Abstract
The discovery of long non-coding RNAs (lncRNAs) and the elucidation of the mechanisms by which they affect different disease states is providing researchers with a better understanding of a wide array of disease pathways. Moreover, lncRNAs are presenting themselves as both unique diagnostic biomarkers as well as novel targets against which to develop new therapeutics. Here we will explore the intricate network of non-coding RNAs associated with infection by the human immunodeficiency virus (HIV). Non-coding RNAs derived from both the human host as well as those from HIV itself are emerging as important regulatory elements. We discuss here the various mechanisms through which both small and long non-coding RNAs impact viral replication, pathogenesis and disease progression. Given the lack of an effective vaccine or cure for HIV and the scale of the current pandemic, a deeper understanding of the complex interplay between non-coding RNAs and HIV will support the development of innovative strategies for the treatment of HIV/acquired immunodeficiency disease (AIDS).
1. HIV
The Human Immunodeficiency Virus Type 1 (HIV-1) epidemic remains a major global health concern. Recent years have seen a worldwide effort to combat this disease, and although this has brought about a concomitant decrease in new infections, UNAIDS reports that a staggering 35 million people are still living with HIV-1. HIV, the causative agent of AIDS, is a lentivirus belonging to the Retroviridae family and was first isolated in 1983 (Barre-Sinoussi et al., 1983; Gallo et al., 1983). More than 30 years following its discovery, scientists are still piecing together the intricacies governing the host-pathogen interactions that enable this virus to stay one step ahead of a preventative vaccine or therapeutic cure. The contributions of ncRNAs, specifically lncRNAs to the biogenesis of HIV have piqued the interest of researchers worldwide and may represent the link to the tools needed to beat this virus. Preliminary studies on viral lncRNAs have even offered clues on potential strategies to purge latent viral reservoirs, which is currently the biggest limitation of antiretroviral drug cocktails which have otherwise had dramatic success in reducing both morbidity and mortality in HIV-infected individuals.
1.1. A simple genome for a complex life cycle
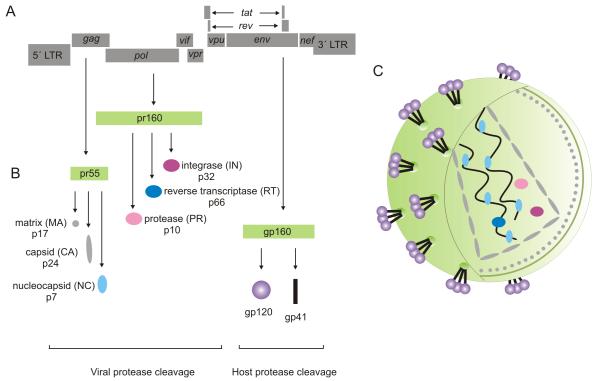
Each virion contains two copies of the RNA genome, which upon being reverse transcribed into cDNA, are then integrated into the human genome for expression and thus long term persistence. The HIV genome is 9.8 kb in length and consists of nine partially overlapping genes (Ratner et al., 1985; Sanchez-Pescador et al., 1985; Wain-Hobson et al., 1985) (Figure 1 A). Complex splicing patterns are responsible for the generation of more than 40 unique HIV mRNA transcripts and post-translational processing enables a rich diversity of protein products (Figure 1 B). The gag (group-specific antigen); pol (polymerase) and env (envelope) genes encode mainly structural and enzymatic proteins. The pr55 Gag protein precursor is processed into the p17 matrix (MA), p24 capsid (CA), and p7 nucleic-acid binding (NC) structural proteins essential for assembly of viral particles (Ganser-Pornillos et al., 2008). The pr160 Gag-Pol protein precursor is processed into the reverse transcriptase (RT), protease (PR), and integrase (IN) enzymes which play critical roles in the viral replicative cycle (Hill et al., 2005). Both the Gag and Gag-Pol polyprotein precursors are cleaved by the viral protease into their respective subunits (Figure 1 B). The gp160 Env protein is cleaved by cellular proteases into the surface gp120 and transmembrane gp41 subunits of the envelope glycoprotein (Figure 1 B) which is necessary for binding to the CD4 primary receptor on host cells (Moulard and Decroly, 2000). The gp120 surface subunit of the Env protein initially engages with the cellular CD4 receptor leading to a conformational change in gp120 which allows it to bind to a co-receptor, either CCR5 or CXCR4 (Salzwedel et al., 2000). Co-receptor binding then triggers the interaction of the gp41 transmembrane subunit of the Env protein with the host cell membrane. Fusion of the cellular and viral membranes ensues (Campbell and Hope, 2008) and the viral core is released into the cellular environment and uncoated, releasing the viral genome (Dvorin and Malim, 2003).
Figure 1. HIV genome organization.
(A) A representation of the nine genes encoded by the HIV-1 genome. (B) The gag, pol and env genes encode protein precursors (pr55, pr160 and gp160) which require further processing by either viral or cellular proteases to generate structural proteins necessary for the formation of a mature virion (C).
The tat (transcriptional transactivator) and rev (regulator of virion gene expression) genes encode two regulatory proteins. Tat, a multiply spliced transcript synthesized early in the viral life cycle, binds to the TAR (transactivation response) element within the R domain of the 5′ long terminal repeat (LTR) to regulate transcription. This interaction between Tat and the TAR loop is required for the synthesis of full-length transcripts. In the absence of Tat, only short attenuated RNA transcripts are produced (Kao et al., 1987). Rev is translated from a completely spliced early phase transcript and is responsible for mediating the transport of singly spliced and unspliced viral transcripts from the nucleus to the cytoplasm. Unspliced transcripts including genomic RNA, Gag-Pol precursors, and incompletely spliced mRNAs encoding Env, Vif, Vpr and Vpu, require the interaction between the regulatory Rev protein and the Rev responsive element present within these transcripts for nuclear export, [reviewed in (Pollard and Malim, 1998)].
In addition to these five genes, HIV also has four accessory genes: nef (negative effector); vif (viral infectivity factor); vpr and vpu (viral proteins r and u) which encode critical virulence factors (Anderson and Hope, 2004; Emerman and Malim, 1998). Flanking the protein coding sequences on either side are the 5′ and 3′ long terminal repeat regions (LTRs), each comprised of 3 domains, namely the U3, R and U5 domains. The U3 region contains the basal promoter as well as an enhancer sequence and multiple transcription factor binding sites (Patarca et al., 1987; Roebuck and Saifuddin, 1999). Transcription initiation occurs at the first nucleotide of the R region and this region also encodes the transcriptional response (TAR) regulatory stem loop structure to which Tat binds. It is apparent that HIV is a complex retrovirus (Figure 1 C) that has evolved a sophisticated, multi-leveled biogenesis pathway that relies on complex splicing and multiple cellular factors for a successful infection cycle.
1.2 The host’s contribution
Limited by a relatively small genome of only 9.8kb and 9 genes, it is no surprise that the virus has evolved sophisticated methods for hijacking cellular machinery for its own benefit. A set of host dependency factors have been identified which is crucial for the completion of HIV’s relatively complex infection cycle. The host factors required for HIV infection and replication are not only limited to host cell receptors. Multiple other host factors have been identified which play an integral role in various aspects of integration, RNA transcription, splicing, cellular localization and protein maturation. These factors include, but are not limited to, integration factors such as BAF1, Emerin and LEDGF/p75 (Jacque and Stevenson, 2006; Llano et al., 2006; Maertens et al., 2003); transcription factors such as NF-kB, PAK-1 and cyclin T1 (Chiu et al., 2004; Nguyen et al., 2006; Surabhi and Gaynor, 2002); and Furin which plays a role in the maturation of the Env protein (Nguyen et al., 2006). Several seminal studies conducting genome-wide siRNA screens were conducted to identify HIV host dependency factors which are implicated in viral infection (Brass et al., 2008; Konig et al., 2008; Zhou et al., 2008). These RNAi screens identified hundreds of potential host factors which are central to the establishment of viral infection however; the overlap of genes identified between these screens was disappointing with only 3 genes overlapping all 3 studies (Bushman et al., 2009; Goff, 2008). Regardless of the number of host dependency factors to be identified, these screens and most other studies to date have focused primarily on protein factors.
A deeper exploration of the transcriptome in the last few years has revealed a complex network of transcripts that are no longer regarded as “noise” or “junk RNA”. Instead, it has become apparent that such non-coding RNAs (ncRNAs) play crucial roles in gene regulation. In addition to protein coding messenger RNA (mRNA) and the well-studied ncRNAs such as transfer RNA (tRNA); ribosomal RNA (rRNA); small nuclear RNA (snRNA); small nucleolar RNA (snoRNA), many other types of ncRNAs are now commonly reported (Morris and Mattick, 2014). These classes of ncRNAs can generally be differentiated upon the basis of their biogenesis pathways, their mechanism, and their cellular roles. Three broad classes of eukaryotic small RNAs have been classified and include microRNAs (miRNAs), endogenous small interfering RNAs (endo-siRNAs) and Piwi-interacting RNAs (piRNAs). Besides the plethora of small ncRNAs identified, considerable attention has been focused on the class of long non-coding RNAs (lncRNAs), generally defined as transcripts longer than 200bp, that have recently been annotated and whose functionality is quickly being established. As a class, lncRNAs play a pivotal role in the regulation of gene expression, acting as epigenetic modulators to an array of genes (Rinn and Chang, 2012).
Many of these ncRNAs may also offer potential as both diagnostic biomarkers and therapeutic targets and in addition, may represent an additional layer of complexity in viral regulation during HIV infection. Considerable attention is therefore owed to both viral-derived and host-derived ncRNAs deregulated during infection. These ncRNA species could provide further pieces of the puzzle and lead to a greater understanding of the mechanisms by which HIV regulates its own transcription and replication and furthermore, could provide additional clues as to how HIV enters and maintains a state of latency.
