Abstract
l-Ascorbic acid (ascorbate, AsA, vitamin C) is essential for animal and plant health. Despite our dependence on fruits and vegetables to fulfill our requirement for this vitamin, the metabolic network leading to its formation in plants is just being fully elucidated. There is evidence supporting the operation of at least four biosynthetic pathways leading to AsA formation in plants. These routes use d-mannose/l-Galactose, l-gulose, d-galacturonate, and myo-inositol as the main precursors. This review focuses on aldonolactone oxidoreductases, a subgroup of the vanillyl alcohol oxidase (VAO; EC 1.1.3.38) superfamily, enzymes that catalyze the terminal step in AsA biosynthesis in bacteria, protozoa, animals, and plants. In this report, we review the properties of well characterized aldonolactone oxidoreductases to date. A shared feature in these proteins is the presence of a flavin cofactor as well as a thiol group. The flavin cofactor in many cases is bound to the N terminus of the enzymes or to a recently discovered HWXK motif in the C terminus. The binding between the flavin moiety and the protein can be either covalent or non-covalent. Substrate specificity and subcellular localization differ among the isozymes of each kingdom. All oxidases among these enzymes possess dehydrogenase activity, however, exclusive dehydrogenases are also found. We also discuss recent evidence indicating that plants have both l-gulono-1,4-lactone oxidases and l-Galactono-1,4-lactone dehydrogenases involved in AsA biosynthesis.
Keywords: Aldonolactone oxidoreductase, ascorbate, galactonolactone dehydrogenase, gulonolactone oxidase, vanillyl alcohol oxidase family
1. Introduction
l-Ascorbic acid (AsA, ascorbate, vitamin C) is a key antioxidant for human and plant health. Humans, non-human primates and guinea pigs cannot produce vitamin C due to the absence of l-gulono-1,4-lactone oxidase (GulLO; EC 1.1.3.8) activity (Burns, 1957). Therefore, we need plant sources to obtain this vitamin. Many approaches have been undertaken to increase vitamin C content in plants yet challenges still remain (reviewed in Gallie, 2013; Locato et al., 2013). In order to successfully engineer elevated AsA content in plants, we need to have a clear understanding about the pathways and enzymes that catalyze the reactions involved in AsA metabolism. In animals, vitamin C biosynthesis proceeds in a single route from d-glucose via d-Glucuronic acid and l-gulono-1,4-lactone (l-GulL), as discovered from the early works of Isherwood and others (1954). In contrast, plants possess an intertwined biochemical network to make this molecule. This network involves myo-inositol, l-gulose, d-mannose/l-Galactose (Man/Gal), and d-galacturonate as entry points. The Man/Gal pathway a.k.a Smirnoff-Wheeler pathway is the best characterized of all and the enzymes involved in this process have been cloned and characterized (Wheeler et al., 1998; Conklin et al., 1999; Laing et al., 2004; Conklin et al., 2006; Laing et al., 2007; Qian et al., 2007; Maruta et al., 2008). Only a few of the enzymes involved in the other three pathways have been studied to date (Agius et al., 2003; Wolucka and van Montagu, 2003; Lorence et al., 2004).
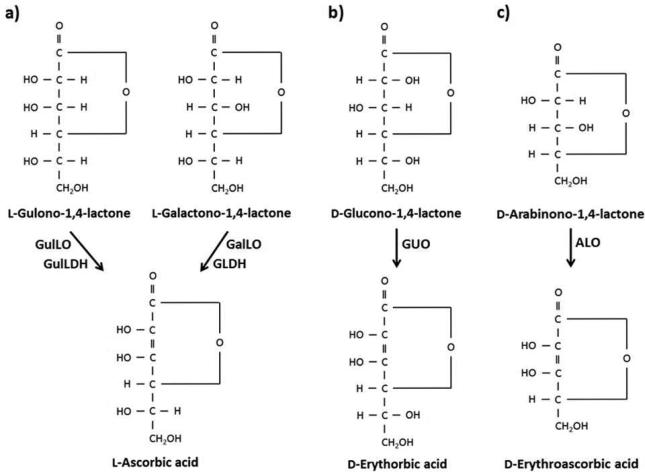
Aldonolactone oxidoreductases catalyze the terminal step in the conversion of an aldonolactone to AsA in plants and animals or its analogues in fungi (Figure 1). Aldonolactone oxidoreductases of different organisms (isoenzymes) are named after the substrate they use, the physiological relevance of substrate and the use of electron acceptors. These two subtopics (substrate specificity and electron acceptors) are discussed in sections 4 and 5 of this paper. There are two immediate precursors to AsA in plants: l-Galactono-1,4-lactone (l-GalL) and l-gulono-1,4-lactone (l-GulL). l-Galactono-1,4-lactone dehydrogenase (GLDH; EC 1.3.2.3) catalyzes the conversion ofl-GalL to AsA, while GulLO catalyzes the transformation of l-GulL into AsA. A detailed study about the existence of aldonolactone oxidoreductases in different prokaryotic and eukaryotic lineages has been recently published (Wheeler et al., 2015). Isoenzymes of GLDH have been characterized from animals, plants, and other organisms. GLDH was suggested to be a good candidate for the control of AsA biosynthesis (Bartoli et al., 2005; Tamaoki et al., 2003; Tokunaga et al., 2005). However, no clear relationship between AsA content and GLDH activity has been found to date (Bartoli et al., 2005). An attempt to increase AsA by overexpressing GLDH in tobacco did not yield positive results (Imai et al., 2009). In contrast one of the earlier successes to increase AsA content in plants, consisted in overexpression of a rat GulLO in lettuce and tobacco that resulted in up to 7-fold increase in foliar AsA content (Jain and Nessler, 2000).
Figure 1.
Aldonolactone oxidoreductases catalyzing the terminal step in the biosynthesis of ascorbate or its analogues in (a) plants, animals, protozoa and bacteria, (b) Penicillia and (c) yeast from their preferred substrates. ALO, d-arabinono-1,4-lactone oxidase; GalLO, l-galactono-1,4-lactone oxidase; GLDH, l-galactono-1,4-lactone dehydrogenase; GulLDH, l-gulono-1,4-lactone dehydrogenase; GulLO, l-gulono-1,4-lactone oxidase; GUO, d-gluconolactone oxidase.
Studies have also been conducted to restore the vitamin C biosynthetic capacity in animal cells by transgenic expression of the rat or mouse GulLO gene in guinea pig cells (Krasnov et al., 1998), human cells (Ha et al., 2004), monkey cells (Yagi et al., 1991), and in fish cells such as the rainbow trout and the Far Eastern catfish (Krasnov et al., 1998; Nam et al., 2002). GulLO is also being studied as a potential therapeutic target in protozoans such as Trypanasoma cruzi (Logan et al., 2007; Kudryashova et al., 2011), T. brucei (Wilkinson et al., 2005), and Leishmania donovani (Biyani and Madhubala, 2011) as well as in the pathogenic yeast Candida albicans (Huh et al., 2001). d-Arabinono-1,4-lactone oxidase (ALO; EC 1.1.3.37), the isoenzyme in yeasts, and d-gluconolactone oxidase (GUO; EC 1.1.3.-) have been also studied for their industrial application to synthesize AsA or its analogues using E. coli or yeasts (Lee et al., 1999; Hancock and Viola, 2001; Salusjärvi et al., 2004; Sauer et al., 2004). In addition, plant GLDH has been studied for its use as a biocatalyst in commercial AsA production (Leferink, 2009a). Isoenzymes from other species in mammals, fungi, and algae have been reported (Takahashi et al., 1976; Nishikimi et al., 1978; Shigeoka et al., 1979; Bleeg and Christensen, 1982; Kiuchi et al., 1982; Okamura, 2001).
A review on recently characterized enzymes from the vanillyl alcohol oxidase (VAO family) including aldonolactone oxidoreductases was published seven years ago (Leferink et al., 2008a). A focused review on the characterization of aldonolactone oxidoreductases was published recently (Leferink and van Berkel, 2014). However, this work does not include any of the plant GulLOs. Therefore, the current review was prepared to expand on the knowledge about the aldonolactone oxidoreductases characterized to date. This paper discusses the history of characterization, cofactor attachment, substrate specificity, electron acceptor, inhibitors, functional residues, and properties of recombinantly expressed GulLO isoenzymes.
2. History of aldonolactone oxidoreductases
l-gulonolactone oxidase (GulLO) activity was first identified in rat liver microsomes by Burns et al. (1956). Eliceiri et al. (1969) partially purified the enzyme from rat liver microsomes. A purification protocol resulting in 8 to15-fold purification was developed and some properties of the enzyme were studied including the finding of a prosthetic group (Nakagawa and Asano, 1970). Later on, Nishikimi et al. (1976) purified the protein to apparent homogeneity; this aided in antisera generation, which further led to the identification of the cDNA sequence (Koshizaka et al., 1988). These tools allowed the study of the molecular mechanism for lack of vitamin C production in scurvy prone animals including humans (Nishikimi and Udenfriend, 1976; Nishikimi and Yagi, 1991). From there on, research moved in the direction of identifying the molecular mechanism(s) for the lack of vitamin C synthesis in scurvy prone animals. Aberrant gene sequences were identified in the human and the guinea pig genomes for GulLO (Nishikimi et al., 1988; Nishikimi et al., 1992; Inai et al., 2003).
In plants, however, a similar activity, l-Galactone-1,4-lactone dehydrogenase (GLDH), was first identified in mitochondria of pea and mung bean seeds (Mapson et al., 1954). This activity was partially purified and characterized from cauliflower floret mitochondria (Mapson and Breslow, 1958). Since then isoforms from spinach (Mutsuda et al., 1995) and white potato tubers (Ôba et al., 1994) have been reported. cDNA sequences encoding GLDH from cauliflower (Østergaard et al., 1997) and sweet potato (Imai et al., 1998) were identified. GLDH and GulLO are both enzymes involved in AsA biosynthesis in plants.
3. Flavin attachment to aldonolactone oxidoreductases
Aldonolactone oxidoreductases are part of the VAO family of flavoenzymes. The flavin group can be either covalently or non-covalently attached to the proteins. Covalent attachment increases the redox power of the enzyme, saturation of the active site with cofactor, protein stability, and prevents flavin modification (Leferink et al., 2008a). Flavin adenine dinucleotide (FAD) is the flavin cofactor associated with all aldonolactone oxidoreductases identified to date, except for the isoform in cauliflower (Mapson and Breslow, 1958), which has flavin mononucleotide (FMN; Table 1).
Table 1.
Properties of flavin characterization in aldonolactone oxidoreductases
| Source | Spectral peaks (nm) | Mode of flavin attachment | Covalent flavin linkage | Residue in N terminus FAD domain | Presence of HWXK motif | FAD or FMN | Fluorescence in SDS-PAGE | References | |
|---|---|---|---|---|---|---|---|---|---|
| Animals | Rat | 346, 443 | Covalent | 8α-N(1)-histidyl | His | Yes | FAD | Yes | Nakagawa et al., 1975 |
| Kenney et al., 1976 | |||||||||
| Nishikimi et al., 1977 | |||||||||
| Chicken | 352, 454 | Covalent | 8α-N(1)-histidyl | ND | ND | FAD | Yes | Kiuchi et al., 1982 | |
| Goat | 350, 455 | Covalent | ND | ND | ND | ND | ND | Nishikimi et al., 1976 | |
| Mouse | ND | Covalent | ND | His | Yes | ND | ND | Ha et al., 2004 | |
| Pig | ND | Covalent | ND | His | Yes | ND | ND | Hasan et al., 2004 | |
| Fungi | S. cerevisiae | 350, 450 | Covalent | 8α-N(1)-histidyl | His | Yes | FAD | Yes | Nishikimi et al., 1978 |
| Kenney et al., 1979 | |||||||||
| Huh et al., 1998 | |||||||||
| C. albicans | 350, 450 | Covalent | 8α-N(3)-histidyl | His | Yes | FAD | Yes | Huh et al., 1994 | |
| Huh et al., 2001 | |||||||||
| Takahashi et al., 1976 | |||||||||
| P. griseoroseum | 350, 450 | Covalent | 8α-N(3)-histidyl | His | Yes | FAD | Yes | Shimizu et al., 1977 | |
| Harada et al., 1979 | |||||||||
| Salusjärvi et al., 2004 | |||||||||
| G. frondosa | 358, 455 | Covalent | ND | ND | ND | ND | Yes | Okamura, 2001 | |
| Bacteria | M. tuberculosis | Nil | Nil | ND | His | Yes | Nil | No | Wolucka and Communi, 2006 |
| G. oxydans | ND | Covalent | ND | ND | ND | ND | Yes | Sugisawa et al., 1995 | |
| Protozoa | T. brucei | ND | Non-covalent | ND | Lys | Yes | FAD (predicted) | ND | Wilkinson et al., 2005 |
| Logan et al., 2007 | |||||||||
| Logan et al., 2007 | |||||||||
| T. cruzi | ND | Non-covalent | ND | Lys | Yes | FAD | ND | Kudryashova et al., 2011 | |
| L. donovani | ND | Non-covalent (predicted) | ND | Lys | Yes | ND | ND | Biyani and Madhubala, 2011 | |
| Plants | Cauliflower | 375, 455 | Non-covalent | ND | Leu | Yes | FMN | ND | Mapson and Breslow, 1958 |
| Østergaard et al., 1997 | |||||||||
| Sweet potato | 373, 448 | Non-covalent | ND | Leu | Yes | ND | ND | Ôba et al., 1995 | |
| Imai et al., 1998 | |||||||||
| Tobacco | ND | Non-covalent | ND | Leu | Yes | ND | ND | Yabuta et al., 2000 | |
| Arabidopsis | 375, 450 | Non-covalent | ND | Leu | Yes | FAD | ND | Leferink et al., 2008b | |
ND: not determined
The flavin attachment is determined from signature spectral analysis of the purified protein, whereas a more detailed analysis usually involves purifying the flavin peptide followed by releasing the cofactor and determining its identity. A review about the methods employed for the identification of flavin linkage is found in Mewies et al (1998). In the spectral analysis, the purified protein is tested for a hypsochromic shift than FAD or FMN at the second maximum (at about 350 nm) indicating covalent flavin attachment to the protein (Nishikimi et al., 1978; Huh et al., 1994). Plant GLDHs with non-covalently bound flavin show the same spectrum as in FAD or FMN or riboflavin (at about 375 nm) without any hypsochromic shift (Table 1). Covalently bound flavin should not be released with trichloroacetic acid, SDS, boiling or acid ammonium sulfate treatment (Nakagawa et al., 1975; Nishikimi et al., 1978; Huh et al., 1994). Trichloroacetic acid treatment may hydrolyze the pyrophosphate bond in FAD to FMN (Nakagawa et al., 1975; Kudryashova et al., 2011) and this should be considered while determining the identity of the flavin. The identity of the flavin cofactor can be determined by TLC, pyrophosphatase treatment (Kiuchi et al., 1982), paper chromatography (Mapson and Breslow, 1958) or HPLC analysis (Logan et al., 2007). SDS and β-mercaptoethanol treated proteins with a covalently bound flavin, when soaked in 7% acetic acid solution, fluoresce under UV illumination (Nishikimi et al., 1977).
