Significance
In mammals, the inequality posed by the difference in the number of X chromosomes between XX females and XY males is remedied by silencing genes along one of the two X chromosomes in females. This process, termed X-chromosome inactivation, is believed to be triggered by X-inactive specific transcript (Xist) RNA. Here we find that Xist RNA can silence X-linked genes efficiently in females but not in males. Thus, Xist RNA is insufficient to inactivate the X chromosome. Our results further suggest that both Xist induction and X-linked gene silencing are orchestrated by the handful of genes that do not undergo X inactivation in females. The increased dosage of one or more such factors in females vs. males may explain why females undergo X inactivation and males do not.
Keywords: Xist, Tsix, X inactivation, embryonic stem cells, epiblast stem cells
Abstract
X-inactive specific transcript (Xist) long noncoding RNA (lncRNA) is thought to catalyze silencing of X-linked genes in cis during X-chromosome inactivation, which equalizes X-linked gene dosage between male and female mammals. To test the impact of Xist RNA on X-linked gene silencing, we ectopically induced endogenous Xist by ablating the antisense repressor Tsix in mice. We find that ectopic Xist RNA induction and subsequent X-linked gene silencing is sex specific in embryos and in differentiating embryonic stem cells (ESCs) and epiblast stem cells (EpiSCs). A higher frequency of XΔTsixY male cells displayed ectopic Xist RNA coating compared with XΔTsixX female cells. This increase reflected the inability of XΔTsixY cells to efficiently silence X-linked genes compared with XΔTsixX cells, despite equivalent Xist RNA induction and coating. Silencing of genes on both Xs resulted in significantly reduced proliferation and increased cell death in XΔTsixX female cells relative to XΔTsixY male cells. Thus, whereas Xist RNA can inactivate the X chromosome in females it may not do so in males. We further found comparable silencing in differentiating XΔTsixY and 39,XΔTsix (XΔTsixO) ESCs, excluding the Y chromosome and instead implicating the X-chromosome dose as the source of the sex-specific differences. Because XΔTsixX female embryonic epiblast cells and EpiSCs harbor an inactivated X chromosome prior to ectopic inactivation of the active XΔTsix X chromosome, we propose that the increased expression of one or more X-inactivation escapees activates Xist and, separately, helps trigger X-linked gene silencing.
X inactivation represents a paradigm of epigenetic regulation and long noncoding RNA (lncRNA) function. In XX female cells, one of the two X chromosomes undergoes transcriptional silencing (1). Moreover, replicated copies of the active and inactive X chromosomes faithfully maintain their respective transcriptional states through many cell division cycles (2–5).
X inactivation requires the X-inactive specific transcript (Xist) (6–8), a lncRNA that is selectively expressed from and physically coats the future inactive X chromosome (9–12). Xist RNA enables X-linked gene silencing by recruiting protein complexes to the inactive X (13–15). Female mouse embryos that inherit a paternal Xist mutation die due to defects in imprinted X inactivation of the paternal X chromosome in extraembryonic tissues (8, 16, 17). Xist is also required in the epiblast-derived embryonic cells, which undergo random X inactivation of either the maternal or the paternal X chromosome. Xist heterozygote fetal cells exhibit inactivation of only the X chromosome with an intact Xist locus, suggesting that Xist is necessary to choose the X chromosome to be inactivated (7, 18, 19). That the Xist-mutant X chromosome is not selected for inactivation, however, precludes assigning to Xist RNA a gene silencing role in the epiblast lineage.
Ectopic expression studies have, however, demonstrated that Xist RNA can silence genes, albeit in a context-dependent manner. Xist transgenes integrated into autosomes can silence neighboring autosomal sequences, but the effect is quite variable. Whereas multicopy Xist transgenes or transgenes driven by artificial promoters often display Xist RNA induction and coating of autosomes in cis accompanied by a degree of silencing of adjacent host sequences (20–28), large single-copy Xist genomic transgenes do not (19, 28, 29). The sequence composition and the chromatin context at the site of transgene integration as well as the level of Xist expression are confounding variables that may influence the ability of transgenic Xist RNA to silence.
We therefore sought to systematically test the impact of Xist RNA on gene silencing by ectopically inducing Xist from the endogenous locus, thus ensuring that the cis-regulatory elements necessary for robust Xist expression are intact. We previously generated male and female embryonic stem cells (ESCs) and epiblast stem cells (EpiSCs) that harbor an X chromosome with a null mutation in the Xist antisense repressor Tsix (XΔTsix) (30). A subset of differentiating XΔTsixY and XΔTsixX cells display ectopic Xist RNA coating of the XΔTsix; thus, male cells harbor a single Xist RNA coat and females possess two Xist coats. These populations enabled us to assess the ability of Xist RNA to silence X-linked genes in males and in females.
Unexpectedly, we observed sex-specific differences in the frequency of cells that induced Xist from the active XΔTsix and silenced X-linked genes once Xist was ectopically induced, both in vitro and in vivo. We found that a higher percentage of XΔTsixY cells displayed ectopic Xist RNA coating compared with XΔTsixX cells. This increase reflected the inability of XΔTsixY cells to efficiently silence X-linked genes upon ectopic Xist induction compared with XΔTsixX cells, despite equivalent levels of Xist expression and RNA coating. We discuss possible underlying reasons for these differences, including the requirement of two X chromosomes to physically interact, epigenetic variation on the XΔTsix between the sexes, and differences in developmental timing between male and female embryos. The comparative analysis compels us to propose that the higher X-chromosomal dose in females, potentially acting through gene(s) that escape X inactivation, induces Xist and, separately, silences X-linked genes once Xist is induced. The increased dosage of such a factor(s) in females compared with males may explain why females undergo X inactivation and males do not.
Results
ESCs Display a Sex-Specific Difference in the Frequency of Ectopic Xist RNA Coating.
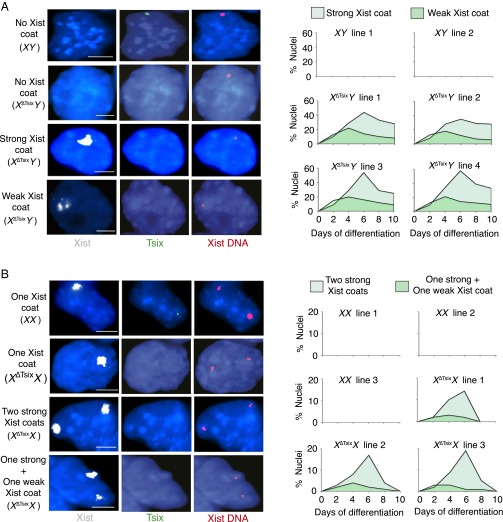
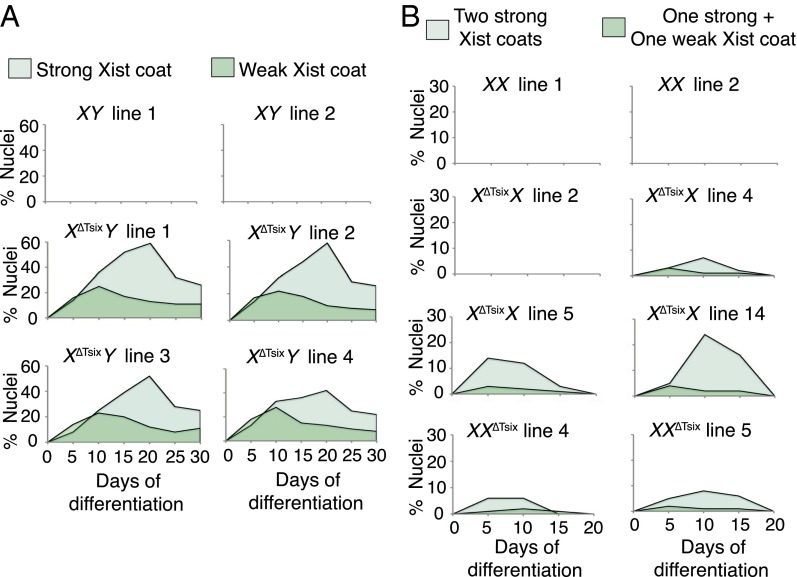
To ectopically induce Xist, we differentiated multiple control wild-type (WT) XY and XX and mutant XΔTsixY and XΔTsixX ESC lines (see SI Appendix, Fig. S1 for map of the ΔTsix mutant allele) (30, 31). We assessed ectopic Xist RNA coating every 2 d over a period of 10 d by RNA fluorescence in situ hybridization (FISH). WT male XY ESC lines did not display Xist RNA-coated nuclei during differentiation. However, mutant male XΔTsixY lines exhibited three classes of nuclei: some had strong Xist RNA coating, resembling Xist RNA coating in female cells, some had weak Xist RNA coating, and some lacked Xist RNA coating altogether. Strong Xist RNA-decorated XΔTsixY nuclei reached a maximum of between 40% and 58% of all nuclei at day 6 (d6) of differentiation, before decreasing to ∼30% at d10 (Fig. 1A). Weak Xist RNA-coated XΔTsixY nuclei peaked at 18–22% at d4 and decreased to ∼8% at d10 of differentiation.
Fig. 1.
Differential Xist RNA coating in X∆TsixY vs. X∆TsixX differentiating ESCs. (A, Left) RNA FISH detection of Xist (white) and Tsix (green) RNAs followed by Xist DNA FISH (red) in representative XY and X∆TsixY differentiated ESCs without or with strong or weak Xist RNA coats. Nuclei are stained blue with DAPI. (Right) Quantification of nuclei with strong or weak Xist RNA coats during differentiation of two XY and four X∆TsixY ESC lines. (B, Left) Xist/Tsix RNA FISH followed by Xist DNA FISH in representative XX and X∆TsixX differentiated ESCs. (Right) Quantification of nuclei with strong or weak ectopic Xist RNA coats during differentiation in three XX and three X∆TsixX ESC lines. Only nuclei with a single Xist locus in males or two Xist loci in females detected by DNA FISH were quantified. n = 100 nuclei per cell line per day of differentiation. (Scale bar, 2 μm.) Related data are included in SI Appendix, Fig. S1.
