Summary
Transcript regulation is essential for cell function, and misregulation can lead to disease. Despite technologies to survey the transcriptome, we lack a comprehensive understanding of transcript kinetics, which limits quantitative biology. This is an acute challenge in embryonic development where rapid changes in gene expression dictate cell fate decisions. By ultra-high frequency sampling of Xenopus embryos and absolute normalization of sequence reads, we present smooth gene expression trajectories in absolute transcript numbers. During a developmental period approximating the first 8 weeks of human gestation, transcript kinetics vary by 8 orders of magnitude. Ordering genes by expression dynamics, we find temporal synexpression predicts common gene function. Remarkably, a single parameter, the characteristic timescale, can classify transcript kinetics globally and distinguish genes regulating development from those involved in cellular metabolism. Overall, our analysis provides unprecedented insight into the reorganization of maternal and embryonic transcripts and redefines our ability to perform quantitative biology.
INTRODUCTION
Gene expression is dynamic and tightly regulated. To build a quantitative understanding of gene regulation, direct measurement of transcript kinetics is necessary. Transcript kinetics describe the rate of change of transcript copy numbers with time. In developing systems such as the embryo, dynamic transcript expression precisely coordinates a sequence of stereotypical events that occur in rapid succession. A quantitative understanding of the transcriptome during embryogenesis will have important applications for congenital malformation research and regenerative medicine (Fakhro et al., 2011; Tebbenkamp et al., 2014; Zaidi et al., 2013). With the ability to measure global transcript kinetics, we can effectively study the impact of different transcript regulation strategies on gene expression, for example, dynamics of chromatin modifications, utilization of cis-regulatory sequences, kinetics of transcription factor binding, and transcript stability. Since the study of gene regulatory networks is limited by the lack of genome wide kinetic data (Karlebach and Shamir, 2008), a kinetic transcriptome dataset will be transformative to build and test gene regulatory network models in development.
The measurement of transcript kinetics has two requirements: 1) measurements of gene expression in absolute numbers of transcripts per cell or embryo and 2) sampling at a sufficiently high temporal resolution to properly calculate rates of change of transcript numbers. While there is great potential to use RNA-seq to study transcript kinetics, the currently available datasets lack one or both of these attributes, and suitable methodologies have yet to be developed (Stegle et al., 2015). In most datasets, sampling timepoints are too widely spaced. Data points are effectively isolated “snapshots” in developmental time, and analysis is restricted to pairwise comparisons of differences in relative gene expression (Aanes et al., 2011; Fang et al., 2010; Kalinka et al., 2010; Paranjpe et al., 2013; Spencer et al., 2011; Tan et al., 2013; Vesterlund et al., 2011; Wang et al., 2004; Yanai et al., 2011). In addition, in each of these datasets, relative normalization precludes kinetic calculations. Relative normalization is ubiquitous within the field and provides expression levels in arbitrary units related to the number of reads that map to a transcript relative to the total number of reads from the library. These relative values do not relate directly to transcript numbers and are not reliable indicators of either the magnitude or the direction of change in gene expression between samples when RNA content changes. For example, as the total amount of RNA increases in the developing embryo, transcripts with a constant copy number will appear to decrease in expression under relative normalization. Therefore, even in those studies where sampling rates are high (Collart et al., 2014; Tomancak et al., 2002), relative normalization combined with varying total mRNA levels confound comparisons such that kinetic calculations cannot be made even in simple fold change terms.
Absolute transcript measurements can be achieved by spiking in RNAs at known abundances for use as quantification standards. Native transcripts are calibrated against RNA standards to calculate absolute transcript copy number estimates. Previous reports have used spike-ins to normalize for sequencing depth, to control for technical variability, and to calculate estimates of absolute transcript numbers (Brennecke et al., 2013; Islam et al., 2014; Junker et al., 2014; Risso et al., 2014). However, recent reports have suggested that such a direct absolute normalization with currently available RNA standards is infeasible due to significant sequencing biases (Risso et al., 2014; SEQC MAQC-III Consortium, 2014). To date, a few RNA-seq datasets have been calibrated indirectly, where a small number of transcripts are first quantified using a non-sequencing technology, which in turn is used to normalize RNA-seq data (Marguerat et al., 2012; Tu et al., 2014). Here, we show that direct absolute quantification using sequence data alone is feasible and yields accurate measurements of embryo RNA content. Using RNA-seq and spike-in RNAs for absolute normalization, we created a genome-wide dataset to precisely measure transcript kinetics, which can be linked to biological processes occurring during embryogenesis (i.e. gastrulation or organogenesis such as eye, muscle, heart). To accomplish this, we sampled rapidly developing embryos at such high temporal resolution that data from neighboring timepoints were found to be as similar as biological replicates, ensuring that we captured precise transcript dynamics.
This rich dataset enables the visualization of gene dynamics of the entire transcriptome over an extended developmental period. Below, we outline a number discoveries enabled by combining absolute normalization and high temporal resolution sampling.
RESULTS
Data Collection, Quality, and Overview of Analysis
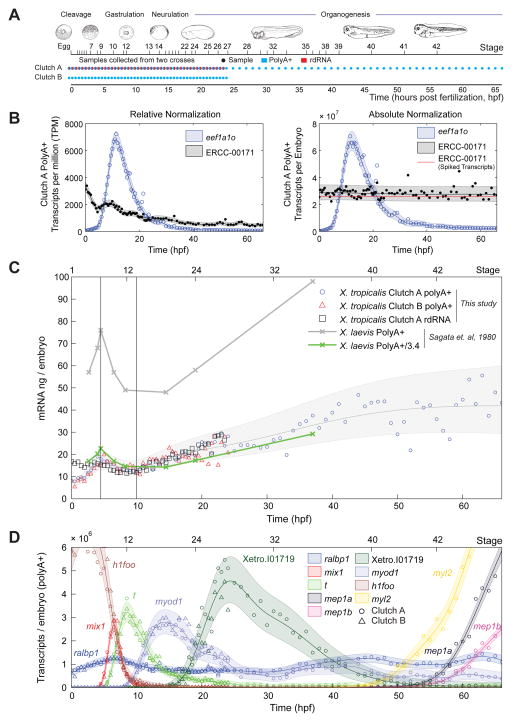
To profile gene expression at high time-resolution in the human disease model, Xenopus tropicalis, we collected eggs (time 0) and then synchronously developing embryos from an in vitro fertilization at 30 minute intervals for the first 24 hours of development, followed by hourly sampling thereafter to 66 hours (Fig 1A). We sampled RNA from two crosses collected in parallel (Fig 1A), Clutch A and B. For Clutch A, we sequenced polyA+ RNA for the full 66 hours and also total RNA depleted of ribosomal RNA (rdRNA; containing both polyA+ and polyA− RNAs) for the first 24 hours. For Clutch B, we sequenced polyA+ RNA for the first 24 hours only.
Figure 1. Data generation, mRNA content of the embryo and gene expression dynamics, see also Fig S1, S2.
A – Experimental design and sample collection
B – Left: eef1a1o and ERCC-00171 abundance in relative normalization (TPM), Clutch A polyA+. RNA standard decreases with time. Right: absolute normalization of left. Red line indicates ERCC-00171 transcripts added based on the manufacturer’s datasheet. The RNA standard is constant at the correct abundance.
C – Total mRNA (PolyA+ and rdRNA) in the embryo in nanograms with time. Gray region and line respectively mark Gaussian process 95% confidence interval (CI) and median of Clutch A & B PolyA+ data.
D – Dynamics of gene expression of PolyA+ RNA in transcripts per embryo. Circles mark Clutch A PolyA+, triangles mark Clutch B PolyA+. The horizontal axis marks time (bottom) and NF developmental stage (top). Shaded regions mark Gaussian process 95% CIs of the data.
To calculate absolute transcript numbers and kinetics, we calibrated read counts of all transcripts using spike-in RNAs as quantification standards. After embryo homogenization and prior to RNA extraction, we added a known amount of spike-in RNAs per embryo to each sample independently. This ensured that RNA standards underwent the same variations in recovery as the endogenous embryonic transcripts during RNA purification, library preparation, and sequencing. To Clutch A and Clutch B, we added the 92 synthetic RNAs available from the External RNA Control Consortium (ERCC) (Baker et al., 2005). In addition, we independently added three of the E. coli derived ArrayControl spike-ins to Clutch B.
