Abstract
The thiolate side chain of cysteine has a unique functionality that drug hunters and chemical biologists have begun to exploit. For example, targeting cysteine residues in the ATP-binding pockets of kinases with thiol-reactive molecules has afforded increased selectivity and potency to drugs like imbrutinib, which inhibits the oncogene BTK, and CO-1686 and AZD9291 that target oncogenic mutant EGFR. Recently, disulfide libraries and targeted GDP-mimetics have been used to selectively label the G12C oncogenic mutation in KRAS. We reasoned that other oncogenes contain mutations to cysteine, and thus screened the Catalog Of Somatic Mutations In Cancer for frequently acquired cysteines. Here, we describe the most common mutations and discuss how these mutations could be potential targets for cysteine-directed personalized therapeutics.
Keywords: covalent drugs, personalized medicine, drug target, cancer, cysteine
Personalized cancer therapeutics
Conventional cancer treatment often comprises a combination of surgery, radiation therapy and/or chemotherapy [1]. Personalized cancer treatment aims to select chemotherapeutics based on the genetic profile of a specific tumor. In many cases, this approach selects drugs based on the upregulation of a pathway in the tumor, but the drug does not necessarily discriminate between the protein expressed in healthy vs tumor tissue. For example, the antibody trastuzumab and the kinase inhibitor lapatinib are used to treat tumors with amplified HER2 [2], but they also target HER2 in healthy tissues. Drugs that selectively target a specific mutant form of an oncogene go one step further, because the wild type tissue should remain unaffected. Of the top ten cancer drugs in the USA in 2014, only Imatinib (Gleevec) predominantly targets a disease-driving mutant protein [3], the Bcr-Abl fusion that is exclusively present in cancer cells positive for the Philadelphia chromosome. Another recent example is vemurafenib, which targets the V600E mutant of the oncogenic kinase BRAF [4]. To make the development of mutant-selective compounds commercially viable, the targeted mutations should occur frequently or should be easily targetable. Ideally, mutations should also be linked to tumor fitness, to reduce the risk of resistance mutations.
Cysteine-directed covalent drugs
Another trend in cancer drug discovery aims to increase the potency and selectivity of small-molecule drugs by introducing a cysteine-targeting element, which covalently links the drug to its target [5,6]. When drugs bind their target covalently, the off-rate is negligible compared to that of a non-covalent drug and therefore these drugs would have a prolonged therapeutic effect. But the same irreversible nature of these compounds potentially increases the severity of off-target effects, leading the pharmaceutical industry to be highly wary of screening programs that include covalent drugs or drug metabolites. However, recent work has shown that this idea should be reevaluated [5]. For example, two of the most widely prescribed cancer drugs - the proteasome inhibitor bortezomib and the CYP17 inhibitor abiraterone acetate - bind their target covalently (for more examples see [7]).
Cysteine contains a redox-sensitive thiol whose special reactivity is often utilized in enzyme active sites. However, non-catalytic cysteine residues can also be found on the surface of proteins, sometimes near active or allosteric sites, and these residues can be targeted with thiol-reactive molecules that contain an electrophilic functionality. For example, kinases represent a large and highly homologous set of targets for cancer drug discovery; starting with the insight that epidermal growth factor receptor (EGFR) kinase contains a cysteine residue poised near the ATP-binding site, several groups have developed inhibitors that gain selectivity by forming a covalent bond with a specific non-conserved cysteine around the active site [6]. The Food and Drug Administration (FDA) awarded “breakthrough drug” status to cysteine-directed covalent inhibitors that target BTK (Bruton’s tyrosine kinase) and a drug-resistant EGFR mutant. Imbrutinib forms a reversible-covalent bond with a cysteine near the active site of BTK (Bruton’s tyrosine kinase) and is successfully used to treat B-cell cancers [8]. Both AZD9291 and CO-1686 form a covalent bond specifically with Cys797 in the EGFR T790M mutant, with minimal activity to wild type EGFR. The latter is of importance because the EGFR T790M mutation confers resistance to other EGFR inhibitors and is often acquired upon treatment [9–11]. Cysteine directed, irreversible and reversible covalent binders of FGFR were also recently reported [12,13]. Additionally, the cyclin-dependent kinase-7 (CDK7) inhibitor THZ1 binds irreversibly through an acrylamide moiety to Cys312 of CDK7, which is located in the proximity of the kinase domain [14] Thus, both irreversible and reversible cysteine-modifying groups have been used. Reversible covalent cysteine directed drugs have the potential advantage that they are less likely to form long-lived covalent adducts with off-targets, thereby increasing specificity and perhaps reducing toxicity [15].
