Abstract
The available drug treatment options for adrenocortical carcinoma (ACC) are limited. In our previous studies, the in vitro activity of 9-cis retinoic acid (9-cisRA) on adrenocortical NCI-H295R cells was shown along with its antitumoral effects in a small pilot xenograft study. Our aim was to dissect the antitumoral effects of 9-cisRA on ACC in a large-scale xenograft study involving mitotane, 9-cisRA and their combination. 43 male SCID mice inoculated with NCI-H295R cells were treated in four groups (i. control, ii. 9-cisRA, iii. mitotane, iv. 9-cisRA + mitotane) for 28 days. Tumor size follow-up, histological and immunohistochemical (Ki-67) analysis, tissue gene expression microarray, quantitative real-time-PCR for the validation of microarray results and to detect circulating microRNAs were performed. Protein expression was studied by proteomics and Western-blot validation. Only mitotane alone and the combination of 9-cisRA and mitotane resulted in significant tumor size reduction. The Ki-67 index was significantly reduced in both 9-cisRA and 9-cisRA+mitotane groups. Only modest changes at the mRNA level were found: the 9-cisRA-induced overexpression of apolipoprotein A4 and down-regulation of phosphodiesterase 4A was validated. The expression of circulating hsa-miR-483-5p was significantly reduced in the combined treatment group. The SET protein was validated as being significantly down-regulated in the combined mitotane+9-cisRA group. 9-cisRA might be a helpful additive agent in the treatment of ACC in combination with mitotane. Circulating hsa-miR-483-5p could be utilized for monitoring the treatment efficacy in ACC patients, and the treatment-induced reduction in protein SET expression might raise its relevance in ACC biology.
Keywords: Adrenocortical cancer, mitotane, 9-cis retinoic acid, xenograft, SET protein, circulating microRNA
Introduction
Adrenocortical carcinoma (ACC) is a rare tumor (incidence is 0.5-2 cases/million people/year) with poor prognosis in its advanced stages [1]. The primary treatment is surgical resection. Even with the most widely used EDPM (etoposide, doxorubicin, cisplatin, mitotane) chemotherapeutic regimen, the median overall survival was only 14.8 months in the recent FIRM-ACT trial [2,3]. Mitotane (o,p-DDD) is the only adrenal specific drug that is registered for the treatment of ACC [4]. Despite its numerous side effects and largely unknown mechanism of action this agent is used in the clinical practice for more than 50 years [5]. Intensive efforts are therefore going on for finding novel, effective drugs with fewer side effects.
In our previous in silico study of adrenocortical tumors genomics data, we have found that retinoic acid signaling via retinoid X receptor is an important pathogenic pathway in ACC [6]. Based on these in silico data, we have performed in vitro studies on the NCI-H295R ACC cell line and found that 9-cis retinoic acid (9-cisRA) was able to diminish hormone synthesis and cell viability in a concentration- and time-dependent manner, and it induced significant gene expression changes [7]. In a pilot in vivo xenograft study in nude mice, 9-cisRA reduced tumor growth, as well [7].
In the present study, we aimed to further dissect the antitumoral effect of 9-cisRA on ACC in a large xenograft study involving both mitotane and 9-cisRA and their combination. We have applied multimodal molecular biological, genomic and proteomic approaches to decipher the molecular way of action of the agents tested.
Materials and methods
Cell culture
The human adrenocortical NCI-H295R cell line was obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in Dulbecco’s modified Eagle’s medium/Nutrient Mixture F-12 Ham (DMEM: F12) supplemented with 3.61*10-8 M selenium (Sigma-Aldrich, St. Louis, MO, USA), 1.92*10-5 M linoleic acid, 0.001 M insulin, 7.81*10-8 M transferrin (Sigma-Aldrich), 1.899*10-5 M bovine serum albumin and adjusted to a final concentration of 1% HEPES, 1% Penicillin/Streptomycin (Sigma-Aldrich), 2.5% Nu-Serum (BD Biosciences, San Jose, CA, USA) and 2.5% L-glutamine (Sigma-Aldrich) at 37°C in a humidified 5% CO2 atmosphere. The medium was changed two or three times a week and subcultured once or twice a week.
Xenograft model
44 male BALB/c SCID (severe combined immunodeficiency) mice aged 6-8 weeks with average weight between 21-23 g were injected subcutaneously with NCI-H295R cell suspension (107 cells/200 µl PBS). Treatment was started when the solid tumor reached 3 mm mean diameter (37 days after the H295R inoculation). The animals have received the treatment by oral gavage for 28 days. Four groups were established: 1. control: 200 µl corn oil/day; 2. mitotane: 200 mg/kg/day/200 µl corn oil; 3. 9-cis retinoic acid: 5 mg/kg/day/200 µl corn oil; and 4. mitotane 200 mg/kg/day/100 µl corn oil + 9-cis retinoic acid 5 mg/kg/day/100 µl corn oil. The tumors were measured twice a week with a caliper by the same investigator. The volumes of the tumors were calculated by the following formula: (a*a*b*π)/6 [8]. On day 29 after starting the treatments, the animals were killed by cervical dislocation in ether anesthesia. Only one animal died before day 29 due to technical problems.
Tumors were removed and their weights were measured. One half of the tumor was fixed in formalin for histological and immunohistochemical examination, the other half was frozen in liquid nitrogen and stored at -80°C until use. Lungs, heart, kidneys, spleen and liver were also removed for histological analysis. Whole blood was collected, then plasma was isolated and stored at -80°C until use. All animal experiments were conducted according to the ethical standards of the animal Health Care and Control Institute, Csongrád County, Hungary, permit No. XVI/02037-2/2008.
Histological and immunohistochemical analysis
Four µm sections of formalin-fixed paraffin-embedded tissues were dewaxed with xylene and ethanol and processed either for hematoxylin-eosin (HE) staining or Ki-67 immunostaining. Ki-67 immunostaining (antibody: Cat. No. M7240, DakoCytomation, Glostrup, Denmark; dilution 1:400) was conducted using a Leica BOND-MAX automated immunostaining system (Leica Biosystems, Wetzlar, Germany). Slides were scanned by Pannoramic Flash II scanner, and digital slides were analyzed by Pannoramic viewer software (3DHistech, Budapest, Hungary). Different regions of the viable xenograft tumor were annotated and Ki-67 positive and negative cells were individually marked on these regions of interests (ROIs). Proliferation index was given in percentages of positive cells. 4-8 slides of each group with good quality were selected for analysis, and 3-5 ROIs/slides were counted with 4498-6273 cells in average. Ki-67 expression was scored by three independent pathologists in a blinded fashion.
RNA isolation from tumor tissue
Total RNA was isolated from the frozen tumors with Qiagen miRNeasy Mini Kit according to the manufacturer’s protocol (Qiagen, Hilden, Germany). RNA concentration was measured with NanoDrop 2000 spectrophotometer. (Thermo Fisher Scientific, Waltham, Mam USA). RNA integrity was determined by Agilent 2100 Bioanalyzer System (Agilent Technologies, Santa Clara, CA, USA). Samples with an RNA integrity number (RIN) above 8.0 were used for further analysis. RNA was stored at -80°C until use.
Messenger RNA (mRNA) expression profiling
Gene expression profiling was performed on 16 samples (4-4 samples from each group) using a single-color array method by 4x44K Agilent Whole Genome Microarray slides (Agilent Technologies).
