Abstract
The neurobiological basis of early life stress (ELS) impact on vulnerability to alcohol use disorder is not fully understood. The effect of ELS, adult ethanol consumption and single housing, on expression of stress and DNA methylation regulatory genes as well as blood corticosterone levels was investigated in the hypothalamus and pituitary of adult out-bred Wistar rats subjected to different rearing conditions. A prolonged maternal separation (MS) of 360 min (MS360) was used to study the effect of ELS, and a short MS of 15 min (MS15) was used as a control. Voluntary ethanol drinking was assessed using a two-bottle free choice paradigm to simulate human episodic drinking. The effects of single housing and ethanol were assessed in conventional animal facility rearing (AFR) conditions. Single housing in adulthood was associated with lower Crhr1 and higher Pomc expression in the pituitary, whereas ethanol drinking was associated with higher expression of Crh in the hypothalamus and Crhr1 in the pituitary, accompanied by lower corticosterone levels. As compared to controls with similar early life handling, rats exposed to ELS displayed lower expression of Pomc in the hypothalamus, and higher Dnmt1 expression in the pituitary. Voluntary ethanol drinking resulted in lower Fkbp5 expression in the pituitary and higher Crh expression in the hypothalamus, independently of rearing conditions. In rats exposed to ELS, water and ethanol drinking was associated with higher and lower corticosterone levels, respectively. The use of conventionally reared rats as control group yielded more significant results than the use of rats exposed to short MS. Positive correlations, restricted to the hypothalamus and ELS group, were observed between the expression of the hypothalamus-pituitary-adrenal receptor and the methylation-related genes. Promoter DNA methylation and expression of respective genes did not correlate suggesting that other loci are involved in transcriptional regulation. Concluding, single housing is a confounding factor to be considered in voluntary ethanol drinking paradigms. ELS and ethanol drinking in adulthood exert independent effects on hypothalamic and pituitary related genes, however, in a manner dependent on the control group used.
Keywords: DNA methylation, early life stress, ethanol, gene expression, housing, hypothalamus, pituitary gland, rat
Introduction
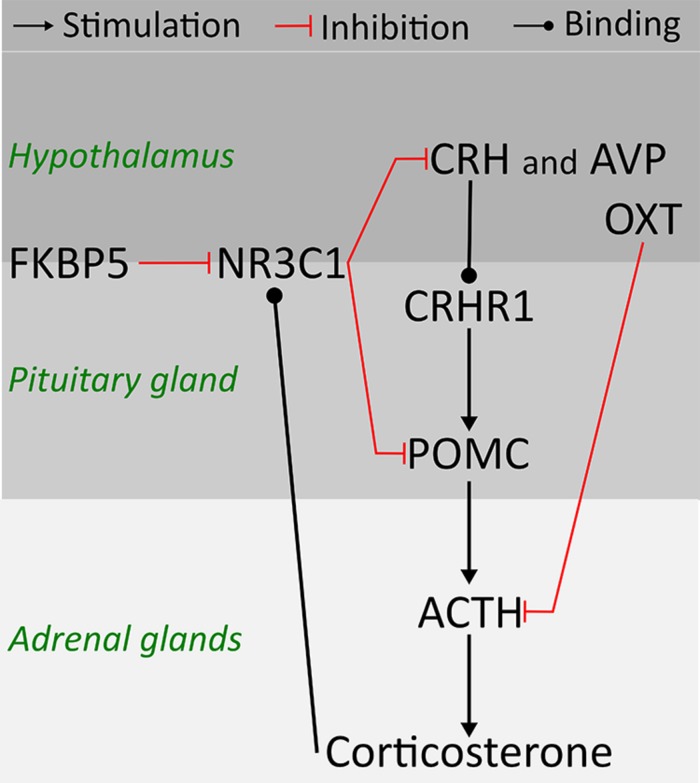
Clinical studies point to early life stress (ELS) as a risk factor for psychiatric disorders, including alcohol use disorder (AUD) (Enoch, 2011). Preclinical studies provide evidence of increased risk for high ethanol consumption later in life as a consequence of ELS (Becker et al., 2011; Nylander and Roman, 2013). Environmentally induced changes in the stress response system and brain plasticity have been suggested to mediate long-term effects of ELS, however, previous findings are not consistent (Pryce and Feldon, 2003; Harrison and Baune, 2014; Maccari et al., 2014). The main system regulating the stress response is the hypothalamus-pituitary-adrenal (HPA) axis, a complex molecular pathway including feedback regulatory interactions between the hypothalamus, the pituitary gland, and the adrenal glands (Figure 1) (Tsigos and Chrousos, 2002). Corticotropin releasing hormone (CRH) and arginine vasopressin (AVP) released from the hypothalamus are essential regulators of the HPA axis. Through synergistic positive reinforcing interaction between CRH and AVP, and CRH actions on the CRH receptor (CRHR1), CRH and AVP stimulate the secretion of adrenocorticotropin hormone (ACTH) from the pituitary (Tsigos and Chrousos, 2002). ACTH is cleaved from proopiomelanocortin (POMC) (Smith and Funder, 1988), and stimulates the adrenal glands to secrete corticosterone, the principal glucocorticoid in rodents. Corticosterone binds to the glucocorticoid receptor (NR3C1) in the brain and inhibits the CRH release through an inhibitory feedback loop (Brunton, 2013). Moreover, the FK506 Binding Protein 5 (FKBP5) by changing the conformation of the receptor complex can reduce the sensitivity of NR3C1 to corticosterone and the efficiency of the negative feedback on the HPA axis (Binder, 2009). In addition to CRH and AVP, oxytocin (OXT) moderates the release of ACTH and corticosterone by either altering sensitivity to the feedback mechanism or by decreasing CRH release (Petersson et al., 1999).
FIGURE 1.
Schematic representation of hypothalamus-pituitary-adrenal (HPA) axis physiology and relationship amongst candidate genes in rodents.
DNA methylation can mediate the crosstalk between genome and environmental cues such as ELS into stable changes of the expression of key-genes of the HPA axis (Szyf, 2013; Maccari et al., 2014). Human as well as experimental studies suggest that ethanol is capable of producing global epigenetic modifications (Contet, 2012). Two important regulators of the DNA methylation machinery are the methyl CpG binding protein 2 (MECP2) and DNA (Cytosine-5)-methyltransferase 1 (DNMT1). MECP2 preferentially binds to methylated DNA and, through the recruitment of other proteins, represses gene expression, but also participates in the maintenance of DNA methylation during replication and reparation by recruiting DNMT1 which transfers methyl groups to hemi-methylated DNA (Moore et al., 2013). The mechanisms forming the relationship between ELS and ethanol consumption behavior in adulthood may involve long-term changes in epigenetic regulation of the HPA system but these mechanisms are largely unknown.
The present study used a rodent model to elucidate how ELS and adult voluntary ethanol drinking affect gene expression, DNA methylation, and corticosterone levels. Also, the effect of using two different but commonly used controls in rodent models of ELS on the outcome was also assessed. The focus was on a set of genes involved in stress response and DNA methylation, based on previous molecular and pharmacological findings (Gehlert et al., 2007; Murgatroyd et al., 2009; Klengel et al., 2013; Wu et al., 2014). The aim was to provide in-depth understanding of the biological mechanisms mediating the impact of stress and ethanol drinking in the hypothalamus and pituitary, which form the up-stream component of the stress system. The specific aims were to examine expression and DNA promoter methylation of genes regulating the HPA axis as well as corticosterone levels, and evaluate: (i) the effect of single housing and voluntary ethanol drinking in adult rats subjected to conventional early rearing conditions; (ii) the long-term effect of ELS alone and in combination with voluntary ethanol drinking; (iii) whether rearing condition and voluntary ethanol drinking affect correlation patterns in gene expression, corticosterone levels and ethanol drinking; and (iv) the results observed when conventionally rearing relative to short maternal separation (MS; 15 min) was used as control condition for prolonged MS (360 min).
Materials and Methods
Animals
Time mated Wistar dams (n = 25; RccHan:WI, Harlan, Europe) arrived at gestation day 15. After birth [postnatal day (PND) 0] the pups were sexed and cross-fostered to avoid the use of biological littermates in the same experimental groups. Each litter contained 10 pups, 6 males and 4 females, and the litters were randomly assigned to the different experimental groups. Only males were used in the present study. The study was approved by the Uppsala Animal Ethical Committee (C32/11) and followed the guidelines of the Swedish Legislation on Animal Experimentation (Animal Welfare Act SFS1998:56) and the European Communities Council Directive (86/609/EEC).
Early Life Rearing Conditions
Pups from the same litters were randomized and subjected to three parallel groups; either MS (15/360 min) or conventional animal facility rearing (AFR) conditions. A rodent MS model was used to simulate ELS during the first three postnatal weeks (Figure 2). Based on previous studies, prolonged daily MS (360 min; MS360) was used to simulate ELS and short MS (15 min; MS15) was used as control to MS360 (Nylander and Roman, 2013). The MS15 and MS360 rats were exposed to the same handling procedures during MS, the only difference being the length of the separation. The separations were performed during the light period and started at 9 AM. The MS procedure has been described in detail elsewhere (Gustafsson and Nylander, 2006). The litters were weighed on PND 0, 3, 7, 10, 13, 16 and the cages were changed on PND 7 and 16. The separations were always performed in the same room and only one person performed all separation and care-taking. The rats subjected to AFR conditions were left undisturbed with the exception of cage change (PND 7, 16) and weighing of the litter (PND 0, 7, 16). These rats were used to assess the possible confounding effect of single housing in the voluntary drinking paradigm. On PND 22 all animals were weaned and then group housed three per cage during adolescence.
FIGURE 2.
