Abstract
One of the hallmark mechanisms activated by type I interferons (IFNs) in human tissues involves cleavage of intracellular RNA by the kinase homology endoribonuclease RNase L. We report 2.8 and 2.1 angstrom crystal structures of human RNase L in complexes with synthetic and natural ligands and a fragment of an RNA substrate. RNase L forms a crossed homodimer stabilized by ankyrin (ANK) and kinase homology (KH) domains, which positions two kinase extension nuclease (KEN) domains for asymmetric RNA recognition. One KEN protomer recognizes an identity nucleotide (U), whereas the other protomer cleaves RNA between nucleotides +1 and +2. The coordinated action of the ANK, KH, and KEN domains thereby provides regulated, sequence-specific cleavage of viral and host RNA targets by RNase L.
Cells of higher vertebrates respond to pathogens and damage by releasing interferons (IFNs), which activate protective programs in surrounding cells. One of the ubiquitous protective programs in mammalian tissues involves cleavage of intracellular RNA by a protein kinase family receptor, RNase L (1). RNase L is a latent endoribonuclease encoded by the hereditary prostate cancer 1 (HPC1) locus and activated by the second messenger, 2–5A (2′,5′-linked oligoadenylates of variable length) (1). In human cells, 2–5As are synthesized by IFN-induced 2–5A synthetases, which serve as sensors of pathogen- and damage-associated double-stranded RNA(dsRNA) (2, 3). The activation of RNase L thus depends on the action of IFNs and accumulation of dsRNA.
Here, we report two crystal structures of human RNase L (table S1). These structures and complementary functional studies reveal the mechanism of 2–5A sensing and RNA cleavage by RNase L and suggest that a similar mechanism of RNA cleavage operates during regulated Ire1-dependent decay (RIDD) (4).We obtained diffracting crystals of nearly full-length human RNase L using cocrystallization with 2–5A, nucleotides, and an RNA 18-nucleotide oligomer 5′-GGCUUUUGACCUUUAUGC-3′ (RNA18). Cocrystallization with RNA18 was enabled by the use of a catalytically inactive RNase L mutant H672N. The final construct includes residues 21 to 719. Aversion of this construct with a wild-type (WT) active site is catalytically active in solution.
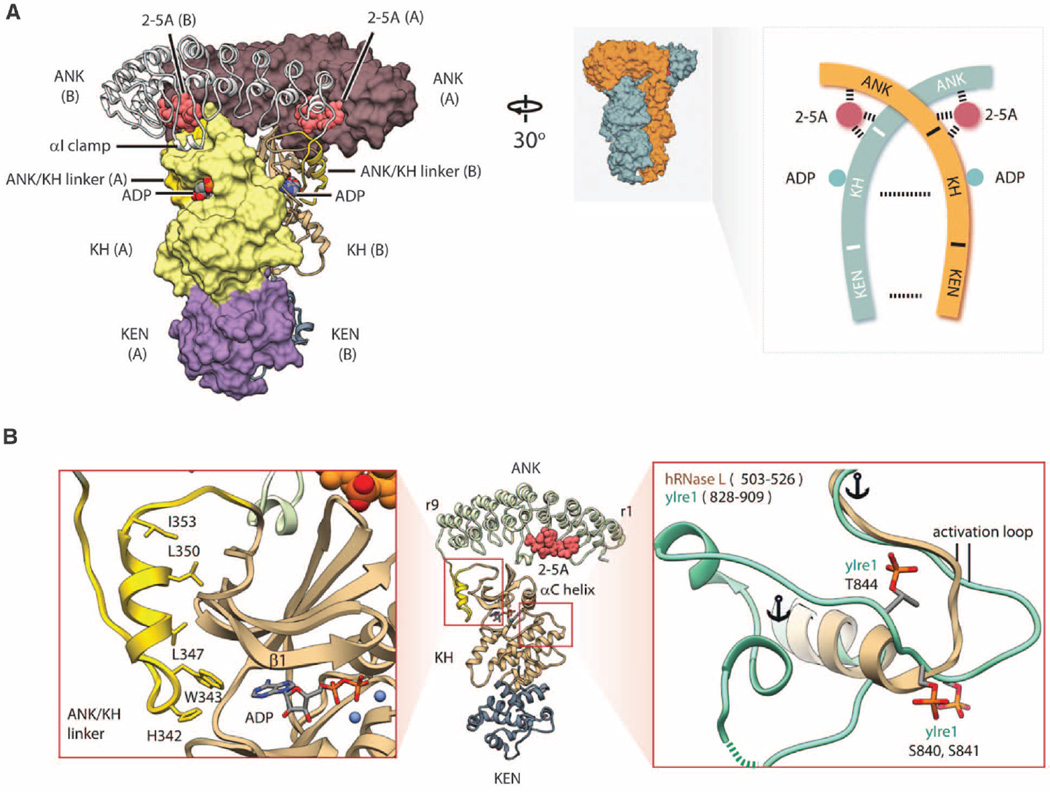
We determined structures of two RNase L complexes, which crystallized in different space groups (table S1). Both complexes reveal the same crossed homodimer that buries >8000Å2 of surface area (Fig. 1A). Previous solution studies indicated that RNase L can form dimers and higher-order oligomers (5).Modeling based on the oligomer of Ire1 (6) predicts that the homodimers of RNase L could form a similar assembly. The kinase homology (KH) domain of RNase L has a typical protein kinase fold with two globular lobes (Fig. 1B). Adenosine diphosphate (ADP) and β,γ-methyleneadenosine triphosphate are bound to the KH domain in the same conformation as ADP in the catalytically active protein kinase Ire1 (fig. S1, A to C). Nonhydrolyzable nucleotides and ATP exhibit the same effect on RNase L (fig. S1D), indicating that ATP hydrolysis is not involved in RNase L regulation, as suggested previously (7).
