Abstract
Legionella spp. are common in various natural and man-made aquatic environments. Recreational hot spring is frequently reported as an infection hotspot because of various factors such as temperature and humidity. Although polymerase chain reaction (PCR) had been used for detecting Legionella, several inhibitors such as humic substances, calcium, and melanin in the recreational spring water may interfere with the reaction thus resulting in risk underestimation. The purpose of this study was to compare the efficiencies of conventional and Taqman quantitative PCR (qPCR) on detecting Legionella pneumophila in spring facilities and in receiving water. In the results, Taqman PCR had much better efficiency on specifying the pathogen in both river and spring samples. L. pneumophila was detected in all of the 27 river water samples and 45 of the 48 hot spring water samples. The estimated L. pneumophela concentrations ranged between 1.0 × 102 and 3.3 × 105 cells/l in river water and 72.1–5.7 × 106 cells/l in hot spring water. Total coliforms and turbidity were significantly correlated with concentrations of L. pneumophila in positive water samples. Significant difference was also found in water temperature between the presence/absence of L. pneumophila. Our results suggest that conventional PCR may be not enough for detecting L. pneumophila particularly in the aquatic environments full of reaction inhibitors.
Keywords: L. pneumophila, Quantitative PCR, Hot spring, River
Introduction
Legionella spp. was first isolated from guinea pigs in 1934, but its potential hazards were not recognized until the first large-scale pneumonia outbreak in Philadelphia, USA in 1976.1,2 At least 59 Legionella species have been identified since, and 26 species are associated with human illness, including Legionnaires' disease and Pontiac fever.3 Legionella pneumophila is the most common pathogenic Legionella species, accounting for 80–90% of Legionnaires' disease cases.4,5
Legionella spp. can be found in natural and man-made water environments such as rivers, hot springs, lakes, cooling towers, and swimming pools. Until recently, many of the Legionella monitoring studies were performed in response to legionellosis outbreaks.6–8 According to previous studies, the detection rate of Legionella spp. ranged between 25.8 and 64% in man-made water facilities,9–12 and the concentration varied from 940 to 1.5 × 106 genome units (GU)/l.12,13 For natural water environment, the detection rate of Legionella spp. was 8.7–100%, and concentrations ranged between 7.4 × 103 and 9.4 × 105 GU/l,14 and 104–108 cells/l.15,16
Polymerase chain reaction assay has been widely used for detection of microbes in environmental samples. Compared to conventional incubation method, PCR is less time-consuming and more sensitive in detecting and identifying specific microorganisms. Although highly specific, PCR is more vulnerable to aquatic pollutants since concentration of bacteria by membrane filtration usually comes with residues of miscellaneous PCR inhibitors including humic substances, metal ions, polysaccharides, and insoluble debris.17–21 Such inhibitors interfere with PCR via inactivating DNA polymerase or sequestering/degrading DNA templates.21 In the recreational spring water, higher concentration of calcium and the impurities such as the melanin from hair and skin, and the urea from human discharges are all potential PCR inhibitors.22 Hot spring facility is a known hotspot of Legionella infection owing to the warmer temperature and humidity, which provides an ideal environment for the bacterial growth. Water swirling and shower nozzles may further increase the pathogens in aerosol which could further increase the infection possibility. However, PCR inhibitors in the recreational spring water may result in underestimation of the Legionella risk.
In this study, qPCR (Taqman) was used for detecting and quantifying L. pneumophila in hot spring and river samples. The efficiencies between the conventional PCR and Taqman qPCR were compared. In addition, relationship between the presence of L. pneumophila and various water quality parameters were evaluated.
