Abstract
Chaperones, nucleosome remodeling complexes, and histone acetyltransferases have been implicated in nucleosome disassembly at promoters of particular yeast genes, but whether these cofactors function ubiquitously, as well as the impact of nucleosome eviction on transcription genome-wide, is poorly understood. We used chromatin immunoprecipitation of histone H3 and RNA polymerase II (Pol II) in mutants lacking single or multiple cofactors to address these issues for about 200 genes belonging to the Gcn4 transcriptome, of which about 70 exhibit marked reductions in H3 promoter occupancy on induction by amino acid starvation. Examining four target genes in a panel of mutants indicated that SWI/SNF, Gcn5, the Hsp70 cochaperone Ydj1, and chromatin-associated factor Yta7 are required downstream from Gcn4 binding, whereas Asf1/Rtt109, Nap1, RSC, and H2AZ are dispensable for robust H3 eviction in otherwise wild-type cells. Using ChIP-seq to interrogate all 70 exemplar genes in single, double, and triple mutants implicated Gcn5, Snf2, and Ydj1 in H3 eviction at most, but not all, Gcn4 target promoters, with Gcn5 generally playing the greatest role and Ydj1 the least. Remarkably, these three cofactors cooperate similarly in H3 eviction at virtually all yeast promoters. Defective H3 eviction in cofactor mutants was coupled with reduced Pol II occupancies for the Gcn4 transcriptome and the most highly expressed uninduced genes, but the relative Pol II levels at most genes were unaffected or even elevated. These findings indicate that nucleosome eviction is crucial for robust transcription of highly expressed genes but that other steps in gene activation are more rate-limiting for most other yeast genes.
Nucleosome disassembly in promoters accompanies gene activation in the yeast Saccharomyces cerevisiae (Boeger et al. 2003; Reinke and Horz 2003) and is thought to enhance transcriptional activation by increasing access of general transcription factors (GTFs) and RNA polymerase II (Pol II) to the promoter and transcription start site (TSS) (Adkins et al. 2007; Zhang and Reese 2007). Genome-wide maps of nucleosome locations reveal a stereotypical pattern of nucleosome-depleted regions (NDRs) of ∼120 bp flanked by the −1 and +1 positioned nucleosomes, with the TSS lying within the +1 nucleosome. Promoter elements and upstream activator binding sites (UAS elements) reside within the NDR and can extend upstream into the −1 nucleosome (Jiang and Pugh 2009b; Wang et al. 2011; Cui et al. 2012; Rando and Winston 2012). The position of the TSS implies that the +1 nucleosome will be displaced or evicted at least transiently by Pol II. Similarly, binding of GTFs should be hindered by nucleosomes in the NDR, and activators will compete with nucleosomes in the NDR or −1 locations. The paucity of nucleosomes in the NDRs appears to be dictated by a combination of poly(dA:dT) tracts thought to destabilize nucleosomes, binding sites for abundant GTFs, and recruitment of chromatin modifying activities by gene-specific activators bound to UAS elements (Cairns 2009; Jiang and Pugh 2009b; Bai et al. 2011).
Nucleosome disassembly may occur by reversing the assembly process catalyzed in vitro by histone chaperones, such that removing H2A-H2B dimers would precede dissociation of the (H3/H4)2 tetrameric core from DNA (Elsasser and D'Arcy 2012). If so, chromatin immunoprecipitation (ChIP) analysis of H3 occupancy is well suited for monitoring eviction of nucleosome cores (henceforth described as nucleosome eviction). Other approaches that measure susceptibility of DNA to cleavage by nucleases may not distinguish between noncanonical nucleosomes, e.g., lacking an H2A-H2B dimer, and histone-free DNA. Importantly, ChIP analysis of H3 has demonstrated that the histone chaperone Asf1 promotes nucleosome eviction at activated PHO5 and PHO8 promoters without affecting UAS occupancy of the activator Pho4 (Adkins et al. 2004, 2007; Korber et al. 2006). Asf1 likely also contributes to eviction of H3/H4 at GAL genes following dissociation of H2A-H2B dimers (Schwabish and Struhl 2006). Yeast FACT complex (Spt16/Pob3) and Nhp6a/Nhp6b, often considered subunits of yFACT, are required for efficient eviction of both H2A-H2B and H3/H4 at PHO5 (Ransom et al. 2009) but could affect UAS binding by Pho4. Both Asf1 and yFACT are required for WT nucleosome eviction at URS2, but not at URS1, of the HO gene, although yFACT and the activator SBF are interdependent for WT URS2 occupancy (Takahata et al. 2009). Interestingly, the general chaperones Hsp90/Hsp70 are required for rapid loss of nucleosomes at GAL genes, without affecting activator Gal4 occupancy, and both proteins are recruited by Gal4 to the UAS (Floer et al. 2008). Thus, among the chaperones, Asf1 and Hsp90/Hsp70 most clearly function downstream from activator binding.
ATP-dependent chromatin remodelers have also been implicated in nucleosome remodeling or eviction (Elsasser and D'Arcy 2012). Eliminating Snf2, a catalytic subunit of SWI/SNF, reduces nucleosome remodeling at PHO8 (Gregory et al. 1999) and impairs remodeling and eviction at PHO5; however, binding of activator Pho4 to the PHO5 UAS is also reduced in snf2Δ cells (Adkins et al. 2007; Barbaric et al. 2007). Reduced Pho4 binding could also apply to overlapping functions described for SWI/SNF, Ino80, RSC, Isw1, and Chd1 in nucleosome remodeling at PHO5 (Musladin et al. 2014). SWI/SNF is needed for efficient nucleosome remodeling (Hirschhorn et al. 1992) and eviction at SUC2 (Schwabish and Struhl 2007), GAL1 (Bryant et al. 2008), HO (Gkikopoulos et al. 2009), RNR1 (Sharma et al. 2003), and CHA1 (Ansari et al. 2014), but whether it functions upstream of or downstream from activator binding is not always clear. SWI/SNF also contributes to nucleosome eviction at heat-shock–induced genes (Shivaswamy and Iyer 2008), but functional redundancy between SWI/SNF and Isw1 in histone eviction at heat-shock promoters involved effects on activator Hsf1 binding (Erkina et al. 2010). Depleting the essential catalytic subunit of RSC, Sth1, was found to increase nucleosome occupancy at a subset of 100–200 yeast promoters in the region of the +1 nucleosome (Parnell et al. 2008) or NDR (Hartley and Madhani 2009), implicating RSC in nucleosome eviction from promoters; however, the opposite conclusion emerged from a reduction in promoter H3 occupancies evoked by Sth1 depletion for the genes enriched for RSC in promoter regions (Spain et al. 2014).
Histone acetylation can promote nucleosome eviction by destabilizing nucleosome structure or promoting recruitment of chromatin remodelers. Hyperacetylated histones promote recruitment of SWI/SNF in vitro (Hassan et al. 2002), consistent with the requirement for both Gcn5, the HAT subunit in SAGA, and Snf2 for nucleosome remodeling at ADH2 (Verdone et al. 2002; Biddick et al. 2008) and PHO8 (Gregory et al. 1999). Gcn5 and Snf2 also promote remodeling at PHO5 under suboptimal induction conditions (Barbaric et al. 2007), although Pho4 binding at PHO5 is impaired in snf2Δ and gcn5Δ cells under certain conditions (Adkins et al. 2007). Interestingly, Gcn5 is required for nucleosome displacement at URS2 but not URS1 at HO, whereas SWI/SNF functions at both sites (Takahata et al. 2009). Acetylation of H3K56 by Rtt109/Asf1 also plays a role in nucleosome eviction, as the deletion of RTT109 or the substitution of H3K56 with nonacetylatable R56 mimics asf1Δ in delaying nucleosome eviction at PHO5 (Williams et al. 2008).
The full complement of chaperones, remodelers, and HATs that participate directly in nucleosome eviction at any yeast promoter is incompletely understood. It is unclear whether every cofactor implicated at one gene also contributes at a majority of other yeast genes. Moreover, the magnitude of changes in transcription that accompany reductions in nucleosome eviction in mutants has not been addressed on a genome-wide scale. To address these issues, we examined the Gcn4 transcriptome, composed of hundreds of genes, including most amino acid biosynthetic genes, that are coordinately induced by amino acid starvation, owing to rapid induction of activator Gcn4 (Hinnebusch 2005). Multiple cofactors have been implicated in modifying promoter nucleosomes and recruiting TATA-binding protein (TBP; Spt15 in yeast) and Pol II to particular Gcn4 target gene promoters (Berger et al. 1992; Georgakopoulos and Thireos 1992; Pina et al. 1993; Drysdale et al. 1998; Grant et al. 1998; Natarajan et al. 1998, 1999; Kuo et al. 2000; Swanson et al. 2003; Qiu et al. 2004; Govind et al. 2005). Induction of Gcn4 evokes strong eviction of H3 from the promoter of one such target gene, ARG1 (Govind et al. 2010), and genome-wide analysis of MNase-resistant nucleosomal DNA revealed extensive nucleosome loss in promoters and coding sequences (CDSs) of many Gcn4 targets (Cole et al. 2011). However, the cofactors required for nucleosome disassembly at ARG1 and other Gcn4 target genes are largely unknown.
Here, we examined a panel of mutants lacking one or more chaperones, remodelers, or HATs for defects in H3 eviction or Gcn4 binding at four canonical Gcn4 target genes and identified the Hsp70 cochaperone Ydj1, Snf2, and Gcn5 as cofactors with important roles in nucleosome disassembly downstream from Gcn4 binding at one or more of these genes. We then examined the consequences of eliminating one or more of these cofactors on nucleosome eviction and transcription for the entire Gcn4 transcriptome, as well as all other yeast genes. The results indicate extensive cooperation by Gcn5, Snf2, and Ydj1 in eviction of promoter nucleosomes genome-wide and in achieving robust transcription of the most highly expressed subset of genes in yeast.
Results
Functional cooperation of SWI/SNF, Gcn5, and Ydj1 at four Gcn4 target promoters
By use of ChIP assays, we showed previously that induction of Gcn4 by limiting cells for isoleucine and valine (Ile/Val) using the drug sulfometuron methyl (SM) evokes reduced H3 occupancy in the ARG1 promoter (Govind et al. 2005). We verified that the SM-induced reduction in H3 occupancy at ARG1 requires Gcn4 (Supplemental Fig. S1A,B) and demonstrated Gcn4-dependent reductions in H3 occupancy in the promoters of ARG4, HIS4, and CPA2 (Supplemental Fig. S1C–E). Subsequently, we conducted H3 ChIP analysis of these four genes in various mutants with defects in the catalytic subunits of remodeling complexes SWI/SNF (Snf2) or RSC (Sth1), Gcn5, cofactors required for H3K56 acetylation (Rtt109/Asf1), or histone chaperones Nap1 or yFACT (Spt16). We also examined mutant chaperones in the Hsp90 family (Hsp82/Hsc82), Hsp70 family (Ssa1/Ssa2), the Hsp40 cochaperone Ydj1 implicated in nucleosome eviction of GAL genes (Floer et al. 2008), and a mutant lacking chromatin-associated AAA-ATPase Yta7 (Gradolatto et al. 2008; Lombardi et al. 2011, 2015). Aiming to identify mutations that impair H3 eviction without reducing Gcn4 binding, we conducted Gcn4 ChIP analysis in the same mutants.
