Abstract
Intracellular components must be recycled for cells to maintain energy and ensure quality control of proteins and organelles. Autophagy is a highly conserved recycling process that involves degradation of cellular constituents in lysosomes. Although autophagy regulates a number of cell functions, it was first found to maintain energy balance in liver cells. As our understanding of autophagy has increased, we have found its connections to energy regulation in liver cells to be tight and complex. We review three mechanisms by which hepatic autophagy monitors and regulates cellular metabolism. Autophagy provides essential components (amino acids, lipids and carbohydrates) required to meet the cell’s energy needs, and it also regulates energy supply by controlling the number, quality, and dynamics of the mitochondria. Lastly, autophagy also modulates levels of enzymes in metabolic pathways. In light of the multiple ways in which autophagy participates to control liver metabolism, it is no surprise that dysregulation of autophagy has been associated with metabolic diseases such as obesity, diabetes, or metabolic syndrome, as well as liver-specific disorders such as fatty liver, non-alcoholic steatohepatitis and hepatocellular carcinoma. We discuss some of these connections and how hepatic autophagy might serve as a therapeutic target in common metabolic disorders.
Keywords: chaperone-mediated autophagy, lipophagy, lysosome, macroautophagy, cancer
Introduction
Recycling is the basis of cellular survival by contributing to reduce cellular waste, to preserve cellular energy and to adapt to environmental challenges by regulating abundance of intracellular components1, 2. Autophagy, the process that mediates degradation of intracellular constituents in lysosomes, is the ultimate example of efficient cellular recycling because it seamlessly couples cellular quality control and cellular energetics2. Autophagy contributes to maintain a positive energetic balance through degradation and recycling of proteins3, 4, glycogen5 or lipids6. Regulated turnover through autophagy also ensures renewal and proper functioning of proteome and intracellular organelles7.
Cellular metabolism has been tightly associated with autophagy since the early days when de Duve demonstrated that starvation and glucagon - a glycogenolytic agent - induced hepatic autophagy8. Since then, many new functions have been linked to autophagic activity such as control of cell cycle, immune response, development, differentiation or cell death9, 10. However, the role of autophagy in cellular metabolism has persisted as one of its main functions because of the identification of new additional ways in which autophagy contributes to cellular energetics. We have recently learnt that, added to the previously demonstrated replenishment of the free pool of amino acids through protein breakdown, autophagy also contributes to mobilization and hydrolysis of lipid stores and glycogen5, 6. Furthermore, a selective form of autophagy known as mitophagy controls number and functionality of the mitochondrial network, responsible for ATP generation from different energy stores11, 12. Lastly, enzymes and other proteins that participate in metabolic pathways such as glycolysis, lipophagy or lipolysis can be timely and selectively degraded by autophagy, contributing in this way to modulate the energetic flux through these metabolic pathways13, 14. In this review, we first provide a brief description of the autophagic pathways in the mammalian liver and then summarize recent findings on the role of these autophagy variants in metabolic process and metabolic disorders in the liver, the organ most tightly related with whole body energetics.
Autophagic Pathways in the Liver
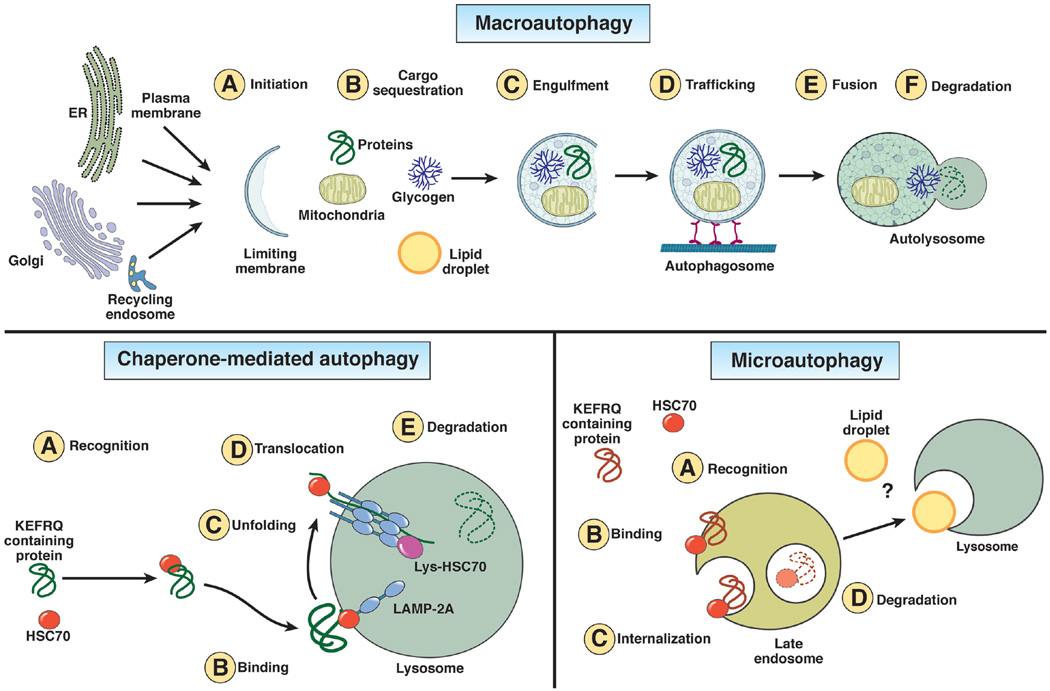
Autophagy stands out among the different post-translational regulatory mechanisms in liver because of its broad range of physiological functions in this organ. The versatility of the two key steps of the autophagy process, breakdown and recycling, along with the co-existence of different autophagic pathways in the same cell, allow for this functional diversity. Three types of autophagy co-exist in hepatocytes, macroautophagy, microautophagy and chaperone-mediated autophagy (CMA) (Fig. 1), and connections with the hepatic energetic balance have been established for all of them. Here, we briefly summarize the main characteristics that differentiate each of these autophagic processes, but readers interested in a more comprehensive description are encouraged to consult recent reviews on this topic9, 15–17.
Figure 1. Scheme of autophagy pathways in the liver.
Schematic depiction of the three types of autophagy that coexist in liver. Macroautophagy is (A) initiated with the formation of the limiting membrane using lipids and proteins from different organelles. Cargo sequestration (B) can occur in bulk or in a selective manner mediated by soluble protein receptors. After engulfment (C) the sealed vesicle (autophagosome) traffics (D) via microtubules and delivers cargo to lysosomes through membrane fusion (E) to form an autolysosome where cargo is degraded by lysosomal hydrolases (F). In chaperone-mediated autophagy (CMA), all substrates carry a pentapeptide (KFERQ-like) recognized (A) by the cytosolic chaperone Hsc70. The substrate-chaperone complex binds (B) to the CMA receptor LAMP-2A at the lysosomal membrane. The substrate must be unfolded (C) before translocation (D) through the multimeric complex formed by LAMP-2A at the lysosomal membrane. A luminal Hsc70 assists in substrate translocation into lysosome where the substrate is finally degraded (E). Microautophagy in liver has been observed in late endosomes where proteins also carrying KFERQ-like motifs are internalized in small microvesicles that form through invagination of the endosomal membrane. As in CMA, the consensus motif allows Hsc70 recognition (A), but in this case the substrate/chaperone complex binds directly to lipids at the endosomal membrane (B). Microvesicles trapping this cargo that form in an ESCRT-dependent manner are internalized (C) into the endosome lumen where degradation takes place (D). Some degradation may also be completed upon endosome/lysosomal fusion. In the case of yeast, direct trapping of lipid droplets by the vacuole (yeast lysosome equivalent) through a microautophagy-like process has been described, but whether or not this process also takes place in mammalian lysosomes requires future investigation.
