Abstract
Changes in the intracellular free calcium concentration ([Ca2 +]i) in neurons regulate many and varied aspects of neuronal function over time scales from microseconds to days. The mystery is how a single signalling ion can lead to such diverse and specific changes in cell function. This is partly due to aspects of the Ca2 + signal itself, including its magnitude, duration, localisation and persistent or oscillatory nature. The transduction of the Ca2 + signal requires Ca2 + binding to various Ca2 + sensor proteins. The different properties of these sensors are important for differential signal processing and determine the physiological specificity of Ca2 + signalling pathways. A major factor underlying the specific roles of particular Ca2 + sensor proteins is the nature of their interaction with target proteins and how this mediates unique patterns of regulation. We review here recent progress from structural analyses and from functional analyses in model organisms that have begun to reveal the rules that underlie Ca2 + sensor protein specificity for target interaction. We discuss three case studies exemplifying different aspects of Ca2 + sensor/target interaction. This article is part of a special issue titled the 13th European Symposium on Calcium.
Abbreviations: CaBP, calcium-binding protein; DREAM, downstream regulatory element antagonist modulator; CDF, Ca2 +-dependent facilitation; CDI, Ca2 +-dependent inhibition; GCAP, guanylyl cyclase activating protein; IL1RAPL1, interleukin 1 receptor accessory protein-like 1 protein 1; KChIP, potassium channel interacting protein; NCS, neuronal calcium sensor; NCX, sodium/calcium exchanger; VGCC, voltage-gated Ca2 + channel; VILIP, visinin-like protein
Keywords: Caenorhabditis elegans, Ca2 +-binding proteins, Ca2 + channel, Ca2 + sensors, NCS-1, Neuronal signalling
Highlights
-
•
Ca2 + regulates multiple aspects of neuronal function over varying time scales.
-
•
The specificity of Ca2 + signalling is determined by the properties of Ca2 + sensors
-
•
Differing properties of closely related Ca2 + sensors contributes to specificity.
-
•
The structure of Ca2 + sensors determines specificity of target protein interactions.
-
•
We discuss three case studies involving calmodulin, NCS proteins and CaBP proteins.
-
•
The examples illustrate the structural–function relationships of Ca2 + sensors.
1. Introduction
The intracellular free calcium concentration ([Ca2 +]i) is tightly regulated through multiple mechanisms in neurons [1], and changes in [Ca2 +]i have crucial roles in the control of normal neuronal function [2]. In addition, abnormalities in Ca2 + signalling have been implicated in many aspects of neuropathology, neurodegeneration [3], [4], [5] and psychiatric disorders [6], [7]. The mechanisms that elevate [Ca2 +]i include entry of extracellular Ca2 + through voltage-gated Ca2 + channels and release from intracellular stores such as the endoplasmic reticulum (ER), lysosomes and mitochondria. [Ca2 +]i can be reduced by sequestration into the same cellular organelles and by extrusion across the plasma membrane and these mechanisms have been well characterised [1], [8]. Large numbers of studies on the localisation, magnitude and time course of [Ca2 +]i fluxes in neurons have been published. Changes in [Ca2 +]i can be local or global, highly transient, oscillatory or persistent. Localisation of [Ca2 +]i changes to particular neuronal compartments is important for the generation of specific neuronal responses. For example, certain stimuli strictly require nuclear rather than cytoplasmic changes in [Ca2 +]i [9] and also highly localised [Ca2 +]i changes restricted to dendritic spines may play an important role in activity-dependent synaptic plasticity [10], [11]. The physiological effects of elevation of [Ca2 +]i can be manifest in microseconds (neurotransmitter release), milliseconds (channel facilitation or inactivation) or over much longer time scales leading to changes in gene expression [12], [13] and effects on synaptic plasticity [14], neuronal development [15], learning [16], neuronal survival and cell death.
Clearly Ca2 + can influence many aspects of neuronal function with the same fundamental signalling ion being used to produce a variety of subtle and distinct changes. It is known, for example, that Ca2 + entry through different types of plasma membrane channels can affect changes in gene expression by different modes of signalling [17], [18]. Of note here is the fact that even different classes of voltage-gated Ca2 + channels (VGCCs) in the same neurons are coupled through distinct signalling pathways to changes in gene expression [19]. Ca2 + signals are translated into changes in cellular function through various types of Ca2 + sensor proteins that in general terms detect increases in [Ca2 +]i by becoming loaded with Ca2 +, undergo a conformational change and then interact with and regulate various target proteins. The subtlety of neuronal Ca2 + signalling is underpinned by differential signal processing by these Ca2 + sensor proteins, which in turn determines the physiological specificity of Ca2 + signalling pathways.
Neurons express a large number of Ca2 + sensor proteins [20] ranging from synaptotagmin I, which is the essential Ca2 +-sensor for fast (microsecond) neurotransmitter release [21], through the annexins [22], to many different EF hand containing proteins [20]. The EF hand is a highly conserved Ca2 +-binding motif [23] which is present, for example, in the ubiquitous Ca2 + sensor protein calmodulin [24]. Calmodulin has numerous targets for regulation and is known to have multiple functions in neurons acting via various targets including Ca2 +/calmodulin-dependent protein kinase II [25]. Other EF hand containing Ca2 + sensors expressed in neurons include the calcium-binding protein (CaBP) family [26], [27] and the neuronal calcium sensor (NCS) proteins [28], [29], [30].
The differential processing of neuronal Ca2 +-signals is affected by multiple aspects of the properties of Ca2 + sensors, and we suggest that they have a key role in determining signalling specificity. Factors that influence differential Ca2 + signalling include varied expression levels of sensors between neuronal cell types [31], differences in subcellular localisation, affinity and dynamics of Ca2 +-binding [32], association with protein signalling complexes [33], [34], regulation and variations in stoichiometry of binding with target proteins [35], [36] and specificity of target protein interaction. Appreciation of the contribution of Ca2 +-sensors to signalling specificity requires analysis of both the Ca2 +-sensors and their target interactions at a structural level [24], [37]. The rules underlying the sensing and specificity of Ca2 + signalling in neurons have begun to emerge in recent years but still remain to be fully understood. In this review, we will present three case studies to illustrate current understanding of the molecular and structural basis of the contribution of the properties of Ca2 + sensors to differential neuronal signal processing.
2. Case Study 1: NCS-1, a Ca2 + sensor with multiple specific targets
NCS-1 is a member of the neuronal calcium sensor family that is encoded by 14 genes in mammals. Of these, recoverin and the guanylyl cyclase activating protein (GCAP) 1–3 have specialised roles in the Ca2 + regulation of phototransduction. One other subfamily, consisting of the visinin-like protein (VILIP) 1–3, neurocalcin δ and hippocalcin has less well-defined functions. Hippocalcin, as suggested by its name, has a very restricted pattern of expression mainly in hippocampal neurons where it shows dynamic membrane association (the Ca2 + myristoyl switch [38]) in response to Ca2 +-elevation [39] and neuronal activity [40], [41], including activity-dependent translocation into dendritic spines. Hippocalcin has been implicated as a Ca2 + sensor in long-term depression [42], [43] and the gating of channels underlying a slow after-hyperpolarisation current [44]. VILIPs also show the Ca2 + myristoyl switch [45] and may have multiple roles, including the regulation of receptor trafficking [46]. The potassium-channel interacting proteins (KChIPs) have all been implicated in the gating [47] and trafficking [48] of A-type potassium channels but show differential expression in different classes of neurons [49], [50]. One of them, KChIP3 also known as calsenilin/DREAM, regulates presenilin function [51] and can act as a transcriptional repressor [52]. The other KChIPs may share the DREAM activity [53], but KChIP3 is specific among the KChIPs in increasing regulated secretion and down regulating the Na+/Ca2 + exchanger NCX3 [54].
NCS-1 appears to be expressed in most neuronal cell types [55], [31], and studies in various organisms have determined that it has multiple physiological functions [29], [56], [57], [58]. Some of the roles of NCS-1 may be specific to certain organisms such as its particular role in temperature-dependent behaviours in Caenorhabditis elegans as a consequence of its restricted neuronal expression [59], [60], [61]. One of two encoded NCS-1 proteins generated through gene duplication (ncs-1a) is required for semi-circular canal formation in the zebrafish inner ear [62] and NCS-1 (frequenin, Frq) is required for the development of synaptic boutons in Drosophila [56], the organism in which NCS-1 was first discovered [63]. In addition, in Saccharomyces cerevisiae NCS-1 (Frq1) is essential for survival as a consequence of its requirement for activation of the phosphatidylinositol-4-kinase Pik1 [64] despite its absence not being lethal in other organisms. In mammalian cells, NCS-1 regulates Ca2 +-dependent exocytosis [65], long-term depression [66], axonal growth and neuronal regeneration [67] and channel function [68]. In mice, knock-out of NCS-1 has relatively subtle effects but results in an increase in anxiety and depressive behaviour [69]. Selective overexpression of NCS-1 in adult mouse dentate gyrus neurons promoted a form of exploratory behaviour and enhanced acquisition of spatial memory [70].
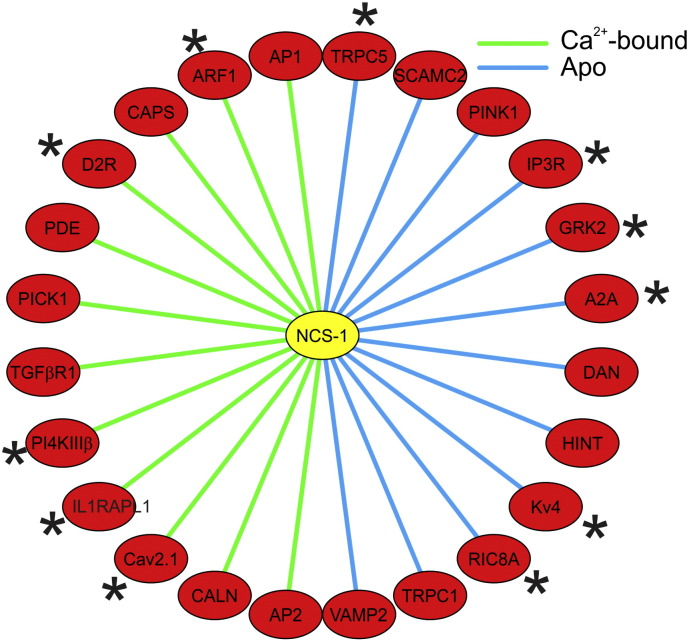
NCS-1 has many known interacting partners [71] (Fig. 1), some of which are unique for NCS-1 but some that are also regulated by other Ca2 + sensors particularly calmodulin [72]. Some of the interactions are known only from in vitro binding assays, and so their biological importance is unclear. It is possible that those binding partners that are also calmodulin targets in vitro [72] are not regulated by NCS-1 under physiological conditions [73]. There are, however, several physiological effects of NCS-1 that can be attributed directly to one of its identified target proteins (Table 1).Two documented NCS-1 interactions are of possible clinical significance. The potential importance of the regulation of dopamine D2 receptors by NCS-1 whereby NCS-1 inhibits D2 receptor internalisation (Fig. 2) [74] stems from the fact that dopamine is of key importance for signalling within the CNS and in addictive behaviour [75], [76]. The regulation of D2 receptors by NCS-1 has been shown to underlie the effect of overexpression of NCS-1 on spatial memory acquisition [70]. Importantly, dopamine D2 receptors are the targets for all known effective antipsychotic drugs [77]. Interestingly, NCS-1 is up-regulated in patients with bipolar disorder or schizophrenia [78] and in response to anti-psychotic drugs [79] and is genetically associated with cocaine addiction [80] believed to be linked to effects of cocaine on dopamine transporters [81]. Recently, NCS-1 has been shown to be required for an adaptive response to dopaminergic agonists in substantia nigra neurons, and coupled with its up-regulation in the substantia nigra from patients with Parkinson's disease, this has resulted in the suggestion that it could be a target for modifying the vulnerability of neurons in the substantia nigra to neurodegeneration [82]. The binding of NCS-1 to the D2 receptor involves the very short cytoplasmic C-terminal domain of the receptor [74]. This interaction has been partially characterised using structural approaches [83] and this may allow exploration of the interaction as a therapeutic drug target.
Fig. 1.
Known target proteins for NCS-1 indicating interactions that require the Ca2 +-bound or the apo form of NCS-1. The interactions shown include ones that are based only in vitro binding assays as well as interactions that have been substantiated and shown to have physiological relevance in functional studies (marked with an asterisk).
Table 1.
