Bi-allelic gene targeting rice plants can be obtained by Agrobacterium-mediated CRISPR/Cas9 transformation and suppression of DNA ligase 4.
Abstract
Sequence-specific nucleases (SSNs) have been used successfully in homology-directed repair (HDR)-mediated gene targeting (GT) in many organisms. However, break-induced GT in plants remains challenging due to inefficient delivery of HDR templates and SSNs into plant nuclei. In many plants, including rice, Agrobacterium-mediated transformation is the most practical means of transformation because this biotic transformation system can deliver longer and more intact DNA payloads with less incorporation of fragmented DNA compared with physical transformation systems such as polyethylene glycol, electroporation, or biolistics. Following infection with Agrobacterium, transfer of transfer DNA (T-DNA) to the nucleus and its integration into the plant genome occur consecutively during cocultivation, thus timing the induction of DNA double-strand breaks (DSBs) on the target gene to coincide with the delivery of the HDR template is crucial. To synchronize DSB induction and delivery of the HDR template, we transformed a Cas9 expression construct and GT vector harboring the HDR template with guide RNAs (gRNAs) targeting the rice acetolactate synthase (ALS) gene either separately or sequentially into rice calli. When gRNAs targeting ALS were transcribed transiently from double-stranded T-DNA containing the HDR template, DSBs were induced in the ALS locus by the assembled Cas9/gRNA complex and homologous recombination was stimulated. Contrary to our expectations, there was no great difference in GT efficiency between Cas9-expressing and nonexpressing cells. However, when gRNA targeting DNA ligase 4 was transformed with Cas9 prior to the GT experiment, GT efficiency increased dramatically and more than one line exhibiting biallelic GT at the ALS locus was obtained.
One of the most powerful and precise ways to introduce specific DNA sequence changes into genomes is to use homology-directed repair (HDR). This approach, referred to as gene targeting (GT), is accomplished by the introduction of DNA fragments encoding a sequence variant of a gene of interest into cells. Since the first report of GT of an integrated antibiotic-tolerant gene in the tobacco genome (Paszkowski et al., 1988), various approaches aimed at HDR-dependent GT have been attempted in plants (for reviews, see Tzfira and White, 2005, Iida and Terada, 2005, and Voytas, 2013). However, in most cases, DNA integrates into random sites in the genome through nonhomologous end joining (NHEJ). A successful approach to enhancing HDR frequency relative to random integration has been to recruit the recombination machinery to the target gene by inducing DNA double-strand breaks (DSBs) at the target gene. Efforts have focused on developing sequence-specific nucleases (SSNs) that can be engineered to create DSBs at a target locus. To date, four classes of SSN—meganucleases (homing endonuclease), zinc finger nucleases (ZFNs), transcriptional activator-like effector nucleases (TALENs), and the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9)—have been developed to cleave genes of interest. Both ZFNs and TALENs have tandem repeats in their DNA-binding domains that can be engineered to recognize specific DNA sequences; the resulting chimeric nucleases can thus be guided to the desired target sequences in the genome to generate DSBs. In these cases, a new chimeric protein must be engineered for each target sequence used. This has been a major hurdle to the wider use of these SSNs because engineering new proteins is no trivial task. In contrast, the CRISPR/Cas9 system uses a single guide RNA (gRNA) to direct the Cas9 endonuclease to the complementary target DNA (Gaj et al., 2013), so only a new gRNA is needed for each new target site. This system thus greatly simplifies the genome-editing process and widens target-site selection.
Since the first publication reporting plant genome editing using CRISPR/Cas9 in 2013, this system has been applied to several plant species, including Arabidopsis, potato, soybean, tobacco, tomato, maize, rice, sorghum, and wheat, suggesting its broad applicability (for a review, see Weeks et al., 2015). Following the discovery that induction of a DSB increases the frequency of HDR by several orders of magnitude, SSNs have emerged as the strategy of choice for improving the efficiency of HDR-mediated genetic alterations. ZFN can be used to increase GT frequency in maize (Shukla et al., 2009). Another study by Townsend et al. (2009) showed that ZFN-mediated GT can be used in a transient expression system in tobacco. Targeted gene insertion with high efficiency using TALEN has been reported in tobacco protoplasts (Zhang et al., 2013). In addition, CRISPR/Cas9 is also useful in HDR-mediated targeted gene insertion in tobacco (Li et al., 2014), rice (Shan et al., 2013), maize (Svitashev et al., 2015), and soybean (Li et al., 2015). Li et al. (2014) and Shan et al. (2013) used tobacco and rice protoplasts as experimental materials, respectively; in these latter studies the donor DNA, which is used as a repair template, was delivered into protoplasts either as a single-stranded oligo DNA (Shan et al., 2013) or as a dsDNA fragment (Li et al., 2014). The next challenge in this strategy is to regenerate whole plants from protoplasts. So far this is possible for only a few plant species (e.g. N. benthamiana and Arabidopsis). Most recently, in maize, immature embryos were bombarded with the oligo or a plasmid to elicit repair templates and Cas9/gRNA expression, respectively (Svitashev et al., 2015). A particle bombardment transformation protocol was also used for donor DNA and Cas9-gRNA DNA delivery in soybean (Li et al., 2015). Transformation efficiency, regeneration efficiency, and issues of regulation must also be taken into consideration when selecting a transformation strategy.
In plants, delivery of SSNs and templates for HDR represent hurdles to the efficient achievement of GT. Literature reports describe the use of either Agrobacterium tumefaciens or physical means to deliver genome engineering reagents (Cai et al., 2009; Shukla et al., 2009), and the highest frequencies of GT have been achieved using protoplasts, i.e. plant cells lacking cell walls (Wright et al., 2005; Townsend et al., 2009; Zhang et al., 2010). Protoplasts can be transformed at high efficiency, but only a handful of plants can be regenerated effectively from protoplasts. Together with the fact that transfer DNA (T-DNA) delivered from Agrobacterium is protected by proteins such as virD2 and virE2 (for review, see Gelvin, 2010), and that intact and long DNA fragments can be delivered directly into plant nuclei, Agrobacterium-mediated transformation is an attractive means of delivering HDR templates. Especially for the positive-negative selection system used in rice GT (Terada et al., 2002, 2007), efficient delivery of long and intact DNA is necessary, because positive and negative selection marker gene expression cassettes make the HDR template long. Using an established Agrobacterium-mediated transformation system (Toki et al., 2006), we succeeded in achieving CRISPR/Cas9-mediated targeted mutagenesis in rice (Endo et al., 2015; Mikami et al., 2015a, 2015b). Here, we report the establishment of an efficient break-induced GT system using Agrobacterium-mediated transformation to deliver both a repair construct and the CRISPR/Cas9 expression system.