Fig. 1. Structure of human RNase L•2–5pA3•ADP•RNA18 quaternary complex.
(A) Crossed homodimer of RNase L. Different protomers are shown as molecular surface and ribbon, respectively. The structured linker connecting the ANK and the KH domains is colored gold. Schematic topology of the RNase L homodimer is shown on the right. (B) Structure of the KH domain. The ATP pocket is flanked by the structured linker (left). The activation loop is short and lacks phosphorylation sites (right). Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys;D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
The KH domain lacks the conserved DFG motif found in most protein kinases and contains the DFD sequence, resembling protein kinases Mnk1/2 (8). RNase L does not carry out autophosphorylation (7), but it remains unknown whether RNase L phosphorylates nonself targets. To examine this possibility, we assayed phosphorylation of a nonspecific substrate, myelin basic protein (MBP), using Ire1 as a control kinase. Ire1 phosphorylated MBP, whereas RNase L was inactive (fig. S1E), supporting the current consensus that RNase L is a pseudokinase. The activation loop of RNase L contains only 13 amino acid residues and is among the shortest in the human protein kinome (fig. S3). The interlobe hinge and the ATP pocket contact a unique helix from the ankyrin (ANK)/KH linker (Fig. 1B and fig. S2). These attributes of the KH domain likely reflect adaptation to autophosphory lation-independent control as a homodimerization scaffold.
Previous structural studies identified two different 2–5A binding sites in the ANK domain (5, 9). The crystal structure of the entire RNase L now reveals an unanticipated third site in the N lobe of the KH domain (Fig. 1A and fig. S4,Ato C). The ANK and KH domains create a composite pocket for 2–5A binding, which exposes the 2′-end to solvent to accommodate long 2–5A molecules and anchors the 5′-end in the ANK domain (fig. S5A). Although RNase L can recognize 5′-p and 5′-ppp groups, at saturating concentrations 2–5pA3 activates RNase L stronger than 2–5pppA3 (fig. S5, B and C). The ANK/ANK homodimer binds 2–5A in a configuration that displays the phosphate p1 for recognition by the KH domain (fig. S6). Mutagenesis of the KH/2–5A interface confirms that the N lobe is functionally involved in 2–5A sensing (fig. S4D). The ANK domain contains a characteristic helix αI, which docks to the KH domain in trans upon homodimerization (fig. S7). The αI/N-lobe interaction and an ANK/N-lobe contact mediated by the residue R238 also facilitate RNase L activation by 2–5A (fig. S4, C and D).
