Abstract
Most living species exploit a limited range of resources. However, little is known on how tight links build up during evolution between specialist species and the hosts they utilize. We examined the dependence of Drosophila pachea on its single host, the senita cactus. Several amino acid changes in the Neverland oxygenase rendered D. pachea unable to transform cholesterol into 7-dehydrocholesterol (first reaction in the steroid hormone biosynthetic pathway in insects) and thus made D. pachea dependent on the uncommon sterols of its host plant. The neverland mutations increase survival on the cactus unusual sterols and are in a genomic region that faced recent positive selection. This study illustrates how relatively few genetic changes in a single gene may restrict the ecological niche of a species.
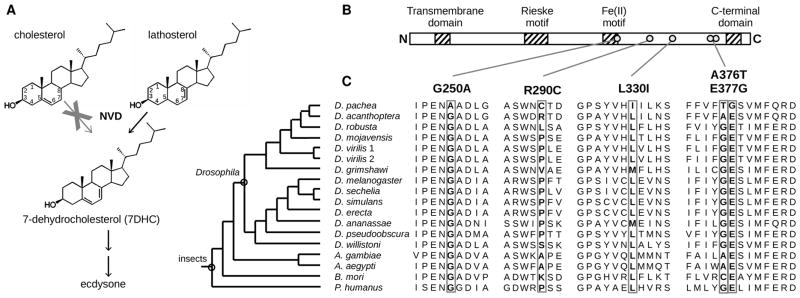
Losses of enzymatic activities are frequent during evolution (1). For example, humans lost the ability to produce nine amino acids and six vitamins, for which we rely on our diet (2). The reasons for such losses are unknown, but it is generally believed that “superfluous” metabolic activities were lost by chance during evolution (3). We examined the dependence of the fly Drosophila pachea on the senita cactus (Lophocereus schottii), a plant species endemic to the Sonoran desert (Northwestern Mexico and Southwestern USA). In insect developmental transitions and egg production are regulated by the steroid hormone ecdysone (4). D. pachea has lost the first metabolic reaction in the ecdysone biosynthetic pathway, i.e. the ability to convert cholesterol into 7-dehydrocholesterol (7DHC) (Fig. 1A, 5–7). The senita cactus, which D. pachea requires as a host (5), does not contain common sterols and is the only plant in the Sonoran desert (7) known to produce Δ7-sterols such as lathosterol (6). D. pachea flies do not reach the adult stage if not raised on senita cactus, but supplementing standard food with senita cactus or with 7DHC fully restores D. pachea viability and fertility (5), indicating that Δ7-sterols are essential compounds required for D. pachea development and survival. Interestingly, D. pachea appears to depend on the senita cactus solely for its sterols as we raised D. pachea on an artificial diet supplemented with 7DHC for more than four years (~60 generations) with no apparent defect (8).
Fig. 1.
Presumed ecdysone biosynthetic pathway (A) and NVD in D. pachea. (B) NVD protein structure. (C) Alignment of multiple NVD protein sequences. Five mutations (boxes) were tested in vitro. For an alignment of full NVD protein sequences with additional insect species see fig. S8.
Conversion of cholesterol into 7DHC is catalyzed by the evolutionary conserved Rieske-domain oxygenase Neverland (NVD) in insects and nematodes (9, 10). To test whether mutation(s) in nvd are responsible for D. pachea dependence upon its host cactus, we sequenced the nvd coding region (8) from D. pachea and the three most closely related species D. nannoptera, D. acanthoptera and D. wassermani, which feed on other cacti (11) (Table S1–2, fig. S1). No stop codon or insertions/deletions were found in the D. pachea sequence, but the ratio of rates of nonsynonymous substitution (dN) over synonymous substitution (dS) is significantly higher in the branch leading to D. pachea (Tables S3, fig. S2). We noticed that several amino acids showing high conservation across insects and vertebrates are different in D. pachea NVD (Fig. 1B–C). We observed that in D. pachea third instar larvae, as in D. melanogaster (9) and D. acanthoptera, nvd is only expressed in the prothoracic gland (fig. S3), an organ whose sole known function is ecdysone production (12). Therefore we conclude that NVD function, if any, should be related to steroid hormone production.
