Abstract
The mitochondrial replicative DNA helicase is essential for animal mitochondrial DNA (mtDNA) maintenance. Deleterious mutations in the gene that encodes it cause mitochondrial dysfunction manifested in developmental delays, defects and arrest, limited life span, and a number of human pathogenic phenotypes that are recapitulated in animals across taxa. In fact, the replicative mtDNA helicase was discovered with the identification of human disease mutations in its nuclear gene, and based upon its deduced amino acid sequence homology with bacteriophage T7 gene 4 protein (T7 gp4), a bi-functional primase-helicase. Since that time, numerous investigations of its structure, mechanism, and physiological relevance have been reported, and human disease alleles have been modeled in the human, mouse, and Drosophila systems. Here, we review this literature and draw evolutionary comparisons that serve to shed light on its divergent features.
Keywords: Animal models, DNA helicase, DNA replication, enzymology, mitochondria
Structure and catalytic activity
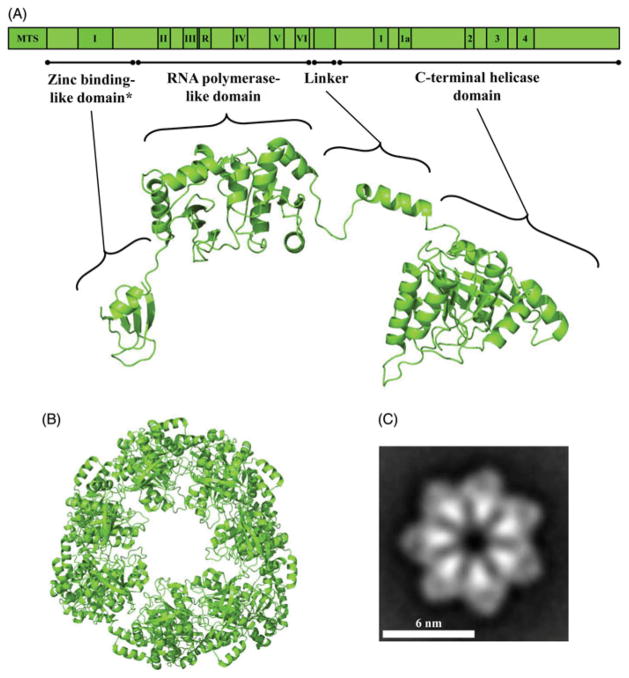
DNA helicases are essential components of the DNA replication, repair and recombination machinery across taxa. The replicative DNA helicases are members of the AAA+ family of ATPases and contain a conserved alpha–beta core structure domain that carries the conserved amino acid sequence motifs required for nucleotide hydrolysis (Caruthers & McKay, 2002; Walker et al., 1982). Additional distinct domains confer functional specificity and together with the core domain, impart a modular architecture. Our group probed the modular nature of a recombinant form of the human mitochondrial DNA (mtDNA) helicase (Ziebarth et al., 2007). We first documented its hexameric quaternary structure by velocity sedimentation and gel filtration, which yielded a sedimentation coefficient of 13.6 S and a Stokes radius of 80 Å, with an estimated native molecular mass of 420 kDa. Limited proteolysis of the hexameric helicase with trypsin resulted in the rapid disappearance of its 72 kDa protomer, giving rise to a stable product of 66 kDa (T66) that we documented to be a C-terminal truncation, and thereafter a relatively stable 57 kDa polypeptide (T57) that proved to have an additional N-terminal deletion. Subsequent products of 50, 34, 20 and 28 kDa were less stable but all were produced by several proteases, reminiscent of the modular architecture of the T7 gp4 and E. coli DnaB proteins (Bird et al., 1997; Frick et al., 1998; Washington et al., 1996). In fact, the C-terminal region of human mtDNA helicase that is removed in T66 represents an extension relative to the T7 gp4 polypeptide, and the further N-terminal deletion in T57 at position K144 is consistent with the removal of the N-terminal zinc-binding domain (ZBD) in T7 gp4 (Ziebarth et al., 2007) (Figure 1A and supplementary Figure 6).
Figure 1.
Schematic representation and structural model of the human mtDNA helicase. (A) Upper image, schematic representation of the conserved amino acid sequence motifs in human mtDNA helicase. MTS, mitochondrial target sequence; I–VI and R, conserved sequence motifs I–IV and RNAP basic motif of prokaryotic primases; 1–4 and 1a, conserved sequence motifs 1–4 and 1a of ring-shaped helicases (Ilyina et al., 1992). The size and position of the conserved sequence motifs are represented to scale. Lower image, structural model of a protomer of the human mtDNA helicase highlighting its modular architecture organized in a zinc binding-like domain (ZBD), RNA polymerase-like domain (RPD), linker region and a C-terminal helicase domain (CTD). *ZBD portion is represented as the polypeptide backbone of the bacteriophage T7 gp4 ZBD. (B) Model of the heptameric human mtDNA helicase, CTD view. The homology models presented in this study were developed using the multiple sequence alignment described in the legend to supplementary Figure 6 and the PDB files 1Q57 (Toth et al., 2003), 1NUI (Kato et al., 2003), 1DDE (Keck et al., 2000), 4E2K (Rymer et al., 2012) and 2AU3 (Corn et al., 2005) as input parameters for the software I-TASSER (Bazzoli et al., 2011). (C) Electron microscopic image of the recombinant human mtDNA helicase at 100 mM NaCl in the presence of Mg2+ and ATPγS, showing its heptameric configuration. This research was originally published in The Journal of Biological Chemistry (Ziebarth et al., 2010). ©The American Society for Biochemistry and Molecular Biology (see color version of this figure at www.informahealthcare.com/bmg).
In subsequent collaborative studies with Carazo and coworkers, we examined the quaternary structure of the human mtDNA helicase by negative-staining electron microscopy (Ziebarth et al., 2010). In doing so, we observed that its solubility is affected greatly by ionic strength, and that at low ionic strength its tendency to aggregate is mitigated by magnesium, nucleoside triphosphate, and temperature. Under conditions of high ionic strength mtDNA helicase forms hexamers and heptamers, the proportion of which is shifted, at low ionic strength, toward the heptameric state in the presence of Mg2+ and ATPγS (Figure 1C). Limited proteolysis with trypsin demonstrated that binding of ATP produces a conformational change that is distinct from that in the presence of ADP. Such conformational changes have also been documented in E. coli DnaB and T7 gp4 (Nakayama et al., 1984; Yong & Romano, 1995), and are likely to facilitate ssDNA translocation and dsDNA unwinding. In support of this, Sola and collaborators have recently reported both oligomeric and conformational changes in human mtDNA helicase (Fernandez-Millan et al., 2015). As with T7 gp4 (Kato et al., 2003; Toth et al., 2003), their combined analysis by negative-staining and cryo-electron microscopy and small angle X-ray scattering supports a structural model in which a hexameric, two-layered ring contains a symmetric, relatively stable C-terminal domain (CTD) ring with a more flexible ring atop that contains the ZBD and the RNA polymerase-like domain (RPD) (Figure 1).
