Abstract
C4.4A, a member of the Ly6/uPAR family of membrane proteins, has been identified as a metastasis-associated molecule, but little is known about its actual expression and possible function in head and neck squamous cell carcinoma (HNSCC). To explore diagnostic and prognostic roles of C4.4A in HNSCC, we investigated the expression of C4.4A in human HNSCC tissue array which contains 43 HNSCC, 6 epithelial dysplasia and 16 normal oral mucosa. Expression of C4.4A was significantly increased in epithelial dysplasia and HNSCC when compared with normal oral mucosa. Moreover, high C4.4A expression indicated a rather poor prognosis of HNSCC patients. To better understand the function of C4.4A in HNSCC progression, we investigated epithelial to mesenchymal transition (EMT) associated proteins including transforming growth factor (TGF-β1), Slug and CD147 in HNSCC. The expression of TGF-β1, Slug, and CD147 was significantly increased in HNSCC when compared with normal oral mucosa. Meanwhile, the expression of C4.4A was significantly correlated with TGF-β1, Slug, and CD147 in HNSCC tissue array. Furthermore, knockdown of C4.4A decreased the cell invasion and migration in CAL27 cell line and suppressed the EMT with increased E-cadherin and decreased N-cadherin and Slug. Our study demonstrated that C4.4A was a potential marker for prognosis of HNSCC, and C4.4A participated in EMT program in HNSCC progression.
Keywords: C4.4A, TGF-β1, slug, CD147, head and neck squamous cell carcinoma, EMT
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer around the world [1]. The main characteristics of HNSCC may be its loco-regional invasion and metastasis to cervical lymph nodes [2]. Despite lots of research efforts, the survival rate of HNSCC has not markedly improved in recent decades because of a high incidence of loco-regional recurrences, distant metastases and second primary tumors [3]. Thus, discovering the biological factor regulating HNSCC invasion and metastasis is important.
The C4.4A protein, a structural homologue to the urokinase-type plasminogen activator receptor [4,5], was regarded as a potential metastasis-associated molecule [6,7]. Pre-vious studies have demonstrated that C4.4A was associated with various human cancers, including bladder cancer [7], esophageal cancer [8], colorectal cancer [9] and lung cancer [10]. Interestingly, C4.4A preferred to highly express at the tumor invasive fronts rather than tumor core, implying a possible connection of C4.4A to the invasive process [11]. These findings suggest the potential role of C4.4A in cancer invasion and metastasis, but further characterization of C4.4A in terms of expression and mechanism in HNSCC remain to be explored.
Emerging evidence demonstrated that Epithelial-mesenchymal transition (EMT) played an important role in HNSCC invasion and metastasis [12,13]. During EMT process, tumor cells lost their epithelial characteristics, including cell-cell adhesion and polarity, and obtained mesenchymal traits, including motility, invasiveness [14]. Transforming growth factor-beta 1 (TGF-β1), a pleiotropic cytokine secreted by cancer cells, has been shown it could process EMT in cancers [15,16]. Continuous research illustrated that the transcription factor Slug is the key molecule regulating EMT program in cancer [17,18]. CD147 is a membrane glycoprotein involved in cell adhesion by regulating matrix metalloproteinases, and participated in EMT of cancer [19].
In this study, to investigate the role of C4.4A in HNSCC progression, we explored the expression of C4.4A, TGF-β1, Slug and CD147 in human HNSCC tissue microarray and analyzed correlation of C4.4A, TGF-β1, Slug and CD147. Furthermore, the impacts of C4.4A on cell migration and EMT of HNSCC were studied by knockdown of C4.4A in vitro.
Material and methods
Chemical and reagents
Chemical and reagents for experiment were purchased from Sigma-Aldrich unless otherwise specified. C4.4A siRNAs were purchased from Sigma-Aldrich. Primary antibody against human C4.4A was purchased from Abcam. TGF-β1, and Slug were purchased from Cell Signaling Technology (Danvers, MA), CD147 was purchased from Proteintech Group, Inc.
Ethics statement, patients’ specimens and human HNSCC tissue microarray
This study was approved by the School and Hospital of Stomatology of Wuhan University Medical Ethics Committee, and informed consent was acquired from the patients before the surgery. All the patients’ tissues histologically confirmed HNSCC were collected from the Hospital of Stomatology of Wuhan University. The clinical stage of HNSCC was classified based on the guidelines of the International Union Against Cancer (UICC 2002), and histological grade was confirmed according to the classification scheme of the World Health Organization. HNSCC tissue array of were constructed with 1.5 mm tissue cylinders from each patient’s specimen mentioned above in collaboration with the Shanghai Biochip Company, Ltd, Shanghai, China.
Immunohistochemical staining
Immunohistochemistry for tissue microarray was performed as follows. Firstly, tissue microarray sections were deparaffinized and hydrated. Antigen retrieval was performed by using citric acid buffer solution (pH 6.0) at high temperature and pressure. Then the sections incubated with 3% hydrogen superoxide to quench endogenous peroxidase activity and 10% normal goat serum to block non-specific binding. Next the sections were incubated overnight at 4°C with polyclonal rabbit anti-human C4.4A (Abcam, 1:200), TGF-β1 (Cell Signaling Technology, 1:200), Slug (Cell Signaling Technology, 1:200), CD147 (Proteintech, 1:400). Then, the sections were incubated with a secondary biotin-labeled antibody solution and streptavidin peroxidase. Finally, the sections were washed with phosphate buffer saline, incubated with 3,3’-diaminobenzidine tetrachloride and then counterstained with Mayer’s haematoxylin.
