Abstract
Besides their use as contrast agents for ultrasound imaging, microbubbles are increasingly studied for a wide range of therapeutic applications. In particular, their ability to enhance the uptake of drugs through the permeabilization of tissues and cell membranes shows great promise. In order to fully understand the numerous paths by which bubbles can interact with cells and the even larger number of possible biological responses from the cells, thorough and extensive work is necessary. In this review, we consider the range of experimental techniques implemented in in vitro studies with the aim of elucidating these microbubble-cell interactions. First of all, the variety of cell types and cell models available are discussed, emphasizing the need for more and more complex models replicating in vivo conditions together with experimental challenges associated with this increased complexity. Second, the different types of stabilized microbubbles and more recently developed droplets and particles are presented, followed by their acoustic or optical excitation methods. Finally, the techniques exploited to study the microbubble-cell interactions are reviewed. These techniques operate over a wide range of timescales, or even off-line, revealing particular aspects or subsequent effects of these interactions. Therefore, knowledge obtained from several techniques must be combined to elucidate the underlying processes.
I. INTRODUCTION
The efficacy of microbubbles as ultrasound contrast agents was discovered by serendipity in the late 1960s.1 They are now used in routine clinical practice for echocardiography and increasingly for other indications2 including the quantification of biological parameters such as blood perfusion rate and hypoxia,3 which are two important indicators of tumor malignancy.4 Advances in the understanding of cell and tissue-specific molecular markers have also led to the development of functionalized microbubbles, which are currently being investigated for targeted molecular imaging.5,6 Upon exposure to ultrasound, microbubbles undergo volumetric oscillations and reradiate a secondary acoustic response of significantly higher amplitude than the scattering that would be produced by a rigid sphere of equivalent size (∼2 μm in diameter). They thus generate much stronger echoes than red blood cells (RBCs), enabling their use as a blood pool agent. In addition, even at moderate ultrasound pressures, microbubble oscillations may be highly nonlinear, and the resulting harmonic content in the radiated signal can be used to achieve even higher contrast to tissue ratios.7–9
Beyond their diagnostic applications, microbubble oscillations have also been shown to enhance the transport and uptake of drugs at both the tissue and cellular levels.10,11 This has led to research across a very broad range of therapeutic applications in which focused ultrasound can be used to locally induce so-called “sonoporation,” leading to spatiotemporally controlled drug and gene delivery. To further improve treatment localization, methods for conjugating drugs and other therapeutic molecules to microbubbles have also been developed. This restricts the release of the drugs to the ultrasound focal zone, thereby increasing the chance that they are taken up in sonoporated areas12 and in these areas only. Microbubbles thus offer considerable promise as a means of improving the therapeutic efficiency, as well as decreasing toxicity in healthy tissue, which is of particular interest for chemotherapeutics. Another exciting application is microbubble-mediated drug delivery across the usually impermeable blood-brain barrier (BBB).13 In addition, microbubbles have also shown potential in immunotherapy: gene-loaded microbubbles have been used to transfect dendritic cells with tumor antigen encoding mRNA to stimulate the immune system and eventually induce an efficient and long-lasting cancer immune response.14 Finally, more complex and indirect methods are also being investigated for the enhanced intracellular delivery of plasmonic nanoparticles through microbubble actuation to increase photothermal therapy.15
Despite some very positive results from animal studies and even preliminary clinical evaluation, the mechanisms underlying microbubble-cell interactions, treatment protocols, and the potential for harmful bioeffects remain poorly defined. Detailed investigation of the fundamental biophysical processes is thus urgently required in order to understand and quantify potential adverse effects on cells and eventually develop efficient protocols to maximize the future impact of the technology in the clinic. Microbubble-cell interactions have been widely investigated in vitro, as evidenced by the extensive literature on this topic, and using the wide range of techniques discussed in this paper. A major challenge is the range of timescales and length scales over which the relevant phenomena occur. For example, microbubble dynamics occur on the tens of nanoseconds to microsecond timescale and on the sub-micrometer to micrometer length scale. Drug diffusion and uptake occurs on the second to minute timescale and over distances ranging from tens of micrometres (tissue level) to nanometres across the cell membrane. The mechanical response of the cell will occur on the same timescales and length scales, as the microbubble dynamics but the subsequent biological effects may extend over hours and throughout a significant volume of tissue. Both this multiscale problem and the extremely large parameter space (physical, chemical, and biological) preclude complete simultaneous control and visualization within a single experiment. Thus, it is necessary to combine the information gathered from multiple techniques during a single experiment.
The aim of this paper is to review the current experimental techniques that have been applied for the in vitro study of specific aspects of bubble-cell interactions. First, an overview of the available cell models is presented, from individual cells to 3D cellular models, and their advantages and disadvantages discussed. This is followed by a review of the different types of microbubbles and related agents that are currently under investigation for therapeutic applications. Finally, the instrumentation and measurement techniques available for studying microbubble-cell interactions are examined and discussed.
II. CHOICE OF THE CELL MODEL
A. Choosing the cell type
A first essential aspect when designing in vitro studies for bubble-cell interactions consists of selecting relevant cell types and cellular models, a choice which is directly motivated by the targeted research question or the ultimate goal of the study. For (bio)medical applications of microbubbles, both bacteria and mammalian cells can be of interest. The majority of in vitro studies is conducted on mammalian cells, since those are the most relevant for applications such as sonoporation for both gene transfection and drug delivery, as well as for imaging purposes. Bacteria are mostly employed in cleaning applications16 and biofilm removal,17,18 as discussed at the end of this section.
Fundamental studies can, for instance, be conducted on large and easy to manipulate cells such as Xenopus oocytes19 for primary proof-of-principle experiments. Alternatively, any cell type (mammalian cell, bacteria, or yeast cell) can be employed for the same purpose of gaining mechanistic information on microbubble-cell interactions at the single cell level. For this purpose, endothelial cells represent the preferred cell type.20,21 They are lining the blood vessels in which microbubbles are injected and therefore constitute the primary targets for both imaging and drug delivery applications. However, the most widely used cell type for microbubble-based drug delivery studies consist of cancer cell lines. Therapeutic applications of microbubbles mostly involve tumorous tissue and cancer cells. Cancer cells are easy to culture and to manipulate and therefore useful for fundamental studies. Finally, dendritic cells14 or lymphocytes22 are also interesting targets, e.g., for immunotherapy, since these can be reached directly from the blood stream or from the lymphatic nodes after subcutaneous injection and drainage of the microbubbles.
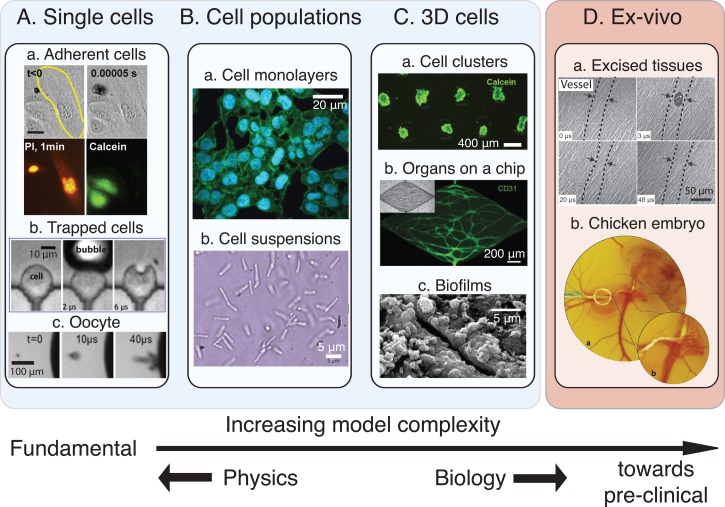
Various cellular models with increasing levels of complexity are available for in vitro experiments, ranging from individual cells and monolayers to complex 3D cellular architectures and organ-on-a-chip platforms. While simple models are particularly interesting to gain fundamental insights into the cell-microbubble interactions, more complex models are closer to the in vivo scenario, so that they provide more physiologically relevant information (Fig. 1).
FIG. 1.
Diverse cell models can be chosen depending on the desired level of complexity and type of information. (A) Models involving the study of single cells for obtaining mechanistic information. Single cells can be adherent (a, reprinted with permission from Fan et al., J. Controlled Release 170, 401 (2013). Copyright 2013 Elsevier), trapped in microfluidic structures (b, reprinted with permission from Li et al., Lab Chip 13, 1144 (2013). Copyright 2013 The Royal Society of Chemistry) and mammalian or not (Xenopus oocyte in c, reprinted with permission from Zhou et al., J. Controlled Release 157, 103 (2012). Copyright 2012 Elsevier). (B) More complete models make use of cell population that can be arranged as a monolayer (a, reprinted with permission from Sridhar et al., PLoS One 9, e93618 (2014). Copyright 2014 Author(s), licensed under a Creative Commons Attribution 4.0 License.79) or be in suspension (b, reprinted with permission from Tandiono et al., Lab Chip 12, 780 (2012). Copyright 2012 The Royal Society of Chemistry) in order to learn about the cell behavior in a collaborative context. (C) More complex bottom-up models are developed in which the cells are placed in a 3D environment, that is more similar to the in vivo situation. These models can be classified in 3 main categories: cell clusters (a), organs-on-a-chip with fully developed blood vessels (b, reprinted with permission from Moya et al., Tissue Eng., Part C 19, 730 (2013). Copyright 2013 Mary Ann Liebert, Inc. publishers), and biofilms (c, reprinted with permission from D. J. Stickler, Nat. Clin. Pract. Urol. 5, 598 (2008). Copyright 2008 Macmillan Publishers Ltd.80), which are particularly used to optimize cleaning processes. (D) Some ex vivo models were also used to study bubble-cell interactions such as excised tissues (a, reprinted with permission from Chen et al., Appl. Phys. Lett. 101, 163704 (2012). Copyright 2012 AIP Publishing LLC) or a chicken egg embryo (b, reprinted with permission from Faez et al., Ultrasound Med. Biol. 38, 1608 (2015). Copyright 2015 Elsevier) to which regulatory restrictions do not apply.
B. Single cell models
Studying interactions between a single microbubble and a single cell yields focused information on the impact of the microbubble on the cell, as well as on the fate of the latter. Large cells such as Xenopus oocytes have been employed as single cell models to study, for instance, cavitation-induced cell poration and to correlate cell-bubble distance with pore size and cytoplasm leakage.19,23 Recent developments in the field of microfluidics have greatly benefited single cell experimentation,24 since microfluidic devices enable the isolation and trapping of individual cells, e.g., with the help of dedicated micrometer-sized structures. This capability not only enables the fate of individual cells to be tracked over a long period of time but it also provides precise control over the cell-bubble distance,25,26 a key-parameter in such bubble interaction studies.27,28
Next to these mechanistic studies on cell membrane poration, other promising applications of single cell-cavitation bubble combinations include single cell lysis before single cell analysis,29 and assessment of the mechanical properties of cells30 and of the rheological properties of their membranes.31 Cavitation-induced cell lysis is particularly advantageous from a timescale point of view to get a precise snapshot of the content of a cell.32 Furthermore, the cell content remains at the same location,33 which prevents any loss of molecular information. When coupled to capillary electrophoresis for on-line separation and analysis of the cellular content, this cell lysis protocol has brought valuable information on targeted intracellular signaling pathways and the activity of specific kinases.34,35 Cavitation-induced cell lysis has also more recently been implemented in a microfluidic format33 and combined with capillary electrophoresis in a flow-through device for high-throughput analysis of individual cells.36 Cell mechanical properties have been probed using microbubbles and a so-called acoustic tweezer cytometer.30 Specifically, microbubbles attached to the membrane of an individual cell were manipulated by acoustic radiation force, and the resulting cell response subsequently recorded using an array of PDMS (Polydimethylsiloxane) posts on which the cell was immobilized. In an alternative approach, RBCs were exposed in a microfluidic channel to a cavitation bubble created by a laser pulse, and their deformability recorded using high-speed imaging.31 This approach, which was tested here on RBCs, with or without chemical treatment to alter their membrane properties, shows great promise for the characterization of the membrane properties of cancer cells which are known to be stiffer than their healthy counterparts.
C. Cell populations: Monolayer models and suspended cells
Interactions between bubbles and cells can also be studied at the level of a cell population, using either monolayers of adherent cells grown in conventional culture dishes or cell suspensions. Cell monolayers are routinely prepared on a multitude of commercially available dishware such as well-plates, Opticells™, or Petri dishes, or alternatively in microchannels for experiments under flow conditions, which explains why this configuration is mostly found in the literature for sonoporation studies.37–40 When exposed to a single jetting cavitation bubble for drug delivery, cell monolayers have provided valuable information on the influence of the distance between the bubble and the substrate on which cells are grown on the amount of cell detachment, the efficiency of molecular delivery in cells, and the cell viability.20,38 Using the same monolayer model, enhanced gene transfection has been reported more recently for human B-cell lines using plasmid-coated microbubbles, which were activated by ultrasound.41 Monolayers formed from endothelial cells can act as artificial blood vessels, and implementing them in a microchannel format allows studying flow-based delivery of microbubbles and recapitulating physiologically relevant cellular shear stress, as would occur in vivo.42 Alternatively, cell population studies rely on the use of suspended cells. A major limitation encountered in this case is that suspension cells freely move with the bubble-induced flow in bulk, which prevents controlled cell exposure to the bubble and from cell tracking over time. This issue has been addressed by restricting the cell suspension in a shallow microfluidic chamber.43 Interestingly, in this confined setting, the bubble-induced flow is mostly planar and was found to have greater effect on cells, this effect being cell-bubble distance dependent. More recently, Tandiono et al. applied the same approach to lyse populations of E. coli (Escherichia Coli) and P. pastoris (Pichia Pastoris) bacteria, confirming the potential of oscillating cavitation bubbles for efficient, fast, and contamination-free lysis of cells, without damaging biomolecules.44
Single cell and cell population models suffer, however, from serious limitations, not only with respect to the bubble dynamics but also from a biological point of view. First, the presence of a substrate even if it is acoustically transparent is known to strongly influence the dynamics of the microbubbles.27,45
So, while these models allow studying bubble-cell interactions in a highly controlled way and have succeeded in bringing insight into bubble-cell interaction mechanisms, the microbubble response to the ultrasound field in these in vitro conditions is not necessarily indicative of that which would occur in vivo. Next to this, single cells and cell monolayers are over-simplified cellular models and are not therefore physiologically relevant. In vivo, cells experience a 3D environment with 3D interactions with neighboring cells, and they are embedded in an extracellular matrix (ECM). As a result, cells cultured under isolated or 2D conditions exhibit differences in their gene and protein expression profiles, as well as in their phenotype and shape.46 Specific differences were found in the expression of cytoskeleton proteins, cell adhesion proteins, and extracellular matrix, in processes such as proliferation and apoptosis, and most importantly, in metabolic processes. This change in phenotype between 2D and 3D was found to correlate with differences in the cell response to therapeutic treatments, which highlights the need to go 3D for drug delivery and efficiency assays.
