Abstract
Midbrain dopamine neurons are an essential component of the basal ganglia circuitry, playing key roles in the control of fine movement and reward. Recently, it has been demonstrated that γ-aminobutyric acid (GABA), the chief inhibitory neurotransmitter, is co-released by dopamine neurons. Here we show that GABA corelease in dopamine neurons does not utilize the conventional GABA synthesizing enzymes, glutamate decarboxylases GAD65 and GAD67. Our experiments reveal an evolutionarily conserved GABA synthesis pathway mediated by aldehyde dehydrogenase 1a1 (ALDH1a1). Moreover, GABA co-release is modulated by ethanol at binge drinking blood alcohol concentrations and diminished ALDH1a1 leads to enhanced alcohol consumption and preference. These findings provide insights into the functional role of GABA co-release in midbrain dopamine neurons, which may be essential for reward-based behavior and addiction.
Midbrain dopamine (DA) neurons are important for fine movement control, motivation and reward-based learning (1, 2). Dysfunction of dopaminergic systems leads to movement disorders, such as Parkinson’s disease, and various forms of addiction and drug abuse (3, 4). DA is the primary neurotransmitter released by DA neurons and activation of DA receptors in post-synaptic neurons can modulate neuronal excitability and circuit output. It has recently been shown that GABA is co-packaged with DA in midbrain DA neurons by the vesicular monoamine transporter 2 (Vmat2) and is subsequently co-released in the striatum (5), where it provides direct and potent inhibition to postsynaptic striatal projection neurons (SPNs) through activation of GABAA receptors.
In the mammalian central nervous system (CNS), GABA biosynthesis is mediated by two glutamate decarboxylases (GAD65 and GAD67, 65 and 67 kDa isoforms, respectively). Expression of either isoform of GAD has traditionally been used to identify GABAergic neurons in the CNS. To identify which subset of midbrain DA neurons is capable of GABA synthesis, we examined GAD expression in DA neurons by coupling immunohistochemistry for tyrosine hydroxylase (TH), the rate-limiting enzyme in DA synthesis with in situ hybridization for Gad1 or Gad2 (which encode GAD67 and GAD65, respectively). Only a small percentage of midbrain DA neurons express Gad in the substantia nigra pars compacta (SNc, ~9%, Fig.1A-K) and the ventral tegmental area (VTA, ~15%, fig.S1) (6, 7).
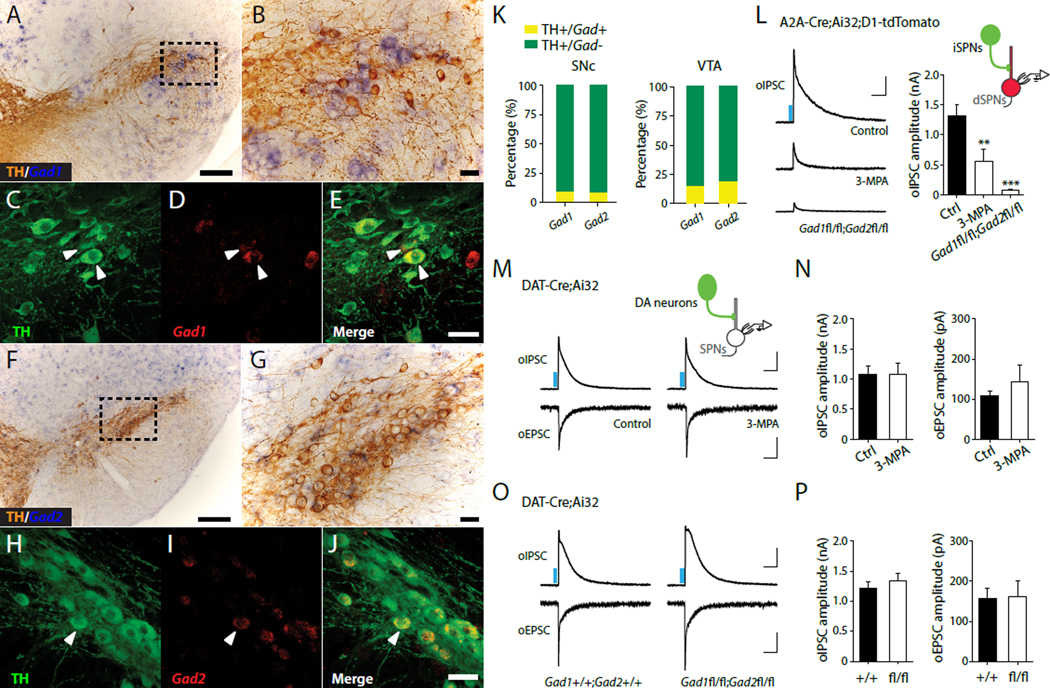
Fig. 1. GABA co-release by midbrain DA neurons does not require GAD.
