Abstract
Although several dozen studies of gene expression in sepsis have been published, distinguishing sepsis from a sterile systemic inflammatory response syndrome (SIRS) is still largely up to clinical suspicion. We hypothesized that a multicohort analysis of the publicly available sepsis gene expression data sets would yield a robust set of genes for distinguishing patients with sepsis from patients with sterile inflammation. A comprehensive search for gene expression data sets in sepsis identified 27 data sets matching our inclusion criteria. Five data sets (n = 663 samples) compared patients with sterile inflammation (SIRS/trauma) to time-matched patients with infections. We applied our multicohort analysis framework that uses both effect sizes and P values in a leave-one-data set-out fashion to these data sets. We identified 11 genes that were differentially expressed (false discovery rate ≤1%, inter–data set heterogeneity P > 0.01, summary effect size >1.5-fold) across all discovery cohorts with excellent diagnostic power [mean area under the receiver operating characteristic curve (AUC), 0.87; range, 0.7 to 0.98]. We then validated these 11 genes in 15 independent cohorts comparing (i) time-matched infected versus noninfected trauma patients (4 cohorts), (ii) ICU/trauma patients with infections over the clinical time course (3 cohorts), and (iii) healthy subjects versus sepsis patients (8 cohorts). In the discovery Glue Grant cohort, SIRS plus the 11-gene set improved prediction of infection (compared to SIRS alone) with a continuous net reclassification index of 0.90. Overall, multicohort analysis of time-matched cohorts yielded 11 genes that robustly distinguish sterile inflammation from infectious inflammation.
INTRODUCTION
Sepsis, a syndrome of systemic inflammation in response to infection, kills about 750,000 people in the United States every year (1). It is also the single most expensive condition treated in the United States, costing the healthcare system more than $20 billion annually (2). Prompt diagnosis and treatment of sepsis is crucial to reducing mortality, with every hour of delay increasing mortality risk (3). Sepsis is defined by the presence of the systemic inflammatory response syndrome (SIRS), in addition to a known or suspected source of infection (1). However, SIRS is not specific for sepsis, because sterile inflammation can arise as a nonspecific response to trauma, surgery, thrombosis, and other non-infectious insults. Thus, sepsis can be difficult to distinguish clinically from systemic inflammation caused by non-infectious sources, such as tissue trauma (4). There is no “gold standard” blood test for distinguishing patients with infections at the time of diagnosis before results become available from standard microbiological cultures. One of the most common biomarkers of infection, procalcitonin, has a summary area under the receiver operating characteristic (ROC) curve (AUC) of 0.78 (range, 0.66 to 0.90) (5). Several groups have evaluated whether cytokine or gene expression arrays can accurately diagnose sepsis; however, because of the highly variable nature of host response and human genetics, no robust diagnostic signature has been found (6–10).
Both infections and tissue trauma activate many of the same innate immune receptor families, such as the Toll-like receptors (TLRs) and NOD-like receptors (NLRs), and consequently activate largely overlapping transcriptional pathways. Thus, distinguishing conserved downstream effects attributable solely to infections has been exceedingly difficult. Recent work has shown that there are pattern recognition receptors potentially specific to pathogen response, including several glycan receptor families (11). Hence, it may be possible that an infection-specific immune response could be differentiated from sterile inflammation.
The ongoing search for new therapies for sepsis, and for new prognostic and diagnostic biomarkers, has generated several dozen microarray-based genome-wide expression studies over the past decade, variously focusing on diagnosis, prognosis, pathogen response, and underlying sepsis pathophysiology (10). Despite tremendous gains in the understanding of gene expression in sepsis, few insights have translated to improvements in clinical practice. Many of these studies have been deposited into public repositories such as the National Institutes of Health Gene Expression Omnibus (GEO) and ArrayExpress, and thus, there is now a wealth of publicly available data in sepsis. In particular, there are several studies comparing patients with sepsis to patients with noninfectious inflammation (such as SIRS) that occurs after major surgery, traumatic injury, or in non–sepsis-related intensive care unit (ICU) admission (thrombosis, respiratory failure, etc.).
One data set in particular, the Inflammation and Host Response to Injury Program (Glue Grant) (6, 12, 13), has yielded several important findings about the effects of time on gene expression after trauma and in sepsis. One part of the Glue Grant longitudinally examined gene expression in patients after severe traumatic injuries. Several groups have examined these data with respect to time; notable findings are that (i) more than 80% of expressed genes show differential expression after traumatic injury (6); (ii) different clusters of genes recover over markedly different time periods (12); (iii) differing scenarios of inflammation such as trauma, burns, and endotoxicosis exhibit similar gene expression changes (12); and (iv) the extent to which posttrauma gene expression profiles differ from those of healthy controls and their degree of gene expression recovery over time are correlated with clinical outcomes (13, 14). There is thus growing understanding of the importance of the changes that underlie recovery from trauma and their impact on specific clinical outcomes.
We hypothesized that only time-matched comparisons, such as those that compare SIRS/trauma to sepsis at the same clinical time points, would yield genes robustly diagnostic of sepsis. We carried out a comprehensive, time-course–based multicohort analysis of the publicly available gene expression data in sepsis to identify a conserved 11-gene set that can robustly distinguish noninfectious inflammation (such as SIRS, trauma, and ICU admissions) from inflammation due to acute infections, as in sepsis. This 11-gene set had excellent diagnostic power in the discovery cohorts and was then validated in 15 independent cohorts.
RESULTS
Comprehensive search and labeled principal components analysis visualizations
We identified 27 independent gene expression data sets that satisfied our criteria in GEO and ArrayExpress, from which we included a total of 2903 microarrays (table S1) (7, 8, 15–40). These 27 data sets comprised only 22 independent cohorts, because the six data sets from the Genomics of Pediatric SIRS/Septic Shock Investigators (GPSSSI) were combined into a single cohort containing 219 patients with SIRS or sepsis at ICU day 1 (15–20). Many of the samples used were from the Glue Grant trauma data sets, which have a total of 333 patients sampled at up to eight time points (1301 samples used here) after traumatic injury. These 27 data sets contain cohorts of children and adults, men and women, with a mix of community- and hospital-acquired sepsis, sampled from whole blood, neutrophils, and peripheral blood mononuclear cells (PBMCs).
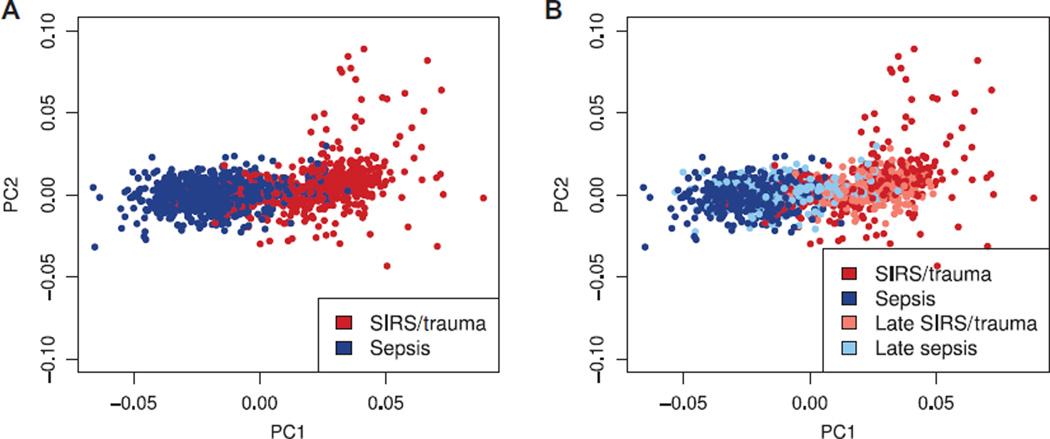
First, we sought to use the simplest possible methods to see whether noninfected SIRS/trauma patients and sepsis/infection patients could be separated by gene expression. We thus co-normalized all available data sets comparing SIRS/trauma with sepsis/infection in a single matrix. Labeled principal components analysis (PCA) (using 168 genes identified by 10-fold cross-validated Lasso-penalized logistic regression) showed that SIRS/trauma patients can be separated from sepsis patients with modest overlap (Fig. 1A). Next, we labeled each sample as “early” (within 48 hours of admission) or “late” (more than 48 hours after admission). Most of the nonseparable samples were the late samples (Fig. 1B). This finding remained true even when we included healthy controls as a separate class (fig. S1). Previous work has shown that gene expression after trauma, burns, or endotoxemia changes nonlinearly over time (6, 12, 14, 35). This continuous change in expression after initial insult could explain the inability to distinguish noninfected SIRS/trauma from sepsis in the late samples if all time points are treated as equal.
Fig. 1. Labeled PCA comparing sterile SIRS/trauma versus sepsis patients.
(A) Sterile SIRS/trauma and sepsis patients appear to be largely separable in the transcriptomic space, with only a minimal non-separable set. (B) The same labeled PCA is shown, with labels updated to reflect patients in recovery from noninfectious SIRS/trauma and patients with hospital-acquired sepsis; the late group (>48 hours after hospital admission) is much harder to separate. n = 1094 combined from 15 studies.
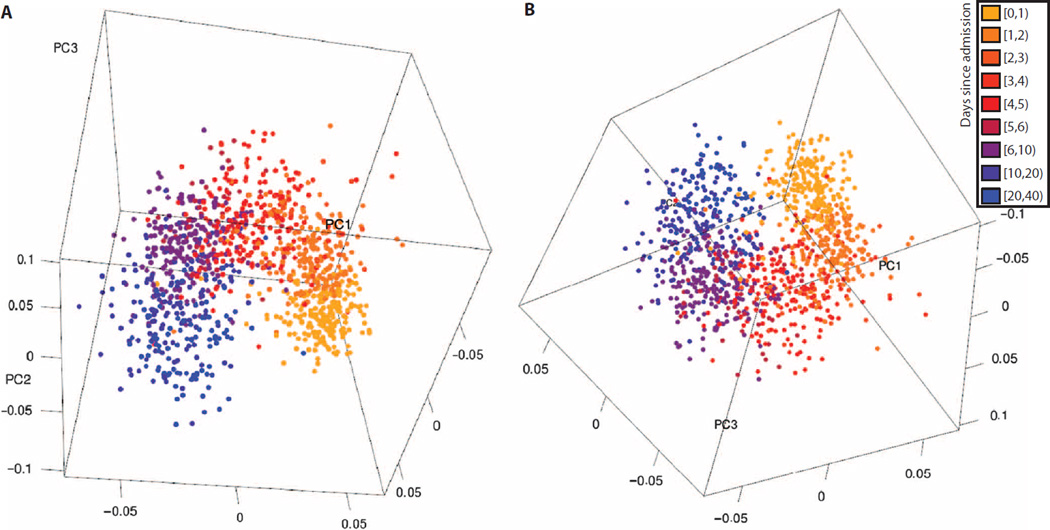
Therefore, we sought to get a qualitative sense of whether gene expression during the hospital course after injury is similar among different cohorts. We included all peripheral blood data sets that examined gene expression longitudinally over time after admission for nonseptic events. We used CUR matrix decomposition to identify the 100 genes that were most orthogonal to each other and used these to perform labeled PCA with classes determined by days after injury. Reassuringly, the gene expression at each time point was closest to the time points by which it was bounded (for example, the days [1,2) group was preceded by days [0,1) and followed by days [2,3); Fig. 2 and movie S1). Furthermore, changes in expression over time explained most variance in the data sets, as evidenced by the different day groups changing in each of the first three labeled principal components. In summary, our analysis showed that the changes in gene expression after trauma/ICU admissions (i) proceed in a nonlinear fashion over time and (ii) show similar changes over time across data sets.
