Abstract
Recently developed ultrasensitive and quantitative methods for detection of Clostridium difficile toxins provide new tools for diagnosis and, potentially, for management of C. difficile infection (CDI). Compared to methods that detect toxigenic organism, ultrasensitive toxin detection may allow diagnosis of CDI with increased clinical specificity, without sacrificing clinical sensitivity; measurement of toxin levels may also provide information relevant to disease prognosis. This minireview provides an overview of these new toxin detection technologies and considers what these new tools might add to the field.
INTRODUCTION
Clostridium difficile is a significant nosocomial and community-acquired pathogen in adults and children, associated with disease ranging from mild diarrhea to severe pseudomembranous colitis resulting in colectomy and even death (1, 2). Transmission is primarily by person-to-person spread of spores (fecal-oral route), and prevention of transmission is significantly complicated by the high prevalence of asymptomatic colonization with C. difficile (e.g., between 5% and 50% in adult inpatient facilities [1]). Since the turn of the millennium, rates of C. difficile infection (CDI) have increased globally, concomitant with increased rates of severe clinical presentations and worsened clinical outcomes (1, 3). A recent U.S. prevalence survey of health care-associated infections (HAI) (4) found that C. difficile was the most commonly reported pathogen, causing 12.1% of HAI. Despite available therapies, treatment failure and relapse are common (1).
C. difficile isolates can be either nontoxigenic or toxigenic (producing toxins A and B); nontoxigenic strains are not considered to be pathogenic. Exposure to antibiotics increases the risk of CDI by disrupting the normal bowel flora and allowing the opportunistic proliferation of toxigenic C. difficile. These high-molecular-mass protein exotoxins (308 and 270 kDa, respectively) are immunologically and biologically distinct; depending on the experimental system used, the activity of each has been described in the literature as proinflammatory, cytotoxic, and enterotoxic (5, 6). Most strains produce both toxins A and B, though a minority of disease-causing strains produce toxin B only (see, e.g., reference 7). Toxins A and B are the primary virulence factors contributing to the pathogenesis of CDI (6, 8, 9), and the genes for these toxins (tcdA and tcdB) are colocated in a pathogenicity locus in toxigenic strains (5, 10). Importantly, these genes are under complex regulatory control and expression of toxin proteins is impacted by numerous environmental factors, including temperature, carbon source/amino acid availability, and antibiotic concentration (10, 11). A recent paper also provided evidence for regulation of toxin production by a quorum sensing system, with toxin synthesis being absent at low bacterial concentrations (12). While each of the two toxins has been shown to be independently capable of causing disease, the relative contributions of the two toxin proteins to disease remain unclear (see, e.g., references 6, 8, 13, and 14), in part due to differences in experimental systems (animals versus humans, purified toxins versus natural infection) and clinical contexts (adults versus children). In short, many complex and important questions remain regarding these toxins and the overall pathogenesis of CDI—questions that could begin to be addressed with a tool with which to sensitively detect and separately quantify toxins A and B in stool.
CURRENT DIAGNOSTIC STRATEGIES AND THEIR LIMITATIONS
Because toxin is necessary for disease, qualitative enzyme immunoassays (EIAs) that detect these toxins in stool were for many years the mainstay of diagnosis, used by more than 90% of U.S. laboratories (1). However, these assays are significantly limited in sensitivity (52% to 75% versus toxigenic culture [TC; see below] [15, 16]). In contrast, the assays have high (96% to 98%) specificity versus TC (15, 16). Attempts have been made to increase sensitivity by combining detection of a more sensitive but less specific target, glutamate dehydrogenase (GDH), with detection of toxin; however, this test must be followed by nucleic acid amplification testing (NAAT; see below) to resolve discordant results (GDH positive [GDH+]/toxin negative [toxin−]), increasing cost and time to results (5).
The test historically used as the laboratory gold standard, TC (in which C. difficile is cultured from stool and isolates are tested for cytotoxin production by cytotoxicity assay [3, 5]), has limited utility for clinical diagnosis. TC methods are slow (requiring 72 to 96 h), nonstandardized, and unsuitable for routine clinical testing. An additional limitation lies in the fact that TC examines toxin production in vitro, which may not reflect the strain's production of toxins in the highly variable in vivo environment. Notably, Akerlund et al. (17) demonstrated no correlation between fecal toxin levels and toxin yields in vitro for given isolates or between in vitro yields and disease severity.