Materials and Methods
Sampling sites, samples collection, and water quality analysis
Water samples were collected from along Puzih River (23˚28′N, 120˚13′E) in southern Taiwan and from three hot spring recreational areas. The three hot spring areas included K.-K. (24˚20′N, 121˚01′E), P.-L. (22˚41′N, 121˚00′E), and Z.-P. (22˚05′N, 120˚44′E), and all are with weak alkaline-carbonated hot springs. The sampling campaign was carried out between August 2011 and July 2012. For each sample, 2,000 ml of water was collected into two sterile 1 lbottles and stored at 4°C before transferring to the laboratory. Three water quality parameters were measured for each water sample at the time of sample collection, including pH level with a portable pH meter (D-24E, Horiba Co., Japan), water temperature with a thermometer, and turbidity with a turbidimeter (HACH Co., Loveland, CO). In addition, microbial water quality was assessed within 24 hours of sample collection, including heterotrophic bacteria by spread method, and total coliforms by membrane filtration and incubation with a differential medium as prescribed in the Standard Method for the Examination of Water and Wastewater (Methods 9215 C and 9222 B).23 Statistical analyses (differences and correlation) on presence and amount of L. pneumophila with the water quality parameters were performed using the STATISTICA® software (StatSoft, Inc., Tulsa, USA).
Sample pretreatment
One liter of each water sample was filtered through 0.22 μm pore size, 47-mm diameter cellulose nitrate membranes (Pall, New York, USA). The number of membranes used for each sample depended on the water turbidity. After filtration, the membranes were collected and swirled with 100 ml of Page's saline solution for 5 min to elute the microbes from the membranes for each sample. Page's saline solution has prepare in 1 l of de-ionized water contained 120 mg of NaCl, 4 mg of MgSO4.7H2O, 4 mg of CaCl2.6H2O, 142 mg of Na2HPO4, and 136 mg of KH2PO4. The resulting eluent was transferred into two conical centrifuge tubes (50 ml each) and centrifuged at 5,800 × g for 30 min. For each centrifuged solution, the top supernatant fluid (about 47.5 ml) was aspirated and discarded. The pellet in the remaining 2.5 ml solution was resuspended by vortexing in a disinfected tube. For each water sample, two tubes of 2.5 ml concentrate were produced. DNA extraction was done with MagNA Pure LC System (Roche, Basel, Switzerland) and MagNA Pure LC DNA Isolation Kit III (Roche). The derived DNA was used for all the PCRs.
Detection and quantification of L. pneumophila with qPCR (Taqman) assay
In this study, mip was used as the target gene for identifying L. pneumophila. The operation conditions of qPCR for L. pneumophila are summarized in Table 1. The qPCR mixtures contained 5 μl template DNA, 0.5 μl of each primer, 0.5 μl of probe, 10 μl of probe mix, and sterile water to 20 ml volume. After qPCR, the reaction product was examined for the presence of specific target genes using electrophoresis separation. Nuclease-free water (Qiagen) was used in all experiments as negative controls. Each qPCR run was conducted using positive control DNA for L. pneumophila ATCC 33823, sample DNA, and negative control.
Table 1.
Summary of primer sequences of polymerase chain reaction (PCR) and quantitative PCR (qPCR) assays for Legionella spp. and L. pneumophila.
| PCR assay | Primer sequences | Predenaturation, denaturation, annealing and extension temperature | Cycling Number | Amplicon size (bp) | Reference | |||
|---|---|---|---|---|---|---|---|---|
| PCR | leg 225 5′-AAGATTAGCCTGCGTCCGAT-3′ | – | 95 | 62 | 72 | 40 | 654 | Miyamoto et al.24 |
| leg 858 5′-GTCAACTTATCGCGTTTGCT-3′ | ||||||||
| mip 920 5′-GCTACAGACAAGGATAAGTTG-3′ | – | 95 | 62 | 72 | 40 | 648 | Bej et al.25 | |
| mip 1548 5′-GTTTTGTATGACTTTAATTCA-3′ | ||||||||
| qPCR | mip F 5′-TTCATTTGYTGYTCGGTTAAAGC-3′ | 95 | 95 | 60 | 72 | 40 | 66 | Behets et al.26 |
| mip R 5′-AWTGGCTAAAGGCATGCAAGAC-3′ | ||||||||
| probe 5′(FAM)-AGCGCCACTCATAG-(BHQ1) 3′ |
leg: Legionella eukaryotic-like genes; leg 225/858 is specific for Legionella species; mip: macrophage infectivity potentiator; mip 920/1548 and mip F/R is specific for Legionella pneumophila.
