Abstract
DNA methylation is a chemical modification that occurs predominantly on CG dinucleotides in mammalian genomes. However, recent studies have revealed that non-CG methylation (mCH) is abundant and nonrandomly distributed in the genomes of pluripotent cells and brain cells, and is present at lower levels in many other human cells and tissues. Surprisingly, mCH in pluripotent cells is distinct from that in brain cells in terms of sequence specificity and association with transcription, indicating the existence of different mCH pathways. In addition, several recent studies have begun to reveal the biological significance of mCH in diverse cellular processes. In reprogrammed somatic cells, mCH marks megabase-scale regions that have failed to revert to the pluripotent epigenetic state. In myocytes, promoter mCH accumulation is associated with the transcriptional response to environmental factors. In brain cells, mCH accumulates during the establishment of neural circuits and is associated with Rett syndrome. In this review, we summarize the current understanding of mCH and its possible functional consequences in different biological contexts.
Keywords: DNA methylation, epigenetics, somatic cell reprogramming, neural development, Rett syndrome
1. Introduction
DNA methylation is an ancient covalent chemical modification of genomic DNA, providing an additional layer of regulatory information on top of the genome sequence (8). This mechanism is present in a variety of organisms, including numerous mammals, plants, fungi, insects, protists, archaebacteria, and eubacteria (19, 24, 25, 48, 55, 56, 59, 72, 81, 91, 102, 108, 110). In mammals, DNA methylation occurs predominantly at cytosine followed by guanine, commonly referred to as CG methylation (mCG). mCG has been extensively studied over the past several decades; it has been found to be involved in transcriptional regulation (47, 91) and biological processes such as cellular differentiation (9, 28, 48, 56, 106) and X-chromosome inactivation (78, 79), and to be associated with diseases such as rheumatoid arthritis (58), neurological disorders (1, 20, 66, 73), and cancer (22, 41, 45). By contrast, methylation at cytosines followed by bases other than guanine, referred to as non-CG methylation (mCH, where H = A, C, or T), was long thought to be an artifact of the methodologies used to detect methylation, until the existence of this DNA modification was finally confirmed in mice (76) and humans (56).
Cytosine methylation can be readily detected at single-base resolution using conventional Sanger sequencing or high-throughput next-generation whole-genome sequencing methods that are coupled to sodium bisulfite conversion of unamplified DNA (19, 26, 55). Sodium bisulfite treatment leads to the deamination of unmethylated cytosine to uracil, whereas methylated cytosine does not undergo this chemical conversion. In this procedure, regulatory information in the form of an epigenetic chemical modification is thus converted to a genetic sequence variant, which can be captured by standard sequencing technologies or other assay methods. Several characteristics of mCH make reliable detection difficult (74). First, mCH is not present in most somatic cells, and only limited types of cells have been found to contain mCH (Figure 1). Second, because it is frequently mingled with the extensive genome-wide mCG, high-resolution methylation assays such as bisulfite sequencing are required to distinguish it from the ubiquitous mCG found in mammalian cells. Finally, for each non-CG site, only a moderate fraction of alleles are methylated in the cell population. Consequently, mCH was dismissed as an artifact of incomplete conversion of unmethylated cytosines during bisulfite treatment.
Figure 1.
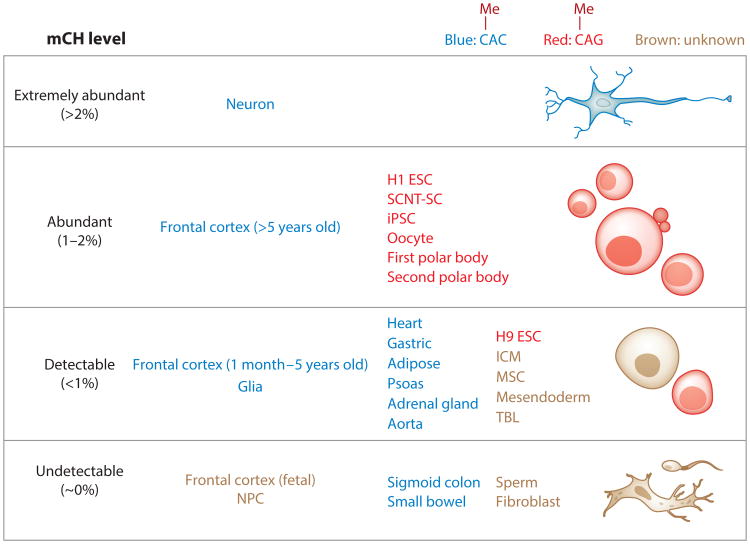
mCH abundance in human cells and tissues. To estimate the abundance, genome-wide mCH levels (83) were calculated as previously described (56) using whole-genome bisulfite sequencing data. Based on these mCH levels, cells and tissues were then divided into four categories: extremely abundant (>2%), abundant (1–2%), detectable (<1%), and undetectable (∼0%). Note that cells and tissues labeled as undetectable have mCH levels that are very close to the background bisulfite nonconversion rate. In the last two categories (detectable and undetectable), only representative cells and tissues are shown, whereas the first two categories (extremely abundant and abundant) contain all qualified cells and tissues based on current knowledge. These cells and tissues are also grouped into three categories by whether the observed mCH occurs primarily in a CAC sequence context (blue) or a CAG sequence context (red) or the sequence preference is unknown (brown). Data for neurons, glia, and frontal cortex are from Lister et al. (54); whole-genome bisulfite sequencing data for H1 ESCs, MSCs, NPCs, TBLs, mesendoderm, and fibroblasts are from Lister et al. (56) and Xie et al. (106); methylome data for H9 ESCs are from Laurent et al. (48); data for SCNT-SCs and iPSCs are from Lister et al. (57) and Ma et al. (61); data for the sigmoid colon, small bowel, heart, gastric system, adipose tissue, psoas, adrenal gland, and aorta are from Schultz et al. (82); and data for the remaining samples (sperm, oocytes, polar bodies, and ICM) are from Guo et al. (31). Abbreviations: ESC, embryonic stem cell; ICM, inner cell mass; iPSC, induced pluripotent stem cell; mCH, non-CG methylation; MSC, mesenchymal stem cell; NPC, neural progenitor cell; SCNT-SC, somatic cell nuclear transfer stem cell; TBL, trophoblast-like cell.
The development of rapid and cost-effective sequencing technologies (65) has enabled accurate genome-wide profiling of mCH at single-base resolution across a large number of mammalian cells and tissues (56, 64, 75, 82, 98, 112, 113). During the past decade, studies have found that numerous human cell and tissue types contain significant amounts of mCH, including embryonic stem cells (ESCs) (48, 56), somatic cell nuclear transfer stem cells (SCNT-SCs) (61), induced pluripotent stem cells (iPSCs) (57, 61), oocytes (31), neurons (54), and glia (54).
Although mCH is present in various cell and tissue types, evidence for its potential function is just beginning to emerge (4, 5, 15, 27, 32, 34, 57, 61, 71), and its true biological role is still unclear and controversial (3, 23, 63, 71, 86, 90). The DNA methylation profile of human ESCs revealed a strong correlation between mCH and adjacent mCG, suggesting that the presence of mCH may be the result of hyperactivity of nonspecific de novo methylation that is simply a tolerated by-product of mCG pathways (90, 113). However, subsequent observations argue against this hypothesis. For example, the level of mCH in the promoter of the peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) gene is higher in type 2 diabetes mellitus (T2DM) patients than in healthy individuals, and is associated with a reduction in the transcription of the PGC-1α gene and fewer mitochondria in T2DM patients (5). In addition, large domains of non-CG hypomethylation were discovered in SCNT-SCs and iPSCs compared with ESCs, corresponding to regions that failed to be reprogrammed at the epigenetic and transcriptional levels (57, 61). Furthermore, in brain cells, gene body mCH is associated with transcriptional repression (32) and can dramatically enhance the binding of methyl-CpG binding protein 2 (MeCP2) (32), a protein critical to the cause of Rett syndrome (14, 36), indicating an association between mCH and this neurological disorder. This association was tested in the mouse model of Rett syndrome, and MeCP2 binding to mCH was suggested to be important for the transcriptional regulation of genes related to neurological function (15, 27).
Given the presence of mCH in various mammalian cell types and its potential functional impact, it is necessary to more deeply understand this specific DNA modification. In this review, we first describe the prevalence of mCH in human cell and tissue types by examining the abundance and sequence specificity of mCH. We next summarize the mechanisms and related proteins in the encoding, decoding, and removal of mCH in the genome. Then, we discuss the genomic features targeted by mCH and the potential functional impact. In the final sections, we review recent efforts toward revealing the roles of mCH in critical biological processes, including somatic cell reprogramming, brain development, and diseases such as diabetes and Rett syndrome. Although this review focuses primarily on studies with human cells, it includes results from mouse models as additional supporting evidence.
