Abstract
Using dilution plating method, 47 fungal isolates were obtained from a soil sample collected from Dokdo in the East Sea of Korea in 2013. In this study, two fungal isolates, EML-MFS30-1 and EML-DDSF4, were confirmed as undescribed species, Metarhizium guizhouense and Mortierella oligospora in Korea based on current classification system using multi loci including rDNA internal transcribed spacer, large subunit, small subunit, and β-tubulin (BTUB) genes. Herein, detailed morphological descriptions on characters of the undescribed fungal species as well as their molecular phylogenetic status are provided with comparisons to related species.
Keywords: Dokdo, Metarhizium guizhouense, Mortierella oligospora, Soil fungi
Dokdo is a rocky island located in the northeastern region of Ulleung Island of Korea and consists of two volcanic islands (Dongdo and Seodo) and 89 small islets. It has a variety of environmental conditions, including drought, strong winds, steep inclinations, soil salinity, high uric acid concentrations in the soil, and low organic matter. Thus, it may offer a distinct marine and fungal biodiversity [1].
Few studies have been done on coastal fungal communities. Such unique ecosystems may provide specific habitat and shelter for diverse organisms including fungi [2]. Mandeel [3] has reported the diversity of Fusarium species community in saline soil habitats. Some new species of Fusarium associated with dieback of Spartina alterniflora plant have been isolated from Atlantic salt marshes [2]. A new genus and species Pileomyces formosanus has been isolated from a rocky shore of Taiwan [4]. High salt tolerant fungal genera have been isolated from mangroves and solar salterns of Goa, India [5], with main genus of Aspergillus, followed by Penicillium, Eurotium, and Hortaea. Rogers [6] has studied fungi from Marshall island of central Pacific ocean. Humber and Rombach [7] have reported a new species of Torrubiella (Pyrenomycetes: Clavicipitales) and other fungi from spiders of Solomon islands. Alias et al. [8] have studied intertidal fungi from Philippines and reported a new species of Acrocordiopsis.
The genus Metarhizium was established in 1883 by Sorokin [9]. Metarhizium species are frequently found in soil or infected insects. The species belonging to this genus are characterized by the production of conidia in long chains, phialides in dense, parallel arrangement, and conidiophores aggregated in with repeated, verticillate branching [10].
The genus Mortierella belongs to the order Mortierellales within the Mortierellaceae family. To date, nearly 100 species has been recognized [11]. Mortierella species can be easily isolated from soil, debris or with living plant.
In Korea, You et al. [12,13] have reported the presence of some endophytic fungi isolated from the roots of six native plants in Dokdo. About 30 bacteria species have been reported from that island, including Virgibacillus dokdonensis [14,15]. However, few studies on the diversity of mitosporic fungi and microfungi on that unique island have been performed. It is important to investigate the fungal diversity of this solitary area that is so far away from the main land of Korea. The aim of this study was to isolate fungi from Dokdo in the East Sea of Korea, using dilution plating method, and to confirm the first records of two fungal species in Korea based on the current classification system using multi loci including rDNA internal transcribed spacer (ITS), large subunit (LSU), small subunit (SSU), and β-tubulin (BTUB) genes with comparisons to related species.
MATERIALS AND METHODS
Sampling and isolation
Soil samples were collected from Dongdo (eastern part) of Dokdo (37°14'21.3" N, 131°52'04.4" E) in the East Sea of Korea in 2013. Three sites of soils were sampled to depths of a couple of cm using a 50-mL conical tube and transferred to the laboratory in sterile plastic containers. In order to isolate fungi from soil samples, serial dilution plating technique was used. Briefly, 1 g of soil sample was mixed with 9mL of sterile distilled water and vortexed for 15 min followed by serial dilution to 10−6 fold of the original suspension. One hundred microliters of each diluted soil solution was then transferred to potato dextrose agar (Difco PDA; 39 g PDA; Becton, Dickinson and Co., Sparks, MD, USA, and 1 L deionized water), and malt extract agar (Difco MEA; 33.6 g MEA; Becton, Dickinson and Co., and 1 L deionized water) plates. Plates were incubated at 25℃ for 2 wk to allow fungal growth. Individual colonies of fungi were transferred to different PDA plates to isolate pure cultures. Pure cultures were selected and isolate numbers assumed to be undescribed species in Korea were assigned as EML-MFS30-1 and EML-DDSF4. Cultures were stored in the 20% glycerol at deep freezer of −80℃ in the EML (Environmental Microbiology Laboratory Fungal Herbarium, Chonnam National University, Gwangju, Korea). The EML strains, EML-MFS30-1 and EML-DDSF4, were also deposited at the culture collection of National Institute of Biological Sciences (NIBR, Incheon, Korea) as ex-types, KOSPGC 1151 and KOSPGC 1123, respectively.