D. More complex cellular models: Toward mimicking the in vivo conditions
The scientific community is becoming more and more aware of the limitations presented by monolayer models and is recognizing the physiological relevance of complex models that reproduce more faithfully the 3D architecture found in vivo. Three-dimensional cellular systems are, in particular, good models for avascular tumors which comprise a hypoxic and/or necrotic core, and to study drug penetration and delivery in tumors.47 Such 3D cellular systems are therefore of utmost importance for the study of microbubble-based drug delivery, where therapeutic agents must penetrate into tumor tissues, that consist of a combination of tumor cells, stromal cells, and ECM.48 Furthermore, such sophisticated and 3D in vitro models appear as attractive alternatives to animal experimentation, which is extremely expensive and time-consuming, while requiring dedicated facilities as well as specific authorization. These complex and 3D cellular models can be classified into three categories, which are discussed in the following paragraphs: artificial cellular aggregates combined with, or without, ECM, excised tissues, and organ-on-a-chip platforms.
A first and fairly simple 3D model consists of a cellular aggregate, which can be prepared from tumor cells only, or from a combination of different cell types (e.g., tumor cells and stromal cells). These cellular aggregates or so-called spheroids can be produced using a variety of techniques such as the hanging drop approach, centrifugation-based techniques,49 or alternatively using microfabricated50 and microfluidic structures51 or even ultrasonic standing waves,52 to control the microtissue size and their size distribution. To produce tissue models, cells can also be mixed with hydrogels, which serve as a surrogate for the ECM and as a “carrier” for growth factors, while presenting mechanical properties similar to those of native tissue.
Alternatively, excised tissues can be employed as 3D cellular models, and this approach has actually already been used in combination with bubbles. For instance, Hossein et al.53 and Chen et al.54 have tested the effect of oscillating cavitation bubbles in capillaries on excised rat tissues. While this model is fairly easy to use and it exhibits the exact architecture of native tissue, reproducibility becomes an issue, and in addition, such experiments require sacrificing animals. Chicken egg embryos which include a vasculature with pulsating flows and for which no authorization is required solve some of these issues.55 However, the latter model is complex to use from an experimental point of view with the injection presenting a particular difficulty.
The last class of in vitro complex models includes organ-on-a-chip platforms.56 These can be defined as hybrid models combining cells and microfabricated structures, aiming at reproducing the physical architecture and cell interactions found in a specific organ. These models are implemented in a microfluidic format, which provides excellent control over the cell microenvironment, while being ideally suited for multiplexed and automated experiments. More importantly, and compared to cellular models alone, they allow the implementation of physiological flow conditions, and they have become an attractive format to create in vitro models of barriers57 (e.g., blood-brain barrier, kidney, intestinal barrier, blood-alveolar barriers, etc.). Organ-on-a-chip platforms are attracting more and more attention for drug screening and toxicity assays and as alternatives to animal experimentation. In a recent review, van Duinen et al.58 mentioned that 87 publications have been reporting organ-on-a-chip platforms since 2012. Of particular interest for microbubble-related studies are vessel-on-a-chip,59 tumor-on-a-chip,60 and barrier models, such as the BBB on a chip.61,62
Microbubbles in vivo are delivered through the blood circulation, and the first barrier they encounter and must cross and/or damage for targeted delivery of imaging or therapeutic agents is the endothelial barrier. A great variety of models for blood vessels have been reported in the literature, since the latter represent an essential element of in vitro engineered tissues for proper and selective delivery of nutrients and oxygen. One particular approach to create in vitro models of blood vessels is worth mentioning for microbubble-related medical applications, since it consists of embedding a functional vascular network in a hydrogel ECM-like matrix, which can act as the surrounding tissue.63 This approach would allow not only the study of the microbubble delivery under physiological flow conditions followed by the rupture of the barrier upon bubble excitation, but also the visualization of the actual release of drugs and the assessment of its penetration into the tissue. Moya et al.64 reported the spontaneous formation of such a complex vasculature in a fibrin matrix. Alternatively, blood vessels were created in channels, which were pre-patterned in a collagen substrate and in which endothelial cells were seeded to yield a functional vasculature.65 In this approach, the integrity of the vascular network is typically evaluated using a leakage assay and fluorescent dextran particles. Interestingly, the same approach would allow the assessment of the opening of the endothelial barrier through the release of fluorescent particles in the hydrogel substrate. Furthermore, the continuous ECM-like phase around the vascular network can easily be supplemented with tumor cells66 or tumor spheroids67 for combined drug efficiency assays.
A highly promising application of microbubbles concerns drug delivery to the brain, which is protected by the so-called BBB, that consists of a densely packed structure formed from endothelial cells on one side, and pericytes and astrocytes on the other side. The BBB prevents the passage of substances larger than small nutrients from the blood. This selective permeability becomes an issue when therapeutic agents must be delivered to the brain to treat diseases. Recent work68,69 has shown that microbubbles upon ultrasound triggering actually allow local and temporal opening of this barrier. The mechanisms of BBB permeation are however not entirely understood, calling about for relevant in vitro models to study cell-microbubble interactions and to monitor variations in the barrier permeability. Recently reported BBB models are of prime interest, since they can reproduce the cellular structure of this biological barrier, including flow on the blood side.61,62
Interestingly, while these more sophisticated three-dimensional models offer exciting possibilities for studying in vitro microbubble-based imaging and drug delivery under in vivo-like conditions, their utilization in combination with microbubbles and cavitation bubbles has been limited so far to excised tissues, for which only a few references are found. This trend could first be accounted for by the fact that these 3D models have recently gained in popularity in the fields of biology and drug screening, where cellular monolayers are still predominant. Furthermore, the field of organ-on-a-chip is still at its infancy, since this concept was introduced in the last decade and does not yet compete with in vivo techniques such as intravital microscopy in terms of biological relevance.70,71 However, since these complex and physiologically relevant models are becoming mature, as evidenced by the existence of commercially available platforms,72–74 they are likely to be adopted for microbubble-based medical applications in the very near future.
E. Biofilms
A different class of applications of (micro)bubbles, related to the medical field, focuses on cleaning and removal of bacterial contamination and biofilms. Biofilms,16,75 which consist of a complex 3D structure allying bacteria, substances secreted by the latter and exogenous proteins coming from the host environment, are found, for instance, on implants and prostheses, in the mouth in the form of dental plaque76 and in the root canals,77 or on medical devices. Simply through their impact on surfaces, bubbles which flow in the vicinity of a biofilm have proven to efficiently remove bacterial contamination. Parini and Pitt76 showed that cleaning of 40–μm thick Streptococcus mutans biofilms, used as models for dental plaque, depended on the bubble streaming velocity, the bubble size, and the gas fraction.76 In more recent work, enhanced cleaning and deeper cleaning penetration were achieved in a root canal model in which a Faecalis biofilm was grown for 7 days, when exposing a microbubble suspension to ultrasonic agitation, most probably through stronger microbubble oscillation (Fig. 1).78
III. MICROBUBBLE TYPES
In the present section, we review the different types of microbubbles that have been used in combination with the aforementioned cell models in order to study bubble-cell interactions. In brief, microbubbles can be used to exert a stress on the cells, either in a direct way by palpating the membrane during its oscillations, by stretching the membrane locally during inertial growth and collapse, or by normal stresses through acoustic radiation forces. Microbubbles can also have indirect action on the cells, by emitting shock waves or generating strong streaming patterns, for example.
A. Microbubbles
1. Stabilized bubbles
One of the first commercial contrast agents, Albunex® (Mallinckrodt Inc., Hazelwood, MO, USA) consisted of air microbubbles coated with a thin stabilizing layer of cross-linked human serum albumin. Its development stemmed from the observation that the relatively short-lived contrast enhancement that could be achieved with agitated saline could be extended through the addition of a small amount of a patient's blood.82,83 While Albunex microbubbles were stable in comparison to uncoated bubbles, they were not yet able to provide a sufficiently long contrast enhancement in vivo, and new agents containing higher-molecular-weight gases were consequently developed. Optison™ (GE Healthcare Inc., Princeton, NJ, USA), for example, contains perfluoropropane with a high molecular weight (MW = 188 g/mol) in order to decrease the diffusivity of the gas across the bubble wall and the solubility in the surrounding liquid. All of the clinical agents currently in use contain gases of this type, although Kwan and Borden84 have shown that even these heavy gases are actually replaced by dissolved blood gasses (mostly N2 and O2) within a few minutes. The improvement in stability is therefore relatively modest. These microbubbles, being a few micrometers in diameter, show limited circulation time in the blood stream. As a consequence, such bubbles interact in the first place with blood cells, which currently attract interest for the targeting of immune cells.85,86 However, most studies using stabilized microbubbles are performed with tumor cells. It must be borne in mind that these microbubbles cannot directly reach these cells, which are located beyond the vascular barrier. Studying the interactions with endothelial cells, however, becomes more and more relevant as the size of the vessels reduces upon nearing the capillary bed, favoring the interaction of the bubbles with the vessel walls. Identifying the type of bubble-cell interactions is crucial for enhancing the permeability of the vessel walls in order to locally improve drug uptake by the interstitial tissue as well as for quantifying the deleterious effects of oscillating microbubbles and the risk of permanent damage. This can be studied best with cell membrane models and 3D complex models that comprise an endothelial cell layer. In more specific cases, such as immunotherapy, immune cells in the blood pool or lymphatic circulation can be a direct target for the microbubbles, which can be studied in an in vitro set-up with direct contact between microbubbles and target cells for fundamental aspects, i.e., single fixed cells, or in a suspension of cells, representative of a blood pool.
To improve convenience of administration, many of the second generation contrast agents were packaged as freeze-dried powders that could be stored and resuspended in saline as required, rather than being prepared immediately prior to injection. Alternative means of administering coated microbubbles have also been developed; e.g., Echovist® and its successor, Levovist® (Schering AG, Berlin, Germany), consisted of suspensions of galactose micro-crystals, which dissolved in the blood following injection, releasing air microbubbles from defects on the crystal surfaces. Levovist also contained palmitic acid to provide additional stability. Levovist microbubbles were found to be more echogenic than Albunex, but considerably less stable on account of the higher diffusivity of their surfactant coatings. Hence, Levovist was soon superseded by agents containing bubbles stabilized by phospholipid monolayers that provided a better compromise between longevity and echogenicity. These included SonoVue® (Bracco, Geneva), Definity® (Bristol-Myers Squibb Medical Imaging Inc., USA), and Sonazoid™ (GE Healthcare Inc., Princeton, NJ, USA). Even greater microbubble stability can be afforded by the use of polymeric coatings (e.g., polylactic(co)glycolic acids, polycaprolactone, or cyanoacrylates).87–91 In selecting the optimum coating material, however, there is a tradeoff between stability and echogenicity and particularly non-linear behavior that is desirable for contrast enhancement.92,93
The majority of microbubble agents used for both for clinical applications and for research purposes are produced by conventional emulsification methods, and most often using a combination of sonication and physical agitation. These methods are efficient and convenient but yield highly polydisperse microbubble size distributions.94 Alternative methods including microfluidics95 and electrospraying96 have been investigated, but systems offering competitive production rates and/or microbubble stability are still under development. Feshitan et al.97 have demonstrated that improvements in the contrast to tissue ratio can be achieved through filtering microbubble suspensions to match their size distribution for a given imaging protocol. Segers and Versluis98 have also recently demonstrated that microbubbles can be sorted according to their acoustic properties similarly offering a significant improvement in imaging performance. Although this smaller size distribution should not influence the circulation time of the microbubbles, it is expected to allow better control over microbubble cavitation behavior, and thus on the mechanical and therapeutic impact on the surrounding cells. The absence of larger microbubbles in the batch might, for instance, reduce the amount of undesired effects caused by cavitation.
2. Functionalized and drug-loaded microbubbles
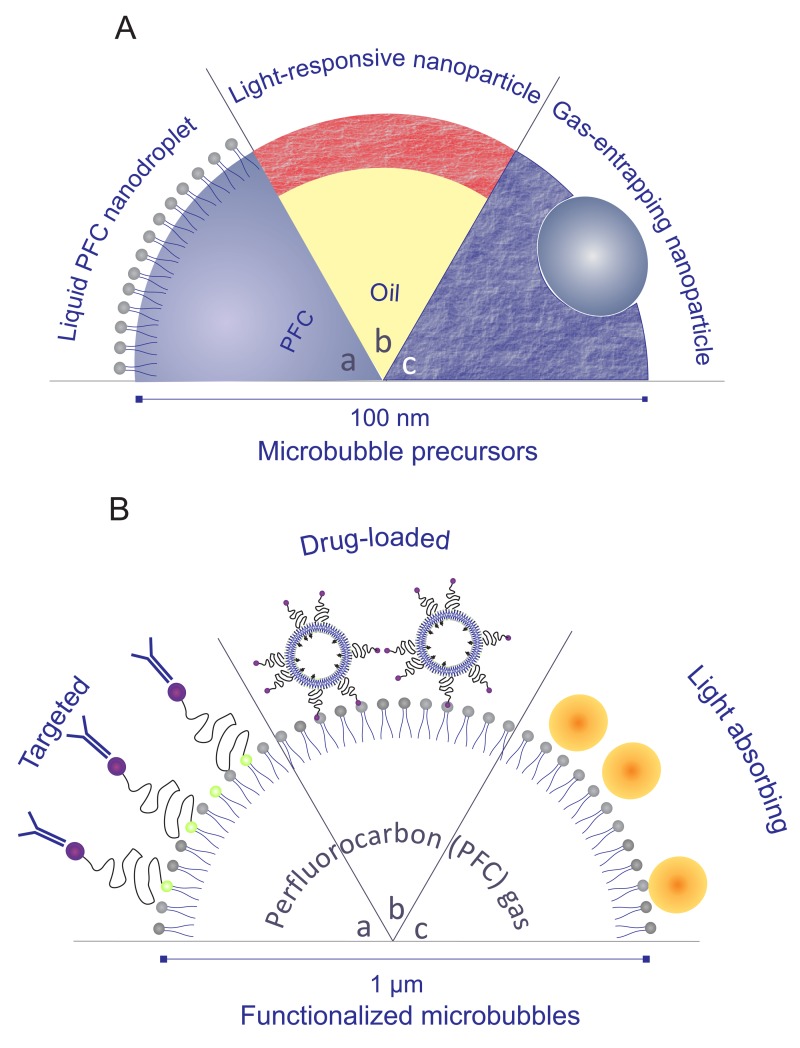
Tissue specificity, e.g., for targeted imaging and therapy, requires functionalization of the microbubble surface (Fig. 2.B). Microbubbles coated with charged material have been shown to locate preferentially in inflamed tissue,99 but a more effective method consists in attaching ligands to the bubble surface that will bind to receptors on particular types of cell.100 Examples include targeting to activated leucocytes by incorporating phosphatidylserine in the microbubble coating;101 angiogenic markers;102 and attaching antibodies to microbubbles targeted to receptors expressed during inflammation (e.g., anti-P-selectin monoclonal antibody, anti-NICAM antibody, and anti-VCAM antibody).103,104 The target here can probably be tumor endothelial markers or neovascular markers on the tumor vessel endothelium. Such systems are most often represented in vitro by cell monolayers, although future research would greatly benefit from the use of organ on chip platforms. These bubbles also present great interest for use in cells suspensions, e.g., for targeting circulating tumor cells (CTCs) with high precision105,106 or to quantify the binding efficiency with white blood cells in a vessel. An alternative method for localizing microbubbles in vivo is to load them with magnetic nanoparticles. This enables the microbubbles to be guided into the target region using an externally applied magnetic field107 either as an alternative to biochemical targeting or as a means of slowing the microbubbles down sufficiently to facilitate binding.