A flavin group is covalently attached to all the identified isoforms from animals, fungi, and bacteria while it is non-covalently attached to the isoforms in protozoa and plants (Table 1). GulLOs of mouse, pig, and Grifola frondosa, a basidiomycete's fungus may have a covalent flavin due to the presence of a histidine residue in the N terminus FAD domain or a weak fluorescence in SDS-PAGE (Table 1). The activity of Euglena gracilis GulLDH was slightly inhibited by riboflavin suggesting flavin involvement in this organism (Shigeoka et al., 1979).
The GalLOs from T. cruzi and T. brucei were shown to have a non-covalently bound FMN (Logan et al., 2007). However, the cofactor in the T. cruzi enzyme was found to be FAD when the factor was extracted with butanol/acetic acid/water system (Kudryashova et al., 2011). The authors point out that the trichloroacetic acid procedure, used to extract the cofactor from the T. cruzi GalLO (Logan et al., 2007), may have hydrolyzed the FAD to FMN giving a false result. It is possible that the true cofactor of the GalLO enzyme in T. brucei may also be FAD (Wilkinson et al., 2005). The isoenzyme from Leishmania donovani seems to have a non-covalently bound FAD (Biyani and Madhubala, 2011).
The GulLDH of Gluconobacter oxydans is reported to contain a cytochrome C component in addition to the flavin group (Sugisawa et al., 1995). The recombinant GulLDH of M. tuberculosis does not have a flavin cofactor, as no flavin spectrum was seen; in addition the enzyme was insensitive to exogenous FAD or riboflavin addition (Wolucka and Communi, 2006).
GLDHs from plants contain a non-covalent flavin prosthetic group as found in cauliflower, sweet potato, tobacco, and Arabidopsis (Table 1). Another GLDH preparation from cauliflower was reported not to have a flavin prosthetic group (Østergaard et al., 1997) and this discrepancy remains unclear. The lack of a shift in the absorption spectrum in the sweet potato GLDH was suggested due to non-covalent flavin attachment (Imai et al., 1998), which was later confirmed in AtGLDH as well (Leferink et al., 2008b). Amino acid sequence analysis of the GLDHs revealed the presence of leucine instead of histidine in the N terminus FAD-binding domain (Imai et al., 1998; Yabuta et al., 2000; Leferink et al., 2008b) and a lysine in the protozoan GalLOs (Biyani and Madhubala, 2011). The replacement of leucine instead of histidine in the FAD-binding domain was proposed as the reason for non-covalent flavin attachment (Imai et al., 1998). Leferink et al (2008b) replaced the leucine with a histidine in AtGLDH but did not observe any covalent FAD attachment. Rather a partial release of the FAD factor, something that did not happen when the wild type enzyme was studied.
4. Substrate specificity of aldonolactone oxidoreductases
4.1. Preferred substrates
The preferred substrate for animal GulLOs is l-GulL and they can also oxidize l-GalL at a rate >70% compared to l-GulL (Table 2). d-Arabinono-1,4-lactone (d-AL) and l-GalL are used at an equally significant rate by the ALOs from ascomycete fungi.However, l-GulL was oxidized only at about one-third rate of d-AL (Table 2).
Table 2.
Substrate specificity of aldonolactone oxidoreductases towards each other's preferred substrates
| Source | l-Gulono-1,4-lactone | d-Mannono-1,4-lactone | l-Galactono-1,4-lactone | d-Altrono-1,4-lactone | d-Arabinono-1,4-lactone | d-Glucono-1,4-lactone | References | |
|---|---|---|---|---|---|---|---|---|
| Animals | Rat | 100 | 68 | 87 | 47 | 22 | NDd | Ashwell et al., 1961 |
| Rat | 100 (0.066 mM) | 61 | 70 | 15 | ND | <3 | Kiuchi et al., 1982 | |
| Nishikimi et al., 1976 | ||||||||
| Chicken | 100 (0.007 mM) | 64 | 90 | 16 | ND | <3 | Kiuchi et al., 1982 | |
| Goat | 100 (0.15 mM) | ND | ND | ND | ND | ND | Nishikimi et al., 1976 | |
| Fungi | S. cerevisiae | 32 | <3 | 100 | 68 | ND | <3 | Nishikimi et al., 1978 |
| S. cerevisiae | 0 | ND | 100 (0.30 mM) | 65 (2.0 mM) | 97 (0.16 mM) | ND | Bleeg and Christensen, 1982 | |
| C. albicans | 24.7 | ND | 87.6 (53.7 mM) | ND | 100 (44.1 mM) | NDd | Huh et al., 1994 | |
| P. griseoroseum | 0 | 0 | ND | ND | ND | 100e (1.7 mM) | Takahashi et al., 1976 | |
| P. notatum | ND | ND | 0 | 0 | ND | 100 | Takahashi, 1969 | |
| G. frondosa | 100 (24 mM) | 25 | 2 | 0 | 0 | NDd | Okamura, 2001 | |
| L. edodes | 100 | 24 | 0 | 0 | 0 | NDd | Okamura, 2001 | |
| Bacteria | M. tuberculosis | 100 (5.5 mM) | ND | 0 | ND | ND | ND | Wolucka and Communi, 2006 |
| G. oxydans | 100 (34.8 mM) | ND | 0 | ND | ND | NDd | Sugisawa et al., 1995 | |
| Protozoa | T. brucei | 3 | ND | 77 (0.154 mM) | ND | 100 (0.055 mM) | ND | Wilkinson et al., 2005 |
| T. cruzi | 0 | ND | 100 (0.161 mM) | ND | 96 (0.285 mM) | ND | Logan et al., 2005 | |
| L. donovani | 0 | ND | ND | ND | 100 (0.039 mM) | ND | Biyani and Madhubala, 2011 | |
| Algae | E. gracilis | 71 | ND | 100 | ND | ND | ND | Shigeoka et al., 1979 |
| Plants | Cauliflower | 0 | 0 | 100 (2 and 4 mM)a | 10 | ND | 0 | Mapson and Breslow, 1958 |
| Cauliflower | 0 | ND | 100 (3.3 mM) | ND | ND | ND | Østergaard et al., 1977 | |
| White potato | 19.5 | ND | 100 (0.08 and 0.23 mM)b | ND | ND | ND | Ôba et al., 1994 | |
| Sweet potato | 1 | ND | 100 (0.12 mM) | ND | ND | ND | Ôba et al., 1995 | |
| Spinach | 0 | ND | 100 (0.192 mM) | ND | ND | ND | Mutsuda et al., 1995 | |
| Tobacco | 7 | ND | 100 (0.06 and 0.08 mM)c | ND | ND | ND | Yabuta et al., 2000 | |
| Strawberry | 0 | ND | 100 | ND | ND | ND | Nascimento et al., 2005 | |
| Arabidopsis | (13.1 mM) | ND | 100 (0.17 mM) | ND | 10.2 mM | ND | Leferink et al., 2008b | |
| Leferink et al., 2009c | ||||||||
with different electron acceptors
with different methods
with native and recombinant enzymes
No or very less activity with Dglucono-1,5-lactone
similar result with d-glucono-1,5-lactone; ND, not determined. Numbers in the parantheses indicate the substrate affinity values (Km).
A GUO partially purified from P. griseroseum is specific for d-glucono-1,4 and d-glucono-1,5-lactones among other hexono acid lactones tested (Takahashi et al., 1976) and that of P. notatum can use d-glucono-1,4-lactone (Takahashi, 1969; Table 2). l-GulL was not a substrate for P. notatum and GUO of P. griseoroseum (Takahashi et al., 1976). l-GalL did not serve as a substrate for the cells of P. notatum (Takahashi et al., 1976) and it may not be a substrate for GUO of P. griseoroseum. GulLO of G. frondosa, a basidiomycetes fungi, is highly specific for l-GulL and it can also use d-mannono-1,4-lactone at 25% efficiency as l-GulL. It appears that this enzyme has substrate specificity for the hydroxyl group at C2 and C3 to be in the “d” form and also only six carbon sugar acid lactones served as substrates (Figure 1; Okamura, 2001). The physiological substrate of this enzyme was thought to be 6-deoxy-GulL because 6-deoxy-5-O-(α-d-glucopyranosyl)-AsA is the naturally occurring AsA analogue in G. frondosa (Okamura, 1994). 6-Deoxy-GalL (l-fucono-1,4-lactone) did not serve as a substrate as expected. Similar results were obtained for crude enzyme preparation from Lentinus edodes (Okamura, 2001). Bacterial GulLDHs from M. tuberculosis and G. oxydans were specific for l-GulL and l-GalL did not serve as a substrate (Sugisawa et al., 1995; Wolucka and Communi, 2006). These enzymes are different from animal, plant, and fungal GulLO isoenzymes in their substrate specificity.
d-Arabinono-1,4-lactone and l-GalL are the preferred substrates for protozoan GalLOs (Table 2). None of the three protozoan enzymes used l-GulL as a substrate (Table 2). The physiologically relevant substrate for T. cruzi was found to be l-GalL (Logan et al., 2007).
Plant GLDHs are highly specific to l-GalL as seen in cauliflower, sweet potato, spinach, tobacco, strawberry, and Arabidopsis (Table 2). l-GalL was found to be the physiological substrate for this enzyme (Østergaard et al., 1997), while l-GulL was not a substrate for these enzymes (Table 2). Recombinant tobacco GLDH used l-GulL as a substrate at 7% as l-GalL but not the native enzyme (Yabuta et al., 2000). Sweet potato GLDH was only 1% efficient towards l-GulL. Recombinant AtGLDH catalyzed l-GulL at a significant rate with a Km of 19.1 mM (Leferink et al., 2008b).
4.2 Stereoisomers
Different stereoisomers among hexono and pentono lactones have been studied for their function as substrates for aldonolactone oxidoreductases. A comprehensive list of substrates was studied for enzymes from G. frondosa, L. edodes (Okamura, 2001) and rat GulLO (Ashwell et al., 1961; Okamura, 2001). Rat GulLO can use substrates, although at low rate, from hexono, pentono and tetrono sugar acid lactones as long as the hydroxyl group at C2 is in the “l” form as in l-GulL and l-GalL (Ashwell et al., 1961; Figure 1). d-mannono-1,4-lactone has a similar chemical configuration as l-GulL except that the hydroxyl group at C5 is opposite to that of l-GulL. The same is the relationship between d-ALtrono-1,4-lactone and l-GalL (Figure 1). d-mannono-1,4-lactone and d-ALtrono-1,4-lactone were was used at low efficiency by some isoforms (Table 2). However, the enantiomer, l-mannono-1,4-lactone was not oxidized by cauliflower and Arabidopsis GLDHs (Østergaard et al., 1997; Leferink et al., 2008b).
d-glucono-1,4 and d-glucono-1,5-lactones, the substrates for P. griseoroseum and P. notatum, were not substrates for any other aldonolactone oxidoreductases tested. Conversely, l-GulL, l-GalL or d-AL, the animal, plant or yeast substrates, were not utilized by GUO of P. notatum and P. griseoroseum (Table 2).
l-fucono (6-deoxy-GalL) and d-threono-1,4-lactones served as substrates for S. cerevisiae ALO (Bleeg and Christensen, 1982). l-Idono, l-talono, and d-ALlono-1,4-lactones are some of the stereoisomers tested and they were not substrates for rat and chicken GulLOs (Kiuchi et al., 1982), S. cerevisiae ALO (Nishikimi et al., 1978), G. frondosa, and L. edodes GulLOs (Okamura, 2001). d-Glucurono-1,4-lactone was used with low efficiency by GulLDH of G. oxydans (Sugisawa et al., 1995) and cauliflower GLDH (Mapson and Breslow, 1958). It was not a substrate for rat GulLO (Ashwell et al., 1961; Nakagawa and Asano, 1970), ALOs (Nishikimi et al., 1978; Huh et al., 1994) or M. tuberculosis GulLDH (Wolucka and Communi, 2006).
Isomers among pentono lactones, d-xylono and d-lyxono-1,4-lactones were not substrates for G. frondosa, L. edodes GulLOs (Okamura, 2001) and S. cerevisiae ALO (Bleeg and Christensen, 1982). d-xylono-1,4-lactone was not oxidized by cauliflower GLDH as well (Østergaard et al., 1997). However, d-lyxono and l-xylono-1,4-lactones were substrates for rat GulLO at 59% and 38% of l-GulL, respectively (Ashwell et al., 1961). C. albicans ALO oxidized l-xylono-1,4-lactone at a rate equal to l-GalL (Huh et al., 1994).
4.3 Sugar acids and sugars
The lactone ring was essential for aldonolactone oxidoreductases as no reaction was observed with precursor acids that are in the AsA biosynthetic pathway of animals and plants. d-Glucuronic acid was found not to be a substrate for rat GulLO (Nakagawa and Asano, 1970) and bacterial GulLDHs (Sugisawa et al., 1995; Wolucka and Communi, 2006). Neither d-Glucuronic acid nor d-galacturonic acid were a substrate for plant GLDHs from spinach (Mutsuda et al., 1995), tobacco (Yabuta et al., 2000) and Arabidopsis (Leferink et al., 2008b). Other precursors such as l-Galactonic acid and l-gulonic acid were also not substrates for rat GulLO (Nakagawa and Asano, 1970), S. cerevisiae ALO (Nishikimi et al., 1978) and cauliflower GLDH (Mapson and Breslow, 1958).
