Abstract
A growing number of studies have highlighted the role of microRNAs (miRNAs or miRs) in the development and progression of cancer. In particular, the aberrant expression of cancer-related proteins, such as oncogenes and tumor suppressors has been shown to correlate with the modulation of the expression of specific miRNAs. In the present study, we aimed to determine which downregulated miRNAs may be involved in modulating the expression of the oncogenic transcription factor, Yin Yang 1 (YY1). YY1 has been reported to be overexpressed in several malignancies and our previous studies have highlighted the significant correlation between the levels of YY1 and aggressive behavior in non-Hodgkin's lymphoma (NHL). A total of 57 miRNAs that are potentially capable of targeting YY1 was identified through in silico approaches. The search of publicly available NHL datasets, including paired mRNA and miRNA data (GSE23026) highlighted a significant correlation (Pearson's correlation, r>0.5) between the expression levels of YY1 and the expression levels of a limited set of miRNAs, including miR-363, miR-200a, miR-23b, miR-15a and miR-15b. Intriguingly, both hsa-miR-363 and hsa-miR-200a belong to the top 20 miRNAs that were found to be downregulated in Burkitt's lymphoma (BL) tissue compared to normal tissue. Although further validation studies are warranted, the identification of these two miRNAs associated with the upregulation of YY1 in BL may provide further insight into the pathogenesis of this tumor and may contribute to more personalized and targeted treatment approaches for patients with this disease.
Keywords: hsa-miR-363, hsa-miR-200a, Yin Yang 1, Burkitt's lymphoma
Introduction
MicroRNAs (miRNAs or miRs) are small non-coding RNAs, comprising 19–25 nucleotides, that reduce the abundance and translational efficiency of mRNAs and play a major role in regulatory networks, influencing diverse biological processes through the effects of individual miRNAs on the translation of multiple mRNAs (1,2). Determining the roles of individual miRNAs in cellular regulatory processes poses a major challenge with the function of the majority of miRNAs that are currently unknown; even for relatively well-studied miRNAs, only a handful of targets have been rigorously characterized and these may differ by tissue type.
miRNAs post-transcriptionally regulate the expression of thousands of genes, including key genes in cell differentiation and cancer pathogenesis (3). A growing number of studies have highlighted the role of miRNAs in the development and progression of cancer (4,5). In particular, the aberrant expression of cancer-related proteins, such as oncogenes and tumor suppressors has been shown to correlate with the modulation of the expression specific miRNAs.
Non-Hodgkin's lymphomas (NHLs) are a heterogeneous group of malignancies of the lymphoid system. NHLs are classified based on cytology, immunophenotype and both genetic and clinical characteristics (6). Genetic aberration and molecular changes are used for a more appropriate diagnostic profile. The most commonly associated genetic abnormality in NHL is the translocation of t(14;18)(q32;q21) (7), mostly detected in low-grade lymphomas, such as follicular lymphoma; while molecular rearrangements involving Bcl-6 or c-Myc are frequently observed in diffuse large B-cell lymphoma (DLBCL) and Burkitt's lymphoma (BL), respectively. Currently, a significant percentage of patients with NHL are successfully cured; however, a fraction of these patients may relapse following treatment due to the development of chemoresitance (8,9).
It has been demonstrated that the transcription factor, Yin Yang 1 (YY1), is involved in the regulation of resistance to chemotherapy and immunotherapy in NHL cell lines. In addition, the regulation of Fas resistance by nuclear factor (NF)-κB is mediated via YY1 expression and activity (10). YY1 is known to play a critical role in normal biological processes, such as embryogenesis, differentiation, replication and cell proliferation. YY1 exerts its effects on genes involved in these processes through its ability to initiate, activate, or repress transcription, depending upon the context in which it binds (11).
It has also been reported that YY1 is overexpressed in several malignancies, including B-cell NHL (12,13), and this overexpression is associated with aggressive behaviour in NHL (14). Based on these findings, in the current study, miRNAs potentially capable of targeting and modulating YY1 were identified through in silico approaches.