Fig. 2. Two views of the first three principal components of labeled PCA of time-course data sets.
Five peripheral whole-blood gene expression data sets were combined and matched for common genes. The genes with the top 100 orthogonality scores were selected via CUR matrix decomposition, and labeled PCA was performed, broken into classes by day. (A and B) The three-dimensional plots of the first three principal components demonstrate that changes by day explain most variance in the data sets, different data sets show similar changes over time, and the changes over time proceed in a nonlinear fashion. Parts (A) and (B) show two different views of the same data; also see movie S1.
Time-matched multicohort analysis
Because changes in gene expression after admission for trauma explain a large amount of variance in the data set, and because these changes proceed nonlinearly, direct comparisons of a patient at admission with that same patient several days later at the time of infection would be confounded by “normal” changes in expression due to recovery from the inciting event, as well as any “abnormal” changes due to the hospital-acquired infection. It would be extremely difficult to disentangle these changes, if not impossible. Consequently, comparisons that do not take clinical time into account will not yield biomarkers that can robustly discriminate infected from noninfected patients (Fig. 1). Therefore, we focused only on infection data sets that also included a time-matched noninfected cohort (to allow for direct time-matched comparisons). We thus separated the data sets into two groups: (i) data sets comparing patients at hospital admission for trauma, surgery, or critical illness versus patients at admission to the hospital for sepsis [GSE28750 (27), GSE32707 (31), GSE40012 (26), and the GPSSSI unique combined data sets (n = 408 samples) (15–20)] and (ii) the Glue Grant data sets containing patients with hospital-acquired infections and day-matched noninfected patients, from which we used only patients in the buffy coat sample cohort (Table 1). The Glue Grant trauma cohorts were sampled at roughly 0.5, 1, 4, 7, 14, 21, and 28 days after injury; these cohorts were thus divided into their sampling time bins, creating subgroups in which patients diagnosed with an infection in a given time bin can be compared to noninfected patients in the same time bin. For the buffy coat samples, there were at least 10 patients present in five time bins, and these were thus taken for further study. Thus, we used a total of nine cohorts comparing time-matched SIRS/trauma to sepsis/infection, comprising 663 samples (326 SIRS/trauma controls and 337 sepsis/ infection cases; Table 2 shows the cohorts in the multicohort analysis; table S2 shows the individual microarray design matrix).
Table 1. Publicly available gene expression data sets comparing SIRS/ICU/trauma to sepsis/infections.
CAP, community-acquired pneumonia; ARDS, acute respiratory distress syndrome.
| Data set | Year | Submitting author |
Paper reference number |
Control used here | Condition used here | Sample type used |
Platform | Time points present in data set (days) |
|---|---|---|---|---|---|---|---|---|
| GPSSSI unique | 2006–2011 | Wong | 15–20 | Pediatric ICU-SIRS | Sepsis and septic shock (bacterial infections only) |
Whole blood |
GPL570 | 1–3 |
| GSE28750 | 2011 | Sutherland | 27 | 24 hours post–”major surgery” |
Community-acquired sepsis |
Whole blood | GPL570 | Admission |
| GSE32707 | 2012 | Dolinay | 31 | MICU patients ± SIRS, nonseptic |
Sepsis, sepsis + ARDS | Whole blood | GPL10558 | 0 and 7 |
| GSE40012 | 2012 | Parnell | 26 | SIRS (66% trauma) | Sepsis from CAP (bacterial infections only) |
Whole blood | GPL6947 | Days 1–5 for both SIRS and sepsis |
|
Glue Grant– Trauma–Buffy Coat |
2004–2006 | Multiple | 6, 12–13 | Trauma patients without infection |
Trauma patients ± 24 hours from diagnosis of infection |
Buffy coat | GPL570 | 0.5, 1, 4, 7, 14, 21, and 28 |
Table 2. All data sets used in the multicohort analysis.
The numbers after the Glue Grant cohort titles indicate days since infection in the given cohort (for instance, [1,3) are patients from 1 to 3 days since injury).
| Cohort | SIRS/ trauma controls (n) |
Sepsis/ infection cases (n) |
Total (n) |
|
|---|---|---|---|---|
|
Admission comparisons |
GSE28750 | 11 | 10 | 21 |
| GSE32707 | 55 | 48 | 103 | |
| GSE40012 | 24 | 41 | 65 | |
| GPSSSI unique | 30 | 189 | 219 | |
|
Hospital-acquired comparisons (Glue Grant buffy coat cohorts) |
Glue Grant buffy coat [1,3) |
65 | 9 | 74 |
|
Glue Grant buffy coat [3,6) |
63 | 17 | 80 | |
|
Glue Grant buffy coat [6,10) |
50 | 15 | 65 | |
|
Glue Grant buffy coat [10,18) |
22 | 4 | 26 | |
|
Glue Grant buffy coat [18,24) |
6 | 4 | 10 | |
| Total used in multicohort analysis | 326 | 337 | 663 | |
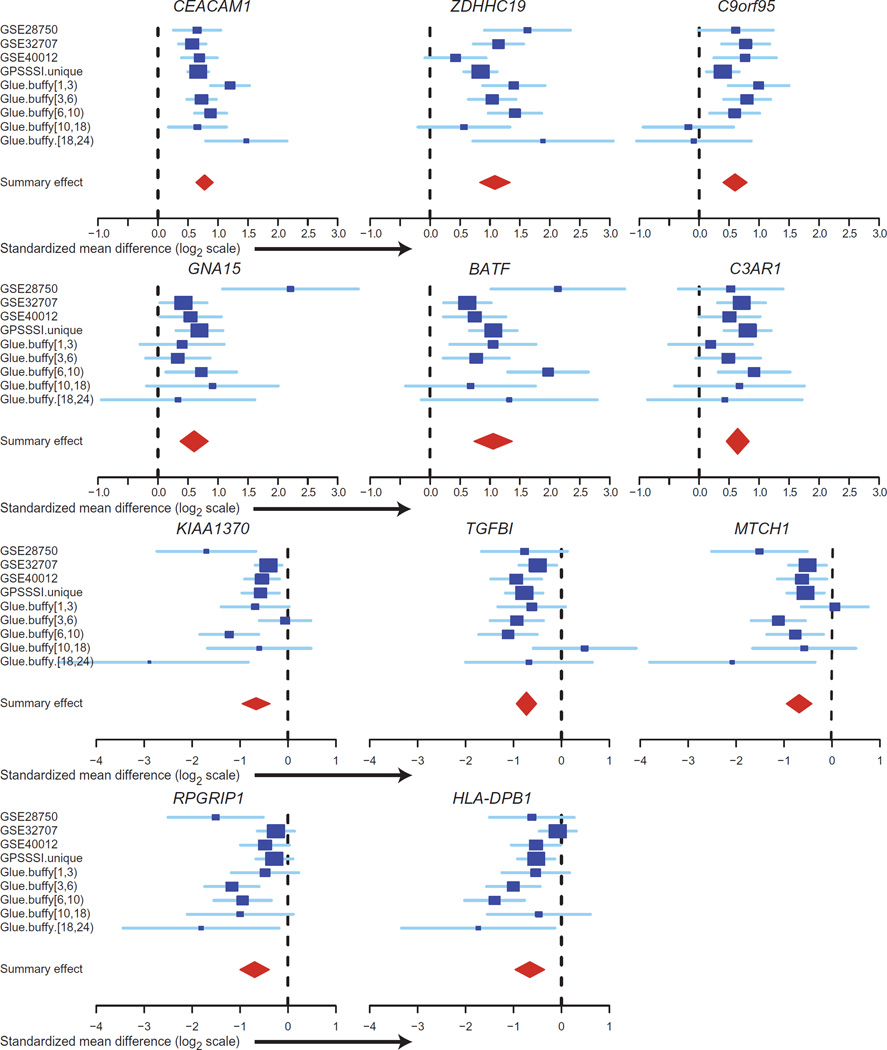
We then applied our previously described (41, 42) multicohort gene expression analysis framework to compare SIRS/trauma with sepsis/ infection, including all nine cohorts in a leave-one-data set-out fashion. The output from this analysis underwent a three-step thresholding process [false discovery rate (FDR) <1% for both pooled effect size and Fischer’s method, inter–data set heterogeneity P > 0.01, and absolute summary effect size fold change > 1.5], which yielded 82 genes differentially expressed between SIRS/trauma and sepsis patients across all time points (summary statistics for all 82 genes shown in table S3). To obtain the most parsimonious set of significant genes that best discriminates between classes, we carried out a greedy forward search to identify which combination of the 82 genes produced the best improvements in AUC across all discovery data sets. Here, discrimination is based on an “infection z score” that combines gene expression levels (using the difference of geometric means between positive and negative genes) into a standardized score for each sample in each data set. This yielded a final set of 11 genes (6 overexpressed and 5 underexpressed in sepsis; Table 3 and Fig. 3). Table S4 shows probe-level expression data for these 11 genes in the discovery cohorts. The mean ROC AUC of this 11-gene set in the nine discovery cohorts was 0.87 (range, 0.70 to 0.98; Fig. 4A and fig. S2).
Table 3. The 11-gene set that separates SIRS/trauma from sepsis.
Included are meta-analysis effect sizes, errors, and heterogeneity analyses.
| Gene symbol | Full name | Pooled effect size |
Pooled effect size, SE |
Effect size, P |
Effect size, Q (FDR) |
Sum of logs, Q (FDR) |
Cochran’s Q |
τ2 | Interstudy heterogeneity, P |
|---|---|---|---|---|---|---|---|---|---|
| CEACAM1 | Carcinoembryonic antigen- related cell adhesion molecule 1 |
0.778 | 0.073 | 1.9 ×10−26 | 6.5 × 10−22 | 7.2 × 10−14 | 16.43 | 0.023 | 0.037 |
| ZDHHC19 | Zinc finger, DHHC-type containing 19 |
1.083 | 0.130 | 6.6 ×10−17 | 3.3 × 10−13 | 7.1 × 10−22 | 18.36 | 0.078 | 0.019 |
| C9orf95 | Nicotinamide riboside kinase 1 |
0.598 | 0.102 | 4.6 × 10−9 | 1.4 × 10−6 | 2.9 × 10−12 | 12.64 | 0.032 | 0.125 |
| GNA15 | Guanine nucleotide binding protein (G protein), α-15 |
0.603 | 0.119 | 4.0 × 10−7 | 4.3 × 10−5 | 4.7 × 10−8 | 10.66 | 0.030 | 0.222 |
| BATF | Basic leucine zipper transcription factor, ATF-like |
1.053 | 0.163 | 9.4 ×10−11 | 6.2 × 10−8 | 1.6 ×10−19 | 17.49 | 0.115 | 0.025 |
| C3AR1 | Complement component 3a receptor 1 |
0.643 | 0.097 | 3.7 ×10−11 | 3.0 × 10−8 | 9.3 × 10−8 | 3.99 | 0.000 | 0.858 |
| KIAA1370 | Family with sequence similarity 214, member A |
−0.664 | 0.148 | 7.8 × 10−6 | 4.5 × 10−4 | 2.1 × 10−10 | 18.79 | 0.095 | 0.016 |
| TGFBI | Transforming growth factor β–induced, 68 kD |
−0.730 | 0.108 | 1.2 ×10−11 | 1.1 × 10−8 | 2.0 ×10−10 | 9.12 | 0.013 | 0.333 |
| MTCH1 | Mitochondrial carrier 1 | −0.686 | 0.135 | 4.1 × 10−7 | 4.3 × 10−5 | 7.9 ×10−10 | 13.04 | 0.058 | 0.111 |
| RPGRIP1 | Retinitis pigmentosa GTPase regulator interacting protein 1 |
−0.694 | 0.156 | 8.5 × 10−6 | 4.7 × 10−4 | 5.2 × 10−9 | 16.93 | 0.103 | 0.031 |
| HLA-DPB1 | Major histocompatibility complex, class II, DP β1 |
−0.659 | 0.157 | 2.6 × 10−5 | 1.1 × 10−3 | 8.6 × 10−9 | 17.38 | 0.107 | 0.026 |
Fig. 3. Effect sizes of the 11-gene set.