Morphological studies
Agar plugs containing stored fungi including EML-MFS30-1 and EML-DDSF4 were transferred to PDA, MEA, yeast malt extract agar (Difco YMA; 1% glucose, 0.5% peptone, 0.3% yeast extract, 0.3% malt extract, 2% agar; Becton, Dickinson and Co. and 1 L deionized water), oatmeal agar (OA; 1.5% oatmeal and 1.5% agar; Junsei, Tokyo, Japan, and 1 L deionized water) and water agar plates for morphological analysis. Plates were incubated at 25℃ in the dark for 1 mon. Colony characteristics (color, size, and texture) were determined at 4~7 days after inoculation. Samples were mounted in lactophenol solution (Junsei) to observe and measure the size and shape of the conidia and conidiophores using light microscope (Leica DMA light microscope; Leica Micriosystems, Jena, Germany). Fine structures of the fungi were observed by scanning electron microscopy (Hitachi S4700 field emission scanning electron microscope; Hitachi, Tokyo, Japan). Samples were cultured on PDA medium in the dark at 27℃ for 7 days. Samples were fixed in 2.5% paraformaldehyde-glutaraldehyde buffer with 0.05 M phosphate (pH 7.2) (Junsei) for 2 hr and washed in carcodylate buffer (Junsei). Cellular membranes were preserved by fixing the samples in 1% osmium tetroxide (diluted in carcodylate buffer; Electron Microscopy Sciences, Hatfield, PA, USA) for 1 hr, washed again in carcodylate buffer, dehydrated in graded ethanol (Emsure, Darmstadt, Germany) and isoamyl acetate (Junsei), and dried under a fume hood. Finally, these samples were covered with gold in a sputter coater and observed at Korea Basic Science Institute, Gwangju, Korea.
Genomic DNA extraction and PCR
Isolates were cultured on PDA plates overlaid with cellophane at 27℃ for 3~5 days. Total genomic DNA was extracted using HiGene Genomic DNA Prep Kit (BIOFACT Corp., Daejeon, Korea). PCR amplification was performed with different primer sets (Table 1) [16,17,18] in a 20 µL reaction using Accupower PCR Premix (Bioneer Corp., Daejeon, Korea) containing Taq DNA polymerase, dNTPs, buffer, and a tracking dye. PCR was conducted using the following conditions: 2 min at 95℃ for the initial denaturation step, followed by 35 cycles of 1min at 94℃ for denaturation, 30 sec at 54℃ for primer annealing, 1 min at 72℃ for extension, and a final extension at 72℃ for 10 min. PCR products were purified using Accuprep PCR Purification Kit (Bioneer Corp.) according to the manufacturer's instructions. Sequencing was performed on an ABI 3700 Automated DNA sequencer (Applied Biosystems Inc., Foster City, CA, USA).
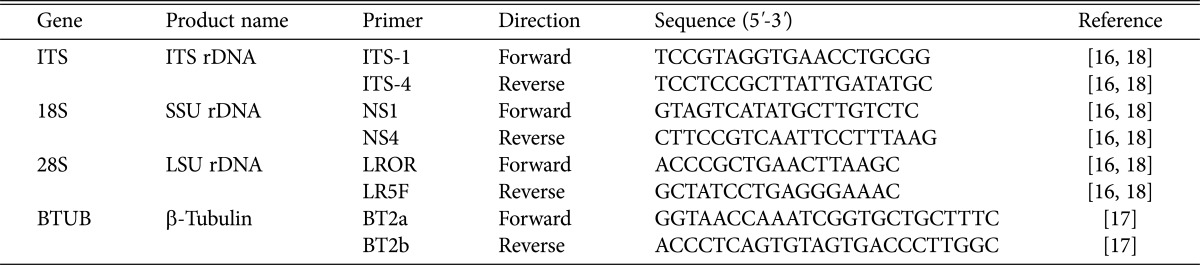
Table 1. List of primers used in this study.
ITS, internal transcribed spacer; SSU, small subunit; LSU, large subunit.
Phylogenetic analysis
Sequences of fungi were submitted for phylogenetic analysis using BioEdit ver. 5.0.9.1 [19], Clustal X ver. 1.83 software [20]. Their phylogenies were assessed using Molecular Evolutionary Genetics Analysis (MEGA) 4 software [21]. Neighbor-joining phylogenetic tree was constructed based on individual ITS, SSU, LSU rDNA, and BTUB sequences as well as their combined sequences. Percent sequence identity (number of matches divided by complete alignment length) was obtained via National Center for Biotechnology Information (NCBI) BLASTn search for each isolate.
Mycelial growth of unrecorded strains
To find the optimum temperature for favorable growth, fungi were incubated for 14 days at three different temperatures (18℃, 27℃, and 32℃). A 6.5 mm diameter plug of inoculum was removed with a cork borer from 7-day-old cultures grown on PDA and placed in the center of OA, MEA, YMA, or PDA media and incubated in the dark for 14 days at 18℃, 27℃, and 32℃. Developing colonies were measured daily for a period of 14 days using a millimeter ruler. Two diameters of the growing colonies were measured at right angles. Growth rates (mm/day) were calculated by linear regression of colony radius against time for each strain grown under each condition.
RESULTS
Using dilution plating method, 47 fungal isolates were obtained from a soil sample collected from Dokdo in the East Sea of Korea in 2013. Out of them, two candidates, EML-MFS30-1 and EML-DDSF4, were primarily identified based on rDNA ITS sequence analysis, and then confirmed as first records in Korea based on the sequence analyses of multi loci including rDNA ITS, LSU, SSU, and β-tubulin (BTUB) genes. The detailed morphological descriptions as well as molecular phylogenetic status of the two undescribed fungal species are presented with comparisons to related species as follows.
Taxonomy of EML-MFS30-1
Metarhizium guizhouense Q. T. Chen & H. L. Guo, Acta Mycol. Sin.: 181 (1986) [MB#130206].
Etymology
This species was isolated from soil sample, Dokdo in the East Sea of Korea.