We align reads to the X. tropicalis genome, known off-genome EST sequences and spike RNA sequences, and calculate relative transcript abundances of an augmented version of X. tropicalis v7.2 models in transcripts per million (TPM). The overall quality of our datasets was excellent according to a number of metrics (Fig S1A, B). Importantly, for capturing transcript kinetics, transcriptome wide comparisons of neighboring time points were as correlated as biological replicates as measured by transcriptome wide Spearman correlation (Fig S1A). Four time points (among 139) and one RNA standard that performed poorly (Qing et al., 2013; SEQC MAQC-III Consortium, 2014) were excluded from all subsequent analysis (Fig S1C,D,E; Supplemental Experimental Procedures). We also found that a small fraction of non-ribosomal RNA transcripts are depleted in the rdRNA dataset (Fig S2F, Table S2) indicating that our ribosomal-depletion protocol may have a small number of off-target effects.
Commonly, in large scale genomics studies, unsupervised machine learning tools such as clustering methods (like WGCNA: weighted correlation network analysis (Langfelder and Horvath, 2008)) or dimensionality reduction methods such as principle components analysis (PCA) (Jolliffe, 2002) can discover relationships within the data. In our case, PCA confirmed a strong temporal correlation inherent in our experimental design (Fig 1A, Fig S1F). Therefore, we opted for a data analysis framework that can explicitly describe these temporal correlations. Gaussian processes are employed commonly in the physical sciences and have received significant attention as general tools for machine learning (Rasmussen and Williams, 2006). They offer a statistical framework for representing and examining the strong temporal correlations present in our data. Gaussian processes have been successfully applied to a range of biological data (Aijo et al., 2014; Heinonen et al., 2014; Honkela et al., 2010; Stegle et al., 2010; wa Maina et al., 2014). Here, Gaussian processes allow us to address multiple topics within a single framework to elucidate transcript dynamics. We employ them in absolute normalization, regression, estimation of confidence intervals, identification of differential expression and the calculation of kinetics.
Direct Absolute Normalization of RNA-seq data
We achieve absolute normalization with a two-step procedure. Our first step is relative normalization for sequencing depth resulting in transcript abundances in TPM. We note that, the relative normalized abundances of our RNA standards decrease with time due to accumulating RNA in the embryo (Fig 1B left, Fig S2A). In our second step, we transform these RNA standard abundances to be constant with time at the known amount spiked into each sample. To minimize the effects of technical noise of the RNA standards, we use Gaussian processes to learn a function that varies smoothly with time to transform the RNA standards. This smooth mapping is critical as it ensures that absolute normalized copy numbers are not contaminated by sample to sample technical noise in the RNA standards.
Recent work has demonstrated that ERCC spikes exhibit significant sequencing biases and their appropriateness for direct absolute normalization in RNA-seq has been questioned (Risso et al., 2014; SEQC MAQC-III Consortium, 2014). To investigate these concerns, we evaluated the consistency of our proposed absolute normalization on our three datasets independently: Clutch A PolyA+, Clutch A rdRNA, and Clutch B PolyA+ (Fig S2B). In agreement with previous observations (Qing et al., 2013; Risso et al., 2014; SEQC MAQC-III Consortium, 2014), we find 1) that ERCC spikes perform poorly in polyA+ sequencing when compared to rdRNA sequencing (Fig S1F, Fig S2B), 2) spike-in variation between the two independently spiked PolyA+ datasets (Clutch A & B) is smaller than between Clutch A PolyA+ and Clutch A rdRNA (Fig S2B), and 3) spike-in performance varies across spike-in species but is consistent over replicates (Fig S2C). We apply corrections for the first two sources of variation (Fig S2B; Supplemental Experimental Procedures) and then calculate absolute transcripts per embryo for native transcripts and RNA standards. We capture the Clutch A/B variation and the spike-in species sequencing biases in a single uncertainty model for the absolute normalization (Fig S2D). Our absolute normalization performs well with R2 = 0.97–0.98 (Fig S2C) and has a 1.11–1.25 fold error when comparing the ERCC and ArrayControl spike-ins that were added independently to Clutch B samples (Fig 2SC). We discuss the limitations of this normalization, the validity of our Gaussian Process models, and influences of gene model quality in Supplemental Experimental Procedures and Table S2.
The detection limit, which is the number of transcripts required to produce a single read on average, increases with time as mRNA accumulates in the embryo (Fig S2E). Averaged over all samples, the detection limit is ~1300 transcripts per embryo, which is less than 1 transcript per cell once the embryo has attained the cell number present in the blastula.
Total mRNA Content of the Embryo Validates Absolute Normalization
We investigated the levels of total mRNA in the embryo during development. We summed the absolute abundance of all transcripts measured to derive the total mRNA in nanograms per embryo at each time point for both polyA+ and rdRNA (Fig 1C). Although there is a notable wave between 0–10 hpf (see below), the total amount of polyA+ mRNA increases with time from 10–15 ng of mRNA/embryo at fertilization to 30–50ng of mRNA/embryo at 66 hpf (swimming tadpole, stage 42). These values validate our absolute normalization, as they agree well with total RNA yields after tissue homogenization (~1.3 μg/embryo and ~2.4 μg/embryo at earliest and latest stages of our timecourse respectively), where ~1–2% of total RNA is polyadenylated (Davidson, 1986). As further validation of the absolute normalization, we compare our X. tropicalis mRNA predictions to experimentally measured polyA+ mRNA yields from X. laevis (Sagata et al., 1980). Accounting for the uncertainty in our absolute normalization, the mean ratio between these X. laevis mRNA yields and our data is 3.38 +/− 0.02 [mean +/− std. dev.] (Fig 1C). This is good agreement with the ratio in volume between X. laevis and X. tropicalis eggs, their respective diameters are 1.19 +/− 0.07 mm and 0.80 +/− 0.05 mm (Crowder et al., 2015), giving a volume ratio of 3.31 +/− 0.76. Therefore, our absolute quantification agrees well with measurements of in vivo mRNA levels determined by an independent experimental method.
Transcript Dynamics in the Developing Embryo
Examining the expression of individual genes, we observe smooth transitions in gene expression (Fig 1D). Our sampling rate is sufficient to capture the dynamics of the expressed genes (see timescale analysis below). We find genes whose expression peaks at different times in development, demonstrating transcript kinetics that are highly dynamic, and suggesting specific roles for these genes during defined developmental periods (Fig 1D). For example, in the case of muscle induction, a series of genes critical for mesoderm and then somite specification accumulate and extinguish at different times as development progresses (Fig S3A). The temporal expression profiles of these genes agree with RNA detection by in situ hybridization in whole mount embryos (Fig S3A). We further examined genes expressed later in development, which also correlated with in situ hybridization data (Fig S3B, C).
There is utility in identifying genes with constant expression as these may serve as optimal loading controls in various experimental regimens (Fig S4). We found 109 genes (<0.7%) in polyA+ (0–66 hpf) and 1078 genes (6.9%) in rdRNA (0–24 hpf) that had less than a two-fold change (Fig S4A, Table S3). Commonly used loading controls (gapdh, odc1, eef1a1) have more variable expression (Fig S4B). Our data would suggest that the RNA helicase ddx3x has the best loading control performance across developmental time and RNA-seq preparations (Fig S4C).