Macroautophagy
Macroautophagy, the better characterized autophagic pathway, starts with the formation of the limiting membrane of the autophagosome, a double membrane vesicle that delivers the cytosolic material targeted for degradation (cargo) to lysosomes. This limiting membrane, or nascent phagophore, forms through the assembly of proteins and lipids from different cellular organelles such as the endoplasmic reticulum (ER), Golgi, mitochondria, endocytic system or plasma membrane (Fig. 1). Cargo sequestration by autophagy in response to starvation occurs, for the most part, in bulk. However, a better characterization of autophagy activated in response to other stimuli has led to the discovery of selective forms of macroautophagy based on the cargo recruited for degradation18. In these instances, formation of the pre-autophagosome structure occurs in close proximity to the cargo thanks to autophagy receptors, such as p62/SQSTM1, NBR1, NDP52 and Optineurin18. These receptors bind simultaneously cargo and key components of the autophagy machinery known as autophagy–related proteins or ATGs18. The elongation of this membrane around the cargo occurs through the coordinated action of a cascade of ATGs that conjugate among themselves and with lipids19. Upon sealing of the autophagosome membrane, this cargo-containing vesicle moves along microtubules to fuse with the lysosome and deliver the sequestered cargo in this hydrolytic organelle16 (Fig. 1). After hydrolysis, the resultant molecules, amino acids, lipids and carbohydrate moieties reach the cytosol through transporters and permeases for recycling16.
How cells sense the need for a boost in autophagy activity is yet to be well defined and may likely occur through multiple mechanisms depending on the type of stimulus. Recent studies have shed some light on one of such mechanisms by showing that nutrient deprivation can be sensed by signaling cascades present on the primary cilia, such as the Sonic Hedgehog pathway20. Signaling by this pathway activates autophagy by recruiting ATGs to the base of the cilia using ciliary trafficking proteins. The unique position of this single ciliary structure that protrudes from the cellular surface and the highly specialized nature of the membrane that surrounds the cilia, supports the idea that primary cilia may also contribute to modulate autophagy by transducing other signals such as those initiated by growth factors, hormone ligands or even physical stimuli that lead to ciliary bending.
This association of ATGs with the plasma membrane may also assist the activation of autophagy in response to changes in the neighboring cells. Thus, for example, under rich nutritional conditions, connexins, the main structural component of intercellular gap junctions, serve as endogenous repressors of autophagy by directly binding to ATG complexes involved in autophagosome formation. During nutrient scarcity, these connexins become platforms for recruitment of additional ATGs (i.e. ATG14 and ATG9), driving the internalization of connexin-ATGs from the plasma membrane to the recycling endocytic compartment for autophagosome formation21.
Once the autophagy-activating signal is transduced, ATGs control each of the steps of the autophagy pathway, from autophagosome formation to cargo degradation and recycling. Consequently, in addition to their reorganization from inactive to active complexes, changes in ATG expression levels in response to different transcriptional programs modulate the magnitude and duration of autophagy activation. One of the master transcriptional regulators of the autophagic program is the transcription factor EB, or TFEB22, that enhances autophagy flux at multiple levels. First, TFEB transcriptionally manages expression of ATGs genes during sustained autophagy to prevent ATG protein depletion. Second, TFEB increases lysosomal biogenesis to avoid overwhelming of this degradative organelle by the arriving autophagosomes. Lastly, TFEB, also controls genes required in other autophagy steps such as molecular motors for trafficking, SNARES for membrane fusion and specific hydrolases, to accommodate the lysosomal enzymatic load to the type of arriving cargo23,24. However, TFEB and the recently described more potent members of this family such as TFE325 are not the only transcriptional regulators of autophagy. The transcriptional autophagy network is also controlled by the sensing nuclear receptor farnesoid X receptor (FXR) in the fed state, guaranteeing physiological proteostasis through basal recycling of proteins26. During fasting, transcriptional regulation of the autophagy program is monitored by cAMP response element-binding protein (CREB) and peroxisome proliferator-activated receptor-alpha (PPARα)26, 27.
Chaperone-mediated autophagy (CMA)
CMA degrades a specific subset of proteins that cross the lysosomal membrane through the CMA receptor, the lysosome-associated membrane protein type 2A (LAMP-2A)17. All CMA substrate proteins contain in their amino acid sequence a pentapeptide motif (KFERQ)28 that is selectively recognized by the cytosolic heat shock cognate protein of 70kDa (hsc70) (Fig. 1). The substrate/chaperone complex docks at the lysosomal membrane through binding to monomeric LAMP-2A proteins that then organize into a multimeric complex required for translocation29. After unfolding and internalization, the substrate protein is rapidly degraded in the lysosomal lumen17 (Fig. 1). LAMP-2A levels, limiting for CMA activity, are controlled both through transcriptional activation, but more frequently, through direct changes in the stability of LAMP-2A at the lysosomal membrane30. CMA was originally described in liver as part of the hepatocyte response to nutritional changes31. Later studies have revealed that CMA also functions as a defense mechanism against cellular insults in an effort to remove damaged proteins and thereby ensuring appropriate hepatic proteostasis reviewed in 32.
The signaling mechanisms that regulate CMA activity are, for the most part, still unknown. Signaling through the nuclear retinoic acid receptor alpha negatively regulates CMA activity33, while recent studies in T-cells have identified the NFAT-calcineurin axis behind CMA induction during T-cell activation34. Metabolites such as ketone bodies activate CMA35 but whether it is because of their pro-oxidizing effects on proteins or by acting as second messengers in signaling pathways requires further investigation. Free fatty acids (FFA) also activate CMA but, their effect is bi-phasic and, as their levels increase, they inhibit CMA through destabilization of LAMP-2A at the lysosomal membrane36. Changes in the lipid composition of the lysosomal membrane with age are also behind the lower LAMP-2A levels and reduced CMA activity in aging36. Whether LAMP-2A instability is secondary to age-related changes in lipid metabolism, or a primary defect behind some of the metabolic changes of aging remains unclear. In any case, this reciprocal interplay between lipid metabolism and CMA may constitute a vicious cycle that contributes to perpetuate the metabolic syndrome associated with aging. Interestingly, in support of the potential anti-aging effect of breaking this vicious circle by enhancing CMA activity, restoration of LAMP-2A levels in livers from old mice improves cellular homeostasis, enhances liver response to stress and preserves normal hepatic function until late in life37.