Calibration for quantification outcome by qPCR (Taqman) assay
The positive control, L. pneumophila ATCC 33823, was transferred onto a BCYE agar plate (buffered charcoal yeast extract with alpha-ketoglutarate, l-cysteine, and ferric pyrophosphate). The inoculated plates were then incubated in a 5% CO2 incubator at 37°C. DNA was extracted from a 5-day culture of L. pneumophila by heating method (heating at 95°C for 10 min, followed by centrifuge at 9,700 × g for 10 min) for qPCR (Taqman) assay. The qPCR products were identified with gel electrophoresis on a 2% agarose gel (Biobasic Inc., New York, USA) with 5 μl reaction solution from qPCR (Taqman) assay. The band of gel in target gene site (66 bp) was cut carefully before gel dissociation. The DNA was purified by Gel/PCR DNA fragments extraction kit (Geneaid, New Taipei City, Taiwan). Subsequently, yT&A clone vector kit (Yeasterm Biotech Corporation, Taipei, Taiwan) was used and the vector will ligate with the insert DNA after overnight reaction at 4°C. The transformed cells were incubated overnight at 37°C for blue/white screening, with white colonies chosen after competent cell transformation. The recombinant plasmid DNA was purified by HiYield™ plasmid mini kit (Real Biotech Corporation, Taipei, Taiwan), and the plasmid DNA amount was determined by Micro-spectrophotometer (CLUBIO CB-4500, Taiwan) at 260 nm in triplicates. The corresponding copy number was calculated using the following equation27
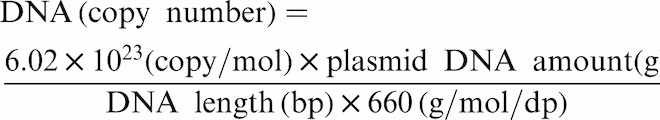 |
Quantification of L. pneumophila was determined with threshold cycle value (Ct) by which the threshold fluorescence level was detected by the ABI StepOne™ Real-Time PCR Systems (Applied Biosystems, Singapore). A detected L. pneumophila cell would be counted as one copy number,28 thus the calculated Ct may be converted to L. pneumophila concentration in cells/l. Plasmid DNA of L. pneumophila from positive control was used with serial 10-fold dilutions by sterile water for qPCR(Taqman) assay, and the resulting copy numbers were compared with observed Ct to construct the calibration curve for quantifying L. pneumophila in the water sample.
PCR analysis
In this study, the PCR method was used for comparison with detection outcomes by qPCR. The PCR solution was prepared with 5 μl of the DNA templates and PCR mixture to a total volume of 50 μl. The PCR mixture contained 5 μl 10 × PCR buffer (20 mM MgCl2), 1 μl dNTP mix (10 mM of each dNTP), 200 pmol each of the oligonucleotide primers and 0.3 μl VioTaqTM DNA Polymerase (Viogene, New Taipei City, Taiwan, 5 U/μl), and DNase-free deionized water. The primer sets and reaction settings for Legionella spp. and L. pneumophila are summarized in Table 1. The target genes (leg and mip) were used to confirm the detection of L. pneumophila. The PCR product was confirmed for the presence of specific target genes using electrophoresis separation. L. pneumophila ATCC 33823 was used as positive control in this study, and negative controls were also included by replacing the DNA template with distilled water for subsequent analyses.
Gel electrophoresis
All positive samples, whether detected by qPCR or PCR method, were subject to gel electrophoresis for species confirmation on a 2% agarose gel (Biobasic Inc.) with 5 μl reaction solution. The DNA fragments were confirmed with ethidium bromide staining (0.5 μg/ml, 10 min). A 100-bp DNA ladder was used as a DNA size marker for image production under UV light.