To date, three catalytic activities have been ascribed to the recombinant human mtDNA helicase: DNA-dependent NTPase, 5′–3′ dsDNA unwinding and nucleotide-independent ssDNA annealing. Notably, DNA primase activity associated with the N-terminal half of the T7 gp4 primase-helicase has not been found, and a role for the evolutionarily conserved sequence elements in mtDNA helicase has been the subject of much investigation. The catalytic activity of a mtDNA helicase was first reported upon DEAE-Sephacel fractionation of sucrose-gradient purified mitochondria derived from bovine brain (Hehman & Hauswirth, 1992). dsDNA unwinding activity was demonstrated on a single-stranded circular DNA to which a short oligonucleotide of complementary or partially complementary sequence was annealed, and was dependent specifically on either ATP or dATP. Nearly a decade later, human mtDNA helicase activity was detected in crude mitochondrial extracts of HEK293 cells, where ATP-dependent DNA unwinding was shown to increase ~50% with the overexpression of a construct containing the full-length product encoded by the Twinkle gene (Spelbrink et al., 2001). Falkenberg and coworkers purified a recombinant form to near-homogeneity upon overexpression in the baculovirus system and demonstrated its DNA-dependent ATPase activity, and 5′–3′ directionality and substantial stimulation by mitochondrial single-stranded DNA-binding protein (mtSSB) in DNA unwinding (Korhonen et al., 2003). With their reconstruction of a minimal mtDNA replisome containing the mitochondrial replicative DNA polymerase, DNA polymerase γ (Pol γ), mtDNA helicase and mtSSB, the functional interactions of mtDNA helicase were demonstrated in unwinding DNA at an active DNA replication fork (Korhonen et al., 2004). Although mtDNA helicase was unable to unwind a duplex region of 55 bp in the absence or presence of mtSSB, it catalyzed extensive NTP-dependent unwinding of a duplex minicircle template with a 5′-ssDNA tail in concert with nucleotide polymerization by Pol γ, which was thus enabled to produce long products of rolling circle replication in a reaction that was stimulated markedly by mtSSB.
We examined further the stimulation of dsDNA unwinding by human mtDNA helicase by wild-type mtSSB and several variant forms, targeting amino acid residues that are conserved among mtSSBs but not present in E. coli SSB by alanine-substitution and deletion mutagenesis (Oliveira & Kaguni, 2011). Whereas maximal stimulation of human mtDNA helicase can be achieved both by human mtSSB and non-cognate SSBs including D. melanogaster mtSSB and E. coli SSB, several of the human variants exhibited substantially reduced stimulation. The variants E33A/G34A/ K35A in loop 1,2 (according to the nomenclature adopted for E. coli SSB (Raghunathan et al., 1997)) and Y100A/G101A/ E102A in loop 4,5–2 showed an ~40% reduced stimulation, while ΔS51–L59 in loop 2,3 showed an ~15% reduction. Notably, several other human mtSSB variants failed differentially to stimulate the activity of human Pol γ. Together, these data argue that specific functional and perhaps physical interactions facilitate dsDNA unwinding by human mtDNA helicase.
A standard DNA unwinding assay on oligonucleotide substrates, the DNA-dependent ATPase assay, several DNA binding assays and the replisome assay have been employed to explore various features of mtDNA helicase activity, and nearly all have focused on human recombinant forms. We examined the DNA-dependent ATPase activity and oligomeric form of several of the stable tryptic forms described earlier, producing recombinant forms of the 66 kDa C-terminal truncation (P66) and the N- and C-terminally truncated 57 kDa protein (P57), showing that they both form hexamers that modulate differentially nucleotide hydrolysis (Ziebarth et al., 2007). P66 reproducibly increased ATPase activity as compared to the similar activities of P57, suggesting that the carboxyl terminus of the intact polypeptide is inhibitory, whereas the amino-terminal zinc-binding region may contribute positively to overall catalytic activity. At the same time, a heterohexamer of the 57 and 50 kDa polypeptides in a 3–4:1 ratio showed dramatically reduced activity. Given that the 50 kDa protein represents a truncated polypeptide terminating at ~amino acid position 590, this would argue that residues between ~590 and ~640 are required for activity. One possibility we considered was the likely involvement of R609 and F621 that are analogous to the arginine finger and base stack, respectively, of T7 gp4, and E. coli DnaB. We tested this hypothesis by producing human alanine substitution variants of them and included another aromatic residue that is conserved among mtDNA helicases, F628 (Matsushima et al., 2008). This demonstrated that each serves a role in nucleotide hydrolysis: DNA-dependent ATPase activity was not detectable in the arginine finger variant R609A, and the F621A and F628A variants showed 10- and 4-fold decreased activity, respectively.
To probe the functional role of the N-terminal domain (NTD) of human mtDNA helicase, Falkenberg and coworkers studied truncation variants and also produced the NTD in the absence of the entire CTD (Farge et al., 2008). The NTD variants were constructed to contain the linker region that separates the NTD from the CTD (Figure 1A). The variants were examined for their ability to bind single- and doublestranded oligonucleotide by electrophoretic mobility shift assay (EMSA). The wild-type helicase was able to bind both substrates, the full NTD construct bound neither, and a NTD-deletion variant (Δ1–314) showed reduced binding to both ssDNA and dsDNA as compared to wild-type, whereas the ZBD-deletion variant showed full dsDNA binding with diminished ssDNA binding. This and the demonstration of diminished DNA-dependent ATPase activity in both N-terminal deletion constructs suggest that the NTD contributes to the DNA-dependent ATPase activity carried in the CTD. At the same time, the NTD only construct (Δ372–684) showed no ATPase activity. In keeping with the requirement for NTP hydrolysis in ssDNA translocation, the DNA unwinding activity of the various forms was shown to be consistent with their DNA-dependent ATPase activities. Finally, in the replisome reconstitution assay, both N-terminal variants were active, but products reflecting lower processivity in DNA strand synthesis by Pol γ were observed.
In the only comparative biochemical analysis so far reported, our group has evaluated NTD-only variants of the D. melanogaster homolog of human mtDNA helicase (Stiban et al., 2014). Modular architecture was documented by limited trypsin proteolysis to produce its N-terminal ZBD and C-terminal RPD, and showed that the full-length NTD and its truncation variants in recombinant form are ssDNA-binding monomers. Remarkably, we found that the ZBD binds an FeS cluster coordinated by the analogous cysteine residues that bind zinc in T7 gp4 and bacterial DnaG primases. These are absent in the mammalian mtDNA helicase, which is thus unlikely to bind a metal in the NTD. As with the human NTD, we were unable to document DNA binding in the Drosophila NTD by EMSA, but using a more sensitive, fluorescence-quenching based assay, both ssDNA and dsDNA binding to a 40-mer oligonucleotide were identified, with an estimated Kd of ~120 nM. This supports the earlier study suggesting a role for the NTD in helicase function and our parallel studies in a Drosophila cell culture model (see the following section).
The most comprehensive analysis reported to date of the biochemical activity of recombinant human mtDNA helicase by Copeland and coworkers (Longley et al., 2010) evaluated the wild-type enzyme and 20 human disease variants, including those in the NTD, linker and CTD. This study demonstrated variable but largely minimal defects in DNA binding affinity, ATP binding and hydrolysis, and DNA helicase activity. Instead, altered protein stability was documented, as measured by thermal inactivation of a number of disease variants, providing a plausible explanation of the late onset of their physiological effects in humans. A more recent study by Patel and coworkers revealed the third catalytic activity of human mtDNA helicase, NTP-independent ssDNA annealing (Sen et al., 2012). The ssDNA annealing activity was found to oppose the dsDNA unwinding activity of human mtDNA helicase in the absence of an SSB or a ssDNA trap, and only ssDNA but not dsDNA serves as a competitive inhibitor of annealing activity even though the helicase was shown to bind dsDNA as reported in the earlier studies. This lead to the proposal that the ssDNA annealing activity might be involved in recombination-mediated replication, a mtDNA replication mechanism supported by physiological studies of mtDNA replication intermediates from human heart (Pohjoismaki et al., 2009), or possibly in replication fork regression during DNA repair as has been described in prokaryotic and nuclear genomes.
