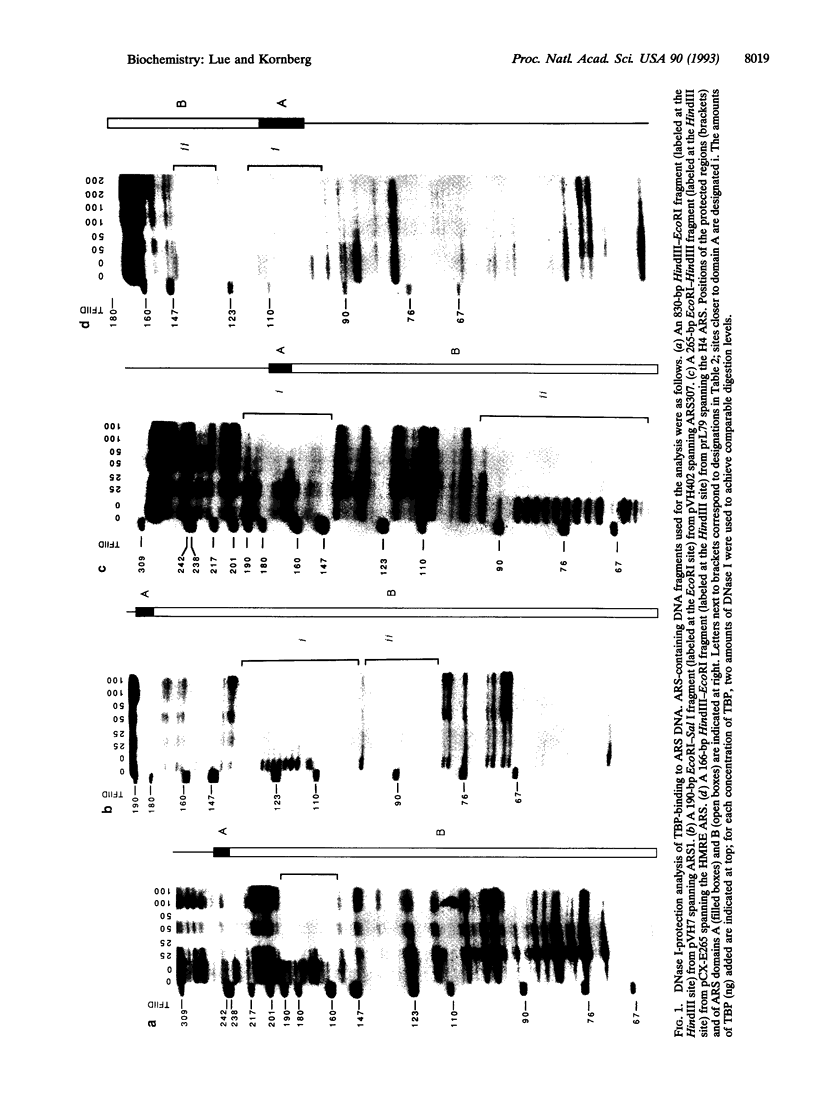
Abstract
The TATA-element-binding protein (TBP) is involved in the initiation of transcription by all three eukaryotic RNA polymerases. The following observations implicate TBP in the initiation of DNA replication at yeast chromosomal origins as well: (i) Recombinant yeast TBP binds specifically to functionally important regions of many yeast replication origins in vitro. (ii) TBP-binding sites from RNA polymerase II promoters can activate defective replication origins in vivo. (iii) Point mutations in TBP-binding sites that diminish their affinity for TBP in vitro reduce their ability to support replication in vivo.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham J., Nasmyth K. A., Strathern J. N., Klar A. J., Hicks J. B. Regulation of mating-type information in yeast. Negative control requiring sequences both 5' and 3' to the regulated region. J Mol Biol. 1984 Jul 5;176(3):307–331. doi: 10.1016/0022-2836(84)90492-3. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992 May 14;357(6374):128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Bouton A. H., Smith M. M. Fine-structure analysis of the DNA sequence requirements for autonomous replication of Saccharomyces cerevisiae plasmids. Mol Cell Biol. 1986 Jul;6(7):2354–2363. doi: 10.1128/mcb.6.7.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Specific protein binding to far upstream activating sequences in polymerase II promoters. Proc Natl Acad Sci U S A. 1985 Jan;82(1):43–47. doi: 10.1073/pnas.82.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987 Nov 6;51(3):463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Kimmerly W. J., Rine J., Kornberg R. D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jan;8(1):210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S., Hahn S., Sharp P. A., Guarente L. Function of a yeast TATA element-binding protein in a mammalian transcription system. Nature. 1988 Jul 7;334(6177):37–42. doi: 10.1038/334037a0. [DOI] [PubMed] [Google Scholar]
- Celniker S. E., Sweder K., Srienc F., Bailey J. E., Campbell J. L. Deletion mutations affecting autonomously replicating sequence ARS1 of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2455–2466. doi: 10.1128/mcb.4.11.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Tanese N., Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992 Mar 6;68(5):965–976. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- Conaway J. W., Hanley J. P., Garrett K. P., Conaway R. C. Transcription initiated by RNA polymerase II and transcription factors from liver. Structure and action of transcription factors epsilon and tau. J Biol Chem. 1991 Apr 25;266(12):7804–7811. [PubMed] [Google Scholar]
- Cormack B. P., Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992 May 15;69(4):685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- Diffley J. F., Cocker J. H. Protein-DNA interactions at a yeast replication origin. Nature. 1992 May 14;357(6374):169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2120–2124. doi: 10.1073/pnas.85.7.