RESULTS
Selection of Cleavage Sites on the OsALS Gene
Previously, we succeeded in inducing two amino acid substitutions (W548L, S627I) in the endogenous rice acetolactate synthase (ALS) gene by GT (Endo et al., 2007). These substitutions confer tolerance to the ALS-inhibiting herbicide bispyribac sodium (BS). To induce DSBs near these two point mutations during the course of CRISPR/Cas9-mediated GT, a highly cleavable target sequence on the ALS gene was selected. Four independent gRNA constructs (gALS-1, gALS-2, gALS-3, and gALS-4) that target near these BS-tolerant mutations (Supplemental Fig. S1A) were designed and transformed with a Cas9 expression construct using the Agrobacterium-mediated method. Because the expected cleavage site lies within the recognition sequence of a restriction enzyme, these restriction sites will be disrupted if CRISPR/Cas9 has cleaved successfully and mutagenized the target sequence; thus, cleaved amplified polymorphic sequences (CAPS) can be used to detect mutations at the target sites. Mutation frequency in calli cultured for one month seemed to be similar in gALS-2, -3, and -4 while that of gALS-1 seemed lower than the others (Supplemental Fig. S1B, Mikami et al., 2015a). To increase cleavage events at the targeted locus, we decided to use both gALS-2 and gALS-3 for DSB induction. When both gALS-2 and gALS-3 were coexpressed with Cas9 in rice calli, the mutation frequency at the target site of gALS-2 was not altered significantly compared to that of calli transformed with a single gRNA (Supplemental Fig. S1B).
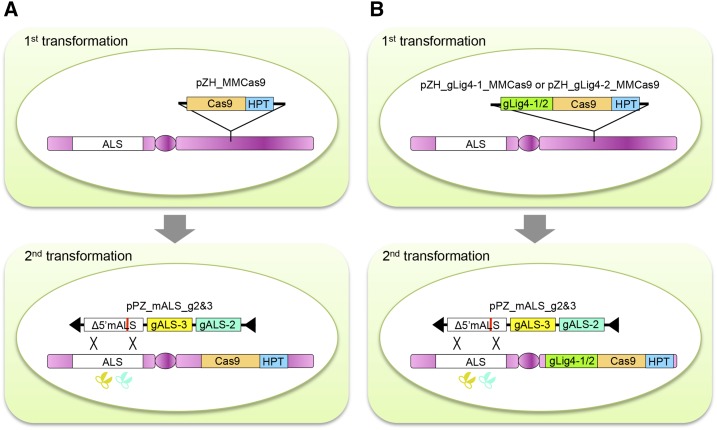
GT System with DSB Induction
Synchronization of DSB induction and delivery of the recombination template is likely to be important for successful GT. However, codelivery of Cas9, gRNA, and the template for recombination simultaneously by Agrobacterium-mediated transformation seemed inadvisable because transformation efficiency drops with increments in the size of the DNA delivered. In addition, it takes time to express the Cas9 protein from the delivered T-DNA. Therefore, we designed a GT system in which the Cas9 expression construct and the GT vector harboring the template for recombination and two gRNA expression constructs were transformed into rice calli sequentially (Fig. 1A). Because the Cas9 expression cassette is stably transformed and expressed in advance in calli, the Cas/gRNA complex becomes functional when gRNAs are transcribed from the T-DNA. In addition, immediate DNA repair at the break sites and random integration of recombination templates should inhibit GT. Thus, gRNAs targeting DNA ligase 4 (Lig4) were transformed with Cas9 at the first transformation, and potential Lig4-disrupted cells were used for GT experiments in a second-round GT transformation (Fig. 1B).
Figure 1.
Strategy for CRISPR/Cas9-mediated GT. A, In the first round of transformation, Cas9 is transformed with the selection marker gene HPT using binary vector pZH_MMCas9 (see Fig. 2). After proliferating transgenic calli, a GT vector harboring mutated the ALS fragment and gALS-2, -3 is transformed using binary vector pPZ_mALS_g2&3 (see Fig. 2). B, To induce mutations in Lig4 before GT experiment, gRNAs targeting Lig4 (gLig4-1 or gLig4-2) are transformed with Cas9.
A two-step transformation strategy might seem complicated. However, we did not have enough seeds of the genetically fixed Cas9 transgenic line because the majority of the transgenic lines were multicopy and transgenes were segregated in the progeny. Since we had already successfully established the sequential transformation system (Nishizawa-Yokoi et al., 2015b; Mikami et al., 2015a, 2015b), we decided to apply it to CRISPR/Cas9-mediated GT.
Sequential Transformation of the Cas9 Expression Construct and the GT Vector Harboring Recombination Template and gRNAs Targeting ALS
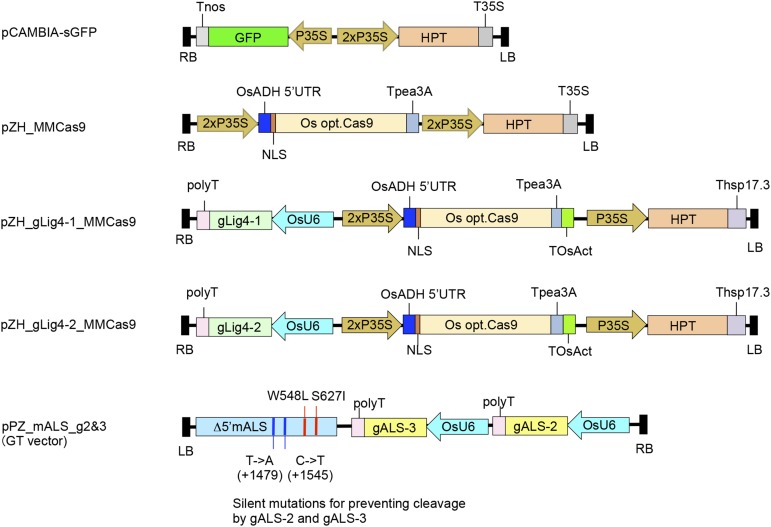
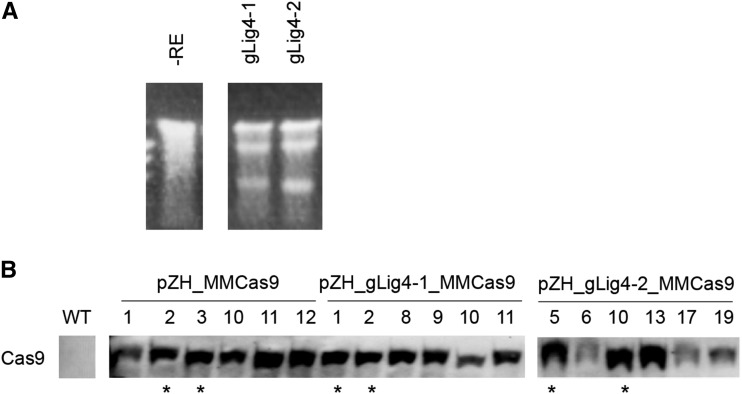
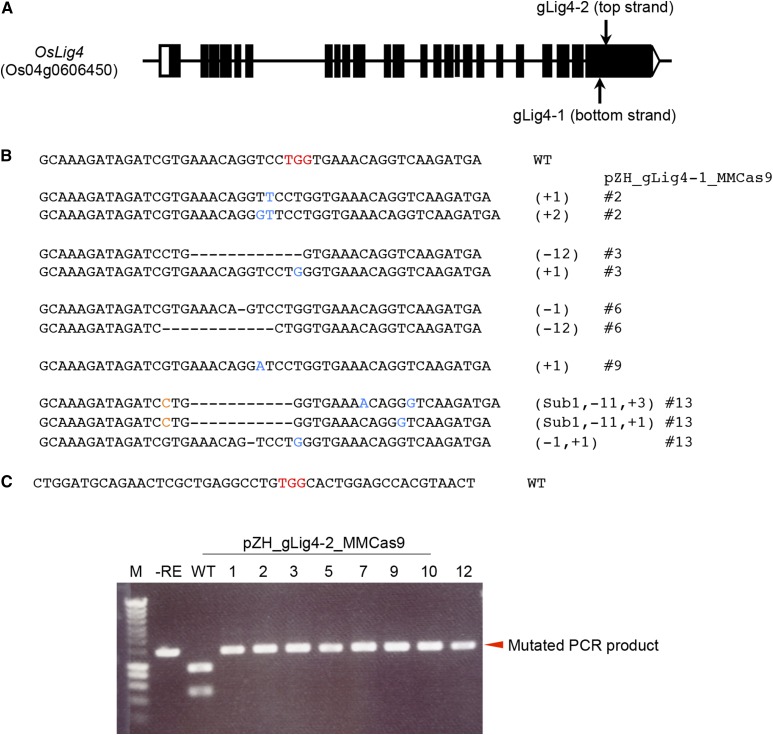