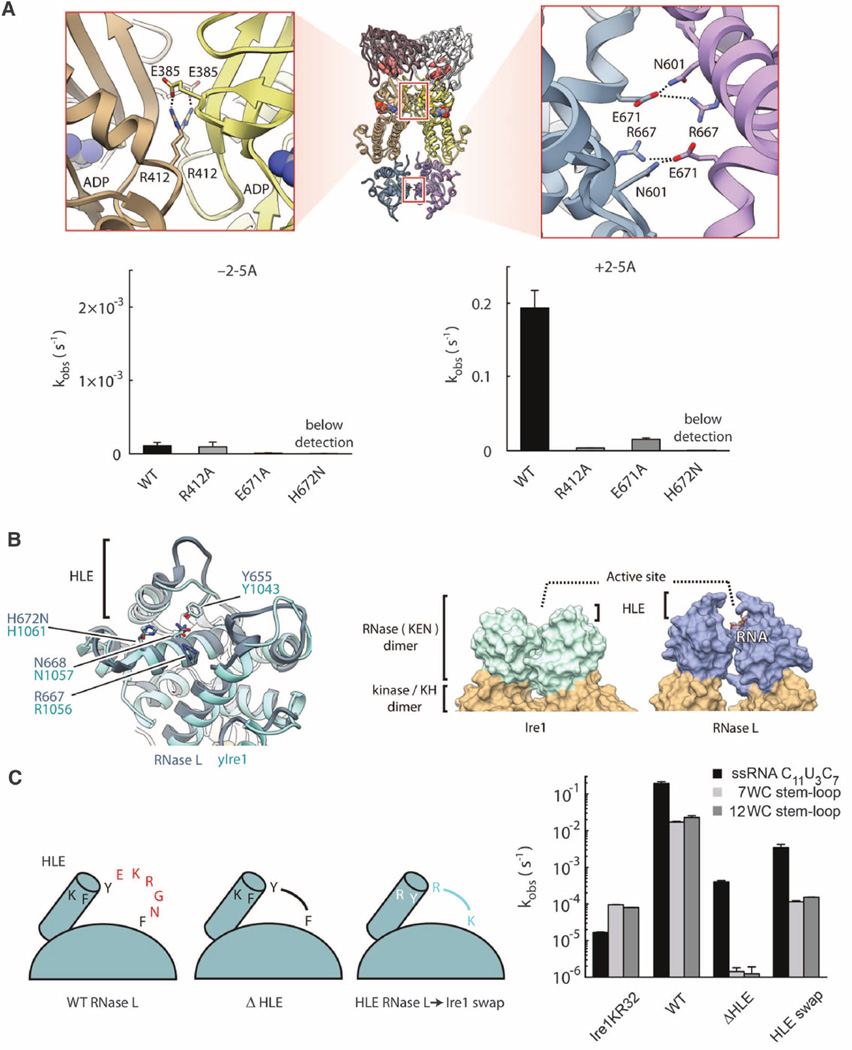
The KH/KH and kinase extension nuclease (KEN)/KEN interfaces resemble those in Ire1 (10). Mutagenesis confirmed that both interfaces are important for 2–5A–dependent RNase L activation (Fig. 2A) and dimerization (fig. S8). The KEN residues involved in catalysis in Ire1 (10, 11) are structurally invariant in RNase L (Fig. 2B), indicating that these enzymes share the same catalytic mechanism. However, the KEN domains contain different α-helix/loop elements (HLE) (6), implicated in RNA specificity (10). To examine the HLE function, we shortened this element in RNase L (ΔHLE). The ΔHLE mutant still cleaved RNA but exhibited a decreased rate and a greater preference for single-stranded RNA (ssRNA) versus stem loops (Fig. 2C). A chimeric RNase L with the HLE sequence from Ire1 (HLE swap) retained the preference for ssRNA (Fig. 2C), revealing that the HLE of Ire1 is insufficient for directing RNase L to stem loops. Thus, the HLE does not determine the substrate specificity of RNase L (and, by extension, of Ire1) and only facilitates RNA cleavage.
Fig. 2. KH and KEN homodimers.