Data collection methods
In order to identify putative miRNAs that target YY1 mRNA, several publicly available miRNA target prediction tools were used, including DIANA-microT (15), miRanda (16), PicTar (17) and TargetScanS (18). Publicly available Gene Expression Omnibus datasets of NHL specimens with the relative normal counterparts were analyzed. Only datasets with both mRNA and miRNA expression levels were considered for the analysis. According to these criteria, the dataset GSE23026 was used in our study (19). This dataset includes 12 samples of BL, 5 tonsil specimens, 7 lymph node specimens and 4 samples of CD19+CD10+ B-cells.
Statistical analysis
The differential analysis of YY1 expression levels was performed using the ANOVA test. Correlation analysis between the miRNA expression and YY1 expression levels was performed using the Pearson correlation test.
Results
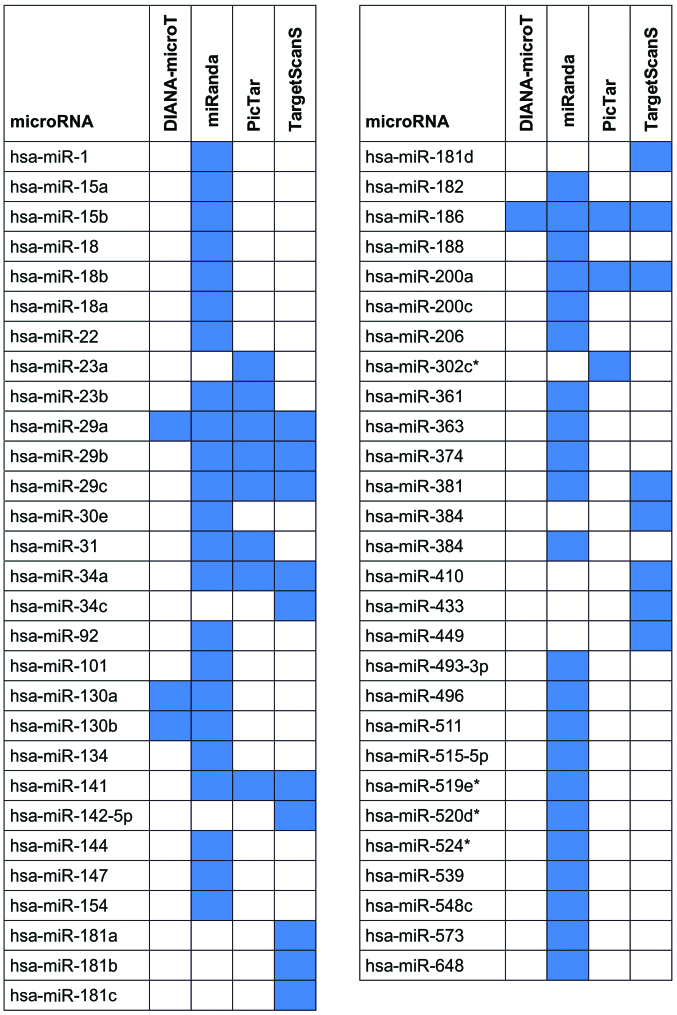
The analysis using the Diana-microT, miRanda, PicTar, TargetScanS tools revealed a total of 57 predicted miRNAs capable of targeting YY1 (Fig. 1).
Figure 1.
Predicted miRNAs capable of targeting YY1 identified using the Diana-microT, miRanda, PicTar and TargetScanS tools. The tools that recognized each predicted miRNA are indicated by the blue-colored boxes.
The search of publicly available NHL datasets, including paired mRNA and miRNA data (GSE23026) highlighted a significant negative correlation (Pearson correlation coefficient ≤-0.5) between the YY1 expression levels and the expression of a limited set of miRNAs, including miR-363, miR-200a, miR-23b, miR-15a and miR-15b among the 57 predicted miRNAs capable of targeting YY1 (Table I).