Total RNA (200 ng) was labeled and amplified using the low Input Quick Amp Labeling Kit according to the instructions of the manufacturer. Labeled RNA was purified and hybridized to Agilent Human Gene Expression Microarray 4x44K array slides, according to the manufacturer’s protocol. After washing, array scanning and feature extraction was performed with default scenario by Agilent DNA Microarray Scanner. Fluorescence intensities of spots were quantified, background subtracted, and dye normalized by Feature Extraction software, version 11.0.1 (Agilent Technologies). The raw data were analyzed with GeneSpring 12.6 software (Agilent Technologies).
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Seven genes were selected for validation by real-time RT-qPCR with Taqman gene expression assays (Thermo Fisher Scientific): MYC (Hs00153408_m1), APOA4 (Hs00166636_m1), CXCR3 (Hs01847760_s1), BAALC (Hs00227249_m1), PDE4A (Hs00183479_m1), PRDM1 (Hs00153357_m1) and TGFBI (Hs00932747_m1). Based on the selection criteria of Cheng et al. [9] and our previous study [10], ZNF625 (Hs00377010_m1) was chosen as reference gene.
Total RNA (10 ng) was reverse transcribed using High-Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific). Quantitative RT-PCR was performed by TaqMan Fast Universal PCR Master Mix (2×) (Thermo Fisher Scientific) on a 7500 Fast Real-Time PCR System (Thermo Fisher Scientific) according to the manufacturer’s protocol. Samples were run in triplicate. For the evaluation of the data we used the ddCT method [11] using Microsoft Excel 2010 (Microsoft Corporation).
RNA isolation from plasma
Total RNA was isolated from 100 µl plasma samples with Qiagen miRNeasy Serum/Plasma Kit according to the attached manufacturer’s protocol (Qiagen) amended with addition of a spike-in control miRNA cel-miR-39 (2594091) (Qiagen). RNA concentration was measured with NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). RNA was stored at -80°C until use.
RT-qPCR for circulating microRNA measurement
Based on our previous studies and literature data, four microRNAs were selected for validation by RT-qPCR with Taqman miRNA assays (Thermo Fisher Scientific): hsa-miR-181b (001098), hsa-miR-184 (000485), hsa-miR-210 (000512) and hsa-miR-483-5p (002338). Cel-mir-39 (000200) was used as reference gene [12].
Total RNA (1 µl) was reverse transcribed using specific TaqMan miRNA assays and TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific) on Proflex Base PCR System (Thermo Fisher Scientific). Quantitative RT-PCR was performed by TaqMan Fast Universal PCR Master Mix (2×) and TaqMan miRNA assays on a 7500 Fast Real-Time PCR System. PCR reactions were run in triplicate. Negative control reactions did not include cDNA templates.
RT-qPCR for tissue microRNA measurement
Considering the results from the circulating microRNA measurements, tissue hsa-miR-483-5p (002338) was studied by RT-qPCR with Taqman miRNA assays according to the instructions of the manufacturer. Four reference genes were used: U6 (001973), RNU6B (001093), RNU44 (001094) and RNU48 (001006).
Proteomics study
Equal amount of protein lysates (20-20 µg of each sample, 3 samples from each group) were separated by SDS-PAGE on a 10% minigel and stained with colloidal Coomassie Brillant Blue. The gel lanes were cut to 10 pieces and their protein content were in-gel digested with trypsin (Promega, Fitchburg, WI, USA) for 4 hours at 37°C after reduction with dithiotreitol and alkylation with iodoacetamide. The digestion reaction was stopped by acidification with formic acid, and the tryptic peptides were extracted from the gel and dried.
Samples were redissolved in 30 µl of 0.1% formic acid and subjected to LC-MSMS (Liquid Chromatography-Mass Spectrometry and Liquid Chromatography-Tandem Mass Spectrometry b, on a Waters nanoAcquity ULC on-line coupled to an Orbitrap Elite Mass spectrometer) analysis. 5 µl sample was loaded onto a Symmetry C18 (5 µm, 180 µm ×20 mm) trap column (Waters, 186003514) in 3% of solvent B and analyzed on a BEH300 C18 (1.7 µm, 075 µm ×250 mm) nanoAcquity UPLC column (Waters 186003815) using a gradient elution (3-40% of solvent B during 37 min, solvent A: 0.1% formic acid in water, solvent B: was 0.1% formic acid in acetonitrile:DMSO (95:5)). The flow rate was 300 nl/min. The LTQ-Orbitrap Elite mass spectrometer operated in data dependent mode, each survey scan (full MS measured in the Orbitrap R=60000, m/z range: 380-1600) was followed with ion-trap collision induced dissociation (CID) scans of the 10 most intense peaks. Dynamic exclusion was used for 30 seconds and single charged ions were not selected for fragmentation.
Mass spectrometry raw data were converted to MSMS peak list files using PAVA script [13]. Peak list files corresponding to the same sample (10 to each) were merged and searched against the human, mouse species specified UniProtKB random concat (06.11.2014) protein database, containing 136244 human and 74540 mouse proteins and their randomized sequences. The search was done with the following parameters: fully specific tryptic peptides with maximum 2 missed cleavages were allowed. All cysteines were considered as carbamidomethylated and some variable modification has been allowed: oxidized methionine, acetylation of protein N-termini and pyroglutamic acid modification at peptide N-terminal glutamine. Parent mass tolerance was set to 10 ppm, while fragment mass tolerance was 0.6 Da. All database search were completed on our in-house ProteinProspector (ver. 5.14.1) (Baker, P.R. and Clauser, K.R. http://prospector.ucsf.edu accessed: July 22. 2015) search engine.
Spectral count was used for semi-quantitation. The number of spectra identified a protein were normalized by the number of all identified spectra in the sample. These relative spectral counts were compared in between the different samples.
Western-blot analysis
Based on our proteomic results and literature data, the SET (SET nuclear proto-oncogene) protein was chosen for Western-blot analysis on 3 samples from each group of the xenograft study and on altogether 6 human adrenocortical samples (2 normal adrenal cortices, 2 ACAs (adrenocortical adenoma) and 2 ACCs (adrenocortical cancer)). Normal human adrenal cortices were obtained from patients operated for hypernephroma [14]. The study on human samples was approved by the Ethical Committee of the Hungarian Health Council and informed consent was obtained from all patients involved.
Tumor tissues were pulverized in liquid nitrogen and lysed in 1 ml of Lysis Buffer (0.02 M Tris, 0.15 M NaCl, 0.002 M EDTA, 0.5% Triton X-100, 0.5% Protease Inhibitor Cocktail (Sigma Aldrich), 0.01 M NaF, 0.002 M Na3VO4 on ice for 30 minutes. Next, samples were centrifuged at 13000 rpm for 15 min. Supernatants were kept and protein concentrations were measured as described before by Bradford [15]. 20 µg of total protein per sample was mixed with loading buffer containing β-mercaptoethanol and was incubated at 99°C for 5 min. Denatured samples were loaded onto a 10% polyacrylamide gel and were run for 30 min at 200 V on a Mini Protean vertical electrophoresis equipment (Bio-Rad, Hercules, CA). Proteins were transferred onto a PVDF membrane (Milipore, Billerca, MA) by blotting overnight at 75 mA. Ponceau-staining was applied to see the loading and blotting efficiency. Membranes were blocked with 5 w/v% non-fat dry milk (Bio-Rad) in TBS (0.15 M NaCl, 0.02 M Tris-HCl pH=7.5) for 1 hour followed by incubation with either anti-SET antibody (ab1183, Abcam, Cambridge, MA, dilution 1:1000) or anti-β-actin antibody (A2228, Sigma Aldrich) for 16 h at 4°C. Membranes were washed 5 times with TBST (TBS+0.05% Tween-20) then incubated with secondary, HRP-conjugated anti-rabbit (P0448, DakoCytomation) or anti-mouse (P0447, DakoCytomation) antibodies for 1 hour. SuperSignal West Pico Chemiluminescent Substrate Kit (Pierce/Thermo Fisher Scientific) and Kodak Image Station 4000 MM Digital Imaging System were used to visualize the signals. β-actin served as loading control. Band density was measured by ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
For the statistical analysis of tumor growth, Mann-Whitney U test was applied (SPSS Statistics 20, IBM). The analysis of Ki-67 index was performed by One-way ANOVA followed by Tukey’s post Hoc test or Kruskal-Wallis ANOVA & Median Test (STATISTICA 7.0) depending on the results of the Shapiro-Wilks normality test. P<0.05 was considered to be significant.