(A) Symmetrical salt bridges stabilize the interfaces between the KH (1364 Å2) and RNase (943 Å2) domains. Mutations of these salt bridges impair RNase L activity. (B) Comparison of KEN domains in RNase L and Ire1. Key active-site residues are indicated. (C) Effect of HLE mutagenesis on cleavage of ssRNA and RNA hairpins derived from human XBP1 mRNA. WC indicates the number of Watson-Crick base pairs in the hairpin stem. Ire1KR32 is yeast Ire1 kinase/RNase. Reactions contained 100 nM Ire1KR32 and 5 nM RNase L. Error bars show mean ± SE.
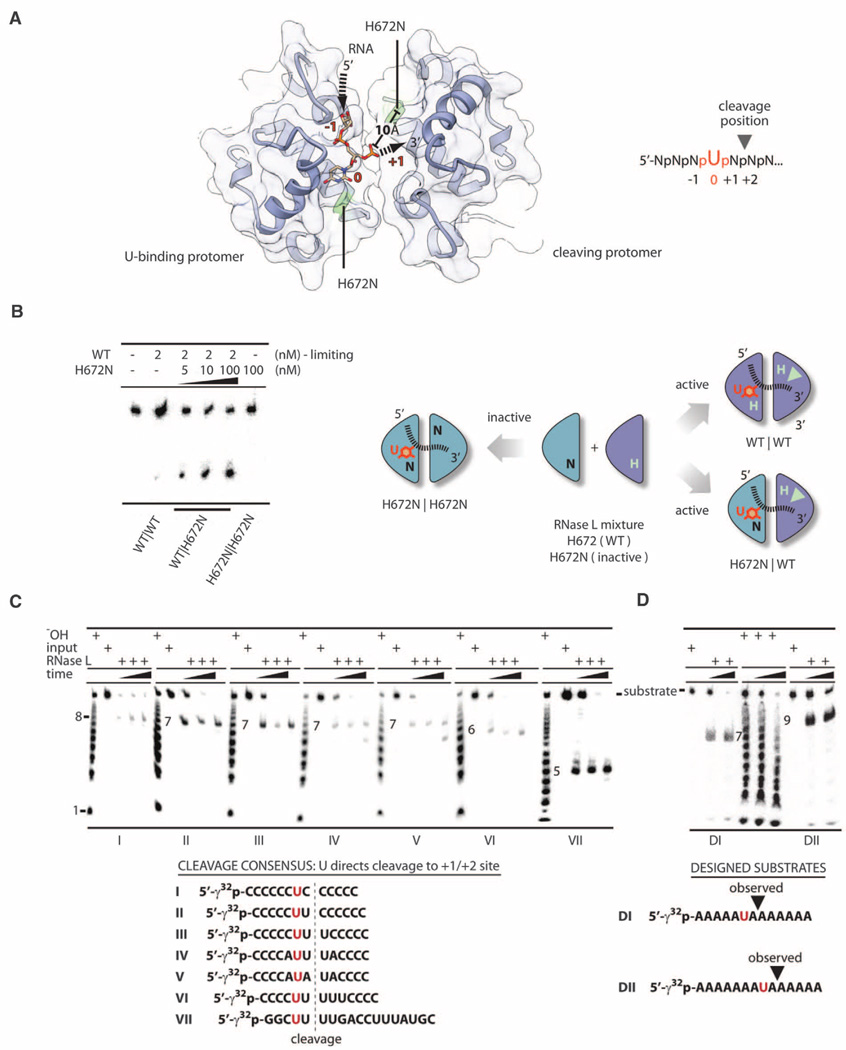
Both complexes of RNase L described here were obtained with an RNA bound in the active site. Two sugar-phosphate groups and one pyrimidine nucleobase are resolved in complex II (fig. S9 and table S1). We modeled uridine based on the preference of RNase L for AU-rich sites (12) and biochemical studies of RNA substrates described below. The U residue blocks access of the nearest catalytic histidine H672 to the phosphodiester backbone. Thus, RNA cleavage should be carried out by the symmetry-related histidine H672 provided by homodimerization (Fig. 3A). Distance constraints suggest that cleavage occurs between the nucleotides +1 and +2 relative to the U residue.