The senita cactus does not contain cholesterol nor 7DHC but does produce three other sterols – lathosterol, campestenol and schottenol (6) – which, if used as precursors for steroid hormone synthesis, are expected to lead to different steroid hormones, respectively 20-hydroxyecdysone, makisterone A and makisterone C (fig. S4), due to the inability of Drosophila to dealkylate phytosterols (13). Steroids from D. pachea extracts were separated by HPLC and fractions of interest were analyzed with mass spectroscopy. We detected ecdysone and 20-hydroxyecdysone but no trace of makisterone A or makisterone C (fig. S5). These results suggest that D. pachea only uses lathosterol and not the other senita cactus sterols as steroid hormone precursors.
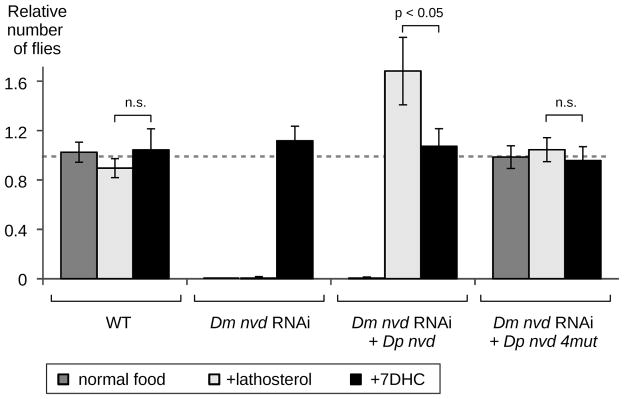
Since conversion of cholesterol into 7DHC biochemically resembles the transformation of lathosterol into 7DHC (Fig. 1A), we hypothesized that D. pachea NVD converts lathosterol rather than cholesterol into 7DHC (14). To test this hypothesis, we generated transgenic D. melanogaster flies in which the endogenous nvd gene is shut down by RNA interference (RNAi) and replaced by D. pachea nvd. The D. melanogaster nvd RNAi flies do not develop on regular fly food (9) nor on food supplemented with lathosterol, yet they reach the adult stage on regular fly food supplemented with 7DHC (9) (Fig. 2, Table S4). As expected, introduction of D. pachea nvd into D. melanogaster nvd RNAi flies rescues development on food supplemented with lathosterol but not with cholesterol (Fig. 2). This demonstrates that D. pachea NVD can use lathosterol but not cholesterol as a substrate (Fig. 1A).
Fig. 2.
Fly survival on various food media. WT: control flies, Dm nvd RNAi: RNAi knockout of wildtype D. melanogaster nvd, +Dp nvd: rescued with wildtype D. pachea nvd, +Dp nvd 4mut: rescued with D. pachea nvd A250G I330L T376A G377E. The proportion of flies of each genotype is indicated relative to the number of UAS-nvd RNAi Sb male siblings (8, Table S4). Bars show average and error bars are SE.
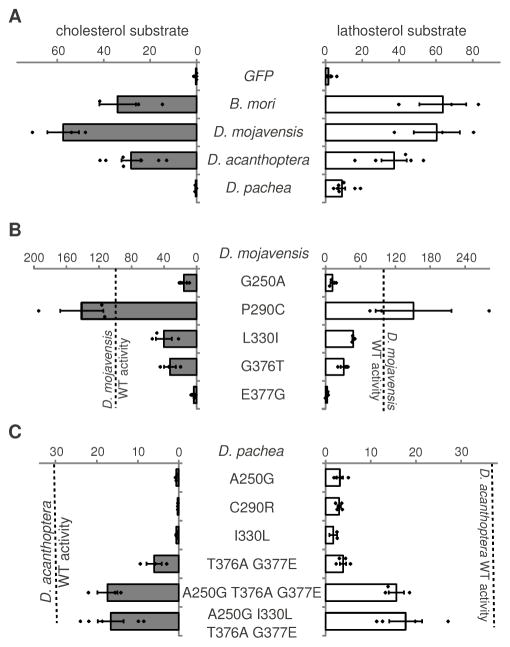
To identify the amino acid changes responsible for the loss of D. pachea NVD activity with cholesterol, we reconstructed ancestral NVD sequences (8) for the entire protein region except for the N-terminal region, which does not show conserved amino acid sequence among insects. Interestingly, we found 19 mutations in the lineage leading to D. pachea, of which five are predicted to affect protein function (Fig. 1C). We sequenced the entire nvd coding region in three D. pachea strains and in two natural population samples. The five predicted functionally relevant amino acids were found in all the 32 individuals. To test whether these five amino acid changes affect NVD activity, we established an in vitro assay of NVD activity with GFP-control and NVD constructs (8, 15). GFP-transfected cells produced no 7DHC whereas cells transfected with nvd from various insects including D. acanthoptera converted cholesterol and lathosterol into 7DHC (Fig. 3A, S6). In accordance with our D. melanogaster transgenic assays, we observed that cells transfected with D. pachea nvd do not convert cholesterol into 7DHC but convert lathosterol into 7DHC (Fig. 3A, S6), although at a lower level relative to other species. Control experiments with HA epitope-tagged NVD constructs revealed that D. pachea NVD accumulates at similar levels as the other NVD homologs in the in vitro assay (fig. S7). These results indicate that the ancestral Drosophila NVD enzyme was likely able to transform both cholesterol and lathosterol into 7DHC and that NVD has subsequently lost the ability to convert cholesterol in the lineage leading to D. pachea.
