBS and then lysed with sterile ice-cold water for 10 min at 37°C (adhesion) or overlaid with fresh prewarmed DMEM containing gentamicin (50 mg/liter) and incubated at 37°C for 45 min (invasion). Following gentamicin treatment, the cell monolayers were washed three times with DPBS and lysed with sterile ice-cold water for 10 min at 37°C. The number of viable bacteria was quantified by spreading a direct inoculum of lysed cells and serial dilutions (10−1 to 10−4) in DPBS onto TSA-YE plates, which were incubated at 37°C for 24 to 48 h. Adhesion was reported as the average log10 CFU per milliliter, where the starting inoculum and recovered cells for each strain were normalized to those of the 08-5578 strain. The invasion efficiency was reported as the percentage of the inoculum recovered by enumeration of intracellular bacteria, normalized to the value for the 08-5578 strain (set at 100%). Listeria monocytogenes BUG5 (Tn1545-induced inlA mutant of EGD-SmR) (29) and 10403S were used as other serotype 1/2a controls. Assays for each isolate were carried out in duplicate and repeated at least three times.
Statistical analysis.
Data analysis was performed using GraphPad Prism 6.0 software (GraphPad Software, Inc., La Jolla, CA). One-way analysis of variance (ANOVA) with Dunnett's multiple-comparison test was used to compare maximum OD600 values for parent and mutant strains exposed to salt stress, as well as adhesion and invasion efficiencies among strains. The unpaired two-tailed t test was used to compare LPD, MGR, and maximum OD600 values between the parent strain and its ΔemrE mutant. The Mann-Whitney test was used to compare LPD, MGR, and maximum OD600 values between strains possessing LGI1 and those without LGI1. For all analyses, differences were considered significant if the P values were <0.05.
RESULTS
LGI1 and emrE genetic content.
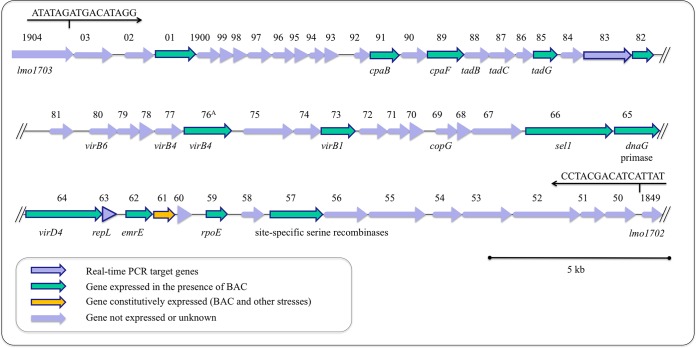
LGI1 is a large, 49.8-kbp contiguous region (39.9% GC content) first described by Gilmour et al. (26) for two Canadian clinical isolates (08-5578 and 08-5923). In L. monocytogenes 08-5578 (accession number CP001602.2), it is inserted at coordinates 1836435 to 1886209 and includes 54 coding sequences (LM5578_1850 to LM5578_1903) oriented in the same direction. The bordering coding sequences LM5578_1849 and LM5578_1904 were 98% identical to contiguous EGDe coding sequences lmo1702 and lmo1703, respectively (26). Compared to other bacterial genomes in the NCBI database, both homology and an organization of coding sequences within LGI1 similar to that for regions present in several environmental firmicutes were observed. These firmicutes included Clostridium saccharolyticum WM1 (accession number CP002109.1; 61% nucleotide [nt] coverage, with 73% identity), Clostridium kluyveri NBRC 12016 (accession number AP009049.1) and DSM 555 (accession number CP000673.1; 57% nt coverage, with 76% identity), and Desulfitobacterium hafniense Y51 (accession number AP008230.1; 50% nt coverage, with 70% identity) (see Fig. S1 in the supplemental material). However, the entire LGI1 sequence was not present within these genomes.
The genetic content of emrELm (36.4% GC content) was closest to those of genes encoding a cationic/cationic drug transporter found in Desulfitobacterium dehalogenans ATCC 51507 (100% coverage, with 74% nt and 77% amino acid [aa] identity) and a small multidrug resistance protein in Desulfitobacterium hafniense DCB-2 (100% coverage, with 72% nt and 73% aa identity) and Y51 (100% coverage, with 71% nt and 72% aa identity) (see Fig. S2 and S3 in the supplemental material). Some homology with predicted multidrug resistance proteins in Clostridium ljungdahlii DSM 13528 (100% coverage, with 68% nt identity [see Fig. S2] and 58% aa identity) and Bacillus subtilis 168 (95% coverage, with 54% aa identity [see Fig. S3]) was also observed.
Growth in the presence of acid, cold, or salt stress.
No differences were observed between the WT parent (08-5578) strain and the LGI1 deletion mutants when strains were grown in BHI adjusted to pH 2.5, 3.5, or 4.5. At pH 4.5, bacterial counts remained constant for all four strains, at approximately 7.6 log10 CFU/ml. When the pH was reduced to 3.5, bacterial counts started to decline after 10 h, with an overall decrease of approximately 1.5 log10 CFU/ml within 24 h. Exposure of cells to pH 2.5 resulted in the largest decrease in bacterial counts for all four strains after approximately 5 h, and no viable bacteria were recovered at 24 h. When exposed to 4°C, L. monocytogenes strain 08-5578 adapted rapidly, and it resumed growth approximately 2 to 2.5 h following cold exposure (Table 5). No differences in adaptation to a cold environment were observed between the parent strain and its LGI1 mutants when LPD, MGR, and maximum CFU/ml were measured at 4°C following a downshift from 37°C (Table 5). All four strains reached the stationary phase within approximately 3 weeks at 4°C. In the presence of salt, the L. monocytogenes Δlmo1851 mutant reached slightly higher maximum OD600 values than those of the parent and other mutant strains in BHI containing 5 and 10% NaCl; however, the differences were not statistically significant (P > 0.05). No growth was observed in the presence of 15 and 20% NaCl for the parent and mutant strains, based on OD600 measurements. Similarly, there was no increase or decrease in viable counts over 24 h in the presence of 15 and 20% NaCl.
TABLE 5.