Animal models of helicase function
Animal models of mtDNA helicase have focused primarily on two key issues, its physiological roles and importance, and the effects of human pathogenic mutations. Initial studies employed both a mouse model and cultured human osteosarcoma 143B cells (Tyynismaa et al., 2004). Here, Suomalainen and collaborators documented that overexpression of the mtDNA helicase in mouse muscle and heart increased mtDNA copy number several fold with no apparent physiological effects. This suggested that mtDNA helicase is limiting for mtDNA replication in vivo, thus potentially exacerbating the mild to moderate biochemical effects reported for disease variants. RNA interference of helicase expression in human 143B cells resulted in a two-fold depletion of mtDNA without apparent effects on mitochondrial number or structure. A subsequent mouse study evaluated the effects of a specific duplication (dup353–365) and point mutation (A360T) that are analogous to those found in patients (dup352–364, A359T) with autosomal dominant progressive external ophthalmoplegia (adPEO), with the finding that the mice recapitulated the histological and molecular results reported in patients, with muscle atrophy accompanied by accumulation of mtDNA deletions and mildly decreased respiratory activity, and yet normal physical function (Tyynismaa et al., 2005). The negative effects developed with age as expected for the late onset of PEO in humans. In later studies, Larsson and collaborators showed embryonic lethality in a conditional mouse knockout that was accompanied by severe and rapid mtDNA depletion in heart and skeletal muscle arguing that the enzyme is essential for mtDNA replication and that its function cannot be compensated by another cellular helicase (Milenkovic et al., 2013).
Our group has pursued a Drosophila model of mtDNA helicase function in cultured Schneider S2 cells and in animals. We have explored the various functions of the NTD, linker, and CTD in S2 cells by a combination of forward mutagenesis and expression of human disease alleles (Matsushima & Kaguni, 2007; Matsushima et al., 2008; Matsushima & Kaguni, 2009). The system was established by evaluating the effects of helicase overexpression and knockdown, with the findings that overexpression increased mtDNA copy number ~1.4 fold while knockdown by RNA interference resulted in ~5-fold decrease in mtDNA, recapitulating in insect cells the results found in the mouse and human systems, and establishing a conserved role for mtDNA helicase in mtDNA replication across taxa (Matsushima & Kaguni, 2007). We also examined catalytic mutants analogous to A318 and D424 in the Walker A and B motifs in the helicase domain of T7 gp4, respectively, and found them to generate a dominant-negative, lethal phenotype resulting from a dramatic mtDNA depletion of 14–20-fold. Interestingly, mitochondrial transcript levels were reduced as a consequence of mtDNA depletion, but only after a two-fold decrease in mtDNA copy number was incurred. Overexpression of variants carrying analogous human-disease alleles in either the linker or CTD showed differential effects. All were able to form hexamers in vivo, the I334P and A442P mutants (I367T, A475P in humans) showed the lethal phenotype and mtDNA depletion found for the catalytic mutants, while A326T, R341Q and W441C (A359T, R734Q, W474C in humans) did not. Notably, I334P maps to the A helix of the linker and A442P to the D3 helix in the helicase domain of T7 gp4; in T7 gp4, the A helix of one protomer interacts with the D2 and D3 helices of the adjacent protomer (Singleton et al., 2000), implicating their involvement in the ssDNA translocation required for dsDNA unwinding (Lee & Richardson, 2004). Garesse and collaborators established a fly model to study further the K388A, A442P and W441C mutations, using ubiquitous overexpression throughout development in the GAL4-UAS system (Sanchez-Martinez et al., 2012). The K338A and A442P mutations cause severe mtDNA depletion and developmental lethality, as might be predicted from the effects in S2 cells. Interestingly, in the animal model, the W441C mutation yielded a modest decrease in mtDNA copy number during the third larval instar stage and a moderate decrease in adult lifespan. Defects in oxidative phosphorylation in the K388A and A442P overexpressing animals were found, which resulted in reduced cell proliferation and the promotion of apoptosis, providing an important link between mitochondrial dysfunction associated with defects in mtDNA metabolism and programed cell death.
Further studies in S2 cells examined the roles of the NTD and CTD by evaluating conserved amino acid residues among mtDNA helicases and both T7 gp4 and bacterial DnaG primases and DnaB helicases, and additional human disease alleles. Surprisingly, overexpression of conserved amino acids in motifs I, IV, V, and VI and the RNA polymerase basic domain (Figure 1A) did not cause mtDNA depletion, while overexpression of the N-terminal variants W282L, R301Q, and P302L that are analogous to human adPEO mutations (W315L, R334Q and P335L) and map within the TOPRIM subdomain comprising motifs IV–VI of the RPD showed deleterious effects (Matsushima & Kaguni, 2009). (These are discussed in the following section on evolutionary comparisons.) In contrast, overexpression of four alanine-substitution variants of conserved amino acids in the CTD, including the putative arginine finger and base stack found in T7 gp4 and DnaB, caused dramatic mtDNA depletion and a dominant-negative lethal phenotype. Interestingly, all of these lie C-terminal to the last conserved motif 4 in superfamily 4 DNA helicases. To evaluate the functional importance of the C-terminal region of the CTD, progressive deletion variants were constructed carrying deletions Δ572–613, Δ586–613, Δ597–613, and Δ607–613; overexpression of Δ527–613 and Δ586–613 led to lethality while cells overexpressing the Δ597–613 and Δ607–613 variants maintained normal mtDNA copy number and growth. These data argue that an ~25 amino acid region beyond motif 4 is required for mtDNA helicase function.
In parallel with studies in mice and insect cells, Spelbrink and collaborators investigated the molecular mechanism of mtDNA depletion and deletion in cultured human HEK293 cells overexpressing both catalytic mutants and human disease alleles of mtDNA helicase (Goffart et al., 2009; Pohjoismaki et al., 2011;Wanrooij et al., 2007). These studies in composite led to the important finding that dose-dependent mtDNA depletion results from replication stalling. Analysis of replicating intermediates (RIs) by two-dimensional neutral/ neutral agarose gel electrophoresis (2DNAGE) and Southern blotting demonstrated that concomitant with the observed mtDNA depletion, both the abundance and nature of the RIs were altered in helicase variants; an accumulation of RIs was observed and these were RNase H resistant indicating full maturation of lagging DNA strands as compared to control cells. Although Pol γ mutants were also shown to cause replication stalling, the features of the RIs they engendered differed with regard to their effects on lagging DNA strand synthesis, likely as a result of the requirement for Pol γ in the replication of both parental DNA strands. Evaluation of the RIs generated in the mouse model where expression of the linker region duplication (dup 353–365) was shown to generate mtDNA deletions with age (9–12 months) (Tyynismaa et al., 2005), produced similar results to those obtained in the human cells, and the impairment of replication appeared as early as six weeks of age (Goffart et al., 2009). This finding suggests that replication stalling predisposes the cells to subsequent mtDNA deletions. This hypothesis is supported by the later finding that replication stalling by catalytically-impaired mtDNA helicase results in multiple mtDNA rearrangements in human cells, which are the possible products of DNA repair of double-stranded breaks (Pohjoismaki et al., 2011).
Evolutionary comparisons and clustering of human pathogenic mutations
The evolutionary origins of the protein components of the animal mtDNA replisome are both intriguing per se and informative relevant to present day function. As predicted by the endosymbiotic theory, mtSSB is a homolog of the homotetrameric SSBs from eubacteria (Mahoungou et al., 1988), but is possibly the only replisome protein that originated from the α-proteobacterium-like mitochondrial ancestor. The mtDNA helicase and Pol γ-α, the catalytic core of Pol γ, share ancestry with the T7 gp4 primase-helicase and gp5 DNA polymerase, respectively (Shutt & Gray, 2006a). On the other hand, in considering the fact that the animal mitochondrial replicase is the only mtDNA polymerase known to function in concert with an accessory factor, Pol γ-β, the accessory subunit of Pol γ, most likely originated from an independent event of lateral class II aminoacyl-tRNA synthetase gene transfer involving eubacterial species and early metazoans (Wolf & Koonin, 2001). How the evolution of proteins with distinct functions and organismal origins has occurred to allow them to function coordinately to promote mtDNA replication remains unclear, but some of the evolutionary routes taken are starting to be unraveled, at least for Metazoa. Here, we evaluate some of the evolutionary changes to provide models of the structural and functional features of the mtDNA helicase.
