Significance
Protein aggregation is a hallmark of neurodegenerative disease and is hypothesized to cause neuron death. Despite extensive study of disease-associated aggregating proteins, mechanisms of neuron death remain a mystery, and no cures or effective treatments yet exist. Here, we demonstrate the toxicity of a small aggregate of the Cu,Zn superoxide dismutase (SOD1) protein, associated with amyotrophic lateral sclerosis (ALS). We present an experimentally verified structural model of this toxic species and show that SOD1 mutants designed to promote formation of this aggregate increase cell death, providing a direct link between aggregate presence and neuron death. Knowledge of toxic species and the ability to manipulate their formation provides a valuable direction for pursuit of therapeutic strategies in ALS.
Keywords: neurodegeneration, protein aggregation, protein misfolding, ALS, structural modeling
Abstract
Since the linking of mutations in the Cu,Zn superoxide dismutase gene (sod1) to amyotrophic lateral sclerosis (ALS) in 1993, researchers have sought the connection between SOD1 and motor neuron death. Disease-linked mutations tend to destabilize the native dimeric structure of SOD1, and plaques containing misfolded and aggregated SOD1 have been found in the motor neurons of patients with ALS. Despite advances in understanding of ALS disease progression and SOD1 folding and stability, cytotoxic species and mechanisms remain unknown, greatly impeding the search for and design of therapeutic interventions. Here, we definitively link cytotoxicity associated with SOD1 aggregation in ALS to a nonnative trimeric SOD1 species. We develop methodology for the incorporation of low-resolution experimental data into simulations toward the structural modeling of metastable, multidomain aggregation intermediates. We apply this methodology to derive the structure of a SOD1 trimer, which we validate in vitro and in hybridized motor neurons. We show that SOD1 mutants designed to promote trimerization increase cell death. Further, we demonstrate that the cytotoxicity of the designed mutants correlates with trimer stability, providing a direct link between the presence of misfolded oligomers and neuron death. Identification of cytotoxic species is the first and critical step in elucidating the molecular etiology of ALS, and the ability to manipulate formation of these species will provide an avenue for the development of future therapeutic strategies.
Protein misfolding and aggregation are linked to cell death and disease progression in neurodegenerative diseases, such as Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis (ALS). In these diseases and others, the formation of amyloid plaques, often observed post mortem, has long been thought to play a role in neurodegeneration, but toxicity has never been confirmed (1–3). Recent research has shown that small, soluble oligomers, rather than insoluble amyloids, are likely to be the cytotoxic species causing neurodegeneration (4–14). These small, soluble oligomers undergo aberrant interactions with cell machinery and activate cell death pathways, but their exact stoichiometry is not known and their properties have yet to be characterized. Recently, metastable soluble Cu,Zn superoxide dismutase (SOD1) oligomers have been identified that contain an epitope associated with disease-linked species of SOD1, mutants of which are implicated in a subset of ALS (15–18). Size exclusion chromatography (SEC) of these oligomers revealed a size range of two to four monomers, consistent with previous findings of potentially cytotoxic SOD1 oligomers (19–21).
Knowledge of the structures of these species would not only allow for definitive testing of their toxicity but could potentially lead to an understanding of disease mechanism and therapeutic strategies against diseases for which no cure or effective treatment exists (22). By their nature, however, these oligomers are only metastable, and therefore difficult to isolate. More importantly, this instability, and consequent transient nature, combined with the proposed size range of the implicated oligomers, means that a high-resolution structure cannot be achieved using traditional methods. Although atomic structures have been explored for folding intermediates of small protein domains (23) or stable oligomeric structures of disease-relevant peptide sequences (24), structural characterization of metastable aggregation products of full-length disease-linked proteins has remained elusive.
Here, we combined experimental and computational approaches to produce a molecular model of a toxic metastable protein oligomer. Using state-of-the-art high-speed atomic force microscopy (HS-AFM), we established the trimeric stoichiometry and highly dynamic nature of the isolated oligomers. We applied limited proteolysis to the isolated SOD1 trimers, allowing us to determine the solvent accessibility and rigidity of the structure. Using this information as constraints in molecular simulations, we evolved a structural model of the SOD1 trimer that is consistent with our experimental data. Using our model, we determined residues critical to trimer formation, and we used rational mutagenesis to verify our model and manipulate trimer formation in vitro and in living cells. We find that mutations that stabilize the trimeric form of SOD1 result in increased cell death over WT SOD1 or SOD1 mutants that inhibit trimer formation, demonstrating the neurotoxicity of the SOD1 trimer and its potential relevance to ALS etiology.
Results
Determination of SOD1 Oligomer Stoichiometry.
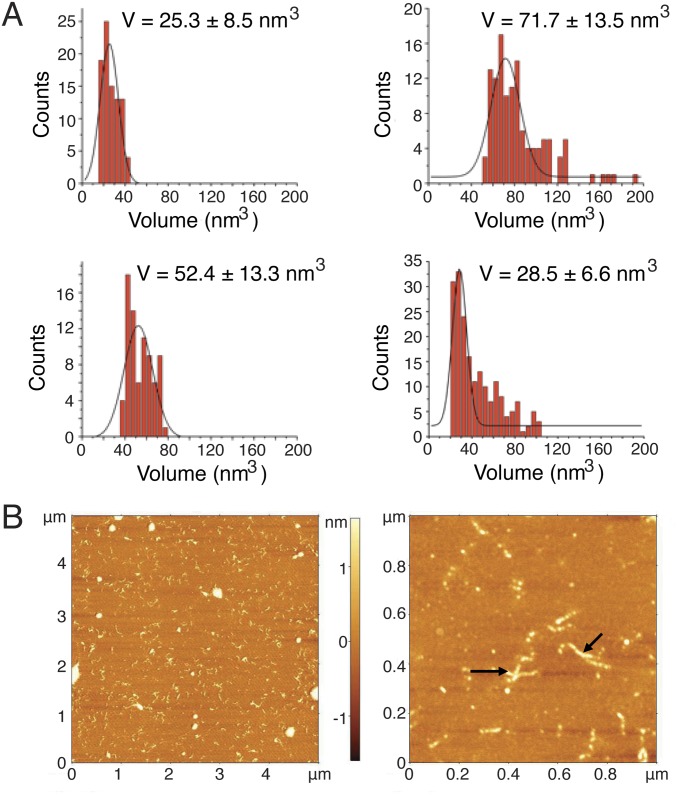
To model potentially toxic SOD1 oligomers accurately, we first determined the number of SOD1 monomers participating in the oligomers using definitive images obtained by both regular AFM and HS-AFM. We find that the metastable SOD1 oligomers are stabilized at pH 3.5, where the SOD1 aggregation pathway has been shown to proceed more rapidly while retaining the same features and characteristics as the physiological process. We have demonstrated previously that although low pH accelerates progression along the aggregation pathway by stabilizing species further down that pathway, it does not affect the pathway itself (25). At pH 3.5, where the SOD1 oligomers are stable, oligomer volume is ∼71.7 nm3, threefold greater than oligomer volume of the monomer (∼25.3 nm3; Fig. S1A). These data are supported by HS-AFM images resolving the three-partite structure of the oligomer (Fig. 1A). We conclude that the primary SOD1 oligomer previously identified by Redler et al. (15) is a trimer. Upon increase of pH to 7.4, where the SOD1 oligomer is less stable, the majority of trimers dissociate into monomers, as confirmed by volumetric AFM measurements (Fig. 1B and Fig. S1A). We find that any larger aggregates encountered at low pH are mainly short fibrillar structures (Fig. S1B).
Fig. S1.
Volume distributions of SOD1 oligomers demonstrate trimeric stoichiometry. (A) At pH 3.5, the volume (V) distribution of SOD1 monomer is narrow, with its peak centered at 25.3 nm3 (Left), whereas the volume distribution of SOD1 oligomer is broad, with its peak centered at 71.7 nm3 (Right). Notably, the volume of oligomer is approximately triple the volume of monomer, denoting that the SOD1 oligomer is trimeric. (Left) At pH 7.4, the peak of the dimeric volume distribution is centered at 52.4 nm3. (Right) Oligomeric volume distribution, although featuring a broad tail leading up to oligomeric values, has a peak centered at 28.5 nm3, comparable to the SOD1 monomer at pH 3.5, denoting that the SOD1 oligomer is primarily dissociated at pH 7.4. Errors are ±SD. Data were obtained with regular AFM of dried samples. (B) Using AFM, under trimer-stabilizing conditions, we observe small populations of short, insoluble fibrils with a molecular mass >600 kDa: 5 μm × 5 μm (Left) and 1 μm × 1 μm (Right) images. The color bar denotes feature height. Arrows indicate fibrils. Data and images were obtained with AFM of dried samples.
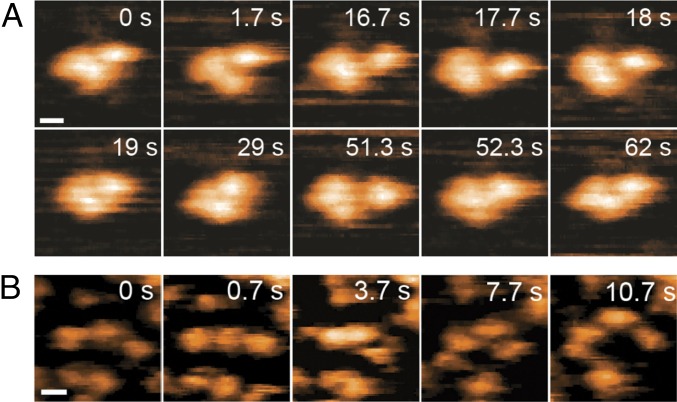
Fig. 1.
SOD1 forms nonnative trimer. (A) HS-AFM analysis reveals that under stabilizing conditions (pH 3.5), SOD1 oligomer features a tripartite structure, in line with volumetric AFM measurements of dry samples, where oligomer volume is ∼71.7 nm3 and monomer volume is ∼25.3 nm3 (Fig. S1A). We observe three distinct monomers in HS-AFM recordings at a SOD1 concentration of 30 nM. The three subunits form a nonspherical compact particle, which is stable over long time periods. (Scale bar: 5 nm.) (B) In destabilizing conditions (pH 7.4), SOD1 trimers dissociate into three distinct monomers, even at the high end of physiological concentration (100 nM). (Scale bar: 5 nm.)