Undifferentiated female WT XX and mutant XΔTsixX ESCs harbor two active X chromosomes, which are randomly inactivated upon differentiation (6, 30, 32). During the 10-d course of differentiation, XX cells either lacked Xist RNA coating, signifying that X inactivation had not yet initiated, or had one Xist RNA coat, characteristic of the inactive X. We did not observe WT XX nuclei with two Xist RNA coats throughout the time course. In contrast, differentiating mutant XΔTsixX ESCs displayed two strong Xist-coated Xs in a significant percentage of XΔTsixX nuclei, peaking at between 14% and 19% at d6. A small percentage of differentiating XΔTsixX nuclei exhibited one strong and one weak Xist RNA-coated X chromosome, with a maximum of 4%. By d10, double Xist RNA-coated XΔTsixX nuclei had rapidly declined and disappeared altogether (Fig. 1B), in marked contrast to the male XΔTsixY ESCs, which showed persistence of ectopic Xist RNA-coated nuclei (Fig. 1A). We previously showed that the second Xist-decorated X chromosome in differentiating XΔTsixX ESCs is the XΔTsix mutant X chromosome (30). Xist is therefore ectopically induced from the XΔTsix in females, as it is in males. Compared with XΔTsixY ESCs, however, differentiating XΔTsixX ESCs appeared to harbor fewer ectopic Xist RNA-decorated nuclei.
Sex-Specific X-Linked Gene Silencing upon Ectopic Xist RNA Coating in ESCs.
We surmised that the difference in the frequency and the kinetics of ectopic Xist RNA-coated nuclei between male XΔTsixY and female XΔTsixX ESC lines may reflect variable silencing of X-linked genes between the sexes. Functional nullizygosity of X-linked genes is expected to be deleterious, leading to selection against these cells (30, 33). Thus, the higher steady-state percentage of XΔTsixY nuclei with Xist RNA coating may reflect inefficient silencing of X-linked genes upon ectopic Xist RNA coating in mutant males compared with females.
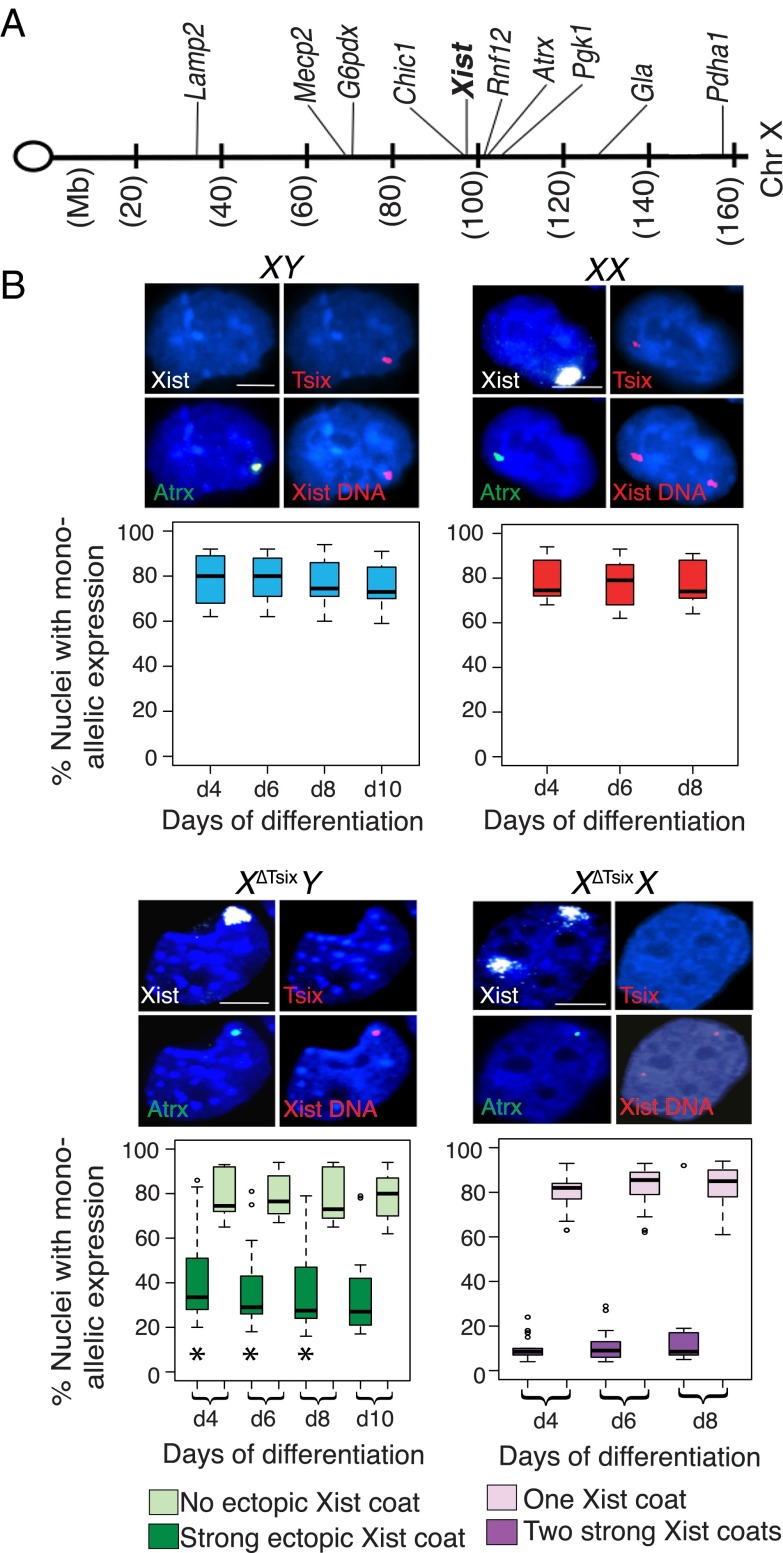
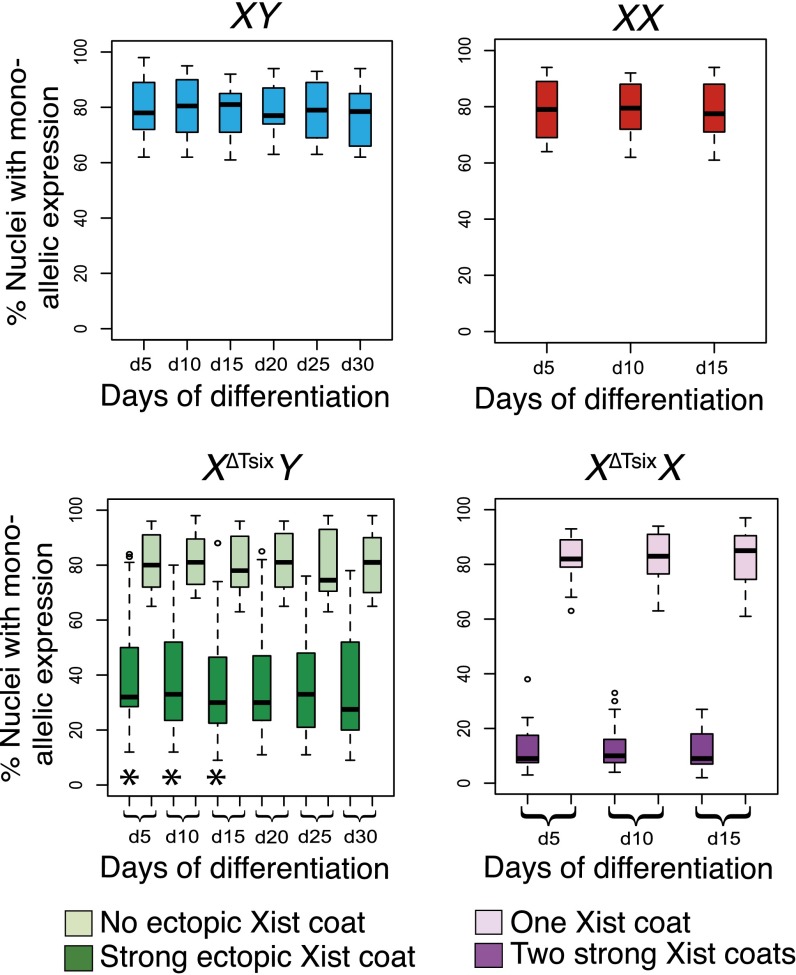
To test the efficiency of X-linked gene silencing in XΔTsixY and XΔTsixX cells, we profiled the expression of Xist RNA together with a panel of genes distributed across the X chromosome in differentiating WT and mutant ESCs by RNA FISH (Fig. 2A). RNA FISH permits detection of nascent transcripts in single cells and is refractory to potentially confounding variables of RNA perdurance and expression heterogeneity in and between cells to which techniques such as RT-PCR or RNA sequencing (RNA-seq) are subject. We assayed nine genes that are subject to X inactivation, Lamp2, Mecp2, G6pdx, Chic1, Rnf12, Atrx, Pgk1, Gla, and Pdha1 (16, 34, 35). In differentiating WT male XY ESCs, all of the genes were expressed in most of the nuclei (61–94%) (Fig. 2B and SI Appendix, Fig. S2). In differentiating WT female XX ESCs, the nine genes were similarly monoallelically expressed (62–92%) (Fig. 2B and SI Appendix, Fig. S2). The remaining cells failed to display expression from the single allele in males or both alleles in females. As a control, we additionally assayed a gene that escapes X inactivation, Smcx, which is expected to be expressed from both the active and the inactive X in females. In XY cells, Smcx was expressed from the single X chromosome in ∼90% of cells; in XX females, Smcx was biallelically expressed in ∼60% of the cells (SI Appendix, Fig. S2B).