An alternative reference standard that detects toxin directly in stool filtrate is the cell culture cytotoxicity assay, which detects characteristic cell rounding in the presence of functional toxin. This qualitative, subjective assay is approximately 86% sensitive compared to TC (16) and primarily detects toxin B, which is far more potent than toxin A in this assay (18); specificity of cytotoxicity is confirmed by neutralizing antitoxin antibodies. Like TC, this assay is slow (requiring 24 to 48 h of incubation), nonstandardized, and not widely used for clinical testing. However, a recent United Kingdom-based study (19) compared TC with cytotoxicity testing on more than 12,000 specimens and correlated results with clinical data. While positive cytotoxicity assay results correlated with increased mortality, the combination of positive TC and negative cytotoxicity assay results did not, indicating that the actual presence of toxin (and not just the presence of toxigenic C. difficile) was of primary importance. The authors concluded that “detection of toxin is an essential step in the diagnosis of C. difficile infection” and proposed a new diagnostic category of “C. difficile excretor” (TC positive but cytotoxicity assay negative) to characterize patients without CDI but with possible colonization.
Given the suboptimal sensitivity of EIAs and the complexities and delayed turnaround times of the cytotoxicity assay (and TC), many laboratories have turned to NAAT for detection of the tcdA and tcdB genes, with its potential for high sensitivity and short turnaround time (despite potentially higher expense). However, despite relatively high sensitivity and specificity versus TC (90% and 96%, respectively, in a large comparison study [20]), the use of NAAT (like that of TC) is confounded by its inability to distinguish disease from colonization (5, 21). The problem remains that positive NAAT results indicate the presence of organisms capable of producing toxin—not whether (or at what levels) they are actually producing it in vivo. While this information may be optimal for determining the need for infection control measures, it is not necessarily optimal for deciding whether or not C. difficile is the cause of the patient's symptoms (22), and patients should be selected appropriately for testing with this highly sensitive method.
IS IT PREFERABLE TO DETECT TOXINS OR TOXIGENIC ORGANISMS?
Whether detection of toxins (EIA, cytotoxicity assay) or detection of toxigenic organisms (NAAT, TC) has higher clinical utility for diagnosis of CDI clearly remains controversial. Arguing for the higher utility of toxin detection, multiple studies comparing the clinical features of patients with different test outcomes have demonstrated that NAAT-positive, toxin-negative patients have milder symptoms than NAAT-positive, toxin-positive patients (see, e.g., references 22, 23, and 24), and others have shown that toxin-positive patients have higher mortality than toxin-negative patients (see, e.g., references 19, 22, 25, 26, and 27). Further arguing for the clinical utility of toxin detection, disease severity has been correlated to stool toxin levels in some preliminary studies (17, 19, 25, 28, 29), suggesting that the ability to quantify toxin levels in stool could potentially be clinically valuable to predict disease and treatment outcomes and in identifying those who need aggressive therapy. Recent data (30) indicate that toxins also may be detectable in blood in some individuals with CDI, providing another potential use for an ultrasensitive toxin detection tool. However, arguing against the utility of toxin detection (and potentially against ultrasensitive detection, in particular), it must be noted that multiple studies have detected toxin in the stool of some asymptomatic individuals (see, e.g., references 31, 32, and 33) and that even after effective therapy (i.e., with clinical improvement) toxin may remain detectable in stool in some patients (see, e.g., references 34 and 35). Clearly, while toxin is necessary for clinical disease, it is not sufficient, in that toxin can be present in stool in the absence of symptoms; the impact of host immunity on detectable toxin levels also remains unclear.
NOVEL APPROACHES TO ULTRASENSITIVE TOXIN DETECTION
Given the limitations of existing diagnostic testing and the building body of evidence that detection of toxins (rather than toxigenic organisms) has the highest clinical utility, the field would seem to be poised for a simple toxin detection test that combines high analytical sensitivity with the clinical specificity of toxin detection. In considering the potential utility of ultrasensitive assay technologies, an important initial question is what analytical sensitivity should be targeted for assay development. The current analytical limits of detection (LODs) for some of the highest-performing EIAs (16) range from 0.8 to 2.5 ng/ml, i.e., ∼1 ng/ml, in stool (36, 37). Ryder et al. (28; discussed below) have described a cell-based assay for quantification of toxin B in stool and calculated toxin concentrations down to as low as 30 pg/ml. Their data indicated that almost half of the toxin-positive specimens in their study would not be detected by EIAs with LODs of ∼1 ng/ml. Conventional cytotoxicity assays have demonstrated analytical LODs far below those of EIA for detection of toxin B in buffer (e.g., 1.5 pg/ml [38]), but achievable LODs for detection of toxins in stool samples appear to be higher (29, 39). Older literature (40) states that “1 pg of toxin B is sufficient to cause rounding of the cells” in this assay format, but how this corresponds to an actual concentration of toxin in stool is unclear.
