Abstract
In this review we discuss the inhibitory effects of dietary polyphenols and amphibian antimicrobial/antitumor peptides on ATP synthase. In the beginning general structural features highlighting catalytic and motor functions of ATP synthase will be described. Some details on the presence of ATP synthase on the surface of several animal cell types, where it is associated with multiple cellular processes making it an interesting drug target with respect to dietary polyphenols and amphibian antimicrobial peptides will also be reviewed. ATP synthase is known to have distinct polyphenol and peptide binding sites at the interface of α/β subunits. Molecular interaction of polyphenols and peptides with ATP synthase at their respective binding sites will be discussed. Binding and inhibition of other proteins or enzymes will also be covered so as to understand the therapeutic roles of both types of molecules. Lastly, the effects of polyphenols and peptides on the inhibition of Escherichia coli cell growth through their action on ATP synthase will also be presented.
Keywords: F1Fo ATP synthase, ATPase, E. coli ATP synthase, dietary polyphenols, amphibian antimicrobial peptides, enzyme inhibitors
INTRODUCTION
ATP synthase is the fundamental means of cellular energy production in animals, plants, and almost all microorganisms. ATP synthase is also the final enzyme in the oxidative phosphorylation pathway and is responsible for ATP synthesis by oxidative or photophosphorylation in the membranes of bacteria, mitochondria, and chloroplasts. It is the smallest known biological nanomotor, found from bacteria to man. In order to synthesize ATP, the cell’s energy currency, a mechanical rotation mechanism is used in which subunits rotate at approximately 100 times per second in order to convert food into energy by oxidation. A typical 70 kg human with a relatively sedentary lifestyle will generate around 2.0 million kg of ATP from ADP and Pi (inorganic phosphate) in a 75-year lifespan [1]. ATP synthase functions in the same way in both prokaryotes and eukaryotes [2]. For different organisms estimates of the number of protons required to synthesize one ATP molecule have ranged from three to four, with the possibility that cells can vary this ratio to suit different conditions [3–5].
STRUCTURE FUNCTION RELATIONSHIP OF ATP SYNTHASE
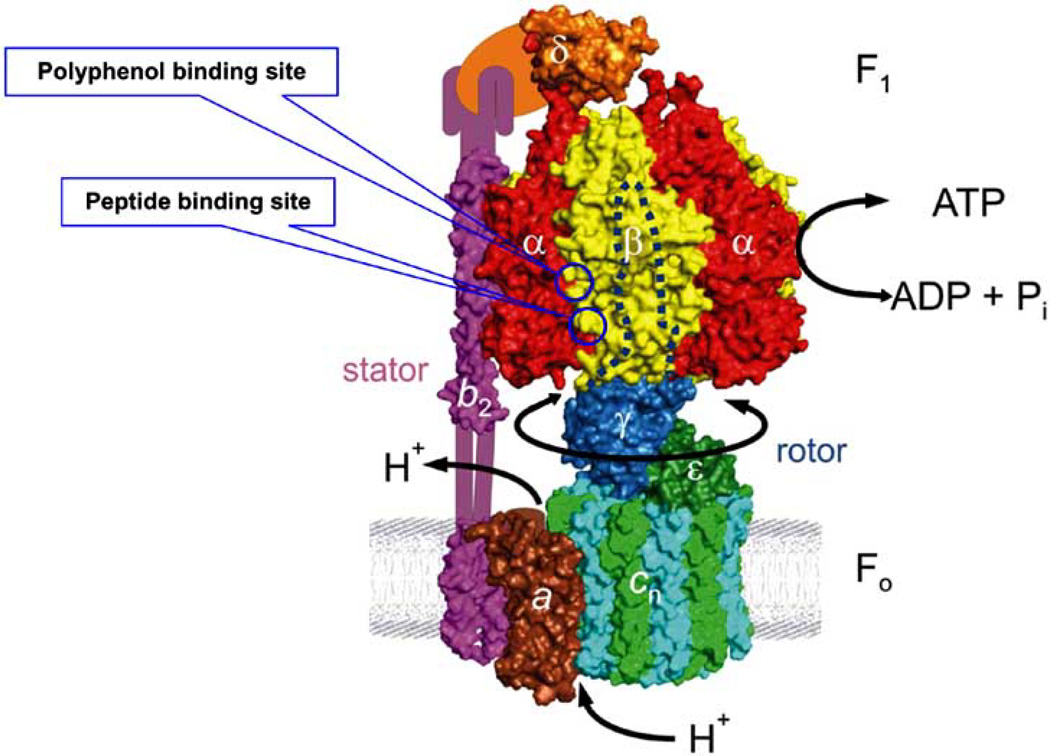
F1Fo-ATP synthase is structurally and functionally similar whatever the source. In its simplest form, as shown in Fig. (1), Escherichia coli ATP synthase contains eight different subunits, namely α3β3γδεab2c10. The total molecular mass is ~530 kDa. F1 corresponds to α3β3γδε and Fo to ab2c10. In chloroplasts, the structure is the same except that there are two isoforms. In mitochondria, there are 7–9 additional subunits, depending on the source, but in toto they contribute only a small fraction of additional mass and may have regulatory functions [6–8]. ATP hydrolysis and synthesis occur on three catalytic sites in the F1 sector, whereas proton transport occurs through the membrane embedded Fo sector. The γ-subunit forms a coiled coil of α-helices that go right up into the central space of the α3β3 hexagon. Proton gradient-driven clockwise rotation of γ (as viewed from the outer membrane) leads to ATP synthesis and anticlockwise rotation of γ results in ATP hydrolysis. In recent nomenclature, the rotor consists of γεcn, and the stator consists of α3β3δab2 [9–11]. The function of the stator is to prevent co-rotation of catalytic sites with the rotor. Current understanding of the F1Fo structure and mechanism has been thoroughly reviewed by Senior’s group and others [1, 11–22].
Fig 1. Structure of Escherichia coli ATP synthase.
In its simplest form in E. coli this enzyme is composed of two sectors, water soluble F1 and membrane bound Fo. Catalytic activity occurs at the interface of α/β subunits of F1 sector which consists of five subunits (α3β3γδε ) and proton conduction occurs at the Fo sector consisting of three subunits (ab2c). In mitochondria and chloroplasts additional subunits are present. Polyphenol and peptide binding sites are identified with circles at the interface of α/β subunits. This model of E. coli ATP synthase is reproduced from Weber [11] with permission; copyright Elsevier.
The three catalytic sites located on the F1 sector of ATP synthase are designated βTP, βDP, and βE by x-ray crystallographers, based on the binding of ATP, ADP, and Pi respectively [23, 24]. βE is the empty site into which Pi (inorganic phosphate) must initially bind for ATP synthesis. It has been proposed that the synthesis reaction in the three catalytic sites does not occur independently but occurs sequentially. In this mechanism, the three catalytic sites have different affinities for nucleotides at any given moment. Each catalytic site undergoes conformational transitions that lead to the following sequence: substrate (ADP+Pi) binding → ATP synthesis → ATP release. Experimental observations of rotation verified the predication made by Boyer [2, 25, 26] that catalysis requires the sequential involvement of three catalytic sites, each of which changes its binding affinity for substrates and products as it proceeds through the cyclical mechanism, hence the term “binding change mechanism.” Proton motive force is converted in Fo to a mechanical rotation of the rotor shaft, which drives conformational changes of the catalytic domains in F1 to synthesize ATP. Conversely, hydrolysis of ATP induces reverse conformational changes of Fo sector and consequently reverses rotation of the shaft. Conformational changes in the catalytic sites are connected to the γ-subunit rotation. γ-Subunit rotation in isolated α3β3γ subcomplex has been observed directly by Yoshida and Kinosita with colleagues in Japan, and subsequently by several other labs [4, 27–31]. The reaction mechanism of ATP hydrolysis and synthesis in F1Fo and their relationship to the γ-subunit mechanical rotation is not the focus of this review, but is a fundamental question that remains to be elucidated. This question also applies to many other ATPases and GTPases, with a relevance to nanotechnology and nanomedicine [32, 33]. Therefore, it is important to summarize the catalytic function of ATP synthase in some detail before describing inhibition chemistry of ATP synthase by polyphenols or peptides [34, 35].
CATALYTIC AND MOTOR FUNCTION OF ATP SYNTHASE
Previous studies were focused on determining the Pi binding residues in the catalytic site so as to better understand the reaction mechanism of ATP synthesis, hydrolysis and their relationship to mechanical rotation. Identification of Pi binding residues and residues surrounding the Pi binding subdomain in the catalytic site is imperative for answering the following two important questions. (I) How does the enzyme bind ADP and Pi rather than ATP at catalytic sites? This is an often overlooked but crucial question in the mechanism of ATP synthesis. In active cells, the cytoplasmic concentrations of ATP and Pi are approximately in the 2–5 mM range, whereas that of ADP is at least 10–50-fold lower. Equilibrium binding assays have established that both ADP and ATP bind to catalytic sites of purified F1 and detergent solubilized F1Fo with relatively similar binding affinities [36–39]. Obviously, a specific mechanism must have evolved for selectively binding ADP into catalytic sites while contemporaneously discouraging access to ATP during proton driven rotation and ATP synthesis. One hypothesis is that during ATP synthesis, proton gradient driven rotation of subunits drives an empty catalytic site to bind Pi tightly, thus stereo-chemically precluding ATP binding and therefore selectively favoring ADP binding [12]. (II) How does subunit rotation affect Pi binding [25, 40, 41]? It was shown that Pi binding appears to be “energy linked”, implying that it is linked directly to subunit rotation [6, 42, 43]. Therefore, for formulating a mechanism for ATP synthesis it is of paramount importance to understand the features that determine Pi binding. Moreover, in the near future it may be possible to use molecular features of Pi binding, derived from mutational and biochemical studies, in the development of potent and novel molecular inhibitors of ATP synthase.
X-ray crystallographic structure of the catalytic sites of ATP synthase shows the following residues αPhe-291, αSer-347, αGly-351, αArg-376, βLys-155, βArg-182, βAsn-243, and βArg-246 in close proximity to bound phosphate analogs AlF3 or SO42− suggesting that these residues are involved in Pi binding [24, 44]. [E. coli residue numbers are used throughout]. Earlier attempts to measure Pi binding in purified E. coli F1 using [32P] Pi [40] or by competition with ATP or AMP-PNP in fluorescence assays of nucleotide binding [37, 45] failed to detect appreciable Pi binding at physiological Pi concentration. An assay devised by Perez et al. [46] in which the protection afforded by Pi against inhibition of ATPase activity induced by covalent reaction with 7-chloro-4-nitrobenzo-2-oxa-1, 3,-diazole (NBD-Cl) provided a measure of Pi binding. Earlier Orris et al. [47] showed by X-ray crystallography that the covalent adduct formed by NBD-Cl is specifically in the βE catalytic site, thus protection afforded by Pi indicates that binding of Pi occurs at the βE catalytic site. By modifying the above assay for use with E. coli purified F1 or membrane bound F1Fo, we have thus far investigated the relationship between Pi binding and catalysis for eight residues, namely αPhe-291, αSer-347, αGly-351, αArg-376, βLys-155, βArg-182, βAsn-243, and βArg-246. We found that the following five residues; αSer-347, ocArg-376, βLys-155, βArg-182, and βArg-246 are grouped in a triangular fashion, and are involved in Pi binding. The other three residues; αPhe-291, αGly-351, and βAsn-243 are not [17, 33, 48–52]. Presence of Pi binding residues in the catalytic site explains the preferential binding of ADP over ATP. The story doesn’t end here as there are many other residues in close proximity to Pi binding subdomain in the catalytic sites which may have a direct or indirect role in Pi binding and thus require further characterization.
ROLE OF ATP SYNTHASE IN HUMAN HEALTH AND DISEASES
ATP synthase is critical to human health. Malfunction of this complex has been implicated in a wide variety of diseases including cancer, tuberculosis, neuropathy, Alzheimer’s, Parkinson’s, and a class of severely debilitating diseases known collectively as mitochondrial myopathies. This enzyme is also a likely therapeutic target in the treatment of diseases such as, cancer, heart disease, mitochondrial diseases, immune deficiency, cystic fibrosis, diabetes, ulcers, tuberculosis, Parkinson’s, and Alzheimer’s ([53] and reference therein). One of the forms of Leigh syndrome, a neurodegenerative disease, is the result of mutation in the a-subunit of ATP synthase [54]. The c-subunit of ATP synthase is involved in both the lysosomal storage diseases, Kufs’ and Battens’. Low expression of β-subunit and accumulation of α-subunit in the cytosol is associated with Alzhimer’s disease. The neuropathy, ataxia, is also caused by dysfunction of ATP synthase. Presence of circulating subunit F6 has been associated with hypertension. Recent studies have also suggested that the presence of ATP synthase on the surface of several animal cell types is correlated with multiple cellular processes including lipid metabolism, angiogenesis, intracellular pH regulation, and programmed cell death [55–59]. Angiostatin is a known inhibitor of angiogenesis, which was shown to bind to ATP synthase on the surface of human endothelial cells. It was shown that angiostatin’s antiproliferative effect on endothelial cells was dependent on its interaction with the α-subunit of ATP synthase. The mechanism behind this is the transport of H across the plasma membrane by mitochondrial ATP synthase causing cytolysis of tumor cells [60].