Experimental outline. ∗One additional group of AFR rats continued to be group-housed during adulthood to assess the effect of housing. AFR, animal facility rearing; MS, Maternal separation (15/360 min).
Voluntary Ethanol Drinking
On postnatal week 10 the MS rats were single housed and randomly assigned to water-drinking (MS15W, n = 10; MS360W, n = 10) or ethanol-drinking groups (MS15E, n = 10; MS360E, n = 20). More rats were included in the MS360 group based on previous findings of subgroups with responding and non-responding rats regarding ethanol intake (Nylander and Roman, 2013). The AFR rats were assigned to three experimental groups; two groups of rats were single housed and assigned to water (AFRW, n = 9) and ethanol drinking (AFRE, n = 11), respectively, whereas one group (n = 7) was group housed during week 10–16.
The rats exposed to ethanol had free choice between non-sweetened ethanol (5 or 20% made from Ethanol 96%; Solveco AB, Rosersberg, Sweden) and water for three consecutive days a week with ethanol-free days in-between. The first week the rats had free access to 5% ethanol for 24 h and the next week limited access to 5% for 2 h; the following 5 weeks they had access to 20% ethanol in 2 h sessions for 3 days. This drinking paradigm is developed to mimic the common human episodic drinking pattern, with repeated drinking days and non-drinking days in-between (Momeni and Roman, 2014; Palm and Nylander, 2014). Various intermittent models are commonly used in voluntary drinking paradigms to increase the ethanol intake, and intermittent ethanol exposure with ethanol-free days in-between has also been shown to be necessary to induce neurobiological alterations similar to those seen in the transition to habitual and compulsive drinking (Carnicella et al., 2014). Since the current study aimed at investigating early neurobiological markers of risk-drinking correlate, rats were sacrificed at a point where the individual ethanol consumption pattern can be discerned but the voluntary intake associates to the non-addicted state. The limited access restricted to 2 h is an optimal choice to ensure less variation in biological parameters due to individual differences in drinking bouts in a 24 h access paradigm (Carnicella et al., 2014). Ethanol and water were changed every session and the bottle position was altered every day to avoid position preference. At the end of each session, the ethanol and water intake was quantified by weighing the bottles. Care was taken to minimize spillage. After 5 weeks of access to 20% ethanol, the rats weight was measured and thereafter they were decapitated (Figure 2). There were no differences in ethanol drinking groups for ethanol intake, as reported in (Comasco et al., 2015). The weekly voluntary ethanol intake in AFR, MS15 and MS360 rats during the last week before decapitation was 1.18 g/kg/2 h (0.62–1.66) in AFR rats, 1.32 g/kg/2 h (0.39–1.77) in MS15 rats, and 1.32 g/kg/2 h (0.60–2.05) in MS360. The ethanol-drinking animals were sacrificed immediately after a 2 h drinking session. Trunk blood was collected from rats randomly chosen from different treatment groups. The hypothalamus and pituitary were removed and immediately frozen on dry ice and stored at -80°C.
Genetic Analyses
RNA and DNA isolation: RNA and DNA were simultaneously isolated from the rat hypothalamus using AllPrep DNA/RNA/miRNA Universal Kit according to the manufacturer’s protocol (Qiagen AB Sollentuna, Sweden). Quantification of both the nucleic acids was carried out using a Nanodrop ND 1000 spectrometer.
Gene Expression Analyses
cDNA synthesis: RNA (700 ng) was converted to cDNA using QuantiTect Reverse: Transcription Kit (Qiagen AB Sollentuna, Sweden). The newly synthesized cDNA was diluted 20x and stored at 20°C until further use. gDNA contamination was controlled at three check-points: (1) On column DNase I treatment during the extraction process; (2) gDNA wipe-out reaction prior to cDNA synthesis; and (3) Designing of primers across two adjacent exons to avoid any unspecific amplification of genomic DNA.
Diluted cDNA (20x) was used to assess the expression of Pomc, Avp, Oxt, Crhr1, Nr3c1, Fkbp5, Mecp2, and Dnmt1 in the hypothalamus, and Pomc, Crhr1, Nr3c1, Fkbp5, Mecp2, and Dnmt1 in the pituitary, as well as Actb, Gapdh and Rpl19, as housekeeping genes, using the CFX96 Touch Real-Time PCR Detection System real time PCR (Supplementary Table S2). The Crh, Avp and Oxt gene expression was studied in the hypothalamus where these genes are known to be expressed but not in the pituitary.
Quantification of Crh mRNA was performed using TaqMan® Universal Master Mix II, and a TaqMan® Gene Expression Assays for Crh (Rn01462137_m1) with a FAM labeled exon spanning probe, and one assay for Actb (Rn00667869_m1) and Gapdh (Rn01775763_g1) which were used as reference genes. PCR conditions were set following the guidelines provided by the manufacturer (Life technologies Europe BV, Stockholm). Primer specificity was checked using melt curve and gel electrophoresis to verify the amplification of a single product (Supplementary Figure S1).
Data Analysis
Data of the relative fluorescence unit (RFU) were collected and PCR efficiency and threshold values were calculated using LinregPCR software (http://www.hartfaalcentrum.nl/index.php?main=files&sub=0) (Ruijter et al., 2009). Cq values were adjusted across the plates using correction factors derived from threshold and PCR efficiency values. Samples with normalized Cq values that had a standard deviation of more than 0.5 were excluded. Relative gene transcripts levels were determined using the ΔCT method (Biorad real time PCR application guide). All the laboratory and pre-processing analyses were performed in a blind manner.
DNA Methylation Analyses
Considering previously shown association of DNA methylation with ELS for Fkbp5, Pomc and Avp (Murgatroyd et al., 2009; Klengel et al., 2013; Wu et al., 2014), as well as the proposed pharmacological relevance of Crhr1 in AUD treatment (Gehlert et al., 2007), these four genes were chosen to investigate potential DNA methylation differences in their promoter region (Supplementary Table S2). The criteria used to select the target amplicons were the presence of Transcription Factor Binding Sites (TFBSs) and/or CpG islands in the promoter region, preferably close to the Transcription Start Site. In addition, previous studies that investigated DNA methylation patterns of the genes of interest were considered. Regarding the Pomc gene, the selected amplicon was a 268 bp long region comprising a CpG island and many TFBSs (Supplementary Figure S2). The target sequences are reported in Supplementary Table S2; Supplementary Figure S2.
The bisulphite quantitative pyrosequencing technique was used to assess the methylation pattern of the four genes using 500 ng of DNA at a concentration of 20 ng/μl (EpigenDx (Hopkinton, MA, USA) (Supplementary Material). To estimate errors, 10% of the samples (9 out of 81) were duplicated in each assay. No significant difference was present between duplicates as assessed by the Wilcoxon signed-rank test. The average difference between the duplicates for any gene was <2%, indicating a 98% replicability of each gene assay. To verify the efficiency of sodium bisulfite DNA conversion, each individual pyrosequencing reaction included a non-CpG cytosine as an internal bisulfite modification control, while low, medium, and high methylated DNA samples were included as controls in each plate. Also, 0, 5, 10, 25, 50, 75 and 100% methylated DNA samples obtained by mixing unmethylated and in vitro methylated DNA were used as standards during the first strand synthesis PCR and pyrosequencing analysis. The percent methylation obtained from the study showed high correlation when compared with expected methylation percentages where correlation coefficient of 0.96 for Avp, 0.97 for Crhr1, 0.98 for Fkbp5, and 0.94 for Pomc, indicating high quality of the data.
Data Analysis
Since the unmethylated CpG sites were converted into uracil, the percentage of methylation was computed by calculating the ratio between both C and T peaks in the pyrogram at the same position using the QCpG software. The calculated percentage was ranging between 100%, indicating the highest methylation, and 0%, indicating no methylation.
Hormone Analysis
Samples were analyzed using the commercial ImmuChemTM Double Antibody Corticosterone 125I RIA kit for rats and mice (MP Biomedicals LLC, Orangeburg, NY, USA) in accordance with the included protocol, with the exception of the addition of one standard (12.5 ng/ml). All samples were analyzed in duplicates. According to the protocol of the RIA kit, the intra-assay variation was 4.4–10.3% and the inter-assay variation 6.5–7.2%. The corticosterone antiserum showed 100% cross-reactivity with corticosterone, while cross-reactivity to other steroids was 0.34% to deoxycorticosterone, 0.10% to testosterone, 0.05% to cortisol, and <0.05% to other tested steroids. Corticosterone levels of the MS rats have been previously reported (Bendre et al., 2015).
Statistical Analyses
To study the effect of ethanol and housing on gene expression, DNA methylation, and corticosterone, between-groups analyses were performed using the Mann–Whitney U test. Mann–Whitney U test was chosen because most of the data for gene expression, DNA methylation (except average methylation) and corticosterone were not normally distributed. Additionally, the GLM two-way ANOVA test with type III sum of squares, which is considered robust to violations of normality (Glass et al., 1972), was performed to assess the interaction and main effects of ELS and ethanol. The Mann–Whitney U test was performed to test group differences between MS groups according to the research questions, i.e., water drinking MS360 vs. water drinking MS15 (effect of ELS), ethanol vs. water drinking MS15 (ethanol-induced effect in MS15 rats), and ethanol vs. water drinking MS360 rats (ethanol-induced effect in MS360 rats). To provide results comparable to the previous literature using AFR animals as controls in MS paradigms, main and interaction effects of MS and ethanol were also assessed in MS360 compared to AFR rats.