Rice calli were transformed with pCAMBIA-sGFP, pZH_MMCas9, pZH_gLig4-1_MMCas9, or pZH_gLig4-2_MMCas9 vectors (Fig. 2A) in a first round of transformation. To confirm induction of mutation in the Lig4 gene in transgenic calli of pZH_gLig4-1_MMCas9 or pZH_gLig4-2_MMCas9, DNA extracted from these calli one month after the first transformation was subjected to Cel I assay. Because half of the amplified DNA was cleaved in this experiment (Fig. 3A), we concluded that half of the Lig4 loci were disrupted. The GT vector, pPZ_mALS_g2&3 (Fig. 2A), was then introduced into these transgenic calli. The GT vector harbors a partial ALS gene with two point mutations (W548L and S627I) that confer tolerance to BS, and silent nucleotide changes to prevent cleavage by the CRISPR/Cas9 system. The other components of the GT vector are gALS-2 and gALS-3 expression constructs. In GT experiments 1 and 3, a mixture of independent transgenic calli of pZH_MMCas9, pCAMBIA-sGFP, pZH_gLig4-1_MMas9, and pZH_gLig4-2_MMCas9 were used for transformation of the GT vector pPZ_mALS_g2&3 (Table I). On the other hand, in GT experiments 2 and 4, independent transgenic lines of the control vector, and pCAMBIA-sGFP and Cas9 constructs with/without gLig4 were devoted to transformation of the GT vector. In GT experiment 4, the expression level of Cas9 in independent transgenic calli of pZH_MMCas9, pZH_gLig4-1_MMas9, and pZH_gLig4-2_MMCas9 was analyzed by western-blot analysis and Cas9-expressing lines were selected for further transformation of GT vector (Fig. 3B).
Figure 2.
Vectors used in this study. pCAMBIA-sGFP is the negative control vector used for the first transformation. pZH_MMCas9, pZH_gLig4-1_MMCas9, and pZH_gLig4-2_MMCas9 harboring the Cas9 expression construct were used for the first transformation. pPZ_mALS_g2&3 is the GT vector used for the second transformation.
Figure 3.
Confirmation of Lig4 mutation and expression level of Cas9 in calli used for GT experiment. A, Cel I assay of Lig4 gene using DNA extracted from one-month cultured pZH_gLig4-1_MMCas9 and pZH_gLig4-2_MMCas9 transgenic calli. B, Immunoblot analysis of Cas9. Transgenic lines used for GT experiment (Exp. 4 in Table I) are marked by asterisk.
Table I. Summary of GT experiments targeting ALS locus.
| TF First | TF Second Transformed Callia | BSR Calli | W548L, S627I Mutations in Endogenous ALS Gene | Ratio of Calli with W548L and S627I (%)b | Recovery of Regenerated Plantsc | ||||
|---|---|---|---|---|---|---|---|---|---|
| W548L and S627I | W548L | None | |||||||
| Homo | Hetero | Homo | Hetero | ||||||
| Exp. 1 | |||||||||
| pCAMBIA-sGFP Mix | 2753 (13.7g) | 3 | 0 | 0 | 0 | 2 | 1 | 0 | 0 |
| pZH_MMCas9 Mix | 3994 (15.5g) | 3 | 0 | 0 | 0 | 2 | 1 | 0 | 0 |
| Exp. 2 | |||||||||
| pCAMBIA-sGFP no. 1 | 674 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| pCAMBIA-sGFP no. 10 | 665 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| pCAMBIA-sGFP no. 12 | 589 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| pZH_MMCas9 no. 1 | 821 | 4 | 0 | 0 | 0 | 2 | 2 | 0 | 0 |
| pZH_MMCas9 no. 2 | 881 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| pZH_MMCas9 no. 3 | 830 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Exp. 3 | |||||||||
| pZH_MMCas9 Mix | 872 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| pZH_gLig4-1_MMCas9 Mix | 985 | 19 | 0 | 8 | 0 | 7 | 4 | 0.812 | 4 |
| pZH_gLig4-2_MMCas9 Mix | 829 | 12 | 1 | 3 | 1 | 4 | 3 | 0.483 | 3 |
| Exp. 4 | |||||||||
| pZH_MMCas9 no. 2 | 1109 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| pZH_MMCas9 no. 3 | 929 | 7 | 0 | 3 | 0 | 3 | 1 | 0.323 | 3 |
| pZH_gLig4-1_MMCas9 no. 1 | 729 | 9 | 0 | 3 | 0 | 3 | 3 | 0.412 | 2 |
| pZH_gLig4-1_MMCas9 no. 2 | 1100 | 24 | 3 | 8 | 1 | 8 | 4 | 1.000 | 7 |
| pZH_gLig4-2_MMCas9 no. 5 | 682 | 5 | 0 | 1 | 0 | 2 | 2 | 0.147 | 1 |
| pZH_gLig4-2_MMCas9 no. 10 | 1003 | 10 | 0 | 4 | 0 | 5 | 1 | 0.399 | 2 |
| Exp. 5 | |||||||||