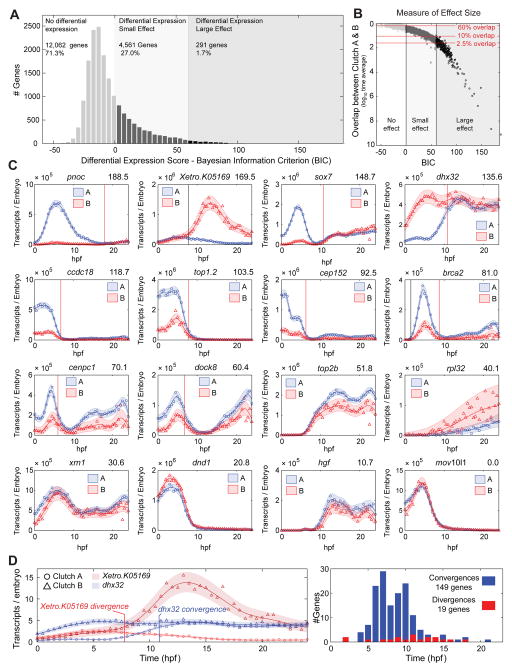
Differential Expression between Biologically Independent Replicates Reveals Precise Regulation of Transcripts
Our dense temporal sampling, offers a unique ability to detect differential expression over the entire time course. We began by exploring differential expression between our Clutch A/B biological replicates (Fig 2, Supplemental Experimental Procedures). We find that 12,062 of 16,914 genes (71%) had identical expression dynamics between the two clutches. Additionally, 4,561 genes (27%) showed differential expression, but with a small statistical effect (Fig 2A,B); their expression profiles show only minor differences (Fig 2C). Thus, expression dynamics of 98% of the transcriptome are similar between two independent biological replicates. This highlights both the high degree of reproducibility in our dataset and the exceptionally robust control of gene expression during development. The remaining 291 genes (1.7%) had a large statistical effect indicating strong differential expression. The majority of these genes have differential maternal expression, but most Clutch A and Clutch B differences converge by 10–12 hpf, displaying identical expression later in development (e.g. dhx32, Fig 2D). Interestingly, this set of genes is enriched for GO terms relating to cell division (Table S4), suggesting roles in the rapid cell division of the early embryo. Notably, a smaller number of genes have expression profiles that are divergent between replicates (Fig 2D). From these data, we conclude that gene expression is tightly regulated throughout embryogenesis and highly reproducible when measured using high-throughput sequencing.
Figure 2. Clutch A vs Clutch B PolyA+ Differential Expression.
A – Clutch A vs Clutch B histogram of differential expression scores as measured by the Bayesian Information Criterion (BIC), larger scores indicate greater differential expression (Supplemental Experimental Procedures). Three regions marked: 1) no differential expression BIC ≤ 0, 2) differential expression small effect 0 < BIC ≤ 60, 3) differential expression large effect BIC > 60. See B for explanation of thresholds.
B – Differential expression effect size as measured by the log10 mean overlap between Clutch A and Clutch B Gaussian process models (Supplemental Experimental Procedures). Mean overlap decreases with increasing BIC. BIC = 0, 60 and mean overlaps of 60%, 10% and 2.5% marked. At BIC = 60 approximately all genes have less than 10% overlap, and all genes with less than 2.5% overlap have BIC > 60.
C – Differential expression examples with decreasing BIC (top right). Genes on the boundary for strong differential expression have highly correlated expression profiles in A and B. Vertical lines mark convergences (red) or divergences (black).
D – Left: Examples of convergence (dhx32) and divergence (Xetro.K05169). Right: Histogram of convergence and divergence times.
Differential Expression and Switching of Transcript Isoforms During Development
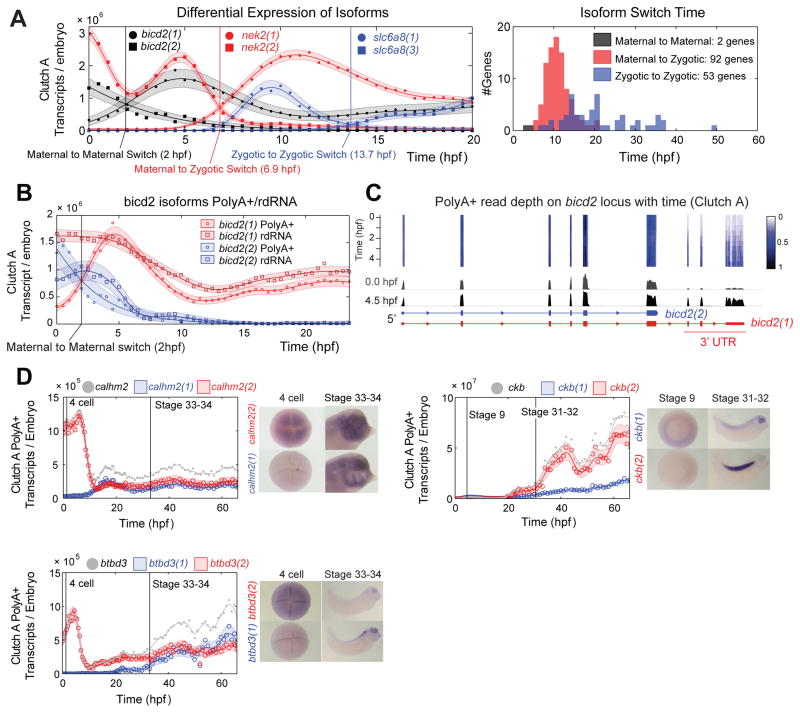
Next, we analyzed the differential expression of alternative transcript isoforms. We can classify differential isoform expression as either differential abundance or differential temporal dynamics. Transcripts are defined to have differential abundance if one transcript is expressed at a different level than the other with a constant ratio between them, and differential dynamics if the ratio varies with time (Fig S5). Here, we restrict our attention to those isoforms that have differential temporal dynamics (Fig 3, Fig S5). We found 761 genes with isoforms showing differential dynamics, with differences in promoter and exon usage, as well as variable 3′ UTRs. Isoform expression profiles for 147 genes show a switch-point, where their expression profiles cross, marking a change in which isoform is most abundant (Fig 3A). The timing of these switch-points was not evenly distributed over development; two thirds are transitions from a maternal to a zygotic isoform concentrated around 10 hpf (Fig 3A). Interestingly, we find examples of isoform switching due to post-transcriptional regulation, two maternal isoforms of bicd2 exhibit differential polyadenylation (Fig 3B). One isoform bicd2(1) has a long 3′ UTR and is polyadenylated upon fertilization, the other bicd2(2) has a short 3′ UTR and is deadenylated upon fertilization (Fig 3C). The 3′ UTR of bicd2(1) must contain control elements to remain deadenylated during oocyte maturation and become polyadenylated after fertilization.
Figure 3. Isoform Differential Expression in Clutch A, see also Fig S5.
A – Isoform switching event examples: maternal to maternal; maternal to zygotic; and zygotic to zygotic switching events (left). Histogram of isoform switching events by category and time (right).
B – bicd2 isoforms switch in PolyA+ data (circles), and do not switch in rdRNA data (squares) indicating differential polyadenylation. PolyA+ and rdRNA abundances agree within uncertainty of the absolute normalization (Supplemental Experimental Procedures).
C – Normalized read depth on bicd2 locus in Clutch A polyA+ between 0–4.5 hpf. Note temporal dynamics shared by the final three bicd2(1) exons. Heatmap and depth of pile ups corrected for changing total mRNA.
D – Spatial expression of three genes with isoforms with differential dynamics. All isoforms show different temporal and spatial expression domains. Grey lines and points mark total expression for each gene (sum of the red and blue isoforms).
We predicted that isoforms with different temporal dynamics may show different spatial as well as temporal expression, implying differential function. We selected three genes where the sequence differences between isoforms were long enough to generate effective in situ hybridization probes. Remarkably, all three showed different spatial expression domains (Fig 3D). This demonstrates the biological importance of isoform dynamics during embryogenesis and suggests specific mechanisms to regulate isoforms in different tissues. Moreover, we demonstrate the power of high-resolution sampling for transcript discovery and gene modeling. The small fragment sizes associated with high-throughput sequencing present challenges for identifying the co-expression of distant exons in long transcripts. The temporal statistics here can aid in the resolution of long-range order; distant exons that are expressed in the same isoform will exhibit related temporal dynamics (Fig 3C).