Key functionally characterised target proteins regulated by NCS-1.
| Target | Ca2 +-bound or apo | Effect on target | Functional consequences | References |
|---|---|---|---|---|
| ARF1 | Ca2 +-bound | Competes for PI4KIIIβ activation | Regulation of TGN to plasma membrane traffic | [117], [173] |
| α1 subunit of CaV2.1 | Ca2 +-bound | Activates Ca2 +-dependent facilitation of channel | Increases facilitation of neurotransmitter release | [145], [174] |
| Dopamine D2 receptor | Ca2 +-bound | Inhibits internalisation of receptor | Promotes spatial memory formation | [70], [74] |
| ILIRAPL1 | Ca2 +-bound | ? | Regulates N-type channels, secretion and neurite elongation | [84], [175] |
| PI4KIIIβ | Ca2 +-bound | Activates the enzyme | Regulation of TGN to plasma membrane traffic | [64], [173] |
| Adenosine A2 receptor | Apo | ? | Increases receptor signalling | [176] |
| GRK2 | Apo | Inhibits kinase activity | Inhibits receptor internalisation | [74] |
| InsP3 receptor | Apo | Enhances receptor activity | Increases calcium signalling in neurons and heart | [177], [178] |
| Ric8A | Apo | ? | Increases synapse number and synaptic release probability | [102] |
| TRPC5 | Apo | Activates channel | Retards neurite growth | [179] |
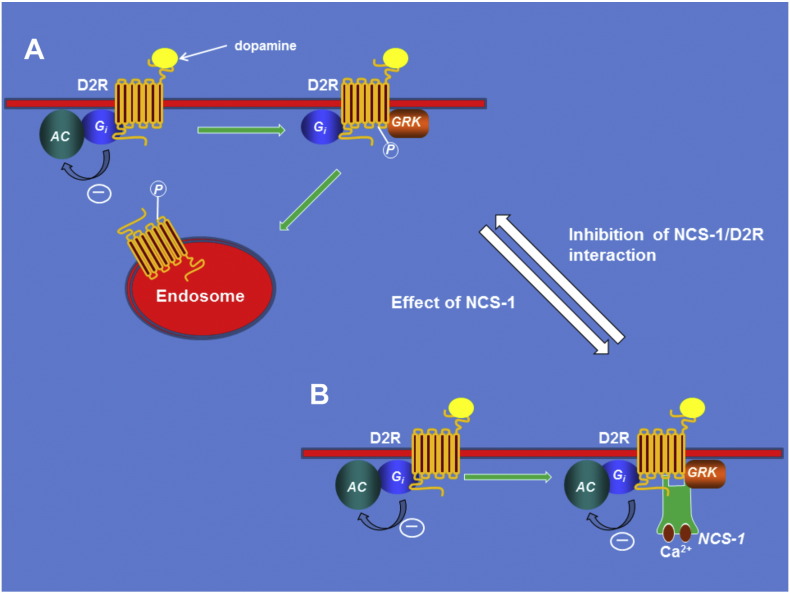
Fig. 2.
The role of NCS-1 in the regulation of dopamine D2 receptors. Agonist occupancy of the receptors results in inhibition of adenylate cyclase (AC) through the inhibitory G-protein Gi. In addition, dopamine D2 receptors undergo desensitisation following agonist binding due to their internalisation by endocytosis. This requires phosphorylation of the receptor by a G-protein coupled receptor kinase (GRK). The role of NCS-1 is to bind both the D2 receptor C-terminal tail and also GRK. Phosphorylation of the receptor is inhibited by NCS-1 and it is not internalised but instead remains at the cell surface. Inhibition of NCS-1/D2R interaction will allow receptor desensitisation.
The other clinically important interaction is with the interleukin 1 receptor accessory protein-like 1 protein (IL1RAPL1), which appears to be specific for NCS-1 [84]. Mutations in ILIRAPL1 have been shown to result in X-linked mental retardation [85], [86] and also have been linked to autistic spectrum disorder (ASD, [87]). Interestingly, the latter study also identified a mutation (R120Q) within NCS-1 in an individual with ASD. This mutation was found to cause a functional deficit in NCS-1 [88] that appeared to be related to a change in the structural dynamics of the C-terminus of the protein [88]. However, the physiological relevance of this mutation and its exact relationship to the disease phenotype remain to be established.
One key question is, what determines the specificity of target recognition for NCS-1 over other Ca2 + sensors and more particularly other members of the NCS protein family which have their own specific targets? The NCS proteins show differences in membrane-association with some such as recoverin and hippocalcin showing transient membrane-association through their Ca2 +/myristoyl switch mechanism [38], [39], [45], whereas others including KChIP3 and KChIP4 are predominantly cytoplasmic [54]. Others including KCIP1 and mammalian NCS-1 appear to be constitutively membrane targeted due to their N-terminal myristoylation [89], [90], but note, however, that Ncs-1 from fission years has been shown to have the Ca2 +/myristoyl switch [91]. The NCS proteins can, however, be very closely related to each other at a protein sequence level (> 60% identity). Structural analyses of several NCS proteins have shown common features with highly similar main chain topologies [37]. The gross tertiary topology of NCS proteins fails therefore to adequately explain target specificity, and so analyses of complexes of NCS proteins with peptides derived from their target proteins have been undertaken in an effort to better address this question. These studies have highlighted both common and distinct aspects of target recognition that help explain both NCS protein promiscuity and specificity. A common feature is that on Ca2 + binding, the NCS proteins undergo a conformational change that exposes a hydrophobic groove within which one or two helices from target proteins can bind. Once Ca2 + loaded the NCS, proteins undergo relatively limited structural changes on target ligand binding unlike the more dramatic changes seen in calmodulin conformation on ligand binding (described below).
Key hydrophobic amino acids within the hydrophobic groove of the NCS proteins that are conserved across the whole NCS family are directly involved in the target interactions in multiple NCS proteins (Fig. 3). Despite this similarity in the mechanism for target protein binding, there are structural differences that will determine specificity [37], [30]. There are differences in the overall size of the exposed hydrophobic groove (which may require some movement of the C-terminal tail of the protein) so that two or only one helix can bind. Also, differences in the distribution of surrounding surface exposed charged residues influences binding of specific ligands. In addition, there are key differences in the role of the C-terminal tail. In the case of KChIP1 [92], [93] and recoverin [94], the C-terminal proximal pocket of the groove is partially occluded by the C-terminal tail of the NCS protein allowing only a single helix to bind. In recoverin, amino acids in the C-terminal tail are actually required to make direct contact with the rhodopsin kinase target peptide for high-affinity binding [95]. In one of the published structures of NCS-1, the C-terminal tail appears to be able to sit in the hydrophobic groove in the absence of ligand [96]. This is likely an auto-regulatory mechanism to control target binding as the groove is fully exposed in another published structure of human NCS-1 [97]. In addition, in the structure of the yeast protein in complex with Pik1, two helices of Pik1 were bound within a fully exposed hydrophobic groove and the C-terminus appeared disordered, suggesting that the C-terminal tail can undergo a conformational change to allow its movement out of the groove on ligand binding [91], [98].
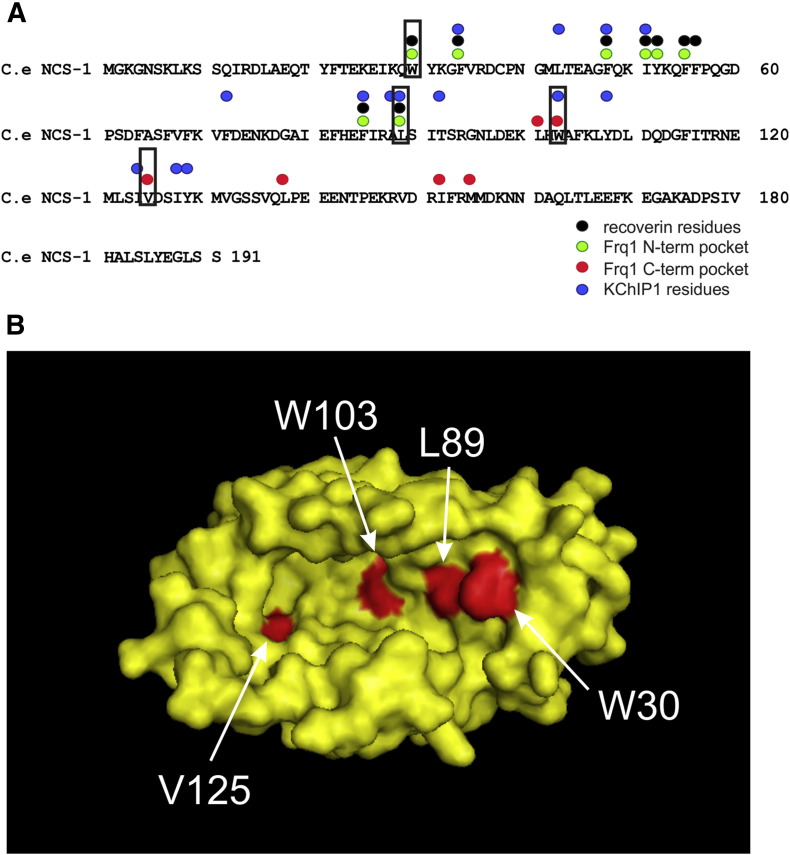
Fig. 3.
Key residues for target protein interactions in the hydrophobic groove of NCS proteins identified from structural studies and amino acids identified for mutagenesis in Caenorhabditis elegans NCS-1. (A) Hydrophobic residues implicated in target protein interactions are conserved in C. elegans NCS-1. Those residues that directly make contact with target proteins in structurally characterised complexes are indicated above the sequence of C. elegans NCS-1. Residues selected for mutagenesis are boxed. (B) Position of residues selected for mutagenesis in the predicted NCS-1 structure. The selected amino acids are within the hydrophobic groove are shown in red in a surface representation of a model structure for C. elegans NCS-1 based on the crystal structure of human NCS-1 (PDB1G8I). Adapted from Martin et al 2013 [60].
The structural studies on NCS protein/target interaction have been based on the use of short peptides derived from the target protein. These studies have provided a consistent picture in which conserved hydrophobic residues are crucial. Nevertheless, it is important to assess the significance of the structural data using a physiological model. This would, for example, allow a test of requirement for the key residues in the hydrophobic groove and also the requirement for the C-terminal tail for NCS-1 function. This was approached using an assay for temperature-dependent locomotion in C. elegans [99] that requires NCS-1 function in the pair of AIY neurons in the worm. Using an NCS-1 null mutant worm strain, an assay was developed for functional reconstitution. Key amino acids were then mutated in the N- and C-terminal parts of the hydrophobic groove (Fig. 3), and the mutant proteins expressed in the null background. The findings from this assay were fully consistent with the structural data and showed that for normal NCS-1 function in this behavioural assay hydrophobic resides in both the N- and C-terminal parts of the hydrophobic groove were required [60]. In addition, full functional reconstitution occurred after truncation of the C-terminus of NCS-1 and indeed truncation of residues 169–191 to remove the entire C-terminal helix resulted in a gain of function [60]. These findings are consistent with the notion that the C-terminal tail of NCS-1 unlike that in for recoverin is not essential for direct target interactions but instead may have an auto-inhibitory role by binding with the C-terminal part of the hydrophobic groove in the absence of a suitable binding ligand [96], and this may prevent binding of inappropriate ligands.
Evidence for a role in specificity determination of residues surrounding the hydrophobic groove has come from analysis of Drosophila frequenins. In Drosophila species, a gene duplication event generated two genes encoding closely similar proteins known as Frq1 (the originally identified frequenin [63]) and Frq2 [100]. Both isoforms affect synaptic transmission and synaptic bouton number [101]. Frq1 and Frq2 differ by only 10 amino acids, and it was unclear whether they would have distinct functions. Recent work has now identified the guanine nucleotide exchange factor Ric8a as a binding partner for Frq2 but not Frq1 and functional analyses suggest that the interaction is involved in the physiological roles of Frq2 in the fly [102]. The interaction with ric8 was conserved in the human proteins [102]. From examination of the crystal structure of Frq2, a small number of solvent exposed amino acids that differed between the two frequenins were highlighted as being potentially important for target recognition, and mutagenesis of Frq1 established that the amino acids R94 and T138 located at the edge of the hydrophobic groove were required for Ric8a binding. Incidentally, the removal of the C-terminal helix of Frq2 increased its binding to Ric8a consistent with the findings on the functional effect of deletion of the C-terminal tail of NCS-1 in C. elegans [60]. These findings provide clear evidence in support of the structural models for NCS protein/target specificity [37].
3. Case Study 2—CaBPs, Ca2 + sensors with targets in common with and distinct from calmodulin
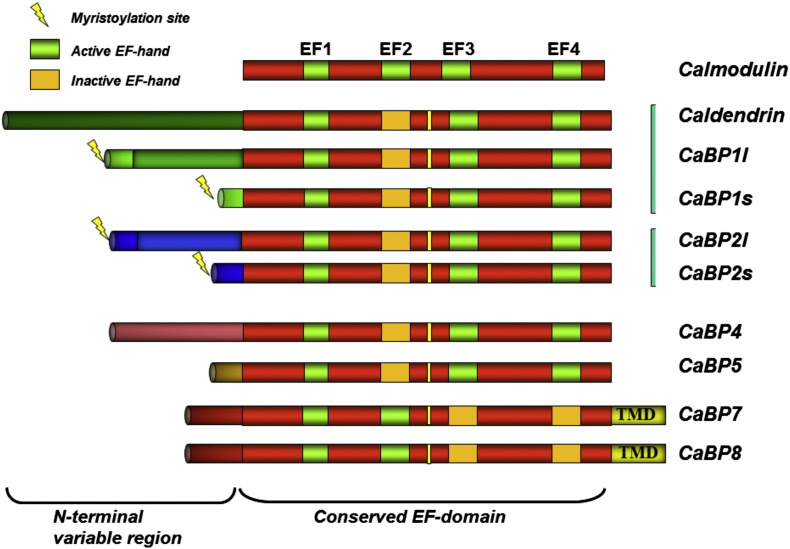
The CaBP proteins in humans are encoded by 6 functional genes (Fig. 4), with CaBP1 and CaBP2 existing as variants derived from alternative splicing [26], [27], [103], [104]. They have a higher level of similarity to calmodulin than do the NCS proteins but have divergent N-terminal domains that differ between the CaBPs and which do not occur in calmodulin and also have a specific larger linker region between the N- and C-terminal EF hand domains. Certain of the CaBPs can be membrane-associated through N-terminal myristoylation or possession of a C-terminal transmembrane domain (Fig. 4). Both CaBP1/caldendrin and calmodulin can interact with VGCCs. In addition, CaBP1 has been shown to bind and regulate InsP3 receptors [105], which also bind calmodulin [106]. Originally, it was suggested that CaBP1 activates the receptor, but subsequent work has established that instead it has an inhibitory effect on receptor function in response to InsP3 [107], [108].