Table I.
Pearson correlation analysis between microRNAs targeting YY1 and YY1 expression levels.
| miRNA | Pearson correlation (r≤-0.5) | miRNA ID |
|---|---|---|
| ahsa-miR-363 | −0.611 | A_25_P00010953 |
| ahsa-miR-200a | −0.608 | A_25_P00010209 |
| hsa-miR-23b | −0.577 | A_25_P00010881 |
| hsa-miR-200a | −0.576 | A_25_P00010208 |
| hsa-miR-15a | −0.552 | A_25_P00010467 |
| ahsa-miR-363 | −0.549 | A_25_P00010954 |
| hsa-miR-15b | −0.517 | A_25_P00011101 |
MicroRNAs (miRNAs or miRs) belonging to the top 20 miRNAs that were found to be differentially expressed in Burkitt's lymphoma samples compared to the normal tissue samples. YY1, Yin Yang 1.
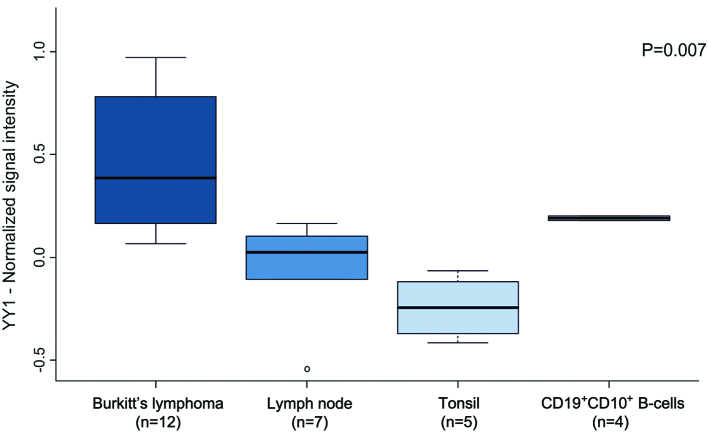
As was expected, the transcript expression levels of YY1 were found to be significantly higher in the BL samples compared to the normal tissue samples (mean value expression for BL=0.458, tonsil=−0.248, lymph node=0.049 and CD19+CD10+ B-cells=0.197; ANOVA, P=0.007) (Fig. 2). Intriguingly, both hsa-miR-363 and hsa-miR-200a belong to the top 20 miRNAs that were found to be downregulated in the BL samples compared to their normal counterparts (Table II).
Figure 2.
Yin Yang 1 transcript levels in Burkitt's lymphoma (GSE23026) are reported as boxed quartiles (median, 25th, and 75th percentile) and whiskers (minimum and maximum). The differences between tumor and normal samples are compared by the ANOVA test.
Table II.
Top 20 microRNAs downregulated in Burkitt's lymphoma samples compared to normal tissue samples.
| miRNA | P-value | FC (log) | miRNA ID |
|---|---|---|---|
| hsa-miR-150 | 2.32E-04 | −3.991 | A_25_P00010490 |
| hsa-miR-150 | 3.13E-04 | −3.869 | A_25_P00010489 |
| hsa-miR-363 | 1.71E-04 | −3.745 | A_25_P00010954 |
| hsa-miR-99a | 1.10E-06 | −3.686 | A_25_P00010471 |
| hsa-miR-10b_v9.1 | 5.03E-05 | −3.657 | A_25_P00010136 |
| hsa-miR-10b_v9.1 | 3.53E-05 | −3.607 | A_25_P00010135 |
| hsa-miR-139_v9.1 | 2.73E-05 | −3.594 | A_25_P00010349 |
| hsa-miR-363 | 9.85E-05 | −3.547 | A_25_P00010953 |
| hsa-let-7g_v9.1 | 7.09E-06 | −3.431 | A_25_P00010306 |
| hsa-miR-31_v9.1 | 6.93E-05 | −3.426 | A_25_P00010510 |
| hsa-let-7g_v9.1 | 5.21E-06 | −3.404 | A_25_P00010305 |
| hsa-miR-200b_v9.1 | 1.01E-03 | −3.389 | A_25_P00010908 |
| hsa-let-7a | 7.41E-07 | −3.37 | A_25_P00011584 |
| hsa-let-7f | 1.59E-06 | −3.335 | A_25_P00010088 |
| hsa-miR-195 | 5.00E-06 | −3.25 | A_25_P00010769 |
| hsa-miR-133b | 7.41E-04 | −3.233 | A_25_P00010963 |
| hsa-miR-195 | 9.00E-06 | −3.198 | A_25_P00010770 |
| hsa-let-7e_v9.1 | 1.04E-06 | −3.192 | A_25_P00010280 |
| hsa-miR-200a | 8.78E-04 | −3.184 | A_25_P00010209 |
| hsa-let-7e_v9.1 | 6.81E-07 | −3.181 | A_25_P00010279 |
Analysis of microRNAs (miRNAs or miRs) was performed using GEO2R tools. FC, fold change.