Forest plots for random effects model estimates of effect size of the positive genes, comparing SIRS/ trauma/ICU to infection/sepsis patients in each of the discovery cohorts.
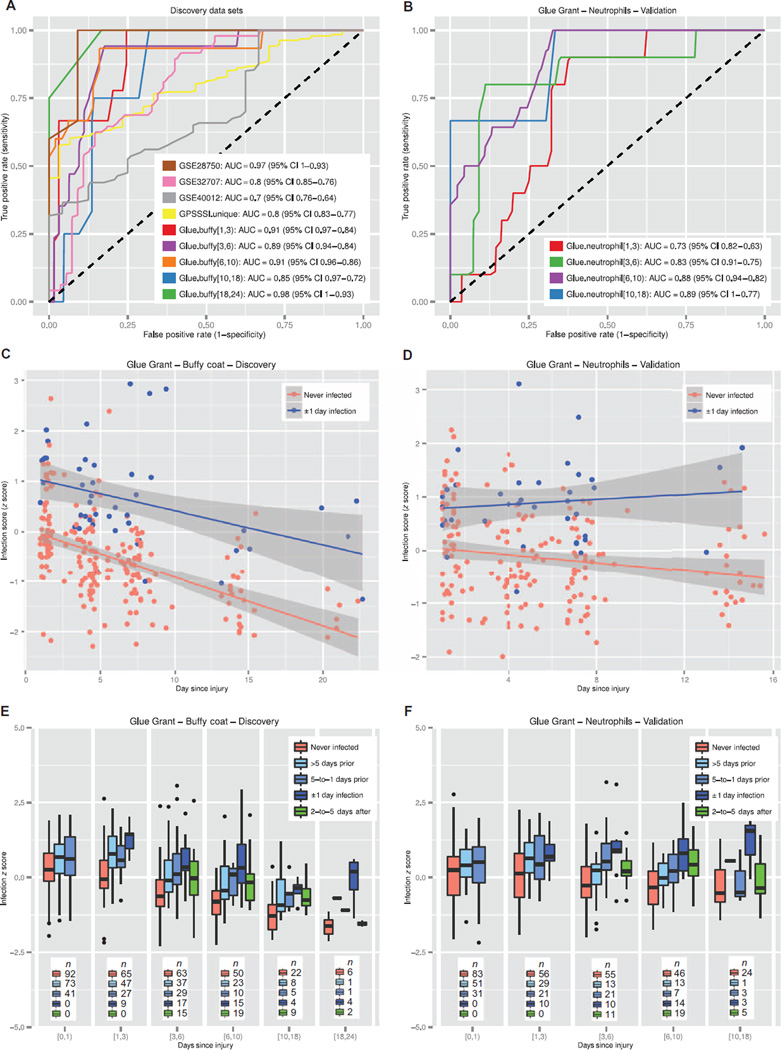
Fig. 4. Results of the 11-gene set in the discovery and neutrophils validation data sets.
(A) ROC curves shown for separating sterile SIRS/ICU/trauma patients from those with sepsis in the discovery data sets. (B) ROC curves shown for separating trauma patients with infections from time-matched trauma patients without infection in the Glue Grant neutrophils validation data sets. (C and D) Glue Grant buffy coat discovery (C) and neutrophils validation samples (D) after >1 day since injury, showing average infection z score in noninfected patients versus patients within ±24 hours of diagnosis. In both cases, there is a significant effect due to both time and infection status. (E and F) Box plots of infection z score by time since injury for buffy coat discovery (E) and neutrophils validation samples (F): patients never infected are compared to patients >5 days before infection, 5-to-1 days before infection, ±1 day of diagnosis (cases), and 2-to-5 days after infection diagnosis. JT trend test was significant (P < 0.01) for an increasing trend from never infected to ±1 day of infection for each time point after admission.
Glue Grant sorted-cells cohort validation
The Glue Grant trauma cohorts have two independent subcohorts: one is the buffy coat cohort (samples processed from 2004 to 2006 on Affymetrix array GPL570), and the other is the sorted-cells cohort, which included neutrophils, monocytes, and T cells [samples processed from 2008 to 2011 on custom Glue Grant Human (GGH) arrays; Table 4]. These cohorts are separate patients, separated in time and profiled using different technologies. Although their inclusion criteria and enrolling sites are largely the same, they are otherwise independent. We thus validated our 11-gene signature in the Glue Grant sorted-cells cohorts. Here, we split the sorted-cells cohorts into the same time bins as the discovery buffy coat cohorts and treated each time bin separately.
Table 4. The Glue Grant sorted-cells cohort.
| Data set | Year | Submitting author |
Paper reference number |
Control used here |
Condition used here |
Sample used here |
Platform | Days since injury |
n control |
n case |
|---|---|---|---|---|---|---|---|---|---|---|
| Glue Grant sorted-cells |
2008–2011 | Multiple | 6 | Trauma patients without infection |
Trauma patients ± 24 hours from diagnosis of infection |
Neutrophils, monocytes, and T cells |
GGH-1, GGH-2 (27) |
[1,3) | 56 | 10 |
| [3,6) | 55 | 10 | ||||||||
| [6,10) | 46 | 14 | ||||||||
| [10,18) | 24 | 3 |
From the sorted-cells subcohort, we expected the neutrophil set to perform most similarly to a whole-blood sample, because neutrophils make up 75 to 85% of the total leukocyte pool after trauma in both infected and noninfected patients (and hence most of the gene expression present in peripheral blood) (fig. S3). Indeed, the 11-gene set performed very well at separating time-matched noninfected trauma patients from septic trauma patients (4 cohorts; 218 samples; mean AUC, 0.83; range, 0.73 to 0.89) (Fig. 4B). Surprisingly, the 11-gene set also showed discriminatory power in the monocytes and T cells from these same patients (monocytes AUC range, 0.71 to 0.97; T cells AUC range, 0.69 to 0.9) (figs. S4 and S5). Because we excluded any sorted-cells data sets from the multicohort analysis, we did not expect diagnostic capability in these cell types. In the sorted-cells cohort, AUC increased with greater time since initial trauma; this may suggest that inflammation due to infection is easier to discriminate as the “genomic storm” of traumatic injury begins to recover.
Examination of the 11-gene set in the Glue Grant cohorts
As expected, in the Glue Grant buffy coat cohort, patients within ±24 hours of diagnosis of infection have significantly higher infection z scores at all time points as compared to time-matched patients with-out infection; this was validated in the neutrophils cohort [repeated-measures analysis of variance (ANOVA) P < 0.0001; Fig. 4, C and D, and table S5A]. Comparison of the infection z score by time since injury in the buffy coat cohort shows a significant decline over time (repeated-measures ANOVA change over time P < 0.0001), but there appears to be a lesser (though still significant) effect in the neutrophils validation cohort (repeated-measures ANOVA change over time P < 0.05) (Fig. 4, C and D, and table S5A). The interaction of group with time since injury was not significant in either discovery or validation cohorts, suggesting that the decline in infection z scores over time for both groups is likely due to recovery from traumatic injury resulting in reduced inflammation (table S5A).
Next, we analyzed how infection z scores changed in infected patients before and after diagnosis of infection (samples that were not included in identifying the 11-gene set). We grouped the samples from patients who were ever diagnosed with infection on the same hospital stay into four groups according to their time from diagnosis of infection (either greater than 5 days before infection, 5-to-1 days before infection, within ±1 day of diagnosis of infection, or 2-to-5 days after diagnosis of infection, where no group besides the ±1 day of diagnosis of infection was included in the multicohort analysis for discovery of the 11-gene set). We further divided these groups into bins according to days since injury. Within each time bin, the infection z scores for the diagnostic groups increased significantly as they progressed toward infection for both the discovery buffy coat cohort and the validation neutrophils cohort [Jonckheere trend (JT) test P < 0.01; Fig. 4, E and F]. Furthermore, in all cohorts, the infection z score declined in the groups that were 2 to 5 days after infection diagnosis, when patients were beginning to recover from infection, presumably due to antibiotic treatment. This may also explain the increase in diagnostic power as time increases since initial injury. We emphasize that the resulting “peak” in infection z score around the time of infection diagnosis validates the association of the infection z score with clinical infection, because neither the >5 days prior cohorts, the 5-to-1 days prior cohorts, nor the 2-to-5 days after cohorts were included in the multicohort analysis, but still shows the hypothesized trends in both the discovery buffy coat cohort and the validation neutrophils cohort. Similar results were seen in the monocytes and T cells samples (same patients as the neutrophils validation cohorts; figs. S4B and S5B).
The infection z scores for patients who were later infected during their hospital stays were significantly higher in buffy coat samples at the time of admission than for those never infected during their hospital admission (P < 0.01; neutrophils validation group P = 0.05; Fig. 4, E and F). One possibility is that there was a baseline difference in injury severity, and that this might influence the infection z score. Severely injured patients are known to be more susceptible to infection (43). To test this hypothesis, we used linear regression of eventual hospital-acquired infection status, injury severity score, and their interaction to predict infection z score as the independent variable (table S5B). Both eventual hospital-acquired infection status and injury severity score were independently significant in predicting infection z score at admission, indicating that injury severity alone does not explain these effects. The interaction term was significant and negative in both the discovery buffy coat cohort and the validation neutrophils cohort samples, perhaps suggesting that higher infection z score at admission may indicate greater susceptibility to later infection. Further studies are needed to examine this observation.
Clinical utility in the Glue Grant
To test whether the infection z score might add to the clinical determinations of infection, we compared logistic regression using SIRS criteria alone to that using SIRS criteria plus our infection z score in discriminating Glue Grant trauma patients (both buffy coat and neutrophils cohorts) with and without infection. The logistic regression model using SIRS criteria alone had an AUC of 0.64, whereas SIRS criteria plus the infection z score had an overall AUC (using a single coefficient for infections at all time points) of 0.81 (fig. S6). The continuous net reclassification index (NRI) is a measure of how many patients would be correctly reclassified by improving a disease marker; here, the continuous NRI of adding the infection z score to SIRS alone was 0.90 (95% confidence interval, 0.62 to 1.17), where a continuous NRI greater than 0.6 is associated with “strong” improvement in prediction (44).