Results and Discussion
Calibration outcome for qPCR (Taqman) assay
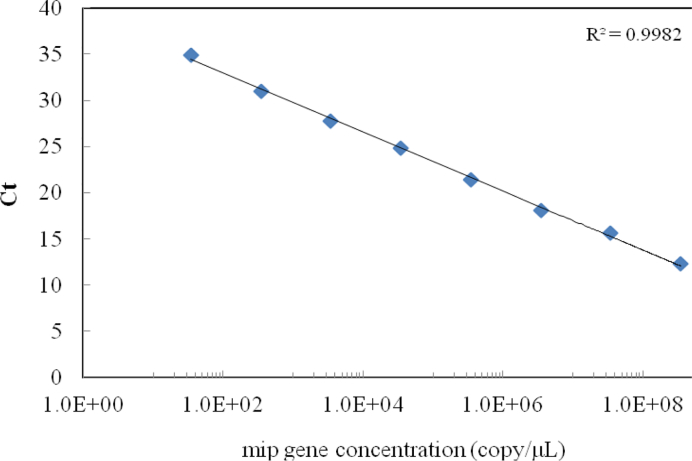
The calibration outcome for qPCR (Taqman) assay is presented in Fig. 1. A linear relationship was observed between Ct values and the logarithmic values of the mip gene copy numbers, and the calibration equation was Ct = –1.39 log10 [copy number] +39.33. The coefficient of determination (r2) was 0.998, with range of detection from 34.3 to 3.5 × 108 cells/l. The calibration results showed that this method would allow a wide range for L. pneumophila concentration.
Figure 1.
The calibration curve of quantitative L. pneumophila mip polymerase chain reaction (PCR).
L. pneumophila monitoring outcomes
A total of 75 water samples were collected from a river and three hot spring recreation areas in this study. Each sample was evaluated with PCR and qPCR, and the detection outcomes are summarized in Table 2. Of the 75 samples analyzed, 72 (96.0%) were found to contain L. pneumophila by qPCR. In contrast, 41 positive samples were detected with leg PCR and 19 samples with mip PCR. As all positive detections were confirmed by electrophoresis, the likelihood of false-positive detection by qPCR was minimized. The estimated concentrations of L. pneumophila in the positive samples ranged between 72.1 and 5.7 × 106 cells/l.
Table 2.
L. pneumophila monitoring for molecular techniques in various types of water samples.
| Detection rate (%)* | ||||
|---|---|---|---|---|
| Type of water sample | leg PCR | mip PCR | mip qPCR | Range of L. pneumophila concentration (cells/l, average concentration)+ |
| Overall | 54.7 (41/75) | 26.7 (19/75) | 96.0 (72/75) | 72.1–5.7 × 106 (1.5 × 104) |
| Puzih river | 70.4 (19/27) | 7.4 (2/27) | 100.0 (27/27) | 1.0 × 102–3.3 × 105 (1.2 × 104) |
| Hot spring | 45.8 (22/48) | 35.4 (17/48) | 93.8 (45/48) | 72.1–5.7 × 106 (2.5 × 104) |
| Source water | 58.3 (7/12) | 41.7 (5/12) | 100.0 (12/12) | 1.7 × 102–5.7 × 106 (8.4 × 105) |
| Hot spring water | 50.0 (5/10) | 40.0 (4/10) | 100.0 (10/10) | 1.7 × 102–5.7 × 106 (1.0 × 106) |
| Cold water | 100.0 (2/2) | 50.0 (1/2) | 100.0 (2/2) | 3.6 × 102–8.0 × 102 (5.8 × 102) |
| Facility water | 42.4 (14/33) | 33.3 (11/33) | 90.9 (30/33) | 72.1–6.5 × 105(4.1 × 104) |
| Public hot tub | 38.9 (7/18) | 33.3 (6/18) | 94.4 (17/18) | 72.1–6.5 × 105 (6.1 × 104) |
| Spa | 66.7 (6/9) | 55.6 (5/9) | 88.9 (8/9) | 1.4 × 102–1.3 × 105 (1.7 × 104) |
| Personal hot tub | 16.7 (1/6) | 0.0 (0/6) | 83.3 (5/6) | 1.6 × 102–6.6 × 104 (1.4 × 104) |
| Wastewater | 33.3 (1/3) | 33.3 (1/3) | 100.0 (3/3) | 2.3 × 103–3.6 × 105 (1.2 × 105) |
Numbers in parentheses indicate positive and total samples, respectively.