The results from the microarray data were analyzed by Genespring 12.6 software (Agilent Technologies). A filter on expression at the 20th percentile of raw signal values, then a 2-fold change filter was used, and One-way ANOVA was performed, followed by Tukey’s post Hoc test without Benjamini-Hochberg false discovery rate calculation. RT-qPCR and proteomics data were analyzed by One-way ANOVA followed by Tukey’s post Hoc test (STATISTICA 7.0).
Western-blot was evaluated by Kruskal-Wallis ANOVA & Median Test (STATISTICA 7.0).
Results
Analysis of xenograft tumor growth
The normalized tumor volume (relative to the first measured volume) was smaller in all treated groups than in controls during the whole experiment. At the end of our experiment, the average values of normalized tumor volumes were 22.48-fold, 9.3-fold, 12.3-fold and 8.22-fold higher in the control, mitotane treated, 9-cisRA-treated, and 9-cisRA+mitotane groups relative to the starting tumor volume, respectively. The reduction in tumor size has been significant in the mitotane only and 9-cisRA+mitotane groups (Figure 1A and 1B).
Figure 1.
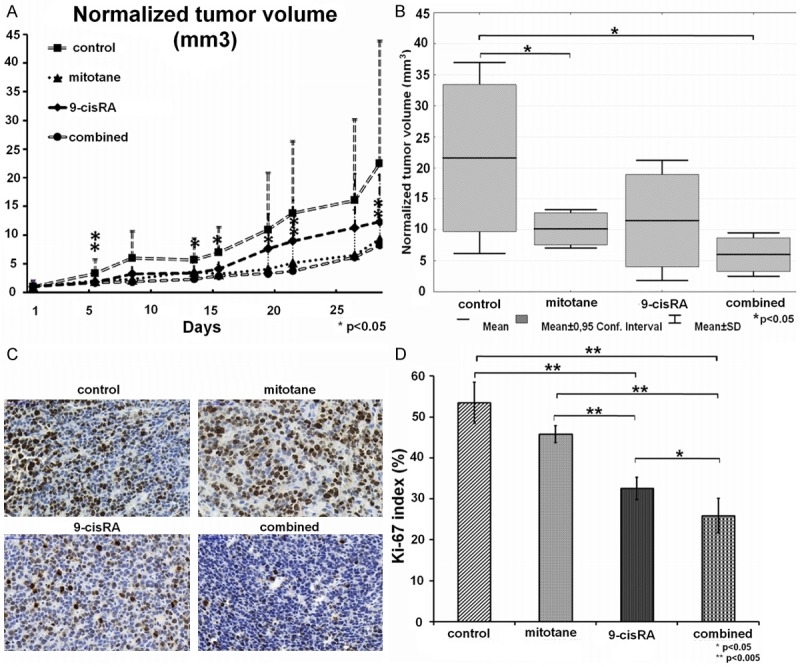
A. Normalized tumor volumes; B. Normalized tumor volumes at the end of the treatments on day 28; C. Representative images of the Ki-67 immunhistochemical analysis from the four treatment groups; D. Numerical values of the Ki-67 immunhistochemical analysis. (Average results by three independent investigators).
Histology, Ki-67 scoring
Tumors in the 9-cis RA receiving groups appeared to be more differentiated compared to the control and mitotane groups by hematoxylin and eosin (H&E) staining. The Ki-67 proliferation index scored blindly by three independent pathologists was significantly lower in the 9-cisRA (32.64 ± 3.07%) group in comparison to the control (54.23 ± 3.71%) and mitotane groups (46.35% ± 2.2%) (P=0.00018 vs. control; P=0.00022 vs. mitotane). The lowest Ki-67 index was noted in the combined 9-cisRA+mitotane group (25.6 ± 4.09%) that was significantly smaller compared to all other groups (P=0.00018 vs. control; P=0.00018 vs. mitotane; P=0.01684 vs. 9-cisRA) (Figure 1C and 1D). Among the other tissues, only moderate fatty lesions were observed in the liver, probably due to the corn oil vehicle. No signs of treatment toxicity have been found.
Microarray analysis
4 samples from each group (altogether 16 samples) have been subjected to microarray. By analyzing the raw data, we have found 483 significant gene expression changes after the statistical analysis, but only by omitting the Benjamini-Hochberg false discovery rate (FDR). By using FDR, only two significantly differentially expressed transcripts emerged whose biological relevance is unknown. Microarray data are accessible at Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo, accession number: GSE73417).
RT-qPCR validation
Seven genes have been selected for validation based on the microarray data chosen from genes with the highest positive and negative fold changes, taking into account literature data, as well. However, only two genes could be validated to be significantly differentially expressed (Figure 2). The well-known 9-cis retinoic acid target gene APOA4 (apolipoprotein A4) turned out to be significantly overexpressed in the combined treated group relative to the control group (P=0.0045), whereas PDE4A (phosphodiesterase 4A) appeared to be significantly underexpressed in the combined treatment group (P=0.0024).
Figure 2.
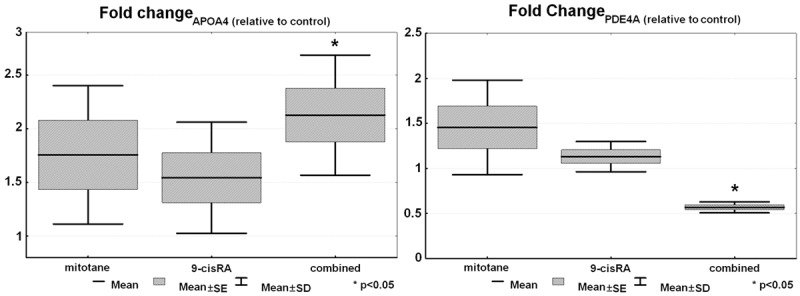
Results of RT-qPCR validation of APOA4 and PDEA4 (n=6).
Proteomics analysis and Western-blot
47 significant protein changes have been found between the groups (Table 1). By considering literature data regarding the tumor biological relevance of these proteins, the SET protein [16-20] was chosen for validation by Western-blotting.
Table 1.