Cold growth adaptation of L. monocytogenes 08-5578 and its LGI1 deletion mutants, based on the lag phase duration, growth rate, and maximum density reached during incubation at 4°C following a downshift from 37°Ca
| L. monocytogenes strain | LPD (h) | Growth rate (Δlog10 CFU/h) | Maximum density (log10 CFU/ml) |
|---|---|---|---|
| 08-5578 (parent) | 2.48 ± 0.74 | 0.47 ± 0.036 | 9.22 ± 0.13 |
| 08-5578 ΔemrE | 1.55 ± 0.56 | 0.40 ± 0.0065 | 9.23 ± 0.19 |
| 08-5578 Δlmo1851 | 2.68 ± 0.59 | 0.49 ± 0.073 | 8.99 ± 0.10 |
| 08-5578 Δsel1 | 2.37 ± 0.17 | 0.43 ± 0.056 | 9.18 ± 0.010 |
Data are means ± SD.
MICs of antimicrobials.
The MICs for L. monocytogenes 08-5578 were 20 μl/ml and 30 μg/ml for E-San and BAC, respectively. The same MICs were observed for the Δlmo1851 and Δsel1 mutants, whereas the ΔemrE mutant had two and three times lower MICs for E-San (10 μl/ml) and BAC (10 μg/ml), respectively. No differences were observed between the MICs for the WT parent strain and those for the deletion mutants for the tested antibiotics (Table 6). All strains were sensitive to CHL, ERY, GEN, and TET and exhibited reduced susceptibility to CIP. Similarly, no differences were observed in the MICs of acriflavine or triclosan (Table 6).
TABLE 6.
MICs of different antimicrobials for the L. monocytogenes 08-5578 wild-type parent strain and its LGI emrELm mutant
| Antimicrobial agent | Strain MIC (μg/ml or μl/ml) |
|||
|---|---|---|---|---|
| 08-5578 | 08-5578 ΔemrELm | 08-5578 Δlmo1851 | 08-5578 Δsel1 | |
| Antibiotics | ||||
| Chloramphenicol | 15 | 15 | 15 | 15 |
| Ciprofloxacin | 5 | 5 | 5 | 5 |
| Erythromycin | 1a | 1 | 1 | 1 |
| Gentamicin | 5 | 5 | 5 | 5 |
| Tetracycline | 5 | 5 | 5 | 5 |
| Sanitizers | ||||
| Benzalkonium chloride | 30 | 10 | 30 | 30 |
| E-San | 20 | 10 | 20 | 20 |
| Other compounds | ||||
| Acriflavine | 18 | 18 | 18 | 18 |
| Triclosan | 8 | 8 | 8 | 8 |
Lowest concentration tested.
Growth in the presence of sublethal concentrations of sanitizers.
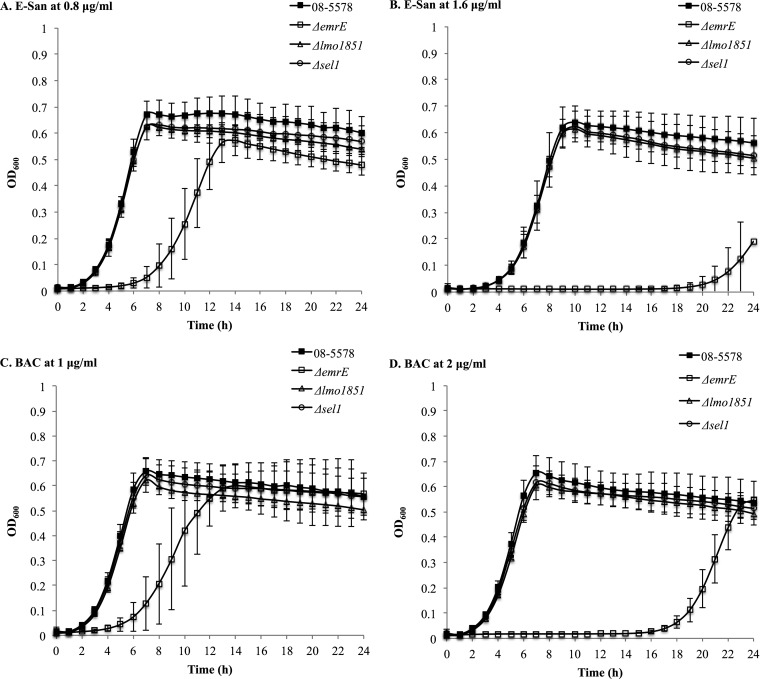
When exposed to sublethal concentrations of QAC sanitizers, the ΔemrELm mutant showed impaired growth compared to that of the other mutants and the parent strain (Fig. 1). This effect was particularly pronounced at concentrations of 1.6 μl/ml and 2 μg/ml for E-San and BAC, respectively (Fig. 1).
FIG 1.
Growth of the L. monocytogenes 08-5578 WT strain (black squares) and its ΔemrELm (white squares), Δlmo1851 (white triangles), and Δsel1 (white circles) mutants in TSB with different concentrations of the E-San (A and B) and benzalkonium chloride (BAC) (C and D) sanitizers at 30°C. The data shown represent the mean OD600 values for five independent cultures.
A significantly longer LPD was observed for the ΔemrELm strain than for the parent strain (P < 0.0001). In particular, it took the ΔemrELm strain 2.6 and 2.4 times longer to resume growth when exposed to E-San and BAC at 0.8 and 1 μg/ml, respectively. The LPD was also 4 and 6 times longer for the strain exposed to E-San and BAC at 1.6 and 2 μg/ml, respectively, than those of the parent strain (Table 7).
TABLE 7.
Average lag phase durations, maximum growth rates, and maximum optical density rates of Listeria monocytogenes 08-5578 and its ΔemrELm mutant exposed to sublethal concentrations of the QAC-based sanitizers E-San and BAC for 24 h at 30°C
| Parameter and treatment | Valuea |
|
|---|---|---|
| 08-5578 (WT) | ΔemrELm mutant | |
| Lag-phase duration (h) | ||
| E-San | ||
| 0.8 μl/ml | 3.31 ± 0.15 | 8.45 ± 1.34*** |
| 1.6 μl/ml | 5.17 ± 0.50 | 21.02 ± 0.79**** |
| BAC | ||
| 1 μg/ml | 2.87 ± 0.39 | 6.80 ± 1.40** |
| 2 μg/ml | 3.16 ± 0.19 | 18.58 ± 0.68**** |
| TSB | 2.68 ± 0.62 | 2.79 ± 0.56 |
| Maximum growth rate (increase in OD600/h) | ||
| E-San | ||
| 0.8 μl/ml | 0.19 ± 0.03 | 0.15 ± 0.02* |
| 1.6 μl/ml | 0.17 ± 0.02 | 0.06 ± 0.04*** |
| BAC | ||
| 1 μg/ml | 0.19 ± 0.01 | 0.15 ± 0.01*** |
| 2 μg/ml | 0.20 ± 0.02 | 0.12 ± 0.01*** |
| TSB | 0.19 ± 0.03 | 0.21 ± 0.03 |
| Maximum OD600 | ||
| E-San | ||
| 0.8 μl/ml | 0.69 ± 0.07 | 0.60 ± 0.06* |
| 1.6 μl/ml | 0.65 ± 0.06 | 0.23 ± 0.18** |
| BAC | ||
| 1 μg/ml | 0.68 ± 0.07 | 0.66 ± 0.11 |
| 2 μg/ml | 0.66 ± 0.07 | 0.56 ± 0.04** |
| TSB | 0.67 ± 0.06 | 0.73 ± 0.07 |
Values represent mean values ± SD for five independent assays, with each sample and treatment measured in duplicate. Statistically significant values are indicated by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001) (P values were obtained using the unpaired two-tailed t test).