In addition to the above mentioned conditions, ATP synthase is also a target enzyme for antimicrobial agents. Streptococcus mutans is a primary microbial agent in the pathogenesis of dental caries through biofilm formation and acid production. Inhibition of ATP synthase of S. mutans inhibits biofilm formation and acid production [61, 62]. Also in Mycobacterium ATP synthase, two mutations in the c-subunit (D32V and A63P) confer resistance to the new tuberculosis drug diarylquinoline [63, 64]. Thus a better understanding of ATP synthase inhibition may aid in the treatment of these diseases.
ATP SYNTHASE INHIBITION
A wide range of natural and synthetic products are known to bind and inhibit both bovine mitochondrial and E. coli ATP synthase [53, 65, 66]. The inhibitory effects and the extent of inhibition on molar scale are variable among different inhibitors. Also, some of the inhibitors are known to inhibit only ATP synthesis and not hydrolysis or vice versa while others inhibit both synthesis and hydrolysis equally. Among the potent inhibitors of ATP synthase are: 7-chloro-4-nitrobenzo-2-oxa-1, 3-diazole (NBD-Cl), sodium azide (NaN3), aluminum fluoride (AlFx), scandium fluoride (ScFx), beryllium fluoride (BeFx), dicyclohexylcarbodiimide (DCCD), several naturally occurring antibiotics such as oligomycin, efrapeptins, aurovertins, leucinostatins, a number of polyphenols like resveratrol, piceatannol, quercetin, morin, epicatechin, and quite a few peptides such as melittin, melittin related peptide (MRP), ascaphin, aurein, caerin, dermaseptin, and maganain II [17, 33–35, 65, 67–75].
The importance of ATP synthase as a promising target for drug development is evident from the fact that many antibiotics such as efrapeptins, aurovertins, and oligomycins inhibit its function. Antibiotics efrapeptins and aurovertins inhibit both synthesis and hydrolysis of ATP by ATP synthase [73, 76]. The efrapeptins bind to ATP synthase at a site extending from the rotor, across the central cavity of the enzyme, into the specific βE-subunit catalytic site. This binding prevents the closure of the βE subunit during the rotary cycle [73, 77]. Aurovertins are known to bind and inhibit mitochondrial ATPase, thereby uncoupling oxidative phosphorylation. Two molecules of aurovertin bind simultaneously to the cleft between nucleotide binding and C-terminal domain of two β subunit domains [76]. Aurovertin was shown to bind non-covalently to wild-type E. coli F1 with Kd of 1µM [78, 79]. Weber and Senior [65] using 10 µM aurovertin found that it inhibited ATPase activity of β-Trp-331 F1 by 87%. Another interesting observation was that aurovertin is an uncompetitive inhibitor of E. coli and mitochondrial F1-ATPase and does not achieve total inhibition of hydrolysis [80].
Oligomycins are macrolides generated by Streptomyces and can be poisonous to other organisms. The macrolides are a group of antibiotic drugs whose activity depends on the presence of a macrolide ring. Macrolides are secondary metabolites from bacteria, fungi, plants, and animals belonging to the polyketide class of natural products [81]. Common macrolide antibiotics in use are Azithromycin, Clarithromycin, Dirithromycin, and Erythromycin. Antibiotic macrolides are used to treat infections such as respiratory tract and soft tissue infections. The antimicrobial spectrum of macrolides is slightly wider than that of penicillin, and therefore macrolides are a common substitute for patients with a penicillin allergy. Beta-hemolytic Streptococci, Pneumococci, Staphylococci and Enterococci are usually susceptible to macrolides. Unlike penicillin, macrolides have been shown to be effective against mycoplasma, mycobacteria, some rickettsia, and chlamydia.
In addition, oligomycin is a potent inhibitor of ATP synthase by binding in the Fo sector and blocking proton conduction. Oligomycin, was shown to induce an apoptotic suicide response in cultured human lymphoblastoid and other mammalian cells within 12–18 hrs, but not in ρo cells that are depleted of a functional mitochondrial respiratory chain [82]. A similar study suggested that interaction with components of mitochondrial pathways by oligomycin may lead to apoptosis of select cells, via CD14 [83]. Thus, it is quite possible that some degree of inhibition, or other interactions between mitochondrial ATP synthase and polyphenols could occur and play a considerable role in apoptosis via mitochondrial pathways [34, 84, 85].
INHIBITORY EFFECTS OF POLYPHENOLS
Polyphenols are naturally occurring plant based phyhtochemicals which possess antioxidant, chemopreventive, and chemotherapeutic properties [84, 86, 87]. Apples, berries, cantaloupe, cherries, grapes, pears, plums, broccoli, cabbages, and onions are rich in polyphenols [88]. A variety of flavonoids or polyphenolic compounds exert a broad range of pharmacological effects, including protection of cells or tissues and invoking multiple responses, including cell death, through their actions on a multitude of targets. A large body of experimental data is available on the effects of dietary polyphenolic compounds and their derivates on human health. Some polyphenols are known to block the action of enzymes and other substances that promote the growth of cancer cells [66, 89–93]. Physiological relevance of dietary polyphenols can be ascribed to their interaction with the mitochondria of eukaryotic cells, while degenerative diseases such as cancer, aging, and neurological disorders are attributed to mitochondrial dysfunction [94, 95]. It was shown that known cardiovascular benefits of dietary polyphenols may derive in part from their inactivation of plasminogen activator inhibitor type 1(PAI-1). PAI-1 has been implicated in variety of pathological processes, such as angiogenesis and tumor metastasis [96, 97].
In vitro studies using rat hepatic mitochondria suggested that flavonoid extracts from Algerian plants have some protective effects against oxidative stress by protecting the mitochondria [98]. Resveratrol (trans-3,4',5,-trihydroxy-stilbene), a phytoalexin, is a toxic antimicrobial compound produced by plants under stress conditions, or in response to pathogen infection or parasitic attack. Phytoalexins are broad spectrum inhibitors that are chemically different among plant species. Phytoalexins are toxic to the pathogens and they may puncture cell walls, delay maturation, disrupt metabolism or prevent reproduction. Inhibition of phytoalexin biosynthesis has been found to result in increased susceptibility of plant tissues to infections [99].
Resveratrol has the potential for multiple uses, with multiple benefits in humans, including but not limited to increased life span, anticancer/antitumor effects, and antimicrobial activities [100]. Administration of resveratrol was shown to increase life span of yeast, Caenorhabditis elegans, Drosophila melanogaster, and mice [101–104]. Resveratrol was also shown to induce apoptosis via mitochondrial pathways [90, 105]. Aziz et al. [93] demonstrated that resveratrol has chemopreventive properties against prostate cancer. They found that treatment with resveratrol concentrations of up to 50 µmol/L/day resulted in stimulation of apoptosis in androgen-responsive human prostate carcinoma cells (LNCaP). At similar concentrations resveratrol had no effect on the rate of cell death in normal human prostate cells.
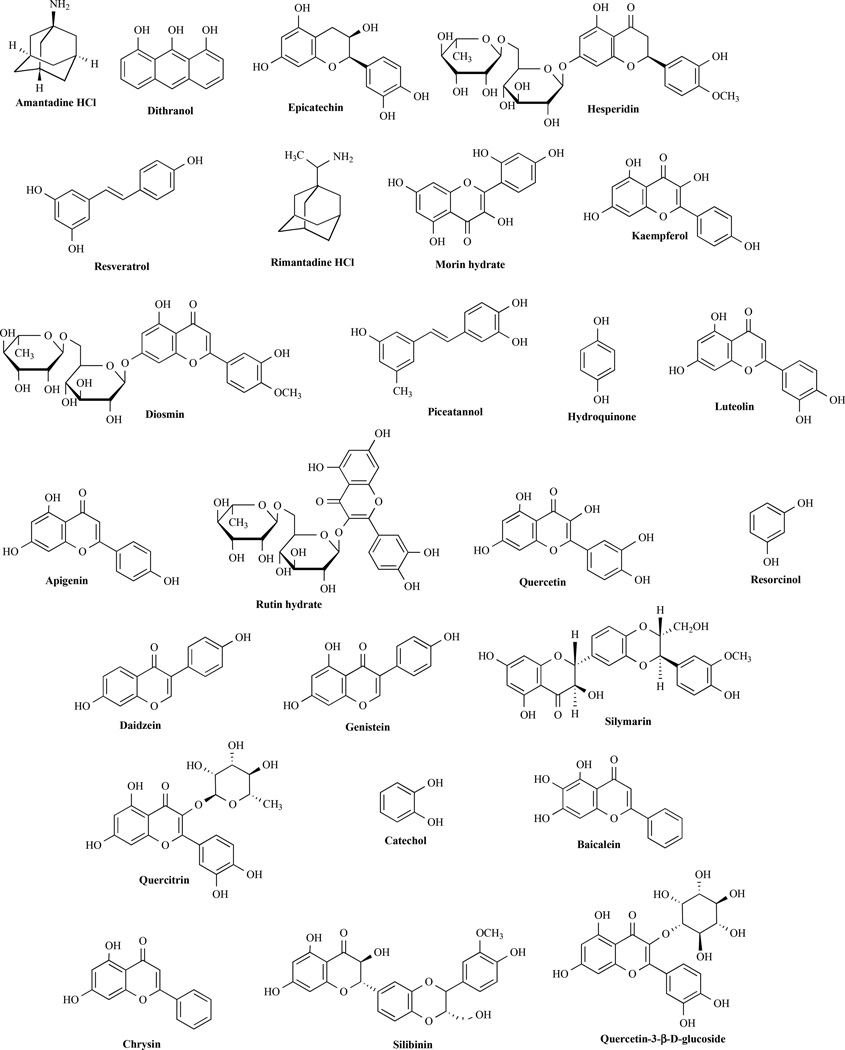
Fig. (2) illustrates some known polyphenol inhibitors of ATP synthase. These polyphenolic compounds have varying degree of inhibitory effects on ATPase activity as well as on E. coli cell growth. One of the best sources of natural polyphenols is tea. Tea has been associated with multiple health benefits that are attributed to the presence of polyphenolic compounds. Catechins are polyphenolic antioxidant plant metabolites that constitute ~25% of the dry weight of fresh tea leaves [106]. The term catechin is also commonly used to refer to the related family of flavonoids. Actual catechin content varies depending on the climate where tea is being grown. Catechins are present in practically all teas made from Camellia sinensis, including white tea, green tea, black tea and Oolong tea. Catechins are also present in chocolate [107], fruits, vegetables and in many other plant species [108, 109]. Catechin gallates are gallic acid esters of the catechins. Epigallocatechin gallate (EGCG) is the most abundant catechin in tea. Catechin and epicatechin are epimers, with (−)-epicatechin and (+)-catechin being the most common optical isomers found in nature. Epigallocatechin and gallocatechin contain an additional phenolic hydroxyl group when compared to epicatechin and catechin, respectively. The health benefits of catechins include reduction in atherosclerotic plaques and reduced carcinogenesis in vitro and in animal models [110, 111]. Green tea polyphenols such as EGCG and epicatechin gallate (ECG) were shown to modulate insulin secretion by inhibiting glutamate dehydrogenase activity [112]. EGCG also causes apoptosis of breast cancer cells by blocking in vitro Fatty acid synthase (FASN) activity that is over expressed in human breast carcinomas [113].
Fig 2. Structure of polyphenolic compounds.
All polyphenolic compound indentified here are known to bind and inhibit ATP synthase.
Mice fed catechins showed decreased levels of aging, lowering of oxidative stress in mitochondria, and an increase in mRNA transcription of mitochondria related proteins [114]. Previous studies indicated concentration dependent effects of epicatechin on biomarkers of oxidative stress in hypertensive and normal patients. The mode of action involves reduction of malondi-aldehyde (MDA) and protein carbonyl content with an increase in glutathione (GSH) and membrane sulfhydryl (−SH) content [115]. Recently we also found that epicatechin is a potent inhibitor of E. coli ATP synthase. Both cell growth and ATPase activity were abrogated (IC50 ~4.0 mM) [85].
