Abstract
The quantitative and qualitative patterns of environmental contamination by Listeria monocytogenes were investigated in the production chain of dry-cured Parma ham. Standard arrays of surfaces were sampled in processing facilities during a single visit per plant in the three compartments of the food chain, i.e., ham production (19 plants) and postproduction, which was divided into deboning (43 plants) and slicing (25 plants) steps. The numbers of sampled surfaces were 384 in ham production, with 25 positive for L. monocytogenes, and 1,084 in postproduction, with 83 positives. Statistical analysis of the prevalence of contaminated surfaces showed that in ham production, contamination was higher at the beginning of processing and declined significantly toward the end, while in postproduction, prevalence rose toward the end of processing. Prevalence was higher in the deboning facilities than in slicing facilities and was dependent on the type of surface (floor/drainage > clothing > equipment). The qualitative pattern of contamination was investigated through an analysis of the survey isolates and a set of isolates derived from routine monitoring, including longitudinal isolations. Pulsed-field gel electrophoresis (PFGE) and whole-genome single-nucleotide polymorphism (SNP) analysis revealed a remarkable clonality of L. monocytogenes within plants, with the detection of 16 plant-specific clones out of 17 establishments with multiple isolates. Repeated detections of clonal isolates >6 months apart were also observed. Six was the maximum number of between-isolate differences in core SNPs observed within these clones. Based on the same six-SNP threshold, three clusters of clonal isolates, shared by six establishments, were also identified. The spread of L. monocytogenes within and between plants, as indicated by its clonal behavior, is a matter of concern for the hygienic management of establishments.
INTRODUCTION
Listeria monocytogenes is a facultative intracellular bacterial pathogen that can infect several species, including humans, and it is one of the most important agents of foodborne disease (1). Human listeriosis, although rarer than other foodborne diseases, can have severe consequences, with a mortality rate of up to 30% (1–4). In particular, susceptible individuals, like pregnant women, newborns, immunocompromised patients, and the elderly, are at risk of developing invasive listeriosis that can arise with septicemia or meningitis and is often complicated by encephalitis, abortion, and perinatal infections (5). A gastrointestinal form of the disease is also reported and referred to as noninvasive listeriosis (6–8). L. monocytogenes is widely diffused in the environment and, like other species of the genus Listeria, its natural habitat is soil, preferably in the presence of decaying vegetation; these bacteria can also be harbored in the intestinal tracts of humans (9), various domestic animals, like ruminants, pigs, and poultry, and wild species (10). Due to its widespread presence in the environment, this pathogen can easily be introduced into food-processing facilities, favored by its resistance to many extreme conditions. In particular, L. monocytogenes can survive and even grow under food-preserving conditions, such as refrigeration temperatures as low as 1°C, extreme pH, and salinity levels of 4.5 and 10%, respectively (11). As a consequence of its widespread presence and environmental resistance, L. monocytogenes can be a frequent contaminant of ready-to-eat foods that have not undergone bactericidal treatment at the end of the production process, unless strict hygiene measures are adopted to prevent their contamination. Many traditional foodstuffs have been developed throughout the centuries by relying on mild conservation conditions, like salting with NaCl, fumigation, or reduction in pH. One of these food products is dry-cured ham, which is prepared by salting fresh swine ham, followed by curing for several months at progressively increasing temperatures from refrigeration to ambient values. The technological details of this basic process vary by different ham types in terms of concentration of NaCl, time-temperature combinations, and specific manipulations, leading to a variety of products worldwide, some of which are produced on a large scale in high-standard industrial facilities. Parma ham is one of these products; it is among the most important protected designations of origin foods in Europe, with a production volume of nearly 9 million pieces in 2014, for a total estimated value at retail of about 1.5 billion euros and 30% of worldwide exports. By regulation, Parma ham is produced exclusively in establishments located in a limited area of the Province of Parma in Italy, in a hill region extending from 5 to about 60 km south of the city of Parma and about 50 km wide. The Parma ham industry is highly specialized and intensive, and due to its important exporting nature, this industry is faced with different regulations posed by the importing countries with regard to the presence of L. monocytogenes in ready-to-eat ham. Some countries, e.g., the European Union Member States (12), Canada, and Japan, tolerate a contamination limit of 100 CFU/g of ready-to-eat foodstuff during its shelf life, while some others, including the United States, apply a zero-tolerance policy and require the complete absence of L. monocytogenes in 25 g of foodstuff. The zero-tolerance limit implies the adoption of strict hygiene procedures by the industry to prevent even low-level contamination by this common bacterial agent. These procedures have been implemented, but contamination of end products sporadically occurs. Therefore, it is important for the industry and the official inspection authority to have an adequate understanding of the extent and pattern of the contamination of plants where ham is processed to better contrast the presence of the pathogen in the end product. A recent study (13) investigated contamination by L. monocytogenes in the Parma ham chain. That study did not consider the environmental contamination of processing facilities but analyzed feces, carcasses, and fresh ham at slaughter and packaged deboned ham as the end product. Furthermore, the study did not investigate the dynamics of prevalence during ham production and postproduction at the processing phase level. Moreover, the relationships between isolates were analyzed by pulsed-field gel electrophoresis (PFGE), but precise clonal patterns could not be identified due to the inadequate resolution of the technique. To fill the knowledge gaps on contamination by L. monocytogenes and to overcome the limitations of this recent study, we assessed the environmental contamination of the production chain of Parma ham by L. monocytogenes to understand its distribution pattern across different compartments and plants. Furthermore, the genetic correlations of the isolates within this study were analyzed with high-resolution methods to investigate the diversity of the contamination across plants. In particular, an analysis of the correlations was performed to ascertain whether different establishments were contaminated by different lineages of L. monocytogenes or if the same clones were shared by different plants.
MATERIALS AND METHODS
Structure of Parma ham production chain and survey scheme.
The production chain of Parma ham is composed of three compartments: (i) production of dry-cured ham, the so-called bone-in ham, (ii) deboning to produce deboned cured ham, and (iii) slicing to produce presliced cured ham. Bone-in ham production (HP) is a process that goes through three main steps. In the first step, referred to as salting, NaCl is added to fresh swine hams. In the second step, referred to as washing and drying, residual salt is washed off after a 3-month rest under refrigeration; after washing, hams are progressively dried at room temperature for 3 to 4 months, and then the muscular surface is sealed with suet to slow down dehydration. In the third step, referred to as shipment, hams are collected from curing rooms and packaged for shipment after 12 to 18 months of curing at ambient temperature. Unlike the long-lasting production time for bone-in ham, deboning and slicing are short processes that last only a few minutes per ham to transform an already-cured product into different forms, namely, deboned ham and presliced ham. For this reason, although deboning and slicing are specialized processes carried out in dedicated environments, within the scope of this study, they were considered two variants of a unique postproduction (PP) phase following ham production. Two processing steps have been identified in PP: (i) the receipt of hams to be processed and (ii) processing of hams (either deboning or slicing).