Scoring system, hierarchical clustering
Tissue array sections were scanned using an Aperio ScanScope CS scanner (Vista, CA, USA) with background subtraction and quantified with Aperio Quantification software (Version 9.1) for membrane, nuclear, or pixel quantification. Histoscore of membrane and nuclear staining was calculated using the formula (3+)×3+(2+)×2+(1+)×1 [20]. Histoscores were translated to scaled values centered at zero in Microsoft Excel, and the hierarchical analysis was performed by the Cluster 3.0 and Java TreeView 1.0.5.
Cell culture, siRNA knockdown assay
Human head neck cancer cell line CAL27, FaDu, SCC-4, SCC-9, SCC-25 were bought from the American Type Culture Collection (Manassas, VA), and cells were maintained in DMEM with 10% fetal bovine serum at 5% CO2 and 37°C. The CAL27 cell line was transfected with C4.4A siRNA (100 nM, Sigma-Aldrich) by using HiPerfect transfection reagent (Qiagen, Germantown, MD).
Cell immunofluorescence
CAL27 cells were seeded on coverglass slide chambers (Millipore). After designed treatment, the cells were fixed by 4% paraformaldehyde at room temperature for 15 min, and then permeabilized with 0.3% triton X-100. After blocked with 2.5% BSA for 1 h, the cells were incubated with primary antibody overnight at 4°C. PerCP-Cy5.5-conjugated secondary antibody (Jackson ImmunoResearch, USA, 1:200) was used for detection and DAPI for nucleus counterstaining. The primary antibodies included the following: C4.4A antibody (Abcam, 1:200), TGF-β1 antibody (Cell Signaling Technology, 1:300), Slug antibody (Cell Signaling Technology, 1:400), CD147 (Proteintech, 1:300). The slides were observed by a fluorescent microscope. Representative cells were selected and photographed.
Western blot
CAL27 cells were treated with indicated concentrations of C4.4A siRNA (100 nM) in DMEM for 48 h. Then the cells were lysed in M-PER Mammalian Protein Extraction Reagent (Pierce, Rockford, IL). Protein concentrations were determined by BCA assay kit (Pierce). The total protein was separated by using 10% SDS-polyacrylamide gel electrophoresis, transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore Corporation, Billerica, MA), blocked with 4% dry milk at room temperature for 1 hour, and incubated with antibodies against C4.4A (Abcam 1:1000), E-cadherin (Cell Signaling Technology, 1:1000), N-cadherin (Cell Signaling Technology, 1:1000), and Slug (Cell Signaling Technology, 1:1000). The blots were then incubated with HRP-conjugated secondary antibody (1:2000; Santa Cruz Biotechnology). Finally, bands were visualized through a chemiluminescent detection system, (West Pico, Thermo) and bands density was calculated with ImageJ software packages.
Wound healing assay
CAL27 cells were seeded in 6-well plates (Corning Life Sciences, USA) at 1.0×105 cells/well. CAL27 cells were treated with C4.4A siRNA and with counterpart control as described above. When cells reached 80% confluence, the center of each well was scratched with a sterile pipette tip to generate a constant gap. After incubated with medium containing no FBS for 24 hours, cells were fixed and photographed under a phase microscope and counted as previously described [21]. Each assay was performed in triplicate.
Transwell assays
In vitro transwell assay was performed using Costar Transwell inserts (#3422, pore size, 8 μm) (Corning, Albany, NY) as described previously. For transwell migration assays, CAL27 cells were added into the upper chamber with a non-coated membrane in each group at a density of 105/well in 100 ul serum-free medium. For transwell invasion assay, CAL27 cells were added into the upper chamber coated with Matrigel (BD Biosciences) in each group at a density of 105/well in 100 ul serum-free medium. Meanwhile 10% FBS medium was added to the bottom chamber to stimulate migration and invasion. After incubation for 24 h at 37°C, cells migrated to the bottom surface of chamber inserts and the cells that had invaded through Matrigel were fixed and stained with Hematoxylin, and then photographed and quantified. CAL27 cells were treated with C4.4A siRNA and with counterpart control as describe above. Each assay was performed in triplicate.
Statistical analysis
Statistical data analysis was performed with GraphPad Prism 5.01 (GraphPad Software, Inc., La Jolla, CA) statistical packages. One-way ANOVA followed by the post-Turkey or Bonferroni multiple comparison tests was used to analyze the differences in immunohisochemical staining. Two-tailed Pearson correlation was used for correlated expression of C4.4A, TGF-β1, Slug and CD147 after confirmation of the sample with a Gaussian distribution. Mann-Whitney U test and student t test was used to analyze differences in Western blotting and immunofluorescence. Mean values ± SEM with P < 0.05 was considered statistically significant.