Acquired cysteines in cancer
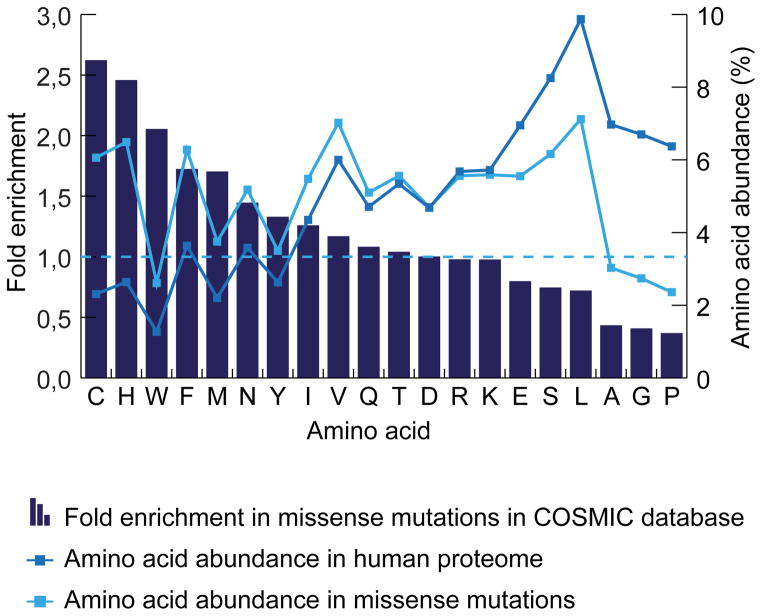
Cancer-associated mutations that give rise to cysteines would combine the above key features of drug discovery: specificity for an oncogenic allele and on-target potency gained through covalent modification. With this idea in mind, we screened the Catalog of Somatic Mutations in Cancer [16] and found that several known oncogenic targets have acquired cysteines. The fifteen most common acquired cysteines in cancer were found to be present in KRAS (2x), FGFR3 (4x), TP53 (3x), IDH1, GNAS, FBXW7, CTNNB1 (2x) and DNMT3. The acquired cysteine is often not the only oncogenic mutation found in these proteins. For instance, acquired cysteines make up 12% of all mutations found in KRAS in cancer, whereas 88% of mutations in FGFR are acquired cysteines, the majority of which are S249C (see Figure 1). Cancer is a mutation-driven disease and, like in evolution, mutations that offer a growth or survival benefit to the tumor cell are selected for and thus are more frequently found mutated in cancer. We can therefore hypothesize that when a certain acquired cysteine is found more often, it plays a role in tumorigenesis. For the purposes of developing a mutation-specific inhibitor, the exact function of the cysteine mutant is not necessarily important, though it may speak to likelihood of generating resistance to a cysteine-dependent inhibitor (as has been observed for EGFR [11]). Interestingly, when looking at all substitution and missense mutations in the COSMIC database, we observed large differences in the rates of acquisition of different amino acids when compared to their relative abundance in the human proteome [17] (see Figure 2). Strikingly, with a fold enrichment of 2.62, cysteine is relatively the most acquired amino acid due to missense mutation in the onco-genome according to the COSMIC database. More information on the structural and functional aspects of this top fifteen acquired cysteines can be found in figure 3 and the supplementary information, which also includes a full list of acquired cysteines in cancer based on the COSMIC database (supplementary table 1).
Figure 1. The most frequently acquired cysteines in cancer.
Pie charts: Percentage of mutations that lead to the acquisition of a cysteine in KRAS, FGFR3, TP53, IDH1, GNAS, FBXW7, CTNNB1 and DNMT3A in cancer based on the COSMIC database. Bars: Number of tumors with a certain acquired cysteine for the 15 most frequently acquired cysteines in cancer in blue. Other acquired cysteines in the selected proteins are shown in grey.