Description
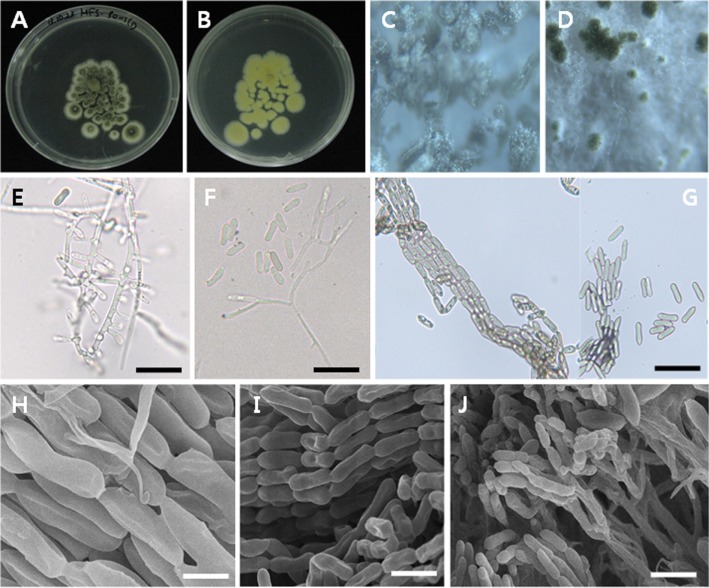
Colonies exhibited slow growth on PDA, attaining a diameter of 10 mm after 5 days at 27℃. The color of colonies was dark green in the center with a lighter margin. The reverse of colony was yellow. A large number of conidia were produced on this medium forming conidial columns. Conidiophores formed a sporulating layer as conidial columns. Basipetal chains were formed in conidiation. The conidia were uninucleate, ovoid to cylindrical, and some more slender in body center. The conidia measured 9.1~10.1 µm wide × 2.4~2.9 µm long (Table 2, Fig. 1).
Table 2. Mycological characteristics of Metarhizium guizhouense EML-MFS30-1 and its closely related reference species.
aFrom description of Bischoff et al. [17].
Fig. 1. Morphology of Metarhizium guizhouense EML-MFS30-1. A, B, Colony on potato dextrose agar (PDA) medium for 5 days at 27℃; C, D, Pustules on PDA under the stereo-microscope (magnification); E, F, Conidiophores, hyphae, phialides and conidia; G, H, Basipetal chains comprising conidia and conidiophores formimg a sporulating layer as conidial columns; I, J, Conidia with basipetal chains under scanning electron microscopy (scale bars: E~I = 20 µm, J = 5 µm).
Molecular phylogeny
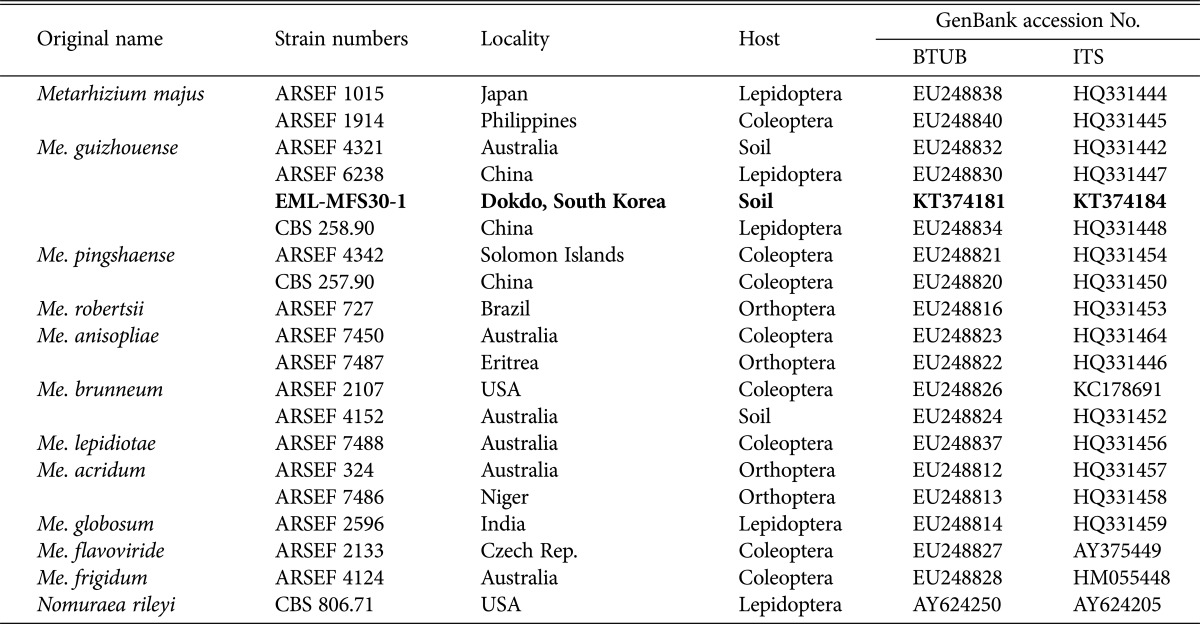
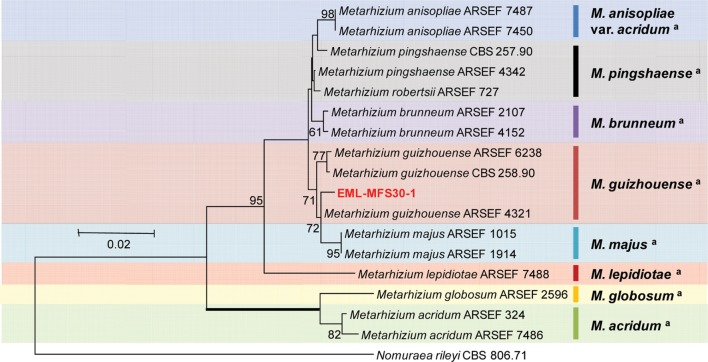
The sequences of EML-MFS30-1 were deposited in the NCBI database under accession numbers KT374181 and KT374184 (Table 3). NCBI BLAST search revealed that EML-MFS30-1 had 99.1% identity (467/471 bp) with Me. guizhouense M335-11 (GenBank accession No. KJ542166, ITS) and 98.8% identity (336/340 bp) with Me. guizhouense ARSEF 4321 (GenBank accession No. EU248832, BTUB). Combined sequence analysis of ITS and BTUB genes produced a guizhouense clade showing that EML-MFS30-1 was closely related to Me. guizhouense ARSEF 4321 (GenBank accession No. EU248832) with a 72% bootstrap value (Fig. 2). Thus, the EML-MFS30-1 isolate was confirmed as Me. guizhouense that has not been previously reported in Korea.