Fig. 4.
Domain structure of the CaBP family of Ca2 + sensors compared to calmodulin. Unlike calmodulin, each of the CaBPs has at least one inactive EF hand. Some are membrane targeted due to N-terminal myristoylation whereas CaBP7 and CaBP8 possess C-terminal transmembrane domains that determine their membrane association [114], [115].
CaBP1 is similar to calmodulin in that it has distinct N-and C-terminal lobes [109], [110], [111]. Each contains two EF hands, but unlike calmodulin, the second EF hand is non-functional in CaBP1. The structure of CaBP1 was derived from analyses of the crystal structure of a form with Ca2 +-bound to EF3 and EF4 [109], [110], [111] and an NMR solution structure of a form with Ca2 + bound to EF hands 1, 3 and 4 [109], [110], [111]. Unlike in the solution structure, CaBP1 in the crystal structure was found to be oligomerised, and there were some differences in the N-lobe between the two structures. The C-terminal but not the N-terminal lobe binds to the InsP3 receptor [110], and the binding interaction and inhibition of receptor function have been shown to require hydrophobic residues that are exposed in the Ca2 +-bound form of the C-lobe and that permit binding to complementary hydrophobic regions on the receptor [112]. In addition to characterising the binding interaction, this structural study also suggested a mechanism whereby CaBP1 inhibited InsP3 receptor gating by limiting inter-subunit movements to stabilise a closed state of the channel.
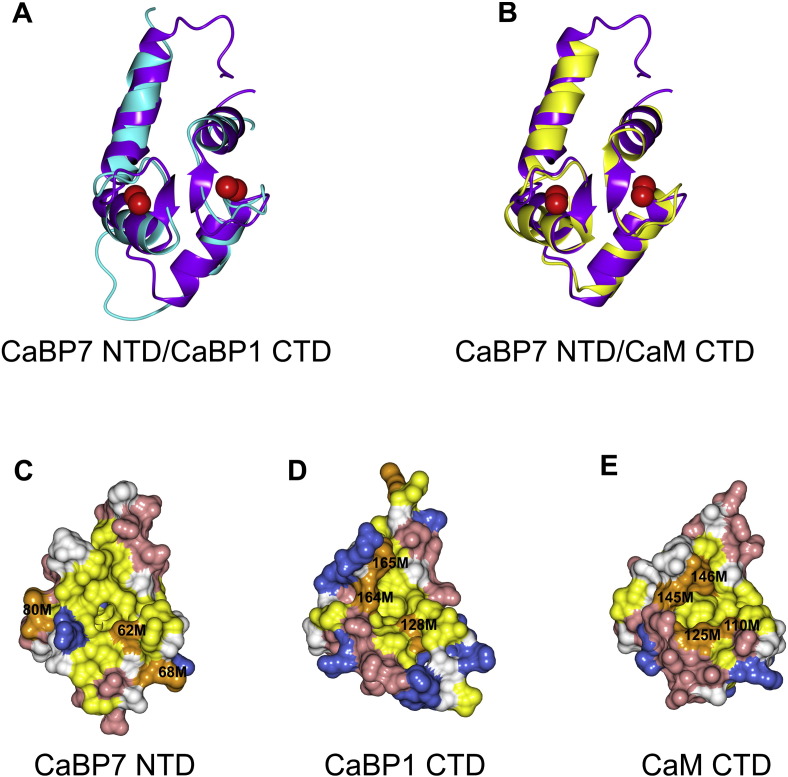
CaBP7 and CaBP8 (also known as calneurons II and I, respectively [104], [113]) are evolutionarily distinct from the other CaBPs [103] and have a unique C-terminal transmembrane domain [114], [115], [116]. Presently, they have only one known target, phosphatidylinositol-4-kinase IIIβ (PI4KIIIβ), which is also regulated by NCS-1 but is not known to be regulated by other Ca2 + sensors. Whereas NCS-1 stimulates the kinase at elevated [Ca2 +]i [117] CaBP7 and CaBP8 inhibit its activity at resting [Ca2 +]I, and this action can fully explain the physiological effects of CaBP7 and CaBP8 on membrane traffic [118]. The active EF hands in CaBP7 and CaBP8 are present in the N-terminal domain (Fig. 4), and this domain from CaBP7 can independently bind PI4KIIIβ [119]. The overall tertiary structure of the Ca2 +-bound N-terminal domain (residues 1–100) of CaBP7 was solved by solution NMR [119]. The structure is most similar to that of the C-terminal lobes of calmodulin and CaBP1 (Fig. 5). Clues to the basis for specificity of target binding come from examination of space-filling representations of the three structures. CaBP7 has an exposed hydrophobic face that is larger in area than that found in calmodulin or CaBP1 with the presence of fewer charged residues [119]. Notably, CaBP7 lacks three of the four methionine residues in the hydrophobic pocket that are present in both the calmodulin N- and C-terminal lobes and that are required for target-binding by calmodulin [24], [120], [121]. Three of these methionines are conserved in the CaBP1 C-terminal lobe, and interestingly, two of these (M164 and M165) make contact with the N-terminal domain of the InsP3 receptor in the docked complex [112]. It may be significant that the methionines in CaBP7 and also CaBP8 have been exchanged for conformationally immobile leucine and isoleucine residues, which might help explain the restricted number of targets available for interaction with CaBP7. The underlying hypothesis is that the CaBP hydrophobic binding surface can be made more flexible by increasing the number of methionines present or more rigid by reducing their number and that the degree of flexibility governs target promiscuity. Consistent with this idea, calmodulin has many more known interaction partners (> 300 [24]) than CaBP1 which in turn exhibits a considerably larger interactome (at least 14 targets) than CaBP7, which is currently known to interact only with PI4KIIIβ. It should be noted, however, that we cannot rule out that further work will identify additional targets for CaBP7 and CaBP8.
Fig. 5.
Comparison of the N-terminal domain of CaBP7 with the C-terminal domains of CaBP1 and calmodulin. Ribbon representation of Ca2 +-bound CaBP7 NTD (PDB code: 2LV7, residues 30–100, purple) superposed with (A) Ca2 +-bound CaBP1 CTD (cyan, PDB code: 2LAP) and (B) Ca2 +-bound CaM CTD (yellow, PDB code 1CLL, residues 80–147). Red spheres represent bound Ca2 +. Space-filling representations of (C) Ca2 +-bound CaBP7 N-terminal domain (PDB code: 2LV7, residues 30–100), (D) Ca2 +-bound CaBP1 C-terminal domain (PDB code: 2LAP) and (E) Ca2 +-bound CaM C-terminal domain (PDB code: 1CLL, residues 80–147), showing the predicted ligand binding face. Acidic residues and basic residues are shown in red and blue, respectively. Hydrophobic residues are shown in yellow with the exception of Met residues which are highlighted in orange. Adapted from McCue et al 2012 [119].
4. Case study 3—regulation of voltage-gated Ca2 + channels by multiple Ca2 + sensors
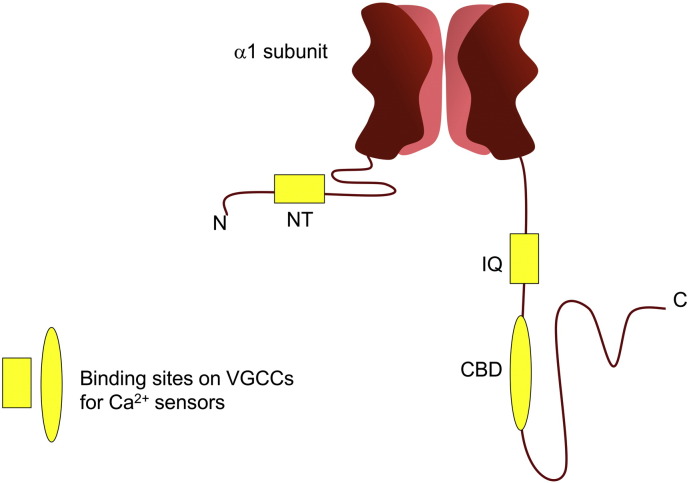
Ca2 +-entry through voltage-gated Ca2 + channels in neurons triggers neurotransmitter release, changes in gene expression, underpins synaptic plasticity and affects neuronal development [122], [123], [124]. The multiple types of VGCCs have been well characterised and found to undergo extensive regulation including feedback by Ca2 + itself leading to rapid Ca2 +-dependent facilitation (CDF) or Ca2 +-dependent inhibition (CDI) on a millisecond timescale. The speed of this regulation is explained by the response of the channels being due to nanodomains of Ca2 + near the mouth of the channel [125]. Both CDF and CDI are believed to contribute to synaptic plasticity [126], [127], [128]. A recent review has summarised the history of the discovery of VGCC regulation by Ca2 + and progress on the characterisation of the mechanisms particularly those involving calmodulin [129]. The regulation of VGCCs by other Ca2 + sensors has also been the subject of a recent review [128]. Pioneering studies identified calmodulin as a Ca2 + sensor required for CDI of L-type (CaV1.2) Ca2 + channels [130], [131]. It was subsequently demonstrated that Ca2 +-free (apo) calmodulin was pre-bound to the pore-forming α1 channel subunit and that this was mediated through the so-called IQ domain (Fig. 6) of the α1 subunit [132], [133], [134]. The fact that calmodulin is already bound to the channel subunit explains how the channels can be regulated so rapidly in response to Ca2 + elevation in local nanodomains.
Fig. 6.
Potential binding sites on VGCCs for Ca2 + sensors that have been identified in the N- or C-terminal cytoplasmic domains of the α1 subunits of various types of VGCCs. These include a site in the N-terminus and the IQ and calmodulin-binding (CBD) domains in the C-terminus. Other regions of the C-terminal domain may also be required for Ca2 + sensor interactions.
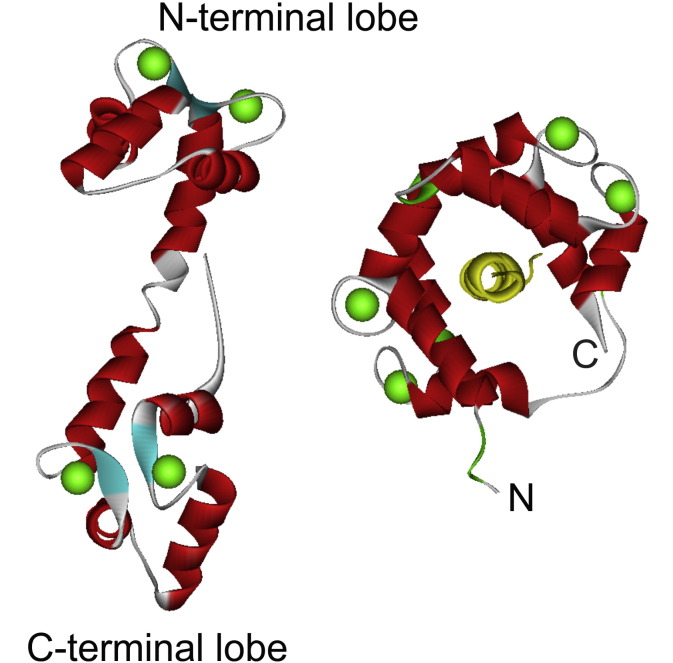
A role for calmodulin was subsequently extended to other VGCC types. The use of Ca2 +-insensitive calmodulin mutants with inactivated EF hands demonstrated its requirement. Calmodulin has independent N- and C-terminal lobes, which each possess 2 active EF hands. The two lobes can show major structural movement relative to each other on ligand binding allowing them to both bind to the same helix (Fig. 7). For CaV1.2 channels, CDI was mediated through the C-lobe [130]. In contrast, for CaV2.1 [135], [136], CaV2.2 [137] and CaV2.3 [137], it is the N-lobe that controls CDI. Of particular interest is the finding that for P/Q-type (CaV2.1) channels calmodulin is required for both CDI and CDF with the N-lobe of calmodulin involved in CDI and the C-lobe in CDF [135], [136]. The majority of studies have focused on the IQ domain in the C-terminal cytoplasmic region of the subunit (Fig. 6) as the binding site for calmodulin on the various channel types [138], [139]. Other studies have implicated an additional calmodulin-binding domain (CBD) in the C-terminus present in CaV2.1 [136], and there are likely to be additional binding sites in the N-terminal domain of the α1 subunit (Fig. 6).
Fig. 7.