Nineteen plants producing bone-in ham (HP survey) and 68 postproduction plants (PP survey) (43 deboning and 25 slicing) were sampled from June 2012 to May 2013. Each plant was sampled upon a single visit during processing operations in the survey period. Specific sampling patterns for HP plants (see Table S1 in the supplemental material) and PP plants (see Table S2 in the supplemental material) were used. Three hundred eighty-four environmental surfaces were sampled in the HP establishments, and 1,084 surfaces were sampled in the PP establishments. The surfaces were selected for sampling so as to represent the diversity of the environmental components of establishments, and for both surveys, they were classified into three categories: (i) floor and drainage, (ii) operator, and (iii) equipment. Some of the surfaces were food contact surfaces and the others were non-food contact surfaces, as specified in Tables S1 and S2 in the supplemental material. Sampling was carried out according to ISO 18593:2004 (14) by swabbing the target surface with a sterile sponge moistened with 10 ml of sterile Dey-Engley neutralizing broth (Biogenetics, Padua, Italy) at approximately 1,000 cm2 or the entire surface if smaller. The sponges were individually put into sterile microbiology bags and transferred to the laboratory inside cooling containers on the day of collection.
Microbiological testing of samples.
Sampling sponges were individually enriched upon arrival in 100 ml of Half-Fraser Listeria enrichment broth (Oxoid, Basingstoke, United Kingdom) for 22 to 26 h at 30 ± 1°C. Two-milliliter aliquots of up to 5 enrichment broths were pooled and tested for L. monocytogenes by real-time PCR with the iQ-Check L. monocytogenes II kit (Bio-Rad Laboratories, Hercules, CA, USA). Positive pools were resolved through testing of the individual enrichment broths according to ISO 11290-1:1996/Amd 1:2004 (15). Only culture-confirmed samples were deemed positive. One isolate (L. monocytogenes LM50) originating from ham was also included in the study as a putative member of a clone colonizing one of the case plants considered (plant S). This isolate was recovered with the microbiological method detailed in USDA FSIS MLG 8.09-2013 (16) from 25 g of ham.
PFGE typing.
The isolates of L. monocytogenes were PFGE typed according to the PulseNet protocol (17) with AscI restriction of DNA. In particular, electrophoresis of DNA was performed using the CHEF Mapper XA system (Bio-Rad Laboratories, Hercules, CA, USA). Pattern identification and comparison were done using the BioNumerics software version 6.6 (Applied Maths, Sint-Martens-Latem, Belgium), with 1% optimization and 1% band matching tolerance. A PFGE pattern (pulsotype) was assigned to each isolate within our laboratory database. In addition to the survey isolates, 20 isolates recovered from Parma ham plants during ad hoc environmental investigations distinct from the survey of this study were PFGE typed and included in the microbiological comparisons of the study. These isolates correspond to five well-defined contamination events that occurred during the period of the study in the products of five independent plants (case plants), with four deboning plants (S, T, U, and Z) and one slicing plant (V). While the primary contamination of the product was detected upon official routine controls at both the facility level and the point of entry of the importing countries, the environmental isolates from the case plants were recovered during the follow-up investigations conducted to track the source of contamination inside the facilities. Multiple isolates per case were recovered, and for one case, the isolate primarily detected in the product was also available and included in the study (plant S). Isolates with AscI-PFGE types shared between different plants, including survey-associated plants (ham production, deboning, and slicing) and case plants, were whole-genome (WG) sequenced, ApaI-PFGE typed, and multilocus sequence typed (MLST) to analyze at the highest detail the potential interrelations between plants with regard to contamination by L. monocytogenes. ApaI-PFGE was performed as described above for AscI-PFGE.
Whole-genome sequencing and assembly.
For whole-genome sequencing (WGS), genomic DNA was extracted from overnight broth cultures in brain heart infusion (Oxoid, Basingstoke, United Kingdom) using the DNeasy blood and tissue kit (Qiagen, Milan, Italy), spectrophotometrically quantified, and quality controlled. Sequencing libraries were prepared with the Nextera XT sample preparation kit (Illumina, Inc., San Diego, CA, USA), and sequencing was performed on the Illumina MiSeq platform with 2 × 250-bp paired-end runs. The sets of sequencing reads were evaluated for sequence quality and read pair length using FastQC (18) and assembled with MIRA 4.0 (19) using accurate settings for de novo assembly mode.
Seven-locus MLST and lineage attribution.
The dedicated Web interface at the Institut Pasteur (http://bigsdb.web.pasteur.fr/perl/bigsdb/bigsdb.pl?db=pubmlst_listeria_seqdef_public&page=sequenceQuery) was used to perform in silico seven-locus multilocus sequence typing (MLST) from assembled genomes according to Ragon et al. (20) on 24 August 2015. The attribution of isolates to genetic lineages was done using the same Web tool.
Genome-based phylogenetic analysis.
Core single-nucleotide polymorphisms (SNPs) were extracted from genome assemblies by kSNP3, with a k-mer length of 21 (21). The core SNPs were all SNPs in positions shared by all genomes under analysis. The data set was used for Bayesian analysis with MRBAYES (22). The analysis was run using the general time-reversible (GTR) substitution model for 2,000,000 generations, sampling the chain at each 1,000th generation. The final tree and parameter values were summarized after 25% of the posterior sample was discarded. The Bayesian tree was displayed and edited with FigTree version 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree) and MEGA 5.2 (23).
Statistical analysis.
The prevalence of positive samples was calculated for the HP and PP surveys, and the relationships between each of the two prevalences and different structural variables were assessed. In particular, for both surveys, generalized linear models (GLM) with binomial error distribution were fitted to test the effect on prevalence of differences in surface category (floor and drainage versus operator versus equipment), contact versus noncontact of the surface with the product, and processing department. In the case of PP, the effect of the type of processing (deboning versus slicing) on prevalence was also tested. The departments identified for the purpose of the study were three in HP (in processing order), i.e., salting, washing, and shipment, and two in PP, i.e., receipt and processing (either deboning or slicing). In particular, no shipment department was considered in PP, as the product at the end of the processing step is hermetically packaged so that no further contamination of the product can occur due to exposure of the packages to the postprocessing environment. The department variable was treated as an ordinal explanatory variable, since the loaf of ham moves along the different departments in both the production and the postproduction steps. Specifically, the department variables were coded through orthogonal polynomials in which dummy regressors provided the trend analysis. In the case of three levels of the categorical variable (as in PP), two dummy regressors were obtained with the orthogonal polynomials, namely, .L and .Q, which correspond to the linear and the quadratic trends, respectively. The response variables (i.e., prevalences) were modeled for dependence on explanatory variables using a forward stepwise selection procedure with log-likelihood ratio tests to define the best model (24). The confidence intervals of the prevalence values were computed with the Wilson binomial approximation (25). Statistical analyses were performed with R 3.2.0 (R Foundation for Statistical Computing, 2010) and the library MASS.