Quantitative assessment outcomes from positive samples only.
Of the 75 water samples, 27 were collected from the same river and 48 were taken from three hot spring recreation areas. The detection rates also varied greatly by type of water source and detection method used (Table 2). With qPCR method, L. pneumophila was detected in all (27/27) river samples and 93.8% (45/48) of the samples from hot spring areas. In the results, L. pneumophila was detected in all river samples, suggesting that the potential human pathogen may be highly prevalent in the natural environment. Further assessment is warranted with respect to potential infection risks associated with recreational use of the water environment. However, it should not be ruled out that qPCR might overestimate cell counts due to nonspecific signals near the detection limits.29,30
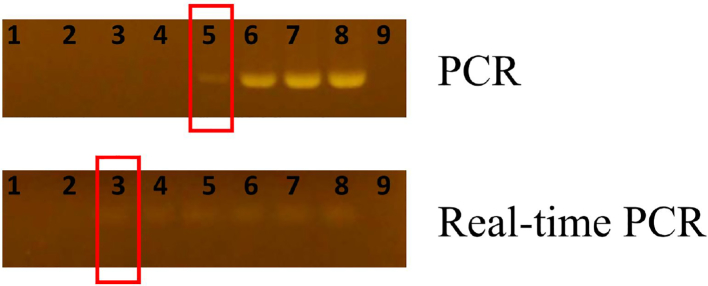
In comparison, the detection rates of conventional PCR are consistently lower than qPCR outcomes. Interestingly, the detection rate of mip PCR is lower than leg PCR especially in the river samples possibly because the primers with higher A/T ratio were more sensitive to PCR inhibitors in receiving waters. On the other hand, evaluation of PCR and TaqMan PCR in artificial samples that inhibitor (Urea) was tested. The results are shown in Fig. 2. TaqMan PCR had much better efficiency on specifying the pathogen in artificial samples. In both Puzih River and hot spring facilities, Taqman qPCR significantly has better efficiency on detecting L. pneumophila. These results suggested that Taqman qPCR is less vulnerable to the environmental PCR inhibitors and may be more suitable for evaluation of the pathogens in recreational spring waters.
Figure 2.
The evaluation of PCR and qPCR inhibitors in artificial samples. DNA concentration: 40 ng/μl, PCR inhibitor: urea. Lane 1:640 mM/μl, lane 2: 320 mM/μl, lane 3: 160 mM/μl, lane 4: 80 mM/μl, lane 5: 40 mM/μl, lane 6: 20 mM/μl, lane 7: 10 mM/μl, lane 8: positive, lane 9: negative.
Quantitative assessment L. pneumophila in hot springs
As shown in Table 2, 45 (93.8%) of the 48 water samples from three hot spring recreation areas were found to contain L. pneumophila and the concentrations ranged from 72.1 to 5.7 × 106 cells/l. Within each hot spring recreation area, separate samples were collected from source water, facilities water, and wastewater. The detection rates ranged from 90.9% in facility water to 100% in source water and wastewater. In addition, the concentrations of L. pneumophila also varied widely by sample source, which ranged up to four orders of magnitude in source and facility water samples. The highest concentration was found among source water samples, but the average concentrations were not significantly different by water type, probably due to large variations and small sample size of the source water (n = 12) and wastewater (n = 3).