Fig. 3. RNA sequence recognition by RNase L.
(A) Structure of RNA bound to the KEN homodimer. Arrows show the RNA trace. RNA cleavage is predicted at UN^N sites. (B) In trans activation of WT RNase L by the catalytically inactive RNase L mutant H672N. (C) RNase L cleavage of substrates I to VII. (D) RNase L cleavage of substrates DI and DII.
To test this model, we employed an in trans complementation assay using a mixture of WT and catalytically inactive RNase L (H672N). The H672N mutation eliminates the proton transfer but preserves the H-bonding and space-filling character of histidine. Thus, the H672N mutant should interact with RNA analogous to the WT protein but without cleaving the RNA. The in trans complementation assay was performed by adding increasing concentrations of the H672N mutant to a fixed and limiting concentration of WT RNase L (2 nM). The H672Nmutant was a potent in trans activator for WT RNase L, whereas double H672N mutant was inactive (Fig. 3B). Thus, a single H672 residue per a KEN/KEN dimer is necessary and sufficient for RNA cleavage. The biochemical and structural data converge on a single mechanism of RNA cleavage by RNase L that involves recognition of a U residue by one KEN protomer and catalytic RNA scission by the second KEN protomer. The size of the nucleotide-binding pocket in RNase L can explain the preference for a pyrimidine, whereas U/C discrimination apparently results from H bonding with H672 (fig. S10).
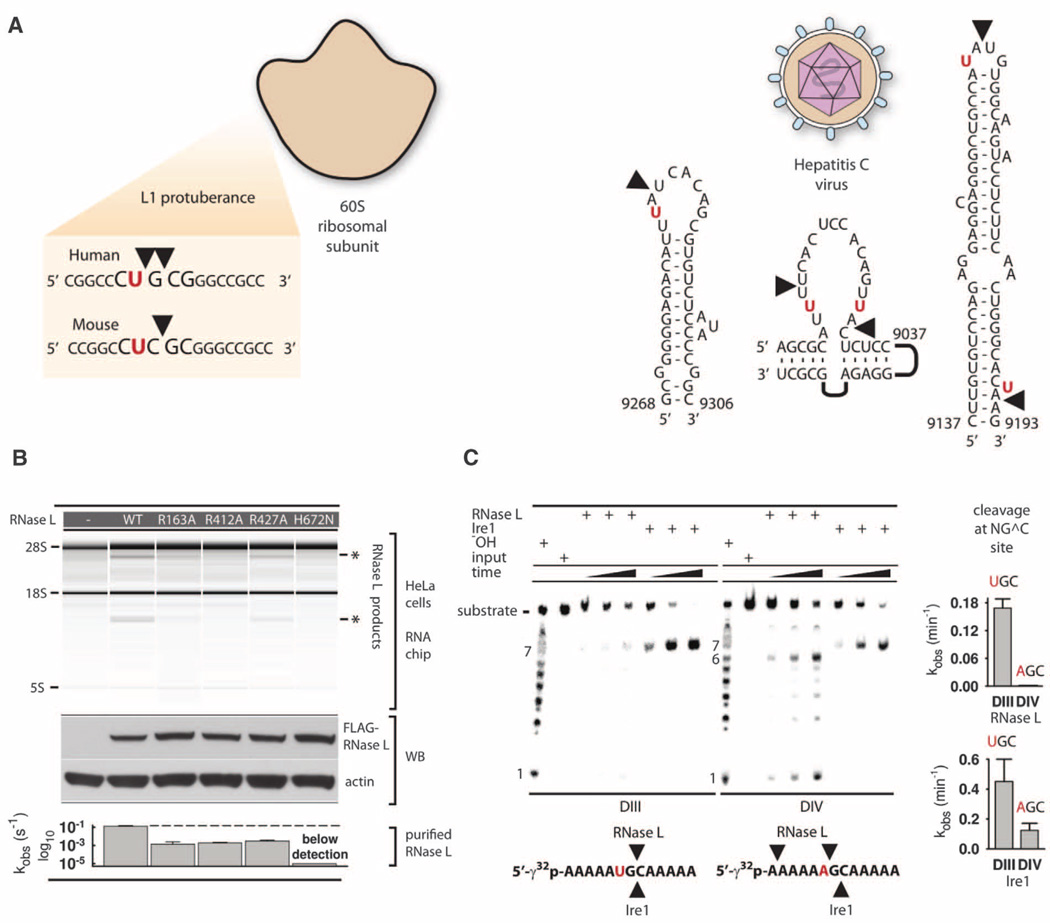
In contrast to other endoribonucleases, such as RNase A and RNase T1, RNase L does not cleave ssRNA randomly. Only a single site is cleaved in 5′-CCCCCCCCCCCUU^UCCCCCCC-3′ ssRNA (RNA21) (^ denotes the cleavage site) (13). RNase L has been described as an enzyme that cleaves RNA at dinucleotides UU and UA (12), as well as after dinucleotides UN (14). Our structural analysis suggests that RNase L recognizes the pattern UN^N, supporting cleavage 3′ of UN sequences. To verify this assignment, we synthesized a series of model RNA substrates. RNase L cleaved these substrates according to the UN^N rule (Fig. 3C). We next designed substrates DI and DII, which use a different RNA backbone and contain the AU, UA and UN^N sequences in mutually exclusive positions. RNase L cleaved these substrates also according to the UN^N pattern (Fig. 3D). The UN^N rule extends to the biological targets of RNase L. Cleavage of 28S rRNA within the L1 protuberance occurs at UG^C, CU^G, and UC^G sites (15) (Fig. 4A), which do not contain the UU/UA dinucleotides conventionally ascribed to RNase L targets (12). However, two of them match the UN^N pattern. Cleavage of hepatitis C virus RNA (12) occurs markedly at the UN^N sequences (Fig. 4A).