Polyphenolic compounds also have promising antimicrobial activities. For example, quercetin (3,3',4',5,7-pentahy-droxyflavone) and apigenin (4',5,7-trihydroxyflavone) both show antibacterial activity against D-Ala:D-Ala ligase (Ddl) and E. coli ATP synthase [34, 116] and [85]. They function as reversible inhibitors of Ddl and compete with its ATP substrate but are non-competitive inhibitors with substrate DAla. Both quercetin and apigenin are reversible inhibitors of E. coli ATP synthase and bind non-covalently at the polyphenol binding site. Quercetin was also found to be a more potent inhibitor on molar scale, having higher affinity than apigenin against Helicobacter pylori Ddl, E. coli Ddl, and E. coli ATP synthase [34, 116] and [85].This can be attributed to the two additional hydroxyl groups on the flavone skeleton of the quercetin, thus facilitating its inhibitory and binding activity to Ddl and ATP synthase (see Fig. 2).
It was shown that polyphenols can also inhibit biofilm formation and acid production by S. mutans. One of the pathways through which polyphenols are active against S. mutans is the inhibition of proton–translocating F1-ATPase activity [61, 62]. The role of mycobacterial ATP synthase is also of interest as tuberculosis (TB) still claims about 2 million lives worldwide yearly. It is interesting that two mutations, D32A and A63P, in the c-subunit of mycobacterium ATP synthase confer resistance to diarylquinoline, a tuberculosis drug [63, 64]. A wide range of polyphenols have been shown to bind at the distinct polyphenol binding site and inhibit ATP synthase partially or maximally. Fig. (3) shows the polyphenol binding pocket of ATP synthase both in unbound and polyphenol bound form [34].
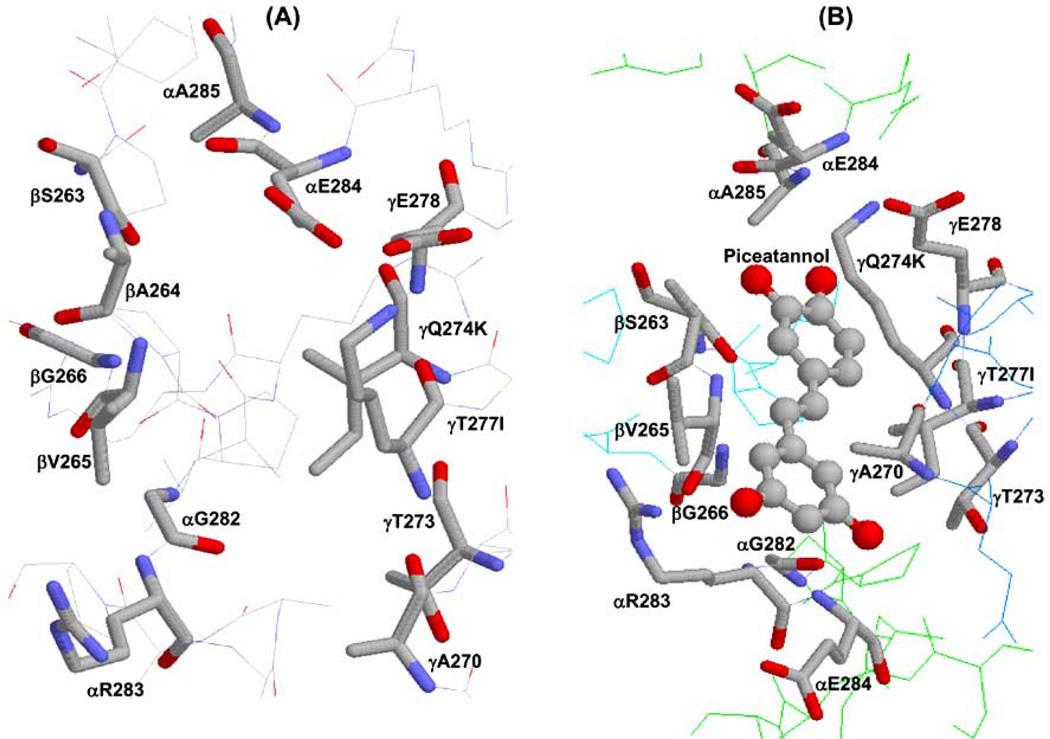
Fig 3. X-ray crystallographic structure of polyphenol binding site of ATP synthase.
(A) Empty and (B) piceatannol bound polyphenol binding pocket. Residues from α, β, and γ subunits involved in interaction with polyphenols are identified. In bovine two variants, Q274K and T277I, occur in the γ subunit and are identified in the figure. PDB file 2jj 1 [84] with RasMol [161] was used to generate this figure.
Among the many polyphenols studied so far, piceatannol is one of the most portent inhibitors of ATP synthase. The polyphenol binding pocket lies at the interface of the α, β, and γ-subunits of F1 sector. In the polyphenol bound ATP synthase crystal structure the polyphenols piceatannol, resveratrol, or quercetin were found to bind in a slightly distorted planar conformation through H-bonds and hydrophobic interactions [84]. As can be seen in Fig. (3B), piceatannol can form hydrophobic interactions with γGln274 (γLys-260), γThr-277 (γIle-263), βAla-264 (βAla-278), or βVal-265 (βVal-279), and an additional non-polar interaction with residues γAla-270 (γAla-256), γThr-273 (γThr-259), γGlu-278 (γGlu-264), αGly-282 (αGly-290), or αGlu-284 (αGlu-292) which are within 4Å of the bound compounds [E. coli residue numbers are used throughout. Bovine residue numbers are shown in parentheses]. Overall, the polyphenol binding pocket residues are highly conserved among different species including human, bovine, rat, and E. coli [117, 118]. The Polyphenol binding pocket residues of E. coli ATP synthase are identical to the bovine polyphenol binding pocket residues, except for two changes, namely γQ274K and γT277I where Gln is replaced by Lys and Thr is replaced by Ile in bovine. Distance measurements using Deep View Swiss-Pdb Viewer, Version 4.01 (http://spdbv.vital-it.ch/) suggested that the –OH group of γThr-277 generates an additional H-bond with the –OH group of γSer-281 [34] and can form additional H-bond with the oxygen or –OH groups of the polyphenol compounds.
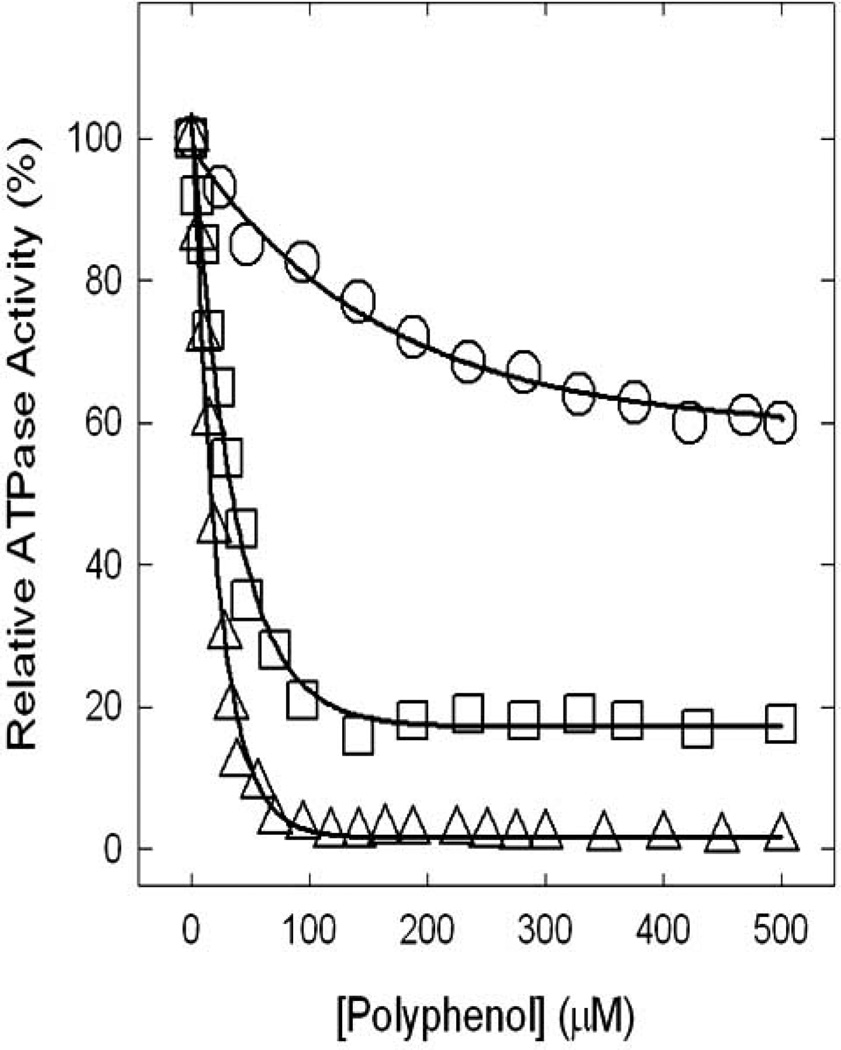
Earlier in this review several potentially relevant effects of natural dietary polyphenolic compounds as antimicrobial and antitumor agents were discussed. Many polyphenols (see Fig. 2) have been shown to bind and inhibit ATP synthase, suggesting that the dietary benefits of these compounds are in part linked to the inhibition of ATP synthesis in tumor cells, thereby leading to apoptosis [34, 119]. All polyphenols illustrated in Fig. (2) cause partial or complete inhibition of ATP synthase and Fig. (4) shows the inhibitory effects of three well known polyphenols (piceatannol, quercetin, or resveratrol) on ATP synthase. On molar scale piceatannol was found to be the most potent inhibitor of ATPase activity and caused 100% inhibition (IC50~ 14µM). Quercetin causes ~80% inhibition (IC50~ 33µM) while resveratrol exerted only ~40% inhibition (IC50~ 94µM) [34]. Our results [85] suggest that all other polyphenols illustrated in Fig. (2) exert partial or maximal inhibition of E. coli ATP synthase. The inhibitory effects of polyphenols have been found to be reversible in all cases. These polyphenols also inhibit intact E. coli cell growth to varied degrees on limiting glucose media [34]. We also found that structural modulation of polyphenols such as resveratrol have the potential to increase inhibitory effects by as much as 100-fold. Our recent unpublished results demonstrate that introduction of an imino group, repositioning of the –OH groups, and or introduction of nitro group enhances inhibitory effects of resveratrol from ~40% to 100% and IC50~ 94µM to IC50~2.25µM.
Fig 4. Inhibitory effects of polyphenols on ATP synthase.
Inhibition profiles induced by resveratrol (ο), quercetin (□), and piceatannol (Δ) resulting in partial or complete inhibition of ATP synthase are shown. For experimental details see Dadi et al. [34].
INHIBITORY EFFECTS OF AMPHIBIAN ANTIMICROBIAL PEPTIDES
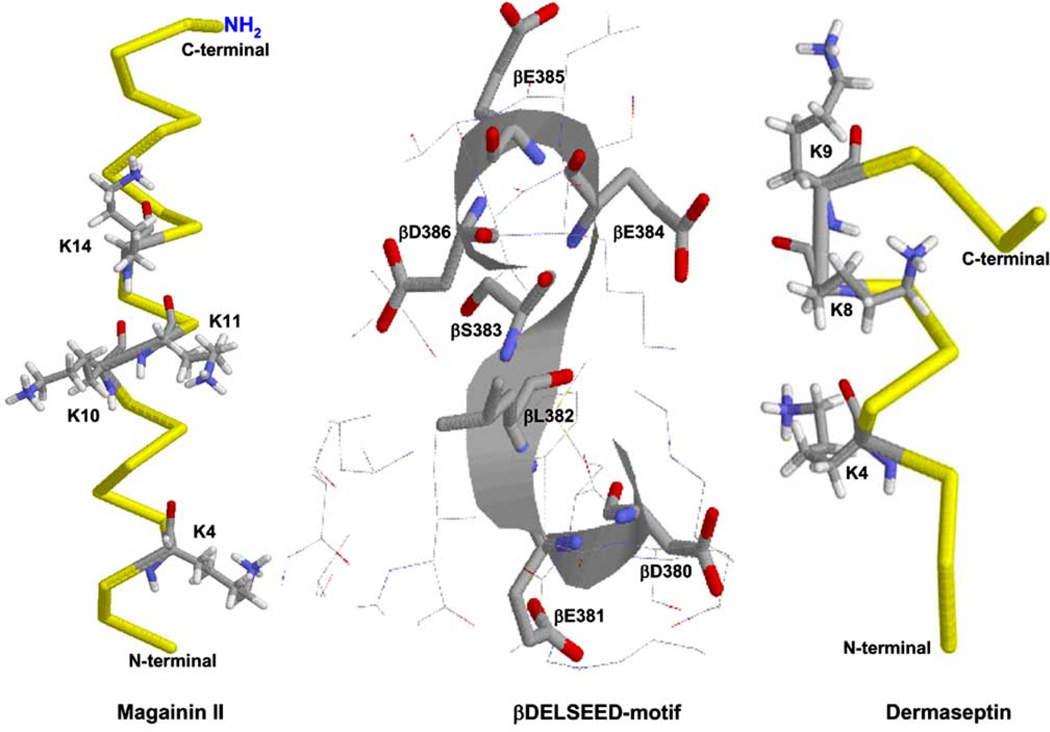
Fig. (5) shows the x-ray crystallographic structure of βDELSEED-loop (residue numbers β380–386) of ATP synthase along with two cationic α-helical peptides maganin II and dermaseptin. Indirect experimental evidence on protection against the inhibition of quinacrine mustard by melittin, the α-helical 26-residue cationic peptide from honey bee venom (Apis mellifera), suggested a common βDELSEED binding site. Several peptides form basic amphiphilic α-helical structures that have been shown to bind at the βDELSEED-loop of E. coli ATP synthase. Peptides such as bacterial/chloroplast ATP synthase ε-subunit, melittin, and the synthetic derivative of cytochrome oxidase SynA2 are known inhibitors of ATP synthase [120–122].