SOD1 Trimer Features Nonnative Secondary, Tertiary, and Quaternary Structure.
After purification (26), aggregation, and isolation of the SOD1 trimer (15), we conducted limited proteolysis, followed by MS, to identify unstructured, solvent-accessible regions (27). Several proteolytic cuts are located in regions that are buried in the dimer interface or contain secondary structure elements in the SOD1 dimer (Fig. 2A), which would make proteolysis impossible or highly unlikely if native secondary and tertiary structure were conserved. Limited proteolysis of SOD1 dimer and isolated SOD1 monomer demonstrate that the pattern of proteolytic cleavage differs significantly (Fig. 2B). In addition to secondary and tertiary structure differences, we conclude from proteolytic data that the interface(s) between monomers in the SOD1 trimer must be located in a different region of the monomer from the native SOD1 dimer interface (i.e., a significant change in quaternary structure). The SOD1 trimer therefore is not simply the result of an addition of a “loose” monomer to a preexisting native-like SOD1 dimer, but features significant rearrangement from the native configuration and fold that necessitates complete dissociation of the native dimer and aggregation of partially unfolded monomers, in agreement with previous findings (25, 28).
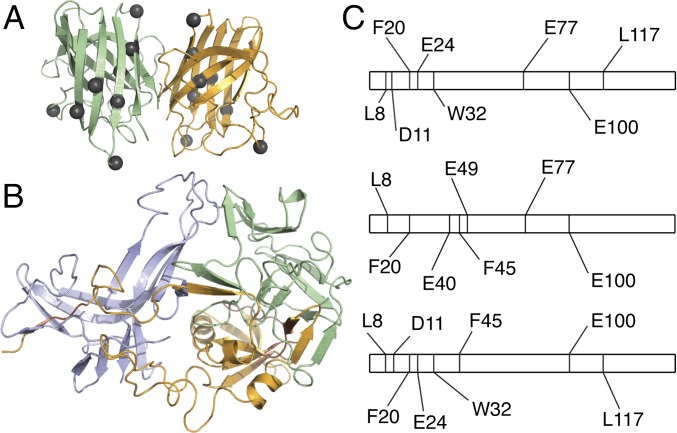
Fig. 2.
Hybrid experimental/computational method leads to a model of metastable SOD1 trimer. (A) SOD1 trimer proteolytic cut sites (gray spheres, shown here on the native dimer structure) in the dimer interface and secondary structural elements suggest significant structural differences between native and trimeric structures. (B) Structural model of the metastable SOD1 trimer obtained using limited proteolysis data as constraints in several rounds of coarse-grained and atomistic DMD simulations (Fig. S2). (C) Representation of the SOD1 linear sequence, residues 1–153. Proteolytic cut sites (vertical lines) determined in limited proteolysis experiments differ significantly in SOD1 monomer (Top), dimer (Middle), and trimer (Bottom), supporting structural rearrangement during the aggregation process and SOD1 trimer formation.
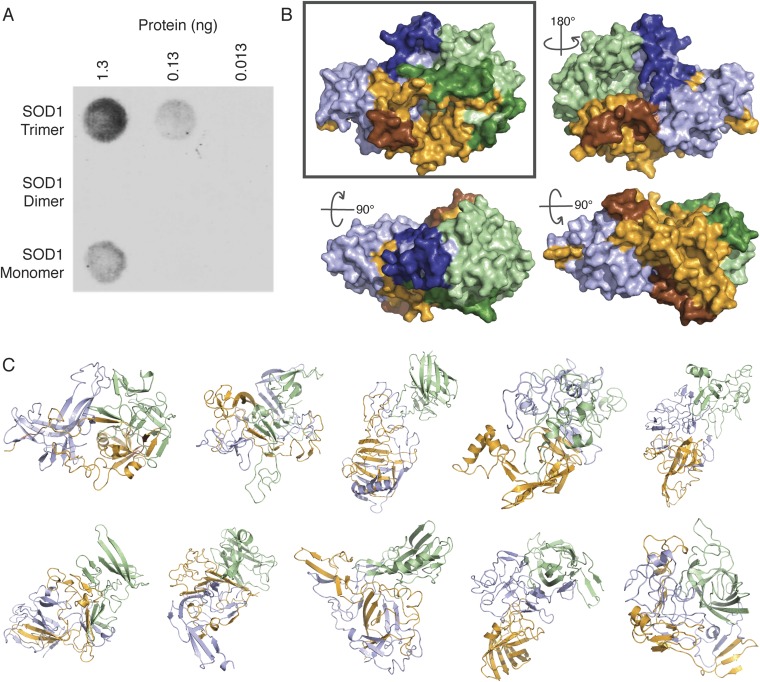
To characterize this rearrangement and obtain a structure of the SOD1 trimer, we performed coarse-grained discrete molecular dynamics (DMD) simulations of three unbound SOD1 monomers with a knowledge-based force field based on the requirement for the experimentally obtained proteolytic cut sites to be unstructured and solvent-exposed (Fig. S2). We designed our force field such that native contacts are attractive to each other and proteolytic cut sites are repulsive to all other residues, fulfilling the requirements for cleavage and introducing competition between the native and nonnative states (Fig. S3). After extensive coarse-grained sampling, followed by equilibration in a physics-based, all-atom force field (29), we obtained a model of the SOD1 trimer that agrees with experimental results (Fig. 2C and Movie S1): The majority of identified proteolytic cut sites are surface-exposed and reside in unstructured regions. Due to competition between the native fold and the artificially imposed, experimentally derived bias, not all proteolytic cut sites are surface-exposed in the set of lowest energy structures, although they are present in the population as a whole (Dataset S1). We find that, consistent with our conclusions from limited proteolysis data, the individual monomers in the SOD1 trimer are severely misfolded and deviate significantly, with rmsds from the native monomer of 12.5 Å, 19.3 Å, and 11.7 Å. More importantly, as predicted from proteolysis data, the potentially cytotoxic SOD1 trimer does not make use of the same monomer–monomer interface as the native SOD1 dimer. Instead, residues comprising the SOD1 dimer interface are spread into discrete regions on each monomer, with a significant fraction solvent-exposed in the SOD1 trimer (Movie S2). Additionally, the recently discovered epitope of the C4F6 Ab (18), which preferentially binds the SOD1 trimer (Fig. S4A) as well as several other disease-linked SOD1 species (15, 16) over the native dimer or monomer, is completely exposed on the surface of the trimer structure (Fig. S4B), further supporting our model and suggesting the potential toxicity of the SOD1 trimer. The atomic coordinates of our SOD1 trimer model are available for download at dokhlab.org.
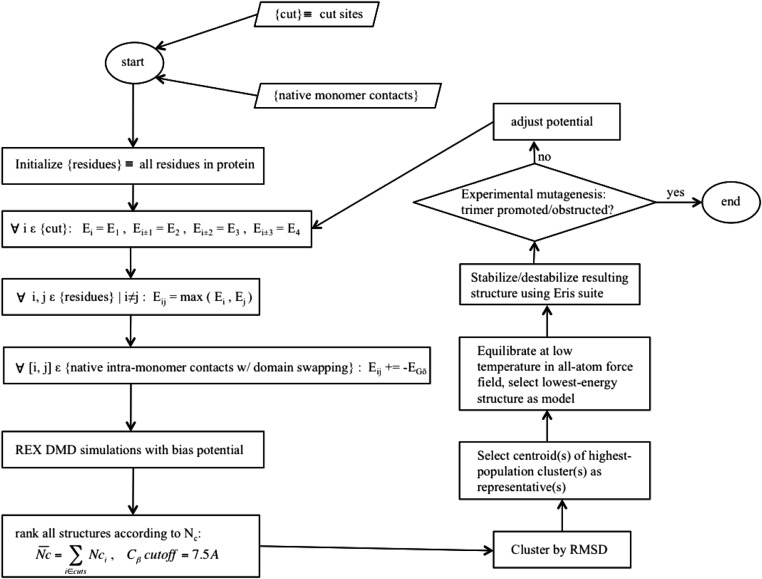
Fig. S2.
Schematic diagram for structural modeling of metastable protein oligomers. The procedure for deriving a bias potential using experimental constraints from limited proteolysis is illustrated. The term {cut} is the set of residues at which a proteolytic cut is made. E1, E2, E3, and E4 are the repulsive potentials applied to (i) the residues that are the site of proteolytic cuts, (ii) residues 1 removed in sequence from cut sites, (iii) residues 2 removed in sequence from cut sites, and (iv) residues 3 removed in sequence from cut sites. Eij are the interresidue interaction energies applied when residues are within contact range. EGō is the interaction energy assigned to native contacts that are in contact range. Nc is the average number of contacts made by each cut site residue.
Fig. S3.