(A to J) Expression of Gad1 and Gad2 mRNA in DA neurons of the SNc. Immunolabeled TH-positive dopaminergic neurons (brown) combined with chromogenic in situ hybridization (ISH) for Gad1 (A and B) and Gad2 (F and G) mRNA. Confocal fluorescence images of ISH for Gad1 (D) and Gad2 (I) mRNA (red) combined with TH immunostaining (green) (panels C-E and H-J) show limited expression of Gad in TH+ cells. (K) Quantification of TH/Gad co-localization in SNc (left) and VTA (right). (L) Left, Representative oIPSC traces in control (upper), 3-MPA-treated (500 μM, middle), and Gad1fl/fl;Gad2fl/fl (lower) A2A-Cre;Ai32;Drd1a-tdTomato mice. Right, summary statistics for oIPSC recordings. Representative traces (M) and summary statistics (N) for oIPSC and oEPSC recorded from DAT-Cre;Ai32 mice treated with ACSF (control) and 3- MPA, respectively. Representative traces (O) and summary statistics (P) for oIPSC and oEPSC recorded from DAT-Cre;Ai32;Gad1+/+;Gad2+/+ and DAT-Cre;Ai32;Gad1fl/fl;Gad2fl/fl mice. Blue bar indicates 450 nm light stimulation. Scale bars: 200 μm for A,F, 50 μm for B-E, G-J, 400 pA, 100 ms for oIPSC and 50 pA, 100 ms for oEPSC. Error bars indicate Mean ± SEM. **P < 0.01, ***P < 0.001.
An individual DA neuron can extend elaborate axonal arbors covering large portions of the striatum (8). Consequently, despite GAD only being expressed in a small subset of DA neurons, it is possible that GAD-expressing neurons can drive sustained GABA co-release throughout the striatum. We thus asked whether GAD is required for GABAergic transmission in the striatum by recording alterations in dopaminergic inhibitory postsynaptic currents (IPSCs) in SPNs resulting from pharmacological inhibition or conditional genetic deletion of GAD. The striatum is comprised of two parallel output pathways arising from distinct groups of ‘direct’ and ‘indirect’ pathway GABAergic SPNs (dSPNs and iSPNs, respectively) that differ in their expression of postsynaptic G-protein coupled DA receptors. SPNs also send collateral inhibitory projections within the striatum. As SPNs express GAD and are considered conventional GABAergic neurons, we used striatal collateral inhibition as an internal control for our experiments. We expressed channelrhodopsin 2 (ChR2) in iSPNs by crossing A2A-Cre mice (in which Cre recombinase is selectively expressed in iSPNs but not in midbrain DA neurons) with transgenic mice containing a conditional floxed allele of ChR2 in the Rosa26 locus (Ai32 mice). Progeny from this cross were bred to Drd1a-tdTomatoexpressing transgenic mice carrying a bacterial artificial chromosome (BAC) transgene that selectively labels dSPNs. We then performed whole-cell voltage-clamp recordings in dSPNs in brain slices of dorsal striatum prepared from A2A-Cre;Ai32;Drd1a-tdTomato mice, in which ChR2 is selectively expressed in A2A adenosine receptor-expressing iSPNs and tdTomato expression is restricted to D1 receptor-expressing dSPNs. Optogenetic stimulation of iSPN axons with brief pulses (0.5 ms) of blue light (450 nm) reliably evoked IPSCs in dSPNs. Optogenetically evoked IPSCs (oIPSCs) recorded in dSPNs were significantly attenuated by GAD inhibitor 3-mercaptopropionic acid (3-MPA, 500 µM, Fig.1L), confirming that local collateral inhibitory transmission arising from iSPNs is dependent on GAD function. We next selectively deleted GAD in iSPNs (9) using Gad1 and Gad2 double conditional knockout mice (A2A-Cre; Gad1fl/fl;Gad2fl/fl). When recording oIPSCs from dSPNs in A2A-Cre; Gad1fl/fl;Gad2fl/fl;Ai32;Drd1a-tdTomato mice, we found that genetic deletion of both GADs in iSPNs nearly abolished nearly all the oIPSCs recorded in dSPNs. These data confirmed that GAD-mediated GABA synthesis is necessary for conventional GABAergic transmission within the striatum (Fig.1L).
To test whether GAD is required for functional GABA co-release by midbrain DA neurons, whose axon terminals project onto SPNs, we used DAT-Cre;Ai32 mice to selectively express ChR2 in DA neurons (5, 10) and recorded oIPSCs and oEPSCs in postsynaptic dorsal striatum SPNs. Monosynaptic oIPSCs and oEPSCs can be abolished by GABAA and glutamate receptor blockers, respectively (fig.S2). Surprisingly, neither incubating brain slices with 3-MPA (Fig.1M-N) nor genetically deleting both GAD isoforms in midbrain DA neurons using DATCre; Ai32;Gad1fl/fl;Gad2fl/fl mice (Fig.1O-P) significantly altered the amplitudes of oIPSCs and oEPSCs in SPNs, indicating that midbrain DA neuron GABA co-released does not require canonical GAD-mediated GABA synthesis. GABA co-release from DA neurons was observed in recordings obtained throughout the striatum, in both dorsal and ventral regions. Notably, the oIPSCs recorded in dorsal striatum SPNs were significantly larger than those recorded from SPNs in the nucleus accumbens (NAc). Within NAc, oIPSC and oEPSC amplitudes were not significantly different between SPNs in the Core or medial Shell, and, as with oIPSCs recorded in dorsal striatum, oIPSCs recorded in the NAc were not blocked by application of 3-MPA (fig.S3). Recent work has suggested that DA can directly activate postsynaptic GABAA receptors (11). To exclude this possibility and test whether dopaminergic oIPSCs were caused by direct activation of GABAA receptors by DA, we locally applied DA and GABA onto individual SPNs and recorded IPSCs. Direct application of DA did not evoke IPSCs, whereas GABA successfully evoked IPSCs in the same SPN, indicating that DA was not likely to be activating GABAA receptors directly in our cells. This idea was further supported by application of the DA transporter (DAT) blocker GBR12935, which elevates extracellular DA concentrations and similarly did not affect oIPSC amplitudes in the striatum (fig. S4). Together, these data suggest that dopaminergic oIPSCs were not caused by direct activation of GABAA receptors by DA.
