Abstract
Whole-genome sequencing (WGS) of 41 patient and environmental sequence type 22 methicillin-resistant Staphylococcus aureus staphylococcal cassette chromosome mec type IV (ST22-MRSA-IV) isolates recovered over 6 weeks in one acute hospital ward in Dublin, Ireland, where ST22-MRSA IV is endemic, revealed 228 pairwise combinations differing by <40 single nucleotide variants corresponding to potential cross-transmission events (CTEs). In contrast, 15 pairwise combinations of isolates representing five CTEs were previously identified by conventional molecular epidemiological typing. WGS enhanced ST22-MRSA-IV tracking and highlighted potential transmission of MRSA via the hospital environment.
TEXT
Sequence type 22 methicillin-resistant Staphylococcus aureus staphylococcal cassette chromosome mec type IV (ST22-MRSA-IV) is endemic in hospitals in Ireland and the United Kingdom and is a predominant cause of nosocomial MRSA infection in several other European countries, Asia, and Australia (1–6). ST22-MRSA-IV is highly clonal and tracking its spread is difficult (6). We previously reported the enhanced discrimination of ST22-MRSA-IV isolates from patients and hospital environmental sites using a combination of spa, dru, and pulsed-field gel electrophoresis (PFGE) typing in combination with key epidemiological data (6–8). Several studies have demonstrated the usefulness of whole-genome sequencing (WGS) for differentiating and tracking MRSA in long-term and global studies and in outbreak settings (2, 9–11). However, no studies have investigated WGS for tracking the spread of ST22-MRSA-IV in an endemic setting. Price et al. investigated the transmission of Staphylococcus aureus in an intensive care unit using WGS over 14 months and reported a low rate of patient-to-patient transmission (12). However, they concluded that important transmission events were probably not identified because environmental sites were not investigated (12). We investigated the usefulness of WGS for tracking ST22-MRSA-IV between patients and environmental sites in an endemic hospital setting and to confirm or disprove cross-transmission events (CTEs) previously identified using conventional molecular epidemiological (CME) typing.
Forty-one ST22-MRSA-IVh isolates recovered from 22 patients (one per patient) and 19 environmental sites (mattresses, bedrails, pillows, and air) in one surgical ward of a 700-bed acute care hospital in Dublin, Ireland, during a 6-week period in 2007 were investigated (6). The 35-bed ward included 6-, 4-, and 2-bed bays and five single rooms as detailed previously by Creamer et al. (8). The 41 isolates were previously characterized using staphylococcal cassette chromosome mec element (SCCmec), spa, and dru typing and PFGE with some isolates undergoing multilocus sequence typing (6).
Among these isolates, CTEs were previously identified using epidemiological information and molecular typing (7). Isolates were deemed to be part of a CTE if they were recovered from ≥2 patients or from a patient and an environmental site within a 3-week period on the same ward bay (a “probable” CTE) or on the same ward but not on the same ward bay (a “possible” CTE) (7). The MRSA status of the patient on admission, the probable source of the patient's MRSA, and the dates of admission, discharge, and first detection of MRSA were also considered (7). Isolates were only included in CTEs if they were deemed to be hospital acquired (HA) or if the patient's MRSA status was determined 72 h after ward admission. The CTEs identified using the epidemiological information were confirmed if the isolates differed by ≤1 typing method, i.e., spa, dru, or PFGE typing. With these criteria, five CTEs were identified (7) and included 5 transmitted isolates from patients with HA MRSA and 14 source isolates, 7 each from patients and environmental sites. Two isolates (M07/0339 and M07/0348, CTEs 2 and 3, respectively) were each implicated in two CTEs (Fig. 1).