Fig. 2.
Differential silencing of X-linked genes upon ectopic Xist RNA coating in differentiating X∆TsixY and X∆TsixX ESCs. (A) X-chromosomal localization of the genes profiled by RNA FISH. (B) A representative nucleus stained to detect Xist RNA (white), Tsix RNA (red), and nascent transcripts of one of the nine genes surveyed (Atrx, green) is shown above boxplots of each genotype. Following RNA FISH, the Xist locus was detected by DNA FISH. Nuclei are stained blue with DAPI. Boxplots show the median percent gene expression (line), second to third quartiles (box), and 1.5 times the interquartile range (whiskers). d, day. In XY and XX cells, nuclei exhibiting monoallelic expression are plotted. In X∆TsixY cells, percent nuclei with monoallelic expression of the genes both without coincident Xist RNA coat and with strong ectopic Xist RNA coats are plotted. In X∆TsixX cells, percent nuclei with monoallelic expression of the genes coincident with a single Xist RNA coat and with two strong ectopic Xist RNA coats are plotted. Two cell lines of each genotype were analyzed. n = 100 nuclei per cell line per day of differentiation for each class of Xist RNA-coated cells. (Scale bar, 2 μm.) *P < 0.003, significant difference in gene expression between X∆TsixY and X∆TsixX nuclei; Welch’s two-sample T test. X-linked gene expression does not differ significantly between X∆TsixY and X∆TsixX nuclei lacking ectopic Xist coats (P > 0.2). Related data are included in SI Appendix, Figs. S2 and S3.
In differentiating XΔTsixY male ESCs, we noticed that all genes were expressed in a significant percentage of nuclei despite strong Xist RNA coating (Fig. 2B and SI Appendix, Fig. S2B). In nuclei with weak Xist RNA coating, all of the genes were expressed more often from the XΔTsix compared with nuclei with strong Xist RNA coating (SI Appendix, Figs. S2B and S3A). By contrast, in differentiating XΔTsixX female ESCs with two strong Xist RNA coats, all X-linked genes were silenced on both Xs in significantly more nuclei than in strong Xist RNA-decorated XΔTsixY male nuclei (Fig. 2B and SI Appendix, Figs. S2B, S3A, and Table S1). In XΔTsixX female nuclei with one strong and one weak Xist RNA coat, all X-linked genes were coincidently expressed with Xist RNA in a greater percentage than in XΔTsixX nuclei with two strong Xist RNA coats (Fig. 2B and SI Appendix, Figs. S2B and S3A). However, X-linked genes were silenced more often in female XΔTsixX nuclei with one strong and one weak Xist RNA coat than in male XΔTsixY nuclei with a weak Xist RNA coat (SI Appendix, Figs. S2B and S3A). In summary, compared with XΔTsixY male cells, differentiating XΔTsixX female ESCs silenced all nine genes in significantly more nuclei upon ectopic Xist RNA coating. The weaker silencing in XΔTsixY males compared with XΔTsixX females may underlie the increased prevalence of Xist RNA-decorated male cells relative to double Xist RNA-coated female cells later in differentiation. Upon ectopic Xist RNA coating, stringent silencing of genes on the second X chromosome potently selects against XΔTsixX cells (see below and ref. 30); by contrast, weaker silencing of X-linked genes in XΔTsixY cells may permit Xist RNA-coated cells to persist.
Xist RNA is thought to potentiate silencing by directly or indirectly recruiting proteins such as the Polycomb group. Thus, as an added indicator of the potency of Xist RNA coating, we tested enrichment of histone H3 trimethylated at lysine 27 (H3-K27me3) on both the strong and weak Xist RNA-coated X chromosomes. H3-K27me3 is catalyzed by the Polycomb repressive complex 2 (PRC2) and is associated with silenced gene expression (36), including on the inactive X chromosome (37, 38). Whereas strong Xist RNA coats displayed robust coincident H3-K27me3 enrichment in both sexes (∼90% of cells), a significant percentage of weak Xist RNA coats also showed overlapping H3-K27me3 enrichment (∼75% of cells), albeit with correspondingly weaker signals in both sexes (SI Appendix, Fig. S3B). The reduced frequency of H3-K27me3 enrichment on the weak Xist RNA-coated Xs correlates with the weaker silencing of genes on that X chromosome in both sexes (SI Appendix, Fig. S3B). In sum, the reduced levels of X-linked gene silencing in XΔTsixY cells is not due to lower frequencies of H3-K27me3 enrichment on the Xist RNA-decorated Xs in comparison with XΔTsixX cells.
Y Chromosome Does Not Protect Against X-Linked Gene Silencing in XΔTsixY ESCs.
To explain the differential X-linked gene silencing in XΔTsixY vs. XΔTsixX cells, we investigated whether the presence of the Y chromosome protected X-linked genes from being silenced. Previous studies have demonstrated that Xist RNA coating can occur in male ESCs with supernumerary X chromosomes (33), but, to our knowledge, whether silencing of individual genes can occur to the same extent in male cells as in corresponding cells without a Y chromosome is not known. We therefore subcloned two 39,XΔTsix (XΔTsixO) ESC lines from XΔTsixX ESCs (ESC line 2 in Fig. 1B) that have lost the WT X chromosome and assessed Xist RNA coating and expression of the 10 X-linked genes by RNA FISH. During differentiation, the frequency of strong and weak Xist RNA-coated nuclei in both of the XΔTsixO ESC lines mimicked XΔTsixY ESCs and not XΔTsixX ESCs, including the parental XΔTsixX ESC line 2 (SI Appendix, Fig. S3C). Moreover, in both strong and weak Xist RNA-coated XΔTsixO nuclei, the expression pattern of all 10 genes matched that of the XΔTsixY cells instead of the parental XΔTsixX cells (SI Appendix, Figs. S2B, S3D, and Table S2). Thus, the absence of the Y chromosome does not explain the greater frequency of X-linked gene silencing in XΔTsixX compared with XΔTsixY cells. The differential silencing between the sexes must therefore be dependent on the X-chromosomal content.
Sex-Specific Difference in Ectopic Xist RNA Coat Frequencies in EpiSCs.
Female ESCs have two active X chromosomes; thus, the higher X-chromosomal dose necessary for efficient X-linked gene silencing could require both Xs to be transcriptionally active. Alternatively, higher X-chromosome dosage may have an effect even if one of the two Xs was inactivated. To distinguish among these two distinct possibilities, we took advantage of Tsix-mutant EpiSCs. Like ESCs, EpiSCs are pluripotent cells of the epiblast lineage (39, 40). However, as opposed to ESCs, undifferentiated female EpiSCs, XX as well as XΔTsixX, harbor a stochastically inactivated X chromosome (30, 41, 42).
We first profiled Xist RNA coating in WT and mutant EpiSCs. Male WT XY as well as mutant XΔTsixY EpiSCs did not display Xist RNA coating in the undifferentiated state (Fig. 3A and SI Appendix, Fig. S4A). WT XY male cells remained devoid of Xist RNA coating throughout differentiation; differentiating mutant XΔTsixY male cells, however, exhibited a significant percentage with Xist RNA coats (Fig. 3A and SI Appendix, Fig. S4A). Undifferentiated XX and XΔTsixX female EpiSCs were also indistinguishable. Both genotypes displayed a single Xist RNA-coated X chromosome (Fig. 3B and SI Appendix, Fig. S4B) (30). Upon differentiation, however, a proportion of XΔTsixX female EpiSCs ectopically induced Xist and coated the second X chromosome (Fig. 3B and SI Appendix, Fig. S4B); XX female EpiSCs continued to exhibit only one Xist RNA coat during the course of differentiation. Xist decoration of the second X chromosome in XΔTsixX female cells is due to ectopic Xist induction from the XΔTsix, as in XΔTsixY male cells (30).
Fig. 3.
Differential ectopic Xist RNA coating in differentiating X∆TsixY vs. X∆TsixX EpiSCs. (A, Left) Quantification of nuclei with strong or weak Xist RNA coats during differentiation of two XY and four X∆TsixY EpiSC lines. (B, Right) Quantification of nuclei with strong or weak ectopic Xist RNA coats during differentiation of two XX and six X∆TsixX EpiSC lines. Only nuclei with a single Xist locus in males or two Xist loci in females detected by DNA FISH were quantified, as shown in SI Appendix, Fig. S4. n = 100 nuclei per cell line per day of differentiation.
Like ESCs, the mutant EpiSCs displayed a sex-specific pattern of Xist RNA coating. In differentiating XΔTsixY male EpiSCs, the percentage of strong Xist RNA-coated nuclei steadily increased up to d20 of differentiation, ranging between 42% and 59% of nuclei, then decreased to between 22% and 26% at d30 (Fig. 3A). Weak Xist RNA-coated XΔTsixY male nuclei peaked between 22% and 28%, at d10, and decreased to 8–11% at d30 (Fig. 3A). By contrast, the percentage of XΔTsixX female nuclei with strong ectopic Xist RNA coating, resulting in two robust Xist RNA-decorated domains, reached a maximum of 24% at d10 and then quickly disappeared by d20 (Fig. 3B). XΔTsixX nuclei with one strong and one weak Xist RNA coats peaked at 4% at d5–d10 and were gone by d20.