Discussion
A growing body of evidence has indicated that deregulated miRNA expression is observed in a variety of human diseases, including lymphomas, suggesting an important role in their pathogenesis (20,21). The data from several experimental studies have supported an important role for miRNAs in lymphomagenesis through systematic miRNA profiling in lymphoma samples using a variety of miRNA expression platforms (22–24).
YY1 is an activator and a repressor transcription factor belonging to the Zinc-finger proteins (25). It is a positive regulator of several oncogenes, such as c-Fos (26), c-Myc (27) and Erb-B2 receptor tyrosine kinase 2 (ERBB2) (28,29). However, it is also a negative regulator of a fraction of tumor suppressor genes, including p27 (30), p16 (31), p73 (32) and p53 (33). YY1 plays a role in the malignant transformation of several diseases, including B-cell lymphomas. Based on these findings, the aim of the present study was to identify, through in silico approaches, which miRNAs are potentially capable of targeting and modulating YY1.
Computational evaluation is an appropriate approach for an initial screening to identify deregulated miRNAs. Based on our results, we speculated that YY1 overexpression may be caused by the downregulation of both hsa-miR-363 and hsa-miR-200a, at least in BL. In fact, the overexpression of YY1 negatively correlated with the expression of hsa-miR-363 and hsa-miR-200a. In turn, the expression levels of hsa-miR-363 and hsa-miR-200a were lower in the BL samples when compared with the normal tissue samples. The increased transcript levels of YY1 in the BL samples is in agreement with the observations of a previous study in which the expression of YY1 was found to be associated with high-grade lymphomas, suggesting that it is linked with post-germinal center transit, where genetic alterations may occur, such as Bcl-6 mutations and/or Bcl-6 translocations (34). These findings may provide further insight into the pathogenesis of BL, and YY1 may thus be considered as a molecular signature of aggressiveness in high-grade lymphomas.
Notably, it has been previously reported that the overexpression of YY1 may promote p53 degradation or inhibit its transcriptional activity (35). Furthermore, the inhibition of YY1 in cancer cells, which lack functional p53, also sensitizes them to cell death in response to apoptotic stimuli (36,37). Although p53 is seldom mutated in NHL, the p53 pathway has been reported to be attenuated in these tumors (38). Among the targets of p53, miR-34a has been found to be repressed as a consequence of promoter methylation (39). These findings support the notion that YY1 may directly control the expression of genes involved in the apoptotic machinery independently of the p53 protein. However, certain drug-resistant hematopoietic disorders that exhibit wild-type p53 may benefit from targeted therapy (40).
In agreement with our results, the loss of miR-200a expression in primary cutaneous melanoma is considered as an additional indicator of aggressiveness, as it was associated with increased thickness and disease progression (41).