Bivariate correlation tests were performed using the Spearman’s rank test. Bonferroni correction for multiple testing was applied. There were performed 105 gene expression vs. gene expression correlations tests; 15 gene expression vs. ethanol intake, and 15 gene expression vs. corticosterone tests, Avp: 5, Crhr1: 14, Fkbp5: 7, and Pomc: 17 gene methylation vs. expression, 50 gene methylation vs. ethanol intake and 50 gene methylation vs. corticosterone correlation tests.
Results
Effects of Single Housing and Voluntary Ethanol Intake in AFR Rats
The effect of single housing and ethanol drinking on gene expression, DNA methylation and corticosterone levels was examined in conventionally reared AFR rats in order to understand how different housing paradigms affect the HPA axis.
Gene Expression (Figure 3)
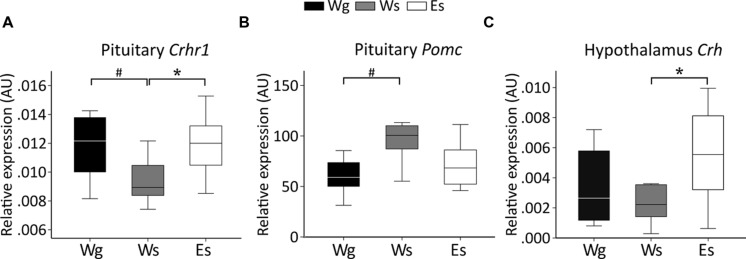
FIGURE 3.
Relative gene expression (median) in animal facility reared rats. Between-group comparisons indicate an effect of voluntary ethanol drinking on (C) Crh in the hypothalamus (U = 20; p = 0.043) and (A) Crhr1 (U = 17; p = 0.012) in the pituitary and an effect of single housing on (A) Crhr1 (U = 14; p = 0.036) and (B) Pomc (U = 14; p = 0.036) in the pituitary. AU, arbitrary units; Crh, corticotropin releasing hormone; Crhr1, corticotropin releasing hormone receptor 1; E, ethanol drinking; g, group housed; Pomc, proopiomelanocortin; s, single housed; W, water drinking.#p ≤ 0.05: effect of single housing; ∗p ≤ 0.05: effect of ethanol.
Single-housed AFR rats had lower Crhr1 expression (Figure 3A) but higher Pomc expression (Figure 3B) in the pituitary gland, compared to group-housed AFR rats. Voluntary ethanol intake was associated with higher Crh expression in the hypothalamus (Figure 3C), and Crhr1 expression in the pituitary (Figure 3A), in comparison with water drinking AFR rats.
Promoter DNA Methylation (Figure 4)
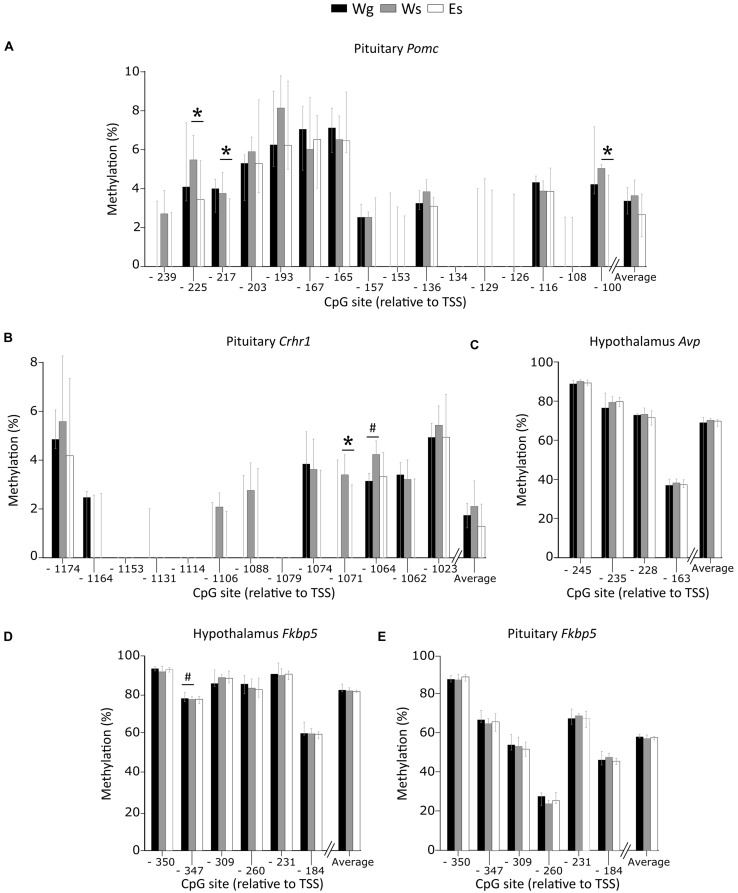
FIGURE 4.
Percent DNA methylation on single CpG sites and average in the AFR rats. Between-group comparisons indicate an effect of voluntary ethanol drinking on methylation of (A) Pomc CpG -225 (U = 15; p = 0.007), -217 (U = 20; p = 0.025), -100 (U = 20.5; p = 0.025), and (B) Crhr1 CpG -1071 (U = 22; p = 0.038) in the pituitary. An effect of single housing was observed on methylation of (B) Crhr1 CpG -1064 in the pituitary (U = 15.5; p = 0.024), and (D) Fkbp5 CpG -347 in the hypothalamus (U = 7; p = 0.029). No effect of ethanol or single housing was observed on the methylation of (C) Avp in the hypothalamus or (E) Fkbp5 in the pituitary. Bars represent median % methylation and 95% confidence interval. Avp, arginine vasopressin; CpG, C—phosphate—G; Crhr1, corticotropin releasing hormone receptor 1; E, ethanol drinking; Fkbp5, FK506 binding protein 5; g, group housed; Pomc, proopiomelanocortin; s, single housed; TSS, transcription start site; W, water drinking #p ≤ 0.05: effect of single housing; ∗p ≤ 0.05: effect of ethanol.
Single housing alone affected methylation patterns with higher methylation of Crhr1 CpG -1064 in the pituitary (Figure 4B), and lower methylation of Fkbp5 CpG -347 in the hypothalamus (Figure 4D), compared to group housed AFR rats. In single housed animals, ethanol drinking AFR rats had lower methylation of Pomc CpG -225, -217, and -100 (Figure 4A) and Crhr1 CpG -1071 (Figure 4B) in the pituitary compared to water drinking AFR rats. Neither single housing nor ethanol intake showed an effect on average DNA methylation of any gene. Promoter DNA methylation of Avp in the hypothalamus (Figure 4C) and Fkbp5 in the pituitary (Figure 4E) were not associated with either ethanol drinking or single housing.
Corticosterone (Figure 5)
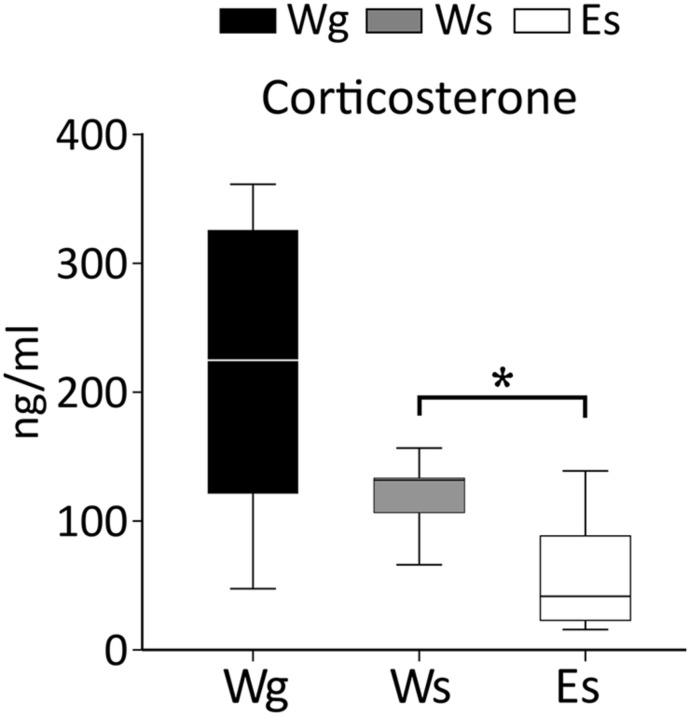
FIGURE 5.
Corticosterone levels in AFR rats. The results indicate an effect of ethanol on corticosterone (U = 14; p = 0.010), E, ethanol drinking; g, group housed; s, single housed; W, water drinking. ∗: Mann–Whitney test p ≤ 0.05.
Single housing in adulthood was not associated with differences in corticosterone levels in AFR rats. Ethanol compared to water drinking AFR rats displayed lower corticosterone. The group housed rats displayed larger within-group variability than the other groups. Such variability was not attributable to diurnal variation because there was no effect of time of day when the rats were decapitated. However, differences in corticosterone levels were observed between the first rat taken from the cage compared to the second and the last rat remaining in the cage.
Effects of ELS and Voluntary Ethanol Intake in MS Rats
To understand how ELS affects genes involved in the HPA axis as well as the interaction with ethanol the main effects of ELS, ethanol drinking, and their interaction, on gene expression, DNA methylation, and corticosterone levels were examined in adult MS rats.
Gene Expression (Figure 6)
FIGURE 6.