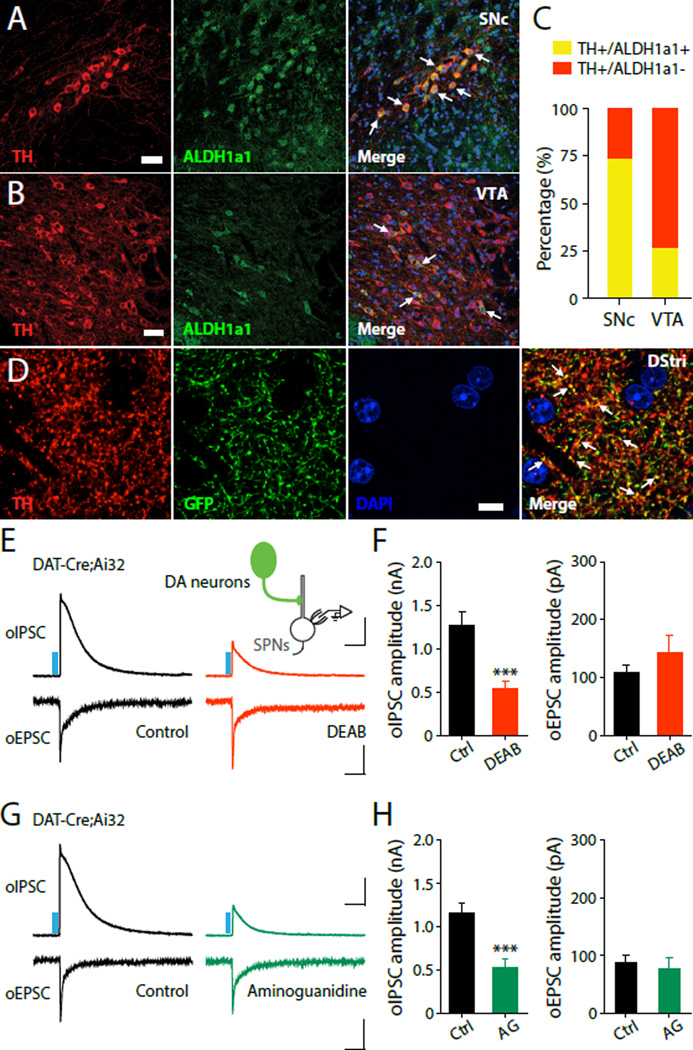
In plants, GABA can be synthesized from putrescine (12) by the enzymes diamine oxidase (DAO) and aldehyde dehydrogenase (ALDH) (fig.S5A) (13, 14). GABA production through this alternative evolutionarily conserved pathway also exists in Xenopus tadpole (15) and mammalian cells (16–19). Glial cells can also utilize putrescine to produce GABA during retinal early development (18, 20). We tested whether ALDH-mediated alternative GABA synthesis drives GABA production in midbrain DA neurons. ALDH1a1 is the most abundant form of cytosolic ALDH (21, 22) and is highly expressed in the ventral midbrain, including the region delineating the SNc (23) (for Aldh1a1 mRNA, see: Allen Brain Atlas, http://mouse.brainmap.org/gene/show/11455). We first examined ALDH expression in midbrain DA neurons by double immunostaining for ALDH1a1 and TH. ALDH1a1 is indeed highly expressed in a subset of DA neurons, co-localizing with TH in the SNc and VTA (Fig.2A-C, and fig.S5B). To examine subcellular localization of ALDH1a1, we injected an adeno-associated virus (AAV) carrying GFP-tagged ALDH1a1 into the midbrain and examined GFP expression in the striatum. We found that GFP strongly co-localizes with TH within axons in the dorsal striatum (Fig.2D), suggesting that ALDH1a1 is highly abundant in dopaminergic terminals (fig.S5C-F). We then tested the involvement of ALDH1a1 in GABA synthesis in these neurons by blocking its activity with the ALDH inhibitors 4-(diethylamino)-benzaldehyde (DEAB, 10 µM), or disulfiram (10 µM). To ensure that intracellular GABA levels were sufficiently depleted, we pretreated brain slices from DAT-Cre;Ai32 mice with ACSF containing these blockers for 2–4 hours (a similar paradigm to our pharmacological treatment with 3-MPA targeting conventional GABA synthesis in SPNs). Treatment with both DEAB and disulfiram dramatically reduced oIPSC amplitude in SPNs following DA axon stimulation (Fig.2E-F, and fig.S6A-B). We also recorded oEPSCs in the same SPNs by stimulating DA fibers. Notably, this treatment did not affect the peak amplitude of oEPSCs (Fig.2E-F, and fig.S6A-B), suggesting that these blockers do not prevent global neurotransmitter release, but rather selectively impair GABA co-release.
Fig. 2. ALDH1a1-mediated non-canonical GABA synthesis in DA neurons.