Nucleotide sequence accession numbers.
De novo-assembled contigs of the 69 newly sequenced isolates from the study were deposited at EBI under project no. PRJEB9682 with individual BioSample identification (ID) numbers ERS762129 to ERS762202. Individual GenBank accession numbers and sequencing statistics are listed in Table S3 in the supplemental material.
RESULTS
Prevalence analysis.
The prevalence of establishments contaminated by L. monocytogenes was 79% (15/19) for ham production, 47% (20/43) for deboning, and 12% (3/25) for slicing. Moreover, among facilities with positive contamination results, 33% (5/15) of the ham production plants, 75% (15/20) of the deboning plants, and 100% (3/3) of the slicing plants had more than one positive sample. In establishments with multiple positive samples, multiple isolates (at least two) with the same AscI-PFGE type were detected in 80% (4/5) of the cases in ham production, 93% (14/15) of the cases in deboning, and 67% (2/3) of the cases in slicing.
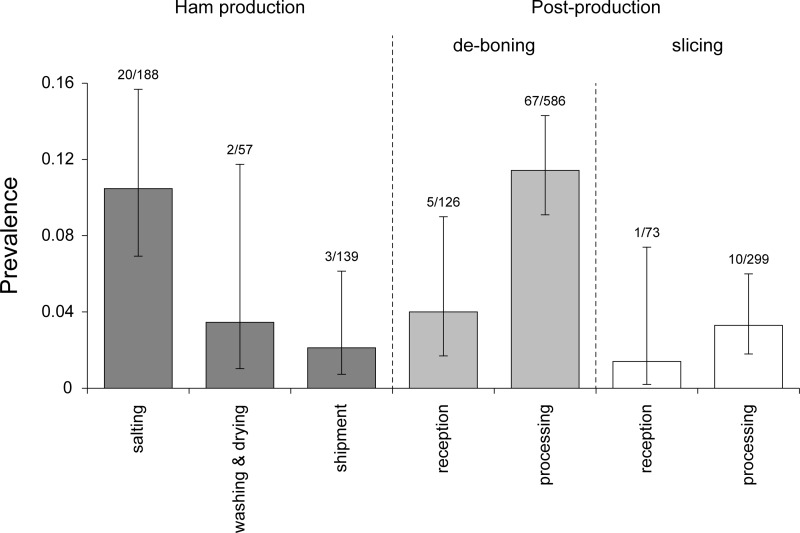
The prevalence of environmental samples contaminated with L. monocytogenes in the different processing compartments and departments is represented in Fig. 1, and the prevalence in the different surface categories across compartments is reported in Table 1. Statistical analysis of the factors affecting prevalence in HP indicated that the department was the only explanatory variable included in the best model from the forward stepwise selection (Table 2). In particular, the best model for describing the prevalence of contamination in HP showed linearly decreasing values moving through the three departments considered (Fig. 1), with maximum prevalence at the beginning (salting department) and progressively lower values in intermediate processing (washing and drying) and shipment (slope [department.L] = −1.1920; standard error [SE] = 0.4453; P = 0.0074). Statistical analysis of the factors affecting prevalence in PP revealed that the surface category (floor and drainage versus operator versus equipment), type of postproduction (deboning versus slicing), and department were included in the best model from the forward stepwise selection (Table 3). In particular, the best model for the description of contamination prevalence in PP showed that there are significant differences between samples collected from floor and drainage, the operator, or the equipment, with significantly higher prevalence of contamination on the floor and drainage than with the operator (P = 0.00031) and equipment (P = 5.24e−10). The best model for PP also showed that prevalence in deboning plants was significantly higher than that in slicing plants (P = 4.46e-05). Moreover, it was apparent in the best model for PP that the prevalence of contamination in processing departments was significantly higher than that in the reception departments (P = 0.001963).
FIG 1.
Prevalence of surfaces contaminated by L. monocytogenes in different departments of ham production and postproduction (i.e., deboning and slicing) establishments. The error bars represent confidence intervals.
TABLE 1.
Distribution of environmental samples across surface categories and production departments
| Production department and parametera | Result by surface category |
|||
|---|---|---|---|---|
| Floor and drainage | Operator | Equipment | Total | |
| Ham production | ||||
| No. of samples | 94 | 74 | 216 | 384 |
| No. of positives | 9 | 7 | 9 | 25 |
| % prevalence (95% CI) | 9.6 (5.1–17.2) | 9.5 (4.7–18.3) | 4.2 (2.2–7.7) | 6.5 (4.4–9.4) |
| Deboning | ||||
| No. of samples | 127 | 171 | 414 | 712 |
| No. of positives | 30 | 20 | 22 | 72 |
| % prevalence (95% CI) | 23.6 (17.1–31.7) | 11.7 (7.7–17.4) | 5.3 (3.5–7.9) | 10.1 (8.1–12.5) |
| Slicing | ||||
| No. of samples | 74 | 75 | 223 | 372 |
| No. of positives | 4 | 3 | 4 | 11 |
| % prevalence (95% CI) | 5.4 (2.1–1.31) | 4.0 (1.4–11.1) | 1.8 (0.7–4.5) | 3.0 (1.7–5.2) |
CI, confidence interval.
TABLE 2.
Forward stepwise model selection for L. monocytogenes prevalence in the ham production survey obtained from a generalized linear model with binomial error distributiona
| Explanatory variable(s) | Loglik | k | P value |
|---|---|---|---|
| 1b | −92.46 | 1 | |
| Department | −86.85 | 3 | 0.00365c |
| Department + contact | −86.71 | 4 | 0.6021 |
The full model is described by prevalence ~ department + contact + surface category. Models were compared using the log-likelihood ratio test. The best models for n explanatory variables are shown, with the log likelihood (Loglik), number of parameters (k), and the P value of the comparison with the n − 1 best model (P value tests: <0.05 as the inclusion criterion and >0.10 as the exclusion criterion).
Null model.
Significant at a P value of <0.01.
TABLE 3.
Forward stepwise model selection for L. monocytogenes prevalence in the postproduction survey obtained from a generalized linear model with binomial error distributiona
| Explanatory variable(s) | Loglik | k | P value |
|---|---|---|---|
| 1b | −292.93 | 1 | |
| Surface category | −276.34 | 3 | 6.25e−08c |
| Surface category + postproduction type | −265.57 | 4 | 3.46e−06c |
| Surface category + postproduction type + department | −259.15 | 5 | 0.00034c |
| Sampling point + postproduction type + department + contact | −258.05 | 6 | 0.1367 |
The full model is described by prevalence ∼ department + contact + surface category + postproduction type. Models were compared using the log-likelihood ratio test. The best models for n explanatory variables are shown, with the log likelihood (Loglik), number of parameters (k), and the P value of the comparison with the n − 1 best model (P value tests: <0.05 as the inclusion criterion and >0.10 as the exclusion criterion).