2. Non-coding RNAs are essential components of HIV’s regulatory network
It has long since been suggested that HIV exploits the host’s gene regulation system to coordinate and direct viral gene expression and replication. Since multiple viral genes are differentially expressed from a single promoter throughout the various stages of the HIV infection cycle, alternative regulatory mechanisms stretching beyond cellular transcription factors must exist to ensure such transcriptional specificity. Recent research studies have pointed to ncRNAs as essential elements in the regulatory network that controls HIV expression (Chen et al., 2014; Farberov et al., 2015; Imam et al., 2015; Patel et al., 2014; Saayman et al., 2014; Schopman et al., 2012; Sung and Rice, 2009; Zhang et al., 2013; Zhang et al., 2014). These regulatory ncRNAs have been demonstrated to originate from both the virus as well as from host cells. While some of these non-coding transcripts arise as part of the host cell’s global response to viral infection, many others have been demonstrated to serve as integral regulatory elements for viral replication, pathogenesis, disease progression and host-pathogen interactions. Regulatory roles are fulfilled by various types of ncRNAs including well-characterized small ncRNAs such as miRNAs, siRNAs, and asRNAs and more recently lncRNAs. Furthermore, it has become readily apparent that HIV manages to actively modulate the expression profiles of host and viral ncRNA during the course of infection, and that through this closely regulated control of the ncRNA expression, HIV may achieve the synchronized gene expression necessary to accomplish a successful replicative cycle. Beyond the regulation of active viral transcription, ncRNAs may also play a role in establishing and maintaining viral latency, wherein the provirus adopts a state of transcriptional shutdown. As is the case with many host genes, suppression of gene expression may often be controlled by ncRNA species. Growing evidence therefore supports an intricate interplay between HIV and host gene regulation machinery directed by various ncRNAs.
2.1 Small non-coding RNAs implicated in HIV infection
2.1.1 Host-derived small ncRNAs
MicroRNAs (miRNAs) are small RNA duplexes 21-24 bp in length that regulate gene expression at the post-transcriptional level. miRNAs are derived from the sequential processing of imperfect RNA stem-loop structures by the Drosha/DGCR8 microprocessor complex and by Dicer. Endogenous host miRNAs have long been implicated in regulatory roles related to viral pathogenesis. This miRNA-based regulation in HIV became obvious after it was demonstrated that HIV gene expression and replication was augmented following the inhibition of two key enzymes in the miRNA biogenesis pathway, Drosha and Dicer (Nathans et al., 2009; Triboulet et al., 2007). Subsequently, a large number of cellular miRNAs differentially expressed during HIV infection have been identified and their roles in viral pathogenesis characterized. These miRNAs are able to function either by directly targeting the viral genomic RNA or mRNA transcripts, or by targeting various host factors which play an integral role in the viral replicative cycle, thereby indirectly impacting viral replication.
Cyclin T1 represents one such critical host factor targeted by a host-derived miRNA during HIV infection, as it serves an integral role in the transcription of full length viral transcripts. The interaction between Tat and the regulatory TAR loop described above is enhanced when Tat binds to the cyclin T1 subunit of the positive elongation factor (P-TEFb). Tat binding recruits the cyclin dependent kinase 9 (CDK9) subunit to the LTR, which in turn phosphorylates RNA Pol II, enabling the transition of initiation to elongation and the consequent synthesis of full-length viral transcripts (Taube et al., 1999). Overexpression of the cellular miR-198 leads to diminished levels of viral replication through the direct targeting of the 3′UTR of cyclinT1 miRNA (Sung and Rice, 2009). Furthermore, while miR-198 was found to be highly expressed in monocytes, its expression is down-regulated upon differentiation to macrophages, offering a potential explanation for the natural restriction of HIV replication in monocytes (Sung and Rice, 2009). The histone acetyltransferase P300/CBP-associated factor (PCAF) is an additional co-factor of Tat and also contributes to the procession of viral transcription. Intriguingly, PCAF is targeted by miR-17-5p and miR-20a, two miRNA components of the polycistronic miR-17/92 cluster which is downregulated in HIV-infected cells (Triboulet et al., 2007).
Upon HIV infection, the host miRNAs let-7c, miR-34a, and miR-124a are upregulated, and this upregulation correlates with the increased ability of HIV to propagate. Working in tandem, miR-34a and miR-124a inhibit TASK1 mRNA (Farberov et al., 2015). TASK1, a mammalian potassium channel, contains high structural homology with the HIV protein Vpu. Due to this homology, these two proteins exhibit strong functional interactions, with TASK overexpression inhibiting Vpu’s ability to enhance the release of HIV virions (Hsu et al., 2004). Let-7c on the other hand serves to inhibit CDKN1A mRNA expression, resulting in decreased p21 protein levels (Farberov et al., 2015). Besides simply being a member of the p53 pathway, p21 protein has been demonstrated to inhibit viral reverse transcriptase and to inhibit the enzymatic activity of the host protein CDK9 described above (Chen et al., 2011). As such, the inhibition of p21 increases both viral integration as well as total viral RNA within an infected cell. It is clear therefore that multiple cellular miRNAs contribute to the elaborate coordination of HIV transcription and thereby indirectly regulate HIV replication.
Two commonly reported cellular miRNAs that appear to play a more direct role in the regulation of viral replication are miR-29a and miR-29b (Ahluwalia et al., 2008; Nathans et al., 2009). Both miR-29a and miR-29b function by directly targeting a conserved site within the viral nef gene, thus inhibiting viral replication (Ahluwalia et al., 2008). More recent studies have also confirmed this ability of miR-29a and miR-29b to play an inhibitory role in viral replication (Sun et al., 2012) while others have gone further to implicate miR-29a in viral latency (Patel et al., 2014). Offering evidence for the role of miR-29a as integral to the latent state, there exists an inverse correlation between HIV replication and levels of miR-29a, with increased miR-29a levels present during HIV latency and reduced levels of miR-29a following activation of HIV replication using PMA (Patel et al., 2014). miR-29a appears to suppress HIV replication through accumulation of viral mRNA in P-bodies (Nathans et al., 2009).
miR-29a is not the only miRNA associated with latency. Further examples support the notion that an intricate relationship existing between HIV and the endogenous RNA-interference (RNAi) pathway contributes to the multifaceted mechanism underlying viral latency. Multiple cellular miRNAs including miR-28; miR-125b; miR-150; miR-223 and miR-382 have all been shown to be upregulated in a latent resting CD4+ T cell population (Huang et al., 2007). These enriched cellular miRNAs inhibit HIV-1 protein translation through interactions with the 3′ end of viral mRNA transcripts and therefore appear to play a pivotal role in HIV latency. A panel of miRNA inhibitors against these miRNAs effectively facilitated viral production in these resting CD4+ T cells and offers the potential to serve as a therapeutic modality to purge the latent reservoirs (Huang et al., 2007; Zhang et al., 2007).
These examples suggest that miRNAs may not only regulate viral transcription, but also serve as key dictators of the fate of the infection cycle. Supporting the view that miRNAs are key regulators of disease progression are observations that cellular miRNAs known to regulate HIV replication are differentially expressed between certain infected populations. A comparison of the plasma miRNA levels between healthy donors and different groups of infected individuals identified a set of miRNAs including miR-29b, miR-33a and miR-146a which are present at significantly higher levels within a population of elite controllers compared to other chronically infected individuals; therefore, the increased expression of these miRNAs has been suggested to influence disease outcome (Reynoso et al., 2014). As nef is a known target of miRNA-29b, it is curious, that modifications in the viral nef gene have been previously associated with elite controllers (Kirchhoff et al., 1995). Nevertheless, it is clear that analyses of the plasma miRNA signature of HIV-infected individuals can thus offer diagnostic and/or prognostic clues in infected individuals.
These are merely a few examples of miRNAs which have already been reported in the literature that directly target HIV thereby manipulating some aspect of the infection cycle. Moreover, it remains highly unlikely that additional novel, HIV-regulating miRNAs will not be discovered in the near future. Already, a study has revealed the presence of up to 22 target sites within the HIV genome for host cellular miRNAs and at least 5 of these synthetically generated miRNAs are capable of decreasing viral replication (Houzet et al., 2012).
Furthermore, recent studies demonstrate that miRNAs can function to modulate HIV infection in the absence of the RISC protein complex thought to be essential for miRNA function. In this novel RNA interference (RNAi)-independent miRNA mechanism, miRNAs have been observed to bind to the Gag protein’s RNA binding nucleocapsid domain. This interaction between miRNA and Gag prevents the assembly of the Gag lattice at the plasma membrane leading to the degradation of incompletely assembled Gag complexes and a reduction in the release of infectious viral particles (Chen et al., 2014). These observations are supported by work which shows that the Gag nucleocapsid domain is dynamic in nature and alters its RNA binding specificity at different stages of virion genesis (Kutluay et al., 2014). Structural changes are essential for the regulation of Gag localization to the plasma membrane and the packaging of the viral RNA genome but may account for the non-specific interactions with cellular miRNAs. These studies demonstrate a new role for miRNAs and add yet another layer of complexity to cellular ncRNA-mediated regulation of HIV infection. Not only is the population of small ncRNAs known to modulate HIV infection growing, but the mechanisms by which these ncRNAs are able to affect HIV biogenesis and infectivity are diversifying too.
2.1.2 Viral-derived small ncRNAs
An abundance of endogenous cellular small ncRNAs have been unquestionably shown to play a role in the replicative life cycle of HIV-1. Until recently however, very few viral derived ncRNAs had been identified, let alone assigned a functional role in infection. Highly coordinated transcriptional regulation and tightly controlled alternative splicing strategies enable HIV to produce more than 40 unique sense transcripts. Therefore, it comes as no surprise that upon closer investigation, HIV generates several additional ncRNAs including antisense transcripts, siRNA duplexes and miRNAs (Schopman et al., 2012). Enrichment of low abundant small ncRNAs using hybridization capture techniques showed that HIV encodes many small ncRNAs of varying lengths spread throughout the viral genome (Althaus et al., 2012). Deep sequencing technologies have also allowed for a more sensitive method to detect virus-derived small RNAs (Schopman et al., 2012) and together these results suggest that numerous small virus-derived RNAs are produced in HIV infected cells which may potentially play a role in viral replication.