(A and B) Confocal images depicting double immunostaining for TH (left, red) and ALDH1a1 (middle, green) in SNc (A) and VTA (B). Scale bar: 40 μm. (C) Quantification of ALDH1a1 expression in TH+ DA neurons in SNc and VTA. (D) Confocal images depicting double immunostaining for TH (red), ALDH1a1–GFP (green), and DAPI (blue) in the dorsal striatum (DStri). Scale bar: 10µm. Representative oIPSC and oEPSC traces (E) and summary statistics (F) recorded from DAT-Cre;Ai32 mice treated with ACSF (control, left) and DEAB (10 µM, right). Representative oIPSC and oEPSC traces (G) and summary statistics (H) recorded from DAT-Cre;Ai32 mice treated with ACSF (control, left) and aminoguanidine (AG, 100 µM, right). Blue bar indicates 450 nm light stimulation. Scale bars: 400 pA, 100 ms for oIPSC and 50 pA, 100 ms for oEPSC. Error bars indicate Mean ± SEM. ***P < 0.001.
If GABA were indeed converted from putrescine, blocking DAO would also reduce GABA production. We thus used DAO blockers (aminoguanidine, 100 µM or amiloride, 10 µM) and examined the effect of this treatment on oIPSCs using the same paradigm as above. Both aminoguanidine and amiloride significantly and selectively reduced oIPSC amplitude in SPNs, with no effect on oEPSC amplitude (Fig.2G-H, and fig.S6C-D) or on conventional GABAergic transmission (fig.S7). Importantly, ALDH1a1 is known to be important for the synthesis of retinoic acid (RA) (24) and breakdown of the DA metabolite 3,4-Dihydroxyphenylacetaldehyde (DOPAL) to 3,4-Dihydroxyphenylacetic acid (DOPAC) (25). It is possible then that deletion of ALDH1a1 may lead to retinoic acid (RA) deficiency and a concomitant increase in extracellular DA concentration, both of which may have effects on synaptic transmission. Application of exogenous RA and a D2 receptor antagonist (sulpiride), however, did not prevent observed reduction of oIPSCs in SPNs and had no effect on oEPSCs, suggesting that these other ALDH1a1-mediated functions, are not important for GABA co-release (fig.S8).
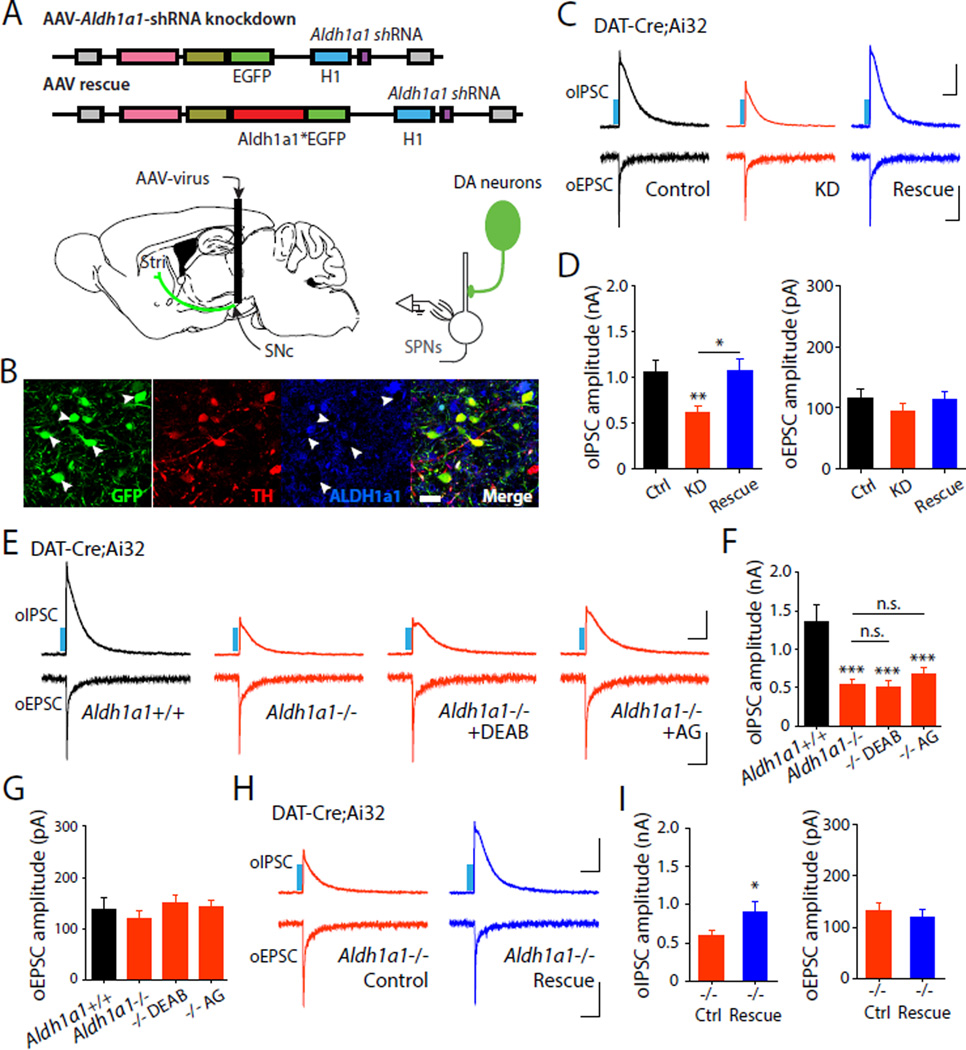
To selectively manipulate ALDH1a1 expression in midbrain DA neurons, we injected an AAV expressing shRNA targeted against Aldh1a1 into the ventral midbrain of DAT-Cre;Ai32 mice to knockdown (KD) Aldh1a1 expression (Fig.3A-B, and fig.S9). Aldh1a1 KD in the midbrain significantly reduced oIPSC amplitude in SPNs (Fig.3C-D) an effect that was fully rescued by simultaneous expression of an shRNA-resistant wildtype Aldh1a1 (Aldh1a1*, Fig.3C-D). Importantly, Aldh1a1 KD and rescue had no effect on oEPSCs recorded in the same neurons. These data suggest that presynaptic expression of ALDH1a1 in DA neurons is critical for GABA synthesis and co-release.