Microautophagy
Microautophagy occurs when small single-walled vesicles form at the lysosomal membrane and invaginate toward the lysosomal lumen while engulfing cytosolic components38. Although the original morphological description of this process was performed in liver, the molecular mechanism and physiological relevance of hepatic microautophagy remains, for the most part, unknown. In fact, most of what is known about lysosomal microautophagy originates from studies in yeast where its selectivity for degradation of organelles, lipids and even nuclear portions has been described39–41. Recently, a variation of this process, termed endosomal microautophagy42, was reported to take place in late endosomes in several cell types including hepatocytes. Endosomal microautophagy involves selection of protein cargo by hsc70 using the same pentapeptide motif described for CMA (Fig. 1). However, in this case, the process relies on the ESCRTI and III machinery for the formation of the small invaginating vesicles in the late endosomes and, instead of to LAMP-2A, hsc70 bringing cargo directly binds to lipids at the late endosome membrane and is internalized and degraded with the cargo42.
Hepatic Metabolism and Autophagy
The liver is essential for maintenance of metabolic homeostasis and thereby, the organismal energetic balance. Dysregulation of hepatic autophagy has been described in severe metabolic disorders such as obesity, fatty liver or diabetes reviewed in 32. Consequently, current efforts are directed to dissect the molecular pathways governing this recycling system and the mechanisms by which hepatic autophagy contributes to liver metabolism. Overall, these mechanisms can be grouped under three main functions as follows:
Energy homeostasis through degradation and recycling by autophagy
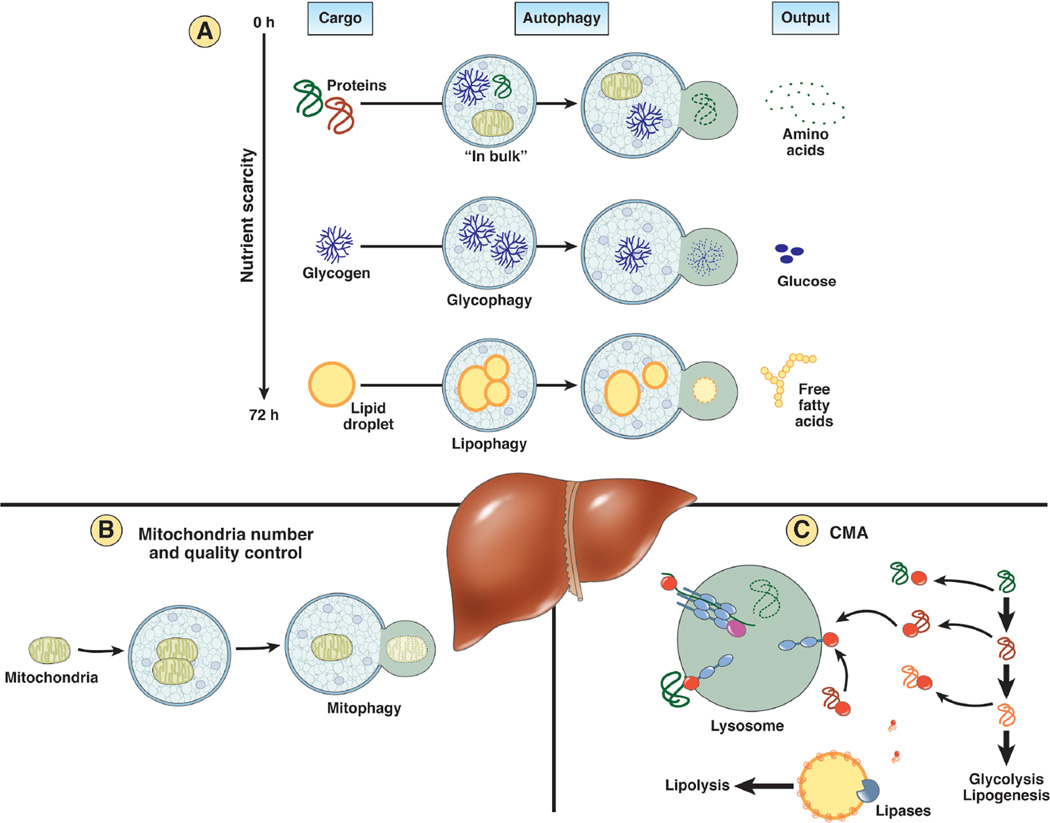
Starvation is the best-characterized trigger for both macroautophagy and CMA activation. In the first 4–6 hours of nutrient shortage inactivation of mechanistic target of rapamycin (mTOR), one of the best-characterized endogenous inhibitors of autophagy, leads to macroautophagy activation43. Amino acids resulting from lysosomal degradation of cytosolic and organelle proteins sustain protein synthesis or directly feed the Krebs cycle in order to produce ATP and/or glucose3 (Fig. 2). Interestingly, some of the released amino acids inhibit autophagy creating an auto-inhibitory feedback.
Figure 2. Three main functions of hepatic autophagy in the control of the energetic balance.
A: Autophagy recycles essential components through degradation of cellular proteins and energy stores. The type of cargo selected by autophagy, at least in liver, changes depending on the duration of nutrient scarcity. While in bulk autophagy of cytosolic proteins and organelles is predominant early in starvation and constitutes an important source of amino acids, if nutrients shortage persists, there is a switch toward glycogen and lipid droplets as preferential cargos. Glycophagy and lipohagy contribute thus glucose and free fatty acids that can be utilized to sustain a positive energetic balance in absence of nutrients. B: Autophagy also manages the cellular energetic balance through the fine-tuned regulation of mitocondrial number and quality control. Mitophagy can eliminate non-functional mitochondria but also controls mitochondrial mass through a coordinated balance with mitochondrial biogenesis. C: Autophagy contributes to accommodation to starvation and other nutritional challenges through selective removal via chaperone-mediated autophagy (CMA) of enzymes that control metabolic pathways such as glycolysis or lipogenesis. In addition, CMA also regulates hepatic lipolytic capacity through degradation of perilipins in the surface of lipid droplets. Removal of these proteins is necessary for cytosolic lipases and autophagy factors to gain access to the lipids in the core of the lipid droplets.
The events connecting mTOR inactivation and autophagy induction have been elucidated. mTOR represses autophagy through direct phosphorylation and sequestration of ULK1, a kinase essential for autophagosome formation. Upon amino acid depletion, inactivation of mTOR is followed by cytosolic release of ULK1 and its relocation to sites of autophagosome biogenesis44. Once amino acids become available again, mTOR reactivation suppresses autophagy. Interestingly, the mechanism for autophagy inhibition seems to be amino acid-specific. For example, changes in leucine levels sensed by the leucyl-tRNA synthetase promote the relocation of the cytosolic mTOR complex to the lysosomal and endosomal membranes through binding to Rag-GTPase and Rheb45. mTOR activation in this location results in inhibitory phosphorylation of TFEB, thus preventing its translocation into the nucleus, and ending the transcriptional autophagic program46. The modulatory effect of other amino acids is less well characterized and apparently more complex. For example, both intracellular uptake of glutamine through SLC1A5 as well as its efflux activate mTOR; this is due to coupling of the exit of this amino acid through the bidirectional transporter SLC7A5/SLC3A2 to intracellular influx of leucine and essential amino acids. Blockage of these transporters results in glutamine depletion and autophagy activation47.