Table 3. List of species within the genus Metarhizium used for molecular phylogenetic analysis.
BTUB, β-tubulin; ITS, internal transcribed spacer; ARSEF, The United States Department of Agriculture Agricultural Research Service (USDA-ARS) Collection of Entomopathogenic Fungi, U.S; EML, Environmental Microbiology Lab Fungal Herbarium, Chonnam National University, Gwangju, South Korea; CBS, Centraalbureau Voor Schimmel cultures, Utrecht, The Netherlands.
Fig. 2. Neighbor-joining tree of alignment based on combined data set of internal transcribed spacer rDNA (GenBank accession No. KT374184) and BTUB (GenBank accession No. KT374181) sequences from Metarhizium guizhouense EML-MFS30-1 and its related species from GenBank database. Nomuraea rileyi CBS 806.71 (GenBank accession No. AY624205) was used as outgroup. Bootstrap values over 50% are shown above the branches supported by 1,000 replications. The tree shows that EML-MFS30-1 strain belongs to guizhouense clade. aClassification by Bischoff et al. [17].
Taxonomy of EML-DDSF4
Mortierella oligospora Björl., Bot. Not. 1936: 121 (1936) [MB#272844].
Etymology
This species was isolated from soil sample, Dokdo in the East Sea of Korea.
Description
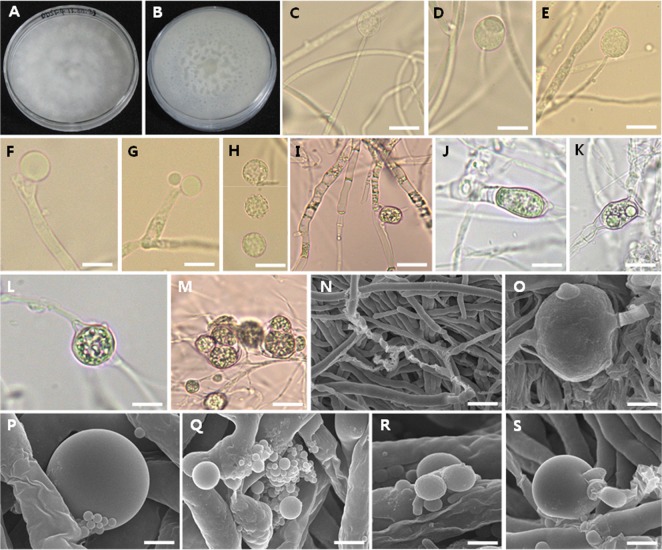
Colonies exhibited fast growth on PDA, attaining a diameter of 70 mm after 7 days at 27℃. The color of colonies was cotton white. The reverse of colony was also white with irregularly zonate. On artificial media, typical sporangia and sporangiospores were not observed although the PDA medium showed good mycelial growth. However, sporangia were produced only on water agar medium. Sporangia were globose to oval, measured 17.4~19.1 × 18.0~21.6 µm. The sporangiospores were globose, roughened, and measured 8.0~14.3 (avr. 12.6) × 8.8~16.0 (avr. 13.4) µm. Intercalary (commonly) or terminal chlamydospores with smooth surface were produced on PDA medium. The structures were mostly globose or subglobose containing different sized vesicles (or spores). Few-spored to 3-spored sporangioles were unclearly observed on PDA (Table 4, Fig. 3).
Table 4. Mycological characteristics of Mortierella oligospora EML-DDSF4 and its closely related reference species.
WA, water agar; OA, oatmeal agar; −, not produced.
aFrom description of Mehrotra et al. [22].
Fig. 3. Morphology of Mortierella oligospora EML-DDSF4. A, B, Culture on potato dextrose agar (PDA) for 7 days at 27℃; C~E, Different stage of sporangia on sporangiophores developed on water agar (WA) medium; F, G, Simple sporangiophore on WA; H, Sporangiospores on WA; I, Irregular and transparent hyphae with septa on PDA; J~M, Chlamydospore structures with intercalary or terminal patterns on PDA; N, Branched hyphae on PDA; O, Magnified chlamydospore structures with intercalary patterns on PDA; P, Globose chlamydospore or chlamydospore-like structure on PDA; Q~S, Different sized vesicles or spores derived from the chlamydospore or chlamydospore-like structures on PDA (scale bars: C~M= 20 µm, N = 30 µm, O~Q = 5 µm, R = 3 µm, S = 10 µm).