In the absence of a crystal structure for the human mtDNA helicase, we developed homology models based on the 3D structures of T7 gp4 and bacterial DNA helicase (DnaBs) and DNA primases (DnaGs) (Figures 1–5), and on a multiple amino acid sequence alignment we derived from 58 plant and animal sequences available in public databases, using the T7 gp4 sequence as the outgroup (Figure 6 of Supplementary material). As with other models reported for the human mtDNA helicase (Fernandez-Millan et al., 2015; Holmlund et al., 2009; Korhonen et al., 2008), we observed that the C-terminal helicase domain (CTD) maintains a structure that is highly similar to those of prokaryotic replicative DNA helicases (Figure 1) (Bailey et al., 2007; Toth et al., 2003). In fact, electron microscopic imaging of the human mtDNA helicase (Fernandez-Millan et al., 2015; Ziebarth et al., 2010) supports this and suggests that the hexameric/heptameric conformation of mtDNA helicase is organized by the CTD and the linker region (Figures 1 and 2), as it is for T7 gp4 and E. coli DnaB (Bailey et al., 2007; Toth et al., 2003). Modeling of the CTD of D. melanogaster mtDNA helicase also provided a structure that resembles the homologous domain of all the DNA helicases mentioned above (data not shown), highlighting the conservation of the mtDNA helicase CTD structure and function across taxa.
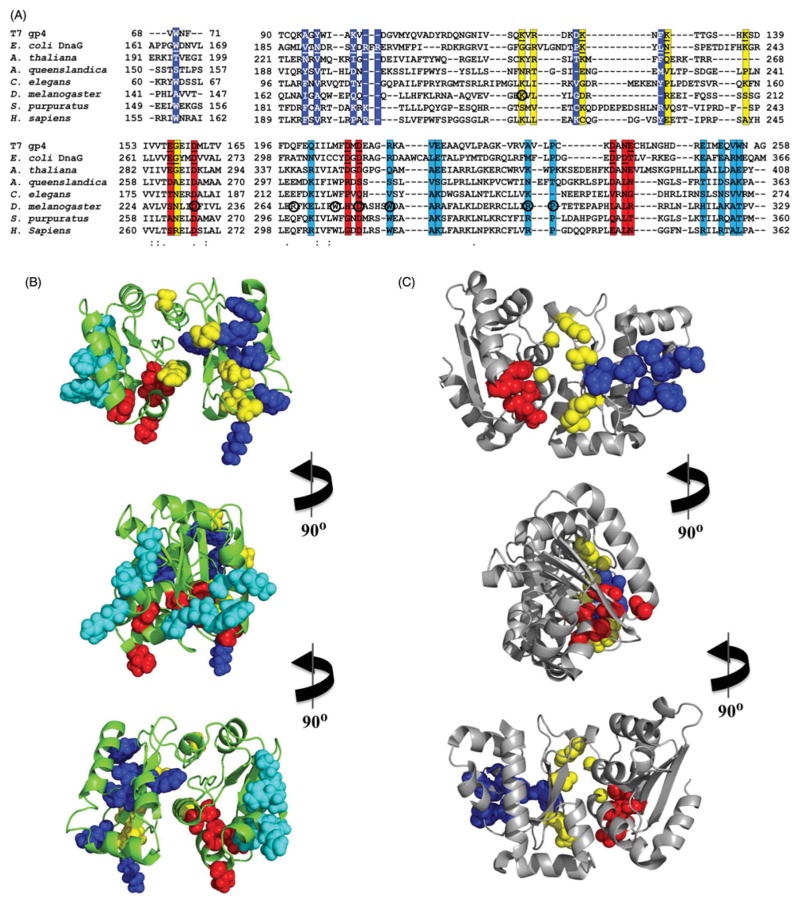
Figure 5.
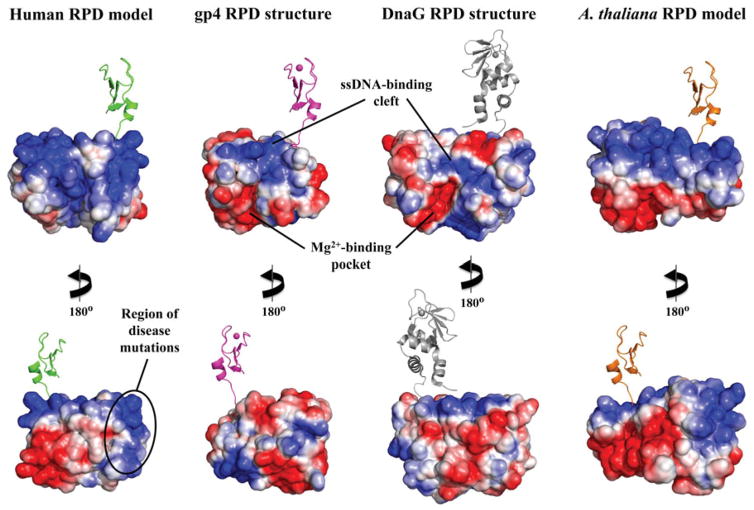
Electrostatic potential surface (EPS) map suggests a new DNA-binding region for the RPD of human mtDNA helicase. EPS maps for the RPD structures of T7 gp4 (residues 64–255, 1NUI), E. coli DnaG (residues 115–366, 1DDE) and the mtDNA helicase models of humans (residues 153–362) and Arabidopsis thaliana (residues 191–408) were calculated using the APBS Tools built into Pymol, with default settings. The solvent accessible surfaces shown are colored according to electrostatic potential (blue≥+10 kT, red≤−10 kT). The ZBD is depicted in cartoon representation to provide a reference for the N-terminal region of these enzymes. The human and A. thaliana ZBD representations are copies of the same domain from T7 gp4 and are positioned as if they provide the same structural arrangements and function as in the bacteriophage enzyme; they are not meant, however, to depict any structural similarities between the ZBDs of mtDNA helicases and the T7 primase–helicase. The bacterial ZBD was retrieved from the crystal structure of the Aquifex aeolicus DnaG (PDB: 2AU3) (see color version of this figure at www.informahealthcare.com/bmg).
Figure 2.
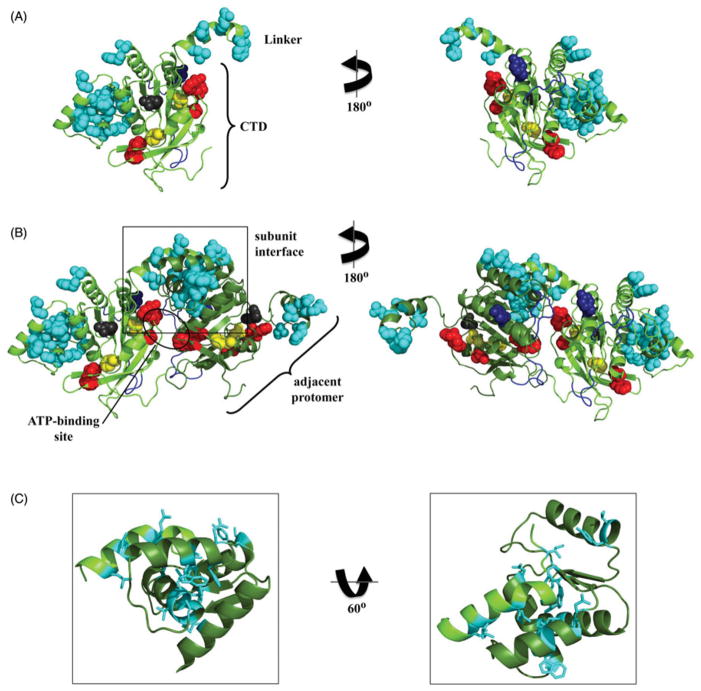
Mapping of active site residues and human pathogenic mutations in the model of the human CTD and linker region. (A) Residues directly important for NTP hydrolysis, including the arginine finger (R609), the putative base stack (F621) and others (K607, F610 and F628) (Matsushima et al., 2008) are shown in red, sphere representation; the loops important for DNA-binding in the CTD of T7 gp4 (Notarnicola et al., 1995; Singleton et al., 2000; Washington et al., 1996) are shown in blue, cartoon representation; and the human disease-related residues (Appendix Table 1) are shown in sphere representation and colored according to their proposed functional properties including subunit interactions (cyan), NTP hydrolysis (yellow) and DNA-binding (dark blue). R391 (dark gray) has not been given a functional assignment (see text for details). (B) Representation of the juxtaposition of two protomers (light and dark green, cartoon representation), highlighting the NTP-binding site (circled) and the subunit interaction region (boxed). (C) Closer view of the subunit interaction region, showing the human disease-related residues in stick representation (see color version of this figure at www.informahealthcare.com/bmg).