Null model.
Significant at a P value of <0.001.
Genotyping and phylogenetic analysis.
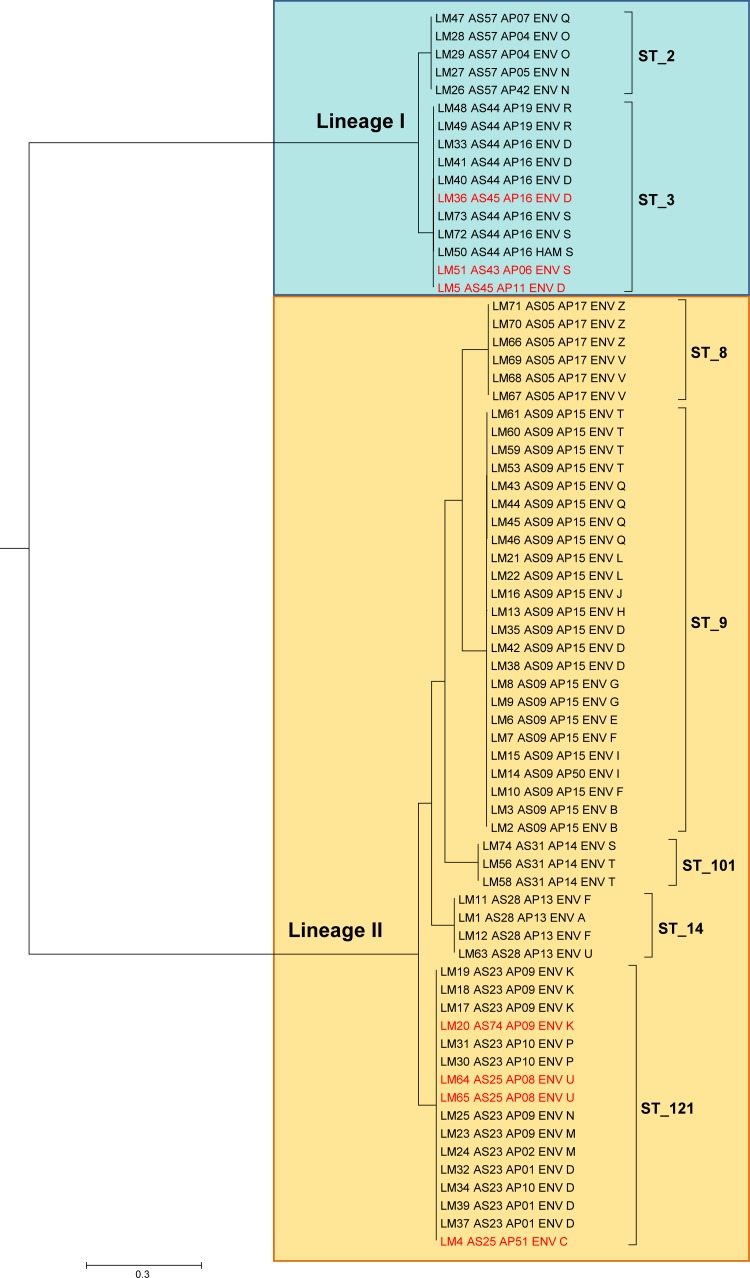
Twenty-five isolates were recovered from HP, and 83 isolates were recovered from PP. The isolates recovered from the surveys in this study (HP and PP) and the 20 case plant isolates recovered during routine controls were treated as a unique set for a comparison of genotypes and phylogenetic analysis. Thirty-nine AscI-PFGE types were identified among the isolates from the surveys, and their distribution among the surveyed plants is represented in Fig. S1 in the supplemental material. Three additional AscI-PFGE types were identified among the case plant isolates, whose remaining types were shared with the survey isolates. All isolates from both the surveys and the case plants belonging to AscI-PFGE types shared by at least 2 plants were subjected to ApaI-PFGE, WGS, and in silico MLST. In total, 69 isolates were selected, including L. monocytogenes LM20, which had a unique AscI-PFGE type in the study but differed by only a single band from 3 other isolates from its plant (plant K). The detailed typing results of these isolates are reported in Table S4 in the supplemental material. In summary, genomic sequencing yielded a median genome coverage of 162-fold (range, 81- to 300-fold), a median of 115 large (>1,000 nucleotides [nt]) contigs (range, 31 to 439 contigs), and a median assembled genome size of 3.07 Mb (range, 2.80 to 3.25 Mb). The sequencing results of the individual genomes are reported in Table S3 in supplemental material. The purpose of this extra typing was twofold: (i) to investigate the possible clonality of contaminations detected in different plants, as suggested by identical AscI-PFGE types, and (ii) to analyze in detail the population structure of L. monocytogenes as disclosed by high-resolution methods. The distance tree generated from the 33,076 core SNPs identified (Fig. 2) divided the isolates into two main clusters corresponding to lineages I and II of L. monocytogenes. Moving down the tree inside lineages I and II, it was revealed that the main 7 branches, those separated by the longest distances, corresponded exactly to 7 distinct sequence types (STs), namely, ST2 and ST3 in lineage I and ST8, ST9, ST14, ST101, and ST121 in lineage II. Furthermore, almost univocal correspondence emerged between the STs and AscI-PFGE types, namely, ST8 and AS05, ST9 and AS09, ST14 and AS28, ST101 and AS31, and ST2 and AS57, while the multiple AscI-PFGE types of ST3 and ST121 differed by a single band within the ST. Greater PFGE heterogeneity emerged with the addition of ApaI profiles to AscI types, specifically within ST2, ST3, and ST121, while AscI-ApaI combined typing still confirmed univocal correspondence within ST8, ST9, ST14, and ST101. Summarizing, PFGE and MLST gave identical clustering for 4 out of 7 STs, all in lineage II, whereas in the remaining 3 STs, the AscI-PFGE types overlapped almost exactly with the STs, while a moderately higher diversity was introduced by ApaI-PFGE typing.
FIG 2.
Maximum clade credibility tree showing genetic distances between isolates of L. monocytogenes based on whole-genome core SNPs. Sequence types (STs) are indicated, while credibility values are omitted for clarity of this figure (reported in Fig. 3). The tree tips list isolate metadata, namely, study ID, AscI- and ApaI-PFGE genotype, source of isolate (environmental or ham), and plant ID (capital letter). AscI-PFGE variants inside STs are in red. The scale bar refers to the branch length representing the number of nucleotide substitutions per site.