A team led by Yi-Wei Tang, in collaboration with ACEA Biosciences (San Diego), has developed a real-time cellular analysis (RTCA) assay for detection of functional C. difficile toxin B directly from stool. This assay applies samples to cultured HS27 cells dispensed in microwells with imbedded electrodes and then measures cell status over time in “cell index” (CI) units, based on changes in electrode impedance with cytotoxic effects (changes in cell number, morphology, and spreading). The first generation of this assay (28) had a LOD for toxin B of 0.2 ng/ml and took >48 h to run. A second-generation assay (“RTCA2” [29]) incorporates a front-end sample processing step (“immunomagnetic separation enrichment process”) in which toxin B is first captured from diluted stool supernatant via magnetic beads coated with toxin B-specific monoclonal antibodies (described as “nonneutralizing”); after elution, captured toxins are inoculated to cultured HS27 cells as described above. The cells are precultured (with CI monitoring, establishing a baseline CI) for 18 to 24 h on the specialized plates prior to addition of sample eluates, after which incubation of up to 36 h (with monitoring at 5-min intervals) is required for CI measurements. The time point at which the normalized cell index (nCI) has dropped by 30% is considered a “positive time step” (PST) which in turn is used to calculate toxin concentrations in samples by comparison to a standard curve generated with buffer spiked with purified toxin B (List Biological Laboratories [Campbell, CA]), also taken through the immunocapture procedure. The calculated LOD for RTCA2 for detection of toxin in stool was 0.12 ng/ml (29). Among the 51 specimens that tested positive by RTCA2 during assay validation, the mean PST ranged from 1.43 to 35.85 h; the total turnaround time was approximately 60 h. Sensitivity and specificity of the RTCA2 versus quantitative toxigenic culture (qTC) were 96.2% and 99.7%, respectively, which were similar to the performance of NAAT (Xpert C. difficile assay [Cepheid, Sunnyvale, CA]) versus qTC (100.0% and 99.7%, respectively).
The authors (29) also performed a retrospective chart review to evaluate CDI severity (broken into 6 clinical categories, including no CDI) in study cohort patients. In the 51 patients whose stools tested positive by RTCA2, comparisons of measured toxin B concentrations across the 5 represented severity score groups indicated a correlation between toxin concentration and clinical CDI severity (R2 = 0.427, P = 0.002), though no significant correlation between clinical CDI severity and threshold cycle (CT) values (Xpert) or toxigenic C. difficile bacterial loads by qTC were observed. Notably, 14/51 patients were determined not to have CDI (severity group 1); the mean stool toxin B concentration in this group as measured by RTCA2 was 2.22 ng/ml (median, 1.59 ng/ml). The fact that this measured concentration is above the LOD of standard EIA, in combination with the very high sensitivity of the RTCA2 assay versus qTC and NAAT in this study (see above), might raise some uncertainty about the accuracy of the assay's calibration curve. However, given that this assay methodology measures concentrations of functional toxin rather than total toxin as detected by immunoassay, it is difficult to directly compare the LODs of the two types of assays. Measuring functional, and thus biologically relevant, toxin is one advantage of this type of assay. However, it should be noted here that cytotoxicity assays by definition measure only one aspect of toxin function and furthermore predominantly detect toxin B, thus potentially underestimating the contributions of toxin A to disease. The authors note the potential disadvantages of prolonged turnaround time and complexity of the RTCA2 assay and are working to improve both. Importantly, differential immunodetection of toxin B from highly virulent strains of C. difficile has recently been demonstrated (41), making it imperative that the front-end immunocapture step of this assay be performed with antibodies that have been shown to detect toxin B from all clinically relevant C. difficile strains.
Investigators from the Feng laboratory (18) developed a cell-based immunocytotoxicity assay similarly based on a real-time cell electronic sensing system (xCELLigence; Roche Applied Science, Indianapolis, IN), with readouts in CI units. These investigators uniquely utilized a mouse monoclonal antibody (A1H3) against toxin A to enhance its cytotoxic effect on mRG1-1 cells (an engineered CHO cell line expressing murine FcγRI-α chain [18]) attached to the bottom of microelectrode-embedded microplate wells. Using this method, the team achieved a sensitivity for toxin A of 0.1 to 1 pg/ml in buffer and also managed to detect toxin activity in a small number of porcine stool samples (the LOD for toxin detection in stool was not reported). Overall turnaround time for this assay was as short as 3 to 4 h, achieved by adding freshly thawed mRG1-1 cells (from cryopreservation) together with toxins to the microplate wells and thus avoiding a prolonged preculture step.