The Transactivating Response (TAR) element of the HIV genome is undoubtedly the most commonly-known source of HIV-derived miRNAs (Bennasser et al., 2004; Klase et al., 2007; Klase et al., 2009; Ouellet et al., 2008; Ouellet et al., 2013). TAR is a structured RNA approximately 50 nt in length found at the 5′ end of all HIV mRNA transcripts and has been reported to be the source of two functional miRNAs. What stands out as interesting is the fact that the 5′ and 3′ TAR regions produce functionally different small RNAs, likely due to the need for differential regulation at various points in the viral life cycle. With respect to the 5′ TAR element, miRNAs are not produced from a complete, full length transcript, but rather as a result of the termination of RNA polymerase II transcription (Harwig et al., 2014; Harwig et al., 2015). At the start of viral transcription at the 5′ LTR, and in the absence of Tat, RNA polymerase II experiences a transcriptional delay, pausing 3 nt following the last paired nucleotide within the 5′ TAR hairpin structure (Palangat et al., 1998). This attenuation of transcription and the formation of a pre-miRNA structure results in further processing of this hairpin by the microprocessor complex in conjunction with the termination factors Setx, Xrn2, and Rrp6. Drosha-mediated cleavage of the TAR hairpin produces a nonadenylated TAR transcript and an uncapped 3′ transcript, which in turn leads to the recruitment of termination factor Xrn2 and the degradation of any potential downstream transcript. Resulting from this process is a single, isolated 60nt TAR transcript from which miRNAs are derived (Wagschal et al., 2012). Binding of the Tat protein to the TAR loop changes the conformation of the TAR hairpin thus removing this transcriptional blockage, and enabling the formation of full length transcripts (Kao et al., 1987). Much effort has been focused on elucidating the role of these TAR-derived miRNAs and a noticeable role in apoptosis is clearly emerging. Two host cellular genes were initially identified as TAR miRNA targets: Excision repair cross complementing-group 1 (ERCC1) and Intermediate early response 3 (IER3). These genes are involved in apoptosis and cell survival and the down-regulation of their expression leads to the protection of HIV-infected cells from apoptosis (Klase et al., 2009). Since then multiple additional genes related to apoptosis have been recognized as TAR miRNA targets and include Caspase 8, Aiolos and Ikaros (Ouellet et al., 2013). The TAR-derived miRNAs therefore play an important role in HIV disease progression by regulating cellular apoptosis and promoting cell survival to ensure the persistence of viral infection.
Another viral-derived miRNA reported early on was miR-N367 which emanates from the viral nef gene (Omoto et al., 2004). This miRNA was believed to target a site at the 3′ end of nef, which overlaps the U3 region of the viral LTR and was thought to play a role in transcriptional regulation of the virus (Omoto and Fujii, 2005; Omoto et al., 2004). As mentioned before, advances in next generation sequencing technology are enabling the identification of further viral derived miRNAs. Recently, deep sequencing led to the discovery of a novel HIV encoded miRNA, miR-H3 (Zhang et al., 2014). The precursor sequence of miR-H3 resides within the RT coding sequence of HIV and the mature miRNA sequence overlaps the conserved catalytic domain of the viral RT. Overexpression of miR-H3 led to upregulation of HIV transcription while the inhibition of miR-H3 through sequence mutations led to diminished viral production thus classifying miR-H3 a viral enhancer. Upon analysis of putative binding sites for miR-H3, it was found that miR-H3 binds to the TATA box within the 5′LTR, thereby targeting the viral core promoter. ChIP analysis showed that miR-H3 binding at the promoter correlated with enhanced association of both RNA Polymerase II as well as the TATA box binding protein at the core promoter (Zhang et al., 2014). This miRNA represents yet another HIV-derived regulatory element capable of activating viral replication.
These small RNAs described above are thought to be processed from hairpin-like secondary structures within the sense RNA strand by Drosha and Dicer in a similar manner to endogenous miRNAs to generate virus-derived miRNAs. In addition to viral miRNAs, it has been demonstrated that antisense transcription from an internal promoter within the HIV genome or from a host promoter downstream of the integrated provirus may lead to the formation of viral siRNAs. A limited number of antisense RNAs emanating from the 3′ LTR region have been identified and these antisense transcripts may form double stranded RNA intermediates with HIV mRNA, which are able to be processed by Dicer to generate viral-derived siRNAs. An early study looking at the potential of HIV-1 to elicit an RNAi response identified a perfectly duplexed 19bp Dicer substrate within the HIV-1 genome. This HIV-derived short interfering RNA (siRNA) was able to specifically target its complementary transcript within the env gene leading to a reduction in both Env mRNA and protein levels (Bennasser et al., 2005). Additional HIV-derived siRNAs were identified in a deep sequencing study of HIV-infected cells and were found to inhibit virus production by mediating cleavage of the viral transcript at the target site (Schopman et al., 2012). Furthermore, in both of the aforementioned studies, inhibition of the viral-derived siRNAs mitigated their inhibitory effects resulting in increased HIV production (Bennasser et al., 2005; Schopman et al., 2012). These viral derived siRNAs appear therefore to function via the endogenous RNAi pathway and are capable of modulating viral production.
To date, only a small number of virally-derived small ncRNAs have been identified and functionally characterized. Nevertheless, evidence suggests that a much larger population of HIV-1 derived small ncRNAs exists that can be detected with improved selection and screening protocols (Althaus et al., 2012; Schopman et al., 2012). Consequently, it is decidedly probable that further viral derived functional ncRNAs will be described in the near future.
2.2 Long non-coding RNAs implicated in HIV infection
Long non-coding RNAs (lncRNAs) have recently emerged as a significant component of the mammalian transcriptome (Morris and Mattick, 2014). The evidence supporting their role in the overall regulation of cellular pathways continues to grow as the dysregulation of these lncRNA transcripts repeatedly correlates with a series of maladies (Wapinski and Chang, 2011). Furthermore, the transcription of lncRNAs has been implicated as a necessary component in numerous differentiation and developmental pathways (Mattick, 2007; Morris and Mattick, 2014). However, to date, the mechanism by which lncRNAs function is not yet completely understood as lncRNA transcripts are capable of impacting gene expression via a myriad of functional mechanisms (Wang and Chang, 2011; Wilusz et al., 2009). Most likely contributing to the breadth by which lncRNAs function is the genomic context from which the transcripts originate. Functional lncRNAs may be transcribed in the sense or antisense direction relative to the protein coding gene, and may stem from intronic regions, long intergenic regions or even pseudogenes (Kung et al., 2013; St Laurent et al., 2015). As research rapidly continues within this field of study, functional roles are being assigned to lncRNAs at an accelerated pace and roles in viral infection are no exception. We have already discussed at length the essential functions performed by small ncRNAs in the replicative cycle of HIV; therefore, it follows that HIV is likely to also act in concert with host lncRNAs to regulate viral infection. Moreover, analysis of the viral transcriptome indicates the presence of viral derived lncRNAs which are expected to contribute to regulatory systems governing viral gene expression.
2.2.1 Host-derived lncRNAs
NEAT1
To investigate lncRNA expression following infection by HIV-1, two independent groups performed RNA-seq on HIV infected cells (Imam et al., 2015; Zhang et al., 2013). Imam et al. identified a set of lncRNAs that are modulated during both HIV infection and the activation of latent virus. In total, two lncRNAs were identified which were downregulated, while eighteen were upregulated in both conditions (Imam et al., 2015). Experimentally, Zhang et al. chose to compare the changes in lncRNA expression across two different T cell lines when infected with HIV. When comparing the infection of these two cell lines, only a total of six lncRNAs displayed either up- or down-regulation (Zhang et al., 2013). When comparing these two data sets, one finds low concordance between the two sets. More disconcerting is the fact that both groups evaluated NL4-3 infection of the Jurkat T cell line and failed to achieve consistent results, suggesting that bioinformatics analysis of deep sequencing results should be experimentally validated. The inconsistent results between these two groups may arise due to the fact that changes in lncRNA expression has been found to be highly sensitive to external stimuli (Tani et al., 2014) as well as the fact that lncRNA transcripts exist in low expression levels across all cells within the population (Cabili et al., 2015), a feature that produces greater variability when analyzing transcript levels. As such, one may find the use of cultured cells lines to have certain limitations in elucidating the role of lncRNAs in HIV. Nevertheless, a mechanistic analysis of two of these unique lncRNAs reveals that they play an integral role in modulating HIV expression.
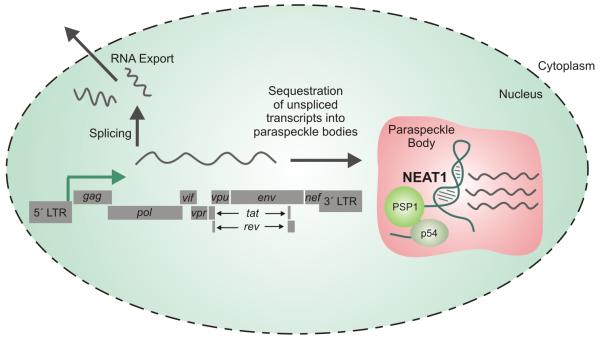
One such lncRNA identified by deep sequencing of host T cells to be upregulated following HIV-1 infection was NEAT1 (Zhang et al., 2013). Within the cell, the primary role of NEAT1 is to regulate and maintain nuclear paraspeckle bodies by acting as a binding scaffold for the proteins PSP1 and p54 (Figure 2). It follows therefore, that upon infection by HIV, upregulation of NEAT1 results in greater formation of paraspeckle bodies within the nucleus (Clemson et al., 2009). As a unit, paraspeckles serve primarily to retain dsRNA molecules in the form of hyperedited A-to-I RNA sequences within the nucleus, enabling cells to discriminate which RNA sequences are exported to the cytoplasm for translation. To perform this function, three proteins, p54, Matrin 3, and PSF, function as a unit to anchor these hyperedited, dsRNA sequences to the nuclear matrix (Zhang and Carmichael, 2001). To counter this repression of nuclear-to-cytoplasmic export, the HIV protein Rev enables the export of both spliceable and non-spliceable pre-mRNA in a manner independent of the mode of nuclear retention, provided that the RNA sequences contain a Rev-response element. This alternative pathway provides a means for the release of viral RNA sequences into the cytoplasm allowing for translation of viral proteins (Fischer et al., 1999). Nevertheless, NEAT1 inhibition during HIV infection and the ensuing loss of paraspeckle bodies results in increased export of unspliced Rev-dependent instability element (INS)-containing HIV mRNAs. The finding that unspliced HIV might be sequestered in these paraspeckles regardless of the Rev protein’s ability for export has led to the conclusion that HIV might have co-opted this paraspeckle machinery to store unspliced RNA transcripts to serve as a means for rapid RNA release upon stress (Zhang et al., 2013).