Fig 5. X-ray crystallographic structure of proposed peptide binding βDELSEED-loop of ATP synthase with amphibian AMPs magainin II and dermaseptin.
Important residue side chains involved in the electrostatic interaction between βDELSEED-loop and peptides are illustrated. PDB files used were 2MAG [162], 2DCX [163], and 1H8E [24]. RasMol [161] was used to generate this figure.
Previous studies indicated that during conditions of high gradients and low ATP concentrations, the C-terminal α-helical domain of the ε-subunit of F1-ATPase undergoes large conformational changes and interacts with the α3β3 hexagon ring, where it then comes in close proximity to the βDELSEED-loop. Electrostatic interactions between basic residues of the ε-subunit and the acidic residues of βDELSEED-motif cause inhibition of ATPase activity [123–125]. Another natural regulatory peptide known to affect ATP synthase activity is IF1 of the mitochondria [126, 127]. IF1 is a natural regulatory peptide of between 56–87 residues in length that inhibits ATPase activity of ATP synthase in a reversible and noncompetitive fashion [128]. Crystal structure of IF1 with the F1- subunit shows the N-terminal domain of IF1 bound at the interface of the α and βF1 subunits [129].
Melittin is an α-helical basic peptide also known to have inhibitory effects on the ATPase activity of F1-ATP synthase [35, 120, 121]. The peptide is composed of 26 residues and is the primary component of honey bee venom (A. mellifera). Melittin is a potent inhibitor of both E. coli and bovine ATP synthase (IC50~5µM) [35, 121]. The inhibitory activity of melittin is suggested to be similar to that of other known α-helical peptide inhibitors, IF1, Wild–type yeast cytochrome oxidase, and synthetic Syn-A2. All the above inhibitors are known to act in a reversible and noncompetitive fashion [120, 121, 130–133].
Defenses against pathogens include a wide variety of systems in both plants and animals. Among these defenses are various types of oligopeptides and peptides [134]. Antimicrobial peptides (AMPs) are a component of vertebrate innate immunity that has been present in most living organisms for over 2.6 billion years [135]. AMPs were first described in insects as an inducible system of protection against bacterial infection [136–138]. Most recently Laughlin and Ahmad [35] showed that several cationic α-helical amphibian AMPs have reversible inhibitory effects on the ATPase activity of E. coli ATP synthase. AMPs are generally cationic and amphipathic molecules that are less than 50 amino acids in length. They have been isolated from all investigated phyla, including microbes, plants, invertebrates and vertebrates. AMPs show potent activity against Gram-positive, Gram-negative bacteria, fungi, parasites, and enveloped viruses [135]. It is noteworthy that several amphibian AMPs are also known to have selective anticancer activity [139]. AMPs are also characterized for having multiple functions with unclear modes of actions. Additionally, AMPs have a neutralizing effect on bacterial endotoxins that are a primary cause of lethality in sepsis [140–142].
Presently there are 1504 entries in the Antimicrobial Database [143], (http://aps.unmc.edu/AP/main.php), of which 1154 (77%) are identified as having antibacterial activity, 438 with antifungal activity, 94 with anticancer activity and 86 with antiviral activity. Amphibian skin is the single largest source of AMPs identified in the database, being 36.4% (N=548) of all listed AMPs. Of all animal AMPs (N=1046) 52.4% are derived from amphibians. The first amphibian AMPs identified were the magainins from skin secretions of the frog Xenopus laevis [144]. Currently known amphibian AMPs in the database were derived from the European toad in the family Bufonidae, South American tree frog species of the family Hylidae and species of frogs in the family Ranidae in Europe, North America or South America [140, 145]. Frogs skin secretions contain a variety of AMPs with up to 100 unique amino acid sequences per species[146]. Based on structural similarity and species of origin there are four identified classes of amphibian AMPs: 1) maginins from Xenopus, 2) dermaseptins from species in the genus Phyllomedusa, 3) bombinins and bombinin H from European toads, and 4) temporins, brevinins, esculentins, ranalexins and ranauerins from species in the genus Rana [140].
The mean length of all peptides in the AMP database is 29.96 residues, and the mean net charge is +3.81. Identification of secondary structure among database AMPs resulted in 14.89% (N=224) as α-helical, 2.92% (N=44) as β-conformation, and 2.32% (N=35) as α+β. Whereas 24.46% (N=368) were found to have disulfide bonds and 5.85% (N=88) were rich in unusual amino acids [143].
Most of the anuran produced AMPs are cationic, between 10 and 50 residues in length, and frequently include a C-terminal amide group. Mode of action studies indicate that AMPs appear to interact with negatively charged phospholipids and then insert into the bacterial cell membrane, or they may also move across the cell membrane by passive transport and there disrupt a number of cellular processes. AMPs are associated with a number of other antimicrobial processes as well, including cell proliferation and angiogenesis [135]. Several mechanisms have been hypothesized regarding the activity of AMPs, including membrane permeabilization and cell death by either a “barrel-stave” model [147] or a “torodial pore” model proposed for magainins from Xenopus skin [148–150]. Dermaseptins appear to cause a non-pore-dependent cytolytic activity that causes membrane bilayer miscellization and disintegration [151]. This leads to the question of whether or not some of the antimicrobial effects of amphibian AMPs could be through their inhibitory actions on ATP synthase.
Following the discovery of AMPs in insects, biochemically active substances in frog skin were identified as bioactive peptides [144, 152, 153]. Frogs and toads secrete AMPs from granular glands of the skin, typically in response to infection or environmental stress [154] and at concentrations as high as mg/g of wet skin [153]. AMPs have become potential sources of compounds with useful pharmacological properties and medical utility in antimicrobial [155–157] and anticancer [158] applications. One probable drawback of the usefulness of these molecules is that they seem to be effective only at very high doses [159]. This may be related to the observed high concentrations in vivo, but may possibly be improved for pharmacological purposes by modifications of amino acid sequences or functional groups, based on molecular modeling studies, as has been observed recently with polyphenolic compounds [P. Dadi and Z. Ahmad unpublished results]. Synergistic effects with AMPs among different dermaseptins have also been observed, which may be related to the large number of different isoforms found within different species [160]. This suggests that evaluation of AMPs potential activity as ATP synthase inhibitory molecules may also be enhanced by combinatorial studies.
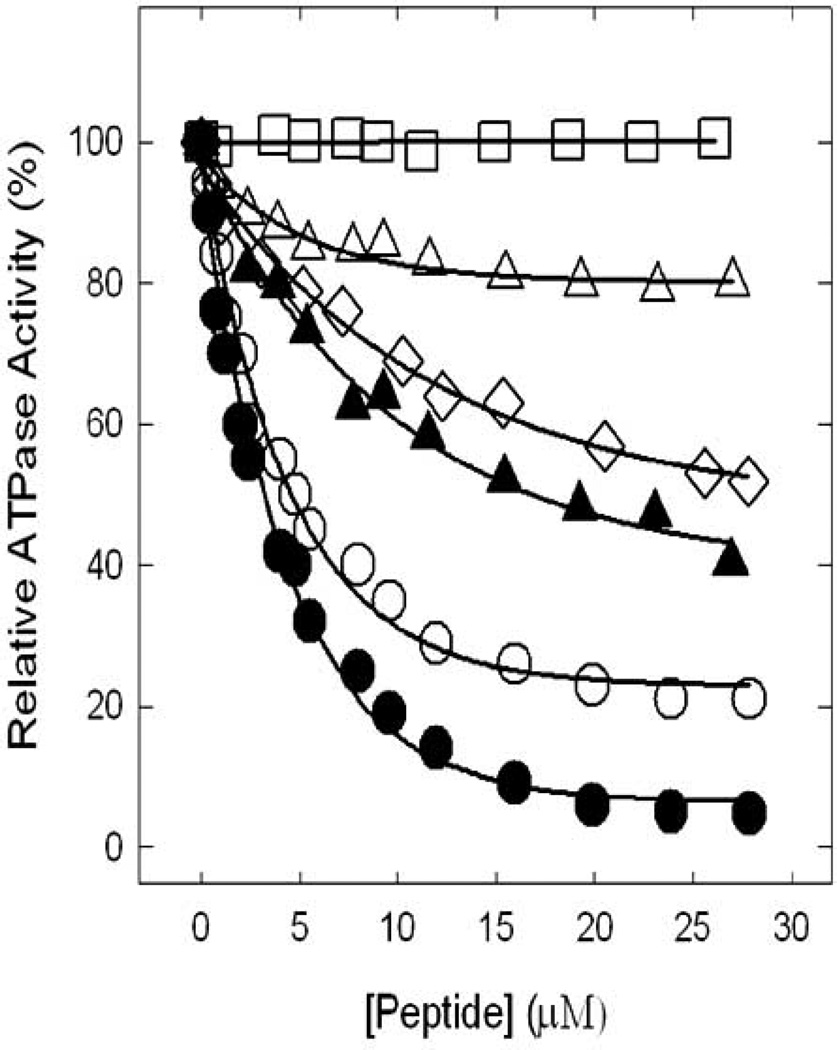
The amphibian AMPs with potential inhibitory effects on ATP synthase through binding at the βDELSEED-loop are expected to be relatively short cationic peptides of approximately 10–30 amino acid residues, with α-helical secondary structure, and having previously identified anti-bacterial or anti-cancer effects. Table 1 identifies a list of sixty candidate amphibian AMPs selected from the AMP database based on an α-helical secondary structure, a net positive charge, a total length of 10–30 residues, and identified antibacterial or anti-cancer activity. We very recently tested fifteen peptides from this candidate list for ATP synthase inhibitory activity [35] based on the previously identified ATP synthase inhibition by the melittin peptide [35, 120, 121]. As shown in Fig. (6) we found that MRP and MRP-amide strongly inhibited the ATPase activity of ATP synthase. However, maginin II, maganin II-amide, or caerin 1.9 only partially inhibited ATPase activity. Other peptides exerting partial inhibition of E. coli ATP synthase but not shown in Fig. (6) were ascaphin-8, aurein 2.2, aurein 2.3, citropin 1.1, and maculatin 1.1. Presence of an amide group at the C-terminal end of MRP and magainin II caused an additional ~20–40% inhibition. All the above amphibian AMPs had varying degrees of effect on E. coli cell growth. Ascaphin-8, aurein 2.2, aurein 2.3, caerin 1.9, citropin 1.1, dermaseptin, magaininII-NH2, MRP, or MRP-NH2 resulted in significant inhibition of cell growth, indicative of complete abrogation of ATP synthesis [35].
Table 1.