Results
C4.4A was over expressed in human HNSCC and served as a risk factor for clinical prognosis
To assess protein expression of C4.4A in human HNSCC, we performed immunohistochemical staining of C4.4A in human HNSCC tissue array including normal oral mucosa (n=16), epithelial dysplasia (n=6) and HNSCC (n=43). We found that C4.4A mostly located in membrane and cytoplasm of HNSCC cells at the invasive front (Figure 1A). Through quantification, we found C4.4A expression in epithelial dysplasia and HNSCC is rather strongly positive than normal oral mucosa (P < 0.01 and P < 0.001, Figure 1B). To further explore whether C4.4A is associated with HNSCC progression, we compared C4.4A expression in different Grades of HNSCC (Figure S1A). Unfortunately, no difference was found between them, same results were found in different T categories and N categories (Figure S1B and S1C). However, when we discuss the prognosis value of C4.4A by using Kaplan-Meier method, we found high C4.4A expression indicated a rather poor prognosis of HNSCC patients, whereas log-Rank analysis indicated that cumulative rate of the patients with high C4.4A (P=0.1176, n=19, Figure 1C) expression did not reach statistical significance.
Figure 1.
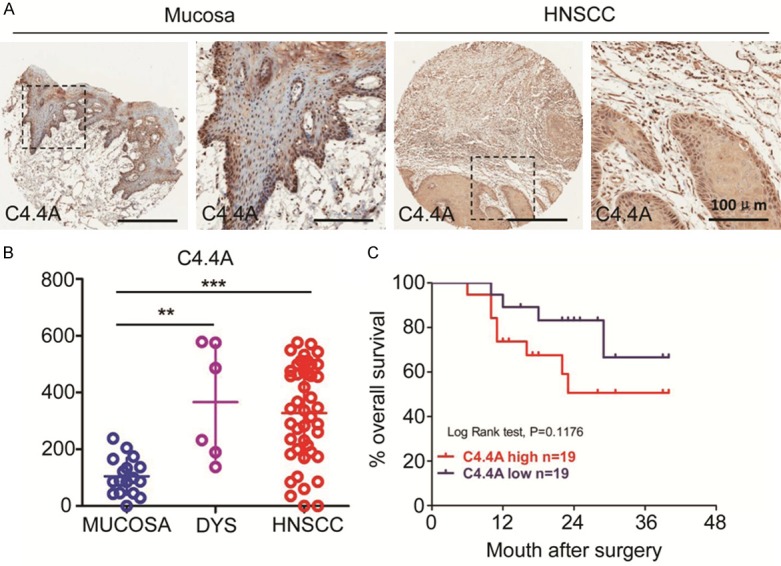
C4.4A expression is up regulated in head and neck squamous cell carcinoma (HNSCC) tissue array and HNSCC cell lines. A. Representative pictures of C4.4A expression in normal oral mucosa and HNSCC by immunohistochemical staining; B. Quantification of Histoscores of C4.4A expression in normal oral mucosa (n=16), epithelial dysplasia (n=6) and HNSCC (n=43). C4.4A expression in oral epithelial dysplasia and HNSCC was higher compared with that in normal oral mucosa (***, P < 0.001; One-way ANOVA); C. Patients with high C4.4A expression have a rather poor prognosis compared with patients with low expression (P=0.1176, n=38).
We also investigated some EMT-associated proteins in the human HNSCC tissue array, we found that TGF-β1 mainly located in cytoplasm and extracellular matrix of HNSCC cells, while Slug was mainly expressed in nucleus, and CD147 is obviously expressed on the membrane of HNSCC cells. To precisely evaluate the differences, we quantified the expression of TGF-β1, Slug, and CD147 (Figure 2A). The quantitative results showed that expression of TGF-β1, Slug, and CD147 was increased in HNSCC when compared with normal oral mucosa and this difference was statistically significant (Figure S1D).
Figure 2.
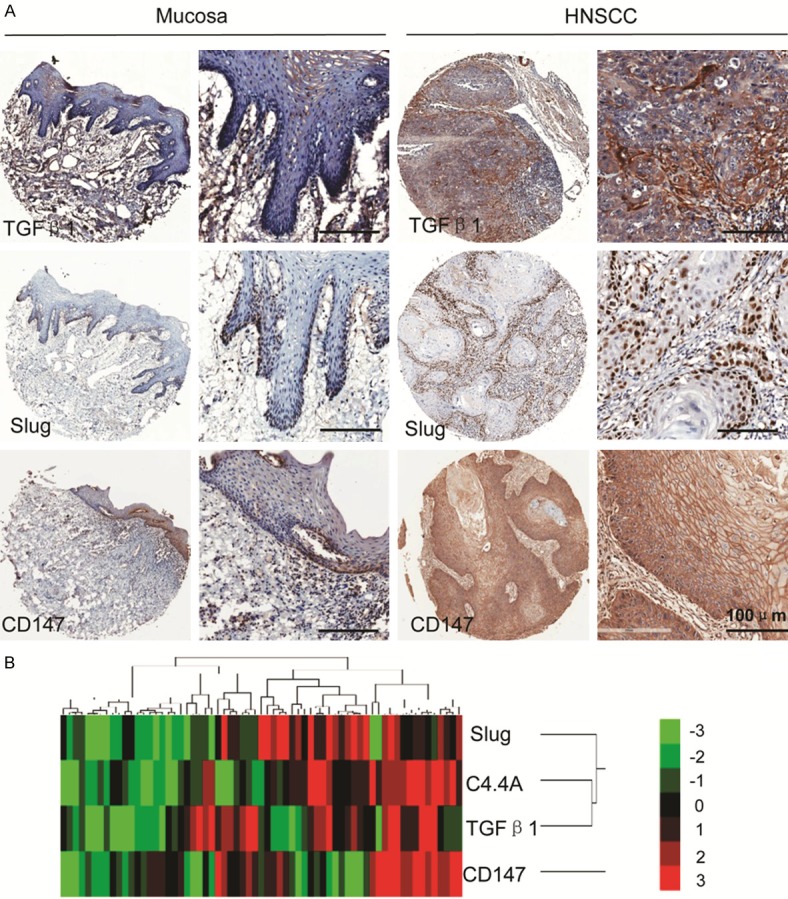
C4.4A expression is correlated with TGF-β1, Slug, and CD147 in human HNSCC tissue. A. Representative IHC staining of TGF-β1, Slug, and CD147 in human HNSCC tissue compared with those of normal oral mucosa (Scale bars =100 μm); B. Hierarchical clustering demonstrated that the correlation between C4.4A, TGF-β1, Slug, and CD147 in human HNSCC tissue array.