Figure 2. NEAT1 lncRNA is upregulated during viral infection.
NEAT1 serves as a scaffold in paraspeckle bodies binding to proteins PSP1 and p54. Paraspeckle bodies, under the control of the NEAT1 lncRNA serve as a reservoir for unspliced and singly spliced HIV transcripts containing cis-acting instability elements (INS) prior to nuclear export.
This hypothesis, wherein paraspeckle bodies under the control of NEAT1 serve as a reservoir for unspliced HIV transcripts prior to nuclear export is further strengthened by the discovery of the protein RBM14, a Rev cofactor demonstrated to localize to paraspeckle bodies. Export of unspliced transcripts to the cytoplasm increases in a RBM14 dependent manner; however, this increase is contingent upon the presence of NEAT1 as overexpression of RBM14 failed to stimulate nuclear export of the unspliced HIV transcripts to the cytoplasm in cells in which NEAT1 expression had been inhibited by siRNAs. Such a finding strengthens the hypothesis that nuclear paraspeckles act as a reservoir for the HIV transcript prior to cytoplasmic release. Furthermore, the RBM14 protein bas been demonstrated to bind to the protein XPO-1, a protein recruited by the Rev protein to mediate nuclear export offering a potential mechanism whereby these unspliced RNAs may be shuttled from the paraspeckles into the cytoplasm via Rev-transport (Budhiraja et al., 2015). The benefit of storing the unspliced HIV transcripts in these paraspeckle bodies can be explained through the finding that increased expression of ADAR1, the protein responsible for the A-to-I conversion, increases the accumulation of overall HIV-1 proteins within the cellular cytoplasm, in turn leading to both a greater release of virions and virions with greater infectivity. While ADAR1 expression also leads to reduced levels of phosphorylated PKR and eIF-2α, the increased infectivity was demonstrated to be directly associated with the A-to-I editing abilities of this protein (Doria et al., 2009).
The NEAT1 sequestration of mRNAs as a regulatory mechanism (Figure 2) is not limited to HIV. In human embryonic stem cells, NEAT1 expression is not detectable, resulting in the loss of paraspeckle formation, which in turn leads to limited nuclear retention of RNA transcripts, despite active A-to-I editing. Cellular differentiation induces NEAT1 expression and the formation of the paraspeckle bodies, and as a result the nuclear retention of various transcripts. Furthermore, sequestration in these paraspeckle bodies is not limited to just RNA. Rather, NEAT1 transcriptional upregulation depletes key paraspeckle proteins from the nucleoplasm by sequestering them into paraspeckle bodies. In terms of HIV infection, one effect of the sequestration of proteins in paraspeckle bodies may be to attenuate the apoptosis pathways upon viral infection as NEAT1 −/− cells are found to be more sensitive to proteasome inhibition which in turn leads to cell death (Hirose et al., 2014). The upregulation of NEAT1, therefore, may serve as a survival mechanism for HIV within the host cell.
Sequestration of proteins has been demonstrated to induce other physiologic changes within these cells too. It appears that overexpression of NEAT1 may be a universal response by T-cells to viral infection as NEAT1 overexpression is found to be induced by the influenza virus, the herpes virus, as well as the immunostimulant Polyinosinic:polycytidylic acid (Poly I:C). Beyond simply retaining dsRNAs within the nucleus, the activation of NEAT1 plays an integral role in the cytokine release upon viral infection. Within an uninfected cell, expression of interleukin-8 (IL-8) expression is transcriptionally regulated by the SFPQ protein, a protein also found within paraspeckle bodies. Upon viral induction of NEAT1 overexpression, SFPQ becomes sequestered within the newly formed paraspeckle bodies, releasing the transcriptional repression of the IL-8 gene. It should be noted that this pathway is not solely under the regulation of the lncRNA NEAT1. Rather, IL-8 activation also occurs through the NFκβ pathway, a pathway that is also upregulated via viral infection (Imamura et al., 2014). While the upregulation of NEAT1 activates the release of cytokines, the ability of HIV to sequester long dsRNA molecules within the nucleus may in turn serve to mute the cell’s innate immune response.
NRON
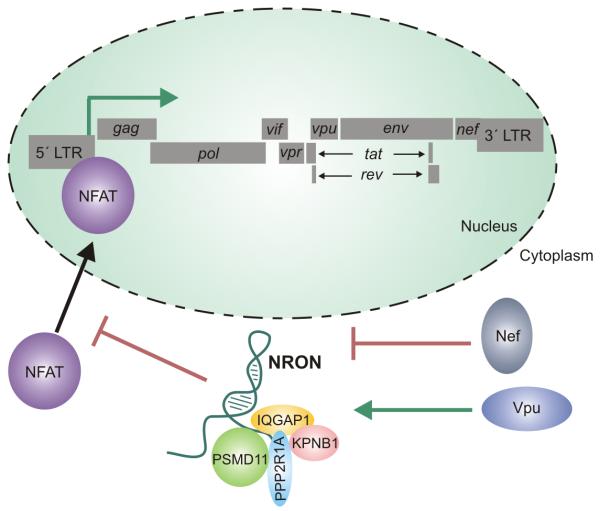
While the regulation of NEAT1 provided a mechanism by which HIV viral RNA could be sequestered within the nucleus until the viral proteins are sufficiently produced in the cytoplasm, the discordant regulation of another lncRNA, NRON (noncoding repressor of Nuclear Factor of T-Cells (NFAT)), by the two HIV proteins, Nef and Vpu, serves to provide a mechanism by which HIV transcription may be differentially regulated at various points of the HIV lifecycle (Imam et al., 2015) (Figure 3). Previous studies demonstrated that NRON acts to regulate the expression of the protein NFAT in active T-cells by binding to multiple proteins including a calmodulin-binding protein (IQGAP1), nuclear transport factor (KPNB1), phosphatase (PPP2R1A), and the proteasome (PSMD11). Interestingly, NRON’s binding of these proteins does not influence the transcription of the NFAT gene. Rather, by sequestering these aforementioned proteins, NRON regulates the subcellular localization of NFAT, most likely through NRON’s interactions with the nuclear import factors, a fact validated by the increased nuclear localization of NFAT following NRON inhibition. The varied expression of NRON throughout the HIV lifecycle therefore regulates NFAT activity by either enabling or inhibiting its nuclear transport (Willingham et al., 2005) (Figure 3).
Figure 3. NRON indirectly regulates the transcription of HIV.
The lncRNA NRON sequesters multiple proteins including a calmodulin-binding protein (IQGAP1), nuclear transport factor (KPNB1), phosphatase (PPP2R1A), and the proteasome (PSMD11), and thereby regulates the subcellular localization of NFAT. NFAT promotes HIV transcriptional activity through its binding to the 5′LTR. NRON is able to regulate NFAT activity by either enabling or inhibiting its nuclear transport. In early stage viral infection, Nef inhibits NRON and thus promotes the activity NFAT which consequently upregulates HIV transcription. Expression of Vpu later in viral infection induces expression of NRON, which then inhibits NFAT activity, and as such, inhibits viral transcription.
The regulation of the protein NFAT has long been known to be essential in the control of HIV transcription within T cells. NFAT promotes HIV transcriptional activity through its binding to the κβ motif of the viral LTR region in proximity to the NFκβ binding site. While in proximity to the NFκβ site, the NFAT binding site remains unique, and NFAT and NFκβ act are able to act in a synergistic manner to increase HIV transcription (Cron et al., 2000; Kinoshita et al., 1997). Interestingly, the two proteins which regulate NRON expression play two unique, yet critical functions during the viral lifecycle. In early stage viral infection, gene profiling suggested that the viral protein Nef functions promiscuously to promote the activity of a wide range of factors that positively promote HIV transcription including NFκβ , Tat-SF1, as well as NFAT, supporting the claim that Nef acts as the primary driving force of early stage viral production (Simmons et al., 2001). The inhibition of NRON by Nef releases NFAT, inducing greater transcriptional activity of HIV found early in the lifecycle (Imam et al., 2015). On the other hand, Vpu is expressed late in the viral lifecycle. During viral replication, the host cell inhibits viral release of retroviral particles by tethering the fully formed viral particles to the cell membrane by CD317. The viral protein Vpu binds to CD317 and inhibits CD317’s ability to retain the fully formed viral particles, enabling virion release (Neil et al., 2008). Furthermore, Vpu contributes to viral induced apoptosis of HIV-infected T cells by inhibiting NFκβ , and in turn promoting caspase activity within the cell (Akari et al., 2001). This late stage expression of Vpu also induces expression of the lncRNA NRON, which serves to inhibit NFAT activity, and as such, inhibits viral transcription prior to viral release and cellular apoptosis (Imam et al., 2015). NRON represents another host lncRNA whose expression changes throughout the lifecycle of HIV. Rather than simply being an artifact of the viral infection, the changes in NRON expression play an integral role in regulating HIV expression through NFAT activity. In addition, the fact that that NRON is discordantly regulated throughout various time points of the HIV life cycle may provide an explanation as to why RNA-seq experiments provided inconsistent results across different studies (Imam et al., 2015; Zhang et al., 2013). If regulation of these lncRNAs vary throughout the lifecycle, it may be difficult to identify significant changes across a large population of cells.