Fig. 4. Cleavage of biological targets.
(A) Site selection by RNase L in mammalian ribosomes and hepatitis C virus RNA. (B) Total RNA from HeLa cells cotransfected with plasmids encoding RNase L mutants or not encoding a protein (–), and 2–5A. Protein expression levels were analyzed by Western blot (WB). The bar graph shows compiled data from in vitro assays in Fig. 2A and fig. S4D. (C) Cleavage of substrates DIII and DIV by RNase L and Ire1KR32. Bar charts show quantification of cleavage. Error bars show mean ± SE of a single-exponential fitting (RNase L) and two time courses (Ire1KR32).
To confirm that key functional residues identified by our structural and biochemical analyses are important in vivo, we tested the activity of several RNase L mutants in human cells using 28S rRNA cleavage readout (12). RNase L mutants R163A (αI clamp), R412A (KH/KH interface), R427A (recognition of 2–5A), and H672N (RNA cleavage) were tested and displayed reduced activity compared with WT RNase L. Western blotting confirmed that all mutants were expressed similarly (Fig. 4B).
The mechanism of RNA cleavage by RNase L identified here has implications for understanding RNA decay by a related receptor, Ire1. In addition to cleaving canonical RNA stem loops within Hac1/XBP1 mRNA, Ire1 has a broader second set of RNA targets, which are cleaved during RIDD (4). Our review of these targets revealed a surprising agreement between the recently mapped RIDD cleavage sites and the consensus UN^N established for RNase L. This is observed with Angptl3 mRNA (16), Cyp1 mRNAs (17), and pre-mir-17microRNA (18), all of which are cleaved at UG^C sites. Moreover, Ire1 in fission yeast, which lacks the Hac1 gene, cleaves mRNA at sites NNUG^CNN (19) that match both the UN^N pattern and the UG^C pattern in RIDD substrates. Thus, structural similarity between RNase L and Ire1 results in a functional similarity, which hitherto escaped notice.
To examine this functional relationship, we tested cleavage of UN^N motifs by Ire1. Ire1 did not cleave UU^U, UU^A, and UA^A sequences, exhibiting a mutually exclusive specificity with RNase L (fig. S11). However, Ire1 and RNase L both cleaved the UG^C sequence (DIII) (Fig. 4C) and were sensitive to a U→A mutation (DIV). Ire1 was still able to cleave the G^C site in substrate DIV, indicating that the catalytic KEN protomer of Ire1 provides G^C recognition. Together, our experiments and the studies of RIDD and fission yeast suggest that RNA decay by RNase L and RIDD use related mechanisms of RNA cleavage (fig. S12).