2. Non-CG Methylation in Human Cells and Tissues
mCH appears in a variety of human cells and tissues, including different pluripotent cells, female germ cells, neurons, and glia. The genome-wide abundance of mCH varies dramatically in different cells and tissues (Figure 1) and can be estimated by computing the genome-wide mCH level. This quantity is determined by calculating the weighted methylation level of all non-CG sites (i.e., the average methylation level of each non-CG site weighted by the coverage at each site) (83). Specifically, the genome-wide mCH level (M) is defined as
where N is the total number of non-CG sites and mi and ci are the number of reads supporting methylation and the number of total reads, respectively, at non-CG site i; mi/ci is the methylation level at non-CG site i (i.e., mCH level at site i), corresponding to the fraction of methylated cytosines out of the total number of cytosines in reads covering this site. Among all sites (including both CG and non-CG sites), a fraction of them are methylated at statistically significantly higher levels than those that might be attributed to the artifacts caused by incomplete bisulfite conversion. These sites are referred to as methylated sites. Methylated CG sites generally have a higher methylation level than methylated non-CG sites. For example, in ESCs, the majority of mCH sites have a methylation level of approximately 20%, whereas the majority of mCG sites have a methylation level of 100% (56).
These metrics enable robust investigation and comparison of the mCH abundances of various human cells and tissues. mCH can be detected in pluripotent cells (e.g., ESCs), female germ cells, brain cells, and cells from skeletal muscle tissue, whereas most of the somatic cells are depleted of mCH. For example, the genome of H1 (male) human ESCs is heavily methylated at non-CG sites: 25% of methylated sites are in a non-CG context, and the genome-wide mCH level is approximately 1% (56). Although the genome-wide mCH level in H1 ESCs is much lower relative to the mCG level (approximately 80%), a significant fraction (25%) of methylated sites are in the non-CG context because the human genome has approximately 20 times more non-CG sites than CG sites. This indicates that mCH is abundant in the genome. Some genomic regions, such as gene bodies, are enriched for mCH and have much higher mCH levels than the rest of the genome (for a detailed discussion, see Section 4). Interestingly, compared with male-derived ESCs, H9 and other female-derived human ESCs are genome-wide hypomethylated in both the CG and non-CG contexts (48, 56, 114), possibly because of lower expression of DNA methyltransferases (DNMTs), which are responsible for the deposition of de novo methylation (114). Similarly to male ESCs, pluripotent cells generated by SCNT (94) or iPSC (95) techniques show high mCH levels (56, 57, 61, 113). Genomes of cell lines that have been differentiated from H1 ESCs into distinct cell types show lower mCH levels (i.e., mCH is not maintained upon differentiation) (106).
Human female germ cells, oocytes, and polar bodies are also highly enriched for mCH (31). By contrast, mCH is not detected in either human (31) or mouse (101) sperm. Mouse studies have shown that mCH is abundant in premitotic prospermatogonia (43, 100) and is then lost during mitotic cell divisions owing to the lack of mCH maintenance, explaining the lack of mCH in sperm (43). After fertilization, only the maternal alleles are non-CG methylated, and the mCH level substantially decreases as the cell begins to divide (37, 97, 101).
Human and mouse adult brain tissues also show genome-wide mCH (32, 54, 98, 105). Although mCH is virtually absent in the genome of the human fetal frontal cortex, the cells accumulate significant mCH levels as this brain area matures (54). Cells isolated from brain show very distinct mCH levels: Glia contain mCH at a similar level as male ESCs, whereas neurons have the highest recorded mCH level, approximately 6% (54).
mCH is not restricted to pluripotent cells, oocytes, and brain cells. It has also been reproducibly detected in human primary myocytes, skeletal muscle tissue, adipose tissue, adrenal glands, the aorta, the gastric system, the heart, and the psoas (4, 5, 82, 98), whereas many somatic cell types seem to lack detectable mCH (48, 56, 57, 61, 98, 113). However, because only a limited number of cell and tissue types have been profiled by single-base-resolution whole-genome bisulfite sequencing, knowledge of the prevalence of mCH in the human body remains incomplete. Figure 1 summarizes the cells and tissues that are known to lack or contain mCH and groups them based on their genome-wide mCH levels calculated from whole-genome bisulfite sequencing data.
Although mCH is present in various cell and tissue types, the sequence context in which it lies is distinct. Specifically, in ESCs it occurs primarily (78% of the methylated CH sites) at cytosines in a 5′CAG3′ context, referred to as mCAG (meaning that the first cytosine is methylated) (48, 56), whereas in neurons the dominant sequence context is 5′CAC3′ (47% of the methylated CH sites, whereas mCAG takes 20%), referred to as mCAC (54, 105). Based on the sequence preference, cells and tissues that contain mCH can be grouped into two categories: The mCAG group consists of pluripotent cells (ESCs, SCNT-SCs, and iPSCs), oocytes, and polar bodies, whereas the mCAC group consists of the frontal cortex, neurons, glia, and several human organs (Figure 1). Intriguingly, the correlation between gene body mCH and gene transcription is tightly associated with the mCH sequence specificity (32, 54, 56, 105). In cells from the mCAG group, intragenic mCH is moderately positively correlated with gene expression (the Pearson correlation coefficient is approximately 0.6), whereas a negative correlation is observed in cells and tissues from mCAC group expression (31, 32, 54, 56, 82, 105). Although mCH occurs at both CAG and CAC sites regardless of the sequence preference, and methylation in these two sequence contexts is moderately correlated genome-wide (56, 82, 105), the distinct associations with transcription imply the existence of at least two different mCH pathways.
3. Non-Cg Methylation Writers, Readers, and Erasers
3.1. Writers
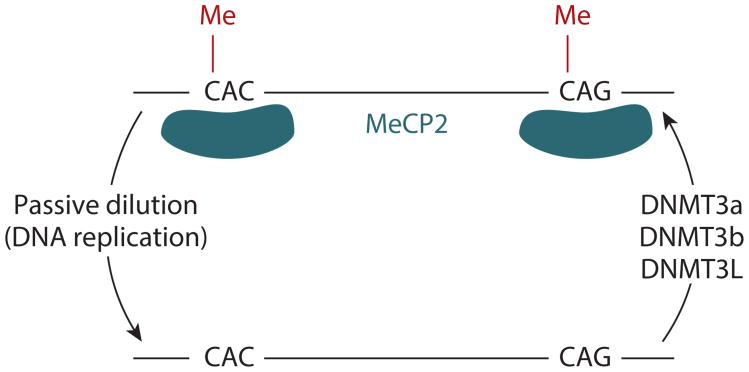
In mammals, DNA methylation is established and maintained by the coordinated activity of three DNMTs (49). mCG patterns are established by the de novo methyltransferases DNMT3a and DNMT3b. In addition, a catalytically inactive DNMT3 family member, DNMT3L, plays a crucial role in methylation establishment specifically during gametogenesis (12, 39, 80). Once established, patterns are transmitted through S phase by the maintenance methyltransferase DNMT1 (10, 85). DNMT1 maintains mCG on newly replicated DNA (10, 85). Biochemical studies indicate that it has much higher activity on hemimethylated oligonucleotides compared with unmethylated oligonucleotides (28a). DNMT1 also strongly prefers CG sites and has almost no activity on non-CG sites, implying that it is not the major enzyme responsible for mCH (29, 92). In line with these results, a knockout of DNMT1 in ESCs had little effect on mCH levels (3, 76), although this type of methylation is relatively rare or absent in many differentiated cell types.
DNMT3a preferentially methylates CG sites as well, but in contrast to DNMT1, non-CG sites (especially CA sites) can also be methylated at lower levels than CG sites by this enzyme (2, 28a, 29, 92). Moreover, introducing DNMT3a into Drosophila leads to both mCG and mCH, demonstrating the enzymes' in vivo ability to methylate non-CG sites (76). This experiment also showed that DNMT3a methylates CA sites with tenfold less preference compared with CG sites. Genetic studies of DNMT3a deficiency and knockdown support its important role in the establishment and maintenance of mCH in ESCs (3, 52, 64, 113), oocytes (87), and neurons (27, 32). Additionally, in vivo DNA-binding sites of DNMT3a in brain tissue are enriched for mCH (32, 54), indicating the in vivo dependency of mCH on DNMT3a.
mCH (primarily at CT and CA sites) has also been attributed to Dnmt3b (2, 92), and it is critical for the formation and maintenance of mCH in ESCs. DNMT3b deletion resulted in a dramatic reduction of mCH in human ESCs (3, 52). DNMT3b partially compensates for the function of DNMT3a in mCH, as knockdown of DNMT3b following DNMT3a deletion leads to a further reduction in mCH in ESCs (113). A recent study revealed that a gene knockout of DNMT3b results in a more striking reduction of mCH compared with a knockout of DNMT3a, indicating that DNMT3b is likely the major enzyme for mCH in human ESCs (52). In brain, however, DNMT3b is expressed at extremely low levels (54). Thus, it seems unlikely to play a major role in the formation of mCH in brain cells (54). In human primary myocytes, DNMT3b is indispensable for the palmitate-induced gain of mCH in the promoter of the PGC-1α locus, supporting its role in the transcriptional response to the environment (5).