Figure 2. Amino acid abundance.
The abundance of each amino acid in the proteome depends on several factors including codon degeneracy. Relative to its abundance in the human proteome (2,31%), cysteine is found most often (6,06%; a 2,62-fold increase) as a result of a missense mutation in cancer in the COSMIC database.
Figure 3. structure and targetability of acquired cysteines.
Local Protein structure for the top fifteen acquired cysteines in cancer. Structural data was available for KRAS G12C and TP53 Y220C. The other structures are based on the structure of the wild type protein or in case of IDH1 on the R132H mutant. The acquired cysteine was modeled in these structures using the mutagenesis tool in the PyMol software package by selecting a rotamer with the lowest hypothetical steric hindrance. The targetability scores given are based on the interpretation of FTMap analysis and includes the number of, distance to and strength (S) of ligand-binding hot spots in the vicinity of the acquired cysteine [19]. The hotspot parameters can be found in supplementary table 2.
Targeting acquired cysteines
A challenge for the design of cysteine-directed covalent drugs is to make them specific for the desired cysteine in the target protein. Cellular proteins and metabolites contain many other thiols; covalent reactions with these molecules could cause side effects and could hamper availability of the drug for its target. It is therefore key that cysteine-directed drugs are designed so that the electrophile reacts with the cysteine of its target only when it is brought into its proximity [18]; noncovalent associations between the drug and target increase the local residency time and thus increase the selectivity of the chemical reaction. Ideally, a cysteine should be surface exposed but in a pocket or cavity that has distinctive features that can bind to functional groups of the small molecule drug. Furthermore, targeting a certain cysteine should make sense in terms of biological function. For example, targeting the acquired cysteine should inhibit the activity of an oncogenic protein and hence it should be in or near an active or allosteric site. By contrast, covalent modification could stabilize a mutant tumor suppressor such as p53. We modeled the top fifteen most acquired cysteines in cancer based on the structure of the mutant or wild type protein to predict whether these cysteines appear to be near a binding pocket. Interestingly, 14 of the 15 cysteine residues appear on the protein surface; TP53 Y163C is the only acquired cysteine that is not surface exposed. Based on FTMap analysis [19] of the structural data, the most promising targets among the top fifteen acquired cysteines seem to be FBXW7 R465C, KRAS G12C and G13C, IDH1 R132C, GNAS R201C, DNMT3a R882C, FGFR3 Y373C and TP53 Y220C (see figure 3). The CTNNB1 S33C and S37C as well as the FGFR3 G370C mutations are surface exposed, but are not in a pocket or cavity. TP53 R273C is also surface exposed, but this mutant has impaired DNA binding and it is unlikely that a covalent modification of this cysteine would restore this function. FBXW7 R465C is very well positioned for targeting, but is adjacent to a wild type cysteine, which could affect selectivity for the mutant.
In principle, two strategies to target acquired cysteines can be distinguished. In the first strategy, an already available non-covalent inhibitor is modified to include a thiol-reactive moiety to tether the inhibitor to an acquired cysteine. Such a strategy has been applied to several kinase inhibitors, such as the CDK7 inhibitor THZ1 [14] with the difference that THZ1 targets a wild-type cysteine. Whether an acquired cysteine can be targeted in this way depends very much on its three-dimensional position relative to the binding-site of the non-covalent inhibitor. A benefit of this approach is that the affinity for the target comes from the non-covalent inhibitor, allowing for a less reactive thiol-directed moiety and thus a lower likelihood of off-target reactivity. Based on the information in figure 3, the acquired cysteines in IDH1 R132C, DNMT3a R882C and KRAS G13C might be targeted by this approach, because non-covalent inhibitors that bind near these cysteines are already available for these proteins. In the second strategy, one screens directly for cysteine-reactive compounds that bind to the site of interest [20–23]. This approach requires a library of compounds that contain cysteine-directed moieties, such as disulfides [24], reversible electrophiles [21,25], or irreversible thio-philes (for an overview see [20]) linked to diverse chemical structures. The wild-type protein should be used as a control for binding selective binding to the cancer-associated acquired cysteine mutant. See figure 4 for a schematic of these two different strategies for covalent targeting.