The physiological relevance of autophagy activation as a source of amino acids is exemplified by the death of autophagy-deficient neonate mice during the period between the interruption at birth of the transplacental-nutrient supply and lactation. Administration of amino acids is sufficient to overcome the negative energetic balance of these autophagy-deficient neonates and guarantee their survival4.
In many cell types, if starvation persists beyond 8h, the contribution of macroautophagy to protein breakdown gradually decreases and it is replaced by activation of CMA, which peaks at 24h and can persist up to 3 days into starvation31. Studies in CMA-incompetent models have confirmed that absence of CMA results in a negative energetic balance during starvation13. However, as described in later sections, CMA does not only supplies amino acids under this conditions, but it is also mediates changes in the proteome that facilitate the metabolic switch required for cellular adaptation to persistent nutrient scarcity.
Beyond protein breakdown
Although protein breakdown has been by far the most studied autophagic catalytic activity, degradation of other cellular energy stores by autophagy has recently gained attention. Lysosomes contain a broad repertoire of hydrolases capable of degrading not only proteins but also carbohydrates, lipids and nucleic acids.
Hepatic glycogen stores become an important source of glucose, when no longer available through dietary intake. Glycogen breakdown during starvation is attained in large extent by the lysosomal enzyme acid alpha-glucosidase48 after glycogen delivery to this organelle by selective forms of macro- and microglycophagy (Fig. 2). How the cell senses the decline in glucose levels and switches its machinery to selective glycophagy remains poorly understood. Nonetheless, improved methods to dynamically track the autophagic process in intact cells and the ability to chemically and genetically modulate autophagy has facilitated the identification of some glycophagy effectors. Starch Binding Domain-containing Protein 1/Genethonin 1 (STBD1) binds to glycogen and drives it to autophagosomes through direct interaction with one of the ATGs (γ-aminobutyric acid receptor-associated protein-like 1 or GABARAPL1)49. Both cAMP and mTOR signaling pathways regulate neonatal hepatic glycophagy50 and a link between growth hormone and hepatic glucose modulation has also been proposed51. Studies in skeletal muscle have highlighted the contribution of microautophagy to mobilization of glycogen stores though microglycophagy5, but the involvement of this pathway in hepatic glycogen turnover is still controversial.
Lysosomal breakdown of hepatic lipids has also gained considerable attention in recent years. In addition to the well-characterized arrival of extracellular lipids to lysosomes through endocytosis, intracellular triglycerides and cholesterol stored in the form of lipid droplets can also reach this compartment through a selective form of macroautophagy known as lipophagy6 (Fig. 2). The FFA resulting from lipid droplet breakdown are used as cellular fuel in case of prolonged nutrient scarcity. In fact, after 8h of starvation there is a gradual shift in the autophagosome cargo from cytosolic proteins and glycogen to lipid droplets6. Identification of lipophagy has promoted a change from a model in which cytosolic lipases were solely responsible for triglyceride catabolism towards a cooperative action of cytosolic and lysosomal lipases in this process. Still, the interconnection between both pathways and the conditions that favor one over the other are under debate.
Although the molecular determinants of autophagic selectivity for lipid droplets are not fully understood, recent studies have shown that RAB7 drives the lipid droplets to the perinuclear region where lipophagy is initiated through kiss-and-run processes52. However, access of the autophagic machinery, and by the same token, of the neutral cytosolic lipases, to the lipid core of lipid droplets requires the CMA-mediated removal of perilipins, the structural proteins that cover the surface of lipid droplets. This step is a prerequisite before lipid breakdown can occur53, thus placing CMA upstream of the two main types of hepatic lipolysis, the one mediated by neutral cytosolic lipases and the other dependent on lipophagy. Degradation of lipid droplets through microautophagy has, to date, only been described in yeast41.
Ongoing investigations have shed light on some of the signaling mechanisms that modulate lipophagy. TFEB exerts an exquisite fine-tuning control on this process by adapting the lysosomal enzymatic load to the arrival of the lipid cargo, and by sustaining the activation of PGC1α and PPARα-dependent programs23,24. Interestingly, lipophagy is activated in the two extremes of nutritional status since, besides starvation, arrival of high levels of dietary lipids to the liver also activates lipophagy to prevent hepatic lipotoxicity6. The well-established FFA sensor PPARα is behind the potent activation of autophagy under these conditions27. Similarly, CREB activation increased lipophagy probably through a mechanism involving AMPK phosphorylation of ULK126, 54. Contrarily, the end-products of lipid metabolism, bile acids, have an inhibitory effect on lipophagy both at the transcriptional level, through FXR sensing in the basal nutritional state27, and by reducing autophagosome-lysosome fusion in hepatocytes through changes in Rab7 association with autophagosomes55. The specific signals that trigger selective removal of perilipins by CMA to initiate lipolysis remain unknown. However, phosphorylation of perilipins in localized regions of the lipid droplets is a pre-requisite for their delivery to lysosomes through this pathway, supporting the possible participation of lipid-sensor kinases in this process53.
Although lipophagy and CMA are activated in response to acute increases in intracellular lipids, pronounced or chronically sustained lipid challenges inhibit both autophagic pathways via changes in the membrane lipid composition of autophagosomes and lysosomes36, 56. In fact, this inhibitory effect along with this newly described role of autophagy in the control of intracellular lipid content may contribute to perpetuate the dysfunctional lipid metabolism characteristic of old organisms. Thus, the decrease in autophagic activity with age may lead to accumulation of intracellular lipids that in turn further inhibits lipophagy, creating a vicious cycle.
Autophagic adjustment of mitochondrial metabolic capacity
Mitochondria, the main source of intracellular ATP, contribute to cellular metabolism through different processes including FFA oxidation, citric acid cycle, biosynthesis of heme and phospholipids and calcium storage. Therefore, adequate maintenance of mitochondria fitness and number is essential for cellular homeostasis. High membrane potential/proton motive force across the inner mitochondrial membrane can promote an excessive production of reactive oxygen species, which in turn results in mitochondrial dysfunction through oxidative damage of membrane proteins and mitochondrial DNA. These events often lead to mitochondrial permeability transition (MPT)11 which causes full and irreversible mitochondrial depolarization57. Selective removal of depolarized and damaged mitochondria through autophagy (mitophagy) under these conditions prevents futile consumption of ATP by the mitochondrial ATP synthase working in reverse and also limits generation of free radicals. Mitophagy contributes in this way to restore the energetic balance and to reduce cellular damage averting in this way activation of cell death pathways (Fig. 2).
The recent detailed dissection of mitophagy has revealed likely co-existent mechanisms for the recognition and elimination of mitochondria via autophagy reviewed in 11, 12, 58. The best described is the PINK1/PARKIN pathway, activated in response to membrane depolarization. Membrane damage leads to PINK1 accumulation on the mitochondria surface which facilitates recruitment of the ubiquitin ligase PARKIN. Ubiquitination of a variety of mitochondrial outer membrane proteins by PARKIN serves as tag for ATG recognition and mitochondria engulfment59. Additional receptors also facilitate mitophagy in response to different environmental and cellular cues, such as hypoxia or nutritional changes. These proteins include BCL2/adenovirus E1B 19-kDa-interacting protein 3 (BNIP3), BNIP3 like (NIX) or FUN14 domain containing 1 (FUNDC1)58. The regulation of mitochondria turnover to accommodate energetic demand occurs at least in part through ULK1 phosphorylation by AMPK60 and by the tight balance between mitochondrial biogenesis and mitophagy orchestrated by the TFEB-PGC1 alpha axis61. Future comparative studies of the different types of mitophagy may help to further understand whether specific forms of mitophagy are more dedicated to basal quality control while others are preferentially engaged to modify the mass of still functional mitochondria.