C4.4A expression is correlated with TGF-β1, Slug, and CD147 in human HNSCC tissue
To evaluate the association of C4.4A with TGF-β1, Slug, and CD147 in HNSCC, we analyzed the quantification of immunehistochemical staining by the Spearman rank correlation coefficient test and linear tendency test. Results showed that expression of C4.4A in HNSCC was significantly correlated with TGF-β1 (P < 0.01, r=0.3623), Slug (P < 0.01, r=0.3276) and CD147 (P < 0.01, r=0.3810), and linear regression demonstrated the positive trendline between C4.4A with TGF-β1, Slug, and CD147 (Figure S2). Meanwhile, cluster results also showed the close relation of C4.4A with TGF-β1, Slug, and CD147. These findings imply the possible function of C4.4A in EMT process of HNSCC.
Knockdown of C4.4A decreases HNSCC cell line migration and invasion
To determine the role of C4.4A in migration and invasion of HNSCC, we detected the expression of C4.4A in HNSCC cell lines. As Figure 3A showed, protein C4.4A expression was up-regulated in HNSCC cell lines (FaDu, SCC-4, SCC-9, SCC-25, CAL27) compared with normal keratinocytes (OKC). CAL27 cell line was selected, which had a higher content of C4.4A. Two different siRNA were designed for knockdown of C4.4A. Based on the suppression efficiency (Figure 3B), siRNA1 was employed to performed wound healing and Boyden chamber invasion assay to detect the migration and invasion potential. As the results showed, knockdown of C4.4A notably decreased the cell mobility of CAL27 cell line, and the cell number of migration after 24 hours is quite different between control group and C4.4A siRNA treatment group (P < 0.01) (Figure 3C and 3D). Then we examined the invasion ability of CAL27, knockdown of C4.4A could also significantly decreased the invasive cell number compared with that in control group (P < 0.001) (Figure 3E). Meanwhile, results of experiments using siRNA2 were showed in Figure S3. Together, the results demonstrated that knockdown of C4.4A decreased the ability of cell migration and invasion of HNSCC.
Figure 3.
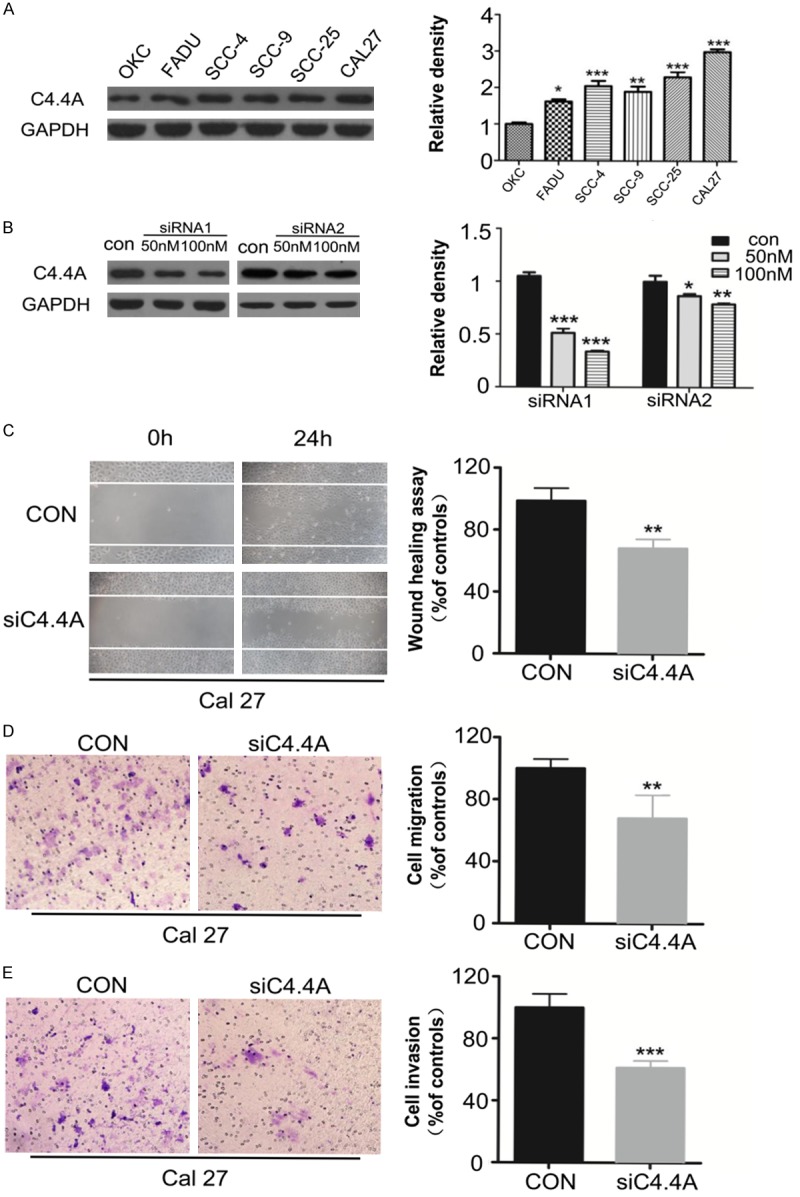