Temporal Synexpression Predicts Common Gene Function
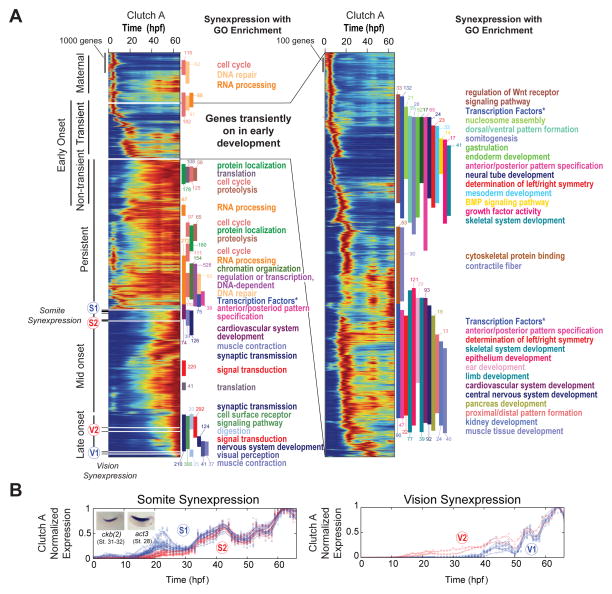
To explore the expression dynamics of the entire dataset, we created a heat map of transcripts organized according to a hierarchical clustering of their normalized expression profiles (Fig 4A). The heat map is ordered such that nearest neighbors have similar expression profiles, which we term temporal synexpression. Synexpression suggests that genes are controlled by shared regulatory networks and may have common function (Niehrs and Pollet, 1999). To investigate temporal synexpression, we used a sliding window to assess local Gene Ontology (GO) enrichment across the heat map (Fig S6A, Table S4). Interestingly, we found many blocks of genes enriched for specific functions ranging from cellular biology to developmental patterning events (Fig 4A). In the set of genes showing early transient expression, we found GO terms associated with early developmental steps including germ layer specification, mesoderm/endoderm development, and axis patterning (Fig 4A right, top of heat map). Genes transiently expressed later in development are enriched for GO terms associated with organogenesis (Fig 4A right, bottom of heat map). Therefore, proximity within the heat map enriches for GO terms that are consistent with developmental events, demonstrating that temporal synexpression is a powerful predictor of gene function (see below).
Figure 4. Temporal Synexpression in Clutch A PolyA+, see also Fig S6.
A – Temporal map of the transcriptome: an enumeration of all gene expression dynamics in the embryo. Heat maps display all genes normalized by maximum expression and ordered by similarity. Inset (right) expands on genes transiently expressed with early onset. Vertical bars mark Gene Ontology (GO) enrichment (colors correspond to GO terms), numbers appended to GO bars indicate the number of genes of given category. All reported GO blocks are enriched with p < 2×10−4 (Fisher Exact) for entire block. S1, S2 and V1, V2 mark the location of somite and vision temporal synexpression genes respectively in B. “Transcription Factors *” labels an annotation of transcription factors separate to GO (Supplemental Experimental Methods).
B – Somite (left) and Vision (right) synexpression groups.
While exploring temporal synexpression, we found two distinct sets of genes (S1 and S2 in Fig 4A, 4B left; Supplemental Experimental Procedures) that have similar oscillatory behaviors between 34 and 66 hours of development. These sets are not immediately adjacent in the overall heat map due to different expression prior to 34 hours. Postulating that there may be more genes across the heat map with similar oscillations, and hence similar function, we used one of these genes, ckb (Fig 3D, right), as a “seed” to identify other similarly oscillating genes and discovered a much larger set (150 genes, Table S5). To determine whether this local temporal synexpression identified common functions, we examined spatial expression patterns. Our seed gene, ckb, has pronounced expression in the somitic mesoderm and lies within a block of genes enriched for muscle contraction GO terms. Remarkably, our larger local temporal synexpression set also has significant enrichment in somitic mesoderm. 38 of these genes have expression patterns available in public databases (Bowes et al., 2010), and 34/38 (89%) have somitic expression (Fig S6B). These genes appear to oscillate with a period of approximately 20 hours, which is significantly longer than the somitic clock (Pourquie, 2003) and more reminiscent of the circadian rhythm. Interestingly, a few circadian clock genes are expressed in the somites (Curran et al., 2014; Curran et al., 2008), and our analysis indicates there may be additional unexplored links between clock genes and somite regulatory networks.
In another case, we found two sets of genes (V1 and V2; Supplemental Experimental Procedures) with similar temporal expression patterns from 40–66 hpf that enrich for visual perception (GO:0007601). With the exception of tmem145, all V1 and V2 genes are well characterized as having roles in rod and cone cells and are associated with human retinal diseases (Table S5). We therefore predicted that the uncharacterized tmem145 also plays a role in vision, and indeed tmem145 does have spatial expression in the eye (XDB3 (Bowes et al., 2010) and XGC EST database (Klein et al., 2002; Morin et al., 2006)).
We conclude that temporal synexpression can be used effectively to predict gene function, and predictions can be refined depending on the local developmental period of interest. Remarkably, we uncovered a large cohort of genes expressed in the somites simply by ranking their similarity in expression to a gene with known somite expression. We anticipate that future investigations of temporal synexpression in this data may be able to assign putative functions to many of the 5,718 genes (~28%) that remain unnamed.
The Reorganization of Maternal and Zygotic Transcriptomes
In our analysis of total mRNA during development (Fig 1C), we noted a difference between total rdRNA levels and polyA+ mRNA levels prior to 4.5 hpf (early stage 9 blastula), the point at which zygotic transcription becomes widespread. From 0–4.5 hpf, there is an increase in polyA+ transcripts while rdRNA levels show little change, reflecting the polyadenylation of maternal transcripts (Collart et al., 2014). Then from 4.5–10 hpf, both polyA+ and rdRNA levels fall as the clearance of maternal mRNA exceeds nascent zygotic transcription. The fall in RNA constitutes a ~20% loss of the embryo’s mRNA content, supporting the notion that very early embryonic development relies heavily on maternally stored transcripts. By 10 hpf (late gastrula stage 12), those maternal mRNAs targeted for clearance are largely extinguished, and zygotic transcription causes mRNA levels to increase for the remainder of our timecourse.
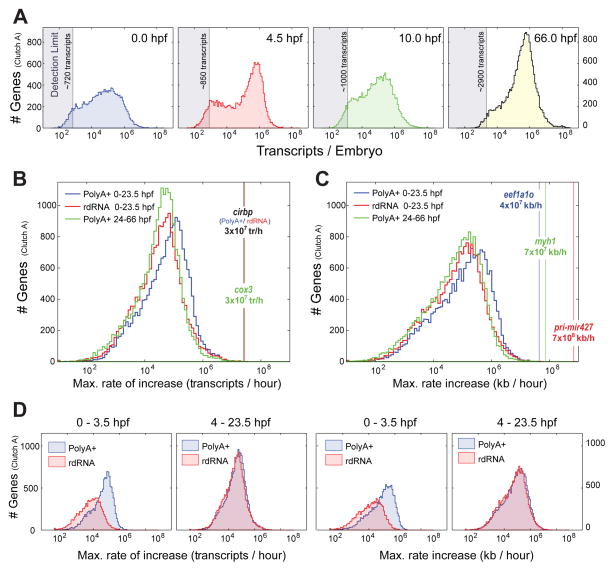
To examine these transitions more closely, we considered the distribution of the number of transcripts per gene at different time points (Fig 5A). Initially, in the egg, the distribution has a broad peak at ~100,000 transcripts, which changes dramatically at 4.5 hpf into a bimodal distribution with a marked peak at ~600,000 transcripts. This change in distribution reflects the rapid increase in polyadenylated maternal transcripts. The distribution is again reorganized at 10 hpf, losing the bimodal appearance and shifting leftwards towards fewer transcripts as maternal transcripts are eliminated. By the end of our timecourse, the peak at ~600,000 transcripts is re-established without the bimodal appearance seen at 4.5 hpf. The coincidence of a mode at approximately ~600,000 transcripts at 4.5 hpf and 66 hpf is unexpected. The tadpole at 66 hpf has twice the mRNA content and is considerably more complex than the 4.5 hpf blastula stage embryo. Therefore, the doubling of the RNA content is achieved by having roughly twice as many genes with at least ~600,000 transcripts/embryo rather than simply shifting the entire distribution (rightward) such that there are twice the copy number of each gene overall.
Figure 5. Transcript copy numbers and kinetics per embryo in Clutch A.
A – Transcript copy number histogram in polyA+ sequencing at 0.0 (egg), 4.5 hpf (early stage 9), 10 hpf (stage 12.5, end of gastrulation), 66 hpf (tadpole stage 42). Detection limits give number of transcripts required to generate a single read averaged over all transcript lengths (Fig S2E). Transcript copy number distributions transition smoothly between these timepoints (data not shown).
B – Histogram of maximum rates of increase in transcripts/hour between any consecutive measurement points calculated from Gaussian process medians for each gene. Blue – PolyA+ 0–23.5 hpf; Red – rdRNA 0–23.5 hpf; Green – PolyA+ 24–66 hpf. Vertical lines mark the gene with max. rate of increase for each category.
C – As B but kb/hour.