FIG. 2.
Schematic representation of microbubbles precursors (A) and functionalized microbubbles (B). (A) Superheated, liquid perfluorocarbon nanodroplets are stabilized by a surfactant layer and can be vaporized with ultrasound or laser pulses (a); fluorescently labeled nanoparticles can generate microbubbles when exposed to laser light (b); and polymeric nanoparticles encapsulating a small gas core can be used as sustainable microbubble precursors (c). (B) Microbubbles can be functionalized by attaching targeting ligands (a); loading them with drug-containing nanoparticles (b) or make them suitable for multimodal imaging by covering them with plasmonic nanoparticles (c).
Surface functionalization can also be exploited for attaching therapeutic components to microbubbles.108,109 Recently developed loading strategies include self-assembly methods for the production of liposome-loaded microbubbles.110 These offer considerable advantages in terms of simplicity and the quantity of drug that can be attached to a single bubble, although it has been shown that the additional loading does modify the microbubble dynamics.111 Nevertheless, such bubbles have been shown to be highly effective in delivering cytotoxic drugs, although the underlying mechanisms still require significant investigation.112 The same method combined with specific binding strategies has also been used to load fluorescent dyes into liposomes attached to phospholipid-coated microbubbles in order to study their dynamic behavior.111 A dye can also be inserted directly within the coating.113 These formulations are most useful to investigate release and uptake mechanisms upon ultrasound exposure.
If contrast enhancement is not the primary consideration, then polymer-coated microbubbles can offer several advantages for drug encapsulation thanks, in particular, to the relative ease with which their size, shell thickness, and drug loading can be controlled.91,114–116 Polymer coatings can also be readily functionalized for molecular targeting.117 The acoustic response of polymeric microbubbles is typically very different from that of a phospholipid-coated microbubble, with negligible volume oscillations followed by rupture of the coating above a threshold acoustic pressure.115,118 The dynamics of the released gas may be comparatively violent, and hence the interactions between polymeric bubbles and cells also require detailed investigation.
3. Echogenic liposomes
Echogenic liposomes are an alternative type of acoustically active drug carrying particle. They are in fact very similar to microbubbles in terms of their composition119 but consist of a multilamellar phospholipid structure entrapping pockets of gas, which also offers the potential for encapsulating both hydrophobic and hydrophilic material.120 They have been shown to offer improved stability compared to microbubbles.121 Larger doses of echogenic liposomes (i.e., particles per unit volume) are required to obtain equivalent levels of contrast enhancement during imaging, on account of their lower gas content per particle, but such high concentrations are well tolerated physiologically.122 Similarly, specific pulse regimes are required to initiate drug release.123
4. Light absorbing microbubbles
A further recent development concerns laser-activated microbubbles. The bubble coating can be loaded with light-absorbing particles (Fig. 2.B.c)124 or a low-viscosity oil containing an absorbing dye.125 Upon exposure to laser light, the heating and subsequent expansion of the coating sets the microbubble into volume oscillation, thereby generating similar acoustic and fluid dynamical effects as ultrasound exposure. These bubbles have the potential to be used for other imaging modalities such as photoacoustic imaging.126 A usual difficulty encountered with laser-based medical imaging techniques such as photoacoustics lies in the limited penetration depth of the laser light combined with the high degree of tissue scattering. Such complications are, however, highly reduced in in vitro models. A typical cell layer thickness of a millimeter, for example, transmits almost 80% of the near-infrared light intensity. The increased specificity of laser light, however, justifies its use in a number of superficial pathologies located in just below the skin, e.g., skin cancers, esophageal cancer, or rheumatoid arthritis in finger joints.
B. Droplets as precursors
Another approach to producing microbubbles utilizes a stabilized emulsion of volatile liquid droplets (Fig. 2.A.a), which vaporize to form microbubbles upon injection into the body or subsequently to exposure to ultrasound. The fluids commonly chosen are octafluoropropane, decafluorobutane, or dodecafluoropentane127 that have boiling points of −37 °C, −4 °C, and 29 °C, respectively. These boiling points are lower than normal body temperature, so that these fluids become superheated in vivo. These perfluorinated liquids present a number of advantages: first, they have a tunable boiling point depending on the carbon chain length that falls precisely in the range that is relevant for medical applications. As for stable microbubbles, the droplets are surfactant-coated to ensure higher stability and better shielding against the immune system and to spontaneous vaporization. Second, these perfluorocarbons are already in clinical use for a number of other applications (including ultrasound contrast agents). Third, this type of agent exhibits improved stability both for storage and administration and, unlike microbubbles, the nanoscale droplets have the potential to extravasate, e.g., through leaky tumor vasculature, before they are vaporized. This facilitates imaging and/or treatment, e.g., within a tumor mass.127,128
The interest of using such precursors lies in the injection in the bloodstream of a nanosized agent that is able to extravasate through large pores as typically found in tumor blood vessels (EPR (enhanced permeability and retention) effect129). Upon vaporization, the resulting micron-sized bubbles can interact with the deeper-lying tissue. In this regard, the collection of mechanistic information on the interaction of stable bubbles with single cells or cell monolayers is most valuable. The dynamics of the penetration process of the precursor, however, can best be studied in vitro using 3D blood vessels or organ-on-a-chip models.
Whilst recent studies have advanced our understanding of the physics of superheated droplets under ultrasound exposure130 extremely little is known about their interactions with cells before, during, and after vaporization. Interestingly, the bubbles generated from superheated droplets exhibit characteristics similar to those of phospholipid-coated bubbles. This implies that the surfactant initially stabilizing the droplet can also stabilize the bubble and support non-linear oscillations.131 Perfluorocarbon droplets also offer the potential to be used as multi-modality agents, since they can be activated using ultrasound127,130,131 but also using light when internally loaded with plasmonic nanoparticles,132 which leads to an increase in photoacoustic contrast and the creation of a ultrasound microbubble contrast agent simultaneously.
C. Solid particles as microbubble nuclei
Notwithstanding the improvements in microbubble stability that can be achieved through selection of appropriate materials and/or surface functionalization, they still offer very limited circulation times. As discussed before, superheated nanodroplets130,133 offer a solution to this problem. Another approach consists in the use of solid functionalized particles on the very same cell models. Here, their surface properties and morphology enable them to act as nuclei for bubble formation.
1. Acoustically responsive particles
As mentioned above, polymeric coatings can significantly enhance microbubble stability, albeit at the cost of echogenicity. Nanoscale polymeric particles can also be produced using standard emulsion and solvent evaporation methods,134 and they either encapsulate a gas core135 or consist of a gas entrapping porous matrix (Fig. 2.A.c).136 Such particles offer excellent stability even under high-intensity ultrasound exposure and have been shown to sustain cavitation over significantly longer periods than microbubbles. Other systems have also been reported in the literature such as the hydrolyzing, gas-generating nanoparticles by Kang et al.,137 able to create bubbles over timescales of several minutes, or carbon nanotubes as ultrasound contrast agents.138 Describing all of these systems, however, would be beyond the scope of this review.
2. Light responsive particles
Just as a drug can be encapsulated within a polymeric particle, so too can a dye that makes the particle light-absorbing (Fig. 2.A.b). Upon irradiation with light of sufficient intensity, such particles can generate bubbles that radiate strong acoustic emissions enabling them to be detected.139 The majority of the research on light-activated bubbles however has made use of plasmonic nanoparticles such as gold nanospheres (AuNS) or nanorods (AuNR). More complex nanoparticles such as gold-coated beads140 have also been investigated for the generation of plasmonic bubbles. Plasmonic nanoparticles have a unique light absorption cross section with respect to their size at the specific wavelength of the plasmon, which itself corresponds to the motion of the electron cloud of the particle. AuNS and AuNR have been used for some years in classical thermoelastic photoacoustics but their capacity for transient microbubble generation has only recently been investigated.132 Although these particles have been used in pre-clinical imaging and/or for inducing hyperthermia, the question of their short and long term toxicity remains unclear. So far, preliminary investigations of the interaction between cells and optically generated bubbles have been reported by Lukianova-Hleb et al.141 but further, more detailed studies are required.
D. Agent behavior and tissue type
Literature related to microbubble-cell interactions focuses on how bubbles act on cells and tissues, and this idea reflects on the organization of Secs. I and II of this review. However, very little is known on how the various tissue types influence the agents as most experiments are simply performed in aqueous fluids. As mentioned before, both the presence of a boundary and the viscosity of the fluid142 are known to have a major effect on the microbubble behavior; therefore, the viscoelastic properties of human tissues can be expected to change the response of the agents to ultrasound in terms of activation threshold, as well as its resonance frequency and amplitude response. This aspect, often forgotten, is nonetheless a crucial part of the intricate problem of bubble-cell interactions. Some very recent numerical work, e.g., that by Gaudron et al.143 and Johnsen and Mancia144 has pioneered microbubble dynamics in a viscoelastic medium. Clearly, more work needs to be done on the topic, in particular, from an experimental point of view, as any smaller number of dimensions than a 3D cell model would fail to reproduce the relevance of the environment. Within the range of available 3D models, the simplest ones, e.g., spheroids, can provide a more fundamental understanding of the effect of the size and type of constructs formed by the cell population. However, different tissues are also known to have different viscoelastic properties,145 one that is exploited in shear rate imaging of tumors, for example. Therefore, studying experimentally the complex two-way coupled problem of agent penetration/activation/forcing on cells requires a well-defined 3D structure that includes the integrative behavior of various cell types and viscous flow, which can be provided by microtissues and organ-on-a-chip models. More advanced 3D models can also add more intricate modifications such as the biochemical activity of the tumor, thereby creating a different tumor micro-environment that induces a higher chemoresistance and, more in general, different cell response.146 Such effects are of prime interest when using bubbles as the local carriers (Fig. 2).
IV. EXPERIMENTAL METHODS
A. Excitation of the microbubbles
1. Optical excitation
Section III has shown that microbubble dynamics may be both excited and interrogated optically and/or acoustically. In terms of optical excitation, current technology already offers pulsed and continuous wave (CW) lasers that provide full control over wavelength, power, and energy distribution in space and time. Therefore and despite the experimental hazards associated with them, lasers are the most suitable light sources for studying the interaction of optically activated or driven microbubbles and cells. Their safe use is already warranted below a medical guidelines threshold that depends on both the duration of the irradiation and the wavelength of the laser light (20 mJ/cm2 at 532 nm and 60 mJ/cm2 at 1064 nm for nanosecond pulsed lasers). These exposure thresholds are directly related to the energy deposited in tissue through light absorption. As mentioned previously, sufficient optical excitation in vivo under this threshold can be difficult for larger penetration depths. Although this limitation is hardly an issue for in vitro models, since even the 3D models have a typical thickness of only a millimeter, it emphasizes the need for highly efficient agents to increase both contrast and imaging depth. Laser light with a 1064 nm wavelength is located right in the biological window,147 i.e., with highest tissue transparency, and thus is less damaging to tissue. In practice, however, a laser operating in the visible range at a wavelength of 532 nm is easier and safer to use. The excitation wavelength of many known absorbers, such as the plasmon resonance of gold nanospheres, is also located near 532 nm. The best choice for the wavelength will therefore also depend on the stage of development of the considered agent. Practically, if the bubbles are equipped with the right fluorophores, light can be used to directly impact on the bubbles. The expansion of the gas core then occurs through thermal effects.124,148 Thus, heat rather than light is generating or driving the bubbles, which in turn generates or scatters the acoustic waves for imaging purposes.
A pre-existing gas microbubble will respond to optical excitation due to thermal expansion.124,149 This phenomenon has been investigated using both pulsed124 and CW lasers.149 The former have been more commonly applied on account of their availability for photoacoustic imaging. A much larger number of studies have used laser exposure and the subsequent heat generation to create vapor and/or gas bubbles. A focused laser beam of sufficient intensity will produce a plasma in water.23,43 Taking advantage of the capability to precisely control the distance between the laser focus and a single cell in a chip, Li et al.26 used an array of cell trapping structures in a PDMS microchamber and found that a shorter cell-bubble distance led to the creation of larger pores. More interestingly, the presence of a trapping structure was found to guide the micro-jet originating from the cavitation bubble collapse towards the cell, thereby enhancing the poration process. Light absorbing precursors, such as described in Sec. III, may also be used to provide better spatiotemporal control over light-induced cavitation processes.132,149 Alternatively, bubbles may be produced through thermally induced vaporization of an injected, initially liquid agent such as perfluorocarbon droplets,150 typically at lower optical intensities.149 In most cases, the laser is directly coupled through an observation microscope using a dichroic filter,124,139,151 allowing for an exact spatial localization of the beam and for a selective excitation of the agents in the sample. The use of CW lasers is still at an early stage and the intensity values reported in literature, as of now, are too high for in-vivo use. Simple cell models such as monolayers are therefore still suitable to evaluate thermal and mechanical damage from pulsed and CW laser-activated agents. Furthermore, laser light offers reduced interference with a potential supporting membrane over ultrasound, thereby overcoming the limitations of the use of monolayer models for laser-activated applications.