Similarly, the ΔemrELm mutant grew 1.3 times slower than the parent strain in the presence of E-San and BAC at 0.8 and 1 μg/ml, respectively, and 2.8 and 1.6 times slower when the sanitizer concentrations were increased to 1.6 and 2 μg/ml, respectively (Table 7). The ΔemrELm mutant also had significantly lower maximum OD600 values over 24 h in the presence of sanitizers than those of the parent strain (Table 7).
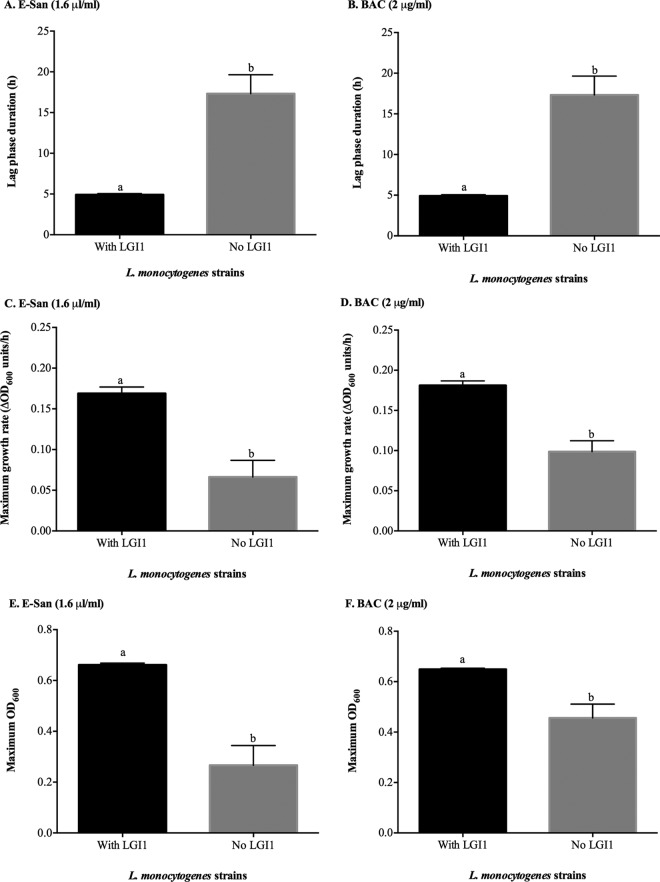
A significantly shorter LPD (P < 0.05) was seen for strains possessing LGI1 than for strains without LGI1 when strains were exposed to sublethal concentrations of E-San and BAC sanitizers (Fig. 2). Similarly, strains possessing LGI1 grew faster (P < 0.001) and reached higher maximum OD600 values (P < 0.05) than those of strains without LGI1 when the strains were grown in the presence of sublethal concentrations of E-San and BAC (Fig. 2).
FIG 2.
Mean lag-phase duration (h), maximum growth rate (ΔOD600/h), and maximum OD600 values for L. monocytogenes isolates possessing LGI1 (n = 9) and isolates without LGI1 (n = 8) grown in the presence of sublethal concentrations of E-San (1.6 μl/ml) (A, C, and E) and benzalkonium chloride (BAC) (2 μg/ml) (B, D, and F) at 30°C for 24 h. Bars represent mean values, and error bars indicate standard errors of the means. Different letters above the bars represent significant differences (P < 0.05) between the groups, determined using the Mann-Whitney test.
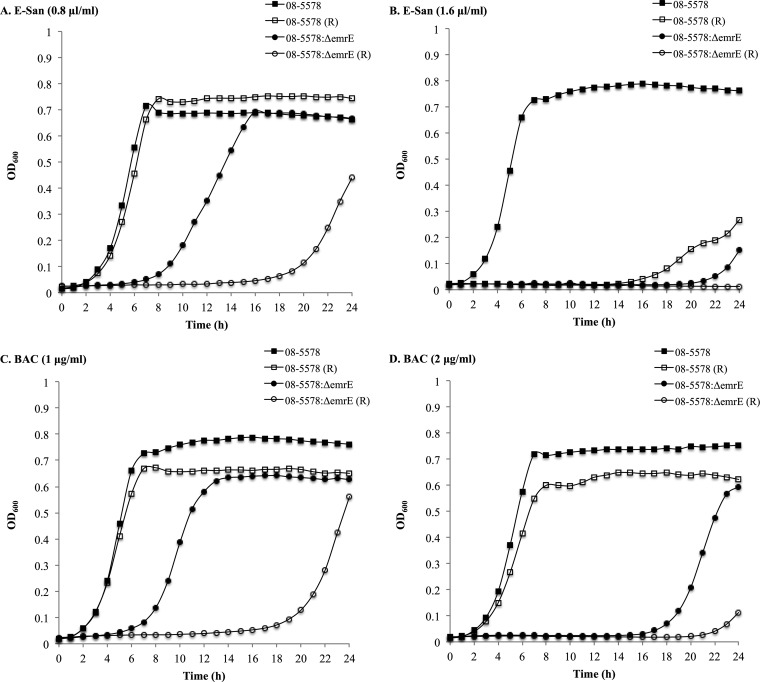
Exposure to sanitizers in the presence of reserpine.