Low levels of miR-200a have also been observed in mucosa-associated lymphoid tissue lymphoma (MALT) lymphoma of the conjunctiva. It was also found that cyclin E2 is a target of miR-200a (42). Accordingly, the putative role of YY1 in tumorigenesis is sustained by its interaction with cell cycle regulation. The downregulated expression of miR-363 can precisely differentiate the high-grade lymphomas into ABC or GCB subtypes, and its expression has been shown to correlate with the clinical outcome (43). Similarly, the downregulation of miR-363 has also been detected in ovarian cancer and has been shown to be associated with the drug-resistance phenomenon (44).
Although further validation studies are warranted, the results of the present study indicate that the negative correlation between miR-200a and miR-363 and YY1 expression, which is observed in BL, may provide further insight into the pathogenesis of BL and may have implications on treatment strategies. Furthermore, our data suggest that miRNA expression profiling provides a useful tool for a better understanding of cancer development and progression and for the identification of molecular predictors of response and outcome.
Acknowledgements
This study was supported in part by Lega Italiana per la Lotta contro i Tumori and by the Jonsson Cancer Research Center.
References
- 1.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: T arget recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 4.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: Diagnostics monitoring and therapeutics. Histopathology. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaffe ES. The 2008 WHO classification of lymphomas: Implications for clinical practice and translational research. Hematology Am Soc Hematol Educ Program. 2009;2009:523–531. doi: 10.1182/asheducation-2009.1.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yunis JJ, Frizzera G, Oken MM, McKenna J, Theologides A, Arnesen M. Multiple recurrent genomic defects in follicular lymphoma. Histopathology. N Engl J Med. 1987;316:79–84. doi: 10.1056/NEJM198701083160204. [DOI] [PubMed] [Google Scholar]
- 8.Ansell SM. Non-HodgkinL ymphoma: Diagnosis and Treatment. Mayo Clin Proc. 2015;90:1152–1163. doi: 10.1016/j.mayocp.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 9.Mehta-Shah N, Younes A. Novel targeted therapies in diffuse large B-cell lymphoma. Semin Hematol. 2015;52:126–137. doi: 10.1053/j.seminhematol.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Vega MI, Jazirehi AR, Huerta-Yepez S, Bonavida B. Rituximab-induced inhibition of YY1 and Bcl-xL expression in Ramos non-Hodgkin's lymphoma cell line via inhibition of NF-kappa B activity, role of YY1 and Bcl-xL in Fas resistance and chemoresistance, respectively. J Immunol. 2005;175:2174–2183. doi: 10.4049/jimmunol.175.4.2174. [DOI] [PubMed] [Google Scholar]
- 11.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: Structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 12.Castellano G, Torrisi E, Ligresti G, Malaponte G, Militello L, Russo AE, McCubrey JA, Canevari S, Libra M. The involvement of the transcription factor Yin Yang 1 in cancer development and progression. Cell Cycle. 2009;8:1367–1372. doi: 10.4161/cc.8.9.8314. [DOI] [PubMed] [Google Scholar]
- 13.Bonavida B, Huerta-Yepez S, Baritaki S, Vega M, Liu H, Chen H, Berenson J. Overexpression of Yin Yang 1 in the pathogenesis of human hematopoietic malignancies. Crit Rev Oncog. 2011;16:261–267. doi: 10.1615/CritRevOncog.v16.i3-4.90. [DOI] [PubMed] [Google Scholar]