p53-dependent lncRNAs
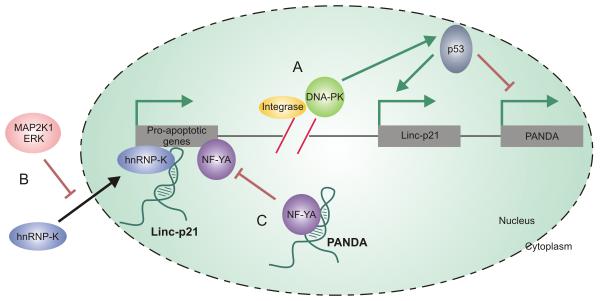
Upon comparison of the lncRNAs that underwent differential expression following HIV infection in two published studies, two unique p53-dependent lncRNAs, lincRNA-p21 and PANDA, were identified (Imam et al., 2015; Zhang et al., 2013). Following reverse transcription of the viral RNA genome and transport into the nucleus, cDNA is integrated into the host genome. To enable integration, the HIV protein integrase cleaves the host cell’s genomic DNA, producing double stranded breaks (Figure 4 A). It is hardly surprising that p53-dependent lncRNAs would be deregulated during viral infection since p53 binding protein plays a critical functional role in the cellular response to double stranded DNA breaks within the genome (Craigie and Bushman, 2012; Schultz et al., 2000). Upon viral integration, two possible outcomes exist for the host cell. Either active viral transcription occurs within the cell, and the cell remains viable, or the cell undergoes apoptosis. Ultimately, the cellular fate of HIV-1 infected CD4 cells is determined by activation of the DNA-dependent protein kinase (DNA-PK) pathway. DNA-PK activation induces apoptosis by phosphorylating p53 (Cooper et al., 2013) (Figure 4 A). Once activated, p53 responds to cellular stress by both activating and repressing a wide range of host genes responsible for DNA repair, apoptosis, cell cycle arrest, and senescence in order to re-establish genomic stability or initiate cellular apoptosis (Riley et al., 2008; Vazquez et al., 2008). Since p53 plays an integral role in host cell fate following HIV infection, dramatic changes in p53-dependent lncRNA expression is not unexpected.
Figure 4. p53-dependent lncRNAs are deregulated upon HIV infection.
(A) Prior to integration, the HIV protein integrase cleaves the host cell’s genomic DNA, producing p53 double stranded breaks. Double stranded DNA breaks result in the activation of DNA-PK which phosphorylates p53, starting the signaling cascade which ultimately results in apoptosis. Two transcripts whose expression is affected by p53 are the lncRNAs lincRNA-p21 and PANDA. (B) LincRNA-p21 expression is induced by p53 and it binds to hnRNP-K enabling its localization at the promoters of pro-apoptotic genes. To prevent this proapoptotic pathway, HIV mediates the initiation of the MAP2K1/ERK2 pathway which causes hnRNP-K to be sequestered in the cytoplasm. MAP2K1/ERK2 activation, therefore, protects HIV-infected cells against p53-induced apoptosis by lincRNA-p21 degradation and hnRNP-K sequestration. (C) Conversely, PANDA acts to inhibit apoptosis by binding to NF-YA and preventing its co-localization at promoter regions of apoptotic genes and is thus downregulated by p53.
Expression of the first of these two lncRNAs, lincRNA-p21, is driven off a p53-dependent promoter, providing an explanation for the upregulation of this particular lncRNA following HIV infection. Furthermore, knockdown of lincRNA-p21 reduces the apoptotic effect of p53 in cells suggesting that this particular lncRNA plays an integral role in p53-induced cellular apoptosis (Huarte et al., 2010). Mechanistically, this lncRNA binds to heterogeneous nuclear ribonucleoprotein K (hnRNP-K), a protein believed necessary for p53-induced repression, enabling its localization at the promoters of p53-repressed genes (Figure 4 B). Without this interaction between the lncRNA and hnRNP-K, p53 fails to induce transcriptional repression, suggesting that lincRNA p-21 plays an integral role in the global p53 response to genomic instability (Huarte et al., 2010). As successful HIV replication requires the evasion of the p53-apoptotic pathway, an alternative pathway must exist whereby HIV suppresses the p53 response. Recent research suggests that HIV succeeds in inhibiting this lincRNA p-21 apoptotic pathway by activating the MAP2K1/ERK2 pathway. Viral initiation of the MAP2K1/ERK2 pathway produces two noticeable effects within the cell: HuR remains localized to the nucleus, and hnRNP-K remains sequestered in the cytoplasm (Barichievy et al., 2015). HuR has long been known to impart stability to RNA transcripts in response to environmental stresses by binding to AU-rich regions, and in doing so, preventing degradation (Brennan and Steitz, 2001). The stabilizing effect of HuR on mRNA transcripts, in response to environment stresses, occurs within the cytoplasm of these cells (Abdelmohsen et al., 2007; Kawai et al., 2006; Wang et al., 2000a; Wang et al., 2000b). While the HuR protein stabilizes mRNA transcripts within the cytoplasm, the localization of HuR to the nucleus has been demonstrated to reduce the overall stability of lincRNA p21 by recruiting let-7/Ago2 to the noncoding transcript (Yoon et al., 2012). The initiation of the MAP2K1/ERK2 pathway by viral infection and the subsequent nuclear sequestration of HuR therefore leads to increased degradation of the lincRNA p21 transcript, which in turn inhibits p53 induced apoptosis. MAP2K1/ERK2 activation, therefore, protects the infected cell against p53-induced apoptosis by lincRNA-p21 degradation and hnRNP-K sequestration (Barichievy et al., 2015).
PANDA (P21 associated noncoding RNA DNA damage activated), the second p53-dependent lncRNA, underwent significant downregulation upon HIV infection (Zhang et al., 2013). As is the case with lincRNA p-21, PANDA expression is regulated in a p53-dependent manner; however, the two appear to have differing roles in regulating the apoptotic response. Whereas lincRNA p-21 activation promoted apoptosis through genetic repression, PANDA inhibits apoptosis by binding to NF-YA and preventing its co-localization at promoter regions of apoptotic genes (Figure 4 C). It follows, therefore, that the reduction of PANDA upon HIV infection supports p53-induced apoptosis within these chronically infected Jurkat T cells (Hung et al., 2011). Upregulation of lincRNA p-21 and the downregulation of PANDA serve as part of the widespread cellular response driven by p53 upon the formation of double strand breaks within the genome. Since HIV induces double stranded breaks in order to integrate into the host genome, changes in the expression of these two lncRNAs are anticipated. As demonstrated with lincRNA-p21, HIV has evolved mechanisms to mitigate the effects of p53 by directly targeting a specific lncRNA pathway, and in doing so, is able to inhibit apoptosis of the host cell.
The importance of lincRNA-p21 in determining the cellular fate of HIV infected cells can be clearly observed when examining the survival of different classes of immune cells. As described before, HIV infection of T lymphocytes activates the DNA-PK pathway and ultimately leads to apoptosis and the depletion of these T cells. HIV infection of macrophages however, results in significantly less apoptosis. Rather, macrophages appear to be less sensitive to the integration of HIV and the formation of double strand breaks within the genome, as these cells are less susceptible to the cytotoxic effects of HIV infection, when compared to T cells (Carter and Ehrlich, 2008). This effect may be best explained by the fact that HIV only activates the MAP2K1/ERK2 pathway, and the subsequent inhibition of lincRNA-p21 and the p53 induced apoptotic pathway, in macrophages and not within T lymphocytes (Barichievy et al., 2015). Such findings demonstrate the importance of lncRNAs in disease progression.
2.2.2 Viral-derived lncRNAs
Given the emerging roles of cellular lncRNAs in viral infection coupled with the notion that HIV may generate viral-derived small ncRNAs, it seems logical that HIV may encode lncRNAs of its own. Indeed reports of antisense lncRNAs emanating from the HIV genome during viral infection have begun to surface. Several independent studies have now described the existence of HIV antisense transcripts (Kobayashi-Ishihara et al., 2012; Landry et al., 2007; Ludwig et al., 2006; Saayman et al., 2014). Promoter activity has clearly been demonstrated in the reverse orientation with transcription start sites identified in the U3 region of the 3′LTR as well as in the nef gene sequence. 5′ RACE analyses identified multiple transcription start sites signifying multiple unique antisense transcripts (Kobayashi-Ishihara et al., 2012; Landry et al., 2007). Supporting these observations, were further experiments characterizing HIV antisense transcripts which specifically identified four unique antisense transcripts with varying termination sites and consequently of varying size (Kobayashi-Ishihara et al., 2012). Such transcripts were found to be localized within the nucleus and upon further exploration of their function found to be natural suppressors of HIV gene expression over extended periods (Kobayashi-Ishihara et al., 2012).
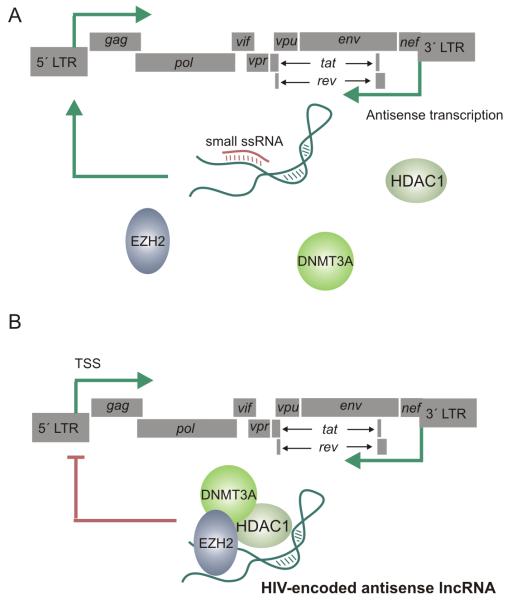
We sought to further characterize and also to elucidate the role of viral-derived antisense lncRNA transcripts and the mechanisms by which their function is achieved. We identified a dominant antisense transcript which spanned the majority of the genome, overlapping env, pol and partially gag, thus confirming the existence of a previously described genome length transcript (Kobayashi-Ishihara et al., 2012), yet extending beyond multiple other reported antisense transcripts. Suppression of this antisense lncRNA by RNAi using small single stranded asRNAs targeted to the lncRNA transcript was shown to result in the activation of viral gene expression (Figure 5 A), thus confirming previous hypotheses that antisense lncRNAs could act to negatively regulate HIV gene expression (Saayman et al., 2014). Furthermore, we demonstrated, using small single stranded asRNAs targeted to the promoter region driving expression of the lncRNA, that we could achieve transcriptional gene silencing (TGS) of a lncRNA transcript. TGS of the HIV lncRNA also resulted in derepression and thus elevated levels of viral gene expression. These phenomena were demonstrated in both latent cell line models as well as in infected primary CD4+ T cells.