Molecular phylogeny
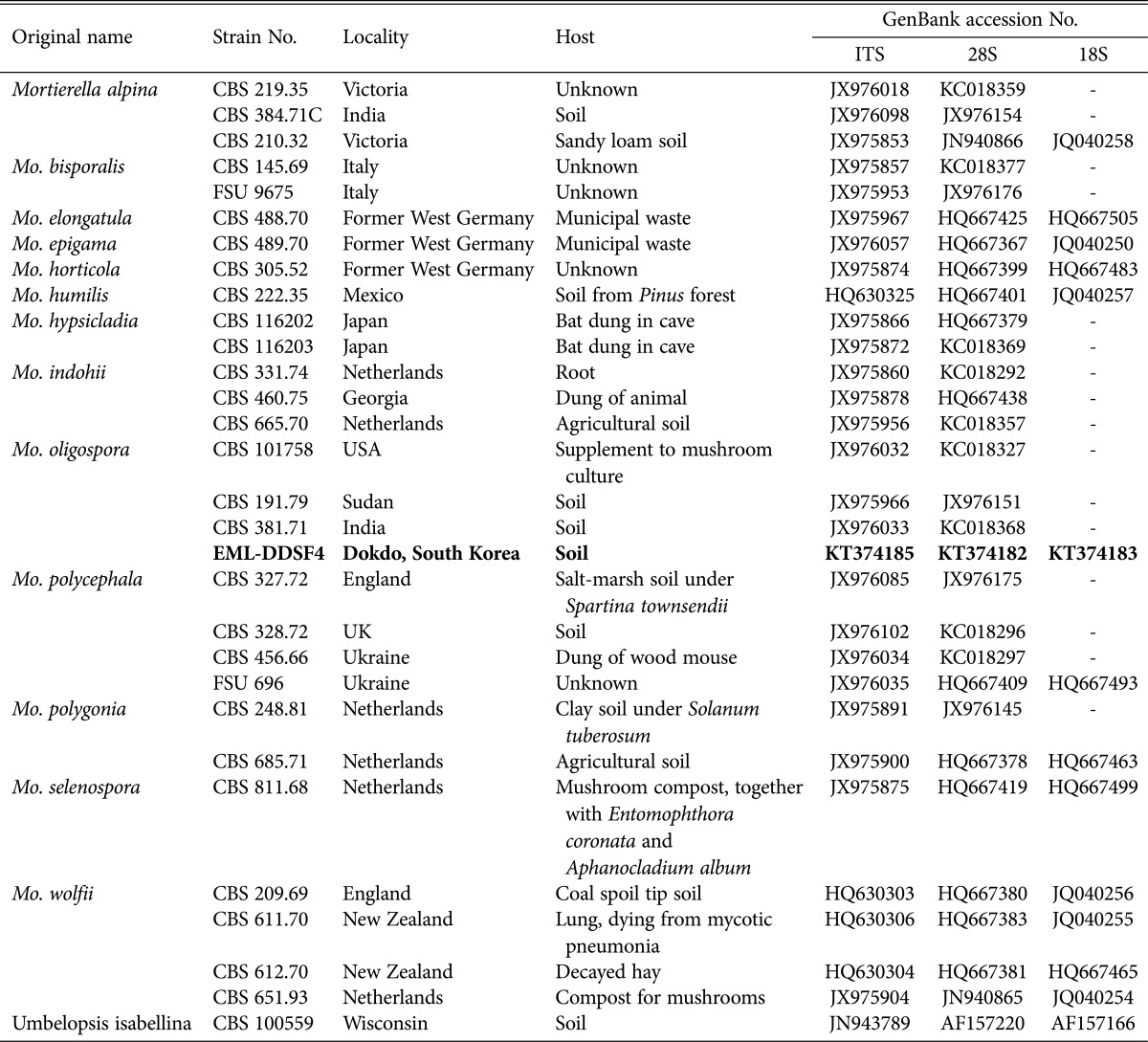
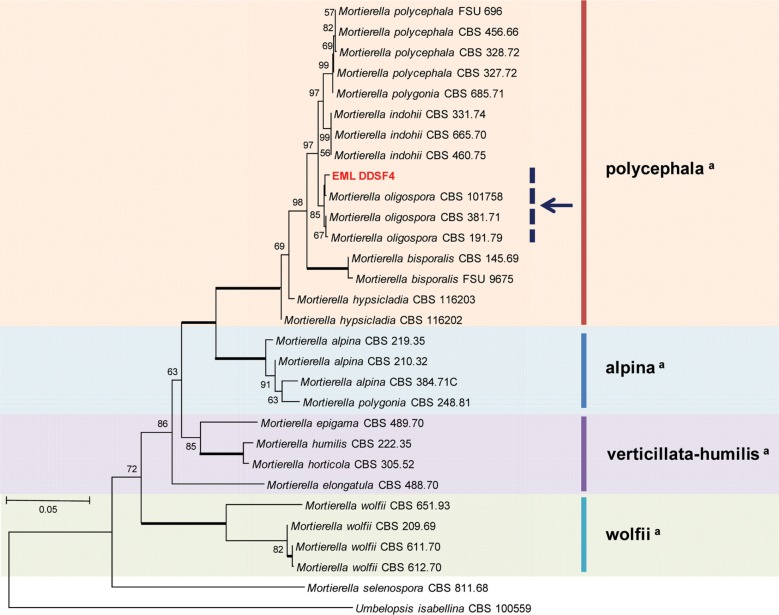
To determine the phylogenetic relationship between EML-DDSF4 isolated from the soil of Ulleung island and its related species, 18S (SSU), ITS, and 28S (LSU) regions were analyzed. The sequences of EML-DDSF4 were deposited in the National Center for Biotechnology Information (NCBI) database under accession numbers KT374185, KT374182, and KT374183 (Table 5). NCBI BLASTn search revealed that EML-DDSF4 shared 100% sequence identity (350/350 bp) with Mo. oligospora CBS 101758 (GenBank accession No. JX976032, ITS), 99.8% identity (810/812 bp) with Mo. polygonia CBS685.71 (GenBank accession No. HQ667463, SSU), and 99.6% identity (967/971 bp) with Mo. oligospora CBS 101758 (GenBank accession No. KC018327, LSU). Sequence analysis revealed 98~100% sequence similarity between EML-DDSF4 isolate and its relevant sequences in GenBank based on NCBI BLASTn searches. The isolate and its related species in GenBank formed a single clade based on combined sequence analysis of the three sequences (ITS, 18S, and 28S). EML-DDSF4, Mo. oligospora CBS 101758 (accession No. JX976032), Mo. oligospora CBS 381.71 (accession No. JX976033), and Mo. oligospora CBS 191.79 (accession No. JX975966) clustered together in a group. The phylogenetic tree shows EML-DDSF4 strain belongs to oligospora subclade (arrow in Fig. 4) within polycephala clade. EML-DDSF4 matched Mo. oligospora CBS 101758 (accession No. JX976032) with 85% bootstrap value (Fig. 4). Based on morphological evaluation and molecular phylogenetic analysis, the isolate EML-DDSF4 was confirmed as Mo. oligospora.