D – Maximum rate of increase in transcripts/hour (left) and kb/hour (right) for 0–3.5 hpf and 4–23.5 hpf time intervals. PolyA+ and rdRNA distributions are discrepant for 0–3.5 hpf reflecting polyadenylation of maternal RNAs. Distributions are identical over 4–23.5 hpf.
Transcript Kinetics Reveal a Rapid Deployment of Maternal Transcripts
To examine these changes in the distribution of transcript numbers in more detail, we calculated transcript kinetics. We determined the maximum rate of increase in transcripts per hour (either as transcripts/hr/embryo or kb/hr/embryo) and examined the distributions of all genes (Fig 5B,C). We compared these maximum rate distributions between rdRNA and polyA+ over 0–24 hpf and then polyA+ over 24–66 hpf. Strikingly, the maximum rates of accumulation vary over 8 orders of magnitude suggesting varied mechanisms for the regulation of transcript accumulation. The distributions for transcripts/hr and kb/hr are similar indicating that the rate distribution is not influenced by transcript length (Pearson correlation of 0.28 between max. accumulation rate and transcript length in Clutch A PolyA+). Both the transcripts/hr and kb/hr distributions are negatively skewed, producing the sharp rise on the right tail of the distribution. This indicates that many genes experience a rate of accumulation that is close to the maximal, and that most genes experience a rate of accumulation that is greater than the mean.
Interestingly, genes measured from 0–24 hpf in polyA+ preparations show faster rates (blue line shifted rightward, Fig 5B,C) than in both 0–24 hpf rdRNA and 24–66 hpf polyA+, which are aligned. Given the reorganization of transcripts from 0–4.5 (Fig 5A), we compared maximum accumulation rates in polyA+ and rdRNA at this time interval. At 0–3.5 hpf, the maximum accumulation rates for polyA+ are faster than for the rdRNA; in contrast, this difference is lost at 4–24 hpf where the polyA+ and rdRNA distributions are coincident (Fig 5D). We conclude that the polyadenylation of maternal transcripts offers the cleavage stage embryo a mechanism by which a large number of transcripts can be deployed for translation very rapidly, overcoming the embryo’s limited capacity for transcription (due to very rapid cell cycling and relatively few nuclei per embryo).
Rapid Kinetics of pri-mir427 and the Clearance of Maternal Transcripts
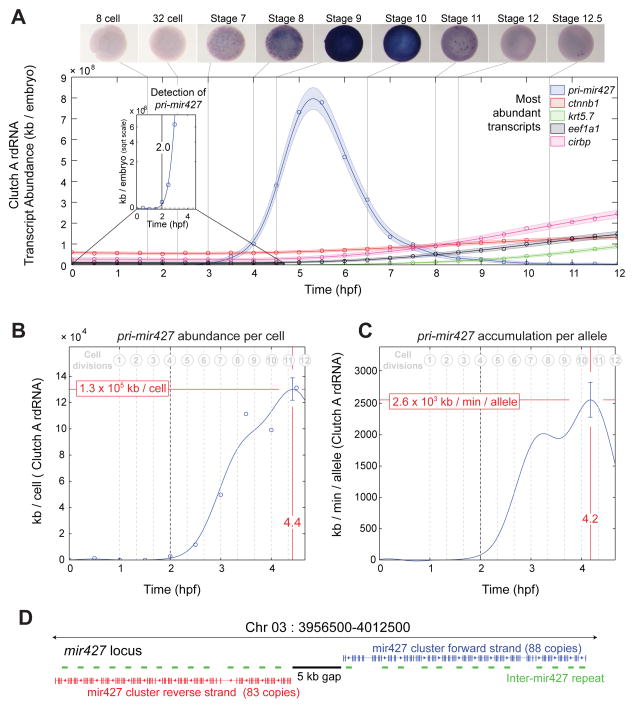
The clearance of maternal RNAs is one of the major embryo-wide transcriptome changes, resulting in a net loss of ~20% of the total mRNA content (Fig 1D). The mir427/430 family, in Xenopus and zebrafish respectively, plays a critical role in clearing maternal RNAs at the onset of zygotic transcription (Giraldez et al., 2006; Lund et al., 2009). Early embryonic phenotypes due to the loss of dicer, and therefore all miRNAs, can be rescued in zebrafish embryos by mir430, highlighting the importance of this microRNA (Giraldez et al., 2005). Interestingly, we find that the pri-mir427 transcript has the fastest kinetics of any transcript in the early frog embryo, at 7×108 kb/hour/embryo (Fig 5C), an order of magnitude faster than the very abundantly expressed ef1a1 (Fig 5C). In addition, we find pri-mir427 transcripts to be ubiquitously expressed, and we first detect their transcription at the 8-cell stage (2 hpf), significantly earlier than the previously identified onset of nodals 3, 5 and 6 and well before the classic midblastula transition (Fig 6A inset, Fig S7) (Kimelman et al., 1987; Newport and Kirschner, 1982; Skirkanich et al., 2011; Yang et al., 2002). Therefore, pri-mir427 is not only transcribed at a prodigious rate but is the earliest detected zygotic transcript in Xenopus.
Figure 6. Accumulation kinetics of pri-mir427 per allele in Clutch A rdRNA, see also Fig S7.
A – pri-mir427 expression over first 12 hpf in rdRNA data and in situ hybridizations. pri-mir427 show ubiquitous expression matching RNA-seq temporal data. Examples of most abundant transcripts shown for comparison. Inset: first detection of pri-mir427 above detection limit at 2.0 hpf (8–16 cell transition).
B – pri-mir427 abundance in kb per cell. Line marks Gaussian process median, error bar is 95% CI (Supplemental Experimental Procedures). Peak per cell abundance occurs after 11th division at 4.4 hpf.
C – pri-mir427 accumulation rate in kb/min/allele with time. Line is median of differential of Gaussian Process, error bar gives 95% CI (Supplemental Experimental Procedures). Peak accumulation rate is achieved at 4.2 hpf just after 10th cell division.
D – Organization of pri-mir427 locus on X. tropicalis Chromosome 3 (X. tropicalis v8 genome, Supplemental Experimental Procedures). Clusters of mir427 hairpins appear in tandem with a repeated sequence arranged symmetrically on opposite strands around a gap in the genome assembly.
As pri-mir427 is expressed during a period where precise cell numbers are known, we can calculate the average rate of transcription per allele from rdRNA data (Supplemental Experimental Procedures). The mir427 locus contains numerous copies of the mir427 hairpin covering ~55kb of transcribed genome (Fig 6D, Data S2). Here, we report the aggregate RNA abundance and accumulation over the entire locus.
The abundance of pri-mir427 transcripts per cell peaks at 4.4 hpf (130 Mb/cell) with the peak accumulation rate per allele occurring at 4.2 hpf (2.6 Mb/min/allele; Fig 6B,C). Performing a similar analysis for other early transcribed genes (Fig S7B), we find that pri-mir427 accumulates nearly 1000-fold more rapidly. We demonstrate that it is the size of the mir427 locus combined with ubiquitous expression that renders such a dramatic accumulation rate possible. We note that pri-mir427 is RNA pol II transcribed (Lund et al., 2009). The maximum RNA pol II elongation rate has been estimated at ~4 kb/min (Ardehali and Lis, 2009). At this elongation rate the locus requires a polymerase density of 11–12 pol II/kb to achieve the measured 2560 (95% CI: 2280 to 2840) kb/min/allele. This is in good agreement with the maximum polymerase densities of ~10 pol II/kb observed on rapidly transcribed genes of amphibian oocyte lampbrush chromosomes (Miller and Hamkalo, 1972). Corroborating this prediction, we examined published X. tropicalis blastula stage 9 RNA pol II ChIP-seq data (van Heeringen et al., 2014), and found high RNA pol II occupancy over the mir427 locus (Fig S7A). In summary, we conclude that the pri-mir427 achieves the highest known rate of transcript production during the very early rapid cell divisions of the frog embryo. To achieve this brisk rate, the size of the locus and the ubiquitous expression (supported by its spatial expression) are essential; however, the RNA pol II density and transcription rate need not have extraordinary values. This does imply that pri-mir427 is transcribed homogenously by all cells; any significant heterogeneity would require some cells to transcribe at rates beyond what has been observed.