Comparison of the structures of Ca2 +-loaded calmodulin with and without bound target peptide. The structures shown are of Ca2 +-bound calmodulin alone ([180]) on the left or in a complex with the IQ-like domain of the Cav1.2 Ca2 +-channel α1-subunit (PDB 2F3Z) [170] on the right with the IQ-like peptide shown in yellow.
The picture of Ca2 +-dependent regulation of VGCCs became more complex when it was discovered that various other Ca2 + sensors could exert differing effects on CDI, CDF (Table 2) or channel current, in some cases apparently also through binding to the IQ domain [128]. The first such indication of an additional regulatory mechanism came from the finding that CaBP1 could elicit CDI itself and also reduce CDF due to calmodulin in CaV2.1 channels [140]. CaBP1, as described above, has been shown to have a bi-lobe structure like calmodulin [109], and it has been suggested that it exerts its effect on CDF by competing-off calmodulin from the IQ domain [109], [141], [142].
Table 2.
Summary of the direct regulation of Ca2 +-dependent facilitation (CDF) or Ca2 +-dependent inhibition (CDI) of neuronal VGCCs by Ca2 + sensors.
| VGCC | Type | CDF | CDI | References |
|---|---|---|---|---|
| CaV1.2 | L-type | CaBP1 | CaM, CaBP1↓ Caldendrin↓ CaBP5↓ |
[130], [131], [150], [151], [154] |
| CaV1.3 | L-type | CaM, CaBP1,2,3,& 4↓ | [155], [156], [157], [158], [167] | |
| CaV1.4 | L-type | CaBP4↓ | [160] | |
| CaV2.1 | P/Q-type | CaM, VILIP-2, NCS-1, CaBP1↓ | CaM, CaBP1, VILIP-2↓ |
[135], [136], [140], [143], [145], [146] |
| CaV2.2 | N-type | CaM | [137] | |
| CaV2.3 | R-type | CaM | [137] |
CaV2.1 channels appear to be under extensive regulatory control by multiple Ca2 + sensors. In addition to calmodulin and CaBP1, these channels are also regulated by the NCS proteins VILIP-2 and NCS-1. VILIP-2 inhibits the inactivation brought about by calmodulin and also enhances facilitation in the same way as calmodulin [143], [144] through interaction with the IQ and the CBD domains. Electrophysiological experiments implicated NCS-1 in CDF of CaV2.1 channels [145] and also in an autocrine pathway that negatively regulates these P/Q-type Ca2 + channels in adrenal chromaffin cells [146], [147]. In addition, functional effects of loss of NCS-1 (Frq) in Drosophila have been investigated using genetic approaches and attributed to the regulation of cacophony [56], [148], which encodes an α1-subunit of a fly VGCC. Despite these studies, a direct interaction of NCS-1 with the CaV2.1 channel or any other VGCC had not been reported. Recently, however, the use of multiple biochemical and structural approaches has consistently demonstrated a direct interaction of NCS-1 with the CaV2.1 channel α1-subunit that requires the IQ domain but does not appear to involve the CBD in the C-terminus of the α1-subunit [149]. NMR analysis also demonstrated direct binding of an IQ domain peptide to NCS-1.
Members of the CaBP family have been found to regulate a range of VGCC types (Table 2). For example, CaBP1 prolongs CaV1.2 currents and inhibits CDI [150], [151]. The gene encoding CaBP1 also gives rise to a longer isoform, caldendrin, which may be the predominant form in brain [152], [153]. Caldendrin also reduces CDI of CaV1.2 channels but to a lesser extent than CaBP1 [150]. Studies of CaBP5 function in the retina have suggested that it acts as an inhibitor of CDI of CaV1.2 channels [154]. Potentially other cell-specific forms of regulation by multiple CaBPs of CaV1.3 channels have also been discovered in auditory hair cells [155], [156], [157], although CaBP1 may be the principal regulator [128]. In photoreceptors, there is a potential function for CaBP4 in the regulation of CaV1.3 channels [158]. CaBP4 may also be important in photoreceptors for the regulation of CaV1.4 channels through an effect on the voltage activation of these channels [159], [160].
Many of the studies on the regulation of CDF or CDI of VGCCs by Ca2 + sensors have been based on expression of the sensors and VGCC subunits in heterologous cell systems. This, therefore, raises questions about the physiological relevance of the interactions of the Ca2 + sensors for neuronal function. Support for a significant role has come from a study in which expression of modified CaV1.2 channels in superior cervical ganglion (SCG) neurons was used to probe the general role for interaction of Ca2 + sensors in synaptic plasticity. In this study, it was shown that mutation of the IQ (IM in Cav2.1) motif or the CBD that would prevent CDF or CDI reduced facilitation or short-term synaptic depression, respectively [161]. In addition, another study using SCG neurons showed that expression of CaBP1 or VILIP-2 had effects on synaptic plasticity [127] consistent with their effects on CDF and CDI in heterologous models [162].
One problem with the originally proposed competitive model of action for CaBPs in which they need to compete endogenous calmodulin from the IQ domain is that calmodulin is expressed in brain at considerably higher levels than CaBPs. CaBP1 has been shown, however, to interact with an additional site in the N-terminus of CaV1.2 [163]. More recently, it has been suggested that apo-CaBP4 and apo-calmodulin may both be associated with CaV1.3 channels as a consequence of CaBP4 binding to regions other than the IQ domain [156] so that the higher concentration of calmodulin would not be a significant barrier to a physiological role for CaBPs. In addition to these considerations, it should also be emphasised that calmodulin interacts with a large number of cellular targets. Its availability for any given interaction is therefore difficult to accurately determine and this also likely impacts on the ability of other calcium sensors to modulate shared targets.
Findings consistent with an important role for CaBP4 in the regulation of phototransduction have come from the study of a CaBP4 mouse knock-out and also the presence of mutations in human CaBP4 [164], [165] and in CaV1.4 [166], which cause retinal disorders including congenital night blindness. Data from the CaBP5 knock-out mouse support a role for CaBP5 in altering retinal sensitivity though its effects on CaV1.2 channels [154]. In addition, a mutation in CaBP2 resulting in its truncation has been found that results in autosomal hearing impairment. CaBP2 is expressed in cochlear hair cells where the effect of the truncation may be due to impaired suppression of CDI of Cav1.3 channels [167].
The structural basis for how calmodulin or other Ca2 + sensors exert their Ca2 +-dependent effects on VGCCs has been a subject of intensive study. Key issues to be resolved include (1) how can the two lobes of calmodulin have divergent functions when both bind to the IQ domain, (2) why does the C-lobe dominate in CDI in CaV1.2 channels but the N-lobe does so in CaV2 channels, (3) how can binding of calmodulin to an intracellular region of the channel α1 subunit affect the gating properties of the channel pore and (4) how can additional Ca2 + sensors that mediate differing effects do so particularly in the presence of calmodulin tightly bound to the IQ domain [129], [168]. A major focus for the calmodulin control of VGCCs has been in analysis of the IQ domain interaction. Initial studies characterised the structure of the complex of Ca2 +-bound calmodulin with an IQ-like peptide from CaV1.2 (Fig. 7). These indicated that calmodulin wrapped around the IQ helix with both the N- and C-terminal lobes binding and with the helix bound in an unusual parallel manner with the N- and C-lobes of calmodulin binding to the N- and C-terminal ends of the IQ helix [169], [170]. It was subsequently found that calmodulin bound the IQ peptides of CaV2.1, CaV2.2 or CaV2.3 channels in an anti-parallel manner giving a possible explanation for the reverse role of the N- and C-lobes in CDI of the two classes of channels [138]. This explanation was not supported by another study, however, that showed that all IQ/calmodulin interactions had a parallel conformation, although this study was based on complexes with a shorter IQ peptide [139]. More recent studies have begun to explore the structural basis for calmodulin interaction with larger constructs from the C-terminal domain of the VGCCs (reviewed in [168]). It is clear that much still remains to be learnt but recent work has suggested that interactions in the C-terminus outside the IQ domain and with the N-terminus of the α1 subunit involving the Ca2 +-loaded N-lobe may be important [171].
A full structural characterisation of the interaction of CaBP1 with VGCCs is not available but one analysis has used chimeras with calmodulin and specific mutants to dissect the requirements for the regulation of CaV1.2 channels. This has demonstrated that both lobes of CaBP1 are functionally important with the C-lobe anchoring to the channel subunit as does the C-lobe of calmodulin. The N-lobe and also the inter-lobe linker specific for the CaBPs are required for modification of channel function and this could contribute to the different effects on the channel of CaBP1 compared to calmodulin [109]. It has been suggested that differential effects of calmodulin and CaBP1 may in part be due to the structurally different interactions with the IQ domain [109]. It is also possible that interactions with other regions of the α1 subunit may be important, but this will require further structural characterisation. There is currently no published data on the structural bases for the interactions of other Ca2 + sensors with VGCCs.
5. Conclusions
One aspect that emerges from consideration of the three case studies above is the variation in level of specificity versus promiscuity of the Ca2 + sensors which is determined by fundamental structural characteristics of the proteins. Calmodulin is the most promiscuous with many target proteins. It consists of two independent lobes joined by a flexible linker that allows calmodulin to wrap around and interact with target proteins in multiple different ways using mutually induced fit to ensure high affinity interaction [172]. Also, calmodulin possesses several residues with flexible side chains on its hydrophobic binding surface that also seems to be important to its promiscuity accommodating binding of numerous different ligands to the same protein surface. The CaBP proteins while superficially similar to calmodulin in having two N- and C-terminal lobes have a reduced range of targets as a consequence of having one or more inactivated EF hands and a reduction to varying levels (see CaBP1 versus CaBP7/8) in the number of methionines presented on their binding surface. They also have specialisations (such as myristoylation or the transmembrane domains of CaBP7 and CaBP8) that will limit their access to potential cytoplasmic target proteins.
On Ca2 + binding, Ca2 + sensors undergo a conformational change that allows them to bind ligands and may then undergo further conformational changes. This in turn will allow them to regulate the function of the target proteins. Most of the available structural characterisation has provided information on the binding mechanism and specificity but so far has provided little information on how the activity of the target protein is regulated. This is particularly notable in the case of VGCCs where even different lobes of calmodulin can have different functional effects after binding to a site on the channel α1 subunit some distance from the channel pore. For VGCC regulation, we need further information on the structural basis of interactions of Ca2 + sensors outside of the IQ domain, with the rest of the C-terminus, with sites in the N-terminus of the α1 subunit and also potentially with other subunits of the holo-channel. The NCS proteins such as NCS-1 have a smaller range of targets and differ from calmodulin in having two lobes that have a very rigid inter-relationship severely reducing the conformational flexibility between the two lobes. Target binding is due to the exposure of a hydrophobic groove the size and surrounding residues of which in conjunction with a variable and more flexible C-terminal tail determine which ligands can bind. A completely un-investigated aspect of NCS-target interaction is how NCS protein binding influences target conformation and structure and how this translates to function. Again, structural insights in this regard are limited by the complexes characterised to date containing only short peptides derived from the target protein. Further progress in this field will, therefore, require characterisation of Ca2 + sensors complexes with larger, ideally full-length, target proteins in order to contribute to an understanding of the mechanistic basis of differential signal processing in response to Ca2 + signals.
Acknowledgements
Work in the authors' laboratory was supported by the Wellcome Trust.
Footnotes
This article is part of a Special Issue entitled: 13th European Symposium on Calcium.