Table 5. List of species within the genus Mortierella and Umbelopsis used for the molecular phylogenetic analysis.
FSU, Friedrich Schiller University, Jena, Germany; EML, Environmental Microbiology Lab Fungal Herbarium, Chonnam National University, Gwangju, South Korea.
Fig. 4. Neighbor-joining tree of combined data set of small subunit rDNA (GenBank accession No. KT374183), internal transcribed spacer rDNA (GenBank accession No. KT374185), and large subunit rDNA (GenBank accession No. KT374182) sequences of Mortierella oligospora EML-DDSF4 and its related species from GenBank databases. Umbelopsis isabellina CBS 100559 (GenBank accession No. JN943789) was used as outgroup. Bootstrap values over 50% are shown above the branches supported by 1,000 replications. The tree shows that EML-DDSF4 strain belongs to oligospora subclade (arrow) within polycephala clade. aClassification by Wagner et al. [16] and Petkovits et al. [18].
Mycelial growth of unrecorded strains
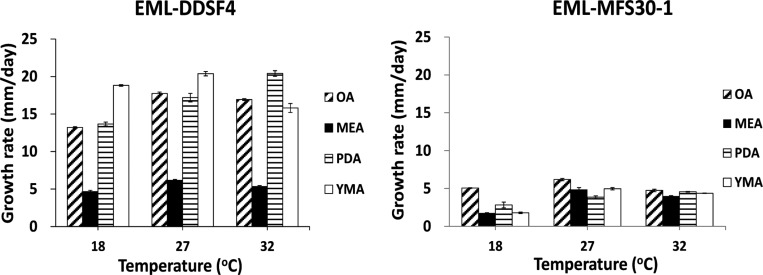
Mycelial growth varied depending on environmental factors such as temperature and growth medium. Mo. oligospora EML-DDSF4 grew fast (avr. 15 mm/day) while Me. guizhouense EML-MFS30-1 grew slowly (avr. 3mm/day) on MEA medium at 18℃. Fungal growth was the slowest in MEA medium (Fig. 5).
Fig. 5. Mycelial growth rates for the newly recorded fungi EML-DDSF4 and EML-MFS30-1. Colonies were grown on oatmeal agar (OA), malt extract agar (MEA), yeast malt extract agar (YMA), and potato dextrose agar (PDA) at different temperatures for 14 days.
DISCUSSION
Most fungal species isolated from Dokdo soil samples belonged to ascomycetes. However, some isolates were classified as zygomycetous fungi including Mo. oligospora and Absidia sp. It was difficult to identify the zygomycetous fungi at the level of species because their detailed descriptions lack in literature. Interestingly, in the case of the species of Mo. oligospora EML-DDSF4, typical sporangia and sporangiospores were produced only on water agar medium. Only different shapes of chlamydospores, sporangiole-like structures and various stages for zygospore formation in EML-DDSF4 strain were observed on PDA medium.
Mortierellales constitutes one of the largest orders in the basal lineages. This group consists of one family and six genera. However, the phylogenetic position of Mortierellales is still controversial. Recently, only one species of Mortierella alpina was reported as new record in Korea [23]. Based on LSU polygram, Mo. oligospora belonged to group 6 including alpina and polycephala. Mo. oligospora, Mo. bisporalis, Mo. hypsicladia, Mo. indohii, Mo. polygonia and Mo. reticulate [22], which was well supported by our SSU, LSU, and ITS rDNA analysis. Many mortierellean species showed potential as very interesting fungi for biotransformations and biotechnological applications [24]. Kataoka et al. [25] have isolated endosulfan-degrading Mortierella species from a soil contaminated with organochlorine pesticides.
On the other hand, Metarhizium is a genus of entomopathogenic fungi in the Clavicipitaceae family in the phylum Ascomycota. The teleomorphs of Metarhizium species appear to be members of the genus Metacordyceps [26]. Metarhizium anisopliae, the type species of the anamorph of genus Metarhizium, contains four varieties, and is closely related to Me. taii, Me. pingshaense, and Me. guizhouense Bischoff et al. [17] has proposed to recognize Me. guizhouense as anamorph of Metacordyceps taii. As in many hyphomycetous genera, it may be still difficult to distinguish them. In this study, we could see that EML-MFS30-1 strain belongs to a guizhouense clade in the phylogenetic tree based on beta-tubulin gene and ITS rDNA sequences. Maniania et al. [27] have studied the potential of Me. anisopliae for the control of Thrips tabaci in onion agroecosystem. New source of chitosanase has been studied from Me. guizhouense under solid state fermentation conditions [28]. Temperature plays an important role in the physiological characteristics of entomopathogenic fungi. In our study, Me. guizhouense EML-MFS30-1 exhibited optimum growth on OA at 27℃ and was unable to grow at 37℃. The results were similar to those by Ouedraogo et al. [29] who studied the optimal temperature for species of Metarhizium. Several species of Metarhizium have shown great potential for the management, as they produce variety of compounds including cyclic peptide, destruxins with insecticidal activity [30,31].