Fig. 3.
NVD enzyme activity with cholesterol (left, grey) or with lathosterol (right, white). (A) wildtype (WT) NVD enzymes. (B) D. mojavensis NVD enzymes containing single mutations. (C) D. pachea enzymes containing reverse mutations. Bars represent average activity, error bars SD, and dots data points. Note that two D. mojavensis nvd wild-type constructs were used in our assays. Enzyme activity is indicated as a percentage relative to the NVD activity obtained with a D. mojavensis nvd construct that includes the nvd gene 5′UTR (9). All the D. mojavensis constructs tested in (B) contained this 5′UTR. The dotted line indicates D. mojavensis NVD wild-type activity (construct containing the 5′UTR) in (B) and D. acanthoptera NVD WT activity in (C).
We tested the effect of the five predicted functionally relevant amino acid changes by introducing each mutation individually in the nvd sequence from D. mojavensis, another cactophilic species endemic to the Sonoran desert, which displayed the highest in vitro NVD activity. With either cholesterol or lathosterol as a substrate, substitution P290C slightly increased the activity, G376T and L300I decreased the activity by half, and substitutions G250A and E377G reduced the activity to less than 18% of the wild-type activity (Fig. 3B, S7). We also performed the reciprocal experiment and reintroduced the predicted ancestral amino acid residues into the D. pachea NVD sequence. We found that a NVD activity close to that of D. acanthoptera is not restored by a single amino acid change but by four amino acid changes in concert (Fig. 3C, S7). Corroborating these in vitro results, introduction of a D. pachea nvd construct containing these four amino acid changes into D. melanogaster nvd RNAi flies rescues development on food supplemented with cholesterol (Fig. 2). We conclude that two to four mutations in the D. pachea nvd coding region have caused the loss of NVD activity with cholesterol substrate. These mutations have turned D. pachea into an obligate specialist dependent on lathosterol, a compound that has been found in a single plant species in the Sonoran desert (5, 6).
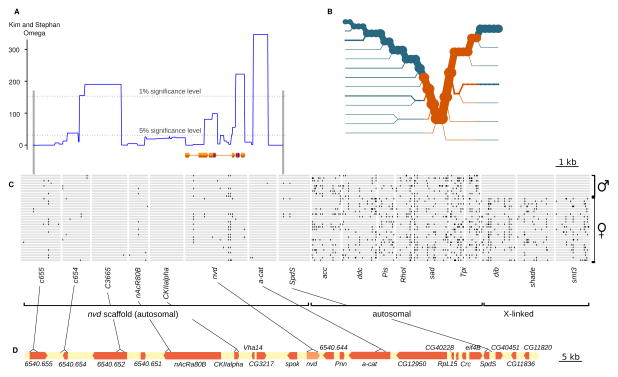
Remarkably, D. melanogaster nvd RNAi flies expressing D. pachea nvd survive significantly better on lathosterol than on cholesterol (t-test, t10,11 = 2.029, p < 0.03, Fig. 2) but no effect on survival was detected with nvd RNAi flies expressing D. pachea nvd with the four ancestral amino acid changes (Fig. 2). This suggests that the mutations that abolished cholesterol conversion during D. pachea evolution provide a fitness advantage on lathosterol. The underlying mechanism remains unclear. Our in vitro assay does not uncover any benefit from the D. pachea nvd mutations: D. pachea NVD in vitro activity with lathosterol is not higher compared to other species (Fig. 3) and the NVD enzymes of related Drosophila species are already able to convert lathosterol into 7DHC. To assess population genetic forces at play on the nvd genomic region, we compared the 3-kb nvd locus and 7 genes on the same 100-kb-scaffold with 9 control genes in 34 individuals from a single natural population. Our analysis reveals that nvd is in a genomic region of low nucleotide diversity, low recombination rate and normal divergence rate (MK test, p > 0.85; MLHKA test, p < 10−5, fig. 4, Tables S5–11). A signature of selective sweep (Kim and Stephan W) is detected over nvd and neighboring loci (fig. 4), but nucleotide polymorphism is too low to infer whether this recent selection acted on the nvd mutations themselves. Tajima’s D and Fu and Li tests are consistent with recovery from selective sweep in the nvd region (Table S6).