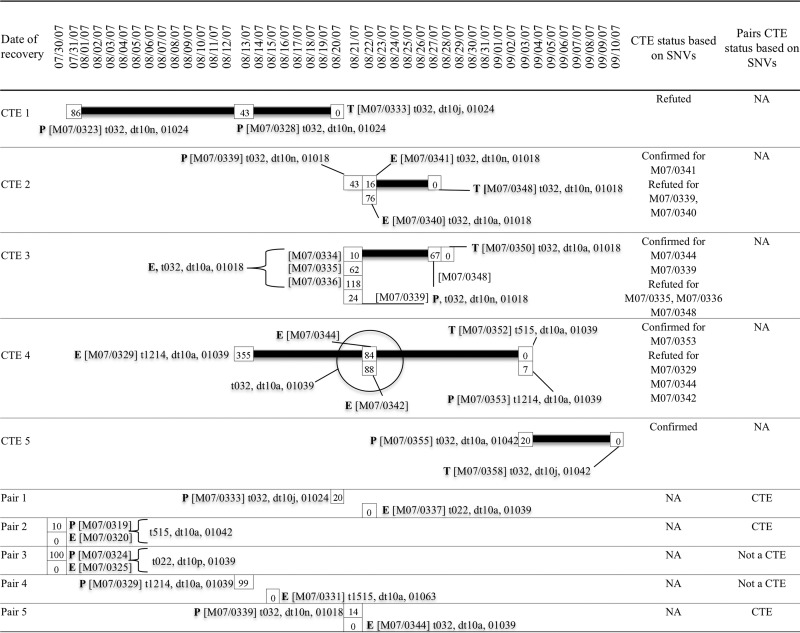
FIG 1.
Timeline showing dates of recovery of ST22-MRSA-IV isolates involved in cross-transmission events (CTEs) previously identified by conventional molecular epidemiological (CME) typing or identified as a pair of isolates recovered from a patient and his or her immediate ward environment. For each CTE, the putative source isolates recovered from patients (P) and the environment (E) as well as putative transmitted isolates (T) are shown, and for each pair of isolates, the patient and environmental isolate are also indicated. Isolate numbers are shown in brackets followed by the spa type, dru type, and PFGE type. The single nucleotide variant (SNV) comparison between each of the source isolates and the transmitted isolates within a CTE or between each pair of isolates is indicated by numerals within a square with the transmitted isolate SNV value denoted by 0 (0 SNVs resulting from self-comparison). CTEs were confirmed by SNV analysis if one or more of the source isolates differed from the transmitted isolate by ≤40 SNVs. For CTEs consisting of multiple source isolates of which some were confirmed and some were refuted as CTEs by SNV analysis, the isolate number for the CTEs either confirmed or refuted are indicated in the second to last column to the right of the figure. Pairs of isolates were confirmed as CTEs if the patient and environmental isolate differed by ≤40 SNVs. Further molecular epidemiological details of isolates implicated in each of the CTEs or as a pair of isolates are provided in Table S1 in the supplemental material and have been published previously (6, 7). NA, not applicable.
Five pairs of isolates, each consisting of one patient isolate and one immediate ward environment isolate, were also previously identified among the 41 isolates (Fig. 1) (6, 7). These included four isolates (M07/0333, M07/0329, M07/0339, and M07/0334) also implicated in CTEs. The isolates associated with 2/5 pairs (pairs 2 and 3) exhibited indistinguishable spa, dru, and PFGE types but were not included in CTEs as the patients concerned were MRSA positive on ward admission. All previously reported molecular epidemiological data for these 41 isolates are summarized in Table S1 in the supplemental material.