Clonality analysis.
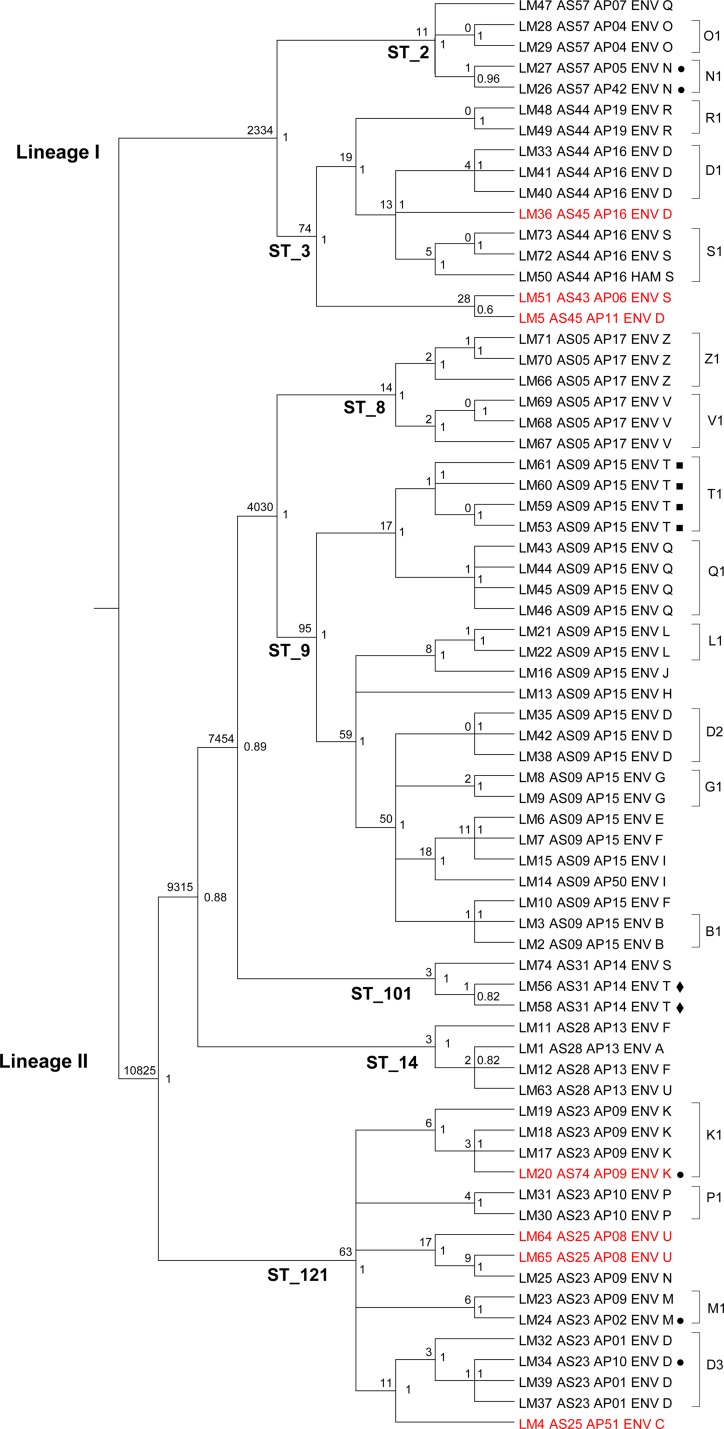
Figure 2 clearly shows the sharp distinction of the isolates in two distant lineages subdivided into a few STs. While the within-ST phylogenetic distances appeared substantially shorter than the between-ST distances, an articulated phylogenetic structure was revealed by the cladogram representation of the SNP-based phylogeny (Fig. 3). In particular, a significant clustering of isolates originating from the same plant became evident. This was the case for 14 out of 17 plants with multiple isolates included in the phylogeny. More specifically, 16 plant-associated clusters were present in these 14 plants, as one of them showed three distinct clones (plant D). The other plant-associated clones were in plants B, G, K, L, M, N, O, P, Q, R, S, V, and Z (Fig. 3). The number of SNPs that differed within the identified plant-associated clones ranged from 0 to 6. Interestingly, in some of these clonal groups, PFGE variants coexisted, always restricted to a single-band polymorphism. In many establishments hosting clones, additional isolates from the same plant were located on the tree as the closest isolates to the plant clone. This is the case for L. monocytogenes isolate LM32 in plant D, LM19 in plant K, LM50 in plant S, LM67 in plant V, and LM66 in plant Z. Furthermore, in establishment S, one of the case plants, LM50 had been isolated 6 months before the sister isolates LM72 and LM73. Similarly, in another case plant, establishment Z, LM66 had been isolated 13 months before the sister isolates LM70 and LM71. Notably, only 2 and 5 core SNPs distinguished these time-separated isolates, respectively. The detection of multiple L. monocytogenes isolates almost indistinguishable by core SNP phylogeny, which were from distinct environmental sites inside facilities and in some cases obtained months apart, supports the hypothesis of persistent clones inside a remarkable proportion of establishments. A particular case was that of plant T, which was not taken into account in the above-mentioned report of clonal evidence. This was another case plant with multiple detected isolates, all simultaneous, in which LM56 and LM58 differing by 1 core SNP were detected on the same surface (a piece of equipment in deboning) immediately before the start of processing, just after preoperative sanitation (LM56) and after the end of daily processing, before postoperative sanitation (LM58). Clearly, LM56 and LM58 were two isolates of the same strain that withstood the sanitation and processing steps, as it persisted throughout. To test the effect of isolation procedures of L. monocytogenes on the repeatability of the WG-SNP approach, multiple colonies from a single microbiological culture of a positive sample were included in the phylogenetic analysis as independent isolates (LM53, LM59, LM60, and LM61). All 4 genomes were closely clustered, as expected. A single core SNP difference was detected between the 4 sister colonies. As a confirmation of its specificity, the genomic SNP-based typing was able to distinguish isolates from plants V and Z, which were identical by PFGE and shared a rare profile that was observed in only two other isolates in our database of about 2,000 isolates. The isolates from the two plants had no known epidemiological relationship and were recovered a year apart from each other. This fact made the finding of the identical PFGE profile unexpected, unless an unknown relationship between the two plants did exist. Indeed, the SNP-based approach confirmed what was epidemiologically meaningful, clearly clustering the isolates from the same facility and distinguishing the two facilities with 14 different core SNPs.
FIG 3.