The source waters used in hot spring recreation areas include cool stream water and hot spring water. The average concentration of L. pneumophila in positive hot spring samples (1.0 × 106 cells/l) was higher than that in cool stream water (5.8 × 102 cells/l). Legionella spp. is known to thrive in hot environment and L. pneumophila has been found to grow in pipelines for hot spring waters. It is likely that L. pneumophila found in the source water samples may have come from soil or other sources during hot spring collection process (Wallis and Robinson 2005).
Typical facilities in the hot spring recreation areas include public hot tubs, spas, and personal hot tubs. According to qPCR results, the average concentration of L. pneumophila in positive samples from public hot tubs (6.1 × 104 cells/l) was slightly higher than those in spas (1.7 × 104 cells/l) and personal hot tubs (1.4 × 104 cells/l). The lower concentrations in water samples from personal hot tubs may have been a result of frequent draining, which prevented proliferation within the facility. For wastewater, only one sample was collected from each hot spring recreational area, and all were found to contain L. pneumophila, with concentrations between 2.3 × 103–3.6 × 105 and averaged 1.2 × 105 cells/l.
Relationships between occurrence of L. pneumophila and water quality parameters
Several water quality parameters were measured at each sampling location during sample collection, including heterotrophic plate counts (HPCs), total coliforms, temperature, pH, and turbidity. The average and standard deviation of these water quality parameters are summarized in Table 3. Significant difference was found between the presence and absence of L. pneumophila for water temperature (P = 0.041, Mann–Whitney U test). The finding was consistent with that reported by Zanetti.31 Correlation between concentration of L. pneumophila and the five water quality parameters were also evaluated using Spearman's rank-order correlation coefficient test. Significant correlations were found between concentration of L. pneumophila and total coliforms and turbidity. The results in this study were in good agreement with those of our previous report, which indicated that total coliforms in the aquatic environment may be from the soil or fecal contamination.32 A previous study also suggested that organic contents in sediment might support microbial growth in the aquatic environment, which may in turn increase water turbidity (Valster et al., 2011).
Table 3.
Comparisons of selected water quality parameters between L. pneumophila detection outcomes and nonparametric test results
| Mean ± SD | p-levels for nonparametric test | |||
|---|---|---|---|---|
| Water quality parameter | L. pneumophila-presence (n = 72) | L. pneumophila-absence (n = 3) | Mann–Whitney U test | Spearman's rank test (n = 72) |
| Heterotrophic plate counts (HPCs) (CFU/ml) | 1.4 × 105 ± 3.6 × 105 | 2.1 × 105 ± 3.0 × 105 | 0.612 | 0.524 |
| Total coliforms (CFU/100 ml) | 5.6 × 103 ± 9.8 × 103 | 44.9 ± 67.5 | 0.069 | 0.022 |
| Turbidity (NTU) | 1.5 × 102 ± 2.2 × 102 | 0.7 ± 0.4 | 0.231 | 0.004 |
| Temperature (°C) | 33.3 ± 11.3 | 47.7 ± 5.1 | 0.041 | 0.116 |
| pH value | 7.5 ± 0.7 | 8.2 ± 1.0 | 0.201 | 0.723 |
Significant difference at alpha = 0.05
In this study, the qPCR (Taqman) combined with electrophoresis is a rapid and highly sensitive procedure for quantitative assessment of L. pneumophila in natural and man-made water environment. The high prevalence and concentration of L. pneumophila in hot spring water may pose a significant health risk. Further assessment is necessary to determine potential health risks associated with recreational water contact. To reduce the infection risk, the devices should be completely disinfected regularly and the hot spring could be mixed with chlorinated water for temperature adjustment. Visitors should avoid getting choked with the hot spring water. Frequent drainage and cleaning of the recreational facility were also required to reduce potential health risks.
Disclaimer Statements
Contributors
SMS, MYC, BMH, WTJ, TKH and YLH contributed to the study concept, design, data collection, analysis, manuscript preparation and approval. PMK, HFT, YCC, ESK and CWF contributed to data collection, analysis, and manuscript preparation and approval.