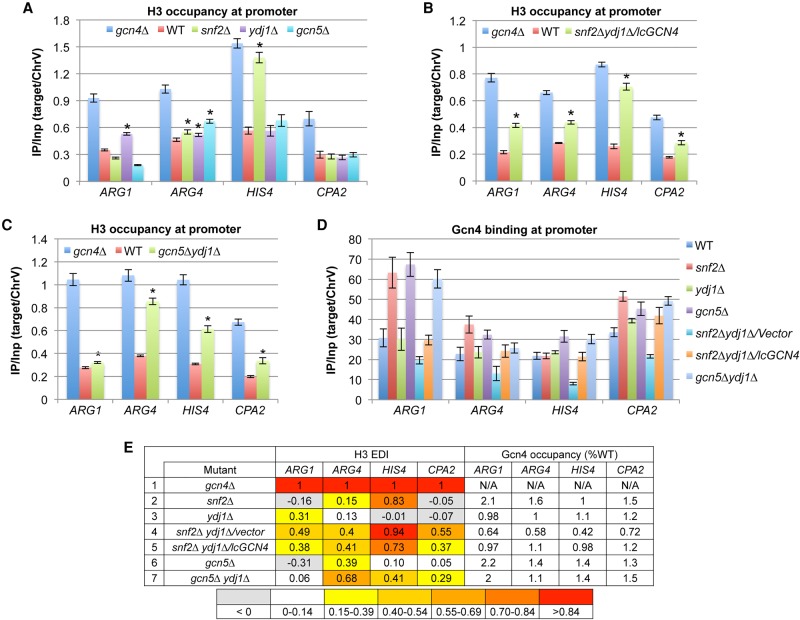
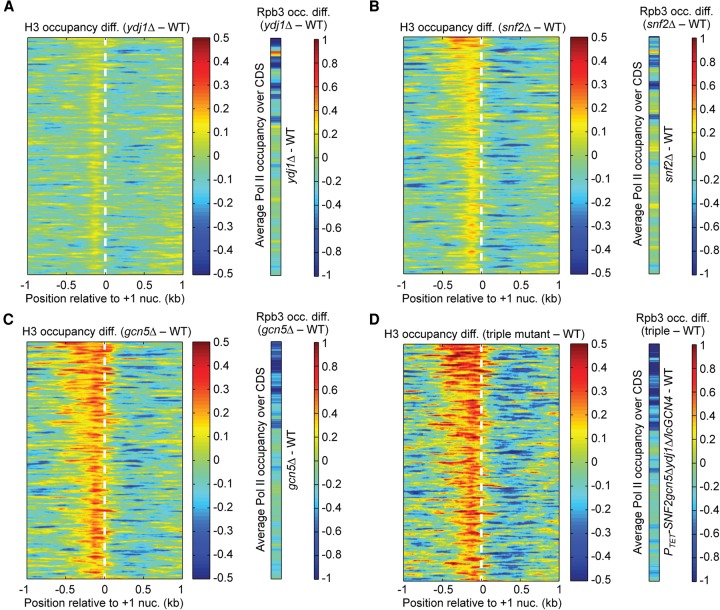
The snf2Δ strain displayed elevated H3 occupancy at HIS4 approaching that seen in gcn4Δ cells, but showed nearly WT H3 occupancies at the other genes (Fig. 1A). The ydj1Δ mutant had a less pronounced defect in H3 eviction, limited primarily to ARG1 (Fig. 1A). WT or greater Gcn4 occupancies occurred at all four genes in both mutants (Fig. 1D). Similar ChIP analyses of gcn5Δ cells revealed a significant defect in H3 eviction only at ARG4 (Fig. 1A), which was not attributable to reduced Gcn4 occupancy (Fig. 1D). Defects in H3 eviction conferred by snf2Δ and gcn5Δ were restricted to inducing conditions (Supplemental Fig. S2).
Figure 1.
Snf2, Gcn5, and Ydj1 cooperate in H3 eviction from promoters of four Gcn4 target genes. (A–D) Conventional ChIP analysis of H3 (A–C) or Gcn4 (D) under inducing conditions, as in Supplemental Figure S1. Mean (±SD) values from three or more biological replicates; Student's t-tests identify significant differences [(*) P < 0.01]. (E) Mean H3 eviction defect indices (EDIs) were calculated as in Supplemental Figure S3A from experiments summarized in Supplemental Figure S4A and color-coded, with all SEMs <20% of mean values. Gcn4 occupancies were determined from replicate experiments conducted as in D and summarized in Supplemental Figure S4B. Occupancies in each mutant were normalized to those in WT cells, and mean normalized occupancies were calculated from three or more replicates, with all SEMs <20% of mean values. (N/A) Not applicable. These data appear in Supplemental Figure S3A,B and are duplicated here for comparison.
The results of ChIP experiments for all single mutants are summarized in Supplemental Figure S3A, using an eviction defect index (EDI) that compares the magnitude of the defect in H3 eviction in each mutant to that observed in the gcn4Δ strain (with the maximum defect in Gcn4-dependent eviction) (Supplemental Fig. S3A). EDI values of zero and 1.0 indicate, respectively, low-level H3 occupancies seen in induced WT cells (WTI) and the high levels observed in gcn4Δ cells. Negative EDI values indicate H3 eviction more extensive than in WTI cells and likely signify defective nucleosome replacement versus eviction. Importantly, none of the mutants lacking only a single cofactor showed marked eviction defects at all four genes (fewer than four yellow to red cells in rows 2–17), and many displayed either no substantial eviction defect (all white cells) or apparent defects in nucleosome replacement (all grey cells) at multiple genes (Supplemental Fig. S3A). The ada1Δ strain, lacking a core subunit of SAGA, displays marked H3 eviction defects at ARG4 and HIS4 (Supplemental Fig. S3A, row 5), and as gcn5Δ strongly affected only ARG4 (row 6), a non-HAT function of SAGA impaired by ada1Δ is likely required at HIS4. The spt16-169 mutant, with an impaired yFACT complex, exhibits moderate H3 eviction defects at ARG1 and ARG4; however, the corresponding Gcn4 occupancies are substantially reduced at both genes (Supplemental Fig. S3A, row 9), precluding the conclusion that yFACT acts downstream from Gcn4 binding.
Eliminating Asf1 or Rtt109 did not impair H3 eviction and, generally, evoked lower H3 occupancies than in WT cells (Supplemental Fig. S3A, rows 7–8), which might reflect the known function of Asf1 in nucleosome reassembly (Das and Tyler 2012). Among mutations affecting HSP chaperones, only ydj1Δ and the ssa1Δssa2Δ double deletion conferred significant defects in H3 eviction with little effect on Gcn4 occupancy, which were limited to ARG1 (Supplemental Fig. S3A, rows 12–16). Deletion of HTZ1, encoding H2A variant H2A.Z, did not markedly alter H3 eviction, neither did inactivation of the RSC ATPase subunit in sth1-3 cells at the restrictive temperature nor elimination of the Med15/Gal11 subunit of Mediator, which reduces Mediator recruitment at ARG1 (Supplemental Fig. S3A; Zhang et al. 2004).
Our finding that H3 eviction was generally impaired at only a single gene in these mutants could indicate distinctive mechanisms of nucleosome eviction at each gene. Alternatively, the cofactors might function broadly, but their contributions are masked by other cofactors at particular genes. To explore this second possibility, we conducted ChIP analysis of various double mutants lacking Snf2, Gcn5, Yta7, or Ydj1, cofactors whose absence in single mutants impaired nucleosome eviction at one or more genes without reducing Gcn4 binding (Supplemental Fig. S3A). H3 eviction in the snf2Δydj1Δ double mutant is substantially defective at all four promoters; however, Gcn4 UAS occupancy was 30%–40% below WT at these genes (Fig. 1E, row 4). Importantly, after introducing additional copies of GCN4 on a low-copy (lc) plasmid, the resulting snf2Δydj1Δ/lcGCN4 strain displayed marked defects in H3 eviction at all four genes (Fig. 1B) with essentially WT Gcn4 binding (Fig. 1D,E). Comparing mean EDI values from multiple replicates confirmed that the snf2Δydj1Δ/lcGCN4 mutant exhibits H3 eviction defects at ARG4 and CPA2 that are significantly greater than those seen in the single mutants, in addition to defects at ARG1 and HIS4 evident in the single mutants (Supplemental Fig. S4A). Thus SWI/SNF and Ydj1 do not act exclusively at HIS4 and ARG1, respectively, and have overlapping functions at ARG4 and CPA2.
Analogous results were obtained for the gcn5Δydj1Δ double mutant, except that extra copies of GCN4 were not required for WT Gcn4 occupancies. In the double mutant, H3 eviction was markedly impaired at ARG4, HIS4, and CPA2 (Fig. 1C), whereas only ARG4 was affected in gcn5Δ cells (Fig. 1A). Moreover, the defect at ARG4 conferred by gcn5Δ was exacerbated by ydj1Δ in the double mutant (Fig. 1E, rows 6,7; Supplemental Fig. S4A). Thus, Gcn5 and Ydj1 have overlapping functions in H3 eviction at ARG4, HIS4, and CPA2. That H3 eviction at ARG1 is impaired in the ydj1Δ mutant but not in the gcn5Δydj1Δ strain can be explained by noting that gcn5Δ confers a defect in nucleosome replacement at ARG1 (negative EDI value) that likely compensates for the eviction defect conferred by ydj1Δ (Fig. 1E, rows 3,6,7).
To determine whether Snf2 and Gcn5 also have overlapping functions, we generated PTET-SNF2 strains, where Snf2 can be depleted by doxycycline (Dox) supplementation (Supplemental Fig. S5), as snf2Δgcn5Δ double mutants are lethal (Roberts and Winston 1997). The PTET-SNF2 single mutant in +Dox medium gave results similar to those described above for snf2Δ (Supplemental Fig. S3B, rows 2,7). The absence of Gcn5 in the PTET-SNF2gcn5Δ double mutant exacerbated the effect of depleting Snf2 at HIS4, producing an eviction defect comparable to that of gcn4Δ cells, and also confers eviction defects at ARG4 and CPA2 that are more severe than those seen in the gcn5Δ or PTET-SNF2 single mutants (Supplemental Figs. S3B, S4A). Comparing the PTET-SNF2gcn5Δydj1Δ triple mutant (also harboring lcGCN4) with the gcn5Δydj1Δ double mutant indicated that Snf2 also functions at ARG1 (Supplemental Figs. S3B, S4A). Thus, Snf2 and Gcn5 have overlapping or additive functions at these genes. While we sought to determine whether Yta7 and Asf1 also cooperate with Snf2 and Gcn5 in nucleosome eviction, the relevant double mutants show reduced Gcn4 occupancies that could not be corrected with extra copies of GCN4 (Supplemental Fig. S6A,B).