The addition of reserpine, a known inhibitor of efflux pumps (31), to TSB containing a low concentration of E-San (0.8 μl/ml) and the two tested BAC concentrations (1 and 2 μg/ml) was observed to only marginally affect the growth of the WT 08-5578 strain (Fig. 3). However, at a higher concentration of E-San (1.6 μl/ml), the effect was more pronounced (Fig. 3B). When reserpine was added to ΔemrELm mutant cultures containing sublethal concentrations of E-San and BAC, longer LPDs were observed at lower concentrations of BAC and E-San (Fig. 3A and C). At the higher concentration of BAC (2 μg/ml), growth was visibly suppressed, whereas the addition of reserpine to cultures with 1.6 μl/ml E-San completely inhibited the growth of the ΔemrELm mutant (Fig. 3B and D).
FIG 3.
Growth of L. monocytogenes 08-5578 (WT) and its ΔemrELm mutant in the presence of E-San at 0.8 μl/ml (A) and 1.6 μl/ml (B) or benzalkonium chloride (BAC) at 1 μg/ml (C) and 2 μg/ml (D) at 30°C, with (white symbols) and without (black symbols) reserpine (R; 20 μg/ml). The data shown represent the mean OD600 values for three independent assays. Standard deviations ranged from 0.0010 to 0.24.
LGI1 gene expression in response to stresses encountered in the food chain.
The expression of LGI1 was analyzed under various growth conditions by using sqRT-PCR with 16 LGI1 gene targets to identify the island's functional roles. Under normal laboratory growth conditions, when the cells were incubated in BHI at 37°C, only one gene (LM5578_1861), encoding a putative MarR family repressor, was expressed (Fig. 4). Similar results were obtained when the cells were subjected to heat or cold shock, and also when they were treated with UV light. In contrast, when the cells were grown at 37°C in BHI supplemented with 5 μg/ml BAC, 14 of the 16 LGI1 gene targets were induced (Fig. 4). This included the putative MarR family repressor gene (LM5578_1861). The two targets for which expression was not detected in the presence of BAC were LM5578_1883, encoding a putative surface/membrane protein, and LM5578_1863, encoding a putative protein involved in DNA processing (Fig. 4).
FIG 4.
Expression profile of selected LGI1 genes when L. monocytogenes 08-5578 was grown in BHI supplemented with 5 μg/ml benzalkonium chloride at 37°C for 18 h.
Gene expression in WT L. monocytogenes 08-5578 exposed to a sublethal concentration of BAC.
To further explore gene expression in the presence of QACs, L. monocytogenes 08-5578 was exposed to BAC at 10 μg/ml for 1 h, and the expression profiles of LGI1 genes and other genes involved in stress responses were investigated. The largest change (82.4-fold) in the expression of LGI1 genes was seen for lmo1861, encoding a putative MarR family transcriptional regulator, followed by emrE (49.6-fold), encoding an SMR efflux pump. The expression of an LGI1-encoded putative response regulator (lmo1851) of a two-component transduction system also increased 2.3-fold, whereas the expression of sigB, encoding a major stress response regulator in L. monocytogenes, increased 4.1-fold. There was no change in the expression of lde and mdrL, encoding proteins that belong to the major facilitator superfamily (MFS) multidrug resistance efflux pumps.
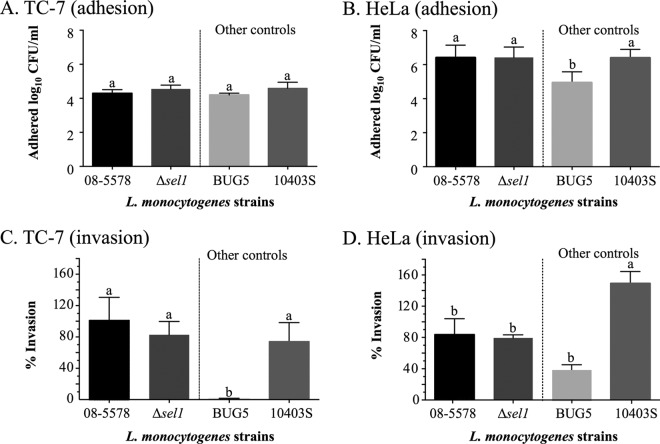
Adhesion and invasion of TC-7 and HeLa cells by the Δsel1 mutant.
There were no differences in the adhesion or invasion efficiencies of the Δsel1 and WT parent (08-5578) strains measured with the TC-7 and HeLa cell lines (Fig. 5). The numbers of adherent cells for the parent and Δsel1 deletion mutant strains were similar to those observed for the control strain 10403S (6.4 log10 CFU/ml). Adhesion efficiencies for all strains were higher for TC-7 cells than for HeLa cells, with the exception of the inlA-deficient mutant BUG5 (Fig. 5A and B). No differences in invasion efficiency were observed between the 10403S control strain, the 08-5578 parent strain, and the Δsel1 mutant by use of TC-7 cells (Fig. 5C). However, significantly (P < 0.05) fewer colonies of 08-5578 and its Δsel1 mutant than of the 10403S control strain invaded HeLa cells (Fig. 5D).
FIG 5.
Adhesion and invasion efficiencies (% bacteria recovered relative to the initial inoculum, normalized to the 08-5578 strain) of the L. monocytogenes WT strain (08-5578) and its isogenic mutant possessing a deletion in the sel1 gene, located on LGI1, compared to those of the clinical isolate 10403S and a Tn1545-induced noninvasive inlA mutant of EGD-SmR (BUG5), using the TC-7 (A and C) and HeLa (B and D) cell lines. Assays for each isolate were carried out in duplicate and repeated four times. Bars show mean adhesion (log10 CFU/ml) and invasion efficiencies (normalized to the 08-5578 strain), and error bars indicate standard errors of the means. Different symbols above the bars indicate significantly different adhesion or invasion efficiencies (P < 0.05; one-way ANOVA with Dunnett's multiple-comparison test).
DISCUSSION
The roles of LGI1 in L. monocytogenes survival in the food chain and in virulence were investigated in the clinical outbreak strain L. monocytogenes 08-5578 and its LGI1 mutant derivatives. LGI1 was first reported for Canadian L. monocytogenes isolates that caused a nationwide listeriosis outbreak in 2008 (26). Since it appears to be absent from all other genomes of L. monocytogenes that are available in the NCBI database, as of August 2015 (see Fig. S1 in the supplemental material), it is highly likely that LGI1 is specific to Canadian isolates. The presence of a number of putative antimicrobial resistance, stress response, and virulence genes on LGI1 indicates that it may be important for L. monocytogenes survival in the food chain and for human listeriosis (26). To investigate this, we deleted three genes located on LGI1, with putative stress response (i.e., the ΔemrE and Δlmo1851 strains) and virulence (i.e., the Δsel1 strain) functions, and exposed the resultant isolates to various conditions relevant to the predicted functions of the deleted genes. Overall, L. monocytogenes 08-5578 possessed high tolerance to acidic, antimicrobial (Table 6), cold (Table 5), and high-salt conditions. Both marked adhesion and invasion of the Caco-2 TC-7 and HeLa cell lines (Fig. 5) were observed, which is consistent with the high mortality rate (40%) and a number of previously reported invasive listeriosis cases (26).