Knockdown of C4.4A decreases migration and invasion of HNSCC cell line. A. Expression of C4.4A protein in HNSCC cell lines was higher than normal keratinocytes cell line (OKC) as the Weston Blot results showed (Mean ± SD; *, P < 0.05, **, P < 0.01, ***, P < 0.001, One-way ANOVA); B. Knockdown of C4.4A by two different siRNA (Mean ± SD; *, P < 0.05, **, P < 0.01, ***, P < 0.001, Two-way ANOVA); C. Wound healing assay showed knockdown of C4.4A suppressed the cell mobility of CAL27 cell line, and quantification of cell numbers with ImageJ “cell counter” module shows the statistical significance of the difference (Mean ± SD; **, P < 0.01, student t-test with GraphPad Prism5.0); D and E. Transwell assay showed the migration and invasion abilities of CAL27 were impaired after knocking down of C4.4A compared with those of control group, and quantification of cell numbers with Image J “cell counter” module (Mean ± SD; **, P < 0.01, ***, P < 0.001, student t-test with GraphPad Prism5.0).
Knockdown of C4.4A suppress EMT process in HNSCC cell line
To better understand the mechanism of how C4.4A affects metastasis of HNSCC cell line, we explored the effect of C4.4A on EMT epithelial marker E-cadherin, N-cadherin and EMT transcriptional factor Slug. As the Weston Blot analysis showed, knockdown of C4.4A could obviously suppress the protein expression of N-cadherin and Slug, and partially suppress Vimentin, while it could increase the expression of E-cadherin in CAL27 cell line (Figure 4A). Furthermore, immunofluorescence results also demonstrated knockdown of C4.4A increased E-cadherin expression and decreased N-cadherin and Slug expression in CAL27 cell line (Figure 4B and 4C). These findings suggest that knockdown of C4.4A could inhibit the EMT progression.
Figure 4.
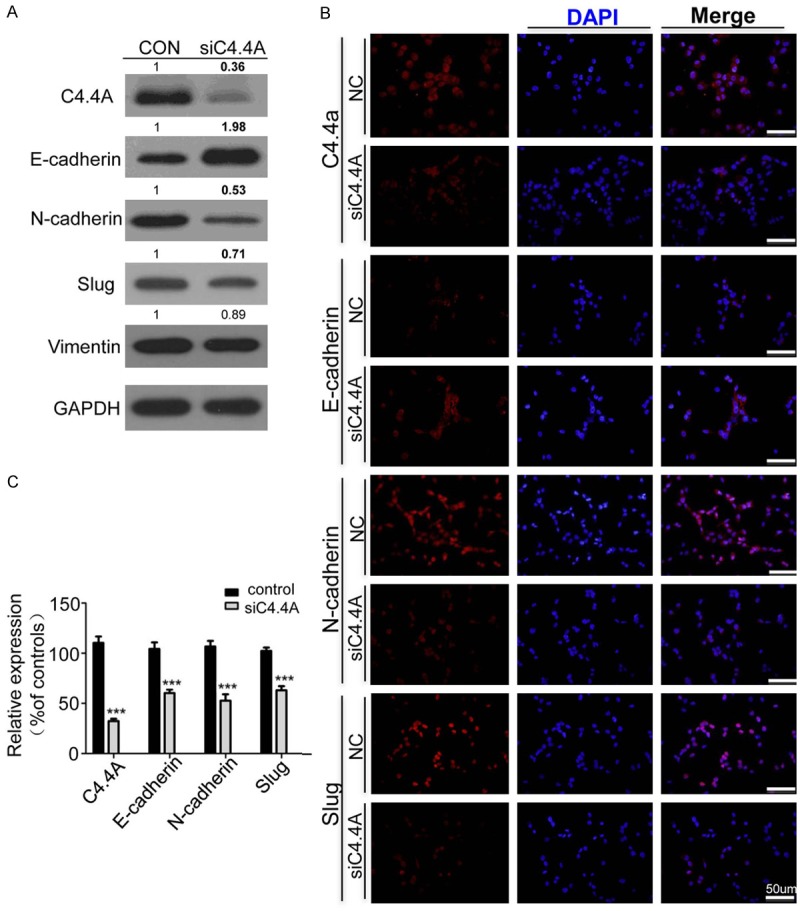
Knockdown of C4.4A suppresses EMT process in HNSCC cell line. A. Weston Blot analysis revealed the up regulation of E-cadherin and down regulation of N-cadherin, Vimentin and Slug after treating CAL27 cell line with C4.4A siRNA (100 nM) for 48 hours. B. Representative immunofluorescence showed knockdown of C4.4A increased E-cadherin and decreased N-cadherin, and Slug, bar =50 μm. C. Quantification of immunofluorescence of CAL27 with Image J, IOD for mean integrated optical density and calculated with total optical density divided by the area (Mean ± SD; ***, P < 0.001, Two-way ANOVA).