Identification of genes exhibiting marked eviction of promoter nucleosomes on SM induction
To extend our analysis to the entire Gcn4 transcriptome, we conducted genome-wide ChIP analysis of H3 and Pol II subunit Rpb3 (ChIP-seq) in WT cells to identify genes exhibiting appreciable eviction of H3 on SM induction. ChIP-seq results for Rpb3 and H3 were highly reproducible in biological replicates (Supplemental Fig. S7A–D). We observed the expected large increase in Rpb3 densities on induction of known Gcn4 targets (Supplemental Fig. S8A–D, upper panels), and identified 223 genes with increased Rpb3 occupancies averaged over the CDSs of greater than or equal to twofold on Gcn4 induction (Supplemental Fig. S9, above dashed diagonal; Supplemental File S1, “211 genes”), of which ∼80% were found by microarray expression analysis (Saint et al. 2014) to exhibit a comparable increase in mRNA abundance on SM treatment (P = 1 × 10−33) (Supplemental File S1, “WT+/−SM”).
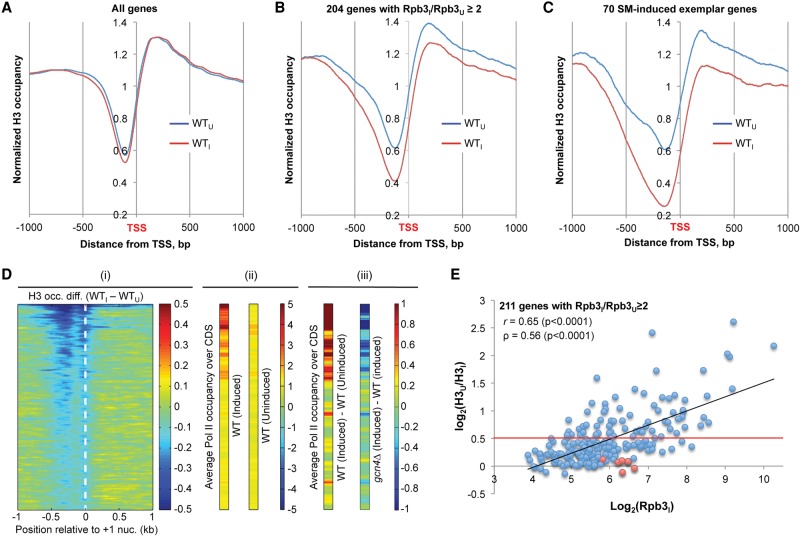
Metagene analysis of H3 occupancies aligned with the TSSs of all approximately 5800 genes (TSS alignment) (Fig. 2A) reveals the expected NDR just upstream of the TSS, as well as H3 occupancy peaks flanking the NDR at the expected positions of the −1 and +1 nucleosomes (Jiang and Pugh 2009a). The locations of additional nucleosomes known to exhibit positional phasing (+2, +3, +4, etc.) are not evident, owing to random fragmentation of chromatin in ChIP-seq. However, we also conducted paired-end sequencing of nucleosomal core particle DNA fragments generated by MNase digestion (Nuc-seq) under the same conditions, and observed the expected nucleosome phasing (Jiang and Pugh 2009a; Cole et al. 2011), with the TSS just inside the 5′ boundary of the +1 nucleosome (Supplemental Fig. S10A). The TSS shows essentially the same location in our H3 ChIP-seq analysis (Fig. 2A). Below, we employed ChIP-seq rather than Nuc-seq for analysis of H3 eviction primarily because the latter technique employs 30-min incubation in sorbitol during preparation of nuclei, during which Gcn4 promoter occupancy was found to decline (H Qiu and AG Hinnebusch, unpubl.).
Figure 2.
Eviction of H3 from promoters correlates with induction of Pol II occupancies at SM-induced genes. (A–C) Plots of average H3 occupancies at each base pair aligned at the TSS, calculated from ChIP-seq data combined from three biological replicates. The relative H3 occupancy at each base pair surrounding the TSS, normalized to the average occupancy on the respective chromosome, was averaged for the indicated gene sets, and the resulting values are plotted for the interval spanning 1000 bp 5′ to 1000 bp 3′ of the TSS. Blue indicates WTU; red, WTI. (D) Heat map depictions of changes in relative H3 and Rpb3 occupancies for 1000 genes with the largest reductions in H3 promoter occupancies on induction of WT cells. (i) Relative H3 occupancy differences between WTI and WTU cells in the region [−1000, +1000] around the +1 nucleosome dyad, sorted in descending order, and color-coded as shown on right. (ii) Relative Rpb3 occupancies, averaged over CDS, in WTI and WTU cells for the same gene order, color-coded as shown on right. (iii) Differences in relative Rpb3 occupancies between WTI and WTU cells (left) or between gcn4Δ and WT cells (right), color-coded as on far right. ChIP-seq data for H3 and Rpb3 were combined from three biological replicates. (E) Scatterplot of Rpb3 occupancies per base pair in CDS in WTI cells versus the ratio of average relative H3 occupancies in the [−1,NDR,+1] intervals of WTU versus WTI cells for 211 SM-induced genes, calculated from ChIP-seq data from three biological replicates. Pearson's (r) and Spearman's (ρ) correlation coefficients are indicated. Seventy-three genes above the red line exhibit a reduction in H3 occupancy of ≥30% on SM-induction. For genes in red exhibiting high-level Rbp3I with little or no H3 eviction (SNZ1, FMN1, PHM8, ARG8, HSP78, MCH1), nucleosome sliding to a new location in the promoter, or eviction of only H2A:H2B dimers, may suffice for PIC assembly.
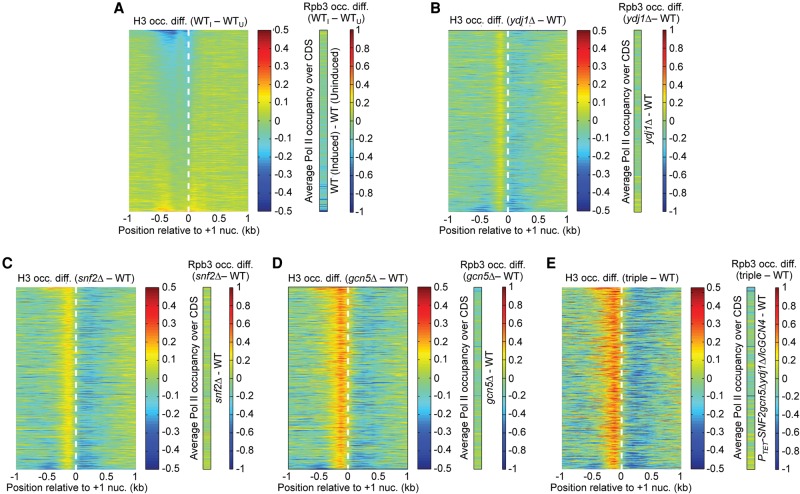
The TSS alignments for all genes were similar under SM-inducing (I) and -uninducing (U) conditions, with only slight reductions in H3 occupancies in the NDR and −1 regions (Fig. 2A). In contrast, alignments for a group of 204 genes with greater than or equal to twofold Pol II induction ratios on SM treatment (Rpb3I/Rpb3U ≥ 2) revealed markedly lower H3 in promoters and CDSs (Fig. 2B), indicating that marked nucleosome depletion occurs primarily at genes transcriptionally induced by SM. (Only 211 of the 223 SM-induced genes identified in Supplemental Fig. S9 contain the annotation required for analyses of promoter H3 occupancies described below, and seven of these genes were excluded from the set of 204 genes analyzed in Fig. 2B because they share a divergent promoter with another of the genes.) The expected correlation between transcription and H3 eviction is illustrated by a heat map depiction of changes in H3 occupancies for 1000 genes exhibiting the greatest nucleosome eviction on SM induction, aligned relative to +1 nucleosome dyads (determined by Nuc-seq data), and sorted on the extent of H3 eviction (Fig. 2D, i). H3 eviction extends upstream of the +1 nucleosome into the NDR and −1 regions for genes at the top of the map with the greatest H3 depletion. Identically sorted maps depicting average Rpb3 occupancies in the CDS of uninduced versus induced cells show that most genes exhibiting appreciable SM induction of Rpb3 are clustered near the top and thus display strong eviction of promoter nucleosomes (Fig. 2D, ii, iii, left). ChIP-seq analysis of Rpb3 in a gcn4Δ mutant revealed that a large fraction of SM-induced genes require Gcn4 for robust induction of Pol II (Fig. 2D, iii, middle).
To quantify the extent of H3 eviction in the −1, NDR, and +1 zones of each gene, we used Nuc-seq data to identify coordinates of the 147 bp surrounding peak positions of the −1 and +1 dyads and intervening NDR at each gene (Supplemental Fig. S10B). We determined the average relative H3 occupancies per nucleotide (Avg. H3 Occ.) across the [−1, NDR, +1] interval for each gene and calculated the ratio of occupancies in uninducing versus inducing conditions (H3U/H3I). Importantly, for the 211 highly induced genes, the H3U/H3I ratio was significantly correlated with both the induced Rpb3 level (Rpb3I) (Fig. 2E) and Rpb3 induction ratio (Rpb3I/Rpb3U) (Supplemental Fig. S11A). A similar correlation was observed for all genes (Supplemental Fig. S11B). Among the highly induced genes, only 70 exhibit reduced H3 occupancies of ≥30% of the uninduced value (above red line in Fig. 2E; Supplemental File S1 “70 genes”), and Figure 2C depicts the profile of H3 eviction for this subset of 70 exemplar induced genes. The correlation between Rpb3 induction and reduction of promoter H3 occupancy on SM treatment (Fig. 2E) supports the idea that eviction of promoter nucleosomes is rate-limiting for transcriptional induction at many genes.