Genomic DNA was extracted from isolates using a Qiagen DNeasy kit according to the manufacturer's instructions (Qiagen, Crawley, United Kingdom). Nextera XT library preparation reagents were used according to the manufacturer's instruction (Illumina, Eindhoven, The Netherlands). Libraries were sequenced on an Illumina MiSeq instrument. Ridom SeqSphere+ software (Munster, Germany), which incorporates the Burrows-Wheeler aligner, was used for assembly with trimmed reads mapped against a previously described ST22-MRSA-IV genome, HO 5096 0412 (GenBank accession number HE681097), recovered in a United Kingdom acute care hospital (2, 12). The assembled genomes were further analyzed against each other with the BioNumerics genome analysis tool (GAT) (version 7.5; Applied Maths, Ghent, Belgium) using the earliest recovered isolate (M07/0319) as a reference genome. Single nucleotide variants (SNVs) were identified and confirmed if they exhibited ≥40× coverage; i.e., each SNV was covered by at least 40 reads, thereby avoiding ambiguous SNVs and increasing confidence in the SNV validity. In fact, >50% of all SNVs exhibited ≥100× coverage. All synonymous and nonsynonymous mutations were included. Insertions and deletions (indels) and repetitive regions were excluded. The genomic SNV data per isolate were compared to those for the other 40 genomes, yielding 861 pairwise comparisons.
Potential CTEs were defined as two isolates recovered at any time during the 6-week period differing by ≤40 SNVs, based on reports of up to 40 SNVs among related S. aureus isolates from outbreaks or among multiple isolates from an individual and studies that used a cutoff of ≤40 SNVs for determining CTEs (12–14).
Whole-genome sequencing of the 41 isolates yielded an average coverage of 189× per genome (range, 100 to 425×) and a total of 20,848 SNVs. Pairwise comparisons across the 41 genomes identified 228/861 pairwise comparisons, involving all 41 genomes in at least one pairwise comparison, where two isolates differed by ≤40 SNVs (range, 0 to 40 SNVs). These included (i) 110 instances, involving 40/41 isolates, where one isolate was recovered from a patient and the other from an environmental source (shaded in Fig. S1A in the supplemental material), (ii) 97 instances, involving 26/41 isolates, where both isolates were recovered from patient sources (shaded in Fig. S1B in the supplemental material), and (iii) 21 instances, involving 11/41 isolates, where both isolates were recovered from an environmental source (shaded in Fig. S1C in the supplemental material). There was no correlation between isolates within pairwise comparisons differing by ≤40 SNVs or >40 SNVs and the CME typing. Isolates differing by one, two, or three of the conventional molecular typing methods were identified among isolates within pairwise comparisons differing by ≤40 and >40 SNVs as were isolates with a range of epidemiological characteristics (see Table S2 in the supplemental material). This may be due to the low correlation between SNV analysis, which detects mutations within the core genome, and PFGE, which is affected by mobile genetic elements (15). Additionally, SNV accumulation within the spa and dru regions may not correlate with the entire genome.
In contrast to the 228 pairwise comparisons implicated as CTEs by SNV analysis, just 15/861 pairwise comparisons were associated with five CTEs using CME typing. The SNV analysis confirmed 4/5 CTEs (CTEs 2, 3, 4, and 5) involving just 5/15 pairwise comparisons as they differed by ≤40 SNVs (Fig. 1). The transmitted and 2/3 source isolates within CTE 2 were indistinguishable based on spa, dru, and PFGE typing, but one isolate (M07/0340) exhibited a different dru type (Fig. 1). However, only 1/3 source isolates (M07/0341) exhibited ≤40 SNVs compared to those for the transmitted isolate (M07/0348) (Fig. 1). Five source isolates within CTE 3 were indistinguishable from the transmitted isolate by spa and PFGE typing, but two isolates (M07/0339 and M07/0348) exhibited a different dru type. However, only two of these isolates (M07/0334 and M07/0339) exhibited ≤40 SNVs compared to those for the transmitted isolate (M07/0350), one of which exhibited the different dru type (Fig. 1). The four CTE 4 source isolates exhibited the same dru and PFGE types but a different spa type from that of the transmitted isolate, and only one of these (M07/0353) exhibited ≤40 SNV differences compared to the transmitted isolate (Fig. 1). The one source isolate within CTE 5 differed in dru type only from that of the transmitted isolate and by 20 SNVs (Fig. 1). The transmitted and source isolates within CTE 1 differed in dru type only and exhibited 43 and 86 SNVs, respectively, compared to the transmitted isolate (Fig. 1).