That the number of mutated residues in the human mtDNA helicase found in association with human diseases (see Appendix Table 1) has increased significantly since its first report 15 years ago (Spelbrink et al., 2001) demonstrates the importance of the enzyme for mitochondrial function and human health, and provides us additional data to map functional regions of the polypeptide, as our group has done for human Pol γ (Euro et al., 2011; Farnum et al., 2014). Figure 2 shows the functional organization of residues for which pathogenic mutations have been identified in the linker region and CTD in our human mtDNA helicase model. We also map the location of catalytic residues that have been identified in biochemical studies and putative DNA-binding regions based upon the T7 gp4 structure (Toth et al., 2003). The disease-related residues were assigned likely functional properties based on their proximity to known sites in the mtDNA helicase and in the homologous T7 gp4 polypeptide including subunit interaction, DNA binding, and ATP binding/ hydrolysis (Figure 2A). Using this criterion, we were unable to make a functional assignment for residue R391, and no biochemical and/or physiological studies using a variant helicase with a mutation at this position that might aid in understanding its role in disease have been reported to date. Moreover, the mutation R391H has been found in compound heterozygosity with N585S and therefore, may not be related to the reported cause of Perrault syndrome (Morino et al., 2014); N585, on the other hand, lies within the conserved amino acid motif 4 of ring-shaped helicases and in close proximity to the arginine finger (R609) and the ATP-binding site (Figure 2B).
Appendix Table 1.
Disease-associated amino acid residues of the human mtDNA helicase. These residues were mapped onto the helicase models presented in Figures 2 and 4.
| Residue | Mutation | Associated disease | References |
|---|---|---|---|
| R303 | R303Q and R303W | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| W315 | W315L and W315S | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| A318 | A318T | Hepatocerebral mtDNA depletion syndrome, infantile-onset spinocerebellar ataxia | (Dundar et al., 2012), (Ji et al., 2014) |
| K319 | K319E and K319T | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| R334 | R334P and R334Q | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| P335 | P335L | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| R354 | R354P | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| R357 | R357P | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| A359 | A359T | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| L360 | L360G | Autosomal dominant progressive external ophthalmoplegia | (Ji et al., 2014) |
| A362 | A362P | Autosomal dominant progressive external ophthalmoplegia | (Ji et al., 2014) |
| W363 | W363L | Autosomal dominant progressive external ophthalmoplegia | (Ji et al., 2014) |
| I367 | I367T | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| S369 | S369P and S369Y | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| F370 | F370C and F370L | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| R374 | R374Q and R374W | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| L378 | L378P | Autosomal dominant progressive external ophthalmoplegia | (Ji et al., 2014) |
| L381 | L381P | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| R391 | R391H/N585S | Perrault syndrome | (Morino et al., 2014) |
| F395 | F395L | Renal tubulopathy | (Prasad et al., 2013) |
| S426 | S426N | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| W441 | W441G/V507I | Perrault syndrome | (Morino et al., 2014) |
| N448 | N448D | Autosomal dominant progressive external ophthalmoplegia | (Morino et al., 2014) |
| M455 | M455T | Autosomal dominant progressive external ophthalmoplegia | (Morino et al., 2014) |
| L456 | L456V | Infantile-onset spinocerebellar ataxia | (Dundar et al., 2012), (Ji et al., 2014) |
| T457 | T457I | Hepatocerebral mtDNA depletion syndrome | (Dundar et al., 2012), (Ji et al., 2014) |
| Q458 | Q458H | Autosomal dominant progressive external ophthalmoplegia | (Ji et al., 2014) |
| A460 | A460G and A460P | Autosomal dominant progressive external ophthalmoplegia | (Ji et al., 2014) |
| R463 | P083S/R463W | Infantile-onset spinocerebellar ataxia | (Hartley et al., 2012) |
| W474 | W474C and W474S | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| A475 | A475D, A475P and A475T | Autosomal dominant progressive external ophthalmoplegia | (Ji et al., 2014) |
| F478 | F478I | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| E479 | E479K | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| F485 | F485L | Autosomal dominant progressive external ophthalmoplegia | (Dundar et al., 2012), (Ji et al., 2014) |
| V507 | W441G/V507I | Perrault syndrome | (Morino et al., 2014) |
| Y508 | Y508C | Infantile-onset spinocerebellar ataxia | (Dundar et al., 2012), (Ji et al., 2014) |
| Y537 | Y537H | Autosomal dominant progressive external ophthalmoplegia | (Ji et al., 2014) |
| N585 | R391H/N585S | Perrault syndrome | (Morino et al., 2014) |
Of the 25 disease-related residues found in the linker region and the CTD, 21 were assigned as important for subunit interaction (Figure 2C). Whereas their putative roles in maintaining the stability and/or conformation of the oligomeric enzyme is not apparent when evaluating the model of a single subunit (Figure 2A), it becomes clear that the linker region of one protomer can interact with a region of the CTD approximately 10.5 Å from the ATP-binding site of the adjacent protomer when two protomers are modeled together (Figure 2B). This is consistent with that suggested previously by Korhonen et al. based on an analysis using a more limited number of pathogenic residues (Korhonen et al., 2008), and by Fernandéz-Millán et al. based on their cryoelectron microscopic study (Fernandez-Millan et al., 2015). A role in subunit interaction does not imply explicit involvement in either hexamer/heptamer formation or stability. On one hand, it has been reported that recombinant forms of the human mtDNA helicase bearing the I367T, R374Q, W474C or A475P substitutions show altered oligomer formation and/ or stability (Korhonen et al., 2008), while other variants, including other linker region and CTD mutants of the human and/or D. melanogaster mtDNA helicase such as A359T, S369P, L381P and F485L (numbering as in the human sequence) do not appear to differ in oligomeric state from that of the wild-type enzyme either in vitro or in vivo (Korhonen et al., 2008; Matsushima & Kaguni, 2007). However, if we assume that the active mtDNA helicase adopts an asymmetric arrangement of protomers that aids in the coupling of ssDNA binding, nucleotide hydrolysis and dsDNA unwinding as has been shown for T7 gp4 (Singleton et al., 2000; Toth et al., 2003), one can argue that most mutations assigned as important for subunit interactions (Figure 2) may disturb this asymmetry and hence influence negatively NTP binding and/or hydrolysis, without affecting the formation of the hexamer/heptamer. This hypothesis is consistent with the biochemical data showing that most mutations in the linker region and CTD, which are located distantly from the ATP-binding site, lead to at least partially defective ATPase and unwinding activities (Korhonen et al., 2008; Longley et al., 2010), even though they most likely form oligomers. Alternatively, these mutations may disrupt the balance between the oligomeric forms; we have shown previously that the recombinant human mtDNA helicase can switch from a hexameric conformation to a heptameric one under low ionic strength and upon binding to Mg2+ and ATPγS (Ziebarth et al., 2010) (Figure 1C). It is also possible that some of the mutations may affect loading of the mtDNA helicase onto DNA rather than ssDNA translocation and dsDNA unwinding, as it has been reported for T7 gp4 carrying the A257T mutation homologous to the human pathogenic mutation A359T, that is located in the linker region (Patel et al., 2011). Notably however, recombinant forms of the human mtDNA helicase bearing A359T, even if hexamers/heptamers, are defective in DNA binding and NTP hydrolysis (Korhonen et al., 2008; Longley et al., 2010). Though, neither the oligomeric forms nor their relative contributions to the mitochondrial replisome in vivo are known, it is tempting to speculate that hexamers and heptamers coexist inside mitochondria, but only heptamers may be active in DNA loading, while hexamers may be involved in nucleotide hydrolysis and dsDNA unwinding. Such a hypothesis would be consistent with models for T7 gp4 in which opening of the heptameric helicase ring occurs after initial DNA contact with the NTD, with the concomitant loss of a protomer (Ahnert et al., 2000; Crampton et al., 2006).