2. Acoustic excitation
a. Acoustic field
Whilst the experimental hazards are usually less significant, accurate spatiotemporal control of an acoustic field may be considerably more challenging than its optical counterpart. Particularly, problematic is the elimination of unwanted reflections from boundaries within the system to avoid the disruption of the sound field incident upon the microbubbles. In the most extreme cases where long pulses are used in combination with an objective lens, for example, it becomes a challenge to suppress the creation of standing waves that were shown to significantly influence the outcome of a sonoporation experiment.152 The calibration of an acoustic field with existing technologies, using, for example, hydrophones, can be done with a typical 10% error at best. With the inclusion of reflections within the setup, the uncertainty of the acoustic pressure can rapidly rise above 50%. In some cases, the transducer is positioned perpendicularly to the cell support, and the occurrence of multiple reflections and ultimately standing waves is unavoidable.153 Hensel et al.154 have also demonstrated that the position of the transducer with respect to the sample holders could greatly influence the acoustic field. They concluded on the necessity to use acoustically transparent materials, e.g., a polymer presenting an acoustic impedance close to that of water. Such materials, however, are not easy to find, in particular, when considering the impact of the angle of incidence. For example, the transmission coefficient of an Opticell membrane drops from over 99% at normal incidence to about 85% at 45° incidence and 1 MHz113 and is strongly frequency dependent. Along the same line, Cochran and Wheatley39 observed that much lower transfection efficiencies were achieved when microbubbles are pushed away from cells by the acoustic wave rather than pushed towards them. This simple observation holds for any choice of cell model and is inherent to in vivo setups. Taking these effects into consideration, optimal control over the acoustic field will be achieved when using a focused ultrasound transducer inserted under an angle that allows for the reflected waves to escape the region of interest. The unknown acoustic field remains in any case an experimental difficulty to consider.
b. Acoustic parameters
Numerous studies have reported that the choice of the acoustic parameters is crucial in achieving cell poration.81,155 When performing a bubble-cell interaction experiment, one can vary the acoustic pressure, the frequency, the number of cycles per bursts, the pulse repetition rate, the total duration of the experiment, and each of these parameters will affect the outcome of the experiment. Some studies have chosen to limit the investigation to a single parameter, usually the acoustic pressure, using short bursts156,157 in order to learn about the precise dynamic phenomena at hand. Others have chosen to maximize the uptake using repeated bursts of a few thousand cycles155 for a total duration of up to several minutes to define the setting that may give the best results in vivo. In addition, the response of a range of microbubbles and microbubble precursors presented in Sec. III, at the very same acoustic setting will be entirely different.90 In order to provide an overview of the state of the art, we classify the acoustic parameters in terms of their effect upon the bubble dynamics, leading to three main categories.
First, “stable” or non-inertial cavitation is observed when microbubbles are driven at relatively low acoustic pressures. They then undergo repetitive volumetric oscillations that are only weakly non-linear. The acoustic pressures used in that regime typically range from 10 to 150 kPa. In this regime, the acoustic emissions from the microbubble will contain some harmonics and potentially also subharmonic and ultraharmonic components. This regime has been shown to induce sonoporation156 usually with relatively low transfection efficiency, but with a high cell viability.81,155
At higher acoustic pressures, inertial cavitation is observed, which is characterized by a much more chaotic microbubble behavior. The term “inertial” is derived from the analysis by Flynn158 and relates to the fact that the compression phase of the microbubbles is driven by the inertia of the surrounding liquid rather than by variations in pressure. In this regime, the bubble usually becomes non-spherical upon collapse, which may lead to micro-jetting.26 In this case, the acoustic pressures typically range from 150 to 500 kPa, leading to stable microbubble cavitation. The corresponding acoustic emissions are broadband in nature.159 Vaporization of liquid precursors of bubble also falls in this category.131 This regime is the most reported one in the study of bubble-cell interactions, for example: Smith et al. reported sonoporation resulting from cavitation of echogenic liposomes,123 Zhou et al. used inertial cavitation to porate oocyte cells,23 and Zhao et al. used cavitation to induce cell apoptosis.11 Using larger bubbles, Ohl et al. observed two regimes of viable porated cell and detached dead cells.20 More specific agents, such as superheated nanodroplets or similar bubble precursor nanoparticles, often require much higher activation pressures, typically over 1 MPa. As such, their subsequent bubble behavior is inherently inertial.
In addition to the dynamics of the microbubbles themselves, there are a number of second order phenomena, induced by the bubbles over a timescale longer than their own oscillations that are likely to play a significant role in microbubble-cell interactions. Most prominent amongst these effects are microstreaming and acoustic radiation forces. Streaming induced by microbubbles under ultrasound irradiation is of interest for the local stress that is generated by local fluid velocities of the order of 0.1 m/s.113 Pommella et al.,160 for example, recently achieved vesicle lysis using the microstreaming generated from a single microbubble (approximately 50 μm diameter) at low acoustic pressures. In this case, the acoustically induced microstreaming could generate stresses as high as 300 Pa. In the absence of cells, Vos et al.161 even reported, based on a parametric analysis, local peak shear stresses as high as 300 kPa. Oh et al.162 used microbubble generated streaming on tissue mimicking samples in order to increase the perfusion distance. The same effect is investigated to improve the efficiency of clot dissolving drugs. Others show interest in the effects of the streaming induced by contrast agents in on-chip vessels.163 Finally, streaming is also suspected to be an important mechanism for the delivery of drugs loaded on microbubbles.113 Moreover, exploiting the acoustic radiation force on microbubbles that arises from the phase difference between the microbubble volumetric oscillations and the acoustic wave164 is an important method for the manipulation of single bubbles, as well as an often undesired effect in vitro. Radiation force was used for the study of the mechanical properties of cells, e.g., their resistance to shear,30 showing that a peak shear stress of a few Pa (corresponding to a maximum shear velocity of approximately 4 mm/s) was sufficient to induce membrane poration. Radiation force was also applied to the non-invasive manipulation of stem cell-bubbles clusters in vitro and in an in vivo CAM (chick embryo chorioallantoic membrane) model to increase their attachment to the vessel wall at the desired location.165 These complexes could be pushed without damage at velocities up to 0.25 mm/s using a pressure of 450 kPa with an off-resonance 1 MHz acoustic wave.
B. Optical imaging
1. Fluorescence imaging
a. Real-time fluorescence imaging
In order to evaluate the effect of microbubbles and ultrasound on drug uptake by cells, fluorescent molecules are almost exclusively used in the literature, since they are easy to use and since there is a wide variety of molecules available. Analysis of fluorescent molecular uptake is often performed by fluorescence microscopy. The most frequently used molecules are propidium iodide (PI),30,166,167 calcein,168–170 and SYTOX-dyes.171–173 These are small molecules (a few nanometer in size) and can serve as model for low molecular weight drugs. Moreover, they are cell-impermeable and can therefore act as a marker for the creation of pores in the cell membrane generated by microbubble-cell interactions. Larger fluorescent molecules such as fluorescent dextrans168,174,175 or fluorescent spheres176,177 represent larger drugs, as, for example, nanoparticles and can be used to determine membrane pore size and to investigate the role of endocytosis.
As an alternative to the small cell-impermeable fluorescent dyes, the influx of calcium can be measured to detect the formation of cell membrane pores. The increase of the intracellular calcium concentration upon membrane poration can be monitored by using probes that bind to intracellular calcium, such as Fluo-4AM169,174,178 and Fura-2AM.153,167,179
The influx of these fluorescent molecules and calcium occurs on a timescale of a few seconds and can therefore be readily imaged by real time fluorescence microscopy. These dyes are also often combined with dyes that stain live or non-viable cells after ultrasound exposure. In this way, these fluorescent molecules are a unique tool to visualize processes occurring in, on, and around the cell. The field of view, and similarly the depth of field, of fluorescence microscopy is inversely proportional to the magnification and is typically a few micrometers. This technique is therefore suitable to image typical in vitro models. The highest imaging precision, however, is achieved on cell monolayers, since the focus in a 3D structure will extend over multiple cell layers, making it difficult to focus on single cells. In the latter case, background noise will increase due to out-of-focus fluorescence and contrast of the deeper layers will decrease due to the turbidity of the tissue. As an example, Luan et al.113 could readily and quantitatively measure fluorescence from released material that was out of focus by tens of micrometers.
b. High-speed fluorescence imaging
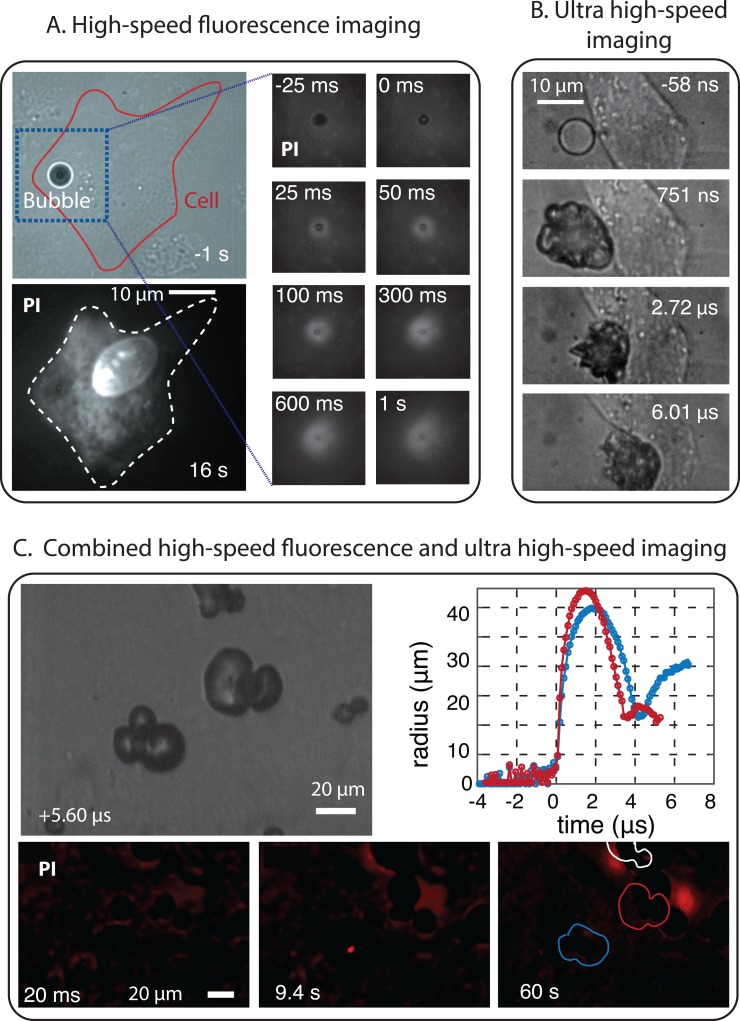
Fluorescence imaging can also be performed at higher frame rates (Fig. 3.A and C). Luan et al.113 were able to monitor on the microsecond time scale the release of model drug from fluorescently labeled microbubbles at up to a frame rate of 150 thousand frames per second, resolving the effects of the microbubble-induced streaming. High-speed fluorescence imaging is thus beneficial for applications involving second-order effects such as streaming or radiation force. However, high-speed fluorescence imaging cannot be used on cells directly, as cell staining has a significantly lower intensity, i.e., membrane staining is hardly imaged faster than at a few tens of frames per second, while PI-uptake can be imaged at a few hundred frames per second.157
FIG. 3.
(A) High-speed fluorescence imaging of the uptake of propidium iodide (PI) used as model drug by a cell as a result of membrane poration by an oscillating microbubble.193 (B) Ultra high-speed recording (interframe time of 58 ns) of the interaction of a vaporizing superheated microdroplet with a cell upon ultrasound exposure at 5 MHz.194 (C) Combined high-speed fluorescence imaging and bright-field ultra high-speed imaging to visualize the sonoporation of cells by short-lived cavitation bubbles. The bubbles are created by laser activation of polymeric microcapsules.195
c. Confocal imaging
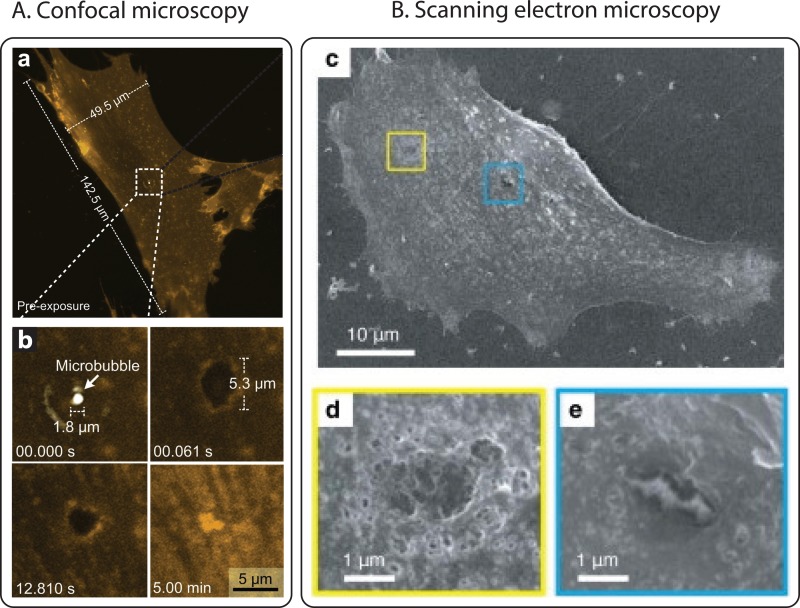
On the same timescales and with the same general motivations than for real-time fluorescence imaging, confocal fluorescence microscopy is used to visualize the cell structures, bubble coatings, and influx following ultrasound exposure.155 Using the high-resolution capability offered by confocal microscopy, Hu et al. could visualize the dynamic recession, or expansion, of single pores in sonoporated cells (Fig. 4.A)180 in order to understand the mechanisms by which a cell can self-heal. Chen et al. also showed how confocal microscopy could be used to image disruptions in the actin cytoskeleton of the cells as a result of their interaction with microbubbles.171 In contrast to fluorescence microscopy, confocal microscopy can also be performed in 3D. This complete spatial information on the cell allows for the assessment of volume information in both the endothelium and the cell nucleus181 or to observe the 3D organization of the cell. Confocal microscopy can display very thin layers (typically a few hundreds of nanometers) and can thus be used to visualize single planes in so-called z-stacks.182 This last aspect is expected to become increasingly important with the development of complex 3D cell model. Finally, confocal imaging of barrier models cells can provide information on bubble-induced gaps within tight-junctions, e.g., the blood-brain barrier by monitoring the location of proteins such as ZO-1.183 Confocal microscopy is very well suited for the imaging of the dynamic cell response180 and to quantify with remarkable detail molecular diffusion processes184–186 within the structure of single cells and cell monolayers. When imaging thicker samples, however, as is the case for 3D cell models, the turbidity of the tissue layers renders high-resolution confocal imaging difficult and calls upon complex image analysis and reconstruction. With such tools, the imaging depth can be increased to beyond 1 mm.187 Alternatively, thicker samples can be imaged with a significant improvement in depth by two-photon or multiphoton microscopy188,189 or prepared post-experiment to offer better imaging capabilities off-line (see Sec. IV D).
FIG. 4.