The variability in ectopic Xist induction between the female EpiSC lines roughly correlates with the number of cells that are eligible to ectopically induce Xist in a given line. Due to random X inactivation, XΔTsixX EpiSCs can inactivate either the mutant XΔTsix or the WT X (30). The greater the percentage of cells in a female EpiSC line in which the XΔTsix is the active X, the higher the percentage of cells that can ectopically express Xist (30). For example, in XΔTsixX EpiSC line 2 the XΔTsix is the inactive X in all cells; this cell line, therefore, entirely lacks cells that can ectopically induce Xist, in agreement with the absolute absence of nuclei with two Xist RNA coats in this cell line during differentiation (Fig. 3B). Conversely, XΔTsixX EpiSC line 14 harbors many cells that have chosen the XΔTsix as the active X chromosome (∼75%), resulting in a relatively high percentage of cells that ectopically induce Xist during differentiation (Fig. 3B). Nevertheless, even in this cell line substantially fewer nuclei displayed ectopic Xist RNA coating (24%) compared with the XΔTsixY EpiSC line with the lowest frequency of ectopic Xist RNA-coated nuclei (cell line 4; 41%). This difference once again suggested diminished silencing of X-linked genes in mutant males compared with females.
Sex-Specific X-Linked Gene Silencing upon Ectopic Xist RNA Coating in EpiSCs.
We therefore assayed expression of the 10 X-linked genes over the course of EpiSC differentiation by RNA FISH (Fig. 4 and SI Appendix, Fig. S5). Similarly to ESCs, significantly more differentiating mutant XΔTsixX female compared with mutant XΔTsixY male EpiSCs exhibited silencing of the 9 genes subject to X inactivation upon ectopic Xist RNA coating (P < 10−6; Fig. 4 and SI Appendix, Fig. S5 and Table S3). We also simultaneously probed pairs of X-linked genes to determine if in the same nucleus the expression of the two genes would concord, or, as implied by the data in SI Appendix, Fig. S5, differ. We tested four different pairs, with 1 gene of each pair exhibiting a greater frequency of silencing than the other when tested individually in XΔTsixY cells (Pgk1/Atrx; Rnf12/Lamp2; Gla/Mecp2; G6pdx/Chic1) (SI Appendix, Figs. S5 and S6). When tested together, the genes in each pair recapitulated the pattern of silencing when assayed individually in both XΔTsixY and XΔTsixX cells (SI Appendix, Fig. S5). One gene was silenced more frequently compared with the other, especially in XΔTsixY cells; thus, the two genes behaved independently in the same nucleus. Notably, 7 of 8 genes were silenced in significantly more XΔTsixX compared with XΔTsixY nuclei (P < 0.03; Pgk1, P < 0.12) (SI Appendix, Fig. S6). Thus, ectopic Xist RNA coating is sufficient to silence X-linked genes in XΔTsixY cells; but, it does not do so as uniformly or as robustly as in XΔTsixX cells.
Fig. 4.
Differential silencing of X-linked genes upon ectopic Xist RNA coating in differentiating X∆TsixY vs. X∆TsixX EpiSCs. Boxplots of expression of the nine X-linked genes surveyed as in Fig. 2 in individual nuclei of differentiating XY, XX, X∆TsixY, and X∆TsixX EpiSC lines (lines 2, 2, 4, and 5, respectively; all EpiSC lines from Fig. 3, with the exception of X∆TsixX EpiSC line 2). d, day. n = 100 nuclei per cell line per day of differentiation for each class of Xist RNA-coated cells. *P < 10−6, significant difference in gene expression between X∆TsixY and X∆TsixX nuclei; Welch’s two-sample T test. X-linked gene expression does not significantly differ between X∆TsixY and X∆TsixX nuclei lacking ectopic Xist coats (P > 0.2). Related data are included in SI Appendix, Figs. S5–11.
We also measured the ability of ectopic Xist RNA coating to recruit the Polycomb PRC2 complex and enrich H3-K27me3 on the X chromosome in differentiating EpiSCs. As in ESCs, both strong and weak Xist coats displayed accumulation of H3-K27me3 in a significant percentage of XΔTsixY and XΔTsixX nuclei in both sexes (∼90%, strong Xist-coated nuclei; ∼75%, weak Xist-coated nuclei) (SI Appendix, Fig. S7). Concurrent detection of H3-K27me3, Xist, and X-linked genes directly showed that despite the robust enrichment of H3-K27me3 on the ectopically Xist-coated XΔTsix, X-linked genes were nevertheless expressed in a significant percentage of differentiating male XΔTsixY EpiSCs (P < 0.001; SI Appendix, Fig. S8). Furthermore, consistent with the relatively stringent silencing upon ectopic Xist RNA coating in differentiating XΔTsixX compared with XΔTsixY cells, mutant female cells displayed a significant reduction in both cell proliferation and viability compared with mutant male cells during differentiation (P < 10−4) (SI Appendix, Fig. S9; see also ref. 30). Reduced proliferation and increased cell death, therefore, potently select against female mutants that have ectopically activated Xist from and silenced genes on the second, i.e., XΔTsix, X chromosome.
In summary, the sex-specific pattern of ectopic Xist induction and X-linked gene silencing occurs in differentiating EpiSCs, as it does in ESCs. Thus, robust silencing of X-linked genes does not require two transcriptionally active Xs and can occur even when one of the two Xs in females is inactivated.
Equivalent Levels of Ectopic Xist Expression in Individual XΔTsixY vs. XΔTsixX EpiSCs.
In principle, the sex-specific X-linked gene silencing may be due to lower levels of ectopic Xist RNA expression in XΔTsixY compared with XΔTsixX cells. We therefore sought to quantify Xist expression in male and female mutant EpiSCs. We took advantage of a single nucleotide polymorphism (SNP) that distinguishes Xist transcripts originating from the XΔTsix vs. the WT X chromosome via pyrosequencing of cDNAs. Whereas the XΔTsix is derived from a M. musculus strain, the WT X is derived from the divergent M. molossinus JF1 strain (XJF1). We first measured Xist expression in XΔTsixY males relative to a reference F1 hybrid female EpiSC line, XΔTsixXJF1 line 15, in which Xist is predominantly expressed from the XJF1 (30). Due to the variability in Xist induction between cells, we profiled Xist expression in individual cells. In the XΔTsixXJF1 EpiSC line 15, Xist was almost exclusively expressed from the WT XJF1 allele (>90% of total Xist expression) in all cells examined (SI Appendix, Fig. S10A). By contrast, in single WT F1 hybrid XJF1XLab EpiSCs, in which the XLab is M. musculus derived, Xist was nearly mutually exclusively expressed from either the XJF1 or the XLab (SI Appendix, Fig. S10B), consistent with stochastic inactivation of either X chromosome in individual cells.
To quantify Xist expression in XΔTsixY male EpiSCs, we combined single XΔTsixY cells (line 3 in Fig. 3A) with single XΔTsixXJF1 line 15 female EpiSCs. Consistent with the lack of Xist RNA coating in undifferentiated XΔTsixY EpiSCs by RNA FISH (Fig. 3A), when single undifferentiated XΔTsixY EpiSCs were combined with single undifferentiated XΔTsixXJF1 line 15 EpiSCs, Xist expression from the XΔTsix did not increase compared with undifferentiated XΔTsixXJF1 line 15 EpiSCs alone (SI Appendix, Fig. S10C). Upon differentiation, based on the RNA FISH data we expected to observe three classes of XΔTsixY cells by RT-PCR (SI Appendix, Fig. S10D). In the first, the cells would be devoid of Xist induction or induced Xist minimally. In the second, Xist would be moderately expressed, thus corresponding to the weak Xist RNA-coated cells by RNA FISH. In the third, Xist would be strongly induced, representing robust Xist RNA-decorated cells. When single d10 differentiated XΔTsixY EpiSCs were combined with single undifferentiated line 15 female EpiSCs, Xist expression from the XΔTsix increased in a substantial percentage of the samples (SI Appendix, Fig. S10E), in agreement with Xist induction in some but not all XΔTsixY cells by RNA FISH (Fig. 3A). Of the three categories, class I cells, which did not induce Xist or induced Xist minimally (≤10% of total Xist expression from the XΔTsix), accounted for 31% of all cells. Class II cells, which induced Xist moderately (30–39% of total Xist expression from the XΔTsix), represented 28% of the cells. We chose 40% as the threshold of expression from the XΔTsix between class II and class III (robust Xist induction from the XΔTsix) because in individual differentiating XΔTsixX female cells, robust ectopic Xist induction from the XΔTsix X chromosome yields values of >40% (see below). The strong Xist-expressing class III cells (40–68% of total Xist expression from the XΔTsix) were 41% of the total cells. In class III XΔTsixY cells, the average Xist expression from the XΔTsix was 57% of total. This level of Xist induction in XΔTsixY male cells matched Xist expression from the XΔTsix in females, measured by combining single cells from female EpiSC lines that express Xist almost exclusively from either the XΔTsix (XΔTsixXJF1 EpiSC line 2) or the XJF1 (XΔTsixXJF1 EpiSC line 15) (30), with an average of 58% of total Xist expression from the XΔTsix (SI Appendix, Fig. S10F). The higher Xist expression from the XΔTsix relative to XJF1 reflects strain-specific differences in Xist levels due to polymorphisms in the X-controlling element (Xce) (43–45).
To gauge ectopic Xist induction later in differentiation, we also similarly profiled d20 differentiated XΔTsixY EpiSCs, when the percentage of Xist-coated cells is at its highest (Fig. 3A). By d20, class I accounted for 19% of all cells, class II 12%, and class III 69% (SI Appendix, Fig. S10G). As with robust Xist-expressing class III cells at d10, on average the d20 class III cells expressed Xist from the XΔTsix at nearly the levels found in females in SI Appendix, Fig. S10F (59% vs. 58%). Of note, at both d10 and d20, a greater percentage of XΔTsixY cells strongly induced Xist (class III) relative to those with robust Xist RNA coats (41% vs. 25% at d10; 69% vs. 52% at d20) (SI Appendix, Fig. S10 E–G and Fig. 3A), suggesting that not all strong Xist expressers display robust Xist RNA coats. The percentage of class II cells with moderate Xist induction more closely approximated the percentage of nuclei displaying weak Xist RNA coats at d10 and at d20 (28% vs. 23% at d10; 19% vs. 12% at d20).