An alternative immunoassay approach to ultrasensitive toxin detection has recently been developed based on single-molecule array (Simoa) technology (39). Simoa technology (Quanterix; Lexington, MA), also known as “digital enzyme-linked immunosorbent assay (ELISA),” is based on efficient capture, labeling, and detection of single protein molecules on paramagnetic beads in arrays of femtoliter-sized wells; in terms of achievable LODs, digital ELISA is typically 1,000-fold more sensitive than conventional ELISA (42). Beth Israel Deaconess Medical Center investigators, in collaboration with Quanterix, developed digital ELISAs for toxins A and B and validated the assays using both culture filtrates prepared from a panel of clinical C. difficile strains (representing the most common strains in circulation) and adult clinical stool specimens submitted to the hospital laboratory for routine testing (NAAT) for C. difficile. The digital ELISAs detected toxins A and B produced by all of the strains in the panel and detected native toxins in stool with LODs of 0.45 pg/ml (toxin A) and 1.50 pg/ml (toxin B), respectively, as calibrated against purified native toxins spiked into NAAT-negative stool samples. Total assay time was 69 min, and sample processing prior to testing was minimal (dilution and filtration); assays were performed on an automated platform (“HD-1”).
For validation of the digital ELISAs, 149 clinical stool samples (previously tested by NAAT [illumigene, Meridian Bioscience, Inc.]) were tested by TC (followed by restriction endonuclease analysis [REA] typing of any C. difficile isolates), cytotoxicity assay, and the digital ELISAs in parallel. A clinical cutoff for positive results for each digital ELISA was established by averaging the Simoa signal values (average enzymes per bead, or AEB) for “true negative” samples (negative by NAAT, TC, and cytotoxicity assay), plus 3 standard deviations of that mean. The calculated assay cutoffs were 29.4 pg/ml (toxin A) and 23.3 pg/ml (toxin B), respectively; with these cutoffs, the specificities of the digital ELISA in the true-negative group were 96% and 98%, respectively. Toxin concentrations in clinical samples as measured by digital ELISAs spanned a >4-log dynamic range (Fig. 1). As expected, despite the low LOD for the toxin B digital ELISA, 16/65 (25%) samples that were positive by NAAT and 14/63 (22%) samples that were positive by TC were negative by the toxin B digital ELISA, consistent with the presence of organism (but minimal or no toxin) in those samples. However, 34/34 (100%) samples positive by cytotoxicity assay were positive by the toxin B digital ELISA (Fig. 1). There were four samples which were positive by TC and cytotoxicity assay but negative by toxin A digital ELISA; REA typing of isolates obtained from TC confirmed that all four were REA type CF (known to produce toxin B but not toxin A). Mean toxin levels (toxin [A], toxin [B], or toxin [A plus B]) in the 5 subjects with CDI-attributable severe outcomes were higher (1.7-fold, 1.5-fold, and 1.6-fold, respectively) than mean toxin levels in the 68 subjects without CDI-attributable severe outcomes, though these trends did not reach statistical significance (P = 0.10, 0.18, and 0.08, respectively). Notably, the sensitivity of the toxin B digital ELISA versus TC (78% [39]) was much lower than the sensitivity reported for the RTCA2 assay versus quantitative TC (96.2% [29]), despite the digital ELISA having a significantly lower reported LOD than the RTCA2 as detailed above. Both toxin B digital ELISA and RTCA2 had high specificity versus TC (97% and 99.7%, respectively), and in both studies, NAAT and TC results were tightly correlated, suggesting similar performances of the TC in both studies. It is difficult to explain this discrepancy without a direct comparison of the two assays, given the different methodologies used for toxin detection, different standards used for assay calibration, and potential differences in sample handling and clinical cohorts studied.
FIG 1.