- 14.Castellano G, Torrisi E, Ligresti G, Nicoletti F, Malaponte G, Traval S, McCubrey JA, Canevari S, Libra M. Yin Yang 1 overexpression in diffuse large B-cell lymphoma is associated with B-cell transformation and tumor progression. Cell Cycle. 2010;9:557–563. doi: 10.4161/cc.9.3.10554. [DOI] [PubMed] [Google Scholar]
- 15.Maragkakis M, Alexiou P, Papadopoulos GL, Reczko M, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, Simossis VA, et al. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics. 2009;10:295. doi: 10.1186/1471-2105-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Correction: Human MicroRNA targets. PLoS Biol. 2005;3:e264. doi: 10.1371/journal.pbio.0030264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 18.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bueno MJ Gómez, de Cedrón M, Gómez-López G, de Pérez Castro I, Di Lisio L, Montes-Moreno S, Martínez N, Guerrero M, Sánchez-Martínez R, Santos J, et al. Combinatorial effects of microRNAs to suppress the Myc oncogenic pathway. Blood. 2011;117:6255–6266. doi: 10.1182/blood-2010-10-315432. [DOI] [PubMed] [Google Scholar]
- 20.Di Lisio L, Sánchez-Beato M, Gómez-López G, Rodríguez ME, Montes-Moreno S, Mollejo M, Menárguez J, Martínez MA, Alves FJ, Pisano DG, et al. MicroRNA signatures in B-cell lymphomas. Blood Cancer J. 2012;2:e57. doi: 10.1038/bcj.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Troppan K, Wenzl K, Deutsch A, Ling H, Neumeister P, Pichler M. MicroRNAs in diffuse large B-cell lymphoma, implications for pathogenesis, diagnosis, prognosis and therapy. Anticancer Res. 2014;34:557–564. [PubMed] [Google Scholar]
- 22.Musilova K, Mraz M. MicroRNAs in B-cell lymphomas: How a complex biology gets more complex. Leukemia. 2015;29:1004–1017. doi: 10.1038/leu.2014.351. [DOI] [PubMed] [Google Scholar]
- 23.Garofalo M, Leva GD, Croce CM. MicroRNAs as anti-cancer therapy. Curr Pharm Des. 2014;20:5328–5335. doi: 10.2174/1381612820666140128211346. [DOI] [PubMed] [Google Scholar]
- 24.Lim EL, Trinh DL, Scott DW, Chu A, Krzywinski M, Zhao Y, Robertson AG, Mungall AJ, Schein J, Boyle M, et al. Comprehensive miRNA sequence analysis reveals survival differences in diffuse large B-cell lymphoma patients. Genome Biol. 2015;16:18. doi: 10.1186/s13059-014-0568-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 26.Lee HY, Chaudhary J, Walsh GL, Hong WK, Kurie JM. Suppression of c-Fos gene transcription with malignant transformation of human bronchial epithelial cells. Oncogene. 1998;16:3039–3046. doi: 10.1038/sj.onc.1201843. [DOI] [PubMed] [Google Scholar]
- 27.Liao WR, Hsieh RH, Hsu KW, Wu MZ, Tseng MJ, Mai RT. WuL ee YH and Yeh TS: The CBF1-independent Notch1 signal pathway activates human c-myc expression partially via transcription factor YY1. Carcinogenesis. 2007;28:1867–1876. doi: 10.1093/carcin/bgm092. [DOI] [PubMed] [Google Scholar]
- 28.Allouche A, Nolens G, Tancredi A, Delacroix L, Mardaga J, Fridman V, Winkler R, Boniver J, Delvenne P, Begon DY. The combined immunodetection of AP-2alpha and YY1 transcription factors is associated with ERBB2 gene overexpression in primary breast tumors. Breast Cancer Res. 2008;10:R9. doi: 10.1186/bcr1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Begon DY, Delacroix L, Vernimmen D, Jackers P, Winkler R. Yin Yang 1 cooperates with activator protein 2 to stimulate ERBB2 gene expression in mammary cancer cells. J Biol Chem. 2005;280:24428–24434. doi: 10.1074/jbc.M503790200. [DOI] [PubMed] [Google Scholar]