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynlacht B. D., Hoey T., Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991 Aug 9;66(3):563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- Eisenberg S., Civalier C., Tye B. K. Specific interaction between a Saccharomyces cerevisiae protein and a DNA element associated with certain autonomously replicating sequences. Proc Natl Acad Sci U S A. 1988 Feb;85(3):743–746. doi: 10.1073/pnas.85.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander S. W., Kallenbach N. R., Heeger A. J., Krumhansl J. A., Litwin S. Nature of the open state in long polynucleotide double helices: possibility of soliton excitations. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7222–7226. doi: 10.1073/pnas.77.12.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman W. L., Hice R. H., Chlebowicz-Sledziewska E. ARS replication during the yeast S phase. Cell. 1983 Mar;32(3):831–838. doi: 10.1016/0092-8674(83)90069-7. [DOI] [PubMed] [Google Scholar]
- Hahn S., Buratowski S., Sharp P. A., Guarente L. Yeast TATA-binding protein TFIID binds to TATA elements with both consensus and nonconsensus DNA sequences. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5718–5722. doi: 10.1073/pnas.86.15.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Bertuccioli C., Takada R., Wang J., Yamamoto T., Roeder R. G. Transcription factor TFIID induces DNA bending upon binding to the TATA element. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):1060–1064. doi: 10.1073/pnas.89.3.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao C. L., Carbon J. High-frequency transformation of yeast by plasmids containing the cloned yeast ARG4 gene. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3829–3833. doi: 10.1073/pnas.76.8.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman J. A., Spotila L. D., Nawotka K. A., el-Assouli S. M., Davis L. R. The in vivo replication origin of the yeast 2 microns plasmid. Cell. 1987 Nov 6;51(3):473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- Kelleher R. J., 3rd, Flanagan P. M., Chasman D. I., Ponticelli A. S., Struhl K., Kornberg R. D. Yeast and human TFIIDs are interchangeable for the response to acidic transcriptional activators in vitro. Genes Dev. 1992 Feb;6(2):296–303. doi: 10.1101/gad.6.2.296. [DOI] [PubMed] [Google Scholar]
- Kimmerly W., Buchman A., Kornberg R., Rine J. Roles of two DNA-binding factors in replication, segregation and transcriptional repression mediated by a yeast silencer. EMBO J. 1988 Jul;7(7):2241–2253. doi: 10.1002/j.1460-2075.1988.tb03064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Torchia T. Structure of the chromosomal copy of yeast ARS1. Biochemistry. 1988 May 31;27(11):3961–3965. doi: 10.1021/bi00411a011. [DOI] [PubMed] [Google Scholar]
- Lorch Y., Kornberg R. D. Near-zero linking difference upon transcription factor IID binding to promoter DNA. Mol Cell Biol. 1993 Mar;13(3):1872–1875. doi: 10.1128/mcb.13.3.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y., Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992 Feb 14;255(5046):817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- Natale D. A., Schubert A. E., Kowalski D. DNA helical stability accounts for mutational defects in a yeast replication origin. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2654–2658. doi: 10.1073/pnas.89.7.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palzkill T. G., Newlon C. S. A yeast replication origin consists of multiple copies of a small conserved sequence. Cell. 1988 May 6;53(3):441–450. doi: 10.1016/0092-8674(88)90164-x. [DOI] [PubMed] [Google Scholar]