Fig. 3. Aldh1a1 knockdown and genetic deletion reduce dopaminergic oIPSCs and are rescued by Aldh1a1 over-expression.
(A) Schematic illustration depicting viral shRNA constructs (Aldh1a1* indicates an shRNA-resistant wild-type Aldh1a1) and experimental configuration. (B) Confocal image showing expression of shRNA (green) in TH+ (red) neurons. Arrowheads indicate absence of ALDH1a1 (blue) in GFP+/TH+ neurons. Scale bar: 20 µm. Representative oIPSC and oEPSC traces (C) and summary statistics (D) in DAT-Cre;Ai32 mice injected with Aldh1a1 knockdown and rescue viruses. Representative oIPSC and oEPSC traces (E) and summary statistics for oIPSC (F) and oEPSC (G) in Aldh1a1+/+;DAT-Cre;Ai32 or Aldh1a1−/−;DAT-Cre;Ai32 mice, or loss of function mice treated with DEAB (10 µM), and aminoguanidine (100 µM). Representative oIPSC and oEPSC traces (H) and summary statistics (I) recorded from Aldh1a1−/−;DAT-Cre;Ai32 mice injected with control and rescue viruses. Blue bar indicates 450 nm light stimulation. Scale bars: 400 pA, 100 ms for oIPSC and 50 pA, 100 ms for oEPSC. Error bars indicate Mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
We next asked if genetic deletion of ALDH1a1 can selectively diminish GABA corelease by midbrain DA neurons (26). We recorded oIPSCs and oEPSCs in SPNs from the dorsal striatum of Aldh1a1−/−;DAT-Cre;Ai32 transgenic mice and Aldh1a1+/+ littermate controls. Dopaminergic oIPSCs were strongly attenuated in Aldh1a1−/− mice, while oEPSCs were unaffected (Fig.3E-G). Because ALDH1a1 is the final enzyme in the conversion process leading to GABA synthesis, the pharmacological effects of DAO and ALDH blockers should be occluded by Aldh1a1 deletion. We treated brain slices from Aldh1a1−/−;DAT-Cre;Ai32 with DAO and ALDH blockers and found that DAO and ALDH blockers did not further reduce oIPSC amplitude in SPNs in Aldh1a1−/− mice (Fig.3E-G). As an additional control, we examined oIPSCs in Aldh1a1−/−;A2ACre; Ai32;Drd1a-tdTomato mice resulting from collateral intrastriatal inhibition and confirmed that conventional GABA transmission is not affected in mutant mice (fig.S10). Lastly, we tested if elevated expression of Aldh1a1 could rescue the reduction of dopaminergic IPSCs observed in SPNs in Aldh1a1−/− mice. To achieve this, we injected AAV-rescue constructs into the midbrain of Aldh1a1−/−;DAT-Cre;Ai32 mice and subsequently recorded oIPSCs in SPNs. Over-expression of Aldh1a1 fully rescued the dopaminergic oIPSCs in SPNs without affecting oEPSCs (Fig.3H-I). GABA transporters also contribute to the accumulation of presynaptic GABA in midbrain DA neurons (fig.S11). Taken together, our data suggest that ALDH1a1-mediated alternative GABA synthesis supports functional GABAergic transmission by DA neurons.
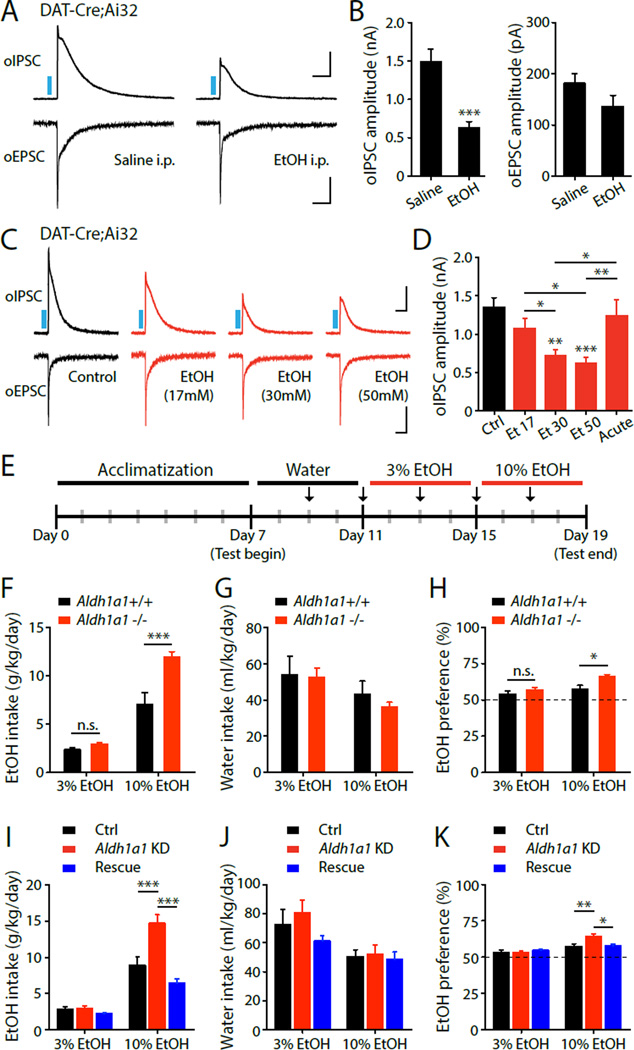
Mutations of aldh1a1 have been linked to alcoholism in human populations (27, 28), suggesting that GABA co-release by DA neurons may be altered by drug abuse. Given the involvement of the dopaminergic system and dorsal striatum in enhanced alcohol consumption, preference and addiction (29–31), we examined GABA co-release in mice exposed to repeated in vivo administration and withdrawal of ethanol (EtOH), a behavioral paradigm approximating binge drinking episodes in humans. Intoxicating levels of EtOH were administered (2 g/kg ETOH, 20%, intraperitoneal injection) daily for consecutive 7 days (31). Two to four hours after the final EtOH injection, we prepared striatal brain slices from DAT-Cre;Ai32 mice and recorded oIPSCs in SPNs. We found that repeated in vivo administration of EtOH significantly decreased oIPSC, but not oEPSC amplitude recorded in SPNs (Fig.4A-B). We then tested if a direct EtOH treatment can affect GABA co-release in brain slices. To mimic binge drinking blood alcohol levels, we pretreated striatal brain slices for 2–4 hours (paradigm comparable to our previous pharmacological manipulations) with EtOH at concentrations comparable to blood alcohol levels during binge drinking episodes (17–50 mM). Prolonged, but not acute, treatment with 17–50 mM EtOH significantly decreased the oIPSCs amplitude recorded in SPNs following DA axon stimulation (Fig.4C-D). The same EtOH treatments did not affect oEPSC amplitude, or oIPSCs recorded from dSPNs in A2ACre; Ai32;Drd1a-tdTomato mice (fig.S12). Reduction of GABA co-release was not observed in Aldh1a1−/− mice (fig.S13), suggesting that EtOH modulation is dependent on ALDH1a1. Our