Independent validation of the infection z score
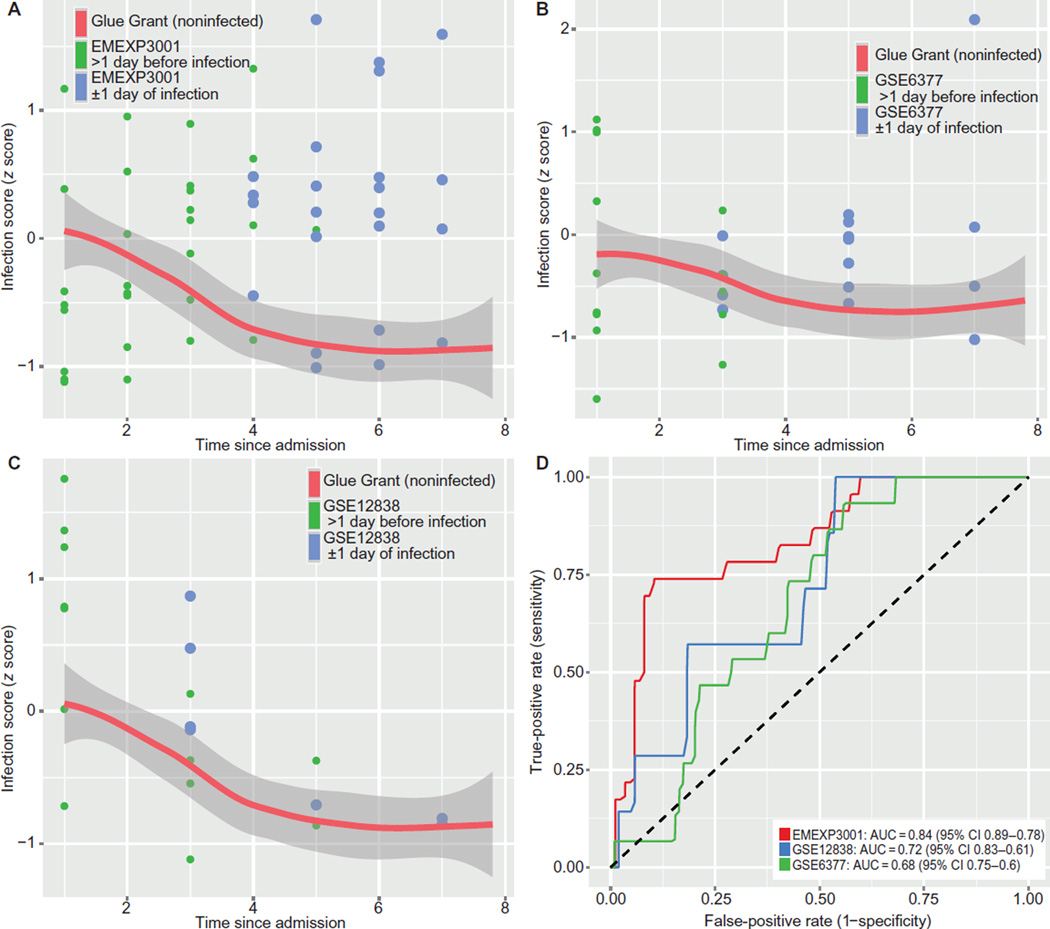
Next, we validated our score in three independent longitudinal cohorts that included only trauma or ICU patients who eventually acquired infections: GSE6377 (35), GSE12838, and EMEXP3001 (23) (Table 5). All three cohorts followed patients from the day of admission at least through the day of infection diagnosis (mostly VAP). Because all patients in each of the three cohorts acquired infections, they did not have time-matched noninfected controls. To compare the validation cohort infection cases with noninfected trauma patients, we used Glue Grant buffy coat noninfected controls. We internally normalized each cohort using housekeeping genes and then co-normalized with the Glue Grant buffy coat patients using empiric Bayes batch correction. Then, we compared the validation cohorts to the Glue Grant noninfected patients at matched time points as a variable reference. Comparing trauma/ICU patients to a time-matched baseline is necessary because our earlier findings (Fig. 4, C to F) showed a change over time in infection z score in the noninfected patients (table S5A). The three independent longitudinal trauma/ICU cohorts show that patients within ±1 day of infection are generally separable from time-matched non-infected Glue Grant patients, with ROC AUCs ranging from 0.68 to 0.84 (Fig. 5).
Table 5. Publicly available gene expression time-course data sets of trauma patients who develop infections.
VAP, ventilator-associated pneumonia; VAT, ventilator-associated tracheobronchitis; N/A, not available.
| Data set | Year | Submitting author |
Paper reference number |
Control used here |
Condition used here |
Sample type used |
Platform |
n control |
n case |
Samples used here |
Time points present in data set (days) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GSE6377 | 2008 | Cobb | 35 | None | ICU patients with eventual VAP |
Buffy coat | GPL201 | 0 | 11 | 99 | 1–21 |
| GSE12838 | 2008 | Cobb | N/A | None | ICU patients with eventual VAP |
Buffy coat and neutrophils |
GPL570 | 0 | 4 | 60 | 1–11 |
| E-MEXP-3001 | 2011 | Lopez | 23 | None | ICU patients with acquired VAP or VAT |
Whole blood | A-Agil-28 (GPL6480) |
0 | 8 | 56 | 1–7 |
Fig. 5. No-controls data sets of trauma/ICU patients who develop VAP.
These data sets did not include noninfected patients, so they were empiric Bayes co-normalized with time-matched Glue Grant patients. Orange line shows Glue Grant loess curve. (A) EMEXP3001. (B) GSE6377. (C) GSE12838, both neutrophils and whole-blood samples. In all cases, only the first 8 days since admission are shown, and patients are censored >1 day after diagnosis of infection. (D) ROC curves compare patients within ±1 day of diagnosis (blue points in A to C) with time-matched noninfected Glue Grant patients. See Table 5 for further data set details.
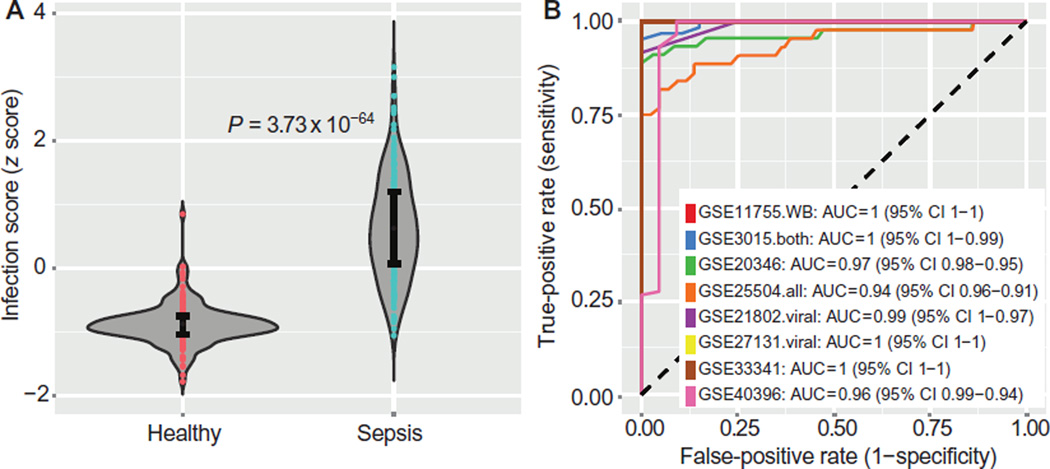
We further validated the 11-gene set in eight additional independent data sets that compared healthy controls to those with bacterial or viral sepsis at admission using whole-blood samples [n = 446: GSE11755 (38), GSE13015 (7), GSE20346 (37), GSE21802 (22), GSE25504 (39), GSE27131 (32), GSE33341 (30), and GSE40396 (25); Table 6]. The infection z scores for all eight data sets were combined in a single violin plot, showing excellent separation (Wilcoxon P < 1 × 10−63; Fig. 6A). The mean ROC for separating healthy and septic patients is 0.98 (range, 0.94 to 1.0; Fig. 6B).
Table 6. Publicly available gene expression data sets in whole blood or neutrophils comparing healthy controls to septic patients.
| Data set | Year | Submitting author |
Paper reference number |
Control used here |
Condition used here |
Sample type used |
Platform |
n control |
n case |
Time points present in data set (days) |
|---|---|---|---|---|---|---|---|---|---|---|
| GSE11755 | 2008 | Emonts | 38 | Healthy children |
Children with meningococcal sepsis |
Whole blood | GPL570 | 3 | 8 | 0, 0.33, 1, and 3 |
| GSE13015 | 2008 | Chaussabel | 7 | Healthy or type 2 diabetic |
Community- acquired or nosocomial sepsis |
Whole blood |
GPL6106 and GPL6947 |
20 | 63 | Mixed: admission or sepsis onset |
| GSE20346 | 2011 | Parnell | 37 | Pre-/post- influenza vaccine |
Bacterial pneumonia and severe influenza |
Whole blood | GPL6947 | 36 | 45 | 1–5 |
| GSE21802 | 2011 | Bermejo-Martin | 22 | Healthy | Severe influenza | Whole blood | GPL6102 | 4 | 12 | Early versus late sepsis |
| GSE25504 | 2014 | Smith | 39 | Healthy neonates |
Neonates with bacterial infection |
Whole blood |
GPL570, GPL6947, and GPL13667 |
44 | 44 | Infection onset |
| GSE27131 | 2011 | Berdal | 32 | Healthy | Severe influenza A | Whole blood | GPL6244 | 7 | 14 | 0–6 |
| GSE33341 | 2011 | Ahn | 30 | Healthy | Sepsis from bacterial CAP |
Whole blood | GPl571 | 43 | 51 | Admission |
| GSE40396 | 2013 | Hu | 25 | Healthy children after surgery |
Children with infection + fever |
Whole blood | GPL10558 | 22 | 30 | Admission |
Fig. 6. Discrimination of healthy versus sepsis.
Eight independent validation data sets that met inclusion criteria (peripheral whole blood or neutrophils, sampled within 48 hours of sepsis diagnosis) were tested with the infection z score. (A) Infection z scores for all patients (n = 446) were combined in a single violin plot; error bars show middle quartiles. P values calculated with Wilcoxon rank-sum test. (B) Separate ROC curves for each of the eight data sets discriminating sepsis patients from healthy controls. Mean ROC AUC = 0.98. See Table 6 for further data set details.
Our results provide strong evidence that the infection z score declines over time since admission/injury in whole blood, buffy coat, neutrophils, and monocytes. We have also shown that non–time-matched comparison yields inaccurate classification of infection, especially for late-acquired infections in SIRS/trauma patients. Hence, comparing infection z scores of SIRS/trauma patients at admission with those of late-acquired sepsis/infection patients would be an inaccurate measure of diagnostic power. However, because the effect of the decrease in infection z score over time is relatively monotonous, comparison of admission SIRS/trauma/surgery patients with late-acquired sepsis/infection would provide a lower limit on detection of ROC AUC for the infection z scores. That is, because the infection z score decreases over time, if the noninfected patients tested at admission had been sampled later (at matched times to the sepsis patients), their infection z scores would be lower at that later time (and hence more easily separable from the higher infection z scores in the septic patients). Using this inference, we examined four independent data sets that compared SIRS/trauma/ surgery patients either to the same patients later in their hospital course at onset of sepsis or to a mixed cohort of patients with community-and hospital-acquired sepsis. These data sets included whole blood [EMTAB1548 (21)], neutrophils [GSE5772 (29)], and PBMCs [GSE9960 (8); EMEXP3621 (40)] (Table 7). In each of these four data sets, the infection z score separated late-acquired infections from admission SIRS or trauma, with ROC AUCs ranging from 0.48 to 0.76 in PBMCs to 0.86 in whole blood (fig. S7). We emphasize that these AUCs are expected to be lower due to their time-mismatched comparison and are essentially the lower limits of what properly time-matched infection z scores would be in each of these cell compartments.