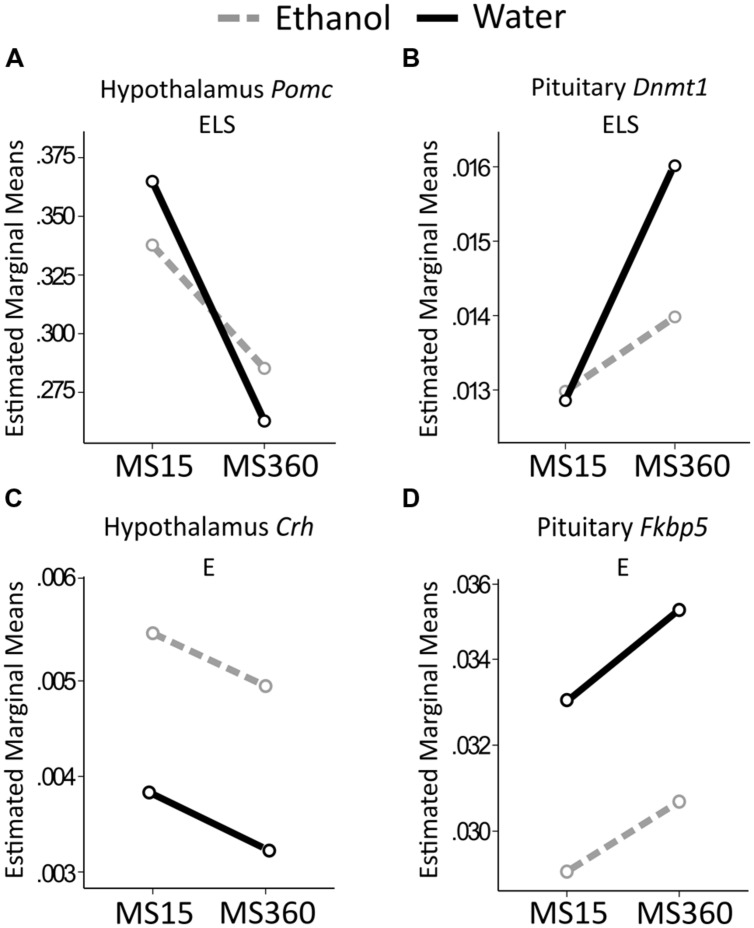
Relative gene expression in the MS rats. The two way ANOVA type III sum of squares test revealed a main effect of ELS on (A) Pomc in the hypothalamus (F = 5.806; p = 0.020; Adj. R2 = 0.060) and (B) Dnmt1 in the pituitary (F = 10.316; p = 0.002; Adj. R2 = 0.175). A main effect of ethanol (E) was found on (C) Crh in the hypothalamus (F = 4.092; p = 0.049; Adj. R2 = 0.026), and (D) Fkbp5 in the pituitary (F = 4.495; p = 0.039; Adj. R2 = 0.039). Crh, Corticotropin releasing hormone; Dnmt1, DNA methyltransferase (cytosine-5) 1; E, ethanol; ELS, early life stress; Fkbp5, FK506 binding protein 5; MS, maternal separation for 15/360 min; Pomc, proopiomelanocortin.
Early life stress had a main effect on Pomc expression in the hypothalamus where MS360 rats had lower expression than MS15 rats, irrespectively of drinking water or ethanol (Figure 6A). Furthermore, a main effect of ELS was present on expression of Dnmt1 in the pituitary (Figure 6B) where MS360 rats had higher Dnmt1 expression than MS15 rats. A focused between-group analysis revealed that water drinking MS360 rats had higher Dnmt1 expression in the pituitary compared to water drinking MS15 rats (U = 21, p = 0.029). Furthermore, ethanol drinking MS360 rats had lower Dnmt1 expression (U = 44, p = 0.013) compared to water drinking MS360 rats whereas ethanol drinking had no impact on expression in the MS15 rats (Figure 6B). A main effect of ethanol intake was associated with higher Crh expression in the hypothalamus (Figure 6C) and lower Fkbp5 expression in the pituitary (Figure 6D) in ethanol drinking rats irrespective of early life rearing condition. No interaction effect was observed on expression of any gene studied.
Promoter DNA Methylation (Figure 7)
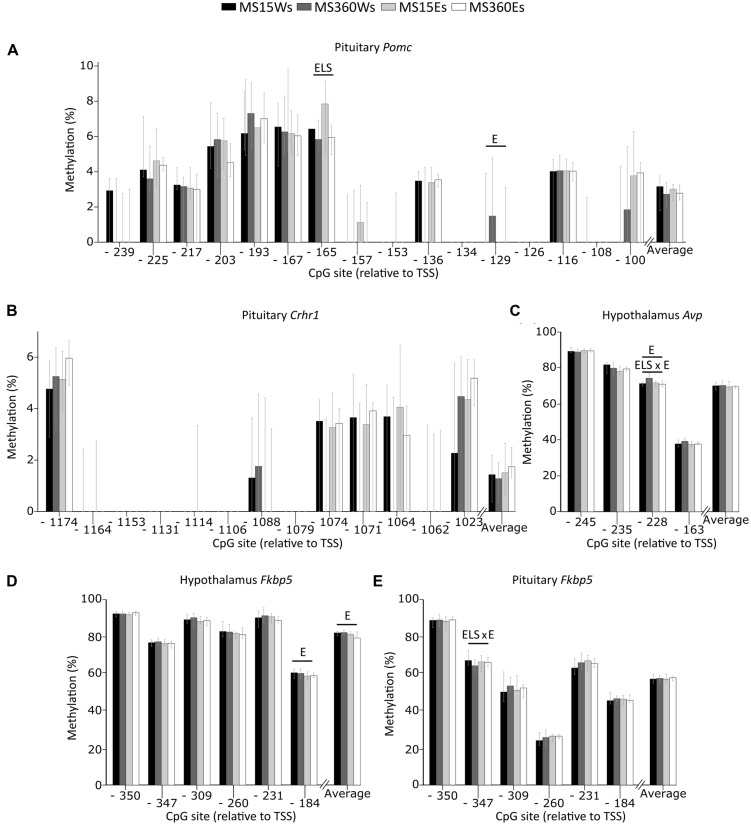
FIGURE 7.
Percent DNA methylation at single CpG sites and average in the MS rats. The two way ANOVA type III sum of squares test revealed a main effect of ELS on methylation of (A) Pomc CpG -165 in the pituitary (F = 7.473; p = 0.009; Adj. R2 = 0.097) as well as a main effect of ethanol (E) on methylation of (A) Pomc CpG -129 (F = 4.766; p = 0.034; Adj. R2 = 0.082) in the pituitary and (C) Avp CpG -228 (F = 4.081; p = 0.049; Adj. R2 = 0.216), (D) Fkbp5 CpG -184 (F = 5.104; p = 0.029 Adj. R2 = 0.043) as well as average (D) Fkbp5 methylation (F = 5.547; p = 0.023; Adj. R2 = 0.060) in the hypothalamus. An interaction between ELS and ethanol effect (ELS × E) was observed on methylation of (C) Avp CpG -228 in the hypothalamus (F = 9.578; p = 0.003; Adj. R2 = 0.216), and (E) Fkbp5 CpG -347 in the pituitary (F = 5.777; p = 0.020; Adj. R2 = 0.082). No effect of stress or ethanol was observed on the methylation of (B) Crhr1 in the pituitary. Bars represent median % methylation and 95% confidence interval Avp, arginine vasopressin; CpG, C—phosphate—G; Crh, corticotropin releasing hormone; Crhr1, corticotropin releasing hormone receptor 1; E, ethanol; Fkbp5, FK506 binding protein 5; Pomc, proopiomelanocortin; ELS, early life stress; TSS, transcription start site.
In the pituitary, a main effect of ELS was associated with lower Pomc CpG -165 DNA methylation in MS360 than MS15 rats (Figure 7A). This effect was mainly driven by the lower methylation on Pomc CpG -165 in ethanol drinking MS360 compared to MS15 rats (U = 51, p = 0.031). Water drinking MS360 rats had higher methylation of Avp -228 compared to MS15 (U = 6, p < 0.001). Furthermore, ethanol intake had a main effect on Pomc CpG -129 where ethanol drinking rats had lower methylation compared to water drinking rats (Figure 7A). A main effect of ethanol intake was present in the hypothalamus on Avp CpG -228, and there was also an interaction between ELS and ethanol intake for Avp CpG -228 methylation driven by lower methylation in ethanol drinking MS360 compared to water drinking MS360 rats (U = 27, p = 0.001) (Figure 7C). Fkbp5 CpG -184 (Figure 7D), and average Fkbp5 methylation in the hypothalamus was associated with ethanol drinking rats having lower methylation compared to water drinking rats (Figure 7D). Furthermore an interaction effect between ELS and ethanol intake was observed on Fkbp5 CpG -347 in the pituitary (Figure 7E) indicating that ethanol drinking results in opposite effects in MS15 and MS360 rats. No difference in the promoter DNA methylation of Crhr1 in the pituitary was observed (Figure 7B).
Corticosterone (Figure 8)
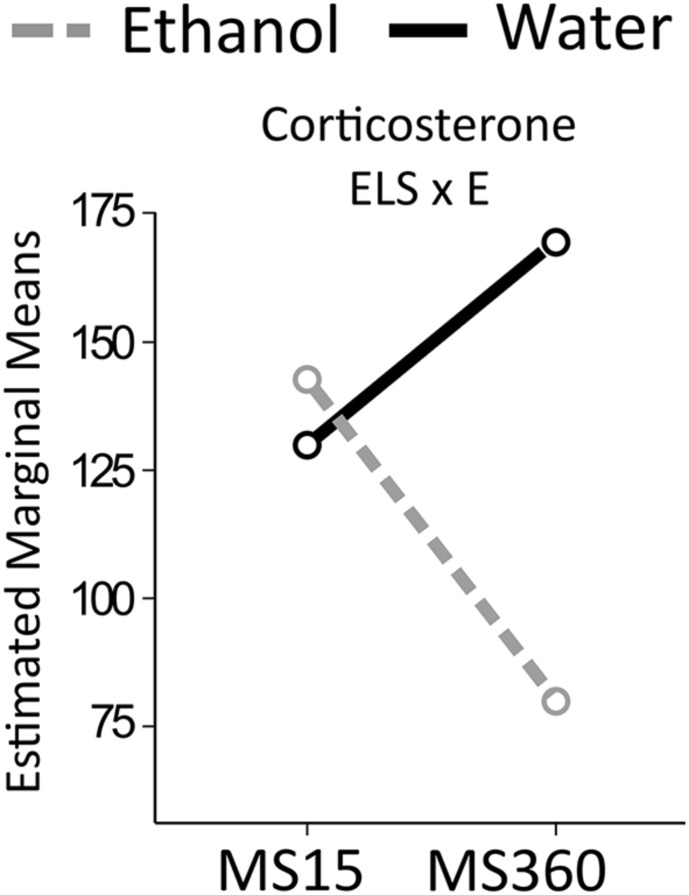
FIGURE 8.