Figure 4. Different strategies for finding compounds to target acquired cysteines.
A. The first strategy is based on modification of an available non-covalent binder to include a cysteine-reactive moiety that cross-links it to an acquired cysteine in or near the binding pocket of the non-covalent binder. B. The second strategy uses a screen with a library of thiophiles as its basis for drug discovery. Thiophiles may include disulfides and irreversible and reversible electrophiles like acrylates and cyano-acrylamides respectively conjugated to different R groups that can provide selectivity for the target.
Exploiting Redox Signaling for targeting Cysteines
The reactivity of the cysteine thiol makes it the lynchpin in cellular redox signaling, a form of signal transduction that is mediated through reversible cysteine oxidation [26]. The redox sensitivity of cysteines can vary over 6 orders of magnitude [27], and it has been suggested that this correlates with electrophilic drug reactivity [28]. When cysteine becomes oxidized to sulfenic acid, it is converted from a nucleophile to an electrophile; these oxidation-prone cysteines could therefore be selectively targeted by nucleophiles that would be unreactive towards highly abundant cellular thiols like glutathione. Furthermore, sulfenic-acid selective inhibitors could modify proteins depending on the cellular redox state, which is often more oxidizing in cancer cells [29]. This concept has been put forward based on the observation that the sulfenylated form of Cys797 in EGFR is selectively reactive to dimedone-based small molecules [30]. It is unknown to what extent acquired cysteines in cancer are redox-sensitive, because mass-spectrometry based screens for redox-sensitivity [31] have used reference databases based on the expected proteolytic fragments from wild-type proteomes and hence have not identified mutant peptides. It would therefore be worthwhile to explore the redox sensitivity of the acquired cysteines.
Covalent allosteric inhibition of KRAS-G12C
Two research groups have recently targeted an acquired cysteine in the oncoprotein KRAS-G12C [22,32]. G12C is an activating mutation that changes the protein structure to favor binding to GTP over GDP and is one of the most common mutations in the KRAS proto-oncogene (see figure 1) and is present in half of Ras-driven lung adenocarcinomas [16]. The Gray lab [32] used the first strategy described above (see also figure 4a), adding an alpha-chloroacetamide to crosslink a GDP-mimetic (‘SML-8-73-1’) to G12C. The Shokat lab [22] used the second strategy (see also figure 4b) and took advantage of a disulfide-containing library to screen for molecules that bound to G12C, and then converted the disulfide to irreversible vinyl sulfonamide or acrylamide based compounds (e.g. ‘compound 12’). They found that these compounds bound in a ‘cryptic binding site’ in the so-called switch II region that is in the vicinity of G12C but is distinct from the GTP binding pocket. (for a review see [33]. Compound 12 allosterically locked KRAS G12C in the GDP - and hence inactive – state and impaired the binding of KRAS to its downstream target RAF. Not surprisingly, compound 12 also inhibited the mutant form of the highly homologous oncoprotein H-RAS(G12C). The covalent inhibitors of both studies were tested for their effectiveness and selectivity in killing various mutant RAS expressing tumor cell lines. Compound 12 decreased viability in KRASG12C mutant cancer cell lines; the effect was specific because it did not compromise viability of wild-type KRAS, KRAS G12D- or NRAS Q61K-expressing cell lines [22]. A membrane permeable analogue of SML-8-73-1 did inhibit proliferation, but relatively high doses were needed and the effect seemed irrespective of RAS mutant status [32]. Although the effectiveness of these compounds have not yet been demonstrated on tumor growth in vivo, these results are certainly promising.
Conclusions
Many oncogenic mutations introduce a novel cysteine that could in principle be used for selective targeting of cancer cells. The special chemistry of this amino acid makes it suitable for covalent binding, which is an up-and-coming trend in drug targeting. The principle of covalent targeting acquired cysteines is especially relevant in cancer because the mutant will only be expressed in the tumor. Nevertheless, this concept might also be applicable to find new drugs for the treatment of genetic diseases that stem from heterozygous missense mutations that yield cysteines.
Supplementary Material
Highlights.
Missense mutations in cancer frequently lead to the gain of a cysteine.