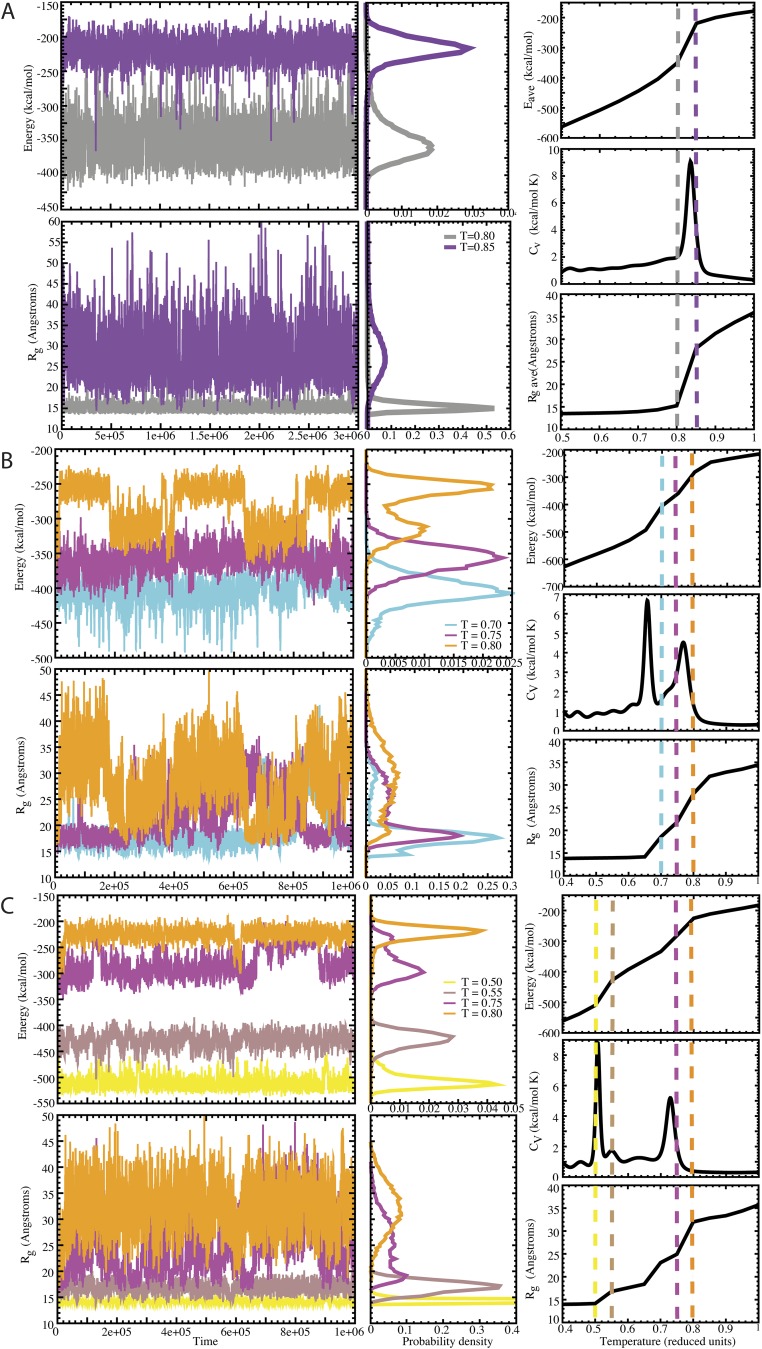
Folding of SOD1 monomer with various values of λ. (A) λ = 0: SOD1 monomer folding features two distinct states with no folding intermediates. Transitions in energy and the radius of gyration are sharp and well-defined. Trajectories from T = 0.80 and T = 0.85 are located at the transition, and exhibit flipping between folded and unfolded structures. (B) λ = 0.66: SOD1 monomer folding features distinct intermediate states. Transitions in energy and the radius of gyration remain well-defined. Trajectories from T = 0.70, T = 0.75, and T = 0.80 are located at the transition, and exhibit flipping between states. (C) λ = 0.99: SOD1 monomer folding features distinct intermediate states, with a greater separation in energy at λ = 0.66. Transitions in energy and the radius of gyration are well-defined. Trajectories from T = 0.50 and T = 0.55 and from T = 0.75 and T = 0.80 are located at the transitions, and exhibit flipping between two respective states. CV, heat capacity at constant volume.
Fig. S4.
Toxic epitope exposed on SOD1 trimer surface. Ensemble of SOD1 trimer models. (A) Conformational Ab C4F6, which binds several disease-associated species of SOD1 (16), selectively binds to WT SOD1 trimer at various concentrations, but does not bind to native WT type SOD1 dimer and exhibits minimal binding to WT SOD1 monomer. (B) Recently identified epitope of the C4F6 Ab (16, 18) is exposed on the surface of our SOD1 trimer model. Individual monomers are depicted in pale green, bright orange, and light blue; residues comprising the C4F6 epitope are highlighted in darker colors: forest, brown, and deep blue, respectively. Rotation angles describe the transformation from the “front” (Upper Left), such that the structures represent the “back” (Upper Right), “top” (Lower Left), and “bottom” (Lower Right). The C4F6 Ab has been shown previously to bind WT and disease-linked mutant SOD1 trimer at physiological pH (15). In addition to experimental data from limited proteolysis, the exposure of the C4F6 epitope further verifies our model, as well as providing support for the toxicity of SOD1 trimer. (C) Replicate SOD1 trimer models resulting from 10 independent repetitions of our protocol (Fig. S3) feature significant variation in tertiary and quaternary structure. Models vary in the degree of domain swapping, as well as in the amount of native tertiary structure maintained in each monomer. We note that despite differences in tertiary and quaternary structure of the 10 SOD1 trimer models, a high level of consensus on the identity of residues involved in trimeric interface interactions is maintained, with at least 77% and as much as 94% identity in interface residue identity between any two models.
Mutations in Predicted Monomer–Monomer Interfaces Affect SOD1 Trimer Formation.
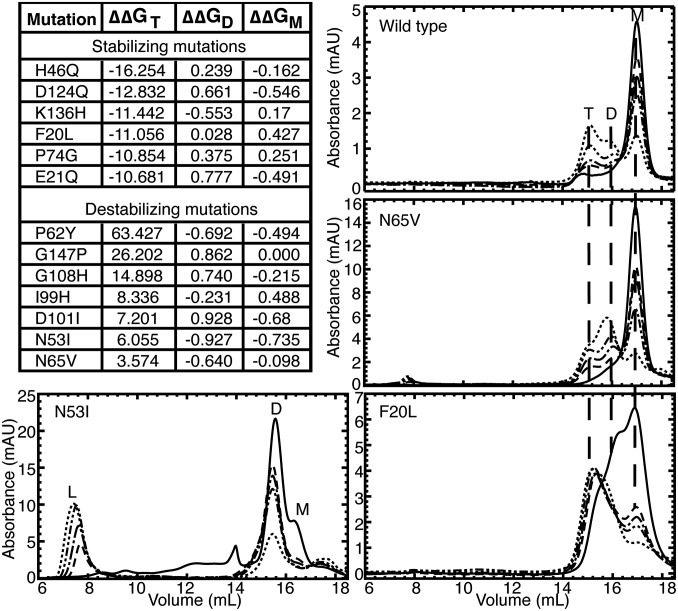
To verify our model of the SOD1 trimer, we designed single-residue mutations to the predicted SOD1 trimer interfaces and tested the effects of these mutations on trimer formation in vitro. Mutations that we predict to stabilize the SOD1 trimer should result in an increased trimer population, whereas mutations that we predict to destabilize the trimer structure should result in a decreased or absent trimer population. Using Monte Carlo energy minimization, we predicted the change in free energy (ΔΔG) of mutation for all interface residues to all possible amino acids, and selected for experimental evaluation those mutations having a significant effect on SOD1 trimer stability (|ΔΔGmut| > 3 kcal/mol). To ascertain that the outcomes of any mutation observed in vitro are caused by directly affecting SOD1 trimer formation and not due to stabilization or destabilization of native SOD1 species, we selected only those mutants that feature a negligible effect (|ΔΔGmut| < 1 kcal/mol) on the native SOD1 dimer and monomer species (Protein Data Bank ID code 1SPD) (Fig. 3).
Fig. 3.
Designed mutations verify the model and demonstrate control of SOD1 aggregation. Mutations to trimer interface residues designed to stabilize or destabilize SOD1 trimer but having no effect on SOD1 monomer or dimer, are shown with ΔΔGmut for the trimer, native dimer, and native monomer structures. Aggregation time courses were measured for each mutant after incubation at physiological concentration (30 μM) and 37 °C for for 0 h (solid line), 2 h (− −), 4 h (− − •), 8 h (− • •), or 24 h (• •). The aggregation of apo-WT SOD1 is shown for comparison, with trimer (T), dimer (D), monomer (M), and large aggregate (L) peaks labeled when present. Vertical dashed lines between panels aid comparison of trimer, dimer, and monomer peaks between mutants. We find that the N65V mutation, predicted to be destabilizing to the SOD1 trimer, results in a smaller population of trimer than WT, shifting the SOD1 population toward dimer and monomer formations. The N53I mutation, also predicted to destabilize SOD1 trimer, results in no detectable SOD1 trimer but, instead, increased populations of large aggregates. The F20L mutation, predicted to stabilize SOD1 trimer, promotes trimer formation early in the aggregation process and maintains high levels of trimer throughout the experiment. Additional aggregation time courses can be found in Fig. S5. mAU, milli-absorbance units.