LGI1 was tightly regulated, with multiple gene targets induced, when strains were grown in the presence of the QAC sanitizer BAC (Fig. 4), but this was not detected with other stresses. When LGI1 mutant strains were exposed to stress conditions, the putative role of the response regulator of a two-component signal transduction system (lmo1851) underlying the increased tolerance to the tested stressors could not be confirmed. Similarly, deletion of the sel1 gene, encoding a putative adhesin, did not affect L. monocytogenes adhesion to and invasion of the TC-7 and HeLa cell lines. These results suggest that sel1 does not affect the virulence potential of L. monocytogenes under the conditions tested herein and that this gene does not contribute to the increased adherence and invasion of L. monocytogenes in vitro. In contrast, deletion of the emrE gene, encoding an SMR efflux pump, resulted in susceptibility to QAC-based sanitizers.
Further analyses revealed that the MICs of two different QAC-based sanitizers against the ΔemrE mutant were up to 3 times lower than those for the WT parent strain (Table 6). When the ΔemrELm mutant was grown in the presence of sublethal concentrations of the BAC and E-San sanitizers, we noted longer lag phases, lower growth rates, and overall lower maximum cell densities (OD600) than those of the parent strain (Table 7). Further evidence that LGI1 improves the survival of L. monocytogenes in the presence of QACs was demonstrated with eight additional clinical and food chain strains possessing LGI1, as these strains exhibited faster adaptation and higher growth rates than those of genetically similar strains without LGI1 (Fig. 2). The addition of reserpine, a known efflux pump inhibitor (31), impaired the growth of both the WT and ΔemrE mutant strains in the presence of different concentrations of QAC sanitizers, thus confirming that increased QAC tolerance is due at least in part to a modified efflux activity. However, definite proof of the role of emrE and the level of its contribution to QAC tolerance would require complementation of the mutants.
The role of emrE in QAC tolerance was also apparent from the significant upregulation of emrE in the presence of the BAC sanitizer. However, the upregulation of other LGI1-carried genes suggests that they may also partially contribute to QAC tolerance. Under the same conditions, increased expression of the lmo1861 gene, encoding a putative MarR family regulator, was also observed. It is tempting to speculate that lmo1861 encodes a repressor of the SMR efflux protein, as the MarR family regulator has been shown to negatively regulate MdrM, a multidrug efflux transporter important in activation of the host cytosolic surveillance system, in L. monocytogenes (40). However, additional experiments are required to confirm this possibility. The expression of mdrL, a gene responsible for the production of an MdrL chromosomal efflux pump belonging to the MFS transporters in L. monocytogenes, was not upregulated in the current study. This result is not surprising, as mdrL was previously shown to be overexpressed only in L. monocytogenes strains that were experimentally adapted to BAC, not in naturally resistant WT strains (21), such as those tested in the current study. The findings obtained here also confirm that lde, encoding an additional efflux pump belonging to the MFS efflux transporters, has no role in L. monocytogenes resistance to QAC-based sanitizers. This conclusion was expected, as the Lde efflux pump has been linked primarily to quinolone resistance and, to some degree, is believed to contribute to the tolerance of L. monocytogenes to dyes, such as ethidium bromide and acridine orange (31, 41).
In WT L. monocytogenes strains, the presence of a plasmid-borne or chromosomally carried bcrABC cassette or a transposon (Tn6188)-based QacH efflux pump has been shown to increase tolerance to QAC-based sanitizers in outbreak, clinical, and food chain L. monocytogenes isolates (22, 23, 42). However, QAC sanitizer resistance in L. monocytogenes due to the activity of the SMR EmrE homolog described here has not been reported before. Efflux pumps belonging to the SMR family of proteins are typically 100 to 140 amino acids long and often confer resistance to aminoglycosides, fluoroquinolones, dyes, and QAC (43–45). The first emrE (i.e., E. coli multidrug resistance E)-encoded efflux pump described was linked to resistance of E. coli to TET, ERY, and sulfadiazine (46). Similar SMR pumps have also been described for Mycobacterium smegmatis (e.g., Mmr), Pseudomonas aeruginosa (e.g., EmrEPae), and Staphylococcus spp. (e.g., QacC/D and QacH); however, the substrates of each efflux pump vary depending on the pump and the bacterial species (44, 45, 47). Li et al. (48) demonstrated that the emrE homolog in P. aeruginosa contributed to resistance to ethidium bromide, acriflavine, and aminoglycosides (i.e., amikacin, GEN, kanamycin, neomycin, and tobramycin), albeit resistance to aminoglycosides was observed only in low-ionic-strength medium. In M. smegmatis, an in-frame deletion in emrE resulted in decreased MICs of ethidium bromide, acriflavine, CIP, and norfloxacin but had no effect on CHL, ERY, GEN, and TET MIC values (49). In L. monocytogenes, emrE did not appear to contribute to resistance to GEN, CHL, CIP, TET, and triclosan. It also did not play a role in L. monocytogenes tolerance to acriflavine dye. This result is not unexpected, since L. monocytogenes emrE did not possess any similarity to other well-characterized SMR efflux pump genes from Gram-negative and Gram-positive bacteria. In fact, its genetic content is closest to that of a small multidrug resistance protein in Desulfitobacterium hafniense (72% similarity) and a cationic/cationic drug transporter found in Desulfitobacterium dehalogenans (74% similarity). Desulfitobacterium spp. are anaerobic, motile, Gram-positive, rod-shaped bacteria that often reside in environments contaminated by halogenated organic compounds (50). Some homology (66 to 68%) also exists among EmrE of L. monocytogenes, a predicted multidrug resistance protein in Clostridium ljungdahlii, and QAC resistance proteins observed in Bacillus thuringiensis serovar kurstaki and Bacillus cereus strains. The presence of these bacteria in soil and effluents, which are also natural environments for L. monocytogenes, may result in sharing of the genetic material that confers survival under harsh conditions, though more research is needed to explore this possibility.