Fig. 4.
The nvd region is under positive selection. (A) Kim and Stephan’s omega statistics across the nvd region. Omega values above the significance level indicate a selective sweep. The nvd coding regions are represented below, with the position of the five tested amino acid changes in purple. (B) Haplotype bifurcation plot. Circles indicate polymorphic sites in the nvd gene (orange) and in neighboring loci (blue). Line thickness is proportional to the number of samples with the indicated haplotype. (C) Representation of the genotypes of 34 individuals. Black bars indicate heterozygote positions. Homozygote sites for rare alleles are not shown. (D) Position of the sequenced loci within the nvd region. Gene annotations are in orange.
A likely scenario is that D. pachea first evolved a resistance towards senita cactus toxic compounds and slowly became restricted to this food source as it escaped competition with other fly species. Evolution of D. pachea’s resistance most likely did not involve NVD since nvd is not expressed in the midgut and fat body (fig. S3), the detoxification organs in insects (16). As lathosterol became D. pachea’s unique source of sterols for steroid hormone synthesis, mutations in nvd that abolished NVD activity on cholesterol appeared and fixed rapidly due to their beneficial effect with lathosterol. As a result, D. pachea became an obligate specialist on the senita cactus. We point out that besides nvd mutations, mutation(s) in other genes might also have contributed to D. pachea dependence on lathosterol. Alternatively, the identified nvd mutations may have spread while D. pachea ancestors were still feeding on various plants and may thus have accelerated its ecological specialization. Our study, which uncovered several mutations underlying the obligate bond between a specialist species and its host, illustrates how a few mutations in a single gene can restrict the ecological niche of a species.
Supplementary Material
Acknowledgments
We thank M. Joron for the LD heat map, M.-A. Félix, N. Gompel and C. Desplan for comments on the manuscript, C. Parada for help in field work, T.A. Markow for D. pachea samples, C.S. Thummel and the DSSC for flies, Y. Hiromi for reagents, T. Blasco for LC-MS/MS analyses and M. Gho for hosting VO in 2009–2010. Supported by CNRS ATIP-AVENIR (to VO), French Foreign Ministry post-doctoral fellowship (to ML), NIH grant AI064950 (to AGC), JSPS Japan-France bilateral Cooperating Program (to HK and CDV), NSF award DEB-1020009 (to LMM), JSPS post-doctoral fellowship (to TYY), Special Coordination Funds for Promoting Science and Technology of the MEXT (to RN) and Amylin Endowment (to T.A. Markow). The nvd sequences were deposited in Genbank under accession numbers JF764559 to JF764595 and JX066807 to JX067384.
Footnotes
The authors declare no conflict of interest.
V.O. conceived the study and wrote the paper. M.L., C.B., R.L., C.D.-V., L.M.M., H.K. and R.N. provided technical support and conceptual advice for designing the experiments. V.O., M.L., G.G., S.M., L.M.M., C.B., E.G., T.Y.-Y., C. D.-V. and R.N. performed the experiments. A.G.C. did the population genetics analysis.
Materials and Methods
References and Notes
- 1.Lwoff A. L’Évolution physiologique: Étude des pertes de fonctions chez les microorganismes. Hermann; Paris: 1944. [Google Scholar]
- 2.Romero P, et al. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2005;6:R2. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voet D. Biochemistry. 2. Wiley; Somerset: 1995. [Google Scholar]
- 4.Huang X, Warren JT, Gilbert LI. New players in the regulation of ecdysone biosynthesis. J Genet Genomics. 2008;35:1–10. doi: 10.1016/S1673-8527(08)60001-6. [DOI] [PubMed] [Google Scholar]
- 5.Heed WB, Kircher HW. Unique sterol in the ecology and nutrition of Drosophila pachea. Science. 1965;149:758–761. doi: 10.1126/science.149.3685.758. [DOI] [PubMed] [Google Scholar]
- 6.Campbell CE, Kircher HW. Senita cactus: a plant with interrupted sterol biosynthetic pathways. Phytochemistry. 1980;19:2777–2779. [Google Scholar]
- 7.Fogleman JC, Danielson PB. Chemical Interactions in the Cactus-Microorganism-Drosophila Model System of the Sonoran Desert. American Zoologist. 2001;41:877–889. [Google Scholar]
- 8.Information on materials and methods is available on Science Online.