Figure 5.
(A) HIV encodes a long non-coding RNA (lncRNA) which is transcribed antisense to the viral genome from a putative promoter within the nef/3′LTR overlapping region. The HIV-encoded lncRNA may be inhibited by small single stranded RNAs targeted to the promoter driving expression of the ncRNA via transcriptional gene silencing, or by small RNAs targeted to the ncRNA transcript via post-transcriptional gene silencing (shown). The inhibition of the antisense lncRNA by small RNAs prevents the recruitment of chromatin remodeling proteins to the viral promoter which remains in a euchromatin state, free of silent state epigenetic marks and ultimately resulting in elevated viral gene expression. (B) The HIV-encoded lncRNA recruits chromatin remodeling proteins DNMT3a, EZH2 and HDAC-1 to the viral promoter resulting in the formation of heterochromatin and the subsequent epigenetic silencing of the virus.
This study suggests that in addition to host lncRNAs, HIV also encodes a lncRNA that contributes to the complex regulatory network governing viral gene expression. Through further mechanistic studies it was revealed that the HIV expressed lncRNA localized to the 5′LTR and interacted directly with DNA methyltransferase 3A (DNMT3a). In addition to DNMT3a, the lncRNA appeared to usurp further components of endogenous cellular pathways that are involved in ncRNA directed epigenetic gene silencing such as EZH2 and HDAC1 (Figure 5 B), observations similar to other studies with lncRNAs regulating endogenous genes (Johnsson et al., 2013). In line with these observations, the increase in viral gene expression and replication following inhibition of the antisense lncRNA correlated to a loss of silent state epigenetic marks at the viral promoter supporting the notion that this lncRNA is functional in the transcriptional and epigenetic regulation of the virus.
Collectively, this body of work demonstrated that HIV-1 expresses an antisense lncRNA, albeit at a low copy number, that emanates from the 3′LTR/nef overlapping region and spans the majority of the HIV genome. This lncRNA localizes to the viral core promoter region and brings about a regulatory effect by recruiting chromatin remodeling proteins and thereby alterating the epigenetic landscape at the viral promoter thus modulating HIV gene expression (Saayman et al., 2014) (Figure 5 B).
3. What lies ahead for lncRNAs and HIV infection?
It has become obvious that ncRNAs, especially lncRNAs, play an important regulatory role in cellular homeostasis. While the functionality of cellular lncRNAs has been quickly established, the existence and functionality of ncRNAs, particularly those that are RNA virus-derived, has remained somewhat controversial. The canonical theory of RNA-based viral regulation supposes that as a class, RNA viruses cannot encode regulatory ncRNAs. This bifurcated theory argues that cytoplasmic RNA viruses cannot express regulatory ncRNAs such as miRNAs because the cellular machinery, specifically the microprocessor complex, functions within the nucleus, and therefore does not come into contact with the viral genome. Retroviruses on the other hand replicate via a DNA intermediate which integrates into the host genome within the nucleus. While miRNA processing is therefore possible, complementary sequences within the viral RNA genome or mRNA transcripts would result in self-targeting and the consequent suppression of viral transcripts (Cullen, 2012; Grundhoff and Sullivan, 2011; Houzet and Jeang, 2011; Skalsky and Cullen, 2010).
Nevertheless, it has since been demonstrated that cytoplasmic RNA viruses have developed non-canonical pathways for miRNA formation (Swaminathan et al., 2014; Usme-Ciro et al., 2013). One mechanism for the generation of pre-miRNA sequences is a Drosha/DGCR8-independent mechanism, first described in Drosophila melanogaster and C. elegans (Ruby et al., 2007) and later discovered in mammals (Babiarz et al., 2008; Berezikov et al., 2007). The miRNA is encoded by an intronic sequence which, following splicing and a debranching, closely resembles the structure of a pre-miRNA, therefore eliminating the need for microprocessor-mediated cleavage. These structures, termed mirtrons, may require exonucleolytic cleavage of extended 5′ or 3′ flanking tails before the convergence of the canonical and mirtronic miRNA biogenesis pathways (Babiarz et al., 2008). Furthermore, the recruitment of Drosha to the cytoplasm has now been demonstrated which allows for efficient miRNA processing within the cytoplasm (Usme-Ciro et al., 2013).
With regards to the HIV, support for the canonical view of viral-derived ncRNAs was corroborated when initial analyses of HIV-infected T-cells failed to identify any virally encoded siRNAs or miRNAs (Lin and Cullen, 2007). As has been discussed throughout this review, however, it is evident that HIV does in fact encode for and transcribe both small and long non-coding RNA moieties, and these ncRNAs are able to regulate various aspects of the viral life cycle. Beyond these isolated descriptions of individual ncRNAs, the strongest support in defense of retrovirus encoded ncRNAs comes from advances in deep sequencing technologies. Improved sensitivity of sequencing has led to the identification of low abundant viral transcripts. These viral-derived small ncRNAs comprise only ~1% of the entirety of the cell’s non-coding profile; therefore, poor sensitivity in earlier studies may explain the failure to previously detect these RNAs (Schopman et al., 2012). Growing evidence supports the expression of retroviral derived ncRNAs, hence one must conclude that HIV has evolved mechanisms to avoid self-targeting and thus auto-inhibition. The primary mechanism by which HIV successfully generates ncRNAs, particularly miRNAs, appears to be via transcriptional regulation leading to the generation of short transcripts rather than full length mRNA transcripts, in the absence of Tat (Harwig et al., 2014; Harwig et al., 2015).
The mechanism of miRNA formation during HIV infection has been best characterized with respect to those derived from the TAR region. Transcription of TAR produces an ~50bp hairpin that has been shown to be processed by Dicer, resulting in mature functional miRNAs that contribute to the maintenance of the latent viral state (Klase et al., 2007) and downregulate apoptotic genes (Ouellet et al., 2013). The detailed mechanism of TAR-derived miRNA biogenesis was described above and provides one possible explanation as to how HIV accomplishes the formation of miRNAs while protecting the full length transcript from degradation (Harwig et al., 2015). Beyond the biogenesis of TAR-derived miRNAs, other mechanisms must also exist whereby the viral genome is protected from self-targeting. To shield viral transcripts from miRNA targeting, HIV appears to have evolved secondary structures within the viral transcripts that inhibit miRNA binding (Whisnant et al., 2013). It is entirely possible therefore that these secondary structures may also confer resistance to self-targeting. The possibility also exists that antisense moieties are the actual targets instead of the canonical predicted sense target.
What is evident is that viral-derived ncRNA transcripts clearly exist and furthermore, a great diversity exists across the population of HIV-derived ncRNA transcripts. In time, as insights are gained into RNA directed gene regulatory systems, additional mechanisms and pathways enabling HIV to encode regulatory ncRNAs without self-targeting are bound to emerge.
The accelerated discovery of ncRNA species, whether derived from the human host or HIV itself, and whether short or long, is providing invaluable insight into the unexplained intricacies that enable a virus with a comparatively small genome to successfully complete a relatively complex life cycle. These ncRNAs appear to play critical roles in pathways governing gene expression, replication, pathogenesis, disease progression and infected cell fate. The significance of ncRNAs in HIV infection has begun to be appreciated; however, these transcripts, in addition to expanding our understanding of the regulatory systems directing the HIV life cycle, denote obvious novel therapeutic targets. Technologies to suppress small ncRNAs have already been developed and extensively optimized and are currently being successfully implemented in pre-clinical studies as a therapeutic for Hepatitis C Virus infection (Lanford et al., 2010). It follows therefore that small RNAs that enhance HIV replication such as TAR-derived miRNAs offer ideal targets for miRNA antagonists. Long non-coding RNAs are no exception. Given that an HIV encoded lncRNA has been found to negatively regulate viral transcription, the role that this lncRNA potentially plays in the establishment and/or maintenance of viral latency is intriguing. The eradication of latent reservoirs of HIV remains the last hurdle in the search for a functional cure for HIV infection. Until now, non-specific histone deacetylase (HDAC) inhibitors have been the primary agents used to reactivate dormant virus in the presence of antiretroviral drugs in order to purge latent reservoirs (Archin et al., 2012; Shirakawa et al., 2013). An activating agent that will specifically activate HIV gene expression is thus highly desirable and the development of a lncRNA inhibitor targeted to the HIV-encoded antisense lncRNA to “de-repress” HIV gene expression could potentially provide an ideal strategy for the elimination of latently-infected populations. Indeed one thing is certain and that is the complexity of HIV infection, and that those non-coding RNAs contributing to this complexity are only now just becoming appreciated.