We speculate that numerous copies are present in Xenopus and zebrafish to maximize production of mature mir427/mir430 microRNAs during the rapid development of these organisms. Interestingly, the mammalian member of the family, mir302, is present in far fewer copies on the genome, presumably because mammalian development is dramatically slower and the embryos smaller (with less maternal mRNA), and thus there is ample time to generate sufficient amounts of mir302 to clear maternal transcripts.
Interestingly, the earliest detection of pri-mir427 occurs at the 8-cell stage when its accumulating transcript breaks our detection limit. The next transcriptional events we detect are from loci that are on average 19 times smaller: nodal3/5/6 and siamois1/2 break our detection limit at 32–256 cell embryos (Fig S7B) which contain 4–32 fold more copies of these loci than 8-cell embryos. It is possible that these shorter loci may be transcribed from the 8-cell stage or earlier, but our expression measurements are insufficiently sensitive to detect this early transcription. Together, we conclude that we are able to detect the onset of zygotic transcription vastly earlier than previously thought.
In principle, the approach taken to quantitate pri-mir427 transcript kinetics can be applied to any gene during any time interval in our timecourse once the numbers of expressing cells/embryo are ascertained.
Characteristic Timescale of Gene Expression Classifies Developmental Gene Function
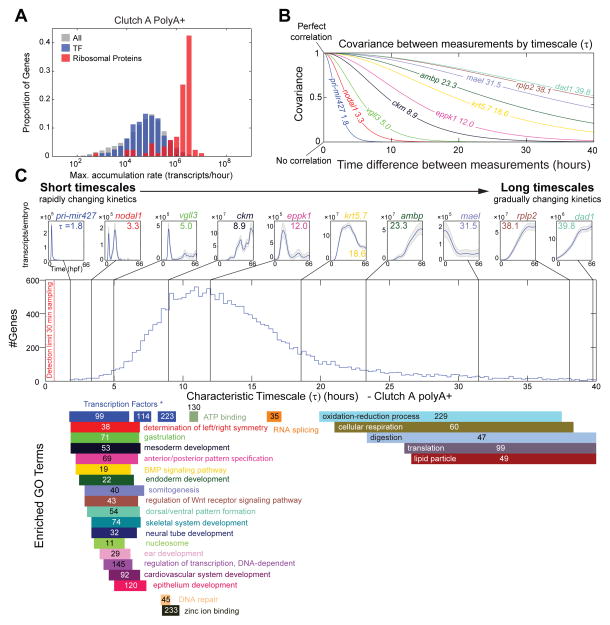
While the rate of accumulation of pri-mir427 is remarkable, transcript accumulation rates may not be optimal for identifying potent developmental regulatory genes, where small changes in transcript levels may be sufficient to alter cell fate and morphogenesis. We note that the distribution of maximum transcript accumulation rate does not differentiate transcription factors from all genes (Fig 7A). Whereas, ribosome related genes accumulate significantly faster. We postulated that developmentally regulated genes would not necessarily be transcribed at maximal rates but would be tightly regulated and transiently expressed, reflecting changing biological events (i.e. their expression would turn on and off rapidly). Using Gaussian processes, we calculate a single parameter to quantify such behavior, a characteristic timescale (τ) for each gene.
Figure 7. Characteristic Timescales of Gene Expression in Clutch A PolyA+.
A – Distribution of max. rate of increase of all transcripts (gray) (as Fig 5), transcription factors (blue), and ribosomal proteins rps* and rpl* (red).
B – Theoretical covariance (Matérn covariance function, Supplemental Experimental Procedures) between two measurements by time interval for example genes in C.
C – Histogram of characteristic timescale over all genes in in Clutch A polyA+. Top row annotates example genes (timescale inset), fast/short timescales are on the left slow/long timescales on the right, Gaussian process 95% CI (gray) and median (blue) marked. Bars below give GO enrichment (calculated as Fig 2, Fig S6A) of histogram, numbers indicate total genes with given category. All enrichments have p < 2.6×10−5 (Table S4). “Transcription Factors *” labels an annotation of transcription factors separate to GO (Supplemental Experimental Methods).
Formally, the timescale parameterizes the Gaussian process covariance function for each gene (Supplemental Experimental Procedures). The covariance function describes the covariance between a gene’s transcript levels at two time points (Fig 7B). Measurements of a gene’s expression that are closer in time will share a greater covariance (be more correlated) than those further apart in time. The timescale reflects how rapidly the covariance decays when considering expression at increasing time intervals (Fig 7B). Genes with rapidly changing expression kinetics lose covariance quickly and have short timescales (Fig 7B, 7C left). Conversely, genes with expression kinetics that vary more smoothly and gradually have covariances that persist in time and are described by long timescales (Fig 7B, 7C right).
To explore the characteristic timescales relevant to development, we plotted the distribution of timescales over all genes (Fig 7C). We find the mass of the timescale distribution to be slower than our detection limit (Supplemental Experimental Methods); in fact, our sampling rate is sufficient to reliably detect very short timescales (for example pri-mir427 with τ = 1.8 hours) (Fig 7C, upper left panel). We investigated the timescale distribution by gene ontology (Fig 7C). Remarkably, GO term enrichments segregated between short and long timescales (Table S4). At short timescales, there was a striking enrichment of genes associated with cell signaling and developmental processes; these genes are involved in rapid, spatially restricted patterning events during embryogenesis. At long timescales, we found enrichment in genes necessary for cellular metabolism, many of which correlate in expression with the changes in total mRNA in the developing embryo. We conclude that timescales are a powerful metric for investigating gene function, and are able to classify novel genes whose kinetics are similar to those with developmental or cell signaling roles, or unidentified roles in cellular metabolism.
DISCUSSION
We present a rich array of findings on transcript kinetics from our ultra-high temporal resolution expression profiling of Xenopus. To establish these biological observations, we overcame the challenges of direct absolute normalization of RNA-seq data using ERCC spikes (SEQC MAQC-III Consortium, 2014). We show: 1) It is essential to spike RNA standards at a constant ratio to embryo/cell numbers directly into homogenates prior to RNA extraction. 2) Relative normalized spikes exhibit a clear decreasing trend that was the result of increasing RNA in the embryo with time. This demonstrated that the ERCC spikes were of suitable fidelity for absolute normalization and identified our absolute normalization strategy. 3) We found it necessary to sequence the same RNA by multiple protocols (PolyA+ and rdRNA) so that protocol specific biases can be understood and minimized. 4) It is essential to estimate the error, or uncertainty, in the measurement of the RNA standards (Fig S2B,C,D) and then average over this uncertainty when making absolute transcript number predictions. We have achieved this latter point using Gaussian processes, and in doing so, we have ensured that our absolute normalization of native transcripts is not unduly influenced by technical noise or sequencing biases of the RNA standards (Fig S2D). 5) Finally, we note that high resolution temporal sampling has a significant advantage over multiple replicates of a low temporal sampling. With high frequency sampling, neighboring timepoints are still as similar as biological replicates, providing the necessary information on reproducibility, but importantly we also capture the transcript dynamics and so obtain more information for the same number of samples.
We have demonstrated that direct absolute normalization of RNA-seq data is feasible; the quality of the RNA standards and the understanding of their sequencing biases are the largest factors limiting progress in the absolute quantification of gene expression data and the accuracy of our predictions (see uncertainty confidence intervals in Fig S2D). Currently, the ERCC spikes are the predominant RNA standards; they are manufactured to log-scale precision and exhibit significant sequencing biases. With log-scale precision, the error is proportional to abundance and doubles for every doubling of target transcript copy numbers. Clearly, transcripts are under much tighter regulation in the embryo and the log-scale precision in RNA standard concentrations is far from ideal. The need for improved RNA quantification standards to advance future absolute quantification efforts is clear.
We demonstrate that high-frequency sampling is critical to define smooth expression trajectories and properly examine transcript dynamics. With such data, we can identify cohorts of temporally co-regulated genes and make accurate predictions on spatial expression and gene function. Our case studies of somite and vision temporal synexpression (Fig 4B) demonstrate the potential of this dataset not only for characterizing genes of unknown function but also uncovering new temporal phenomena in gene expression. High frequency sampling combined with Gaussian process analysis is a powerful tool for capturing the temporal evolution of differential expression, including identifying convergence and divergence points. Importantly, our analysis of timescales confirmed that our sampling rate was sufficient to capture the transcript behavior for even the most dynamic genes. The concept of the characteristic timescale will be critical for future transcriptome time series studies to ensure that dynamic behaviors are not missed or misinterpreted.