Cladogram of the maximum clade credibility tree of isolates of L. monocytogenes based on whole-genome core SNPs. Main lineages and sequence types (STs) are reported on the corresponding branches. Credibility values are reported on the right side of each node, and the numbers of core SNPs differing between isolates of each node are on the left. The tree tips list isolate metadata, namely, study ID, AscI- and ApaI-PFGE genotype, source of isolate (environmental or ham), and plant ID (capital letter). AscI-PFGE variants inside STs are in red. ■, multiple colonies from the culture of a single environmental sample; ◆, preoperative and postoperative sanitation samples; ●, AscI- and ApaI-PFGE variants inside plant-specific clones.
DISCUSSION
Pattern of contamination shows fluctuating trend over processing steps.
The results of this study showed that the environmental contamination of processing facilities by L. monocytogenes, represented by both between-plant prevalence and within-plant prevalence, was significant although varied across the components of the production chain. It declined significantly moving from the first phase of ham production, i.e., salting, to the intermediate and final phases. This is consistent with the continuous introduction of relatively contaminated fresh hams from slaughterhouses into the salting departments, where they are intensively manipulated for the salting process, as opposed to the reduced manipulation the hams go through during subsequent phases. Furthermore, in addition to reduced manipulation, the postsalting phases are characterized by the presence of hams that are virtually free of Listeria due to the combined action of salt and reduced water activity following months of curing. This combination of reduced manipulation and absence of product contamination can explain the observed decline in environmental contamination moving toward the final phases of ham processing. Consistent with this observation was the further finding that in both of the postproduction compartments, i.e., deboning and slicing, the receipt departments were significantly less contaminated than the respective processing departments, where contamination reappeared. This finding results in a v-shaped pattern of contamination moving through ham production and postproduction, with higher contamination at the beginning, a significant decline at the end of curing, and the reappearance of contamination during deboning and slicing. This pattern indicates that postproduction processing implies some level of contamination. At the same time, the observed quantitative difference in contamination between phases is indicative of the existence of limitations to cross-contamination between compartments and departments, as required by the hygiene standards. The reappearance of contamination during postproduction processing is probably due to an increased chance of introduction associated with working procedures, like movement of materials, staff, etc., and to the establishment of persistent populations of L. monocytogenes in processing environments, as discussed below.
Varied contamination levels across processing types reflect their operative features.
With regard to postproduction processes, deboning turned out to be a significantly more contaminated process than slicing. This corresponds to the invasive mechanical operation needed to extract the bone by means of several pieces of semiautomatic equipment used in a series. The operation causes the spread of organic debris on the equipment, floor, and operators' clothing. Conversely, slicing is a high-precision process with very limited spread of organic matter.
Different contamination prevalences on different environmental surfaces have practical implications for prevention and monitoring strategies in processing facilities.
Interestingly, in postproduction, the floor and drainage categories of sampled surfaces were more contaminated than the operator category, which was in turn more contaminated than the equipment category. This evidence has some practical implications. First, floors and drainage systems should be regarded as important sources of contamination for the hams being processed, unless adequate separation is put in place between the environmental and food compartments during the handling of foodstuffs and sanitation procedures. Second, the operators might be effective carriers of cross-contamination through their clothes, gloves, and footwear. Third, floors and drainage systems should be regularly included in routine monitoring programs to maximize the probability of detecting environmental contamination. The findings in this study on the contamination of floors and drainage systems confirm the results of previous work that found these environmental compartments to be a significant source of L. monocytogenes in other food-processing facilities, including meat- and fish-processing plants (26–28). The ability of this pathogen to colonize the same environmental niches in different industrial establishments is indicative of its tendency to persist inside plants.
PFGE results were suggestive of both within-plant clonality and between-plant cross-contamination.
The high frequency of establishments with multiple surfaces contaminated by isolates with identical AscI-PFGE types indicated possible colonization by clonal populations of plant-specific L. monocytogenes. At the same time, some AscI-PFGE types were shared by different plants, suggesting possible cross-contamination. Considering the nonnegligible level of operative interlink between establishments, this might actually be the case.
In-depth genomic analysis identifies general and context-specific features of L. monocytogenes.
Whole-genome SNP (WG-SNP) analysis of the isolates belonging to AscI-PFGE types shared by two or more establishments confirmed the sharp distinction of L. monocytogenes into distant genomic lineages, as previously described (20, 29–34). In our study, only lineages I and II were detected, with a predominance of lineage II. Another recent study on the contamination pattern of food-associated environments (35) observed the exclusive presence of lineages I and II but with a substantial predominance of lineage I (179 out of 188 isolates). The two studies investigated different food compartments, namely, dry-cured ham establishments versus various retail delicatessen establishments in distant geographical areas, Italy, and the United States. The different target environments might underpin the observed lineage reversal in the two populations of L. monocytogenes, corresponding to limited overlap of STs from the two populations, with only 2 STs shared out of 17 STs cumulatively identified in the two studies. The phylogenetic distance tree generated from the core SNPs in our isolates showed that the fundamental clades inside lineages corresponded to different STs, which in turn corresponded to different PFGE types with limited PFGE diversity inside STs.
Whole-genome SNP analysis detects diffused clonality of contamination.
Beyond the ST/PFGE level of discrimination, the high-resolution power of WG-SNP analysis, already evidenced in recent outbreak investigations (36–39), showed an articulated structure of well-supported branches in the tree. Although the phylogenetic distances corresponding to these branches were virtually negligible compared to those at the ST and lineage I/II levels, the branching structure revealed a wealth of informative insights on the qualitative pattern of contamination among facilities. In particular, while the high-resolution WG-SNP analysis had been done primarily to detect the occurrence of clone sharing between plants, the remarkable outcome was the detection of plant-specific clones (highly similar isolates from different surfaces inside single establishments) in the majority of plants, with a case of three distinct clones coexisting inside a single establishment (plant D). In two cases, within-plant clonality was also confirmed by the longitudinal detection of clonal isolates 6 and 13 months apart. Overall, 13 out of 16 plant-specific clusters were composed of isolates from different departments or surface categories, or they were recovered months apart from each other, confirming the independence of the isolates. The simultaneous existence of the same microorganism in distinct points of an establishment, its reisolation after several months, and the coexistence of close genomic neighbors with just a few differing core SNPs indicate the tendency of L. monocytogenes to become established and persist inside food-processing facilities, as already observed (40, 41). Besides within-plant clonality, our study also evidenced a few clusters of highly similar isolates across establishments. The threshold of six core SNPs was used as an empirical discriminant for the definition of between-plant clonality, and the threshold of six was chosen for consistency with the limit observed in within-plant clones. Three between-plant clones were detected; these clones involved the establishment groups F and B, S and T, and F, A, and U, as displayed in the cladogram in Fig. 3. The finding of between-plant clones is consistent with the moderate grade of interlink existing between establishments of the Parma ham compartments. This interlink consists of sharing of suppliers, including slaughterhouses for fresh hams, sharing of other processing materials, like salt, suet, etc., and occasional sharing of services, like transportation, cleaning, etc. These results confirm, at the level of an important food chain, like the Parma ham industry, the outcomes of the recent study by Stasiewicz et al. (35), who reported similar clonal and persistent patterns of environmental contamination in delicatessen retail establishments in the United States (35). Interestingly, the two studies revealed remarkably similar behaviors of L. monocytogenes in very different food-processing contexts and geographical areas, which is the tendency to establish clonal populations both inside and across facilities.