Mortierellean fungi have important biotechnological applications as producers of polyunsaturated fatty acids, lipoxygenase enzyme [32,33]. In particular, polyunsaturated fatty acids as arachidonic acid (5,8,11,14-ciseicosatetraenoic acid) is very important for maintaining biofunctions in mammalians [34]. However, the genus Mortierella also contains an animal pathogen, Mo. wolfii [35].
Fungal diversity may be affected by different climate conditions, soil conditions, and collection seasons. Thus, it is important to conduct more studies on seasonal distribution of fungi in Korea. The weather on the island is characterized by mostly snow in winter season and an oceanic climate that is affected by warm ocean currents. It is foggy and cloudy for more than 160 days a year [36]. Our soil samples were collected in August. However, the distribution of fungi may be different in other months. More studies on seasonal diversity of (dominant) fungi, their ecophysiological characteristics and bioactivities are merited for future studies.
ACKNOWLEDGEMENTS
This work was supported by the Project (NIBR201401205) on Survey and Discovery of Indigenous Fungal Species of Korea and by the Graduate Program for the Undiscovered Taxa of Korea (NIBR201524202) by NIBR of the Ministry of Environment (MOE), Republic of Korea.
References
- 1.Ryu SH, Jang KH, Choi EH, Kim SK, Song SJ, Cho HJ, Ryu JS, Kim YM, Sagong J, Lee JH, et al. Biodiversity of marine invertebrates on rocky shores of Dokdo, Korea. Zool Stud. 2012;51:710–726. [Google Scholar]
- 2.Elmer WH, Marra RE. New species of Fusarium associated with dieback of Spartina alterniflora in Atlantic salt marshes. Mycologia. 2011;103:806–819. doi: 10.3852/10-155. [DOI] [PubMed] [Google Scholar]
- 3.Mandeel QA. Biodiversity of the genus Fusarium in saline soil habitats. J Basic Microbiol. 2006;46:480–494. doi: 10.1002/jobm.200510128. [DOI] [PubMed] [Google Scholar]
- 4.Pang KL, Jheng JS. Pileomyces formosanus gen. et sp. nov. (Halosphaeriaceae, Ascomycota) from a rocky shore of Taiwan. Bot Stud. 2012;53:535–539. [Google Scholar]
- 5.Nayak SS, Gonsalves V, Nazareth SW. Isolation and salt tolerance of halophilic fungi from mangroves and solar salterns in Goa-India. Indian J Geomarine Sci. 2012;41:164–172. [Google Scholar]
- 6.Rogers DP. Fungi of the Marshall islands, central Pacific Ocean. Pac Sci. 1947;1:92–107. [Google Scholar]
- 7.Humber RA, Rombach MC. Torrubiella ratticaudata sp. nov. (Pyrenomycetes: Clavicipitales) and other fungi from spiders on the Solomon Islands. Mycologia. 1987;79:375–382. [Google Scholar]
- 8.Alias SA, Jones EB, Torres J. Intertidal fungi from the Philippines, with a description of Acrocordiopsis sphaerica sp. nov. (Ascomycota) Fungal Divers. 1999;2:35–41. [Google Scholar]
- 9.Sorokin N. Rastitelnye parazity cheloveka i zhivotnykn' kak' prichina zaraznykn' boleznei (Plant parasites causing infectious diseases of man and animals). Vyp. II.Pervoe prilozenie k Voenno-Meditsinskomu Zhurnalu za 1883 g (First supplement to J Military Med 1883) St. Petersburg: Izdanie glavnogo Voenno-Meditsinskago Upraveleneia; 1883. pp. 168–198. [Google Scholar]
- 10.Tulloch M. The genus Metarhizium. Trans Br Mycol Soc. 1976;66:407–411. [Google Scholar]
- 11.Benny GL. Zygomycetes [Internet] Gerald L. Benny; 2009. [cited 2015 Oct 1]. Available from: http://www.zygomycetes.org. [Google Scholar]
- 12.You YH, Yoon H, Lee GS, Woo JR, Shin JH, Lee IJ, Rim SO, Choo YS, Kim JG. Diversity and plant growth-promotion of endophytic fungi isolated from the roots of plants in Dokdo islands. J Life Sci. 2011;21:992–996. [Google Scholar]
- 13.You YH, Yoon H, Kim H, Lim SH, Shin JH, Lee IJ, Choo YS, Kim JG. Plant growth-promoting activity and genetic diversity of endophytic fungi isolated from native plants in Dokdo islands for restoration of a coastal ecosystem. J Life Sci. 2013;23:95–101. [Google Scholar]
- 14.Yoon JH, Kang SJ, Lee SY, Lee MH, Oh TK. Virgibacillus dokdonensis sp. nov., isolated from a Korean island, Dokdo, located at the edge of the East Sea in Korea. Int J Syst Evol Microbiol. 2005;55(Pt 5):1833–1837. doi: 10.1099/ijs.0.63613-0. [DOI] [PubMed] [Google Scholar]
- 15.Lee CM, Weon HY, Yoon SH, Kim SJ, Koo BS, Kwon SW. Burkholderia denitrificans sp. nov., isolated from the soil of Dokdo Island, Korea. J Microbiol. 2012;50:855–859. doi: 10.1007/s12275-012-1554-2. [DOI] [PubMed] [Google Scholar]