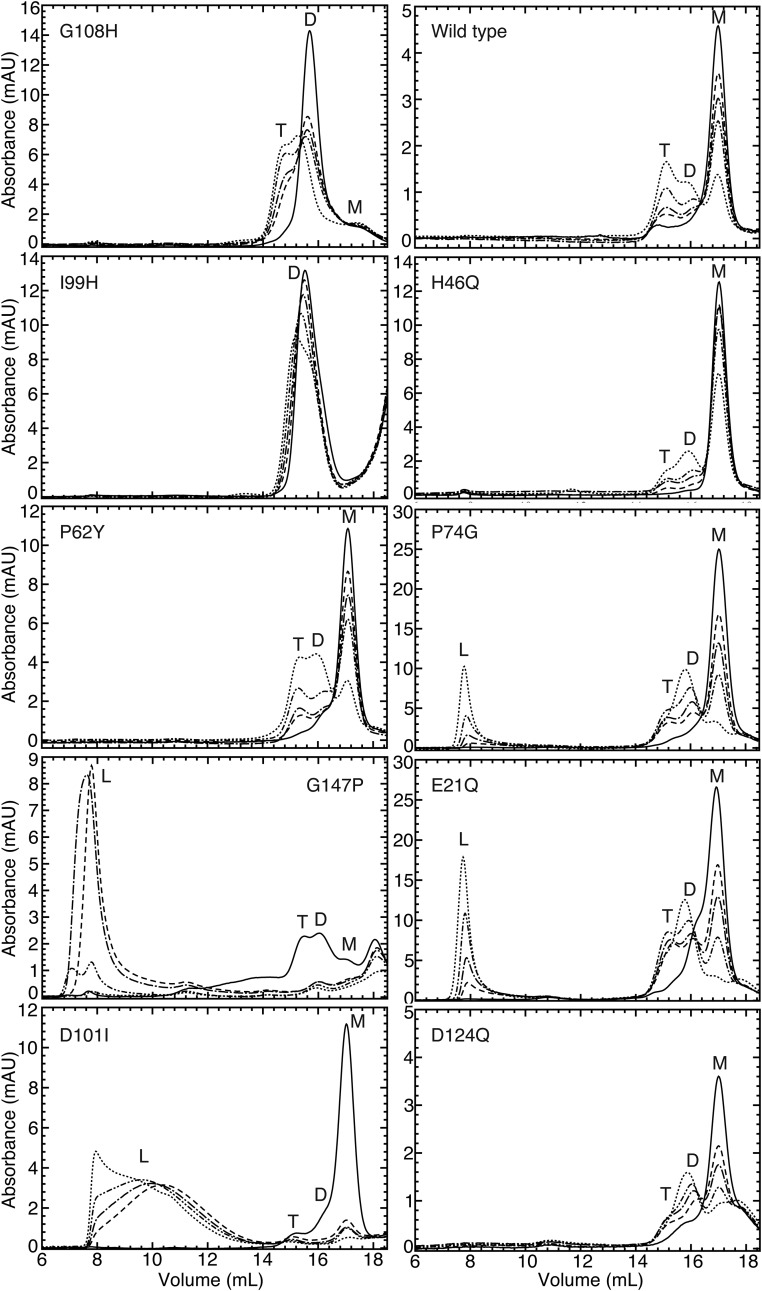
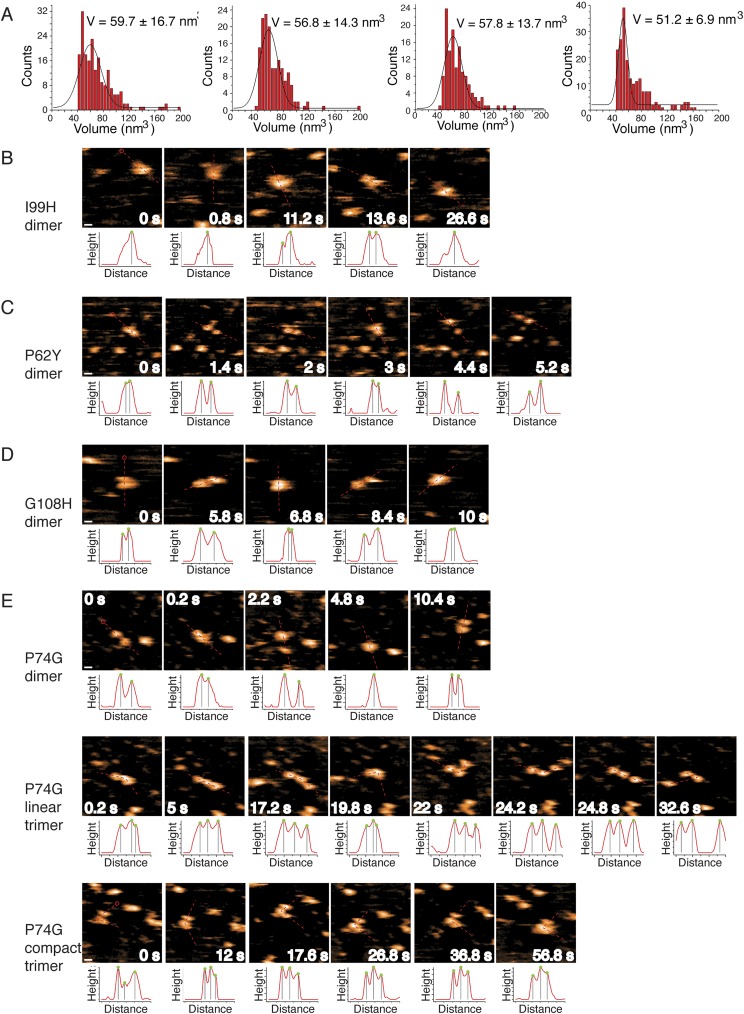
We observed the aggregation time course of our designed SOD1 mutants using SEC. All seven of our designed destabilizing mutations successfully decrease or eliminate the population of SOD1 trimer. Upon destabilization of the trimer interfaces, we observe one of two outcomes: (i) inhibited trimer formation with resulting increased populations of SOD1 monomer and dimer species or (ii) inhibited trimer formation with a resulting increase in large aggregates. Exhibiting the first outcome, in four cases (P62Y-SOD1, G108H-SOD1, I99H-SOD1, and N65V-SOD1), we observe the formation of a nonnative extended dimer species, confirmed by AFM (Fig. 3 and Figs. S5 and S6). In the second possible outcome of trimer-destabilizing mutations, we observe the immediate formation of large aggregates in D101I-SOD1, G147P-SOD1, and N53I-SOD1 (Fig. 3 and Figs. S5 and S6). In these high-aggregation mutants, some SOD1 trimer is formed in the early hours of the time course, quickly followed by the appearance of large aggregates. None of these three trimer-destabilizing mutations form SOD1 trimer, instead quickly aggregating into large, potentially insoluble aggregates, supporting the hypothesis that large, insoluble aggregates, such as fibrils, lie on a different, competing pathway from the formation of small, soluble aggregates.
Fig. S5.
Designed mutations verify the model and demonstrate control of SOD1 aggregation. Mutations to trimer interface residues are designed to stabilize or destabilize SOD1 trimer but to have no effect on SOD1 monomer or dimer. ΔΔGmut for the trimer, native dimer, and native monomer structures can be found in Fig. 3. Aggregation time courses were measured for each mutant after incubation at physiological concentration (30 μM) and 37 °C for 0 h (solid line), 2 h (− −), 4 h (− − •), 8 h (− • •), or 24 h (• •). The aggregation of apo-WT SOD1 is shown for comparison, with trimer (T), dimer (D), monomer (M), and large aggregate (L) peaks labeled when present. We find that the G108H, I99H, and P62Y mutations, predicted to be destabilizing to the SOD1 trimer, result in smaller populations of trimer than WT, shifting the SOD1 population toward dimer and monomer formations. The G147P and D101I mutations, also predicted to destabilize SOD1 trimer, result in no or very little detectable SOD1 trimer after 24 h, but instead increased populations of large aggregates, especially in G147P-SOD1. In four additional mutants that we predict to be trimer-stabilizing, H46Q-SOD1, D124Q-SOD1, P74G-SOD1, and E21Q-SOD1, we observe the formation of trimer that is overcome by a nonnative extended dimer species (Fig. S6), which (as discussed in the main text) we could not predict computationally. mAU, milli-absorbance units.
Fig. S6.
Mutations to SOD1 trimer interfaces stabilize formation of nonnative extended dimer conformations. (A) Volume distributions of (left to right) I99H, P74G, P62Y, and G108H after 24 h of aggregation at 30 μM SOD1 at 37 °C indicate dominance of a nonnative dimer species. Solid lines are Gaussian fits, with most probable volume designated. Errors are ±SD. (B) I99H-SOD1, a trimer-destabilizing mutant, exists almost exclusively as an extended dimer, as demonstrated by HS-AFM images. (C) P62Y-SOD1, a trimer-destabilizing mutant, exists as an extended dimer, as demonstrated by HS-AFM images. (D) G108H-SOD1, a trimer-destabilizing mutant, exists as an extended dimer with a small trimeric population (not shown), as demonstrated by HS-AFM images. (E) P74G-SOD1, a trimer-stabilizing mutant, exists as an extended dimer and linear or compact trimer. In B–E, the scan area is 50 nm × 50 nm. (Scale bar: B–E, 5 nm.) Dashed lines on the image represent the 1D cross-section used for height profiling below each image, with red circles indicating the initial point of measurement. Green dots on the height profiles indicate peak positions, with the distance between the two gray lines representing the distance of two adjacent subunits. Data were obtained with regular AFM imaging of dried samples.
Because the SOD1 trimer is a nonnative, metastable, intermediate aggregation state, stabilizing its formation is a much more challenging task than destabilizing it. In stabilizing one transient species, we may unknowingly stabilize, destabilize, or even create another transient species. Although we can account for the stabilization or destabilization of known structures, such as the native SOD1 dimer and monomer, aggregation is a complex process, and we cannot account for other metastable species that may be perturbed during our mutation of trimer interface residues, which may affect whether or not we observe increased levels of SOD1 trimer. Despite these difficulties, we succeeded in predicting and designing a SOD1 trimer-stabilizing mutant. The F20L-SOD1 mutant demonstrates a clear stabilization of the SOD1 trimer species: monomer, dimer, and trimer species are all present at time t = 0, but the trimer species quickly dominates and stabilizes over t = 24 h, with no larger aggregates forming (Fig. 3). Five additional mutations predicted to stabilize the SOD1 trimer, while exhibiting a trimer population, result in stabilization of other nonnative species ranging from misfolded monomer to protofibrils (Fig. S5), with stoichiometry confirmed by AFM (Fig. S6). This finding suggests that the nonnative monomer–monomer interfaces we identify in the SOD1 trimer are also exploited by other potentially toxic species.
Trimer-Stabilizing SOD1 Mutants Promote Cell Death.
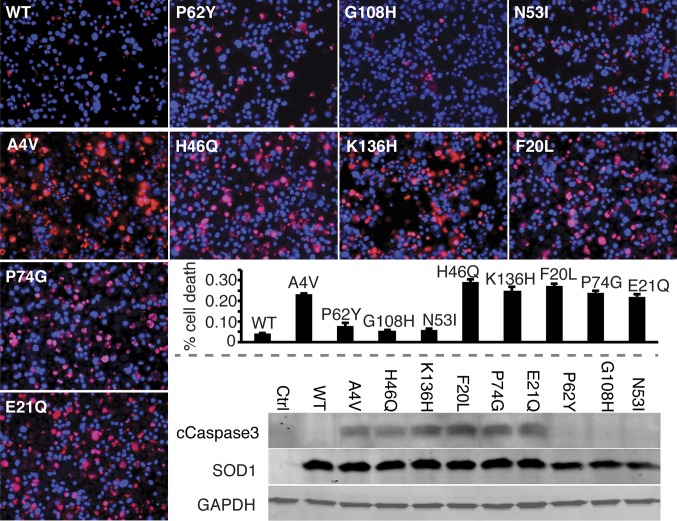
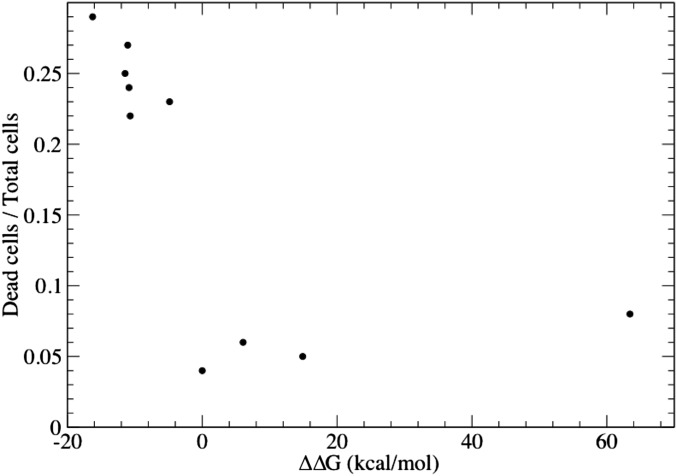
Because mutations can cause conformational changes in proteins, especially when located in a structurally important region like an interface, SOD1 mutations clinically linked to ALS could cause an entirely different trimeric species to form, which we would not capture in our model. Computational testing of the stabilizing effect of each mutation on all possible SOD1 trimer structures would be prohibitive, and still may not yield an answer to the crucial question: Do disease mutations cause an increase in the population of trimeric SOD1? Instead, we address the question of SOD1 trimer disease relevance from another direction: by directly testing the toxicity of SOD1 trimer in motor neuron-like cells (NSC-34 cells). We transfected cells with expression constructs for each of our designed SOD1 mutants, WT SOD1, and A4V-SOD1, an aggressive ALS-linked mutation responsible for the majority of familial ALS cases in North America (30, 31), which we use as a positive control for ALS-relevant cell death. The A4V mutation has previously been observed to promote the formation nonnative SOD1 trimer (15). We observed significantly increased cell death in NSC-34 hybrid motor neuron cells expressing A4V-SOD1 (positive control) or our designed trimer-stabilizing SOD1 mutants (Fig. 4). Conversely, our trimer-destabilizing mutants do not cause cell death, with cell viability comparable to cells expressing WT SOD1 (Fig. 4). Similarly, expression of A4V-SOD1 or trimer-stabilizing SOD1 mutants, but not trimer-destabilizing SOD1 mutants, resulted in elevated levels of cleaved caspase-3 compared with WT SOD1 (Fig. 4). Finally, we note that the amount of cell death present was correlated to the stability of the SOD1 mutant trimer (Fig. S7).