Resistance to QACs is a particular concern in regard to L. monocytogenes, as these compounds are often used as sanitizers in food processing facilities due to their noncorrosive properties. Even though the risk of selecting for sanitizer-resistant microorganisms when sanitizers are used at the concentrations recommended by manufacturers is low, it should not be overlooked that inadequate cleaning and sanitation practices can result in exposure of L. monocytogenes to sublethal concentrations of sanitizers, which in turn can lead to selection pressure for progeny possessing increasing sanitizer tolerance (51). This is especially likely to occur if equipment and niches that are difficult to clean and sanitize are encountered, allowing bacteria to persist in food processing environments (52, 53). A number of listeriosis outbreaks implicating isolates with increased tolerance to sanitizers have been noted (23, 42, 52). The L. monocytogenes 08-5578 isolate characterized in the current study was implicated in one of the largest listeriosis outbreaks in Canada, with the source of contamination suspected to be a large commercial slicer harboring the bacterium (54). The presence of LGI1-carried emrE likely contributed to survival of this isolate in the food processing environment.
An additional concern with isolates possessing efflux pumps that enhance L. monocytogenes tolerance to QAC-based sanitizers is the potential for these isolates to develop enhanced tolerance to antibiotics due to similar mechanisms of action (45). Rakic-Martinez et al. (51) demonstrated that L. monocytogenes strains selected on sublethal concentrations of CIP (2 μg/ml) or BAC (10 μg/ml) exhibited higher MICs not only of these agents but also of several other toxic compounds, including GEN, the dye ethidium bromide, and the chemotherapeutic drug tetraphenylphosphonium chloride. While the research performed in this study did not show that emrE increased tolerance to antibiotics relevant to listeriosis treatment (e.g., aminoglycosides), the adaptation to high concentrations of QAC sanitizers and the antibiotic coselection phenomenon were not investigated. In future research, it would be interesting to investigate whether the coselection phenomenon between QACs and antibiotics can occur via the EmrELm efflux pump. While the coselection phenomenon occurring in L. monocytogenes is not well understood, a growing body of evidence suggests that pressures occurring at food processing facilities may contribute to the selection of isolates with enhanced tolerance to different antimicrobials (32, 55). As only a small number of antibiotics were tested in the experiments described here, future studies should include a larger panel of antimicrobials comprising different classes of antibiotics and sanitizer compounds.
Conclusions.
Collectively, our data demonstrate a role for LGI1-carried emrE in QAC tolerance, as evidenced by the following: (i) adaptation and growth of L. monocytogenes strains possessing LGI1-carried emrE are significantly improved in the presence of QACs compared to the adaptation and growth of genetically similar strains without LGI1, (ii) the expression of emrE and several other genes on LGI1 is induced in the presence of BAC, and (iii) deletion of emrELm results in reduced MICs and impaired isolate growth and survival in the presence of QACs. While, to date, there has been no evidence that proper use of sanitizers in food processing will lead to the development of resistant microorganisms, exposure of microorganisms to concentrations below the recommended levels required to eradicate L. monocytogenes is not an unlikely scenario. Since QACs are commonly used in the food industry, the presence of LGI1-carried emrELm may provide a survival advantage and lead to the persistence of L. monocytogenes in food processing environments, though more work is required to fully understand the mechanisms afforded by LGI1 that underlie persistence. In particular, further studies are needed to determine the extent of the contribution of EmrELm to QAC tolerance, the role of this efflux pump in resistance to other antimicrobials via the coselection phenomenon, the genetic regions involved in the regulation of emrE, and the potential contributions of additional LGI1-carried genes to antimicrobial resistance and/or virulence. The identification and characterization of emrELm provide evidence for a novel mechanism for L. monocytogenes to resist injury when exposed to sanitizers commonly encountered in food processing, reaffirming the resilient nature of this foodborne pathogen.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by the National Sciences and Engineering Research Council of Canada.
We acknowledge the Canadian National Microbiology Laboratory for donating bacterial isolates; Cornell University (Ithaca, NY) and Kendra Nightingale (Texas Tech University) for donating the pKSV7 vector and strain 10403S, respectively; and Monique Rousset (Centre de Recherche des Cordeliers, Paris, France) and Pascale Cossart (Institut Pasteur, France) for donating Caco-2 TC-7 cells and the EGD-SmR strain, respectively, via B. Brett Finlay. We thank Kristie Keeney and B. Brett Finlay for their guidance and critical reviews of this work and for financial support, Kevin Allen for providing laboratory space and financial support, and Lili R. Mesak and Jessica Chen for technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03741-15.
