Abstract
The surface of eukaryotic cells is a multi-component fluid bilayer in which glycosylphosphatidylinositol (GPI)-anchored proteins are an abundant constituent. In this review, we discuss the complex nature of the organization and dynamics of GPI-anchored proteins at multiple spatial and temporal scales. Different biophysical techniques have been utilized for understanding this organization, including fluorescence correlation spectroscopy, fluorescence recovery after photobleaching, single particle tracking, and a number of super resolution methods. Major insights into the organization and dynamics have also come from exploring the short-range interactions of GPI-anchored proteins by fluorescence (or Förster) resonance energy transfer microscopy. Based on the nanometer to micron scale organization, at the microsecond to the second time scale dynamics, a picture of the membrane bilayer emerges where the lipid bilayer appears inextricably intertwined with the underlying dynamic cytoskeleton. These observations have prompted a revision of the current models of plasma membrane organization, and suggest an active actin-membrane composite.
Keywords: lipid rafts, glycosylphosphatidylinositol-anchored protein, nanoscale, diffusion, homo-fluorescence resonance energy transfer, super-resolution, signal transduction, microdomains, actomyosin, activity
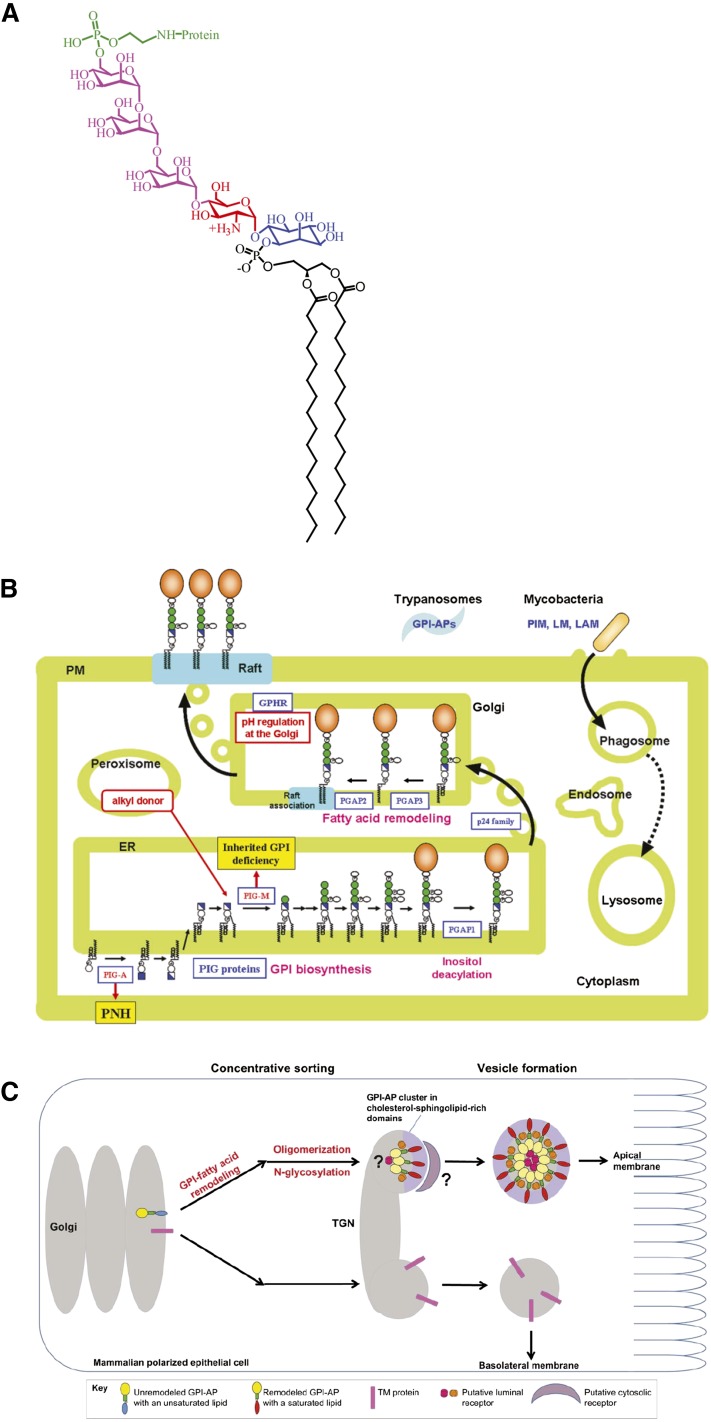
The plasma membrane of the cell is comprised of a diverse set of proteins, many of which are transmembrane (TM) proteins spanning the entire bilayer, but a significant proportion of proteins are also lipid tethered, containing a complex glycan core attached to the C terminus of the protein called glycosylphosphatidylinositol (GPI)-anchored proteins. The basic structure of a GPI-anchored protein consists of phosphatidylinositol (PI) linked to an unusual non-N-acetyl glucosamine, which, in turn, is linked to three mannose residues followed by an ethanolamine covalently linked to the protein via an amide linkage (EtNP-6Manα1-2Manα1-6Manα1-4GlcNα1-6myoinositol-phospholipid, Fig. 1A). Depending on the species and functional context, there may exist variations in the side chain associated with the glycan core. These have been summarized in (1). The lipid moiety is necessary for the incorporation of GPI-anchored proteins into so-called lipid rafts/microdomains (2, 3), which can serve as a sorting station for a number of cell signaling molecules, thereby functioning as a reaction center. GPI-anchored proteins can exist in different forms depending on the context and the tissue in which they are expressed. Alternate splicing can cause the same protein to exhibit TM, soluble, or GPI-anchored forms; for example, neural cell adhesion molecule (NCAM) can exist in its GPI-anchored and soluble form when expressed in muscles; whereas, it takes up a TM form instead of the soluble form in brain. GPI anchoring of proteins occurs at the luminal face of the endoplasmic reticulum (ER), following which, these proteins also undergo a number of posttranslational modifications at the lipid and glycan parts, which play a crucial role in sorting them at the secretory level (4). Use of synthetic GPI analogs have greatly improved our understanding of the structural contribution of the anchor toward the protein’s membrane behavior (5, 6), because perturbing the biosynthetic enzymes that are key to glycan modifications might result in complete disorganization of the structure.
Fig. 1.
GPI-anchored protein structure, biosynthesis, and trafficking. A: Schematic depicts chemical structure of full-length GPI-ethanolamine linked to terminal mannose of a trimannose oligosaccharide via phosphodiester linkage, in turn coupled to glucosamine PI. Reproduced with permission from (5). B: Schematic represents biosynthesis and trafficking of remodeled GPI-anchored proteins, their association to membrane rafts, and diseases caused by defective GPI synthesis. Adapted from T. Kinoshita, unpublished observations. PNH, paroxysmal nocturnal hemoglobinuria. C: Schematic shows the sorting mechanism of GPI-anchored proteins at the trans-Golgi network in polarized epithelial cells. The primary level of sorting involves the segregation of remodeled GPI-anchored proteins into cholesterol- and sphingolipid-enriched domains and other TM proteins. The next level of sorting occurs mainly due to oligomerization effected by the interaction of different receptors with GPI-anchored proteins, which, in turn, might help in vesicle formation and budding. LAM, lipoarabinomannan; LM, lipomannan; PIG, phosphatidylinositol glycan; PIM, phosphatidylinositol mannoside; PM, plasma membrane. Adapted from (132).
GPI-anchored proteins are known to play crucial roles in various cellular processes, such as cell signaling and cell adhesion. This has implications in health and disease, because GPI-anchored proteins carry out diverse functions in different organisms by acting as surface hydrolases, coat proteins, protozoan antigens, toxin binders, receptors, and so on (7). Moreover, impairment of GPI anchoring is also implicated in a large number of diseases, such as the formation of the scrapie form of the prion protein, the causative agent for Creutzfeldt-Jakob disease. Paroxysmal nocturnal hemoglobinuria is caused by the absence of GPI on the membrane. The role of GPI anchoring is necessary for embryonic development (8), and its perturbation is the cause of several neurological disorders (9), abnormal cell growth in yeast (10), and the survival of many protozoan parasites (11). A study of the cell surface dynamics and the organization is crucial to provide an understanding of their many roles.
DELIVERY OF GPI-ANCHORED PROTEINS TO THE PLASMA MEMBRANE
The GPI anchor plays an important role in delivering the attached membrane protein to the plasma membrane, and this has been addressed in a review from Muniz and Reizman also in this thematic review series. The plasma membrane domain to which the GPI-anchored protein localizes also depends on the cell type; for example, in Madin-Darby canine kidney (MDCK) cells, GPI-anchored proteins are present mainly on the apical surface. On the contrary, in Fisher rat thyroid cells, they are sorted to the basolateral domain; the information for targeting the proteins to these distinct locations lies in the GPI anchor sequence (12, 13). The mechanism by which sorting takes place from the Golgi to the appropriate membrane domain also depends on the cell type (13).
Posttranslational modification of the GPI anchor also affects its sorting at the level of the ER. Unlike TM proteins, GPI-anchored proteins do not have a cytoplasmic domain that can directly interact with coat proteins. Hence, the modified GPI anchor efficiently interacts with protein complexes, like the p24 family of proteins, which selectively sort them into COPII-containing vesicles for anterograde transport, thereby establishing quality control at the site of protein attachment itself (14). Knocking down Sec24, an interactor of the p24-p23 complex, was shown to impede the trafficking of GPI-anchored proteins from the ER to the Golgi, while the other cargo molecules remained unaffected (15). In yeast, the p24 protein complex acts as an adaptor between GPI-anchored proteins and the COPII coat by recognizing the remodeled GPI-anchored proteins. On the contrary, in mammalian cells, the p24 protein complex mediates the concentration of GPI-anchored proteins to ER exit sites, because the lipid remodeling occurs much later at the Golgi (14). Posttranslational modifications of GPI-anchored proteins, which involve remodeling enzymes like post GPI attachment to protein (PGAP)1 and PGAP5, are also known to tightly regulate ER-Golgi transport (Fig. 1B) of several GPI-anchored proteins in mammalian cells (14, 16, 17).
The ability of GPI anchor to incorporate proteins into membrane domains helps in sorting these proteins all along the secretory pathway (18). There exist different mechanisms by which sorting can take place in the secretory pathway (Fig. 1C). The sorting takes place at the ER exit site, in the case of yeast, in a sphingolipid-dependent manner; whereas in mammalian cells, this event primarily occurs at the Golgi (14, 19, 20). The sorting process is preceded by posttranslational modification of the lipid tail, so that GPI-anchored proteins are incorporated into specific membrane domains and can be transported efficiently to the cell surface. Depending on the cell type, for example, in MDCK or Fisher rat thyroid cells, GPI-anchored proteins are sorted and routed to the apical or basal membrane, respectively. For the apical sorting of GPI-anchored protein oligomerization, N-glycosylation and raft association have been shown to be important (13); although the underlying mechanism for basolateral sorting of GPI-anchor proteins is not fully understood (21). Synthetic chemistry approaches have led to the development of modified GPI analogs and, hence, allowed studying the trafficking of analogs with modified glycan cores, which showed that despite exhibiting a minor difference in the efficiency of membrane incorporation, the trafficking and endocytic fates of these analogs with differing glycan core does not show an appreciable change (22). However, there is compelling evidence that these analogs differ hugely in their diffusion properties, possibly due to their differential engagement with membrane molecules (6). Further studies utilizing this powerful synthetic tool would provide answers to several questions prevailing in the field.
THE BEHAVIOR OF GPI-ANCHORED PROTEINS AT THE CELL SURFACE
Once at the cell surface, GPI-anchored proteins exhibit a rich diversity of dynamic behaviors in terms of their diffusion, organization, and interactions with other membrane-resident proteins. This has indeed been an active area of research for the past three decades. Studying these behaviors has been instrumental in shaping the modern understanding of how specialized domains in the cell membrane could be organized, maintained, and utilized for signal transduction. In the next sections, we provide a chronological perspective of how the study of GPI-anchored proteins has led to a complex understanding of cell membrane organization and structure.
GPI-ANCHORED PROTEINS AND LATERAL HETEROGENEITIES IN THE CELL MEMBRANE
The plasma membrane has been thought of as a fluid mosaic, as proposed by Singer and Nicholson (23) in 1972. The fluid-mosaic model was a simple, yet elegant, model, rooted in the principles of equilibrium thermodynamics of lipid-protein interactions. Here the cell membrane essentially behaves like a “fluid,” where membrane lipids act as “solvents” for the TM and peripheral membrane proteins. The proposed fluid-like nature of membrane lipids implied the absence of local lateral heterogeneities in the distribution of lipids. However, studies on the lateral heterogeneity of membrane lipid and protein composition (24–26) and their polarized distribution in epithelia (27) suggested that the cell surface and internal membranes were not a well-mixed membrane, and this led to a rethinking of the fluid-mosaic model.