Table 7. Publicly available gene expression data sets comparing sterile SIRS/trauma patients to later or non–time-matched sepsis/infection patients.
| Data set | Year | Submitting author |
Paper reference number |
Control used here |
Condition used here |
Sample type used |
Platform |
n control |
n case |
Time points present in data set |
|---|---|---|---|---|---|---|---|---|---|---|
| GSE5772 | 2007 | Tang | 29 | ICU nonsepsis (43% trauma) |
Sepsis— early + late |
Neutrophils | GPL4274 | 23 | 70 | Mixed: admission or sepsis onset |
| GSE9960 | 2009 | Tang | 8 | ICU nonsepsis | Sepsis | Monocytes | GPL570 | 16 | 54 | Mixed: admission or sepsis onset |
| E-MEXP-3621 | 2012 | Harokopos | 40 | Trauma at admission |
Trauma at onset of sepsis |
Monocytes | A-Affy-37 (GPL571) |
5 | 5 | Admission versus onset of sepsis |
| E-MTAB-1548 | 2014 | Almansa | 21 | Postsurgery (average 2 days) |
Sepsis after surgery (average 4 days) |
Whole blood | A-MEXP-2183 (GPL10332) |
34 postsurgery |
39 | Postsurgery (average 2 days), onset of sepsis (average 4 days) |
Finally, we examined our 11-gene set in one data set comparing healthy controls or those with autoimmune inflammation to acute bacterial infections after diagnosis confirmation (GSE22098, n = 274) (33). Exact sampling times are not available, but typically, confirmation of infection takes 24 to 72 hours, so these infection samples are expected to show lower z scores than at the time of diagnosis. Still, the infection z score was able to discriminate healthy and autoimmune inflammation patients from those with acute infections (ROC AUC, 0.72; fig. S8). Considering that cohorts with autoimmune inflammation were not included in our discovery set, this provides validation of the specificity of the infection z score for infectious inflammation.
The effect of infection type on infection z score
To examine whether there were any infection type–specific differences in the infection z score, we compared patients infected with Gram-positive versus Gram-negative bacteria, as well as those with viral infections to those with bacterial infections. The Glue Grant patients were not analyzed, because there were too few time-matched infection patients in each subcohort. Four data sets had information on Gram-positive versus Gram-negative infection, and four had data on bacterial versus viral infections; in neither case was there a clear trend of differences in infection z score based on infection subtype (table S6).
Gene set pathway evaluation and transcription factor analysis
Having validated the 11-gene set, we examined whether any mechanism might explain why these genes were acting in concert. We analyzed the 11-gene set with Ingenuity Pathway Analysis, which showed that several of the genes are downstream of IL-6 and JUN (fig. S9). All 11 genes identified by the multicohort analysis were tested with both EncodeQT and PASTAA (chosen for a mix of experimental results and in silico transcription factor predictions). EncodeQT found only one significant transcription factor among the positive genes (MAX) and none for the negative genes (EncodeQT Q ≤ 0.01, table S7A). PASTAA showed enrichment for well-known proinflammatory transcription factors, such as nuclear factor κB (NF-κB) member c-REL, STAT5, and interferon response factors (IRF) 1 and 10 (table S7B).
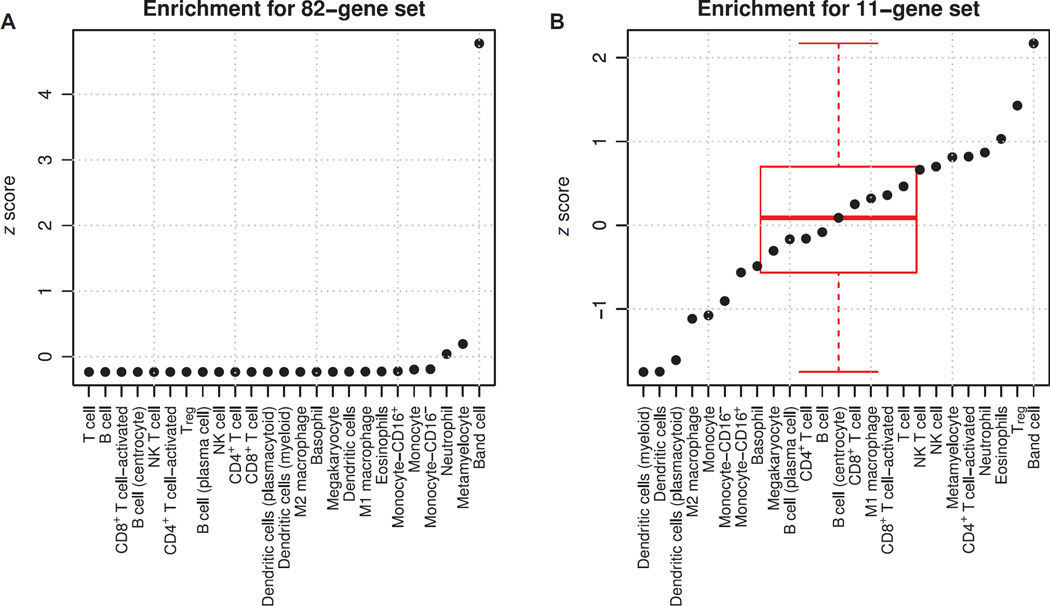
Because we did not find an obvious network driver, we next studied whether the genes were enriched in certain immune cell types that might explain their relation to sepsis. We searched GEO for human immune cell type–specific gene expression profiles and found 277 samples from 18 data sets matching our criteria (table S8). We aggregated these into broad immune cell type signatures using mean gene expression scores. We then calculated standardized enrichment scores using the same method as the infection z score (difference of geometric means between positive and negative genes). We did this both for the initial set of 82 genes found to be significantly enriched in the multicohort analysis and for the 11-gene set found after forward search (the genes included in the infection z score) (Fig. 7). The set of all 82 significant genes was found to be highly enriched in band cells only (>4 SDs above the mean; P < 1 × 10−6). The 11-gene set was significantly enriched (>2 SDs above the mean; P = 0.015) in band cells but also showed up-regulation in regulatory T cells (Tregs) and down-regulation in dendritic cells. This suggests that one driving force in differential gene expression between sterile SIRS and sepsis is the presence of band cells; however, the best set of genes for diagnosis contains information that may incorporate multiple cell type shifts at once. Finally, we checked whether there was a difference in band counts (where present) between acutely infected and noninfected patients in the Glue Grant trauma cohort, but there was no significant difference (mean noninfected = 2.13; mean infected = 2.74; P = 0.49).
Fig. 7. Cell type enrichment analyses.
(A and B) Standardized enrichment scores (z scores, dots) for human immune cell types for both (A) the entire set of 82 genes found to be significant in multicohort analysis and (B) the 11-gene set found after forward search (subset of the 82 genes). Part (B) also shows a box plot of distributions of z scores.
DISCUSSION
The dozens of studies that we examined here have reported valuable insights into changes in gene expression that occur in response to SIRS, trauma, surgery, and sepsis; one key insight is that time after injury is an important factor in gene expression after injury (6, 12, 14, 35). Across multiple independent cohorts, we showed that changes in gene expression over time during recovery are nonlinear but follow a similar trajectory (Fig. 2). Therefore, a comparison of gene expression across early and late time points in the same patient will yield a large number of differentially expressed genes solely due to the recovery process. It is thus very difficult to identify relatively small changes in gene expression due to late complication such as infection from the large changes caused by recovery. Therefore, we separated longitudinal studies into subcohorts of patients at matched time points. We used an integrated, time-course–based multicohort analysis (41, 42) to evaluate differential gene expression between sterile SIRS/trauma and sepsis/infection patients. We then used a forward search to select a parsimonious set of differentially expressed genes optimized for discriminatory power for sepsis. An infection z score, derived from the geometric mean of the 11-gene set, had a mean ROC AUC of 0.87 in the discovery cohorts for distinguishing SIRS/trauma from sepsis/ infection patients.
We validated the 11-gene set in an independent group of patients from the Glue Grant. The mean AUC for distinguishing sepsis from noninfectious inflammation was 0.83 in the neutrophils validation cohort, with a trend toward better diagnostic power with greater time since initial injury, when initial traumatic inflammation wanes and hospital-acquired infections manifest (43). Although we expect the whole-blood transcriptional profiles to be largely driven by neutrophils, the signal in sorted cells will certainly differ from whole blood. Despite this limitation, the infection z scores performed comparably in validation cohorts. We further validated the infection z score in several additional external data sets, which included three longitudinal cohorts of ICU/trauma patients who developed VAP/VAT; eight cohorts of healthy controls compared to patients with bacterial or viral sepsis; four cohorts of admission SIRS/trauma patients compared to patients at mixed or later time points using whole blood, neutrophils, and PBMCs; and one cohort of patients with autoimmune inflammation compared to patients with acute infection. Finally, we showed that the infection z score does not have systematic trends with regard to infection type (Gram-positive versus Gram-negative and bacterial versus viral) across those data sets for which infection type information is available.
Using the extensive clinical phenotype data available for patients in the Glue Grant, we illustrated two important points about the application of the infection z score. First, the infection z score showed a decline over time since injury that was similar in both infected and non-infected patients. Thus, for maximal discriminatory power, if the infection z score were to be tested in a longitudinal study, the diagnostic thresholds would need to be a function of the time since initial injury/ event. Second, the infection z scores increased over the days before infection, peaked within 1 day of diagnosis, and decreased afterwards (presumably due to treatment of infection). This observation raises the possibility that earlier diagnosis or stratification of patients at risk of developing sepsis may be possible using the 11-gene set. In particular, we note that the early rise in infection z score that precedes a clinical diagnosis of infection is not a false positive but an “early positive” result.
In the Glue Grant buffy coat cohort, SIRS binary parameters alone performed poorly in discriminating patients at time of infection from noninfected patients. SIRS criteria plus the infection z score with a global cutoff (that is, not broken into separate time bins) increased the discriminatory power with a continuous NRI of 0.9. However, SIRS is only one of several criteria used to diagnose sepsis. Procalcitonin is a well-studied biomarker for differentiating sepsis from SIRS, with a summary ROC AUC of 0.78 (range, 0.66 to 0.90) (5). The average AUC in our discovery cohorts was 0.87 and the time-matched neutrophils validation cohort had a mean AUC of 0.83, both of which are thus at least comparable to procalcitonin. None of the publicly available data sets included procalcitonin levels, so no direct comparison is available. We emphasize, however, that each of these markers need not be used separately; any prospective study of the infection z score should also include known biomarkers to test for better diagnostic performance using biomarker combinations and for head-to-head comparisons.