Gcn5, SWI/SNF, and Ydj1 function broadly in the Gcn4 transcriptome
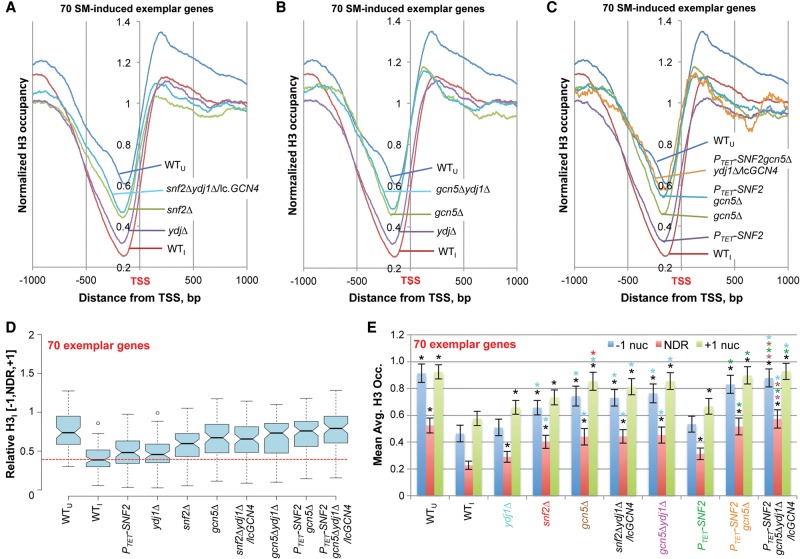
We examined the effects of eliminating Snf2 in cells containing or lacking Ydj1 on H3 eviction at the 70 exemplar genes by H3 ChIP-seq analysis of replicate cultures of SM-treated snf2Δ, ydj1Δ, and snf2Δydj1Δ/lcGCN4 mutants. As a group, the 70 genes displayed substantial defects in H3 eviction in snf2Δ cells, stronger than in the ydj1Δ mutant and nearly as great as seen in the snf2Δydj1Δ/lcGCN4 double mutant (Fig. 3A,D, ydj1Δ, snf2Δ, and snf2Δydj1Δ/lcGCN4 vs. WTI). In all three mutants, the largest increases in mean H3 occupancy occurred in the NDR, approximately twofold in the double mutant (Fig. 3E; tabulated in Supplemental File S2, sheet 1). That H3 occupancies in the double mutant remained lower than in WTU cells (Fig. 3A,D,E) implies that only a portion of SM-induced nucleosome eviction at these 70 genes requires Ydj1 and Snf2.
Figure 3.
Snf2, Ydj1, and Gcn5 cooperate in H3 eviction from promoters of 70 exemplar induced genes. (A–C) TSS plots of average relative H3 occupancies calculated as in Figure 2A–C, except for only the 70 exemplar genes, for WTU, WTI, and the indicated mutants under inducing conditions. (D) Notched-box plots of average relative H3 occupancies in the [−1,NDR,+1] region for the 70 exemplar genes calculated from ChIP-seq data from three replicates for each mutant. Each box depicts the interquartile range containing 50% of the data, intersected by the median; the notch indicates a 95% confidence interval (CI) around the median. (E) Mean Avg. H3 Occ. per base pair (±95% CI) calculated for the 70 exemplar genes for the three promoter zones using the relative H3 occupancies determined by ChIP-seq analysis of three replicates. Asterisks indicating significant differences in Student's t-tests (P < 0.05) are color-coded to indicate strains being compared, e.g., red asterisk above the gcn5Δ +1 nuc bar indicates a mean significantly different than the corresponding +1 nuc mean for the snf2Δ strain (labeled red).
H3 ChIP-seq of mutants containing gcn5Δ revealed that eliminating Gcn5 alone evokes a marked H3 eviction defect slightly greater than in snf2Δ cells, and combining gcn5Δ with ydj1Δ produced an insignificant further increase in H3 occupancy for the group of 70 genes (Fig. 3B,D,E, ydj1Δ, gcn5Δ, and gcn5Δydj1Δ vs. WTI). However, the H3 eviction defect in the PTET-SNF2gcn5Δydj1Δ/lcGCN4 triple mutant is significantly larger than seen in gcn5Δ cells, indicating cooperation between Gcn5, Snf2, and Ydj1 at the exemplar genes (Fig. 3C–E). Essentially identical conclusions were reached by analyzing the larger group of 204 SM-induced genes (Supplemental Fig. S12). The fact that the triple mutation increases the mean H3 occupancies in the −1, NDR, and +1 zones to the level observed in WTU cells (Fig. 3C–E; Supplemental Fig. S12) indicates that Ydj1, Snf2, and Gcn5 mediate a large measure of the nucleosome eviction from promoters at SM-induced genes. Interestingly, the mutations do not increase H3 occupancies in the adjacent CDS relative to that seen in WTI cells.
Gene-specific patterns of cooperation among SWI/SNF, Gcn5, and Ydj1 in H3 eviction
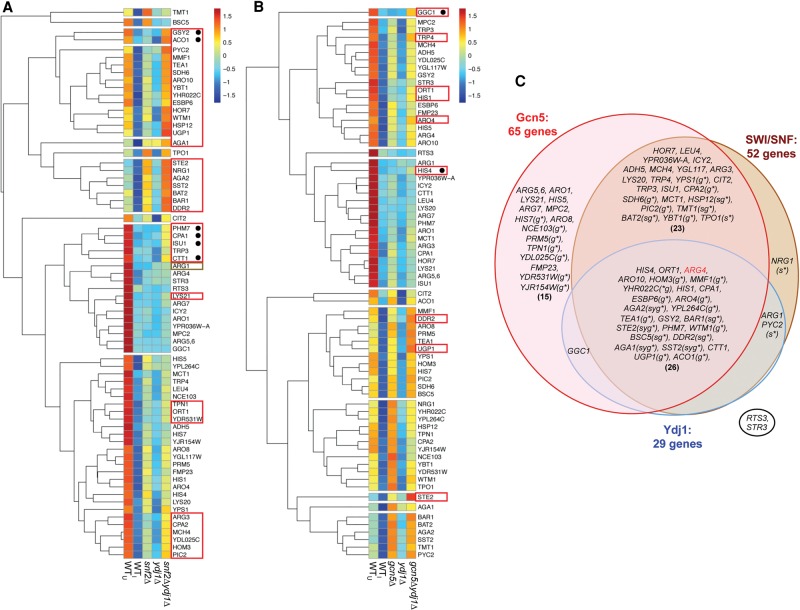
To examine the effects of snf2Δ, ydj1Δ, and gcn5Δ on H3 eviction at each of the 70 genes individually, the Avg. H3 Occ. for the [−1,NDR,+1] window was calculated from three biological replicates, averaged, and mean values compared between SM-treated mutant and WTI cells. Results for the snf2Δ, ydj1Δ, and snf2Δydj1Δ/lcGCN4 mutants are presented in Supplemental Figure S13, A–E (with significant changes summarized in Supplemental File S3, sheet 1) and analyzed by hierarchical clustering in Figure 4A. Cluster analysis reveals that a majority of genes exhibit higher H3 levels in snf2Δ versus WT cells, but the magnitude of the increases varies considerably at different genes, and statistically significant changes were observed for only 25 of 70 genes (P < 0.05) (Supplemental File S3, sheet 1, col. 3). Relatively few genes exhibit pronounced increases in H3 levels in ydj1Δ cells (Fig. 4A), and only 18 genes display significant increases compared with the WT (P < 0.05) (Supplemental File S3, sheet 1, col. 4). In fact, ARG1 is the only gene with a significantly greater eviction defect in ydj1Δ versus snf2Δ cells (Fig. 4A, gold box). Numerous genes show stronger eviction defects in the snf2Δydj1Δ/lcGCN4 double mutant versus either single mutant (Fig. 4A, red boxes), and consistent with this, a large set of 52 genes showed significantly increased H3 levels in this double mutant compared with WT cells (P < 0.05) (Supplemental File S3, sheet 1, col. 5). Thus, Snf2 and Ydj1 cooperate in H3 eviction at a sizeable fraction of the exemplar genes. Their functions appear to be highly redundant in a subset of genes (black bullets in Fig. 4A) where a marked defect occurred only in the double mutant, including CPA1 (Supplemental Fig. S13B), GSY2, ISU1, PHM7 (Supplemental Fig. S13C), CTT1, and ACO1 (Supplemental Fig. S13E). Genes ARO10 (Supplemental Fig. S13A) and MMF1 (Supplemental Fig. S13B) provide contrasting examples of additive contributions by Ydj1 and Snf2, as both single mutations confer lesser eviction defects than seen in the double mutant.
Figure 4.
Snf2, Gcn5, and Ydj1 cooperate in promoter H3 eviction at many of the 70 exemplar induced genes. (A,B) Hierarchical clustering analysis of the average relative H3 occupancies in the [−1, NDR, +1] regions was performed in R using the “pheatmap” function, with the following parameters: scale = “row,” clustering_method = “average,” clustering_distance_rows = “euclidean,” cluster_rows = TRUE, cluster_cols = FALSE, cutree_row = 10. The avg. H3 occ. values for each gene given in Supplemental Figures S13 and S14 were transformed to z-scores. Boxed genes display additive effects of mutations; those with bullets show strong eviction defects only in double mutants. (C) Venn diagram depicting involvement of Snf2, Gcn5, or Ydj1 at the 70 exemplar genes deduced from effects of single, double, or triple mutations eliminating or depleting these cofactors on H3 promoter occupancies in SM-treated cells, quantified in Supplemental Figures S13–S15 and summarized in Supplemental Figure S18. (g*, s*, or y*) Strong defect in H3 eviction in gcn5Δ, snf2Δ, or ydj1Δ single mutants, respectively, with H3 occupancies similar to those in WTU cells. As described in Supplemental Results, there was generally good agreement between conclusions reached from ChIP-seq (Supplemental Figs. S13–S16) and conventional ChIP (Supplemental Fig. S3B) for ARG1, ARG4, HIS4, and CPA2. However, the lower variance of conventional ChIP data for ARG4 (Supplemental Fig. S4A) led us to place it among the group of genes employing all three cofactors.
H3 promoter occupancies measured in the gcn5Δ, ydj1Δ, and gcn5Δydj1Δ mutants are presented in Supplemental Figure S14, A–E, and Supplemental File S3, sheet 2, and analyzed by clustering in Figure 4B. As observed for snf2Δ, the majority of genes exhibit increased H3 occupancies in the gcn5Δ mutant, covering a broad range of eviction defects (Fig. 4B). As gcn5Δ has comparatively stronger effects than snf2Δ, there are relatively fewer genes where cooperation between Gcn5 and Ydj1 is revealed by a marked exacerbation of eviction defects in the double mutant (boxed red in Fig. 4B). STE2 (Supplemental Fig. S14C) represents a clear case where defects in the single mutants appear to be additive in the double mutant, whereas GGC1 (Supplemental Fig. S14E) and HIS4 (Supplemental Fig. S14A) provide examples of pronounced functional redundancy. Similar analyses of the gcn5Δ, PTET-SNF2, and PTET-SNF2gcn5Δ strains (Supplemental Fig. S15A–E) uncovered a sizeable group of genes where the eviction defect conferred by gcn5Δ was exacerbated by combining it with PTET-SNF2 or both PTET-SNF2 and ydj1Δ (red boxes in clustergrams of Supplemental Fig. S16A,B), consistent with functional cooperation of Gcn5 with Snf2 and Ydj1.