In 1988, Simons and van Meer (27) argued that polarized membrane composition could be achieved by sorting GPI-anchored proteins and sphingolipids into lipidic microdomains at the level of Golgi membranes, followed by their polarized trafficking. However, a biochemical correlate of such membrane domains was lacking. In 1992, Brown and Rose (2) showed that placental alkaline phosphatase, a GPI-anchored protein destined for apical transport in MDCK cells, was enriched in membrane extracts insoluble in cold nonionic detergents, such as Triton X-100 (1% at 4°C). These detergent-resistant membrane (DRM) fractions had very high lipid content and exhibited an enrichment of sphingolipids and cholesterol along with GPI-anchored proteins. The DRMs hence appeared as a biochemical correlate of the functional membrane domains proposed by Simons and van Meer (27). This laid the foundations of the “lipid raft” hypothesis (28, 29) in cellular and functional context, as well as from a biochemical perspective, wherein these “rafts” were envisaged as lateral sorting platforms for specific lipids (sphingolipids), lipid-linked proteins (like GPI-anchored proteins), and other TM proteins (e.g., CD44), which form distinct domains at the cell surface (schematic in Fig. 2D). Moreover, it was shown that the partitioning of GPI-anchored proteins and other raft markers into DRMs depended on cholesterol levels. For example, the depletion of cholesterol led to nearly complete solubilization of GPI-anchored proteins by the same detergents (30). Hence, the ability to partition into DRMs and its sensitivity to cholesterol depletion gained ground as a working definition of lipid rafts in cells. Rafts were envisaged as domains formed by the phase segregation of lipids, as observed in artificial membranes of composition resembling the plasma membrane (31, 32).
Fig. 2.
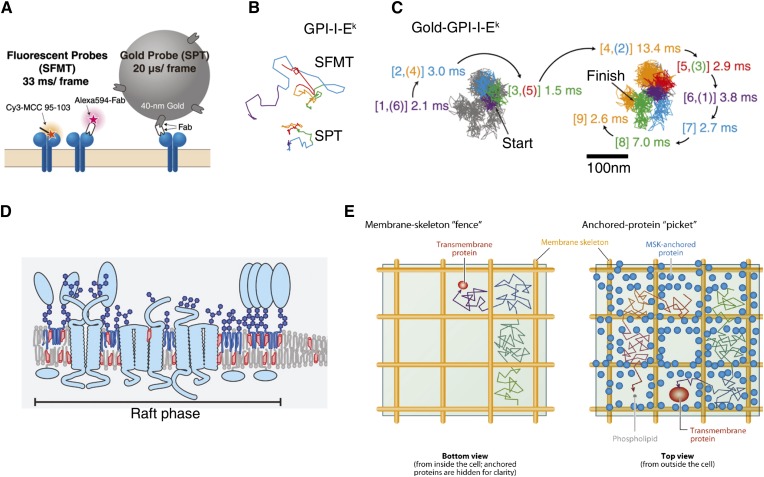
SPT of GPI-anchored protein. A: Probes for single fluorescent-molecule tracking (SFMT, left) and SPT (right) as used in (56). SFMT provides the effective diffusion coefficient on the timescale of 100 ms. GPI-anchored MHC-IIs (GPI-I-Ek) were labeled with either Alexa594-conjugated anti-I-Ek Fab fragments or the Cy3-tagged peptide at its N terminus (left). SPT provides the compartment size sensed by the diffusant. GPI-I-Eks were labeled with anti-I-Ek Fab fragments and then labeled with 40 nm colloidal gold nanoparticle probes coated with anti-mouse IgG antibodies’ Fab fragments (right). B: Representative trajectories for GPI-I-Ek observed at a 33 ms resolution. Representative trajectories in the CHO cell plasma membrane for 3 s (total number of frames, n = 90). The colors (purple, cyan, green, orange, and red) represent the passage of time (every 600 ms or 18 video frames). C: GPI-I-Ek tagged with gold probe and observed at a 20 μs resolution exhibited hop diffusion. Representative 40 ms trajectories (containing 2,000 determined coordinates) of GPI-I-Ek. The residency time within each compartment is shown and is color coordinated with the respective compartment. The numbers in the square brackets indicate the order of the compartments the molecules entered. When repeated passages across the same compartment took place in these trajectories, the compartment is numbered by two numbers [(A–C) have been reproduced with permission from (55)]. D: Schematic showing lipid rafts in cell membrane, which are envisaged as cholesterol- and sphingolipid-rich liquid-ordered domains enriching GPI-anchored proteins and raft-partitioning TM receptors. Adapted from (131). E: A contrasting picture of the membrane structure arose from the SPT studies of Kusumi et al. (133). Schematic of the bottom view (left) of the plasma membrane depicting the MSK fence model and the top view (right) of the membrane representing the anchored TM protein picket model. This model shows that the membrane constituents can undergo short-term confined diffusion within the domains and long-term hop diffusion between neighboring compartments. The actin meshwork-based MSK (fence) and the TM proteins anchored and aligned across the actin fences (pickets) create the compartmental boundaries. Reproduced with permission from (133).
These initial developments gave an impetus to using GPI-anchored proteins as a marker to study the organization and dynamics of the raft domains, using both fluorescence and electron microscopy in intact cell membranes. In one such early study (33), fluorescence microscopy of labeled folate receptor (FR)-GPI in CHO cells showed a diffuse and uniform distribution at the cell membranes. Upon detergent extraction, FR clearly redistributed to detergent-insoluble patches on the membrane. By this time, it was recognized that the process of detergent extraction could induce artifacts by coalescence of microdomains, differential removal of specific lipid species, and inducing changes in fluidity and phase behavior of the native lipids in the membrane (34, 35). Hence, while the direct visualization of native raft domains remained elusive, the focus shifted to other noninvasive or minimally perturbing approaches to study GPI-anchored protein organization under native (-like) conditions.
In the next sections, we focus on the biophysical and microscopy studies on cell-surface dynamics of GPI-anchored proteins, which have given pointers regarding the development of a new model of membrane organization that has been driven by these studies.
DIFFUSIONAL DYNAMICS OF GPI-ANCHORED PROTEINS
Fluorescence recovery after photobleaching (FRAP), single particle tracking (SPT), and fluorescence correlation spectroscopy (FCS) are three widely used tools to study the diffusion of GPI-anchored proteins. Although all three techniques monitor diffusion, they do so over different spatial-temporal regimes, which are in the order of SPT (10–20 nm, ~1 ms) < FCS (∼40 nm–1 μm, ∼100 μs–100 ms) < FRAP (>1 μm, ~1 s) (36). This has indeed provided complementary insights on the scale-dependent diffusion behavior of GPI-anchored proteins, as well as the local organization of the membrane.
Pioneering FRAP studies (37, 38) on GPI-anchored proteins from Jacobson’s group reported that GPI-anchored proteins, such as Thy-1, placental alkaline phosphatase, and Ly6E, exhibit diffusion coefficients comparable to membrane lipids, but ∼2- to 5-fold faster than TM proteins like VSV-G. However, the GPI molecules showed a mobile fraction of only ∼50% across different cell types (38). The origin of this reduced lateral mobility was not well-understood. It was speculated that the size, glycosylation, and interactions of the ecto-domain of the GPI-anchored proteins with the extracellular matrix, other TM receptors, or cortical cytoskeletal corrals could be a potential explanation (37, 39). FRAP-based studies also showed that apically sorted GPIs are immobile just after reaching the cell surface, and their mobility increases as they become long-term residents at the cell surface, suggesting that GPI-anchored proteins are delivered to the membrane in a clustered form (40). Studying the dynamics of GPI-anchored proteins (41) provided a scale dependence to GPI-anchored protein diffusion, which suggested that they diffuse freely as individual molecules over a larger length scale, but may dynamically partition into raft domains. However, the poor spatial resolution, the inherent averaging of dynamics of multiple individual molecules, and a possible convolution of diffusion, turnover, and interactions (like trapping or binding) in a typical FRAP measurement precluded further investigation (42). Hence, studying the dynamics at (or near) the scale of single molecules (or a small group) was necessary to gain a more resolved picture regarding the microscopics of the diffusion process.
The advent of fast (video-rate and higher) and sensitive “low” light detectors in the early 90s coupled with the ability to specifically tag membrane molecules led to the emergence of SPT as a popular method in studying membrane dynamics (42, 43). A unique feature about SPT is the ability to classify the observed particle trajectory into several categories/modes of diffusive behavior like free diffusion, anomalous diffusion, directed motion, confined diffusion, and trapping/stalling. Early SPT studies (44, 45) on GPI-anchored proteins, like Thy-1 and NCAM-125 (a GPI-linked isoform of NCAM), showed that the individual trajectories of both proteins exhibit all the different modes, like stationary, free, anomalous sub-diffusive, and transiently confined diffusion. The most striking observation was that ∼35% of all tracked molecules showed compartmentalized/confined diffusion in compartments of sizes ∼250–300 nm in C3H fibroblasts. These regions/domains were called “transient confinement zones” (TCZs), and GPI-anchored proteins and other glycosphingolipids, like GM-1, were found to be trapped for ∼5–10 s in these cholesterol- and sphingolipid-sensitive domains (44, 46). In parallel, SPT data from the Kusumi group on TM proteins, like cadherin, EGF-receptor, and transferrin receptor, showed similar signatures of diffusional trapping for these proteins into compartments of ∼300 nm in NRK cells (43, 47). To explain these findings, Kusumi and Sako (48) proposed the “membrane-skeleton (MSK) fence” model, wherein the diffusion of TM receptors could be corralled into confined zones by the barrier effects of the membrane underlying the cortical matrix, which include the actin cytoskeleton and spectrin meshwork (Fig. 2E, left). This presented a conundrum about the true nature of TCZs, do they represent MSK fence-bound compartments or cholesterol-sphingolipid-rich “raft domains” (49). Moreover, it was unclear how MSKs could affect the diffusion of outer-leaflet lipid-linked proteins, like GPI-anchored proteins, which, unlike TM receptors, cannot directly interact with the underlying cortex, but show distinctly confined diffusion.
Two new lines of observation attempted to resolve this. The first approach combined particle tracking with optical trapping, wherein an individual (or a small group of) membrane protein (here GPI-anchor protein probes) could be bound to a bead held by the laser trap. The fluctuations of the bead position (50) and the resistance felt by the bead in its movement during stage scanning (51) provided a measure of local viscous drag (50) and the nature and abundance of resistive barriers in diffusion (51), respectively. In conjunction, they showed that the viscous drag and the density of these domains were sensitive to cholesterol levels. The local viscous drag measurements yielded a size of ∼26 nm for individual domains and the force measurements from scanning of beads estimated ∼50–100 domains for every square micron. However, the inherent noise in these measurements precluded any definitive conclusion.
The second approach, pioneered by Kusumi et al. (52), involved enhancing the temporal resolution of SPT measurements to an unprecedented 40,000 frames per second for colloidal gold nanoparticle-labeled species and 10,000 frames per second for fluorescently labeled molecules. In 2002, Fujiwara et al. (53) reported that outer-leaflet phospholipids also showed the signature of compartmentalized diffusion. The high temporal resolution of the particle trajectories showed that particles tend to diffuse freely for ∼1–5 ms in a given compartment before “hopping” onto a neighboring compartment. In a series of SPT-based studies (54, 55), the Kusumi group subsequently showed that GPI-anchored proteins (like GPI-linked MHC-II) exhibit this distinct hop-diffusion behavior (Fig. 2A–C), like other phospholipids and TM proteins (56). These SPT studies were also able to estimate the membrane compartment sizes in many cell lines (55), which closely correlate with the sizes of the juxtamembrane cortical actin (CA) meshwork (57). It was suggested that the diffusion of the outer-leaflet components could be corralled due to the presence of TM-protein “pickets,” which can directly sense the MSK “fences” underneath (52). This led to the more generalized form of MSK concept, the “anchored picket fence” model. These SPT studies clearly implicated the CA network as a major regulator of membrane structure.