(A) Confocal images of membrane perforation and resealing in microbubble-induced sonoporation (Reprinted with permission from Hu et al., Ultrasound Med. Biol. 39, 2393 (2013). Copyright 2013 Elsevier). Upon the application of an ultrasound pulse, a disruption of 5.3 m is created in the cell, exactly at the site where the microbubble was. The cell membrane is stained with an orange dye (CellMask Orange). After ultrasound exposure, the pore progressively reseals within a few minutes. (B) SEM images of cell membrane damage after ultrasound radiation (Reprinted with permission from Kudo et al., Biophys. J. 96, 4866 (2009). Copyright 2009 Elsevier). The high magnification views show perforations in the cell membrane surrounded by a rough surface.
2. High-speed bright-field imaging
High-speed, bright field microscopy allows for the study of microbubble-cell interactions on the timescale of the ultrasound cycle or faster (Fig. 3.B and C). In particular, the Brandaris 128 ultra-high-speed camera was designed 15 years ago as a unique tool to record at up to 25 × 106 frames per second190 and has been widely used to study microbubble related problems. The ability to record at sufficiently high frame rates to resolve the ultrasound contrast microbubbles oscillation has facilitated the obtention of invaluable insight into fundamental bubble behavior.191 More recently, recordings made with the Brandaris camera have also revealed that bubble oscillations can result in the pulling and pushing of the cell membrane.21,192 More violent bubble behavior such as microjetting towards the cell can also occur and be recorded by such a system, as observed by Prentice et al.25 Ultra high-speed imaging is also the only existing method that allows for relating the details of the microbubble oscillation dynamics with the impact on the cells,21 or the microbubble behavior to the release of drugs loaded on their surface.113 This represents a critical aspect of the investigation of bubble-cell interactions. Other microbubbles such as plasmonic nano-bubbles have a very short lifetime (a fraction of microsecond) and therefore can be directly characterized only at such high frame rates. Other methods based on light scattering, for example, can detect transient bubbles but with limited possibilities to characterize them.151
Most commercial high-speed cameras cannot record faster than a few hundred thousand frames per second. This is insufficient to resolve microbubbles oscillations, but can prove instrumental in learning about the events on the timescale of the ultrasound bursts, e.g., the effect of microbubble cavitation on capillary blood vessel excised from rats.53,54 These camera systems were also used to study the impact of larger bubbles, with a slower dynamics, on a collection of cells.38
High-speed bright-field imaging can however not bring structural information on the cells at these times scale, which limits its relevance mostly to the observation and quantification of the bubble behavior itself and thus requires the simultaneous use of complementary techniques in order to extract information from the cells (Fig. 3).
C. Sensing
Aside from imaging methods, sensing techniques can be used to probe the nature of bubble-cell interactions and their consequences. Sensing methods are important for the study of bubble-cell interaction as they offer a direct measure of cell membranes and/or intercellular matrix integrity on the timescale of the ultrasound, e.g., using electrical sensing methods. Moreover, they give a remote feedback on the bubble activity in relation with the cell response. This feedback can be used later to quantify the resulting effects in vivo where the aforementioned imaging techniques cannot be used.
1. Electrical methods
a. Transepithelial electrical resistance (TEER)
Bubbles have proven to be an attractive active means to pass foreign substances, such as drugs, across physiological barriers like the BBB. The permeation of barrier models following their exposure to bubbles can first be assessed by evaluating the integrity of the barrier. A conventional approach to this end consists in measuring the TEER of the barrier model,196,197 which corresponds to the electrical resistance of the cell monolayer grown on a porous substrate/membrane (Transwell inserts, typically). Two main approaches are reported to determine TEER values: the first one directly assesses the electrical resistance of the cell monolayers. This principle is employed by the commercially available systems for TEER measurement (EVOM, World Precision Instruments, Sarasota, FL). The second method is based on impedance spectroscopy, which provides additional information on the electrical capacitance of the cell monolayer. When the cell monolayer reaches confluency, cells establish tight junctions with each other, and the monolayer is characterized by a high TEER value. On the contrary, the creation of gaps between cells after bubble actuation, for instance, is accompanied by a decrease in the resistance of the monolayer. Interestingly, the TEER can be monitored continuously and in a non-invasive manner, yielding real-time information on the barrier characteristics.
b. Trans-membrane current (TMC)
TMC measurement is a sensing technique that measures the electrical intensity carried by the ions that passes across the membrane of a single cell. The technique involves a pair of electrodes, which are placed in the intracellular and extracellular medium, respectively. This technique give real-time information on the cell membrane porosity as the flux of ions directly relates to the number and size of the pores in the membrane. TMC can be directly correlated to the cell pore size.23
2. Ultrasound sensing
Ultrasound was largely discussed in Sec. IV A of this review as an excitation method. Ultrasound detection is also one of the main sensing methods and is very often used for the detection of broadband cavitation noise159 as a measure for the energy release by the bubbles in the vicinity of the cells. Although ultrasound sensing is typically performed with a single element transducer with sensitivity on the order of 1 Pa, it can also be achieved with a commercial ultrasound system in order to obtain, in addition, the spatial information of the occurring events.198 Ultrasound is also a crucial sensing method to quantify the response of a photoacoustic agent132 or to obtain direct information on the physical mechanisms underlying the cavitation events.139
D. Off-line techniques
Some techniques such as described hereafter cannot be performed simultaneously with the experiment or ultrasound exposure. These methods, termed here off-line techniques, can be used to access precise details of the cell structure after ultrasound irradiation or to obtain quantitative statistical information (Fig. 4).
1. Off-line confocal imaging
In the fluorescence imaging section, we mentioned the difficulties encountered in studying 3D samples using fluorescence techniques. As an alternative, these 3D samples can be studied off-line to yield high-resolution information on the impact of the bubbles on the tissues, e.g., in terms of tissue damage, drug penetration, and efficiency of intracellular drug delivery. The most commonly used approach relies on the fixation of the tissue using, for instance, paraformaldehyde followed by the embedding of the tissue in paraffin, to then perform cryosectioning of the tissue to yield thin slices that can readily be imaged using confocal microscopy. Another more recent and highly promising strategy, also known as tissue clearance or CLARITY,199,200 consists of embedding the tissue or 3D cellular constructs in an acrylamide gel matrix, followed by removal of the lipid molecules to yield a transparent 3D structure. This 3D structure is not only suitable for 3D imaging by allowing deep optical penetration but it also facilitates the penetration of all probes required for tissue staining, another concern encountered with the thicker 3D cellular aggregates.
2. Electron and atomic force microscopy
Other forms of microscopy have also been used to examine single fixed cells or cell monolayers. These include scanning electron microscopy (SEM, Fig. 4.B),23 transmission electron microscopy (TEM),201 and atomic force microscopy (AFM).25,202,203 Using these techniques, the direct effects of microbubble cavitation on cell morphology and topography can be studied. Cell size has been shown to decrease after ultrasound radiation,176 and smoother and flatter cell surfaces have also been observed.21 Moreover, clear perforations in the cell membrane could be imaged.25,91,166,176 The size of these pores has been estimated to be several hundred nanometres up to a few micrometres. These techniques, however, are applied after the actual experiment in which the bubble-cell interaction occurs. Therefore, small pores can have already resealed by the time, the sample preparation is completed, and the sample preparation itself can also cause artifacts in the image. Thus, these microscopy techniques give great details on the cell itself, but little on the mechanism by which the observed effects are caused.
3. Flow cytometry: High-throughput technique
A common limitation of the techniques presented above is their throughput. Flow cytometry can provide statistical information on a large number155,204 of cells, simultaneously. It makes use of similar dyes as these added for fluorescence imaging. Flow cytometry, however, suffers from some of the same limitations as SEM, TEM, or AFM: it provides a snapshot of the cell properties at a given time point after the experiment. It requires a sample preparation step that may alter the cells, and its results are difficult to directly correlate to the local microscopy observations.
Similar methods exist that make use of non-fluorescent dyes for viability examinations of a cell population.204,205
V. SUMMARY AND OUTLOOK
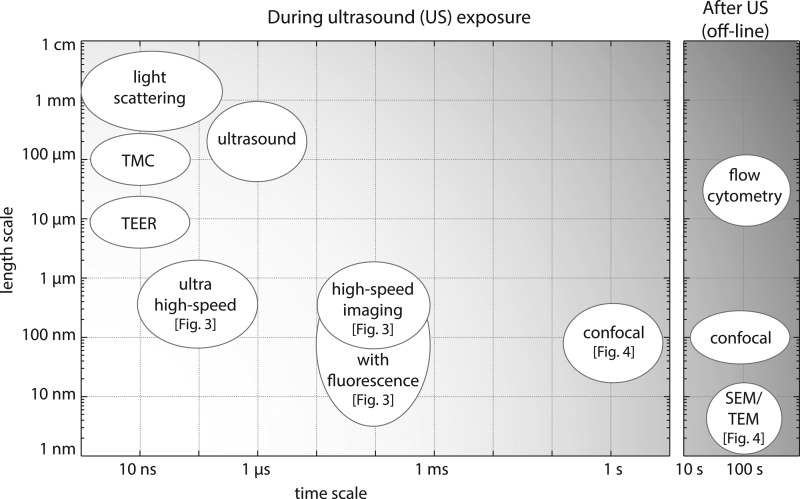
There is a growing scientific literature demonstrating the unique potential of microbubbles for targeted therapeutic delivery. However, a much more complete understanding of the fundamental mechanisms underlying microbubble-cell interactions is required to translate this potential into clinical benefit. Elucidating these interactions represents a significant challenge owing to the complexity of the interactions and the range of timescales and length scales involved, and consequently the range of different methods and techniques required to study them (Fig. 5).
FIG. 5.
Summary of the diverse techniques used for the study of bubble-cell interaction in relation with the accessible time scales and length scales. The left panel presents techniques that can be used simultaneously with the ultrasound exposure, while the right panel shows the techniques that cannot be applied during ultrasound exposure and thus require a 2-step experiment.
In this paper, we have reviewed the available experimental methods. First, we have described various cell models exhibiting different levels of complexity, and ranging from individual cells up to more complex 3D cellular models, typically implemented in an organ-on-a-chip format. Each model carries specific interest for the understanding of either the fundamentals of bubble-cell interactions or that of the chain of events occurring on a larger scale. Following this, we have reviewed various types of microbubbles and precursors that are currently used in research and in preclinical setting. We have presented their differences in activation, their behavior, and their potential. These bubbles can also target specific cells and are therefore mostly relevant in combination with these specific cell models. Finally, based on an appropriate combination of a cell model and a bubble type, we have reviewed the great variety of experimental methods which have been reported in literature for studying bubble-cell interactions, to yield specific information on the various physical and biological time scales and length scales.
The development of ultra high-speed imaging systems has offered unparalleled insight into microbubble dynamics, including the release and transport of model drugs and physical interactions with biological structures. Similarly, real-time fluorescence microscopy techniques combined with novel molecular probes have enabled membrane dynamics and sub-cellular processes to be studied, revealing hitherto unsuspected mechanisms of interaction and with significant implications for both treatment efficacy, outcome, as well as safety. Technology in both of these areas continues to advance rapidly and will undoubtedly facilitate further and deeper understanding in the near future. Recent examples include the use of ultra high-speed fluorescence microscopy to reveal the details of drug transport and uptake around oscillating microbubbles and of live confocal microscopy to observe the dynamics of individual cell membranes following cavitation events.206 Miniaturization of ultrasound devices and improved control over microbubble and microbubble precursor fabrication will similarly enable the design of more versatile experimental systems to investigate these phenomena.
Each of the aforementioned techniques has the ability to elucidate specific aspects of microbubble-cell interactions, and an important challenge of future experimental work will be the ability to integrate these methods to be able to relate and correlate the different time and length scales. This in combination with the newly developed cell models will greatly increase the relevance of in vitro research on the interaction of bubbles with tissue in vivo. Similarly, advances in endoscopy and intravital microscopy will ultimately enable these studies to bridge to in vivo models so that microbubble-cell interactions can be studied in situ.
A detailed understanding of the underlying mechanisms will translate into the design of more effective delivery agents, treatment protocols, and monitoring strategies and ultimately the realization of microbubble-mediated therapy as a powerful new clinical approach.
ACKNOWLEDGMENTS
This work was made possible by the funding of the Dutch national NanoNextNL project, a micro and nanotechnology consortium of the Government of the Netherlands and 130 partners, and the UK Engineering and Physical Sciences Research Council (Grant No. EP/I021795/1). Ine De Cock is a doctoral fellow of the Institute for the Promotion of Innovation through Science and Technology in Flanders, Belgium (IWT-Vlaanderen). Ine Lentacker is a postdoctoral fellow of the Research Foundation Flanders, Belgium (FWO-Vlaanderen). The support of both these institutions is gratefully acknowledged. Finally, Séverine Le Gac would like to thank the MESA+ Institute for Nanotechnology for its financial support via the Strategic Research Orientation “Nanotechnology for Innovative Medicine”. We also thank Adithya Sridhar for providing the image of Fig. 1.C.a.