REFERENCES
- 1.Clark CG, Farber J, Pagotto F, Ciampa N, Doré K, Nadon C, Bernard K, Ng L-K. 2010. Surveillance for Listeria monocytogenes and listeriosis, 1995–2004. Epidemiol Infect 138:559–572. doi: 10.1017/S0950268809990914. [DOI] [PubMed] [Google Scholar]
- 2.Farber JM, Peterkin PI. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol Mol Biol Rev 55:476–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vàzquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, Gonzalez-Zorn B, Wehland J, Kreft J. 2001. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev 14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Coillie E, Hadewig W, Marc H, Lieve H, Nancy R. 2004. Prevalence and typing of Listeria monocytogenes in ready-to-eat food products on the Belgian market. J Food Prot 67:2480–2487. [DOI] [PubMed] [Google Scholar]
- 5.Little CL, Sagoo SK, Gillespie IA, Grant K, McLauchlin J. 2009. Prevalence and level of Listeria monocytogenes and other Listeria species in selected retail ready-to-eat foods in the United Kingdom. J Food Prot 72:1869–1877. [DOI] [PubMed] [Google Scholar]
- 6.Gianfranceschi M, Gattuso A, Tartaro S, Aureli P. 2003. Incidence of Listeria monocytogenes in food and environmental samples in Italy between 1990 and 1999: serotype distribution in food, environmental and clinical samples. Eur J Epidemiol 18:1001–1006. [DOI] [PubMed] [Google Scholar]
- 7.Low JC, Donachie W. 1997. A review of Listeria monocytogenes and listeriosis. Vet J 153:9–29. doi: 10.1016/S1090-0233(97)80005-6. [DOI] [PubMed] [Google Scholar]
- 8.Orsi RH, den Bakker HC, Wiedmann M. 2011. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol 301:79–96. doi: 10.1016/j.ijmm.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Roche SM, Gracieux P, Milohanic E, Albert I, Virlogeux-Payant I, Temoin S, Grepinet O, Kerouanton A, Jacquet C, Cossart P, Velge P. 2005. Investigation of specific substitutions in virulence genes characterizing phenotypic groups of low-virulence field strains of Listeria monocytogenes. Appl Environ Microbiol 71:6039–6048. doi: 10.1128/AEM.71.10.6039-6048.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonquieres R, Bierne H, Mengaud J, Cossart P. 1998. The inlA gene of Listeria monocytogenes LO28 harbors a nonsense mutation resulting in release of internalin. Infect Immun 66:3420–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rousseaux S, Olier M, Lemaitre JP, Piveteau P, Guzzo J. 2004. Use of PCR-restriction fragment length polymorphism of inlA for rapid screening of Listeria monocytogenes strains deficient in the ability to invade Caco-2 cells. Appl Environ Microbiol 70:2180–2185. doi: 10.1128/AEM.70.4.2180-2185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovacevic J, Arguedas-Villa C, Wozniak A, Tasara T, Allen KJ. 2013. Examination of food chain-derived Listeria monocytogenes of different serotypes reveals considerable diversity in inlA genotypes, mutability, and adaptation to cold temperature. Appl Environ Microbiol 79:1915–1922. doi: 10.1128/AEM.03341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nightingale K, Windham K, Martin K, Yeung M, Wiedmann M. 2005. Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl Environ Microbiol 71:8764–8772. doi: 10.1128/AEM.71.12.8764-8772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nightingale KK, Ivy RA, Ho AJ, Fortes ED, Njaa BL, Peters RM, Wiedmann M. 2008. inlA premature stop codons are common among Listeria monocytogenes isolates from foods and yield virulence-attenuated strains that confer protection against fully virulent strains. Appl Environ Microbiol 74:6570–6583. doi: 10.1128/AEM.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Handa-Miya S, Kimura B, Takahashi H, Sato M, Ishikawa T, Igarashi K, Fujii T. 2007. Nonsense-mutated inlA and prfA not widely distributed in Listeria monocytogenes isolates from ready-to-eat seafood products in Japan. Int J Food Microbiol 117:312–318. doi: 10.1016/j.ijfoodmicro.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Roche SM, Grepinet O, Kerouanton A, Ragon M, Leclercq A, Temoin S, Schaeffer B, Skorski G, Mereghetti L, Le Monnier A, Velge P. 2012. Polyphasic characterization and genetic relatedness of low-virulence and virulent Listeria monocytogenes isolates. BMC Microbiol 12:304. doi: 10.1186/1471-2180-12-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tasara T, Stephan R. 2006. Cold stress tolerance of Listeria monocytogenes: a review of molecular adaptive mechanisms and food safety implications. J Food Prot 69:1473–1484. [DOI] [PubMed] [Google Scholar]
- 18.Arguedas-Villa C, Kovacevic J, Allen KJ, Stephan R, Tasara T. 2014. Cold growth behaviour and genetic comparison of Canadian and Swiss Listeria monocytogenes strains associated with the food supply chain and human listeriosis cases. Food Microbiol 40:81–87. doi: 10.1016/j.fm.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Arguedas-Villa C, Stephan R, Tasara T. 2010. Evaluation of cold growth and related gene transcription responses associated with Listeria monocytogenes strains of different origins. Food Microbiol 27:653–660. doi: 10.1016/j.fm.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Casey A, Fox EM, Schmitz-Esser S, Coffey A, McAuliffe O, Jordan K. 2014. Transcriptome analysis of Listeria monocytogenes exposed to biocide stress reveals a multi-system response involving cell wall synthesis, sugar uptake, and motility. Front Microbiol 5:68. doi: 10.3389/fmicb.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romanova NA, Wolffs PFG, Brovko LY, Griffiths MW. 2006. Role of efflux pumps in adaptation and resistance of Listeria monocytogenes to benzalkonium chloride. Appl Environ Microbiol 72:3498–3503. doi: 10.1128/AEM.72.5.3498-3503.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dutta V, Elhanafi D, Kathariou S. 2013. Conservation and distribution of the benzalkonium chloride resistance cassette bcrABC in Listeria monocytogenes. Appl Environ Microbiol 79:6067–6074. doi: 10.1128/AEM.01751-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elhanafi D, Dutta V, Kathariou S. 2010. Genetic characterization of plasmid-associated benzalkonium chloride resistance determinants in a Listeria monocytogenes strain from the 1998-1999 outbreak. Appl Environ Microbiol 76:8231–8238. doi: 10.1128/AEM.02056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.To MS, Favrin S, Romanova N, Griffiths MW. 2002. Postadaptational resistance to benzalkonium chloride and subsequent physicochemical modifications of Listeria monocytogenes. Appl Environ Microbiol 68:5258–5264. doi: 10.1128/AEM.68.11.5258-5264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knabel SJ, Reimer A, Verghese B, Lok M, Ziegler J, Farber J, Pagotto F, Graham M, Nadon CA, Gilmour MW. 2012. Sequence typing confirms that a predominant Listeria monocytogenes clone caused human listeriosis cases and outbreaks in Canada from 1988 to 2010. J Clin Microbiol 50:1748–1751. doi: 10.1128/JCM.06185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilmour M, Graham M, Van Domselaar G, Tyler S, Kent H, Trout-Yakel K, Larios O, Allen V, Lee B, Nadon C. 2010. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 11:120. doi: 10.1186/1471-2164-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovačević J, Mesak LR, Allen KJ. 2012. Occurrence and characterization of Listeria spp. in ready-to-eat retail foods from Vancouver, British Columbia. Food Microbiol 30:372–378. doi: 10.1016/j.fm.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 28.Althaus D, Lehner A, Brisse S, Maury M, Tasara T, Stephan R. 2014. Characterization of Listeria monocytogenes strains isolated during 2011–2013 from human infections in Switzerland. Foodborne Pathog Dis 11:753–758. doi: 10.1089/fpd.2014.1747. [DOI] [PubMed] [Google Scholar]