DNMT3L, although catalytically inactive, interacts with DNMT3a and DNMT3b, functioning as a targeting factor and allosteric regulator (40, 46, 93). DNMT3L is highly expressed in ESCs, prospermatogonia, and growing oocytes, and its deletion results in almost complete loss of mCH (3, 43, 87), likely resulting from the disruption of the DNMT3a/b-DNMT3L complex and the later dramatic reduction of de novo methylation activity. More specifically, in mouse prospermatogonia, the ATRX-Dnmt3-Dnmt3L (ADD) domain in the Dnmt3L protein is essential for the establishment of mCH in the genome (100). The ADD domain is responsible for recognizing histone H3, and disrupting this interaction is sufficient to eliminate mCH, suggesting that the ability of Dnmt3L to bind to histone H3 is critical for mCH in mouse prospermatogonia (100). Surprisingly, a recent study showed that DNMT3L knockdown in human embryonic carcinoma cells leads to increased mCH (96), which contradicts the observations in germ cells and ESCs (3, 43, 87, 100). This result suggests the possibility that DNMT3L plays different roles in the mCH pathway in different cells, although it remains to be validated in follow-up studies. DNMT3L expression has not been observed in brain cells, suggesting that it is not involved in mCH in this tissue. Collectively, DNMT3a, DNMT3b, and the cofactor DNMT3L are responsible for the mCH in various cell and tissue types, but the presence of all three is not necessary for the mCH in every cell or tissue type (Figure 2).
Figure 2.
Proteins and mechanism responsible for establishing, recognizing, and removing mCH. DNMTs, including DNMT3a, DNMT3b, and DNMT3L, are part of the complex responsible for the establishment and maintenance of mCH. MeCP2 is able to recognize non-CG-methylated regions. A non-CG demethylase has not yet been identified, and it is speculated that passive dilution (resulting from DNA replication and lack of maintenance of mCH) is the mechanism underlying the loss of mCH in dividing cells. Abbreviations: DNMT, DNA methyltransferase; mCH, non-CG methylation; MeCP2, methyl-CpG binding protein 2.
Although DNMT3a and DNMT3b are capable of methylating cytosines in a non-CG context, it is not fully understood how they target specific regions in the genome to establish the nonrandom mCH landscape (described in Section 4). Nucleosome positioning, which affects the accessibility of DNA by DNMTs, may account for the observed ∼180-base-pair periodicity of mCH (6, 32). Also, histone modifications on genomic regions can either recruit or exclude DNMTs (13), which may determine the genomic distribution of mCH. For example, histone H3 with unmethylated lysine 4 can be recognized by the DNMT3a-DNMT3L complex (69, 70) and alter its de novo methylation activity (35). By contrast, the complex can neither bind nor be activated by histone H3 with methylated lysine 4 (H3K4me3), which protects the H3K4me3-marked regions from de novo methylation (35, 69, 70). Moreover, actively transcribed genes are enriched in mCH in ESCs (56) (for details, see Section 4.2), raising the question of whether transcription results in intragenic mCH, or mCH is deposited to regulate transcription. It has been suggested that the binding and activity of DNMT3s can be instructed by histone H3 with trimethylated lysine 36 (H3K36me3), the histone modification that marks the bodies of actively expressed genes (6, 21), which might be one mechanism of intragenic mCH deposition. A recent study by Baubec et al. (6) in mouse ESCs demonstrated that Dnmt3b can recognize H3K36me3 in vivo, whereas gene transcription alone is unable to recruit Dnmt3b. This finding implies that intragenic mCH (possibly deposited by Dnmt3b) may be an indirect result of transcription, and it also provides a possible explanation of the positive correlation between mCH in the gene body and transcription in human ESCs (56).
However, the models discussed above account only for the inclusion and exclusion of DNMT3a/b, and direct evidence that histone modifications affect mCH deposition is still limited. Furthermore, the negative correlation in brain between intragenic mCH and gene transcription cannot be explained by these models, indicating that the mechanisms of mCH deposition may be different in brain cells and that components such as mCH readers (discussed below) are likely missing (in current models).
3.2. Readers
Several proteins whose binding is affected by the methylation state of non-CG sites have recently been identified, and these proteins are referred to as the readers of mCH. MeCP2 is a DNA-binding protein that recognizes CG-methylated DNA, and mutations in the MeCP2 gene can lead to Rett syndrome (36). Guo et al. (32) showed that MeCP2 also binds to mCH-containing DNA. They discovered through an in vitro experiment that MeCP2 is able to bind to methylated non-CG sites in oligonucleotides with lower-affinity binding than to oligonucleotides methylated at CG sites. However, the combined presence of mCH and mCG on the same oligonucleotide molecule greatly enhanced the binding affinity of MeCP2. Additionally, chromatin immunoprecipitation of MeCP2 followed by high-throughput sequencing confirmed that MeCP2 in vivo binding sites are enriched for both mCG and mCH (32). Together, these results imply that the genome-wide binding landscape of MeCP2 can be shaped by both mCH and mCG profiles (Figure 2). Several recent studies have confirmed the in vitro binding of MeCP2 to mCH (15, 27). Furthermore, a study by Gabel et al. (27) revealed that MeCP2 has significantly higher affinity for mCA than for other mCH contexts. However, whether MeCP2 has different binding affinities for mCAC and mCAG remains to be studied.
Considering that MeCP2 can function as a transcriptional repressor (67), its binding may link mCH and mCG to transcriptional repression, which would partly explain the negative correlation between gene transcription and intragenic mCH in brain (32). In line with this, deleting MeCP2 in mouse brain resulted in upregulation of genes that are typically longer than 100 kb, which tended to be highly non-CG methylated (27). However, it was also shown that mutations of MeCP2 in neurons led to a global decrease in transcription, implying that MeCP2 plays a role as a global transcriptional activator (51), although these effects could also be indirect. In addition, a recent study found that when MeCP2 is depleted in mouse brain, both upregulated and downregulated genes are enriched in mCH, suggesting that MeCP2 may function differently in different sets of genes (15). The contradictory evidence indicates that more studies are required to elucidate the actual effect of MeCP2 binding, especially by teasing out the direct and indirect effects. In addition to MeCP2, the binding of methyl-CpG binding protein 2b (MBD2b) can be enhanced by the co-occurrence of mCG and mCH (32), whereas the binding of Sp-family transcription factors is reduced by mCH within their binding sites (44).
3.3. Erasers
Active and passive mechanisms have been proposed in which a cell (or cell lineage) can lose DNA methylation (103). Passive demethylation accounts for the loss of mCH through dilution in dividing cells when the maintenance by de novo mCH is less active, because it is likely that there is not a replication-dependent mechanism for mCH maintenance similar to the one by which DNMT1 maintains mCG (10, 85). Active demethylation would involve a chemical process that selectively removes cytosine methylation. Currently, there is little evidence about active non-CG demethylases.
One mechanism of active mCH removal may involve the recently discovered ten-eleven translocation (TET) methylcytosine dioxygenases (103). The TET enzymes are able to oxidize 5-methylcytosine (5mC) to 5-hydroxymethyl cytosine (5hmC) in vitro regardless of the sequence context (33, 42, 107). However, Tet-assisted bisulfite sequencing (TAB-seq) (107) detected almost no 5hmC at non-CG sites in ESCs and brain cells (33, 107). The absence of 5hmC at non-CG sites may simply be due to its rapid turnover, which is supported by the observation that TET1 has higher activity at methylated non-CG sites than at CG sites (33). An alternative explanation is that at the time when mCH appears in these cells, TETs are not active at methylated non-CG sites owing to the presence of unknown cofactors that modulate the specificity of TETs. Other active demethylation mechanisms (such as DNA repair and activation-induced cytosine deaminase–dependent pathways) are possible candidates for non-CG demethylation in human cells, but they are either restricted to certain biological conditions or not fully demonstrated (103). Because of the lack of a known active demethylation mechanism for non-CG sites, it is speculated that the reduction of mCH in dividing cells is exclusively due to the lack of constitutive de novo methylation activity to maintain the existing methylation patterns during DNA replication, which leads to the passive dilution of mCH when these enzymes are not active, i.e., passive demethylation (Figure 2). There is currently no evidence for TET-dependent demethylation on non-CG sites in postmitotic cells such as neurons (54), possibly explaining why these contain abundant mCH.