Despite these studies, the link between these compartments and the lipid raft-like (cholesterol-sphingolipid-rich liquid-ordered) microdomains remained unclear. An explanation for the cholesterol sensitivity of the TCZs (or any microdomains brought about by lipidic interactions) also comes from the fact that the perturbation of cholesterol can lead to the loss of the PI 4,5-bis phosphate (PIP2) pool from the inner leaflet, effectively disrupting both the membrane-cortex coupling and the organization of the actin cortex (58). This key observation not only linked the TCZs to anchored picket fences, but also hinted at far-reaching implications, that the spatial architecture and topology of the membrane cytoskeleton interactions could dictate the diffusion behavior of TM components, as well as inner and outer leaflet lipids.
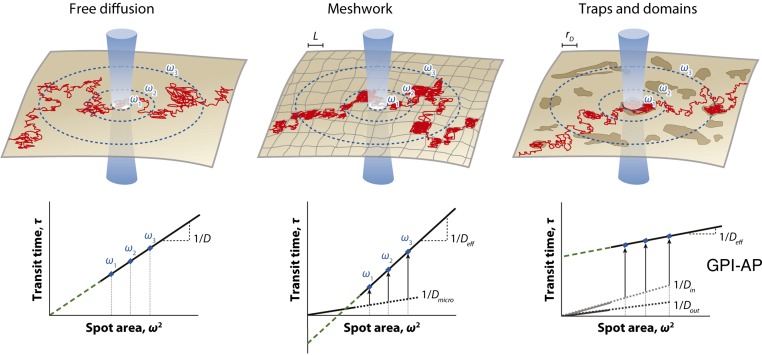
FCS is the third major technique to study the dynamics of GPI-anchored proteins on the cell surface. In FCS, a confocal spot is parked on the cell membrane and the emitted fluorescence is recorded at a very high temporal resolution using avalanche photo diodes over a few seconds (typically 5–10 s). The fluctuation of fluorescence intensity is then autocorrelated to generate the autocorrelation decay over time. This can then be fitted with suitable models describing dynamics (like single component or multi-component diffusion/mobilities/flow, etc.) to extract quantitative measures of diffusion timescales (and their amplitudes) and the number of molecules. Unlike FRAP, FCS monitors equilibrium diffusion and, hence, the measurements are made at sparse molecular density (∼10–100 molecules per square micrometer) and low illumination intensities. This allows monitoring dynamics without the deleterious effects of photobleaching and overexpression, often associated with FRAP studies on cellular physiology. While FCS can readily measure the local density and the diffusion coefficients of membrane molecules, just the value of the diffusion constant is not sufficient to infer on the local membrane environment, which the diffusing probe encounters. However, measuring the diffusion properties across a range of observation areas provided more resolved understanding. In fact, FRAP studies had already exploited the scale-dependence of the mobile fraction and diffusion coefficient to infer on the size of membrane domains (59). Marguet and coworkers implemented this approach in FCS and developed a simulation-based approach to interpret the scale-dependence data. This FCS-based technique, also called spot-variation FCS (sv-FCS), and the scale-dependence of the diffusion timescales (τD) as a function of the spot size were analyzed using the “FCS Diffusion Law” (60). According to this framework (Fig. 3), linear fits of τD versus confocal spot area plots, which pass through the origin (hence have zero y-intercept), were interpreted as simple free diffusion; a positive y-intercept would reflect transient dynamic association with raft-like microdomains and finally a negative y-intercept would signify meshwork compartmentalized diffusion (60, 61). The authors used this to show that Thy1, a GPI-anchored protein, shows a positive y-intercept, suggesting dynamic partitioning into membrane domains of ∼120 nm in size (Fig. 3, right panel) (62). The diffusion behavior of GPI-anchored proteins switched to free diffusion upon cholesterol and sphingomyelin depletion, while actin perturbations did not show any discernible effect (62). Subsequently, the spatial resolution of FCS measurements was improved using two different approaches. Marguet and colleagues also reported the use of nanometric apertures (63) for FCS measurements on the cell membrane, while Stephan Hell’s group used stimulated emission depletion (STED) beam for excitation (64), thereby bringing down the resolution of FCS measurements to ∼40 nm. Both approaches confirmed the cholesterol-sensitive positive y-intercept for GPI-anchored proteins. Further STED-FCS studies suggested that GPI-anchored proteins and sphingolipids could be trapped into lipid shell-like domains of ∼20 nm size, which were likely to be held together by engaging the CA cytoskeleton (65).
Fig. 3.
Diffusion behavior of GPI-anchored protein. sv-FCS is another approach to categorize the diffusion behavior of membrane molecules (top panels) based on the y-intercept of the linear fit to the diffusion time versus confocal spot area data (bottom panels). The schematic represents membrane organization arising from free diffusion, meshwork barriers, and trap/domain confinements with the trajectory drawn for a single molecule (red). Blue circles depict the membrane intersection of the laser beam of waist ω. (bottom panels). FCS diffusion laws represented by plotting the diffusion time τd as a function of the squared radius ω2. D, lateral diffusion coefficient for Brownian motion; Deff, effective diffusion coefficient; Dmicro, microscopic diffusion coefficient; Din, diffusion coefficient inside domains; Dout, diffusion coefficient outside domains; L, size of the side of a square domain; rD, radius of a circular domain. FCS diffusion law scaling from sv-FCS measurements has suggested that GPI-anchored proteins follow the “traps and domains” mode of membrane organization. Reproduced with permission from (135).
In summary, studying diffusion dynamics of membrane molecules such as GPI-anchored proteins has contributed significantly to the evolving picture of cell membrane structure and organization. However, the picture is not complete without understanding the molecular scale organization of GPI-anchored proteins at the cell surface. In the next section, we summarize the studies focused on the nanoscale organization of GPI-anchored proteins and its functional role in cellular physiology.
CELL-SURFACE ORGANIZATION OF GPI-ANCHORED PROTEINS
Different microscopy approaches have been employed to unravel the organization of GPI-anchored proteins in their native environment. Direct visualization of the membrane domains of these proteins also posed unique challenges on multiple fronts because living cells at (or near) physiological temperatures do not show phase-segregated domains, unlike model membranes. Instead, fluorescence imaging of GPI-anchored proteins shows a uniform distribution under both native conditions and in DRMs (33). Moreover, it was only upon antibody-mediated cross-linking of GPI-anchored proteins that optically resolvable patches could be generated (66), possibly by bringing together the sub-resolution native domains of GPI-anchored proteins. Early studies of GPI-anchored protein organization using electron microscopy also pointed out that GPI-anchored proteins primarily show diffused distribution and clustering only upon cross-linking (66) and in DRMs (33). Although electron microscopy can provide the ultimate resolution, its application is limited to fixed cells only. Moreover, potential artifacts of the fixation and EM preparation process might alter the native organization of the molecule studied. However, the early electron microscopy studies hinted of the possible nanoscale organization of GPI-anchored proteins (33, 66, 67).
Proximity measurements based on fluorescence (or Förster) resonance energy transfer (FRET) [reviewed in (67–69)] and, more recently, super-resolution imaging (71, 72) have been utilized to explore the nanoscale organization of the GPI-anchored proteins at the cell surface.
FRET imaging of GPI-anchored protein organization
FRET is a photophysical phenomenon involving the nonradiative energy transfer between a donor fluorophore in excited state and an acceptor fluorophore in ground state (73, 74). The efficiency of energy transfer is inversely proportional to the sixth power of distances between the fluorophores, rendering FRET an ideal “spectroscopic ruler” (75, 76) to probe distances at the biomolecular scales (1–10 nm). Energy transfer can also occur between like fluorophores (homo-FRET) with low Stokes shift and, hence, a significant overlap between their absorption and emission spectra (69, 75). Homo-FRET is estimated by measuring the loss of polarization of emitted fluorescence, which in turn is monitored by determining fluorescence emission anisotropy. Homo-FRET can be estimated from steady-state anisotropy images or from time-resolved anisotropy decays (69). These approaches have been extensively used to study the nanoscale organization of multiple cell surface constituents, such as fluorescent lipid analogs, GPI-anchored proteins, and the EGF receptor (5, 70, 78).
Over the years, homo-FRET (68) has been employed to study the nanoscale clustering of GPI-anchored proteins. Using this technique with steady-state anisotropy as a readout, Varma and Mayor (3) showed that FR-GPI labeled with a fluorescent analog of folate undergoes homo-FRET, suggesting for the first time that these proteins can form small submicron-sized domains at the surface of the living cell. Subsequently, in collaboration with Madan Rao, a theoretical physicist, and his colleagues, the structure and dynamics of these submicron-sized domains were elucidated (79). Perturbation of cholesterol levels leads to a loss of homo-FRET between labeled FR-GPI, suggesting that cholesterol plays a key role in their formation and maintenance. Moreover, cells from a wide range of surface density of FR-GPI showed similar levels of homo-FRET, suggesting that they could be organized into submicron domains (<70 nm), whose dimensions are not dictated by the overall concentration of proteins. The fact that a TM form of FR does not exhibit concentration-independent homo-FRET, underlines the importance of GPI-anchor in dictating the nature of clustering. Although these measurements were made at lower resolution scale of whole cells, which precluded direct observation of domains, these results complemented the chemical cross-linking studies on GPI-anchored proteins, which demonstrated that these proteins could form oligomeric clusters on the cell surface (80).
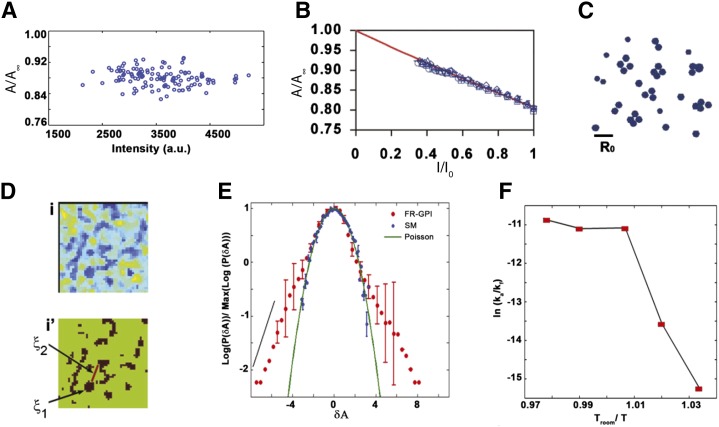
Subsequently, GPI-anchored green fluorescent protein (GFP) and monomeric-yellow fluorescent protein were also shown to form cholesterol-sensitive sub-resolution clusters (79). Time-resolved anisotropy decays of these probes (77) allowed a more direct estimation of the fraction of molecules undergoing FRET and their intermolecular distances. These measurements clearly showed that at least 10–20% of the cell surface pool of both probes are organized into very high density clusters with intermolecular distances of ∼4 nm (79). To estimate cluster size at the nanoscale, a methodology that is based on how fluorescence anisotropy changes when local fluorophore density is reduced via photobleaching was adopted (79, 81). If, indeed, robust homo-FRET occurs in specific arrangements, this should predictably increase distances between fluorophores, thereby reducing FRET probabilities and leading to a systematic increase in anisotropy (Fig. 4A, B). Photobleaching and/or chemical quenching can reduce fluorophore densities. The change in the fluorescence anisotropy upon photobleaching was analyzed by Monte-Carlo-based simulations of a number of expected models of organizing the fluorophores, wherein the expected FRET changes from different nanoscale configurations of probes are able to distinguish the nature of the cluster. This analysis suggested about 20–40% of the cell surface FR-GPIs form oligomers or nanoclusters of two to four molecules each (Fig. 4C) (79).
Fig. 4.
GPI-anchored protein organization by homo-FRET. A: Fluorescence emission anisotropy profile of GPI-anchored proteins plotted against a wide range of intensities (A) or normalized intensities (I/I0) after photobleaching (B) from the membrane of live CHO cells (79, 97). The data clearly show that nanoclustering of GPI-anchored proteins is concentration independent (A) and the change in the anisotropy with decreased intensity upon photobleaching reflects the change in homo-FRET due to GPI-anchored proteins nanoclustering (B). Theoretical modeling of the FRET changes upon photobleaching suggested that GPI-anchored proteins form nanoscale clusters of two to four molecules (C). D: The top panel shows the presence of GPI-anchored protein cluster-rich regions (blue) from the flat surfaces of CHO cells expressing FR-GPI labeled with PLF (fluorescein-conjugated folate analog ligand for FR). The bottom panel represents the thresholded binary map of the region in [D (top)] to clearly show the cluster size ξ1 (≤450 nm) and inter cluster distance ξ2 (∼800–1,250 nm). E: Pixel anisotropy distributions from regions mentioned in [D (top)] show a slower exponentially decaying tail (red dots) which appears as a linear decay (black line) in contrast to the simulated Poisson distribution (green line) or the measured distribution of exogenously incorporated NBD-SM at levels that give rise to homo-FRET, which coincides with a quadratic decay profile (green line). F: Ratio of inter-conversion rates of GPI-anchored protein clustering (represented in an Arrhenius plot) shows a sharp cross-over from temperature dependence to independence at 24°C, implying that cluster remodeling is an active process. Reproduced with permission from (83).