- Reinberg D., Horikoshi M., Roeder R. G. Factors involved in specific transcription in mammalian RNA polymerase II. Functional analysis of initiation factors IIA and IID and identification of a new factor operating at sequences downstream of the initiation site. J Biol Chem. 1987 Mar 5;262(7):3322–3330. [PubMed] [Google Scholar]
- Samuels M., Fire A., Sharp P. A. Separation and characterization of factors mediating accurate transcription by RNA polymerase II. J Biol Chem. 1982 Dec 10;257(23):14419–14427. [PubMed] [Google Scholar]
- Schultz M. C., Reeder R. H., Hahn S. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III promoters. Cell. 1992 May 15;69(4):697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- Singer V. L., Wobbe C. R., Struhl K. A wide variety of DNA sequences can functionally replace a yeast TATA element for transcriptional activation. Genes Dev. 1990 Apr;4(4):636–645. doi: 10.1101/gad.4.4.636. [DOI] [PubMed] [Google Scholar]
- Snyder M., Buchman A. R., Davis R. W. Bent DNA at a yeast autonomously replicating sequence. Nature. 1986 Nov 6;324(6092):87–89. doi: 10.1038/324087a0. [DOI] [PubMed] [Google Scholar]
- Stinchcomb D. T., Struhl K., Davis R. W. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979 Nov 1;282(5734):39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- Strubin M., Struhl K. Yeast and human TFIID with altered DNA-binding specificity for TATA elements. Cell. 1992 Feb 21;68(4):721–730. doi: 10.1016/0092-8674(92)90147-5. [DOI] [PubMed] [Google Scholar]
- Umek R. M., Kowalski D. The ease of DNA unwinding as a determinant of initiation at yeast replication origins. Cell. 1988 Feb 26;52(4):559–567. doi: 10.1016/0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Van Houten J. V., Newlon C. S. Mutational analysis of the consensus sequence of a replication origin from yeast chromosome III. Mol Cell Biol. 1990 Aug;10(8):3917–3925. doi: 10.1128/mcb.10.8.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S. S., Francesconi S. C., Eisenberg S. A DNA replication enhancer in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4665–4669. doi: 10.1073/pnas.87.12.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S. S., Malik A. K., Eisenberg S. Analysis of the interactions of functional domains of a nuclear origin of replication from Saccharomyces cerevisiae. Nucleic Acids Res. 1991 Nov 25;19(22):6255–6262. doi: 10.1093/nar/19.22.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley S. R., Kraus R. J., Mertz J. E. Functional binding of the "TATA" box binding component of transcription factor TFIID to the -30 region of TATA-less promoters. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5814–5818. doi: 10.1073/pnas.89.13.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Lieberman P. M., Boyer T. G., Berk A. J. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992 Oct;6(10):1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]