4. Genome-Wide Distribution of Non-CG Methylation
In cells and tissues where mCH is present, this form of DNA methylation is prevalent throughout the genome. The genome-wide distribution of mCH is neither uniform nor random but is greatly enriched in several genomic features, including gene bodies (31, 32, 105), repeat elements (3, 34, 113), and inactive enhancers (60), and it tends to be absent in active enhancers (56), promoters (31, 48, 56, 105), transcription factor binding sites (56), and mCH deserts, which are megabase-scale regions that have been recently identified in brain (54) (Figure 3). In addition, mCH preferentially occurs in regions of low CG dinucleotide density (32). Given that CG dinucleotides are enriched in CG islands and generally depleted in the rest of the genome (47), mCH may complement mCG in regions with low CG density. Furthermore, single mCH sites have been shown to be conserved between biological replicates, indicating that mCH establishment and maintenance are tightly regulated events: High concordance was observed when comparing, at single cytosines, mCH in neurons from two different individuals (54), and in human ESCs, although a high variation of genome-wide mCH was reported, highly methylated non-CG sites are conserved in different ESC lines (16, 113). These reproducible nonrandom patterns of mCH serve as clues about how mCH is regulated as well as its functional consequences in cellular processes. In the following sections, we summarize mCH patterns in different genomic contexts and discuss their potential functional impacts on various biological processes.
Figure 3.
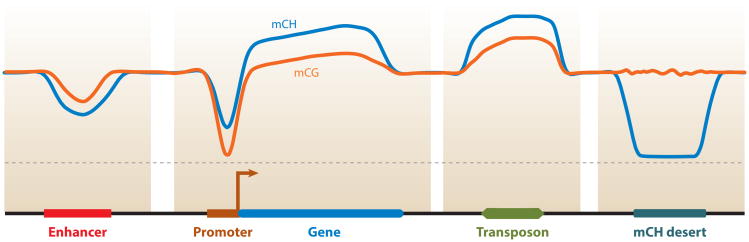
Average mCH and mCG levels reported at enhancers, promoters, genes, transposons, and a recently identified mCH desert (54). The scale is schematic, and intergenic mCH and mCG levels have been scaled to be the same level. Abbreviations: mCG, CG methylation; mCH, non-CG methylation.
4.1. Promoters and Enhancers
mCH levels are generally lower in promoters than in surrounding regions (31, 48, 56, 105) (Figure 3). The depletion of mCH in promoters may be explained by the presence of unmethylated promoter-proximal CG islands. To maintain the unmethylated state of CG islands, DNMT3a and DNMT3b are constantly excluded from these regions by competitive binding of transcription factors such as FBLX10 (11), removal of nucleosomes, recruitment of H3K4 methyltransferases, and possibly active transcription (90). Consequently, lack of de novo methylation activity in CG islands results in the exclusion of mCH. The promoters that lack CG islands are also depleted of mCH (48), which may be due to the exclusion of the de novo methylation complex by the H3K4me3 in these regions (13, 70). Although mCH levels in promoters are generally low, high levels of promoter mCH are able to repress transcription (4, 5, 44). The repressive effect of promoter mCH may be due to its influence on protein-DNA interactions. By assessing the interaction between proteins and methylated oligonucleotides, Inoue & Oishi (44) showed that mCH can reduce the binding of certain transcription factors, which results in decreased transcription.
Similar to promoters, active enhancers in H1 ESCs are non-CG hypomethylated at CH sites compared with upstream and downstream regions, appearing as a valley of unmethylated CH sites (56) (Figure 3). Similar mCH profiles were also observed in the binding sites of the embryonic transcription factors (56). Inactive enhancers instead are enriched for mCH (60). These results indicate that mCH is closely correlated with the activity of enhancers, but whether mCH has a regulatory role is unclear.
4.2. Genes
mCH is more abundant in gene bodies than in intergenic regions (31, 32, 56, 105), and this intragenic mCH is closely correlated with gene transcription (31, 54, 56, 105). In human ESCs, mCH levels are higher in exons than in introns (56). In addition, mCH is highly enriched at splice sites owing to the presence of CAG in the canonical acceptor sequence (16, 48). The distinctive distribution of mCH between exons and introns and the presence of mCH at splice sites imply that a relationship may exist between alternative splicing and mCH, which remains to be elucidated and validated (16, 48). One possible explanation is that the different mCH levels in exons and introns are due to the different nucleosome positioning patterns (17). Another interesting observation in ESCs is that gene body mCH does not occur evenly in both strands; the antisense strand is non-CG hypermethylated (56). A recent study showed that this phenomenon derives from the skewed frequency of methylation-prone motifs (5′ACA3′) on the two strands (34). The functional consequences of this strand-specific mCH in the gene body remain unclear.
Intragenic mCH patterns were also found in human brain cells. Lister et al. (54) found that the relative abundance of mCH in gene bodies compared with flanking regions was closely related to gene expression patterns and was informative about gene function. For example, in neurons, intragenic mCG and mCH hypermethylation was observed in genes associated with glial function, whereas in glia, the bodies of these genes tended to be mCG and mCH hypomethylated (54). In addition, Lister et al. (54) identified hundreds of genes enriched for neuronal and synaptic development and function that had higher gene body mCH levels in glia than in neurons, indicating the distinct gene body mCH patterns in different cell types. Lister et al. (54) also discovered that the genes that escape X chromosome inactivation showed higher intragenic mCH in female brains compared with male brains. This female-specific higher intragenic mCH is speculated to compensate for the failure of repression.
A recent study (82) revealed that the findings from Lister et al. (54)—e.g., that tissue-specific intragenic CH hypomethylation is associated with tissue function—also held true for multiple nonbrain tissues that contain mCH, although the mCH abundance is much lower in these tissues. In addition to the mCH patterns in normal tissues, it was found that the bodies of genes misregulated in brains with Rett syndrome (MeCP2 mutations) were highly non-CG methylated and were bound by the mCH-binding protein MeCP2, highlighting the potential functional role of intragenic mCH in this neurological disease (15, 27) (for more details, see Section 8).
4.3. Transposons and Non-CG Methylation Deserts
Transposable elements are also heavily non-CG methylated (3, 34). In human ESCs, the highest mCH levels were observed in short interspersed element (SINE) repeats (113). In addition, abundant mCH was found in long interspersed nuclear element 1 (LINE1) and intracisternal A particle (IAP) elements in mouse prospermatogonia (100). Similar to gene bodies, the bodies of transposons are strand-specific non-CG methylated owing to differences in sequence motif (5′ACA3′) frequencies between the sense and antisense strand (34). The mCH at transposons is dependent on DNMT3a, DNMT3b, and DNMT3L, indicating that these regions are under constitutively de novo methylation (3, 100).
mCH deserts are megabase-scale regions in which mCH is depleted but mCG remains present (54). This mCH feature was identified in both human and mouse frontal cortex, but whether this pattern is unique to brain cells is still unknown. The chromatin state of mCH deserts shows a high degree of inaccessibility (54), which is hypothesized to prevent the entrance of the de novo methylation complex and thus prevent the establishment of mCH in these regions. Olfactory receptor genes and immunoglobulin genes are enriched in mCH deserts, which may provide the inaccessible chromatin state that may be required for the monoallelic expression of these genes (18, 62, 99).
5. Non-CG Methylation in Somatic Cell Reprogramming
Reprogramming differentiated somatic cells into pluripotent cells has immense potential in regenerative medicine. To fully regain pluripotency, somatic cells are required to correctly reconfigure the epigenome so that it resembles the ESC state (63). Restoring the ESC-like mCH landscape may be important in obtaining high-quality pluripotent cells. iPSCs generated by overexpressing a defined set of embryonic transcription factors (95) successfully restore the abundant genome-wide mCH observed in ESCs, but certain regional patterns of mCH are not faithfully restored (57).