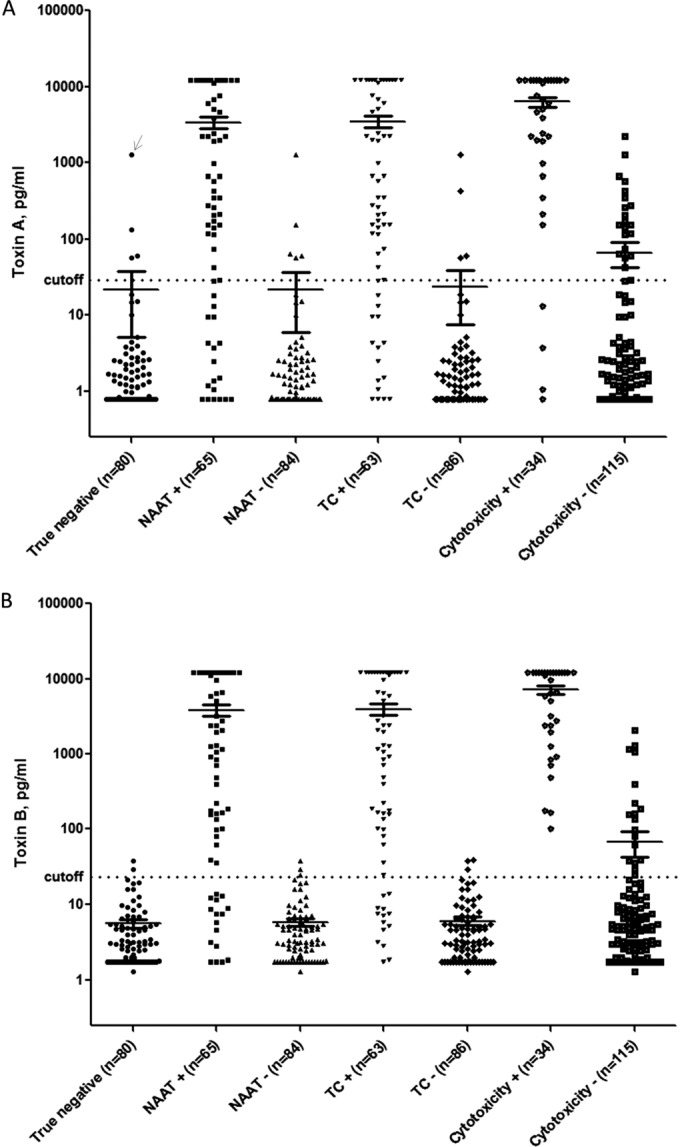
Toxin A and B digital ELISA results (39). Toxin A and B digital ELISA results for groups of samples testing positive versus negative on other assays (i.e., NAAT+ versus NAAT−, TC+ versus TC−, and cytotoxicity+ versus cytotoxicity−) are shown in Fig. 1A and B, respectively. Mean signals in each group are indicated by horizontal lines. The calculated clinical cutoffs for each digital ELISA (29.4 pg/ml for the toxin A assay and 23.3 pg/ml for the toxin B assay) are shown as dotted lines spanning each panel. The arrow in Fig. 1A indicates a sample that was excluded from calculation of the cutoff because it was an extreme outlier and substantially distorted the mean for that assay. NAAT, nucleic acid amplification testing; TC, toxigenic culture. (Reprinted from reference 39.)
Alternative immunoassay approaches to ultrasensitive toxin detection are currently in early development, including two based on sandwich-type electrochemical immunosensor methodology (43, 44). Preliminarily, both methods appear to be able to detect toxins in buffer with analytical LODs of ≤1 pg/ml; definition of LOD for toxin in stool samples and, ultimately, assay validation using well-characterized clinical samples will provide important detail as to the potential clinical utility of these approaches.
In summary, a tool capable of sensitive detection and quantification of C. difficile toxins in stool offers significant potential for improvements to the current paradigm for diagnosis of CDI. Compared to methods that detect toxigenic organism, ultrasensitive toxin detection may allow diagnosis of CDI with increased clinical specificity, without sacrificing clinical sensitivity. For such a tool to be suitable for clinical use, it must improve on the complexity and lengthy turnaround times limiting cytotoxicity assays, while also improving on the sensitivity limitations of currently available EIAs; optimally, the new tool will be rapid, robust, and simple to use. Future studies should focus on determining the clinical diagnostic and prognostic value of ultrasensitive detection and quantification of stool toxins (both A and B) in symptomatic patients, as well as the clinical significance of TC or NAAT positivity in the absence of detectable toxin. Optimization of clinical cutoffs for these ultrasensitive assays may be refined by analysis of toxin presence and quantity in asymptomatic hosts and the potential impact of host factors (particularly host antitoxin antibodies) on disease expression. If a direct and definitive correlation between toxin quantities and clinical course were shown, this new tool would have not only diagnostic but also prognostic value, allowing toxin measurements made at the time of diagnosis to influence management decisions—a rational yet entirely new direction for the field.