Steady-state anisotropy-based homo-FRET assays also indicated that multiple GPI-anchored proteins, when expressed on the same cell, could intermix and co-cluster at nanoscale (79). The small cluster configuration and the low fraction of the GPI-anchored proteins forming nanoscale clusters can explain why earlier hetero-FRET measurements (82) were not sensitive enough to detect the GPI-anchored protein nanoclusters.
More recently, the implementation of homo-FRET measurement on several microscopy modalities/platforms allowed studying GPI-anchored protein clustering at superior spatiotemporal resolution (68, 83). Homo-FRET imaging at a higher spatial resolution (∼300 nm) showed that GPI-anchored protein nanoclusters exhibit spatially nonrandom distributions (Fig. 4E). Interestingly, cell membrane regions with uniform fluorescence intensity (and hence concentration of FR-GPI) mapped to intricate spatial anisotropy patterns of variegated membrane patches, which were strongly enriched (or devoid) of GPI clusters. This clearly suggests a hierarchical organization wherein nanoscale FR-GPI clusters (∼10 nm) are collected into mesoscale optically resolvable cluster-enriched domains or hotspots (∼450 nm) (Fig. 4D). The clustering of GPI-anchored proteins was also found to be sensitive to the perturbations of cortical actomyosin, as clustering was completely abrogated on membrane blebs devoid of functional actin cortex (83).
Homo-FRET-based photobleaching recovery assays provide a window into cluster remodeling and report on dynamics of cluster remodeling. Using assays at different length scale (FRAP-based, as well as single point photolysis), GPI-anchored protein clusters were ascertained to be relatively immobile, but underwent dynamic remodeling by formation/fragmentation more readily at 37°C than at 20°C. This suggested that the cluster remodeling is an activity-dependent process. Indeed, it was also shown that the dynamics of GPI-anchored protein cluster remodeling are also sensitive to acute perturbations of actin polymerization, myosin activity, and cholesterol (83).
In a more dynamical cellular context, like spontaneously generated membrane blebs which undergo both detachment (during bleb growth) and reattachment (during bleb retraction) to the CA mesh, the anisotropy of EGFP-GPI was monitored, along with the distribution of the C-terminal fragment of ezrin (an F-actin binding protein), to address the interplay between CA activity and local clustering. This assay showed that growing blebs (detached from the underlying cortex) are devoid of EGFP-GPI clusters and clustering recovers gradually on the retracting blebs with reformed functional membrane-cortex links (84). Together these observations show that the nanometer scale organization exhibits unusual properties and is dependent on the CA mesh.
Super-resolution imaging of GPI-anchored protein clustering
The last decade has witnessed significant advances in the field of super-resolution microscopy techniques. Technical breakthroughs in super-resolution imaging have been closely followed by their application to diverse biological contexts (85–87). This has opened up the new possibility of directly studying the molecular clustering of membrane molecules at or near scales (10–100 nm) over which membrane domains are most likely to form and operate (71, 72). An elegant near-field scanning optical microscopy (NSOM)-based study from the Garcia-Parajo group directly demonstrated that endogenous GPI-anchored proteins in monocyte apical membrane form cholesterol-sensitive nanoclusters (88). The inherent single molecule sensitivity of NSOM (Fig. 5A) allowed the authors to precisely calculate the presence of three to five molecules per cluster. The clusters were also found to form cluster-rich “hotspots” (Fig. 5B). These results agreed well with the homo-FRET measurements described above and provided an independent confirmation of GPI-anchored protein nanoclustering. Recently, STED imaging by the same group also detected the presence of actin-sensitive FR-GPI clusters on CHO cells (89). Pair-correlation analysis in conjunction with photo-activation localization microscopy (PC-PALM) imaging also confirmed that GPI-anchored proteins form nanoscale clusters of three to five molecules in Cos-7 cells (Fig. 5D) (90). Perturbation of cholesterol, sphingolipids, and CA led to a loss of clustering, while antibody-mediated cross-linking of GPI-anchored proteins redistributed the GPI-clusters to co-localize strongly with CA. Together, the super-resolution data on GPI-anchored protein nanoscale organization not only confirms the presence and configuration of cholesterol- and actin-sensitive nanoclusters, but also suggests that these clusters can (re)organize to form larger cluster-rich hotspots of ∼100 nm size (Fig. 5E, F). Arguably, this also clarifies our understanding of the mesoscale organization of GPI-anchored proteins and supports the notion of hierarchical organization proposed earlier (83).
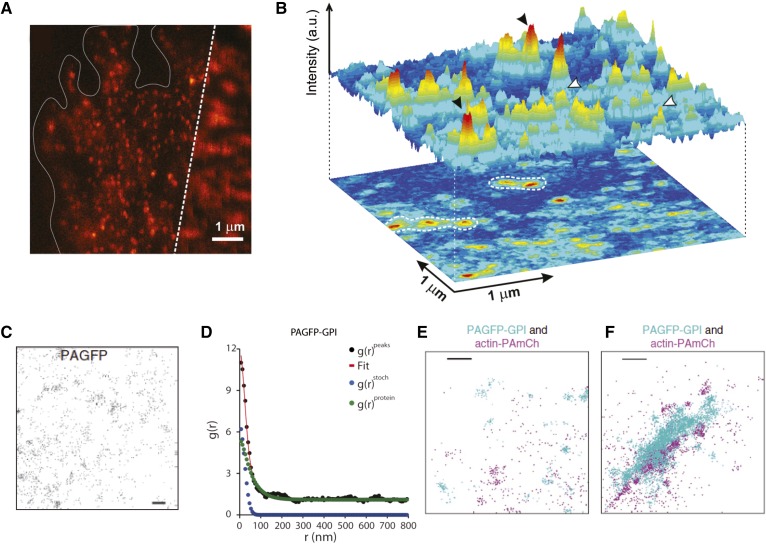
Fig. 5.
GPI-anchored protein organization by super-resolution microscopy. A: Combined confocal (right) and NSOM (left) image of GPI-anchored protein distribution at the cell surface of a fixed monocyte. NSOM imaging provided a direct measure of the cluster configuration of the GPI-anchored proteins. B: 3D projection of an NSOM image with nanodomains (black arrowheads) and monomers (white arrowheads) of GPI-anchored proteins show that these nanoclusters concentrate on specific sites as hotspots (white contour dashed lines). Reproduced with permission from (88). Super-resolution techniques like PALM have been used to study GPI-anchored protein nanoclustering. C: PC-PALM detects the spatial distribution of peak centers of photoactivatable GFP (PAGFP) molecules immobilized on a glass coverslip. D: Plot of calculated autocorrelation function [g(r)peaks] of PAGFP-GPI molecules. The correlation due to multiple appearances of a single protein [g(r)stoch] and the protein correlation [g(r)protein] were evaluated from the fit to clustering model. E, F: Cos-7 cells co-expressing PAGFP-GPI and actin-PAmCh in the absence (E) or presence (F) of cross-linking antibodies. This data shows that actin-PAmCh localizes with clusters of antibody-cross-linked PAGFP-GPI (C–F). Reproduced with permission from (90).
GPI-ANCHORED PROTEIN NANOCLUSTERING REQUIRES TRANSBILAYER ACYL CHAIN COUPLING TO PHOSPHATIDYLSERINE
CA dynamics and cholesterol play a critical role in the clustering and diffusion of GPI-anchored proteins. If CA dynamics are necessary for the creation and maintenance of the nanoclusters, it is imperative that the outer-leaflet GPI-anchored proteins need to sense and interact with the underlying CA. Two possibilities emerge for this connection: either via TM proteins that (in)directly bind CA or by harnessing lipidic interactions laterally (to cholesterol and sphingolipids) and across the bilayer (to inner-leaflet lipids). While TM anchors can certainly corral GPI diffusion, it is unlikely that protein-protein interaction can account for the extremely small and high density GPI clustering, as seen by homo-FRET and other super-resolution microscopies.
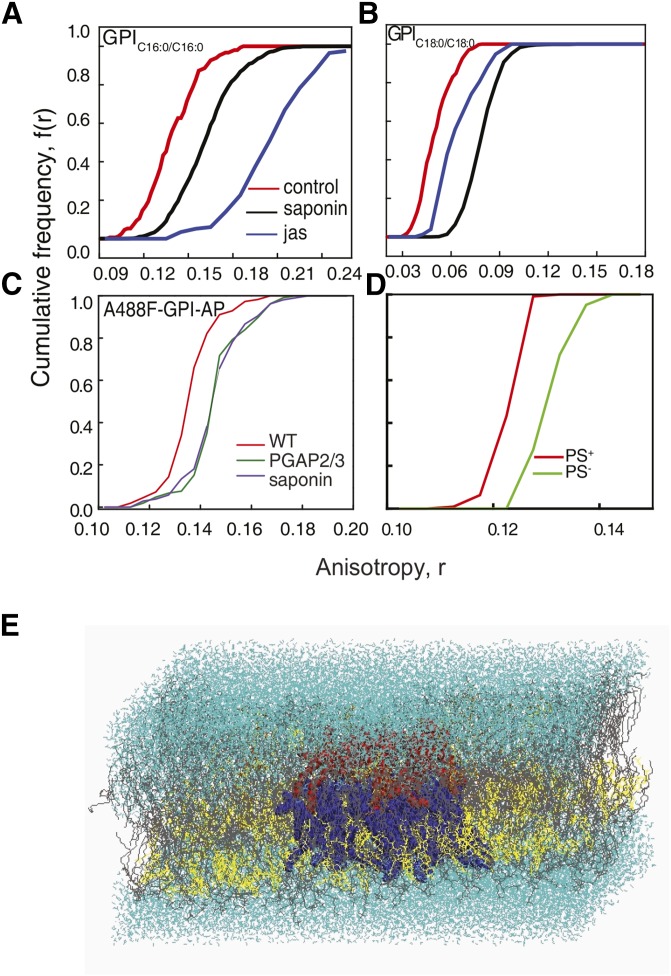
The idea of transbilayer coupling has been proposed previously. Experimental studies on symmetric membranes and phospholipid monolayers supported on immobile hydrocarbon films have hinted toward how lipid composition in the outer leaflet influences the inner-leaflet distribution of lipids (91, 92). Recent studies using free-standing model membrane vesicles (93) and atomistic molecular dynamic simulations (94) show that transbilayer asymmetry, lateral heterogeneity, acyl chain length, and saturation are key prerequisites for transbilayer coupling (Fig. 6E). This makes the GPI anchor’s structure viable for establishing transbilayer coupling between the outer-leaflet protein and the actin cytoskeleton. We find that the chemistry of the fatty acid chain of the GPI anchor is a prerequisite for nanoclustering to occur. Mutation of enzymes affecting lipid remodeling of the GPI anchor, inhibiting the replacement of short and unsaturated acyl chains with long saturated ones, leads to disrupted nanoclustering at the plasma membrane (Fig. 6C). The chemistry of the GPI anchor, particularly the nature of the lipid moiety, is extremely important for organizing into cholesterol-sensitive nanoclusters at the plasma membrane (5) (Fig. 6A, B). Though the underlying mechanism of nanocluster formation is not fully understood, recent work has shown the significance of acyl chain interactions between GPI-anchored proteins and phosphatidylserine (PS) at the inner leaflet that ultimately leads to the formation of clustered domains at the plasma membrane (5). Cells incapable of producing a sufficient amount of PS, due to mutations in enzymes in the biosynthetic pathway of PS, lack the ability to create GPI nanoclusters at the plasma membrane (Fig. 6D). This ability could be restored only on the addition of PS with a long saturated fatty acid chain (18:0). The addition of short (12:0) and unsaturated PS (18:1) into mutant cells failed to rescue nanoclustering of endogenous GPI-anchored proteins.
Fig. 6.