Practical implications of the study encompass hygienic management of establishments and careful interpretation of molecular methods in the epidemiology of L. monocytogenes.
The clonality and persistence of contamination in food-associated environments, as demonstrated by WG-SNP analysis in independent studies and uncorrelated contexts, have two main practical implications for the industry and control authorities. First, the evidence emphasizes the potential of WGS-based molecular epidemiology in source tracking of food contamination. Second, it is an important warning to hygiene managers about the ability of L. monocytogenes to persistently colonize processing environments, prompting the need for adequate microbiological monitoring of plants. A practical consequence of our results is also apparent on the methodological grounds of molecular epidemiology. In fact, while the discriminatory power of PFGE appeared to be essentially not higher than that of MLST and, likewise, that of MLST far lower than that of WG-SNP analysis, some concerns derive from the epidemiological use of PFGE in light of the misleading diversity detected within highly clonal clusters of isolates, like those identified inside establishments. In our study, the PFGE type diversity observed within plant-specific clones was limited to single-band variants; nevertheless, these variants cannot easily be interpreted for epidemiological purposes. A strict interpretation of PFGE differences would classify the variants as nonclonal, leading to epidemiological mistakes. On the other hand, a tolerant interpretation, e.g., one using the criteria of Tenover et al. (42), would render PFGE of limited usefulness due to a significant reduction in discriminatory power. Our results provide field evidence that PFGE should be interpreted with particular care during detailed epidemiological investigations because of its limited discriminatory power and, more importantly, its instability and uncertain meaning. In fact, these single-band variants in PFGE can be the effect of very limited genetic changes, like single-nucleotide mutations that commonly arise during clonal expansion, or the consequence of more significant modifications in the genome, like the insertion of mobile genetic elements. Indeed, more significant modifications, unlike point mutations, can constitute useful markers for tracking epidemic clones of L. monocytogenes in outbreak investigations, as was recently reported (39). Also in this case, WGS, with its ability to ascertain the presence of mobile genetic elements, is the key to overcoming the ambiguity posed by PFGE single-band variants.
Supplementary Material
ACKNOWLEDGMENT
This study was supported by Italian Health Ministry grant PRC2010_013 for a postdoctoral fellowship to Erika Scaltriti.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03103-15.
REFERENCES
- 1.EFSA, ECDC. 2015. The European Union summary report on trends and sources of zoonoses, zoonotic agents and foodborne outbreaks in 2013. EFSA J 13:3991. [Google Scholar]
- 2.Pinner RW, Schuchat A, Swaminathan B, Hayes PS, Deaver KA, Weaver RE, Plikaytis BD, Reeves M, Broome CV, Wenger JD. 1992. Role of foods in sporadic listeriosis. II. Microbiologic and epidemiologic investigation. The Listeria Study Group. JAMA 267:2046–2050. [PubMed] [Google Scholar]
- 3.Schuchat A, Deaver KA, Wenger JD, Plikaytis BD, Mascola L, Pinner RW, Reingold AL, Broome CV. 1992. Role of foods in sporadic listeriosis. I. Case-control study of dietary risk factors. The Listeria Study Group. JAMA 267:2041–2045. [PubMed] [Google Scholar]
- 4.Schuchat A, Swaminathan B, Broome CV. 1991. Epidemiology of human listeriosis. Clin Microbiol Rev 4:169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, Gonzalez-Zorn B, Wehland J, Kreft J. 2001. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev 14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pichler J, Much P, Kasper S, Fretz R, Auer B, Kathan J, Mann M, Huhulescu S, Ruppitsch W, Pietzka A, Silberbauer K, Neumann C, Gschiel E, de Martin A, Schuetz A, Gindl J, Neugschwandtner E, Allerberger F. 2009. An outbreak of febrile gastroenteritis associated with jellied pork contaminated with Listeria monocytogenes. Wien Klin Wochenschr 121:149–156. doi: 10.1007/s00508-009-1137-3. [DOI] [PubMed] [Google Scholar]
- 7.Dalton CB, Austin CC, Sobel J, Hayes PS, Bibb WF, Graves LM, Swaminathan B, Proctor ME, Griffin PM. 1997. An outbreak of gastroenteritis and fever due to Listeria monocytogenes in milk. N Engl J Med 336:100–105. doi: 10.1056/NEJM199701093360204. [DOI] [PubMed] [Google Scholar]
- 8.Aureli P, Fiorucci GC, Caroli D, Marchiaro G, Novara O, Leone L, Salmaso S. 2000. An outbreak of febrile gastroenteritis associated with corn contaminated by Listeria monocytogenes. N Engl J Med 342:1236–1241. doi: 10.1056/NEJM200004273421702. [DOI] [PubMed] [Google Scholar]
- 9.Grif K, Hein I, Wagner M, Brandl E, Mpamugo O, McLauchlin J, Dierich MP, Allerberger F. 2001. Prevalence and characterization of Listeria monocytogenes in the feces of healthy Austrians. Wien Klin Wochenschr 113:737–742. [PubMed] [Google Scholar]
- 10.Farber JM, Peterkin PI. 1991. Listeria monocytogenes, a foodborne pathogen. Microbiol Rev 55:476–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClure PJ, Kelly TM, Roberts TA. 1991. The effects of temperature, pH, sodium chloride and sodium nitrite on the growth of Listeria monocytogenes. Int J Food Microbiol 14:77–91. doi: 10.1016/0168-1605(91)90039-R. [DOI] [PubMed] [Google Scholar]
- 12.Commission Regulation (EC). 2005. Commission regulation (EC) no. 2073/2005 of 15 November 2005 on microbiological criteria for foodstuffs. European Union, Brussels, Belgium: http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2005:338:0001:0026:EN:PDF. [Google Scholar]
- 13.Prencipe VA, Rizzi V, Acciari V, Iannetti L, Giovannini A, Serraino A, Rossi A, Morelli D, Marino L, Migliorati G, Caporale V. 2012. Listeria monocytogenes prevalence, contamination levels and strains characterization throughout the Parma ham processing chain. Food Control 25:150–158. doi: 10.1016/j.foodcont.2011.10.018. [DOI] [Google Scholar]
- 14.International Organization for Standardization (ISO). 2004. ISO 18593:2004. Microbiology of food and animal feeding stuffs. Horizontal methods for sampling techniques from surfaces using contact plates and swabs. International Organization for Standardization, Geneva, Switzerland: http://www.iso.org/iso/iso_catalogue/catalogue_tc/catalogue_detail.htm?csnumber=39849. [Google Scholar]