Selective regulation of enzymes
Autophagy can also regulate the cellular energetic balance through degradation of enzymes that catalyze different cellular metabolic cycles. This function requires a high degree of selectivity for individual proteins and consequently, it was first described for CMA. The first evidence of physiological relevance of the degradation of glycolytic enzymes by CMA was obtained in the context of cancer metabolism as CMA was recently described to modulate degradation of pyruvate kinase, muscle isoform 2 (PKM2), a key enzyme in glucose metabolism that is usually expressed only in embryonic cells62. The M2 isoform is also more abundant in cancer cells because it favors the switch toward aerobic glycolysis rather than oxidative phosphorylation for the production of ATP, a process known as the Warburg effect. This switch provides metabolic advantage to rapidly growing transformed cells and also increases abundance of growth promoting glycolytic intermediates since PKM2 has lower activity than PKM1 (ubiquitously expressed in most cells)63. Acetylation of PKM2 contributes to its turnover via CMA and further tumor growth because of the accumulation of intermediates62.
Later studies demonstrated a complex interplay between CMA and cancer metabolism. Thus, CMA has been found upregulated in more than 14 different cancer types and its blockage slows down growth of solid tumors and markedly reduces metastasis frequency64. Active CMA is necessary to preserve the Warburg effect in different types of cancer cells in a p53-dependent manner64. Blockage of CMA in cancer cells increases p53, which in turn transcriptionally represses the subset of glycolytic enzymes required to sustain the Warburg effect. Although the reasons behind the increase in p53 observed upon CMA blockage in cancer cells requires further investigation, it is possible that its increase is result of the inefficient DNA repair and accumulation of DNA damage observed in CMA-incompetent cells65.
Despite the first connection between CMA and cellular metabolism was established in cancer cells, recent studies in animals with hepatic CMA blockage have revealed an important physiological role for this pathway both in basal and starvation-induced turnover of enzymes involved in lipid, carbohydrate and amino acid metabolism13. Comparative proteomics of lysosomes isolated from livers with functional or ablated CMA revealed that more than 40% of the proteins identified as CMA substrates are involved in metabolism, with almost 2/3 of them involved in carbohydrate and lipid metabolism13. Accordingly, livers unable to perform CMA exhibit elevated basal levels of glycolytic enzymes such as GAPDH, PK, aldolase A, malate dehydrogenase 1 or enolase 1, leading to elevated basal glycolysis. This continuous consumption of glucose by the liver leads to defective gluconeogenesis and scarcity of glycogen hepatic stores. CMA deficiency becomes even more obvious under starvation conditions, when CMA degrades glycolytic enzymes to reduce the usage of hepatic glucose and favor its mobilization to other organs (Fig. 2).
In absence of CMA, the continuous glycolytic flux in the liver leads to a negative body energetic balance and depletion of the peripheral adipose tissues13. The reduced content of peripheral fat is in part also due to the failure to mobilize hepatic lipids under these conditions and their subsequent accumulation as LD in this organ. Although elevated levels of lipogenic enzymes that normally undergo degradation via CMA could contribute to the pronounced steatosis of CMA-deficient livers, most of the lipid accumulation has been recently shown due to their failure to perform lipolysis53. In this case, the CMA substrate responsible for the control of the lipolytic flux is not an enzyme but instead the peripheral proteins that cover lipid droplets in this organ known as perilipin 2 and 3. These proteins need to be partially removed upon lipolysis stimulation to allow access of cytosolic lipases (responsible for neutral lipolysis) and ATGs (responsible for lipophagy) to the lipids in the core of lipid droplets. This discrete removal of perilipins involves their recognition by hsc70 and phosphorylation that delivers them for lysosomal degradation via CMA53 (Fig. 2). If the CMA lysosomal receptor is ablated, hsc70 and perilipins persist bound to the surface of lipid droplets interfering with the initiation of the lipolytic process. CMA degradation of perilipins both during starvation, to facilitate mobilization of hepatic lipids for usage by peripheral tissues and during sustained lipogenic challenges (i.e. diets rich in fat), when lipid mobilization prevents hepatic lipotoxicity53. All together, these findings highlight CMA importance in hepatic metabolic homeostasis and as an essential component for the adaptation to nutritional changes.
Recently published studies in yeast support regulated degradation by macroautophagy of fatty acid synthase, an enzyme key for lipid metabolism and for adaptation to starvation14. The determinants of selective degradation in this case are unknown and future studies are needed to discriminate if degradation of fatty acid synthase by macroautophagy occurs as single isolated units or along with other cytosolic cargo. However, since CMA is not present in yeast or invertebrates, it is appealing to speculate that evolutionary pressure to separate degradation of enzymes with regulatory purposes from in bulk degradation contributed to the development of CMA in mammals.
Hepatic Autophagy and Metabolic Diseases
The tight connections between autophagy and cellular and organism metabolism described in previous sections, make it easy to infer why altered autophagic function underlies the basis of important metabolic disorders. A detailed description of each of these disorders and the causative or aggravating role that autophagy dysregulation plays in them is beyond the scope of this review. Instead, we just highlight here representative examples of different types of autophagic failure and include references that review in more detail the interplay between autophagy and specific metabolic disorders32, 66.
Fatty liver/Non-alcoholic steatohepatitis
Obesity and type two diabetes are often associated with non-alcoholic fatty liver disease (NAFLD) or non-alcoholic steatohepatitis (NASH). Increased lipid flux into the liver, augmented de novo lipid synthesis or decreased lipid catabolism all contribute to the hepatic accumulation of lipids and NAFLD. Chronic persistence of this lipid overload leads to liver injury with inflammation, cell death and fibrosis characteristic of NASH. As mentioned in the previous section, reduced macroautophagy and CMA in liver both result in marked hepatosteatosis although through different mechanisms.
In the case of macroautophagy, part of the alterations in lipid metabolism are at the level of lipid mobilization since hepatic ATG7 deletion, decreases TGs breakdown resulting in lipid droplets accumulation6. Furthermore, macroautophagy blockage also leads to deficiencies in proteostasis (increased polyubiquitinated protein content67) and in organelle 68,69. ER stress and defective ER functioning in the context of the chronic metabolic dysfunction that associates with NASH contributes to exacerbate the defects in glucose metabolism68. Failure of mitochondria quality control, due to their reduced turnover through mitophagy, can promote oxidative stress through ROS production and activation of downstream inflammatory pathways such as the NOD-like receptor family, pyrin domain containing 3 (NLRP3) inflammasome and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB)69. The combination of lipotoxicity, oxidative stress and chronic activation of the inflammatory response upon macroautophagy failure often leads to hepatocyte cell death, thus recapitulating the hallmarks of NASH (inflammation, oxidative stress, cell death and fibrosis).