- 30.Wan M, Huang W, Kute TE, Miller LD, Zhang Q, Hatcher H, Wang J, Stovall DB, Russell GB, Cao PD, et al. Yin Yang 1 plays an essential role in breast cancer and negatively regulates p27. Am J Pathol. 2012;180:2120–2133. doi: 10.1016/j.ajpath.2012.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. Int J Cancer. 2012;130:1715–1725. doi: 10.1002/ijc.27316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu S, Murai S, Kataoka K, Miyagishi M. Cooperative regulation og p73 promoter by Yin Yang 1 and E2F1. Nucleic Acids Symp Ser (Oxf) 2007;51:347–348. doi: 10.1093/nass/nrm174. [DOI] [PubMed] [Google Scholar]
- 33.Grönroos E, Terentiev AA, Punga T, Ericsson J. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc Natl Acad Sci USA. 2004;101:12165–12170. doi: 10.1073/pnas.0402283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Migliazza A, Martinotti S, Chen W, Fusco C, Ye BH, Knowles DM, Offit K, Chaganti RS, Dalla-Favera R. Frequent somatic hypermutation of the 5′ noncoding region of the BCL6 gene in B-cell lymphoma. Proc Natl Acad Sci USA. 1995;92:12520–12524. doi: 10.1073/pnas.92.26.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Libra M, Torrisi E, Castellano G, Nicoletti F, Malaponte G, Mazzarino MC, Marconi A, Proietti L, Militello L, Bonavida B, et al. Computational Evaluation of Yin Yang 1 Transcript Levels in the Spectrum of B-cell Neoplasia. Immunopathol Dis Therap. 2010;1:115–125. [Google Scholar]
- 36.Sui G, Affar B, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 37.Affar B, Gay F, Shi Y, Liu H, Huarte M, Wu S, Collins T, Li E, Shi Y. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol. 2006;26:3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maestro R, Gloghini A, Doglioni C, Piccinin S, Vukosavljevic T, Gasparotto D, Carbone A, Boiocchi M. Human non-Hodgkin's lymphomas overexpress a wild-type form of p53 which is a functional transcriptional activator of the cyclin-dependent kinase inhibitor p21. Blood. 1997;89:2523–2528. [PubMed] [Google Scholar]
- 39.Chim CS, Wong KY, Qi Y, Loong F, Lam WL, Wong LG, Jin DY, Costello JF, Liang R. Epigenetic inactivation of the miR-34a in hematological malignancies. Carcinogenesis. 2010;31:745–750. doi: 10.1093/carcin/bgq033. [DOI] [PubMed] [Google Scholar]
- 40.McCubrey JA, Abrams SL, Ligresti G, Misaghian N, Wong EW, Steelman LS, Bäsecke J, Troppmair J, Libra M, Nicoletti F, et al. Involvement of p53 and Raf/MEK/ERK pathways in hematopoietic drug resistance. Leukemia. 2008;22:2080–2090. doi: 10.1038/leu.2008.207. [DOI] [PubMed] [Google Scholar]
- 41.van Kempen LC, van den Hurk K, Lazar V, Michiels S, Winnepenninckx V, Stas M, Spatz A, van den Oord JJ. Loss of microRNA-200a and c, and microRNA-203 expression at the invasive front of primary cutaneous melanoma is associated with increased thickness and disease progression. Virchows Arch. 2012;461:441–448. doi: 10.1007/s00428-012-1309-9. [DOI] [PubMed] [Google Scholar]
- 42.Cai J, Liu X, Cheng J, Li Y, Huang X, Li Y, Ma X, Yu H, Liu H, Wei R. MicroRNA-200 is commonly repressed in conjunctival MALT lymphoma, and targets cyclin E2. Graefes Arch Clin Exp Ophthalmol. 2012;250:523–531. doi: 10.1007/s00417-011-1885-4. [DOI] [PubMed] [Google Scholar]
- 43.Sandhu SK, Croce CM, Garzon R. Micro-RNA expression and function in lymphomas. Adv Hematol. 2011;2011:347137. doi: 10.1155/2011/347137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou J, Yin F, Wang Q, Zhang W, Li L. Analysis of microarray-identified genes and microRNAs associated with drug resistance in ovarian cancer. Int J Clin Exp Pathol. 2015;8:6847–6858. [PMC free article] [PubMed] [Google Scholar]