- 9.Yoshiyama T, Namiki T, Mita K, Kataoka H, Niwa R. Neverland is an evolutionally conserved Rieske-domain protein that is essential for ecdysone synthesis and insect growth. Development. 2006;133:2565–2574. doi: 10.1242/dev.02428. [DOI] [PubMed] [Google Scholar]
- 10.Rottiers V, et al. Hormonal control of C. elegans dauer formation and life span by a Rieske-like oxygenase. Dev Cell. 2006;10:473–482. doi: 10.1016/j.devcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Pitnick S, Heed WB. New species of cactus-breeding Drosophila (Diptera: Drosophilidae) in the Nannoptera species group. Ann Entomol Soc Am. 1994;87:307–310. [Google Scholar]
- 12.Lafont R, Dauphin-Villemant C, Warren JT, Rees HH. In: Comprehensive Molecular Insect Science. Gilbert LI, Iatrou K, Gill S, editors. Vol. 3. Oxford: 2005. pp. 125–195. [Google Scholar]
- 13.Blais C, Blasco T, Maria A, Dauphin-Villemant C, Lafont R. Characterization of ecdysteroids in Drosophila melanogaster by enzyme immunoassay and nano-liquid chromatography-tandem mass spectrometry. J Chromatogr B. 2010;878:925–932. doi: 10.1016/j.jchromb.2010.02.018. [DOI] [PubMed] [Google Scholar]
- 14.Goodnight KC, Kircher HW. Metabolism of lathosterol by Drosophila pachea. Lipids. 1971;6:166–169. doi: 10.1007/BF02533031. [DOI] [PubMed] [Google Scholar]
- 15.Yoshiyama-Yanagawa T, et al. The conserved Rieske oxygenase DAF-36/Neverland is a novel cholesterol metabolizing enzyme. J Biol Chem. 2011;286:25756–62. doi: 10.1074/jbc.M111.244384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mittapalli O, et al. Tissue-Specific Transcriptomics of the Exotic Invasive Insect Pest Emerald Ash Borer (Agrilus planipennis) PLoS ONE. 2010;5:e13708. doi: 10.1371/journal.pone.0013708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldlaufer MF, Weirich GF, Imberski RB, Svoboda JA. Ecdysteroid production in Drosophila melanogaster reared on defined diets. Insect Biochem Mol Biol. 1995;25:709–712. doi: 10.1016/0965-1748(95)00009-k. [DOI] [PubMed] [Google Scholar]
- 18.Timmons L, et al. Green fluorescent protein/β-galactosidase double reporters for visualizing Drosophila gene expression patterns. Developmental Genetics. 1997;20:338–347. doi: 10.1002/(SICI)1520-6408(1997)20:4<338::AID-DVG5>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 19.Huang AM, Rehm EJ, Rubin GM. Recovery of DNA sequences flanking P-element insertions in Drosophila: inverse PCR and plasmid rescue. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.prot5199. pdb.prot5199. [DOI] [PubMed] [Google Scholar]
- 20.Rutherford K, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 21.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 23.Yang Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Molecular Biology and Evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 24.Wessner M, et al. Ecdysteroids from Ajuga iva. Phytochemistry. 1992;31:3785–3788. [Google Scholar]
- 25.Girault JP, et al. Ecdysteroids from Leuzea carthamoides. Phytochemistry. 1988;27:737–741. [Google Scholar]
- 26.Niwa R, et al. CYP306A1, a cytochrome P450 enzyme, is essential for ecdysteroid biosynthesis in the prothoracic glands of Bombyx and Drosophila. J Biol Chem. 2004;279:35942–35949. doi: 10.1074/jbc.M404514200. [DOI] [PubMed] [Google Scholar]
- 27.Warren JT, et al. Molecular and biochemical characterization of two P450 enzymes in the ecdysteroidogenic pathway of Drosophila melanogaster. Proc Natl Acad Sci USA. 2002;99:11043–11048. doi: 10.1073/pnas.162375799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roth GE, et al. The Drosophila gene Start1: a putative cholesterol transporter and key regulator of ecdysteroid synthesis. Proc Natl Acad Sci USA. 2004;101:1601–1606. doi: 10.1073/pnas.0308212100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christian AE, Haynes MP, Phillips MC, Rothblat GH. Use of cyclodextrins for manipulating cellular cholesterol content. J Lipid Res. 1997;38:2264–2272. [PubMed] [Google Scholar]
- 30.Lavrov S, Déjardin J, Cavalli G. Combined immunostaining and FISH analysis of polytene chromosomes. Methods Mol Biol. 2004;247:289–303. doi: 10.1385/1-59259-665-7:289. [DOI] [PubMed] [Google Scholar]
- 31.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson JT, et al. Integrative genomics viewer. Nature Biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drummond A, et al. Geneious. 2011 http://www.geneious.com/