References
- 1.Brini M., Cali T., Ottolini D., Carafoli E. Neuronal calcium signaling: function and dysfunction. Cell. Mol. Life Sci. 2014;71:2787–2814. doi: 10.1007/s00018-013-1550-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berridge M.J. Neuronal calcium signalling. Neuron. 1998;21:13–26. doi: 10.1016/s0896-6273(00)80510-3. [DOI] [PubMed] [Google Scholar]
- 3.Berridge M.J. Calcium hypothesis of Alzheimer's disease. Pflugers Arch. 2010;459:441–449. doi: 10.1007/s00424-009-0736-1. [DOI] [PubMed] [Google Scholar]
- 4.Stutzmann G.E., Caccamo A., LaFerla F.M., Parker I. Dysregulated IP3 signaling in cortical neurons of knock-in mice expressing an Alzheimer's-linked mutation in presenilin1 results in exaggerated Ca2 + signals and altered membrane excitability. J. Neurosci. 2004;24:508–513. doi: 10.1523/JNEUROSCI.4386-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Popugaeva E., Bezprozvanny I. Role of endoplasmic reticulum Ca2 + signaling in the pathogenesis of Alzheimer disease. Front. Mol. Neurosci. 2013;6:29. doi: 10.3389/fnmol.2013.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berridge M.J. Calcium signalling and psychiatric disease: bipolar disorder and schizophrenia. Cell Tissue Res. 2014;357:477–492. doi: 10.1007/s00441-014-1806-z. [DOI] [PubMed] [Google Scholar]
- 7.C. Cross-Disorder Group of the Psychiatric Genomics Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berridge M.J., Lipp P., Bootman M.D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 9.Bading H. Nuclear calcium signalling in the regulation of brain function. Nat. Rev. Neurosci. 2013;14:593–608. doi: 10.1038/nrn3531. [DOI] [PubMed] [Google Scholar]
- 10.Takechi H., Eilers J., Konnerth A. A new class of synaptic response involving calcium release in dendritic spines. Nature. 1998;396:757–760. doi: 10.1038/25547. [DOI] [PubMed] [Google Scholar]
- 11.Segal M., Korkotian E. Endoplasmic reticulum calcium stores in dendritic spines. Front. Neuroanat. 2014;8:64. doi: 10.3389/fnana.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan J.I., Curran T. Role of ion flux in the control of c-fos expression. Nature. 1986;322:552–555. doi: 10.1038/322552a0. [DOI] [PubMed] [Google Scholar]
- 13.Bito H., Deisseroth K., Tsien R.W. Ca2 +-dependent regulation in neuronal gene expression. Curr. Opin. Neurobiol. 1997;7:419–429. doi: 10.1016/s0959-4388(97)80072-4. [DOI] [PubMed] [Google Scholar]
- 14.Yang S.-N., Tang Y.-G., Zucker R.S. Selective induction of LTP and LTD by postsynaptic [Ca2 +]i elevation. J. Neurophysiol. 1999;81:781–787. doi: 10.1152/jn.1999.81.2.781. [DOI] [PubMed] [Google Scholar]
- 15.Spitzer N.C., Lautermilch N.J., Smith R.D., Gomez T.M. Coding of neuronal differentiation by calcium transients. Bioessays. 2000;22:811–817. doi: 10.1002/1521-1878(200009)22:9<811::AID-BIES6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 16.Dash P.K., Moore A.N., Kobori N., Runyan J.D. Molecular activity underlying working memory. Learn. Mem. 2007;14:554–563. doi: 10.1101/lm.558707. [DOI] [PubMed] [Google Scholar]
- 17.Bading H., Ginty D.D., Greenberg M.E. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science. 1993;260:181–186. doi: 10.1126/science.8097060. [DOI] [PubMed] [Google Scholar]
- 18.Deisseroth K., Heist E.K., Tsien R.W. Translocation of calmodulin to the nucleus supports CREB phosphorylation in hippocampal neurons. Nature. 1998;392:198–202. doi: 10.1038/32448. [DOI] [PubMed] [Google Scholar]
- 19.Wheeler D.G., Groth R.D., Ma H., Barrett C.F., Owen S.F., Safa P., Tsien R.W. CaV1 and CaV2 channels engage distinct modes of Ca2 + signaling to control CREB-dependent gene expression. Cell. 2012;149:1112–1124. doi: 10.1016/j.cell.2012.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCue H.V., Haynes L.P., Burgoyne R.D. The diversity of calcium sensor proteins in the regulation of neuronal function. Cold Spring Harb. Perspect. Biol. 2010;2:a004085. doi: 10.1101/cshperspect.a004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez-Chacon R., Konigstorfer A., Gerber S.H., Garcia J., Matos M.F., Stevens C.F., Brose N., Rizo J., Rosenmund C., Sudhof T.C. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 22.Gerke V., Creutz C.E., Moss S.E. Annexins: linking Ca2 + signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 23.Moncreif N.D., Kretsinger R.H., Goodman M. Evolution of EF-hand calcium-modulated proteins. 1. Relationships based on amino-acid sequences. J. Mol. Evol. 1990;30:522–562. doi: 10.1007/BF02101108. [DOI] [PubMed] [Google Scholar]
- 24.Ikura M., Ames J.B. Genetic polymorphism and protein conformational plasticity in the calmodulin superfamily: two ways to promote multifunctionality. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1159–1164. doi: 10.1073/pnas.0508640103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamauchi T. Neuronal Ca2 +/calmodulin-dependent protein kinase II–discovery, progress in a quarter of a century, and perspective: implication for learning and memory. Biol. Pharm. Bull. 2005;28:1342–1354. doi: 10.1248/bpb.28.1342. [DOI] [PubMed] [Google Scholar]
- 26.Haeseleer F., Sokal I., Verlinde C.L.M.J., Erdjument-Bromage H., Tempst P., Pronin A.N., Benovic J.L., Fariss R.N., Palczewski K. Five members of a novel Ca2 + binding protein (CABP) subfamily with similarity to calmodulin. J. Biol. Chem. 2000;275:1247–1260. doi: 10.1074/jbc.275.2.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haynes L.P., McCue H.V., Burgoyne R.D. Evolution and functional diversity of the calcium binding proteins (CaBPs) Front. Mol. Neurosci. 2012;5:9. doi: 10.3389/fnmol.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burgoyne R.D., Weiss J.L. The neuronal calcium sensor family of Ca2 +-binding proteins. Biochem. J. 2001;353:1–12. [PMC free article] [PubMed] [Google Scholar]
- 29.Burgoyne R.D. Neuronal calcium sensor proteins: generating diversity in neuronal Ca2 + signalling. Nat. Rev. Neurosci. 2007;8:182–193. doi: 10.1038/nrn2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgoyne R.D., Haynes L.P. Understanding the physiological roles of the neuronal calcium sensor proteins. Mol. Brain. 2012;5:2. doi: 10.1186/1756-6606-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paterlini M., Revilla V., Grant A.L., Wisden W. Expression of the neuronal calcium sensor protein family in the rat brain. Neuroscience. 2000;99:205–216. doi: 10.1016/s0306-4522(00)00201-3. [DOI] [PubMed] [Google Scholar]
- 32.Raghuram V., Sharma Y., Kreutz M.R. Ca2 + sensor proteins in dendritic spines: a race for Ca2 + Front. Mol. Neurosci. 2012;5:61. doi: 10.3389/fnmol.2012.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson D., Engbers J.D., Heath N.C., Bartoletti T.M., Mehaffey W.H., Zamponi G.W., Turner R.W. The Cav3-Kv4 complex acts as a calcium sensor to maintain inhibitory charge transfer during extracellular calcium fluctuations. J. Neurosci. 2013;33:7811–7824. doi: 10.1523/JNEUROSCI.5384-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navedo M.F., Hell J.W. AKAP5 keeps L-type channels and NFAT on their toes. Cell Rep. 2014;7:1341–1342. doi: 10.1016/j.celrep.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitazawa M., Kubo Y., Nakajo K. The stoichiometry and biophysical properties of the Kv4 potassium channel complex with K+ channel-interacting protein (KChIP) subunits are variable, depending on the relative expression level. J. Biol. Chem. 2014;289:17597–17609. doi: 10.1074/jbc.M114.563452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kunjilwar K., Qian Y., Pfaffinger P.J. Functional stoichiometry underlying KChIP regulation of Kv4.2 functional expression. J. Neurochem. 2013;126:462–472. doi: 10.1111/jnc.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ames J.B., Lim S. Molecular structure and target recognition of neuronal calcium sensor proteins. Biochim. Biophys. Acta. 2012;1820:1205–1213. doi: 10.1016/j.bbagen.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ames J.B., Ishima R., Tanaka T., Gordon J.I., Stryer L., Ikura M. Molecular mechanics of calcium-myristoyl switches. Nature. 1997;389:198–202. doi: 10.1038/38310. [DOI] [PubMed] [Google Scholar]
- 39.O'Callaghan D.W., Tepikin A.V., Burgoyne R.D. Dynamics and calcium-sensitivity of the Ca2 +-myristoyl switch protein hippocalcin in living cells. J. Cell Biol. 2003;163:715–721. doi: 10.1083/jcb.200306042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Markova O., Fitzgerald D., Stepanyuk A., Dovgan A., Cherkas V., Tepikin A., Burgoyne R.D., Belan P. Hippocalcin signaling via site-specific translocation in hippocampal neurons. Neurosci. Lett. 2008;442:152–157. doi: 10.1016/j.neulet.2008.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dovgan A.V., Cherkas V.P., Stepanyuk A.R., Fitzgerald D.J., Haynes L.P., Tepikin A.V., Burgoyne R.D., Belan P.V. Decoding glutamate receptor activation by the Ca sensor protein hippocalcin in rat hippocampal neurons. Eur. J. Neurosci. 2010;32:347–358. doi: 10.1111/j.1460-9568.2010.07303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmer C.L., Lim W., Hastie P.G., Toward M., Korolchuk V.I., Burbidge S.A., Banting G., Collingridge G.L., Isaac J.T., Henley J.M. Hippocalcin functions as a calcium sensor in hippocampal LTD. Neuron. 2005;47:487–494. doi: 10.1016/j.neuron.2005.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jo J., Son G.H., Winters B.L., Kim M.J., Whitcomb D.J., Dickinson B.A., Lee Y.B., Futai K., Amici M., Sheng M., Collingridge G.L., Cho K. Muscarinic receptors induce LTD of NMDAR EPSCs via a mechanism involving hippocalcin, AP2 and PSD-95. Nat. Neurosci. 2010;13:1216–1224. doi: 10.1038/nn.2636. [DOI] [PubMed] [Google Scholar]
- 44.Tzingounis A.V., Kobayashi M., Takamatsu K., Nicoll R.A. Hippocalcin gates the calcium activation of the slow after hyperpolarization in hippocampal pyramidal cells. Neuron. 2007;53:487–493. doi: 10.1016/j.neuron.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spilker C., Dresbach T., Braunewell K.-H. Reversible translocation and activity-dependent localisation of the calcium-myristoyl switch protein VILIP-1 to different membrane compartments in living hippocampal neurons. J. Neurosci. 2002;22 doi: 10.1523/JNEUROSCI.22-17-07331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braunewell K.H., Klein-Szanto A.J. Visinin-like proteins (VSNLs): interaction partners and emerging functions in signal transduction of a subfamily of neuronal Ca2 +-sensor proteins. Cell Tissue Res. 2009;335:301–316. doi: 10.1007/s00441-008-0716-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.An W.F., Bowlby M.R., Bett M., Cao J., Ling H.P., Mendoza G., Hinson J.W., Mattsson K.I., Strassle B.W., Trimmer J.S., Rhodes K.J. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403:553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- 48.Hasdemir B., Fitzgerald D.J., Prior I.A., Tepikin A.V., Burgoyne R.D. Traffic of Kv4 K+ channels mediated by KChIP1 is via a novel post-ER vesicular pathway. J. Cell Biol. 2005;171:459–469. doi: 10.1083/jcb.200506005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rhodes K.J., Carroll K.I., Sung M.A., Doliveira L.C., Monaghan M.M., Burke S.L., Strassle B.W., Buchwalder L., Menegola M., Cao J., An F.W., Trimmer J.S. KChIPs and Kv4α subunits as integral components of A-type potassium channels in mammalian brain. J. Neurosci. 2004;24:7903–7915. doi: 10.1523/JNEUROSCI.0776-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dabrowska J., Rainnie D.G. Expression and distribution of Kv4 potassium channel subunits and potassium channel interacting proteins in subpopulations of interneurons in the basolateral amygdala. Neuroscience. 2010;171:721–733. doi: 10.1016/j.neuroscience.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buxbaum J.D., Choi E.K., Luo Y.X., Lilliehook C., Crowley A.C., Merriam D.E., Wasco W. Calsenilin: a calcium-binding protein that interacts with the presenilins and regulates the levels of a presenilin fragment. Nat. Med. 1998;4:1177–1181. doi: 10.1038/2673. [DOI] [PubMed] [Google Scholar]
- 52.Carrion A.M., Link W.A., Ledo F., Mellstrom B., Naranjo J.R. DREAM is a Ca2 +-regulated transcriptional repressor. Nature. 1999;398:80–84. doi: 10.1038/18044. [DOI] [PubMed] [Google Scholar]