GulLDH of G. oxydans, however, can use d-xylose in addition to l-GulL at high efficiency. d-glucose can also be used at about 25% efficiency. d-Mannose was used less efficiently but d-galactose and l-gulose were not substrates for this enzyme (Sugisawa et al., 1995). d-Arabinose and d-xylose did not serve as substrates for M. tuberculosis GulLDH (Wolucka and Communi, 2006).
4.4 Enantiomers
The enantiomers of preferred substrates for animals (l-GulL), plants, protozoa (l-GalL) and yeast (d-AL) enzymes are d-GulL, d-GalL and l-AL, respectively (Figure 1). The enantiomers were never a substrate for any of the aldonolactone oxidoreductases tested.
The enantiomers, d-GalL and d-GulL were not substrates to rat GulLO (Ashwell et al., 1961), ALOs of C. albicans (Huh et al., 1994) and S. cerevisiae (Nishikimi et al., 1978), G. frondosa, L. edodes GulLOs (Okamura, 2001), GLDHs of cauliflower (Østergaard et al., 1997; Mapson and Breslow, 1958), sweet potato (Ôba et al., 1995), spinach (Mutsuda et al., 1995), tobacco (Yabuta et al., 2000), strawberry (Nascimento et al., 2005) or Arabidopsis (Leferink et al., 2008b). P. griseoroseum GUO (Takahashi et al., 1976) was not tested with d-GulL but it cannot oxidize d-GalL. Isoforms of GulLDHs from bacteria and E. gracilis were not tested for d-GalL and d-GulL. l-AL was not a substrate for C. albicans (Huh et al., 1994).
4.5 Configurational specificity
Animal GulLOs have a configurational specificity for the hydroxyl group at C2 and C4 in the substrates and the C5 hydroxyl group being in the “d” arrangement makes it more efficient (Figure 1). Ascomycete enzymes have a configurational specificity for the hydroxyl group at C2, C3, and C4, (Figure 1). Protozoan enzymes are similar to ascomycete enzymes with a substrate specificity for C2 and C3 hydroxyl group (Figure 1). In plants, the specificity is for the hydroxyl group at C2, C3, C4 and C5. Basidiomycete enzymes from G. frondosa and L. edodes and bacterial enzymes are similar to plant enzymes but the difference is the hydroxyl group at C2 is opposite to that of l-GalL.
S. cerevisiae and C. albicans ALOs (Nishikimi et al., 1978; Huh et al., 1994) use substrates as five or six carbon sugar acid lactones with hydroxyl groups in “d” form at C2 and C3. Molecules with hydroxyl groups on the opposite side in C3 to the above can also be substrates. However, GulLO of G. frondosa and L. edodes are specific for both the hydroxyl groups.
4.6 Product formed
GulLOs of animals such as the ones from rat, goat (Nishikimi et al., 1976), chicken (Kiuchi et al., 1982), mouse (Ha et al., 2004) and pig (Hasan et al., 2004) and a basidiomycetes fungus, G. frondosa (Okamura, 2001) produce AsA when l-GulL or l-GalL are used as a substrate (Kiuchi et al., 1982; Okamura, 2001; Figure 1). Similarly, GulLDHs from bacteria (Sugisawa et al., 1995; Wolucka and Communi, 2006) produce AsA as well, when l-GulL is the substrate. The enzyme products of G. oxydans were d-xylosone and d-glucosone when d-xylose and d-glucose were used as substrates, respectively (Sugisawa et al., 1995). Plant GLDHs also produced AsA as product with l-GalL as the starting reactant (Mapson and Breslow, 1958; Leferink et al., 2008b; Figure 1).
The reaction product of ascomycete fungal ALOs from S. cerevisiae and C. albicans was d-erythroascorbic acid when d-AL was used as the substrate (Bleeg and Christensen, 1982; Huh et al., 1994; Figure 1). d-erythroascorbic acid was found to be the naturally occurring AsA analogue in C. albicans (Huh et al., 1994). These enzymes can also make AsA when l-GalL was used as the substrate (Nishikimi et al., 1978; Huh et al., 1994). Interestingly, this property was exploited to make a yeast strain to produce AsA (Hancock et al., 2000). GUO enzymes from P. notatum and P. griseorseum yield d-araboascorbic acid (d-erythorbic acid) as their enzyme reaction products (Takahashi, 1969; Takahashi et al., 1976; Figure 1).
5. Electron acceptor of aldonolactone oxidoreductases
Aldonolactone oxidoreductases are called “oxidases” when the oxidation of an aldonolactone uses molecular oxygen as electron acceptor. Hydrogen peroxide is the byproduct of this reaction. These enzymes are called “dehydrogenases”, if the reaction involves alternative electron acceptors such as cytochrome C, phenazine methosulfate, 2,6-dichloroindophenol or benzoquinone.
Almost all characterized aldonolactone oxidoreductases reported as oxidases have also dehydrogenase activity. Various enzyme assays exist to determine oxidase activity. The activity staining technique in native gel electrophoresis is based on the dehydrogenase property of the enzyme using phenazine methosulfate (Nishikimi et al., 1976). Animal GulLOs, fungal ALOs and GUO have both oxidase and dehydrogenase activities. Animal enzymes from rat, goat (Nishikimi et al., 1976), chicken (Kiuchi et al., 1982) and fungal enzymes: C. albicans (Huh et al., 1994), S. cerevisiae (Nishikimi et al., 1978), P. griseoroseum (Takahshi et al., 1976; Salusjärvi et al., 2004) and G. frondosa (Okamura, 2001) were all reported to use oxygen and alternative electron acceptors, i.e, to have both oxidase and dehydrogenase activities.
Plant GLDHs have been reported to be exclusive dehydrogenases in cauliflower (Mapson and Breslow, 1958; Østergaard et al., 1997), spinach (Mutsuda et al., 1995), tobacco (Yabuta et al., 2000) and Arabidopsis (Leferink et al., 2008b). Recently, an alanine residue was found in AtGLDH to functions as a gatekeeper in not allowing oxygen to reach the reduced flavin (Leferink et al., 2009b; Figure 2). A glycine or proline is present in nearly all oxidases of the VAO family near the C4a locus of the isoalloxazine ring of flavin, whereas other residues are present in dehydrogenases (Leferink et al., 2009b). Exceptions to this rule are also found in the analysis of different VAO family members. For example, alditol oxidase from Streptomyces coelicolor has an alanine and it functions as an oxidase while eugenol hydroxylase from Pseudomonas sp. strain HR199 functions as a dehydrogenase although it has a proline residue (Leferink et al., 2009b). Among aldonolactone oxidoreductases, all members obey this rule except M. tuberculosis GulLDH (Wolucka and Communi, 2006) and AtGulLO5 (At2g46740; Aboobucker et al., submitted; Figure 2). Nevertheless, changing the Ala-113 to Gly in AtGLDH gained oxidase activity without any loss in the dehydrogenase activity (Leferink et al., 2009b; amino acid numbered based on mature peptide as in Leferink et al., 2008b). AtGLDH modified to use oxygen is the first enzyme to use both oxygen and cytochrome C as electron acceptors among the plant GLDHs.
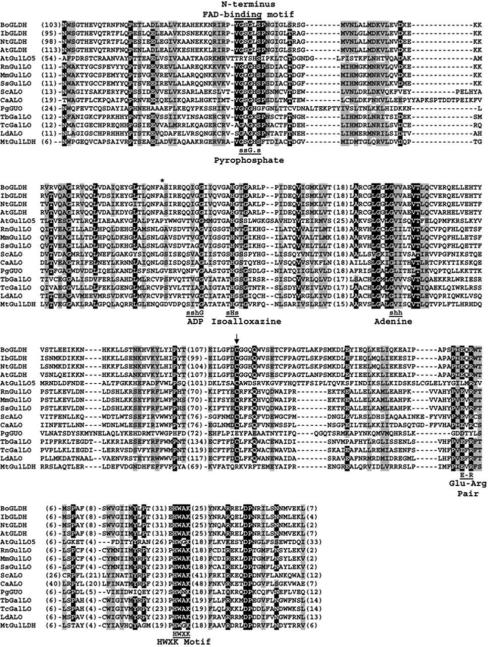
Figure 2. Multiple sequence alignment of all characterized aldonolactone oxidoreductases including Arabidopsis GulLO (AtGulLO5 or At2g46740).
Conserved residues interacting with the parts of FAD in the alignment are shown as in Fraaije et al (1998). (G, glycine; H, histidine; s, small: G, A, S, T; h, hydrophobic: I, L, V; -, any residue). Numbers in parenthesis indicate the residues present in gaps and termini. The residue indicating oxygen reactivity is marked by asterisk (*; Leferink et al. 2009b). Arrow indicates the cysteine residue known to be involved with thiol modifying agents in AtGLDH (Leferink et al. 2009d). The Glu-Arg pair identified in the catalytic site of AtGLDH (Leferink et al. 2009c) is also highlighted. The sequences used are: BoGLDH (cauliflower; Swiss-Prot: O47881), IbGLDH (sweet potato; GenBank: BAA34995), NtGLDH (tobacco; GenBank: BAB13368), AtGLDH (Arabidopsis; Swiss-Prot: Q9SU56), AtGulLO5 (Arabidopsis; GenBank: AEC10747), RnGulLO (rat; Swiss-Prot: P10867), MmGulLO (mouse; GenBank: AAR15891), SsGulLO (pig; GenBank: AAN63634), ScALO (S. cerevisiae; Swiss-Prot: P54783), CaALO (C. albicans; GenBank: AAC98913), PgGUO (P. griseoroseum; Swiss-Prot: Q671X8), TbGalLO (T. brucei; Swiss-Prot: Q57ZU1), TcGalLO (T. cruzi; Swiss-Prot: Q4DPZ5), LdALO (L. donovani; GenBank: ACV04074), MtGulLDH (M. tuberculosis; Swiss-Prot: O06804).
Enzymes that function exclusively as dehydrogenases have also been reported in bacteria such as G. oxydans (Sugisawa et al., 1995) and M. tuberculosis (Wolucka and Communi, 2006). The amino acid sequence for M. tuberculosis GulLDH also has a glycine at this position (Leferink et al., 2009b), but it was reported not to use molecular oxygen (Wolucka and Communi, 2006). It is worth noting that the recombinant M. tuberculosis GulLDH did not have a flavin and this could be the reason for its non-reactivity to molecular oxygen. G. oxydans GulLDH is an interesting protein as it has a covalently bound FAD and functions exclusively as a dehydrogenase (Sugisawa et al., 1995). However, its amino acid sequence is unknown to date.
Protozoan enzymes from T. brucei, T. cruzi and L. donovani were reported to have dehydrogenase activity but their oxidase activity was not tested, even though they were named “oxidases” (Biyani and Madhubala, 2011; Logan et al., 2007; Wilkinson et al., 2005). Recently, a GalLO from T. cruzi was tested for its oxidase activity and found to operate as an oxidase as well (Kudryashova et al., 2011). The authors reported the absence of an alanine residue in T. cruzi, similar to AtGLDH (Figure 2; Kurdyashova et al., 2011). Similarly, this alanine residue is absent in T. brucei GalLO and L. donovani ALO and they both may function as oxidases as well (Figure 2).
A single protein component is responsible for both the activities in all cases examined. The presence of two components for oxidase and dehydrogenase activity in rat GulLO and their separation was reported (Isherwood et al., 1960; Bublitz, 1961). Later studies of the enzyme found the presence of both the activities, but were unable to separate them (Eliceiri et al., 1969; Nakagawa and Asano, 1970). However, the presence of two active sites in one component for both oxidase and dehydrogenase activities was postulated (Nakagawa and Asano, 1970). Nevertheless, the above studies were based on partial purification of the enzyme. It was later confirmed in a homogeneous preparation that only one component was responsible for both oxidase and dehydrogenase activities in rat GulLO (Nishikimi et al., 1976).
One interesting observation is that rat GulLO (Eliceiri et al., 1969), GUO from P. cyaneo-fulvum (Takahashi et al., 1976) and GulLO from G. frondosa (Okamura, 2001) cannot take cytochrome C but use molecular oxygen and phenazine methosulfate as electron acceptors. The use of cytochrome C as an electron acceptor was not tested in other animal or yeast enzymes. GLDH from plants, on the other hand, can use cytochrome C and phenazine methosulfate but not molecular oxygen as acceptors. Enzyme catalysis occurs via flavin when cytochrome and oxygen are acceptors as demonstrated by inhibitor studies, but the inhibitors do not interfere in the reaction via phenazine (Mapson and Breslow, 1958). Cytochrome C was used as an electron acceptor by plant enzymes but not animal isoforms. The residue in charge of allowing the molecular oxygen to enter the active site is known (Leferink et al., 2009b). However, the interaction between cytochrome C and AtGLDH is dynamic, involving low affinity and electrostatic protein-protein interaction rather than a single residue (Hervás et al., 2013).
The GulLDH from E. gracilis uses only phenazine methosulfate among other alternative electron acceptors tested including cytochrome C (Shigeoka et al., 1979) differing from animal, yeast and plant isoenzymes. The GulLDH did not have oxidase activity either, similar to plants and bacterial enzymes.
6. Active site, inhibitors, and functional residues of aldonolactone oxidoreductases
A flavin group or cofactor is found to be involved in the enzyme catalysis of many characterized aldonolactone oxidoreductases. Its involvement is observed by the disappearance of 450 nm peak when the substrate is added to the enzyme solution such as in GulLOs of rat (Nakagawa et al., 1975), chicken (Kiuchi et al., 1982) and G. frondosa (Okamura, 2001) as well as im the GUO of P. griseoroseum (Takahshi et al., 1976), S. cerevisiae ALO (Nishikimi et al., 1980; Bleeg and Christensen, 1982), T. brucei GalLO (Wilkinson et al., 2005) and in the GLDHs ofcauliflower (Mapson and Breslow, 1958) and Arabidopsis (Leferink et al., 2008b). Flavin involved reactions proceed as two half reactions. In the first reductive half, flavin is reduced by the substrate and the latter is oxidized. The flavin is reoxidized, in the second half, by electron acceptors such as oxygen (oxidase) or alternative electron acceptors (dehydrogenases). Hydrogen peroxide is produced as a result of the transfer of electrons from flavin to oxygen. Both half reactions can be studied separately due to characteristic spectral properties exhibited during oxidation and reduction cycles of the flavin group (Leferink, 2009a). Flavin involvement in the catalysis of l-GalL by AtGLDH was studied in detail. During the first reductive half, the flavin was reduced by l-GalL to a two electron reduced state. The second oxidative half proceeds in two steps as cytochrome C is a one-electron acceptor. The appearance of a semiquinone radical intermediate was monitored during the oxidative half (Leferink et al., 2008b).