Amphibian Cationic, Alpha-Helical, Antimicrobial/Anticancer Peptides
| Amphibian peptides | Sequence | Length | Charge | Species of origin |
|---|---|---|---|---|
| Ascaphin-8 a, [164] | GFKDLLKGAAKALVKTVLF | 19 | 3 | Ascaphus truei |
| Aurein 2.2 a, [165] | GLFDIVKKVVGALGSL-NH2 | 16 | 1 | Littoria aurea, L. raniformis |
| Aurein 2.3 a, [165] | GLFDIVKKVVGAIGSL-NH2 | 16 | 1 | Littoria aurea |
| Bombinin H2b, [166] | IIGPVLGLVGSALGGLLKKI | 20 | 2 | Bombina variegata |
| Bombinin H4b, [166] | LIGPVLGLVGSALGGLLKKI | 21 | 2 | Bombina. variegata |
| Bombinin-like pep3b, [167] | GIGAAILSAGKSALKGLAKGLAEHF | 25 | 3 | Bombina orientalis |
| Bombinin-like pep4b, [167] | GIGAAILSAGKSIIKGLANGLAEHF | 25 | 2 | Bombina orientalis |
| Buforin II a, [168] | TRSSRAGLQFPVGRVHRLLRK | 31 | 7 | Bufo bufo gargarizans |
| Caerin 1.1 a, [169] | GLLSVLGSVAKHVLPHVVPVIAEHL | 25 | 3 | Littoria splendida |
| Caerin 1.2 b, [170] | GLLGVLGSVAKHVLPHVVPVIAEHL | 25 | 3 | Litoria caerula |
| Caerin 1.3 a, [170] | GLLSVLGSVAQHVLPHVVPVIAEHL | 25 | 2 | Littoria caerula |
| Caerin 1.4 b, [170] | GLLSSLSSVAKHVLPHVVPVIAEHL | 25 | 3 | Litoria caerula |
| Caerin 1.7 a, [171] | GLFKVLGSVAKHLLPHVAPVIAEK | 24 | 4 | Littoria chloris |
| Caerin 1.8 a, [171] | GLFKVLGSVAKHLLPHVVPVIAEK | 24 | 4 | Littoria chloris |
| Caerin 1.9 b, [171] | GLFGVLGSIAKHVLPHVVPVIAEK | 24 | 3 | Littoria chloris |
| Citropin 1.1a, [172] | GLFDVIKKVASVIGGL | 16 | 1 | Littoria citropa |
| D-1CDYa b, [173] | IIPLPLGYFAKKT | 13 | 2 | Rana chensinensis |
| Dermaseptin-B4b, [174] | ALWKDILKNVGKAAGKAVLNTVTDMVNQ | 28 | 2 | Phyllomedusa bicolor |
| Dermaseptin-B5b, [174] | GLWNKIKEAASKAAGKAALGFVNEMV | 26 | 2 | Phyllomedusa bicolor |
| Dermaseptin-B9b, [174] | ALWKTIIKGAGKMIGSLAKNLLGSQAQPES | 30 | 3 | Phyllomedusa bicolor |
| Dermaseptin S1a, [175] | ALWFTMLKKLGTMALHAGKAALGAAANTISQGTQ | 34 | 4 | Phyllomedusa sauvagei |
| Dermaseptin-S3b, [175] | ALWKNMLKGIGKLAGKAALGAVKKLVGAES | 30 | 5 | Phyllomedusa sauvagei |
| Dermaseptin-S4b, [175] | ALWMTLLKKVLKAAAKALNAVLVGANA | 27 | 4 | Phyllomedusa sauvagei |
| Dermaseptin-S5b, [175] | GLWSKIKTAGKSVAKAAAKAAVKAVTNAV | 29 | 6 | Phyllomedusa sauvagei |
| Dermaseptin-S9b, [176] | GLRSKIWLWVLLMIWQESNKFKKM | 24 | 4 | Phyllomedusa sauvagei |
| Distinctinb, [177] | NLVSGLIEARKYLEQLHRKLKNCKV | 25 | 5 | Phyllomedusa disticta |
| Fallaxidin 4.1b, [178] | GLLSFLPKVIGVIGHLIHPPS | 21 | 3 | Litoria fallax |
| Frenatin 3b, [179] | GLMSVLGHAVGNVLGGLFKS | 20 | 2 | Litoria infrafrenata |
| Hylaseptin P1b, [180] | GILDAIKAIAKAAG | 14 | 1 | Hyla punctata |
| Hylin a1b, [181] | IFGAILPLALGALKNLIK-NH2 | 18 | 2 | Hypsiboas albopunctatus |
| Japonicin-CDYab, [173] | FFPLALLCKVFKKC | 14 | 3 | Rana chensinensis |
| Kassinatuerin-1b, [182] | GFMKYIGPLIPHAVKAISDLI- NH2 | 21 | 2 | Kassina senegalensi |
| Maculatin 1.1 a, [183] | GLFVGVLAKVAAHVVPAIAEHF- NH2 | 22 | 2 | Litorria genimaculate |
| Magainin II a, [144] | GIGKFLHSAKKFGKAFVGEIMNS | 23 | 4 | Xenopus laevis |
| Magainin II amide a, [144] | GIGKFLHSAKKFGKAFVGEIMNS-NH2 | 23 | 4 | Xenopus laevis |
| MRP b, [184] | AIGSILGALAKGLPTLISWIKNR | 23 | 3 | Rana tagoi |
| MRP amide b, [184] | AIGSILGALAKGLPTLISWIKNR-NH2 | 23 | 3 | Rana tagoi |
| Nigrocin 2b, [185] | GLLSKVLGVGKKVLCGVSGLC | 21 | 3 | Rana nigromaculata |
| Ocellatin-F1b, [186] | GVVDILKGAAKDIAGHLASKVMNKL | 25 | 3 | Leptodactylus fallax |
| Odorranain-B1b, [187] | AALKGCWTKSIPPKPCFGKR | 20 | 5 | Odorrana grahami |
| Odorranain-F1b, [187] | GFMDTAKNVAKNVAVTLIDNLKCKITKAC | 29 | 3 | Odorrana grahami |
| Odorranain-G1b, [187] | FMPILSCSRFKRC | 13 | 2 | Odorrana grahami |
| Odorranain-H1b, [187] | GIFGKILGVGKKVLCGLSGWC | 21 | 3 | Odorrana grahami |
| Odorranain-T1b, [187] | TSRCYIGYRRKVVCS | 15 | 4 | Odorrana grahami |
| PGLab, [188] | GMASKAGAIAGKIAKVALKAL | 21 | 4 | Xenopus laevis |
| Phylloseptin-H1b, [189] | FLSLIPHAINAVSAIAKHN | 19 | 3 | Phyllomedusa hypochondrialis |
| Phylloseptin-H2b, [189] | FLSLIPHAINAVSTLVHHF- NH2 | 19 | 3 | Phyllomedusa hypochondrialis |
| Phylloseptin-H3b, [189] | FLSLIPHAINAVSALANHG- NH2 | 19 | 2 | Phyllomedusa hypochondrialis |
| Phylloxinb, [190] | GWMSKIASGIGTFLSGMQQ | 19 | 1 | Phyllomedusa bicolor |
| Pseudin 1b, [191] | GLNTLKKVFQGLHEAIKLINNHVQ | 24 | 4 | Pseudis paradoxa |
| Pseudin 3b, [191] | GINTLKKVIQGLHEVIKLVSNHE | 23 | 3 | Pseudis paradoxa |
| Pseudin 4b, [191] | GINTLKKVIQGLHEVIKLVSNHA | 23 | 4 | Pseudis paradoxa |
| Ranalexinb, [192] | FLGGLIKIVPAMICAVTKKC | 20 | 3 | Rana catesbeiana |
| Temporin Ab, [193] | FLPLIGRVLSGIL | 13 | 1 | Rana temporaria |
| Temporin Bb, [193] | LLPIVGNLLKSLL-NH2 | 13 | 1 | Rana temporaria |
| Temporin Lb, [193] | FVQWFSKFLGRIL | 13 | 2 | Rana temporaria |
| Temporin-SHab, [194] | FLSGIVGMLGKLF | 13 | 1 | Pelophylax saharica |
| Temporin-SHcb, [194] | FLSHIAGFLSNLF | 13 | 1 | Pelophylax saharica |
| Uperin 3.6b, [195] | GVIDAAKKVVNVLKNLF-NH2 | 17 | 2 | Uperoleia mjobergii |
| XT-7 a, [196] | GLLGPLLKIAAKVGSNLL | 18 | 2 | Xenopus tropicalis |
Peptides known for both antibacterial and anticancer activity.
Peptides known for antibacterial activity only.
Fig. 6. Inhibitory effects of amphibian AMPs on ATP synthase.
Inhibition profiles induced by MRP (ο), MRP-NH2 (●), maginin II (Δ), maginin II-NH2 (▲), caerin 1.8 (□), and caerin 1.9 (◊) resulting in partial or complete inhibition of ATP synthase are shown. For experimental details see Laughlin and Ahmad [35].
It seems probable that there will be variable results in terms of ATP synthase and cell growth inhibition among different dietary polyphenols and amphibian AMPs, depending on the target molecules and organisms. We also expect that various modifications of naturally occurring dietary polyphenols and amphibian AMPs may be used to modulate their effectiveness on a molar scale with regard to both ATP synthase inhibition and cytotoxicity. By virtue of the great variability in the structures and potential functions exhibited by both dietary polyphenols and amphibian AMPs, these relatively simple molecules may constitute a formidable natural resource of new drug compounds that may be targeted at microorganisms and neoplasms through their inhibitory actions on ATP synthase.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health Grant GM085771 to ZA, Dr. William R Duncan, and the Office of Research and Sponsored Programs Administration, East Tennessee State University.