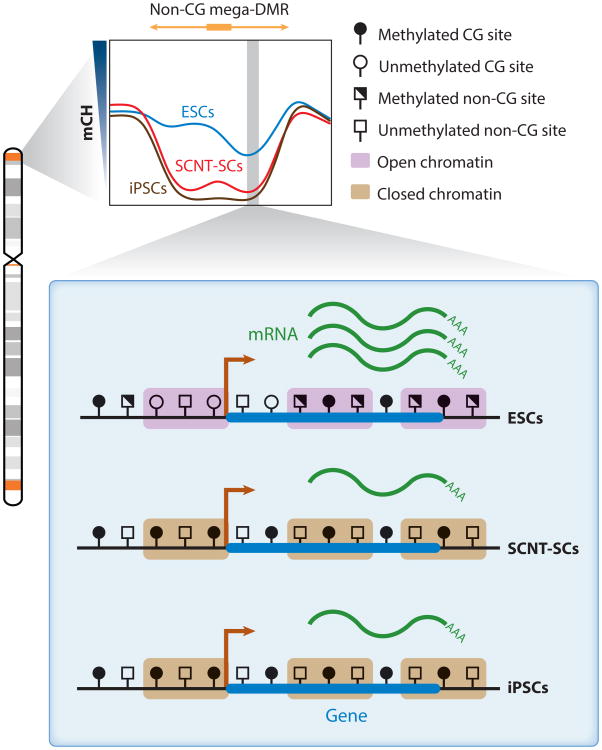
Lister et al. (57) identified non-CG megabase-scale differentially methylated regions (non-CG mega-DMRs) where mCH was not properly reestablished; in these regions, they observed a much lower mCH abundance in iPSCs than in ESCs (Figure 4). The disruption of mCH coincided with a dramatic increase of a heterochromatin histone mark (H3K9me3), whereas the transcription of genes in these regions was repressed (57). Many of the non-CG mega-DMRs observed in iPSCs lie within so-called partially (CG) methylated domains of the parental fibroblast (57), which coincide with lamina-associated domains that are late replicating (7). Moreover, the locations of non-CG mega-DMRs in iPSCs overlapped with regions of the genome that acquired dense H3K9me3 at later stages of passage after primary cells were put in a culture environment that contained growth stimuli such as serum (111). These results imply that the culture condition of the iPSCs may contribute to the aberrant reprogramming caused by a “memory” (lack of erasure) of stress-induced chromatin accumulation in the parental cells. In a more recent study, Ma et al. (61) compared the methylation landscape of SCNT-SCs (94) with that of ESCs. Non-CG mega-DMRs were also identified in SCNT-SCs, and similarly to iPSCs, transcription was disrupted in these regions (61) (Figure 4). Although the role of mCH in the reprogramming process is still unclear, these two studies demonstrate its potential as a marker for regions that are resistant to epigenetic and transcriptional reprogramming, a measurement for reprogramming completeness, and a metric for the quality of pluripotent cells generated by SCNT or iPSC approaches.
Figure 4.
Non-CG mega-DMRs in ESCs, SCNT-SCs, and iPSCs (57, 61). The non-CG mega-DMRs are shown in orange in the chromosome diagram and tend to be proximal to telomeres and centromeres (64, 76); the gray shades are cytogenetic bands. The vast majority of non-CG mega-DMRs are hypomethylated in SCNT-SCs and iPSCs compared with ESCs (shown in the mCH profile of non-CG mega-DMRs). In SCNT-SCs and iPSCs, the heterochromatin mark H3K9me3 is enriched in these regions. In addition, the transcription of genes within non-CG mega-DMRs is disrupted, and the promoters of these genes are typically CG hypermethylated in SCNT-SCs and iPSCs compared with ESCs. Abbreviations: ESC, embryonic stem cell; H3K9me3, histone H3 with trimethylated lysine 9; iPSC, induced pluripotent stem cell; mCH, non-CG methylation; mega-DMR, megabase-scale differentially methylated region; SCNT-SC, somatic cell nuclear transfer stem cell.
6. Non-CG Methylation in Diabetes and Obesity
Growing evidence suggests that chromatin factors are involved in the incidence of T2DM and may mediate the complex interaction between genetic variants, environment, and gene expression (53). Barrès et al. (5) identified mCH as one of the critical factors involved in diabetes. They examined the methylation patterns in muscle tissue from a cohort of T2DM, impaired glucose-tolerant, and normal glucose-tolerant individuals. They discovered that the T2DM and impaired glucose-tolerant subjects had significantly higher mCH in the promoter of the PGC-1α gene compared with normal glucose-tolerant subjects, whereas the number of CG sites was limited in the promoter regions and their methylation states were not significantly altered. They did not observe methylation alteration in the promoters of nearby genes, indicating that the PGC-1α promoter's non-CG hypermethylation was not a result of increased mCH in a large region. PGC-1α is a critical gene involved in the regulation of mitochondria biogenesis (104). The non-CG hypermethylation of its promoter was accompanied by lower transcription and a dramatic (approximately 50%) reduction of mitochondrial DNA content.
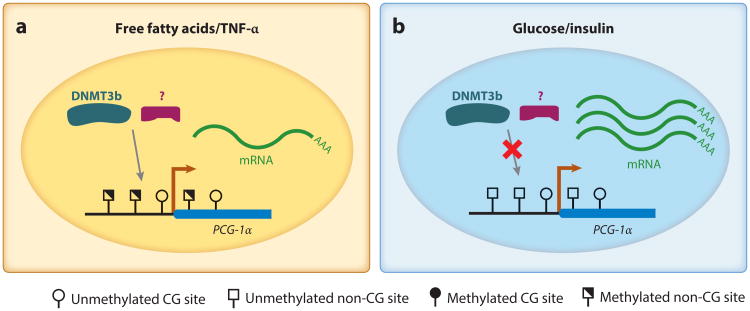
Barrès et al. (5) further studied the environmental factors that may induce non-CG hypermethylation. They found that in human primary myocytes, the mCH in the PGC-1α promoter can be induced by a high concentration of free fatty acids, such as palmitate and oleate or cytokine tumor necrosis factor α (TNF-α), all of which were present at high levels in diabetic individuals. By contrast, neither a high concentration of glucose nor the addition of insulin induced mCH in this region (Figure 5). A genome-wide methylation assay showed that the palmitate treatment on human primary myocytes induced an increase of mCH levels across the genome, indicating that under this treatment the mCH in the PGC-1α promoter is a result of the genome-wide increased methylation at non-CG sites (5). They also found that this induction is mediated by DNMT3b but not by DNMT1 or DNMT3a. Similar to the observation in the cohort study, palmitate-induced mCH was associated with a reduction of mitochondria content. In summary, these results indicate that a rapid increase of mCH may mediate transient gene repression as a response to certain environmental cues.
Figure 5.
mCH in the promoter of the PGC-1α gene. In human primary myocytes, mCH can be induced by free fatty acids and TNF-α(left), both of which are elevated in type 2 diabetes mellitus patients compared with healthy individuals. Under normal culture conditions and glucose or insulin treatment, the promoter of PGC-1α has a low mCH level (right). The induced non-CG hypermethylation is associated with a reduction of gene expression and a further decrease in the number of mitochondria. The induction of mCH is mediated by DNMT3b and potentially other unknown proteins or cofactors of DNMT3b. Abbreviations: DNMT3b, DNA methyltransferase 3b; mCH, non-CG methylation; PGC-1α, peroxisome proliferator-activated receptor γ coactivator 1α; TNF-α, tumor necrosis factor α.
Barrès et al. (4) observed that mCH alteration is also associated with obesity. This study compared the methylation patterns in skeletal muscle from normal-weight subjects and obese subjects before and after Roux-en-Y gastric bypass surgery. The comparison revealed that, similarly to the T2DM subjects, the PGC-1α promoter is non-CG hypermethylated in obese individuals compared with normal-weight individuals, and this is accompanied by a lower transcription level. After the surgery, obese individuals lost weight and the mCH level of the PGC-1α promoter was reduced to levels similar to those of healthy controls, associated with an increase of transcription. A similar restoration of promoter mCH and transcription patterns was found in PDK4, a gene involved in mitochondrial function. The study further assessed the activity of an in vitro non-CG-methylated PDK4 promoter by luciferase assay and confirmed that the mCH is sufficient to repress transcription in vivo. Collectively, these observations support the hypothesis that promoter mCH can be affected by changes in physiological conditions and mCH alterations can directly regulate the expression of specific genes.
7. Non-CG Methylation in Brain Development
The genomes of adult brain cells are highly non-CG methylated (54, 98, 105). However, the developmental stage at which mCH appears in brain was previously unknown. Following the trajectory of frontal cortex development, Lister et al. (54) discovered that mCH is barely detectable in the human fetal frontal cortex, but its abundance in neurons dramatically increases in the early years of postnatal brain life and reaches a plateau as the frontal cortex reached maturity. This developmental process is not restricted to the frontal cortex, having also been observed in mouse dentate gyrus by Guo et al. (32). The accumulation of mCH coincides with the developmental increase of synapse density and later synaptic pruning, but whether there is a causative relationship remains obscure (54). Lister et al. (54) further profiled the methylomes of purified neurons and glia, providing insights into the brain cell type–specific patterns of mCH. Neurons were found to have the highest recorded mCH level in the human body (Figure 1). In glia, mCH was also present, but its levels were lower and more similar to those observed in the frontal cortex between one month and five years of age (54) (Figure 1).