| T1 (lig4, +1(A) homo; pZH_gLig4-1_MMCas9, segregated out) | 877 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Molecular Characterization of the ALS Locus in BS-Tolerant Calli
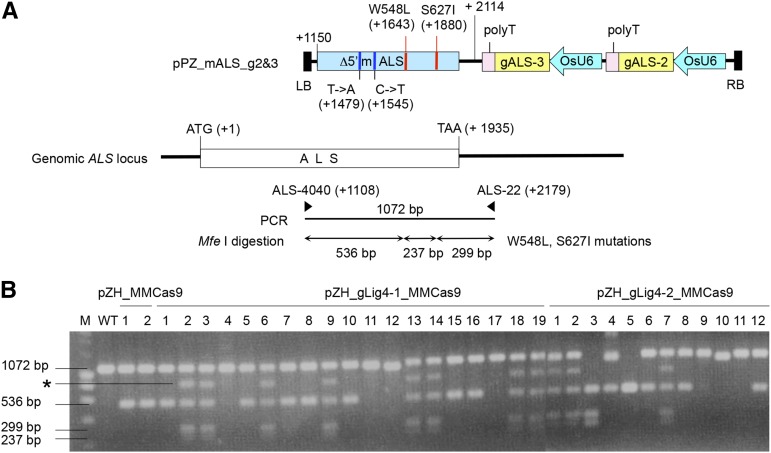
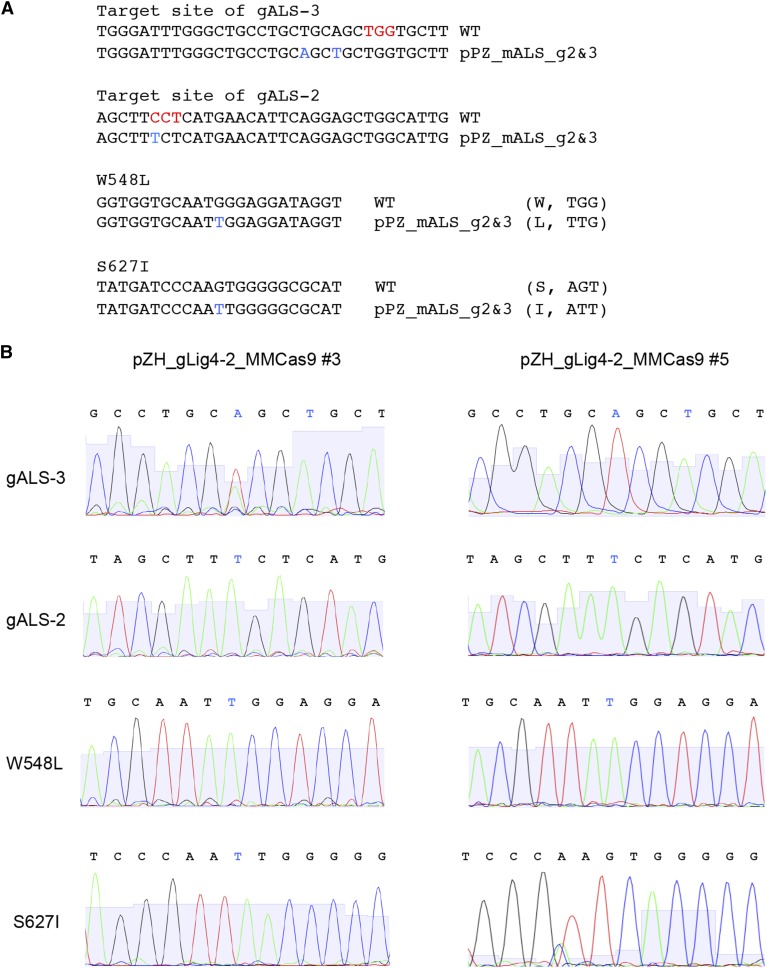
Because both W548L and S627I mutations generate recognition sites for the restriction enzyme Mfe I (CAATTG), CAPS analysis can be used to screen GT cells from BS-tolerant calli. PCR amplification using primer OsALS-4040, which anneals to the ALS coding region located outside the GT vector (not present on the targeting construct) and primer ALS-22, which exists downstream of ALS, yield a PCR product of 1070 bp (Fig. 4A). When W548L and S627I mutations are introduced into the ALS locus by GT, this PCR product was split into three subfragments (536, 237, and 299 bps) by Mfe I digestion. Figure 4B shows an example of PCR-Mfe I digestion analysis of GT experiment 3. Patterns indicating GT (536, 299, and 237 bps) were found in DNA products derived from eight individual BS-tolerant calli out of 19 BS-tolerant pZH_gLig4-1_MMCas9 transformed calli. In the case of pZH_gLig4-2_MMCas9 transformed calli, 4 out of 12 BS-tolerant calli showed the positive band pattern, indicating that W548L and S627I mutations were induced in the endogenous ALS gene. In some DNA samples, not only the expected 536-, 299-, and 237-bp bands, but also an additional band of approximately 700 bps (shown by an asterisk in Fig. 4), was observed. We think this 700-bp fragment is due to partially digested heteroduplex PCR products of wild-type ALS and ALS with W548L and S627I mutations generated artificially during PCR (see Supplemental Fig. S2). Although BS-tolerant mutations might occur spontaneously, a combination of the W548L mutation and the S627I mutation in the rice ALS gene could be discovered only after two years of selection of BS-tolerant rice calli in liquid culture medium (Shimizu et al., 2005). Thus, the possibility that these two point mutations were acquired spontaneously during only one-and-a-half months of cell culture before BS selection is minimal. Furthermore, direct sequencing of the same PCR product revealed the presence of nonselectable silent mutations: T−>A at +1479, preventing digestion by gALS-3; and C−>T at +1545, preventing digestion by gALS-2, in these BS-tolerant rice calli. These results demonstrate that W548L and S627I mutations were integrated into the rice genome via GT rather than via spontaneously occurring mutation.
Figure 4.
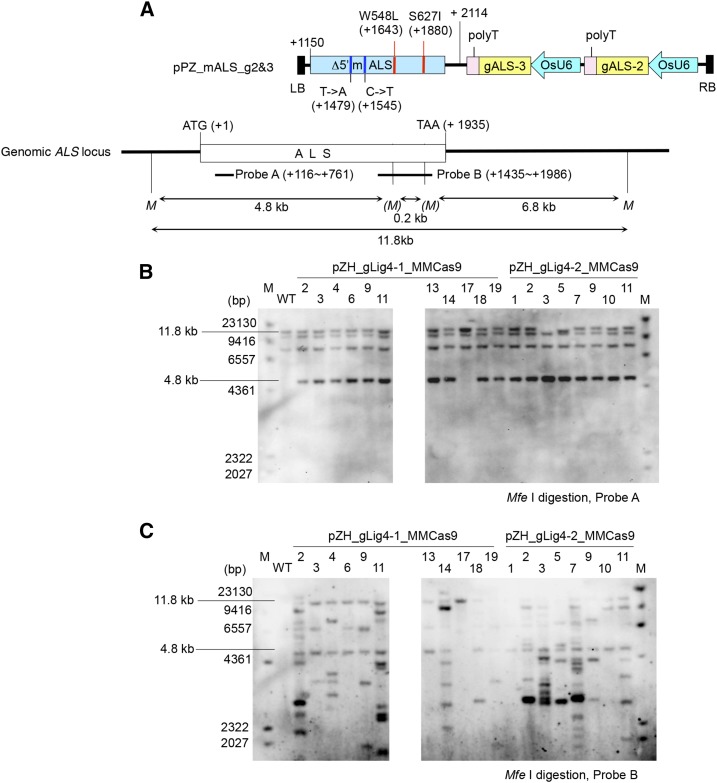
Analysis of GT events in BS-tolerant calli. A, Schematic representation of PCR-Mfe I digestion analysis. The blue boxes represent the coding region of the ALS gene. Partial ALS coding sequence and the 3′ region of the ALS gene (179 bps) exist on the GT vector, pPZ_mALS_g2&3. A sequence encoding 383 amino acids, including the chloroplast-targeting signal, is deleted in the GT vector, rendering the gene nonfunctional. The two point mutations (W548L and S627I), which confer BS tolerance and two silent mutations (T−>A at +1479, C−>T at +1545) are marked by blue and red vertical lines, respectively. The W548L and S627I mutations create novel Mfe I restriction sites (CAATTG). The positions of primers (ALS-4040 and ALS-22) used for PCR, and the expected size of PCR-amplified fragments and their Mfe I endonuclease digestion products, are shown. B, Example of an initial DNA analysis of BS-tolerant calli by PCR-Mfe I digestion analysis. When GT occurred in a single ALS locus in a monoallelic state, three fragments, 536, 299, and 237 bps, appear in addition to the nondigested wild-type band of 1072 bps. Callus with a band corresponding to 536 bps indicates introduction of the W548L mutation but not the S627I mutation into the ALS locus. Heteroduplex of mutated and nonmutated amplicons generated by PCR reaction is partially tolerant to Mfe I-HF and imperfect digested products at W548L appeared as an approximately 700-bp fragment (*).