results indicate that GABA co-release is attenuated by EtOH at binge drinking blood alcohol concentrations. As excessive alcohol drinking enhances the inclination for alcohol drinking behavior (32), we used the home cage continuous twobottle choice test to test behavioral consequences of ALDH1a1 deletion on EtOH intake (33). Basal locomotion, as assessed by open-field test, remains intact in Aldh1a1−/− mice (fig.S14). When given continuous access to EtOH, Aldh1a1−/− mice significantly increased their intake of and preference for EtOH, with no significant difference in daily water intake, compared with WT littermates (Fig.4E-H). To more conclusively demonstrate that loss of ALDH1a1 specifically in midbrain neurons is responsible for enhanced EtOH consumption, we injected an AAV expressing Aldh1a1-shRNA into the ventral midbrain of wild-type mice to knockdown Aldh1a1 expression (fig.S9). Aldh1a1 KD in the midbrain recapitulated the increased intake of and preference for EtOH behavioral phenotype observed in Aldh1a1−/− mice. This behavioral effect was fully rescued by simultaneous expression of an shRNA-resistant wild-type Aldh1a1* (Fig.4IK). . Together, our studies suggest that diminished GABA co-release may serve as potential determinant for enhanced alcohol consumption and preference.
Fig. 4. Altered GABA co-release in conditions related to alcohol binge drinking.
Representative oIPSC and oEPSC traces (A) and summary statistics (B) in DAT-Cre;Ai32 mice in control or following repeated in vivo administration of EtOH. Representative oIPSC and oEPSC traces (C) and summary statistics (D) in DAT-Cre;Ai32 mice in control, or treated with EtOH (17–50 mM). (E) Shematic illustration depicting the timeline of the two-bottle choice behavioral assay. (F) Quantification of average daily EtOH intake, (G) average daily water intake, and (H) average EtOH preference. (I) Quantification of average daily EtOH intake in Aldh1a1 KD or overespression mice. (J) average daily water intake. (K) average EtOH preference. Blue bar indicates 450 nm light stimulation. Scale bars: 400 pA, 100 ms for oIPSC and 50 pA, 100 ms for oEPSC. Error bars indicate Mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Midbrain DA neurons are known to act through slow neuromodulatory mechanisms. However, GABA co-release in DA neurons demonstrates a rapid and potent inhibitory control employed by DA neurons. A variety of neuronal subtypes can co-release multiple neurotransmitters in different neural circuits (34–36). How an individual neuron releases and regulates multiple neurotransmitters remains unclear. We found that DA neurons utilize an alternative GABA synthesis pathway to support functional GABAergic neurotransmission. Co-released GABA can permit very local inhibition of dendritic excitability, a key mechanism controlling synaptic plasticity. Moreover, as this pathway is evolutionarily conserved, and given the widespread expression of ALDH1 in a variety of cell types, including cells of the retina (19, 20) and hippocampus (21), and subset of SPNs (fig.S5), our findings suggest that GABA alternative synthesis may represent a more fundamental mechanism employed by broader classes of GABAergic neurons.
The dorsal striatum plays important roles in the transitioning from initial voluntary drug use to habitual, and ultimately compulsive, drug abuse (29, 37, 38). The GABAergic synapse has also been the focus of extensive study for its role in the behavioral consequences of EtOH exposure. In particular, acute and chronic EtOH exposure has been suggested to modulate GABAergic release and synaptic GABAA receptors through pre- and postsynaptic mechanisms, respectively (39). Our studies indicate that EtOH attenuates GABA co-release by inhibiting GABA bio-synthesis at binge drinking alcohol concentrations, providing an additional mechanism through which EtOH exposure can modulate the activity of GABAergic synapses. Diminished GABA co-release and ALDH1a1 activity may directly contribute to enhancement of alcohol intake and preference behavior, reminiscent of humans with Aldh1a1 mutations. In evaluating genetic factors associated with risk of alcohol abuse then, we will likely need to consider pre- and post-synaptic components that converge on the GABAergic system. Together, our data propose that GABA co-release, in addition to DA, may serve an essential function in the regulation of the development of addictive behaviors.