Fig. 4.
SOD1 trimer is cytotoxic. Expression of A4V-SOD1 (positive control) and trimer-stabilizing mutant SOD1s in NSC-34 motor neuron-like cells increases the incidence of cell death compared with WT SOD1 (negative control), whereas trimer-destabilizing mutants have no effect on viability. Applied 3 d posttransfection, red stain [propidium iodide (PI)] identifies nuclei of dead cells, whereas blue stain (Hoechst) identifies nuclei of all cells. Incidence of cell death is measured as the percentage of total cells exhibiting PI staining (left to right from Upper Left): WT, 4%; P62Y, 8%; G108H, 5%; N53I, 6%; A4V, 23%; H46Q, 29%; K136H, 25%; F20L, 27%; P74G, 24%; and E21Q, 22%. (Magnification: 20×.) Cell death correlates with trimer stability with a P value of 0.0476, or a P value of 0.000966 with exclusion of the outlier P62Y, which is extremely destabilizing (Fig. S7). Elevated levels of the apoptotic marker cleaved caspase 3 in NSC-34 motor neuron hybrid cells expressing A4V-SOD1 (positive control) and trimer-stabilizing SOD1 mutants (demonstrated by Western blot, Lower Right) confirm increased cytotoxicity of these mutants compared with WT SOD1 (negative control) and trimer-destabilizing SOD1 mutants.
Fig. S7.
Stabilization of trimer correlates with cell death. The size of the SOD1 trimer population is proportional to the stability of that species in relation to other SOD1 species. We find that the stability of the SOD1 trimer is highly indicative of the incidence of cell death, with the stability of the mutants corresponding to cell death with a P value of 0.0476 (as calculated from the Fisher test) when all points are included. With removal of the outlier P62Y-SOD1, which mutation is extremely destabilizing at +62 kcal/mol (Fig. 3), the P value of the relation between SOD1 trimer stability and cell death becomes 0.001.
Discussion
We conclude that the SOD1 trimer causes cell death in motor neuron-like cells, and that the nonnative trimeric interfaces appear to be shared by multiple cytotoxic SOD1 species. Studies from many groups have suggested potential mechanisms of SOD1 oligomer neurotoxicity but were unable to characterize these oligomers fully. Excellent reviews of these mechanisms can be found elsewhere (32–34). Many of these studies resulted in such broad characterization that we may only conclude that the toxic SOD1 species was soluble; however, in those studies that determine a more limited size range, we note that the size of these oligomers closely mirrors the size of the SOD1 trimer, and we propose that the trimeric form of SOD1 that we describe in this work is a likely candidate for the aberrant and detrimental interactions with cellular machinery observed in these previous studies.
Given that SOD1 has been shown to undergo aberrant aggregation in both familial and sporadic ALS (35–37), we propose that formation of SOD1 trimer is a common pathogenic mechanism in ALS, causing the death of motor neurons and progression of the disease. At the time of writing, 62% of residues known to feature disease-relevant point mutations are located in the proposed SOD1 trimer interfaces (Table S1 and Dataset S1). Moreover, at least two-thirds of disease mutations located in the trimer interfaces are overall stabilizing to trimer formation; the effect of the remaining third cannot currently be established because of the metastable nature of these oligomers. Due to metastability, the SOD1 trimer exists as an ensemble of diverse structures (Fig. S4C and Dataset S1), and although the effect of a mutation on one trimeric structure may be destabilizing, the same mutation could promote formation of another trimeric form, resulting in a higher fraction of disease mutants stabilizing a trimeric form of SOD1.
Table S1.
ALS-linked mutations stabilize SOD1 trimer
| Original AA | Residue | Mutation | ΔΔGT | ΔΔGD | ΔΔGM |
| A | 4 | S | −9.51 | 5.728 | −2.256 |
| A | 4 | T | −12.996 | 9.072 | −2.741 |
| A | 4 | V | −4.844 | 8.659 | 0.62 |
| V | 7 | E | −3.711 | 12.873 | −4.104 |
| L | 8 | Q | −1.052 | 12.078 | 1.658 |
| L | 8 | V | −6.481 | 1.131 | 4.981 |
| G | 10 | R | 15.741 | 20 | 2.724 |
| G | 10 | V | 16.084 | 20 | −2.917 |
| G | 12 | R | −2.968 | 20 | 2.05 |
| G | 16 | S | −5.171 | 19.269 | 6.765 |
| N | 19 | S | −7.77 | 2.324 | −2.961 |
| E | 21 | K | −10.52 | 4.360 | 1.798 |
| Q | 22 | L | −11.609 | 4.624 | −6.264 |
| G | 37 | R | −7.104 | 7.437 | 22.474 |
| L | 38 | V | −2.494 | −0.416 | 6.446 |
| G | 41 | D | 2.055 | 0.098 | −3.873 |
| G | 41 | S | −1.33 | 1.254 | −1.104 |
| H | 46 | R | −1.715 | 7.855 | −1.624 |
| H | 48 | Q | −5.294 | 6.819 | 0.665 |
| H | 48 | R | −2.546 | 5.670 | −5.052 |
| N | 65 | S | 3.241 | 1.199 | −0.813 |
| D | 76 | V | −6.563 | 12.791 | 2.922 |
| D | 76 | Y | −6.816 | 2.921 | −1.19 |
| G | 93 | A | −3.804 | 12.803 | 0.707 |
| G | 93 | D | 22.897 | 20 | −0.728 |
| G | 93 | R | 33.12 | 20 | −0.087 |
| G | 93 | S | 2.8 | 20 | 9.944 |
| G | 93 | V | 20.699 | 14.997 | 15.008 |
| A | 95 | T | −15.237 | 16.537 | −0.281 |
| D | 101 | G | −0.759 | 4.967 | −1.86 |
| D | 101 | H | 0.31 | 9.361 | 0.142 |
| D | 101 | N | −8.335 | −1.371 | −1.119 |
| D | 101 | Y | 0.921 | 14.853 | −1.064 |
| I | 104 | F | 19.486 | 20 | 6.935 |
| S | 105 | L | −14.356 | −7.769 | −6.383 |
| L | 106 | F | 13.187 | 20 | 2.706 |
| L | 106 | V | 7.353 | 4.349 | −2.924 |
| I | 112 | M | −3.703 | −7.888 | −1.988 |
| I | 112 | T | −4.586 | 6.172 | 2.826 |
| G | 114 | A | −10.812 | 11.700 | −1.375 |
| R | 115 | G | −3.484 | −0.425 | 0.439 |
| V | 118 | L | 2.858 | 8.345 | −4.0 |
| D | 124 | G | −6.602 | 7.756 | −4.152 |
| D | 125 | H | −6.848 | 5.791 | −3.271 |
| L | 127 | R | −2.293 | 20 | 24.563 |
| S | 134 | N | −8.502 | 3.170 | 6.118 |
| G | 141 | E | 21.177 | 7.283 | 1.218 |
| A | 145 | T | −8.669 | 5.199 | 4.399 |
| G | 147 | R | 24.122 | 20 | 20.585 |
| V | 148 | G | 1.548 | 13.447 | −5.315 |
| V | 148 | I | −11.136 | −2.324 | 2.884 |
Predicted ΔΔG (in kilocalories per mole) for known ALS disease mutations on structures of the SOD1 trimer, dimer, and monomer. Dimer and monomer structures are from Protein Data Bank ID code 1SPD; trimer structure, as in the main text, is model 1 described in Dataset S1. We note that our identified mutations for stabilizing or destabilizing the SOD1 trimer are not known disease mutants, likely because we specially selected them to affect only the trimeric form of SOD1 and not the native monomer or dimer species, artificially limiting our choices. Our purpose in choosing such mutations was for clean, unambiguous results verifying the identity of residues in the trimer interfaces, whereas the disease is most likely caused by mutations that affect the stability of all forms of SOD1, including the native forms for which we deselect here. AA, amino acid; ΔΔGD, change in free energy upon mutation for the structure of SOD1 native dimer; ΔΔGM, change in free energy upon mutation for the structure of SOD1 native monomer; ΔΔGT, change in free energy upon mutation for the structure of SOD1 trimer.
Recognizing and sequestering SOD1 trimer or preventing its formation is therefore a potential strategy for preventing neurodegeneration in ALS. Because of the high concentration of SOD1 in motor neurons (30–100 μM), stoichiometric binding of a drug compound to native SOD1 may not be a viable option to abolish trimer formation completely. One option is to stabilize larger, nontoxic aggregation products; this strategy has been adopted for amyloid-β aggregates in Alzheimer’s disease, to positive effect in a cellular model (9). However, another strategy would involve identifying and blocking toxic interactions of SOD1 trimer in the cell. Alternatively, the structural knowledge that we uncover in this work can be used for the design of recognition and sequestering strategies. The characterization of structural features, such as the identification of nonnative aggregation interfaces in this work, provides a key step in the understanding of ALS disease etiology and development of therapeutic or preventive strategies.