- 35.Dmitriev DA, Rakitov RA. Decoding of superimposed traces produced by direct sequencing of heterozygous indels. PLoS Comput Biol. 2008;4:e1000113. doi: 10.1371/journal.pcbi.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reich D, Price AL, Patterson N. Principal component analysis of genetic data. Nat Genet. 2008;40:491–492. doi: 10.1038/ng0508-491. [DOI] [PubMed] [Google Scholar]
- 39.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 40.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 41.Wright SI, Charlesworth B. The HKA test revisited: a maximum-likelihood-ratio test of the standard neutral model. Genetics. 2004;168:1071–1076. doi: 10.1534/genetics.104.026500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hudson RR, Kreitman M, Aguadé M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim Y, Nielsen R. Linkage disequilibrium as a signature of selective sweeps. Genetics. 2004;167:1513–1524. doi: 10.1534/genetics.103.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim Y, Stephan W. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics. 2002;160:765–777. doi: 10.1093/genetics/160.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavlidis P, Alachiotis N. OmegaPlus 2.0.0 software. 2012 doi: 10.1093/bioinformatics/bts419. available at http://sco.h-its.org/exelixis/software/OmegaPlus_Manual.pdf. [DOI] [PubMed]
- 46.Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 47.Sabeti P. Sweep software. available at http://www.broadinstitute.org/mpg/sweep/index.html.
- 48.Zaykin DV, Pudovkin A, Weir BS. Correlation-based inference for linkage disequilibrium with multiple alleles. Genetics. 2008;180:533–545. doi: 10.1534/genetics.108.089409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin J-H, Blay S, McNeney B, Graham J, Dheatmap L. An R Function for Graphical Display of Pairwise Linkage Disequilibria Between Single Nucleotide Polymorphisms. Journal of Statistical Software. :16. available at http://rgm2.lab.nig.ac.jp/RGM2/func.php?rd_id=LDheatmap:LDheatmap.
- 50.Nishi S, Nishino H, Ishibashi T. cDNA cloning of the mammalian sterol C5-desaturase and the expression in yeast mutant. Biochim Biophys Acta. 2000;1490:106–108. doi: 10.1016/s0167-4781(99)00248-1. [DOI] [PubMed] [Google Scholar]
- 51.Vinci G, Xia X, Veitia RA. Preservation of genes involved in sterol metabolism in cholesterol auxotrophs: facts and hypotheses. PLoS ONE. 2008;3:e2883. doi: 10.1371/journal.pone.0002883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schaeffer SW, et al. Polytene chromosomal maps of 11 Drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics. 2008;179:1601–1655. doi: 10.1534/genetics.107.086074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 54.Filion GJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoskins RA, et al. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science. 2007;316:1625–1628. doi: 10.1126/science.1139816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mount SM, et al. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 1992;20:4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yasuhara JC, Wakimoto BT. Oxymoron no more: the expanding world of heterochromatic genes. Trends Genet. 2006;22:330–338. doi: 10.1016/j.tig.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 58.Yasuhara JC, DeCrease CH, Wakimoto BT. Evolution of heterochromatic genes of Drosophila. Proc Natl Acad Sci USA. 2005;102:10958–10963. doi: 10.1073/pnas.0503424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schulze SR, et al. Heterochromatic genes in Drosophila: a comparative analysis of two genes. Genetics. 2006;173:1433–1445. doi: 10.1534/genetics.106.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mackay TFC, et al. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482:173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kulathinal RJ, Bennett SM, Fitzpatrick CL, Noor MAF. Fine-scale mapping of recombination rate in Drosophila refines its correlation to diversity and divergence. Proc Natl Acad Sci USA. 2008;105:10051–10056. doi: 10.1073/pnas.0801848105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clark AG, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 63.Ward BL, Heed WB. Chromosome phylogeny of Drosophila pachea and related species. J Hered. 1970;61:248–258. doi: 10.1093/oxfordjournals.jhered.a108095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.