- 53.Link W.A., Ledo F., Torres B., Palczewski M., Madsen T.M., Savignac M., Albar J.P., Mellstrom B., Naranjo J.R. Day-night changes in downstream regulatory element antagonist aodulator/potassium channel interacting protein activity contribute to circadian gene expression in pineal gland. J. Neurosci. 2004;24:5346–5355. doi: 10.1523/JNEUROSCI.1460-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venn N., Haynes L.P., Burgoyne R.D. Specific effects of KChIP3/calsenilin/DREAM but not KChIPs1, 2 and 4 on calcium signalling and regulated secretion in PC12 cells. Biochem. J. 2008;413:71–80. doi: 10.1042/BJ20080441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gierke P., Zhao C., Brackmann M., Linke B., Heinemann U., Braunewell K.-H. Expression analysis of members of the neuronal calcium sensor protein family: combining bioinformatics and Western Blot analysis. Biochem. Biophys. Res. Commun. 2004;323:38–43. doi: 10.1016/j.bbrc.2004.08.055. [DOI] [PubMed] [Google Scholar]
- 56.Dason J.S., Romero-Pozuelo J., Atwood H.L., Ferrus A. Multiple roles for frequenin/NCS-1 in synaptic function and development. Mol. Neurobiol. 2012;45:388–402. doi: 10.1007/s12035-012-8250-4. [DOI] [PubMed] [Google Scholar]
- 57.Mikhaylova M., Hradsky J., Kreutz M.R. Between promiscuity and specificity: novel roles of EF-hand calcium sensors in neuronal Ca2 + signalling. J. Neurochem. 2011;118:695–713. doi: 10.1111/j.1471-4159.2011.07372.x. [DOI] [PubMed] [Google Scholar]
- 58.Kerrigan T.L., Daniel J.W., Regan P.L., Cho K. The role of neuronal calcium sensors in balancing synaptic plasticity and synaptic dysfunction. Front. Mol. Neurosci. 2012;5:57. doi: 10.3389/fnmol.2012.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gomez M., De Castro E., Guarin E., Sasakura H., Kuhara A., Mori I., Bartfai T., Bargmann C.I., Nef P. Ca2 + signalling via the neuronal calcium sensor-1 regulates associative learning and memory in C. elegans. Neuron. 2001;30:241–248. doi: 10.1016/s0896-6273(01)00276-8. [DOI] [PubMed] [Google Scholar]
- 60.Martin V.M., Johnson J.R., Haynes L.P., Barclay J.W., Burgoyne R.D. Identification of key structural elements for neuronal calcium sensor-1 function in the regulation of the temperature-dependency of locomotion in C. elegans. Mol. Brain. 2013;6:39. doi: 10.1186/1756-6606-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang D., O'Halloran D., Goodman M.B. GCY-8, PDE-2, and NCS-1 are critical elements of the cGMP-dependent thermotransduction cascade in the AFD neurons responsible for C. elegans thermotaxis. J. Gen. Physiol. 2013;142:437–449. doi: 10.1085/jgp.201310959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blasiole B., Kabbani N., Boehmler W., Thisse B., Thisse C., Canfield V., Levenson R. Neuronal calcium sensor-1 gene ncs-1 is essential for semicircular canal formation in zebrafish inner ear. J. Neurobiol. 2005;64:285–297. doi: 10.1002/neu.20138. [DOI] [PubMed] [Google Scholar]
- 63.Pongs O., Lindemeier J., Zhu X.R., Theil T., Endelkamp D., Krah-Jentgens I., Lambrecht H.-G., Koch K.W., Schwemer J., Rivosecchi R., Mallart A., Galceran J., Canal I., Barbas J.A., Ferrus A. Frequenin—a novel calcium-binding protein that modulates synaptic efficacy in the drosophila nervous system. Neuron. 1993;11:15–28. doi: 10.1016/0896-6273(93)90267-u. [DOI] [PubMed] [Google Scholar]
- 64.Hendricks K.B., Wang B.Q., Schnieders E.A., Thorner J. Yeast homologue of neuronal frequenin is a regulator of phosphatidylinositol-4-OH kinase. Nat. Cell Biol. 1999;1:234–241. doi: 10.1038/12058. [DOI] [PubMed] [Google Scholar]
- 65.McFerran B.W., Graham M.E., Burgoyne R.D. NCS-1, the mammalian homologue of frequenin is expressed in chromaffin and PC12 cells and regulates neurosecretion from dense-core granules. J. Biol. Chem. 1998;273:22768–22772. doi: 10.1074/jbc.273.35.22768. [DOI] [PubMed] [Google Scholar]
- 66.Jo J., Heon S., Kim M.J., Son G.H., Park Y., Henley J.M., Weiss J.L., Sheng M., Collingridge G.L., Cho K. Metabotropic glutamate receptor-mediated LTD involves two interacting Ca2 + sensors, NCS-1 and PICK1. Neuron. 2008;60:1095–1111. doi: 10.1016/j.neuron.2008.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yip P.K., Wong L.F., Sears T.A., Yanez-Munoz R.J., McMahon S.B. Cortical overexpression of neuronal calcium sensor-1 induces functional plasticity in spinal cord following unilateral pyramidal tract injury in rat. PLoS Biol. 2010;8:e1000399. doi: 10.1371/journal.pbio.1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weiss J.L., Hui H., Burgoyne R.D. Neuronal calcium sensor-1 regulation of calcium channels, secretion, and neuronal outgrowth. Cell. Mol. Neurobiol. 2010;30:1283–1292. doi: 10.1007/s10571-010-9588-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Rezende V.B., Rosa D.V., Comim C.M., Magno L.A., Rodrigues A.L., Vidigal P., Jeromin A., Quevedo J., Romano-Silva M.A. NCS-1 deficiency causes anxiety and depressive-like behavior with impaired non-aversive memory in mice. Physiol. Behav. 2014;130:91–98. doi: 10.1016/j.physbeh.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 70.Saab B.J., Georgiou J., Nath A., Lee F.J., Wang M., Michalon A., Liu F., Mansuy I.M., Roder J.C. NCS-1 in the dentate gyrus promotes exploration, synaptic plasticity, and rapid acquisition of spatial memory. Neuron. 2009;63:643–656. doi: 10.1016/j.neuron.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 71.Haynes L.P., Fitzgerald D.J., Wareing B., O'Callaghan D.W., Morgan A., Burgoyne R.D. Analysis of the interacting partners of the neuronal calcium-binding proteins L-CaBP1, hippocalcin, NCS-1 and neurocalcin. Proteomics. 2006;6:1822–1832. doi: 10.1002/pmic.200500489. [DOI] [PubMed] [Google Scholar]
- 72.Schaad N.C., De Castro E., Nef S., Hegi S., Hinrichsen R., Martone M.E., Ellisman M.H., Sikkink R., Sygush J., Nef P. Direct modulation of calmodulin targets by the neuronal calcium sensor NCS-1. Proc. Natl. Acad. Sci. U. S. A. 1996;93:9253–9258. doi: 10.1073/pnas.93.17.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fitzgerald D.J., Burgoyne R.D., Haynes L.P. Neuronal calcium sensor proteins are unable to modulate NFAT activation in mammalian cells. Biochim. Biophys. Acta. 2008;1780:240–248. doi: 10.1016/j.bbagen.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kabbani N., Negyessy L., Lin R., Goldman-Rakic P., Levenson R. Interaction with the neuronal calcium sensor NCS-1 mediates desensitization of the D2 dopamine receptor. J. Neurosci. 2002;22:8476–8486. doi: 10.1523/JNEUROSCI.22-19-08476.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dagher A., Robbins T.W. Personality, addiction, dopamine: insights from Parkinson's disease. Neuron. 2009;61:502–510. doi: 10.1016/j.neuron.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 76.Koob G.F. The neurobiology of addiction: a neuroadaptational view relevant for diagnosis. Addiction. 2006;101(Suppl. 1):23–30. doi: 10.1111/j.1360-0443.2006.01586.x. [DOI] [PubMed] [Google Scholar]
- 77.Seeman P. Dopamine receptor sequences. Therapeutic levels of neuroleptics occupy D2 receptors, clozapine occupies D4. Neuropsychopharmacology. 1992;7:261–284. [PubMed] [Google Scholar]
- 78.Koh P.O., Undie A.S., Kabbani N., Levenson R., Goldman-Rakic P.S., Lidow M.S. Up-regulation of neuronal calcium sensor-1 (NCS-1) in the prefrontal cortex of schizophrenic and bipolar patients. Proc. Natl. Acad. Sci. U. S. A. 2003;100:313–317. doi: 10.1073/pnas.232693499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kabbani N., Levenson R. Antipsychotic-induced alterations in D2 dopamine receptor interacting proteins within the cortex. Neuroreport. 2006;17:299–301. doi: 10.1097/01.wnr.0000199460.24412.04. [DOI] [PubMed] [Google Scholar]
- 80.Multani P.K., Clarke T.K., Narasimhan S., Ambrose-Lanci L., Kampman K.M., Pettinati H.M., Oslin D.W., O'Brien C.P., Berrettini W.H., Lohoff F.W. Neuronal calcium sensor-1 and cocaine addiction: a genetic association study in African-Americans and European Americans. Neurosci. Lett. 2012;531:46–51. doi: 10.1016/j.neulet.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ritz M.C., Lamb R.J., Goldberg S.R., Kuhar M.J. Cocaine receptors on dopamine transporters are related to self-administration of cocaine. Science. 1987;237:1219–1223. doi: 10.1126/science.2820058. [DOI] [PubMed] [Google Scholar]
- 82.Dragicevic E., Poetschke C., Duda J., Schlaudraff F., Lammel S., Schiemann J., Fauler M., Hetzel A., Watanabe M., Lujan R., Malenka R.C., Striessnig J., Liss B. Cav1.3 channels control D2-autoreceptor responses via NCS-1 in substantia nigra dopamine neurons. Brain. 2014;137:2287–2302. doi: 10.1093/brain/awu131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lian L.Y., Pandalaneni S.R., Patel P., McCue H.V., Haynes L.P., Burgoyne R.D. Characterisation of the Interaction of the C-Terminus of the Dopamine D2 Receptor with Neuronal Calcium Sensor-1. PLoS One. 2011;6:e27779. doi: 10.1371/journal.pone.0027779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bahi N., Friocourt G., Carrié A., Graham M.E., Weiss J.L., Chafey P., Fauchereau F., Burgoyne R.D., Chelly J. IL1 receptor accessory protein like, a protein involved in X-linked mental retardation, interacts with Neuronal Calcium Sensor-1 and regulates exocytosis. Hum. Mol. Genet. 2003;12:1415–1425. doi: 10.1093/hmg/ddg147. [DOI] [PubMed] [Google Scholar]
- 85.Zhang Y.H., Huang B.L., Niakan K.K., McCabe L.L., McCabe E.R., Dipple K.M. IL1RAPL1 is associated with mental retardation in patients with complex glycerol kinase deficiency who have deletions extending telomeric of DAX1. Hum. Mutat. 2004;24:273. doi: 10.1002/humu.9269. [DOI] [PubMed] [Google Scholar]
- 86.Tabolacci E., Pomponi M.G., Pietrobono R., Terracciano A., Chiurazzi P., Neri G. A truncating mutation in the IL1RAPL1 gene is responsible for X-linked mental retardation in the MRX21 family. Am. J. Med. Genet. A. 2006;140:482–487. doi: 10.1002/ajmg.a.31107. [DOI] [PubMed] [Google Scholar]
- 87.Piton A., Michaud J.L., Peng H., Aradhya S., Gauthier J., Mottron L., Champagne N., Lafreniere R.G., Hamdan F.F., Joober R., Fombonne E., Marineau C., Cossette P., Dube M.P., Haghighi P., Drapeau P., Barker P.A., Carbonetto S., Rouleau G.A. Mutations in the calcium-related gene IL1RAPL1 are associated with autism. Hum. Mol. Genet. 2008;17:3965–3974. doi: 10.1093/hmg/ddn300. [DOI] [PubMed] [Google Scholar]
- 88.Handley M.T., Lian L.Y., Haynes L.P., Burgoyne R.D. Structural and functional deficits in a neuronal calcium sensor-1 mutant identified in a case of autistic spectrum disorder. PLoS One. 2010;5:e10534. doi: 10.1371/journal.pone.0010534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O'Callaghan D.W., Ivings L., Weiss J.L., Ashby M.C., Tepikin A.V., Burgoyne R.D. Differential use of myristoyl groups on neuronal calcium sensor proteins as a determinant of spatio-temporal aspects of Ca 2 +-signal transduction. J. Biol. Chem. 2002;277:14227–14237. doi: 10.1074/jbc.M111750200. [DOI] [PubMed] [Google Scholar]
- 90.O'Callaghan D.W., Hasdemir B., Leighton M., Burgoyne R.D. Residues within the myristoylation motif determine intracellular targeting of the neuronal Ca2 + sensor protein KChIP1 to post-ER transport vesicles and traffic of Kv4 K + channels. J. Cell Sci. 2003;116:4833–4845. doi: 10.1242/jcs.00803. [DOI] [PubMed] [Google Scholar]
- 91.Lim S., Strahl T., Thorner J., Ames J.B. Structure of a Ca2 +-myristoyl switch protein that controls activation of a phosphatidylinositol 4-kinase in fission yeast. J. Biol. Chem. 2011;286:12565–12577. doi: 10.1074/jbc.M110.208868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pioletti M., Findeisen F., Hura G.L., Minor D.L. Three-dimensional structure of the KChIP1–Kv4.3 T1 complex reveals a cross-shaped octamer. Nat. Struct. Mol. Biol. 2006;13:987–995. doi: 10.1038/nsmb1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang H., Yan Y., Liu Q., Huang Y., Shen Y., Chen L., Chen Y., Yang Q., Hao Q., Wang K., Chai J. Structural basis for modulation of Kv4 K+ channels by auxiliary KChIP subunits. Nat. Neurosci. 2007;10:32–39. doi: 10.1038/nn1822. [DOI] [PubMed] [Google Scholar]