Materials and Methods
Purification of SOD1.
For determination of surface-accessible sites in the SOD1 trimer in limited proteolysis studies, we isolated and purified SOD1 from human erythrocytes, as previously described (26). To generate SOD1 mutants for aggregation time courses using SEC, and WT SOD1 for comparison of comparable species, we performed cloning, expression, and purification of human recombinant SOD1 in Saccharomyces cerevisiae as described elsewhere (15, 26, 38). We determined the concentration of purified SOD1 species by measuring the A at 280 nm with an extinction coefficient of 10,800 M−1⋅cm−1. We stored samples at −80 °C after flash-freezing in liquid nitrogen.
Formation and Isolation of SOD1 Species.
We formed SOD1 trimer by incubating SOD1 dimer at a concentration of 100–120 μM in 50 mM acetate, 150 mM NaCl, and 10 mM EDTA at pH 3.5 for 24 h. We then isolated trimer and monomer using an AKTA Purifier with a Superdex 200 10/300 GL column (GE Healthcare) equilibrated in either 50 mM phosphate and 150 mM NaCl at pH 3.5 (for use with V8 protease) or 50 mM acetate, 150 mM NaCl, and 10 mM EDTA at pH 3.5 (for all other proteases). We isolated monomer species fresh daily. We obtained trimer fractions from gel filtration chromatography, concentrated using an Amicon Ultra filter (Millipore) and used fresh or stored at 4 °C overnight and used the next day.
AFM.
For AFM imaging of dried samples, we immersed freshly cleaved mica stripes (5.0 × 1.5 cm; Asheville Schoonmaker Mica Co.) in plastic cuvettes containing 167 μM 1-(3-aminopropyl)silatrane (APS) solution for 30 min (39), followed by deionized water rinsing and drying in argon flow. We stored the APS mica in a vacuum chamber for use over the next few weeks (39). We cut the strips into ∼0.75 × 0.75-cm pieces for sample preparation. We diluted the SOD1 samples to the appropriate concentrations (3.3–10 nM) with the corresponding buffers before deposition onto APS mica [50 mM sodium acetate, 150 mM NaCl, and 10 mM EDTA (pH 3.5) or 20 mM Tris⋅HCl and 150 mM NaCl (pH 7.4)]. We deposited 20 mL of diluted sample onto APS mica surfaces for 2 min, followed by thorough rinsing with deionized water and drying with argon flow. We kept the specimens in a vacuum chamber for at least 3 h, after which we mounted the specimens onto metal discs with double-sided tape (carbon-coated; Ted Pella, Inc.). We performed AFM imaging on a MultiMode 8 atomic force microscope (Bruker NanoA, Bruker Co., Santa Barbara, CA) equipped with PeakForce modulus with microlever (MSNL; Bruker) probes. We acquired AFM images with 512 × 512 pixels at a scan rate of 4–6 Hz. We conducted volume and height analysis using Femtoscan (Advanced Technologies Center). We used “Enum features,” a functional tool provided by the software, to measure the heights and volumes of the protein in AFM images (40). We measured the background over the noncovered surface area. We performed Gaussian fitting of the volume and height values using Origin 6.0 software (Microcal Software, Inc.).
We acquired HS-AFM images using an HS-AFM instrument, operating in tapping mode in liquid (RIBM). We prepared samples as reported by Miyagi et al. (41). Briefly, we glued freshly cleaved 1.5-mm mica discs to the glass rods and modified with 167 μM APS for 30 min. After rinsing with 20 μL Milli-Q water (AquaMax-Ultra, LabWater.com, a division of APS water Services Corporation, Van Nuys, CA) and then three times with 20 μL of pH 3.5 buffer (50 mM sodium acetate with an additional 150 mM NaCl and 10 mM EDTA), we deposited the SOD1 samples onto the APS mica in the same buffer. We performed measurements at two different concentrations of SOD1: 100 nM and 30 nM. After 5 min of incubation, we rinsed the surface with the same buffer used for imaging. To compare the effect of pH on sample dynamics, we removed the pH 3.5 buffer by pipette and injected the pH 7.4 buffer (20 mM Tris with 150 mM NaCl) to continue sample scanning. We used BL-AC10DS-A2 cantilevers (Olympus) with carbon tips obtained by electron beam deposition for imaging (39). The spring constant of the AFM probes is between 0.1 and 0.2 N/m, with resonance frequency between 400 and 1,000 kHz in water. We performed continued scanning over the selected area (50 nm × 50 nm) with a scan rate of approximately three frames per second. Images contain 128 × 128 pixels. We processed HS-AFM images using home-built software written by Atsushi Miyagi, Y.L.L. laboratory, University of Nebraska Medical Center, Omaha, NE.
Limited Proteolysis.
We performed limited proteolysis at 25 °C in 100 mM buffer (acetate, phosphate, or Tris) at the appropriate pH for each enzyme: V8 protease reactions in 100 mM phosphate buffer (pH 4.0), chymotrypsin reactions in 100 mM Tris 50 mM CaCl2 buffer (pH 7.8), and pepsin reactions in 100 mM acetate buffer (pH 3.5), quenched by addition of 1 M Tris buffer (pH 8.0) to a final concentration of 150 mM. We performed 1- to 50-min time course proteolysis reactions for trimer, allowing the reaction to proceed for a sufficient time to allow a single proteolytic cut and no more than four proteolytic cuts: 1 min for V8 and pepsin reactions and 2 min for chymotrypsin reactions. This technique preserves the structural integrity of the trimer during the period of the limited proteolytic reaction, ensuring relevance of the obtained structural information. Time course reactions for monomer and dimer extended longer, as is necessary to obtain SOD1 cleavage. The proteolysis reaction for dimer contained an equimolar SOD1 concentration to the trimer reaction (7–15 μM), but we performed reactions with SOD1 monomer at the concentration eluted from the FPLC (2.6–5 μM), because we could not concentrate the monomer without significant aggregation. We determined protease/SOD1 ratios empirically. We quenched samples by the addition of 100 mM PMSF (1 mM final) and froze them until ready for further use. We desalted samples using Ziptip C18 pipette tips (Millipore) and eluted the peptides in 20 μL of MS grade solvent [50% acetonitrile, 50% water, and 0.1% TFA (vol/vol); Fisher].
MS.
We performed MS on an AB SCIEX 4800 PLUS MALDI-TOF-TOF instrument at the University of North Carolina Michael Hooker Proteomics Center using α-cyano-4-hydroxycinnamic acid as the matrix. Data output was analyzed in Data Explorer, with the signal-to-noise ratio set to 20–30, MS/MS tolerance set to ±1 Da, and peptide charge set to 1+. We identified peptides using Mascot (Matrix Science) (42) with the UniProt database, with the search limited to the taxonomy Homo sapiens. We used the data from the earliest time points that resulted in cleavage (1–2 min) for constraints in modeling (Table S2).
Table S2.
Human SOD1 limited proteolytic digest peptides identified by MS
| Form of SOD | Observed | Mr(expt) | Mr(calc) | Peptide | Mascot assignment | Notes |
| V8 (100 mM phosphate, pH 4.0, t = 1 min) | ||||||
| Monomer | No matches | First peptide identified at t = 5 min | ||||
| Dimer | No matches | No peptides out to t = 10 min | ||||
| Trimer | 1,459.0857 | 1,458.0784 | 1,457.7514 | D.GPVQGIINFEQKE.S | SOD1 G12-E24 | |
| 2,396.6919 | 2,395.6846 | 2,395.1616 | E.ERHVGDLGNVTADKDGVADVSIE.D | SOD1 E78-E100 | ||
| Chymotrypsin (100 mM Tris 50 mM CaCl2, pH 7.8, t = 2 min) | ||||||
| Monomer | No matches | First peptide identified at t = 10 min | ||||
| Dimer | No matches | No peptides out to t = 10 min | ||||
| Trimer | 1,400.4094 | 1,399.4021 | 1,399.7096 | F.EQKESNGPVKVW.G | SOD1 E21-W32 | |
| 3,580.9929 | 3,579.9856 | 3,580.7904 | L.VVHEKADDLGKGGNEESTKTGNAGSRLACGVIGIAQ.- | SOD1 V118-Q153 | ||
| Pepsin (100 mM acetate, pH 3.5, t = 1 min) | ||||||
| Monomer | 1,244.4987 | 1,243.4914 | 1,243.6561 | L.KGDGPVQGIINF.E | SOD1 K9-F20 | |
| 2,696.9983 | 2,695.9910 | 2,696.3922 | F.EQKESNGPVKVWGSIKGLTEGLHGF.H | SOD1 E21-F45 | ||
| Dimer | 1,244.7208 | 1,243.7135 | 1,243.6561 | L.KGDGPVQGIINF.E | SOD1 K9-F20 | |
| 2,697.4995 | 2,696.4922 | 2,696.3922 | F.EQKESNGPVKVWGSIKGLTEGLHGF.H | SOD1 E21-F45 | ||
| Trimer | 1,244.6426 | 1,243.6353 | 1,243.6561 | L.KGDGPVQGIINF.E | SOD1 K9-F20 | |
We used three proteases with different specificity: V8 cuts after D and E; chymotrypsin cuts after F, W, Y, H, M, and L (unless followed by P); and pepsin cuts before L, F, W, and Y (unless preceded by P). Buffer, pH, and digest time are listed for each limited proteolytic reaction with the purified SOD1 monomer, dimer, or trimer. SOD1 monomer and dimer required longer digest times with V8 or chymotrypsin to observe proteolytic cleavage products; we collected time points of proteolytic cleavage reactions out to t = 10 min. Notably, we did not observe proteolytic cleavage of the SOD1 dimer with V8 or chymotrypsin even after 10 min of incubation, in agreement with the known highly stable structure of the native SOD1 dimer. Mr(calc) is the calculated molecular mass from the matched peptide sequence, Mr(expt) is the experimental m/z transformed to a relative molecular mass, and Observed is the experimental m/z from the mass spectrometer. Definitions are from Matrix Science (www.matrixscience.com/help/pmf_summaries_help.html#PROTSUM).
Incorporation of Experimental Constraints.