The gene-specific nature of dependencies on Snf2 and Gcn5 is illustrated by evaluating the proportions of exemplar genes displaying different H3 eviction defects (Supplemental Fig. S17). In both gcn5Δ and snf2Δ cells, the 70 exemplars exhibit a broad distribution of EDI values ranging from almost none (EDI = 0.0–0.1) to apparent loss of H3 eviction (EDI ≥ 1.0), while in ydj1Δ cells, the 70 genes exhibit a narrower distribution of less severe eviction defects.
Evaluation of statistically significant increases in H3 occupancy in the gcn5Δ and gcn5Δydj1Δ strains (Supplemental File S3, sheet 2) identified 42 genes where Gcn5 appeared to function alone and 18 genes where Gcn5 and Ydj1 cooperate in H3 eviction (Supplemental File S3, sheet 2, cols. 8,9). Comparing results for the PTET-SNF2, gcn5Δ, and PTET-SNF2gcn5Δ strains increased the Gcn5-dependent genes from 60 to 65 by uncovering defects conferred by gcn5Δ in the PTET-SNF2 background and, likewise, increased the number of Snf2-dependent genes from 44 to 51. Similarly compiling effects of ydj1Δ in the two double mutants containing snf2Δ or gcn5Δ, as well as in the triple mutant, revealed 29 genes clearly dependent on Ydj1 (Supplemental Fig. S18). The dependencies of all 70 genes deduced from combining results from all of the mutants are summarized in Figure 4C. This analysis provides a minimal estimate, as contributions of Gcn5, Snf2, or Ydj1 might be too small, or the variance in data too large, to reveal a significant defect in H3 occupancy at particular genes in a given mutant. It is instructive, however, that 93% and 74% of the exemplar genes were scored as dependent on Gcn5 or Snf2, respectively, indicating a broad role for both cofactors in the Gcn4 transcriptome.
Defects in H3 eviction are associated with reduced transcription of induced genes
We analyzed the consequences of impaired H3 eviction on transcription by ChIP-seq analysis of Rpb3 on the same biological replicates of the mutants analyzed above (Supplemental File S4). Heat maps of changes in H3 and Rpb3 occupancies in different mutants are depicted in Figure 5, A–D, and Supplemental Figure S19, A–C, for the 1000 genes with the greatest nucleosome eviction in induced WT cells. As noted in Figure 2D for this gene set, those with the greatest Rpb3 induction in WT cells are clustered near the top of each map. Consistent with the results described above, among single mutants, ydj1Δ has the smallest effect, while gcn5Δ confers the greatest increases in H3 occupancy compared with WTI cells (Fig. 5A–C). Combining ydj1Δ and snf2Δ or combining gcn5Δ and PTET-SNF2 in double mutants (Supplemental Fig. S19A,C) evokes greater increases in H3 occupancies versus the corresponding single mutants (Fig. 5A–C), and the triple mutation confers the largest increases, extending across the entire promoter regions of highly induced genes at the top of the map (Fig. 5D). As discussed below, genes in the lower portions of the maps also show increased H3 occupancies that are largely restricted to the NDRs, and many genes throughout the maps exhibit decreased H3 levels in the adjacent CDS in cofactor mutants (Fig. 5A–D). Examining Rpb3 difference maps for each of the mutants shows that a substantial fraction of the highly induced genes at the top of each map exhibit decreased Rpb3 occupancies, with the gcn5Δ, double and triple mutants displaying the most pervasive reductions (Fig. 5A–D; Supplemental S19A–C, maps at far right). Thus, increases in promoter H3 occupancies in the mutants are associated with decreased Pol II levels at many induced genes.
Figure 5.
Reductions in Pol II occupancy are associated with increased H3 promoter occupancy for 1000 genes with the greatest eviction of promoter nucleosomes in WT induced cells. (A–D) Heat maps for the identically ordered 1000 genes shown in Figure 2D, constructed as described there, for the indicated mutants.
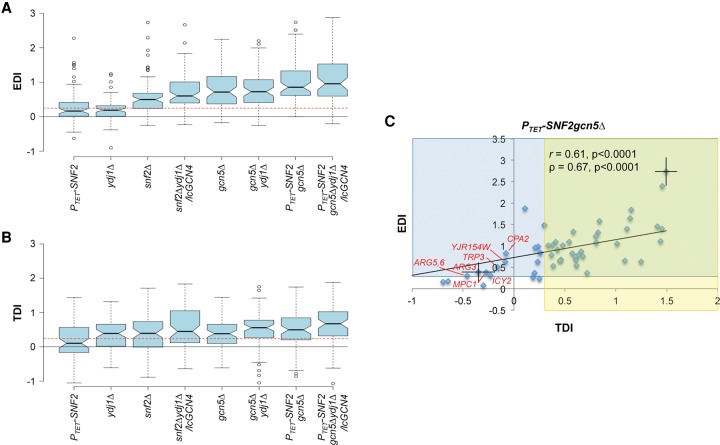
This relationship was examined in greater detail for the 70 exemplar genes by comparing their EDI values to a comparable measure of the transcription defect, the TDI, which expresses the reduction in Rpb3 occupancy in an SM-induced mutant as a proportion of the total Rpb3 induction seen in WT cells. Consistent with findings above, the EDI values are generally lower for single mutants, higher for double mutants, and greatest for the triple mutant (Fig. 6A; Supplemental File S5, sheet 2 for EDI and TDI values). A similar trend was observed for TDIs, but of a smaller degree (Fig. 6B; Supplemental File S5, sheet 2 for EDI and TDI values), and the mean TDI and mean EDI values for the group of 70 genes are highly correlated among different strains (Supplemental Fig. S20A). Thus, progressive impairment of histone eviction is associated with increasing reductions in transcription in the eight mutants for the group of 70 exemplar genes.
Figure 6.
Defects in promoter H3 eviction are associated with defects in Pol II induction for 70 induced exemplar genes. (A,B) Notched-box plots of EDI and TDI values for the 70 exemplar genes in eight mutants (tabulated in Supplemental File S5, sheet 2). EDIs were calculated for each gene from H3 ChIP-seq data from three replicates as (H3I(mut) − H3I(WT))/(H3U(WT) − H3I(WT)), where H3I(mut) and H3U(WT) signify the Avg. H3 Occs. per base pair in the [−1,NDR,+1] regions in induced mutant or WTU cells, respectively. TDIs were calculated similarly for each gene as (RI(WT) − RI(mut))/(RI(WT) − RU(WT)), where R designates average Rpb3 occupancy per base pair in CDS. (C) EDI versus TDI scatterplot for 54 of the group of 70 exemplar genes in PTET-SNF2gcn5Δ cells. Mean EDI and TDI values (±SEM) were calculated for each gene by averaging the EDI or TDI values calculated from H3 or Rpb3 ChIP-seq data from each of three biological replicates (tabulated in Supplemental File S5, sheet 1), and genes with both SEM values <0.25 of the mean EDI and with SEM values <0.125 of the mean TDI values (thus excluding genes CIT2, ACO1, SDH6, LYS21, LYS20, YDR531W, ESBP6, BSC5, UGP1, YBT1, CTT1, HOR7, BAR1, STE2, BAT2, and NRG1) were subjected to correlation analyses to obtain the indicated Pearson's and Spearman's coefficients and P-values. Error bars (±SD) shown for two genes (TPO1 and ARG3) are the largest observed for any of the 54 genes. Yellow and blue zones indicate TDI or EDI values ≥0.3, respectively, and exceptional genes with EDI > 0.3/TDI < 0 are labeled red. Data for RTS3 were not plotted because its large negative EDI value would confine the majority of data points to a small portion of the x-axis.
This conclusion was supported by analyzing EDI-TDI relationships for individual genes. In correlations of EDIs with TDIs for each gene across eight different mutants, the Pearson's coefficient (r) was ≥0.45 for more than two-thirds of the 70 genes (Supplemental Fig. S20B). Moreover, the EDIs and TDIs for the majority of exemplar genes are highly correlated in each of the different mutants, as shown for the PTET-SNF2gcn5Δ strain in Figure 6C (Supplemental File S5, sheet 1 for EDI and TDI values) and for the other mutants in Supplemental Figure S21A–F. Deviations from the trendline observed for certain genes in Figure 6C do not arise from experimental error, as indicated by errors bars for the two genes with the largest standard deviations in the EDI or TDI values calculated from replicate measurements. Despite the variance in EDI-TDI association, two-thirds of plotted genes with EDIs ≥ 0.3 (≥30% reduction in H3 eviction relative to WTI cells) exhibit an equally strong transcription defect, with TDI ≥ 0.3 (Fig. 6C; intersection of blue and yellow sectors). That the increased H3 occupancy is associated with decreased transcription in mutant cells for most exemplar genes suggests that Gcn5, SWI/SNF, and Ydj1 generally cooperate to stimulate transcription at least partly by their roles in evicting promoter nucleosomes.
Thirteen genes exhibit appreciable H3 eviction defects but little transcriptional defect (Fig. 6C, in blue sector only), and seven (red labels) actually show increased Rpb3 levels in the mutant to yield negative TDI values. Similar analyses of other mutants (Supplemental S21A–F) revealed a set of six genes exhibiting this behavior (EDI ≥ 0.3 and TDI < 0), including CPA1, LYS20, LYS21, CIT2, LEU4, and ICY2 and was also observed for ARG5,6, and ARG3 in PTET-SNF2gcn5Δ cells (Fig. 6C). These genes might undergo transcriptional repression in SM-induced cells that requires one or more cofactors, so that transcription is elevated in the mutants despite increased H3 occupancies. Indeed, ARG5,6, and ARG3 are repressed by the ArgR/Mcm1 repressor in arginine-replete cells (Hinnebusch 1992).
Interestingly, all mutants containing ydj1Δ exhibit larger mean TDIs than expected from the magnitude of their mean EDIs for the 70 exemplars (Supplemental Fig. S20A). This is also evident in heat map analyses of Figure 5, A and B, revealing comparable reductions in Rpb3 in snf2Δ and ydj1Δ cells despite greater increases in H3 occupancies in snf2Δ cells. Thus, Ydj1 likely enhances transcription by an additional mechanism independent of its role in nucleosome eviction.