Transbilayer coupling is brought about by long acyl chain interactions of GPI and PS. A: CFD of fluorescence anisotropy measurements performed on GPI analogs, GPIC16:0/C16:0 tagged with BODIPY TMR (A) and GPIC18:0/C18:0 tagged with fluorescein (B), incorporated into cells shows the effect of cholesterol depletion by saponin treatment (black line) or on blebs treated with jasplankinolide (jas; blue line) compared with control cells (red line). C: Fluorescence anisotropy measurements of fluorescently tagged FLAER [Alexa-488-FLAER (A488F), which is a bacterial toxin that binds to the glycan core of GPI-anchored proteins in a monovalent manner) in wild-type and PGAP2/3 double mutant CHO cells: CFD plots of anisotropy measurements for wild-type (red line), PGAP2/3 double-mutant (green line), and saponin-treated (violet line) cells (5). The data clearly show that unremodeled GPI-anchored proteins fail to form nanoclusters. D: CFDs of anisotropy measurements of A488F-labeled PS mutant CHO cells grown with [PS replete (PS+; red line)] or without [PS deplete (PS−; green line)] ethanolamine. The data indicate that endogenous GPI-anchored proteins do not form nanoclusters in the absence of PS. E: Equilibrium snapshot of asymmetric bilayer: upper leaflet [POPC (gray) + 4% PSM (orange), and cholesterol (yellow) + few GPI (red) (Ld phase)]; lower leaflet [POPC (gray) + 35% of cholesterol (yellow) + few PS (blue)]; water in cyan. These data show strong bilayer registry between GPI and PS across the bilayer.
Results from atomistic molecular dynamics simulations done on the asymmetric bilayer have indicated that, along with the adequate levels of cholesterol and the appropriate lipid chemistry on either leaflet, it is also necessary that either of these components in the outer or inner leaflets needs to be immobilized. The actin cytoskeleton, along with myosin, gets reoriented and aids in immobilizing the lipids at the inner leaflet. We hypothesized that PS can form functional links to the underlying actin cytoskeleton via actin-adaptor proteins capable of binding both the membrane lipids and actin, thus unraveling the basic mechanism that leads to membrane domains.
UNIQUE FEATURES OF GPI-ANCHORED PROTEIN CLUSTERING SUGGESTIVE OF ACTIVE ORGANIZATION
Here we summarize the key results obtained from the homo-FRET and super-resolution imaging of GPI clustering. These quantitative approaches have unraveled unique (and peculiar) features of the clustering behavior of GPI proteins, which suggest some rethinking of the existing paradigms (lipid-raft, critical fluctuations, and picket-fence) for the organization of membrane microdomains. 1) GPI-anchored proteins are organized as monomers and nanoscale clusters, whose fraction is independent of the total protein concentration (Fig. 4A), suggesting an organization maintained away from chemical equilibrium (3, 79, 88, 90). 2) The nanoclusters are enriched in optically resolvable cluster-rich domains at steady state; the statistical distribution of nanoclusters and the statistics and spatial distribution of the optically resolvable domains are affected by acute perturbations of cholesterol levels and CA activity (83). 3) The single-point distribution of the local density of GPI-anchored proteins shows prominent exponential tails at both high and low density. This suggests a nonrandom patterning, where the GPI proteins are held at concentrations well beyond equilibrium mixing and segregation (83). 4) The monomers and nanoclusters are highly dynamic and the rates of aggregation-fragmentation (∼0.1 s) are strongly non-Arrhenius (hence they disobey thermal equilibrium). The ratio of the rates is insensitive to temperature in the range 24–37°C (Fig. 4F), and shows a sharp fall below this range coinciding with a sharp reduction in CA activity (83). 5) The rates of fragmentation and reformation of nanoclusters show a wide spatial variation, consistent with the spatial heterogeneity of CA activity. The remodeling of the GPI clustering is also sensitive to CA activity and cholesterol levels (83). 6) The nanoclusters are immobile, whereas the monomers exhibit diffusive motion on the cell surface (56, 83, 95). 7) The nanoclusters fragment into monomers as soon as blebs are spontaneously created on the cell surface, where the cell membrane detaches from the CA (96). The dynamic nanoclusters reform when the bleb retracts due to repolymerization of actin on the cell membrane, recruitment of myosin, and subsequent actomyosin contractility (83). 8) Transbilayer acyl chain coupling with inner-leaflet PS is critical for the clustering of GPI-anchored proteins. This presumably allows the exoplasmic GPI-anchored proteins to link with the actin cortex (5). 9) Local activation and reorganization of actin machinery by integrin activation triggers enhanced clustering of GPI-anchored proteins (88). Conversely, cross-linking of GPI-anchored proteins presumably recruits and hence colocalizes strongly with CA (90).
These features suggest that GPI-anchored protein clustering is an active “non-equilibrium” process, which requires functional links (via PS) between GPIs to cortical actomyosin activity. Recently, an active composite membrane model (97, 98) of the cell surface as a general framework has been proposed to explain the active organization of cell surface molecules (schematic in Fig. 7).
Fig. 7.
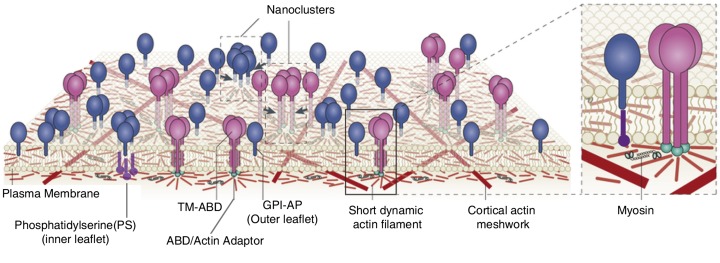
Clustering in active composite membrane. Active composite membrane model proposes that myosin activity (zoomed inset, right) can organize short dynamic actin filaments into “aster-like” configurations. The “aster” can, in turn, drive nanoclustering of outer membrane proteins and lipids, which can couple (in)directly to the short dynamic actin filaments. Outer leaflet GPI-anchored proteins (blue) with long acyl chains do so via transbilayer coupling (5) with inner-leaflet PS connecting to aster via actin adaptors (green). On the other hand, TM-ABD (pink) can couple directly to the underlying actin asters to form distinct nanoclusters. Adapted from (96, 125).
ACTIVE COMPOSITE MEMBRANE MODEL: A CONCEPTUAL FRAMEWORK
The active composite membrane model (97, 98) regards the cell surface as a composite of the multi-component plasma membrane and the membrane juxtaposed CA configuration that it rests on. The CA has a wide distribution of filament length and is expected to be composed of at least two species of actin filaments. The longer filaments form a static cross-linked meshwork (57) along with a pool of short and polar active filaments (97). Active energy-consuming processes, like filament treadmilling and myosin contractility, can drive these filaments. These dynamic short filaments can also couple with the membrane transiently via membrane-actin linkers. This coupling, in turn, can dictate the local organization, dynamics, and environment of the membrane molecules, which can interact with this dynamic CA. Hence, in an active composite membrane, cell surface molecules, based on their coupling to this active CA, are classified into three types: inert, passive, and active. Inert molecules (like short chain lipids) do not interact with CA, while passive (GPI-anchored proteins) and active molecules interact with the dynamic CA. While passive molecules cannot regulate local actin architecture, active molecules (like signaling receptors, integrin and T (or B) cell receptors) can do so via triggering nucleation, (de)polymerization, and cross-linking locally. The model also proposes that passive molecules, like GPI-anchored proteins, are driven into nanoclusters by localized and dynamically remodeling platforms of short actin filaments (Fig. 7). These dynamic platforms, called asters, are emergent structures resulting from the active hydrodynamics of the short filaments driven by filament treadmilling and motor activity (Fig. 7). The active composite model has provided a consistent explanation for the different peculiar features of GPI-anchored protein nanoclustering mentioned earlier (97).
The active composite model offers a paradigm shift in the way we understand membrane organization, because it gives equal importance to inter-molecular interactions as it does to the interaction of cell membrane molecules with the CA.
ACTIVE COMPOSITE MEMBRANE: EXPECTATIONS AND EXPERIMENTS
The active composite model is a framework to understand membrane organization and has faithfully accounted for the clustering behavior of GPI-anchored proteins. However, the framework also assumes a two-component CA architecture and makes general predictions regarding the organization, cell surface dynamics, and distribution of passive molecules. A systematic experimental verification of these predictions, in turn, can reveal newer qualitative and quantitative properties of the membrane and its resident molecules. Indeed, recent experiments have now verified a number of these predictions.
Considerable evidence now exists for the existence of short dynamic actin filaments from FCS measurements and SPT measurements (97). These data are in agreement with earlier biochemical- (99) and electron microscopy-based (100, 101) studies on short actin filaments. Furthermore, the active composite model predicts that a model passive TM probe with an actin binding domain (TM-ABD) will couple to actin asters and, hence, can be nanoclustered. Homo-FRET (97) and STED imaging (89) have confirmed that a model TM-ABD probe can exhibit actin-dependent clustering.
The dynamic CA activity can also impart active fluctuations to drive the diffusion of these molecules. This leads to temperature independence of membrane diffusion of GPI-anchored proteins at length scales similar to those of “actin asters,” ∼200 nm. The active fluctuations can be quelled by perturbations of actin polymerization, myosin activity, and cholesterol, resulting in a reversal of temperature-dependent diffusion akin to inert particles (102). Similarly, active anomalous density fluctuations were also detected in the density (or intensity) distribution of GPI-anchored proteins on the cell surface (97).
PHYSIOLOGICAL RELEVANCE OF GPI-ANCHORED PROTEIN STRUCTURE AND MEMBRANE DYNAMICS
Functional roles of GPI-anchored proteins in cellular physiology
GPI-anchored proteins are known to critically regulate a wide range of physiological functions, from nutrient uptake to cell migration to immune recognition. Therefore, it is obvious that perturbations resulting in altered GPI-anchored protein trafficking, as well as plasma membrane organization, can lead to several diseases. Recent studies have shown that the genes involved in the biosynthesis of GPI-anchored proteins are linked with various diseases because the fully processed GPI anchor is essential for the proper functioning, as well as targeting, of the protein (9, 103–106). Mutations in GPI remodeling enzymes, like PGAP3, are known to generate intellectual disorders, like hyperphosphatasia (9), and deletion of this gene results in impaired T cell receptor signaling (107). Interestingly GPI-anchored proteins are also known to function as cargo transporters, where GPIHBP1, a lipoprotein lipase transporter, carries LPL from subendothelial spaces to the capillary lumen in endothelial cells, thereby initiating lipolysis. The absence of GPIHBP1 leads to diseases such as chylomicronemia, due to elevated triglyceride levels in the plasma (108).
Ras signaling, which is usually associated with plasma membrane, plays a crucial role in the biosynthesis of GPI-anchored proteins in yeast. Experiments done in yeast show that downregulating GPI-anchored protein expression at the plasma membrane leads to hyperactive Ras phenotypes (109). This is an interesting aspect, which connects cell signaling to expression and organization of GPI-anchored proteins at the membrane. Recent studies show that subunits of GPI transamidase can act as oncogenes. For example, overexpression of PIG U leads to an increased cell division rate and a good correlation between the overexpression of these genes and several cancers, like bladder, colon, and ovarian cancers, was also observed (110). Though the direct functional relevance of GPI anchor is not known, it is essential for localizing the proteins into membrane domains, which might have implications in processes like endocytosis (111, 112) or ER-Golgi cargo sorting (113). Formation of domains at the plasma membrane or at the endosomal level might help concentrate the cargo, thereby increasing endocytic and cargo sorting efficiency, respectively. This can even help in determining the fate of the cargo by preferentially sorting them into different domains. A clear example, which links GPI-anchored protein clustering at the plasma membrane and endocytosis, is in the context of MA104 cells where clustering of the FR, which is a GPI-anchored protein, is found necessary for ligand uptake (114). Synthetic GPI analogs, which have minimal structural similarity to endogenous GPI anchor, have been used in studies to dissect out the contribution of the glycan core in protein trafficking (22). Similar studies would prove to be useful for understanding the structural and functional relationship of GPI-anchored proteins.
GPI-hotspots and signal transduction
While many cell-surface GPI-anchored proteins are implicated in diverse signaling roles, it is unclear how an outer-leaflet signaling protein can stimulate intracellular downstream effectors. Studying the membrane organization, interactions, and diffusion of these proteins have also contributed to our understanding of GPI-anchored protein signaling. GPI hotspots are regions on the cell surface that have a high local concentration of clusters (83, 88) or show transiently confined and compartmentalized diffusion (44, 56). Such regions, in turn, can act as signaling platforms for either GPI-anchored signaling proteins or TM signaling receptors, which associate with GPI microdomains. Early biochemical studies (115) showed that antibody-mediated cross-linking of several GPI-anchored proteins, like DAF, CD59, Thy-1, CD14, and Ly-6, led to the association and activation of Src family kinases. This classical assay has been revisited using high-resolution SPT and imaging tools on live cells to uncover the dynamics of the signaling process. Recent studies have shown that antibody-conjugated gold bead-mediated cross-linking of CD 59 leads to the formation of stabilized clusters, which exhibit stimulation-induced temporary arrest of lateral diffusion (also transient anchorage) and recruit signaling effectors, like Gαi2, Lyn kinase, and PLCγ, to elicit a transient IP3-calcium signal (116, 117). The transient anchorage of Thy-1 clusters requires Src family kinase activity and cholesterol (118). The same group identified Csk-binding protein (CBP) as a TM anchor which connects the Thy-1 clusters to CA via ezrin (119).