Inhibitor studies have been conducted on many aldonolactone oxidoreductases to determine the functional groups and they are summarized in Table 3. The involvement of flavin through cytochrome C had also been determined using flavoprotein inhibitors such as atebrin, riboflavin, and acriflavine in cauliflower, sweet potato, and tobacco GLDHs, E. gracilis GulLDH, and G. frondosa GulLO (Table 3). In another observation, riboflavin did not inhibit any activity of GLDH in cauliflower (Østergaard et al., 1997). Interestingly, the reaction proceeding through phenazine methosulfate of cauliflower GLDH was not inhibited by flavoprotein inhibitors (Mapson and Breslow, 1958). Rat GulLO was not inhibited by flavoprotein inhibitors and notably the authors were unable to find flavin as a prosthetic group in this particular preparation (Nakagwa and Asano, 1970). GulLDH of G. oxydans (Sugisawa et al., 1995) and cauliflower GLDH (Mapson and Breslow, 1958) were reported to have a heme component in addition to flavin.
Table 3.
A comparison of the effects of different classes of inhibitors on aldonolactone oxidoreductases
| Source | Sulfhydryl group inhibitors | Metal ions | Metal Chelating agents | Substrate analogs | Flavin inhibitors | Reference | ||
|---|---|---|---|---|---|---|---|---|
| Animals | Rat | Sufide, sulfite, PCMS |
1-Hg2+ 2-Cd2+ |
Nil | ND | Nil | Nakagawa and Asano, 1970 | |
| Fungi | S. cerevisiae | PCMS, iodoacetamide, NEM, sulfite, sulfide | 1 -Hg2+, Cd2+ | Nil | l-GulL, d-GalL | ND | Bleeg and Christensen, 1982 | |
| S. cerevisiae | PCMB | ND | Nil | Nil | ND | Nishikimi et al., 1978 | ||
| C. albicans | PCMB, NEM, iodoacetic acid, iodoacetamide |
1-Cd2+, Hg2+, Mn2+, Zn2+ 2-Cu2+, Mg2+ |
Nil | d-Glucono-1,5-lactone, l-AL, d-GalL, d-GulL | ND | Huh et al., 1994 | ||
| P. griseoroseum | Nil |
1-Hg2+, Ag2+, Fe3+, Fe2+, Cu2+ 2- Co2+, Mn2+, Zn2+, Cd2+, Mg2+, Ca2+, As2+ |
ND | ND | ND | Takahashi et al., 1976 | ||
| P. notatum | PCMB | ND | Nil | ND | ND | Takahashi, 1969 | ||
| G. frondosa | Nil | ND | ND | ND | Acriflavine | Okamura, 2001 | ||
| Bacteria | M. tuberculosis | NEM |
1-Cu2+, Zn2+ 2-Mg2+, Ca2+ 3-Mn2+ |
EDTA, KCN | ND | Nil | Wolucka and Communi, 2006 | |
| G. oxydans | Nil |
1 -Cu2+, Mn2+ 3 -Fe6+, Mo6+, Ti+, Zn2+, Ni2+ |
Nil | ND | ND | Sugisawa et al., 1995 | ||
| Protozoa | T. brucei | ND | 3 -Cu2+ | ND | Substrate >4mM | ND | Wilkinson et al., 2005 | |
| T. cruzi | PCMB, NEM | 1 -Hg2+, Zn2+ | ND | ND | ND | Logan et al., 2007 | ||
| L. donovani | NEM | 1 -Zn2+, Hg2+ | ND | ND | ND | Biyani and Madhubala, 2011 | ||
| Algae | E. gracilis | PCMB, NEM | 1 -Cu2+, Zn2+, Fe2+ | Nil | ND | Riboflavin | Shigeoka et al., 1979 | |
| Plants | Cauliflower | PCMB, o-Iodosobenzoate |
1-Cu2+ 2-Mn2+, Zn2+, Fe3+, MoO42− |
o-phenanthroline | d-GulL, d-GalL | Atebrin, Riboflavin | Mapson and Breslow, 1958 | |
| Cauliflower | PHMB, NEM, monoiodoacetate | Nil | Nil | l-GalL – 32.6 mM | Nil | Østergaard et al., 1997 | ||
| Sweet potato | PCMB, NEM* | 1- Cu2+, Zn2+* | ND | l-GalL ≥ 4 mM | Acriflavine | Ôba et al.,1995; * Imai et al.,1998 | ||
| Spinach | PCMB, NEM | ND | ND | l-GalL > 1 mM | ND | Mutsuda et al., 1995 | ||
| Tobacco | PCMB, NEM | Cu2+, Zn2+ | ND | ND | Acriflavine | Yabuta et al., 2000 | ||
| Strawberry | ND | ND | ND | l-GalL > 10 mM | ND | Nascimento et al., 2005 | ||
| Arabidopsis | DTNB, NEM | ND | ND | l-GalL > 2 mM* | ND | * Leferink et al., 2008b; Leferink, 2009a | ||
1: inhibitory, 2: not inhibitory, 3: slightly inhibitory, Nil: no inhibition found, DTNB: 5,5′ -dithiobis(2-nitrobenzoic acid), NEM: N-ethylmaleimide, PCMB: p-chloromercuribenzoate, PCMS: p-chloromercurisulfate, PHMB: p-hydroxymercuribenzoate. ND: not determined
Another important feature of aldonolactone oxidoreductases is the participation of a thiol group. Enzyme activity of all members of this family was inhibited by sulfhydryl group modifiers with two exceptions (Table 3). P. griseoroseum GUO is one of the exceptions whose sequence has been deduced (Salusjärvi et al., 2004). Interestingly, a Cys residue, conserved among other isoenzymes, is not present in P. griseoroseum GUO, which was the lead to identify the functional Cys residue in AtGLDH (Figure 2; Leferink et al., 2009d) as will be discussed later. p-Chloromercuribenzoate (PCMB) or p-chloromercurisulfate (PCMS) inhibition was reversible with glutathione, 2-mercaptoethanol or potassium cyanide. Reversal of enzyme activity with glutathione was observed in rat GulLO (Nakagawa and Asano, 1970), P. notatum GUO (Takahashi, 1969) and cauliflower GLDH (Mapson and Breslow, 1958). 2-Mercaptoethanol reversal was observed in ALOs of S. cerevisiae (Nishikimi et al., 1978), C. albicans (Huh et al., 1994) and sweet potato GLDH (Ôba et al., 1995). Potassium cyanide reversed inhibition was seen in E. gracilis GulLDH (Shigeoka et al., 1979) and ALOs of C. albicans (Huh et al., 1994) and S. cerevisiae (Bleeg and Christensen, 1982). N-Ethylmaleimide (NEM) inhibition, on the other hand, was reversible with glutathione in sweet potato GLDH (Imai et al., 1998) but irreversible in C. albicans ALO (Huh et al., 1994) and tobacco GLDH (Yabuta et al., 2000). Prior incubation with glutathione decreased inhibition by Hg2+ or Ag2+ in P. griseoroseum GUO (Takahashi et al., 1976).
The inhibition of rat GulLO activity by PCMS and Hg2+ was through inhibition of flavin reduction as observed by the disappearance of the peak at 450 nm corresponding to flavin. Sulfite, however, did not inhibit flavin reduction although it inhibited the oxidase activity. Therefore, the authors postulated the presence of two active sites for oxidase and dehydrogenase activities (Nakagawa and Asano, 1970). This idea may be correct as indeed, an alanine residue (Ala-113) was found to prevent oxygen from entering the active site and thereby lacking oxidase activity in AtGLDH (Leferink et al., 2009b). When replaced with glycine, AtGLDH was able to utilize oxygen without any reduction in the dehydrogenase activity (Leferink et al., 2009b) supporting the previous finding. The same was observed in GalLO of T. cruzi, which had both oxidase and dehydrogenase activities and a gatekeeper alanine residue is absent (Kudryashova et al., 2011).
Sulfhydryl group inhibition was studied in detail in S. cerevisiae ALO (Noguchi et al., 1981) and in AtGLDH (Leferink et al., 2009d). Both the studies found that the sulfhydrl group is not playing a role in the catalysis itself but it was involved in substrate binding. The same was observed in cauliflower GLDH (Mapson and Breslow, 1958). Also, only one sulfhydrl group was involved in this function in both the studies and it was identified as Cys-340 in AtGLDH (Leferink et al., 2009d). This residue is conserved in all aldonolactone oxidoreductases except P. griseoroseum GUO and M. tuberculosis GulLDH (Figure 2). In AtGLDH, a Cys-340 was found to be buried away from the flavin and not directly involved in the catalysis as the flavin spectrum remain unchanged (Leferink et al., 2009d). Noguchi et al. (1981) suggested the sulfhydryl group may be situated near flavin and that the sulfhydryl group is provoked during catalytic process to be more reactive with the sulfhydryl modifying reagents. Replacing Cys-340 with Ser or Ala strongly weakened substrate binding in AtGLDH, while the catalytic activity was retained. In support of this, pre-incubation with the substrate slowed down inhibition by thiol modifying agents, as was observed in AtGLDH (Leferink et al., 2009d), S. cerevisiae ALO (Noguchi et al., 1981) and also in cauliflower GLDH (Mapson and Breslow, 1958). The sulfhydrl group is required for phenazine activity in addition to substrate binding in cauliflower GLDH (Mapson and Breslow, 1958).
In addition to the two active sites for oxidase and dehydrogenase activities, reactions proceeding with phenazine methosulfate may have a different active site and flavin is probably not involved. In cauliflower GLDH, cytochrome C activity was inhibited with flavoprotein inhibitors such as atebrin or riboflavin while phenazine activity was not inhibited (Mapson and Breslow, 1958). Similarly, in M. tuberculosis, GulLDH activity was higher with phenazine methosulfate than with cytochrome C and it did not have a flavin prosthetic group (Wolucka and Communi, 2006). A contrasting observation, in GulLDH of E. gracilis, is that only phenazine was used as an electron acceptor but not cytochrome C and flavin participation was also indicated by riboflavin inhibition (Shigeoka et al., 1979).
Metal ions inhibited most of the characterized aldonolactone oxidoreductases (Table 3). Hg2+, Zn2+ and Cu2+ were found as common inhibitors and they were implicated to inhibit through sulfhydryl groups (Mapson and Breslow, 1958; Biyani and Madhubala, 2011). None of these enzymes are considered metalloproteins as chelating agents had no effect on their activity except M. tuberculosis GulLDH and cauliflower GLDH (Table 3). KCN inhibition suggests the presence of iron-like groups in the GulLDH enzyme of M. tuberculosis (Wolucka and Communi, 2006). The involvement of an iron-sulfur cluster in catalysis was also suggested for S. cerevisiae ALO (Bleeg and Christensen, 1982).
Substrate analogs were also inhibitory in some cases (Table 3) and they were mainly the “d” forms of the aldonolactone substrates. Preferred substrates were also found to be inhibitory for T. brucei GalLO and many plant GLDHs at above certain concentrations (Table 3). Quinoprotein inhibitors were tested on GulLDH of G. oxydans and found not to be inhibitory (Sugisawa et al., 1995). Anions were inhibitory to cytochrome activity of cauliflower GLDH but not to phenazine activity (Mapson and Breslow, 1958). Lycorine is a specific inhibitor for GLDH (Imai et al., 1998), but no inhibition was found with AtGLDH (Leferink et al., 2008b) or cauliflower GLDH (Østergaard et al., 1997).
Amino acid residues involved in flavin binding have also been identified. A two-domain folding topology is common in VAO family members and they are FAD-binding domain (PFAM Id: 01565) and a catalytic domain (PFAM Id: 04030) found at the interface of the two domains is the active site. The catalytic domain in the C terminus is where the substrate binds (Fraaije et al., 1998; Leferink et al., 2008a). Although the sequence similarity among aldonolactone oxidoreductases from different species is low, the presence of these two characteristic features is shared. His-61 of VAO was shown to promote covalent flavin binding as no covalent FAD was found in His-61Thr mutant (Fraaije et al., 2000). The flavin is covalently linked to His-104 in the N-terminus in berberine bridge enzyme (Kutchan and Dittrich, 1995) and 6-hydroxy-d-nicotine oxidase (Brandsch et al., 1987), two members of VAO family. The N terminus histidine of FAD-binding domain is found in a stretch of VGSGHS sequences in aldonolactone oxidoreductases (Imai et al., 1998; Wolucka and Communi, 2006). All covalently bound FAD containing isoenzymes have a histidine, whereas a leucine or lysine is present in enzymes with non-covalent FAD (Figure 2). An exception to this is GulLDH of M. tuberculosis that has a histidine residue but it was found to be without flavin (Table 1; Wolucka and Communi, 2006). It was thought that FAD is covalently linked to this histidine residue in VAO (Fraaije et al., 1998). However, it is not known if the flavin is linked to the histidine in N or C termini in aldonolactone oxidoreductases.
Attempts were made to generate enzymes with covalently bound FAD by replacing leucine with histidine in AtGLDH (Leferink et al., 2008) and lysine with histidine in T. cruzi GalLO (Logan et al., 2007). No covalent linkage was observed, rather FAD was loosely bound to AtGLDH (Leferink et al., 2008b) and flavin binding was dramatically reduced in T. cruzi GalLO (Logan et al., 2007). Leferink et al. (2008b) explained that the His-422 is activated by His-61 in VAO for covalent flavinylation. The presence of similar His-61 is not known in AtGLDH due to the lack of structural information for aldonolactone oxidoreductases. However, it is not known if flavin binding in these enzymes is in the C terminus, similar to VAO. Attempts to solve the crystal structure of AtGLDH failed as all crystals for AtGLDH, including variants, diffracted to 3.5-4 Å and it was not enough to solve the structure (Leferink, 2009a).