Our analysis of the N-terminal region of the human mtDNA helicase reveals a story different from that of the CTD and linker region. We found that modeling of ZBD was unreliable using in silico methods based on sequence homology, because the amino acid sequence similarities between the mtDNA helicase, T7 gp4 and bacterial DnaGs are too low. Although several homology models have been reported for the ZBD (Fernandez-Millan et al., 2015; Holmlund et al., 2009), in our opinion, this domain likely adopts a novel structural fold, different from its counterpart in the prokaryotic enzymes. Evidence supporting this derives from our recent report demonstrating that the D. melanogaster ZBD binds an FeS cluster, instead of zinc (Stiban et al., 2014) and it appears that the human ZBD binds neither (Shutt & Gray, 2006b).
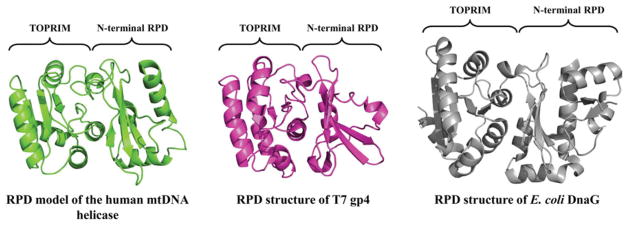
In contrast, in the C-terminal half of the NTD of human mtDNA helicase, we show the RPD adopts a conformation comprising two subdomains: an N-terminal region and a TOPRIM fold (Figure 3). These are highly similar to that of prokaryotic primases (Kato et al., 2003; Keck et al., 2000), although in this case the human RPD resembles more closely that of bacteriophage T7. Though the human ZBD has apparently diverged from a structural conformation that binds zinc, the RPD appears to have the backbone of a primase, and presents some of the conserved amino acid residues that are important for primase function in T7 gp4 (Figure 4). Interestingly, however, the electrostatic potential surface (EPS) of the human RPD model, as compared to those of the RPD structures of T7 gp4 and E. coli DnaG, shows that the negatively-charged Mg2+-binding pocket that is so evident in the prokaryotic structures is absent in the human RPD (Figure 5), arguing that human mtDNA helicase is unable to coordinate the bivalent cations to catalyze primer synthesis. Rather, the homologous aspartate and glutamate residues that form the Mg2+-binding pocket in the T7 gp4 and DnaG RPDs are buried inside the human RPD model (Figure 4), perhaps involved in electrostatic interactions with positively charged residues in the TOPRIM subdomain (data not shown). In agreement, no primase activity has been detected in vitro for the human mtDNA helicase ((Farge et al., 2008) and our unpublished data) and in vivo, alanine substitutions of the homologous negatively-charged residues caused no effect on mtDNA copy number in Drosophila S2 cells (Matsushima & Kaguni, 2009). On the other hand, a comparative RPD model of the Arabidopsis thaliana mtDNA helicase, for which primase activity has been demonstrated biochemically (Diray-Arce et al., 2013), clearly presents a Mg2+-binding pocket similar to those in the crystal structures of T7 gp4 and E. coli DnaG (Figure 5).
Figure 3.
Structural model for the RNA polymerase-like domain (RPD) of human mtDNA helicase. The polypeptide backbone of the human RPD (residues 153–362) is highly similar to those of T7 gp4 (residues 64–255, PDB: 1NUI; (Kato et al., 2003)) and E. coli DnaG (residues 115–366, PDB: 1DDE; (Keck et al., 2000)), even though no primase activity is apparently present in the human enzyme (see color version of this figure at www.informahealthcare.com/bmg).
Figure 4.
Mapping of conserved amino acid residues and human pathogenic mutations in the human RPD model. (A) Amino acid sequence alignment among T7 gp4, E. coli DnaG and selected mtDNA helicases, showing important residues for ssDNA (highlighted in blue), NTP-(yellow) and Mg2+-binding (red), and human disease mutations (cyan). The residues in T7 gp4 and bacterial DnaG that were shown experimentally to bind ssDNA, NTP or Mg2+ (Corn et al., 2008; Kato et al., 2003; Keck et al., 2000; Lee & Richardson, 2001; Rymer et al., 2012) are underlined, and the residues in the D. melanogaster mtDNA helicase that have been mutated in S2 cells (Matsushima & Kaguni, 2009) are circled. These residues have been positioned on the human RPD model (B) and in the E. coli RPD structure (PDB: 1DDE, (Keck et al., 2000)) (C) to illustrate (1) that the homologous residues in the human RPD, important for primase activity in DnaG, are displaced from an active configuration and (2) that the human disease mutations (Appendix Table 1) are clustered in a region of the TOPRIM fold of the human RPD (see color version of this figure at www.informahealthcare.com/bmg).
In addition to the lack of a Mg2+-binding pocket, the EPS of the human RPD model shows a relatively large, positively-charged area within the TOPRIM subdomain (Figure 5). Truncated forms of the human mtDNA helicase that lack the RPD, either partially or completely, exhibit diminished ssDNA-binding properties, which is accompanied by decreased ATPase and unwinding activities in vitro and decreased mtDNA copy number in cultured cells (Farge et al., 2008; Wanrooij et al., 2007). Because the positively-charged area in the human RPD is distinct from the ssDNA-binding groove of prokaryotic RPDs that allows proper positioning of the template DNA for the clamping by the ZBD (Corn et al., 2008; Kato et al., 2003), we propose that the human RPD binds ssDNA in a new configuration to impart alternate functionalities. One such feature might be to bind ssDNA complementary to the DNA template during dsDNA unwinding on a surface-exposed site of the protein to promote NTP-independent ssDNA annealing that is not found in the prokaryotic counterparts. Another possibility may be that this region of the human RPD facilitates ssDNA loading by the mtDNA helicase.
Interestingly, when we mapped amino acid residues associated with human disease mutations on the RPD model, we found that they cluster in a region of the TOPRIM subdomain that forms part of the positively-charged surface area (Figure 4). We suggest then that these changes interfere with ssDNA-binding to this region, arguing for its functional importance in the mtDNA replisome, consequently leading to mitochondrial dysfunction. Support for an important role for the proposed ssDNA-binding region derives from our studies modeling mutations found in human patients in the D. melanogaster mtDNA helicase, which results in severe mtDNA depletion in S2 cells, whereas alanine-substitutions of the amino acids that are conserved in prokaryotic RPDs have no effect on mtDNA copy number (Matsushima & Kaguni, 2009). Because a similar positively-charged area is also present in RPD models of other distantly related animals, such as D. melanogaster (Arthropoda) and Strongylocentrotus purpuratus (Echinodermata) (Figure 7 of Supplementary material), and Amphimedon queenslandica (Porifera) and Caenorhabditis elegans (Nematoda) (data not shown), this suggests conservation of function in animal mtDNA helicases.