Discussion
In this study, we investigated the protein expression of C4.4A in human HNSCC tissue array. We found that C4.4A tended to express in invasive front of HNSCC, which suggested the possible function of C4.4A in the invasive process. Quantification results showed that C4.4A expression was significantly increased in human HNSCC compared with normal oral mucosa. And C4.4A expression in epithelial dysplasia was increased compared with normal oral mucosa, while C4.4A expression in epithelial dysplasia was similar to that in HNSCC. Meanwhile, the analysis of clinical data of HNSCC tissue array suggested that C4.4A was not associated with pathological grades. This indicated that the appearance of C4.4A might be an early event in HNSCC progression and was constitutively activated. It has been reported that C4.4A expression increased in metastatic lymph nodes and metastatic skin lesions compared with primary malignant melanoma [22]. However, in this study, no difference was found between node-negative and node-positive HNSCC, for which more cases may be needed.
Evidence showed that C4.4A was associated with a poor prognosis in colorectal cancer, esophageal squamous cell carcinoma and lung adenocarcinama [23-25]. In present study, high C4.4A expression indicated a rather poor prognosis of HNSCC patients, whereas no statistical significance was found. More cases and longer follow-up period may be needed in future research.
To further explore the possible role of C4.4A in HNSCC, we investigated TGF-β1, Slug and CD147, which correlated with EMT process of cancers. It has been proved that EMT played an important role in HNSCC metastasis [26,27]. TGF-β1 is an epithelial-to-mesenchymal inducer [28], and has been reported participating in HNSCC progression [29]. Slug is an epithelial-to-mesenchymal transition-inducing transcription factor [30], and overexpression of Slug in HNSCC played an important role in EMT [31]. It has been proved that CD147 could promote cell proliferation and metastasis of HNSCC [32]. An early report indicated that CD147 promoted EMT through TGF-β signaling and was regulated by Slug [33]. Increased expression of these proteins was found in HNSCC compared with that in normal mucosa in the tissue array. Furthermore, expression of C4.4A was significantly correlated with that of TGF-β1, Slug and CD147. Cluster results suggested the close relation of C4.4A TGF-β1, Slug, and CD147. The evidence implies that C4.4A may be a downstream target of the EMT inducer TGF-β1, and could participate in the EMT process along with Slug and CD147.
A recent report suggested that the blockade of AGR2-C4.4A pathway reduced growth and metastasis of pancreatic tumor [34]. Similarly, our previous research showed that AGR2 could promote HNSCC progression by regulating cancer stem cell and EMT signaling pathways in HNSCC [35]. However, the function of C4.4A in HNSCC progression has not been investigated. We knocked down C4.4A by siRNA in CAL27 cell line. Inhibition of C4.4A decreased the metastasis and invasion of HNSCC cells. Moreover, blockade of C4.4A induced the up-regulation of epithelial marker E-cadherin and the down regulation of N-cadherin and transcriptional factor Slug, but CD147 expression was invariant. These results showed that knocking down of C4.4A reduced the EMT program in HNSCC cell line, which indicated that C4.4A took part in EMT in HNSCC. Some other reporters demonstrated that C4.4A promoted migration by associating with alpha6beta4 integrin [36]. C4.4A was also reported bind with LN1 and LN5 promoted transition of epithelial cells to a migratory phenotype [37]. In addition, in accordance with our findings, an early report suggested that C4.4A was associated with epithelial-to-mesenchymal transition with alterations of EMT markers including E-cadherin, Vimentin and N-cadherin in colorectal cancer [38].
In summary, we studied the expression of C4.4A in human HNSCC tissue microarray and function of C4.4A in vitro, and demonstrated that the aberrant expression of C4.4A promoted the invasion and metastasis of HNSCC. We further found that blockade of C4.4A reduced the EMT program in HNSCC. Thus, C4.4A may be a potential therapeutic target for HNSCC.