Both infectious and noninfectious inflammation can lead to SIRS through activation of the same innate immune pathways [TLRs, RIG-like receptor (RLRs), NLRs, etc.], so the “typical” proinflammatory genes and cytokines (such as tumor necrosis factor and the inter-leukins) are generally expressed in both sterile and infectious inflammation (45). For instance, one recent study showed high correlation in gene expression between sterile inflammation (Glue Grant burns cohort) and four independent sepsis data sets, with as much as 93% of the genes changing in the same direction in the two conditions (12). Thus, a standard hypothesis-driven approach in the search of biomarkers specifically differentially expressed between sterile SIRS and sepsis is unlikely to succeed, given that the “standard” suite of cytokines and chemokines known to be expressed in sepsis is mostly also activated in sterile SIRS. However, several protein families have been shown to have specificity for pathogen-associated molecular patterns, thus giving rise to the possibility of infection-specific innate immune signaling pathways (11). Our data-driven, unbiased approach searched specifically for genes that are homogeneously statistically differentially expressed between sterile SIRS/trauma patients and sepsis patients across multiple cohorts.
Some of the genes in the sepsis-specific 11-gene set, such as CEACAM1, C3AR1, GNA15, and HLA-DPB1, have been previously associated with sepsis or infections (46, 47). The regulatory control of these genes may be enriched for several proinflammatory factors, but no single common factor explained the network. The gene sets found here may be better explained by cell type enrichment analyses. We show that band cells and the myeloid cell line are highly enriched for the gene sets found to be significantly differentially expressed between sterile SIRS and sepsis. The finding of enrichment in band cells is particularly intriguing, because bands have previously been shown to help differentiate sterile SIRS and sepsis (48). However, there is very high variability in band counts both by automatic blood counters and by hand (49), and no good serum marker exists. The 11-gene set may distinguish sepsis from sterile SIRS at least in part because it also includes information on increased Tregs and decreased dendritic cells, both of which have previously been implicated in sepsis (50, 51). In particular, the joint findings that the 11-gene set is overexpressed in bands but underexpressed in adaptive immune cells are remarkably similar to the phenotype of increased immature granulocytes and decreased adaptive immunity caused by myeloid-derived suppressor cells in infection and chronic critical illness (52, 53). The connection between the 11-gene set and different immune cell types may help explain some sepsis biology, but certainly, these 11 genes require further study.
Our study has some limitations. First, although we validated the 11-gene set in all available independent data sets, prospective validation is required. Second, the Glue Grant buffy coat and neutrophils cohorts were incorporated in a way that treated different periods of time since injury as different data sets, though the noninfected controls came from the same patient cohort at different sampling times (with some dropouts due to injury, recovery, or missed sampling). These time-based control subsets are thus not independent of one another, which may lead to underestimation of effect size variance; however, this was only the case for the two Glue Grant data sets and not the rest of the data sets.
This work presents several future directions. First, both the 11-gene set and the protein products of these genes will need to be tested prospectively in a time-matched manner. Although protein assays are faster than transcript quantitation assays, a number of advances in polymerase chain reaction technology have brought assay times down toward the range of clinical applicability (54). Second, our results showed that the changes in gene expression due to normal recovery from a traumatic event (such as injury or surgery) mean that time must be properly accounted for in any gene expression study of acute illness. Our search found several studies that examine time course after SIRS/trauma (GSE6377, GSE12838, GSE40012, and EMEXP3001) and several that examine the time course since onset of sepsis/infection (GSE20346, GSE2713, GSE40012, and EMEXP3850). However, we found only one publicly available microarray study (the Glue Grant) that examined a cohort of patients over time, where some of the cohorts develop infection and some do not. Thus, on the basis of our results, we recommend that future studies of sepsis diagnostics should be designed with longitudinal cohorts both with and without infection to enable appropriate time-matched comparisons (9, 10).
Overall, our comprehensive analysis of publicly available gene expression data in SIRS/trauma and sepsis has yielded a parsimonious 11-gene set with excellent discriminatory power in both the discovery cohorts and in 15 independent cohorts. Optimizing a clinical assay for this gene set to get results within a window of clinical relevance should be feasible. Further study will be needed both to confirm our clinical findings in a prospective manner and to investigate the molecular pathways upstream of these genes.
Supplementary Material
Acknowledgments
We would like to thank T. Roth for the help in curating the public immune cell profiles; T. Chakraborty, J. P. Cobb, H. Hossain, M. Lissauer, G. Parnell, and B. Tang for helpful discussions concerning their publicly available data; P. Mason for assistance with accessing the Glue Grant data; and the Glue Grant investigators for sharing their data publicly; they are supported in this by National Institute of General Medical Sciences Glue Grant Legacy Award R24GM102656.
Funding: T.E.S is funded by National Library of Medicine grant 2T15LM007033, a Stanford Child Health Research Institute Young Investigator Award (through the Institute for Immunity, Transplantation and Infection), and the Stanford Department of Surgery. P.K. is funded by National Institute of Allergy and Infectious Diseases grants 1U19AI109662, U19AI057229, U54I117925, and U01AI089859.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/7/287/287ra71/DC1
Materials and Methods
Fig. S1. Labeled PCA comparing healthy controls, SIRS/trauma patients, and sepsis patients.
Fig. S2. Violin plots for the data sets included in the discovery multicohort analysis.
Fig. S3. Neutrophil percentages for the Glue Grant patients with both complete blood count and microarray data.
Fig. S4. Performance of the infection z score in the sorted monocytes from the Glue Grant cohort.
Fig. S5. Performance of the infection z score in the sorted T cells from the Glue Grant cohort.
Fig. S6. Linear models of SIRS criteria and the infection z score.
Fig. S7. The infection z score in non-time-matched data sets.
Fig. S8. Comparison of the infection z scores in patients with acute infections to healthy controls and patients with autoimmune diseases.
Fig. S9. Ingenuity Pathway Analysis results for the 11-gene set.
Fig. S10. Schematic of the entire integrated multicohort analysis.
Table S1. Summary spreadsheet of all data sets referenced in the manuscript.
Table S2. Design matrix of individual phenotypes for multicohort analysis.
Table S3. Summary statistics for the 82 genes that passed significance, heterogeneity, and effect size filtering after multicohort analysis.
Table S4. Probe-level data for all 11 genes in the diagnostic set for all patients in the multi-cohort analysis.
Table S5. Linear models of infection score in the Glue Grant data.
Table S6. Comparison of infection z score across infection types.
Table S7. In silico transcription factor binding analyses for the 11-gene set.
Table S8. Design matrix for cell type enrichment analyses.
Movie S1. Rotation of a time-course-labeled PCA of trauma patients.
References (55–79)
Author contributions: Study conception and design: T.E.S. and P.K.; contributed materials and methods: A.S. and H.R.W.; performed the analysis: T.E.S. and P.K.; drafted manuscript: T.E.S. and P.K.; and critical revision: T.E.S., A.S., H.R.W., and P.K.
Competing interests: The 11-gene set has been disclosed for possible patent protection to Stanford Office of Technology and Licensing by T.E.S. and P.K.
Data and materials availability: Links to the data sets used here are available in table S1; patient design matrix and probe-level data are available in tables S2 and S3. The GPSSSI unique data set has been deposited in GEO at accession number GSE66099. The Glue Grant data are publicly available, pending Institutional Review Board approval per the instructions found at https://www.gluegrant.org/. The data and code necessary to recreate the multicohort analysis have been deposited online at http://khatrilab.stanford.edu/sepsis; access to the data will be granted after approval by the Glue Grant consortium.
REFERENCES AND NOTES
- 1.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: Analysis of incidence, outcome, and associated costs of care. Crit. Care Med. 2001;29:1303–1310. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Torio CA, Andrews RA. Agency for Healthcare Research and Quality, Rockville, MD. 2013 [Google Scholar]
- 3.Ferrer R, Martin-Loeches I, Phillips G, Osborn TM, Townsend S, Dellinger RP, Artigas A, Schorr C, Levy MM. Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour: Results from a guideline-based performance improvement program. Crit. Care Med. 2014;42:1749–1755. doi: 10.1097/CCM.0000000000000330. [DOI] [PubMed] [Google Scholar]
- 4.Coburn B, Morris AM, Tomlinson G, Detsky AS. Does this adult patient with suspected bacteremia require blood cultures? JAMA. 2012;308:502–511. doi: 10.1001/jama.2012.8262. [DOI] [PubMed] [Google Scholar]
- 5.Tang BM, Eslick GD, Craig JC, McLean AS. Accuracy of procalcitonin for sepsis diagnosis in critically ill patients: Systematic review and meta-analysis. Lancet Infect. Dis. 2007;7:210–217. doi: 10.1016/S1473-3099(07)70052-X. [DOI] [PubMed] [Google Scholar]
- 6.Xiao W, Mindrinos MN, Seok J, Cuschieri J, Cuenca AG, Gao H, Hayden DL, Hennessy L, Moore EE, Minei JP, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Brownstein BH, Mason PH, Baker HV, Finnerty CC, Jeschke MG, López MC, Klein MB, Gamelli RL, Gibran NS, Arnoldo B, Xu W, Zhang Y, Calvano SE, McDonald-Smith GP, Schoenfeld DA, Storey JD, Cobb JP, Warren HS, Moldawer LL, Herndon DN, Lowry SF, Maier RV, Davis RW, Tompkins RG. Inflammation and Host Response to Injury Large-Scale Collaborative Research Program, A genomic storm in critically injured humans. J. Exp. Med. 2011;208:2581–2590. doi: 10.1084/jem.20111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pankla R, Buddhisa S, Berry M, Blankenship DM, Bancroft GJ, Banchereau J, Lertmemongkolchai G, Chaussabel D. Genomic transcriptional profiling identifies a candidate blood biomarker signature for the diagnosis of septicemic melioidosis. Genome Biol. 2009;10:R127. doi: 10.1186/gb-2009-10-11-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang BM, McLean AS, Dawes IW, Huang SJ, Lin RC. Gene-expression profiling of peripheral blood mononuclear cells in sepsis. Crit. Care Med. 2009;37:882–888. doi: 10.1097/CCM.0b013e31819b52fd. [DOI] [PubMed] [Google Scholar]