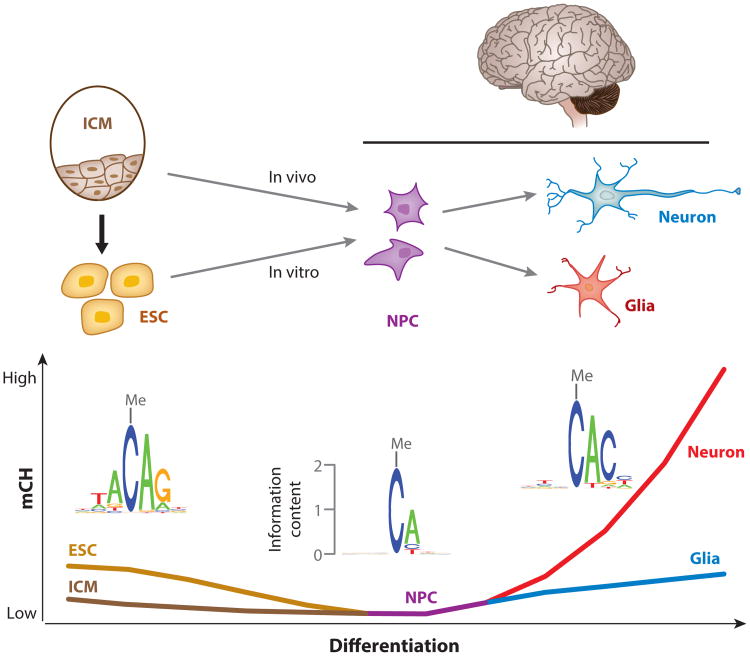
In contrast to the high mCH levels in neurons, neural progenitor cells (NPCs) in vitro derived from human ESCs are depleted of mCH (106). In line with this, the fetal frontal cortex, which contains abundant NPCs (38), also lacks mCH (54). The high mCH levels in both pluripotent cells (56) and neurons (54), together with the depletion of mCH in NPCs (106), imply that the mCH landscape undergoes a reconfiguration during neural differentiation. In vivo, NPCs are differentiated from inner cell mass (ICM) cells (Figure 6). ICM cells initially show low levels of mCH (31) that then disappear as they differentiate to NPCs, although it is unclear whether mCH is abundant in the intermediate cell states. At the next stage, NPCs further differentiate into neurons and glia, in which new mCH patterns are established (54). Neurons can also be generated from ESCs via in vitro differentiation (77, 109) (Figure 6). During this process, presumable passive loss of mCH takes place when ESCs differentiate to NPCs, resulting in the depletion of mCH in NPCs (106), and these NPCs can further differentiate into neurons and glia (77, 109). However, because methylation data for ESC-derived neurons are not available, it is unclear whether similar mCH patterns are established in these cells, as in the neurons isolated from brain tissue. Collectively, although the non-CG reconfiguration during in vitro neural differentiation has not been fully demonstrated owing to the lack of methylation data for in vitro differentiated neurons, recent evidence suggests that the reconfiguration occurs during in vivo neural differentiation, when the mCH present in ICM cells becomes depleted in NPCs, and the methylation is then reestablished in neurons and glia.
Figure 6.
Hypothesized reconfiguration of mCH patterns during neural differentiation. Abundant mCH is present in ESCs derived from ICM cells, which show low levels of mCH. During the in vitro differentiation process from ESCs to NPCs, non-CG demethylation takes place, and genome-wide mCH is lost in NPCs. When NPCs differentiate into neurons and glia, the genome-wide mCH could possibly be reestablished in in vitro differentiated neurons and glia, but methylation measurements to support this hypothesis are lacking. The absence of mCH in NPCs and the reestablishment of mCH in neurons and glia during in vivo neural differentiation are supported by methylation data (54), but whether mCH is absent in intermediate cells between ICM cells and NPCs is unclear. In addition, the fact that mCH occurs primarily at CAG sites in ESCs but at CAC sites in both neurons and glia implies that an interesting switch of mCH pathways occurs during neural differentiation. Abbreviations: ESC, embryonic stem cell; ICM, inner cell mass; mCH, non-CG methylation; NPC, neural progenitor cell.
In addition to the change in mCH levels, the pathways to establish and maintain mCH may be distinct because mCH occurs primarily at CAC sites in neurons and glia but at CAG sites in ESCs, which are derived from ICM cells (Figure 6). The difference in sequence specificity indicates that during the reestablishment of mCH in neurons and glia, the new patterns are not simply regenerated by copying and amplifying those in ESCs. Furthermore, despite the extremely similar sequence specificity of mCH in neurons and glia, differential mCH was identified in the bodies of hundreds of genes that are enriched for synapse function (54). Although the biological significance of this mCH reconfiguration is still obscure, the differences in mCH patterns among ESCs, neurons, and glia indicate that de novo methylation at non-CG sites is tightly regulated by restricting the activity to specific cytosines in a unique sequence context and to a set of selected regions in the genome.
The abundant mCH in neurons raises the question of whether this base modification has functional consequences. To gain deeper mechanistic insight into the regulatory role of mCH, Guo et al. (32) in vitro methylated the non-CG sites in a plasmid and showed that mCH alone can repress transcription as effectively as mCG. Furthermore, they discovered that mCH can be recognized by MeCP2, a protein whose mutations lead to Rett syndrome (14). They also showed that the reduction in intragenic mCH resulting from Dnmt3a knockdown (by injecting adeno-associated viruses expressing short hairpin RNAs targeting Dnmt3a) in adult mouse dentate gyrus increased transcription of mCH-enriched genes but not that of mCH-depleted genes, supporting the notion of mCH as a repressive chromatin modification. Xie et al. (105) studied the allelic mCH patterns in imprinted regions in mouse brain, and in line with the above findings, they found that the silenced allele has higher levels of mCH than the active allele. Although these observations support the repressive influence of mCH on gene transcription in brain cells, the exact mechanism remains to be elucidated.
8. Non-CG Methylation in Rett Syndrome
The observations that (a) mCH can be recognized and bound by MeCP2, (b) mCH accumulates as the brain matures, (c) disrupting MeCP2 leads to Rett syndrome, and (d) a brain with Rett syndrome is normal in the early development stage raise the possibility that MeCP2 binding to mCH is critical for neurological function and that the late appearance of mCH in the brain may partly explain the delayed onset of Rett syndrome (15, 27). Two recent studies have tested this hypothesis (15, 27). Chen et al. (15) profiled the MeCP2 binding landscape in the hypothalamus of adult mouse and found that MeCP2 preferentially binds genes with high mCH. Disrupting or overexpressing MeCP2 led to the misregulation of these highly non-CG-methylated genes, whereas the transcription of genes with low mCH remained unchanged under these conditions. By contrast, little mCG difference was observed between the two sets of genes, suggesting that the MeCP2 binding to mCH rather than mCG in these genes is important for their transcription regulation.
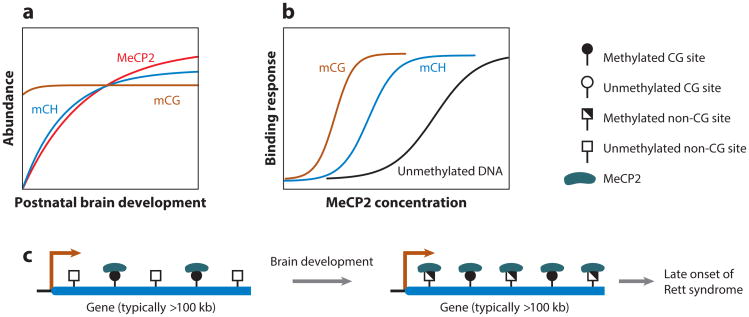
Based on these findings about the functionality of mCH and MeCP2 interaction, Chen et al. (15) proposed a model to explain the functionality of mCH and the delayed onset of Rett syndrome (Figure 7). During brain development, the abundance of mCH and MeCP2 increases dramatically, whereas that of mCG remains approximately the same (32, 54, 84, 88) (Figure 7). As the amount of both mCH and MeCP2 increases in mature brain, the bodies of some genes begin to accumulate mCH and are then bound by the highly abundant MeCP2, which regulates the transcription. Because the mCH- and MeCP2-dependent gene regulation starts only in adult brain where both components are present, this model is able to at least partly explain the delayed onset of Rett syndrome. By contrast, mCG in gene bodies is bound by MeCP2 in both immature and mature brain owing to the constantly high level of mCG and high binding affinity of MeCP2 to it.