On the other hand, two (nos. 1 and 2), seven (nos. 1, 5, 7, 8, 10, 15, and 16), and four (nos. 4, 6, 8, and 12) BS-tolerant calli obtained from pZH_MMCas9, pZH_gLig4-1_MMCas9, and pZH_gLig4-2_MMCas9 transgenic calli, respectively, generated fragments of 1072 bps and 536 bps upon PCR-Mfe I digestion analysis. This result could be explained by spontaneous mutation at W548L. Alternatively, HDR might have occurred at a point between the W548L and S627I mutations. Interestingly, no nondigested 1072-bp band was found in pZH_gLig4-2_MMCas9 no. 3 and no. 5. These results mean that W548L, S627I double mutations (no. 3) or only the W548L mutation (no. 5) were induced in the ALS locus in the biallelic state. The number of BS-tolerant calli and details of mutations in independent GT experiments are shown in Table I.
Detailed Analysis of the ALS Locus in GT Candidate Calli Harboring Induction of Biallelic Mutations
In previous GT experiments using OsALS as a target gene for modification, we could not obtain GT plants with biallelic mutation despite identifying more than 60 GT plants with monoallelic mutations (Endo et al., 2007). Suppose that a GT event occurs at each homologous chromosome independently; biallelic GT events in single cells should be rare. To reveal such rare events in pZH_gLig4-2_MMCas9 no. 3, the PCR fragments used for PCR-Mfe I digestion analysis were sequenced directly. Direct sequencing of PCR fragments ALS-4040–ALS-22 (Fig. 4A) revealed that all silent mutations at the target sites of gALS-3 and gALS-2 and W548L and S627I mutations were integrated into the endogenous ALS locus in a biallelic manner (Fig. 5, A and B). On the other hand, PCR-Mfe I digestion analysis of pZH_gLig4-2_MMCas9 no. 5 indicated that W548L but not S627I was introduced into the ALS locus as a biallelic mutation (Fig. 4B). In fact, direct sequencing of the PCR fragment used for PCR-Mfe I digestion analysis revealed that, in the case of the W548L mutation, silent mutations at the target sites of gALS-3 and gALS-2 existed in the ALS locus in a biallelic manner but the S627I mutation was not introduced in pZH_gLig4-2_MMCas9 no. 5 (Fig. 5B). Because the S627I mutation was farthest from the expected cleavage sites of gALS-2 and gALS-3, crossover between W548L and S627I failed to induce the S627I mutation. Another possibility is the occurrence of mismatch correction of the heteroduplex molecules formed between genomic DNA and the recombination template (Johzuka-Hisatomi et al., 2008). Considering the results found with pZH_gLig4-2_MMCas9 no. 5, we checked for the existence of silent mutations in 13 BS-tolerant calli without the S627I mutation. When Mfe I-digested PCR fragments corresponding to 536 bp were purified and sequenced, we found one GT line, pZH_gLig4-2_MMCas9 no. 4, that possessed silent mutations at the target site of gALS-2 and gALS-3.
Figure 5.
Sequence analysis of calli in which biallelic modification of the ALS gene was expected. A, The TGG and CCT shown in red are the PAM sequences recognized by Cas9. SNPs existing on the partial ALS fragment in GT vector pPZ_mALS_g2&3 are shown in blue. B, Sequencing chromatograms of the mutation sites. Direct sequencing of the PCR product amplified using ALS-4040 and ALS-22 revealed that all SNPs derived from the GT vector were induced in the ALS locus as a homozygous state in pZH_gLig4-2_MMCas9 no. 3. In the case of pZH_gLig4-2_MMCas9 no. 5, all SNPs except S627I exist in the ALS locus in the homozygous state.
Confirmation of GT Event by Southern-Blot Analysis
In the case of true GT, no modification of the ALS locus other than the two point mutations should have occurred. Southern-blot analysis was conducted using genomic DNA extracted from calli of GT candidates. When Mfe I-digested DNA was hybridized with probe A (Fig. 6A), three bands existing in the wild type plus a 4.8-kb band caused by the W548L mutation were detected in all GT candidates (Fig. 6B). In pZH_gLig4-2_MMCas9 no. 3 and no. 5, in which disappearance of the wild-type ALS locus was suggested by PCR-Mfe I digestion analysis (Fig. 4B), no 11.8-kb band corresponding to the wild-type ALS locus was detected (Fig. 6B). To analyze the copy number of randomly integrated GT vectors, probe B was hybridized to Mfe I-digested DNA (Fig. 6C). Bands other than 11.8 kb, 6.8 kb, and 4.8 kb seemed to represent hybridization at the randomly integrated GT vector, pPZ_mALS_g2&3. Because calli no. 3, no. 6, no. 13, and no. 18 of pZH_gLig4-1_MMCas9 and no. 1 of pZH_gLig4-2_MMCas9 did not show the extra band, random integration of the GT vector was thought not to have occurred in these calli.
Figure 6.
Southern-blot analysis of the ALS locus. A, Diagram showing the location of probes and expected band size in Southern-blot analysis using probes A and B. B, Southern-blot analysis of Mfe I-digested DNA with probe A. An 11.8-kb band indicates the existence of the ALS gene without W548L and S627I mutations. A 4.8-kb band indicates the ALS gene with the W548L mutation. The two bands other than 11.8 kb and 4.8 kb are due to nonspecific hybridization, since these additional bands are also seen in the wild-type lane. C, Southern-blot analysis of Mfe I-digested DNA with probe B. Bands other than 11.8 kb and 4.8 kb indicate the occurrence of random integration of GT or unexpected rearrangements of the ALS locus. Abbreviations: LB, left border; RB, right border; M, Mfe I.
Effect on GT of Lig4 Knockout
When pZH_MMCas9 was used for the first transformation, calli with W548L/S627I double mutations were obtained only in GT experiment 4 (Table I); the ratio of these double-mutated calli in this GT experiment was 0.323% (3/929). In other independent GT experiments (experiments 1, 2, and 3), rice cells with W548L and S627I mutations in ALS were not obtained although over 7000 calli were used for transformation of the GT vector. On the other hand, several calli with double mutations were consistently obtained from pZH_gLig4-1_MMCas9 or pZH_gLig4-2_MMCas9 transgenic calli in GT experiments 3 and 4, and the GT frequency was 0.147 to 1%. Because only a few GT calli were obtained from pZH_MMCas9 transgenic calli, and a relatively large number of GT calli were obtained from pZH_gLig4-1_MMCas9 or pZH_gLig4-2_MMCas9 transgenic calli, depletion of Lig4 seems to increase GT. To confirm whether GT events had indeed occurred in Lig4 mutated cells, genotyping of Lig4 in GT cells was conducted. When cloned PCR products covering the target site of gLig4-1 (Fig. 7A) were sequenced, no wild-type sequences were found (Fig. 7B). Because the expected cleavage site of gLig4-2 lies within the recognition sequence of the restriction enzyme Stu I, CAPS analysis can be used to detect mutation at the target site of gLig4-2. PCR products covering this site were not digested by Stu I in all GT calli (Fig. 7C). We cannot rule out the possibility that Lig4 mutations were induced after GT because Cas9 and gRNA targeting Lig4 expression cassettes were stably transformed into the rice genome. However, considering the result of the surveyor nuclease assay conducted prior to transformation of the GT vector (Fig. 3A), and the significant difference in the rate of appearance of GT calli between pZH_MMCas9 transgenic calli and pZH_gLig4-1/2_MMCas9 transgenic calli, it is reasonable to assume that Lig4 was mutated before transformation of the GT vector.