In relation to the five pairs of patient and environmental isolates, SNV analysis indicated that 3/5 pairs of isolates, i.e., pairs 1, 2, and 5, differed by ≤40 SNVs compared to 0/5 pairs which were assigned to CTEs by CME typing (Fig. 1). Among those that differed by ≤40 SNVs, different molecular typing results were detected in pairs 1 (differences in spa, dru, and PFGE types) and 5 (differences in dru and PFGE types) only (Fig. 1). Among the two pairs of isolates that differed by >40 SNVs, one pair exhibited identical spa, dru, and PFGE types (pair 3) and one pair differed in spa and PFGE type (pair 4) (Fig. 1).
This study highlights the increased sensitivity of WGS over CME typing for tracking the highly clonal ST22-MRSA-IV in an endemic setting. The involvement of all isolates in at least one potential CTE using WGS and the identification of 228 pairwise comparisons differing by ≤40 SNVs compared to 15 pairwise comparisons representing CTEs by CME typing highlights ST22-MRSA-IVh transmissibility and the way that MRSA transmission may be significantly underestimated or incorrectly designated by CME approaches. The hospital environment had a significant role in ST22-MRSA-IV transmission with the identification of 110 instances of isolates differing by ≤40 SNVs from patients and their immediate ward environment, a further 21 instances involving environmental sites only, and 3/5 pairs of isolates from patients and their surrounding environment that were potential CTEs. However, health care workers should also be considered a reservoir for nosocomial MRSA transmission. Further in vivo and in vitro investigations are required with SNV accumulation rates in particular MRSA clones to enable accurate inference of CTEs. This will allow more accurate assignment of SNV thresholds for defining strain relatedness as other studies used different thresholds (15). Indels were excluded from SNV analysis and might be considered in future investigations.
Supplementary Material
Funding Statement
This work was supported by the Microbiology Research Unit, Dublin Dental University Hospital, Dublin, Ireland.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02662-15.
REFERENCES
- 1.Grundmann H, Schouls LM, Aanensen DM, Pluister GN, Tami A, Chlebowicz M, Glasner C, Sabat AJ, Weist K, Heuer O, Friedrich AW. 2014. The dynamic changes of dominant clones of Staphylococcus aureus causing bloodstream infections in the European region: results of a second structured survey. Euro Surveill 19(49):pii=20987 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20987. [DOI] [PubMed] [Google Scholar]
- 2.Holden MT, Hsu LY, Kurt K, Weinert LA, Mather AE, Harris SR, Strommenger B, Layer F, Witte W, de Lencastre H, Skov R, Westh H, Zemlickova H, Coombs G, Kearns AM, Hill RL, Edgeworth J, Gould I, Gant V, Cooke J, Edwards GF, McAdam PR, Templeton KE, McCann A, Zhou Z, Castillo-Ramirez S, Feil EJ, Hudson LO, Enright MC, Balloux F, Aanensen DM, Spratt BG, Fitzgerald JR, Parkhill J, Achtman M, Bentley SD, Nubel U. 2013. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res 23:653–664. doi: 10.1101/gr.147710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim KT, Hanifah YA, Yusof MY, Goering RV, Thong KL. 2012. Temporal changes in the genotypes of methicillin-resistant Staphylococcus aureus strains isolated from a tertiary Malaysian hospital based on MLST, spa, and mec-associated dru typing. Diagn Microbiol Infect Dis 74:106–112. doi: 10.1016/j.diagmicrobio.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 4.Teo J, Tan TY, Hon PY, Lee W, Koh TH, Krishnan P, Hsu LY. 2013. ST22 and ST239 MRSA duopoly in Singaporean hospitals: 2006-2010. Epidemiol Infect 141:153–157. doi: 10.1017/S0950268812000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coombs GW, Nimmo GR, Daly DA, Le TT, Pearson JC, Tan HL, Robinson JO, Collignon PJ, McLaws ML, Turnidge JD. 2014. Australian Staphylococcus aureus Sepsis Outcome Programme annual report, 2013. Commun Dis Intell Q Rep 38:E309–E319. [DOI] [PubMed] [Google Scholar]