Serum corticosterone levels in the MS rats. The two way ANOVA type III sum of squares test revealed an interaction effect between ELS and ethanol (ELS × E) on corticosterone (F = 5.314; p = 0.027; Adj. R2 = 0.178). MS, maternal separation for15/360 min.
An interaction effect between ELS and ethanol was found on corticosterone levels. Ethanol drinking MS360 rats had lower corticosterone levels (U = 20, p = 0.003) compared to water drinking MS360 rats, whereas no difference was noted between water and ethanol drinking MS15 rats. Ethanol drinking MS360 rats also had lower levels than ethanol drinking MS15 rats (U = 22, p = 0.033).
Body weight of the water drinking rats exposed to ELS was lower compared to MS15 rats (U = 6, p < 0.001). However, body weight as a covariate did not influence the results.
Correlations
Correlation analyses were performed separately in each group to assess the effect of ELS or voluntary drinking on (i) correlations amongst expression of the genes, and (ii) correlations between gene expression, promoter methylation, ethanol intake and corticosterone.
Expression vs. Expression (Table 1)
Table 1.
Group-wise bivariate correlation between expression of genes.
| Gene 1 | Gene 2 | Region | Group | rs | p | |
|---|---|---|---|---|---|---|
| HPA axis - HPA axis | Crhr1 | Nr3c1 | Hypothalamus | MS360 Es | 0.878 | 3.57E-07 |
| Crhr1 | Nr3c1 | Hypothalamus | MS360 Ws | 0.903 | 3.44E-04 | |
| Crhr1 | Oxt | Hypothalamus | MS15 Ws | 0.903 | 3.44E-04 | |
| Crhr1 | Oxt | Hypothalamus | MS360 Ws | 0.964 | 7.32E-06 | |
| Oxt | Avp | Hypothalamus | MS360 Es | 0.753 | 2.01E-04 | |
| HPA axis - DNA methylation | Crhr1 | Dnmt1 | Hypothalamus | MS15 Ws | 0.952 | 2.28E-05 |
| Crhr1 | Dnmt1 | Hypothalamus | MS360 Es | 0.798 | 2.44E-05 | |
| Crhr1 | Dnmt1 | Hypothalamus | MS360 Ws | 0.952 | 2.28E-05 | |
| Nr3c1 | Dnmt1 | Hypothalamus | AFR Es | 0.927 | 1.12E-04 | |
| Nr3c1 | Dnmt1 | Hypothalamus | MS15 Ws | 0.952 | 2.28E-05 | |
| Nr3c1 | Dnmt1 | Hypothalamus | MS360 Es | 0.949 | 1.88E-10 | |
| Nr3c1 | Dnmt1 | Hypothalamus | MS360 Ws | 0.964 | 7.32E-06 | |
| Crhr1 | Mecp2 | Hypothalamus | AFR Es | 0.927 | 3.97E-05 | |
| Crhr1 | Mecp2 | Hypothalamus | MS360 Ws | 0.939 | 5.48E-05 | |
| Oxt | Mecp2 | Hypothalamus | MS360 Ws | 0.939 | 5.48E-05 | |
| Nr3c1 | Mecp2 | Pituitary | MS360 Es | 0.805 | 1.90E-05 | |
AFR, animal facility reared; E, ethanol drinking; g, group housed; MS, maternal separation (15 or 360 min); rs, spearman’s correlation coefficient; s, single housed; W, water drinking.
After Bonferroni correction, strong positive correlations, mainly restricted to the hypothalamus and ELS group, were observed amongst expression of HPA receptor (Nr3c1, Crhr1) and methylation-related genes (Dnmt1 and Mecp2).
Expression vs. Methylation, Ethanol Intake and Corticosterone
After Bonferroni correction, gene expression was not correlated with average gene methylation, except in water drinking MS15 rats where Pomc expression in the pituitary was positively correlated with its average methylation (rs = 0.903; p < 0.001). Regarding methylation at single CpG sites, in group housed AFR rats methylation of Fkbp5 CpG -335 in the hypothalamus (rs = 0.943, p = 0.005) and Fkbp5 CpG -184 in the pituitary (rs = 0.881, p = 0.004) was positively correlated with Fkbp5 expression in the respective brain regions. In ethanol drinking MS360 rats, methylation of Fkbp5 CpG -376 positively correlated with its expression in the pituitary (rs = 0.636, p = 0.003). Gene expression was not correlated with ethanol intake at postnatal week 15 for any gene studied, after Bonferroni correction. A strong positive correlation surviving correction for multiple testing was noted between Dnmt1 expression in the hypothalamus and corticosterone levels only in ethanol drinking MS15 rats (rs = 1.000; p < 0.001).
Methylation vs. Ethanol Intake and Corticosterone
There were no significant correlations, after Bonferroni correction, in any of experimental group between average DNA methylation of any gene and ethanol intake at postnatal week 15, likewise with corticosterone levels.
Comparison of Main and Interaction Effects Using Different Reference Groups
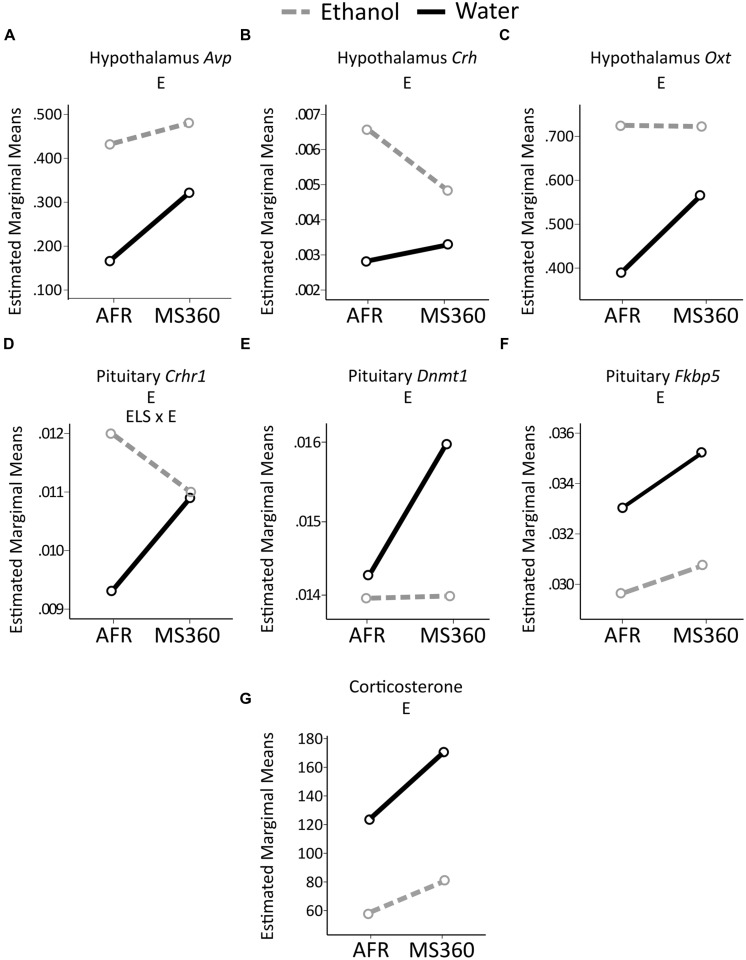
In the literature, MS paradigms with either AFR or MS15 as controls to prolonged MS can be found. Therefore, ELS and ethanol drinking effects were also tested in MS360 compared to AFR rats. Irrespectively of reference group (i.e., MS15 or AFR) a main effect of ethanol intake was found on expression of Fkbp5 in the pituitary and Crh in the hypothalamus as well as no main effect of ELS on corticosterone levels (Table 2). The rest of the results differed depending on the control group used, with more significant associations being present when testing the comparison MS360 vs. AFR rats (Table 2, Figures 6 and 9).
Table 2.
Comparison between group differences using two different reference groups, MS15 or AFR conditions.
| Effect | Measurement | Gene (Region) | MS15 and MS360 Direction | AFR and MS360 Direction |
|---|---|---|---|---|
| ELS | Expression | Dnmt1 (pit) | ↑ in MS360 | – |
| Pomc (hyp) | ↓ in MS360 | – | ||
| Hormone | Corticosterone | – | – | |
| Ethanol | Expression | Fkbp5 (pit) | ↓in ethanol drinking | ↓in ethanol drinking |
| Crh (hyp) | ↑ in ethanol drinking | ↑ in ethanol drinking | ||
| Avp (hyp) | – | ↑ in ethanol drinking | ||
| Dnmt1 (pit) | – | ↓ in ethanol drinking | ||
| Oxt (hyp) | – | ↑ in ethanol drinking | ||
| Crhr1 (pit) | – | ↑ in ethanol drinking (in AFR) | ||
| Hormone | Corticosterone | – | ↓ in ethanol drinking | |
| ELS × ethanol | Expression | Crhr1 (pit) | – | ↑ in AFR, ethanol drinking (no difference in MS360) |
| Hormone | ↓ in MS360, ethanol drinking (no difference in MS15) | – | ||
The bold represents consistent results across the two comparisons. AFR, animal facility reared; E, ethanol drinking; ELS, early life stress; g, group housed; hyp, hypothalamus; MS, maternal separation; pit, pituitary; W, water drinking.