To incorporate information from limited proteolysis experiments into DMD simulations, we created an algorithm for converting knowledge of the sequence positions of proteolytic cleavage sites into pairwise simulation constraints (Fig. S2). The formation of trimeric SOD1 is made possible by a perturbation to the native state, which must occur as local unfolding in a background of native interactions. To account for this phenomenon, we first represent the native background as a Gō potential (43), which rewards contacts that are present in the native structure, assigning a pairwise attraction of 1 kcal/mol between pairs of residues with a distance between beta-carbons that is less than 7.5 Å. Next, we define a bias potential based on the location of proteolytic cleavage sites. Because each cleavage site must necessarily be (i) solvent-exposed and (ii) unstructured, we assign a repulsive interaction of energy E0 between the cleavage site j and every other residue in the system. Because residues near a solvent-exposed site are also likely to be solvent-exposed, and because an unstructured region of ∼12 residues surrounding the cleavage site is necessary for the proteolytic enzyme to gain access to the site (27, 44), we apply a stepwise, decreasingly repulsive potential to the two residues on either side of each cleavage site, such that the pairwise repulsive interaction of the respective residue with each residue in the system has energy:
For each identified cleavage site, we thus generate 5n pairwise constraints, where n is the total number of residues in the system. All Gō and experimental constraint potentials are additive. Thus, the combined potential function is:
where Δij is the native contact matrix of the conformation, δij is the contact matrix of the current conformation, and λ is a scaling parameter discussed in the next section.
Parameterization of Bias Potential.
To scale the two terms of our bias potential, we tested the effects of various values of λ from 0.0 to 30 at intervals of 0.3 (from 0.0 to 3.0) or 3 (from 3 to 30) in coarse-grained (45) replica exchange simulations of apo-SOD1 monomer, which features a well-defined folding pathway. We examined the resulting energetic profiles and folding trajectories, and selected values of λ for which folding intermediates are introduced by the competition between Gō and experimental energy terms but distinct folding transitions are evident. At λ = 0 (Gō potential only), we find a single, sharp folding transition with no intermediates, as expected from experimental findings (46). At λ ≥ 1.5, we cannot resolve individual thermodynamic transitions, and protein unfolding occurs with increasing linearity as λ increases. We find that that λ values 0.33 ≤ λ ≤ 1.2 fit our criteria for selection, and that values in this range produce nearly identical final structures in our test apo-monomer system (Fig. S3). We therefore selected λ = 0.99 for use in modeling of trimeric SOD1.
Coarse-Grained DMD.
To obtain initial trimeric SOD1 structures that agree with experimental data, we use DMD (47, 48) replica exchange (REX) (49) simulations. We apply our bias potential to a coarse-grained model of three natively folded SOD1 apo-monomers placed in proximity to each other but not initially bound. We use a four-bead protein model (45) for increased sampling and for simplified application of the bias potential to the Cβ bead of each residue, with the exception of Ala, for which the bias potential is applied to the Cα bead. For each value of λ, we determined the ideal number of replicas and spread of replica temperatures such that we sample energetic space spanning the entire melting transition and that exchange of replicas occurs with an acceptance rate between 0.2 and 0.7, with exchange attempted every 1,000 time steps. For λ = 0.99, we used 27 replicas, with temperatures of 0.435, 0.445, 0.455, 0.462, 0.470, 0.480, 0.490, 0.503, 0.513, 0.523, 0.538, 0.552, 0.566, 0.581, 0.595, 0.610, 0.625, 0.639, 0.654, 0.674, 0.694, 0.713, 0.733, 0.753, 0.768, 0.788, and 0.815 kcal⋅mol−1⋅kB−1. We performed simulations for 106 time steps.
We evaluated the experimental agreement of each structure snapshot from simulation by calculating the average number of contacts made by each proteolytic cut site residue (Nc). Our rationale is that to demonstrate agreement with limited proteolysis results, each proteolytic cut site residue should make as few contacts with other residues as possible, because its ability to be cleaved by proteolytic enzymes denotes its solvent accessibility and lack of participation in secondary structure interactions. However, although the proteolytic cleavage sites should demonstrate a minimum of structural interaction, SEC results (15) indicate that the structure should be a compact, associated trimer. We therefore use a combination of proteolytic cut site contacts and trimer radius of gyration (Rg) to select a pool of candidate structures from all REX simulation trajectories. When we examined these two criteria together, we found a cluster of structures having Nc < 1.5 and Rg < 30 Å. We clustered this pool of candidate structures by pairwise rmsd, and selected the centroid of the largest cluster for further simulation and structural refinement.
All-Atom DMD.
We reconstructed the four-bead centroid structure obtained from the DMD REX simulations described above to an all-atom model (29) and performed structural minimization using Chiron (50) to remove clashes introduced by reconstruction. To equilibrate our structure in a physical force field, we first performed low-temperature (below the melting transition) all-atom REX simulations using 26 replicas with temperatures of 0.350, 0.360, 0.370, 0.380, 0.390, 0.400, 0.410, 0.420, 0.430, 0.440, 0.450, 0.460, 0.470, 0.480, 0.490, 0.500, 0.510, 0.520, 0.530, 0.540, 0.550 0.560, 0.570, 0.580, 0.590, and 0.600 kcal⋅mol−1⋅kB−1. We selected the ideal number of replicas and spread of replica temperatures such that exchange of replicas occurs with an acceptance rate between 0.2 and 0.7, with exchange attempted every 1,000 time steps. We performed simulations for 106 time steps. We isolated the lowest energy structure from all simulation replicas, and continued simulation of that structure at a temperature of 0.350 kcal⋅mol−1⋅kB−1 for an additional 106 time steps. Finally, we selected the lowest energy structure from this single-temperature trajectory as our final model, and verified the quality of the model using Gaia (51).
Computational Mutagenesis.
Using our final SOD1 trimer model, we performed computational mutagenesis of every residue residing in an interface between monomer chains. We designate a residue as residing in an interface if any atom of that residue is within 4.5 Å of any atom of a residue belonging to a different monomer chain. We performed computational mutagenesis of each interface residue to every possible residue and calculated the change in free energy upon mutation (ΔΔGmut) using Eris (52, 53). Due to complexity in their interactions, we did not perform mutations to Cys.
SOD1 Aggregation Time Course Experiments.
We used time-resolved analytical SEC to measure the relative population sizes of the various SOD1 species over time. We diluted purified mutant and WT SOD1 to 30 μM and incubated them at 37 °C at time 0 (t = 0). At designated time points (t = 0, t = 2, t = 4, t = 8, t = 24), we withdrew and filtered aliquots of each sample, and separated SOD1 species by SEC at 4 °C using an AKTA Purifier with a Superdex 200 10/300 GL column (GE Healthcare) equilibrated in 50 mM acetate, 150 mM NaCl, and 10 mM EDTA (pH 3.5).
Measurement of Cytotoxicity of SOD1 Trimer.
We maintained neuroblastoma spinal cord hybrid cell lines (NSC-34) in DMEM containing 10% (vol/vol) FBS. We plated ∼5,000 NSC-34 cells in one well of a 24-well plate with differentiation medium [1% FBS, 1% nonessential amino acid supplement, (vol/vol) 10 μM of RA]. After a 2-d differentiation period in DMEM, we transfected cells with 2 μg of plasmid encoding WT SOD1 or SOD1 mutants (within the pCI-Neo vector) using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. To verify that SOD1 had been properly transfected, we performed immunostaining. We fixed cells 24 h posttransfection using 4% (vol/vol) PFA for 10 min at room temperature. We blocked with 3% (vol/vol) goat serum, followed by Abs against SOD1 (Sigma) and Hoechst 33342 counterstain. We performed propidium iodide (red) and Hoechst (blue) staining in culture 3 d posttransfection without fixation. We performed a Western blot with ∼20,000 cells per well, collecting cells 2 d posttransfection. We lysed cells in PBS with 0.5% Triton X-100, 0.5 mM EDTA, and EDTA-free protease inhibitor (Roche Life Sciences). We denatured cell lysates and resolved using 18% (vol/vol) SDS/PAGE, and incubated with Abs against cleaved caspase-3 (Cell Signaling Technology), SOD1 (Calbiochem), and GAPDH (Chemcon), followed by secondary Ab incubation. We visualized protein bands using an Odyssey IR imaging system (LI-COR Biosciences). Cell viability was not assayed in all mutants because we were unable to verify SOD1 transfection in five of our predicted mutants, four of which are trimer-destabilizing mutations: D124Q-SOD1, G147P-SOD1, I99H-SOD1, D101I-SOD1, and N65V-SOD1.
Supplementary Material
Acknowledgments
We thank Dr. Feng Ding and Dr. Michael Caplow for helpful discussions, and Dr. Joan S. Valentine for graciously providing the EG118 yeast strain and the yEP351:hwtSOD1 vector. MS data were collected at the University of North Carolina Michael Hooker Proteomics Center with help from Nedyalka Dicheva. This work was supported by NIH Grants R01GM080742 (to N.V.D.), R01GM096039 (to Y.L.L.), and R01GM078366 (to M.D.). E.A.P. was supported by a Ruth L. Kirschstein National Research Service Award (F31AG039266) from the National Institute on Aging.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1516725113/-/DCSupplemental.