The His-422 residue in the C terminus of VAOs is where the FAD is covalently bound (Fraaije et al., 1999). This histidine is conserved among all aldonolactone oxidoreductases in a HWXK motif present in the catalytic domain (Figure 2; Logan et al., 2007). The functional role of the HWXK motif was studied in T. cruzi GalLO by site-directed mutagenesis (Logan et al., 2007). His-447 and Trp-448 were individually mutated to glycine and absence of flavin binding and enzyme activity were observed. When, Lys-450 was mutated to Gly, the enzyme retained half of the flavin but not the activity (Logan et al., 2007). The authors proposed that His-447 and Trp-448 are involved directly or indirectly in interactions with FMN while Lys-450 has a role in activity independent of flavin binding. Kudryashova et al. (2011) reported after examining T. cruzi GalLO with alditol oxidase (AldO) that His-447 and Lys-450 are located close enough to the flavin for direct interaction. They also proposed that the flavin moiety could be binding to His-447. Also, the lysine residue is part of the active site of AldO and Lys-450 in T. cruzi may have a similarly important role. Kudryashova et al. (2011) suggested that loss of flavin binding and activity in the Trp448Gly mutant could be due to incorrect protein folding. The X in this motif is an alanine in all of the enzymes except AtGulLO5, M. tuberculosis GulLDH and P. griseoroseum GUO (Figure 2). It is interesting to note that l-GalL is not a substrate for AtGulLO5, M. tuberculosis GulLDH, and the resting cells of P. notatum (Aboobucker et al., submitted; Takahashi et al., 1976; Wolucka and Communi, 2006). The substrate, l-GalL was not tested with P. griseoroseum GUO.
Comparison of the structure of VAO with several of its family members including rat GulLO, S. cerevisiae ALO and cauliflower GLDH enabled the identification of residues interacting with parts of the FAD molecule (Fraaije et al., 1998). These residues are conserved in all aldonolactone oxidoreductases whose sequences are known (Figure 2). AtGulLO5 may not have the complete stretch of sequences that interact with pyrophosphate but part of it is still present (Figure 2). A similar analysis was done for the sequence of T. cruzi and T. brucei GalLOs and FAD was predicted to be the cofactor in T. cruzi (Kudryashova et al., 2011). Experimental evidence also proved that the cofactor was FAD (Kudryashova et al., 2011), which was previously reported as FMN (Logan et al., 2007).
The functional role of the Glu-Arg pair in the catalytic domain, conserved among aldonolactone oxidoreductases, was studied in AtGLDH by site-directed mutagenesis (Leferink et al., 2009c). It was demonstrated that this pair is essential for optimal catalysis. Glu-386 is involved in substrate binding and determining substrate specificity. Glu386Asp variant lost its specificity for l-GalL but used l-GulL with more efficiency. The Arg-388 residue is essential for the stabilization of negative charge resulting from flavin reduction. An Arg388Lys variant showed some activity while the Arg388Ala variant was essentially inactive (Leferink et al., 2009c). This pair is also widely conserved among all aldonolactone oxidoreductases with the exception of AtGulLO5 and P. griseoroseum GUO (Figure 2).
7. Flavin binding in recombinantly expressed aldonolactone oxidoreductases
There is a discrepancy about covalent flavin attachment in recombinantly produced GulLO isoenzymes in E. coli. Three enzymes were expressed in E. coli, viz. ALO from S. cerevisiae (Lee et al., 1999), GUO from P. griserosoeum (Salusjärvi, 2006) and GulLDH from M. tuberculosis (Wolucka and Communi, 2006). Native ALO (Kenney et al., 1979) and GUO (Harada et al., 1979) were experimentally found to have a covalently bound FAD as well as a histidine residue in the FAD-binding domain in N terminus. GulLDH from M. tuberculosis was predicted to have a covalently bound FAD, as a histidine is found in the N terminal FAD-binding domain. The recombinant enzymes from S. cerevisiae (ALO) and M. tuberculosis (GulLDH) were active but did not have a flavin prosthetic group. The GUO from P. griseoroseum was not active and Salusjärvi (2006) suggested it could be due to the absence of covalent flavin attachment. However, GUO was found to have covalently linked FAD when expressed in Pichia pastoris (Salusjärvi et al., 2004). Rat GulLO and AtGulLO2, 3, 4 and 5 were not able to be successfully expressed in E. coli (Maruta et al., 2010).
The rat GulLO has been expressed in monkey cells (Yagi et al., 1991) and BmN4 silkworm cells (Nishikimi et al., 1994) and found to be covalently linked to FAD. A similar observation was made with mouse GulLO expressed in guinea pig cells (Ha et al., 2004). Rat GulLO was expressed as both holo and apo forms in a riboflavin rich and free media, respectively. The apo form only made non-covalent interaction when FAD was added to it (Nishikimi et al., 1994), but the activity was as in the holo form with covalent FAD. This finding is in contrast to 6-hydroxy-d-nicotine-oxidase (Brandsch and Bichler, 1991), a VAO family member, wherein the apo form made covalent FAD, when exogenous FAD was added.
On the other hand, protozoan GulLO isoenzymes from T. brucei (Wilkinson et al., 2005) and T. cruzi (Logan et al., 2007; Kudryashova et al., 2011) as well as AtGLDH from Arabidopsis (Leferink et al., 2008b) and GLDH from tobacco (Yabuta et al., 2000) were expressed in E. coli and found to have a non-covalent flavin as predicted from their amino acid sequences. It is interesting to note that T. cruzi GalLO was expressed in the inclusion bodies of E. coli and successfully refolded in a reverse micellar system to a native-like structure in the presence of FAD but not with FMN (Kudryashova et al., 2011). L. donovani GalLO (Biyani and Madhubala, 2011), predicted to have non-covalently bound FAD, was found to be in the apo form when expressed in E. coli as no activity was detected without FAD addition.
Expression of aldonolactone oxidoreductases in E. coli did not produce an enzyme with covalently linked FAD. This could be because the recombinant proteins may not have correctly folded, since several VAO members were successfully expressed as holo proteins in E. coli, including VAO (Benen et al., 1998), such as eugenol oxidase (Jin et al., 2007), alditol oxidase (Heuts et al., 2007a), and chitooligosaccharide oxidase (Heuts et al., 2007b). Incorrect folding of recombinant M. tuberculosis GulLDH, expressed in E. coli may have been the reason for producing a non flavoprotein.
8. Subcellular localization of aldonolactone oxidoreductases
Aldonolactone oxidoreductases from animals such as rat (Eliceiri et al., 1969), goat (Nishikimi et al., 1976), mouse (Braun et al., 1999), chicken (Kiuchi et al., 1982), and pig (Hasan et al., 2004) were localized to microsomes and they all are integral membrane proteins. Several possible membrane associating regions were identified in rat GulLO amino acid sequence (Koshizaka et al., 1988). The active site of rat GulLO was found to be intraluminal and the enzyme releases its products towards the lumen of endoplasmic reticulum (Puskás et al., 1998).
Plant GLDHs are localized in the mitochondria in cauliflower (Mapson and Breslow, 1958; Østergaard et al., 1997), spinach leaves (Mutsuda et al., 1995), tobacco (Yabuta et al., 2000), white potato tubers and leaves (Ôba et al., 1994; Bartoli et al., 2000). About one-fifth of GLDH activity was also observed in microsomes of white potato tubers (Ôba et al., 1994) and leaves (Bartoli et al., 2000). A mitochondrial transit peptide is found in all plant GLDHs whose cDNA sequences have been identified in cauliflower (Østergaard et al., 1997), tobacco (Yabuta et al., 2000), Arabidopsis (Tamaoki et al., 2003), sweet potato (Imai et al., 1998), strawberry (Nascimento et al., 2005), muskmelon (Pateraki et al., 2004), wheat (Bartoli et al., 2005), Rosa roxburghii (An et al., 2007), tomato (Alhagdow et al., 2007) and rice (Yu et al., 2010).
Detailed localization studies of GLDH in white potato (Bartoli et al., 2000) and kidney beans (Siendones et al., 1999) found that GLDH is located at the inner mitochondrial membrane. Bartoli et al. (2000) proposed that the catalytic site is on the outside of the inner mitochondrial membrane by predicting the location of the FAD-binding site, which was thought to be Leu-137. It is worth noting that the Leu-137 was only thought to be a putative FAD binding site (Imai et al., 1998). Recent identification of the HWXK motif towards the C terminus of T. cruzi GalLO suggests that the FAD binding site could be the histidine in this motif (Logan et al., 2007; Kudryashova et al., 2011). Nevertheless, the prediction by Bartoli et al. (2000) could still be correct as the HWXK motif is also present on the outside of the inner mitochondrial membrane in the GLDH from white potato leaves.
S. cerevisiae (Nishikimi et al., 1978; Bleeg and Christensen, 1982) and C. albicans (Huh et al., 1994) ALOs are localized in the mitochondria, where they are predicted to be integral membrane proteins (Huh et al., 1998; Huh et al., 2001). The subcellular localization of G. frondosa GulLO was not determined but it may be a membrane protein (Okamura, 2001). Interestingly, P. griseoroseum GUO was isolated as a soluble enzyme (Takahashi et al., 1976) and the native signal peptide was sufficient to be secreted when expressed in P. pastoris (Salusjärvi et al., 2004). The G. oxydans GulLDH was also isolated from the soluble fraction (Sugisawa et al., 1995) and that of E. gracilis was localized to the cytosol by sucrose density gradient centrifugation, although about 12% of the activity was also found in the mitochondria (Shigeoka et al., 1979). GulLDH of M. tuberculosis (Mawuenyega et al., 2005) and Arabidopsis AtGulLO5 (Aboobucker et al., submitted), however, are localized to the cell wall.
Trypanasomal enzymes are localized in glycosome and they have a characteristic SHL motif at the C terminus of their amino acid sequence which may be targeted to the glycosomes (Wilkinson et al., 2005; Logan et al., 2007). L. donovani ALO is also localized to the glycosomes as well as to the cytosol. The amino acid sequence lacks the SHL motif (Biyani and Madhubala, 2011).
9. Plants have GulLOs, in addition to GLDHs
It is absolutely clear from the characterization of GLDHs of many plant species that they are highly specific to l-GalL and cannot use l-GulL as a substrate, except at a very low efficiency in some cases (Table 2). In a recent article, Wheeler et al. (2015) conclude that plants have been replaced with the GLDH enzyme instead of GulLO. However, the following evidences demonstrate the existence of a GulLO enzyme in plants in addition to GLDH.
First, multiple precursor feeding studies have reported the use of l-GulL as a substrate since 1954 (Isherwood et al., 1954) including our observation in the detached leaves of Nicotiana benthamiana (wood tobacco; Lisko et al., 2014). The increase in AsA when fed with l-GulL is different than with l-GalL or l-Gal feeding. The substrates l-GalL or l-Gal are readily converted to AsA while l-GulL is converted less efficiently. This activity of converting l-GulL to AsA can be attributed to the existence of a GulLO enzyme since GLDH is strictly specific to l-GalL (Table 2). The possibilities of differential substrate uptake (Baig et al., 1970; Davey et al., 1999) and the stimulation of AsA synthesis by a general increase in carbohydrate pool (Loewus, 1963; Baig et al., 1970) have been ruled out. Iinterestingly, when Wheeler et al. (2015) fed l-Gal or l-GulL to the rhodophyte algae Galdieria sulphuraria, the phenomenon of AsA increase observed was just as it has been observed in many plant species as mentioned above (Isherwood et al., 1954; Lisko et al., 2014). Rightfully so, Wheeler et al. (2015) concluded that G. sulphuraria has a GulLO enzyme instead of a GLDH. For this same reason, we argue the idea of a GulLO enzyme in plants in addition to GLDH based on the precursor feeding experiments with l-GulL. Second, the enzyme activity responsible for this catalysis has been observed in hypocotyl homogenates of kidney beans (Siendones et al., 1999), cytosolic and mitochondrial fractions of Arabidopsis cell culture (Davey et al., 1999) and potato tubers (Ôba et al., 1994; Wolucka and van Montagu, 2003). Third, rat GulLO, the enzyme known to use both l-GalL and l-GulL, has been successfully expressed in multiple plant species including lettuce, tobacco (Jain and Nessler, 2000), potato (Hemavathi et al., 2010), Arabidopsis (Radzio et al., 2003), and tomato (Lim et al., 2012) resulting in elevated AsA levels. It may be possible that these observed increases were due to rat GulLO using l-GalL, as the enzyme can also use this as a substrate (Table 2). However, it might not have been the case as the level of l-GalL is reduced in the vitamin C deficient (vtc) mutants and expression of rat GulLO rescued AsA level in these mutants (Radzio et al., 2003; Gallie, 2014). Overexpression of GLDH has failed to increase AsA in tobacco plants (Imai et al., 2009). Bartoli et al. (2005) did not find an increase in AsA level with increase in GLDH activity in wild type wheat. This further strengthens the idea for the requirement of an enzyme to use l-GulL as a substrate. Multiple authors (Davey et al., 1999; Wolucka and van Montagu, 2003; Leferink et al., 2008b) have insisted on the existence of a GulLO isoenzyme in plants. Baig et al. (1970) went on to predict that this isoenzyme will be specific to l-GulL. Seven putative GulLOs were predicted in Arabidopsis based on the homology to rat GulLO (Wolucka and van Montagu, 2003) and two ESTs were identified for GulLOs in kiwi (Crowhurst et al., 2008). Therefore, we and others strongly support the idea of a GulLO enzyme using l-GulL as a substrate in plants, distinct from GLDH.