Gcn5, SWI/SNF, and Ydj1 cooperate in NDR formation at virtually all genes but enhance transcription of only highly expressed genes
Inspection of heat maps in Figure 5, A–D, suggested that ydj1Δ, snf2Δ, gcn5Δ, and the triple mutation confer increased promoter H3 occupancy at many genes beyond those at the top of the maps with the greatest H3 eviction in WTI cells. To examine this further, we constructed a heat map for a group of 4744 genes with Rpb3I/Rpb3U less than 1.5-fold in WT cells, which excludes the approximately 600 genes most highly induced by SM, aligned at the +1 dyad and sorted on H3 eviction in WTI cells (Fig. 7A). Excluding the most highly induced genes eliminates the majority of genes displaying strong H3 eviction, leaving only a small group at the top showing modest H3 eviction in WTI cells. A group of genes at the bottom display increased rather than decreased H3 occupancies on SM induction and also tend to exhibit reduced Rpb3 occupancies in WTI cells (Fig. 7A, right). Additional heat maps in Figure 7, B–E, compare the effects of the single and triple mutations on H3 and Rpb3 occupancies for the same order of 4744 genes. Regardless of position in the map, all genes show a fairly uniform increase in H3 occupancy in the NDR that is the least in ydj1Δ cells, progressively greater in snf2Δ and gcn5Δ cells, and greatest in the triple mutant (Fig. 7B–E, left). Comparable heat maps for the double mutants (Supplemental Fig. S22A–C) reveal increases in H3 occupancies intermediate in degree between those in Figure 7 for the ydj1Δ or snf2Δ single mutants and the triple mutant. Thus, it appears that Ydj1, Snf2, and Gcn5 cooperate to promote nucleosome depletion in the NDRs of nearly all genes. Interestingly, however, the corresponding Rpb3 difference maps do not indicate marked decreases in transcription, even for the triple mutant (Fig. 7B–E, maps on right). Thus, it appears that significant increases in NDR H3 occupancy can occur at many genes with little effect on transcription.
Figure 7.
Elimination of Ydj1, Snf2, and Gcn5 increases H3 occupancies in the NDRs of most genes without substantially altering Pol II occupancies. (A) Heat map depictions of changes in H3 and Rpb3 relative occupancies on SM treatment of WT cells for 4744 genes not appreciably induced by SM, displayed as in Figure 2D. (B–E) Heat maps for the identically ordered 4744 genes shown in A, constructed as described in Figure 5 for the indicated mutants.
To examine this further, we divided the 4744 genes into 10 deciles based on Rpb3 occupancies in WTI cells. As might be expected, decile 1, with highest Rpb3 levels, shows the lowest median H3 occupancy in the [−1, NDR, +1] among all 10 deciles (Fig. 8A, WT). Median H3 occupancy is relatively higher in decile 2 but increases only slightly across deciles 2 to 10 (Fig. 8A). The 70 exemplar genes exhibit even lower median H3 occupancies and higher median Rpb3 levels than decile 1 (Fig. 8A,B, WT, 70 exemplars vs. decile 1). These findings support our conclusion that strong eviction of promoter nucleosomes is restricted to only the most highly transcribed genes, found in decile 1 and among the 70 exemplars.
Figure 8.
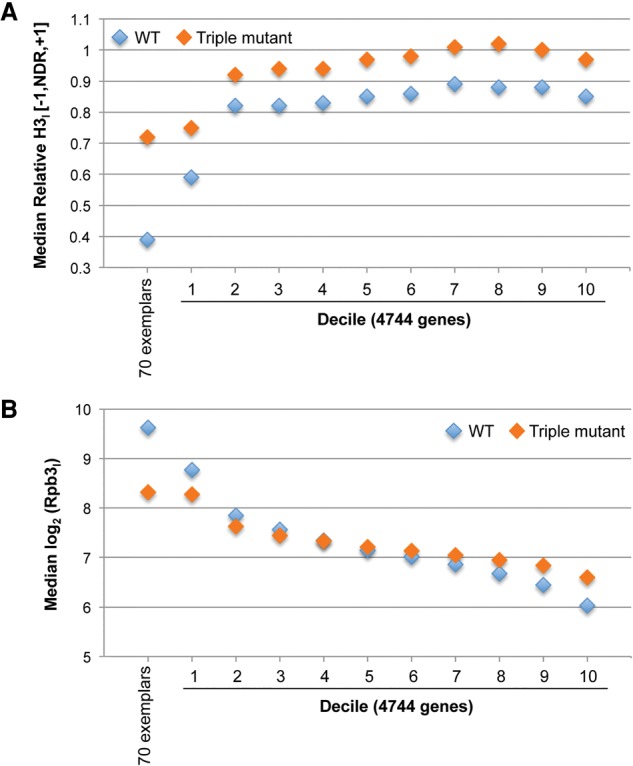
Defects in promoter H3 eviction are associated with marked reductions in Pol II levels only for the most highly expressed subset of genes. (A,B) Median relative H3 occupancies per base pair in the [−1,NDR,+1] regions (A) or median log2 values of Rpb3I occupancies per base pair in CDS (B) for the 70 exemplar genes or for each Rpb3 decile. The 4744 genes not appreciably induced by SM were divided into 10 deciles according to Rpb3 occupancies in SM-induced WT cells.
In the triple cofactor mutant, only the 70 exemplar genes and first decile display reduced median Rpb3 levels versus WTI cells, with a substantially larger effect for the 70 exemplars (Fig. 8B). Remarkably, the triple mutation evokes increased H3 occupancies for deciles 4–10 comparable to that seen for decile 1 (Fig. 8A) with little change (deciles 4–8) or even increased Rpb3 occupancies (deciles 9–10) compared with WTI cells (Fig. 8B). This phenomenon is also evident in Supplemental Figure S23, A–C, for genes in deciles 1, 5, and 10 that exhibit increased H3 occupancies but decreased (decile 1), unchanged (decile 5), or increased (decile 10) Rpb3 levels in cofactor mutants; with only decile 1 resembling the response of the 70 exemplar genes to the mutations (Supplemental Fig. S24A–B). Thus, it appears that defects in promoter H3 eviction are associated with reduced Rpb3 levels in the cofactor mutants only for genes with the highest Pol II levels in WTI cells, which includes highly SM-induced genes and those in decile 1 of the set of 4744 genes. The more than 3000 genes in deciles 4–10, in contrast, exhibit no change or even increased relative Rpb3 occupancies in the face of increased promoter H3 density in mutant strains. Note that because Rpb3 occupancies for each gene are expressed relative to all other genes in each strain, the absolute transcription rate of genes in deciles 9 and 10 might actually be reduced in some or all cofactor mutants compared with WT cells but decreased to a much smaller degree than occurs for other genes. Similarly, if total H3 levels were reduced in cofactor mutants, this would diminish the absolute increase in promoter H3 occupancies relative to those in WTI cells but would not alter the fact that increased promoter H3 levels in the mutants seem to reduce transcription to a greater extent at highly expressed genes than at the majority of other genes.
Discussion
Our results demonstrate that SWI/SNF, Gcn5, and Ydj1 function broadly in the yeast transcriptome, acting cooperatively at many genes, and in many cases redundantly, to evict promoter nucleosomes and that defects in this process confer marked reductions in transcription only for the most highly expressed genes. By conventional H3 ChIP, we found that eliminating Snf2, Gcn5, either Ydj1 or the Hsp70 proteins Ssa1/Ssa2, or AAA-ATPase Yta7 all impaired H3 eviction in SM-induced cells for at least one of four target genes, and mutants lacking different combinations of Ydj1, Snf2, and Gcn5 displayed H3 eviction defects at three or all four genes, with WT or higher Gcn4 UAS occupancies. ChIP-seq analysis of the same mutants implicated these three cofactors in nucleosome eviction throughout the Gcn4 transcriptome. Based on statistically significant increases in H3 occupancies conferred by each cofactor mutation in single, double, or triple mutants, which undoubtedly underestimates involvement of the mutated cofactors, we determined that Gcn5, Snf2, and Ydj1 function at 93%, 74%, and 42%, respectively, of the 70 exemplar genes showing the greatest H3 eviction in WTI cells. Functional cooperation among Ydj1, Snf2, and Gcn5 is widespread, as about two-thirds of these 70 genes utilize both Gcn5 and Snf2, and more than one-third employ all three cofactors in nucleosome eviction (Fig. 4C).
Nucleosome eviction is frequently overdetermined, such that removal of one cofactor confers a strong H3 eviction defect only in a mutant lacking another cofactor. Indeed, the H3 occupancies at many of the 70 exemplar genes and the larger group of 204 highly SM-induced genes increased progressively with elimination of two or three cofactors and approached the high levels observed in uninduced WT cells in the triple mutant lacking all three cofactors. It is likely that these are not the only cofactors functioning at Gcn4 targets, and combinations of other cofactor mutations might well produce equally strong defects in H3 eviction. Indeed, that appreciable H3 eviction remains intact in the triple mutant for a subset of the 70 exemplars (e.g., LEU4, YPR036W-A, ICY2, and RTS3 in Supplemental Fig. S16B) implicates other cofactors at these genes. The clustergrams in Figure 4A,B reveal a wide range of defects in H3 eviction among Gcn4 target genes in the snf2Δ and gcn5Δ single mutants. Perhaps overlapping functions of other chaperones, remodelers, or HATs can explain why certain genes are affected less than others by elimination of Snf2 or Gcn5 in otherwise WT cells. However, we have discovered significant correlations between the A/T and T/A dinucleotide frequencies in DNA and the increase in H3 occupancy in the [−1,NDR,+1] regions of the 204 highly induced genes observed in snf2Δ and gcn5Δ mutants, with Pearson's correlation coefficients of 0.51 and 0.61, respectively (P-values <0.0001). This might suggest that Gcn5 and Snf2 preferentially promote eviction of nucleosomes that are relatively unstable (Jiang and Pugh 2009b), with other cofactors being needed to evict more stable nucleosomes, although it could also reflect the presence of an AT-hook motif in Snf2 (Aravind and Landsman 1998).
Gcn5 and Snf2 probably act directly to evict nucleosomes at Gcn4 targets, as we showed that SAGA and SWI/SNF are recruited by Gcn4 to promoters (Qiu et al. 2005). Considering that med15Δ reduces Pol II recruitment to Gcn4 targets (Qiu et al. 2004) but had no effect on H3 eviction (Supplemental Fig. S3A), it is unlikely that gcn5Δ and snf2Δ impair nucleosome eviction merely by their deleterious effects on PIC assembly (Qiu et al. 2004), although reduced Pol II recruitment could exacerbate the effects of diminished histone acetylation (gcn5Δ) or nucleosome remodeling (snf2Δ) on H3 eviction in these mutants (Ansari et al. 2014).
Our analysis of 4744 genes that are either weakly induced by SM, constitutively expressed, or nonexpressed unveiled a nearly ubiquitous cooperation among Ydj1, Snf2, and Gcn5 in evicting promoter nucleosomes from NDRs, albeit to a smaller degree than observed for highly induced genes. The relative contributions of these cofactors, with Ydj1 contributing the least and Gcn5 the most, and their functional cooperation at the 4744 genes was essentially identical to our findings for the highly induced genes, implying that these cofactors act similarly throughout much of the genome.