Biography

Nira Pollock completed her M.D./Ph.D. at the University of California, San Francisco, an internal medicine residency at Brigham and Women’s Hospital (Boston, MA), and infectious diseases and clinical microbiology training at Beth Israel Deaconess Medical Center (BIDMC; Boston, MA). She is currently the associate medical director of the Infectious Diseases Diagnostic Laboratory at Boston Children's Hospital and a member of the Division of Infectious Diseases at BIDMC. She has an active research program focused on the development and evaluation of diagnostic tests for infectious diseases and related applications. Her diagnostics research has spanned a range of diseases, including C. difficile infection, active and latent tuberculosis, Ebola virus disease, influenza, and Lyme disease, and has involved many different technologies, ranging from simple paper-based lateral-flow and microfluidic platforms to automated single-molecule array (Simoa) technology.
REFERENCES
- 1.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect Control Hosp Epidemiol 31:431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 2.Schutze GE, Willoughby RE. 2013. Clostridium difficile infection in infants and children. Pediatrics 131:196–200. doi: 10.1542/peds.2012-2992. [DOI] [PubMed] [Google Scholar]
- 3.Gilligan PH. 2015. Optimizing the laboratory diagnosis of Clostridium difficile infection. Clin Lab Med 35:299–312. doi: 10.1016/j.cll.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK. 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burnham CA, Carroll KC. 2013. Diagnosis of Clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin Microbiol Rev 26:604–630. doi: 10.1128/CMR.00016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyras D, O'Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, Poon R, Adams V, Vedantam G, Johnson S, Gerding DN, Rood JI. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson S, Kent SA, O'Leary KJ, Merrigan MM, Sambol SP, Peterson LR, Gerding DN. 2001. Fatal pseudomembranous colitis associated with a variant Clostridium difficile strain not detected by toxin A immunoassay. Ann Intern Med 135:434–438. doi: 10.7326/0003-4819-135-6-200109180-00012. [DOI] [PubMed] [Google Scholar]
- 8.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713. doi: 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 9.Carter GP, Chakravorty A, Pham Nguyen TA, Mileto S, Schreiber F, Li L, Howarth P, Clare S, Cunningham B, Sambol SP, Cheknis A, Figueroa I, Johnson S, Gerding D, Rood JI, Dougan G, Lawley TD, Lyras D. 2015. Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during Clostridium difficile infections. mBio 6:e00551. doi: 10.1128/mBio.00551-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antunes A, Dupuy B. 2010. Molecular methods to study transcriptional regulation of Clostridium difficile toxin genes. Methods Mol Biol 646:93–115. doi: 10.1007/978-1-60327-365-7_7. [DOI] [PubMed] [Google Scholar]
- 11.Karlsson S, Burman LG, Akerlund T. 2008. Induction of toxins in Clostridium difficile is associated with dramatic changes of its metabolism. Microbiology 154:3430–3436. doi: 10.1099/mic.0.2008/019778-0. [DOI] [PubMed] [Google Scholar]
- 12.Darkoh C, DuPont HL, Norris SJ, Kaplan HB. 2015. Toxin synthesis by Clostridium difficile is regulated through quorum signaling. mBio 6:e02569. doi: 10.1128/mBio.02569-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kader HA, Piccoli DA, Jawad AF, McGowan KL, Maller ES. 1998. Single toxin detection is inadequate to diagnose Clostridium difficile diarrhea in pediatric patients. Gastroenterology 115:1329–1334. doi: 10.1016/S0016-5085(98)70009-5. [DOI] [PubMed] [Google Scholar]
- 14.Riegler M, Sedivy R, Pothoulakis C, Hamilton G, Zacherl J, Bischof G, Cosentini E, Feil W, Schiessel R, LaMont JT. 1995. Clostridium difficile toxin B is more potent than toxin A in damaging human colonic epithelium in vitro. J Clin Invest 95:2004–2011. doi: 10.1172/JCI117885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crobach MJ, Dekkers OM, Wilcox MH, Kuijper EJ. 2009. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): data review and recommendations for diagnosing Clostridium difficile-infection (CDI). Clin Microbiol Infect 15:1053–1066. doi: 10.1111/j.1469-0691.2009.03098.x. [DOI] [PubMed] [Google Scholar]