GPI hotspots also exhibit proximity to nanoclusters of LFA1, an integrin, possibly creating a membrane niche poised for the initiation of cell adhesion (88). Ligand-mediated integrin activation resulted in a dramatic increase in nanoclustering of GPI-anchored proteins, locally driving the formation of nascent adhesion sites (88), which can eventually stabilize into lipid ordered domains in stable focal adhesions (120). In the same context, ECM components can also modulate the dimerization and diffusion of uPAR, the GPI-linked receptor for urokinase (121). Interestingly, uPAR also interacts with integrin and uses integrin as a coreceptor for its signaling (122), pointing toward yet another functional link between GPI-clusters and integrin signaling.
FUTURE DIRECTIONS AND PERSPECTIVE
GPI-anchored proteins are a diverse set of cell surface molecules with unique structural features that modulate the transport and function of the proteins. Here, we have provided an overview of the contemporary understanding of membrane diffusion and organizational features of the GPI-anchored proteins. We have focused primarily on the biophysical measurements of these properties of GPI-anchored proteins. We have also highlighted the bearing of the structure and membrane dynamics of GPI-anchored proteins in a functional context. It is clear that studying GPI-anchored protein organization and dynamics has greatly contributed to the evolving picture of membrane structure. This has now led to the Active Composite Model of the plasma membrane (97, 98), which incorporates the contribution from the association of the dynamic actin filaments to the membrane, along with the structuring influence of the static meshwork on membrane organization (from the picket fence model). Arguably, studying GPI-anchored proteins has been fundamental to the evolving landscape of conceptual models describing membrane structure and organization.
However, many unresolved issues remain in GPI-anchored protein cell surface dynamics, organization, and function. GPI-anchored proteins are unique molecules because they require both lipidic interaction and actin activity for their membrane organization and dynamics (83). This very fact makes them an ideal candidate probe for mechanistic studies on membrane-CA interaction and how the cell might leverage such interactions to create membrane domains or regulate mobilities. To strengthen the role of dynamic actin in organizing GPIs, it will be crucial to study the clustering or diffusion of GPI-anchored proteins in conjunction with the monitoring of the local actin dynamics. This will entail the coupling of proximity or super-resolution microscopy of native GPI-anchored proteins or synthetic analogs to the particle tracing approaches for the actin markers. Imaging assays based on dual-color SPT-PALM (123) or spatiotemporal image correlation spectroscopy (124) might provide crucial clues.
Moreover, genetic and biochemical approaches would be necessary to unravel the nature of the molecular linker(s) (and their regulation) between the GPI-anchored proteins and actin machinery (125). Understanding such functional links and their bearing on the membrane dynamics will naturally allow us to probe the properties of the membrane domains GPI-anchored protein’s form at the cell surface and how such domains can be endocytosed (126). Addressing this will necessitate better assays to relate the local composition of the membrane milieu to the clustering of GPI-anchored proteins. Multimodal approaches combining the measure of local lipid order (127–129) and clustering or imaging-mass spectroscopy tools like NanoSIMS (130, 131) could be key.
Finally, many GPI-anchored proteins have well-defined roles in cellular signaling (7, 132); hence allowing study of the dynamics of membrane proximal signaling events in physiological contexts (133). This also extends the possibility of making progress in our fundamental understanding of the biology of GPI-related diseases (134).
Acknowledgments
The authors thank Prof. Madan Rao for stimulating interactions and other members of the Mayor Laboratory for their support and discussions. The authors also thank the Central Imaging Facility at the National Centre for Biological Sciences for microscopy infrastructure.
Footnotes
Abbreviations:
- CA
- cortical actin
- CFD
- cumulative frequency distribution
- DRM
- detergent-resistant membrane
- ER
- endoplasmic reticulum
- FCS
- fluorescence correlation spectroscopy
- FR
- folate receptor
- FRAP
- fluorescence recovery after photobleaching
- FRET
- fluorescence (or Förster) resonance energy transfer
- GFP
- green fluorescent protein
- GPI
- glycosylphosphatidylinositol
- MDCK
- Madin-Darby canine kidney
- MSK
- membrane-skeleton
- NCAM
- neural cell adhesion molecule
- NSOM
- near-field scanning optical microscopy
- PALM
- photo-activation localization microscopy
- PC
- pair-correlation
- PGAP
- post glycosylphosphatidylinositol attachment to protein
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- SPT
- single particle tracking
- STED
- stimulated emission depletion
- sv-FCS
- spot-variation fluorescence correlation spectroscopy
- TCZ
- transient confinement zone
- TM
- transmembrane
- TM-ABD
- transmembrane probe with an actin binding domain
- τD
- diffusion timescale
S.M. is a J. C. Bose Fellow (Department of Science and Technology, Government of India) and acknowledges support from a Human Frontier Science Program Grant (RGP0027/2012). S.S. and A.A.A. acknowledge fellowship support from the NCBS-TIFR graduate program and Council of Scientific and Industrial Research, Government of India, respectively.
REFERENCES
- 1.Orlean P., and Menon A. K.. 2007. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J. Lipid Res. 48: 993–1011. [DOI] [PubMed] [Google Scholar]
- 2.Brown D. A., and Rose J. K.. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 68: 533–544. [DOI] [PubMed] [Google Scholar]
- 3.Varma R., and Mayor S.. 1998. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 394: 798–801. [DOI] [PubMed] [Google Scholar]
- 4.Kinoshita T. 2014. Biosynthesis and deficiencies of glycosylphosphatidylinositol. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 90: 130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raghupathy R., Anilkumar A. A., Polley A., Singh P. P., Yadav M., Johnson C., Suryawanshi S., Saikam V., Sawant S. D., Panda A., et al. 2015. Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell. 161: 581–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paulick M. G., Wise A. R., Forstner M. B., Groves J. T., and Bertozzi C. R.. 2007. Synthetic analogues of glycosylphosphatidylinositol-anchored proteins and their behavior in supported lipid bilayers. J. Am. Chem. Soc. 129: 11543–11550. [DOI] [PubMed] [Google Scholar]
- 7.Paulick M. G., and Bertozzi C. R.. 2008. The glycosylphosphatidylinositol anchor: a complex membrane-anchoring structure for proteins. Biochemistry. 47: 6991–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murata D., Nomura K. H., Dejima K., Mizuguchi S., Kawasaki N., Matsuishi-Nakajima Y., Ito S., Gengyo-Ando K., Kage-Nakadai E., Mitani S., et al. 2012. GPI-anchor synthesis is indispensable for the germline development of the nematode Caenorhabditis elegans. Mol. Biol. Cell. 23: 982–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howard M. F., Murakami Y., Pagnamenta A. T., Daumer-Haas C., Fischer B., Hecht J., Keays D. A., Knight S. J. L., Kölsch U., Krüger U., et al. 2014. Mutations in PGAP3 impair GPI-anchor maturation, causing a subtype of hyperphosphatasia with mental retardation. Am. J. Hum. Genet. 94: 278–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leidich S. D., Drapp D. A., and Orlean P.. 1994. A conditionally lethal yeast mutant blocked at the first step in glycosyl phosphatidylinositol anchor synthesis. J. Biol. Chem. 269: 10193–10196. [PubMed] [Google Scholar]
- 11.Nagamune K., Nozaki T., Maeda Y., Ohishi K., Fukuma T., Hara T., Schwarz R. T., Sutterlin C., Brun R., Riezman H., et al. 2000. Critical roles of glycosylphosphatidylinositol for Trypanosoma brucei. Proc. Natl. Acad. Sci. USA. 97: 10336–10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lisanti M. P., Sargiacomo M., Graeve L., Saltiel A. R., and Rodriguez-Boulan E.. 1988. Polarized apical distribution of glycosyl-phosphatidylinositol-anchored proteins in a renal epithelial cell line. Proc. Natl. Acad. Sci. USA. 85: 9557–9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imjeti N. S. S., Lebreton S., Paladino S., de la Fuente E., Gonzalez A., and Zurzolo C.. 2011. N-Glycosylation instead of cholesterol mediates oligomerization and apical sorting of GPI-APs in FRT cells. Mol. Biol. Cell. 22: 4621–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujita M., Watanabe R., Jaensch N., Romanova-Michaelides M., Satoh T., Kato M., Riezman H., Yamaguchi Y., Maeda Y., and Kinoshita T.. 2011. Sorting of GPI-anchored proteins into ER exit sites by p24 proteins is dependent on remodeled GPI. J. Cell Biol. 194: 61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonnon C., Wendeler M. W., Paccaud J-P., and Hauri H-P.. 2010. Selective export of human GPI-anchored proteins from the endoplasmic reticulum. J. Cell Sci. 123: 1705–1715. [DOI] [PubMed] [Google Scholar]
- 16.Fujita M., Maeda Y., Ra M., Yamaguchi Y., Taguchi R., and Kinoshita T.. 2009. GPI glycan remodeling by PGAP5 regulates transport of GPI-anchored proteins from the ER to the Golgi. Cell. 139: 352–365. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka S., Maeda Y., Tashima Y., and Kinoshita T.. 2004. Inositol deacylation of glycosylphosphatidylinositol-anchored proteins is mediated by mammalian PGAP1 and yeast Bst1p. J. Biol. Chem. 279: 14256–14263. [DOI] [PubMed] [Google Scholar]
- 18.Surma M. A., Klose C., and Simons K.. 2012. Lipid-dependent protein sorting at the trans-Golgi network. Biochim. Biophys. Acta. 1821: 1059–1067. [DOI] [PubMed] [Google Scholar]
- 19.Castillon G. A., Watanabe R., Taylor M., Schwabe T. M. E., and Riezman H.. 2009. Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic. 10: 186–200. [DOI] [PubMed] [Google Scholar]
- 20.Rivier A. S., Castillon G. A., Michon L., Fukasawa M., Romanova-Michaelides M., Jaensch N., Hanada K., and Watanabe R.. 2010. Exit of GPI-anchored proteins from the ER differs in yeast and mammalian cells. Traffic. 11: 1017–1033. [DOI] [PubMed] [Google Scholar]
- 21.Zurzolo C., Lisanti M. P., Caras I. W., Nitsch L., and Rodriguez-Boulan E.. 1993. Glycosylphosphatidylinositol-anchored proteins are preferentially targeted to the basolateral surface in fischer rat thyroid epithelial cells. J. Cell Biol. 121: 1031–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhagatji P., Leventis R., Comeau J., Refaei M., and Silvius J. R.. 2009. Steric and not structure-specific factors dictate the endocytic mechanism of glycosylphosphatidylinositol-anchored proteins. J. Cell Biol. 186: 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singer S. J., and Nicolson G. L.. 1972. The fluid mosaic model of the structure of cell membranes. Science. 175: 720–731. [DOI] [PubMed] [Google Scholar]
- 24.Edidin M. 1974. Rotational and translational diffusion in membranes. Annu. Rev. Biophys. Bioeng. 3: 179–201. [DOI] [PubMed] [Google Scholar]
- 25.Searls D. B., and Edidin M.. 1981. Lipid composition and lateral diffusion in plasma membranes of teratocarcinoma-derived cell lines. Cell. 24: 511–517. [DOI] [PubMed] [Google Scholar]
- 26.Yechiel E., and Edidin M.. 1987. Micrometer-scale domains in fibroblast plasma membranes. J. Cell Biol. 105: 755–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simons K., and van Meer G.. 1988. Lipid sorting in epithelial cells. Biochemistry. 27: 6197–6202. [DOI] [PubMed] [Google Scholar]
- 28.Simons K., and Ikonen E.. 1997. Functional rafts in cell membranes. Nature. 387: 569–572. [DOI] [PubMed] [Google Scholar]
- 29.Lingwood D., and Simons K.. 2010. Lipid rafts as a membrane-organizing principle. Science. 327: 46–50. [DOI] [PubMed] [Google Scholar]