- 94.Ames J.B., Levay K., Wingard J.N., Lusin J.D., Slepak V.Z. Structural basis for calcium-induced inhibition of rhodopsin kinase by recoverin. J. Biol. Chem. 2006;281:37237–37245. doi: 10.1074/jbc.M606913200. [DOI] [PubMed] [Google Scholar]
- 95.Zernii E., Komolov K., Permyakov S., Kolpakova T., Dell Orco D., Poetzsch A., Knyazeva E., Grigoriev I., Permyakov E., Senin I., Philippov P., Koch K.W. Involvement of recoverin C-terminal segment in recognition of the target enzyme rhodopsin kinase. Biochem. J. 2011;435:441–450. doi: 10.1042/BJ20110013. [DOI] [PubMed] [Google Scholar]
- 96.Heidarsson P.O., Bjerrum-Bohr I.J., Jensen G.A., Pongs O., Finn B.E., Poulsen F.M., Kragelund B.B. The C-terminal tail of human neuronal calcium sensor 1 regulates the conformational stability of the Ca2 +-activated state. J. Mol. Biol. 2012;417:51–64. doi: 10.1016/j.jmb.2011.12.049. [DOI] [PubMed] [Google Scholar]
- 97.Bourne Y., Dannenberg J., Pollmann V., Marchot P., Pongs O. Immunocytochemical localisation and crystal structure of human frequenin (neuronal calcium sensor 1) J. Biol. Chem. 2001;276:11949–11955. doi: 10.1074/jbc.M009373200. [DOI] [PubMed] [Google Scholar]
- 98.Strahl T., Huttner I.G., Lusin J.D., Osawa M., King D., Thorner J., Ames J.B. Structural insights into activation of phosphatidylinositol 4-kinase (pik1) by yeast frequenin (Frq1) J. Biol. Chem. 2007;282:30949–30959. doi: 10.1074/jbc.M705499200. [DOI] [PubMed] [Google Scholar]
- 99.Edwards M.R., Johnson J.R., Rankin K., Jenkins R.E., Maguire C., Morgan A., Burgoyne R.D., Barclay J.W. PKC-2 phosphorylation of UNC-18 Ser322 in AFD neurons regulates temperature dependency of locomotion. J. Neurosci. 2012;32:7042–7051. doi: 10.1523/JNEUROSCI.4029-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sanchez-Gracia A., Romero-Pozuelo J., Ferrus A. Two frequenins in Drosophila: unveiling the evolutionary history of an unusual neuronal calcium sensor (NCS) duplication. BMC Evol. Biol. 2010;10:54. doi: 10.1186/1471-2148-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Romero-Pozuelo J., Dason J.S., Atwood H.L., Ferrus A. Chronic and acute alterations in the functional levels of Frequenins 1 and 2 reveal their roles in synaptic transmission and axon terminal morphology. Eur. J. Neurosci. 2007;26:2428–2443. doi: 10.1111/j.1460-9568.2007.05877.x. [DOI] [PubMed] [Google Scholar]
- 102.Romero-Pozuelo J., Dason J.S., Mansilla A., Banos-Mateos S., Sardina J.L., Chaves-Sanjuan A., Jurado-Gomez J., Santana E., Atwood H.L., Hernandez-Hernandez A., Sanchez-Barrena M.J., Ferrus A. The guanine-exchange factor Ric8a binds the calcium sensor NCS-1 to regulate synapse number and probability of release. J. Cell Sci. 2014;127:4246–4259. doi: 10.1242/jcs.152603. [DOI] [PubMed] [Google Scholar]
- 103.McCue H.V., Haynes L.P., Burgoyne R.D. Bioinformatic analysis of CaBP/calneuron proteins reveals a family of highly conserved vertebrate Ca2 +- binding proteins. BMC Res. Notes. 2010;3:118. doi: 10.1186/1756-0500-3-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mikhaylova M., Sharma Y., Reissner C., Nagel F., Aravind P., Rajini B., Smalla K.H., Gundelfinger E.D., Kreutz M.R. Neuronal Ca2 + signaling via caldendrin and calneurons. Biochim. Biophys. Acta. 2006;1763:1229–1237. doi: 10.1016/j.bbamcr.2006.08.047. [DOI] [PubMed] [Google Scholar]
- 105.Yang J., McBride S., Mak D.-O.D., Vardi N., Palczewski K., Haeseleer F., Foskett J.K. Identification of a family of calcium sensors as protein ligands of inositol trisphosphate receptor Ca2 + release channels. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7711–7716. doi: 10.1073/pnas.102006299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Patel S., Morris S.A., Adkins C.E., O'Beirne G., Taylor C.W. Ca2 +-independent inhibition of inositol trisphosphate receptors by calmodulin: redistribution of calmodulin as a possible means of regulating Ca2 + mobilization. Proc. Natl. Acad. Sci. U. S. A. 1997;94:11627–11632. doi: 10.1073/pnas.94.21.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Haynes L.P., Tepikin A.V., Burgoyne R.D. Calcium binding protein 1 is an inhibitor of agonist-evoked, inositol 1,4,5-trisphophate-mediated calcium signalling. J. Biol. Chem. 2004;279:547–555. doi: 10.1074/jbc.M309617200. [DOI] [PubMed] [Google Scholar]
- 108.Kasri N.N., Holmes A.M., Bultynck G., Parys J.B., Bootman M.D., Rietdorf K., Missiaen L., McDonald F., Smedt H.D., Conway S.J., Holmes A.B., Berridge M.J., Roderick H.L. Regulation of InsP3 receptor activity by neuronal Ca2 +-binding proteins. EMBO J. 2004;23:1–10. doi: 10.1038/sj.emboj.7600037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Findeisen F., Minor D.L., Jr. Structural basis for the differential effects of CaBP1 and calmodulin on Ca(V)1.2 calcium-dependent inactivation. Structure. 2010;18:1617–1631. doi: 10.1016/j.str.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li C., Chan J., Haeseleer F., Mikoshiba K., Palczewski K., Ikura M., Ames J.B. Structural insights into Ca2 +-dependent regulation of inositol 1,4,5-trisphosphate receptors by CaBP1. J. Biol. Chem. 2009;284:2472–2481. doi: 10.1074/jbc.M806513200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Park S., Li C., Ames J.B. Nuclear magnetic resonance structure of calcium-binding protein 1 in a Ca2 +-bound closed state: implications for target recognition. Protein Sci. 2011;20:1356–1366. doi: 10.1002/pro.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li C., Enomoto M., Rossi A.M., Seo M.D., Rahman T., Stathopulos P.B., Taylor C.W., Ikura M., Ames J.B. CaBP1, a neuronal Ca2 + sensor protein, inhibits inositol trisphosphate receptors by clamping intersubunit interactions. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8507–8512. doi: 10.1073/pnas.1220847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu Y.Q., Lin X., Liu C.M., Jamrich M., Shaffer L.G. Identification of a human brain-specific gene, calneuron 1, a new member of the calmodulin superfamily. Mol. Genet. Metab. 2001;72:343–350. doi: 10.1006/mgme.2001.3160. [DOI] [PubMed] [Google Scholar]
- 114.McCue H.V., Burgoyne R.D., Haynes L.P. Membrane targeting of the EF-hand containing calcium-sensing proteins CaBP7 and CaBP8. Biochem. Biophys. Res. Commun. 2009;380:825–831. doi: 10.1016/j.bbrc.2009.01.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McCue H.V., Burgoyne R.D., Haynes L.P. Determination of the membrane topology of the small EF-hand Ca-sensing proteins CaBP7 and CaBP8. PLoS One. 2011;6:e17853. doi: 10.1371/journal.pone.0017853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hradsky J., Raghuram V., Reddy P.P., Navarro G., Hupe M., Casado V., McCormick P.J., Sharma Y., Kreutz M.R., Mikhaylova M. Post-translational membrane insertion of tail-anchored transmembrane EF-hand Ca2 + sensor calneurons requires the TRC40/Asna1 protein chaperone. J. Biol. Chem. 2011;286:36762–36776. doi: 10.1074/jbc.M111.280339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Haynes L.P., Sherwood M.W., Dolman N.J., Burgoyne R.D. Specificity, promiscuity and localization of ARF protein interactions with NCS-1 and phosphatidylinositol-4 kinase-IIIβ. Traffic. 2007;8:1080–1092. doi: 10.1111/j.1600-0854.2007.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mikhaylova M., Reddy P.P., Munsch T., Landgraf P., Suman S.K., Smalla K.H., Gundelfinger E.D., Sharma Y., Kreutz M.R. Calneurons provide a calcium threshold for trans-Golgi network to plasma membrane trafficking. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9093–9098. doi: 10.1073/pnas.0903001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McCue H.V., Patel P., Herbert A.P., Lian L.Y., Burgoyne R.D., Haynes L.P. Solution NMR structure of the Ca2 +-bound N-terminal domain of CaBP7: a regulator of Golgi trafficking. J. Biol. Chem. 2012;287:38231–38243. doi: 10.1074/jbc.M112.402289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang M., Li M., Wang J.H., Vogel H.J. The effect of Met– > Leu mutations on calmodulin's ability to activate cyclic nucleotide phosphodiesterase. J. Biol. Chem. 1994;269:15546–15552. [PubMed] [Google Scholar]
- 121.O'Neil K.T., DeGrado W.F. How calmodulin binds its targets: sequence independent recognition of amphiphilic alpha-helices. Trends Biochem. Sci. 1990;15:59–64. doi: 10.1016/0968-0004(90)90177-d. [DOI] [PubMed] [Google Scholar]
- 122.Catterall W.A., Few A.P. Calcium channel regulation and presynaptic plasticity. Neuron. 2008;59:882–901. doi: 10.1016/j.neuron.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 123.Dolphin A.C. A short history of voltage-gated calcium channels. Br. J. Pharmacol. 2006;147(Suppl. 1):S56–S62. doi: 10.1038/sj.bjp.0706442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Catterall W.A. Structure and regulation of voltage-gated Ca2 + channels. Ann. Rev. Cell. Dev. Biol. 2000;16:521–555. doi: 10.1146/annurev.cellbio.16.1.521. [DOI] [PubMed] [Google Scholar]
- 125.Tadross M.R., Tsien R.W., Yue D.T. Ca2 + channel nanodomains boost local Ca2 + amplitude. Proc. Natl. Acad. Sci. U. S. A. 2013;110:15794–15799. doi: 10.1073/pnas.1313898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Adams P.J., Rungta R.L., Garcia E., van den Maagdenberg A.M., MacVicar B.A., Snutch T.P. Contribution of calcium-dependent facilitation to synaptic plasticity revealed by migraine mutations in the P/Q-type calcium channel. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18694–18699. doi: 10.1073/pnas.1009500107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Leal K., Mochida S., Scheuer T., Catterall W.A. Fine-tuning synaptic plasticity by modulation of CaV2.1 channels with Ca2 + sensor proteins. Proc. Natl. Acad. Sci. U. S. A. 2012;109:17069–17074. doi: 10.1073/pnas.1215172109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Christel C., Lee A. Ca2 +-dependent modulation of voltage-gated Ca2 + channels. Biochim. Biophys. Acta. 2012;1820:1243–1252. doi: 10.1016/j.bbagen.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ben-Johny M., Yue D.T. Calmodulin regulation (calmodulation) of voltage-gated calcium channels. J. Gen. Physiol. 2014;143:679–692. doi: 10.1085/jgp.201311153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Peterson B.Z., DeMaria C.D., Yue D.T. Calmodulin is the Ca2 + sensor for Ca2 +-dependent inactivation of L-type calcium channels. Neuron. 1999;22:549–558. doi: 10.1016/s0896-6273(00)80709-6. [DOI] [PubMed] [Google Scholar]
- 131.Zuhlke R.D., Pitt G.S., Deisseroth K., Tsien R.Y., Reuter H. Calmodulin supports both inactivation and facilitation of L-type calcium channels. Nature. 1999;399:159–162. doi: 10.1038/20200. [DOI] [PubMed] [Google Scholar]
- 132.Erickson M.G., Alseikhan B.A., Peterson B.Z., Yue D.T. Preassociation of calmodulin with voltage-gated Ca2 + channels revealed by FRET in single living cells. Neuron. 2001;31:973–985. doi: 10.1016/s0896-6273(01)00438-x. [DOI] [PubMed] [Google Scholar]
- 133.Erickson M.G., Liang H., Mori M.X., Yue D.T. FRET two-hybrid mapping reveals function and location of L-type Ca2 + channel CaM preassociation. Neuron. 2003;39:97–107. doi: 10.1016/s0896-6273(03)00395-7. [DOI] [PubMed] [Google Scholar]
- 134.Pitt G.S., Zuhlke R.D., Hudmon A., Schulman H., Reuter H., Tsien R.W. Molecular basis of calmodulin tethering and Ca2 +-dependent inactivation of L-type Ca2 + channels. J. Biol. Chem. 2001;276:30794–30802. doi: 10.1074/jbc.M104959200. [DOI] [PubMed] [Google Scholar]
- 135.DeMaria C.D., Soong T.W., Alselkhan B.A., Alvania R.S., Yue D.T. Calmodulin bifurcates the local Ca2 + signal that modulates P/Q-type Ca2 + channels. Nature. 2001;411:484–489. doi: 10.1038/35078091. [DOI] [PubMed] [Google Scholar]
- 136.Lee A., Zhou H., Scheuer T., Catterall W.A. Molecular determinants of Ca2 +/calmodulin-dependent regulation of Ca(v)2.1 channels. Proc. Natl. Acad. Sci. U. S. A. 2003;100:16059–16064. doi: 10.1073/pnas.2237000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liang H., DeMaria C.D., Erickson M.G., Mori M.X., Alseikhan B.A., Yue D.T. Unified mechanisms of Ca2 + regulation across the Ca2 + channel family. Neuron. 2003;39:951–960. doi: 10.1016/s0896-6273(03)00560-9. [DOI] [PubMed] [Google Scholar]