Treatment-induced significant protein changes (47 proteins) based on the results of proteomic analysis (P<0.05)
| UniProt ID | Gene Name | Protein Name |
|---|---|---|
| P30050 | RPL12 | 60S ribosomal protein L12 |
| Q13765 | NACA | Nascent polypeptide-associated complex subunit alpha |
| B1AK87 | CAPZB | Capping protein (Actin filament) muscle Z-line, beta |
| Q01105 | SET | Phosphatase 2A inhibitor I2PP2A |
| Q86SZ7 | PSME2 | PSME2 protein |
| P14314 | PRKCSH | Protein kinase C substrate 60.1 kDa protein heavy chain |
| Q5U8W9 | HRMT1L2 | Protein arginine methyltransferase 1 isoform 4 |
| Q6DC98 | LMNB1 | LMNB1 protein |
| M0QZL1 | BLVRB | Flavin reductase (NADPH) |
| P61158 | ACTR3 | Actin-related protein 3 |
| Q9UBQ7 | GRHPR | Glyoxylate reductase/hydroxypyruvate reductase |
| O75347 | TBCA | Tubulin-specific chaperone A |
| Q6NVY0 | CACYBP | Calcyclin binding protein |
| K7ES63 | TUBB6 | Tubulin beta-6 chain |
| P42126 | ECI1 | Enoyl-CoA delta isomerase 1 |
| Q5T1M5 | FKBP15 | FK506-binding protein 15 |
| O75477 | ERLIN1 | Erlin-1 |
| H7BZ09 | ANP32A | Acidic leucine-rich nuclear phosphoprotein 32 family member A |
| E9PK45 | FTH1 | Ferritin heavy chain |
| P62714 | PPP2CB | Serine/threonine-protein phosphatase 2A catalytic subunit beta |
| P31040 | SDHA | Succinate dehydrogenase |
| H7C333 | GBAS | Protein NipSnap homolog 2 |
| Q15404 | RSU1 | Ras suppressor protein 1 |
| Q92882 | OSTF1 | Osteoclast-stimulating factor 1 |
| D6RC06 | HINT1 | Histidine triad nucleotide-binding protein 1 |
| P62888 | RPL30 | 60S ribosomal protein L30 |
| Q969G3 | SMARCE1 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 |
| P61326 | MAGOH | Protein mago nashi homolog |
| Q16539 | MAPK14 | Mitogen-activated protein kinase 14 |
| P10599 | TXN | Thioredoxin |
| Q9Y333 | LSM2 | U6 snRNA-associated Sm-like protein LSm2 |
| P26885 | FKBP2 | Peptidyl-prolyl cis-trans isomerase FKBP2 |
| Q9UJV8 | PURG | Purine-rich element-binding protein gamma |
| P04080 | CSTB | Cystatin-B |
| E5RJH5 | ENO3 | Beta-enolase |
| P28062 | PSMB8 | Proteasome subunit beta type-8 |
| P67775 | PPP2CA | Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform |
| Q53G26 | DNAJA3 | DnaJ (Hsp40) homolog, subfamily A, member 3 variant |
| Q9UBI6 | GNG12 | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-12 |
| Q8NHP8 | PLBD2 | Putative phospholipase B-like 2 |
| D6R9R5 | SAR1B | GTP-binding protein SAR1b |
| Q00169 | PITPNA | Phosphatidylinositol transfer protein alpha isoform |
| P63173 | RPL38 | 60S ribosomal protein L38 |
| Q5T6V5 | C9orf64 | UPF0553 protein C9orf64 |
| A4D2P2 | RAC1 | Ras-related C3 botulinum toxin substrate 1 |
SET protein expression was smaller in all treated groups relative to control, but only reached the level of significance in the combined treated group (Figure 3A and 3C). To look at the potential relevance of SET in human adrenocortical tumors, we have analyzed some human tissues in a preliminary study, and found that SET protein is undetectable in normal and ACA tissues, whereas it is expressed in ACC (Figure 3B).
Figure 3.
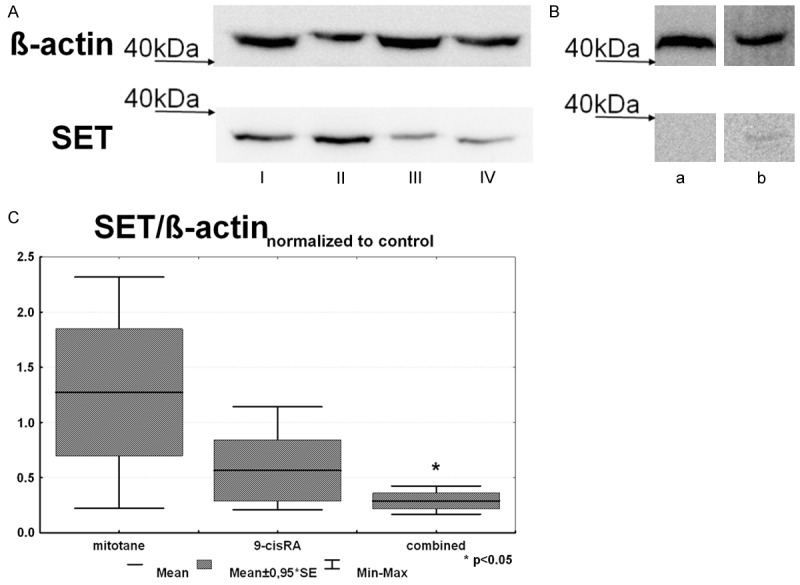
Results of Western-blot analysis of SET expression. A: Representative image SET expression in the xenograft model: I: Mitotane, II: Control, III: 9-cisRA, IV: Combined treatment group; B: Representative image of SET expression from a normal human adrenocortical cortex (a) and ACC tissue (b); C: Results of the densitometry analysis of SET expression in the xenograft model (n=3).
microRNA expression
From the four selected circulating miRNAs, hsa-miR-483-5p has been significantly underexpressed in the combined group relative to control (P=0.028) (Figure 4). The expression of the other three tested circulating microRNAs was unchanged. However, we have not found significant changes in the expression of tissue hsa-miR-483-5p. From the four reference genes tested for tissue microRNAs, RNU44 and RNU6B turned out to be the best for normalizing the results.
Figure 4.
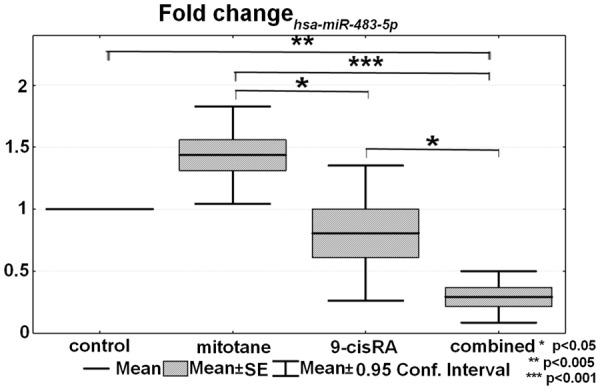
The expressional difference of circulating hsa-miR-483-5p (n=6).
Discussion
Mitotane is the only available adrenal cortex specific agent in the therapy of adrenocortical cancer. Despite being used for more than 50 years, its precise mechanism of action is largely unknown [5]. The degeneration of mitochondria, free radicals, inhibition of steroid biosynthetic enzymes, lipid-mediated endoplasmic reticulum stress have all been suggested to take part in the action of mitotane [5,21]. The narrow therapeutic range and frequent side effects of mitotane constitute serious problems in clinical practice. Based on our previous in vitro and pilot xenograft study [7], 9-cisRA appeared to be a promising agent in ACC treatment. To further dissect its actions and to compare it to mitotane, we have performed a large-scale xenograft study involving 9-cisRA, mitotane and their combination. We have observed that these agents act together in a synergistic manner on tumor growth.