FIGURE 9.
Relative gene expression in the MS360 and AFR rats. The two way ANOVA type III sum of squares test indicates a main effect of ethanol (E) on expression of (A) Avp (F = 8.44, p = 0.006 Adj. R2 = 0.138), (B) Crh (F = 9.17, p = 0.004, Adj. R2 = 0.129) and (C) Oxt (F = 4.39, p = 0.042 Adj. R2 = 0.043) in the hypothalamus as well as on expression on (D) Crhr1 (F = 6.00, p = 0.018, Adj. R2 = 0.114), (E) Dnmt1 (F = 4.79, p = 0.034, Adj. R2 = 0.115) and (F) Fkbp5 (F = 4.71, p = 0.035, Adj. R2 = 0.046) in the pituitary (G) corticosterone levels (F = 22.26, p < 0.001, Adj. R2 = 0.330) and an interaction effect between ELS and ethanol (ELS × E) on expression of (D) Crhr1 (F = 4.58, p = 0.038, Adj. R2 = 0.114). AFR, animal facility reared; MS, maternal separation for 360 min.
Discussion
Effects of Single Housing and Voluntary Ethanol Intake in AFR Rats
The effect of housing was assessed by comparing single and group housed water drinking rats, whereas the effect of voluntary ethanol drinking was assessed by comparing single housed ethanol vs. water drinking AFR rats. Single housing was associated with down-regulation of Crhr1 and up-regulation of Pomc in the pituitary whereas ethanol drinking was associated with up-regulation of Crhr1. Single housing stress during adolescence associates with increased basal ACTH levels, which is derived from Pomc (Weiss et al., 2004), and increased ethanol intake (Wolffgramm, 1990). Furthermore, housing dependent effects of alcohol have been observed on opioid peptides in both adolescent and adult rats (Palm and Nylander, 2014).
On the other hand, single housing has been previously associated with lower ethanol intake in Sprague Dawley rats (Doremus et al., 2005) or no effect on ethanol intake in adult Long Evans or Wistar rats (Schenk et al., 1990; Thorsell et al., 2005). Moreover, single housing has been shown to influence molecular profiles despite having no effect on the ethanol intake (Thorsell et al., 2005). The up-regulation of Crhr1 expression in the pituitary in ethanol drinking rats is in line with the proposed use of Crhr1 antagonism as a treatment for AUD (Funk et al., 2007; Gehlert et al., 2007; Spanagel et al., 2014). However, a recent clinical trial observed no effect of a Crhr1 antagonist on neuroendocrine correlates or behavior in AUD patients (Kwako et al., 2015).
The higher Crh expression observed in the hypothalamus in ethanol drinking rats is likely a result of adaptive mechanisms, suggested by the observed lower corticosterone levels. The finding is in agreement with previous reports showing increased Crh expression in the hypothalamus following acute or short-term ethanol exposure in adult rats (Madeira and Paula-Barbosa, 1999).
Neither single housing nor ethanol drinking was associated with Avp or Oxt expression in the hypothalamus. Chronic ethanol vapor exposure has been associated with both lower (Sanna et al., 1993) and higher Avp expression (Logrip et al., 2013), whereas prolonged oral ethanol did not affect mRNA expression in the paraventricular nucleus (Silva et al., 2002). Lower Oxt peptide levels have been detected in the hypothalamus of mice exposed to single compared to group housing in adulthood (Liu et al., 2013).
Regarding Fkbp5, Nr3c1, Mecp2, and Dnmt1, no effect of ethanol or housing was found. Finally, both ethanol and single housing affected methylation at single CpG sites; however, whether these small differences (<5%) have a biological impact remains unknown.
Effects of ELS and Voluntary Ethanol Intake in MS Rats
Prolonged MS induces long-term changes in various neurotransmitter systems (Gudsnuk and Champagne, 2012; Harrison and Baune, 2014; Maccari et al., 2014), and relates to propensity for excessive voluntary ethanol intake (Roman and Nylander, 2005). Although previous studies support long-term alterations of the HPA axis after MS, the exact nature of the impact of ELS is not understood (Lehmann and Feldon, 2000; Pryce and Feldon, 2003).
Effect of ELS in MS Rats
Down-regulation of Pomc in the hypothalamus was associated with ELS, but no effect was observed on Crh or Crhr1 expression. MS has been associated with increased expression of Crh in the hypothalamus of rats (Plotsky and Meaney, 1993; Huot et al., 2004; Plotsky et al., 2005) and Pomc in the pituitary of mice (Wu et al., 2014). The contradictory results for Crh and Pomc might be due to difference in the duration of MS (180 vs. 360 min), group vs. single housing in adulthood, and species (mice vs. rats) or strains (Long-Evans vs. Wistar). As the product of Pomc mRNA is a prohormone which is cleaved into tissue specific peptides (e.g., ACTH, beta-endorphin) with diverse physiological functions (Smith and Funder, 1988), changes in Pomc gene expression can relate to other physiological conditions than stress. In the present study, ELS had no effect on Avp expression and on methylation of the Avp promoter region, in line with findings on mRNA (Veenema et al., 2006) and peptide levels (Oreland et al., 2010). However, Murgatroyd et al. (2009) reported that mice subjected to MS180 had higher mRNA expression of Avp and Nr3c1 as well as hypomethylation of an enhancer region of Avp in the hypothalamus compared to AFR animals. Combination of methodological differences discussed earlier for Crh and Pomc as well as different genomic loci assessed for methylation herein might explain discordant results. Regarding Fkbp5 or Oxt no effect of ELS was observed.
Effects of Voluntary Ethanol Drinking in MS Rats
All voluntary ethanol drinking rats had higher Crh expression in the hypothalamus. Acute ethanol exposure has been reported to increase Crh expression in the hypothalamus of conventionally reared rats (Madeira and Paula-Barbosa, 1999). This finding is consistent with what observed in AFR rats, indicating a robust effect of ethanol on Crh expression irrespective of rearing condition.
Ethanol drinking rats also had lower Fkbp5 expression in the pituitary irrespective of MS condition. However, the absence of an effect of ethanol drinking in AFR rats suggests Fkbp5 mediated mechanism following exposure to ethanol in the pituitary to be restricted to handling of pups performed in the MS procedure. Moreover, a main effect of ethanol was found on average Fkbp5 promoter methylation in the hypothalamus, but Fkbp5 promoter methylation was not correlated with its gene expression. Reports of ethanol-induced effects on Fkbp5 in HPA axis are lacking. Corticosterone levels in MS360, but not in MS15, rats were sensitive to ethanol as evidenced by lower levels detected only in MS360 rats.
No main effect of ethanol was found on Avp, Crhr1, Oxt, Nr3c1, and Pomc or DNA methylation genes, Dnmt1 and Mecp2. However, between-group analyses revealed that rats exposed to ELS displayed higher Dnmt1 expression in the pituitary compared to controls and that ethanol drinking in these rats resulted in lower Dnmt1 expression, i.e., more close to MS15 rats. This indicates an opposite effect of ethanol drinking and ELS on the methylation machinery. Moreover, in ethanol drinking MS15 rats, Dnmt1 expression in the hypothalamus was positively correlated with corticosterone levels. Corticosterone treatment of pituitary cells has been reported to decrease Dnmt1 expression in a dose dependent manner (Yang et al., 2012) thus a negative correlation can be expected. MS15 is suggested to be protective against risk of AUD (Nylander and Roman, 2013; Nishi et al., 2014). Whether the positive correlation observed herein is underlying a protective mechanism against AUD in MS15 rats needs further investigation. Also, the positive correlation between average methylation and Pomc expression only in water drinking MS15 rats might also indicate a possible underlying mechanism against behavioral abnormalities in adulthood.
DNA Methylation and HPA Axis
In the current study ELS was associated with higher Dnmt1 expression in the pituitary. In vitro mice hippocampal cells challenged with corticosterone showed a dose-dependent decrease in Dnmt1 expression indicating a possible regulatory role in stress physiology (Yang et al., 2012). No effect of ELS was observed on Mecp2. Finally, most of the correlations between genes were found in the hypothalamus and in the MS360 group. The correlations between the expression of HPA receptors (Nr3c1, Crhr1) and DNA methylation regulatory genes (Dnmt1, Mecp2) in the hypothalamus of M360 rats indicate possible epigenetic regulation of the two receptor genes upon exposure to ELS. These correlations thus call for further studies on the molecular mechanisms underlying ELS mediated vulnerability to ethanol drinking.
Methodological Considerations
A well validated MS model was employed to investigate long-term consequences of ELS (Nylander and Roman, 2013). Herein, MS15 was the control based on the notion that short periods of MS are associated with adult low ethanol consumption (Moffett et al., 2007; Nylander and Roman, 2013) as well as it controls for the handling effect. However, the outcome of prolonged separations are depending on the control group (Lehmann and Feldon, 2000; Pryce and Feldon, 2003; Nylander and Roman, 2013), and therefore AFR was also included to improve the comparability with previous studies. When AFR was used as control, more effects of ethanol and ELS were found on gene expression, and only two findings were consistent irrespectively of the control group. The higher number of significant differences in gene expression when AFR animals were used as controls was expected due to the different handling conditions in the two controls (AFR and MS15) and the constant presence of the dam (AFR) vs. the more naturalistic short periods of maternal absence (MS15). The comparison was performed to test a methodological consideration regarding the choice of the control group. These results thus highlight the importance of choosing the control group, and confirm previous reported effects of handling per se (Pryce and Feldon, 2003; Jaworski et al., 2005; Nylander and Roman, 2013). The lower body weight after MS accompanied by higher corticosterone levels observed in the present study has been previously demonstrated (Kalinichev et al., 2002). However, body weight increase was similar between groups over time and it had no effect on the gene expression or corticosterone levels when used as a covariate in the general linear model. Corticosterone levels depend on circadian rhythm and single measurement is a limitation of the present study. On the other hand, the sampling of blood for the measurement of corticosterone levels at the same time of the sampling of the brain for gene expression and DNA methylation analyses makes the correlation tests of relevance.