RNase L is expressed in most human tissues, which imposes the requirement for a delicate regulation of this protein to prevent uncontrolled RNA cleavage, growth inhibition, and apoptosis. Here, we describe the mechanism of this regulation. Our findings further the understanding of the IFN response and RIDD and provide a structural basis for designing synthetic 2–5A analogs as molecular probes and modulators of IFN action and adjuvants for treating infections and inflammatory diseases.
Supplementary Material
Acknowledgments
We thank F. Hughson (Princeton University) and A. Korostelev (University of Massachusetts, Worcester) for reading the manuscript and making valuable suggestions. We thank the staff of Brookhaven National Laboratory for providing access to a synchrotron x-ray source. We are grateful to our colleagues at the Department of Molecular Biology for stimulating discussions and an excellent research environment. We thank Princeton University for supporting and funding our work. Coordinates have been deposited to the Protein Data Bank with accession codes 4OAU and 4OAV.
Footnotes
Note added in proof: After this manuscript was submitted, groups of F. Sicheri and R. Silverman published a structural analysis of porcine RNase L. This work reports a similar crossed homodimer but observes different KEN/KEN interactions (20) due to a conformational difference presumably caused by RNA binding.
Author contributions: Y.H., J.D., and A.K. determined the structures of RNase L. S.R. carried out biochemical studies of model RNA substrates. G.W. and A.C. carried out studies in HeLa cells. Y.H. and A.K. wrote the manuscript. A.K. supervised the work.
Supplementary Materials
www.sciencemag.org/content/343/6176/1244/suppl/DC1
Materials and Methods
Figs. S1 to S12
Table S1
References (21–30)
References and Notes
- 1.Chakrabarti A, Jha BK, Silverman RH. J. Interferon Cytokine Res. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donovan J, Dufner M, Korennykh A. Proc. Natl. Acad. Sci. U.S.A. 2013;110:1652–1657. doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin R-J, et al. J. Immunol. 2009;183:8035–8043. doi: 10.4049/jimmunol.0902728. [DOI] [PubMed] [Google Scholar]
- 4.Hollien J, Weissman JS. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 5.Han Y, Whitney G, Donovan J, Korennykh A. Cell Rep. 2012;2:902–913. doi: 10.1016/j.celrep.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Korennykh AV, et al. Nature. 2009;457:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong B, Silverman RH. Nucleic Acids Res. 1999;27:439–445. doi: 10.1093/nar/27.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turowec JP, et al. Methods Enzymol. 2010;484:471–493. doi: 10.1016/B978-0-12-381298-8.00023-X. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka N, et al. EMBO J. 2004;23:3929–3938. doi: 10.1038/sj.emboj.7600420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee KPK, et al. Cell. 2008;132:89–100. [Google Scholar]
- 11.Korennykh AV, et al. BMC Biol. 2011;9:47. doi: 10.1186/1741-7007-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Washenberger CL, et al. Virus Res. 2007;130:85–95. doi: 10.1016/j.virusres.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll SS, et al. J. Biol. Chem. 1996;271:4988–4992. doi: 10.1074/jbc.271.9.4988. [DOI] [PubMed] [Google Scholar]
- 14.Wreschner DH, McCauley JW, Skehel JJ, Kerr IM. Nature. 1981;289:414–417. doi: 10.1038/289414a0. [DOI] [PubMed] [Google Scholar]
- 15.Iordanov MS, et al. Mol. Cell. Biol. 2000;20:617–627. doi: 10.1128/mcb.20.2.617-627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.So JS, et al. Cell Metab. 2012;16:487–499. doi: 10.1016/j.cmet.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hur KY, et al. J. Exp. Med. 2012;209:307–318. doi: 10.1084/jem.20111298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Upton JP, et al. Science. 2012;338:818–822. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimmig P, et al. eLife. 2012;1:e00048. doi: 10.7554/eLife.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang H, et al. Mol. Cell. 2014;53:221–234. doi: 10.1016/j.molcel.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.