To quantify ectopic Xist expression in mutant females, we used an F1 hybrid XΔTsixXJF1 female EpiSC line that exhibits ectopic Xist RNA coating of the XΔTsix in a significant percentage of cells at d10 of differentiation (22%; XΔTsixX EpiSC line 14 in Fig. 3B). Undifferentiated line 14 XΔTsixXJF1 EpiSCs displayed a slightly biased pattern of X inactivation; two-thirds of the individual undifferentiated cells surveyed displayed Xist induction from the XJF1 X chromosome and one-third from the XΔTsix X chromosome (SI Appendix, Fig. S10H). This distribution was expected to change during differentiation based on the RNA FISH data (Fig. 3B; see also ref. 30), with the cells once again expected to be stratified into three classes (SI Appendix, Fig. S10I). Class I would express Xist exclusively or almost exclusively from the XJF1 and lack much ectopic Xist induction from the XΔTsix due to the lack of differentiation. Class II would correspond to cells that originally inactivated the WT XJF1 and robustly ectopically induced Xist from the XΔTsix during differentiation. Class III represents cells that initially inactivated the XΔTsix and therefore are not eligible to ectopically induce Xist from the second X (i.e., the WT XJF1); these cells would therefore only express Xist from the XΔTsix throughout differentiation. At d10 of differentiation, class I (≤10% of total Xist expression from the XΔTsix) accounted for 18% of the cells; class II (48–66% of total Xist expression from the XΔTsix) represented 23% of all cells; and class III (≥90% of total Xist expression from the XΔTsix), 59% of the cells (SI Appendix, Fig. S10J). Class II female cells, therefore, are most informative with respect to ectopic Xist induction, because only this group of cells expresses both Xist alleles. Notably, ectopic Xist expression in XΔTsixXJF1 females is almost always robust, consistent with RNA FISH detecting very few mutant female nuclei with weak ectopic Xist coats (<4%) during EpiSC differentiation (Fig. 3B). The average expression of the two Xist alleles in individual class II cells was 57% from the XΔTsix and 43% from the XJF1 X chromosome (SI Appendix, Fig. S10J). This distribution matched allelic Xist expression in single-cell mixtures of female EpiSCs that display preferential inactivation of either the XΔTsix or the XJF1 (58% from the XΔTsix; 42% from the XJF1) (SI Appendix, Fig. S10F). Moreover, class II percentage (23%) agrees well with the percentage of cells displaying two Xist RNA coats in this EpiSC line at d10 of differentiation by RNA FISH (22%) (Fig. 3B). Thus, in both XΔTsixY and XΔTsixX EpiSCs, the XΔTsix is able to ectopically induce Xist at levels matching Xist normally expressed from the WT inactive X in females.
By d20 of differentiation, XΔTsixXJF1 cells only exhibited Xist RNA originating from the XΔTsix (SI Appendix, Fig. S10K). The uniform expression of Xist from the XΔTsix is consistent with selection against cells that had originally inactivated the XJF1. During differentiation, all of these cells ectopically induced Xist robustly from the previously active XΔTsix and became deficient in X-linked gene expression (Figs. 3B and 4 and SI Appendix, Fig. S5), leading to reduced proliferation and induced cell death (SI Appendix, Fig. S9) (see also ref. 30). By contrast, a significant fraction of male XΔTsixY d20 differentiated cells either failed to induce Xist (19%) or induced Xist only moderately (12%) (SI Appendix, Fig. S10G). And, at d10, when some differentiating female XΔTsixXJF1 cells had not yet ectopically induced Xist, a greater percentage of XΔTsixY compared with XΔTsixX cells either lacked ectopic Xist induction (31% vs. 18%) or induced Xist moderately (28% vs. 0%) (SI Appendix, Fig. S10 E and J).
Equivalent Ectopic Xist RNA Coating in XΔTsixY vs. XΔTsixX EpiSCs.
Although Xist RNA can be equivalently robustly expressed between the sexes, reduced Xist RNA coating could explain the diminished frequency of X-linked gene silencing in differentiated XΔTsixY compared with XΔTsixX cells. We therefore quantified the volume as well as the intensity of the Xist RNA coats in XΔTsixY, XΔTsixX, and XX EpiSCs, using an automated voxel-based analysis (Materials and Methods). We focused these measurements on nuclei with strong Xist RNA coats quantified in Figs. 3 and 4. Neither the volume nor the intensity of the Xist RNA coats was significantly different between male and female mutants (SI Appendix, Fig. S11). Moreover, neither measurement correlated with silencing of the five X-linked genes tested (Lamp2, Mecp2, Atrx, Gla, and Pdha1) (SI Appendix, Fig. S11); cells that expressed the X-linked genes did not have smaller or less intense coats compared with cells in which the genes were silenced. Together, these data demonstrate that, in EpiSC nuclei with robust ectopic Xist RNA coating, the strength of the Xist RNA coats in differentiating XΔTsixY is equivalent to that of XΔTsixX EpiSCs. The difference in X-linked gene silencing between the sexes, therefore, is not due to weaker Xist RNA coating in XΔTsixY compared with XΔTsixX cells.
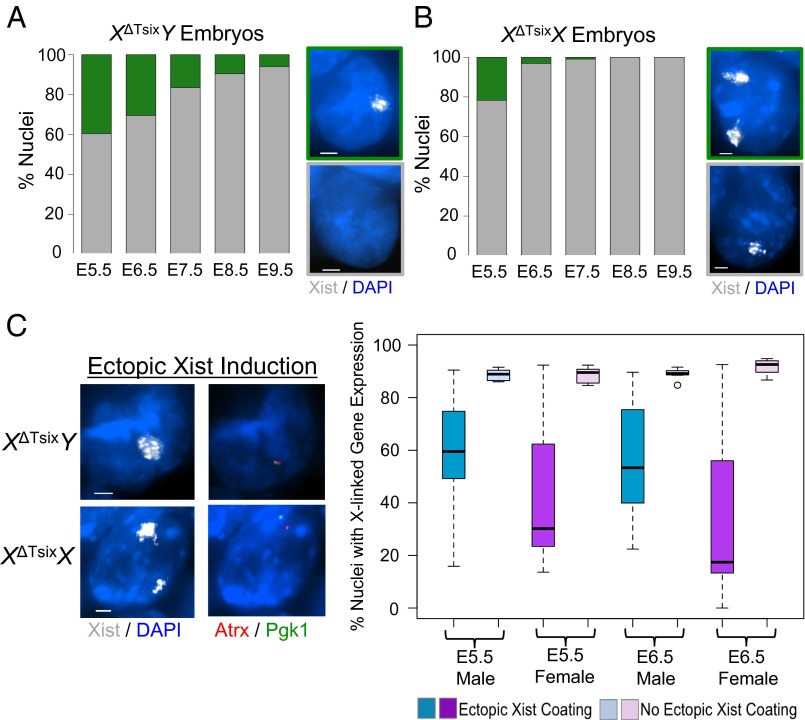
Embryos Display Sex-Specific Differences in Ectopic Xist Induction and X-Linked Gene Silencing.
We next interrogated Tsix-mutant embryonic epiblasts to test whether the sex-specific pattern of Xist induction and X-linked silencing observed in differentiating ESCs and EpiSCs is also observed in vivo. We first assayed the percentage of ectopic Xist RNA-coated epiblast cells from embryonic day 5.5 (E5.5), E6.5, E7.5, E8.5, and E9.5 XΔTsixY male and XΔTsixX female embryos. In XΔTsixY male embryos, a significant percentage of epiblast nuclei of E5.5 embryos, a stage shortly after random X-inactivation initiates (30, 46), exhibited ectopic Xist RNA coating (39%), which decreased at E6.5 (30%) and continued declining thereafter but were still found at E9.5 (Fig. 5A). In XΔTsixX embryos, ectopic Xist RNA-coated epiblast nuclei with two Xist RNA coats were almost exclusively observed at E5.5 (22%), with very few at E6.5 (3%), and none at the later stages (Fig. 5B). Thus, XΔTsixY and XΔTsixX embryos also display the sex-specific pattern of ectopic Xist induction observed in ESCs and EpiSCs. The difference in the kinetics of ectopic Xist induction in embryos compared with ESCs and EpiSCs reflects the relatively rapid rate of differentiation of embryonic epiblasts (30).
Fig. 5.
Ectopic Xist induction and X-linked gene silencing in postimplantation XΔTsixY and XΔTsixX embryos. (A) Quantification of XΔTsixY E5.5–E9.5 embryonic nuclei with and without Xist RNA coats. (B) Quantification of XΔTsixX E5.5–E9.5 embryonic nuclei with one and two Xist RNA coats. (C) Analysis of expression of the X-linked genes Lamp2, Mecp2, G6pdx, Chic1, Rnf12, Atrx, Pgk1, and Gla in XΔTsixY and XΔTsixX E5.5 and E6.5 embryonic epiblasts by RNA FISH. (Left) Representative images of nuclei stained to detect Xist, Atrx, and Pgk1 RNAs. (Scale bar, 2 μm.) (Right) Boxplots of expression of all eight X-linked genes surveyed. Boxplots show the median percent gene expression (line), second to third quartiles (box), and 1.5 times the interquartile range (whiskers). Related data are included in SI Appendix, Fig. S12.