References
- 1. Gramiak R. and Shah P., Invest. Radiol. 3, 356 (1968). 10.1097/00004424-196809000-00011 [DOI] [PubMed] [Google Scholar]
- 2. Cosgrove D. and Harvey C., Med. Biol. Eng. Comput. 47, 813 (2009). 10.1007/s11517-009-0434-3 [DOI] [PubMed] [Google Scholar]
- 3. Hoyt K., Umphrey H., Lockhart M., Robbin M., and Forero-Torres A., Ultrasound Med. Biol. 41, 2292 (2015). 10.1016/j.ultrasmedbio.2015.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Klotz L. V., Gürkov R., Eichhorn M. E., Siedek V., Krause E., Jauch K.-W., Reiser M. F., and Clevert D.-A., Eur. J. Radiol. 82, 2227 (2013). 10.1016/j.ejrad.2013.08.033 [DOI] [PubMed] [Google Scholar]
- 5. Abou-Elkacem L., Bachawal S. V., and Willmann J. K., Eur. J. Radiol. 84, 1685 (2015). 10.1016/j.ejrad.2015.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang H., Tam S., Ingham E. S., Mahakian L. M., Lai C.-Y., Tumbale S. K., Teesalu T., Hubbard N. E., Borowsky A. D., and Ferrara K. W., Biomaterials 56, 104 (2015). 10.1016/j.biomaterials.2015.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hope Simpson D., Chin C. T., and Burns P. N., IEEE Trans. Ultrason., Ferroelectr., Freq. Control 46, 372 (1999). 10.1109/58.753026 [DOI] [PubMed] [Google Scholar]
- 8. Miller D. L., Ultrasonics 19, 217 (1981). 10.1016/0041-624X(81)90006-8 [DOI] [Google Scholar]
- 9. Daeichin V., Bosch J. G., Needles A., Foster F. S., van der Steen A., and de Jong N., Ultrasound Med. Biol. 41, 486 (2015). 10.1016/j.ultrasmedbio.2014.10.003 [DOI] [PubMed] [Google Scholar]
- 10. Marmottant P., Biben T., and Hilgenfeldt S., Proc. R. Soc. London A 464, 1781 (2008). 10.1098/rspa.2007.0362 [DOI] [Google Scholar]
- 11. Zhao L., Feng Y., Shi A., Zong Y., and Wan M., Ultrasound Med. Biol. 41, 2755 (2015). 10.1016/j.ultrasmedbio.2015.05.021 [DOI] [PubMed] [Google Scholar]
- 12. Lentacker I., Geers B., Demeester J., Smedt S. C. D., and Sanders N. N., J. Controlled Release 148, e113 (2010). 10.1016/j.jconrel.2010.07.085 [DOI] [PubMed] [Google Scholar]
- 13. Alonso A., Reinz E., Leuchs B., Kleinschmidt J., Fatar M., Geers B., Lentacker I., Hennerici M. G., de Smedt S. C., and Meairs S., Mol. Ther.—Nucl. Acids 2, e73 (2013). 10.1038/mtna.2012.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dewitte H., Lint S. V., Heirman C., Thielemans K., Smedt S. C. D., Breckpot K., and Lentacker I., J. Controlled Release 194, 28 (2014). 10.1016/j.jconrel.2014.08.011 [DOI] [PubMed] [Google Scholar]
- 15. Tarapacki C., Kumaradas C., and Karshafian R., Ultrasonics 53, 793 (2013). 10.1016/j.ultras.2012.11.006 [DOI] [PubMed] [Google Scholar]
- 16. Fernández Rivas D., Verhaagen B., Seddon J. R. T., Zijlstra A. G., Jiang L.-M., van der Sluis L. W. M., Versluis M., Lohse D., and Gardeniers H. J. G. E., Biomicrofluidics 6, 034114 (2012). 10.1063/1.4747166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parini M. R., Eggett D. L., and Pitt W. G., J. Clin. Periodontol. 32, 1151 (2005). 10.1111/j.1600-051X.2005.00836.x [DOI] [PubMed] [Google Scholar]
- 18. van der Sluis L., Boutsioukis C., Jiang L., Macedo R., Verhaagen B., and Versluis M., in The Root Canal Biofilm, edited by de Paz L. E. C., Sedgley C. M., and Kishen A. ( Springer-Verlag, Berlin Heidelberg, 2015). [Google Scholar]
- 19. Deng C. X., Sieling F., Pan H., and Cui J., Ultrasound Med. Biol. 30, 519 (2004). 10.1016/j.ultrasmedbio.2004.01.005 [DOI] [PubMed] [Google Scholar]
- 20. Ohl C.-D., Arora M., Ikink R., de Jong N., Versluis M., Delius M., and Lohse D., Biophys. J. 91, 4285 (2006). 10.1529/biophysj.105.075366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Wamel A., Kooiman K., Harteveld M., Emmer M., ten Cate F. J., Versluis M., and de Jong N., J. Controlled Release 112, 149 (2006). 10.1016/j.jconrel.2006.02.007 [DOI] [PubMed] [Google Scholar]
- 22. Kato S., Shirai Y., Kanzaki H., Sakamoto M., Mori S., and Kodama T., Ultrasound Med. Biol. 41, 1411 (2015). 10.1016/j.ultrasmedbio.2014.12.014 [DOI] [PubMed] [Google Scholar]
- 23. Zhou Y., Yang K., Cui J., Ye J., and Deng C., J. Controlled Release 157, 103 (2012). 10.1016/j.jconrel.2011.09.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Le Gac S. and van den Berg A., Trends Biotechnol. 28, 55 (2010). 10.1016/j.tibtech.2009.10.005 [DOI] [PubMed] [Google Scholar]
- 25. Prentice P., Cuschieri A., Dholokia K., Prausnitz M., and Campbell P., Nat. Phys. 1, 107 (2005). 10.1038/nphys148 [DOI] [Google Scholar]
- 26. Li Z. G., Liu A. Q., Klaseboer E., Zhang J. B., and Ohl C. D., Lab Chip 13, 1144 (2013). 10.1039/c3lc41252k [DOI] [PubMed] [Google Scholar]
- 27. Garbin V., Cojoc D., Ferrari E., Di Fabrizio E., Overvelde M. L. J., Van Der Meer S. M., De Jong N., Lohse D., and Versluis M., Appl. Phys. Lett. 90, 114103 (2007). 10.1063/1.2713164 [DOI] [Google Scholar]
- 28. Helfield B. L., Leung B. Y. C., and Goertz D. E., Phys. Med. Biol. 59, 1721 (2014). 10.1088/0031-9155/59/7/1721 [DOI] [PubMed] [Google Scholar]
- 29. Li H., Sims C. E., Wu H. Y., and Allbritton N. L., Anal. Chem. 73, 4625 (2001). 10.1021/ac0105235 [DOI] [PubMed] [Google Scholar]
- 30. Fan Z., Sun Y., Chen D., Tay D., Chen W., Deng C. X., and Fu J., Sci. Rep. 3, 2176 (2013). 10.1038/srep02176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Quinto-Su P. A., Kuss C., Preiser P. R., and Ohl C.-D., Lab Chip 11, 672 (2011). 10.1039/C0LC00182A [DOI] [PubMed] [Google Scholar]
- 32. Sims C. E., Meredith G. D., Krasieva T. B., Berns M. W., Tromberg B. J., and Allbritton N. L., Anal. Chem. 70, 4570 (1998). 10.1021/ac9802269 [DOI] [PubMed] [Google Scholar]
- 33. Quinto-Su P. A., Lai H.-H., Yoon H. H., Sims C. E., Allbritton N. L., and Venugopalan V., Lab Chip 8, 408 (2008). 10.1039/b715708h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meredith G. D., Sims C. E., Soughayer J. S., and Allbritton N. L., Nat. Biotechnol. 18, 309 (2000). 10.1038/73760 [DOI] [PubMed] [Google Scholar]
- 35. Phillips R. M., Bair E., Lawrence D. S., Sims C. E., and Allbritton N. L., Anal. Chem. 85, 6136 (2013). 10.1021/ac401106e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Phillips K. S., Lai H. H., Johnson E., Sims C. E., and Allbritton N. L., Lab Chip 11, 1333 (2011). 10.1039/c0lc00370k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Compton J. L., Hellman A. N., and Venugopalan V., Biophys. J. 105, 2221 (2013). 10.1016/j.bpj.2013.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dijkink R. J. and Ohl C.-D., Lab Chip 8, 1676 (2008). 10.1039/b806912c [DOI] [PubMed] [Google Scholar]
- 39. Cochran M. and Wheatley M. A., Ultrasound Med. Biol. 39, 1102 (2013). 10.1016/j.ultrasmedbio.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qin P., Xu L., Hu Y., Zhong W., Cai P., Du L., Jin L., and Yu A. C., Ultrasound Med. Biol. 40, 979 (2014). 10.1016/j.ultrasmedbio.2013.11.024 [DOI] [PubMed] [Google Scholar]
- 41. Ling Yong C. L., Siak-Wei Ow D., Tandiono T., Mei Heng L. L., Kwok-Keung Chan K., Ohl C.-D., Klaseboer E., Ohl S.-W., and Boon-Hwa Choo A., Biotechnol. J. 9, 1081 (2014). 10.1002/biot.201300507 [DOI] [PubMed] [Google Scholar]
- 42. Park J., Fan Z., and Deng C. X., J. Biomech. 44, 164 (2011). 10.1016/j.jbiomech.2010.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gac S. L., Zwaan E., Berg A. V. D., and Ohl C.-D., Lab Chip 7, 1666 (2007). 10.1039/b712897p [DOI] [PubMed] [Google Scholar]
- 44. Tandiono T., Siak-Wei Ow D., Driessen L., Sze-Hui Chin C., Klaseboer E., Boon-Hwa Choo A., Ohl S.-W., and Ohl C.-D., Lab Chip 12, 780 (2012). 10.1039/C2LC20861J [DOI] [PubMed] [Google Scholar]
- 45. Overvelde M., Garbin V., Dollet B., de Jong N., Lohse D., and Versluis M., Ultrasound Med. Biol. 37, 1500 (2011). 10.1016/j.ultrasmedbio.2011.05.025 [DOI] [PubMed] [Google Scholar]
- 46. Ramaiahgari S., den Braver M., Herpers B., Terpstra V., Commandeur J., van de Water B., and Price L., Arch. Toxicol. 88, 1083 (2014). 10.1007/s00204-014-1215-9 [DOI] [PubMed] [Google Scholar]
- 47. Mehta G., Hsiao A. Y., Ingram M., Luker G. D., and Takayama S., J. Controlled Release 164, 192 (2012). 10.1016/j.jconrel.2012.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Young E. W. K., Integr. Biol. 5, 1096 (2013). 10.1039/c3ib40076j [DOI] [PubMed] [Google Scholar]
- 49. Breslin S. and O'Driscoll L., Drug Discovery Today 18, 240 (2013). 10.1016/j.drudis.2012.10.003 [DOI] [PubMed] [Google Scholar]
- 50. Torisawa Y.-S., Chueh B.-H., Huh D., Ramamurthy P., Roth T. M., Barald K. F., and Takayama S., Lab Chip 7, 770 (2007). 10.1039/b618439a [DOI] [PubMed] [Google Scholar]
- 51. Rivron N. C., Vrij E. J., Rouwkema J., Le Gac S., van den Berg A., Truckenmüller R. K., and van Blitterswijk C. A., Proc. Natl. Acad. Sci. U.S.A. 109, 6886 (2012). 10.1073/pnas.1201626109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Christakou A. E., Ohlin M., Onfelt B., and Wiklund M., Lab Chip 15, 3222 (2015). 10.1039/C5LC00436E [DOI] [PubMed] [Google Scholar]
- 53. Hosseinkhah N., Chen H., Matula T. J., Burns P. N., and Hynynen K., J. Acoust. Soc. Am. 134, 1875 (2013). 10.1121/1.4817843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen H., Brayman A. A., and Matula T. J., Appl. Phys. Lett. 101, 163704 (2012). 10.1063/1.4761937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Faez T., Skachkov I., Versluis M., Kooiman K., and de Jong N., Ultrasound Med. Biol. 38, 1608 (2015). 10.1016/j.ultrasmedbio.2012.05.014 [DOI] [PubMed] [Google Scholar]
- 56. Bhatia S. N. and Ingber D. E., Nat. Biotechnol. 32, 760 (2014). 10.1038/nbt.2989 [DOI] [PubMed] [Google Scholar]
- 57. Bischel L. L., Young E. W., Mader B. R., and Beebe D. J., Biomaterials 34, 1471 (2013). 10.1016/j.biomaterials.2012.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. van Duinen V., Trietsch S. J., Joore J., Vulto P., and Hankemeier T., Curr. Opin. Biotechnol. 35, 118 (2015). 10.1016/j.copbio.2015.05.002 [DOI] [PubMed] [Google Scholar]
- 59. Franco C. and Gerhardt H., Nature 488, 465 (2012). 10.1038/488465a [DOI] [PubMed] [Google Scholar]
- 60. Albanese A., Lam A. K., Sykes E. A., Rocheleau J. V., and Chan W. C. W., Nat. Commun. 4, 2718 (2013). 10.1038/ncomms3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Prabhakarpandian B., Shen M.-C., Nichols J. B., Mills I. R., Sidoryk-Wegrzynowicz M., Aschner M., and Pant K., Lab Chip 13, 1093 (2013). 10.1039/c2lc41208j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Griep L. M., Wolbers F., de Wagenaar B., ter Braak P. M., Weksler B. B., Romero I. A., Couraud P. O., Vermes I., van der Meer A. D., and van den Berg A., Biomed. Microdevices 15, 145–150 (2013). 10.1007/s10544-012-9699-7 [DOI] [PubMed] [Google Scholar]
- 63. Song H.-H. G., Park K. M., and Gerecht S., Adv. Drug Delivery Rev. 79–80, 19 (2014). 10.1016/j.addr.2014.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Moya M. L., Hsu Y.-H., Lee A. P., Hughes C. C. W., and George S. C., Tissue Eng., Part C 19, 730 (2013). 10.1089/ten.tec.2012.0430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zheng Y., Chen J., Craven M., Choi N. W., Totorica S., Diaz-Santana A., Kermani P., Hempstead B., Fischbach-Teschl C., López J., and Stroock A. D., Proc. Natl. Acad. Sci. U.S.A. 109, 9342 (2012). 10.1073/pnas.1201240109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Buchanan C. F., Voigt E. E., Szot C. S., Freeman J. W., Vlachos P. P., and Rylander M. N., Tissue Eng., Part C 20, 64 (2014). 10.1089/ten.tec.2012.0731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Moya M., Tran D., and George S., Stem Cell Res. Ther. 4, S15 (2013). 10.1186/scrt376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Choi J. J., Wang S., Brown T. R., Small S. A., Duff K. E. K., and Konofagou E. E., Ultrason. Imaging 30, 189 (2008). 10.1177/016173460803000304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Choi J. J., Pernot M., Small S. A., and Konofagou E. E., Ultrasound Med. Biol. 33, 95 (2007). 10.1016/j.ultrasmedbio.2006.07.018 [DOI] [PubMed] [Google Scholar]
- 70. Lindner J. R., Cardiovasc. Res. 84, 182 (2009). 10.1093/cvr/cvp302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Villanueva F. S., Lu E., Bowry S., Kilic S., Tom E., Wang J., Gretton J., Pacella J. J., and Wagner W. R., Circulation 115, 345 (2007). 10.1161/CIRCULATIONAHA.106.633917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.See http://www.insphero.com/ for Insphero (2015).
- 73.See http://mimetas.com/ for Mimetas (2015).
- 74.See http://www.tissuse.com/ for TissUse (2015).