REFERENCES
- 1.Senior AE, Nadanaciva S, Weber J. The molecular mechanism of ATP synthesis by F1F0-ATP synthase. Biochim. Biophys. Acta. 2002;1553:188–211. doi: 10.1016/s0005-2728(02)00185-8. [DOI] [PubMed] [Google Scholar]
- 2.Boyer PD. The ATP synthase-a splendid molecular machine. Annu. Rev. Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 3.Van Walraven HS, Strotmann H, Schwarz O, Rumberg B. The H+/ATP coupling ratio of the ATP synthase from thiol-modulated chloroplasts and two cyanobacterial strains is four. FEBS Lett. 1996;379:309–313. doi: 10.1016/0014-5793(95)01536-1. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida M, Muneyuki E, Hisabori T. ATP synthase--a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2001;2:669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- 5.Schemidt RA, Qu J, Williams JR, Brusilow WS. Effects of carbon source on expression of F0 genes and on the stoichiometry of the c subunit in the F1F0 ATPase of Escherichia coli. J. Bacteriol. 1998;180:3205–3208. doi: 10.1128/jb.180.12.3205-3208.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Senior AE. ATP synthesis by oxidative phosphorylation. Physiol. Rev. 1988;68:177–231. doi: 10.1152/physrev.1988.68.1.177. [DOI] [PubMed] [Google Scholar]
- 7.Karrasch S, Walker JE. Novel features in the structure of bovine ATP synthase. J. Mol. Biol. 1999;290:379–384. doi: 10.1006/jmbi.1999.2897. [DOI] [PubMed] [Google Scholar]
- 8.Devenish RJ, Prescott M, Roucou X, Nagley P. Insights into ATP synthase assembly and function through the molecular genetic manipulation of subunits of the yeast mitochondrial enzyme complex. Biochim. Biophys. Acta. 2000;1458:428–442. doi: 10.1016/s0005-2728(00)00092-x. [DOI] [PubMed] [Google Scholar]
- 9.Diez M, Zimmermann B, Borsch M, Konig M, Schweinberger E, Steigmiller S, Reuter R, Felekyan S, Kudryavtsev V, Seidel CAM, Graber P. Proton-powered subunit rotation in single membrane-bound F0F1-ATP synthase. Nat. Struct. Mol. Biol. 2004;11:135–141. doi: 10.1038/nsmb718. [DOI] [PubMed] [Google Scholar]
- 10.Itoh H, Takahashi A, Adachi K, Noji H, Yasuda R, Yoshida M, Kinosita K. Mechanically driven ATP synthesis by F1-ATPase. Nature. 2004;427:465–468. doi: 10.1038/nature02212. [DOI] [PubMed] [Google Scholar]
- 11.Weber J. ATP synthase: subunit-subunit interactions in the stator stalk. Biochim. Biophys. Acta. 2006;1757:1162–1170. doi: 10.1016/j.bbabio.2006.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber J, Senior AE. ATP synthase: what we know about ATP hydrolysis and what we do not know about ATP synthesis. Biochim. Biophys. Acta. 2000;1458:300–309. doi: 10.1016/s0005-2728(00)00082-7. [DOI] [PubMed] [Google Scholar]
- 13.Senior AE, Nadanaciva S, Weber J. Rate acceleration of ATP hydrolysis by F(1)F(o)-ATP synthase. J. Exp. Biol. 2000;203:35–40. doi: 10.1242/jeb.203.1.35. [DOI] [PubMed] [Google Scholar]
- 14.Frasch WD. The participation of metals in the mechanism of the F(1)-ATPase. Biochim. Biophys. Acta. 2000;1458:310–325. doi: 10.1016/s0005-2728(00)00083-9. [DOI] [PubMed] [Google Scholar]
- 15.Nakamoto RK, Ketchum CJ, al-Shawi MK. Rotational coupling in the F0F1 ATP synthase. Annu. Rev. Biophys. Biomol. Struct. 1999;28:205–234. doi: 10.1146/annurev.biophys.28.1.205. [DOI] [PubMed] [Google Scholar]
- 16.Pedersen PL. Transport ATPases into the year 2008: a brief overview related to types, structures, functions and roles in health and disease. J. Bioenerg. Biomembr. 2007;39:349–355. doi: 10.1007/s10863-007-9123-9. [DOI] [PubMed] [Google Scholar]
- 17.Ahmad Z, Senior AE. Identification of phosphate binding residues of Escherichia coli ATP synthase. J. Bioenerg. Biomembr. 2005;37:437–440. doi: 10.1007/s10863-005-9486-8. [DOI] [PubMed] [Google Scholar]
- 18.Senior AE. ATP synthase: motoring to the finish line. Cell. 2007;130:220–221. doi: 10.1016/j.cell.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Noji H, Yoshida M. The rotary machine in the cell, ATP synthase. J. Biol. Chem. 2001;276:1665–1668. doi: 10.1074/jbc.R000021200. [DOI] [PubMed] [Google Scholar]
- 20.Weber J, Senior AE. ATP synthesis driven by proton transport in F1F0-ATP synthase. FEBS Lett. 2003;545:61–70. doi: 10.1016/s0014-5793(03)00394-6. [DOI] [PubMed] [Google Scholar]
- 21.Khan S. Rotary chemiosmotic machines. Biochim. Biophys. Acta. 1997;1322:86–105. doi: 10.1016/s0005-2728(97)00075-3. [DOI] [PubMed] [Google Scholar]
- 22.Ren H, Allison WS. On what makes the gamma subunit spin during ATP hydrolysis by F(1) Biochim. Biophys. Acta. 2000;1458:221–233. doi: 10.1016/s0005-2728(00)00075-x. [DOI] [PubMed] [Google Scholar]
- 23.Leslie AG, Walker JE. Structural model of F1-ATPase and the implications for rotary catalysis. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2000;355:465–471. doi: 10.1098/rstb.2000.0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menz RI, Walker JE, Leslie AG. Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell. 2001;106:331–341. doi: 10.1016/s0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- 25.Boyer PD. A perspective of the binding change mechanism for ATP synthesis. FASEB J. 1989;3:2164–2178. doi: 10.1096/fasebj.3.10.2526771. [DOI] [PubMed] [Google Scholar]
- 26.Boyer PD. A research journey with ATP synthase. J. Biol. Chem. 2002;277:39045–39061. doi: 10.1074/jbc.X200001200. [DOI] [PubMed] [Google Scholar]
- 27.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 28.Kinosita K, Jr, Yasuda R, Noji H, Ishiwata S, Yoshida M. F1-ATPase: a rotary motor made of a single molecule. Cell. 1998;93:21–24. doi: 10.1016/s0092-8674(00)81142-3. [DOI] [PubMed] [Google Scholar]
- 29.Nishizaka T, Oiwa K, Noji H, Kimura S, Muneyuki E, Yoshida M, Kinosita K., Jr Chemomechanical coupling in F1-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nat. Struct. Mol. Biol. 2004;11:142–148. doi: 10.1038/nsmb721. [DOI] [PubMed] [Google Scholar]
- 30.Senior AE, Weber J. Happy motoring with ATP synthase. Nat. Struct. Mol. Biol. 2004;11:110–112. doi: 10.1038/nsmb0204-110. [DOI] [PubMed] [Google Scholar]
- 31.Yasuda R, Noji H, Kinosita K, Jr, Yoshida M. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120 degree steps. Cell. 1998;93:1117–1124. doi: 10.1016/s0092-8674(00)81456-7. [DOI] [PubMed] [Google Scholar]
- 32.Whitesides GM. The ‘right’ size in nanobiotechnology. Nat. Biotechnol. 2003;21:1161–1165. doi: 10.1038/nbt872. [DOI] [PubMed] [Google Scholar]
- 33.Li W, Brudecki LE, Senior AE, Ahmad Z. Role of {alpha}-subunit VISIT-DG sequence residues Ser-347 and Gly-351 in the catalytic sites of Escherichia coli ATP synthase. J. Biol. Chem. 2009;284:10747–10754. doi: 10.1074/jbc.M809209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dadi PK, Ahmad M, Ahmad Z. Inhibition of ATPase activity of Escherichia coli ATP synthase by polyphenols. Int. J. Biol. Macromol. 2009;45:72–79. doi: 10.1016/j.ijbiomac.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Laughlin TF, Ahmad Z. Inhibition of Escherichia coli ATP synthase by amphibian antimicrobial peptides. Int. J. Biol. Macromol. 2010 doi: 10.1016/j.ijbiomac.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber J, Wilke-Mounts S, Lee RS, Grell E, Senior AE. Specific placement of tryptophan in the catalytic sites of Escherichia coli F1-ATPase provides a direct probe of nucleotide binding: maximal ATP hydrolysis occurs with three sites occupied. J. Biol. Chem. 1993;268:20126–20133. [PubMed] [Google Scholar]
- 37.Lobau S, Weber J, Senior AE. Catalytic site nucleotide binding and hydrolysis in F1F0-ATP synthase. Biochemistry. 1998;37:10846–10853. doi: 10.1021/bi9807153. [DOI] [PubMed] [Google Scholar]
- 38.Weber J, Hammond ST, Wilke-Mounts S, Senior AE. Mg2+ coordination in catalytic sites of F1-ATPase. Biochemistry. 1998;37:608–614. doi: 10.1021/bi972370e. [DOI] [PubMed] [Google Scholar]
- 39.Dou C, Fortes PA, Allison WS. The alpha 3(beta Y341W)3 gamma subcomplex of the F1-ATPase from the thermophilic Bacillus PS3 fails to dissociate ADP when MgATP is hydrolyzed at a single catalytic site and attains maximal velocity when three catalytic sites are saturated with MgATP. Biochemistry. 1998;37:16757–16764. doi: 10.1021/bi981717q. [DOI] [PubMed] [Google Scholar]
- 40.al-Shawi MK, Senior AE. Effects of dimethyl sulfoxide on catalysis in Escherichia coli F1-ATPase. Biochemistry. 1992;31:886–891. doi: 10.1021/bi00118a034. [DOI] [PubMed] [Google Scholar]
- 41.Al-Shawi MK, Ketchum CJ, Nakamoto RK. The Escherichia coli FOF1 gammaM23K uncoupling mutant has a higher K0.5 for Pi. Transition state analysis of this mutant and others reveals that synthesis and hydrolysis utilize the same kinetic pathway. Biochemistry. 1997;36:12961–12969. doi: 10.1021/bi971478r. [DOI] [PubMed] [Google Scholar]
- 42.Rastogi VK, Girvin ME. Structural changes linked to proton translocation by subunit c of the ATP synthase. Nature. 1999;402:263–268. doi: 10.1038/46224. [DOI] [PubMed] [Google Scholar]
- 43.Gibbons C, Montgomery MG, Leslie AG, Walker JE. The structure of the central stalk in bovine F(1)-ATPase at 2.4 A resolution. Nat. Struct. Biol. 2000;7:1055–1061. doi: 10.1038/80981. [DOI] [PubMed] [Google Scholar]
- 44.Braig K, Menz RI, Montgomery MG, Leslie AG, Walker JE. Structure of bovine mitochondrial F(1)-ATPase inhibited by Mg(2+) ADP and aluminium fluoride. Structure. 2000;8:567–573. doi: 10.1016/s0969-2126(00)00145-3. [DOI] [PubMed] [Google Scholar]
- 45.Weber J, Senior AE. Location and properties of pyrophosphate-binding sites in Escherichia coli F1-ATPase. J. Biol. Chem. 1995;270:12653–12658. doi: 10.1074/jbc.270.21.12653. [DOI] [PubMed] [Google Scholar]
- 46.Perez JA, Greenfield AJ, Sutton R, Ferguson SJ. Characterisation of phosphate binding to mitochondrial and bacterial membrane-bound ATP synthase by studies of inhibition with 4-chloro-7-nitrobenzofurazan. FEBS Lett. 1986;198:113–118. doi: 10.1016/0014-5793(86)81195-4. [DOI] [PubMed] [Google Scholar]
- 47.Orriss GL, Leslie AG, Braig K, Walker JE. Bovine F1-ATPase covalently inhibited with 4-chloro-7-nitrobenzofurazan: the structure provides further support for a rotary catalytic mechanism. Structure. 1998;6:831–837. doi: 10.1016/s0969-2126(98)00085-9. [DOI] [PubMed] [Google Scholar]
- 48.Ahmad Z, Senior AE. Mutagenesis of residue betaArg-246 in the phosphate-binding subdomain of catalytic sites of Escherichia coli F1-ATPase. J. Biol. Chem. 2004;279:31505–31513. doi: 10.1074/jbc.M404621200. [DOI] [PubMed] [Google Scholar]
- 49.Ahmad Z, Senior AE. Role of betaAsn-243 in the phosphate-binding subdomain of catalytic sites of Escherichia coli F(1)-ATPase. J. Biol. Chem. 2004;279:46057–46064. doi: 10.1074/jbc.M407608200. [DOI] [PubMed] [Google Scholar]
- 50.Ahmad Z, Senior AE. Involvement of ATP synthase residues alphaArg-376, betaArg-182, and betaLys-155 in Pi binding. FEBS Lett. 2005;579:523–528. doi: 10.1016/j.febslet.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 51.Ahmad Z, Senior AE. Modulation of charge in the phosphate binding site of Escherichia coli ATP synthase. J. Biol. Chem. 2005;280:27981–27989. doi: 10.1074/jbc.M503955200. [DOI] [PubMed] [Google Scholar]
- 52.Ahmad Z, Senior AE. Inhibition of the ATPase activity of Escherichia coli ATP synthase by magnesium fluoride. FEBS Lett. 2006;580:517–520. doi: 10.1016/j.febslet.2005.12.057. [DOI] [PubMed] [Google Scholar]
- 53.Hong S, Pedersen PL. ATP synthase and the actions of inhibitors utilized to study its roles in human health, disease, and other scientific areas. Microbiol. Mol. Biol. Rev. 2008;72:590–641. doi: 10.1128/MMBR.00016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Vries DD, van Engelen BG, Gabreels FJ, Ruitenbeek W, van Oost BA. A second missense mutation in the mitochondrial ATPase 6 gene in Leigh’s syndrome. Ann. Neurol. 1993;34:410–412. doi: 10.1002/ana.410340319. [DOI] [PubMed] [Google Scholar]
- 55.Arakaki N, Nagao T, Niki R, Toyofuku A, Tanaka H, Kuramoto Y, Emoto Y, Shibata H, Magota K, Higuti T. Possible role of cell surface H+ -ATP synthase in the extracellular ATP synthesis and proliferation of human umbilical vein endothelial cells. Mol. Cancer Res. 2003;1:931–939. [PubMed] [Google Scholar]