- 29.Gaillard JL, Berche P, Sansonetti P. 1986. Transposon mutagenesis as a tool to study the role of hemolysin in the virulence of Listeria monocytogenes. Infect Immun 52:50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell AD. 2004. Whither triclosan? J Antimicrob Chemother 53:693–695. doi: 10.1093/jac/dkh171. [DOI] [PubMed] [Google Scholar]
- 31.Godreuil S, Galimand M, Gerbaud G, Jacquet C, Courvalin P. 2003. Efflux pump Lde is associated with fluoroquinolone resistance in Listeria monocytogenes. Antimicrob Agents Chemother 47:704–708. doi: 10.1128/AAC.47.2.704-708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kovacevic J, Sagert J, Wozniak A, Gilmour MW, Allen KJ. 2013. Antimicrobial resistance and co-selection phenomenon in Listeria spp. recovered from food and food production environments. Food Microbiol 34:319–327. doi: 10.1016/j.fm.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Baranyi J, Roberts TA. 1994. A dynamic approach to predicting bacterial growth in food. Int J Food Microbiol 23:277–294. doi: 10.1016/0168-1605(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 34.Ells TC, Truelstrup Hansen L. 2011. Increased thermal and osmotic stress resistance in Listeria monocytogenes 568 grown in the presence of trehalose due to inactivation of the phosphotrehalase-encoding gene treA. Appl Environ Microbiol 77:6841–6851. doi: 10.1128/AEM.00757-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oliver HF, Orsi RH, Wiedmann M, Boor KJ. 2010. Listeria monocytogenes σB has a small core regulon and a conserved role in virulence but makes differential contributions to stress tolerance across a diverse collection of strains. Appl Environ Microbiol 76:4216–4232. doi: 10.1128/AEM.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camilli A, Tilney LG, Portnoy DA. 1993. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol Microbiol 8:143–157. doi: 10.1111/j.1365-2958.1993.tb01211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith K, Youngman P. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spollM gene. Biochimie 74:705–711. doi: 10.1016/0300-9084(92)90143-3. [DOI] [PubMed] [Google Scholar]
- 38.Tasara T, Stephan R. 2007. Evaluation of housekeeping genes in Listeria monocytogenes as potential internal control references for normalizing mRNA expression levels in stress adaptation models using real-time PCR. FEMS Microbiol Lett 269:265–272. doi: 10.1111/j.1574-6968.2007.00633.x. [DOI] [PubMed] [Google Scholar]
- 39.Chantret I, Rodolosse A, Barbat A, Dussaulx E, Brot-Laroche E, Zweibaum A, Rousset M. 1994. Differential expression of sucrase-isomaltase in clones isolated from early and late passages of the cell line Caco-2: evidence for glucose-dependent negative regulation. J Cell Sci 107:213–225. [DOI] [PubMed] [Google Scholar]
- 40.Crimmins GT, Herskovits AA, Rehder K, Sivick KE, Lauer P, Dubensky TW Jr, Portnoy DA. 2008. Listeria monocytogenes multidrug resistance transporters activate a cytosolic surveillance pathway of innate immunity. Proc Natl Acad Sci U S A 105:10191–10196. doi: 10.1073/pnas.0804170105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lismond A, Tulkens PM, Mingeot-Leclercq MP, Courvalin P, Van Bambeke F. 2008. Cooperation between prokaryotic (Lde) and eukaryotic (MRP) efflux transporters in J774 macrophages infected with Listeria monocytogenes: studies with ciprofloxacin and moxifloxacin. Antimicrob Agents Chemother 52:3040–3046. doi: 10.1128/AAC.00105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Müller A, Rychli K, Muhterem-Uyar M, Zaiser A, Stessl B, Guinane CM, Cotter PD, Wagner M, Schmitz-Esser S. 2013. Tn6188—a novel transposon in Listeria monocytogenes responsible for tolerance to benzalkonium chloride. PLoS One 8:e76835. doi: 10.1371/journal.pone.0076835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bay DC, Rommens KL, Turner RJ. 2008. Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim Biophys Acta 1778:1814–1838. doi: 10.1016/j.bbamem.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 44.Piddock LJ. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat Rev Microbiol 4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 45.Poole K. 2005. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother 56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 46.Yerushalmi H, Lebendiker M, Schuldiner S. 1995. EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J Biol Chem 270:6856–6863. doi: 10.1074/jbc.270.12.6856. [DOI] [PubMed] [Google Scholar]
- 47.Piddock LJ. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev 19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li XZ, Poole K, Nikaido H. 2003. Contributions of MexAB-OprM and an EmrE homolog to intrinsic resistance of Pseudomonas aeruginosa to aminoglycosides and dyes. Antimicrob Agents Chemother 47:27–33. doi: 10.1128/AAC.47.1.27-33.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li XZ, Zhang L, Nikaido H. 2004. Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob Agents Chemother 48:2415–2423. doi: 10.1128/AAC.48.7.2415-2423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Villemur R, Lanthier M, Beaudet R, Lepine F. 2006. The Desulfitobacterium genus. FEMS Microbiol Rev 30:706–733. doi: 10.1111/j.1574-6976.2006.00029.x. [DOI] [PubMed] [Google Scholar]
- 51.Rakic-Martinez M, Drevets DA, Dutta V, Katic V, Kathariou S. 2011. Listeria monocytogenes strains selected on ciprofloxacin or the disinfectant benzalkonium chloride exhibit reduced susceptibility to ciprofloxacin, gentamicin, benzalkonium chloride, and other toxic compounds. Appl Environ Microbiol 77:8714–8721. doi: 10.1128/AEM.05941-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lundén JM, Autio TJ, Korkeala HJ. 2002. Transfer of persistent Listeria monocytogenes contamination between food-processing plants associated with a dicing machine. J Food Prot 65:1129–1133. [DOI] [PubMed] [Google Scholar]
- 53.Lundén JM, Autio TJ, Markkula A, Hellstrom S, Korkeala HJ. 2003. Adaptive and cross-adaptive responses of persistent and non-persistent Listeria monocytogenes strains to disinfectants. Int J Food Microbiol 82:265–272. doi: 10.1016/S0168-1605(02)00312-4. [DOI] [PubMed] [Google Scholar]
- 54.Weatherill S. 2009. Listeriosis investigative review. Report of the independent investigator into the 2008 listeriosis outbreak. Government of Canada, Ottawa, Canada. [Google Scholar]
- 55.Lungu B, O'Bryan CA, Muthaiyan A, Milillo SR, Johnson MG, Crandall PG, Ricke SC. 2011. Listeria monocytogenes: antibiotic resistance in food production. Foodborne Pathog Dis 8:569–578. doi: 10.1089/fpd.2010.0718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.