Acknowledgements
This work was supported by National Natural Science Foundation of China (81272963, 81472528) to Z.J. Sun, (81272946, 81472529) to W.F. Zhang and (81402241) to C.F. Huang. This work was also supported by program for new century excellent talents in university (NCET-13-0439), Ministry of Education of China to Z.J. Sun.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Markwell SM, Weed SA. Tumor and stromal-based contributions to head and neck squamous cell carcinoma invasion. Cancers (Basel) 2015;7:382–406. doi: 10.3390/cancers7010382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 4.Hansen LV, Gårdsvoll H, Nielsen BS, Lund LR, Danø K, Jensen ON, Ploug M. Structural analysis and tissue localization of human C4.4A: a protein homologue. Biochem J. 2004;380:845–857. doi: 10.1042/BJ20031478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobsen B, Ploug M. The urokinase receptor and its structural homologue C4.4A in human cancer. Curr Med Chem. 2008;15:2559–2573. doi: 10.2174/092986708785909012. [DOI] [PubMed] [Google Scholar]
- 6.Rösel M, Claas C, Seiter S, Herlevsen M, Zöller M. Cloning and functional characterization of a new phosphatidyl-inositol anchored. Oncogene. 1998;17:1989–2002. doi: 10.1038/sj.onc.1202079. [DOI] [PubMed] [Google Scholar]
- 7.Smith BA, Kennedy WJ, Harnden P, Selby PJ, Trejdosiewicz LK, Southgate J. Identification of genes involved in human urothelial cell-matrix interactions. Cancer Res. 2001;61:1678–1685. [PubMed] [Google Scholar]
- 8.Hansen LV, Laerum OD, Illemann M, Nielsen BS, Ploug M. Altered expression of the urokinase receptor homologue, C4.4A, in invasive areas of human esophageal squamous cell carcinoma. Int J Cancer. 2008;122:734–741. doi: 10.1002/ijc.23082. [DOI] [PubMed] [Google Scholar]
- 9.Konishi K, Yamamoto H, Mimori K, Takemasa I, Mizushima T, Ikeda M, Sekimoto M, Matsuura N, Takao T, Doki Y, Mori M. Expression of C4.4A at the invasive front is a novel prognostic marker for disease recurrence of colorectal cancer. Cancer Sci. 2010;101:2269–2277. doi: 10.1111/j.1349-7006.2010.01674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen LV, Skov BG, Ploug M, Pappot H. Tumour cell expression of C4.4A, a structural homologue of the urokinase. Lung Cancer. 2007;58:260–266. doi: 10.1016/j.lungcan.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 11.Jacobsen B, Kriegbaum MC, Santoni-Rugiu E, Ploug M. C4.4A as a biomarker in pulmonary adenocarcinoma and squamous cell carcinoma. World J Clin Oncol. 2014;5:621–632. doi: 10.5306/wjco.v5.i4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S, Ye D, Guo W, Yu W, He Y, Hu J, Wang Y, Zhang L, Liao Y, Song H, Zhong S, Xu D, Yin H, Sun B, Wang X, Liu J, Wu Y, Zhou BP, Zhang Z, Deng J. G9a is essential for EMT-mediated metastasis and maintenance of cancer stem cell-like characters in head and neck squamous cell carcinoma. Oncotarget. 2015;6:6887–6901. doi: 10.18632/oncotarget.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin Y, Mallen-St Clair J, Luo J, Sharma S, Dubinett S, John MS. p53 modulates NFkappaB mediated epithelial-to-mesenchymal transition in head and neck squamous cell carcinoma. Oral Oncol. 2015;51:921–928. doi: 10.1016/j.oraloncology.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 14.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han J, Alvarez-Breckenridge CA, Wang QE, Yu J. TGF-beta signaling and its targeting for glioma treatment. Am J Cancer Res. 2015;5:945–955. [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao ZL, Ma SR, Wang WM, Huang CF, Yu GT, Wu TF, Bu LL, Wang YF, Zhao YF, Zhang WF, Sun ZJ. Notch signaling induces epithelialmesenchymal transition to promote invasion and metastasis in adenoid cystic carcinoma. Am J Transl Res. 2015;7:162–174. [PMC free article] [PubMed] [Google Scholar]
- 17.Xie J, Gao H, Peng J, Han Y, Chen X, Jiang Q, Wang C. Hispidulin prevents hypoxia-induced epithelial-mesenchymal transition in human colon carcinoma cells. Am J Cancer Res. 2015;5:1047–1061. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer-Hardi U, Bell G, Tam WL, Mani SA, van Oudenaarden A, Weinberg RA. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez-Teja M, Gronau JH, Minamidate A, Darby S, Gaughan L, Robson C, Mauri F, Waxman J, Sturge J. Survival Outcome and EMT Suppression Mediated by a Lectin Domain Interaction of Endo180 and CD147. Mol Cancer Res. 2015;13:538–547. doi: 10.1158/1541-7786.MCR-14-0344-T. [DOI] [PubMed] [Google Scholar]
- 20.Sun ZJ, Zhang L, Hall B, Bian Y, Gutkind JS, Kulkarni AB. Chemopreventive and chemotherapeutic actions of mTOR inhibitor in genetically defined head and neck squamous cell carcinoma mouse model. Clin Cancer Res. 2012;18:5304–5313. doi: 10.1158/1078-0432.CCR-12-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Sun ZJ, Bian Y, Kulkarni AB. MicroRNA-135b acts as a tumor promoter by targeting the hypoxia-inducible factor pathway in genetically defined mouse model of head and neck squamous cell carcinoma. Cancer Lett. 2013;331:230–238. doi: 10.1016/j.canlet.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seiter S, Stassar M, Rappl G, Reinhold U, Tilgen W, Zoller M. Upregulation of C4.4A expression during progression of melanoma. J Invest Dermatol. 2001;116:344–347. doi: 10.1046/j.1523-1747.2001.01230.x. [DOI] [PubMed] [Google Scholar]