Funding
This work was supported by research grants from National Science Council of Taiwan, ROC (NSC 100-2116-M-194-004-MY2) and Cheng Hsin General Hospital, Taiwan, ROC (CH104-2).
Conflicts of interest
All authors have agreed to publish the results.
Ethics approval
No ethics concern in this study.
References
- 1.Tatlock H. A Rickettsia-like organism recovered from guinea pigs. Proc Soc Exp Biol Med. 1944;57:95–9. [Google Scholar]
- 2.Fraser DW, Tsai TR, Orenstein W, Parkin WE, Beecham HJ. Legionnaires' disease: description of an epidemic of pneumonia. N Engl J Med. 1977;297:1189–97. [DOI] [PubMed] [Google Scholar]
- 3.Gomez-Valero L, Rusniok C, Buchrieser C. Legionella pneumophila: population genetics, phylogeny and genomics. Infect Genet Evol. 2009;9(5):727–39. [DOI] [PubMed] [Google Scholar]
- 4.Yu VL, Plouffe JF, Pastoris MC, Stout JE, Schousboe M, Widmer A, et al. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: An international collaborative survey. J Infect Dis. 2002;186(1):127–8. [DOI] [PubMed] [Google Scholar]
- 5.Yanez MA, Nocker A, Soria-Soria E, Murtula R, Martinez L, Catalan V. Quantification of viable Legionella pneumophila cells using propidium monoazide combined with quantitative PCR. J Microbiol Meth. 2011;85(2):124–30. [DOI] [PubMed] [Google Scholar]
- 6.Watson JM, Mitchell E, Gabbay J, Maguire H, Boyle M, Bruce J, et al. Piccadilly Circus legionnaires' disease outbreak. J Public Health Med. 1994;16(3):341–7. [PubMed] [Google Scholar]
- 7.Joseph CA, Harrison TG, Ilijic-Car D, Bartlett CL. Legionnaires' disease in residents of England and Wales: 1996. Commun Dis Rep CDR Rev. 1997;7(11):R153–9. [PubMed] [Google Scholar]
- 8.Marston BJ, Lipman HB, Breiman RF. Surveillance for Legionnaires' disease. Risk factors for morbidity and mortality. Arch Intern Med. 1994;154(21):2417–22. [PubMed] [Google Scholar]
- 9.Hsu BM, Lin CL, Shih FC. Survey of pathogenic free-living amoebae and Legionella spp. in mud spring recreation area. Water Res. 2009;43(11):2817–28. [DOI] [PubMed] [Google Scholar]
- 10.Huang SW, Hsu BM, Wu SF, Fan CW, Shih FC, Lin YC, et al. Water quality parameters associated with prevalence of Legionella in hot spring facility water bodies. Water Res. 2010;44(16):4805–11. [DOI] [PubMed] [Google Scholar]
- 11.Huang SW, Hsu BM, Chen NH, Huang CC, Huang KH, Chen JS, et al. Isolation and identification of Legionella and their host amoebae from weak alkaline carbonate spring water using a culture method combined with PCR. Parasitol Res. 2011;109(5):1233–41. [DOI] [PubMed] [Google Scholar]
- 12.Lee JV, Lai S, Exner M, Lenz J, Gaia V, Casati S, et al. An international trial of quantitative PCR for monitoring Legionella in artificial water systems. J Appl Microbiol. 2011;110(4):1032–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wery N, Bru-Adan V, Minervini C, Delgenes JP, Garrelly L, Godon JJ. Dynamics of Legionella spp. and bacterial populations during the proliferation of L. pneumophila in a cooling tower facility. Appl Environ Microbiol. 2008;74(10):3030–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parthuisot N, West NJ, Lebaron P, Baudart J. High diversity and abundance of Legionella spp. in a Pristine River and impact of seasonal and anthropogenic effects. Appl Environ Microbiol. 2010;76(24):8201–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ortiz-Roque CM, Hazen TC. Abundance and distribution of Legionellaceae in Puerto Rican waters. Appl Environ Microbiol. 1987;53(9):2231–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Declerck P, Behets J, van Hoef V, Ollevier F. Detection of Legionella spp. and some of their amoeba hosts in floating biofilms from anthropogenic and natural aquatic environments. Water Res. 2007;41(14):3159–67. [DOI] [PubMed] [Google Scholar]