- 16.Eastwood K, Else P, Charlett A, Wilcox M. 2009. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol 47:3211–3217. doi: 10.1128/JCM.01082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akerlund T, Svenungsson B, Lagergren A, Burman LG. 2006. Correlation of disease severity with fecal toxin levels in patients with Clostridium difficile-associated diarrhea and distribution of PCR ribotypes and toxin yields in vitro of corresponding isolates. J Clin Microbiol 44:353–358. doi: 10.1128/JCM.44.2.353-358.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He X, Wang J, Steele J, Sun X, Nie W, Tzipori S, Feng H. 2009. An ultrasensitive rapid immunocytotoxicity assay for detecting Clostridium difficile toxins. J Microbiol Methods 78:97–100. doi: 10.1016/j.mimet.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Planche TD, Davies KA, Coen PG, Finney JM, Monahan IM, Morris KA, O'Connor L, Oakley SJ, Pope CF, Wren MW, Shetty NP, Crook DW, Wilcox MH. 2013. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C difficile infection. Lancet Infect Dis 13:936–945. doi: 10.1016/S1473-3099(13)70200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deshpande A, Pasupuleti V, Rolston DD, Jain A, Deshpande N, Pant C, Hernandez AV. 2011. Diagnostic accuracy of real-time polymerase chain reaction in detection of Clostridium difficile in the stool samples of patients with suspected Clostridium difficile Infection: a meta-analysis. Clin Infect Dis 53:e81–90. doi: 10.1093/cid/cir505. [DOI] [PubMed] [Google Scholar]
- 21.Wilcox MH, Planche T, Fang FC, Gilligan P. 2010. What is the current role of algorithmic approaches for diagnosis of Clostridium difficile infection? J Clin Microbiol 48:4347–4353. doi: 10.1128/JCM.02028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polage CR, Gyorke CE, Kennedy MA, Leslie JL, Chin DL, Wang S, Nguyen HH, Huang B, Tang YW, Lee LW, Kim K, Taylor S, Romano PS, Panacek EA, Goodell PB, Solnick JV, Cohen SH. 2015. Overdiagnosis of Clostridium difficile infection in the molecular test era. JAMA Intern Med 175:1792–1801. doi: 10.1001/jamainternmed.2015.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Longtin Y, Trottier S, Brochu G, Paquet-Bolduc B, Garenc C, Loungnarath V, Beaulieu C, Goulet D, Longtin J. 2013. Impact of the type of diagnostic assay on Clostridium difficile infection and complication rates in a mandatory reporting program. Clin Infect Dis 56:67–73. doi: 10.1093/cid/cis840. [DOI] [PubMed] [Google Scholar]
- 24.Beaulieu C, Dionne LL, Julien AS, Longtin Y. 2014. Clinical characteristics and outcome of patients with Clostridium difficile infection diagnosed by PCR versus a three-step algorithm. Clin Microbiol Infect 20:1067–1073. doi: 10.1111/1469-0691.12676. [DOI] [PubMed] [Google Scholar]
- 25.Polage CR, Chin DL, Leslie JL, Tang J, Cohen SH, Solnick JV. 2012. Outcomes in patients tested for Clostridium difficile toxins. Diagn Microbiol Infect Dis 74:369–373. doi: 10.1016/j.diagmicrobio.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker I, Leeming JP, Reynolds R, Ibrahim I, Darley E. 2013. Clinical relevance of a positive molecular test in the diagnosis of Clostridium difficile infection. J Hosp Infect 84:311–315. doi: 10.1016/j.jhin.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 27.Walker AS, Eyre DW, Wyllie DH, Dingle KE, Griffiths D, Shine B, Oakley S, O'Connor L, Finney J, Vaughan A, Crook DW, Wilcox MH, Peto TE. 2013. Relationship between bacterial strain type, host biomarkers, and mortality in Clostridium difficile infection. Clin Infect Dis 56:1589–1600. doi: 10.1093/cid/cit127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryder AB, Huang Y, Li H, Zheng M, Wang X, Stratton CW, Xu X, Tang YW. 2010. Assessment of Clostridium difficile infections by quantitative detection of tcdB toxin by use of a real-time cell analysis system. J Clin Microbiol 48:4129–4134. doi: 10.1128/JCM.