- 30.Cerneus D. P., Ueffing E., Posthuma G., Strous G. J., and Van Der Ende A.. 1993. Detergent insolubility of alkaline phosphatase during biosynthetic transport and endocytosis: role of cholesterol. J. Biol. Chem. 268: 3150–3155. [PubMed] [Google Scholar]
- 31.Edidin M. 2003. The state of lipid rafts: from model membranes to cells. Annu. Rev. Biophys. Biomol. Struct. 32: 257–283. [DOI] [PubMed] [Google Scholar]
- 32.Simons K., and Vaz W. L. C.. 2004. Model systems, lipid rafts, and cell membranes. Annu. Rev. Biophys. Biomol. Struct. 33: 269–295. [DOI] [PubMed] [Google Scholar]
- 33.Mayor S., and Maxfield F. R.. 1995. Insolubility and redistribution of GPI-anchored proteins at the cell surface after detergent treatment. Mol. Biol. Cell. 6: 929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heerklotz H. 2002. Triton promotes domain formation in lipid raft mixtures. Biophys. J. 83: 2693–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heerklotz H., Szadkowska H., Anderson T., and Seelig J.. 2003. The sensitivity of lipid domains to small perturbations demonstrated by the effect of Triton. J. Mol. Biol. 329: 793–799. [DOI] [PubMed] [Google Scholar]
- 36.Chen Y., Lagerholm B. C., Yang B., and Jacobson K.. 2006. Methods to measure the lateral diffusion of membrane lipids and proteins. Methods. 39: 147–153. [DOI] [PubMed] [Google Scholar]
- 37.Zhang F., Crise B., Rose J. K., Jacobson K., Hill C., Biology C., Haven N., and Haven N.. 1991. Lateral diffusion of membrane-spanning and glycosylphosphatidylinositol- linked proteins: toward establishing rules governing the lateral mobility of membrane proteins. J. Cell Biol. 115: 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishihara A., Hou Y., and Jacobson K.. 1987. The Thy-1 antigen exhibits rapid lateral diffusion in the plasma membrane of rodent lymphoid cells and fibroblasts. Proc. Natl. Acad. Sci. USA. 84: 1290–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang F., Lee G. M., and Jacobson K.. 1993. Protein lateral mobility as a reflection of membrane microstructure. BioEssays. 15: 579–588. [DOI] [PubMed] [Google Scholar]
- 40.Hannan L. A., Lisanti M. P., Rodriguez-Boulan E., and Edidin M.. 1993. Correctly sorted molecules of a GPI-anchored protein are clustered and immobile when they arrive at the apical surface of MDCK cells. J. Cell Biol. 120: 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kenworthy A. K., Nichols B. J., Remmert C. L., Hendrix G. M., Kumar M., Zimmerberg J., and Lippincott-Schwartz J.. 2004. Dynamics of putative raft-associated proteins at the cell surface. J. Cell Biol. 165: 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saxton M. J., and Jacobson K.. 1997. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 26: 373–399. [DOI] [PubMed] [Google Scholar]
- 43.Kusumi A., Sako Y., and Yamamoto M.. 1993. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys. J. 65: 2021–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheets E. D., Lee G. M., Simson R., and Jacobson K.. 1997. Transient confinement of a glycosylphosphatidylinositol-anchored protein in the plasma membrane. Biochemistry. 36: 12449–12458. [DOI] [PubMed] [Google Scholar]
- 45.Sheets E. D., Simson R., and Jacobson K.. 1995. New insights into membrane dynamics from the analysis of cell surface interactions by physical methods. Curr. Opin. Cell Biol. 7: 707–714. [DOI] [PubMed] [Google Scholar]
- 46.Dietrich C., Yang B., Fujiwara T., Kusumi A., and Jacobson K.. 2002. Relationship of lipid rafts to transient confinement zones detected by single particle tracking. Biophys. J. 82: 274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sako Y., and Kusumi A.. 1994. Compartmentalized structure of the plasma membrane for receptor movements as revealed by a nanometer-level motion analysis. J. Cell Biol. 125: 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kusumi A., and Sako Y.. 1996. Cell surface organization by the membrane skeleton. Curr. Opin. Cell Biol. 8: 566–574. [DOI] [PubMed] [Google Scholar]
- 49.Simson R., Yang B., Moore S. E., Doherty P., Walsh F. S., and Jacobson K. A.. 1998. Structural mosaicism on the submicron scale in the plasma membrane. Biophys. J. 74: 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pralle A., Keller P., Florin E. L., Simons K., and Hörber J. K.. 2000. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J. Cell Biol. 148: 997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki K., and Sheetz M. P.. 2001. Binding of cross-linked glycosylphosphatidylinositol-anchored proteins to discrete actin-associated sites and cholesterol-dependent domains. Biophys. J. 81: 2181–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kusumi A., Nakada C., Ritchie K., Murase K., Suzuki K., Murakoshi H., Kasai R. S., Kondo J., and Fujiwara T.. 2005. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 34: 351–378. [DOI] [PubMed] [Google Scholar]
- 53.Fujiwara T., Ritchie K., Murakoshi H., Jacobson K., and Kusumi A.. 2002. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 157: 1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritchie K., Iino R., Fujiwara T., Murase K., and Kusumi A.. 2003. The fence and picket structure of the plasma membrane of live cells as revealed by single molecule techniques (Review). Mol. Membr. Biol. 20: 13–18. [DOI] [PubMed] [Google Scholar]
- 55.Murase K., Fujiwara T., Umemura Y., Suzuki K., Iino R., Yamashita H., Saito M., Murakoshi H., Ritchie K., and Kusumi A.. 2004. Ultrafine membrane compartments for molecular diffusion as revealed by single molecule techniques. Biophys. J. 86: 4075–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Umemura Y. M., Vrljic M., Nishimura S. Y., Fujiwara T. K., Suzuki K. G. N., and Kusumi A.. 2008. Both MHC class II and its GPI-anchored form undergo hop diffusion as observed by single-molecule tracking. Biophys. J. 95: 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morone N., Fujiwara T., Murase K., Kasai R. S., Ike H., Yuasa S., Usukura J., and Kusumi A.. 2006. Three-dimensional reconstruction of the membrane skeleton at the plasma membrane interface by electron tomography. J. Cell Biol. 174: 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwik J., Boyle S., Fooksman D., Margolis L., Sheetz M. P., and Edidin M.. 2003. Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin. Proc. Natl. Acad. Sci. USA. 100: 13964–13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edidin M., and Stroynowski I.. 1991. Differences between the lateral organization of conventional and inositol phospholipid-anchored membrane proteins. A further definition of micrometer scale membrane domains. J. Cell Biol. 112: 1143–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wawrezinieck L., Rigneault H., Marguet D., and Lenne P-F.. 2005. Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys. J. 89: 4029–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He H.-T., and Marguet D.. 2011. Detecting nanodomains in living cell membrane by fluorescence correlation spectroscopy. Annu. Rev. Phys. Chem. 62: 417–436. [DOI] [PubMed] [Google Scholar]
- 62.Lenne P.-F., Wawrezinieck L., Conchonaud F., Wurtz O., Boned A., Guo X.-J., Rigneault H., He H.-T., and Marguet D.. 2006. Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. EMBO J. 25: 3245–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wenger J., Conchonaud F., Dintinger J., Wawrezinieck L., Ebbesen T. W., Rigneault H., Marguet D., and Lenne P-F.. 2007. Diffusion analysis within single nanometric apertures reveals the ultrafine cell membrane organization. Biophys. J. 92: 913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eggeling C., Ringemann C., Medda R., Schwarzmann G., Sandhoff K., Polyakova S., Belov V. N., Hein B., von Middendorff C., Schonle A., et al. 2009. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 457: 1159–1162. [DOI] [PubMed] [Google Scholar]
- 65.Mueller V., Ringemann C., Honigmann A., Schwarzmann G., Medda R., Leutenegger M., Polyakova S., Belov V. N., Hell S. W., and Eggeling C.. 2011. STED nanoscopy reveals molecular details of cholesterol- and cytoskeleton-modulated lipid interactions in living cells. Biophys. J. 101: 1651–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mayor S., Rothberg K. G., and Maxfield F. R.. 1994. Sequestration of GPI-anchored proteins in caveolae triggered by cross-linking. Science. 264: 1948–1951. [DOI] [PubMed] [Google Scholar]
- 67.Prior I. A., Muncke C., Parton R. G., and Hancock J. F.. 2003. Direct visualization of ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 160: 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ghosh S., Saha S., Goswami D., Bilgrami S., and Mayor S.. 2012. Dynamic imaging of homo-FRET in live cells by fluorescence anisotropy microscopy. Methods Enzymol. 505: 291–327. [DOI] [PubMed] [Google Scholar]
- 69.Rao M., and Mayor S.. 2005. Use of Forster’s resonance energy transfer microscopy to study lipid rafts. Biochim. Biophys. Acta. 1746: 221–233. [DOI] [PubMed] [Google Scholar]
- 70.Saha S., Raghupathy R., and Mayor S.. 2015. Homo-FRET imaging highlights the nanoscale organization of cell surface molecules. Methods Mol. Biol. 1251: 151–173. [DOI] [PubMed] [Google Scholar]
- 71.Garcia-Parajo M. F., Cambi A., Torreno-Pina J. A., Thompson N., and Jacobson K.. 2014. Nanoclustering as a dominant feature of plasma membrane organization. J. Cell Sci. 127: 4995–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Owen D. M., Magenau A., Williamson D., and Gaus K.. 2012. The lipid raft hypothesis revisited - new insights on raft composition and function from super-resolution fluorescence microscopy. BioEssays. 34: 739–747. [DOI] [PubMed] [Google Scholar]
- 73.Jares-Erijman E. A., and Jovin T. M.. 2003. FRET imaging. Nat. Biotechnol. 21: 1387–1395. [DOI] [PubMed] [Google Scholar]
- 74.Förster T. 1948. Intermolecular energy migration and fluorescence. Ann. Phys. 2: 55–75. [Google Scholar]
- 75.Stryer L., and Haugland R. P.. 1967. Energy transfer: a spectroscopic ruler. Proc. Natl. Acad. Sci. USA. 58: 719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stryer L. 1978. Fluorescence energy transfer as a spectroscopic ruler. Annu. Rev. Biochem. 47: 819–846. [DOI] [PubMed] [Google Scholar]
- 77.Gautier I., Tramier M., Durieux C., Coppey J., Pansu R. B., Nicolas J. C., Kemnitz K., and Coppey-Moisan M.. 2001. Homo-FRET microscopy in living cells to measure monomer-dimer transition of GFT-tagged proteins. Biophys. J. 80: 3000–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bader A. N., Hofman E. G., Voortman J., en Henegouwen P. M., and Gerritsen H. C.. 2009. Homo-FRET imaging enables quantification of protein cluster sizes with subcellular resolution. Biophys. J. 97: 2613–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sharma P., Varma R., Sarasij R. C. , , Gousset K., Krishnamoorthy G., Rao M., and Mayor S.. 2004. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 116: 577–589. [DOI] [PubMed] [Google Scholar]
- 80.Friedrichson T., and Kurzchalia T. V.. 1998. Microdomains of GPI-anchored proteins in living cells revealed by crosslinking. Nature. 394: 802–805. [DOI] [PubMed] [Google Scholar]
- 81.Yeow E. K. L., and Clayton A. H. A.. 2007. Enumeration of oligomerization states of membrane proteins in living cells by homo-FRET spectroscopy and microscopy: theory and application. Biophys. J. 92: 3098–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kenworthy A. K. K., and Edidin M.. 1998. Distribution of a glycosylphosphatidylinositol-anchored protein at the apical surface of MDCK cells examined at a resolution of <100 A using imaging fluorescence resonance energy transfer. J. Cell Biol. 142: 69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goswami D., Gowrishankar K., Bilgrami S., Ghosh S., Raghupathy R., Chadda R., Vishwakarma R., Rao M., and Mayor S.. 2008. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell. 135: 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Charras G. T., Hu C. K., Coughlin M., and Mitchison T. J.. 2006. Reassembly of contractile actin cortex in cell blebs. J. Cell Biol. 175: 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang B., Babcock H., and Zhuang X.. 2010. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 143: 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Eggeling C., Willig K. I., Sahl S. J., and Hell S. W.. 2015. Lens-based fluorescence nanoscopy. Q. Rev. Biophys. 48: 178–243. [DOI] [PubMed] [Google Scholar]