Perspectives
Nearly 15 years of research on the mitochondrial replicative DNA helicase has elucidated its basic physical properties, subunit composition and catalytic activities, and documented its central role in mtDNA replication. The specific role(s) of the N-terminal domain remain to be elucidated, including those in ssDNA and dsDNA binding relevant to DNA loading, translocation, unwinding, and annealing, the formation and stability of hexamers and heptamers, and in replisome interactions. The mechanism of dsDNA unwinding by mtDNA helicase and the involvement of the hexameric and heptameric forms remains to be determined, as do its three-dimensional structure, and specific contacts with nucleotide and DNA substrates and replisome partners. The possibility that the enzyme may unwind RNA–DNA duplexes should be explored to help understand the mechanism(s) by which RNA associated with mtDNA is removed. In addition, the possible roles of mtDNA helicase in mtDNA repair and recombination are important areas for future studies. Furthermore, the possible roles of the replicative helicase in the regulation of mtDNA replication, and in inter-organellar communication with the nucleus and possibly the endoplasmic reticulum, perhaps mediated by membrane association, are emerging areas of research.
The importance of the mitochondrial replicative DNA helicase in human health and disease remains a key area of interest. A critical area for future research will be to establish genotype–phenotype relationships for human pathogenic alleles such that despite the widely variable clinical presentations observed, age of onset and severity of symptoms can be predicted in both homozygous and heterozygous forms, and the tissue specificity, and molecular basis and outcomes relevant to mtDNA metabolism can be understood.
Intriguing and broader questions still remain concerning the N-terminal region in mtDNA helicase. Why does the mtDNA helicase retain the NTD and RPD domains whereas bacterial DnaBs function without their equivalents? What might have driven this evolutionary and functional rewiring, and why is the enzyme of some animal species now able to bind an iron-sulfur cluster (and perhaps other cofactors)? A road as long as that taken by research on the bacteriophage T7 and E. coli DNA replisomes appears to lie ahead to understand the roles of the mtDNA helicase in mtDNA replication, repair and recombination.
Acknowledgments
We thank Jani Haukka for his contribution to the multiple amino acid sequence alignment shown in Supplementary material as Figure 6. We are grateful to Dr. Grzegorz Ciesielski for critical reading of the manuscript.
Footnotes
Declaration of interest
We acknowledge support from the National Institutes of Health (Grant GM45295) and the Academy of Finland to L.S.K. and the Fundação de Amparo à Pesquisa do Estado de São Paulo (Grant 2014/02253-6) to M.T.O. The authors report no conflicts of interest.
References
- Ahnert P, Picha KM, Patel SS. A ring-opening mechanism for DNA binding in the central channel of the T7 helicase-primase protein. EMBO J. 2000;19:3418–27. doi: 10.1093/emboj/19.13.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bailey S, Eliason WK, Steitz TA. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science. 2007;318:459–63. doi: 10.1126/science.1147353. [DOI] [PubMed] [Google Scholar]
- Bazzoli A, Tettamanzi AG, Zhang Y. Computational protein design and large-scale assessment by I-TASSER structure assembly simulations. J Mol Biol. 2011;407:764–76. doi: 10.1016/j.jmb.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird LE, Hakansson K, Pan H, Wigley DB. Characterization and crystallization of the helicase domain of bacteriophage T7 gene 4 protein. Nucleic Acids Res. 1997;25:2620–6. doi: 10.1093/nar/25.13.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruthers JM, Mckay DB. Helicase structure and mechanism. Curr Opin Struct Biol. 2002;12:123–33. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- Corn JE, Pease PJ, Hura GL, Berger JM. Crosstalk between primase subunits can act to regulate primer synthesis in trans. Mol Cell. 2005;20:391–401. doi: 10.1016/j.molcel.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Corn JE, Pelton JG, Berger JM. Identification of a DNA primase template tracking site redefines the geometry of primer synthesis. Nat Struct Mol Biol. 2008;15:163–9. doi: 10.1038/nsmb.1373. [DOI] [PubMed] [Google Scholar]
- Crampton DJ, Ohi M, Qimron U, et al. Oligomeric states of bacteriophage T7 gene 4 primase/helicase. J Mol Biol. 2006;360:667–77. doi: 10.1016/j.jmb.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Diray-Arce J, Liu B, Cupp JD, et al. The Arabidopsis At1g30680 gene encodes a homologue to the phage T7 gp4 protein that has both DNA primase and DNA helicase activities. BMC Plant Biol. 2013;13:36. doi: 10.1186/1471-2229-13-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundar H, Ozgul RK, Yalnizoglu D, et al. Identification of a novel Twinkle mutation in a family with infantile onset spinocerebellar ataxia by whole exome sequencing. Pediatr Neurol. 2012;46:172–7. doi: 10.1016/j.pediatrneurol.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euro L, Farnum GA, Palin E, et al. Clustering of Alpers disease mutations and catalytic defects in biochemical variants reveal new features of molecular mechanism of the human mitochondrial replicase, Pol gamma. Nucleic Acids Res. 2011;39:9072–84. doi: 10.1093/nar/gkr618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farge G, Holmlund T, Khvorostova J, et al. The N-terminal domain of TWINKLE contributes to single-stranded DNA binding and DNA helicase activities. Nucleic Acids Res. 2008;36:393–403. doi: 10.1093/nar/gkm1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnum GA, Nurminen A, Kaguni LS. Mapping 136 pathogenic mutations into functional modules in human DNA polymerase gamma establishes predictive genotype–phenotype correlations for the complete spectrum of POLG syndromes. Biochim Biophys Acta. 2014;1837:1113–21. doi: 10.1016/j.bbabio.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Millan P, Lazaro M, Cansiz-Arda S, et al. The hexameric structure of the human mitochondrial replicative helicase Twinkle. Nucleic Acids Res. 2015;43:4284–95. doi: 10.1093/nar/gkv189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick DN, Baradaran K, Richardson CC. An N-terminal fragment of the gene 4 helicase/primase of bacteriophage T7 retains primase activity in the absence of helicase activity. Proc Natl Acad Sci U S A. 1998;95:7957–62. doi: 10.1073/pnas.95.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffart S, Cooper HM, Tyynismaa H, et al. Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum Mol Genet. 2009;18:328–40. doi: 10.1093/hmg/ddn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley JN, Booth FA, Del Bigio MR, Mhanni AA. Novel autosomal recessive c10orf2 mutations causing infantile-onset spinocerebellar ataxia. Case Rep Pediatr. 2012;2012:303096. doi: 10.1155/2012/303096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehman GL, Hauswirth WW. DNA helicase from mammalian mitochondria. Proc Natl Acad Sci USA. 1992;89:8562–6. doi: 10.1073/pnas.89.18.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmlund T, Farge G, Pande V, et al. Structure-function defects of the twinkle amino-terminal region in progressive external ophthalmoplegia. Biochim Biophys Acta. 2009;1792:132–9. doi: 10.1016/j.bbadis.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Ilyina TV, Gorbalenya AE, Koonin EV. Organization and evolution of bacterial and bacteriophage primase–helicase systems. J Mol Evol. 1992;34:351–7. doi: 10.1007/BF00160243. [DOI] [PubMed] [Google Scholar]
- Ji K, Liu K, Lin P, et al. Twinkle mutations in two Chinese families with autosomal dominant progressive external ophthalmoplegia. Neurol Sci. 2014;35:443–8. doi: 10.1007/s10072-013-1557-8. [DOI] [PubMed] [Google Scholar]
- Kato M, Ito T, Wagner G, et al. Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol Cell. 2003;11:1349–60. doi: 10.1016/s1097-2765(03)00195-3. [DOI] [PubMed] [Google Scholar]
- Keck JL, Roche DD, Lynch AS, Berger JM. Structure of the RNA polymerase domain of E. coli primase. Science. 2000;287:2482–6. doi: 10.1126/science.287.5462.2482. [DOI] [PubMed] [Google Scholar]