- 56.Berger K, Winzell MS, Mei J, Erlanson-Albertsson C. Enterostatin and its target mechanisms during regulation of fat intake. Physiol. Behav. 2004;83:623–630. doi: 10.1016/j.physbeh.2004.08.040. [DOI] [PubMed] [Google Scholar]
- 57.Champagne E, Martinez LO, Collet X, Barbaras R. Ecto-F1Fo ATP synthase/F1 ATPase: metabolic and immunological functions. Curr. Opin. Lipidol. 2006;17:279–284. doi: 10.1097/01.mol.0000226120.27931.76. [DOI] [PubMed] [Google Scholar]
- 58.Kenan DJ, Wahl ML. Ectopic localization of mitochondrial ATP synthase: a target for anti-angiogenesis intervention? J. Bioenerg. Biomembr. 2005;37:461–465. doi: 10.1007/s10863-005-9492-x. [DOI] [PubMed] [Google Scholar]
- 59.Moser TL, Kenan DJ, Ashley TA, Roy JA, Goodman MD, Misra UK, Cheek DJ, Pizzo SV. Endothelial cell surface F1-F0 ATP synthase is active in ATP synthesis and is inhibited by angiostatin. Proc. Natl. Acad. Sci. USA. 2001;98:6656–6661. doi: 10.1073/pnas.131067798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moser TL, Stack MS, Asplin I, Enghild JJ, Hojrup P, Everitt L, Hubchak S, Schnaper HW, Pizzo SV. Angiostatin binds ATP synthase on the surface of human endothelial cells. Proc. Natl. Acad. Sci. USA. 1999;96:2811–2816. doi: 10.1073/pnas.96.6.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Percival RS, Devine DA, Duggal MS, Chartron S, Marsh PD. The effect of cocoa polyphenols on the growth, metabolism, and biofilm formation by Streptococcus mutans and Streptococcus sanguinis . Eur. J. Oral Sci. 2006;114:343–348. doi: 10.1111/j.1600-0722.2006.00386.x. [DOI] [PubMed] [Google Scholar]
- 62.Duarte S, Gregoire S, Singh AP, Vorsa N, Schaich K, Bowen WH, Koo H. Inhibitory effects of cranberry polyphenols on formation and acidogenicity of Streptococcus mutans biofilms. FEMS Microbiol. Lett. 2006;257:50–56. doi: 10.1111/j.1574-6968.2006.00147.x. [DOI] [PubMed] [Google Scholar]
- 63.Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis . Science. 2005;307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 64.Cole ST, Alzari PM. Microbiology: Enhanced: TB-A New Target, a New Drug. Science. 2005;307:214–215. doi: 10.1126/science.1108379. [DOI] [PubMed] [Google Scholar]
- 65.Weber J, Senior AE. Effects of the inhibitors azide, dicyclohexylcarbodiimide, and aurovertin on nucleotide binding to the three F1-ATPase catalytic sites measured using specific tryptophan probes. J. Biol. Chem. 1998;273:33210–33215. doi: 10.1074/jbc.273.50.33210. [DOI] [PubMed] [Google Scholar]
- 66.Zheng J, Ramirez VD. Inhibition of mitochondrial proton F0F1-ATPase/ATP synthase by polyphenolic phytochemicals. Br. J. Pharmacol. 2000;130:1115–1123. doi: 10.1038/sj.bjp.0703397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frasch AC, Cazzulo JJ, Stoppani AO. Solubilization and some properties of the Mg2+-activated adenosine triphosphatase from Trypanosoma cruzi . Comp. Biochem. Physiol. B. 1978;61:207–212. doi: 10.1016/0305-0491(78)90162-1. [DOI] [PubMed] [Google Scholar]
- 68.Bowler MW, Montgomery MG, Leslie AG, Walker JE. How azide inhibits ATP hydrolysis by the F-ATPases. Proc. Natl. Acad. Sci. USA. 2006;103:8646–8649. doi: 10.1073/pnas.0602915103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zharova TV, Vinogradov AD. Energy-dependent transformation of F0.F1-ATPase in Paracoccus denitrificans plasma membranes. J. Biol. Chem. 2004;279:12319–12324. doi: 10.1074/jbc.M311397200. [DOI] [PubMed] [Google Scholar]
- 70.Yoshida M, Allison WS, Esch FS, Futai M. The specificity of carboxyl group modification during the inactivation of the Escherichia coli F1-ATPase with dicyclohexyl[14C]carbodiimide. J. Biol. Chem. 1982;257:10033–10037. [PubMed] [Google Scholar]
- 71.Hermolin J, Fillingame RH. H+-ATPase activity of Escherichia coli F1F0 is blocked after reaction of dicyclohexylcarbodiimide with a single proteolipid (subunit c) of the F0 complex. J. Biol. Chem. 1989;264:3896–3903. [PubMed] [Google Scholar]
- 72.Tommasino M, Capaldi RA. Effect of dicyclohexylcarbodiimide on unisite and multisite catalytic activities of the adenosinetriphosphatase of Escherichia coli. Biochemistry. 1985;24:3972–3976. doi: 10.1021/bi00336a026. [DOI] [PubMed] [Google Scholar]
- 73.Abrahams JP, Buchanan SK, Van Raaij MJ, Fearnley IM, Leslie AG, Walker JE. The structure of bovine F1-ATPase complexed with the peptide antibiotic efrapeptin. Proc. Natl. Acad. Sci. USA. 1996;93:9420–9424. doi: 10.1073/pnas.93.18.9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lardy H, Reed P, Lin CH. Antibiotic inhibitors of mitochondrial ATP synthesis. Fed. Proc. 1975;34:1707–1710. [PubMed] [Google Scholar]
- 75.Yarlett N, Lloyd D. Effects of inhibitors on mitochondrial adenosine triphosphatase of Crithidia fasciculata: an unusual pattern of specificities. Mol. Biochem. Parasitol. 1981;3:13–17. doi: 10.1016/0166-6851(81)90073-6. [DOI] [PubMed] [Google Scholar]
- 76.van Raaij MJ, Abrahams JP, Leslie AG, Walker JE. The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B. Proc. Natl. Acad. Sci. USA. 1996;93:6913–6917. doi: 10.1073/pnas.93.14.6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gledhill JR, Montgomery MG, Leslie AG, Walker JE. How the regulatory protein, IF(1), inhibits F(1)-ATPase from bovine mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15671–15676. doi: 10.1073/pnas.0707326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wise JG, Duncan TM, Latchney LR, Cox DN, Senior AE. Properties of F1-ATPase from the uncD412 mutant of Escherichia coli. Biochem. J. 1983;215:343–350. doi: 10.1042/bj2150343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Satre M, Bof M, Vignais PV. Interaction of Escherichia coli adenosine triphosphatase with aurovertin and citreoviridin: inhibition and fluorescence studies. J. Bacteriol. 1980;142:768–776. doi: 10.1128/jb.142.3.768-776.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Linnett PE, Beechey RB. Inhibitors of the ATP synthethase system. Methods Enzymol. 1979;55:472–518. doi: 10.1016/0076-6879(79)55061-7. [DOI] [PubMed] [Google Scholar]
- 81.Robinson JA. Polyketide synthase complexes: their structure and function in antibiotic biosynthesis. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1991;332:107–114. doi: 10.1098/rstb.1991.0038. [DOI] [PubMed] [Google Scholar]
- 82.Wolvetang EJ, Johnson KL, Krauer K, Ralph SJ, Linnane AW. Mitochondrial respiratory chain inhibitors induce apoptosis. FEBS Lett. 1994;339:40–44. doi: 10.1016/0014-5793(94)80380-3. [DOI] [PubMed] [Google Scholar]
- 83.Mills KI, Woodgate LJ, Gilkes AF, Walsh V, Sweeney MC, Brown G, Burnett AK. Inhibition of mitochondrial function in HL60 cells is associated with an increased apoptosis and expression of CD14. Biochem. Biophys. Res. Commun. 1999;263:294–300. doi: 10.1006/bbrc.1999.1356. [DOI] [PubMed] [Google Scholar]
- 84.Gledhill JR, Montgomery MG, Leslie AG, Walker JE. Mechanism of inhibition of bovine F1-ATPase by resveratrol and related polyphenols. Proc. Natl. Acad. Sci. USA. 2007;104:13632–13637. doi: 10.1073/pnas.0706290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chinnam N, Dadi PK, Sabri SA, Ahmad M, Kabir MA, Ahmad Z. Dietary bioflavonoids inhibit Escherichia coli ATP synthase in a differential manner. Int. J. Biol. Macromol. 2010;46:478–486. doi: 10.1016/j.ijbiomac.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Barta I, Smerak P, Polivkova Z, Sestakova H, Langova M, Turek B, Bartova J. Current trends and perspectives in nutrition and cancer prevention. Neoplasma. 2006;53:19–25. [PubMed] [Google Scholar]
- 87.Nishino H, Murakoshi M, Mou XY, Wada S, Masuda M, Ohsaka Y, Satomi Y, Jinno K. Cancer Prevention by Phytochemicals. Oncology. 2005;69:38–40. doi: 10.1159/000086631. [DOI] [PubMed] [Google Scholar]
- 88.Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CWW, Fong HHS, Farnsworth NR, Kinghorn AD, Mehta RG, Moon RC, Pezzuto JM. Cancer Chemopreventive Activity of Resveratrol, a Natural Product Derived from Grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 89.Stahl W, Sies H. Carotenoids and flavonoids contribute to nutritional protection against skin damage from sunlight. Mol. Biotechnol. 2007;37:26–30. doi: 10.1007/s12033-007-0051-z. [DOI] [PubMed] [Google Scholar]
- 90.Pervaiz S. Resveratrol: from grapevines to mammalian biology. FASEB J. 2003;17:1975–1985. doi: 10.1096/fj.03-0168rev. [DOI] [PubMed] [Google Scholar]
- 91.Athar M, Back JH, Kopelovich L, Bickers DR, Kim AL. Multiple molecular targets of resveratrol: Anti-carcinogenic mechanisms. Arch. Biochem. Biophys. 2009;486:95–102. doi: 10.1016/j.abb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Athar M, Back JH, Tang X, Kim KH, Kopelovich L, Bickers DR, Kim AL. Resveratrol: a review of preclinical studies for human cancer prevention. Toxicol. Appl. Pharmacol. 2007;224:274–283. doi: 10.1016/j.taap.2006.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Aziz MH, Nihal M, Fu VX, Jarrard DF, Ahmad N. Resveratrol-caused apoptosis of human prostate carcinoma LNCaP cells is mediated via modulation of phosphatidylinositol 3'-kinase/Akt pathway and Bcl-2 family proteins. Mol. Cancer Ther. 2006;5:1335–1341. doi: 10.1158/1535-7163.MCT-05-0526. [DOI] [PubMed] [Google Scholar]
- 94.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 95.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McMahon GA, Petitclerc E, Stefansson S, Smith E, Wong MK, Westrick RJ, Ginsburg D, Brooks PC, Lawrence DA. Plasminogen activator inhibitor-1 regulates tumor growth and angiogenesis. J. Biol. Chem. 2001;276:33964–33968. doi: 10.1074/jbc.M105980200. [DOI] [PubMed] [Google Scholar]
- 97.Leik CE, Su EJ, Nambi P, Crandall DL, Lawrence DA. Effect of pharmacologic plasminogen activator inhibitor-1 inhibition on cell motility and tumor angiogenesis. J. Thromb. Haemost. 2006;4:2710–2715. doi: 10.1111/j.1538-7836.2006.02244.x. [DOI] [PubMed] [Google Scholar]
- 98.Lahouel M, Amedah S, Zellagui A, Touil A, Rhouati S, Benyache F, Leghouchi E, Bousseboua H. The interaction of new plant flavonoids with rat liver mitochondria: relation between the anti- and pro-oxydant effect and flavonoids concentration. Therapie. 2006;61:347–355. doi: 10.2515/therapie:2006025. [DOI] [PubMed] [Google Scholar]
- 99.Glazebrook J, Ausubel FM. Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. USA. 1994;91:8955–8959. doi: 10.1073/pnas.91.19.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pirola L, Frojdo S. Resveratrol: one molecule, many targets. IUBMB Life. 2008;60:323–332. doi: 10.1002/iub.47. [DOI] [PubMed] [Google Scholar]
- 101.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 102.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 103.Valenzano DR, Terzibasi E, Genade T, Cattaneo A, Domenici L, Cellerino A. Resveratrol prolongs lifespan and retards the onset of age-related markers in a short-lived vertebrate. Curr. Biol. 2006;16:296–300. doi: 10.1016/j.cub.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 104.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Clement MV, Hirpara JL, Chawdhury SH, Pervaiz S. Chemopreventive agent resveratrol, a natural product derived from grapes, triggers CD95 signaling-dependent apoptosis in human tumor cells. Blood. 1998;92:996–1002. [PubMed] [Google Scholar]
- 106.Spiller GA. Caffeine. Boca Raton, Fla: CRC Press; 1998. [Google Scholar]
- 107.Hammerstone JF, Lazarus SA, Schmitz HH. Procyanidin content and variation in some commonly consumed foods. J. Nutr. 2000;130:2086S–2092S. doi: 10.1093/jn/130.8.2086S. [DOI] [PubMed] [Google Scholar]
- 108.Ruidavets J, Teissedre P, Ferrieres J, Carando S, Bougard G, Cabanis J. Catechin in the Mediterranean diet: vegetable, fruit or wine? Atherosclerosis. 2000;153:107–117. doi: 10.1016/s0021-9150(00)00377-4. [DOI] [PubMed] [Google Scholar]
- 109.Harborne JB, Mabry TJ, Mabry H. The Flavonoids. London: Chapman & Hall; 1975. [Google Scholar]