- 87.Schermelleh L., Heintzmann R., and Leonhardt H.. 2010. A guide to super-resolution fluorescence microscopy. J. Cell Biol. 190: 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van Zanten T. S., Cambi A., Koopman M., Joosten B., Figdor C. G., and Garcia-Parajo M. F.. 2009. Hotspots of GPI-anchored proteins and integrin nanoclusters function as nucleation sites for cell adhesion. Proc. Natl. Acad. Sci. USA. 106: 18557–18562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Manzo C., van Zanten T. S., Saha S., Torreno-Pina J. A., Mayor S., and Garcia-Parajo M. F.. 2014. PSF decomposition of nanoscopy images via Bayesian analysis unravels distinct molecular organization of the cell membrane. Sci. Rep. 4: 4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sengupta P., Jovanovic-Talisman T., Skoko D., Renz M., Veatch S. L., and Lippincott-Schwartz J.. 2011. Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat. Methods. 8: 969–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schram V., and Thompson T. E.. 1995. Interdigitation does not affect translational diffusion of lipids in liquid crystalline bilayers. Biophys. J. 69: 2517–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schmidt C. F., Barenholz Y., Huang C., and Thompson T. E.. 1978. Monolayer coupling in sphingomyelin bilayer systems. Nature. 271: 775–777. [DOI] [PubMed] [Google Scholar]
- 93.Chiantia S., and London E.. 2012. Acyl chain length and saturation modulate interleaflet coupling in asymmetric bilayers: Effects on dynamics and structural order. Biophys. J. 103: 2311–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Polley A., Mayor S., and Rao M.. 2014. Bilayer registry in a multicomponent asymmetric membrane: dependence on lipid composition and chain length. J. Chem. Phys. 141: 064903. [DOI] [PubMed] [Google Scholar]
- 95.Lenne P-F., Wawrezinieck L., Conchonaud F., Wurtz O., Boned A., Guo X-J., Rigneault H., He H-T., and Marguet D.. 2006. Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. EMBO J. 25: 3245–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Charras G. T., Coughlin M., Mitchison T. J., and Mahadevan L.. 2007. Life and times of a cellular bleb. Biophys. J. 94: 1836–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gowrishankar K., Ghosh S., Saha S., Rumamol C., Mayor S., and Rao M.. 2012. Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell. 149: 1353–1367. [DOI] [PubMed] [Google Scholar]
- 98.Rao M., and Mayor S.. 2014. Active organization of membrane constituents in living cells. Curr. Opin. Cell Biol. 29: 126–132. [DOI] [PubMed] [Google Scholar]
- 99.Cano M. L., Lauffenburger D. A., and Zigmond S. H.. 1991. Kinetic analysis of F-actin depolymerization in polymorphonuclear leukocyte lysates indicates that chemoattractant stimulation increases actin filament number without altering the filament length distribution. J. Cell Biol. 115: 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reichl E. M., Ren Y., Morphew M. K., Delannoy M., Effler J. C., Girard K. D., Divi S., Iglesias P. A., Kuo S. C., and Robinson D. N.. 2008. Interactions between Myosin and Actin Crosslinkers Control Cytokinesis Contractility Dynamics and Mechanics. Curr. Biol. 18: 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Puthenveedu M. A., Lauffer B., Temkin P., Vistein R., Carlton P., Thorn K., Taunton J., Weiner O. D., Parton R. G., and von Zastrow M.. 2010. Sequence-dependent sorting of recycling proteins by actin-stabilized endosomal microdomains. Cell. 143: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Saha S., Lee I-H., Polley A., Groves J. T., Rao M., and Mayor S.. Diffusion of GPI-anchored proteins is influenced by the activity of dynamic cortical actin. Mol. Biol. Cell. Epub ahead of print. September 16, 2015; doi:10.1091/mbc.E15-06-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lakhan S. E., Sabharanjak S., and De A.. 2009. Endocytosis of glycosylphosphatidylinositol-anchored proteins. J. Biomed. Sci. 16: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chatterjee S., and Mayor S.. 2001. The GPI-anchor and protein sorting. Cell. Mol. Life Sci. 58: 1969–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Almeida A. M., Murakami Y., Layton D. M., Hillmen P., Sellick G. S., Maeda Y., Richards S., Patterson S., Kotsianidis I., Mollica L., et al. 2006. Hypomorphic promoter mutation in PIGM causes inherited glycosylphosphatidylinositol deficiency. Nat. Med. 12: 846–851. [DOI] [PubMed] [Google Scholar]
- 106.Ng B. G., Hackmann K., Jones M. A., Eroshkin A. M., He P., Wiliams R., Bhide S., Cantagrel V., Gleeson J. G., Paller A. S., et al. 2012. Mutations in the glycosylphosphatidylinositol gene PIGL cause CHIME syndrome. Am. J. Hum. Genet. 90: 685–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Murakami H., Wang Y., Hasuwa H., Maeda Y., Kinoshita T., and Murakami Y.. 2012. Enhanced response of T lymphocytes from Pgap3 knockout mouse: Insight into roles of fatty acid remodeling of GPI anchored proteins. Biochem. Biophys. Res. Commun. 417: 1235–1241. [DOI] [PubMed] [Google Scholar]
- 108.Young S. G., Davies B. S. J., Voss C. V., Gin P., Weinstein M. M., Tontonoz P., Reue K., Bensadoun A., Fong L. G., and Beigneux P.. 2011. GPIHBP1, an endothelial cell transporter for lipoprotein lipase. J. Lipid Res. 52: 1869–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sobering A. K., Watanabe R., Romeo M. J., Yan B. C., Specht C. A., Orlean P., Riezman H., and Levin D. E.. 2004. Yeast Ras regulates the complex that catalyzes the first step in GPI-anchor biosynthesis at the ER. Cell. 117: 637–648. [DOI] [PubMed] [Google Scholar]
- 110.Nagpal J. K., Dasgupta S., Jadallah S., Chae Y. K., Ratovitski E. A., Toubaji A., Netto G. J., Eagle T., Nissan A., Sidransky D., et al. 2008. Profiling the expression pattern of GPI transamidase complex subunits in human cancer. Mod. Pathol. 21: 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sabharanjak S., Sharma P., Parton R. G., and Mayor S.. 2002. GPI-anchored proteins are delivered to recycling endosomes via a distinct cdc42-regulated clathrin-independent pinocytic pathway. Dev. Cell. 2: 411–423. [DOI] [PubMed] [Google Scholar]
- 112.Mayor S., and Pagano R. E.. 2007. Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell Biol. 8: 603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kinoshita T., Maeda Y., and Fujita M.. 2013. Transport of glycosylphosphatidylinositol-anchored proteins from the endoplasmic reticulum. Biochim. Biophys. Acta. 1833: 2473–2478. [DOI] [PubMed] [Google Scholar]
- 114.Kamen B. A., Wang M. T., Streckfuss A. J., Peryea X., and Anderson R. G.. 1988. Delivery of folates to the cytoplasm of MA104 cells is mediated by a surface membrane receptor that recycles. J. Biol. Chem. 263: 13602–13609. [PubMed] [Google Scholar]
- 115.Stefanová I., Horejsí V., Ansotegui I. J., Knapp W., and Stockinger H.. 1991. GPI-anchored cell-surface molecules complexed to protein tyrosine kinases. Science. 254: 1016–1019. [DOI] [PubMed] [Google Scholar]
- 116.Suzuki K. G., Fujiwara T. K., Sanematsu F., Iino R., Edidin M., and Kusumi A.. 2007. GPI-anchored receptor clusters transiently recruit Lyn and G alpha for temporary cluster immobilization and Lyn activation: single-molecule tracking study 1. J. Cell Biol. 177: 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Suzuki K. G. N., Fujiwara T. K., Edidin M., and Kusumi A.. 2007. Dynamic recruitment of phospholipase Cγ at transiently immobilized GPI-anchored receptor clusters induces IP3-Ca2+ signaling: Single-molecule tracking study 2. J. Cell Biol. 177: 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen Y., Thelin W. R., Yang B., Milgram S. L., and Jacobson K.. 2006. Transient anchorage of cross-linked glycosyl-phosphatidylinositol-anchored proteins depends on cholesterol, Src family kinases, caveolin, and phosphoinositides. J. Cell Biol. 175: 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen Y., Veracini L., Benistant C., and Jacobson K.. 2009. The transmembrane protein CBP plays a role in transiently anchoring small clusters of Thy-1, a GPI-anchored protein, to the cytoskeleton. J. Cell Sci. 122: 3966–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gaus K., Le Lay S., Balasubramanian N., and Schwartz M. A.. 2006. Integrin-mediated adhesion regulates membrane order. J. Cell Biol. 174: 725–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Caiolfa V. R., Zamai M., Malengo G., Andolfo A., Madsen C. D., Sutin J., Digman M. A., Gratton E., Blasi F., and Sidenius N.. 2007. Monomer-dimer dynamics and distribution of GPI-anchored uPAR are determined by cell surface protein assemblies. J. Cell Biol. 179: 1067–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Smith H. W., and Marshall C. J.. 2010. Regulation of cell signalling by uPAR. Nat. Rev. Mol. Cell Biol. 11: 23–36. [DOI] [PubMed] [Google Scholar]
- 123.Manley S., Gillette J. M., Patterson G. H., Shroff H., Hess H. F., Betzig E., and Lippincott-Schwartz J.. 2008. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods. 5: 155–157. [DOI] [PubMed] [Google Scholar]
- 124.Kolin D. L., and Wiseman P. W.. 2007. Advances in image correlation spectroscopy: measuring number densities, aggregation states, and dynamics of fluorescently labeled macromolecules in cells. Cell Biochem. Biophys. 49: 141–164. [DOI] [PubMed] [Google Scholar]
- 125.Doherty G. J., and McMahon H. T.. 2008. Mediation, modulation, and consequences of membrane-cytoskeleton interactions. Annu. Rev. Biophys. 37: 65–95. [DOI] [PubMed] [Google Scholar]
- 126.Johannes L., Parton R. G., Bassereau P., and Mayor S.. 2015. Building endocytic pits without clathrin. Nat. Rev. Mol. Cell Biol. 16: 311–321. [DOI] [PubMed] [Google Scholar]
- 127.Owen D. M., Williamson D. J., Magenau A., and Gaus K.. 2012. Sub-resolution lipid domains exist in the plasma membrane and regulate protein diffusion and distribution. Nat. Commun. 3: 1256. [DOI] [PubMed] [Google Scholar]
- 128.Kwiatek J. M., Hinde E., and Gaus K.. 2014. Microscopy approaches to investigate protein dynamics and lipid organization. Mol. Membr. Biol. 31: 141–151. [DOI] [PubMed] [Google Scholar]
- 129.Sanchez S. A., Tricerri M. A., and Gratton E.. 2012. Laurdan generalized polarization fluctuations measures membrane packing micro-heterogeneity in vivo. Proc. Natl. Acad. Sci. USA. 109: 7314–7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Frisz J. F., Lou K., Klitzing H. A., Hanafin W. P., Lizunov V., Wilson R. L., Carpenter K. J., Kim R., Hutcheon I. D., Zimmerberg J., et al. 2013. Direct chemical evidence for sphingolipid domains in the plasma membranes of fibroblasts. Proc. Natl. Acad. Sci. USA. 110: E613–E622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Frisz J. F., Klitzing H. A., Lous K., Hutcheon I. D., Weber P. K., Zimmerberg J., and Kraft M. L.. 2013. Sphingolipid domains in the plasma membranes of fibroblasts are not enriched with cholesterol. J. Biol. Chem. 288: 16855–16861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ferguson M. A. 1999. The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J. Cell Sci. 112: 2799–2809. [DOI] [PubMed] [Google Scholar]
- 133.Kusumi A., Fujiwara T. K., Chadda R., Xie M., Tsunoyama T. A., Kalay Z., Kasai R. S., and Suzuki K. G. N.. 2012. Dynamic organizing principles of the plasma membrane that regulate signal transduction: commemorating the fortieth anniversary of Singer and Nicolson’s fluid-mosaic model. Annu. Rev. Cell Dev. Biol. 28: 215–250. [DOI] [PubMed] [Google Scholar]
- 134.Paladino S., Lebreton S., and Zurzolo C.. 2015. Trafficking and membrane organization of GPI-anchored proteins in health and diseases. Curr. Top. Membr. 75: 269–303. [DOI] [PubMed] [Google Scholar]
- 135.Muñiz M., and Zurzolo C.. 2014. Sorting of GPI-anchored proteins from yeast to mammals - common pathways at different sites? J. Cell Sci. 127: 2793–2801. [DOI] [PubMed] [Google Scholar]