- Korhonen JA, Gaspari M, Falkenberg M. TWINKLE Has 5′->3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J Biol Chem. 2003;278:48627–32. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- Korhonen JA, Pande V, Holmlund T, et al. Structure–function defects of the TWINKLE linker region in progressive external ophthalmoplegia. J Mol Biol. 2008;377:691–705. doi: 10.1016/j.jmb.2008.01.035. [DOI] [PubMed] [Google Scholar]
- Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–9. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Richardson CC. Essential lysine residues in the RNA polymerase domain of the gene 4 primase–helicase of bacteriophage T7. J Biol Chem. 2001;276:49419–26. doi: 10.1074/jbc.M108443200. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Richardson CC. The linker region between the helicase and primase domains of the gene 4 protein of bacteriophage T7. Role in helicase conformation and activity. J Biol Chem. 2004;279:23384–93. doi: 10.1074/jbc.M400857200. [DOI] [PubMed] [Google Scholar]
- Longley MJ, Humble MM, Sharief FS, Copeland WC. Disease variants of the human mitochondrial DNA helicase encoded by C10orf2 differentially alter protein stability, nucleotide hydrolysis, and helicase activity. J Biol Chem. 2010;285:29690–702. doi: 10.1074/jbc.M110.151795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoungou C, Ghrir R, Lecaer JP, et al. The amino-terminal sequence of the Xenopus laevis mitochondrial SSB is homologous to that of the Escherichia coli protein. FEBS Lett. 1988;235:267–70. doi: 10.1016/0014-5793(88)81276-6. [DOI] [PubMed] [Google Scholar]
- Matsushima Y, Farr CL, Fan L, Kaguni LS. Physiological and biochemical defects in carboxyl-terminal mutants of mitochondrial DNA helicase. J Biol Chem. 2008;283:23964–71. doi: 10.1074/jbc.M803674200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima Y, Kaguni LS. Differential phenotypes of active site and human autosomal dominant progressive external ophthalmoplegia mutations in Drosophila mitochondrial DNA helicase expressed in Schneider cells. J Biol Chem. 2007;282:9436–44. doi: 10.1074/jbc.M610550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima Y, Kaguni LS. Functional importance of the conserved N-terminal domain of the mitochondrial replicative DNA helicase. Biochim Biophys Acta. 2009;1787:290–5. doi: 10.1016/j.bbabio.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic D, Matic S, Kuhl I, et al. TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum Mol Genet. 2013;22:1983–93. doi: 10.1093/hmg/ddt051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morino H, Pierce SB, Matsuda Y, et al. Mutations in Twinkle primase–helicase cause Perrault syndrome with neurologic features. Neurology. 2014;83:2054–61. doi: 10.1212/WNL.0000000000001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama N, Arai N, Kaziro Y, Arai K. Structural and functional studies of the dnaB protein using limited proteolysis. Characterization of domains for DNA-dependent ATP hydrolysis and for protein association in the primosome. J Biol Chem. 1984;259:88–96. [PubMed] [Google Scholar]
- Notarnicola SM, Park K, Griffith JD, Richardson CC. A domain of the gene 4 helicase/primase of bacteriophage T7 required for the formation of an active hexamer. J Biol Chem. 1995;270:20215–24. doi: 10.1074/jbc.270.34.20215. [DOI] [PubMed] [Google Scholar]
- Oliveira MT, Kaguni LS. Reduced stimulation of recombinant DNA polymerase gamma and mitochondrial DNA (mtDNA) helicase by variants of mitochondrial single-stranded DNA-binding protein (mtSSB) correlates with defects in mtDNA replication in animal cells. J Biol Chem. 2011;286:40649–58. doi: 10.1074/jbc.M111.289983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel G, Johnson DS, Sun B, et al. A257T linker region mutant of T7 helicase–primase protein is defective in DNA loading and rescued by T7 DNA polymerase. J Biol Chem. 2011;286:20490–9. doi: 10.1074/jbc.M110.201657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjoismaki JL, Goffart S, Spelbrink JN. Replication stalling by catalytically impaired Twinkle induces mitochondrial DNA rearrangements in cultured cells. Mitochondrion. 2011;11:630–4. doi: 10.1016/j.mito.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Pohjoismaki JL, Goffart S, Tyynismaa H, et al. Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J Biol Chem. 2009;284:21446–57. doi: 10.1074/jbc.M109.016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad C, Melancon SB, Rupar CA, et al. Exome sequencing reveals a homozygous mutation in TWINKLE as the cause of multisystemic failure including renal tubulopathy in three siblings. Mol Genet Metab. 2013;108:190–4. doi: 10.1016/j.ymgme.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Raghunathan S, Ricard CS, Lohman TM, Waksman G. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multi-wavelength X-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proc Natl Acad Sci U S A. 1997;94:6652–7. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymer RU, Solorio FA, Tehranchi AK, et al. Binding mechanism of metalNTP substrates and stringent-response alarmones to bacterial DnaG-type primases. Structure. 2012;20:1478–89. doi: 10.1016/j.str.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Martinez A, Calleja M, Peralta S, et al. Modeling pathogenic mutations of human twinkle in Drosophila suggests an apoptosis role in response to mitochondrial defects. PLoS One. 2012;7:e43954. doi: 10.1371/journal.pone.0043954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen D, Nandakumar D, Tang GQ, Patel SS. Human mitochondrial DNA helicase TWINKLE is both an unwinding and annealing helicase. J Biol Chem. 2012;287:14545–56. doi: 10.1074/jbc.M111.309468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutt TE, Gray MW. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet. 2006a;22:90–5. doi: 10.1016/j.tig.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Shutt TE, Gray MW. Twinkle, the mitochondrial replicative DNA helicase, is widespread in the eukaryotic radiation and may also be the mitochondrial DNA primase in most eukaryotes. J Mol Evol. 2006b;62:588–99. doi: 10.1007/s00239-005-0162-8. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- Spelbrink JN, Li FY, Tiranti V, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet. 2001;28:223–31. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- Stiban J, Farnum GA, Hovde SL, Kaguni LS. The N-terminal domain of the Drosophila mitochondrial replicative DNA helicase contains an iron–sulfur cluster and binds DNA. J Biol Chem. 2014;289:24032–42. doi: 10.1074/jbc.M114.587774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth EA, Li Y, Sawaya MR, et al. The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol Cell. 2003;12:1113–23. doi: 10.1016/s1097-2765(03)00442-8. [DOI] [PubMed] [Google Scholar]
- Tyynismaa H, Mjosund KP, Wanrooij S, et al. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc Natl Acad Sci U S A. 2005;102:17687–92. doi: 10.1073/pnas.0505551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynismaa H, Sembongi H, Bokori-Brown M, et al. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum Mol Genet. 2004;13:3219–27. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–51. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanrooij S, Goffart S, Pohjoismaki JL, et al. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–51. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington MT, Rosenberg AH, Griffin K, et al. Biochemical analysis of mutant T7 primase/helicase proteins defective in DNA binding, nucleotide hydrolysis, and the coupling of hydrolysis with DNA unwinding. J Biol Chem. 1996;271:26825–34. doi: 10.1074/jbc.271.43.26825. [DOI] [PubMed] [Google Scholar]
- Wolf YI, Koonin EV. Origin of an animal mitochondrial DNA polymerase subunit via lineage-specific acquisition of a glycyl-tRNA synthetase from bacteria of the Thermus-Deinococcus group. Trends Genet. 2001;17:431–3. doi: 10.1016/s0168-9525(01)02370-8. [DOI] [PubMed] [Google Scholar]
- Yong Y, Romano LJ. Nucleotide and DNA-induced conformational changes in the bacteriophage T7 gene 4 protein. J Biol Chem. 1995;270:24509–17. doi: 10.1074/jbc.270.41.24509. [DOI] [PubMed] [Google Scholar]
- Ziebarth TD, Farr CL, Kaguni LS. Modular architecture of the hexameric human mitochondrial DNA helicase. J Mol Biol. 2007;367:1382–91. doi: 10.1016/j.jmb.2007.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebarth TD, Gonzalez-Soltero R, Makowska-Grzyska MM, et al. Dynamic effects of cofactors and DNA on the oligomeric state of human mitochondrial DNA helicase. J Biol Chem. 2010;285:14639–47. doi: 10.1074/jbc.M109.099663. [DOI] [PMC free article] [PubMed] [Google Scholar]