Figure 7.
Genotyping of Lig4 gene in GT calli. A, Target sites of gLig4-1 and gLig4-2. gLig4-1 and gLig4-2 recognize the bottom and top strands of the Lig4 gene, respectively. B, Sequences of the gLig4-1 target site in GT calli. Sequences without mutations were not detected in all sequenced samples. C, CAPS analysis of the target site of gLig4-2 in GT calli. Expected cleavage site is located on Stu I site (AGGCCT). Biallelic mutation seemed to be introduced into Lig4 gene in all GT calli.
Considering the possibility that Lig4 knockout alone stimulates GT, we used for our GT experiment progeny of pZH_gLig4-1_MMCas9 transgenic plants in which a 1-bp “A” insertion in the Lig4 gene was fixed in a homozygote state but in which Cas9 and gRNA expression constructs were segregated out. When the GT vector pPZ_mALS_g2&3 was transformed into one-month-cultured calli of this lig4 mutant, no BS-tolerant calli were obtained. This result suggested that Lig4 deficiency alone is not enough to enhance GT (Table I).
In our GT system, Cas9 with gRNA targeting Lig4 (pZH_gLig4-1/2_MMCas9) and HDR template with gRNAs targeting ALS (pPZ_mALS_g2&3) were transformed sequentially to rice calli and the total culture period of calli was approximately 3.5 months. Furthermore, almost all GT events occurred in cells with the Lig4 mutation. In addition, more than half of the GT cell lines could be regenerated (Table I). As expected, all T2 progenies of biallelic GT plants, ZH_gLig4-2_MMCas9 no. 3, contained W548L and S627I mutations in the ALS gene and showed a BS-tolerant phenotype (Supplemental Fig. S3, A and B).
DISCUSSION
GT using the CRISPR/Cas9 system has been achieved in plants using an Agrobacterium-mediated system (Schiml et al., 2014). In this report, an all-in-one vector harboring Cas9, gRNA expression constructs, an HDR template, and a selection marker gene was stably transformed into the Arabidopsis genome. In this case, the target sequence and stably integrated HDR template were cleaved simultaneously by the Cas9/gRNA complex, thus leading to successful GT in planta in a similar manner to in planta GT using I-SceI (Fauser et al., 2012). Furthermore, Baltes et al. (2014) demonstrated that DNA carried by geminiviruses can be used as a template for HDR.
In our study, the HDR template was supplied transiently from Agrobacterium as T-DNA. Because transgenic calli expressing Cas9 were used for the GT experiment, and gRNAs could be supplied from extrachromosomal T-DNA harboring a HDR template, delivery of the HDR template, and timing of DSB induction might be synchronized temporally and spatially. However, no positive effect of this CRISPR/Cas9 system on GT was apparent. Several hypotheses could explain why the CRISPR/Cas9 system does not stimulate GT in our study. (1) Expression levels of Cas9 and gRNA in this study might have been insufficient to induce DSBs for stimulating GT, although two cleavage sites were prepared near BS-tolerant mutations. (2) Amounts of HDR template may have been insufficient to stimulate recombination. (3) NHEJ, rather than HDR, might operate to repair break sites.
On the other hand, when gRNAs targeting Lig4 were transformed with Cas9 in the first round of transformation, GT cells were obtained efficiently (Table I). This result means that, by manipulating DNA repair pathways, CRISPR/Cas9-induced Agrobacterium-mediated GT can work efficiently. Notably, HDR activity in Lig4 knockdown rice calli was two- to three-fold higher than that in control calli, and suppression of Lig4 leads to decreased stable transformation in rice (Nishizawa-Yokoi et al., 2012). Furthermore, Lig4 deficiency increases mutation frequency and produce longer indels at the target site of TALENs (Nishizawa-Yokoi et al., 2015a). Another study reported that HDR-based GT was 5- to 16-fold and 3- to 4-fold enhanced in Arabidopsis ku70 and lig4 mutants, respectively (Qi et al., 2013). These latter authors also showed that NHEJ mutagenesis frequency was not changed significantly in ku70 and lig4 mutants but that DNA repair was shifted to the alternative, microhomology-dependent, NHEJ. As a result, mutations in both Ku70 and Lig4 were predominantly large deletions (Qi et al., 2013). Considering these results, the effect of Lig4 knockout on GT can be explained as follows: in our GT system, DSBs may be induced at the ALS locus to some extent even though expression levels of Cas9 and gRNA are somewhat lower than those seen in polyethylene glycol-mediated protoplast transformation. However, DSBs induced by CRISPR/Cas9 are repaired by NHEJ due to low HDR activity and/or insufficient HDR template. When Lig4 is mutated prior to transformation of the GT vector, NHEJ repair at DSB sites in ALS is inhibited, and the competitive homologous recombination pathway can be recruited to the DSB site. In addition, suppression of random integration of the GT vector due to Lig4 deficiency might have a positive effect on GT. In fact, it has been reported that suppression of Ku70/80 or Lig4 function can increase the frequency of GT in several organisms, such as fungi (de Boer et al., 2010; Ushimaru et al., 2010; Nakazawa et al., 2011), bacteria (Zhang et al., 2012), vertebrates (Bertolini et al., 2009; Fattah et al., 2008; Iiizumi et al., 2008), and plants (Tanaka et al., 2010; Jia et al., 2012).
Thus, although Lig4 deficiency appears effective in enhancing GT, NHEJ is an important DNA repair pathway and somaclonal mutations might be increased in a Lig4 knockout background. Indeed, Lig4-deficient mutants in Arabidopsis are sensitive to DNA damage such as ionizing radiation (Friesner and Britt, 2003). So, instead of knockout, transient suppression of Lig4 at the time of DSB induction with template delivery, or utilization of a Lig4 inhibitor, will be the next step toward efficient GT without undesirable mutations in rice.
A noteworthy result of this study is the generation of biallelic GT plants. Possible mechanisms leading to biallelic mutation include the following: (1) simultaneous or consecutive GT events at both alleles; or (2) GT occurring first on one allele with consecutive HDR of the second allele using the modified homologous chromosome as a template. Both these mechanisms would be favored by blocking NHEJ. It is difficult to obtain homozygous mutated plants by self-pollination in the case of vegetatively propagated and self-incompatible plants. Thus, biallelic modification of a gene of interest will be necessary in some cases. Even with seed-propagated plants, biallelic GT has the advantage of saving time for fixing gene modification. Improvements in HDR template delivery, and expression of Cas9 and gRNA, suppression of the NHEJ pathway and in the selection system of GT cells, will all contribute to the establishment of an efficient GT system in plants.