Our finding of functional cooperation between Gcn5 and Snf2 in nucleosome eviction is consistent with previous results implicating them in nucleosome remodeling at PHO8 (Gregory et al. 1999) and PHO5 (Adkins et al. 2007; Barbaric et al. 2007). It also fits with in vitro studies indicating that SWI/SNF binding to reconstituted nucleosomes, as well as attendant octamer displacement, are stimulated by acetylation by SAGA (Gcn5) (Hassan et al. 2001), dependent on the bromodomain in Snf2 (Hassan et al. 2002, 2006). Thus, Gcn5 might stimulate nucleosome eviction indirectly at some genes by enhancing SWI/SNF recruitment via the Snf2 bromodomain. However, there was no effect of gcn5Δ on SWI/SNF recruitment by Gcn4 at ARG1 (Yoon et al. 2004) or a modified PHO5 promoter (Syntichaki et al. 2000). Similarly, histone acetylation promotes nucleosome remodeling independently of SWI/SNF recruitment at RNR3 (Sharma et al. 2003) and PHO5 (Barbaric et al. 2007). A SWI/SNF-independent role for Gcn5 is also supported by our finding that H3 eviction defects are generally greater in gcn5Δ versus snf2Δ cells (Figs. 5, 7) and our identification of SM-induced genes with a greater requirement for Gcn5 versus Snf2 for nucleosome eviction (Fig. 4). Histone acetylation by Gcn5 could stimulate the recruitment or function of a remodeler besides SWI/SNF, as there is evidence that RSC, Isw1, and Chd1 act with SWI/SNF and INO80 in nucleosome remodeling at PHO5 (Musladin et al. 2014), and functional overlap among different remodeling complexes occurs in mammalian cells (Morris et al. 2014).
Our results implicating Ydj1 in nucleosome eviction at Gcn4 target genes follow on previous findings implicating Ydj1, Hsp70 orthologs Ssa1/Ssa2, and Hsp90 chaperones (Hsp82 and Hsc82) in rapid nucleosome eviction and induction of GAL genes by Gal4. Ssa1 and Hsc82 were recruited to GAL1 under inducing conditions (Floer et al. 2008), consistent with a direct role in nucleosome eviction. Although we observed no defects in nucleosome eviction in the hsc82Δhsp82 double mutants, the ydj1Δ and ssa1Δssa2Δ mutations had the strongest effects on GAL1 induction (Floer et al. 2008). Hence, Hsp70 and cochaperone Ydj1 might be generally more important than Hsp90 in nucleosome eviction.
As noted above, the cofactor mutations increase H3 occupancies in the [−1,NDR,+1] regions but not in the adjacent CDS of the 70 exemplar genes, even in the triple mutant (Fig. 3C). One explanation might be that because Pol II levels at these genes remain much higher in the triple mutant compared with the WT levels at most other genes (Fig. 8B), transcription-coupled nucleosome eviction from CDSs (Schwabish and Struhl 2004) should still operate at the 70 exemplar genes in the triple mutant. However, it is noteworthy that the heat maps in Figures 5, A–D, and 7, B–E, reveal that the cofactor mutations reduce H3 occupancies in promoter-proximal CDS (blue shading) while increasing H3 levels in the NDR (orange-red shading) throughout the genome. We calculated the decrease in average H3 occupancy throughout the CDS in the triple mutant versus WTI cells to be 6.7% for all yeast genes, and similar decreases were found for the 204 highly induced genes (4.6%) and for the most highly or weakly expressed genes among the set of 4744 genes (decile 1, 4.5%; decile 10, 6.7%). Given that the NDRs of all genes exhibit increased H3 occupancies in the cofactor mutants (Figs. 5A–D, 7B–E), at least a portion of the reduced H3 levels in CDSs could involve redistribution of nucleosomes from the CDS into the NDR in cofactor mutants, although it could also reflect a general defect in nucleosome replacement in CDSs. Regardless of the mechanism, the general decrease in H3 occupancy in CDSs conferred by the cofactor mutations might contribute to the moderately reduced H3 levels in the CDSs of the 70 exemplar genes seen in the triple mutant compared with WTU cells (Fig. 3C).
Another important conclusion of our study is that defects in nucleosome eviction are tightly associated with defects in transcription, but this coupling is evident only for the subset of genes with the highest transcription rates. Combining mutations that eliminate or deplete Snf2, Gcn5, or Ydj1 produced increasingly severe transcription defects that paralleled progressive increases in promoter H3 occupancies for the 70 exemplar induced genes and the first decile of the 4744 gene set with highest Rpb3 levels in WT cells. Thus, it appears that steady-state removal of (H3/H4)2 tetramers across the promoter, as opposed to limited nucleosome sliding or eviction of only H2A-H2B dimers, is important for maximal transcription of the most highly expressed genes, presumably to facilitate cofactor recruitment, PIC assembly, and initiation of transcription. Unexpectedly, a progressive increase in promoter H3 occupancies was not associated with reduced transcription for the majority of the 4744 genes, and the deciles with lowest Rpb3 occupancies in WT cells showed increased, rather than decreased, Rpb3 levels in the triple mutant (Fig. 8B; Supplemental Fig. S23B,C). This is consistent with previous observations of increased nucleosome occupancy at many promoters in an abf1 mutant that did not alter mRNA expression (Ganapathi et al. 2011).
To explain these findings, we propose that nucleosome eviction is crucial to achieve WT levels of transcription at highly expressed genes, whereas recruitment of cofactors, GTFs, or Pol II (or another step of initiation) is more rate-limiting at weakly expressed genes. Accordingly, increased H3 occupancies conferred by cofactor mutations evoke the greatest reductions in transcription at the most highly expressed genes. The resulting increased availability of cofactors, GTFs, or Pol II could mitigate the effects of increased nucleosome density at genes with inherently lower transcription rates, such that their relative Pol II occupancies are increased, rather than decreased, in the cofactor mutants. Thus, although eviction of nucleosomes probably enhances the transcription of all genes by increasing access of the transcription machinery to promoter DNA, the magnitude of the transcription defect evoked by global attenuation of histone eviction in cofactor mutants (or possibly an environmental state mimicking such mutations) likely depends on the particular step of transcriptional activation that is most rate-limiting at each gene.
Methods
Yeast strain and plasmid constructions
Yeast strains used in this study are listed in Supplemental Table S1, and strain and plasmid constructions are described in the Supplemental Material.
Conventional ChIP, ChIP-seq, and Nuc-seq analyses
Yeast cells were grown in synthetic complete medium lacking isoleucine and valine (SC-Ilv) to log-phase (OD600 = 0.6–0.8), and SM was added at 1 µg/mL for 30 min to induce Gcn4. Conventional ChIP experiments were conducted as described previously (Qiu et al. 2012) using anti-H3 (abcam, ab1791) and anti-Gcn4 (Zhang et al. 2008) antibodies and primers summarized in Supplemental Figure S1. For ChIP-seq analysis, DNA libraries for Illumina paired-end sequencing of chromatin immunoprecipitates (PESCI) were prepared as described previously (Cole et al. 2014) with modifications indicated in the Supplemental Material. For Nuc-seq, nucleosomal DNA was obtained by MNase digestion of nuclei and prepared for Illumina paired-end sequencing (Cole et al. 2011; Cole et al. 2012). Paired-end sequencing (50 nt from each end) was conducted by the DNA Sequencing and Genomics core facility of the NHLBI, NIH.
Sequence data were aligned to the SacCer2 version of the genome sequence using alignment software Bowtie 2 (Langmead and Salzberg 2012). Numbers of aligned paired reads from H3 and Rpb3 PESCI, as well as Nuc-seq, are summarized in Supplemental Tables S2 though S4, respectively. H3 (Supplemental Figs. S7, S8), and Rpb3 occupancy profiles were obtained from the alignment (.sam) files using custom Perl programs written by Bruce Howard and James Iben (for Perl scripts used in this study, see Supplemental File S6). The chromosomal coordinates of the 5′-ends of both reads in each pair were extracted and saved in chromosome-specific (“cpsOut”) files, which were used to construct occupancy plots in zero-notation format (“.znt4” files) (Cole et al. 2011). The occupancy at each nucleotide is defined as the number of sequenced molecules containing that nucleotide, such that the occupancy plots in Supplemental Figure S8 represent the number of sequenced molecules containing the nucleotide at coordinate n plotted against coordinate n.
Positions of NDRs and flanking nucleosomes at each gene (Supplemental Fig. S10B) were determined using a custom MATLAB script (Supplemental File S6; also at https://github.com/rchereji/Detect_NDRs_and_flanking_nucleosomes) using two replicate Nuc-seq experiments from WTU cells, providing coordinates of 147-bp regions centered on the +1 and −1 dyads, and intervening NDR for each gene. Some pairs of divergent genes share the same NDR and flanking nucleosomes, with exchanged roles; i.e., +1 nucleosome of one gene becomes −1 nucleosome of the divergent gene.
As different chromosomes frequently have slightly different sequencing depths, the H3 occupancy profiles are normalized so that the average occupancy for every chromosome is unity; i.e., the occupancy at each base pair is expressed relative to the average occupancy on that chromosome. The genome-wide average H3 distributions relative to TSS and the average H3 occupancy in the three zones (−1, NDR, +1) for every gene were computed using TSS data (Xu et al. 2009) and custom MATLAB scripts (Supplemental File S6, also at https://github.com/rchereji/Compute_H3_averages). Heat maps were generated in MATLAB using the heatmap function (http://www.mathworks.com/matlabcentral/fileexchange/24253-customizable-heat-maps).
Data access
Sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE74787.
Supplementary Material
Acknowledgments
We thank Krishnamurthy Natarajan for sharing expression microarray data on SM-induced WT cells, James Iben for valuable assistance with bioinformatics, and Chhabi Govind and members of our laboratories for helpful suggestions. This work was supported by the Intramural Research Program of the National Institutes of Health (NIH) and utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.196337.115.