- 23.Paret C, Hildebrand D, Weitz J, Kopp-Schneider A, Kuhn A, Beer A, Hautmann R, Zoller M. C4.4A as a candidate marker in the diagnosis of colorectal cancer. Br J Cancer. 2007;97:1146–1156. doi: 10.1038/sj.bjc.6604012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohtsuka M, Yamamoto H, Masuzawa T, Takahashi H, Uemura M, Haraguchi N, Nishimura J, Hata T, Yamasaki M, Miyata H, Takemasa I, Mizushima T, Takiguchi S, Doki Y, Mori M. C4.4A expression is associated with a poor prognosis of esophageal squamous cell carcinoma. Ann Surg Oncol. 2013;20:2699–2705. doi: 10.1245/s10434-013-2900-2. [DOI] [PubMed] [Google Scholar]
- 25.Jacobsen B, Muley T, Meister M, Dienemann H, Christensen IJ, Santoni-Rugiu E, Laerum OD, Ploug M. Ly6/uPAR-related protein C4.4A as a marker of solid growth pattern and poor prognosis in lung adenocarcinoma. J Thorac Oncol. 2013;8:152–160. doi: 10.1097/JTO.0b013e318279d503. [DOI] [PubMed] [Google Scholar]
- 26.Way TD, Huang JT, Chou CH, Huang CH, Yang MH, Ho CT. Emodin represses TWIST1-induced epithelial-mesenchymal transitions in head and neck squamous cell carcinoma cells by inhibiting the beta-catenin and Akt pathways. Eur J Cancer. 2014;50:366–378. doi: 10.1016/j.ejca.2013.09.025. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen PT, Tsunematsu T, Yanagisawa S, Kudo Y, Miyauchi M, Kamata N, Takata T. The FGFR1 inhibitor PD173074 induces mesenchymal-epithelial transition through the transcription factor AP-1. Br J Cancer. 2013;109:2248–2258. doi: 10.1038/bjc.2013.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeisberg M, Hanai J, Sugimoto H, Mammoto T, Charytan D, Strutz F, Kalluri R. BMP-7 counteracts TGF-beta1-induced epithelial-tomesenchymal transition and reverses chronic renal injury. Nat Med. 2003;9:964–968. doi: 10.1038/nm888. [DOI] [PubMed] [Google Scholar]
- 29.Bian Y, Hall B, Sun ZJ, Molinolo A, Chen W, Gutkind JS, Waes CV, Kulkarni AB. Loss of TGF-beta signaling and PTEN promotes head and neck squamous cell carcinoma through cellular senescence evasion and cancer-related inflammation. Oncogene. 2012;31:3322–3332. doi: 10.1038/onc.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, Weinberg RA. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–260. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katafiasz D, Smith LM, Wahl JK 3rd. Slug (SNAI2) expression in oral SCC cells results in altered cell-cell adhesion and increased motility. Cell Adh Migr. 2011;5:315–322. doi: 10.4161/cam.5.4.17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sweeny L, Liu Z, Bush BD, Hartman Y, Zhou T, Rosenthal EL. CD147 and AGR2 expression promote cellular proliferation and metastasis of head and neck squamous cell carcinoma. Exp Cell Res. 2012;318:1788–1798. doi: 10.1016/j.yexcr.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J, Ru NY, Zhang Y, Li Y, Wei D, Ren Z, Huang XF, Chen ZN, Bian H. HAb18G/CD147 promotes epithelial-mesenchymal transition through TGF-beta signaling and is transcriptionally regulated by Slug. Oncogene. 2011;30:4410–4427. doi: 10.1038/onc.2011.149. [DOI] [PubMed] [Google Scholar]
- 34.Arumugam T, Deng D, Bover L, Wang H, Logsdon CD, Ramachandran V. New blocking antibodies against novel AGR2-C4.4A pathway reduce growth and metastasis of pancreatic tumors and increase survival in mice. Mol Cancer Ther. 2015;14:941–951. doi: 10.1158/1535-7163.MCT-14-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma SR, Wang WM, Huang CF, Zhang WF, Sun ZJ. Anterior gradient protein 2 expression in high grade head and neck squamous cell carcinoma correlated with cancer stem cell and epithelial mesenchymal transition. Oncotarget. 2015;6:8807–8821. doi: 10.18632/oncotarget.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thuma F, Ngora H, Zoller M. The metastasis-associated molecule C4.4A promotes tissue invasion and anchorage independence by associating with the alpha6beta4 integrin. Mol Oncol. 2013;7:917–928. doi: 10.1016/j.molonc.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paret C, Bourouba M, Beer A, Miyazaki K, Schnolzer M, Fiedler S, Zoller M. Ly6 family member C4.4A binds laminins 1 and 5, associates with galectin-3 and supports cell migration. Int J Cancer. 2005;115:724–733. doi: 10.1002/ijc.20977. [DOI] [PubMed] [Google Scholar]
- 38.Oshiro R, Yamamoto H, Takahashi H, Ohtsuka M, Wu X, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Sekimoto M, Matsuura N, Doki Y, Mori M. C4.4A is associated with tumor budding and epithelial-mesenchymal transition of colorectal cancer. Cancer Sci. 2012;103:1155–1164. doi: 10.1111/j.1349-7006.2012.02263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