Figure 7.
mCH in Rett syndrome. (a) As the brain matures, the abundances of mCH (blue) and MeCP2 (red) increase dramatically, while the abundance of mCG (brown) remains stable. (b) The binding affinity of MeCP2 to mCH (blue), mCG (brown), and unmethylated DNA (black) varies under different MeCP2 protein concentrations. The scales of panels a and b are schematic. (c) In immature brain, MeCP2 binds only to mCG sites owing to both a low mCH level and a low MeCP2 concentration. In mature brain, the increased mCH level and MeCP2 concentration enable the binding of (extra) MeCP2 at mCH sites in the bodies of genes that typically have a length greater than 100 kb and are enriched for neurological function (27). Based on this model, only in mature brain does disrupting MeCP2 function or concentration result in misregulation of long genes that is severe enough to cause a neurological disorder. The prototype of this model was proposed by Chen et al. (15), and the gene length feature of MeCP2-regulated genes was found by Gabel et al. (27). Abbreviations: mCG, CG methylation; mCH, non-CG methylation; MeCP2, methyl CpG binding protein 2. Adapted from figure 6 of Reference 15.
Evidence from Gabel et al. (27) also supports this model. The authors found that in the cortexes of patients with Rett syndrome, long genes (length greater than 100 kb) rather than short genes tended to be misregulated (specifically, upregulated). They further found that in the cortexes and cerebellums of mice, long genes are bound by MeCP2 and are highly CA methylated (the major form of mCH recognized by MeCP2). Deleting the Dnmt3a gene during early brain development or disrupting MeCP2 led to the misregulation of long genes, indicating that mCA and MeCP2 are essential for their normal expression. Furthermore, they showed that only long genes with high mCH (rather than short genes with high mCH or long genes with low mCH) tended to be misregulated in the brain where MeCP2 was disrupted. This observation demonstrates that both long gene length and high mCH are essential features of genes regulated by MeCP2 binding, which adds one feature (gene length) to the current model of mCH gene regulation in adult brain (Figure 7).
Embryonic deletion of Dnmt3a affected the accumulation of both mCH and mCG (27) and produced brain malformations that resulted in early postnatal death (68). These confounding effects prevent examination of the effect of specific loss of mCH on animal behavior. Future studies that create postnatal deletions of Dnmt3a at precisely the time of mCH accumulation may allow investigators to assess the role of this epigenetic mark on synapse formation during brain development as well as its role in complex behaviors and diseases such as Rett syndrome.
9. Perspectives
In this review, we have summarized recent efforts to understand the biological significance of mCH. Many questions remain unanswered:
Our understanding of the prevalence of mCH in human cell types is far from complete. Although the human methylome repertoire is growing rapidly, many cells and tissues remain uncharacterized. Furthermore, even if tissues or cell mixtures are found to contain mCH, the exact cell types enriched for mCH are yet to be pinpointed (e.g., neurons are known to be enriched in mCH, but the abundance of mCH in specific types of neurons is unknown). At the single-cell level, we still do not know how much the mCH landscape varies within cells from the same tissue. Is mCH mosaic in the genome but present in every cell, or does it appear only in a subset of cells of that cell type? Or is it a combination of both? These questions may be resolved by the recently developed single-cell bisulfite sequencing technique (23a, 30, 89).
The mechanisms that determine the existence and shape of the mCH landscape are still obscure. It is speculated that abundant DNMT3a/b (high de novo methylation activity) in nondividing cells might be sufficient to add mCH to the genome. This hypothesis is supported by results observed from overexpressing Dnmt3a in Drosophila (76), but in human cells, direct evidence is still limited. Moreover, although recent studies have revealed that histone modifications are able to direct the binding of the de novo methylation complex (13, 35, 69, 70), whether these events directly contribute to mCH deposition remains to be uncovered.
The direct functional consequences of mCH remain to be demonstrated. Currently, the interaction between mCH and transcription factor binding has only been tested for a limited number of transcription factors (44). Systematic screening is needed to reveal the effect (if any) of mCH on the binding of human transcription factors, especially the neuronal and pluripotent transcription factors, as these are the contexts in which mCH seems to be the most active. Moreover, mCH in brain cells, as shown by an in vitro experiment, has the capacity to repress gene transcription (32), but whether this result was a direct effect of mCH or the repression was mediated by proteins that recognize mCH is unknown. Additionally, the fact that mCG has been shown to change the shape of DNA (50) leaves open the possibility that mCH may also have an effect on chromatin structure, but the function remains to be investigated.
Because of our incomplete understanding of mCH and its absence in most somatic cells, the clinical utility of knowing the variance in mCH levels or distribution is unknown. However, its presence has been found to be associated with obesity and diabetes (4, 5). mCH may also serve as an indicator of the degree of reprogramming and extent of epigenetic memory of iPSCs (57, 61). Furthermore, several recent studies have begun to demonstrate the association between mCH and Rett syndrome (15, 27, 32). These studies hint at the possible future applications of mCH as a biomarker and highlight the need for further investigations of this poorly understood DNA modification.
Acknowledgments
We thank Chongyuan Luo, Robert J. Schmitz, Eran A. Mukamel, M. Margarita Behrens, Matthew D. Schultz, Ryan O'Neil, Manoj Hariharan, and Mark Zander for their critical comments and helpful discussions. Y.H. is supported by the H.A. and Mary K. Chapman Charitable Trust. J.R.E. is an Investigator of the Howard Hughes Medical Institute and is supported by grants from the Gordon and Betty Moore Foundation (GBMF3034), the National Institutes of Health (R01 MH094670 and U01 MH105985), and the California Institute for Regenerative Medicine (GC1R-06673-B).
Footnotes
The Annual Review of Genomics and Human Genetics is online at genom.annualreviews.org
Disclosure Statement: The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Yupeng He, Email: yuhe@salk.edu.
Joseph R. Ecker, Email: ecker@salk.edu.
Literature Cited
- 1.Aberg KA, McClay JL, Nerella S, Clark S, Kumar G, et al. Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71:255–64. doi: 10.1001/jamapsychiatry.2013.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki A, Suetake I, Miyagawa J, Fujio T, Chijiwa T, et al. Enzymatic properties of de novo-type mouse DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 2001;29:3506–12. doi: 10.1093/nar/29.17.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arand J, Spieler D, Karius T, Branco MR, Meilinger D, et al. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLOS Genet. 2012;8:e1002750. doi: 10.1371/journal.pgen.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrès R, Kirchner H, Rasmussen M, Yan J, Kantor FR, et al. Weight loss after gastric bypass surgery in human obesity remodels promoter methylation. Cell Rep. 2013;3:1020–27. doi: 10.1016/j.celrep.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 5.Barrès R, Osler ME, Yan J, Rune A, Fritz T, et al. Non-CpG methylation of the PGC-1α promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10:189–98. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520:243–47. doi: 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- 7.Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2012;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 9.Bock C, Beerman I, Lien WH, Smith ZD, Gu H, et al. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell. 2012;47:633–47. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–64. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 11.Boulard M, Edwards JR, Bestor TH. FBXL10 protects Polycomb-bound genes from hypermethylation. Nat Genet. 2015;47:479–85. doi: 10.1038/ng.3272. [DOI] [PubMed] [Google Scholar]
- 12.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–39. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 13.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 14.Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–37. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Chen L, Chen K, Lavery LA, Baker SA, Shaw CA, et al. MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. PNAS. 2015;112:5509–14. doi: 10.1073/pnas.1505909112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen PY, Feng S, Joo JW, Jacobsen SE, Pellegrini M. A comparative analysis of DNA methylation across human embryonic stem cell lines. Genome Biol. 2011;12:R62. doi: 10.1186/gb-2011-12-7-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–92. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, et al. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–37. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–19. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20:4786–96. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, et al. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem. 2010;285:26114–20. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–53. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dyachenko OV, Schevchuk TV, Kretzner L, Buryanov YI, Smith SS. Human non-CG methylation: Are human stem cells plant-like? Epigenetics. 2010;5:569–72. doi: 10.4161/epi.5.7.12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Farlik M, Sheffield NC, Nuzzo A, Datlinger P, Schönegger A et al. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 2015;10:1386–97. doi: 10.1016/j.celrep.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, et al. Conservation and divergence of methylation patterning in plants and animals. PNAS. 2010;107:8689–94. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foret S, Kucharski R, Pellegrini M, Feng S, Jacobsen SE, et al. DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. PNAS. 2012;109:4968–73. doi: 10.1073/pnas.1202392109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. PNAS. 1992;89:1827–31. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabel HW, Kinde B, Stroud H, Gilbert CS, Harmin DA, et al. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015;522:89–93. doi: 10.1038/nature14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–63. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Gowher H, Jeltsch A. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: The enzyme modifies DNA in a non-processive manner and also methylates non-CpA sites. J Mol Biol. 2001;309:1201–8. doi: 10.1006/jmbi.2001.4710. [DOI] [PubMed] [Google Scholar]
- 29.Gowher H, Jeltsch A. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J Biol Chem. 2002;277:20409–14. doi: 10.1074/jbc.M202148200. [DOI] [PubMed] [Google Scholar]