- 17.Rossen L, Norskov P, Holmstrom K, Rasmussen OF. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int J Food Microbiol. 1992;17(1):37–45. [DOI] [PubMed] [Google Scholar]
- 18.Kreader CA. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Applied and Environmental Microbiology. 1996;62(3):1102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson IG. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol. 1997;63(10):3741–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burtscher C, Wuertz S. Evaluation of the use of PCR and reverse transcriptase PCR for detection of pathogenic bacteria in biosolids from anaerobic digestors and aerobic composters. Appl Environ Microbiol. 2003;69(8):4618–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kermekchiev MB, Kirilova LI, Vail EE, Barnes WM. Mutants of Taq DNA polymerase resistant to PCR inhibitors allow DNA amplification from whole blood and crude soil samples. Nucleic Acids Res. 2009;37(5):e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radstrom P, Knutsson R, Wolffs P, Lovenklev M, Lofstrom C. Pre-PCR processing – strategies to generate PCR-compatible samples. Mol Biotechnol. 2004;26(2):133–46. [DOI] [PubMed] [Google Scholar]
- 23.APHA Standard method for the examination of water and wastewater. 21st edn Washington, DC: APHA, WEF and AWWA; 2005. [Google Scholar]
- 24.Miyamoto H, Yamamoto H, Arima K, Fujii J, Maruta K, Izu K, et al. Development of a new seminested PCR method for detection of Legionella species and its application to surveillance of Legionellae in hospital cooling tower water. Appl Environ Microb. 1997;63(7):2489–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bej AK, Mahbubani MH, Atlas RM. Detection of viable Legionella pneumophila in water by polymerase chain reaction and gene probe methods. Appl Environ Microbiol. 1991;57(2):597–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Behets J, Declerck P, Delaedt Y, Creemers B, Ollevier F. Development and evaluation of a Taqman duplex real-time PCR quantification method for reliable enumeration of Legionella pneumophila in water samples. J Microbiol Methods. 2007;68(1):137–44. [DOI] [PubMed] [Google Scholar]
- 27.Whelan JA, Russell NB, Whelan MA. A method for the absolute quantification of cDNA using real-time PCR. J Immunol Methods. 2003;278(1–2):261–9. [DOI] [PubMed] [Google Scholar]
- 28.Koide M, Saito A, Kusano N, Higa F. Detection of Legionella spp. in cooling tower water by the polymerase chain reaction method. Appl Environ Microbiol. 1993;59(6):1943–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rawsthorne H, Dock CN, Jaykus LA. PCR-based method using propidium monoazide to distinguish viable from nonviable bacillus subtilis spores. Applied and environmental microbiology. 2009;75(9):2936–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen NT, Chang CW. Rapid quantification of viable legionellae in water and biofilm using ethidium monoazide coupled with real-time quantitative PCR. J Appl Microbiol. 2010;109(2):623–34. [DOI] [PubMed] [Google Scholar]
- 31.Zanetti F, Stampi S, De Luca G, Fateh-Moghadam P, Antonietta M, Sabattini B, et al. Water characteristics associated with the occurrence of Legionella pneumophila in dental units. Eur J Oral Sci. 2000;108(1):22–8. [DOI] [PubMed] [Google Scholar]
- 32.Hsu BM, Chen CH, Wan MT, Cheng HW. Legionella prevalence in hot spring recreation areas of Taiwan. Water Res. 2006;40(17):3267–73. [DOI] [PubMed] [Google Scholar]