Recently, Maruta et al. (2010) expressed four of the seven putative GulLOs from Arabidopsis in BY2 cell suspension cultures. They found that transgenic cell cultures expressing three of the GulLOs had increased AsA content when l-GulL was fed for 48 h. This marks the first observation of plant genes encoding proteins with GulLO activity. While enzyme activity was present, no detailed characterization was reported. Recently, we characterized, for the first time, a recombinant AtGulLO5 (TAIR: At2g46740) expressed in a Nicotiana benthamiana based transient expression system (Aboobucker et al. submitted). AtGulLO5 has an absolute specificity for l-GulL and it cannot use l-GalL as predicted by Baig et al. (1970). The enzyme did not have oxidase activity and it behaved more like bacterial GulLDHs in terms of its substrate specificity and the use of electron acceptor. AtGulLO5 is localized in the cell wall (Boudart et al., 2005; Aboobucker and Lorence, unpublished observation). This provides further evidence for the existence of a GulLO in addition to GLDH in plants thereby to the existence of alternate routes for AsA biosynthesis in plants.
10. Conclusions and perspectives
Animal and fungal GulLO and ALO have covalently bound FAD while plants and protozoans have non-covalently bound FAD. There is a strong correlation between the presence of histidine and lysine/leucine in amino acid sequences in the N terminus of FAD-binding domain and covalent and non-covalent FAD interactions, respectively. A second histidine residue, involved in FAD binding in VAO, is also found in a widely conserved HWXK motif among all aldonolactone oxidoreductases. Flavin is assumed to be bound to aldonolactone oxidoreductases in the N terminus as in two of VAO family members, berberine bridge enzyme and 6-hydroxy-d-nicotine oxidase. Rather, the flavin group may be binding to the HWXK motif in the C terminus as in VAO. Resolving the crystal structure of aldonolactone oxidoreductases will provide a clear answer.
All animal and yeast aldonolactone oxidoreductases are oxidases, but in addition they also possess dehydrogenase-like activity utilizing phenazine methosulfate but not cytochrome C as electron acceptor. Protozoan enzymes are both oxidases and dehydrogenases as well and they use both cytochrome C and oxygen. Plant GLDHs are exclusive dehydrogenases using cytochrome C and phenazine methosulfate. An amino acid responsible for oxygen reactivity is found in AtGLDH which is present in all plant GLDHs. Bacterial enzymes are exclusive dehydrogenases as well. An isoenzyme from algae is unique to use only phenazine methosulfate and neither cytochrome C nor oxygen as an electron acceptor in vitro.
The flavin moiety is involved in enzyme catalysis in many aldonolactone oxidoreductases. The thiol group is essential for these enzymes as all isoenzymes except GUO of P. griseoroseum were inhibited by sulfhydryl groups. The thiol group participates in substrate binding but not in the catalysis itself. Only one sulfhydryl group is involved in this function and it was identified as Cys-340 in AtGLDH. M. tuberculosis GulLDH was the only enzyme identified as a metalloenzyme among all. Site-directed mutagenesis studies assisted to understand the roles of some amino acid residues in these enzymes. Although, multiple studies have been done to understand the roles of residues involved in enzyme function in these families, molecular determinants for substrate specificity are yet to be found.
Animal GulLOs and an algal GulLDH use l-GulL as well as l-GalL as substrates. Plant GLDHs are highly specific to l-GalL and cannot use l-GulL. Similarly, protozoan enzymes can use l-GalL and/or d-AL but not l-GulL. Ascomycete fungal ALOs prefer d-AL, in addition they catalyze l-GalL at a significant rate and l-GulL is also a substrate although at a low rate. An exception to this is GUO of Penicillia that is specific to d-gluconolactone and doesn't oxidize l-GalL, l-GulL or d-AL. Conversely, animal, yeast, plant and bacterial enzymes don't catalyze d-gluconolactone. A basidiomycete GulLO isoenzyme and bacterial GulLDHs are very specific to l-GulL and l-GalL is not a substrate for them. So, three kinds of enzymes exist, one can use only l-GulL, the second can use both l-GulL and l-GalL, and the third can use only l-GalL.
Animal GulLOs are localized to microsomes. Plant GLDHs and ascomycete fungal ALOs are localized to the mitochondria except GUO of P. griseoroseum that was isolated from the soluble fraction. Bacterial GulLDHs were found to be localized in the soluble fraction in G. oxydans and in cell wall in M. tuberculosis while the algal GulLDH was localized in both cytosol and mitochondria.
Plants, in addition to GLDH, have GulLO with an absolute specificity for l-GulL, similar to bacterial enzymes. It is not known why plants have both GLDH and GulLO. The differential expression of these genes spatially and temporally may be involved in regulating the synthesis of AsA, which is yet to be studied. Also, GLDH is a single copy gene in many species whereas GulLO exists as a gene family at least in Arabidopsis and the reason for this is unclear.
In the past, enzymes from the AsA biosynthetic and recycling pathways have been overexpressed to increase AsA in plants (reviewed in Lisko et al., 2014). Ascorbate biosynthetic pathways in plants converge to either l-GulL or l-GalL as the immediate precursor. Substrate availability is the limiting factor for AsA synthesis through l-GalL in plants (Imai et al., 2009). The l-GulL pool may be exploited to increase AsA in the plants. Understanding the regulation of GulLO enzyme(s) in plants may provide opportunitites to increase AsA through l-GulL.
Highlights.
A flavin cofactor is a characteristic feature present in almost all aldonolactone oxidoreductases.
An HWXK motif is present in all aldonolactone oxidoreductases for which amino acid sequences are known.
All aldonolactone oxidoreductases that are oxidases also have dehydrogenase activity however, exclusive dehydrogenases do exist.
Aldonolactone oxidoreductases with absolute specificity to l-galactono lactone or l-gulono lactone and promiscuous enzymes are found.
Plants have both l-gulono-1,4-lactone oxidases (GulLOs) and l-galactono-1,4-lactone dehydrogenases (GLDHs) involved in ascorbate biosynthesis
Acknowledgements
This study was supported at AL Laboratory with funds from the Arkansas Biosciences Institute, the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000, and a sub-award from the Arkansas INBRE program [National Center for Research Resources (5P20RR016460-11) and the National Institute of General Medical Sciences (8P20GM103429-11) from the National Institutes of Health]. SIA thanks the Molecular Biosciences PhD program at Arkansas State University for a scholarship.
Abbreviations
- AtGLDH
Arabidopsis thaliana l-galactono-1,4-lactone dehydrogenase
- AtGulLO
A. thaliana l-gulono-1,4-lactone oxidase
- d-AL
d-Arabinono-1,4-lactone
- l-AL
l-Arabinono-1,4-lactone
- ALO
d-Arabinono-1,4-lactone oxidase
- FAD
Flavin adenine dinucleotide
- FMN
Flavin mononucleotide
- GalLO
l-galactono-1,4-lactone oxidase
- l-GalL
l-Galactono-1,4-lactone
- d-GalL
d-Galactono-1,4-lactone
- GLDH
l-Galactono-1,4-lactone dehydrogenase
- d-GulL
d-Gulono-1,4-lactone
- l-GulL
l-Gulono-1,4-lactone
- GulLDH
l-Gulono-1,4-lactone dehydrogenase
- GulLO
l-Gulono-1,4-lactone oxidase
- GUO
d-Gluconolactone oxidase
- Man/Gal
d-Mannose/l-Galactose
- VAO
Vanillyl alcohol oxidase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agius F, González-Lamonthe R, Caballero JL, Muñoz-Blanco J, Botella MA, Valpuesta V. Engineering increased vitamin C levels in plants by over-expression of a d-galacturonic acid reductase. Nat Biotechnol. 2003;21:177–181. doi: 10.1038/nbt777. [DOI] [PubMed] [Google Scholar]
- Alhagdow M, Mounet F, Gilbert L, Nunes-Nesi A, Garcia V, Just D, Petit J, Beauvoit B, Fernie AR, Rothan C, Baldet P. Silencing of the mitochondrial ascorbate synthesizing enzyme l-galactono-1,4-lactone dehydrogenase affects plant and fruit development in tomato. Plant Physiol. 2007;145:1408–1422. doi: 10.1104/pp.107.106500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An HM, Fan WG, Chen LG, Asghar S, Liu QL. Molecular characterization and expression of l-galactono-1,4-lactone dehydrogenase and l-ascorbic acid accumulation during fruit development in Rosa Roxburghii. J Hortic Sci Biotech. 2007;82:627–635. [Google Scholar]
- Ashwell G, Kanfer J, Smiley JD, Burns JJ. Metabolism of ascorbic acid and related uronic acids, aldonic acids, and pentoses. Ann NY Acad Sci. 1961;92:105–114. doi: 10.1111/j.1749-6632.1961.tb46110.x. [DOI] [PubMed] [Google Scholar]
- Baig MM, Kelly S, Loewus FA. l-Ascorbic acid biosynthesis in higher plants from l-gulono-1,4- lactone and l-galactono-1,4-lactone. Plant Physiol. 1970;46:277–280. doi: 10.1104/pp.46.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli CG, Pastori GM, Foyer CH. Ascorbate biosynthesis in mitochondria is linked to the electron tansport chain between complexes III and IV. Plant Physiol. 2000;123:335–343. doi: 10.1104/pp.123.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli CG, Guiamet JJ, Kiddle G, Pastori GM, Di Cagno R, Theodoulou FL, Foyer CH. Ascorbate content of wheat leaves is not determined by maximal l-galactono-1,4-lactone dehydrogenase (GalLDH) activity under drought stress. Plant Cell Environ. 2005;28:1073–1081. [Google Scholar]
- Benen JAE, Sanchez-Torres P, Wagemaker MJM, Fraaije MW, van Berkel WJH, Visser J. Molecular cloning, sequencing, and heterologous expression of the vaoA gene from Penicillium simplicissimum CBS 170.90 encoding vanillyl-alcohol oxidase. J Biol Chem. 1998;273:7865–7872. doi: 10.1074/jbc.273.14.7865. [DOI] [PubMed] [Google Scholar]
- Biyani N, Madhubala R. Leishmania donovani encodes a functional enzyme involved in vitamin C biosynthesis: Arabino-1,4-lactone oxidase. Mol Biochem Parasitol. 2011;180:76–85. doi: 10.1016/j.molbiopara.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Bleeg HS, Christensen F. Biosynthesis of ascorbate in yeast. Purification of l-galactono-1,4-lactone oxidase with properties different from mammalian l-gulonolactone oxidase. Eur J Biochem. 1982;127:391–396. doi: 10.1111/j.1432-1033.1982.tb06884.x. [DOI] [PubMed] [Google Scholar]
- Boudart G, Jamet E, Rossignol M, Lafitte C, Borderies G, Jauneau A, Esquerré-Tugayé MT, Pont-Lezica R. Cell wall proteins in apoplastic fluids of Arabidopsis thaliana rosettes: Identification by mass spectrometry and bioinformatics. Proteomics. 2005;5:212–221. doi: 10.1002/pmic.200400882. [DOI] [PubMed] [Google Scholar]
- Brandsch R, Hinkkanen AE, Mauch L, Nagursky H. 6-Hydroxy-d-nicotine oxidase of Arthrobacter oxidans. Eur J Biochem. 1987;167:315–320. doi: 10.1111/j.1432-1033.1987.tb13338.x. [DOI] [PubMed] [Google Scholar]
- Brandsch R, Bichler V. Autoflavinylation of apo6-hydroxy-d-nicotine oxidase. J Biol Chem. 1991;266:19056–19062. [PubMed] [Google Scholar]
- Braun L, Mile V, Schaff Z, Csala M, Kardon T, Mandl J, Bánhegyi G. Induction and peroxisomal appearance of gulonolactone oxidase upon clofibrate treatment in mouse liver. FEBS Lett. 1999;458:359–362. doi: 10.1016/s0014-5793(99)01184-9. [DOI] [PubMed] [Google Scholar]
- Bublitz C. l-Gulono-γ-lactone oxidase and dehydrogenase. Biochim Biophys Acta. 1961;48:61–70. [Google Scholar]
- Burns JJ. Missing step in man, monkey and guinea pig required for the biosynthesis of l-ascorbic acid. Nature. 1957;180:553. doi: 10.1038/180553a0. [DOI] [PubMed] [Google Scholar]
- Burns JJ, Peyser P, Moltz A. Missing step in guinea pigs required for the biosynthesis of l-ascorbic acid. Science. 1956;7:1148–1149. doi: 10.1126/science.124.3232.1148-a. [DOI] [PubMed] [Google Scholar]
- Conklin PL, Norris SR, Wheeler GL, Williams EH, Smirnoff N, Last RL. Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc Natl Acad Sci USA. 1999;96:4198–4203. doi: 10.1073/pnas.96.7.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Gatzek S, Wheeler GL, Dowdle J, Raymond MJ, Rolinski S, Isupov M, Littlechild JA, Smirnoff N. Arabidopsis thaliana VTC4 encodes l-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme. J Biol Chem. 2006;281:15662–15670. doi: 10.1074/jbc.M601409200. [DOI] [PubMed] [Google Scholar]