We next probed pairs of X-linked genes to test whether they were concordantly or discordantly silenced in individual embryonic nuclei upon ectopic Xist RNA coating in Tsix-mutant E5.5 and E6.5 epiblast cells of both sexes, as in the EpiSCs (SI Appendix, Fig. S6). The X-linked genes were variably silenced within and between the sexes, with a greater average frequency of silencing in XΔTsixX females compared with XΔTsixY males (Fig. 5C). For three pairs of genes exhibiting differential levels of silencing (Atrx/Pgk1; Lamp2/Rnf12; Mecp2/Gla), one gene of each pair was silenced significantly less often than the other gene in the pair in XΔTsixY nuclei (P < 0.05), as compared to the XΔTsixX nuclei (SI Appendix, Fig. S12), consistent with the EpiSC results (SI Appendix, Fig. S6). The fourth gene pair, G6pdx/Chic1, was rarely discordantly silenced in either sex. At E5.5, for six of the eight genes tested significantly fewer XΔTsixY male nuclei displayed silencing of the X-linked genes compared with XΔTsixX female nuclei upon ectopic Xist coating (P < 0.01); only silencing of Pgk1 and Rnf12 did not differ significantly between the sexes (P > 0.05) (SI Appendix, Fig. S12). Similarly, at E6.5, upon ectopic Xist RNA coating all X-linked genes except Pgk1 and Rnf12 were silenced in significantly fewer XΔTsixY compared with XΔTsixX nuclei (P < 0.05).
Discussion
Xist RNA is believed to be both necessary and sufficient to initiate X inactivation. To test Xist function, in this study we analyzed differentiating ESCs, EpiSCs, and embryonic epiblast cells harboring a mutation in the Xist antisense repressor Tsix. The XΔTsix X chromosome offered a sensitized background in which to assess the impact of Xist RNA on X-linked gene silencing, because Xist is ectopically induced from the active XΔTsix in the epiblast lineage of both males and females. We found that a higher frequency of XΔTsixY and XΔTsixO cells displayed ectopic Xist RNA coating compared with XΔTsixX cells. This increase reflected the inability of XΔTsixY and XΔTsixO cells to efficiently silence X-linked genes upon ectopic Xist induction. Silencing of genes on both Xs due to ectopic Xist induction from the XΔTsix resulted in significantly reduced proliferation and increased cell death in XΔTsixX female cells relative to XΔTsixY male cells. The rapid loss of this population of XΔTsixX female cells leaves behind only cells or descendants of cells that had originally inactivated the XΔTsix and which could not induce Xist from the second, i.e., WT, X chromosome during differentiation (see also ref. 30). Therefore, despite a lower steady-state frequency of ectopic Xist RNA coating, all XΔTsixX female mutant cells in which the XΔTsix was the active X ultimately ectopically induce Xist from the XΔTsix. By contrast, a significant percentage of differentiating XΔTsixY male mutant epiblast cells do not induce Xist. Thus, XΔTsixY mutants not only display lower frequencies of X-linked gene silencing upon ectopic Xist induction, but also exhibit a reduced number of cells with ectopic Xist induction compared with XΔTsixX female cells.
The X-chromosome:autosome ratio determines whether X inactivation occurs and how many X chromosomes undergo inactivation, ensuring that only one X remains active per diploid genome (47–51). In the Tsix mutants, the differences in Xist induction and X-linked gene silencing must be genetically attributed to the sex chromosomes, as both XΔTsixX and XΔTsixY cells have identical complements of autosomes. Because differentiating XΔTsixO ESCs behave similarly to XΔTsixY ESCs, we exclude the Y chromosome as the source of the sex-specific differences by, for example, Y-linked genes functioning to prevent X-linked gene silencing in males. Instead, the data implicate the presence of the second X chromosome—i.e., the WT X—as the cause of the increased frequencies of ectopic Xist induction and X-linked gene silencing in females compared with males.
In addition to truncating Tsix transcription, the ΔTsix mutation deletes the critical DXPas34 repeat sequence close to the Xist–Tsix topological associated domain (TAD) boundary (30, 31, 52–56). Consistent with the broadly coordinated regulation of genes within each of the two adjacent TADs, a 58-kb deletion encompassing the TAD boundary changes the transcription of multiple genes within the X-inactivation center (Xic) (33, 57, 58). The ΔTsix mutation may result in a similar long-range dysregulation of X-linked genes in cis by perturbing the Xist–Tsix TAD boundary. However, the requirement for the second X chromosome implies that the observed sex-specific differences are a trans-effect, rather than due to a cis-limited sex-specific transcriptional defect on the XΔTsix imparted by the ΔTsix mutation.
Another potential explanation for the sex-specific effects is differential epigenetic marking of the XΔTsix in the two sexes. In females, the second X chromosome may alter the XΔTsix chromatin in a manner that later facilitates ectopic Xist induction from and X-linked gene silencing on the XΔTsix. The observation that XΔTsixO ESCs, which were derived from XΔTsixX female ESCs and thus previously harbored two Xs, ectopically induce Xist and undergo X-linked gene silencing at frequencies similar to XΔTsixY male ESCs suggests that the presence of two active X chromosomes does not mark the XΔTsix differently in females compared with males. Further arguing against such epigenetic differences is the propensity of XΔTsixX ESCs to undergo random X inactivation, a pattern similar to that of WT XX cells (30). If the XΔTsix was especially prone to inducing Xist and undergoing inactivation in females, the expectation is that it would preferentially be chosen for inactivation in XΔTsixX heterozygotes.
Developmental differences between male and female embryos may also explain the sex-specific differences in Xist induction and X-linked gene silencing in the Tsix mutants. XY male embryos develop slightly faster compared with their XX siblings, owing both to the absence of the Y chromosome and the presence of the second X chromosome (59). Moreover, previous observations have suggested that Xist RNA is competent to silence X-linked genes in a defined developmental window (26). If cells in male embryos exceed this developmental window due to their faster development, then they may become refractory to X inactivation. Several observations, however, argue against the faster development of male embryos underlying the lower frequencies of X-linked gene silencing in XΔTsixY compared with XΔTsixX embryos. For example, any difference in the rate of development of embryonic epiblasts between the sexes at E5.25 is not expected to be as large as the difference between E5.25 and E6.5 epiblasts. E6.5 epiblasts harbor more than five times the number of cells as in E5.25 epiblasts (60, 61). Upon ectopic Xist RNA coating, epiblasts in E6.5 XΔTsixX females silenced each of the eight genes surveyed significantly more frequently than did E5.25 XΔTsixY males. Thus, female embryos older by >1 d are nevertheless more competent to silence X-linked genes than their younger male counterparts.
Cultured XΔTsixY and XΔTsixX ESCs and EpiSCs recapitulate the sex-specific patterns observed in embryos, also arguing against developmental timing differences as the underlying cause of the sex-specific differences. Any developmental timing differences would be normalized by capturing cells of both sexes at equivalent stages of differentiation. Ectopic Xist induction and X-linked gene silencing occur at the same stage of ESC differentiation in both sexes (30). Xist is ectopically induced just after the ESCs differentiate beyond the epiblast-like cell (EpiLC) stage in both sexes (30). EpiLCs molecularly and morphologically mimic EpiSCs (30). In agreement, Xist and X-linked gene silencing are ectopically induced only when XΔTsixY male and XΔTsixX female EpiSCs differentiate (30). In fact, XΔTsixY and XΔTsixX embryonic epiblasts, ESCs, and EpiSCs all display ectopic Xist induction and X-linked gene silencing as a function of differentiation, rather than developmental timing (30).
The X-chromosome dosage effect in the Tsix mutants may be intimately linked to the mechanism that senses, or “counts,” the cellular X-chromosomal complement. The counting mechanism ensures that only if the X-chromosomal ploidy is sufficiently high does an X become targeted for inactivation. One prominent X-counting model invokes physical pairing of the two X chromosomes in females (62), via sequences within the Xic, including the Tsix locus, at the onset of X inactivation (63–65). As a consequence of this coupling, Xist is believed to be selectively upregulated from one of the two Xs (63–65), presumably through a transvection-like mechanism (62). However, deletions of all Xic elements thought to take part in X homolog pairing nevertheless result in Xist induction and inactivation of one of the two Xs in female cells (30, 33, 66–68).
Another mode of X-chromosomal dose sensing is the higher expression in XX cells of specific X-linked genes that lie within the Xic. The Xic-encoded Ftx and Jpx/Enox lncRNAs, both of which are expressed from the active and the inactive X chromosomes, are believed to facilitate X inactivation by activating Xist (19, 69, 70). Similarly, the Rnf12 protein-coding gene, also encoded within the Xic but subject to X inactivation, is also posited to induce Xist through its higher expression in females before inactivation (67, 71, 72). However, a deletion of the Xic segment encompassing all three of these factors does not prevent Xist induction or gene silencing, since the mutant X chromosome is able to undergo Xist RNA coating and inactivation in differentiating ESCs (68).