- 6.Shore AC, Rossney AS, Kinnevey PM, Brennan OM, Creamer E, Sherlock O, Dolan A, Cunney R, Sullivan DJ, Goering RV, Humphreys H, Coleman DC. 2010. Enhanced discrimination of highly clonal ST22-methicillin-resistant Staphylococcus aureus IV isolates achieved by combining spa, dru, and pulsed-field gel electrophoresis typing data. J Clin Microbiol 48:1839–1852. doi: 10.1128/JCM.02155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Creamer E, Shore AC, Rossney AS, Dolan A, Sherlock O, Fitzgerald-Hughes D, Sullivan DJ, Kinnevey PM, O'Lorcain P, Cunney R, Coleman DC, Humphreys H. 2012. Transmission of endemic ST22-MRSA-IV on four acute hospital wards investigated using a combination of spa, dru and pulsed-field gel electrophoresis typing. Eur J Clin Microbiol Infect Dis 31:3151–3161. doi: 10.1007/s10096-012-1678-7. [DOI] [PubMed] [Google Scholar]
- 8.Creamer E, Shore AC, Deasy EC, Galvin S, Dolan A, Walley N, McHugh S, Fitzgerald-Hughes D, Sullivan DJ, Cunney R, Coleman DC, Humphreys H. 2014. Air and surface contamination patterns of meticillin-resistant Staphylococcus aureus on eight acute hospital wards. J Hosp Infect 86:201–208. doi: 10.1016/j.jhin.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris SR, Cartwright EJ, Torok ME, Holden MT, Brown NM, Ogilvy-Stuart AL, Ellington MJ, Quail MA, Bentley SD, Parkhill J, Peacock SJ. 2013. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis 13:130–136. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Köser CU, Holden MT, Ellington MJ, Cartwright EJ, Brown NM, Ogilvy-Stuart AL, Hsu LY, Chewapreecha C, Croucher NJ, Harris SR, Sanders M, Enright MC, Dougan G, Bentley SD, Parkhill J, Fraser LJ, Betley JR, Schulz-Trieglaff OB, Smith GP, Peacock SJ. 2012. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med 366:2267–2275. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price JR, Golubchik T, Cole K, Wilson DJ, Crook DW, Thwaites GE, Bowden R, Walker AS, Peto TE, Paul J, Llewelyn MJ. 2014. Whole-genome sequencing shows that patient-to-patient transmission rarely accounts for acquisition of Staphylococcus aureus in an intensive care unit. Clin Infect Dis 58:609–618. doi: 10.1093/cid/cit807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azarian T, Cook RL, Johnson JA, Guzman N, McCarter YS, Gomez N, Rathore MH, Morris JG, Salemi M. 2015. Whole-genome sequencing for outbreak investigations of methicillin-resistant Staphylococcus aureus in the neonatal intensive care unit: time for routine practice? Infect Control Hosp Epidemiol 36:777–785. doi: 10.1017/ice.2015.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golubchik T, Batty EM, Miller RR, Farr H, Young BC, Larner-Svensson H, Fung R, Godwin H, Knox K, Votintseva A, Everitt RG, Street T, Cule M, Ip CL, Didelot X, Peto TE, Harding RM, Wilson DJ, Crook DW, Bowden R. 2013. Within-host evolution of Staphylococcus aureus during asymptomatic carriage. PLoS One 8:e61319. doi: 10.1371/journal.pone.0061319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindsay JA. 2014. Evolution of Staphylococcus aureus and MRSA during outbreaks. Infect Genet Evol 21:548–553. doi: 10.1016/j.meegid.2013.04.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.