- 9.Johnson SB, Lissauer M, Bochicchio GV, Moore R, Cross AS, Scalea TM. Gene expression profiles differentiate between sterile SIRS and early sepsis. Ann. Surg. 2007;245:611–621. doi: 10.1097/01.sla.0000251619.10648.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maslove DM, Wong HR. Gene expression profiling in sepsis: Timing, tissue, and translational considerations. Trends Mol. Med. 2014;20:204–213. doi: 10.1016/j.molmed.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Kooyk Y, Rabinovich GA. Protein-glycan interactions in the control of innate and adaptive immune responses. Nat. Immunol. 2008;9:593–601. doi: 10.1038/ni.f.203. [DOI] [PubMed] [Google Scholar]
- 12.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, López CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG. Inflammation and Host Response to Injury, Large Scale Collaborative Research Program, Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. U.S.A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warren HS, Elson CM, Hayden DL, Schoenfeld DA, Cobb JP, Maier RV, Moldawer LL, Moore EE, Harbrecht BG, Pelak K, Cuschieri J, Herndon DN, Jeschke MG, Finnerty CC, Brownstein BH, Hennessy L, Mason PH, Tompkins RG. Inflammation and the Host Response to Injury Large Scale Collaborative Research Program, A genomic score prognostic of outcome in trauma patients. Mol. Med. 2009;15:220–227. doi: 10.2119/molmed.2009.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai KH, Tan CS, Leek JT, Maier RV, Tompkins RG, Storey JD. Inflammation and the Host Response to Injury Large Scale Collaborative Research Program, Dissecting inflammatory complications in critically injured patients by within-patient gene expression changes: A longitudinal clinical genomics study. PLOS Med. 2011;8:e1001093. doi: 10.1371/journal.pmed.1001093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cvijanovich N, Shanley TP, Lin R, Allen GL, Thomas NJ, Checchia P, Anas N, Freishtat RJ, Monaco M, Odoms K, Sakthivel B, Wong HR. Genomics of Pediatric SIRS/Septic Shock Investigators, Validating the genomic signature of pediatric septic shock. Physiol. Genomics. 2008;34:127–134. doi: 10.1152/physiolgenomics.00025.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shanley TP, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, Kalyanaraman M, Tofil NM, Penfil S, Monaco M, Odoms K, Barnes M, Sakthivel B, Aronow BJ, Wong HR. Genome-level longitudinal expression of signaling pathways and gene networks in pediatric septic shock. Mol. Med. 2007;13:495–508. doi: 10.2119/2007-00065.Shanley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong HR, Shanley TP, Sakthivel B, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, Kalyanaraman M, Tofil NM, Penfil S, Monaco M, Tagavilla MA, Odoms K, Dunsmore K, Barnes M, Aronow BJ. Genomics of Pediatric SIRS/Septic Shock Investigators, Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol. Genomics. 2007;30:146–155. doi: 10.1152/physiolgenomics.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong HR, Cvijanovich N, Allen GL, Lin R, Anas N, Meyer K, Freishtat RJ, Monaco M, Odoms K, Sakthivel B, Shanley TP. Genomics of Pediatric SIRS/Septic Shock Investigators, Genomic expression profiling across the pediatric systemic inflammatory response syndrome, sepsis, and septic shock spectrum. Crit. Care Med. 2009;37:1558–1566. doi: 10.1097/CCM.0b013e31819fcc08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong HR, Freishtat RJ, Monaco M, Odoms K, Shanley TP. Leukocyte subset-derived genomewide expression profiles in pediatric septic shock. Pediatr. Crit. Care Med. 2010;11:349–355. doi: 10.1097/PCC.0b013e3181c519b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong HR, Cvijanovich NZ, Allen GL, Thomas NJ, Freishtat RJ, Anas N, Meyer K, Checchia PA, Lin R, Shanley TP, Bigham MT, Wheeler DS, Doughty LA, Tegtmeyer K, Poynter SE, Kaplan JM, Chima RS, Stalets E, Basu RK, Varisco BM, Barr FE. Validation of a gene expression-based subclassification strategy for pediatric septic shock. Crit. Care Med. 2011;39:2511–2517. doi: 10.1097/CCM.0b013e3182257675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Almansa R, Tamayo E, Heredia M, Gutierrez S, Ruiz P, Alvarez E, Gomez-Sanchez E, Andaluz-Ojeda D, Ceña R, Rico L, Iglesias V, Gomez-Herreras JI, Bermejo-Martin JF. Transcriptomic evidence of impaired immunoglobulin G production in fatal septic shock. J. Crit. Care. 2014;29:307–309. doi: 10.1016/j.jcrc.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 22.Bermejo-Martin JF, Martin-Loeches I, Rello J, Antón A, Almansa R, Xu L, Lopez-Campos G, Pumarola T, Ran L, Ramirez P, Banner D, Ng DC, Socias L, Loza A, Andaluz D, Maravi E, Gómez-Sánchez MJ, Gordón M, Gallegos MC, Fernandez V, Aldunate S, León C, Merino P, Blanco J, Martin-Sanchez F, Rico L, Varillas D, Iglesias V, Marcos M, Gandía F, Bobillo F, Nogueira B, Rojo S, Resino S, Castro C, Ortiz de Lejarazu R, Kelvin D. Host adaptive immunity deficiency in severe pandemic influenza. Crit. Care. 2010;14:R167. doi: 10.1186/cc9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin-Loeches I, Papiol E, Almansa R, López-Campos G, Bermejo-Martin JF, Rello J. Intubated patients developing tracheobronchitis or pneumonia have distinctive complement system gene expression signatures in the pre-infection period: A pilot study. Med. Intensiva. 2012;36:257–263. doi: 10.1016/j.medin.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Tamayo E, Fernández A, Almansa R, Carrasco E, Goncalves L, Heredia M, Andaluz-Ojeda D, March G, Rico L, Gómez-Herreras JI, de Lejarazu RO, Bermejo-Martin JF. Beneficial role of endogenous immunoglobulin subclasses and isotypes in septic shock. J. Crit. Care. 2012;27:616–622. doi: 10.1016/j.jcrc.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Hu X, Yu J, Crosby SD, Storch GA. Gene expression profiles in febrile children with defined viral and bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 2013;110:12792–12797. doi: 10.1073/pnas.1302968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parnell GP, McLean AS, Booth DR, Armstrong NJ, Nalos M, Huang SJ, Manak J, Tang W, Tam O-Y, Chan S, Tang BM. A distinct influenza infection signature in the blood transcriptome of patients with severe community-acquired pneumonia. Crit. Care. 2012;16:R157. doi: 10.1186/cc11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sutherland A, Thomas M, Brandon RA, Brandon RB, Lipman J, Tang B, McLean A, Pascoe R, Price G, Nguyen T, Stone G, Venter D. Development and validation of a novel molecular bio-marker diagnostic test for the early detection of sepsis. Crit. Care. 2011;15:R149. doi: 10.1186/cc10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang Y, Xu H, Du X, Lit L, Walker W, Lu A, Ran R, Gregg JP, Reilly M, Pancioli A, Khoury JC, Sauerbeck LR, Carrozzella JA, Spilker J, Clark J, Wagner KR, Jauch EC, Chang DJ, Verro P, Broderick JP, Sharp FR. Gene expression in blood changes rapidly in neutrophils and mono-cytes after ischemic stroke in humans: A microarray study. J. Cereb. Blood Flow Metab. 2006;26:1089–1102. doi: 10.1038/sj.jcbfm.9600264. [DOI] [PubMed] [Google Scholar]
- 29.Tang BM, McLean AS, Dawes IW, Huang SJ, C Lin R. The use of gene-expression profiling to identify candidate genes in human sepsis. Am. J. Respir. Crit. Care Med. 2007;176:676–684. doi: 10.1164/rccm.200612-1819OC. [DOI] [PubMed] [Google Scholar]
- 30.Ahn SH, Tsalik EL, Cyr DD, Zhang YC, van Velkinburgh J, Langley RJ, Glickman SW, Cairns CB, Zaas AK, Rivers EP, Otero RM, Veldman T, Kingsmore SF, Lucas JC, Woods W, Ginsburg GS, Fowler VG., Jr Gene expression-based classifiers identify Staphylococcus aureus infection in mice and humans. PLOS One. 2013;8:e48979. doi: 10.1371/journal.pone.0048979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dolinay T, Kim YS, Howrylak J, Hunninghake GM, An CH, Fredenburgh L, Massaro AF, Rogers A, Gazourian L, Nakahira K, Haspel JA, Landazury R, Eppanapally S, Christie JD, Meyer NJ, Ware LB, Christiani DC, Ryter SW, Baron RM, Choi AM. Inflammasome-regulated cytokines are critical mediators of acute lung injury. Am. J. Respir. Crit. Care Med. 2012;185:1225–1234. doi: 10.1164/rccm.201201-0003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berdal J-E, Mollnes TE, Wæhre T, Olstad OK, Halvorsen B, Ueland T, Laake JH, Furuseth MT, Maagaard A, Kjekshus H, Aukrust P, Jonassen CM. Excessive innate immune response and mutant D222G/N in severe A (H1N1) pandemic influenza. J. Infect. 2011;63:308–316. doi: 10.1016/j.jinf.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, Quinn C, Blankenship D, Dhawan R, Cush JJ, Mejias A, Ramilo O, Kon OM, Pascual V, Banchereau J, Chaussabel D, O’Garra A. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fredriksson K, Tjäder I, Keller P, Petrovic N, Ahlman B, Schéele C, Wernerman J, Timmons JA, Rooyackers O. Dysregulation of mitochondrial dynamics and the muscle transcriptome in ICU patients suffering from sepsis induced multiple organ failure. PLOS One. 2008;3:e3686. doi: 10.1371/journal.pone.0003686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDunn JE, Husain KD, Polpitiya AD, Burykin A, Ruan J, Li Q, Schierding W, Lin N, Dixon D, Zhang W, Coopersmith CM, Dunne WM, Colonna M, Ghosh BK, Cobb JP. Plasticity of the systemic inflammatory response to acute infection during critical illness: Development of the riboleukogram. PLOS One. 2008;3:e1564. doi: 10.1371/journal.pone.0001564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung TP, Laramie JM, Meyer DJ, Downey T, Tam LH, Ding H, Buchman TG, Karl I, Stormo GD, Hotchkiss RS, Cobb JP. Molecular diagnostics in sepsis: From bedside to bench. J. Am. Coll. Surg. 2006;203:585–598. doi: 10.1016/j.jamcollsurg.2006.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parnell G, McLean A, Booth D, Huang S, Nalos M, Tang B. Aberrant cell cycle and apoptotic changes characterise severe influenza A infection—A meta-analysis of genomic signatures in circulating leukocytes. PLOS One. 2011;6:e17186. doi: 10.1371/journal.pone.0017186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emonts M. thesis, Erasmus University Rotterdam. 2008 [Google Scholar]