MATERIALS AND METHODS
Vector Construction
All-in-one vectors of Cas9, gALS-1 to -4 and HPT (pZH_OsU6gALS_MMCas9) were constructed according to the method described in Mikami et al. (2015a, 2015b). The binary vector pCAMBIA-sGFP (Toki 1997; Toki et al., 2006) was used as a control vector in GT experiments as it contains the HPT gene. pZH_MMCas9 is described in Mikami et al. (2015a, 2015b). pZH_gLig4-1_MMCas9 and pZH_gLig4-2_MMCas9 were assembled according to the strategy shown in Mikami et al. (2015a, 2015b). The GT vector pPZ_mALS_g2&3 was constructed as follows: (1) a partial ALS fragment with silent mutations W548L and S627I was synthesized by Life Technologies, Thermo Fisher Scientific (Carlsbad, CA) and cloned into the Sal I/BamHI site of the binary vector pPZP2028, which is a derivative of pPZP202 (Hajdukiewicz et al., 1994) with the addition of rare restriction enzyme (Asc I and Pac I) sites to both ends of the multicloning site of pPZP202. (2) pU6gRNA-oligo (Mikami et al., 2015a) has two Bbs I sites between the OsU6 promoter and the gRNA scaffold sequence. This vector was linearized using Bbs I and the 20-bp target sequences of gLig4-1 and gLig4-2 were ligated into this site. (3) To connect the two gRNA expression cassettes, a gALS-3 expression construct containing the OsU6 promoter, gRNA scaffold sequence, and polyT sequence was extracted from pU6gRNA using Pvu II and Asc I and cloned into the downstream region of the gALS-2 expression construct using EcoRV and Asc I sites. (4) Tandemly repeated gALS-2 and gALS-3 were eliminated using Puv II and cloned into the Sma I site next to ALS fragments in pPZP2028.
Rice Transformation
Agrobacterium-mediated transformation of rice (O. sativa L. cv Nipponbare) was performed as described previously (Toki, 1997; Toki et al., 2006). To select promising target sites for CRISPR/Cas9 on the ALS gene, 3-week-old cultured rice scutellum-derived calli were used for transformation of all-in-one vector of Cas9, gRNA, and HPT expression constructs (pZH_OsU6gALS_MMCas9) and mutation frequency was analyzed 1 month after transformation. For GT experiments, 2-week-old cultured calli were used for the transformation of pCAMBIA-sGFP, pZH_MMCas9, pZH_gLig4-1_MMCas9, and pZH_gLig4-2_MMCas9 at the first transformation. Transgenic calli were selected on callus-induction medium (CIM, Toki, 1997) containing 50 mg/L hygromycin B (Wako Pure Chemicals, Osaka, Japan) and 25 mg/L meropenem (Wako Pure Chemicals) to remove Agrobacterium for three weeks. One-week before the second transformation, calli were transferred to CIM without meropenem. Then, the GT vector pPZ_mALS_g2&3 was transformed in a second transformation. After a 3-d cocultivation, infected calli were transferred to fresh CIM containing 0.75 μM BS (Kumiai Chemical Industry, Tokyo, Japan) to select GT calli.
PCR-Mfe I Digestion Analysis
Genomic DNA was extracted from calli using an Agencourt Chloropure kit (Beckman Coulter, Brea, CA) for PCR. A 1072-bp ALS fragment was amplified using the following primers: ALS-4040 5′-ATTGATCCAGCAGAGATTGGAAAG-3′, ALS-22 5′-ACATGATATCTTGTGATGCATATGCCTAC-3, and Prime STAR GXL DNA Polymerase (Takara Clontech, Shiga, Japan). To confirm the occurrence of GT in BS-tolerant rice plants, we performed PCR analysis coupled with Mfe I digestion using Mfe I-HF; a high fidelity version of Mfe I was supplied by New England Biolabs (Ipswich, MA).
Sequencing Analysis
PCR products were subjected to direct sequence analysis or cloned into pCR-Blunt II-TOPO (Invitrogen, San Diego, CA) and then subjected to sequencing analysis using an ABI3130 sequencer (Applied Biosystems, Foster City, CA).
Southern-Blot Analysis
Genomic DNA was extracted from calli using a Nucleon Phytopure extraction kit (GE Healthcare Life Sciences, Little Chalfont, Buckinghamshire, UK) according to the manufacturer’s instructions. After Mfe I digestion and electrophoresis on a 1% agarose gel, DNA fragments were transferred onto a positively charged nylon membrane (Roche, Basel, Switzerland). Probes were prepared using a PCR DIG probe synthesis kit (Roche) and the following primers: Asc-ALS 5′-GGCGCGCCGCCGGCCACGCCGCTCCGGCCGT-3′ and OsALS-14 5′-AGGCGTGCGATGTACCCTGGTAGATT-3′ for probe A; OsALS-4340 5′-TGTCTTCGGCTGGTCTGGGCGCA-3′ and OsALS-4860 5′-TCAGGTCAAACATAGGCCGGGCTGGT-3′ for probe B. Hybridization was performed according to the DIG Application Manual.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. CRISPR/Cas9-mediated target mutagenesis in OsALS gene.
Supplemental Figure S2. Demonstration of the fact that artificial heteroduplex of the wild type and mutated ALS creased by PCR is not digested by a high fidelity version of Mfe I.
Supplemental Figure S3. T1 progenies of biallelic GT plant.
Supplementary Material
Acknowledgments
We thank K. Amagai, R. Aoto, C. Furusawa, A. Nagashii, R. Takahashi, and F. Suzuki for general experimental technical support.
Glossary
- ALS
acetolactate synthase
- Cas9
CRISPR-associated protein 9
- BS
bispyribac sodium
- CIM
callus-induction medium
- CRISPR
clustered regularly interspaced short palindromic repeat
- DSB
double-strand break
- gRNA
guide RNA
- GT
gene targeting
- HDR
homology-directed repair
- Lig4
DNA ligase 4
- NHEJ
nonhomologous end joining
- SSN
sequence-specific nuclease
- TALEN
transcriptional activator-like effector nuclease
- T-DNA
transfer DNA
- ZFN
zinc finger nuclease
References
- Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF (2014) DNA replicons for plant genome engineering. Plant Cell 26: 151–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolini LR, Bertolini M, Maga EA, Madden KR, Murray JD (2009) Increased gene targeting in Ku70 and Xrcc4 transiently deficient human somatic cells. Mol Biotechnol 41: 106–114 [DOI] [PubMed] [Google Scholar]
- Cai CQ, Doyon Y, Ainley WM, Miller JC, Dekelver RC, Moehle EA, Rock JM, Lee YL, Garrison R, Schulenberg L, Blue R, Worden A, et al. (2009) Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Mol Biol 69: 699–709 [DOI] [PubMed] [Google Scholar]
- de Boer P, Bastiaans J, Touw H, Kerkman R, Bronkhof J, van den Berg M, Offringa R (2010) Highly efficient gene targeting in Penicillium chrysogenum using the bi-partite approach in deltalig4 or deltaku70 mutants. Fungal Genet Biol 47: 839–846 [DOI] [PubMed] [Google Scholar]
- Endo M, Mikami M, Toki S (2015) Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol 56: 41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Osakabe K, Ono K, Handa H, Shimizu T, Toki S (2007) Molecular breeding of a novel herbicide-tolerant rice by gene targeting. Plant J 52: 157–166 [DOI] [PubMed] [Google Scholar]
- Fattah FJ, Lichter NF, Fattah KR, Oh S, Hendrickson EA (2008) Ku70, an essential gene, modulates the frequency of rAAV-mediated gene targeting in human somatic cells. Proc Natl Acad Sci USA 105: 8703–8708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauser F, Roth N, Pacher M, Ilg G, Sánchez-Fernández R, Biesgen C, Puchta H (2012) In planta gene targeting. Proc Natl Acad Sci USA 109: 7535–7540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner J, Britt AB (2003) Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant J 34: 427–440 [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF III (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31: 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin SB. (2010) Plant proteins involved in Agrobacterium-mediated genetic transformation. Annu Rev Phytopathol 48: 45–68 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Iida S, Terada R (2005) Modification of endogenous natural genes by gene targeting in rice and other higher plants. Plant Mol Biol 59: 205–219 [DOI] [PubMed] [Google Scholar]
- Jia Q, Bundock P, Hooykaas P, de Pater S (2012) Agrobacterium tumefaciens T-DNA integration and gene targeting in Arabidopsis thaliana non-homologous end-joining mutants. J Bot 2012: 989272.