- 138.Kim E.Y., Rumpf C.H., Fujiwara Y., Cooley E.S., Van Petegem F., Minor D.L., Jr. Structures of CaV2 Ca2 +/CaM-IQ domain complexes reveal binding modes that underlie calcium-dependent inactivation and facilitation. Structure. 2008;16:1455–1467. doi: 10.1016/j.str.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mori M.X., Vander Kooi C.W., Leahy D.J., Yue D.T. Crystal structure of the CaV2 IQ domain in complex with Ca2 +/calmodulin: high-resolution mechanistic implications for channel regulation by Ca2 + Structure. 2008;16:607–620. doi: 10.1016/j.str.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee A., Westenbroek R.E., Haeseleer F., Palczewski K., Scheuer T., Catterall W.A. Differential modulation of Cav2.1 channels by calmodulin and Ca2 +-binding protein 1. Nat. Neurosci. 2002;5:210–217. doi: 10.1038/nn805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Oz S., Benmocha A., Sasson Y., Sachyani D., Almagor L., Lee A., Hirsch J.A., Dascal N. Competitive and non-competitive regulation of calcium-dependent inactivation in CaV1.2 L-type Ca2 + channels by calmodulin and Ca2 +-binding protein 1. J. Biol. Chem. 2013;288:12680–12691. doi: 10.1074/jbc.M113.460949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Findeisen F., Rumpf C.H., Minor D.L., Jr. Apo States of Calmodulin and CaBP1 Control CaV1 Voltage-Gated Calcium Channel Function through Direct Competition for the IQ Domain. J. Mol. Biol. 2013;425:3217–3234. doi: 10.1016/j.jmb.2013.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lautermilch N.J., Few A.P., Scheuer T., Catterall W.A. Modulation of Cav2.1 channels by the neuronal calcium-binding protein visinin-like protein-2. J. Neurosci. 2005;25:7062–7070. doi: 10.1523/JNEUROSCI.0447-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nanou E., Martinez G.Q., Scheuer T., Catterall W.A. Molecular determinants of modulation of CaV2.1 channels by visinin-like protein 2. J. Biol. Chem. 2012;287:504–513. doi: 10.1074/jbc.M111.292581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tsujimoto T., Jeromin A., Satoh N., Roder J.C., Takahashi T. Neuronal calcium sensor 1 and activity-dependent facilitation of P/Q-type calcium channel currents at presynaptic nerve terminals. Science. 2002;295:2276–2279. doi: 10.1126/science.1068278. [DOI] [PubMed] [Google Scholar]
- 146.Weiss J.L., Archer D.A., Burgoyne R.D. NCS-1/frequenin functions in an autocrine pathway regulating Ca2 + channels in bovine adrenal chromaffin cells. J. Biol. Chem. 2000;275:40082–40087. doi: 10.1074/jbc.M008603200. [DOI] [PubMed] [Google Scholar]
- 147.Weiss J.L., Burgoyne R.D. Voltage-independent inhibition of P/Q-type Ca2 + channels in adrenal chromaffin cells via a neuronal Ca2 + sensor-1-dependent pathway involves Src-family tyrosine kinase. J. Biol. Chem. 2001;276:44804–44811. doi: 10.1074/jbc.M103262200. [DOI] [PubMed] [Google Scholar]
- 148.Dason J.S., Romero-Pozuelo J., Marin L., Iyengar B.G., Klose M.K., Ferrus A., Atwood H.L. Frequenin/NCS-1 and the Ca2 +-channel α-subunit co-regulate synaptic transmission and nerve-terminal growth. J. Cell Sci. 2009;122:4109–4121. doi: 10.1242/jcs.055095. [DOI] [PubMed] [Google Scholar]
- 149.Lian L.Y., Pandalaneni S.R., Todd P.A., Martin V.M., Burgoyne R.D., Haynes L.P. Demonstration of neuronal calcium sensor-1 binding to the Cav2.1 P/Q-type calcium channel. Biochemistry. 2014;53:6052–6062. doi: 10.1021/bi500568v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tippens A.L., Lee A. Caldendrin, a neuron-specific modulator of Cav/1.2 (L-type) Ca2 + channels. J. Biol. Chem. 2007;282:8464–8473. doi: 10.1074/jbc.M611384200. [DOI] [PubMed] [Google Scholar]
- 151.Zhou H., Kim S.-A., Kirk E.A., Tippens A.L., Sun H., Haeseleer F., Lee A. Ca2 +-binding protein-1 facilitates and forms a postsynaptic complex with Cav1.2 (L-type) Ca2 + channels. J. Neurosci. 2004;24:4698–4708. doi: 10.1523/JNEUROSCI.5523-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Reddy P.P., Raghuram V., Hradsky J., Spilker C., Chakraborty A., Sharma Y., Mikhaylova M., Kreutz M.R. Molecular dynamics of the neuronal EF-hand Ca2 +-sensor caldendrin. PLoS One. 2014;9:e103186. doi: 10.1371/journal.pone.0103186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kim K.Y., Scholl E.S., Liu X., Shepherd A., Haeseleer F., Lee A. Localization and expression of CaBP1/caldendrin in the mouse brain. Neuroscience. 2014;268:33–47. doi: 10.1016/j.neuroscience.2014.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rieke F., Lee A., Haeseleer F. Characterization of Ca2 +-binding protein 5 knockout mouse retina. Investig. Ophthalmol. Vis. Sci. 2008;49:5126–5135. doi: 10.1167/iovs.08-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yang P.S., Alseikhan B.A., Hiel H., Grant L., Mori M.X., Yang W., Fuchs P.A., Yue D.T. Switching of Ca2 +-dependent inactivation of Ca(V)1.3 channels by calcium binding proteins of auditory hair cells. J. Neurosci. 2006;26:10677–10689. doi: 10.1523/JNEUROSCI.3236-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yang P.S., Johny M.B., Yue D.T. Allostery in Ca2 + channel modulation by calcium-binding proteins. Nat. Chem. Biol. 2014;10:231–238. doi: 10.1038/nchembio.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cui G., Meyer A.C., Calin-Jageman I., Neef J., Haeseleer F., Moser T., Lee A. Ca2 +-binding proteins tune Ca2 +-feedback to Cav1.3 channels in mouse auditory hair cells. J. Physiol. 2007;585:791–803. doi: 10.1113/jphysiol.2007.142307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lee A., Jimenez A., Cui G., Haeseleer F. Phosphorylation of the Ca2 +-binding protein CaBP4 by protein kinase C zeta in photoreceptors. J. Neurosci. 2007;27:12743–12754. doi: 10.1523/JNEUROSCI.4264-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Haeseleer F., Imanishi Y., Maeda T., Possin D.E., Maeda A., Lee A., Rieke F., Palczewski K. Essential role of Ca2 +-binding protein 4, a Cav1.4 channel regulator in photoreceptor synaptic function. Nat. Neurosci. 2004;7:1079–1087. doi: 10.1038/nn1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shaltiel L., Paparizos C., Fenske S., Hassan S., Gruner C., Rotzer K., Biel M., Wahl-Schott C.A. Complex regulation of voltage-dependent activation and inactivation properties of retinal voltage-gated Cav1.4 L-type Ca2 + channels by Ca2 +-binding protein 4 (CaBP4) J. Biol. Chem. 2012;287:36312–36321. doi: 10.1074/jbc.M112.392811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mochida S., Few A.P., Scheuer T., Catterall W.A. Regulation of presynaptic Ca(V)2.1 channels by Ca2 + sensor proteins mediates short-term synaptic plasticity. Neuron. 2008;57:210–216. doi: 10.1016/j.neuron.2007.11.036. [DOI] [PubMed] [Google Scholar]
- 162.Catterall W.A., Leal K., Nanou E. Calcium channels and short-term synaptic plasticity. J. Biol. Chem. 2013;288:10742–10749. doi: 10.1074/jbc.R112.411645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhou H., Yu K., McCoy K.L., Lee A. Molecular mechanism for divergent regulation of Cav1.2 Ca2 + channels by calmodulin and Ca2 +-binding protein-1. J. Biol. Chem. 2005;280:29612–29619. doi: 10.1074/jbc.M504167200. [DOI] [PubMed] [Google Scholar]
- 164.Zeitz C., Kloeckener-Gruissem B., Forster U., Kohl S., Magyar I., Wissinger B., Matyas G., Borruat F.X., Schorderet D.F., Zrenner E., Munier F.L., Berger W. Mutations in CABP4, the gene encoding the Ca2 +-binding protein 4, cause autosomal recessive night blindness. Am. J. Hum. Genet. 2006;79:657–667. doi: 10.1086/508067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Littink K.W., van Genderen M.M., Collin R.W., Roosing S., de Brouwer A.P., Riemslag F.C., Venselaar H., Thiadens A.A., Hoyng C.B., Rohrschneider K., den Hollander A.I., Cremers F.P., van den Born L.I. A novel homozygous nonsense mutation in CABP4 causes congenital cone-rod synaptic disorder. Investig. Ophthalmol. Vis. Sci. 2009;50:2344–2350. doi: 10.1167/iovs.08-2553. [DOI] [PubMed] [Google Scholar]
- 166.Hoda J.C., Zaghetto F., Koschak A., Striessnig J. Congenital stationary night blindness type 2 mutations S229P, G369D, L1068P, and W1440X alter channel gating or functional expression of Ca(v)1.4 L-type Ca2 + channels. J. Neurosci. 2005;25:252–259. doi: 10.1523/JNEUROSCI.3054-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Schrauwen I., Helfmann S., Inagaki A., Predoehl F., Tabatabaiefar M.A., Picher M.M., Sommen M., Seco C.Z., Oostrik J., Kremer H., Dheedene A., Claes C., Fransen E., Chaleshtori M.H., Coucke P., Lee A., Moser T., Van Camp G. A Mutation in CABP2, expressed in cochlear hair cells, causes autosomal-recessive hearing impairment. Am. J. Hum. Genet. 2012;91:636–645. doi: 10.1016/j.ajhg.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Minor D.L., Jr., Findeisen F. Progress in the structural understanding of voltage-gated calcium channel (CaV) function and modulation. Channels (Austin) 2010;4:459–474. doi: 10.4161/chan.4.6.12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Van Petegem F., Chatelain F.C., Minor D.L., Jr. Insights into voltage-gated calcium channel regulation from the structure of the CaV1.2 IQ domain-Ca2 +/calmodulin complex. Nat. Struct. Mol. Biol. 2005;12:1108–1115. doi: 10.1038/nsmb1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fallon J.L., Halling D.B., Hamilton S.L., Quiocho F.A. Structure of calmodulin bound to the hydrophobic IQ domain of the cardiac Ca(v)1.2 calcium channel. Structure. 2005;13:1881–1886. doi: 10.1016/j.str.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 171.Ben Johny M., Yang P.S., Bazzazi H., Yue D.T. Dynamic switching of calmodulin interactions underlies Ca2 + regulation of CaV1.3 channels. Nat. Commun. 2013;4:1717. doi: 10.1038/ncomms2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang Q., Zhang P., Hoffman L., Tripathi S., Homouz D., Liu Y., Waxham M.N., Cheung M.S. Protein recognition and selection through conformational and mutually induced fit. Proc. Natl. Acad. Sci. U. S. A. 2013;110:20545–20550. doi: 10.1073/pnas.1312788110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Haynes L.P., Thomas G.M.H., Burgoyne R.D. Interaction of neuronal calcium sensor-1 and ARF1 allows bidirectional control of phosphatidylinositol 4-kinase beta and TGN-plasma membrane traffic. J. Biol. Chem. 2005;280:6047–6054. doi: 10.1074/jbc.M413090200. [DOI] [PubMed] [Google Scholar]
- 174.Sippy T., Cruz-Martin A., Jeromin A., Schweizer F.E. Acute changes in short-term plasticity at synapses with elevated levels of neuronal calcium sensor-1. Nat. Neurosci. 2003;6:1031–1038. doi: 10.1038/nn1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gambino F., Pavlowsky A., Begle A., Dupont J.L., Bahi N., Courjaret R., Gardette R., Hadjkacem H., Skala H., Poulain B., Chelly J., Vitale N., Humeau Y. IL1-receptor accessory protein-like 1 (IL1RAPL1), a protein involved in cognitive functions, regulates N-type Ca2 +-channel and neurite elongation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:9063–9068. doi: 10.1073/pnas.0701133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Navarro G., Hradsky J., Lluis C., Casado V., McCormick P.J., Kreutz M.R., Mikhaylova M. NCS-1 associates with adenosine A(2A) receptors and modulates receptor function. Front. Mol. Neurosci. 2012;5:53. doi: 10.3389/fnmol.2012.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schlecker C., Boehmerle W., Jeromin A., Degray B., Varshney A., Sharma Y., Szigeti-Buck K., Ehrlich B.E. Neuronal calcium sensor-1 enhancement of InsP(3) receptor activity is inhibited by therapeutic levels of lithium. J. Clin. Invest. 2006;116:1668–1674. doi: 10.1172/JCI22466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Nakamura T.Y., Jeromin A., Mikoshiba K., Wakabayashi S. Neuronal calcium sensor-1 promotes immature heart function and hypertrophy by enhancing Ca2 + signals. Circ. Res. 2011;109:512–523. doi: 10.1161/CIRCRESAHA.111.248864. [DOI] [PubMed] [Google Scholar]
- 179.Hui H., McHugh D., Hannan M., Zeng F., Xu S.Z., Khan S.U., Levenson R., Beech D.J., Weiss J.L. Calcium-sensing mechanism in TRPC5 channels contributing to retardation of neurite outgrowth. J. Physiol. 2006;572:165–172. doi: 10.1113/jphysiol.2005.102889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Chattopadhyaya R., Meador W.E., Means A.R., Quiocho F.A. Calmodulin structure refined at 1.7 A resolution. J. Mol. Biol. 1992;228:1177–1192. doi: 10.1016/0022-2836(92)90324-d. [DOI] [PubMed] [Google Scholar]