- 75. Verhaagen B. and Fernández Rivas D., Ultrason. Sonochem. 29, 619 (2016). 10.1016/j.ultsonch.2015.03.009 [DOI] [PubMed] [Google Scholar]
- 76. Parini M. R. and Pitt W. G., Colloids Surf., B 52, 39 (2006). 10.1016/j.colsurfb.2006.06.005 [DOI] [PubMed] [Google Scholar]
- 77. Macedo R. G., Robinson J. P., Verhaagen B., Walmsley A. D., Versluis M., Cooper P. R., and van der Sluis L. W. M., Int. Endod. J. 47, 1040 (2014). 10.1111/iej.12246 [DOI] [PubMed] [Google Scholar]
- 78. Halford A., Ohl C.-D., Azarpazhooh A., Basrani B., Friedman S., and Kishen A., J. Endod. 38, 1530 (2012). 10.1016/j.joen.2012.07.007 [DOI] [PubMed] [Google Scholar]
- 79. Sridhar A., de Boer H. L., van den Berg A., and Gac S. L., PLoS One 9, e93618 (2014). 10.1371/journal.pone.0093618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Stickler D. J., Nat. Clin. Pract. Urol. 5, 598 (2008). 10.1038/ncpuro1231 [DOI] [PubMed] [Google Scholar]
- 81. Fan Z., Chen D., and Deng C., J. Controlled Release 170, 401 (2013). 10.1016/j.jconrel.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Feinstein S. B., Ten Cate F. J., Zwehl W., Ong K., Maurer G., Tei C., Shah P. M., Meerbaum S., and Corday E., J. Am. Coll. Cardiol. 3, 14 (1984). 10.1016/S0735-1097(84)80424-6 [DOI] [PubMed] [Google Scholar]
- 83. Faez T., Emmer M., Kooiman K., Versluis M., van der Steen A. F. W., and de Jong N., IEEE Trans. Ultrason., Ferroelectr., Freq. Control 60, 7–20 (2013). 10.1109/TUFFC.2013.2533 [DOI] [PubMed] [Google Scholar]
- 84. Kwan J. J. and Borden M. A., Adv. Colloid Interface Sci. 183–184, 82 (2012). 10.1016/j.cis.2012.08.005 [DOI] [PubMed] [Google Scholar]
- 85. Un K., Kawakami S., Suzuki R., Maruyama K., Yamashita F., and Hashida M., Hum. Gene Ther. 21, 65 (2010). 10.1089/hum.2009.106 [DOI] [PubMed] [Google Scholar]
- 86. De Temmerman M.-L., Dewitte H., Vandenbroucke R. E., Lucas B., Libert C., Demeester J., De Smedt S. C., Lentacker I., and Rejman J., Biomaterials 32, 9128 (2011). 10.1016/j.biomaterials.2011.08.024 [DOI] [PubMed] [Google Scholar]
- 87. Forsberg F., Lathia J. D., Merton D. A., Liu J.-B., Le N. T., Goldberg B. B., and Wheatley M. A., Ultrasound Med. Biol. 30, 1281 (2004). 10.1016/j.ultrasmedbio.2004.07.006 [DOI] [PubMed] [Google Scholar]
- 88. Wheatley M. A., Forsberg F., Oum K., Ro R., and El-Sherif D., Ultrasonics 44, 360 (2006). 10.1016/j.ultras.2006.04.003 [DOI] [PubMed] [Google Scholar]
- 89. Fokong S., Siepmann M., Liu Z., Schmitz G., Kiessling F., and Gätjens J., Ultrasound Med. Biol. 37, 1622 (2011). 10.1016/j.ultrasmedbio.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 90. Chlon C., Guédon C., Verhaagen B., Shi W. T., Hall C. S., Lub J., and Böhmer M. R., Biomacromolecules 10, 1025 (2009). 10.1021/bm801243u [DOI] [PubMed] [Google Scholar]
- 91. Kooiman K., Böhmer M. R., Emmer M., Vos H. J., Chlon C., Shi W. T., Hall C. S., de Winter S. H., Schroën K., Versluis M., de Jong N., and van Wamel A., J. Controlled Release 133, 109 (2009). 10.1016/j.jconrel.2008.09.085 [DOI] [PubMed] [Google Scholar]
- 92. Helfield B. L., Cherin E., Foster F. S., and Goertz D. E., Ultrasound Med. Biol. 38, 846 (2012). 10.1016/j.ultrasmedbio.2012.01.011 [DOI] [PubMed] [Google Scholar]
- 93. van Rooij T., Luan Y., Renaud G., van der Steen A. F. W., Versluis M., de Jong N., and Kooiman K., Ultrasound Med. Biol. 41, 1432 (2015). 10.1016/j.ultrasmedbio.2015.01.004 [DOI] [PubMed] [Google Scholar]
- 94. Goertz D. E., de Jong N., and van der Steen A. F., Ultrasound Med. Biol. 33, 1376 (2007). 10.1016/j.ultrasmedbio.2007.03.009 [DOI] [PubMed] [Google Scholar]
- 95. Hettiarachchi K., Talu E., Longo M. L., Dayton P. A., and Lee A. P., Lab Chip 7, 463 (2007). 10.1039/b701481n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Farook U., Stride E., and Edirisinghe M., J. R. Soc. Interface 6, 271 (2009). 10.1098/rsif.2008.0225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Sirsi S., Feshitan J., Kwan J., Homma S., and Borden M., Ultrasound Med. Biol. 36, 935 (2010). 10.1016/j.ultrasmedbio.2010.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Segers T. and Versluis M., Lab Chip 14, 1705 (2014). 10.1039/c3lc51296g [DOI] [PubMed] [Google Scholar]
- 99. Fisher N. G., Christiansen J. P., Klibanov A., Taylor R. P., Kaul S., and Lindner J. R., J. Am. Coll. Cardiol. 40, 811 (2002). 10.1016/S0735-1097(02)02038-7 [DOI] [PubMed] [Google Scholar]
- 100. Unnikrishnan S. and Klibanov A. L., Am. J. Roentgenol. 199, 292 (2012). 10.2214/AJR.12.8826 [DOI] [PubMed] [Google Scholar]
- 101. Lindner J. R., Song J., Xu F., Klibanov A. L., Singbartl K., Ley K., and Kaul S., Circulation 102, 2745 (2000). 10.1161/01.CIR.102.22.2745 [DOI] [PubMed] [Google Scholar]
- 102. Deshpande N., Ren Y., Foygel K., Rosenberg J., and Willmann J. K., Radiology 258, 804 (2011). 10.1148/radiol.10101079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lindner J. R., Song J., Christiansen J., Klibanov A. L., Xu F., and Ley K., Circulation 104, 2107 (2001). 10.1161/hc4201.097061 [DOI] [PubMed] [Google Scholar]
- 104. Klibanov A. L., Rychak J. J., Yang W. C., Alikhani S., Li B., Acton S., Lindner J. R., Ley K., and Kaul S., Contrast Media Mol. Imaging 1, 259 (2006). 10.1002/cmmi.113 [DOI] [PubMed] [Google Scholar]
- 105. Rao C. G., Chianese D., Doyle G. V., Miller M. C., Russell T., Sanders R. A., and Terstappen L. W. M. M., Int. J. Oncol. 27, 49 (2005). 10.3892/ijo.27.1.49 [DOI] [PubMed] [Google Scholar]
- 106. Plaks V., Koopman C. D., and Werb Z., Science 341, 1186 (2013). 10.1126/science.1235226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Owen J., Pankhurst Q., and Stride E., Int. J. Hyperthermia 28, 362 (2012). 10.3109/02656736.2012.668639 [DOI] [PubMed] [Google Scholar]
- 108. Gao Z., Kennedy A. M., Christensen D. A., and Rapoport N. Y., Ultrasonics 48, 260 (2008). 10.1016/j.ultras.2007.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shih R., Bardin D., Martz T. D., Sheeran P. S., Dayton P. A., and Lee A. P., Lab Chip 13, 4816 (2013). 10.1039/c3lc51016f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Geers B., Lentacker I., Sanders N. N., Demeester J., Meairs S., and De Smedt S. C., J. Controlled Release 152, 249 (2011). 10.1016/j.jconrel.2011.02.024 [DOI] [PubMed] [Google Scholar]
- 111. Luan Y., Faez T., Gelderblom E. C., Skachkov I., Geers B., Lentacker I., van der Steen A. F. W., Versluis M., and de Jong N., Ultrasound Med. Biol. 38, 2174 (2012). 10.1016/j.ultrasmedbio.2012.07.023 [DOI] [PubMed] [Google Scholar]
- 112. Lentacker I., Geers B., Demeester J., De Smedt S. C., and Sanders N. N., Mol. Ther. 18, 101 (2010). 10.1038/mt.2009.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Luan Y., Lajoinie G., Gelderblom E., Skachkov I., van der Steen A. F. W., Vos H. J., Versluis M., and De Jong N., Ultrasound Med. Biol. 40, 1834 (2014). 10.1016/j.ultrasmedbio.2014.02.031 [DOI] [PubMed] [Google Scholar]
- 114. Bouakaz A., Versluis M., and de Jong N., Ultrasound Med. Biol. 31, 391 (2005). 10.1016/j.ultrasmedbio.2004.12.004 [DOI] [PubMed] [Google Scholar]
- 115. Lensen D., Gelderblom E. C., Vriezema D. M., Marmottant P., Verdonschot N., Versluis M., de Jong N., and van Hest J. C., Soft Matter 7, 5417 (2011). 10.1039/c1sm05324h [DOI] [Google Scholar]
- 116. Yang F., Li Y., Chen Z., Zhang Y., Wu J., and Gu N., Biomaterials 30, 3882 (2009). 10.1016/j.biomaterials.2009.03.051 [DOI] [PubMed] [Google Scholar]
- 117. Lathia J. D., Leodore L., and Wheatley M. A., Ultrasonics 42, 763 (2004). 10.1016/j.ultras.2003.12.018 [DOI] [PubMed] [Google Scholar]
- 118. Marmottant P., Bouakaz A., De Jong N., and Quillet C., J. Acoust. Soc. Am. 129, 1231 (2011). 10.1121/1.3543943 [DOI] [PubMed] [Google Scholar]
- 119. Raymond J. L., Luan Y., van Rooij T., Kooiman K., Huang S.-L., McPherson D. D., Versluis M., de Jong N., and Holland C. K., J. Acoust. Soc. Am. 137, 1693 (2015). 10.1121/1.4916277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Huang S.-L., McPherson D. D., and MacDonald R. C., Ultrasound Med. Biol. 34, 1272 (2008). 10.1016/j.ultrasmedbio.2008.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kee P. H., Abruzzo T. A., Smith D. A., Kopechek J. A., Wang B., Huang S. L., MacDonald R. C., Holland C. K., and McPherson D. D., J. Liposome Res. 18, 263 (2008). 10.1080/08982100802354558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Coussios C.-C., Holland C. K., Jakubowska L., Huang S.-L., MacDonald R. C., Nagaraj A., and McPherson D. D., Ultrasound Med. Biol. 30, 181 (2004). 10.1016/j.ultrasmedbio.2003.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Smith D. A. B., Porter T. M., Martinez J., Huang S., MacDonald R. C., McPherson D. D., and Holland C. K., Ultrasound Med. Biol. 33, 797 (2007). 10.1016/j.ultrasmedbio.2006.11.017 [DOI] [PubMed] [Google Scholar]
- 124. Dove J. D., Borden M. A., and Murray T. W., Opt. Lett. 39, 3732 (2014). 10.1364/OL.39.003732 [DOI] [PubMed] [Google Scholar]
- 125. Lajoinie G., Lee J. Y., Kruizinga P., Owen J., van Soest G., De Jong N., Stride E., and Versluis M., “ Laser-driven resonance of dye-doped oil-coated microbubbles” (unpublished). [DOI] [PubMed]
- 126. Firouzi K., Stride E., and Saffari N., J. Acoust. Soc. Am. 133, 3853 (2013). 10.1121/1.4802939 [DOI] [PubMed] [Google Scholar]
- 127. Sheeran P. S., Rojas J. D., Puett C., Hjelmquist J., Arena C. B., and Dayton P. A., Ultrasound Med. Biol. 41, 814 (2015). 10.1016/j.ultrasmedbio.2014.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Rapoport N., Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 4, 492 (2012). 10.1002/wnan.1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Maeda H., Wu J., Sawa T., Matsumura Y., and Hori K., J. Controlled Release 65, 271 (2000). 10.1016/S0168-3659(99)00248-5 [DOI] [PubMed] [Google Scholar]
- 130. Shpak O., Verweij M., Vos H. J., de Jong N., Lohse D., and Versluis M., Proc. Natl. Acad. Sci. U.S.A. 111, 1697 (2014). 10.1073/pnas.1312171111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Reznik N., Lajoinie G., Shpak O., Gelderblom E. C., Williams R., de Jong N., Versluis M., and Burns P. N., Ultrasound Med. Biol. 40, 1379 (2014). 10.1016/j.ultrasmedbio.2013.11.025 [DOI] [PubMed] [Google Scholar]
- 132. Wilson K., Homan K., and Emelianov S., Nat. Commun. 3, 618 (2012). 10.1038/ncomms1627 [DOI] [PubMed] [Google Scholar]
- 133. Rapoport N. Y., Gao Z., and Kennedy A., J. Natl. Cancer Inst. 99, 1095 (2007). 10.1093/jnci/djm043 [DOI] [PubMed] [Google Scholar]
- 134. Song C., Labhasetwar V., Guzman L., Topol E., and Levy R. J., Proc. Controlled Release Soc. 22, 444 (1995). [Google Scholar]
- 135. Wang S., Dai Z., Ke H., Qu E., Qi X., Zhang K., and Wang J., Eur. J. Radiol. 83, 117 (2014). 10.1016/j.ejrad.2013.09.010 [DOI] [PubMed] [Google Scholar]
- 136. Straub J. A., Chickering D. E., Church C. C., Shah B., Hanlon T., and Bernstein H., J. Controlled Release 108, 21 (2005). 10.1016/j.jconrel.2005.07.020 [DOI] [PubMed] [Google Scholar]
- 137. Kang E., Min H., Lee J., Han M., Ahn H., Yoon I.-C., Choi K., Kim K., Park K., and Kwon I., Angew. Chem., Int. Ed. 49, 524 (2010). 10.1002/anie.200903841 [DOI] [PubMed] [Google Scholar]
- 138. Delogu L. G., Vidili G., Venturelli E., Ménard-Moyon C., Zoroddu M. A., Pilo G., Nicolussi P., Ligios C., Bedognetti D., Sgarrella F., Manetti R., and Bianco A., Proc. Natl. Acad. Sci. U.S.A. 109, 16612 (2012). 10.1073/pnas.1208312109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Lajoinie G., Gelderblom E., Chlon C., Böhmer M., Steenbergen W., de Jong N., Manohar S., and Versluis M., Nat. Commun. 5, 3671 (2014). 10.1038/ncomms4671 [DOI] [PubMed] [Google Scholar]
- 140. Arnal B., Perez C., Wei C.-W., Xia J., Lombardo M., Pelivanov I., Matula T. J., Pozzo L. D., and O'Donnell M., Photoacoustics 3, 3 (2015). 10.1016/j.pacs.2014.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Lukianova-Hleb E. Y., Wagner D. S., Brenner M. K., and Lapotko D. O., Biomaterials 33, 5441 (2012). 10.1016/j.biomaterials.2012.03.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Helfield B., Black J. J., Qin B., Pacella J., Chen X., and Villanueva F. S., “ Fluid Viscosity Affects the Fragmentation and Inertial Cavitation Threshold of Lipid-Encapsulated Microbubbles,” Ultrasound Med. Biol. (to be published 2015). 10.1016/j.ultrasmedbio.2015.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Gaudron R., Warnez M. T., and Johnsen E., J. Fluid Mech. 766, 54 (2015). 10.1017/jfm.2015.7 [DOI] [Google Scholar]
- 144. Johnsen E. and Mancia L., J. Phys.: Conf. Ser. 656, 012022 (2015). 10.1088/1742-6596/656/1/012022 [DOI] [Google Scholar]
- 145. Culjat M. O., Goldenberg D., Tewari P., and Singh R. S., Ultrasound Med. Biol. 36, 861 (2010). 10.1016/j.ultrasmedbio.2010.02.012 [DOI] [PubMed] [Google Scholar]
- 146. Milane L., Ganesh S., Shah S., Duan Z.-F., and Amiji M., J. Controlled Release 155, 237 (2011). 10.1016/j.jconrel.2011.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Phan T. G. and Bullen A., Immunol. Cell Biol. 88, 438 (2010). 10.1038/icb.2009.116 [DOI] [PubMed] [Google Scholar]
- 148. Dove J. D., Murray T. W., and Borden M. A., Soft Matter 9, 7743 (2013). 10.1039/c3sm51690c [DOI] [Google Scholar]
- 149. Lajoinie G., Visscher M., Blazejewski E., Lathuile A., De Jong N., Veldhuis G., and Versluis M., “ Laser-activated microcapsules: three phase theory and single capsule behavior” (unpublished).