- Crowhurst RN, Gleave AP, MacRae EA, et al. Analysis of expressed sequence tags from Actinidia: applications of a cross species EST database for gene discovery in the areas of flavor, health, color and ripening. BMC Genomics. 2008;9:351. doi: 10.1186/1471-2164-9-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MW, Gilot C, Persiau G, Ostergaard J, Han Y, Bauw GC, Van Montagu MC. Ascorbate biosynthesis in Arabidopsis cell suspension culture. Plant Physiol. 1999;121:535–543. doi: 10.1104/pp.121.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliceiri GL, Lai EK, McCay PB. Gulonolactone oxidase solubilization, properties and partial purification. J Biol Chem. 1969;244:2641–2645. [PubMed] [Google Scholar]
- Fraaije MW, van Berkel WJH, Benen JAE, Visser J, Mattevi A. A novel oxidoreductase family sharing a conserved FAD-binding domain. Trends Biochem Sci. 1998;23:206–207. doi: 10.1016/s0968-0004(98)01210-9. [DOI] [PubMed] [Google Scholar]
- Fraaije MW, van den Heuvel RHH, van Berkel WJH, Mattevi A. Covalent flavinylation is essential for efficient redox catalysis in vanillyl-alcohol oxidase. J Biol Chem. 1999;274:35514–35520. doi: 10.1074/jbc.274.50.35514. [DOI] [PubMed] [Google Scholar]
- Fraaije MW, van den Heuvel RHH, van Berkel WJH, Mattevi A. Structural analysis of flavinylation in vanillyl-alcohol oxidase. J Biol Chem. 2000;275:38654–38658. doi: 10.1074/jbc.M004753200. [DOI] [PubMed] [Google Scholar]
- Gallie DR. Increasing vitamin C content in plant foods to improve their nutritional value - successes and challenges. Nutrients. 2013;5:3424–3446. doi: 10.3390/nu5093424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. l-Ascorbic acid: A multifunctional molecule supporting plant growth and development. Scientifica. 2014;2014:795964. doi: 10.1155/2013/795964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha MN, Graham FL, D'Souza CK, Muller WJ, Igdoura SA, Schellhorn HE. Functional rescue of vitamin C synthesis deficiency in human cells using adenoviral-based expression of murine l-gulono-γ-lactone oxidase. Genomics. 2004;83:482–492. doi: 10.1016/j.ygeno.2003.08.018. [DOI] [PubMed] [Google Scholar]
- Hancock RD, Galpin JR, Viola R. Biosynthesis of l-ascorbic acid (vitamin C) by Saccharomyces cerevisiae. FEMS Microbiol Lett. 2000;186:245–250. doi: 10.1111/j.1574-6968.2000.tb09112.x. [DOI] [PubMed] [Google Scholar]
- Hancock RD, Viola R. The use of micro-organisms for l-ascorbic acid production: current status and future perspectives. Appl Microbiol Biotechnol. 2001;56:567–576. doi: 10.1007/s002530100723. [DOI] [PubMed] [Google Scholar]
- Harada Y, Shimizu M, Murakawa S, Takahashi T. Identification of FAD of d-glucono-lactone dehydrogenase: d-erythorbic acid producing enzyme of Penicillium cyaneo-fulvum. Agric Biol Chem. 1979;43:2635–2636. [Google Scholar]
- Hasan L, Vögeli P, Stoll P, Kramer SS, Stranzinger G, Neuenschwander S. Intragenic deletion in the gene encoding l-gulonolactone oxidase causes vitamin C deficiency in pigs. Mamm Genome. 2004;15:323–333. doi: 10.1007/s00335-003-2324-6. [DOI] [PubMed] [Google Scholar]
- Hemavathi, Upadhyaya CP, Akula N, Young KE, Chun SC, Kim DH, Park SW. Enhanced ascorbic acid accumulation in transgenic potato confers tolerance to various abiotic stresses. Biotechnol Lett. 2010;32:321–330. doi: 10.1007/s10529-009-0140-0. [DOI] [PubMed] [Google Scholar]
- Hervás M, Bashir Q, Leferink NGH, Ferreira P, Moreno-Beltrán B, Westphal AH, Dίaz-Moreno I, Medina M, de la Rosa MA, Ubbink M, Navarro JA, van Berkel WJH. Communication between l-galactono-1,4-lactone dehydrogenase and cytochrome C. FEBS J. 2013;280:1830–1840. doi: 10.1111/febs.12207. [DOI] [PubMed] [Google Scholar]
- Heuts DPHM, van Hellemond EW, Janssen DB, Fraaije MW. Discovery, characterization, and kinetic analysis of an alditol oxidase from Streptomyces coelicolor. J Biol Chem. 2007a;282:20283–20291. doi: 10.1074/jbc.M610849200. [DOI] [PubMed] [Google Scholar]
- Heuts DPHM, Janssen DB, Fraaije MW. Changing the substrate specificity of a chitooligosaccharide oxidase from Fusarium graminearum by model-inspired site-directed mutagenesis. FEBS Lett. 2007b;581:4905–4909. doi: 10.1016/j.febslet.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Huh WK, Kim ST, Yang KS, Seok YJ, Hah YC, Kang SO. Characterisation of d-arabinono-1,4-lactone oxidase from Candida albicans ATCC 10231. Eur J Biochem. 1994;225:1073–1079. doi: 10.1111/j.1432-1033.1994.1073b.x. [DOI] [PubMed] [Google Scholar]
- Huh WK, Lee BH, Kim ST, Kim YR, Rhie GE, Baek YW, Hwang CS, Lee JS, Kang SO. d-Erythroascorbic acid is an important antioxidant molecule in Saccharomyces cerevisiae. Mol Microbiol. 1998;30:895–903. doi: 10.1046/j.1365-2958.1998.01133.x. [DOI] [PubMed] [Google Scholar]
- Huh WK, Kim ST, Kim H, Jeong G, Kang SO. Deficiency of d-erythroascorbic acid attenuates hyphal growth and virulence of Candida albicans. Infect Immun. 2001;69:3939–3946. doi: 10.1128/IAI.69.6.3939-3946.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Karita S, Shratori G, Hattori M, Nunome T, Ôba K, Hirai M. l-Galactono-γ-lactone dehydrogenase from sweet potato: Purification and cDNA sequence analysis. Plant Cell Physiol. 1998;39:1350–1358. doi: 10.1093/oxfordjournals.pcp.a029341. [DOI] [PubMed] [Google Scholar]
- Imai T, Niwa M, Ban Y, Hirai M, Ôba K, Moriguchi T. Importance of the l-galactonolactone pool for enhancing the ascorbate content revealed by l-galactonolactone dehydrogenase-overexpressing tobacco plants. Plant Cell Tiss Organ Cult. 2009;96:105–112. [Google Scholar]
- Inai Y, Ohta Y, Nishikimi M. The whole structure of the human nonfunctional l-gulono-γ-lactone oxidase gene – the gene responsible for scurvy – and the evolution of repetitive sequences thereon. J Nutr Sci Vitaminol. 2003;49:315–319. doi: 10.3177/jnsv.49.315. [DOI] [PubMed] [Google Scholar]
- Isherwood FA, Chen YT, Mapson LW. Synthesis of l-ascorbic acid in plants and animals. Biochem J. 1954;56:1–14. doi: 10.1042/bj0560001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isherwood FA, Mapson LW, Chen YT. Synthesis of l-ascorbic acid in rat liver homogenates. Biochem J. 1960;76:157–171. doi: 10.1042/bj0760157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AK, Nessler CL. Metabolic engineering of an alternative pathway for ascorbic acid biosynthesis in plants. Mol Breed. 2000;6:73–78. [Google Scholar]
- Jin J, Mazon H, van den Heuvel RHH, Janssen DB, Fraaije MW. Discovery of a eugenol oxidase from Rhodococcus sp.strain RHA1. FEBS J. 2007;274:2311–2321. doi: 10.1111/j.1742-4658.2007.05767.x. [DOI] [PubMed] [Google Scholar]
- Kenney WC, Edmondson DE, Singer TP, Nakagawa H, Asano A, Sato R. Identification of the covalently bound flavin of l-gulono-γ-lactone oxidase. Biochem Biophys Res Comm. 1976;71:1194–1200. doi: 10.1016/0006-291x(76)90780-4. [DOI] [PubMed] [Google Scholar]
- Kenney WC, Edmondson DE, Singer TP, Nishikimi M, Noguchi E, Yagi K. Identification of the covalently-bound flavin of l-galactonolactone oxidase from yeast. FEBS Lett. 1979;97:40–42. doi: 10.1016/0014-5793(79)80047-2. [DOI] [PubMed] [Google Scholar]
- Kiuchi K, Nishikimi M, Yagi K. Purification and characterization of l-gulonolactone oxidase from chicken kidney microsomes. Biochem. 1982;21:5076–5082. doi: 10.1021/bi00263a035. [DOI] [PubMed] [Google Scholar]
- Koshizaka T, Nishikimi M, Ozawa T, Yagi K. Isolation and sequence analysis of a complementary DNA encoding rat liver l-gulono-γ-lactone oxidase, a key enzyme for l-ascorbic acid biosynthesis. J Biol Chem. 1988;263:1619–1621. [PubMed] [Google Scholar]
- Krasnov A, Reinisalo M, Pitkänen TI, Nishikimi M, Mölsä H. Expression of rat gene for l-gulono-γ-lactone oxidase, the key enzyme of l-ascorbic acid biosynthesis, in guinea pig cells and in teleost fish rainbow trout (Oncorhynchus mykiss). Biochim Biophys Acta. 1998;1381:241–248. doi: 10.1016/s0304-4165(98)00037-3. [DOI] [PubMed] [Google Scholar]
- Kudryashova EV, Leferink NGH, Slot IGM, van Berkel WJH. Galactonolactone oxidoreductase from Trypanosoma cruzi employs a FAD cofactor for the synthesis of vitamin C. Biochim Biophys Acta. 2011;1814:545–552. doi: 10.1016/j.bbapap.2011.03.001. [DOI] [PubMed] [Google Scholar]
- Kutchan TM, Dittrich H. Characterization and mechanism of the berberine bridge enzyme, a covalently flavinated oxidase of benzophenanthridine alkaloid biosynthesis in plants. J Biol Chem. 1995;270:24475–24481. doi: 10.1074/jbc.270.41.24475. [DOI] [PubMed] [Google Scholar]
- Laing WA, Bulley Sy, Wright My, Cooney Jyy, Jensen Dy, Barraclough D, MacRae E. A highly specific l-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis. Proc Natl Acad Sci USA. 2004;101:16976–16981. doi: 10.1073/pnas.0407453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing WA, Wright MA, Cooney J, Bulley SM. The missing step of the l-galactose pathway of ascorbate biosynthesis in plants, an l-galactose guanyltransferase, increases leaf ascorbate content. Proc Natl Acad Sci USA. 2007;104:9534–9539. doi: 10.1073/pnas.0701625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Huh WK, Kim ST, Lee JS, Kang SO. Bacterial production of d-erythroascorbic acid and l-ascorbic acid through functional expression of Saccharomyces cerevisiae d-arabinono-1,4-lactone oxidase in Escherichia coli. Appl Environ Microbiol. 1999;65:4685–4687. doi: 10.1128/aem.65.10.4685-4687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leferink NGH, Heuts DPHM, Fraaije MW, van Berkel WJH. The growing VAO flavoprotein family. Arch Biochem Biophys. 2008a;474:292–301. doi: 10.1016/j.abb.2008.01.027. [DOI] [PubMed] [Google Scholar]
- Leferink NGH, van den Berg WAM, van Berkel WJH. l-Galactono-γ-lactone dehydrogenase from Arabidopsis thaliana, a flavoprotein involved in vitamin C biosynthesis. FEBS J. 2008b;275:713–726. doi: 10.1111/j.1742-4658.2007.06233.x. [DOI] [PubMed] [Google Scholar]
- Leferink NGH. PhD thesis. Wageningen University; The Netherlands: 2009a. Characterization and redesign of galactonolactone dehydrogenase, a flavoprotein producing vitamin C. [Google Scholar]
- Leferink NGH, Fraaije MW, Joosten HJ, Schaap PJ, Mattevi A. Identification of a gatekeeper residue that prevents dehydrogenases from acting as oxidases. J Biol Chem. 2009b;284:4392–4397. doi: 10.1074/jbc.M808202200. [DOI] [PubMed] [Google Scholar]
- Leferink NGH, Jose MDF, van den Berg WAM, van Berkel WJH. Functional assignment of Glu386 and Arg388 in the active site of l-galactono-γ-lactone dehydrogenase. FEBS Lett. 2009c;583:3199–3203. doi: 10.1016/j.febslet.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Leferink NGH, van Duijn E, Barendregt A, Heck AJR, van Berkel WJH. Galactonolactone dehydrogenase requires a redox-sensitive thiol for optimal production of vitamin C. Plant Physiol. 2009d;150:596–605. doi: 10.1104/pp.109.136929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leferink NGH, van Berkel WJH. Aldonolactone oxidoreductases. In: Weber S, Schleicher E, editors. Flavins and Flavoproteins: Methods and Protocols, Methods in Molecular Biology. Vol. 1146. Springer; New York: 2014. pp. 95–111. [DOI] [PubMed] [Google Scholar]
- Lim MY, Pulla RK, Park JM, Harn CH, Jeong BR. Over-expression of l-gulono-γ-lactone oxidase (GLOase) gene leads to ascorbate accumulation with enhanced abiotic stress tolerance in tomato. In Vitro Cell Dev Biol. 2012;48:453–461. [Google Scholar]
- Lisko KA, Aboobucker SI, Torres R, Lorence A. Engineering elevated vitamin C in plants to improve their nutritional content, growth, and tolerance to abiotic stress. In: Jetter R, editor. Phytochemicals – Biosynthesis, Function and Application, Recent Advances in Phytochemistry. Vol. 44. Springer; Switzerland: 2014. pp. 109–128. [Google Scholar]
- Locato V, Cimini S, De Gara L. Strategies to increase vitamin C in plants: from plant defense perspective to food biofortification. Front Plant Sci. 2013;4:152. doi: 10.3389/fpls.2013.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewus FA. Tracer studies of ascorbic acid formation in plants. Phytochem. 1963;2:109–128. [Google Scholar]