Contrary to the several human in vivo studies applying mitotane, there are only few studies on mitotane-treated xenograft models to date [22-27]. Since there was no uniformly defined dose of mitotane given to mice, we have chosen the average amount used in published studies. The results of these studies on the effectiveness of mitotane in animal models are also contradictory. Lindhe et al. described that mitotane has antitumoral effect given simultaneously with the tumor cells inoculation [24], whereas Doghman et al. did not find any long-lasting effect in a similar study [25]. Here, we have observed the antitumoral activity of mitotane.
Retinoids, the natural and synthetic derivates of vitamin A play crucial roles in cell differentiation, growth, and death. The anti-proliferative and antitumoral effects [28] of 9-cisRA acting via the retinoid X receptor are well documented in many different types of tumors in vitro and in vivo e.g. gastric cell line [29], head and neck squamous cell line [30], neuroblastoma [31] and breast cancer [32].
The dose of 9-cis retinoic acid (5 mg/kg) was much less than the documented toxic dose [30] and it was comparable to other in vivo doses used in the literature [31,33-35].
We have observed antitumoral effects of both 9-cisRA and mitotane when used alone, but significant reduction in tumor volume could only be achieved by groups receiving mitotane alone and the combination of mitotane and 9-cisRA. The Ki-67 index was the lowest in the group receiving the combined treatment. Despite mitotane was more effective than 9-cisRA as a single agent for reducing tumor size, the Ki-67 index was lower in the 9-cisRA-treated group than in the mitotane only group (Figure 1). The background for this phenomenon is unclear.
To decipher the molecular mechanisms behind the observed effects on tumor growth and proliferation, we have performed molecular analysis at mRNA and protein levels. In our previous in vitro studies on the adrenocortical cell line NCI-H295R, we have observed that 9-cisRA affected the expression of more than 2000 genes [7] whereas the effect of mitotane on gene expression was more limited [10]. In contrast to these in vitro data and the observed changes in tumor size and Ki-67 levels, we could observe only modest changes in gene expression in the present study. By omitting the Benjamini-Hochberg False Discovery Rate, 483 genes were found to be significantly genes, and from the 7 selected genes only 2 (APOA4, PDEA4) was validated to be significantly differentially expressed.
APOA4, a member of apolipoprotein family is a known 9-cisRA target gene that has important roles in fat and glucose metabolism [36], in antioxidant [37], and anti-atherogenic processes [38], but it has been suggested also as a predictive factor in inflammatory bowel disease [39,40], and as a novel serum biomarker in ovarian, cervical cancer, and acute lymphocytic leukemia [41-44]. There are no data, however, on the potential relevance of APOA4 in adrenocortical tumors.
PDE4A, as a member of PDE (phosphodiesterase) family, act as crucial cAMP level regulator [45], and it is involved in the basic cell pathways and also takes part in cancer progression. Its inhibition has been associated with many antitumoral actions e.g. regression of brain tumors [46], reduced proliferation and angiogenesis in lung cancer [47], reduced motility and invasion in colon [48] and breast cancer cell line [49], decreased growth of malignant melanoma [50]. In line with these observations, we have also observed reduced expression of PDE4A in the combined treatment group, thus PDE4A might also be involved in ACC pathogenesis or progression.
We hypothesize that after the long treatment period of 28 days, mRNA expression does not really reflects the altered tumor behavior. We have therefore turned to the protein level, where we have found 47 significantly differentially expressed proteins by proteomics analysis. We have chosen the SET protein from the significantly differentially expressed proteins for validation by Western-blot. As expected, SET expression was down-regulated by the treatments, showing the lowest expression in the combined treatment group.
Protein SET is an inhibitor of tumor suppressor PP2A (protein phosphatase 2A) that has widespread actions in the cellular functions implying cell cycle [18], apoptosis [51] and cell migration [19]. Due its target PP2A, it also influences ß-catenin [20], c-Myc [17] and Akt [16] pathways, and via inhibition of nm23-H1 (NME/NM23 nucleoside diphosphate kinase 1) it also promotes metastatic potential [52], and thus it is associated with tumor progression. SET overexpression is described in many tumors e.g. Wilm’s tumor [53], acute lymphoblastic leukaemia [54], chronic lymphocytic leukemia and non-Hodgkin lymphoma [55], lung [56], colon [57], pancreatic [58], prostate [59] and ovarian cancer [60], as well. To the best of our knowledge, our study is the first to raise the potential relevance of SET in ACC biology.
We have performed a preliminary evaluation of SET protein expression in human adrenocortical tumor samples, as well. SET was weakly expressed in ACC, but absent from benign adenomas and normal adrenocortical samples. This finding that certainly awaits validation on a larger cohort appears to support our xenograft findings where SET expression was the highest in the untreated xenograft, and suppressed in the treated samples parallel to tumor shrinkage.
In addition to our mechanistic efforts aimed at deciphering the molecular way of action of 9-cisRA and mitotane in our xenograft model, we have also studied some circulating microRNAs as potential markers for treatment efficacy monitoring based on our previous studies in humans. Circulating hsa-miR-483-5p appears to be the best circulating microRNA marker of adrenocortical malignancy [61-63]. From the four analyzed circulating microRNAs, only hsa-miR-483-5p expression was modulated by the treatments, and its expression was significantly suppressed by the combined 9-cisRA + mitotane treatment. Circulating hsa-miR-483-5p thus appears to be a marker of treatment efficacy, and this could be relevant in the clinical setting, as well. Regarding the tissue expression of hsa-miR-483-5p, however, no significant differences have been observed. We can conclude that the circulating and tissue hsa-miR-483-5p alter independently. There are data that microRNAs can be changed parallel, contrary, or independently between the circulation and tissues [64,65], but the molecular mechanisms underlying these discrepancies and the regulation of adrenocortical hsa-miR-483-5p expression are largely unknown, yet.
In conclusion, 9-cisRA might represent an alternative additive treatment option in ACC that seems to be most efficient when combined with mitotane. The tumor size, Ki-67, SET protein, and circulating hsa-miR-483-5p were all lowest in the group receiving the combined treatment that suggest synergistic action of 9-cisRA and mitotane in our xenograft model. The molecular background of the observed synergistic mitotane-9-cisRA action is, however, unclear. We have found only modest gene expression changes, and although the number of affected proteins is higher, their list is not extensive, either. The decrease of SET protein expression by the treatments might be noteworthy, as paralleled by its expression in human adrenocortical tumors. The potential applicability of circulating hsa-miR-483-5p for monitoring ACC treatment efficacy might be relevant in the clinical setting, as there is no reliable blood-based tumor marker of ACC at present. Circulating microRNA might be exploited for treatment monitoring in other diseases, as well.
Acknowledgements
This study has been supported by grants from the Hungarian National Research, Development and Innovation Office-NKFIH (grants K100295 and K115398) to Dr. Peter Igaz.
Disclosure of conflict of interest
None.