Reduced hepatic CMA promotes hepatosteatosis due to an increase in lipogenic enzymes and failure in the timely removal of perilipins13,53. Added to lipotoxicity, oxidative damage can also contribute to hepatic injury upon CMA blockage. Thus, CMA is upregulated in liver in response to oxidative stress to selectively remove damaged proteins70. Defects in proteostasis upon persistent blockage of hepatic CMA promote accumulation of oxidized protein aggregates and thus contribute to perpetuate chronic oxidative stress in this organ71.
Most of the changes that characterize the metabolic syndrome and that include obesity, hyperglycemia, dyslipidemia and high blood pressure, have also shown to exert a negative effect on autophagy. In the presence of insulin resistance and hyperinsulinemia, the regulatory effect of forkhead box O1 (FoxO1) on the expression of several Atg genes is lost resulting in autophagy malfunction72. The increase in intracellular lipids also reduces both macroautophagy and CMA due to changes in intracellular membrane composition36,56. High dietary lipids alter the lysosomal stability of the CMA receptor and lead to reduced activity of this pathway36. Likewise, lipid changes in the autophagosome limiting membrane, reduce the ability of these vesicles to fuse with lysosomes and leads to a decrease in macroautophagic flux56. This reduced clearance of autophagosomes could explain the accumulation in LC3-II and p62 observed in patients diagnosed with non-alcoholic steatosis (NAS) and NASH and that positively correlate with the severity of the disease73. Studies in obese mice and NALD patients have also demonstrated that fatty liver decreases lysosome activity through inhibition of cathepsin expression which reduces lysosomal degradation of cargo delivered by all types of autophagy and endocytosis74.
Given the above-described essential role of the different types of autophagy in liver metabolism, their dysregulation during dietary challenges or abnormal metabolic conditions can further aggravate the metabolic malfunction, thus creating a deleterious vicious cycle. This continuous negative feedback makes it often difficult to determine if the autophagic failure is primary or secondary.
Hepatocarcinogenesis
Numerous studies have revealed complex interconnections between autophagy and oncogenesis in different organs reviewed in 75. In the liver, where one of the late consequences of metabolic malfunction is development of hepatocarcinomas, changes in autophagy activity have also been linked to tumorogenesis.
Hepatic macroautophagy exerts an anti-oncogenic function, demonstrated by the fact that lowering ATG5 or ATG7 levels triggers occurrence of multiple liver tumors. Accordingly, human hepatocellular carcinomas (HCC) show reduced levels of autophagic proteins and activity which is associated with malignancy and bad prognosis76. Accumulation of the autophagy adaptor protein p62 that occurs upon autophagy failure is in part responsible for the observed increase in liver tumors, since deletion of p62 in autophagy-deficient livers counteracts tumorogenesis77. In fact, p62 accumulation promotes development and growth of hepatocellular adenomas in these mice78 through overactivation of the nuclear factor erythroid 2–related factor 2 (Nrf2)79. P62 binds to the Nrf2-binding site on kelch-like ECH-associated protein 1 (Keap1), the ligase that mediates Nrf2 degradation, resulting in Nrf2 release, nuclear translocation and upregulation of Nrf2-mediated expression of cytoprotective and detoxifying enzymes that support tumor growth 79. Furthermore, Nrf2 activates the expression of multi-drug efflux pumps conferring liver tumors resistance against oxidative stress and chemotherapeutic agents80. Interestingly, liver tumors in autophagy deficient mice lack the malignant phenotype indicating the requirement of other genetic changes coupled to macroautophagy malfunction for hepatic carcinogenesis. Growing evidence support that macroautophagy can also be utilized by hepatic cancer cells for tumor progression and, in fact, increased levels of macroautophagy markers such as LC3 in hepatocarcinoma has been associated with bad prognosis and higher rates of recurrence after surgery81. Hence, it might possible that, at least in mice, deficient autophagy only promotes the development of benign tumors but halts their progression into cancer.
CMA has also a dual role in liver tumor development. In support of an anti-tumoral function of CMA, mice with defective hepatic CMA develop hepatocellular adenomas spontaneously by middle age. This enhanced tumorogenesis could be a combination of the hepatosteatosis, poor quality control, and increased oxidative damage associated with chronic CMA deficiency71. However, once malignant transformation occurs, cancer cells display a marked increase in CMA activity that is key to sustain their unique metabolic requirements64. As described in previous sections, blockage of CMA in cancer cells attenuates their characteristic high rates of aerobic glycolysis in a p53-dependent manner leading to reduced tumorogenic and metastastatic capacities64. Further investigations are needed to determine if the recent identified role for CMA in DNA repair65 also contributes to cancer cell survival and resistance to anti-oncogenic interventions.
Autophagy as an Attractive Target for Metabolic Diseases
Multiple genetic interventions have provided proof-of-principle to the value of autophagy as a therapeutic target in multiple liver diseases and metabolic disorders. For example, ATG7 adenoviral delivery has been proven successful to restore insulin/glucose homeostasis as well as ER homeostasis in obese mice68. Similarly, overexpression of TFEB through similar approaches displays remarkable protection against lipotoxicity induced by high fat diet in mice23. Chemical upregulation of macroautophagy with compounds such as resveratrol has recently shown efficient in reducing NAFLD-caused hepatic injury82. Even in the case of CMA, regulated expression of LAMP-2A in old mice, to counteract the decrease in endogenous levels of this receptor with aging, reduces liver proteotoxicity and improves hepatic function37.
Although activation of autophagy in liver is considered protective, modulation of autophagy for the treatment of NASH may require a more targeted approach since enhanced macroautophagy activity in hepatic stellate cells has been shown to underlie hepatic fibrosis83. In fact, lipophagy provides the fueling required for the switch of these cells from their normal quiescent phenotype towards activated myofibroblast-like cells that synthesize extracellular matrix, triggering liver fibrosis and NASH
An added current challenge, is how to translate these genetic triumphs into effective chemical modulation of the autophagic pathways. Very promising results have been obtained in obese mice with some non-autophagy specific drugs such as blockers of calcium channels, where the protective effect against hepatosteatosis and insulin resistance has been directly linked to the activation of autophagy by these drugs84. Other drugs known to activate autophagy such as carbamazepine and rapamycin are able to decrease fatty liver, reduce circulating triglycerides and improve insulin homeostasis85. However, the direct application of many of these reagents in clinic is not without limitations due to the non-autophagy effects of many of them. Selective chemical modulation of CMA has also been challenging. However, the recent development of compounds that activate CMA through selective blockage of a subset of the retinoic-acid alpha signaling33 may open now new options for the treatment of metabolic disorders in which CMA downregulation is involved.