Acknowledgements
The work was supported by the following grants: NCI R01 CA151574, NIH R01 CA153124, NIAID P01 AI099783-01, R01 DK104681-01, and ARC Future Fellow FT1300100572 to KVM
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelmohsen K, Pullmann R, Jr., Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, Furneaux H, Gorospe M. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Molecular cell. 2007;25(4):543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahluwalia JK, Khan SZ, Soni K, Rawat P, Gupta A, Hariharan M, Scaria V, Lalwani M, Pillai B, Mitra D, Brahmachari SK. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology. 2008;5:117. doi: 10.1186/1742-4690-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akari H, Bour S, Kao S, Adachi A, Strebel K. The human immunodeficiency virus type 1 accessory protein Vpu induces apoptosis by suppressing the nuclear factor kappaB-dependent expression of antiapoptotic factors. The Journal of experimental medicine. 2001;194(9):1299–1311. doi: 10.1084/jem.194.9.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althaus CF, Vongrad V, Niederost B, Joos B, Di Giallonardo F, Rieder P, Pavlovic J, Trkola A, Gunthard HF, Metzner KJ, Fischer M. Tailored enrichment strategy detects low abundant small noncoding RNAs in HIV-1 infected cells. Retrovirology. 2012;9:27. doi: 10.1186/1742-4690-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JL, Hope TJ. HIV accessory proteins and surviving the host cell. Curr HIV/AIDS Rep. 2004;1(1):47–53. doi: 10.1007/s11904-004-0007-x. [DOI] [PubMed] [Google Scholar]
- Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487(7408):482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22(20):2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barichievy S, Naidoo J, Mhlanga MM. Non-coding RNAs and HIV: viral manipulation of host dark matter to shape the cellular environment. Frontiers in genetics. 2015;6:108. doi: 10.3389/fgene.2015.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220(4599):868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22(5):607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Bennasser Y, Le SY, Yeung ML, Jeang KT. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology. 2004;1:43. doi: 10.1186/1742-4690-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28(2):328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319(5865):921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58(2):266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhiraja S, Liu H, Couturier J, Malovannaya A, Qin J, Lewis DE, Rice AP. Mining the Human Complexome Database Identifies RBM14 as an XPO1-Associated Protein Involved in HIV-1 Rev Function. Journal of virology. 2015;89(7):3557–3567. doi: 10.1128/JVI.03232-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD, Malani N, Fernandes J, D’Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, Konig R, Bandyopadhyay S, Ideker T, Goff SP, Krogan NJ, Frankel AD, Young JA, Chanda SK. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS pathogens. 2009;5(5):e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, Rinn JL, Raj A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015;16:20. doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EM, Hope TJ. Live cell imaging of the HIV-1 life cycle. Trends Microbiol. 2008;16(12):580–587. doi: 10.1016/j.tim.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CA, Ehrlich LS. Cell biology of HIV-1 infection of macrophages. Annu Rev Microbiol. 2008;62:425–443. doi: 10.1146/annurev.micro.62.081307.162758. [DOI] [PubMed] [Google Scholar]
- Chen AK, Sengupta P, Waki K, Van Engelenburg SB, Ochiya T, Ablan SD, Freed EO, Lippincott-Schwartz J. MicroRNA binding to the HIV-1 Gag protein inhibits Gag assembly and virus production. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(26):E2676–2683. doi: 10.1073/pnas.1408037111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Li C, Huang J, Cung T, Seiss K, Beamon J, Carrington MF, Porter LC, Burke PS, Yang Y, Ryan BJ, Liu R, Weiss RH, Pereyra F, Cress WD, Brass AL, Rosenberg ES, Walker BD, Yu XG, Lichterfeld M. CD4+ T cells from elite controllers resist HIV-1 infection by selective upregulation of p21. J Clin Invest. 2011;121(4):1549–1560. doi: 10.1172/JCI44539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YL, Cao H, Jacque JM, Stevenson M, Rana TM. Inhibition of human immunodeficiency virus type 1 replication by RNA interference directed against human transcription elongation factor P-TEFb (CDK9/CyclinT1) J Virol. 2004;78(5):2517–2529. doi: 10.1128/JVI.78.5.2517-2529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Molecular cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A, Garcia M, Petrovas C, Yamamoto T, Koup RA, Nabel GJ. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature. 2013;498(7454):376–379. doi: 10.1038/nature12274. [DOI] [PubMed] [Google Scholar]
- Craigie R, Bushman FD. HIV DNA integration. Cold Spring Harb Perspect Med. 2012;2(7):a006890. doi: 10.1101/cshperspect.a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cron RQ, Bartz SR, Clausell A, Bort SJ, Klebanoff SJ, Lewis DB. NFAT1 enhances HIV-1 gene expression in primary human CD4 T cells. Clinical immunology. 2000;94(3):179–191. doi: 10.1006/clim.1999.4831. [DOI] [PubMed] [Google Scholar]
- Cullen BR. MicroRNA expression by an oncogenic retrovirus. Proc Natl Acad Sci U S A. 2012;109(8):2695–2696. doi: 10.1073/pnas.1200328109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria M, Neri F, Gallo A, Farace MG, Michienzi A. Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic acids research. 2009;37(17):5848–5858. doi: 10.1093/nar/gkp604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorin JD, Malim MH. Intracellular trafficking of HIV-1 cores: journey to the center of the cell. Curr Top Microbiol Immunol. 2003;281:179–208. doi: 10.1007/978-3-642-19012-4_5. [DOI] [PubMed] [Google Scholar]
- Emerman M, Malim MH. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science. 1998;280(5371):1880–1884. doi: 10.1126/science.280.5371.1880. [DOI] [PubMed] [Google Scholar]
- Farberov L, Herzig E, Modai S, Isakov O, Hizi A, Shomron N. MicroRNA-mediated regulation of p21 and TASK1 cellular restriction factors enhances HIV-1 infection. Journal of cell science. 2015;128(8):1607–1616. doi: 10.1242/jcs.167817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Pollard VW, Luhrmann R, Teufel M, Michael MW, Dreyfuss G, Malim MH. Rev-mediated nuclear export of RNA is dominant over nuclear retention and is coupled to the Ran-GTPase cycle. Nucleic acids research. 1999;27(21):4128–4134. doi: 10.1093/nar/27.21.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo RC, Sarin PS, Gelmann EP, Robert-Guroff M, Richardson E, Kalyanaraman VS, Mann D, Sidhu GD, Stahl RE, Zolla-Pazner S, Leibowitch J, Popovic M. Isolation of human T-cell leukemia virus in acquired immune deficiency syndrome (AIDS) Science. 1983;220(4599):865–867. doi: 10.1126/science.6601823. [DOI] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol. 2008;18(2):203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SP. Knockdown screens to knockout HIV-1. Cell. 2008;135(3):417–420. doi: 10.1016/j.cell.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. 2011;411(2):325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwig A, Das AT, Berkhout B. Retroviral microRNAs. Curr Opin Virol. 2014;7:47–54. doi: 10.1016/j.coviro.2014.03.013. [DOI] [PubMed] [Google Scholar]
- Harwig A, Das AT, Berkhout B. HIV-1 RNAs: sense and antisense, large mRNAs and small siRNAs and miRNAs. Curr Opin HIV AIDS. 2015;10(2):103–109. doi: 10.1097/COH.0000000000000135. [DOI] [PubMed] [Google Scholar]
- Hill M, Tachedjian G, Mak J. The packaging and maturation of the HIV-1 Pol proteins. Curr HIV Res. 2005;3(1):73–85. doi: 10.2174/1570162052772942. [DOI] [PubMed] [Google Scholar]
- Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Benard M, Fox AH, Pierron G. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Molecular biology of the cell. 2014;25(1):169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzet L, Jeang KT. MicroRNAs and human retroviruses. Biochim Biophys Acta. 2011;1809(11-12):686–693. doi: 10.1016/j.bbagrm.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzet L, Klase Z, Yeung ML, Wu A, Le SY, Quinones M, Jeang KT. The extent of sequence complementarity correlates with the potency of cellular miRNA-mediated restriction of HIV-1. Nucleic acids research. 2012;40(22):11684–11696. doi: 10.1093/nar/gks912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu K, Seharaseyon J, Dong P, Bour S, Marban E. Mutual functional destruction of HIV-1 Vpu and host TASK-1 channel. Molecular cell. 2004;14(2):259–267. doi: 10.1016/s1097-2765(04)00183-2. [DOI] [PubMed] [Google Scholar]
- Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med. 2007;13(10):1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Wang Y, Kong B, Langerod A, Borresen-Dale AL, Kim SK, van de Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y, Wong DJ, Chang HY. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43(7):621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam H, Bano AS, Patel P, Holla P, Jameel S. The lncRNA NRON modulates HIV-1 replication in a NFAT-dependent manner and is differentially regulated by early and late viral proteins. Scientific reports. 2015;5:8639. doi: 10.1038/srep08639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, Kai C, Yada T, Suzuki Y, Yamada T, Ozawa T, Kaneki K, Inoue T, Kobayashi M, Kodama T, Wada Y, Sekimizu K, Akimitsu N. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Molecular cell. 2014;53(3):393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- Jacque JM, Stevenson M. The inner-nuclear-envelope protein emerin regulates HIV-1 infectivity. Nature. 2006;441(7093):641–645. doi: 10.1038/nature04682. [DOI] [PubMed] [Google Scholar]
- Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grander D, Morris KV. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nature structural & molecular biology. 2013;20(4):440–446. doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SY, Calman AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330(6147):489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol Cell Biol. 2006;26(8):3295–3307. doi: 10.1128/MCB.26.8.3295-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita S, Su L, Amano M, Timmerman LA, Kaneshima H, Nolan GP. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity. 1997;6(3):235–244. doi: 10.1016/s1074-7613(00)80326-x. [DOI] [PubMed] [Google Scholar]
- Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. The New England journal of medicine. 1995;332(4):228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, McCaffrey T, Kashanchi F. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol. 2007;8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klase Z, Winograd R, Davis J, Carpio L, Hildreth R, Heydarian M, Fu S, McCaffrey T, Meiri E, Ayash-Rashkovsky M, Gilad S, Bentwich Z, Kashanchi F. HIV-1 TAR miRNA protects against apoptosis by altering cellular gene expression. Retrovirology. 2009;6:18. doi: 10.1186/1742-4690-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi-Ishihara M, Yamagishi M, Hara T, Matsuda Y, Takahashi R, Miyake A, Nakano K, Yamochi T, Ishida T, Watanabe T. HIV-1-encoded antisense RNA suppresses viral replication for a prolonged period. Retrovirology. 2012;9:38. doi: 10.1186/1742-4690-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135(1):49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutluay SB, Zang T, Blanco-Melo D, Powell C, Jannain D, Errando M, Bieniasz PD. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell. 2014;159(5):1096–1109. doi: 10.1016/j.cell.2014.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry S, Halin M, Lefort S, Audet B, Vaquero C, Mesnard JM, Barbeau B. Detection, characterization and regulation of antisense transcripts in HIV-1. Retrovirology. 2007;4:71. doi: 10.1186/1742-4690-4-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Cullen BR. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. Journal of virology. 2007;81(22):12218–12226. doi: 10.1128/JVI.01390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314(5798):461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- Ludwig LB, Ambrus JL, Jr., Krawczyk KA, Sharma S, Brooks S, Hsiao CB, Schwartz SA. Human Immunodeficiency Virus-Type 1 LTR DNA contains an intrinsic gene producing antisense RNA and protein products. Retrovirology. 2006;3:80. doi: 10.1186/1742-4690-3-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem. 2003;278(35):33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- Mattick JS. A new paradigm for developmental biology. J Exp Biol. 2007;210(Pt 9):1526–1547. doi: 10.1242/jeb.005017. [DOI] [PubMed] [Google Scholar]
- Morris KV, Mattick JS. The rise of regulatory RNA. Nature reviews. Genetics. 2014;15(6):423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulard M, Decroly E. Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim Biophys Acta. 2000;1469(3):121–132. doi: 10.1016/s0304-4157(00)00014-9. [DOI] [PubMed] [Google Scholar]
- Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Molecular cell. 2009;34(6):696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451(7177):425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Nguyen DG, Wolff KC, Yin H, Caldwell JS, Kuhen KL. “UnPAKing” human immunodeficiency virus (HIV) replication: using small interfering RNA screening to identify novel cofactors and elucidate the role of group I PAKs in HIV infection. J Virol. 2006;80(1):130–137. doi: 10.1128/JVI.80.1.130-137.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto S, Fujii YR. Regulation of human immunodeficiency virus 1 transcription by nef microRNA. J Gen Virol. 2005;86(Pt 3):751–755. doi: 10.1099/vir.0.80449-0. [DOI] [PubMed] [Google Scholar]
- Omoto S, Ito M, Tsutsumi Y, Ichikawa Y, Okuyama H, Brisibe EA, Saksena NK, Fujii YR. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1:44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, Flamand L, Tremblay MJ, Provost P. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic acids research. 2008;36(7):2353–2365. doi: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet DL, Vigneault-Edwards J, Letourneau K, Gobeil LA, Plante I, Burnett JC, Rossi JJ, Provost P. Regulation of host gene expression by HIV-1 TAR microRNAs. Retrovirology. 2013;10:86. doi: 10.1186/1742-4690-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palangat M, Meier TI, Keene RG, Landick R. Transcriptional pausing at +62 of the HIV-1 nascent RNA modulates formation of the TAR RNA structure. Molecular cell. 1998;1(7):1033–1042. doi: 10.1016/s1097-2765(00)80103-3. [DOI] [PubMed] [Google Scholar]
- Patarca R, Heath C, Goldenberg GJ, Rosen CA, Sodroski JG, Haseltine WA, Hansen UM. Transcription directed by the HIV long terminal repeat in vitro. AIDS Res Hum Retroviruses. 1987;3(1):41–55. doi: 10.1089/aid.1987.3.41. [DOI] [PubMed] [Google Scholar]
- Patel P, Ansari M, Bapat S, Thakar M, Gangakhedkar R, Jameel S. The microRNA miR-29a is associated with human immunodeficiency virus latency. Retrovirology. 2014;11(1):108. doi: 10.1186/s12977-014-0108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard VW, Malim MH. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- Ratner L, Haseltine W, Patarca R, Livak KJ, Starcich B, Josephs SF, Doran ER, Rafalski JA, Whitehorn EA, Baumeister K, et al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313(6000):277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- Reynoso R, Laufer N, Hackl M, Skalicky S, Monteforte R, Turk G, Carobene M, Quarleri J, Cahn P, Werner R, Stoiber H, Grillari-Voglauer R, Grillari J. MicroRNAs differentially present in the plasma of HIV elite controllers reduce HIV infection in vitro. Scientific reports. 2014;4:5915. doi: 10.1038/srep05915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9(5):402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roebuck KA, Saifuddin M. Regulation of HIV-1 transcription. Gene Expr. 1999;8(2):67–84. [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448(7149):83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saayman S, Ackley A, Turner AM, Famiglietti M, Bosque A, Clemson M, Planelles V, Morris KV. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Molecular therapy : the journal of the American Society of Gene Therapy. 2014;22(6):1164–1175. doi: 10.1038/mt.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzwedel K, Smith ED, Dey B, Berger EA. Sequential CD4-coreceptor interactions in human immunodeficiency virus type 1 Env function: soluble CD4 activates Env for coreceptor-dependent fusion and reveals blocking activities of antibodies against cryptic conserved epitopes on gp120. J Virol. 2000;74(1):326–333. doi: 10.1128/jvi.74.1.326-333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pescador R, Power MD, Barr PJ, Steimer KS, Stempien MM, Brown-Shimer SL, Gee WW, Renard A, Randolph A, Levy JA, et al. Nucleotide sequence and expression of an AIDS-associated retrovirus (ARV-2) Science. 1985;227(4686):484–492. doi: 10.1126/science.2578227. [DOI] [PubMed] [Google Scholar]
- Schopman NC, Willemsen M, Liu YP, Bradley T, van Kampen A, Baas F, Berkhout B, Haasnoot J. Deep sequencing of virus-infected cells reveals HIV-encoded small RNAs. Nucleic acids research. 2012;40(1):414–427. doi: 10.1093/nar/gkr719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151(7):1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa K, Chavez L, Hakre S, Calvanese V, Verdin E. Reactivation of latent HIV by histone deacetylase inhibitors. Trends in microbiology. 2013;21(6):277–285. doi: 10.1016/j.tim.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons A, Aluvihare V, McMichael A. Nef triggers a transcriptional program in T cells imitating single-signal T cell activation and inducing HIV virulence mediators. Immunity. 2001;14(6):763–777. doi: 10.1016/s1074-7613(01)00158-3. [DOI] [PubMed] [Google Scholar]
- Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends in genetics : TIG. 2015 doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Li H, Wu X, Covarrubias M, Scherer L, Meinking K, Luk B, Chomchan P, Alluin J, Gombart AF, Rossi JJ. Interplay between HIV-1 infection and host microRNAs. Nucleic acids research. 2012;40(5):2181–2196. doi: 10.1093/nar/gkr961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung TL, Rice AP. miR-198 inhibits HIV-1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of cyclin T1. PLoS pathogens. 2009;5(1):e1000263. doi: 10.1371/journal.ppat.1000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surabhi RM, Gaynor RB. RNA interference directed against viral and cellular targets inhibits human immunodeficiency Virus Type 1 replication. J Virol. 2002;76(24):12963–12973. doi: 10.1128/JVI.76.24.12963-12973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan G, Navas-Martin S, Martin-Garcia J. MicroRNAs and HIV-1 infection: antiviral activities and beyond. J Mol Biol. 2014;426(6):1178–1197. doi: 10.1016/j.jmb.2013.12.017. [DOI] [PubMed] [Google Scholar]
- Tani H, Onuma Y, Ito Y, Torimura M. Long non-coding RNAs as surrogate indicators for chemical stress responses in human-induced pluripotent stem cells. PloS one. 2014;9(8):e106282. doi: 10.1371/journal.pone.0106282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube R, Fujinaga K, Wimmer J, Barboric M, Peterlin BM. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology. 1999;264(2):245–253. doi: 10.1006/viro.1999.9944. [DOI] [PubMed] [Google Scholar]
- Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, Reynes J, Corbeau P, Jeang KT, Benkirane M. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315(5818):1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- Usme-Ciro JA, Campillo-Pedroza N, Almazan F, Gallego-Gomez JC. Cytoplasmic RNA viruses as potential vehicles for the delivery of therapeutic small RNAs. Virology journal. 2013;10:185. doi: 10.1186/1743-422X-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7(12):979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- Wagschal A, Rousset E, Basavarajaiah P, Contreras X, Harwig A, Laurent-Chabalier S, Nakamura M, Chen X, Zhang K, Meziane O, Boyer F, Parrinello H, Berkhout B, Terzian C, Benkirane M, Kiernan R. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell. 2012;150(6):1147–1157. doi: 10.1016/j.cell.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain-Hobson S, Sonigo P, Danos O, Cole S, Alizon M. Nucleotide sequence of the AIDS virus, LAV. Cell. 1985;40(1):9–17. doi: 10.1016/0092-8674(85)90303-4. [DOI] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Molecular cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000a;19(10):2340–2350. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000b;20(3):760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Whisnant AW, Bogerd HP, Flores O, Ho P, Powers JG, Sharova N, Stevenson M, Chen CH, Cullen BR. In-depth analysis of the interaction of HIV-1 with cellular microRNA biogenesis and effector mechanisms. mBio. 2013;4(2):e000193. doi: 10.1128/mBio.00193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309(5740):1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Molecular cell. 2012;47(4):648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Li H, Zhou G, Zhang Q, Zhang T, Li J, Zhang J, Hou J, Liew CT, Yin D. Transcriptional silencing of the TMS1/ASC tumour suppressor gene by an epigenetic mechanism in hepatocellular carcinoma cells. The Journal of pathology. 2007;212(2):134–142. doi: 10.1002/path.2173. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Chen CY, Yedavalli VS, Jeang KT. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. mBio. 2013;4(1):e00596–00512. doi: 10.1128/mBio.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fan M, Geng G, Liu B, Huang Z, Luo H, Zhou J, Guo X, Cai W, Zhang H. A novel HIV-1-encoded microRNA enhances its viral replication by targeting the TATA box region. Retrovirology. 2014;11:23. doi: 10.1186/1742-4690-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106(4):465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. Genome-scale RNAi screen for host factors required for HIV replication. Cell host & microbe. 2008;4(5):495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]