References
- 1.Fassnacht M, Kroiss M, Allolio B. Update in adrenocortical carcinoma. J Clin Endocrinol Metab. 2013;98:4551–64. doi: 10.1210/jc.2013-3020. [DOI] [PubMed] [Google Scholar]
- 2.Else T, Kim AC, Sabolch A, Raymond VM, Kandathil A, Caoili EM, Jolly S, Miller BS, Giordano TJ, Hammer GD. Adrenocortical Carcinoma. Endocr Rev. 2014;35:282–326. doi: 10.1210/er.2013-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fassnacht M, Terzolo M, Allolio B, Baudin E, Haak H, Berruti A, Welin S, Schade-Brittinger C, Lacroix A, Jarzab B, Sorbye H, Torpy DJ, Stepan V, Schteingart DE, Arlt W, Kroiss M, Leboulleux S, Sperone P, Sundin A, Hermsen I, Hahner S, Willenberg HS, Tabarin A, Quinkler M, de la Fouchardière C, Schlumberger M, Mantero F, Weismann D, Beuschlein F, Gelderblom H, Wilmink H, Sender M, Edgerly M, Kenn W, Fojo T, Müller HH, Skogseid B. Combination chemotherapy in advanced adrenocortical carcinoma. N Engl J Med. 2012;366:2189–97. doi: 10.1056/NEJMoa1200966. [DOI] [PubMed] [Google Scholar]
- 4.Terzolo M, Angeli A, Fassnacht M, Daffara F, Tauchmanova L, Conton PA, Rossetto R, Buci L, Sperone P, Grossrubatscher E, Reimondo G, Bollito E, Papotti M, Saeger W, Hahner S, Koschker AC, Arvat E, Ambrosi B, Loli P, Lombardi G, Mannelli M, Bruzzi P, Mantero F, Allolio B, Dogliotti L, Berruti A. Adjuvant mitotane treatment for adrenocortical carcinoma. N Engl J Med. 2007;356:2372–80. doi: 10.1056/NEJMoa063360. [DOI] [PubMed] [Google Scholar]
- 5.Igaz P, Tombol Z, Szabo P, Liko I, Racz K. Steroid Biosynthesis Inhibitors in the Therapy of Hypercortisolism: Theory and Practice. Curr Med Chem. 2008;15:2734–47. doi: 10.2174/092986708786242921. [DOI] [PubMed] [Google Scholar]
- 6.Szabó PM, Tamási V, Molnár V, Andrásfalvy M, Tömböl Z, Farkas R, Kövesdi K, Patócs A, Tóth M, Szalai C, Falus A, Rácz K, Igaz P. Metaanalysis of adrenocortical tumour genomics data: novel pathogenic pathways revealed. Oncogene. 2010;29:3163–72. doi: 10.1038/onc.2010.80. [DOI] [PubMed] [Google Scholar]
- 7.Szabó DR, Baghy K, Szabó PM, Zsippai A, Marczell I, Nagy Z, Varga V, Éder K, Tóth S, Buzás EI, Falus A, Kovalszky I, Patócs A, Rácz K, Igaz P. Antitumoral effects of 9-cis retinoic acid in adrenocortical cancer. Cell Mol Life Sci. 2014;71:917–32. doi: 10.1007/s00018-013-1408-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doghman M, El Wakil A, Cardinaud B, Thomas E, Wang J, Zhao W, Peralta-Del Valle MH, Figueiredo BC, Zambetti GP, Lalli E. Regulation of insulin-like growth factor-mammalian target of rapamycin signaling by microRNA in childhood adrenocortical tumors. Cancer Res. 2010;70:4666–75. doi: 10.1158/0008-5472.CAN-09-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng WC, Chang CW, Chen CR, Tsai ML, Shu WY, Li CY, Hsu IC. Identification of reference genes across physiological states for qRT-PCR through microarray meta-analysis. PLoS One. 2011;6:e17347. doi: 10.1371/journal.pone.0017347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zsippai A, Szabó DR, Tömböl Z, Szabó PM, Éder K, Pállinger É, Gaillard RC, Patócs A, Tóth S, Falus A, Rácz K, Igaz P. Effects of mitotane on gene expression in the adrenocortical cell line NCI-H295R: a microarray study. Pharmacogenomics. 2012;13:1351–61. doi: 10.2217/pgs.12.116. [DOI] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan S, Price JC, Prusiner SB, Ghaemmaghami S, Burlingame AL. A Data Processing Pipeline for Mammalian Proteome Dynamics Studies Using Stable Isotope Metabolic Labeling. Mol Cell Proteomics. 2011;10:M111.010728–M111.010728. doi: 10.1074/mcp.M111.010728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tömböl Z, Szabó PM, Molnár V, Wiener Z, Tölgyesi G, Horányi J, Riesz P, Reismann P, Patócs A, Likó I, Gaillard RC, Falus A, Rácz K, Igaz P. Integrative molecular bioinformatics study of human adrenocortical tumors: microRNA, tissue-specific target prediction, and pathway analysis. Endocr Relat Cancer. 2009;16:895–906. doi: 10.1677/ERC-09-0096. [DOI] [PubMed] [Google Scholar]
- 15.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 16.Leopoldino AM, Squarize CH, Garcia CB, Almeida LO, Pestana CR, Polizello AC, Uyemura SA, Tajara EH, Gutkind JS, Curti C. Accumulation of the SET protein in HEK293T cells and mild oxidative stress: Cell survival or death signaling. Mol Cell Biochem. 2012;363:65–74. doi: 10.1007/s11010-011-1158-x. [DOI] [PubMed] [Google Scholar]
- 17.Arnold HK, Sears RC. A tumor suppressor role for PP2A-B56alpha through negative regulation of c-Myc and other key oncoproteins. Cancer Metastasis Rev. 2008;27:147–58. doi: 10.1007/s10555-008-9128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canela N, Rodriguez-Vilarrupla A, Estanyol JM, Diaz C, Pujol MJ, Agell N, Bachs O. The SET protein regulates G2/M transition by modulating cyclin B-cyclin-dependent kinase 1 activity. J Biol Chem. 2003;278:1158–64. doi: 10.1074/jbc.M207497200. [DOI] [PubMed] [Google Scholar]
- 19.Ten Klooster JP, Leeuwen Iv, Scheres N, Anthony EC, Hordijk PL. Rac1-induced cell migration requires membrane recruitment of the nuclear oncogene SET. EMBO J. 2007;26:336–45. doi: 10.1038/sj.emboj.7601518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Götz J, Probst A, Mistl C, Nitsch RM, Ehler E. Distinct role of protein phosphatase 2A subunit Cα in the regulation of E-cadherin and β-catenin during development. Mech Dev. 2000;93:83–93. doi: 10.1016/s0925-4773(00)00267-7. [DOI] [PubMed] [Google Scholar]
- 21.Sbiera S, Leich E, Liebisch G, Sbiera I, Schirbel A, Wiemer L, Matysik S, Eckhardt C, Gardill F, Gehl A, Kendl S, Weigand I, Bala M, Ronchi CL, Deutschbein T, Schmitz G, Rosenwald A, Allolio B, Fassnacht M, Kroiss M. Mitotane inhibits Sterol-O-Acyl Transferase 1 triggering lipidmediated endoplasmic reticulum stress and apoptosis in adrenocortical carcinoma cells. Endocrinology. 2015;156:3895–908. doi: 10.1210/en.2015-1367. [DOI] [PubMed] [Google Scholar]
- 22.Stacpoole PW, Varnado CE, Island DP. Stimulation of rat liver 3-hydroxy-3-methylglutarylcoenzyme A reductase activity by o,p’-DDD. Biochem Pharmacol. 1982;31:857–60. doi: 10.1016/0006-2952(82)90474-9. [DOI] [PubMed] [Google Scholar]