Beyond chemical modulation, the close relationship between autophagy and the nutritional status justifies recent attempts to use dietary interventions to modulate this catabolic process with therapeutic purposes. Extreme dietary regimes such as caloric restriction, proven to be one of the most effective methods to increase health span in many species, exert their effect in part through activation of autophagy86. Although the ability of caloric restriction to increase life-span in non-human primates is still controversial, there is general agreement on the protective effect of caloric restriction against metabolic stress and associated disorders even in these species87, 88. These positive results justify current efforts to chemically mimic the effects of calorie restriction which in large part has been based on modification of cytosolic levels of acetyl-coenzyme A, recently described to regulate autophagy86. Interestingly, most of the caloric restriction mimetic drugs, that include among others, resveratrol, metformin, spermidine, rapamycin, nicotinamide or curcumin, have all been shown to activate autophagy reviewed in 86, further supporting the value of autophagy regulation in metabolic disorders. However, for most of the currently available drugs that activate autophagy, their action is not limited to this catabolic pathway and the coexistence of non-desirable additional effects limits their use in therapeutics.
Concluding Remarks and Open Questions
Studies during the last 10 years have broadly expanded our understanding of the multiple points of interaction between autophagy and cellular and organism metabolism. Autophagy has proven essential for the adaptation of nutritional availability, both during nutrient excess and paucity. There are however many pending questions about the newly identified connections between autophagy and the cellular energetic balance. Although organelle and protein selectivity during autophagy is convincingly demonstrated, the determinants of selectivity for lipids or glycogen are poorly understood. Also pending is the analysis of the possible interplay among the different functions of autophagy in the control of cellular energetics, the relative contribution of different forms of autophagy to the energetic balance and the basis for cell- and tissue- differences. From a translational point of view, although the connections between autophagy malfunctioning and metabolic disorders and their bidirectional nature are unquestionable, the therapeutic advancement is on hold until more selective activators of autophagy become available. Nutrient interventions along with other changes in life-style such as physical exercise, recently shown to improve glucose homeostasis through activation of BCl2-mediated autophagy89, could substitute for the lack of chemical modulators in the prevention against metabolic disorders. However, in patients already affected by these severe conditions, it is likely that only acute and prominent chemical activation of autophagy will be able to disrupt the negative vicious cycles established between metabolic and autophagy dysfunction.
ACKNOWLEDGMENTS
We thank Drs. Susmita Kaushik for critical reading of the manuscript. We apologize to those whose work could not be cited owing to space limitations.
FUNDING SOURCES: Work in our laboratory is supported by National Institutes of Health grants AG021904, AG031782 and DK098408 and the generous support of R&R Belfer (to A.M.C.). JMM is supported by a postdoctoral fellowship from the American Diabetes Association.
Abbreviations
- ATG
autophagy-related genes
- BNIP3
BCL2/adenovirus E1B 19kDa-interacting protein 3
- BNIP3L
BCL2/adenovirus E1B 19kDa-interacting protein 3 like (NIX)
- CMA
chaperone-mediated autophagy
- CREB
cAMP response element-binding protein
- FFA
free fatty acids
- FXR
farnesoid X receptor
- FOXO1
forkhead box protein O1
- GAA
acid alpha-glucosidase
- GABARAPL1
γ-aminobutyric acid receptor-associated protein-like 1
- HCC
hepatocellular carcinoma
- Keap1
kelch-like ECH-associated protein 1
- mTOR
mechanistic target of rapamycin
- NAFLD
non-alcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- NAS
non-alcoholic steatosis
- Nrf2
nuclear factor erythroid 2–related factor 2
- HSCs
hepatic stellate cells
- PPARα
peroxisome proliferator-activated receptor alpha
- PKM2
pyruvate kinase muscle isoform 2
- STBD1
starch binding domain-containing protein 1/genethonin 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR’S CONTRIBUTION: JMM performed bibliographic search, summarized and analyzed the content of the selected manuscripts, wrote a complete draft of the review and figures and revised the edited version; AMC contributed the design of sections and organization of the original outline, performed bibliographic searches, edited and added to the original draft and review the final version of the manuscript.
DISCLOSURE: The authors have no conflict of interest.
References
- 1.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 2005;12:1535–1541. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- 3.Ezaki J, Matsumoto N, Takeda-Ezaki M, et al. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy. 2011;7:727–736. doi: 10.4161/auto.7.7.15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 5.Raben N, Hill V, Shea L, et al. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet. 2008;17:3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh R, Kaushik S, Wang Y, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizushima N, Levine B, Cuervo AM, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deter RL, Baudhuin P, De Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol. 1967;35:C11–C16. doi: 10.1083/jcb.35.2.c11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Baehrecke EH. Eaten alive: novel insights into autophagy from multicellular model systems. Trends Cell Biol. 2015;25:376–387. doi: 10.1016/j.tcb.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15:344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemasters JJ. Variants of mitochondrial autophagy: Types 1 and 2 mitophagy and micromitophagy (Type 3) Redox Biol. 2014;2:749–754. doi: 10.1016/j.redox.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu J, Wang KZ, Chu CT. After the banquet: mitochondrial biogenesis, mitophagy, and cell survival. Autophagy. 2013;9:1663–1676. doi: 10.4161/auto.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider JL, Suh Y, Cuervo AM. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 2014;20:417–432. doi: 10.1016/j.cmet.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shpilka T, Welter E, Borovsky N, et al. Fatty acid synthase is preferentially degraded by autophagy upon nitrogen starvation in yeast. Proc Natl Acad Sci U S A. 2015;112:1434–1439. doi: 10.1073/pnas.1409476112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Y, He D, Yao Z, et al. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen HM, Mizushima N. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem Sci. 2014;39:61–71. doi: 10.1016/j.tibs.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 19.Mizushima N, Yoshimori T, Ohsumi Y. The Role of Atg Proteins in Autophagosome Formation. Annual Review of Cell and Developmental Biology. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 20.Pampliega O, Orhon I, Patel B, et al. Functional interaction between autophagy and ciliogenesis. Nature. 2013;502:194–200. doi: 10.1038/nature12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bejarano E, Yuste A, Patel B, et al. Connexins modulate autophagosome biogenesis. Nat Cell Biol. 2014;16:401–414. doi: 10.1038/ncb2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Settembre C, Di Malta C, Polito VA, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Settembre C, De Cegli R, Mansueto G, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol. 2013;15:668–676. doi: 10.1038/ncb2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martina JA, Diab HI, Lishu L, et al. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal. 2014;7:ra9. doi: 10.1126/scisignal.2004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seok S, Fu T, Choi SE, et al. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014;516:108–111. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JM, Wagner M, Xiao R, et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 29.Bandyopadhyay U, Kaushik S, Varticovski L, et al. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuervo AM, Dice JF. Regulation of lamp2a levels in the lysosomal membrane. Traffic. 2000;1:570–583. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- 31.Cuervo AM, Knecht E, Terlecky SR, et al. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- 32.Schneider JL, Cuervo AM. Liver autophagy: much more than just taking out the trash. Nat Rev Gastroenterol Hepatol. 2014;11:187–200. doi: 10.1038/nrgastro.