The chemistry of the cysteine thiol allows for covalent modification.
Covalent drugs have a prolonged or permanent interaction with their targets.
Covalent targeting of acquired cysteines could be a new personalized cancer therapy.
Acknowledgments
We would like to thank Dr. Michael Hadders for help with the PyMol software. MV and TBD were supported by grants from the Dutch Cancer Society (KWF Kankerbestrijding).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Michelle R. Arkin, Email: Michelle.arkin@ucsf.edu.
Tobias B. Dansen, Email: t.b.dansen@umcutrecht.nl.
References
- 1.DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 2.Rimawi MF, Schiff R, Osborne CK. Targeting HER2 for the treatment of breast cancer. Annu Rev Med. 2015;66:111–128. doi: 10.1146/annurev-med-042513-015127. [DOI] [PubMed] [Google Scholar]
- 3.Mulcahy N. Top 10 Best-Selling Cancer Drugs Globally. Medscape 2014 Jun; [Google Scholar]
- 4.Bollag G, Tsai J, Zhang J, Zhang C, Ibrahim P, Nolop K, Hirth P. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat Rev Drug Discov. 2012;11:873–886. doi: 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- 5.Bauer RA. Covalent inhibitors in drug discovery: from accidental discoveries to avoided liabilities and designed therapies. Drug Discov Today. 2015;20:1061–1073. doi: 10.1016/j.drudis.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Liu Q, Sabnis Y, Zhao Z, Zhang T, Buhrlage SJ, Jones LH, Gray NS. Developing irreversible inhibitors of the protein kinase cysteinome. Chem Biol. 2013;20:146–159. doi: 10.1016/j.chembiol.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh J, Petter RC, Baillie TA, Whitty A. The resurgence of covalent drugs. Nat Rev Drug Discov. 2011;10:307–317. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- 8.Tucker DL, Rule SA. A critical appraisal of ibrutinib in the treatment of mantle cell lymphoma and chronic lymphocytic leukemia. Ther Clin Risk Manag. 2015;11:979–990. doi: 10.2147/TCRM.S73559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finlay MR, Anderton M, Ashton S, Ballard P, Bethel PA, Box MR, Bradbury RH, Brown SJ, Butterworth S, Campbell A, et al. Discovery of a potent and selective EGFR inhibitor (AZD9291) of both sensitizing and T790M resistance mutations that spares the wild type form of the receptor. J Med Chem. 2014;57:8249–8267. doi: 10.1021/jm500973a. [DOI] [PubMed] [Google Scholar]
- 10.Walter AO, Sjin RT, Haringsma HJ, Ohashi K, Sun J, Lee K, Dubrovskiy A, Labenski M, Zhu Z, Wang Z, et al. Discovery of a mutant-selective covalent inhibitor of EGFR that overcomes T790M-mediated resistance in NSCLC. Cancer Discov. 2013;3:1404–1415. doi: 10.1158/2159-8290.CD-13-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, Cortot AB, Chirieac L, Iacob RE, Padera R, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •12.Bradshaw JM, McFarland JM, Paavilainen VO, Bisconte A, Tam D, Phan VT, Romanov S, Finkle D, Shu J, Patel V, et al. Prolonged and tunable residence time using reversible covalent kinase inhibitors. Nat Chem Biol. 2015;11:525–531. doi: 10.1038/nchembio.1817. This study reversibly crosslinked inhibitors of BTK and FGFR to a non-catalytic cysteine, thereby enhancing drug residence time up to seven days. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan L, Wang J, Tanizaki J, Huang Z, Aref AR, Rusan M, Zhu SJ, Zhang Y, Ercan D, Liao RG, et al. Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors. Proc Natl Acad Sci U S A. 2014;111:E4869–4877. doi: 10.1073/pnas.1403438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •14.Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, Dastur A, Amzallag A, Ramaswamy S, Tesar B, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511:616–620. doi: 10.1038/nature13393. The authors modify a known CDK7 inhibitor to include a cysteine reactive moiety that crosslinks the inhibitor to a cysteine near its binding site. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson MH. Reversible Michael additions: covalent inhibitors and prodrugs. Mini Rev Med Chem. 2012;12:1330–1344. doi: 10.2174/13895575112091330. [DOI] [PubMed] [Google Scholar]