- 23.Barker EN, Campbell S, Tebb AJ, Neiger R, Herrtage ME, Reid SWJ, Ramsey IK. A comparison of the survival times of dogs treated with mitotane or trilostane for pituitary-dependent hyperadrenocorticism. J Vet Intern Med. 2005;19:810–5. doi: 10.1892/0891-6640(2005)19[810:acotst]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 24.Lindhe Ö, Skogseid B. Mitotane Effects in a H295R Xenograft Model of Adjuvant Treatment of Adrenocortical Cancer. Horm Metab Res. 2010;42:725–30. doi: 10.1055/s-0030-1261923. [DOI] [PubMed] [Google Scholar]
- 25.Doghman M, Lalli E. Lack of long-lasting effects of mitotane adjuvant therapy in a mouse xenograft model of adrenocortical carcinoma. Mol Cell Endocrinol. 2013;381:66–9. doi: 10.1016/j.mce.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 26.Arenas C, Melián C, Pérez-Alenza MD. Longterm survival of dogs with adrenal-dependent hyperadrenocorticism: A comparison between mitotane and twice daily trilostane treatment. J Vet Intern Med. 2014;28:473–80. doi: 10.1111/jvim.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hantel C, Jung S, Mussack T, Reincke M, Beuschlein F. Liposomal polychemotherapy improves adrenocortical carcinoma treatment in a preclinical rodent model. Endocr Relat Cancer. 2014;21:383–94. doi: 10.1530/ERC-13-0439. [DOI] [PubMed] [Google Scholar]
- 28.Sun SY, Lotan R. Retinoids and their receptors in cancer development and chemoprevention. Crit Rev Oncol Hematol. 2002;41:41–55. doi: 10.1016/s1040-8428(01)00144-5. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Zhu Z, Zhang SN, Mou J, Liu L, Cui T, Pei DS. Combinational effect of PPARγ agonist and RXR agonist on the growth of SGC7901 gastric carcinoma cells in vitro. Tumour Biol. 2013;34:2409–18. doi: 10.1007/s13277-013-0791-2. [DOI] [PubMed] [Google Scholar]
- 30.Shalinsky DR, Bischoff ED, Gregory ML, Gottardis MM, Hayes JS, Lamph WW, Heyman RA, Shirley MA, Cooke TA, Davies PJ. Retinoidinduced suppression of squamous cell differentiation in human oral squamous cell carcinoma xenografts (line 1483) in athymic nude mice. Cancer Res. 1995;55:3183–91. [PubMed] [Google Scholar]
- 31.Ponthan F, Kogner P, Bjellerup P, Klevenvall L, Hassan M. Bioavailability and dose-dependent anti-tumour effects of 9-cis retinoic acid on human neuroblastoma xenografts in rat. Br J Cancer. 2001;85:2004–9. doi: 10.1054/bjoc.2001.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maeng S, Kim GJ, Choi EJ, Yang HO, Lee DS, Sohn YC. 9-Cis-retinoic acid induces growth inhibition in retinoid-sensitive breast cancer and sea urchin embryonic cells via retinoid X receptor α and replication factor C3. Mol Endocrinol. 2012;26:1821–35. doi: 10.1210/me.2012-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCormick DL, Rao KV, Steele VE, Lubet RA, Kelloff GJ, Bosland MC. Chemoprevention of rat prostate carcinogenesis by 9-cis-retinoic acid. Cancer Res. 1999;59:521–4. [PubMed] [Google Scholar]
- 34.Shalinsky DR, Bischoff ED, Gregory ML, Lamph WW, Heyman RA, Hayes JS, Thomazy V, Davies PJ. Enhanced antitumor efficacy of cisplatin in combination with ALRT1057 (9-cis retinoic acid) in human oral squamous carcinoma xenografts in nude mice. Clin Cancer Res. 1996;2:511–20. [PubMed] [Google Scholar]
- 35.Wu K, Kim H, Rodriquez JL, Munoz-medellin D, Mohsin SK, Hilsenbeck SG, Lamph WW, Gottardis MM, Shirley MA, Kuhn JG, Green JE, Brown PH. 9-cis-Retinoic Acid Suppresses Mammary Tumorigenesis in C3(1)-Simian Virus 40 T Antigen-transgenic Mice 9-cis-Retinoic Acid Suppresses Mammary Tumorigenesis in. Clin Cancer Res. 2000;3:3696–704. [PubMed] [Google Scholar]
- 36.Wang F, Kohan AB, Kindel TL, Corbin KL, Nunemaker CS, Obici S, Woods SC, Davidson WS, Tso P. Apolipoprotein A-IV improves glucose homeostasis by enhancing insulin secretion. Proc Natl Acad Sci U S A. 2012;109:9641–6. doi: 10.1073/pnas.1201433109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spaulding HL. Apolipoprotein A-IV attenuates oxidant-induced apoptosis in mitotic competent, undifferentiated cells by modulating intracellular glutathione redox balance. Am J Physiol Cell Physiol. 2005;290:C95–103. doi: 10.1152/ajpcell.00388.2005. [DOI] [PubMed] [Google Scholar]
- 38.Culnan DM, Cooney RN, Stanley B, Lynch CJ. Apolipoprotein A-IV, a putative satiety/antiatherogenic factor, rises after gastric bypass. Obesity (Silver Spring) 2009;17:46–52. doi: 10.1038/oby.2008.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Broedl UC, Schachinger V, Lingenhel A, Lehrke M, Stark R, Seibold F, Goke B, Kronenberg F, Parhofer KG, Konrad-Zerna A. Apolipoprotein A-IV is an independent predictor of disease activity in patients with inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:391–7. doi: 10.1002/ibd.20078. [DOI] [PubMed] [Google Scholar]
- 40.Orsó E, Moehle C, Boettcher A, Szakszon K, Werner T, Langmann T, Liebisch G, Buechler C, Ritter M, Kronenberg F, Dieplinger H, Bornstein SR, Stremmel W, Schmitz G. The satiety factor apolipoprotein A-IV modulates intestinal epithelial permeability through its interaction with α-catenin: Implications for inflammatory bowel diseases. Horm Metab Res. 2007;39:601–11. doi: 10.1055/s-2007-984466. [DOI] [PubMed] [Google Scholar]
- 41.Timms JF, Arslan-Low E, Kabir M, Worthington J, Camuzeaux S, Sinclair J, Szaub J, Afrough B, Podust VN, Fourkala EO, Cubizolles M, Kronenberg F, Fung ET, Gentry-Maharaj A, Menon U, Jacobs I. Discovery of serum biomarkers of ovarian cancer using complementary proteomic profiling strategies. Proteomics Clin Appl. 2014;8:982–93. doi: 10.1002/prca.201400063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo X, Hao Y, Kamilijiang M, Hasimu A, Yuan J, Wu G, Reyimu H, Kadeer N, Abudula A. Potential predictive plasma biomarkers for cervical cancer by 2D-DIGE proteomics and Ingenuity Pathway Analysis. Tumour Biol. 2014;36:1711–20. doi: 10.1007/s13277-014-2772-5. [DOI] [PubMed] [Google Scholar]
- 43.Li L, Xu Y, Yu CX. Proteomic analysis of serum of women with elevated Ca-125 to differentiate malignant from benign ovarian tumors. Asian Pac J Cancer Prev. 2012;13:3265–70. doi: 10.7314/apjcp.2012.13.7.3265. [DOI] [PubMed] [Google Scholar]