- 30.Guo H, Zhu P, Wu X, Li X, Wen L, Tang F. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 2013;23:2126–35. doi: 10.1101/gr.161679.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo H, Zhu P, Yan L, Li R, Hu B, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–10. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- 32.Guo JU, Su Y, Shin JH, Shin J, Li H, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci. 2014;17:215–22. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–34. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo W, Chung WY, Qian M, Pellegrini M, Zhang MQ. Characterizing the strand-specific distribution of non-CpG methylation in human pluripotent cells. Nucleic Acids Res. 2014;42:3009–16. doi: 10.1093/nar/gkt1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo X, Wang L, Li J, Ding Z, Xiao J, et al. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature. 2014;517:640–44. doi: 10.1038/nature13899. [DOI] [PubMed] [Google Scholar]
- 36.Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annu Rev Cell Dev Biol. 2011;27:631–52. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- 37.Haines TR, Rodenhiser DI, Ainsworth PJ. Allele-specific non-CpG methylation of the Nf1 gene during early mouse development. Dev Biol. 2001;240:585–98. doi: 10.1006/dbio.2001.0504. [DOI] [PubMed] [Google Scholar]
- 38.Hansen DV, Lui JH, Flandin P, Yoshikawa K, Rubenstein JL, et al. Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat Neurosci. 2013;16:1576–87. doi: 10.1038/nn.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–93. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 40.Holz-Schietinger C, Reich NO. The inherent processivity of the human de novo methyltransferase 3A (DNMT3A) is enhanced by DNMT3L. J Biol Chem. 2010;285:29091–100. doi: 10.1074/jbc.M110.142513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hovestadt V, Jones DT, Picelli S, Wang W, Kool M, et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature. 2014;510:537–41. doi: 10.1038/nature13268. [DOI] [PubMed] [Google Scholar]
- 42.Hu L, Li Z, Cheng J, Rao Q, Gong W, et al. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell. 2013;155:1545–55. doi: 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 43.Ichiyanagi T, Ichiyanagi K, Miyake M, Sasaki H. Accumulation and loss of asymmetric non-CpG methylation during male germ-cell development. Nucleic Acids Res. 2013;41:738–45. doi: 10.1093/nar/gks1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inoue S, Oishi M. Effects of methylation of non-CpG sequence in the promoter region on the expression of human synaptotagmin XI (syt11) Gene. 2005;348:123–34. doi: 10.1016/j.gene.2004.12.044. [DOI] [PubMed] [Google Scholar]
- 45.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–51. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 48.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–31. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–20. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lazarovici A, Zhou T, Shafer A, Dantas Machado AC, Riley TR, et al. Probing DNA shape and methylation state on a genomic scale with DNase I. PNAS. 2013;110:6376–81. doi: 10.1073/pnas.1216822110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, Wang H, Muffat J, Cheng AW, Orlando DA, et al. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell. 2013;13:446–58. doi: 10.1016/j.stem.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao J, Karnik R, Gu H, Ziller MJ, Clement K, et al. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat Genet. 2015;47:469–78. doi: 10.1038/ng.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ling C, Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58:2718–25. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–36. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31:142–47. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lodwick D, Ross HN, Harris JE, Almond JW, Grant WD. dam methylation in the Archaebacteria. J Gen Microbiol. 1986;132:3055–59. doi: 10.1099/00221287-132-11-3055. [DOI] [PubMed] [Google Scholar]
- 60.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–13. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 61.Ma H, Morey R, O'Neil RC, He Y, Daughtry B, et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature. 2014;511:177–83. doi: 10.1038/nature13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–70. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol. 2010;28:1079–88. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- 64.Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–77. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 66.Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–81. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 68.Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev Dyn. 2007;236:1663–76. doi: 10.1002/dvdy.21176. [DOI] [PubMed] [Google Scholar]
- 69.Ooi SKT, Qiu C, Bernstein E, Li K, Jia D, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–17. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, Shirakawa M. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10:1235–41. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patil V, Ward RL, Hesson LB. The evidence for functional non-CpG methylation in mammalian cells. Epigenetics. 2014;9:823–28. doi: 10.4161/epi.28741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peoples OP, Hardman N. An abundant family of methylated repetitive sequences dominates the genome of Physarum polycephalum. Nucleic Acids Res. 1983;11:7777–88. doi: 10.1093/nar/11.22.7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pidsley R, Viana J, Hannon E, Spiers HH, Troakes C, et al. Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biol. 2014;15:483. doi: 10.1186/s13059-014-0483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pinney SE. Mammalian non-CpG methylation: stem cells and beyond. Biology. 2014;3:739–51. doi: 10.3390/biology3040739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Plongthongkum N, Diep DH, Zhang K. Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nat Rev Genet. 2014;15:647–61. doi: 10.1038/nrg3772. [DOI] [PubMed] [Google Scholar]
- 76.Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. PNAS. 2000;97:5237–42. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 78.Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 79.Riggs AD. X chromosome inactivation, differentiation, and DNA methylation revisited, with a tribute to Susumu Ohno. Cytogenet Genome Res. 2002;99:17–24. doi: 10.1159/000071569. [DOI] [PubMed] [Google Scholar]
- 80.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–40. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 81.Schmitz RJ, He Y, Valdes-Lopez O, Khan SM, Joshi T, et al. Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 2013;23:1663–74. doi: 10.1101/gr.152538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schultz MD, He Y, Whitaker JW, Hariharan M, Mukamel EA, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015 doi: 10.1038/nature14465. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schultz MD, Schmitz RJ, Ecker JR. Leveling” the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 2012;28:583–85. doi: 10.1016/j.tig.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY. Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum Mol Genet. 2002;11:115–24. doi: 10.1093/hmg/11.2.115. [DOI] [PubMed] [Google Scholar]
- 85.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–12. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 86.Shin J, Ming GL, Song H. DNA modifications in the mammalian brain. Philos Trans R Soc B. 2014;369:20130512. doi: 10.1098/rstb.2013.0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shirane K, Toh H, Kobayashi H, Miura F, Chiba H, et al. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. PLOS Genet. 2013;9:e1003439. doi: 10.1371/journal.pgen.1003439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–68. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods. 2014;11:817–20. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–20. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 91.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–95. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 92.Suetake I, Miyazaki J, Murakami C, Takeshima H, Tajima S. Distinct enzymatic properties of recombinant mouse DNA methyltransferases Dnmt3a and Dnmt3b. J Biochem. 2003;133:737–44. doi: 10.1093/jb/mvg095. [DOI] [PubMed] [Google Scholar]
- 93.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279:27816–23. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 94.Tachibana M, Amato P, Sparman M, Gutierrez NM, Tippner-Hedges R, et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell. 2013;153:1228–38. doi: 10.1016/j.cell.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 96.Tiedemann RL, Putiri EL, Lee JH, Hlady RA, Kashiwagi K, et al. Acute depletion redefines the division of labor among DNA methyltransferases in methylating the human genome. Cell Rep. 2014;9:1554–66. doi: 10.1016/j.celrep.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tomizawa S, Kobayashi H, Watanabe T, Andrews S, Hata K, et al. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development. 2011;138:811–20. doi: 10.1242/dev.061416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–67. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vettermann C, Schlissel MS. Allelic exclusion of immunoglobulin genes: models and mechanisms. Immunol Rev. 2010;237:22–42. doi: 10.1111/j.1600-065X.2010.00935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vlachogiannis G, Niederhuth CE, Tuna S, Stathopoulou A, Viiri K, et al. The Dnmt3L ADD domain controls cytosine methylation establishment during spermatogenesis. Cell Rep. 2015;10:944–56. doi: 10.1016/j.celrep.2015.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang L, Zhang J, Duan J, Gao X, Zhu W, et al. Programming and inheritance of parental DNA methylomes in mammals. Cell. 2014;157:979–91. doi: 10.1016/j.cell.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wilson GG, Murray NE. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 103.Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–24. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 105.Xie W, Barr CL, Kim A, Yue F, Lee AY, et al. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–31. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–48. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–80. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–19. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 109.Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–33. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 110.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 111.Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–54. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–81. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ziller MJ, Muller F, Liao J, Zhang Y, Gu H, et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLOS Genet. 2011;7:e1002389. doi: 10.1371/journal.pgen.1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zvetkova I, Apedaile A, Ramsahoye B, Mermoud JE, Crompton LA, et al. Global hypomethylation of the genome in XX embryonic stem cells. Nat Genet. 2005;37:1274–79. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]