References
- 1.Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: Deciphering selective motor neuron death in ALS. Nat Rev Neurosci. 2001;2(11):806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 2.Rothstein JD. Current hypotheses for the underlying biology of amyotrophic lateral sclerosis. Ann Neurol. 2009;65(Suppl 1):S3–S9. doi: 10.1002/ana.21543. [DOI] [PubMed] [Google Scholar]
- 3.Walker LC, LeVine H., 3rd Corruption and spread of pathogenic proteins in neurodegenerative diseases. J Biol Chem. 2012;287(40):33109–33115. doi: 10.1074/jbc.R112.399378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redler RL, Dokholyan NV. The complex molecular biology of amyotrophic lateral sclerosis (ALS) Prog Mol Biol Transl Sci. 2012;107:215–262. doi: 10.1016/B978-0-12-385883-2.00002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zetterström P, et al. Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. Proc Natl Acad Sci USA. 2007;104(35):14157–14162. doi: 10.1073/pnas.0700477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirkitadze MD, Bitan G, Teplow DB. Paradigm shifts in Alzheimer’s disease and other neurodegenerative disorders: The emerging role of oligomeric assemblies. J Neurosci Res. 2002;69(5):567–577. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- 7.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431(7010):805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 8.Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: Separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 9.Bieschke J, et al. Small-molecule conversion of toxic oligomers to nontoxic β-sheet-rich amyloid fibrils. Nat Chem Biol. 2012;8(1):93–101. doi: 10.1038/nchembio.719. [DOI] [PubMed] [Google Scholar]
- 10.Rakhit R, et al. An immunological epitope selective for pathological monomer-misfolded SOD1 in ALS. Nat Med. 2007;13(6):754–759. doi: 10.1038/nm1559. [DOI] [PubMed] [Google Scholar]
- 11.Jonsson PA, et al. Minute quantities of misfolded mutant superoxide dismutase-1 cause amyotrophic lateral sclerosis. Brain. 2004;127(Pt 1):73–88. doi: 10.1093/brain/awh005. [DOI] [PubMed] [Google Scholar]
- 12.Israelson A, et al. Misfolded mutant SOD1 directly inhibits VDAC1 conductance in a mouse model of inherited ALS. Neuron. 2010;67(4):575–587. doi: 10.1016/j.neuron.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedrini S, et al. ALS-linked mutant SOD1 damages mitochondria by promoting conformational changes in Bcl-2. Hum Mol Genet. 2010;19(15):2974–2986. doi: 10.1093/hmg/ddq202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishitoh H, et al. ALS-linked mutant SOD1 induces ER stress- and ASK1-dependent motor neuron death by targeting Derlin-1. Genes Dev. 2008;22(11):1451–1464. doi: 10.1101/gad.1640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Redler RL, Fee L, Fay JM, Caplow M, Dokholyan NV. Non-native soluble oligomers of Cu/Zn superoxide dismutase (SOD1) contain a conformational epitope linked to cytotoxicity in amyotrophic lateral sclerosis (ALS) Biochemistry. 2014;53(14):2423–2432. doi: 10.1021/bi500158w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosco DA, et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat Neurosci. 2010;13(11):1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brotherton TE, et al. Localization of a toxic form of superoxide dismutase 1 protein to pathologically affected tissues in familial ALS. Proc Natl Acad Sci USA. 2012;109(14):5505–5510. doi: 10.1073/pnas.1115009109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rotunno MS, et al. Identification of a misfolded region in superoxide dismutase 1 that is exposed in amyotrophic lateral sclerosis. J Biol Chem. 2014;289(41):28527–28538. doi: 10.1074/jbc.M114.581801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banci L, et al. Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: A possible general mechanism for familial ALS. Proc Natl Acad Sci USA. 2007;104(27):11263–11267. doi: 10.1073/pnas.0704307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banci L, et al. SOD1 and amyotrophic lateral sclerosis: mutations and oligomerization. PloS One. 2008;3(2):e1677. doi: 10.1371/journal.pone.0001677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luchinat E, et al. In-cell NMR reveals potential precursor of toxic species from SOD1 fALS mutants. Nat Commun. 2014;5:5502. doi: 10.1038/ncomms6502. [DOI] [PubMed] [Google Scholar]
- 22.Auclair JR, Boggio KJ, Petsko GA, Ringe D, Agar JN. Strategies for stabilizing superoxide dismutase (SOD1), the protein destabilized in the most common form of familial amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2010;107(50):21394–21399. doi: 10.1073/pnas.1015463107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korzhnev DM, Religa TL, Banachewicz W, Fersht AR, Kay LE. A transient and low-populated protein-folding intermediate at atomic resolution. Science. 2010;329(5997):1312–1316. doi: 10.1126/science.1191723. [DOI] [PubMed] [Google Scholar]
- 24.Laganowsky A, et al. Atomic view of a toxic amyloid small oligomer. Science. 2012;335(6073):1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khare SD, Caplow M, Dokholyan NV. The rate and equilibrium constants for a multistep reaction sequence for the aggregation of superoxide dismutase in amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2004;101(42):15094–15099. doi: 10.1073/pnas.0406650101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilcox KC, et al. Modifications of superoxide dismutase (SOD1) in human erythrocytes: A possible role in amyotrophic lateral sclerosis. J Biol Chem. 2009;284(20):13940–13947. doi: 10.1074/jbc.M809687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hubbard SJ, Campbell SF, Thornton JM. Molecular recognition. Conformational analysis of limited proteolytic sites and serine proteinase protein inhibitors. J Mol Biol. 1991;220(2):507–530. doi: 10.1016/0022-2836(91)90027-4. [DOI] [PubMed] [Google Scholar]
- 28.Rakhit R, et al. Monomeric Cu,Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J Biol Chem. 2004;279(15):15499–15504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- 29.Ding F, Tsao D, Nie H, Dokholyan NV. Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure. 2008;16(7):1010–1018. doi: 10.1016/j.str.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng HX, et al. Amyotrophic lateral sclerosis and structural defects in Cu,Zn superoxide dismutase. Science. 1993;261(5124):1047–1051. doi: 10.1126/science.8351519. [DOI] [PubMed] [Google Scholar]
- 31.Juneja T, Pericak-Vance MA, Laing NG, Dave S, Siddique T. Prognosis in familial amyotrophic lateral sclerosis: Progression and survival in patients with glu100gly and ala4val mutations in Cu,Zn superoxide dismutase. Neurology. 1997;48(1):55–57. doi: 10.1212/wnl.48.1.55. [DOI] [PubMed] [Google Scholar]
- 32.Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 33.Joyce PI, Fratta P, Fisher EMC, Acevedo-Arozena A. SOD1 and TDP-43 animal models of amyotrophic lateral sclerosis: Recent advances in understanding disease toward the development of clinical treatments. Mamm Genome. 2011;22(7-8):420–448. doi: 10.1007/s00335-011-9339-1. [DOI] [PubMed] [Google Scholar]
- 34.Rotunno MS, Bosco DA. An emerging role for misfolded wild-type SOD1 in sporadic ALS pathogenesis. Front Cell Neurosci. 2013;7:253. doi: 10.3389/fncel.2013.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shibata N, Asayama K, Hirano A, Kobayashi M. Immunohistochemical study on superoxide dismutases in spinal cords from autopsied patients with amyotrophic lateral sclerosis. Dev Neurosci. 1996;18(5-6):492–498. doi: 10.1159/000111445. [DOI] [PubMed] [Google Scholar]
- 36.Shibata N, et al. Cu/Zn superoxide dismutase-like immunoreactivity in Lewy body-like inclusions of sporadic amyotrophic lateral sclerosis. Neurosci Lett. 1994;179(1-2):149–152. doi: 10.1016/0304-3940(94)90956-3. [DOI] [PubMed] [Google Scholar]
- 37.Gruzman A, et al. Common molecular signature in SOD1 for both sporadic and familial amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2007;104(30):12524–12529. doi: 10.1073/pnas.0705044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goscin SA, Fridovich I. The purification and properties of superoxide dismutase from Saccharomyces cerevisiae. Biochim Biophys Acta. 1972;289(2):276–283. doi: 10.1016/0005-2744(72)90078-2. [DOI] [PubMed] [Google Scholar]
- 39.Shlyakhtenko LS, Gall AA, Lyubchenko YL. Mica functionalization for imaging of DNA and protein-DNA complexes with atomic force microscopy. Methods Mol Biol. 2013;931:295–312. doi: 10.1007/978-1-62703-056-4_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Portillo AM, Krasnoslobodtsev AV, Lyubchenko YL. Effect of electrostatics on aggregation of prion protein Sup35 peptide. J Phys Condens Matter. 2012;24(16):164205. doi: 10.1088/0953-8984/24/16/164205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyagi A, Ando T, Lyubchenko YL. Dynamics of nucleosomes assessed with time-lapse high-speed atomic force microscopy. Biochemistry. 2011;50(37):7901–7908. doi: 10.1021/bi200946z. [DOI] [PubMed] [Google Scholar]
- 42.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 43.Go N, Abe H. Noninteracting local-structure model of folding and unfolding transition in globular proteins. I. Formulation. Biopolymers. 1981;20(5):991–1011. doi: 10.1002/bip.1981.360200511. [DOI] [PubMed] [Google Scholar]
- 44.Hubbard SJ, Eisenmenger F, Thornton JM. Modeling studies of the change in conformation required for cleavage of limited proteolytic sites. Protein Sci. 1994;3(5):757–768. doi: 10.1002/pro.5560030505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding F, Borreguero JM, Buldyrey SV, Stanley HE, Dokholyan NV. Mechanism for the alpha-helix to beta-hairpin transition. Proteins. 2003;53(2):220–228. doi: 10.1002/prot.10468. [DOI] [PubMed] [Google Scholar]
- 46.Stroppolo ME, Malvezzi-Campeggi F, Mei G, Rosato N, Desideri A. Role of the tertiary and quaternary structures in the stability of dimeric copper, zinc superoxide dismutases. Arch Biochem Biophys. 2000;377(2):215–218. doi: 10.1006/abbi.2000.1780. [DOI] [PubMed] [Google Scholar]
- 47.Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Discrete molecular dynamics studies of the folding of a protein-like model. Fold Des. 1998;3(6):577–587. doi: 10.1016/S1359-0278(98)00072-8. [DOI] [PubMed] [Google Scholar]
- 48.Zhou Y, Karplus M. Folding thermodynamics of a model three-helix-bundle protein. Proc Natl Acad Sci USA. 1997;94(26):14429–14432. doi: 10.1073/pnas.94.26.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okamoto Y. Generalized-ensemble algorithms: enhanced sampling techniques for Monte Carlo and molecular dynamics simulations. J Mol Graph Model. 2004;22(5):425–439. doi: 10.1016/j.jmgm.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 50.Ramachandran S, Kota P, Ding F, Dokholyan NV. Automated minimization of steric clashes in protein structures. Proteins. 2011;79(1):261–270. doi: 10.1002/prot.22879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kota P, Ding F, Ramachandran S, Dokholyan NV. Gaia: Automated quality assessment of protein structure models. Bioinformatics. 2011;27(16):2209–2215. doi: 10.1093/bioinformatics/btr374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin S, Ding F, Dokholyan NV. Modeling backbone flexibility improves protein stability estimation. Structure. 2007;15(12):1567–1576. doi: 10.1016/j.str.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 53.Yin S, Ding F, Dokholyan NV. Eris: An automated estimator of protein stability. Nat Methods. 2007;4(6):466–467. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.