- 16.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tekaia F, Yeramian E, Dujon B. Amino acid composition of genomes, lifestyles of organisms, and evolutionary trends: a global picture with correspondence analysis. Gene. 2002;297:51–60. doi: 10.1016/s0378-1119(02)00871-5. [DOI] [PubMed] [Google Scholar]
- 18.Flanagan ME, Abramite JA, Anderson DP, Aulabaugh A, Dahal UP, Gilbert AM, Li C, Montgomery J, Oppenheimer SR, Ryder T, et al. Chemical and computational methods for the characterization of covalent reactive groups for the prospective design of irreversible inhibitors. J Med Chem. 2014;57:10072–10079. doi: 10.1021/jm501412a. [DOI] [PubMed] [Google Scholar]
- 19.Kozakov D, Grove LE, Hall DR, Bohnuud T, Mottarella SE, Luo L, Xia B, Beglov D, Vajda S. The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat Protoc. 2015;10:733–755. doi: 10.1038/nprot.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kathman SG, Xu Z, Statsyuk AV. A fragment-based method to discover irreversible covalent inhibitors of cysteine proteases. J Med Chem. 2014;57:4969–4974. doi: 10.1021/jm500345q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller RM, Paavilainen VO, Krishnan S, Serafimova IM, Taunton J. Electrophilic fragment-based design of reversible covalent kinase inhibitors. J Am Chem Soc. 2013;135:5298–5301. doi: 10.1021/ja401221b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••22.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. The authors use a chemical screen to identify compounds that covalently and selectively bind to acquired cysteine G12C in the KRAS oncogene. This is the first example of covalent targeting of an acquired cysteine in cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson CG, Arkin MR. Probing structural adaptivity at PPI interfaces with small molecules. Drug Discov Today Technol. 2013;10:e501–508. doi: 10.1016/j.ddtec.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Erlanson DA, Braisted AC, Raphael DR, Randal M, Stroud RM, Gordon EM, Wells JA. Site-directed ligand discovery. Proc Natl Acad Sci U S A. 2000;97:9367–9372. doi: 10.1073/pnas.97.17.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnan S, Miller RM, Tian B, Mullins RD, Jacobson MP, Taunton J. Design of reversible, cysteine-targeted Michael acceptors guided by kinetic and computational analysis. J Am Chem Soc. 2014;136:12624–12630. doi: 10.1021/ja505194w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 27.Nagy P, Karton A, Betz A, Peskin AV, Pace P, O’Reilly RJ, Hampton MB, Radom L, Winterbourn CC. Model for the exceptional reactivity of peroxiredoxins 2 and 3 with hydrogen peroxide: a kinetic and computational study. J Biol Chem. 2011;286:18048–18055. doi: 10.1074/jbc.M111.232355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan LB, Chandel NS. Mitochondrial reactive oxygen species and cancer. Cancer Metab. 2014;2:17. doi: 10.1186/2049-3002-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •30.Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, Carroll KS. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat Chem Biol. 2012;8:57–64. doi: 10.1038/nchembio.736. In this study the redox sensitivty of a cysteine in EGFR is exploited to target it with a nucleophile rather than an electrophile, thereby reducing interference of high-abundant thiols in the cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furdui CM, Poole LB. Chemical approaches to detect and analyze protein sulfenic acids. Mass Spectrom Rev. 2014;33:126–146. doi: 10.1002/mas.21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •32.Lim SM, Westover KD, Ficarro SB, Harrison RA, Choi HG, Pacold ME, Carrasco M, Hunter J, Kim ND, Xie T, et al. Therapeutic targeting of oncogenic K-Ras by a covalent catalytic site inhibitor. Angew Chem Int Ed Engl. 2014;53:199–204. doi: 10.1002/anie.201307387. The authors modify a GTP mimetic to include a thiol reactive moiety that crosslinks it to acquired cysteine G12C in the KRAS oncogene. Like in reference 22, this is an example of covalent targeting of an acquired cysteine in cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudolph J, Stokoe D. Selective inhibition of mutant Ras protein through covalent binding. Angew Chem Int Ed Engl. 2014;53:3777–3779. doi: 10.1002/anie.201400233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.