- Logan FJ, Taylor MC, Wilkinson SR, Kaur H, Kelly JM. The terminal step in vitamin C biosynthesis in Trypanosoma cruzi is mediated by a FMN-dependent galactonolactone oxidase. Biochem J. 2007;407:419–426. doi: 10.1042/BJ20070766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorence A, Chevone BI, Mendes P, Nessler CL. Myo-Inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol. 2004;134:1200–1205. doi: 10.1104/pp.103.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapson LW, Isherwood FA, Chen YT. Biological synthesis of l-ascorbic acid: The conversion of l-galactono-γ-lactone into l-ascorbic acid by plant mitochondria. Biochem J. 1954;56:21–28. doi: 10.1042/bj0560021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapson LW, Breslow E. Biological synthesis of ascorbic acid: l-Galactono-γ-lactone dehydrogenase. Biochem J. 1958;68:395–406. doi: 10.1042/bj0680395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawuenyega KG, Forst CV, Dobos KM, Belisle JT, Chen J, Bradbury EM, Bradbury AR, Chen X. Mycobacterium tuberculosis functional network analysis by global subcellular protein profiling. Mol Biol Cell. 2005;16:396–404. doi: 10.1091/mbc.E04-04-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta T, Yonemitsu M, Yabuta Y, Tamoi M, Ishikawa T, Shigeoka S. Arabidopsis phosphomannose isomerase 1, but not phosphomannose isomerase 2, is essential for ascorbic acid biosynthesis. J Biol Chem. 2008;283:28842–28851. doi: 10.1074/jbc.M805538200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta T, Ichikawa Y, Mieda T, Takeda T, Tamoi M, Yabuta Y, Ishikawa T, Shigeoka S. The contribution of Arabidopsis homologs of l-gulono,1,4-lactone oxidase to the biosynthesis of ascorbic acid. Biosci Biotechnol Biochem. 2010;74:1494–1497. doi: 10.1271/bbb.100157. [DOI] [PubMed] [Google Scholar]
- Mawuenyega KG, Forst CV, Dobos KM, Belisle JT, Chen J, Bradbury EM, Bradbury AR, Chen X. Mycobacterium tuberculosis functional network analysis by global subcellular protein profiling. Mol Biol Cell. 2005;16:396–404. doi: 10.1091/mbc.E04-04-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewies M, McIntire WS, Scrutton NS. Covalent attachment of flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) to enzymes: the current state of affairs. Protein Sci. 1998;7:7–20. doi: 10.1002/pro.5560070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsuda M, Ishikawa T, Takeda T, Shigeoka S. Subcellular localization and properties of l-galactono-γ-lactone dehydrogenase in spinach leaves. Biosci Biotechnol Biochem. 1995;59:1983–1984. [Google Scholar]
- Nakagawa H, Asano A. Ascorbate synthesizing system in rat liver microsomes. I. Gulonolactone-reducible pigment as a prosthetic group of gulonolactone oxidase. J Biochem. 1970;68:737–746. doi: 10.1093/oxfordjournals.jbchem.a129408. [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Asano A, Sato R. Ascorbate-synthesizing system in rat liver microsomes. II. A peptide-bound flavin as the prosthetic group of l-gulono-γ-lactone oxidase. J Biochem. 1975;77:221–232. [PubMed] [Google Scholar]
- Nam YK, Cho YS, Douglas SE, Gallant JW, Reith ME, Kim DS. Isolation and transient expression of a cDNA encoding l-gulono-γ-lactone oxidase, a key enzyme for l-ascorbic acid biosynthesis, from the tiger shark Scyliorhinus torazame. Aquaculture. 2002;209:271–284. [Google Scholar]
- Nascimento JRO, Higuchi BK, Gomez ML, Oshiro RA, Lajolo FM. l-Ascorbate biosynthesis in strawberries: l-galactono-1,4-lactone dehydrogenase expression during fruit development and ripening. Postharvest Biol Tech. 2005;38:34–42. [Google Scholar]
- Nishikimi M, Udenfriend S. Immunologic evidence that the gene for l-gulono-γlactone oxidase is not expressed in animals subject to scurvy. Proc Natl Acad Sci USA. 1976;73:2066–2068. doi: 10.1073/pnas.73.6.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikimi M, Yagi K. Molecular basis for the deficiency in humans of gulonolactone oxidase, a key enzyme for ascorbic acid biosynthesis. Am J Clin Nutr. 1991;54:1203S–1208S. doi: 10.1093/ajcn/54.6.1203s. [DOI] [PubMed] [Google Scholar]
- Nishikimi M, Tolbert BM, Udenfriend S. Purification and characterization of l-gulono-γ-lactone oxidase from rat and goat liver. Arch Biochem Biophys. 1976;175:427–435. doi: 10.1016/0003-9861(76)90530-0. [DOI] [PubMed] [Google Scholar]
- Nishikimi M, Kiuchi K, Yagi K. Detection of l-gulono-γ-lactone oxidase on SDS-polyacrylamide gels by the fluorescence of its covalently bound flavin. FEBS Lett. 1977;81:323–325. doi: 10.1016/0014-5793(77)80545-0. [DOI] [PubMed] [Google Scholar]
- Nishikimi M, Noguchi E, Yagi K. Occurrence in yeast of l-galactonolactone oxidase which is similar to a key enzyme for ascorbic acid biosynthesis in animals, l-gulonolactone oxidase. Arch Biochem Biophys. 1978;191:479–486. doi: 10.1016/0003-9861(78)90386-7. [DOI] [PubMed] [Google Scholar]
- Nishikimi M, Noguchi E, Yagi K. Redox properties of l-galactonolactone oxidase purified from Baker's yeast. Biochem Int. 1980;1:155–161. [Google Scholar]
- Nishikimi M, Koshizaka T, Ozawa T, Yagi K. Occurrence in humans and guinea pigs of the gene related to their missing enzyme l-gulono-γ-lactone oxidase. Arch Biochem Biophys. 1988;267:842–846. doi: 10.1016/0003-9861(88)90093-8. [DOI] [PubMed] [Google Scholar]
- Nishikimi M, Kawai T, Yagi K. Guinea pigs possess a highly mutated gene for l-gulono-γ-lactone oxidase, the key enzyme for l-ascorbic acid biosynthesis missing in this species. J Biol Chem. 1992;267:21967–21972. [PubMed] [Google Scholar]
- Nishikimi M, Kobayashi J, Yagi K. Production by a baculovirus expression system of the apoprotein of l-gulono-γ-lactone oxidase, a flavoenzyme possessing a covalently-bound FAD. Biochem Mol Biol Int. 1994;33:313–320. [PubMed] [Google Scholar]
- Noguchi E, Nishikimi M, Yagi K. Studies on the sulfhydryl group of l-galactonolactone oxidase. J Biochem. 1981;90:33–38. doi: 10.1093/oxfordjournals.jbchem.a133467. [DOI] [PubMed] [Google Scholar]
- Ôba K, Fukui M, Imai Y, Iriyama S, Nogami K. l-Galactono-γ-lactone dehydrogenase: partial characterization, induction of activity and role in the synthesis of ascorbic acid in wounded white potato tuber tissue. Plant Cell Physiol. 1994;35:473–478. [Google Scholar]
- Ôba K, Ishikawa S, Nishikawa M, Mizuno H, Yamamoto T. Purification and properties of l-galactono-γ-lactone dehydrogenase, a key enzyme for ascorbic acid biosynthesis, from sweet potato roots. J Biochem. 1995;117:120–124. doi: 10.1093/oxfordjournals.jbchem.a124697. [DOI] [PubMed] [Google Scholar]
- Okamura M. Distribution of ascorbic acid analogs and associated glycosides in mushrooms. J Nutr Sci Vitaminol. 1994;40:81–94. doi: 10.3177/jnsv.40.81. [DOI] [PubMed] [Google Scholar]
- Okamura M. Purification and properties of l-gulono-1,4-lactone oxidase from Grifola frondosa. J Nutri Sci Vitaminol. 2001;47:258–262. doi: 10.3177/jnsv.47.258. [DOI] [PubMed] [Google Scholar]
- Østergaard J, Persiau G, Cavey MW, Bauw G, Van Montagu M. Isolation and cDNA cloning for l-galactono-γ-lactone dehydrogenase, an enzyme involved in the biosynthesis of ascorbic acid in plants. J Biol Chem. 1997;272:30009–30016. doi: 10.1074/jbc.272.48.30009. [DOI] [PubMed] [Google Scholar]
- Pateraki I, Sanmartin M, Kalamaki MS, Gerasopoulos D, Kanellis AK. Molecular characterization and expression studies during melon fruit development and ripening of l-galactono-1,4-lactone dehydrogenase. J Exp Bot. 2004;55:1623–1633. doi: 10.1093/jxb/erh186. [DOI] [PubMed] [Google Scholar]
- Puskás F, Braun L, Csala M, Kardon T, Marcolongo P, Benedetti A, Mandl J, Bánhegyi G. Gulonolactone oxidase activity-dependent intravesicular glutathione oxidation in rat liver microsomes. FEBS Lett. 1998;430:293–296. doi: 10.1016/s0014-5793(98)00678-4. [DOI] [PubMed] [Google Scholar]
- Qian W, Yu C, Qin H, Liu X, Zhang A, Johansen AE, Wang D. Molecular and functional analysis of phosphomannomutase (PMM) from higher plants and genetic evidence for the involvement of PMM in ascorbic acid biosynthesis in Arabidopsis and Nicotiana benthamiana. Plant J. 2007;49:399–413. doi: 10.1111/j.1365-313X.2006.02967.x. [DOI] [PubMed] [Google Scholar]
- Radzio JA, Lorence A, Chevone BI, Nessler CL. l-Gulono-1,4-lactone oxidase expression rescues vitamin C-deficient Arabidopsis (vtc) mutants. Plant Mol Biol. 2003;53:837–844. doi: 10.1023/B:PLAN.0000023671.99451.1d. [DOI] [PubMed] [Google Scholar]
- Salusjärvi T, Kalkkinen N, Miasnikov AN. Cloning and characterization of gluconolactone oxidase of Penicillium cyaneo-fulvum ATCC 10431 and evaluation of its use for production of d-erythorbic acid in recombinant Pichia pastoris. Appl Environ Microbiol. 2004;70:5503–5510. doi: 10.1128/AEM.70.9.5503-5510.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salusjärvi . PhD thesis. University of Helsinki; Finland: 2006. Characterisation, cloning and production of industrially interesting enzymes: gluconolactone oxidase of Penicillium cyaneofulvum and gluconate 5-dehydrogenase of Gluconobacter suboxydans. [Google Scholar]
- Sauer M, Branduardi P, Valli M, Porro D. Production of l-ascorbic acid by metabolically engineered Saccharomyces cerevisiae and Zygosaccharomyces bailii. Appl Environ Microbiol. 2004;70:6086–6091. doi: 10.1128/AEM.70.10.6086-6091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka S, Yokota A, Nakano Y, Kitaoka S. The effect of illumination on the l-ascorbic acid content in Euglena gracilis Z. Agric Biol Chem. 1979;43:2053–2058. [Google Scholar]
- Shimizu M, Murakawa S, Takahashi T. The covalently bound flavin prosthetic group of d-gluconolactone dehydrogenase of Penicillium cyaneo-fulvum. Agric Biol Chem. 1977;41:2107–2108. [Google Scholar]
- Siendones E, González-Reyes JA, Santos-Ocaña C, Navas P, Córdoba F. Biosynthesis of ascorbic acid in kidney bean. l-galactono-γ-lactone dehydrogenase is an instrinsic protein located at the mitochondrial inner membrane. Plant Physiol. 1999;120:907–912. doi: 10.1104/pp.120.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugisawa T, Setsuko O, Matzinger PK, Hoshino T. Isolation and characterization of a new vitamin C producing enzyme (l-gulono-γ-lactone dehydrogenase) of bacterial origin. Biosci Biotech Biochem. 1995;59:190–196. [Google Scholar]
- Takahashi T. Erythorbic acid fermentation. Biotechnol Bioeng. 1969;11:1157–1171. [Google Scholar]
- Takahashi T, Yamashita H, Kato E, Mitsumoto M, Murakawa S. Purification and properties of d-glucono-γ-lactone dehydrogenase d-erythorbic acid producing enzyme of Penicillium cyaneo-fulvum. Agric Biol Chem. 1976;40:121–129. [Google Scholar]
- Tamaoki M, Mukai F, Asai N, Nakajima N, Kubo A, Aono M, Saji H. Light-controlled expression of a gene encoding l-galactono-γ-lactone dehydrogenase which affects ascorbate pool size in Arabidopsis thaliana. Plant Sci. 2003;164:1111–1117. [Google Scholar]
- Tokunaga T, Miyahara K, Tabata K, Esaka M. Generation and properties of ascorbic acid-overproducing transgenic tobacco cells expressing sense RNA for l-galactono-1,4-lactone dehydrogenase. Planta. 2005;220:854–863. doi: 10.1007/s00425-004-1406-3. [DOI] [PubMed] [Google Scholar]
- Wheeler GL, Jones MA, Smirnoff N. The biosynthetic pathway of vitamin C in higher plants. Nature. 1998;393:365–369. doi: 10.1038/30728. [DOI] [PubMed] [Google Scholar]
- Wheeler G, Ishikawa T, Pornsaksit V, Smirnoff N. Evolution of alternative biosynthetic pathways for vitamin C following plastid acquisition in photosynthetic eukaryotes. eLife. 2015;4:e06369. doi: 10.7554/eLife.06369. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson SR, Prathalingam SR, Taylor MC, Horn D, Kelly JM. Vitamin C biosynthesis in trypanosomes:A role for the glycosome. Proc Natl Acad Sci USA. 2005;102:11645–11650. doi: 10.1073/pnas.0504251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolucka BA, van Montagu M. GDP-mannose-3’,5’-epimerase forms GDP-l-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. J Biol Chem. 2003;278:47483–47490. doi: 10.1074/jbc.M309135200. [DOI] [PubMed] [Google Scholar]
- Wolucka BA, Communi D. Mycobacterium tuberculosis possesses a functional enzyme for the synhesis of vitamin C, l-gulono-1,4-lactone dehydrogenase. FEBS J. 2006;273:4435–4445. doi: 10.1111/j.1742-4658.2006.05443.x. [DOI] [PubMed] [Google Scholar]
- Yabuta Y, Yoshimura K, Takeda T, Shigeoka S. Molecular characterization of tobacco mitochondrial l-galactono-γ-dehydrogenase and its expression in Escherichia coli. Plant Cell Physiol. 2000;41:666–675. doi: 10.1093/pcp/41.6.666. [DOI] [PubMed] [Google Scholar]
- Yagi K, Koshizaka T, Kito M, Ozawa T, Nishikimi M. Expression in monkey cells of the missing enzyme in l-ascorbic acid biosynthesis, l-gulono-gamma-lactone oxidase. Biochem Biophys Res Commun. 1991;177:659–663. doi: 10.1016/0006-291x(91)91839-5. [DOI] [PubMed] [Google Scholar]
- Yu L, Jiang JZ, Zhang C, Jiang LR, Ye NH, Lu YS, Yang GZ, Liu EE, Peng CL, He ZH, Peng XX. Glyoxylate rather than ascorbate is an efficient precursor for oxalate biosynthesis in rice. J Exp Bot. 2010;61:1625–1634. doi: 10.1093/jxb/erq028. [DOI] [PMC free article] [PubMed] [Google Scholar]