References
- Adkins MW, Howar SR, Tyler JK. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell 14: 657–666. [DOI] [PubMed] [Google Scholar]
- Adkins MW, Williams SK, Linger J, Tyler JK. 2007. Chromatin disassembly from the PHO5 promoter is essential for the recruitment of the general transcription machinery and coactivators. Mol Cell Biol 27: 6372–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari SA, Paul E, Sommer S, Lieleg C, He Q, Daly AZ, Rode KA, Barber WT, Ellis LC, LaPorta E, et al. 2014. Mediator, TATA-binding protein, and RNA polymerase II contribute to low histone occupancy at active gene promoters in yeast. J Biol Chem 289: 14981–14995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Landsman D. 1998. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res 26: 4413–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Ondracka A, Cross FR. 2011. Multiple sequence-specific factors generate the nucleosome-depleted region on CLN2 promoter. Mol Cell 42: 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaric S, Luckenbach T, Schmid A, Blaschke D, Horz W, Korber P. 2007. Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J Biol Chem 282: 27610–27621. [DOI] [PubMed] [Google Scholar]
- Berger SL, Pina B, Silverman N, Marcus GA, Agapite J, Regier JL, Triezenberg SJ, Guarente L. 1992. Genetic isolation of ADA2: a potential transcription adaptor required for function of certain acidic activation domains. Cell 70: 251–265. [DOI] [PubMed] [Google Scholar]
- Biddick RK, Law GL, Chin KK, Young ET. 2008. The transcriptional coactivators SAGA, SWI/SNF, and mediator make distinct contributions to activation of glucose-repressed genes. J Biol Chem 283: 33101–33109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell 11: 1587–1598. [DOI] [PubMed] [Google Scholar]
- Bryant GO, Prabhu V, Floer M, Wang X, Spagna D, Schreiber D, Ptashne M. 2008. Activator control of nucleosome occupancy in activation and repression of transcription. PLoS Biol 6: 2928–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR. 2009. The logic of chromatin architecture and remodelling at promoters. Nature 461: 193–198. [DOI] [PubMed] [Google Scholar]
- Cole HA, Howard BH, Clark DJ. 2011. Activation-induced disruption of nucleosome position clusters on the coding regions of Gcn4-dependent genes extends into neighbouring genes. Nucleic Acids Res 39: 9521–9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole HA, Howard BH, Clark DJ. 2012. Genome-wide mapping of nucleosomes in yeast using paired-end sequencing. Methods Enzymol 513: 145–168. [DOI] [PubMed] [Google Scholar]
- Cole HA, Ocampo J, Iben JR, Chereji RV, Clark DJ. 2014. Heavy transcription of yeast genes correlates with differential loss of histone H2B relative to H4 and queued RNA polymerases. Nucleic Acids Res 42: 12512–12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F, Cole HA, Clark DJ, Zhurkin VB. 2012. Transcriptional activation of yeast genes disrupts intragenic nucleosome phasing. Nucleic Acids Res 40: 10753–10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C, Tyler JK. 2012. Histone exchange and histone modifications during transcription and aging. Biochim Biophys Acta 1819: 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale CM, Jackson BM, McVeigh R, Klebanow ER, Bai Y, Kokubo T, Swanson M, Nakatani Y, Weil PA, Hinnebusch AG. 1998. The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID, and the Adap-Gcn5p coactivator complex. Mol Cell Biol 18: 1711–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser SJ, D'Arcy S. 2012. Towards a mechanism for histone chaperones. Biochim Biophys Acta 1819: 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkina TY, Zou Y, Freeling S, Vorobyev VI, Erkine AM. 2010. Functional interplay between chromatin remodeling complexes RSC, SWI/SNF and ISWI in regulation of yeast heat shock genes. Nucleic Acids Res 38: 1441–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M, Bryant GO, Ptashne M. 2008. HSP90/70 chaperones are required for rapid nucleosome removal upon induction of the GAL genes of yeast. Proc Natl Acad Sci 105: 2975–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi M, Palumbo MJ, Ansari SA, He Q, Tsui K, Nislow C, Morse RH. 2011. Extensive role of the general regulatory factors, Abf1 and Rap1, in determining genome-wide chromatin structure in budding yeast. Nucleic Acids Res 39: 2032–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos T, Thireos G. 1992. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J 11: 4145–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkikopoulos T, Havas KM, Dewar H, Owen-Hughes T. 2009. SWI/SNF and Asf1p cooperate to displace histones during induction of the Saccharomyces cerevisiae HO promoter. Mol Cell Biol 29: 4057–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Yoon S, Qiu H, Govind S, Hinnebusch AG. 2005. Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol Cell Biol 25: 5626–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. 2010. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell 39: 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradolatto A, Rogers RS, Lavender H, Taverna SD, Allis CD, Aitchison JD, Tackett AJ. 2008. Saccharomyces cerevisiae Yta7 regulates histone gene expression. Genetics 179: 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Sterner DE, Duggan LJ, Workman JL, Berger SL. 1998. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol 8: 193–197. [DOI] [PubMed] [Google Scholar]
- Gregory PD, Schmid A, Zavari M, Munsterkotter M, Horz W. 1999. Chromatin remodelling at the PHO8 promoter requires SWI–SNF and SAGA at a step subsequent to activator binding. EMBO J 18: 6407–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD. 2009. Mechanisms that specify promoter nucleosome location and identity. Cell 137: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL. 2001. Histone acetyltransferase complexes stabilize SWI/SNF binding to promoter nucleosomes. Cell 104: 817–827. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111: 369–379. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Awad S, Prochasson P. 2006. The Swi2/Snf2 bromodomain is required for the displacement of SAGA and the octamer transfer of SAGA-acetylated nucleosomes. J Biol Chem 281: 18126–18134. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. 1992. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae. In The molecular and cellular biology of the yeast Saccharomyces: gene expression (ed. Broach JR, et al. ), pp. 319–414. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Hinnebusch AG. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Brown SA, Clark CD, Winston F. 1992. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev 6: 2288–2298. [DOI] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. 2009a. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol 10: R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. 2009b. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet 10: 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber P, Barbaric S, Luckenbach T, Schmid A, Schermer UJ, Blaschke D, Horz W. 2006. The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J Biol Chem 281: 5539–5545. [DOI] [PubMed] [Google Scholar]
- Kuo MH, vom Bauer E, Struhl K, Allis CD. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters Independently of transcription. Mol Cell 6: 1309–1320. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi LM, Ellahi A, Rine J. 2011. Direct regulation of nucleosome density by the conserved AAA-ATPase Yta7. Proc Natl Acad Sci 108: E1302–E1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi LM, Davis MD, Rine J. 2015. Maintenance of nucleosomal balance in cis by conserved AAA-ATPase Yta7. Genetics 199: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Baek S, Sung MH, John S, Wiench M, Johnson TA, Schiltz RL, Hager GL. 2014. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat Struct Mol Biol 21: 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musladin S, Krietenstein N, Korber P, Barbaric S. 2014. The RSC chromatin remodeling complex has a crucial role in the complete remodeler set for yeast PHO5 promoter opening. Nucleic Acids Res 42: 4270–4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K, Jackson BM, Rhee E, Hinnebusch AG. 1998. yTAFII61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol Cell 2: 683–692. [DOI] [PubMed] [Google Scholar]
- Natarajan K, Jackson BM, Zhou H, Winston F, Hinnebusch AG. 1999. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and SRB/mediator. Mol Cell 4: 657–664. [DOI] [PubMed] [Google Scholar]
- Parnell TJ, Huff JT, Cairns BR. 2008. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J 27: 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina B, Berger S, Marcus GA, Silverman N, Agapite J, Guarente L. 1993. ADA3: a gene, identified by resistance to GAL4-VP16, with properties similar to and different from those of ADA2. Mol Cell Biol 13: 5981–5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Yoon S, Natarajan K, Swanson MJ, Hinnebusch AG. 2004. An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol Cell Biol 24: 4104–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Zhang F, Hwang GJ, Swanson MJ, Boonchird C, Hinnebusch AG. 2005. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol Cell Biol 25: 3461–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Gaur NA, Hinnebusch AG. 2012. Pol II CTD kinases Bur1 and Kin28 promote Spt5 CTR-independent recruitment of Paf1 complex. EMBO J 31: 3494–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Winston F. 2012. Chromatin and transcription in yeast. Genetics 190: 351–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom M, Williams SK, Dechassa ML, Das C, Linger J, Adkins M, Liu C, Bartholomew B, Tyler JK. 2009. FACT and the proteasome promote promoter chromatin disassembly and transcriptional initiation. J Biol Chem 284: 23461–23471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke H, Horz W. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell 11: 1599–1607. [DOI] [PubMed] [Google Scholar]
- Roberts SM, Winston F. 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147: 451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint M, Sawhney S, Sinha I, Singh RP, Dahiya R, Thakur A, Siddharthan R, Natarajan K. 2014. The TAF9 C-terminal conserved region domain is required for SAGA and TFIID promoter occupancy to promote transcriptional activation. Mol Cell Biol 34: 1547–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol 24: 10111–10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. 2006. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell 22: 415–422. [DOI] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. 2007. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol Cell Biol 27: 6987–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VM, Li B, Reese JC. 2003. SWI/SNF-dependent chromatin remodeling of RNR3 requires TAFIIs and the general transcription machinery. Genes Dev 17: 502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S, Iyer VR. 2008. Stress-dependent dynamics of global chromatin remodeling in yeast: dual role for SWI/SNF in the heat shock stress response. Mol Cell Biol 28: 2221–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain MM, Ansari SA, Pathak R, Palumbo MJ, Morse RH, Govind CK. 2014. The RSC complex localizes to coding sequences to regulate Pol II and histone occupancy. Mol Cell 56: 653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MJ, Qiu H, Sumibcay L, Krueger A, Kim S-J, Natarajan K, Yoon S, Hinnebusch AG. 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol Cell Biol 23: 2800–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syntichaki P, Topalidou I, Thireos G. 2000. The Gcn5 bromodomain co-ordinates nucleosome remodeling. Nature 404: 414–417. [DOI] [PubMed] [Google Scholar]
- Takahata S, Yu Y, Stillman DJ. 2009. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol Cell 34: 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdone L, Wu J, van Riper K, Kacherovsky N, Vogelauer M, Young ET, Grunstein M, Di Mauro E, Caserta M. 2002. Hyperacetylation of chromatin at the ADH2 promoter allows Adr1 to bind in repressed conditions. EMBO J 21: 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bai L, Bryant GO, Ptashne M. 2011. Nucleosomes and the accessibility problem. Trends Genet 27: 487–492. [DOI] [PubMed] [Google Scholar]
- Williams SK, Truong D, Tyler JK. 2008. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc Natl Acad Sci 105: 9000–9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Govind CK, Qiu H, Kim SJ, Dong J, Hinnebusch AG. 2004. Recruitment of the ArgR/Mcm1p repressor is stimulated by the activator Gcn4p: a self-checking activation mechanism. Proc Natl Acad Sci 101: 11713–11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Reese JC. 2007. Exposing the core promoter is sufficient to activate transcription and alter coactivator requirement at RNR3. Proc Natl Acad Sci 104: 8833–8838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Sumibcay L, Hinnebusch AG, Swanson MJ. 2004. A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol Cell Biol 24: 6871–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gaur NA, Hasek J, Kim SJ, Qiu H, Swanson MJ, Hinnebusch AG. 2008. Disrupting vesicular trafficking at the endosome attenuates transcriptional activation by Gcn4. Mol Cell Biol 28: 6796–6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.