Moreover, the investigation of the effect of housing allowed detecting possible effect of single housing, a factor often overlooked. Another strength of the present study is the gene set based approach which provides a comprehensive view of effects of ELS and ethanol consumption on the HPA axis. To gain a deeper understanding, proteomic analyses as well as analysis of hypothalamic subnuclei would have been an optimal complement.
Regarding the ethanol intake, the majority of the rats consumed 1–2 g/kg, during the 2 h session. Even though this level of consumption is moderate compared to that of genetically alcohol-preferring rats or rats in the addictive state, it relates to a binge intake that is defined as “a pattern of drinking that brings blood alcohol concentration levels to 0.08 g/dL in about 2 h” according to NIAAA and that clearly has neurobiological effects. However, the purpose of the present study was to investigate early neurobiological signatures related to voluntary alcohol intake in habitual drinking non-preferring rats. In fact, molecular changes concomitant to initial ethanol drinking are likely to contribute to the development of AUD (Koob, 2013). The voluntary and episodic drinking paradigm used herein simulates human episodic drinking patterns (Crabbe et al., 2011; Carnicella et al., 2014). The use of outbred rats gives further strength for translational interpretations since they represent a heterogeneous population compared to alcohol-preferring strains of rodents.
Conclusion
This study provides a comprehensive investigation of stress and DNA methylation regulatory genes. Ethanol displayed opposite effects compared to single housing indicating ethanol drinking to alleviate stress, which is a core factor in AUD. Thus, single housing is a potential confounding factor to be considered in voluntary ethanol drinking paradigms.
The few effects of ELS alone, compared to what expected, might be explained by allostatic mechanisms during development or confounding due to single housing in adulthood. ELS and ethanol drinking in adulthood exerted independent effects on hypothalamic and pituitary related genes, in a manner dependent on the control group used. As no regulatory effect of methylation in the targeted promoter regions was observed, further investigations is needed since correlative relationships between genes regulating DNA methylation and receptor genes in HPA axis were observed in rats exposed to ELS and ethanol drinking. The present study deepens understanding of the ELS mediated changes in genes related to the HPA axis and how they can affect the ELS-mediated propensity to ethanol drinking in adulthood. Moreover, it provides insight into the importance of the choice of the control group while studying the effects of ELS and indicates single housing as a potential confounding factor in ethanol drinking paradigms.
Author Contributions
IN: study concept and design; LG: performing of animal experiment; AT: genetic analyses; AT, MA: epigenetic analyses; AT, EC, IN, KN: statistical analyses; IN, AT, EC, KN: interpretation of findings; AT, EC, IN: writing of the first draft; all authors: critical revision of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to sincerely thank Professor Georgy Bakalkin, Dr. Igor Bazov and Dr. Daniil Sarkisyan for helpful discussion on the methods for epigenetic analyses.
Footnotes
Funding. The study was partially supported by funds from the Swedish Council for Working Life and Social Research (2011-0627), Fredrik and Ingrid Thuring foundation (2012, 2013), Lars Hierta’s Minne foundation (2013), Swedish Brain foundation (PS2013-0052), and Lundberg’s and Karlsson’s foundation (2013) to EC; from the Alcohol Research Council of the Swedish Alcohol Retailing Monopoly, the European Foundation for Alcohol Research (EA 11 30), the Swedish Research Council (K2012-61X-22090-01-3; IN) to IN, and from the Swedish Brain Foundation, the Swedish Alcohol Monopoly Research Council (SRA), the Swedish Council for Working Life and Social Research (FAS/FORTE), the Uppsala and Örebro Regional Research Council, the Fredrik and Ingrid Thurings Foundation, the County Council of Västmanland, the König-Söderströmska Foundation, and the Svenska Spel Research Foundation to KN.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fnmol.2015.00090
References
- Becker H. C., Lopez M. F., Doremus-Fitzwater T. L. (2011). Effects of stress on alcohol drinking: a review of animal studies. Psychopharmacology 218 131–156. 10.1007/s00213-011-2443-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendre M., Comasco E., Nylander I., Nilsson K. W. (2015). Effect of voluntary alcohol consumption on Maoa expression in the mesocorticolimbic brain of adult male rats previously exposed to prolonged maternal separation. Transl. Psychiatry 5:e690 10.1038/tp.2015.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder E. B. (2009). The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology 34(Suppl. 1), S186–S195. 10.1016/j.psyneuen.2009.05.021 [DOI] [PubMed] [Google Scholar]
- Brunton P. J. (2013). Effects of maternal exposure to social stress during pregnancy: consequences for mother and offspring. Reproduction 146 R175–R189. 10.1530/REP-13-0258 [DOI] [PubMed] [Google Scholar]
- Carnicella S., Ron D., Barak S. (2014). Intermittent ethanol access schedule in rats as a preclinical model of alcohol abuse. Alcohol 48 243–252. 10.1016/j.alcohol.2014.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comasco E., Todkar A., Granholm L., Nilsson K. W., Nylander I. (2015). Alphalpha 2a-adrenoceptor gene expression and early life stress-mediated propensity to alcohol drinking in outbred rats. Int. J. Environ. Res. Public Health 12 7154–7171. 10.3390/ijerph120707154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contet C. (2012). Gene expression under the influence: transcriptional profiling of ethanol in the brain. Curr. Psychopharmacol. 1 301–314. 10.2174/2211556011201040301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe J. C., Harris R. A., Koob G. F. (2011). Preclinical studies of alcohol binge drinking. Ann. N. Y. Acad. Sci. 1216 24–40. 10.1111/j.1749-6632.2010.05895.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doremus T. L., Brunell S. C., Rajendran P., Spear L. P. (2005). Factors influencing elevated ethanol consumption in adolescent relative to adult rats. Alcohol. Clin. Exp. Res. 29 1796–1808. 10.1097/01.alc.0000183007.65998.aa [DOI] [PubMed] [Google Scholar]
- Enoch M.-A. (2011). The role of early life stress as a predictor for alcohol and drug dependence. Psychopharmacology 214 17–31. 10.1007/s00213-010-1916-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk C. K., Zorrilla E. P., Lee M. J., Rice K. C., Koob G. F. (2007). Corticotropin-releasing factor 1 antagonists selectively reduce ethanol self-administration in ethanol-dependent rats. Biol. Psychiatry 61 78–86. 10.1016/j.biopsych.2006.03.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehlert D. R., Cippitelli A., Thorsell A., Le A. D., Hipskind P. A., Hamdouchi C., et al. (2007). 3-(4-Chloro-2-morpholin-4-yl-thiazol-5-yl)-8-(1-ethylpropyl)-2,6-dimethyl-imidazo [1,2-b]pyridazine: a novel brain-penetrant, orally available corticotropin-releasing factor receptor 1 antagonist with efficacy in animal models of alcoholism. J. Neurosci. 27 2718–2726. 10.1523/JNEUROSCI.4985-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass G. V., Peckham P. D., Sanders J. R. (1972). Consequences of failure to meet assumptions underlying the fixed effects analyses of variance and covariance. Rev. Educ. Res. 42 237–288. 10.3102/00346543042003237 [DOI] [Google Scholar]
- Gudsnuk K., Champagne F. A. (2012). Epigenetic influence of stress and the social environment. ILAR J. 53 279–288. 10.1093/ilar.53.3-4.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson L., Nylander I. (2006). Time-dependent alterations in ethanol intake in male wistar rats exposed to short and prolonged daily maternal separation in a 4-bottle free-choice paradigm. Alcohol. Clin. Exp. Res. 30 2008–2016. 10.1111/j.1530-0277.2006.00247.x [DOI] [PubMed] [Google Scholar]
- Harrison E. L., Baune B. T. (2014). Modulation of early stress-induced neurobiological changes: a review of behavioural and pharmacological interventions in animal models. Transl. Psychiatry 4:e390 10.1038/tp.2014.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot R. L., Gonzalez M. E., Ladd C. O., Thrivikraman K. V., Plotsky P. M. (2004). Foster litters prevent hypothalamic-pituitary-adrenal axis sensitization mediated by neonatal maternal separation. Psychoneuroendocrinology 29 279–289. 10.1016/S0306-4530(03)00028-3 [DOI] [PubMed] [Google Scholar]
- Jaworski J. N., Francis D. D., Brommer C. L., Morgan E. T., Kuhar M. J. (2005). Effects of early maternal separation on ethanol intake, GABA receptors and metabolizing enzymes in adult rats. Psychopharmacology (Berl) 181 8–15. 10.1007/s00213-005-2232-4 [DOI] [PubMed] [Google Scholar]