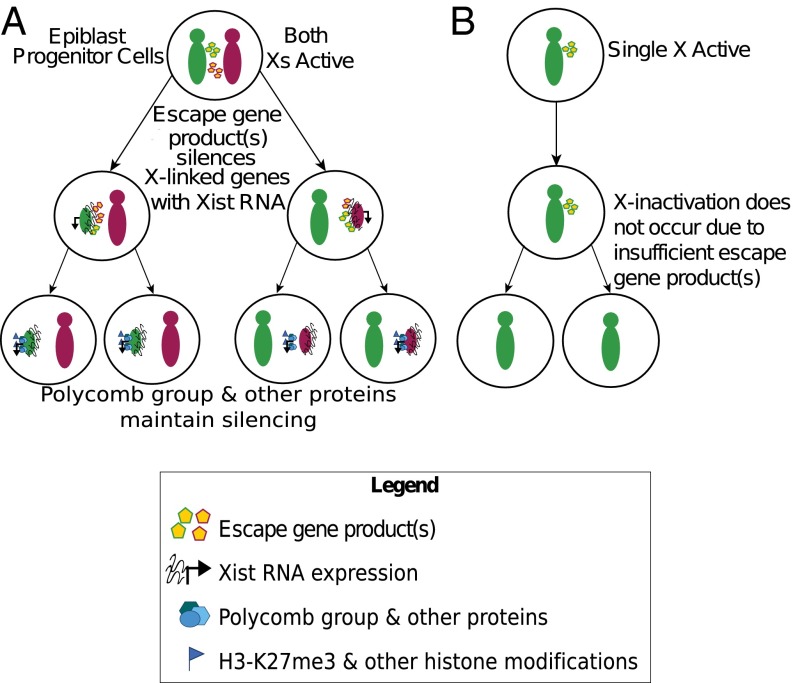
These observations open the possibility of an alternate X-linked dosage-sensing mechanism. Most X-linked genes in females with an inactive X chromosome are expressed at levels equal to that in males (73–75). A subset of X-linked genes, however, escape X inactivation in female cells and are capable of being expressed from both X chromosomes despite inactivation of one of the two Xs (76, 77). Due to expression from both alleles, these X-inactivation escapees are expressed at higher levels in females compared with males (78). We therefore suggest that the relatively higher expression of one or more X-inactivation escapees in females ectopically activates Xist expression as well as induces X-linked gene silencing in XΔTsixX cells. The lower dosage of such factors in males may explain the reduced frequency of Xist induction and X-linked gene silencing in XΔTsixY cells. Similarly, female XΔTsixO cells lack a second X chromosome, and, like males, would have a lower dosage of X-inactivation escapees compared with XΔTsixX females. The dose-dependent effect of the X-inactivation escapees implies that they function as diffusible/trans-acting factors.
The escapees in XΔTsixX mutants are also expected to escape X inactivation in WT XX cells. In the Tsix mutants, we propose that Xist is induced in both males and females by the escapees due to the lower threshold conferred by Tsix absence (30). In WT cells, the same dose of the escapees activates Xist only in females and not in males due to an intact Tsix locus (Fig. 6). Thus, we postulate that one or more X-inactivation escapees normally induces Xist from the future inactive X in WT XX females.
Fig. 6.
A model of random X-inactivation initiation by X-inactivation escapee(s). XX female pluripotent epiblast progenitor cells (A) have two active X chromosomes and express the products of the escape gene(s) at equal levels from both. Upon differentiation, the 2× dose of the escape gene product(s) robustly induces Xist from the future inactive X. Once Xist is induced, the same or different escape gene product(s) cooperates with Xist to initiate silencing of genes on the inactive X chromosome. Polycomb group and other proteins then maintain silencing on the inactive X in part by depositing repressive histone marks. In XY males (B), the lower dose of the escape gene product(s) is insufficient to induce Xist and to silence X-linked genes.
Once Xist is induced, the same, or a different, escape gene product may silence X-chromosomal genes cooperatively with Xist, either by interacting with Xist RNA or through a parallel pathway. Recent reports of proteins bound to Xist found two X-encoded proteins, NONO and RBM3, as direct Xist RNA partners (14, 15). Neither gene, however, escapes random X inactivation (77, 78). The critical escapee proteins may therefore only indirectly or transiently interact with Xist, or may indirectly induce Xist and X-linked gene silencing. Alternatively, the catalog of Xist-binding proteins may be incomplete (14, 15). Consistent with the dose-dependent function of the escapee(s) in triggering X inactivation, ESCs with supernumerary X chromosomes display faster kinetics of Xist induction and X inactivation, commensurate with the number of extra X chromosomes (33). By inducing Xist and X-linked gene silencing in a dose-dependent manner, the escapee(s) thus also serve as X-chromosomal counting factor(s).
The increased dose of one or more X-chromosomal genes is believed to underlie DNA methylation differences between male and female ESCs. Bulk DNA in XX ESCs is hypomethylated relative to XY ESCs (79). The same X-linked factor(s) may contribute to sex-specific differences in Xist induction and X-linked gene silencing. However, XX female somatic cells with an inactive X chromosome do not display reduced DNA methylation levels compared with XY male cells (79), suggesting that the effect is not modulated by X-inactivation escapees. Nevertheless, early X-inactivated cells such as female EpiSCs may be hypomethylated compared with male EpiSCs, potentially implicating the same X-inactivation escapee(s) in regulating both DNA hypomethylation and X inactivation.
The Xic is an obvious X-chromosomal segment where the candidate escapee genes may reside. Classical mouse and human studies of X-chromosome truncations, translocations, and deletions, pinpointed the Xic as necessary for initiating random X inactivation (19, 32, 80–83). Expectedly, the Xist locus maps to the Xic (9). The Xic, however, may not be sufficient to recapitulate the various steps underlying random X inactivation, including the sensing of the X-chromosomal dose, as suggested by the inability of large single-copy YAC Xic transgenes to induce Xist (28, 29). A recent report delineating X-inactivation escapees in ESCs via allele-specific RNA-seq may yield candidate X-inactivation regulators (77). It is also plausible that the escapees indirectly control Xist induction and gene silencing by up-regulating the active allele of a gene(s) that is subject to X inactivation, inducing other escapees, or triggering the sex-specific expression of autosomal factors. We are currently defining the repertoire of X-inactivation escapees in EpiSCs, to investigate which escape genes function as dosage-sensitive factors that induce Xist and trigger X-linked gene silencing.
Materials and Methods
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (84). All animals were handled according to protocols approved by the University Committee on Use and Care of Animals (UCUCA) at the University of Michigan (protocol #PRO00004007).
ESC and EpiSC derivation, RNA/DNA FISH, immunofluorescence (IF), RT-PCR, pyrosequencing, cell proliferation, viability assays, microscopy, and the mice used in this study have previously been described in ref. 30 and are detailed in SI Appendix.
Supplementary Material
Acknowledgments
We thank members of the S.K. laboratory for discussions and critical review of the manuscript; Shigeki Iwase and Jacob Mueller for critically evaluating the manuscript; Angela Andersen of Pickersgill and Andersen, Life Science Editors (lifescienceeditors.com/), for editing services; and we acknowledge the services of the University of Michigan Sequencing Core Facility, supported in part by the University of Michigan Comprehensive Cancer Center. This work was funded by an NIH National Research Service Award 5-T32-GM07544 from the National Institute of General Medicine Sciences (to E.M.), a University of Michigan Reproductive Sciences Program training grant, NIH National Research Service Award 1F31HD080280-01 from the National Institute of Child Health and Human Development (to E.M.), a Rackham Predoctoral Fellowship from the University of Michigan (to E.M.), an NIH Director’s New Innovator Award (DP2-OD-008646-01) (to S.K.), a March of Dimes Basil O’Connor Starter Scholar Research Award (5-FY12-119) (to S.K.), and the University of Michigan Endowment for Basic Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515971113/-/DCSupplemental.
References
- 1.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 2.Morey C, Avner P. The demoiselle of X-inactivation: 50 years old and as trendy and mesmerising as ever. PLoS Genet. 2011;7(7):e1002212. doi: 10.1371/journal.pgen.1002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152(6):1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 4.Barakat TS, Jonkers I, Monkhorst K, Gribnau J. X-changing information on X inactivation. Exp Cell Res. 2010;316(5):679–687. doi: 10.1016/j.yexcr.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol. 2009;21(3):359–366. doi: 10.1016/j.ceb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 6.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379(6561):131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 7.Marahrens Y, Loring J, Jaenisch R. Role of the Xist gene in X chromosome choosing. Cell. 1998;92(5):657–664. doi: 10.1016/s0092-8674(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 8.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11(2):156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 9.Brown CJ, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 10.Brown CJ, et al. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 11.Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132(3):259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panning B, Jaenisch R. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 1996;10(16):1991–2002. doi: 10.1101/gad.10.16.1991. [DOI] [PubMed] [Google Scholar]
- 13.Lee JT. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol. 2011;12(12):815–826. doi: 10.1038/nrm3231. [DOI] [PubMed] [Google Scholar]
- 14.Chu C, et al. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161(2):404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalantry S, Purushothaman S, Bowen RB, Starmer J, Magnuson T. Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature. 2009;460(7255):647–651. doi: 10.1038/nature08161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256(5519):640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- 18.Gribnau J, Luikenhuis S, Hochedlinger K, Monkhorst K, Jaenisch R. X chromosome choice occurs independently of asynchronous replication timing. J Cell Biol. 2005;168(3):365–373. doi: 10.1083/jcb.200405117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maclary E, Hinten M, Harris C, Kalantry S. 2013. Long nonoding RNAs in the X-inactivation center. Chromosome Res 21(6–7):601–614. [DOI] [PMC free article] [PubMed]
- 20.Jiang J, et al. Translating dosage compensation to trisomy 21. Nature. 2013;500(7462):296–300. doi: 10.1038/nature12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bala Tannan N, et al. DNA methylation profiling in X;autosome translocations supports a role for L1 repeats in the spread of X chromosome inactivation. Hum Mol Genet. 2014;23(5):1224–1236. doi: 10.1093/hmg/ddt553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cotton AM, et al. Spread of X-chromosome inactivation into autosomal sequences: Role for DNA elements, chromatin features and chromosomal domains. Hum Mol Genet. 2014;23(5):1211–1223. doi: 10.1093/hmg/ddt513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herzing LB, Romer JT, Horn JM, Ashworth A. Xist has properties of the X-chromosome inactivation centre. Nature. 1997;386(6622):272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- 24.Lee JT, Jaenisch R. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature. 1997;386(6622):275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- 25.Tang YA, et al. Efficiency of Xist-mediated silencing on autosomes is linked to chromosomal domain organisation. Epigenetics Chromatin. 2010;3(1):10. doi: 10.1186/1756-8935-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5(4):695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 27.Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30(2):167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 28.Heard E, Mongelard F, Arnaud D, Avner P. Xist yeast artificial chromosome transgenes function as X-inactivation centers only in multicopy arrays and not as single copies. Mol Cell Biol. 1999;19(4):3156–3166. doi: 10.1128/mcb.19.4.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heard E, et al. Transgenic mice carrying an Xist-containing YAC. Hum Mol Genet. 1996;5(4):441–450. doi: 10.1093/hmg/5.4.441. [DOI] [PubMed] [Google Scholar]