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang B, Jin D, Zhang J, Sun JY, Wang X, Stiles J, Xu X, Kamboj M, Babady NE, Tang YW. 2014. Real-time cellular analysis coupled with a specimen enrichment accurately detects and quantifies Clostridium difficile toxins in stool. J Clin Microbiol 52:1105–1111. doi: 10.1128/JCM.02601-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu H, Chen K, Wu J, Yang Z, Shi L, Barlow LL, Aronoff DM, Garey KW, Savidge TC, von Rosenvinge EC, Kelly CP, Feng H. 2015. Identification of toxemia in patients with Clostridium difficile infection. PLoS One 10:e0124235. doi: 10.1371/journal.pone.0124235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McFarland LV, Mulligan ME, Kwok RY, Stamm WE. 1989. Nosocomial acquisition of Clostridium difficile infection. N Engl J Med 320:204–210. doi: 10.1056/NEJM198901263200402. [DOI] [PubMed] [Google Scholar]
- 32.Kyne L, Warny M, Qamar A, Kelly CP. 2000. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N Engl J Med 342:390–397. doi: 10.1056/NEJM200002103420604. [DOI] [PubMed] [Google Scholar]
- 33.Koo HL, Van JN, Zhao M, Ye X, Revell PA, Jiang ZD, Grimes CZ, Koo DC, Lasco T, Kozinetz CA, Garey KW, Dupont HL. 2014. Real-time polymerase chain reaction detection of asymptomatic Clostridium difficile colonization and rising C. difficile-associated disease rates. Infect Control Hosp Epidemiol 35:667–673. doi: 10.1086/676433. [DOI] [PubMed] [Google Scholar]
- 34.Wullt M, Odenholt I. 2004. A double-blind randomized controlled trial of fusidic acid and metronidazole for treatment of an initial episode of Clostridium difficile-associated diarrhoea. J Antimicrob Chemother 54:211–216. doi: 10.1093/jac/dkh278. [DOI] [PubMed] [Google Scholar]
- 35.Louie TJ, Miller MA, Mullane KM, Weiss K, Lentnek A, Golan Y, Gorbach S, Sears P, Shue YK. 2011. Fidaxomicin versus vancomycin for Clostridium difficile infection. N Engl J Med 364:422–431. doi: 10.1056/NEJMoa0910812. [DOI] [PubMed] [Google Scholar]
- 36.Meridian Bioscience Inc. 2009. Premier toxins A & B package insert. Meridian Bioscience, Inc., Nice, France: http://www.meridianbioscience.com/Content/Assets/Files/2.1%20%20C.%20difficile%20Products/Package-Insert-Premier-Toxins-A-and-B.pdf. [Google Scholar]
- 37.TechLab. 2008. C. difficile Tox A/B II package insert. TechLab, Inc., Blacksburg, VA: http://www.techlab.com/wp-content/uploads/2013/06/t5015insert_rev_0308.pdf. [Google Scholar]
- 38.Kelly CP, Pothoulakis C, Vavva F, Castagliuolo I, Bostwick EF, O'Keane JC, Keates S, LaMont JT. 1996. Anti-Clostridium difficile bovine immunoglobulin concentrate inhibits cytotoxicity and enterotoxicity of C. difficile toxins. Antimicrob Agents Chemother 40:373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song L, Zhao M, Duffy DC, Hansen J, Shields K, Wungjiranirun M, Chen X, Xu H, Leffler DA, Sambol SP, Gerding DN, Kelly CP, Pollock NR. 22 July 2015. Development and validation of digital ELISAs for ultrasensitive detection and quantification of C. difficile toxins in stool. J Clin Microbiol doi: 10.1128/JCM.01334-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyerly DM, Krivan HC, Wilkins TD. 1988. Clostridium difficile: its disease and toxins. Clin Microbiol Rev 1:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollock NR, Song L, Zhao M, Duffy DC, Chen X, Sambol SP, Gerding DN, Kelly CP. 2015. Differential immunodetection of toxin B from highly virulent Clostridium difficile BI/NAP-1/027. J Clin Microbiol 53:1705–1708. doi: 10.1128/JCM.03419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, Ferrell EP, Randall JD, Provuncher GK, Walt DR, Duffy DC. 2010. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol 28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Z, Shi L, Feng H, Zhou HS. 2015. Single domain antibody coated gold nanoparticles as enhancer for Clostridium difficile toxin detection by electrochemical impedance immunosensors. Bioelectrochemistry 101:153–158. doi: 10.1016/j.bioelechem.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang YS, Chen SY, Huang XJ, Wang LS, Wang HY, Wang JF. 2014. Simple approach for ultrasensitive electrochemical immunoassay of Clostridium difficile toxin B detection. Biosens Bioelectron 53:238–244. doi: 10.1016/j.bios.2013.09.063. [DOI] [PubMed] [Google Scholar]