- Iiizumi S, Kurosawa A, So S, Ishii Y, Chikaraishi Y, Ishii A, Koyama H, Adachi N (2008) Impact of non-homologous end-joining deficiency on random and targeted DNA integration: implications for gene targeting. Nucleic Acids Res 36: 6333–6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka-Hisatomi Y, Terada R, Iida S (2008) Efficient transfer of base changes from a vector to the rice genome by homologous recombination: involvement of heteroduplex formation and mismatch correction. Nucleic Acids Res 36: 4727–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Zhang D, Sheen J (2014) Cas9-based genome editing in Arabidopsis and tobacco. Methods Enzymol 546: 459–472 [DOI] [PubMed] [Google Scholar]
- Li Z, Liu ZB, Xing A, Moon BP, Koellhoffer JP, Huang L, Ward RT, Clifton E, Falco SC, Cigan AM (2015) Cas9-guide RNA directed genome editing in soybean. Plant Physiol 169: 960–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami M, Toki S, Endo M (2015a) Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol Biol 88: 561–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami M, Toki S, Endo M (2015b) Parameters affecting frequency of CRISPR/Cas9 mediated targeted mutagenesis in rice. Plant Cell Rep 34: 1807–1815 [DOI] [PubMed] [Google Scholar]
- Nakazawa T, Ando Y, Kitaaki K, Nakahori K, Kamada T (2011) Efficient gene targeting in ΔCc.ku70 or ΔCc.lig4 mutants of the agaricomycete Coprinopsis cinerea. Fungal Genet Biol 48: 939–946 [DOI] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A, Cermak T, Hoshino T, Sugimoto K, Saika H, Mori A, Osakabe K, Hamada M, Katayose Y, Starker C, et al. (2015a) A defect in DNA ligase 4 enhances the frequency of TALEN-mediated targeted mutagenesis in rice. Plant Physiol http://dx.doi.org/10.1104/pp.15.01542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A, Endo M, Ohtsuki N, Saika H, Toki S (2015b) Precision genome editing in plants via gene targeting and piggyBac-mediated marker excision. Plant J 81: 160–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A, Nonaka S, Saika H, Kwon YI, Osakabe K, Toki S (2012) Suppression of Ku70/80 or Lig4 leads to decreased stable transformation and enhanced homologous recombination in rice. New Phytol 196: 1048–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski J, Baur M, Bogucki A, Potrykus I (1988) Gene targeting in plants. EMBO J 7: 4021–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Zhang Y, Zhang F, Baller JA, Cleland SC, Ryu Y, Starker CG, Voytas DF (2013) Increasing frequencies of site-specific mutagenesis and gene targeting in Arabidopsis by manipulating DNA repair pathways. Genome Res 23: 547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiml S, Fauser F, Puchta H (2014) The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J 80: 1139–1150 [DOI] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Chen K, Liang Z, Li J, Zhang Y, Zhang K, Liu J, Voytas DF, Zheng X, Zhang Y, Gao C (2013) Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol Plant 6: 1365–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Kaku K, Kawai K, Miyazawa T, Tanaka Y (2005) Molecular characterization of acetolactate synthase in resistant weeds and crops. In Clark, J M, Ohkawa, H, eds, ACS Symposium Series 899: Environmental Fate and Safety Management of Agrochemicals. American Chemical Society, Washington, DC, pp 255–271 [Google Scholar]
- Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, Choi VM, Rock JM, et al. (2009) Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459: 437–441 [DOI] [PubMed] [Google Scholar]
- Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM (2015) Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol 169: 931–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Ishii C, Hatakeyama S, Inoue H (2010) High efficient gene targeting on the AGAMOUS gene in an ArabidopsisAtLIG4 mutant. Biochem Biophys Res Commun 396: 289–293 [DOI] [PubMed] [Google Scholar]
- Terada R, Johzuka-Hisatomi Y, Saitoh M, Asao H, Iida S (2007) Gene targeting by homologous recombination as a biotechnological tool for rice functional genomics. Plant Physiol 144: 846–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada R, Urawa H, Inagaki Y, Tsugane K, Iida S (2002) Efficient gene targeting by homologous recombination in rice. Nat Biotechnol 20: 1030–1034 [DOI] [PubMed] [Google Scholar]
- Toki S. (1997) Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol Biol Rep 15: 16–21 [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47: 969–976 [DOI] [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF (2009) High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 459: 442–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, White C (2005) Towards targeted mutagenesis and gene replacement in plants. Trends Biotechnol 23: 567–569 [DOI] [PubMed] [Google Scholar]
- Ushimaru T, Terada H, Tsuboi K, Kogou Y, Sakaguchi A, Tsuji G, Kubo Y (2010) Development of an efficient gene targeting system in Colletotrichum higginsianum using a non-homologous end-joining mutant and Agrobacterium tumefaciens-mediated gene transfer. Mol Genet Genomics 284: 357–371 [DOI] [PubMed] [Google Scholar]
- Voytas DF. (2013) Plant genome engineering with sequence-specific nucleases. Annu Rev Plant Biol 64: 327–350 [DOI] [PubMed] [Google Scholar]
- Weeks DP, Spalding MH, Yang B (2015) Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol J http://dx.doi.org/10.1111/pbi.12448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DA, Townsend JA, Winfrey RJ. , Jr., Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF (2005) High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J 44: 693–705 [DOI] [PubMed] [Google Scholar]
- Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyon D, Christian M, Li X, Pierick CJ, Dobbs D, Peterson T, Joung JK, Voytas DF (2010) High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc Natl Acad Sci USA 107: 12028–12033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang F, Li X, Baller JA, Qi Y, Starker CG, Bogdanove AJ, Voytas DF (2013) Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol 161: 20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen W, Zhang Y, Jiang L, Chen Z, Wen Y, Li J (2012) Deletion of ku homologs increases gene targeting frequency in Streptomyces avermitilis. J Ind Microbiol Biotechnol 39: 917–925 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.