- 44.Braoudaki M, Lambrou GI, Vougas K, Karamolegou K, Tsangaris GT, Tzortzatou-Stathopoulou F. Protein biomarkers distinguish between high- and low-risk pediatric acute lymphoblastic leukemia in a tissue specific manner. J Hematol Oncol. 2013;6:52. doi: 10.1186/1756-8722-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conti M, Richter W, Mehats C, Livera G, Park JY, Jin C. Cyclic AMP-specific PDE4 phosphodiesterases as critical components of cyclic AMP signaling. J Biol Chem. 2003;278:5493–6. doi: 10.1074/jbc.R200029200. [DOI] [PubMed] [Google Scholar]
- 46.Goldhoff P, Warrington NM, Limbrick DD, Hope A, Woerner BM, Jackson E, Perry A, Piwnica-Worms D, Rubin JB. Targeted inhibition of cyclic AMP phosphodiesterase-4 promotes brain tumor regression. Clin Cancer Res. 2008;14:7717–25. doi: 10.1158/1078-0432.CCR-08-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pullamsetti SS, Banat GA, Schmall A, Szibor M, Pomagruk D, Hänze J, Kolosionek E, Wilhelm J, Braun T, Grimminger F, Seeger W, Schermuly RT, Savai R. Phosphodiesterase-4 promotes proliferation and angiogenesis of lung cancer by crosstalk with HIF. Oncogene. 2013;32:1121–34. doi: 10.1038/onc.2012.136. [DOI] [PubMed] [Google Scholar]
- 48.Murata K, Sudo T, Kameyama M, Fukuoka H, Mukai M, Doki Y, Sasaki Y, Ishikawa O, Kimura Y, Imaoka S. No Title. Clin Exp Metastasis. 2000;18:599–604. doi: 10.1023/a:1011926116777. [DOI] [PubMed] [Google Scholar]
- 49.Dong H, Claffey KP, Brocke S, Epstein PM. Inhibition of breast cancer cell migration by activation of cAMP signaling. Breast Cancer Res Treat. 2015;152:17–28. doi: 10.1007/s10549-015-3445-9. [DOI] [PubMed] [Google Scholar]
- 50.Narita M, Murata T, Shimizu K, Nakagawa T, Sugiyama T, Inui M, Hiramoto K, Tagawa T. A role for cyclic nucleotide phosphodiesterase 4 in regulation of the growth of human malignant melanoma cells. Oncol Rep. 2007;17:1133–9. [PubMed] [Google Scholar]
- 51.Madeira A, Pommet JM, Prochiantz A, Allinquant B. SET protein (TAF1beta, I2PP2A) is involved in neuronal apoptosis induced by an amyloid precursor protein cytoplasmic subdomain. FASEB J. 2005;19:1905–7. doi: 10.1096/fj.05-3839fje. [DOI] [PubMed] [Google Scholar]
- 52.Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein set is its inhibitor. Cell. 2003;112:659–72. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 53.Carlson SG, Eng E, Kim EG, Perlman EJ, Copeland TD, Ballermann BJ. Expression of SET, an inhibitor of protein phosphatase 2A, in renal development and Wilms’ tumor. J Am Soc Nephrol. 1998;9:1873–80. doi: 10.1681/ASN.V9101873. [DOI] [PubMed] [Google Scholar]
- 54.Sirma Ekmekci S, G Ekmekci C, Kandilci A, Gulec C, Akbiyik M, Emrence Z, Abaci N, Karakas Z, Agaoglu L, Unuvar A, Anak S, Devecioglu O, Ustek D, Grosveld G, Ozbek U. SET oncogene is upregulated in pediatric acute lymphoblastic leukemia. Tumori. 2012;98:252–6. doi: 10.1177/030089161209800212. [DOI] [PubMed] [Google Scholar]
- 55.Christensen DJ, Chen Y, Oddo J, Matta KM, Neil J, Davis ED, Volkheimer AD, Lanasa MC, Friedman DR, Goodman BK, Gockerman JP, Diehl LF, de Castro CM, Moore JO, Vitek MP, Weinberg JB. SET oncoprotein overexpression in B-cell chronic lymphocytic leukemia and non-Hodgkin lymphoma: a predictor of aggressive disease and a new treatment target. Blood. 2011;118:4150–8. doi: 10.1182/blood-2011-04-351072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu H, Gu Y, Yin J, Zheng G, Wang C, Zhang Z, Deng M, Liu J, Jia X, He Z. SET-mediated NDRG1 inhibition is involved in acquisition of epithelial-to-mesenchymal transition phenotype and cisplatin resistance in human lung cancer cell. Cell Signal. 2014;26:2710–20. doi: 10.1016/j.cellsig.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 57.Jiang Q, Zhang C, Zhu J, Chen Q, Chen Y. The set gene is a potential oncogene in human colorectal adenocarcinoma and oral squamous cell carcinoma. Mol Med Rep. 2011;4:993–9. doi: 10.3892/mmr.2011.526. [DOI] [PubMed] [Google Scholar]
- 58.Bhutia YD, Hung SW, Krentz M, Patel D, Lovin D, Manoharan R, Thomson JM, Govindarajan R. Differential processing of let-7a precursors influences RRM2 expression and chemosensitivity in pancreatic cancer: role of LIN-28 and SET oncoprotein. PLoS One. 2013;8:e53436. doi: 10.1371/journal.pone.0053436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anazawa Y, Nakagawa H, Furihara M, Ashida S, Tamura K, Yoshioka H, Shuin T, Fujioka T, Katagiri T, Nakamura Y. PCOTH, a Novel Gene Overexpressed in Prostate Cancers, Promotes Prostate Cancer Cell Growth through Phosphorylation of Oncoprotein TAF-Iβ/SET. Cancer Res. 2005;65:4578–86. doi: 10.1158/0008-5472.CAN-04-4564. [DOI] [PubMed] [Google Scholar]
- 60.Ouellet V, Page CL, Guyot MC, Lussier C, Tonin PN, Provencher DM, Mes-Masson AM. SET complex in serous epithelial ovarian cancer. Int J Cancer. 2006;119:2119–26. doi: 10.1002/ijc.22054. [DOI] [PubMed] [Google Scholar]
- 61.Patel D, Boufraqech M, Jain M, Zhang L, He M, Gesuwan K, Gulati N, Nilubol N, Fojo T, Kebebew E. MiR-34a and miR-483-5p are candidate serum biomarkers for adrenocortical tumors. Surgery. 2013;154:1224–8. doi: 10.1016/j.surg.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chabre O, Libe R, Assie G, Barreau O, Bertherat J, Bertagna X, Feige JJ, Cherradi N. Serum miR-483-5p and miR-195 are predictive of recurrence risk in adrenocortical cancer patients. Endocr Relat Cancer. 2013;20:579–94. doi: 10.1530/ERC-13-0051. [DOI] [PubMed] [Google Scholar]
- 63.Szabó DR, Luconi M, Szabó PM, Tóth M, Szücs N, Horányi J, Nagy Z, Mannelli M, Patócs A, Rácz K, Igaz P. Analysis of circulating microRNAs in adrenocortical tumors. Lab Investig. 2014;94:331–9. doi: 10.1038/labinvest.2013.148. [DOI] [PubMed] [Google Scholar]
- 64.Waters PS, McDermott AM, Wall D, Heneghan HM, Miller N, Newell J, Kerin MJ, Dwyer RM. Relationship between Circulating and Tissue microRNAs in a Murine Model of Breast Cancer. PLoS One. 2012;7:1–8. doi: 10.1371/journal.pone.0050459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J, Zhang KY, Liu SM, Sen S. Tumor-Associated circulating micrornas as biomarkers of cancer. Molecules. 2014;19:1912–38. doi: 10.3390/molecules19021912. [DOI] [PMC free article] [PubMed] [Google Scholar]