We also analyzed the global properties and kinetics of the transcriptome during our 66 hour timecourse. Interestingly, we find that much of the transcriptome accumulates to ~600,000 transcripts/embryo at different developmental stages. Examinations of transcript kinetics revealed that different kinetic signatures can be associated with the accumulation of transcripts via polyadenylation or transcription. We expect that further examination of kinetics may reveal additional signatures associated with different mechanisms of mRNA regulation.
The maternal to zygotic transition constitutes the most dramatic changes in the transcriptome that we observe in our timecourse. It begins with the polyadenylation of maternal transcripts, followed by onset of zygotic transcription and the clearance of maternal transcripts. The absolute nature of our data provides new quantitative insights into this period. For example, zygotic transcription starts early during the cleavage stages with the activation of pri-mir427, which plays an important role in maternal RNA clearance. The reduction in maternal mRNA exceeds transcription as the cleavage stages are completed (blastula stage 9; 4.5 hpf) and mRNA levels continue to fall until the end of gastrulation (stage 12.5; 10 hpf). During this time window, the majority of transcript level differences between Clutch A and Clutch B converge (Fig 2D), and many zygotic isoforms are activated whose abundance will exceed their maternal counterpart by ~10 hpf (Fig 3A).
Our data demonstrate that transcript levels during embryonic development are exquisitely controlled with only 2% of transcripts showing large differences in expression between replicates. Although the biological importance of these differences is not certain, these 2% of genes are nevertheless tightly regulated within the embryos of each clutch. This differential expression appears to be interesting and worthy of further exploration. Tight control over gene expression occurs at multiple levels, including chromatin modifications, cis-regulatory elements, transcription factors, and transcript stability. Our experimental approach can be used to investigate the effects of perturbation of these regulatory processes on transcript kinetics, which is particularly difficult without measurements of absolute transcript levels. Such real measurements have the potential to transform our understanding of the transcriptional control that is fundamental to gene regulatory networks. Our approach affords many opportunities to build gene regulatory networks whose foundation lies on much needed quantitative data. The potential for using Gaussian processes with time series expression data for uncovering gene regulatory interactions has been demonstrated (Honkela et al., 2010). Combining these approaches with our timecourse data holds exciting prospects for the elucidation of gene regulatory networks in early vertebrate development.
A major goal in developmental biology is to determine the temporal and spatial expression patterns of all genes during development. Our work is a step towards that goal. By combining absolute normalization and high frequency sampling with the recent advances in RNA tomography (Junker et al., 2014) and/or the spatial reconstruction of single cell RNA-seq data (Satija et al., 2015), there is an exciting opportunity to create a 4D atlas of developmental gene expression especially in vertebrate human disease models such as Xenopus.
EXPERIMENTAL PROCEDURES
Embryo Collection
To obtain RNA samples for RNA-seq, we performed 2 parallel in vitro fertilizations of siblings of 12th generation inbred Nigerian Xenopus tropicalis. All fertilizations and subsequent culturing of embryos were performed in a temperature-controlled room maintained at ~24°C, with fluctuations +/− ~1°C over the entire timecourse. All samples are reported in hours post fertilization (hpf) and developmental stage. Mapping from hpf to Nieuwkoop and Faber stage Is given in Table S1. We labelled the progeny of the two crosses Clutch A and Clutch B. All collections from the two clutches occurred concurrently.
RNA isolation and Spike ins
For most samples, 10 embryos/timepoint were homogenized in 200 μl Trizol® and frozen at −80°C, with the exception of egg, 0.5, 1, 1.5, 2, 2.5 and 3 hpf timepoints, which were sampled at 25, 30, 20, 20, 20, 15 and 15 embryos respectively. ERCC Spike In Mix 1 was added on a per embryo basis to Clutch A samples. For the Clutch B timecourse, ERCC spike in Mix 1 was also added along with the independent adding of Ambion Array Control RNA spikes. See Supplemental Experimental Procedures for additional details.
Data Analysis
All libraries were sequenced with 76bp paired-ends on an Illumina HiSeq 2000. Read pairs were aligned using TopHat v2.0.10 (Trapnell et al., 2009) to the X. tropicalis version 7.1 genome (Hellsten et al., 2010; Karpinka et al., 2015) along with our ERCC & ArrayControl exogenous spike sequences and known off-assembly gene sequences used in a previous study (Collart et al., 2014). Alignment was guided by the v7.2 X. tropicalis gene models, to which we made improvements and corrections (Data S1). We estimated the relative abundance of all transcripts in fragments per kilobase per million (FPKM) with Cufflinks v2.1.1 (Trapnell et al., 2010), and converted to transcripts per million (TPM). Absolute normalization was achieved by calibrating relative normalized transcript abundances against spikes to achieve absolute normalization. Corrections for polyA+ bias and Clutch A/B variation are applied to absolute normalization of native transcripts. All subsequent analysis employs Gaussian processes. See Supplemental Experimental Procedures for details.
Supplementary Material
Acknowledgments
We thank Sarah Kubek and Michael Slocum for animal husbandry. We are indebted to the support and sequencing at the Yale Center for Genome Analysis especially the assistance of Shrikant Mane. The authors also wish to thank Joe Gall and Margaret Fish for stimulating discussions. The authors thank James Noonan, Xiaohui Xie, Sarah Teichmann, and George Gentsch and the members of the Gilchrist, Khokha and Cho labs for helpful discussion of the manuscript. This work was supported by NIH/NIGMS 5R01GM099149 to MJG, KWYC, and MKK. MKK is a Mallinckrodt Scholar. This work was supported in part by the Francis Crick Institute, which receives its core funding from Cancer Research UK, the UK Medical Research Council, and the Wellcome Trust.
Footnotes
Accession Numbers
Sequencing reads are available in GEO GSE65785.
AUTHOR CONTRIBUTIONS
ILB and MKK collected the embryos for the timecourse while ILB prepared all RNAs. JDO supervised and provided critical expertise on RNA-seq at YCGA. NDLO performed all of the bioinformatics analysis. MAL did all of the in situ hybridizations. IP helped build visualization tools for the data. NDLO, ILB, MJG, KWYC, and MKK conceived the work, designed the experiments, and interpreted all experimental data. All authors contributed to the writing of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aanes H, Winata CL, Lin CH, Chen JP, Srinivasan KG, Lee SG, Lim AY, Hajan HS, Collas P, Bourque G, et al. Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome research. 2011;21:1328–1338. doi: 10.1101/gr.116012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aijo T, Butty V, Chen Z, Salo V, Tripathi S, Burge CB, Lahesmaa R, Lahdesmaki H. Methods for time series analysis of RNA-seq data with application to human Th17 cell differentiation. Bioinformatics. 2014;30:i113–120. doi: 10.1093/bioinformatics/btu274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardehali MB, Lis JT. Tracking rates of transcription and splicing in vivo. Nature structural & molecular biology. 2009;16:1123–1124. doi: 10.1038/nsmb1109-1123. [DOI] [PubMed] [Google Scholar]
- Baker SC, Bauer SR, Beyer RP, Brenton JD, Bromley B, Burrill J, Causton H, Conley MP, Elespuru R, Fero M, et al. The External RNA Controls Consortium: a progress report. Nat Methods. 2005;2:731–734. doi: 10.1038/nmeth1005-731. [DOI] [PubMed] [Google Scholar]