- 16.Wagner L, Stielow B, Hoffmann K, Petkovits T, Papp T, Vágvölgyi C, de Hoog GS, Verkley G, Voigt K. A comprehensive molecular phylogeny of the Mortierellales (Mortierellomycotina) based on nuclear ribosomal DNA. Persoonia. 2013;30:77–93. doi: 10.3767/003158513X666268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bischoff JF, Rehner SA, Humber RA. A multilocus phylogeny of the Metarhizium anisopliae lineage. Mycologia. 2009;101:512–530. doi: 10.3852/07-202. [DOI] [PubMed] [Google Scholar]
- 18.Petkovits T, Nagy LG, Hoffmann K, Wagner L, Nyilasi I, Griebel T, Schnabelrauch D, Vogel H, Voigt K, Vágvölgyi C, et al. Data partitions, Bayesian analysis and phylogeny of the zygomycetous fungal family Mortierellaceae, inferred from nuclear ribosomal DNA sequences. PLoS One. 2011;6:e27507. doi: 10.1371/journal.pone.0027507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 20.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 22.Yadav DR, Kim SW, Babu AG, Adhikari M, Kim C, Lee HB, Lee YS. First report of Mortierella alpina (Mortierellaceae, Zygomycota) isolated from crop field soil in Korea. Mycobiology. 2014;42:401–404. doi: 10.5941/MYCO.2014.42.4.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mehrotra BS, Mehrotra BR, Baijal U. Species of Mortierella from India: V. Mortierella oligospora Bjorling and M. oligospora var. indica var. nov. Sydowia. 1963;17:186–189. [Google Scholar]
- 24.Walter M, Ford CI, Guthrie JV, Boyd-Wilson KS, Di HJ, Cameron KC, Parker E. Bioremediation of aged pentachlorophenol residues by white-rot fungi. In: Rai M, editor. Mycotechnology: present status and future prospects. New Delhi: I. K. International Publishing House Pvt. Ltd.; 2007. pp. 18–54. [Google Scholar]
- 25.Kataoka R, Takagi K, Sakakibara F. A new endosulfan-degrading fungus, Mortierella species, isolated from a soil contaminated with organochlorine pesticides. J Pestic Sci. 2010;35:326–332. [Google Scholar]
- 26.Sung GH, Hywel-Jones NL, Sung JM, Luangsa-ard JJ, Shrestha B, Spatafora JW. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud Mycol. 2007;57:5–59. doi: 10.3114/sim.2007.57.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniania NK, Sithanantham S, Ekesi S, Ampong-Nyarko K, Baumgärtner J, Löhr B, Matoka CM. A field trial of the entomogenous fungus Metarhizium anisopliae for control of onion thrips, Thrips tabaci. Crop Prot. 2003;22:553–559. [Google Scholar]
- 28.Yang L, Zhang T, Li XH, Yu R. Purification of a novel chitosanase from Metarhizium guizhouense in solid-state fermentation with affinity chromatography. Sichuan Da Xue Xue Bao Yi Xue Ban. 2007;38:713–716. [PubMed] [Google Scholar]
- 29.Ouedraogo A, Fargues J, Goettel MS, Lomer CJ. Effect of temperature on vegetative growth among isolates of Metarhizium anisopliae and M. flavoviride. Mycopathologia. 1997;137:37–43. doi: 10.1023/A:1006882621776. [DOI] [PubMed] [Google Scholar]
- 30.Wang C, Skrobek A, Butt TM. Investigations on the destruxin production of the entomopathogenic fungus Metarhizium anisopliae. J Invertebr Pathol. 2003;85:168–174. doi: 10.1016/j.jip.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Lomer CJ, Prior C, Kooyman C. Development of Metarhizium spp. for the control of grasshopper and locusts. Mem Entomol Soc Can. 1997;129(Suppl S171):265–286. [Google Scholar]
- 32.Filippovich SY, Rybakov YA, Afanasieva TP, Bachurina GP, Lukina GP, Ezhova IE, Nosova AV, Artjushkina TV, Sineokii SP, Kritskii MS. Characterization of lipoxygenase from fungi of the genus Mortierella. Appl Biochem Microbiol. 2001;37:473–479. [Google Scholar]
- 33.Nagao K, Yanagita T. Bioactive lipids in metabolic syndrome. Prog Lipid Res. 2008;47:127–146. doi: 10.1016/j.plipres.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Botha A, Paul I, Roux C, Kock JL, Coetzee DJ, Strauss T, Maree C. An isolation procedure for arachidonic acid producing Mortierella species. Antonie Van Leeuwenhoek. 1999;75:253–256. doi: 10.1023/a:1001848709005. [DOI] [PubMed] [Google Scholar]
- 35.Davies JL, Ngeleka M, Wobeser GA. Systemic infection with Mortierella wolfii following abortion in a cow. Can Vet J. 2010;51:1391–1393. [PMC free article] [PubMed] [Google Scholar]
- 36.Dokdo [Internet] Daegu: Gyeongsangbuk-do Province; 2015. [cited 2015 Oct 1]. Available from: http://en.dokdo.go.kr/pages/s03/page.html?mc=7180. [Google Scholar]