- 15.International Organization for Standardization (ISO). 2004. ISO 11290-1:1996/Amd.1:2004. Microbiology of food and animal feeding stuffs—horizontal method for the detection and enumeration of Listeria monocytogenes—part 1: detection method amendment 1: modification of the isolation media and the haemolysis test, and inclusion of precision data. International Organization for Standardization, Geneva, Switzerland: https://saz.isolutions.iso.org/obp/ui#iso:std:iso:11290:-1:ed-1:v1:amd:1:v1:en. [Google Scholar]
- 16.US Department of Agriculture, Food Safety and Inspection Service, Office of Public Health Science Laboratory Quality Assurance Division. 2013. Isolation and identification of Listeria monocytogenes from red meat, poultry and egg products, and environmental samples. USDA FSIS MLG 8.09, revision 9 US Department of Agriculture, Athens GA: http://www.fsis.usda.gov/wps/wcm/connect/1710bee8-76b9-4e6c-92fc-fdc290dbfa92/MLG-8.pdf?MOD=AJPERES. [Google Scholar]
- 17.Centers for Disease Control and Prevention. 2013. Standard operating procedure for PulseNet PFGE of Listeria monocytogenes. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/pulsenet/PDF/listeria-pfge-protocol-508c.pdf. [Google Scholar]
- 18.Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 19.Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information, p 45–46. In Computer science and biology: Proceedings of the German Conference on Bioinformatics (GCB), GCB '99. GCB, Hannover, Germany. [Google Scholar]
- 20.Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, Le Monnier A, Brisse S. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog 4:e1000146. doi: 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardner SN, Hall BG. 2013. When whole-genome alignments just won't work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One 8:e81760. doi: 10.1371/journal.pone.0081760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venables WN, Ripley BD. 2002. Modern applied statistics with S. Springer, New York, NY. [Google Scholar]
- 25.Brown L, Cat TT, DasGupta A. 2001. Interval estimation for a proportion. Stat Sci 16:101–133. doi: 10.1214/ss/1009213286. [DOI] [Google Scholar]
- 26.Autio T, Hielm S, Miettinen M, Sjöberg AM, Aarnisalo K, Björkroth J, Mattila-Sandholm T, Korkeala H. 1999. Sources of Listeria monocytogenes contamination in a cold-smoked rainbow trout processing plant detected by pulsed-field gel electrophoresis typing. Appl Environ Microbiol 65:150–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gudbjörnsdóttir B, Suihko M-L, Gustavsson P, Thorkelsson G, Salo S, Sjöberg A-M, Niclasen O, Bredholt S. 2004. The incidence of Listeria monocytogenes in meat, poultry and seafood plants in the Nordic countries. Food Microbiol 21:217–225. doi: 10.1016/S0740-0020(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 28.Zhao T, Podtburg TC, Zhao P, Schmidt BE, Baker DA, Cords B, Doyle MP. 2006. Control of Listeria spp. by competitive-exclusion bacteria in floor drains of a poultry processing plant. Appl Environ Microbiol 72:3314–3320. doi: 10.1128/AEM.72.5.3314-3320.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brosch R, Chen J, Luchansky JB. 1994. Pulsed-field fingerprinting of listeriae: identification of genomic divisions for Listeria monocytogenes and their correlation with serovar. Appl Environ Microbiol 60:2584–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graves LM, Swaminathan B, Reeves MW, Hunter SB, Weaver RE, Plikaytis BD, Schuchat A. 1994. Comparison of ribotyping and multilocus enzyme electrophoresis for subtyping of Listeria monocytogenes isolates. J Clin Microbiol 32:2936–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piffaretti JC, Kressebuch H, Aeschbacher M, Bille J, Bannerman E, Musser JM, Selander RK, Rocourt J. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc Natl Acad Sci U S A 86:3818–3822. doi: 10.1073/pnas.86.10.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasmussen OF, Skouboe P, Dons L, Rossen L, Olsen JE. 1995. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology 141(Pt 9):2053–2061. doi: 10.1099/13500872-141-9-2053. [DOI] [PubMed] [Google Scholar]
- 33.Orsi RH, den Bakker HC, Wiedmann M. 2011. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol 301:79–96. doi: 10.1016/j.ijmm.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Doijad S, Weigel M, Barbuddhe S, Blom J, Goesmann A, Hain T, Chakraborty T. 2015. Phylogenomic grouping of Listeria monocytogenes. Can J Microbiol 9:1–10. doi: 10.1139/cjm-2015-0281. [DOI] [PubMed] [Google Scholar]
- 35.Stasiewicz MJ, Oliver HF, Wiedmann M, den Bakker HC. 2015. Whole-genome sequencing allows for improved identification of persistent Listeria monocytogenes in food-associated environments. Appl Environ Microbiol 81:6024–6037. doi: 10.1128/AEM.01049-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilmour MW, Graham M, Van Domselaar G, Tyler S, Kent H, Trout-Yakel KM, Larios O, Allen V, Lee B, Nadon C. 2010. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 11:120. doi: 10.1186/1471-2164-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rychli K, Müller A, Zaiser A, Schoder D, Allerberger F, Wagner M, Schmitz-Esser S. 2014. Genome sequencing of Listeria monocytogenes “Quargel” listeriosis outbreak strains reveals two different strains with distinct in vitro virulence potential. PLoS One 9:e89964. doi: 10.1371/journal.pone.0089964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmid D, Allerberger F, Huhulescu S, Pietzka A, Amar C, Kleta S, Prager R, Preussel K, Aichinger E, Mellmann A. 2014. Whole genome sequencing as a tool to investigate a cluster of seven cases of listeriosis in Austria and Germany, 2011–2013. Clin Microbiol Infect 20:431–436. doi: 10.1111/1469-0691.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Q, Holmes N, Martinez E, Howard P, Hill-Cawthorne G, Sintchenko V. 2015. It is not all about SNPs: comparison of mobile genetic elements and deletions in Listeria monocytogenes genomes links cases of hospital-acquired listeriosis to the environmental source. J Clin Microbiol 53:3492–3500. doi: 10.1128/JCM.00202-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orsi RH, Borowsky ML, Lauer P, Young SK, Nusbaum C, Galagan JE, Birren BW, Ivy RA, Sun Q, Graves LM, Swaminathan B, Wiedmann M. 2008. Short-term genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genomics 9:539. doi: 10.1186/1471-2164-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vongkamjan K, Roof S, Stasiewicz MJ, Wiedmann M. 2013. Persistent Listeria monocytogenes subtypes isolated from a smoked fish processing facility included both phage susceptible and resistant isolates. Food Microbiol 35:38–48. doi: 10.1016/j.fm.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 42.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.