- Kalinichev M., Easterling K. W., Plotsky P. M., Holtzman S. G. (2002). Long-lasting changes in stress-induced corticosterone response and anxiety-like behaviors as a consequence of neonatal maternal separation in Long-Evans rats. Pharmacol. Biochem. Behav. 73 131–140. 10.1016/S0091-3057(02)00781-5 [DOI] [PubMed] [Google Scholar]
- Klengel T., Mehta D., Anacker C., Rex-Haffner M., Pruessner J. C., Pariante C. M., et al. (2013). Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat. Neurosci. 16 33–41. 10.1038/nn.3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob G. F. (2013). Theoretical frameworks and mechanistic aspects of alcohol addiction: alcohol addiction as a reward deficit disorder. Curr. Top Behav. Neurosci. 13 3–30. 10.1007/7854_2011_129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwako L. E., Spagnolo P. A., Schwandt M. L., Thorsell A., George D. T., Momenan R., et al. (2015). The corticotropin releasing hormone-1 (CRH1) receptor antagonist pexacerfont in alcohol dependence: a randomized controlled experimental medicine study. Neuropsychopharmacology 40 1053–1063. 10.1038/npp.2014.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann J., Feldon J. (2000). Long-term biobehavioral effects of maternal separation in the rat: consistent or confusing? Long-term biobehavioral effects of maternal separation in the rat: consistent or confusing? Rev. Neurosci. 11 383–408. [DOI] [PubMed] [Google Scholar]
- Liu X., Wu R., Tai F., Ma L., Wei B., Yang X., et al. (2013). Effects of group housing on stress induced emotional and neuroendocrine alterations. Brain Res. 1502 71–80. 10.1016/j.brainres.2013.01.044 [DOI] [PubMed] [Google Scholar]
- Logrip M. L., Rivier C., Lau C., Im S., Vaughan J., Lee S. (2013). Adolescent alcohol exposure alters the rat adult hypothalamic–pituitary–adrenal axis responsiveness in a sex-specific manner. Neuroscience 235 174–186. 10.1016/j.neuroscience.2012.12.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccari S., Krugers H. J., Morley-Fletcher S., Szyf M., Brunton P. J. (2014). The Consequences of early-life adversity: neurobiological, behavioural and epigenetic adaptations. J. Neuroendocrinol. 26 707–723. 10.1111/jne.12175 [DOI] [PubMed] [Google Scholar]
- Madeira M. D., Paula-Barbosa M. M. (1999). Effects of alcohol on the synthesis and expression of hypothalamic peptides. Brain Res. Bull. 48 3–22. 10.1016/S0361-9230(98)00131-2 [DOI] [PubMed] [Google Scholar]
- Moffett M. C., Vicentic A., Kozel M., Plotsky P., Francis D. D., Kuhar M. J. (2007). Maternal separation alters drug intake patterns in adulthood in rats. Biochem. Pharmacol. 73 321–330. 10.1016/j.bcp.2006.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momeni S., Roman E. (2014). Subgroup-dependent effects of voluntary alcohol intake on behavioral profiles in outbred Wistar rats. Behav. Brain Res. 275 288–296. 10.1016/j.bbr.2014.08.058 [DOI] [PubMed] [Google Scholar]
- Moore L. D., Le T., Fan G. (2013). DNA methylation and its basic function. Neuropsychopharmacology 38 23–38. 10.1038/npp.2012.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C., Patchev A. V., Wu Y., Micale V., Bockmuhl Y., Fischer D., et al. (2009). Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat. Neurosci. 12 1559–1566. 10.1038/nn.2436 [DOI] [PubMed] [Google Scholar]
- Nishi M., Horii-Hayashi N., Sasagawa T. (2014). Effects of early life adverse experiences on the brain: implications from maternal separation models in rodents. Front. Neurosci. 8:166 10.3389/fnins.2014.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander I., Roman E. (2013). Is the rodent maternal separation model a valid and effective model for studies on the early-life impact on ethanol consumption? Psychopharmacology 229 555–569. 10.1007/s00213-013-3217-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreland S., Gustafsson-Ericson L., Nylander I. (2010). Short- and long-term consequences of different early environmental conditions on central immunoreactive oxytocin and arginine vasopressin levels in male rats. Neuropeptides 44 391–398. 10.1016/j.npep.2010.06.001 [DOI] [PubMed] [Google Scholar]
- Palm S., Nylander I. (2014). Alcohol-induced changes in opioid peptide levels in adolescent rats are dependent on housing conditions. Alcohol. Clin. Exp. Res. 38 2978–2987. 10.1111/acer.12586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson M., Hulting A. L., Uvnas-Moberg K. (1999). Oxytocin causes a sustained decrease in plasma levels of corticosterone in rats. Neurosci. Lett. 264 41–44. 10.1016/S0304-3940(99)00159-7 [DOI] [PubMed] [Google Scholar]
- Plotsky P. M., Meaney M. J. (1993). Early, postnatal experience alters hypothalamic corticotropin-releasing factor (CRF) mRNA, median eminence CRF content and stress-induced release in adult rats. Brain Res. Mol. Brain Res. 18 195–200. 10.1016/0169-328X(93)90189-V [DOI] [PubMed] [Google Scholar]
- Plotsky P. M., Thrivikraman K. V., Nemeroff C. B., Caldji C., Sharma S., Meaney M. J. (2005). Long-term consequences of neonatal rearing on central corticotropin-releasing factor systems in adult male rat offspring. Neuropsychopharmacology 30 2192–2204. 10.1038/sj.npp.1300769 [DOI] [PubMed] [Google Scholar]
- Pryce C. R., Feldon J. (2003). Long-term neurobehavioural impact of the postnatal environment in rats: manipulations, effects and mediating mechanisms. Neurosci. Biobehav. Rev. 27 57–71. 10.1016/S0149-7634(03)00009-5 [DOI] [PubMed] [Google Scholar]
- Roman E., Nylander I. (2005). The impact of emotional stress early in life on adult voluntary ethanol intake-results of maternal separation in rats. Stress 8 157–174. 10.1080/10253890500188666 [DOI] [PubMed] [Google Scholar]
- Ruijter J. M., Ramakers C., Hoogaars W. M., Karlen Y., Bakker O., Van Den Hoff M. J., et al. (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37 e45. 10.1093/nar/gkp045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna P. P., Folsom D. P., Barizo M. J., Hirsch M. D., Melia K. R., Maciejewski-Lenoir D., et al. (1993). Chronic ethanol intake decreases vasopressin mRNA content in the rat hypothalamus: a PCR study. Mol. Brain Res. 19 241–245. 10.1016/0169-328X(93)90035-N [DOI] [PubMed] [Google Scholar]
- Schenk S., Gorman K., Amit Z. (1990). Age-dependent effects of isolation housing on the self-administration of ethanol in laboratory rats. Alcohol 7 321–326. 10.1016/0741-8329(90)90090-Y [DOI] [PubMed] [Google Scholar]
- Silva S. M., Madeira M. D., Ruela C., Paula-Barbosa M. M. (2002). Prolonged alcohol intake leads to irreversible loss of vasopressin and oxytocin neurons in the paraventricular nucleus of the hypothalamus. Brain Res. 925 76–88. 10.1016/S0006-8993(02)03346-2 [DOI] [PubMed] [Google Scholar]
- Smith A. I., Funder J. W. (1988). Proopiomelanocortin processing in the pituitary. Central nervous system, and peripheral tissues. Endocr. Rev. 9 159–179. [DOI] [PubMed] [Google Scholar]
- Spanagel R., Noori H. R., Heilig M. (2014). Stress and alcohol interactions: animal studies and clinical significance. Trends Neurosci. 37 219–227. 10.1016/j.tins.2014.02.006 [DOI] [PubMed] [Google Scholar]
- Szyf M. (2013). How do environments talk to genes? Nat. Neurosci. 16 2–4. 10.1038/nn.3286 [DOI] [PubMed] [Google Scholar]
- Thorsell A., Slawecki C. J., Khoury A., Mathe A. A., Ehlers C. L. (2005). Effect of social isolation on ethanol consumption and substance P/neurokinin expression in Wistar rats. Alcohol 36 91–97. 10.1016/j.alcohol.2005.07.003 [DOI] [PubMed] [Google Scholar]
- Tsigos C., Chrousos G. P. (2002). Hypothalamic-pituitary-adrenal axis, neuroendocrine factors and stress. J. Psychosom. Res. 53 865–871. 10.1016/S0022-3999(02)00429-4 [DOI] [PubMed] [Google Scholar]
- Veenema A. H., Blume A., Niederle D., Buwalda B., Neumann I. D. (2006). Effects of early life stress on adult male aggression and hypothalamic vasopressin and serotonin. Eur. J. Neurosci. 24 1711–1720. 10.1111/j.1460-9568.2006.05045.x [DOI] [PubMed] [Google Scholar]
- Weiss I. C., Pryce C. R., Jongen-Rêlo A. L., Nanz-Bahr N. I., Feldon J. (2004). Effect of social isolation on stress-related behavioural and neuroendocrine state in the rat. Behav. Brain Res. 152 279–295. 10.1016/j.bbr.2003.10.015 [DOI] [PubMed] [Google Scholar]
- Wolffgramm J. (1990). Free choice ethanol intake of laboratory rats under different social conditions. Psychopharmacology (Berl.) 101 233–239. 10.1007/BF02244132 [DOI] [PubMed] [Google Scholar]
- Wu Y., Patchev A. V., Daniel G., Almeida O. F., Spengler D. (2014). Early-life stress reduces DNA methylation of the Pomc gene in male mice. Endocrinology 155 1751–1762. 10.1210/en.2013-1868 [DOI] [PubMed] [Google Scholar]
- Yang X., Ewald E. R., Huo Y., Tamashiro K. L., Salvatori R., Sawa A., et al. (2012). Glucocorticoid-induced loss of DNA methylation in non-neuronal cells and potential involvement of DNMT1 in epigenetic regulation of Fkbp5. Biochem. Biophys. Res. Commun. 420 570–575. 10.1016/j.bbrc.2012.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.