- Bowes JB, Snyder KA, Segerdell E, Jarabek CJ, Azam K, Zorn AM, Vize PD. Xenbase: gene expression and improved integration. Nucleic Acids Res. 2010;38:D607–612. doi: 10.1093/nar/gkp953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke P, Anders S, Kim JK, Kolodziejczyk AA, Zhang X, Proserpio V, Baying B, Benes V, Teichmann SA, Marioni JC, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- Collart C, Owens ND, Bhaw-Rosun L, Cooper B, De Domenico E, Patrushev I, Sesay AK, Smith JN, Smith JC, Gilchrist MJ. High-resolution analysis of gene activity during the Xenopus mid-blastula transition. Development. 2014;141:1927–1939. doi: 10.1242/dev.102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder ME, Strzelecka M, Wilbur JD, Good MC, von Dassow G, Heald R. A comparative analysis of spindle morphometrics across metazoans. Curr Biol. 2015;25:1542–1550. doi: 10.1016/j.cub.2015.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran KL, Allen L, Porter BB, Dodge J, Lope C, Willadsen G, Fisher R, Johnson N, Campbell E, VonBergen B, et al. Circadian genes, xBmal1 and xNocturnin, modulate the timing and differentiation of somites in Xenopus laevis. PloS one. 2014;9:e108266. doi: 10.1371/journal.pone.0108266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran KL, LaRue S, Bronson B, Solis J, Trow A, Sarver N, Zhu H. Circadian genes are expressed during early development in Xenopus laevis. PloS one. 2008;3:e2749. doi: 10.1371/journal.pone.0002749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. Gene activity in early development. 3. Orlando: Academic Press; 1986. [Google Scholar]
- Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Yang Y, Li C, Fu S, Yang Z, Jin G, Wang K, Zhang J, Jin Y. Transcriptome analysis of early organogenesis in human embryos. Developmental cell. 2010;19:174–184. doi: 10.1016/j.devcel.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Heinonen M, Guipaud O, Milliat F, Buard V, Micheau B, Tarlet G, Benderitter M, Zehraoui F, d’Alche-Buc F. Detecting time periods of differential gene expression using Gaussian processes: an application to endothelial cells exposed to radiotherapy dose fraction. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu699. [DOI] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkela A, Girardot C, Gustafson EH, Liu YH, Furlong EE, Lawrence ND, Rattray M. Model-based method for transcription factor target identification with limited data. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7793–7798. doi: 10.1073/pnas.0914285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lonnerberg P, Linnarsson S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- Jolliffe IT. Principal component analysis. 2. New York: Springer; 2002. [Google Scholar]
- Junker JP, Noel ES, Guryev V, Peterson KA, Shah G, Huisken J, McMahon AP, Berezikov E, Bakkers J, van Oudenaarden A. Genome-wide RNA Tomography in the zebrafish embryo. Cell. 2014;159:662–675. doi: 10.1016/j.cell.2014.09.038. [DOI] [PubMed] [Google Scholar]
- Kalinka AT, Varga KM, Gerrard DT, Preibisch S, Corcoran DL, Jarrells J, Ohler U, Bergman CM, Tomancak P. Gene expression divergence recapitulates the developmental hourglass model. Nature. 2010;468:811–814. doi: 10.1038/nature09634. [DOI] [PubMed] [Google Scholar]
- Karlebach G, Shamir R. Modelling and analysis of gene regulatory networks. Nat Rev Mol Cell Biol. 2008;9:770–780. doi: 10.1038/nrm2503. [DOI] [PubMed] [Google Scholar]
- Karpinka JB, Fortriede JD, Burns KA, James-Zorn C, Ponferrada VG, Lee J, Karimi K, Zorn AM, Vize PD. Xenbase, the Xenopus model organism database; new virtualized system, data types and genomes. Nucleic Acids Res. 2015;43:D756–763. doi: 10.1093/nar/gku956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimelman D, Kirschner M, Scherson T. The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell. 1987;48:399–407. doi: 10.1016/0092-8674(87)90191-7. [DOI] [PubMed] [Google Scholar]
- Klein SL, Strausberg RL, Wagner L, Pontius J, Clifton SW, Richardson P. Genetic and genomic tools for Xenopus research: The NIH Xenopus initiative. Dev Dyn. 2002;225:384–391. doi: 10.1002/dvdy.10174. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Liu M, Hartley RS, Sheets MD, Dahlberg JE. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. Rna. 2009;15:2351–2363. doi: 10.1261/rna.1882009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, Bahler J. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–683. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OL, Jr, Hamkalo BA. Visualization of RNA synthesis on chromosomes. International review of cytology. 1972;33:1–25. doi: 10.1016/s0074-7696(08)61446-1. [DOI] [PubMed] [Google Scholar]
- Morin RD, Chang E, Petrescu A, Liao N, Griffith M, Chow W, Kirkpatrick R, Butterfield YS, Young AC, Stott J, et al. Sequencing and analysis of 10,967 full-length cDNA clones from Xenopus laevis and Xenopus tropicalis reveals post-tetraploidization transcriptome remodeling. Genome research. 2006;16:796–803. doi: 10.1101/gr.4871006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- Niehrs C, Pollet N. Synexpression groups in eukaryotes. Nature. 1999;402:483–487. doi: 10.1038/990025. [DOI] [PubMed] [Google Scholar]
- Paranjpe SS, Jacobi UG, van Heeringen SJ, Veenstra GJ. A genome-wide survey of maternal and embryonic transcripts during Xenopus tropicalis development. BMC genomics. 2013;14:762. doi: 10.1186/1471-2164-14-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourquie O. Vertebrate somitogenesis: a novel paradigm for animal segmentation? Int J Dev Biol. 2003;47:597–603. [PubMed] [Google Scholar]
- Qing T, Yu Y, Du T, Shi L. mRNA enrichment protocols determine the quantification characteristics of external RNA spike-in controls in RNA-Seq studies. Science China Life sciences. 2013;56:134–142. doi: 10.1007/s11427-013-4437-9. [DOI] [PubMed] [Google Scholar]
- Rasmussen CE, Williams CKI. Gaussian processes for machine learning. Cambridge, Mass: MIT Press; 2006. [Google Scholar]
- Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nature biotechnology. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagata N, Shiokawa K, Yamana K. A study on the steady-state population of poly(A)+RNA during early development of Xenopus laevis. Developmental biology. 1980;77:431–448. doi: 10.1016/0012-1606(80)90486-8. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nature biotechnology. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEQC MAQC-III Consortium. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nature biotechnology. 2014;32:903–914. doi: 10.1038/nbt.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirkanich J, Luxardi G, Yang J, Kodjabachian L, Klein PS. An essential role for transcription before the MBT in Xenopus laevis. Developmental biology. 2011;357:478–491. doi: 10.1016/j.ydbio.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer WC, Zeller G, Watson JD, Henz SR, Watkins KL, McWhirter RD, Petersen S, Sreedharan VT, Widmer C, Jo J, et al. A spatial and temporal map of C. elegans gene expression. Genome research. 2011;21:325–341. doi: 10.1101/gr.114595.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegle O, Denby KJ, Cooke EJ, Wild DL, Ghahramani Z, Borgwardt KM. A robust Bayesian two-sample test for detecting intervals of differential gene expression in microarray time series. Journal of computational biology: a journal of computational molecular cell biology. 2010;17:355–367. doi: 10.1089/cmb.2009.0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nature reviews Genetics. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- Tan MH, Au KF, Yablonovitch AL, Wills AE, Chuang J, Baker JC, Wong WH, Li JB. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome research. 2013;23:201–216. doi: 10.1101/gr.141424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebbenkamp AT, Willsey AJ, State MW, Sestan N. The developmental transcriptome of the human brain: implications for neurodevelopmental disorders. Current opinion in neurology. 2014;27:149–156. doi: 10.1097/WCO.0000000000000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome biology. 2002;3:RESEARCH0088. doi: 10.1186/gb-2002-3-12-research0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Q, Cameron RA, Davidson EH. Quantitative developmental transcriptomes of the sea urchin Strongylocentrotus purpuratus. Developmental biology. 2014;385:160–167. doi: 10.1016/j.ydbio.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heeringen SJ, Akkers RC, van Kruijsbergen I, Arif MA, Hanssen LL, Sharifi N, Veenstra GJ. Principles of nucleation of H3K27 methylation during embryonic development. Genome research. 2014;24:401–410. doi: 10.1101/gr.159608.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesterlund L, Jiao H, Unneberg P, Hovatta O, Kere J. The zebrafish transcriptome during early development. BMC developmental biology. 2011;11:30. doi: 10.1186/1471-213X-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- wa Maina C, Honkela A, Matarese F, Grote K, Stunnenberg HG, Reid G, Lawrence ND, Rattray M. Inference of RNA polymerase II transcription dynamics from chromatin immunoprecipitation time course data. PLoS computational biology. 2014;10:e1003598. doi: 10.1371/journal.pcbi.1003598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Developmental cell. 2004;6:133–144. doi: 10.1016/s1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Developmental cell. 2011;20:483–496. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Tan C, Darken RS, Wilson PA, Klein PS. Beta-catenin/Tcf-regulated transcription prior to the midblastula transition. Development. 2002;129:5743–5752. doi: 10.1242/dev.00150. [DOI] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.