Supplementary Material
Acknowledgments
The authors thank Amy Du, Dr. Yu Liu for technical assistance, Dr. Dorit Ron for suggestions on 2-bottle choice behavior test, and Drs. Tom Sudhof, Georgia Panagiotakos, Wei Wei, Zayd Khaliq and members of Ding laboratory for helpful discussions. Supported by grants from the NINDS/NIH NS075136, NS091144 (J.B.D.) and the Klingenstein Foundation (J.B.D.), NIMH MH086403 (L.C.), MH091193 (L.C.). All primary electrophysiological and immunohistochemical data are archived on a server in the Department of Neurosurgery at Stanford University School of Medicine.
Footnotes
The authors declare no conflicts of interest.
SUPPLEMENTARY MATERIALS
Materials and Methods
figs. S1 to S14
Table. S1
References (40, 50)
References
- 1.Schultz W. Multiple dopamine functions at different time courses. Annual review of neuroscience. 2007;30:259. doi: 10.1146/annurev.neuro.28.061604.135722. [DOI] [PubMed] [Google Scholar]
- 2.Tritsch NX, Sabatini BL. Dopaminergic modulation of synaptic transmission in cortex and striatum. Neuron. 2012 Oct 4;76:33. doi: 10.1016/j.neuron.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen BT, Hopf FW, Bonci A. Synaptic plasticity in the mesolimbic system: therapeutic implications for substance abuse. Annals of the New York Academy of Sciences. 2010 Feb;1187:129. doi: 10.1111/j.1749-6632.2009.05154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redgrave P, et al. Goal-directed and habitual control in the basal ganglia: implications for Parkinson's disease. Nature reviews. Neuroscience. 2010 Nov;11:760. doi: 10.1038/nrn2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tritsch NX, Ding JB, Sabatini BL. Dopaminergic neurons inhibit striatal output through non-canonical release of GABA. Nature. 2012 Oct 11;490:262. doi: 10.1038/nature11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez-Hernandez T, Barroso-Chinea P, Acevedo A, Salido E, Rodriguez M. Colocalization of tyrosine hydroxylase and GAD65 mRNA in mesostriatal neurons. The European journal of neuroscience. 2001 Jan;13:57. [PubMed] [Google Scholar]
- 7.Tritsch NX, Oh WJ, Gu C, Sabatini BL. Midbrain dopamine neurons sustain inhibitory transmission using plasma membrane uptake of GABA, not synthesis. eLife. 2014;3:e01936. doi: 10.7554/eLife.01936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuda W, et al. Single nigrostriatal dopaminergic neurons form widely spread and highly dense axonal arborizations in the neostriatum. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009 Jan 14;29:444. doi: 10.1523/JNEUROSCI.4029-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heusner CL, Beutler LR, Houser CR, Palmiter RD. Deletion of GAD67 in dopamine receptor-1 expressing cells causes specific motor deficits. Genesis. 2008 Jul;46:357. doi: 10.1002/dvg.20405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lammel S, et al. Diversity of transgenic mouse models for selective targeting of midbrain dopamine neurons. Neuron. 2015 Jan 21;85:429. doi: 10.1016/j.neuron.2014.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoerbelt P, Lindsley TA, Fleck MW. Dopamine directly modulates GABAA receptors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015 Feb 25;35:3525. doi: 10.1523/JNEUROSCI.4390-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams K. Modulation and block of ion channels: a new biology of polyamines. Cellular signalling. 1997 Jan;9:1. doi: 10.1016/s0898-6568(96)00089-7. [DOI] [PubMed] [Google Scholar]
- 13.Xing SG, Jun YB, Hau ZW, Liang LY. Higher accumulation of gamma-aminobutyric acid induced by salt stress through stimulating the activity of diamine oxidases in Glycine max (L.) Merr. roots. Plant physiology and biochemistry : PPB / Societe francaise de physiologie vegetale. 2007 Aug;45:560. doi: 10.1016/j.plaphy.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Shelp BJ, et al. Hypothesis/review: contribution of putrescine to 4-aminobutyrate (GABA) production in response to abiotic stress. Plant science : an international journal of experimental plant biology. 2012 Sep;193–194:130. doi: 10.1016/j.plantsci.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Bell MR, Belarde JA, Johnson HF, Aizenman CD. A neuroprotective role for polyamines in a Xenopus tadpole model of epilepsy. Nature neuroscience. 2011 Apr;14:505. doi: 10.1038/nn.2777. [DOI] [PubMed] [Google Scholar]
- 16.Sequerra EB, Gardino P, Hedin-Pereira C, de Mello FG. Putrescine as an important source of GABA in the postnatal rat subventricular zone. Neuroscience. 2007 May 11;146:489. doi: 10.1016/j.neuroscience.2007.01.062. [DOI] [PubMed] [Google Scholar]
- 17.Noto T, et al. The effect of L-erythro-dihydroxyphenylserine injected into the lateral ventricle and the hypothalamus on the locomotor activity. Pharmacology, biochemistry, and behavior. 1986 Aug;25:411. doi: 10.1016/0091-3057(86)90017-1. [DOI] [PubMed] [Google Scholar]
- 18.Yamasaki EN, Barbosa VD, De Mello FG, Hokoc JN. GABAergic system in the developing mammalian retina: dual sources of GABA at early stages of postnatal development. International journal of developmental neuroscience : the official journal of the International Society for Developmental Neuroscience. 1999 Jun;17:201. doi: 10.1016/s0736-5748(99)00002-7. [DOI] [PubMed] [Google Scholar]