- 30.Gayen S, Maclary E, Buttigieg E, Hinten M, Kalantry S. A primary role for the Tsix lncRNA in maintaining random X-chromosome inactivation. Cell Reports. 2015;11(8):1251–1265. doi: 10.1016/j.celrep.2015.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sado T, Wang Z, Sasaki H, Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. 2001;128(8):1275–1286. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- 32.Rastan S, Robertson EJ. X-chromosome deletions in embryo-derived (EK) cell lines associated with lack of X-chromosome inactivation. J Embryol Exp Morphol. 1985;90:379–388. [PubMed] [Google Scholar]
- 33.Monkhorst K, Jonkers I, Rentmeester E, Grosveld F, Gribnau J. X inactivation counting and choice is a stochastic process: Evidence for involvement of an X-linked activator. Cell. 2008;132(3):410–421. doi: 10.1016/j.cell.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 34.Namekawa SH, Payer B, Huynh KD, Jaenisch R, Lee JT. Two-step imprinted X inactivation: Repeat versus genic silencing in the mouse. Mol Cell Biol. 2010;30(13):3187–3205. doi: 10.1128/MCB.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patrat C, et al. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc Natl Acad Sci USA. 2009;106(13):5198–5203. doi: 10.1073/pnas.0810683106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300(5616):131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 38.Silva J, et al. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4(4):481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 39.Tesar PJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448(7150):196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 40.Brons IG, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448(7150):191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 41.Pasque V, Gillich A, Garrett N, Gurdon JB. Histone variant macroH2A confers resistance to nuclear reprogramming. EMBO J. 2011;30(12):2373–2387. doi: 10.1038/emboj.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bao S, et al. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461(7268):1292–1295. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chadwick LH, Pertz LM, Broman KW, Bartolomei MS, Willard HF. Genetic control of X chromosome inactivation in mice: Definition of the Xce candidate interval. Genetics. 2006;173(4):2103–2110. doi: 10.1534/genetics.105.054882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnston PG, Cattanach BM. Controlling elements in the mouse. IV. Evidence of non-random X-inactivation. Genet Res. 1981;37(2):151–160. doi: 10.1017/s0016672300020127. [DOI] [PubMed] [Google Scholar]
- 45.Ohhata T, Hoki Y, Sasaki H, Sado T. Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development. 2008;135(2):227–235. doi: 10.1242/dev.008490. [DOI] [PubMed] [Google Scholar]
- 46.Kalantry S, Magnuson T. The Polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS Genet. 2006;2(5):e66. doi: 10.1371/journal.pgen.0020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grumbach MM, Morishima A, Taylor JH. Human sex chromosome abnormalities in relation to DNA replication and heterochromatinization. Proc Natl Acad Sci USA. 1963;49(5):581–589. doi: 10.1073/pnas.49.5.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lyon MF. Sex chromatin and gene action in the mammalian X-chromosome. Am J Hum Genet. 1962;14:135–148. [PMC free article] [PubMed] [Google Scholar]
- 49.Speirs S, Cross JM, Kaufman MH. The pattern of X-chromosome inactivation in the embryonic and extra-embryonic tissues of post-implantation digynic triploid LT/Sv strain mouse embryos. Genet Res. 1990;56(2-3):107–114. doi: 10.1017/s0016672300035175. [DOI] [PubMed] [Google Scholar]
- 50.Webb S, de Vries TJ, Kaufman MH. The differential staining pattern of the X chromosome in the embryonic and extraembryonic tissues of postimplantation homozygous tetraploid mouse embryos. Genet Res. 1992;59(3):205–214. doi: 10.1017/s0016672300030494. [DOI] [PubMed] [Google Scholar]
- 51.Monkhorst K, et al. The probability to initiate X chromosome inactivation is determined by the X to autosomal ratio and X chromosome specific allelic properties. PLoS One. 2009;4(5):e5616. doi: 10.1371/journal.pone.0005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohen DE, et al. The DXPas34 repeat regulates random and imprinted X inactivation. Dev Cell. 2007;12(1):57–71. doi: 10.1016/j.devcel.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 53.Navarro P, et al. Molecular coupling of Tsix regulation and pluripotency. Nature. 2010;468(7322):457–460. doi: 10.1038/nature09496. [DOI] [PubMed] [Google Scholar]
- 54.Stavropoulos N, Rowntree RK, Lee JT. Identification of developmentally specific enhancers for Tsix in the regulation of X chromosome inactivation. Mol Cell Biol. 2005;25(7):2757–2769. doi: 10.1128/MCB.25.7.2757-2769.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vigneau S, Augui S, Navarro P, Avner P, Clerc P. An essential role for the DXPas34 tandem repeat and Tsix transcription in the counting process of X chromosome inactivation. Proc Natl Acad Sci USA. 2006;103(19):7390–7395. doi: 10.1073/pnas.0602381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maclary E, et al. Differentiation-dependent requirement of Tsix long non-coding RNA in imprinted X-chromosome inactivation. Nat Commun. 2014;5:4209. doi: 10.1038/ncomms5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485(7398):381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsai CL, Rowntree RK, Cohen DE, Lee JT. Higher order chromatin structure at the X-inactivation center via looping DNA. Dev Biol. 2008;319(2):416–425. doi: 10.1016/j.ydbio.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burgoyne PS, et al. 1995. The genetic basis of XX-XY differences present before gonadal sex differentiation in the mouse. Philos Trans R Soc Lond B Biol Sci 350(1333):253–260, discussion 260–251.
- 60.Nagy A, Gertsenstein M, Vintersten K, Behringer RR. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 2003. p 71. [Google Scholar]
- 61.Snow MHL. Gastrulation in the mouse: Growth and regionalization of the epiblast. J Embryol Exp Morphol. 1977;42:293–303. [Google Scholar]
- 62.Marahrens Y. X-inactivation by chromosomal pairing events. Genes Dev. 1999;13(20):2624–2632. doi: 10.1101/gad.13.20.2624. [DOI] [PubMed] [Google Scholar]
- 63.Augui S, et al. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science. 2007;318(5856):1632–1636. doi: 10.1126/science.1149420. [DOI] [PubMed] [Google Scholar]
- 64.Bacher CP, et al. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8(3):293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 65.Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311(5764):1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 66.Sun S, Fukue Y, Nolen L, Sadreyev R, Lee JT. Characterization of Xpr (Xpct) reveals instability but no effects on X-chromosome pairing or Xist expression. Transcription. 2010;1(1):46–56. doi: 10.4161/trns.1.1.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jonkers I, et al. RNF12 is an X-encoded dose-dependent activator of X chromosome inactivation. Cell. 2009;139(5):999–1011. doi: 10.1016/j.cell.2009.10.034. [DOI] [PubMed] [Google Scholar]
- 68.Barakat TS, et al. The trans-activator RNF12 and cis-acting elements effectuate X chromosome inactivation independent of X-pairing. Mol Cell. 2014;53(6):965–978. doi: 10.1016/j.molcel.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 69.Chureau C, et al. Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum Mol Genet. 2011;20(4):705–718. doi: 10.1093/hmg/ddq516. [DOI] [PubMed] [Google Scholar]
- 70.Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143(3):390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barakat TS, et al. RNF12 activates Xist and is essential for X chromosome inactivation. PLoS Genet. 2011;7(1):e1002001. doi: 10.1371/journal.pgen.1002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gontan C, et al. RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature. 2012;485(7398):386–390. doi: 10.1038/nature11070. [DOI] [PubMed] [Google Scholar]
- 73.Deng X, Disteche CM. Genomic responses to abnormal gene dosage: The X chromosome improved on a common strategy. PLoS Biol. 2010;8(2):e1000318. doi: 10.1371/journal.pbio.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Disteche CM. Dosage compensation of the sex chromosomes. Annu Rev Genet. 2012;46:537–560. doi: 10.1146/annurev-genet-110711-155454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deng X, et al. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat Genet. 2011;43(12):1179–1185. doi: 10.1038/ng.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Berletch JB, Yang F, Disteche CM. Escape from X inactivation in mice and humans. Genome Biol. 2010;11(6):213. doi: 10.1186/gb-2010-11-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marks H, et al. Dynamics of gene silencing during X inactivation using allele-specific RNA-seq. Genome Biol. 2015;16(1):149. doi: 10.1186/s13059-015-0698-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berletch JB, et al. Escape from X inactivation varies in mouse tissues. PLoS Genet. 2015;11(3):e1005079. doi: 10.1371/journal.pgen.1005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zvetkova I, et al. Global hypomethylation of the genome in XX embryonic stem cells. Nat Genet. 2005;37(11):1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
- 80.Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet. 2011;12(6):429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- 81.Brown CJ, et al. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature. 1991;349(6304):82–84. doi: 10.1038/349082a0. [DOI] [PubMed] [Google Scholar]
- 82.Takagi N. Primary and secondary nonrandom X chromosome inactivation in early female mouse embryos carrying Searle’s translocation T(X; 16)16H. Chromosoma. 1980;81(3):439–459. doi: 10.1007/BF00368155. [DOI] [PubMed] [Google Scholar]
- 83.Rastan S. Non-random X-chromosome inactivation in mouse X-autosome translocation embryos—location of the inactivation centre. J Embryol Exp Morphol. 1983;78:1–22. [PubMed] [Google Scholar]
- 84.Committee on Care and Use of Laboratory Animals . Guide for the Care and Use of Laboratory Animals. Natl Inst Health; Bethesda: 1996. DHHS Publ No (NIH) 85-23. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.