- 39.Smith CL, Dickinson P, Forster T, Craigon M, Ross A, Khondoker MR, France R, Ivens A, Lynn DJ, Orme J, Jackson A, Lacaze P, Flanagan KL, Stenson BJ, Ghazal P. Identification of a human neonatal immune-metabolic network associated with bacterial infection. Nat. Commun. 2014;5:4649. doi: 10.1038/ncomms5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vassiliou AG, Maniatis NA, Orfanos SE, Mastora Z, Jahaj E, Paparountas T, Armaganidis A, Roussos C, Aidinis V, Kotanidou A. Induced expression and functional effects of aquaporin-1 in human leukocytes in sepsis. Crit. Care. 2013;17:R199. doi: 10.1186/cc12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khatri P, Roedder S, Kimura N, De Vusser K, Morgan AA, Gong Y, Fischbein MP, C Robbins R, Naesens M, Butte AJ, Sarwal MM. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J. Exp. Med. 2013;210:2205–2221. doi: 10.1084/jem.20122709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen R, Khatri P, Mazur PK, Polin M, Zheng Y, Vaka D, Hoang CD, Shrager J, Xu Y, Vicent S, Butte AJ, Sweet-Cord ero EA. A meta-analysis of lung cancer gene expression identifies PTK7 as a survival gene in lung adenocarcinoma. Cancer Res. 2014;74:2892–2902. doi: 10.1158/0008-5472.CAN-13-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hietbrink F, Koenderman L, Althuizen M, Pillay J, Kamp V, Leenen LP. Kinetics of the innate immune response after trauma: Implications for the development of late onset sepsis. Shock. 2013;40:21–27. doi: 10.1097/SHK.0b013e318295a40a. [DOI] [PubMed] [Google Scholar]
- 44.Pencina MJ, D’Agostino RB, Demler OV. Novel metrics for evaluating improvement in discrimination: Net reclassification and integrated discrimination improvement for normal variables and nested models. Stat. Med. 2012;31:101–113. doi: 10.1002/sim.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newton K, Dixit VM. Signaling in innate immunity and inflammation. Cold Spring Harb. Perspect. Biol. 2012;4:a006049. doi: 10.1101/cshperspect.a006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madsen-Bouterse SA, Romero R, Tarca AL, Kusanovic JP, Espinoza J, Kim CJ, Kim J-S, Edwin SS, Gomez R, Draghici S. The transcriptome of the fetal inflammatory response syndrome. Am. J. Reprod. Immunol. 2010;63:73–92. doi: 10.1111/j.1600-0897.2009.00791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwan A, Hubank M, Rashid A, Klein N, Peters MJ. Transcriptional instability during evolving sepsis may limit biomarker based risk stratification. PLOS One. 2013;8:e60501. doi: 10.1371/journal.pone.0060501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cavallazzi R, Bennin CL, Hirani A, Gilbert C, Marik PE. Is the band count useful in the diagnosis of infection? An accuracy study in critically ill patients. J. Intensive Care Med. 2010;25:353–357. doi: 10.1177/0885066610377980. [DOI] [PubMed] [Google Scholar]
- 49.Cornbleet PJ. Clinical utility of the band count. Clin. Lab. Med. 2002;22:101–136. doi: 10.1016/s0272-2712(03)00069-6. [DOI] [PubMed] [Google Scholar]
- 50.Venet F, Chung C-S, Monneret G, Huang X, Horner B, Garber M, Ayala A. Regulatory T cell populations in sepsis and trauma. J. Leukoc. Biol. 2008;83:523–535. doi: 10.1189/jlb.0607371. [DOI] [PubMed] [Google Scholar]
- 51.Grimaldi D, Louis S, Pène F, Sirgo G, Rousseau C, Claessens YE, Vimeux L, Cariou A, Mira JP, Hosmalin A, Chiche JD. Profound and persistent decrease of circulating dendritic cells is associated with ICU-acquired infection in patients with septic shock. Intensive Care Med. 2011;37:1438–1446. doi: 10.1007/s00134-011-2306-1. [DOI] [PubMed] [Google Scholar]
- 52.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanzant EL, Lopez CM, Ozrazgat-Baslanti T, Ungaro R, Davis R, Cuenca AG, Gentile LF, Nacionales DC, Cuenca AL, Bihorac A, Leeuwenburgh C, Lanz J, Baker HV, McKinley B, Moldawer LL, Moore FA, Efron PA. Persistent inflammation, immunosuppression, and catabolism syndrome after severe blunt trauma. J. Trauma Acute Care Surg. 2014;76:21–30. doi: 10.1097/TA.0b013e3182ab1ab5. discussion 29–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park S, Zhang Y, Lin S, Wang T-H, Yang S. Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnol. Adv. 2011;29:830–839. doi: 10.1016/j.biotechadv.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu W, Seok J, Mindrinos MN, Schweitzer AC, Jiang H, Wilhelmy J, Clark TA, Kapur K, Xing Y, Faham M, Storey JD, Moldawer LL, Maier RV, Tompkins RG, Wong WH, Davis RW, Xiao W. Inflammation and Host Response to Injury Large-Scale Collaborative Research Program, Human transcriptome array for high-throughput clinical studies. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3707–3712. doi: 10.1073/pnas.1019753108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koren Y, Carmel L. Robust linear dimensionality reduction. IEEE Trans. Vis. Comput. Graph. 2004;10:459–470. doi: 10.1109/TVCG.2004.17. [DOI] [PubMed] [Google Scholar]
- 57.Bodor A, Csabai I, Mahoney MW, Solymosi N. rCUR: An R package for CUR matrix decomposition. BMC Bioinformatics. 2012;13:103. doi: 10.1186/1471-2105-13-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034–research0034.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Auerbach RK, Chen B, Butte AJ. Relating genes to function: Identifying enriched transcription factors using the ENCODE ChIP-Seq significance tool. Bioinformatics. 2013;29:1922–1924. doi: 10.1093/bioinformatics/btt316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roider HG, Manke T, O’Keeffe S, Vingron M, Haas SA. PASTAA: Identifying transcription factors associated with sets of co-regulated genes. Bioinformatics. 2009;25:435–442. doi: 10.1093/bioinformatics/btn627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jeffrey KL, Brummer T, Rolph MS, Liu SM, Callejas NA, Grumont RJ, Gillieron C, Mackay F, Grey S, Camps M, Rommel C, Gerondakis SD, Mackay CR. Positive regulation of immune cell function and inflammatory responses by phosphatase PAC-1. Nat. Immunol. 2006;7:274–283. doi: 10.1038/ni1310. [DOI] [PubMed] [Google Scholar]
- 63.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: New molecules and patterns of gene expression. J. Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 64.Radom-Aizik S, Zaldivar F, Jr, Leu S-Y, Galassetti P, Cooper DM. Effects of 30 min of aerobic exercise on gene expression in human neutrophils. J. Appl. Physiol. 2008;104:236–243. doi: 10.1152/japplphysiol.00872.2007. [DOI] [PubMed] [Google Scholar]
- 65.He F, Chen H, Probst-Kepper M, Geffers R, Eifes S, Del Sol A, Schughart K, Zeng A-P, Balling R. PLAU inferred from a correlation network is critical for suppressor function of regulatory T cells. Mol. Syst. Biol. 2012;8:624. doi: 10.1038/msb.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Giefing M, Winoto-Morbach S, Sosna J, Döring C, Klapper W, Küppers R, Böttcher S, Adam D, Siebert R, Schütze S. Hodgkin-Reed-Sternberg cells in classical Hodgkin lymphoma show alterations of genes encoding the NADPH oxidase complex and impaired reactive oxygen species synthesis capacity. PLOS One. 2013;8:e84928. doi: 10.1371/journal.pone.0084928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meyers JA, Su DW, Lerner A. Chronic lymphocytic leukemia and B and T cells differ in their response to cyclic nucleotide phosphodiesterase inhibitors. J. Immunol. 2009;182:5400–5411. doi: 10.4049/jimmunol.0804255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eckerle S, Brune V, Döring C, Tiacci E, Bohle V, Sundström C, Kodet R, Paulli M, Falini B, Klapper W, Chaubert AB, Willenbrock K, Metzler D, Bräuninger A, Küppers R, Hansmann M-L. Gene expression profiling of isolated tumour cells from anaplastic large cell lymphomas: Insights into its cellular origin, pathogenesis and relation to Hodgkin lymphoma. Leukemia. 2009;23:2129–2138. doi: 10.1038/leu.2009.161. [DOI] [PubMed] [Google Scholar]
- 69.Stegmann KA, Björkström NK, Veber H, Ciesek S, Riese P, Wiegand J, Hadem J, Suneetha PV, Jaroszewicz J, Wang C, Schlaphoff V, Fytili P, Cornberg M, Manns MP, Geffers R, Pietschmann T, Guzmán CA, Ljunggren HG, Wedemeyer H. Interferon-a-induced TRAIL on natural killer cells is associated with control of hepatitis C virus infection. Gastroenterology. 2010;138:1885–1897. doi: 10.1053/j.gastro.2010.01.051. [DOI] [PubMed] [Google Scholar]
- 70.Vinh DC, Patel SY, Uzel G, Anderson VL, Freeman AF, Olivier KN, Spalding C, Hughes S, Pittaluga S, Raffeld M, Sorbara LR, Elloumi HZ, Kuhns DB, Turner ML, Cowen EW, Fink D, Long-Priel D, Hsu AP, Ding L, Paulson ML, Whitney AR, Sampaio EP, Frucht DM, DeLeo FR, Holland SM. Autosomal dominant and sporadic monocytopenia with susceptibility to mycobacteria, fungi, papillomaviruses, and myelodysplasia. Blood. 2010;115:1519–1529. doi: 10.1182/blood-2009-03-208629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ancuta P, Liu K-Y, Misra V, Wacleche VS, Gosselin A, Zhou X, Gabuzda D. Transcriptional profiling reveals developmental relationship and distinct biological functions of CD16+ and CD16 monocyte subsets. BMC Genomics. 2009;10:403. doi: 10.1186/1471-2164-10-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, Frampton GM, Drake AC, Leskov I, Nilsson B, Preffer F, Dombkowski D, Evans JW, Liefeld T, Smutko JS, Chen J, Friedman N, Young RA, Golub TR, Regev A, Ebert BL. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allantaz F, Cheng DT, Bergauer T, Ravindran P, Rossier MF, Ebeling M, Badi L, Reis B, Bitter H, D’Asaro M, Chiappe A, Sridhar S, Pacheco GD, Burczynski ME, Hochstrasser D, Vonderscher J, Matthes T. Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression. PLOS One. 2012;7:e29979. doi: 10.1371/journal.pone.0029979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsitsiou E, Williams AE, Moschos SA, Patel K, Rossios C, Jiang X, Adams O-D, Macedo P, Booton R, Gibeon D, Chung KF, Lindsay MA. Transcriptome analysis shows activation of circulating CD8+ T cells in patients with severe asthma. J. Allergy Clin. Immunol. 2012;129:95–103. doi: 10.1016/j.jaci.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 75.Frankenberger M, Hofer TP, Marei A, Dayyani F, Schewe S, Strasser C, Aldraihim A, Stanzel F, Lang R, Hoffmann R, Prazeres da Costa O, Buch T, Ziegler-Heitbrock L. Transcript profiling of CD16-positive monocytes reveals a unique molecular fingerprint. Eur. J. Immunol. 2012;42:957–974. doi: 10.1002/eji.201141907. [DOI] [PubMed] [Google Scholar]
- 76.Huen N-Y, Pang AL, Tucker JA, Lee T-L, Vergati M, Jochems C, Intrivici C, Cereda V, Chan W-Y, Rennert OM, Madan RA, Gulley JL, Schlom J, Tsang KY. Up-regulation of proliferative and migratory genes in regulatory T cells from patients with metastatic castration-resistant prostate cancer. Int. J. Cancer. 2013;133:373–382. doi: 10.1002/ijc.28026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Malcolm KC, Nichols EM, Caceres SM, Kret JE, Martiniano SL, Sagel SD, Chan ED, Caverly L, Solomon GM, Reynolds P, Bratton DL, Taylor-Cousar JL, Nichols DP, Saavedra MT, Nick JA. Mycobacterium abscessus induces a limited pattern of neutrophil activation that promotes pathogen survival. PLOS One. 2013;8:e57402. doi: 10.1371/journal.pone.0057402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rapin N, Bagger FO, Jendholm J, Mora-Jensen H, Krogh A, Kohlmann A, Thiede C, Borregaard N, Bullinger L, Winther O, Theilgaard-Mönch K, Porse BT. Comparing cancer vs normal gene expression profiles identifies new disease entities and common transcriptional programs in AML patients. Blood. 2014;123:894–904. doi: 10.1182/blood-2013-02-485771. [DOI] [PubMed] [Google Scholar]
- 79.Mabbott NA, Baillie JK, Brown H, Freeman TC, Hume DA. An expression atlas of human primary cells: Inference of gene function from coexpression networks. BMC Genomics. 2013;14:632. doi: 10.1186/1471-2164-14-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.