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anguiano J, Garner TP, Mahalingam M, et al. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat Chem Biol. 2013;9:374–382. doi: 10.1038/nchembio.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valdor R, Mocholi E, Botbol Y, et al. Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nat Immunol. 2014;15:1046–1054. doi: 10.1038/ni.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finn PF, Dice JF. Ketone bodies stimulate chaperone-mediated autophagy. J Biol Chem. 2005;280:25864–25870. doi: 10.1074/jbc.M502456200. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez-Navarro JA, Kaushik S, Koga H, et al. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2012;109:E705–E714. doi: 10.1073/pnas.1113036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mortimore GE, Hutson NJ, Surmacz CA. Quantitative correlation between proteolysis and macro- and microautophagy in mouse hepatocytes during starvation and refeeding. Proc Natl Acad Sci U S A. 1983;80:2179–2183. doi: 10.1073/pnas.80.8.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki K. Selective autophagy in budding yeast. Cell Death Differ. 2013;20:43–48. doi: 10.1038/cdd.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uttenweiler A, Mayer A. Microautophagy in the yeast Saccharomyces cerevisiae. Methods Mol Biol. 2008;445:245–259. doi: 10.1007/978-1-59745-157-4_16. [DOI] [PubMed] [Google Scholar]
- 41.van Zutphen T, Todde V, de Boer R, et al. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2014;25:290–301. doi: 10.1091/mbc.E13-08-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahu R, Kaushik S, Clement CC, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanazawa T, Taneike I, Akaishi R, Yoshizawa F, Furuya N, Fujimura S, Kadowaki M. Amino acids and insulin control autophagic proteolysis through different signaling pathways in relation to mTOR in isolated rat hepatocytes. J Biol Chem. 2004;279:8452–8459. doi: 10.1074/jbc.M306337200. [DOI] [PubMed] [Google Scholar]
- 44.Egan D, Kim J, Shaw RJ, et al. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy. 2011;7:643–644. doi: 10.4161/auto.7.6.15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han JM, Jeong SJ, Park MC, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 46.Settembre C, Zoncu R, Medina DL, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicklin P, Bergman P, Zhang B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hers HG. alpha-Glucosidase deficiency in generalized glycogenstorage disease (Pompe's disease) Biochem J. 1963;86:11–16. doi: 10.1042/bj0860011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang S, Heller B, Tagliabracci VS, et al. Starch binding domain-containing protein 1/genethonin 1 is a novel participant in glycogen metabolism. J Biol Chem. 2010;285:34960–34971. doi: 10.1074/jbc.M110.150839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kondomerkos DJ, Kalamidas SA, Kotoulas OB, et al. Glycogen autophagy in the liver and heart of newborn rats. The effects of glucagon, adrenalin or rapamycin. Histol Histopathol. 2005;20:689–696. doi: 10.14670/HH-20.689. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Fang F, Goldstein JL, et al. Reduced autophagy in livers of fasted, fat-depleted, ghrelin-deficient mice: Reversal by growth hormone. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1423643112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schroeder B, Schulze RJ, Weller SG, et al. The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology. 2015;61:1896–1907. doi: 10.1002/hep.27667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol. 2015;17:759–770. doi: 10.1038/ncb3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koo SH, Flechner L, Qi L, et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 55.Manley S, Ni HM, Kong B, et al. Suppression of autophagic flux by bile acids in hepatocytes. Toxicol Sci. 2014;137:478–490. doi: 10.1093/toxsci/kft246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu H, Chen Q. Hypoxia activation of mitophagy and its role in disease pathogenesis. Antioxid Redox Signal. 2014 doi: 10.1089/ars.2014.6204. [DOI] [PubMed] [Google Scholar]
- 59.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Egan DF, Shackelford DB, Mihaylova MM, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma X, Liu H, Murphy JT, et al. Regulation of TFEB-PGC1alpha Axis by BECLIN-1 Controls Mitochondrial Quality and Cardiomyocyte Death under Stress. Mol Cell Biol. 2015 doi: 10.1128/MCB.01091-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lv L, Li D, Zhao D, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 64.Kon M, Kiffin R, Koga H, et al. Chaperone-mediated autophagy is required for tumor growth. Sci Transl Med. 2011;3:109ra117. doi: 10.1126/scitranslmed.3003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park C, Suh Y, Cuervo AM. Regulated Degradation of Chk1 by chaperone-mediated autophagy in response to DNA damage. Nat. Comm. 2015 doi: 10.1038/ncomms7823. E-Pub ahead of printng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang X, Overholtzer M, Thompson CB. Autophagy in cellular metabolism and cancer. J Clin Invest. 2015;125:47–54. doi: 10.1172/JCI73942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang L, Li P, Fu S, et al. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou R, Yazdi AS, Menu P, et al. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 70.Kiffin R, Christian C, Knecht E, et al. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schneider JL, Villarroya J, Diaz-Carretero A, et al. Loss of hepatic chaperone-mediated autophagy accelerates proteostasis failure in aging. Aging Cell. 2015;14:249–264. doi: 10.1111/acel.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu HY, Han J, Cao SY, et al. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284:31484–31492. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gonzalez-Rodriguez A, Mayoral R, Agra N, et al. Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis. 2014;5:e1179. doi: 10.1038/cddis.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Inami Y, Yamashina S, Izumi K, et al. Hepatic steatosis inhibits autophagic proteolysis via impairment of autophagosomal acidification and cathepsin expression. Biochem Biophys Res Commun. 2011;412:618–625. doi: 10.1016/j.bbrc.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 75.Galluzzi L, Pietrocola F, Bravo-San Pedro JM, et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34:856–880. doi: 10.15252/embj.201490784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ding ZB, Shi YH, Zhou J, et al. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Res. 2008;68:9167–9175. doi: 10.1158/0008-5472.CAN-08-1573. [DOI] [PubMed] [Google Scholar]
- 77.Takamura A, Komatsu M, Hara T, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ni HM, Woolbright BL, Williams J, et al. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J Hepatol. 2014;61:617–625. doi: 10.1016/j.jhep.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Inami Y, Waguri S, Sakamoto A, et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shibata T, Ohta T, Tong KI, et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci U S A. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang JD, Seol SY, Leem SH, et al. Genes associated with recurrence of hepatocellular carcinoma: integrated analysis by gene expression and methylation profiling. J Korean Med Sci. 2011;26:1428–1438. doi: 10.3346/jkms.2011.26.11.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li L, Hai J, Li Z, et al. Resveratrol modulates autophagy and NF-kappaB activity in a murine model for treating non-alcoholic fatty liver disease. Food Chem Toxicol. 2014;63:166–173. doi: 10.1016/j.fct.2013.08.036. [DOI] [PubMed] [Google Scholar]
- 83.Hernandez-Gea V, Ghiassi-Nejad Z, Rozenfeld R, et al. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142:938–9346. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Park HW, Park H, Semple IA, et al. Pharmacological correction of obesity-induced autophagy arrest using calcium channel blockers. Nat Commun. 2014;5:4834. doi: 10.1038/ncomms5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin CW, Zhang H, Li M, et al. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J Hepatol. 2013;58:993–999. doi: 10.1016/j.jhep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marino G, Pietrocola F, Madeo F, et al. Caloric restriction mimetics: natural/physiological pharmacological autophagy inducers. Autophagy. 2014;10:1879–1882. doi: 10.4161/auto.36413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Colman RJ, Anderson RM, Johnson SC, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mattison JA, Roth GS, Beasley TM, et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489:318–321. doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He C, Bassik MC, Moresi V, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]