- 150. Mountford P. A., Thomas A. N., and Borden M. A., Langmuir 31, 4627 (2015). 10.1021/acs.langmuir.5b00399 [DOI] [PubMed] [Google Scholar]
- 151. Lapotko D., Shnip A., and Lukianova E., J. Biomed. Opt. 10, 014006 (2005). 10.1117/1.1854685 [DOI] [PubMed] [Google Scholar]
- 152. Hassan M. A., Buldakov M. A., Ogawa R., Zhao Q.-L., Furusawa Y., Kudo N., Kondo T., and Riesz P., J. Controlled Release 141, 70 (2010). 10.1016/j.jconrel.2009.08.020 [DOI] [PubMed] [Google Scholar]
- 153. Kumon R., Aehle M., Sabens D., Parikh P., Han Y., Kourennyi D., and Deng C., Ultrasound Med. Biol. 35, 494 (2009). 10.1016/j.ultrasmedbio.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Hensel K., Mienkina M. P., and Schmitz G., Ultrasound Med. Biol. 37, 2105 (2011). 10.1016/j.ultrasmedbio.2011.09.007 [DOI] [PubMed] [Google Scholar]
- 155. De Cock I., Zagato E., Braeckmans K., Luan Y., de Jong N., Smedt S. C. D., and Lentacker I., J. Controlled Release 197, 20 (2015). 10.1016/j.jconrel.2014.10.031 [DOI] [PubMed] [Google Scholar]
- 156. Kooiman K., Foppen-Harteveld M., van der Steen A. F., and de Jong N., J. Controlled Release 154, 35 (2011). 10.1016/j.jconrel.2011.04.008 [DOI] [PubMed] [Google Scholar]
- 157. Gelderblom E. C., “ Ultra-high-speed fluorescence imaging,” Ph.D. dissertation ( University of Twente, The Netherlands, 2012). [Google Scholar]
- 158. Flynn H. G., J. Acoust. Soc. Am. 57, 1379 (1975). 10.1121/1.380624 [DOI] [Google Scholar]
- 159. Tu J., Matula T. J., Brayman A. A., and Crum L. A., Ultrasound Med. Biol. 32, 281 (2006). 10.1016/j.ultrasmedbio.2005.10.001 [DOI] [PubMed] [Google Scholar]
- 160. Pommella A., Brooks N. J., Seddon J. M., and Garbin V., Sci. Rep. 5, 13163 (2015). 10.1038/srep13163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Vos J., Dollet H. B., Versluis M., and de Jong N., Ultrasound Med. Biol. 37, 935 (2011). 10.1016/j.ultrasmedbio.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 162. Oh J. S., Kwon Y. S., Lee K. H., Jeong W., Chung S. K., and Rhee K., Comput. Biol. Med. 44, 37 (2014). 10.1016/j.compbiomed.2013.10.020 [DOI] [PubMed] [Google Scholar]
- 163. Cho E., Chung S. K., and Rhee K., Ultrasonics 62, 66 (2015). 10.1016/j.ultras.2015.05.002 [DOI] [PubMed] [Google Scholar]
- 164. Garbin V., Dollet B., Overvelde M., Cojoc D., di Fabrizio E., van Wijngaarden L., Prosperetti A., de Jong N., Lohse D., and Versluis M., Phys. Fluids 21, 092003 (2009). 10.1063/1.3227903 [DOI] [Google Scholar]
- 165. Kokhuis T., Skachkov I., Naaijkens B., Juffermans L., Kamp O., Kooiman K., van der Steen A., Versluis M., and de Jong N., Biotechnol. Bioeng. 112, 220 (2015). 10.1002/bit.25337 [DOI] [PubMed] [Google Scholar]
- 166. Kudo N., Okada K., and Yamamoto K., Biophys. J. 96, 4866 (2009). 10.1016/j.bpj.2009.02.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Fan Z., Liu H., Mayer M., and Deng C. X., Proc. Natl. Acad. Sci. U.S.A. 109, 16486 (2012). 10.1073/pnas.1208198109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Schlicher R. K., Radhakrishna H., Tolentino T. P., Apkarian R. P., Zarnitsyn V., and Prausnitz M. R., Ultrasound Med. Biol. 32, 915 (2006). 10.1016/j.ultrasmedbio.2006.02.1416 [DOI] [PubMed] [Google Scholar]
- 169. Hutcheson J., Schlicher R., Hicks H., and Prausnitz M., Ultrasound Med. Biol. 36, 1008 (2010). 10.1016/j.ultrasmedbio.2010.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Guzman H. R., Nguyen D. X., Khan S., and Prausnitz M. R., J. Acoust. Soc. Am. 110, 588 (2001). 10.1121/1.1376131 [DOI] [PubMed] [Google Scholar]
- 171. Chen X., Leow R. S., Hu Y., Wan J. M. F., and Yu A. C. H., J. R. Soc. Interface 11, 1–11 (2014). 10.1098/rsif.2014.0071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Yudina A., Lepetit-Coiffé M., and Moonen C., Mol. Imaging Biol. 13, 239 (2011). 10.1007/s11307-010-0346-5 [DOI] [PubMed] [Google Scholar]
- 173. Derieppe M., Yudina A., Lepetit-Coiffé M., de Senneville B., Bos C., and Moonen C., Mol. Imaging Biol. 15, 3 (2013). 10.1007/s11307-012-0568-9 [DOI] [PubMed] [Google Scholar]
- 174. Meijering B. D., Juffermans L. J., van Wamel A., Henning R. H., Zuhorn I. S., Emmer M., Versteilen A. M., Paulus W. J., van Gilst W. H., Kooiman K., de Jong N., Musters R. J., Deelman L. E., and Kamp O., Circ. Res. 104, 679 (2009). 10.1161/CIRCRESAHA.108.183806 [DOI] [PubMed] [Google Scholar]
- 175. Karshafian R., Samac S., Bevan P. D., and Burns P. N., Ultrasonics 50, 691 (2010). 10.1016/j.ultras.2010.01.009 [DOI] [PubMed] [Google Scholar]
- 176. Mehier-Humbert S., Bettinger T., Yan F., and Guy R. H., J. Controlled Release 104, 213 (2005). 10.1016/j.jconrel.2005.01.007 [DOI] [PubMed] [Google Scholar]
- 177. Lum A. F. H., Borden M. A., Dayton P. A., Kruse D. E., Simon S. I., and Ferrara K. W., J. Controlled Release 111, 128 (2006). 10.1016/j.jconrel.2005.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Juffermans L. J. M., Dijkmans P. A., Musters R. J. P., Visser C. A., and Kamp O., Am. J. Physiol.: Heart Circ. Physiol. 291, 1595 (2006). 10.1152/ajpheart.01120.2005 [DOI] [PubMed] [Google Scholar]
- 179. Zhou Y., Kumon R. E., Cui J., and Deng C. X., Ultrasound Med. Biol. 35, 1756 (2009). 10.1016/j.ultrasmedbio.2009.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Hu Y., Wan J. M., and Yu A. C., Ultrasound Med. Biol. 39, 2393 (2013). 10.1016/j.ultrasmedbio.2013.08.003 [DOI] [PubMed] [Google Scholar]
- 181. Hu Y., Wan J. M., and Yu A. C., Ultrasound Med. Biol. 40, 1587 (2014). 10.1016/j.ultrasmedbio.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 182. Hsu Y.-H., Moya M. L., Hughes C. C. W., George S. C., and Lee A. P., Lab Chip 13, 2990 (2013). 10.1039/c3lc50424g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Takeshita Y., Obermeier B., Cotleur A., Sano Y., Kanda T., and Ransohoff R. M., J. Neurosci. Methods 232, 165 (2014). 10.1016/j.jneumeth.2014.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Schwille P., Korlach J., and Webb W. W., Cytometry 36, 176 (1999). [DOI] [PubMed] [Google Scholar]
- 185. Douglass A. D. and Vale R. D., Cell 121, 937 (2005). 10.1016/j.cell.2005.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Wawrezinieck L., Rigneault H., Marguet D., and Lenne P.-F., Biophys. J. 89, 4029 (2005). 10.1529/biophysj.105.067959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Susaki E. A., Tainaka K., Perrin D., Kishino F., Tawara T., Watanabe T. M., Yokoyama C., Onoe H., Eguchi M., Yamaguchi S., Abe T., Kiyonari H., Shimizu Y., Miyawaki A., Yokota H., and Ueda H. R., Cell 157, 726 (2014). 10.1016/j.cell.2014.03.042 [DOI] [PubMed] [Google Scholar]
- 188. Hoover E. E. and Squier J. A., Nat. Photonics 7, 93 (2013). 10.1038/nphoton.2012.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Ducourthial G., Leclerc P., Mansuryan T., Fabert M., Brevier J., Habert R., Braud F., Batrin R., Vever-Bizet C., Bourg-Heckly G., Thiberville L., Druilhe A., Kudlinski A., and Louradour F., Sci. Rep. 5, 18303 (2015). 10.1038/srep18303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Chin C. T., Lancée C., Borsboom J., Mastik F., Frijlink M. E., de Jong N., Versluis M., and Lohse D., Rev. Sci. Instrum. 74, 5026 (2003). 10.1063/1.1626013 [DOI] [Google Scholar]
- 191. Gelderblom E. C., Vos H. J., Mastik F., Faez T., Luan Y., Kokhuis T. J. A., van der Steen A. F. W., Lohse D., de Jong N., and Versluis M., Rev. Sci. Instrum. 83, 103706 (2012). 10.1063/1.4758783 [DOI] [PubMed] [Google Scholar]
- 192. van Wamel A., Bouakaz A., Versluis M., and de Jong N., Ultrasound Med. Biol. 30, 1255 (2004). 10.1016/j.ultrasmedbio.2004.07.015 [DOI] [PubMed] [Google Scholar]
- 193. Gelderblom E., Wolbers F., de Jong N., van den Berg A., and Versluis M., “ Time-resolved high-speed fluorescence imaging of microbubble-induced sonoporation” (unpublished).
- 194. Shpak O., Adan M., de Jong N., Fowlkes B., Fabiilli M., and Kripfgans O., “ Ultra high-speed imaging of bubble-cell interactions in acoustic droplet vaporization” (unpublished).
- 195. Lajoinie G., van Rooij T., Kooiman K., de Jong N., and Versluis M., “ Bubble-cell interactions with laser activated polymeric microcapsules” (unpublished).
- 196. Kooiman K., Emmer M., Foppen-Harteveld M., van Wamel A., and de Jong N., IEEE Trans. Biomed. Eng. 57, 29 (2010). 10.1109/TBME.2009.2030335 [DOI] [PubMed] [Google Scholar]
- 197. Kooiman K., van der Steen A. F. W., and de Jong N., IEEE Trans. Ultrason., Ferroelectr., Freq. Control 60, 1811 (2013). 10.1109/TUFFC.2013.2767 [DOI] [PubMed] [Google Scholar]
- 198. Cui W., Tavri S., Benchimol M. J., Itani M., Olson E. S., Zhang H., Decyk M., Ramirez R. G., Barback C. V., Kono Y., and Mattrey R. F., Biomaterials 34, 4926 (2013). 10.1016/j.biomaterials.2013.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Chung K., Wallace J., Kim S.-Y., Kalyanasundaram S., Andalman A. S., Davidson T. J., Mirzabekov J. J., Zalocusky K. A., Mattis J., Denisin A. K., Pak S., Bernstein H., Ramakrishnan C., Grosenick L., Gradinaru V., and Deisseroth K., Nature 497, 332 (2013). 10.1038/nature12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Underwood E., Science 340, 131 (2013). 10.1126/science.340.6129.131 [DOI] [PubMed] [Google Scholar]
- 201. Zeghimi A., Uzbekov R., Arbeille B., Escoffre J. M., and Bouakaz A., in 2012 IEEE International Ultrasonics Symposium (IUS) (2012), p. 2045. [Google Scholar]
- 202. Duvshani-Eshet M., Baruch L., Kesselman E., Shimoni E., and Machluf M., Gene Ther. 13, 163 (2006). 10.1038/sj.gt.3302642 [DOI] [PubMed] [Google Scholar]
- 203. Zhao Y.-Z., Luo Y.-K., Lu C.-T., Xu J.-F., Tang J., Zhang M., Zhang Y., and Liang H.-D., J. Drug Targeting 16, 18 (2008). 10.1080/10611860701637792 [DOI] [PubMed] [Google Scholar]
- 204. Leung K. S., Chen X., Zhong W., Yu A. C. H., and Lee C.-Y. J., Chem. Phys. Lipids 180, 53 (2014). 10.1016/j.chemphyslip.2014.02.004 [DOI] [PubMed] [Google Scholar]
- 205. Lai C.-Y., Wu C.-H., Chen C.-C., and Li P.-C., Ultrasound Med. Biol. 32, 1931 (2006). 10.1016/j.ultrasmedbio.2006.06.020 [DOI] [PubMed] [Google Scholar]
- 206. Leow R. S., Wan J. M. F., and Yu A. C. H., J. R. Soc. Interface 12, 1–10 (2015). 10.1098/rsif.2015.0029 [DOI] [PMC free article] [PubMed] [Google Scholar]