- 19.Laschet J, Grisar T, Bureau M, Guillaume D. Characteristics of putrescine uptake and subsequent GABA formation in primary cultured astrocytes from normal C57BL/6J and epileptic DBA/2J mouse brain cortices. Neuroscience. 1992;48:151. doi: 10.1016/0306-4522(92)90345-3. [DOI] [PubMed] [Google Scholar]
- 20.Barres BA, Koroshetz WJ, Swartz KJ, Chun LL, Corey DP. Ion channel expression by white matter glia: the O-2A glial progenitor cell. Neuron. 1990 Apr;4:507. doi: 10.1016/0896-6273(90)90109-s. [DOI] [PubMed] [Google Scholar]
- 21.Aoto J, Nam CI, Poon MM, Ting P, Chen L. Synaptic signaling by all-trans retinoic acid in homeostatic synaptic plasticity. Neuron. 2008 Oct 23;60:308. doi: 10.1016/j.neuron.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu G, et al. Aldehyde dehydrogenase 1 defines and protects a nigrostriatal dopaminergic neuron subpopulation. The Journal of clinical investigation. 2014 Jul;124:3032. doi: 10.1172/JCI72176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCaffery P, Drager UC. High levels of a retinoic acid-generating dehydrogenase in the meso-telencephalic dopamine system. Proceedings of the National Academy of Sciences of the United States of America. 1994 Aug 2;91:7772. doi: 10.1073/pnas.91.16.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasiliou V, Pappa A, Petersen DR. Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism. Chemico-biological interactions. 2000 Dec 1;129:1. doi: 10.1016/s0009-2797(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 25.Goldstein DS, et al. Determinants of buildup of the toxic dopamine metabolite DOPAL in Parkinson's disease. Journal of neurochemistry. 2013 Sep;126:591. doi: 10.1111/jnc.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan X, et al. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Molecular and cellular biology. 2003 Jul;23:4637. doi: 10.1128/MCB.23.13.4637-4648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherva R, et al. Associations and interactions between SNPs in the alcohol metabolizing genes and alcoholism phenotypes in European Americans. Alcoholism, clinical and experimental research. 2009 May;33:848. doi: 10.1111/j.1530-0277.2009.00904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, et al. Haplotype-based study of the association of alcohol-metabolizing genes with alcohol dependence in four independent populations. Alcoholism, clinical and experimental research. 2011 Feb;35:304. doi: 10.1111/j.1530-0277.2010.01346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Everitt BJ, Robbins TW. Neural systems of reinforcement for drug addiction: from actions to habits to compulsion. Nature neuroscience. 2005 Nov;8:1481. doi: 10.1038/nn1579. [DOI] [PubMed] [Google Scholar]
- 30.Chen G, et al. Striatal involvement in human alcoholism and alcohol consumption, and withdrawal in animal models. Alcoholism, clinical and experimental research. 2011 Oct;35:1739. doi: 10.1111/j.1530-0277.2011.01520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, et al. Ethanol-mediated facilitation of AMPA receptor function in the dorsomedial striatum: implications for alcohol drinking behavior. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012 Oct 24;32:15124. doi: 10.1523/JNEUROSCI.2783-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Dell LE, Roberts AJ, Smith RT, Koob GF. Enhanced alcohol self-administration after intermittent versus continuous alcohol vapor exposure. Alcoholism, clinical and experimental research. 2004 Nov;28:1676. doi: 10.1097/01.alc.0000145781.11923.4e. [DOI] [PubMed] [Google Scholar]
- 33.Ben Hamida S, et al. Protein tyrosine phosphatase alpha in the dorsomedial striatum promotes excessive ethanol-drinking behaviors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013 Sep 4;33:14369. doi: 10.1523/JNEUROSCI.1954-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hnasko TS, et al. Vesicular glutamate transport promotes dopamine storage and glutamate co-release in vivo. Neuron. 2010 Mar 11;65:643. doi: 10.1016/j.neuron.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Root DH, et al. Single rodent mesohabenular axons release glutamate and GABA. Nature neuroscience. 2014 Nov;17:1543. doi: 10.1038/nn.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shabel SJ, Proulx CD, Piriz J, Malinow R. Mood regulation. GABA/glutamate co-release controls habenula output and is modified by antidepressant treatment. Science. 2014 Sep 19;345:1494. doi: 10.1126/science.1250469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lovinger DM. Neurotransmitter roles in synaptic modulation, plasticity and learning in the dorsal striatum. Neuropharmacology. 2010 Jun;58:951. doi: 10.1016/j.neuropharm.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pennartz CM, et al. Corticostriatal Interactions during Learning, Memory Processing, and Decision Making. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009 Oct 14;29:12831. doi: 10.1523/JNEUROSCI.3177-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiner JL, Valenzuela CF. Ethanol modulation of GABAergic transmission: the view from the slice. Pharmacology & therapeutics. 2006 Sep;111:533. doi: 10.1016/j.pharmthera.2005.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.