- 110.Chyu KY, Babbidge SM, Zhao X, Dandillaya R, Rietveld AG, Yano J, Dimayuga P, Cercek B, Shah PK. Differential effects of green tea-derived catechin on developing versus established atherosclerosis in apolipoprotein E-null mice. Circulation. 2004;109:2448–2453. doi: 10.1161/01.CIR.0000128034.70732.C2. [DOI] [PubMed] [Google Scholar]
- 111.Mittal A, Pate MS, Wylie RC, Tollefsbol TO, Katiyar SK. EGCG down-regulates telomerase in human breast carcinoma MCF-7 cells, leading to suppression of cell viability and induction of apoptosis. Int. J. Oncol. 2004;24:703–710. [PubMed] [Google Scholar]
- 112.Li C, Allen A, Kwagh J, Doliba NM, Qin W, Najafi H, Collins HW, Matschinsky FM, Stanley CA, Smith TJ. Green tea polyphenols modulate insulin secretion by inhibiting glutamate dehydrogenase. J. Biol. Chem. 2006;281:10214–10221. doi: 10.1074/jbc.M512792200. [DOI] [PubMed] [Google Scholar]
- 113.Puig T, Turrado C, Benhamu B, Aguilar H, Relat J, Ortega-Gutierrez S, Casals G, Marrero PF, Urruticoechea A, Haro D, Lopez-Rodriguez ML, Colomer R. Novel inhibitors of fatty acid synthase with anticancer activity. Clin. Cancer Res. 2009;15:7608–7615. doi: 10.1158/1078-0432.CCR-09-0856. [DOI] [PubMed] [Google Scholar]
- 114.Murase T, Haramizu S, Ota N, Hase T. Tea catechin ingestion combined with habitual exercise suppresses the aging-associated decline in physical performance in senescence-accelerated mice. Am. J. Physiol. Regu. Integr. Comp. Physiol. 2008;295:R281–R289. doi: 10.1152/ajpregu.00880.2007. [DOI] [PubMed] [Google Scholar]
- 115.Kumar N, Kant R, Maurya PK. Concentration-dependent effect of (−) epicatechin in hypertensive patients. Phytother. Res. 2010 doi: 10.1002/ptr.3119. [DOI] [PubMed] [Google Scholar]
- 116.Wu D, Kong Y, Han C, Chen J, Hu L, Jiang H, Shen X. D-Alanine:D-alanine ligase as a new target for the flavonoids quercetin and apigenin. Int. J. Antimicrob. Agents. 2008;32:421–426. doi: 10.1016/j.ijantimicag.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 117.Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, Williams RL. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol. Cell. 2000;6:909–919. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- 118.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 119.Zheng J, Ramirez VD. Piceatannol, a stilbene phytochemical, inhibits mitochondrial F0F1-ATPase activity by targeting the F1 complex. Biochem. Biophys. Res. Commun. 1999;261:499–503. doi: 10.1006/bbrc.1999.1063. [DOI] [PubMed] [Google Scholar]
- 120.Gledhill JR, Walker JE. Inhibition sites in F1-ATPase from bovine heart mitochondria. Biochem. J. 2005;386:591–598. doi: 10.1042/BJ20041513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bullough DA, Ceccarelli EA, Roise D, Allison WS. Inhibition of the bovine-heart mitochondrial F1-ATPase by cationic dyes and amphipathic peptides. Biochim. Biophys. Acta. 1989;975:377–383. doi: 10.1016/s0005-2728(89)80346-9. [DOI] [PubMed] [Google Scholar]
- 122.Kato-Yamada Y, Bald D, Koike M, Motohashi K, Hisabori T, Yoshida M. Epsilon subunit, an endogenous inhibitor of bacterial F(1)-ATPase, also inhibits F(0)F(1)-ATPase. J. Biol. Chem. 1999;274:33991–33994. doi: 10.1074/jbc.274.48.33991. [DOI] [PubMed] [Google Scholar]
- 123.Hara KY, Kato-Yamada Y, Kikuchi Y, Hisabori T, Yoshida M. The role of the betaDELSEED motif of F1-ATPase: propagation of the inhibitory effect of the epsilon subunit. J. Biol. Chem. 2001;276:23969–23973. doi: 10.1074/jbc.M009303200. [DOI] [PubMed] [Google Scholar]
- 124.Suzuki T, Murakami T, Iino R, Suzuki J, Ono S, Shirakihara Y, Yoshida M. F0F1-ATPase/synthase is geared to the synthesis mode by conformational rearrangement of epsilon subunit in response to proton motive force and ADP/ATP balance. J. Biol. Chem. 2003;278:46840–46846. doi: 10.1074/jbc.M307165200. [DOI] [PubMed] [Google Scholar]
- 125.Tsunoda SP, Rodgers AJ, Aggeler R, Wilce MC, Yoshida M, Capaldi RA. Large conformational changes of the epsilon subunit in the bacterial F1F0 ATP synthase provide a ratchet action to regulate this rotary motor enzyme. Proc. Natl. Acad. Sci. USA. 2001;98:6560–6564. doi: 10.1073/pnas.111128098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lebowitz MS, Pedersen PL. Regulation of the mitochondrial ATP synthase/ATPase complex: cDNA cloning, sequence, overexpression, and secondary structural characterization of a functional protein inhibitor. Arch. Biochem. Biophys. 1993;301:64–70. doi: 10.1006/abbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- 127.Lebowitz MS, Pedersen PL. Protein inhibitor of mitochondrial ATP synthase: relationship of inhibitor structure to pH-dependent regulation. Arch. Biochem. Biophys. 1996;330:342–354. doi: 10.1006/abbi.1996.0261. [DOI] [PubMed] [Google Scholar]
- 128.Van Heeke G, Deforce L, Schnizer RA, Shaw R, Couton JM, Shaw G, Song PS, Schuster SM. Recombinant bovine heart mitochondrial F1-ATPase inhibitor protein: overproduction in Escherichia coli, purification, and structural studies. Biochemistry. 1993;32:10140–10149. doi: 10.1021/bi00089a033. [DOI] [PubMed] [Google Scholar]
- 129.Cabezon E, Montgomery MG, Leslie AG, Walker JE. The structure of bovine F1-ATPase in complex with its regulatory protein IF1. Nat. Struct. Biol. 2003;10:744–750. doi: 10.1038/nsb966. [DOI] [PubMed] [Google Scholar]
- 130.Iwadate M, Asakura T, Williamson MP. The structure of the melittin tetramer at different temperatures--an NOE-based calculation with chemical shift refinement. Eur. J. Biochem. 1998;257:479–487. doi: 10.1046/j.1432-1327.1998.2570479.x. [DOI] [PubMed] [Google Scholar]
- 131.Roise D, Horvath SJ, Tomich JM, Richards JH, Schatz G. A chemically synthesized pre-sequence of an imported mitochondrial protein can form an amphiphilic helix and perturb natural and artificial phospholipid bilayers. EMBO J. 1986;5:1327–1334. doi: 10.1002/j.1460-2075.1986.tb04363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Terwilliger TC, Eisenberg D. The structure of melittin. I. Structure determination and partial refinement. J. Biol. Chem. 1982;257:6010–6015. doi: 10.2210/pdb1mlt/pdb. [DOI] [PubMed] [Google Scholar]
- 133.Roise D, Theiler F, Horvath SJ, Tomich JM, Richards JH, Allison DS, Schatz G. Amphiphilicity is essential for mitochondrial presequence function. EMBO J. 1988;7:649–653. doi: 10.1002/j.1460-2075.1988.tb02859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Powers JP, Hancock RE. The relationship between peptide structure and antibacterial activity. Peptides. 2003;24:1681–1691. doi: 10.1016/j.peptides.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 135.Jakubke H-D, Sewald N. Peptides from A to Z: a concise encyclopedia. Weinheim: Wiley-VCH; 2008. [Google Scholar]
- 136.Boman HG, Nilsson I, Rasmuson B. Inducible antibacterial defence system in Drosophila. Nature. 1972;237:232–235. doi: 10.1038/237232a0. [DOI] [PubMed] [Google Scholar]
- 137.Hultmark D, Steiner H, Rasmuson T, Boman HG. Insect immunity. Purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of Hyalophora cecropia. Eur. J. Biochem. 1980;106:7–16. doi: 10.1111/j.1432-1033.1980.tb05991.x. [DOI] [PubMed] [Google Scholar]
- 138.Steiner H, Hultmark D, Engstrom A, Bennich H, Boman HG. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature. 1981;292:246–248. doi: 10.1038/292246a0. [DOI] [PubMed] [Google Scholar]
- 139.Hoskin DW, Ramamoorthy A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta. 2008;1778:357–375. doi: 10.1016/j.bbamem.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mangoni ML, Shai Y. Temporins and their synergism against Gram-negative bacteria and in lipopolysaccharide detoxification. Biochim. Biophys. Acta. 2009;1788:1610–1619. doi: 10.1016/j.bbamem.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 141.Martin GS, Mannino DM, Eaton S, Moss M. The epidemiology of sepsis in the United States from 1979 through 2000. N. Engl. J. Med. 2003;348:1546–1554. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- 142.Rosenfeld Y, Papo N, Shai Y. Endotoxin (lipopolysaccharide) neutralization by innate immunity host-defense peptides. Peptide properties and plausible modes of action. J. Biol. Chem. 2006;281:1636–1643. doi: 10.1074/jbc.M504327200. [DOI] [PubMed] [Google Scholar]
- 143.Wang Z, Wang G. APD: the Antimicrobial Peptide Database. Nucleic Acids Res. 2004;32:D590–D592. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. USA. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Amiche M, Ladram A, Nicolas P. A consistent nomenclature of antimicrobial peptides isolated from frogs of the subfamily Phyllomedusinae. Peptides. 2008;29:2074–2082. doi: 10.1016/j.peptides.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 146.Mangoni ML. Temporins, anti-infective peptides with expanding properties. Cell. Mol. Life Sci. 2006;63:1060–1069. doi: 10.1007/s00018-005-5536-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Oren Z, Shai Y. Mode of action of linear amphipathic alpha-helical antimicrobial peptides. Biopolymers. 1998;47:451–463. doi: 10.1002/(SICI)1097-0282(1998)47:6<451::AID-BIP4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 148.Ludtke SJ, He K, Heller WT, Harroun TA, Yang L, Huang HW. Membrane pores induced by magainin. Biochemistry. 1996;35:13723–13728. doi: 10.1021/bi9620621. [DOI] [PubMed] [Google Scholar]
- 149.Matsuzaki K, Murase O, Fujii N, Miyajima K. An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation. Biochemistry. 1996;35:11361–11368. doi: 10.1021/bi960016v. [DOI] [PubMed] [Google Scholar]
- 150.Matsuzaki K, Nakamura A, Murase O, Sugishita K, Fujii N, Miyajima K. Modulation of magainin 2-lipid bilayer interactions by peptide charge. Biochemistry. 1997;36:2104–2111. doi: 10.1021/bi961870p. [DOI] [PubMed] [Google Scholar]
- 151.Shai Y. Mode of action of membrane active antimicrobial peptides. Biopolymers. 2002;66:236–248. doi: 10.1002/bip.10260. [DOI] [PubMed] [Google Scholar]
- 152.Erspamer V. Half a century of comparative research on biogenic amines and active peptides in amphibian skin and molluscan tissues. Comp. Biochem. Physiol. C. 1984;79:1–7. doi: 10.1016/0742-8413(84)90153-1. [DOI] [PubMed] [Google Scholar]
- 153.Erspamer V, Falconieri Erspamer G, Cei JM. Active peptides in the skins of two hundred and thirty American amphibian species. Comp. Biochem. Physiol. C. 1986;85:125–137. doi: 10.1016/0742-8413(86)90063-0. [DOI] [PubMed] [Google Scholar]
- 154.Davidson C, Benard MF, Shaffer HB, Parker JM, O’Leary C, Conlon JM, Rollins-Smith LA. Effects of chytrid and carbaryl exposure on survival, growth and skin peptide defenses in foothill yellow-legged frogs. Environ. Sci. Technol. 2007;41:1771–1776. doi: 10.1021/es0611947. [DOI] [PubMed] [Google Scholar]
- 155.Rinaldi AC. Antimicrobial peptides from amphibian skin: an expanding scenario. Curr. Opin. Chem. Biol. 2002;6:799–804. doi: 10.1016/s1367-5931(02)00401-5. [DOI] [PubMed] [Google Scholar]
- 156.Conlon JM, Kolodziejek J, Nowotny N. Antimicrobial peptides from ranid frogs: taxonomic and phylogenetic markers and a potential source of new therapeutic agents. Biochim. Biophys. Acta. 2004;1696:1–14. doi: 10.1016/j.bbapap.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 157.Conlon JM, Al-Ghaferi N, Abraham B, Leprince J. Strategies for transformation of naturally-occurring amphibian antimicrobial peptides into therapeutically valuable anti-infective agents. Methods. 2007;42:349–357. doi: 10.1016/j.ymeth.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 158.Papo N, Shai Y. Host defense peptides as new weapons in cancer treatment. Cell. Mol. Life Sci. 2005;62:784–790. doi: 10.1007/s00018-005-4560-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 160.Mor A, Hani K, Nicolas P. The vertebrate peptide antibiotics dermaseptins have overlapping structural features but target specific microorganisms. J. Biol. Chem. 1994;269:31635–31641. [PubMed] [Google Scholar]
- 161.Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 162.Gesell J, Zasloff M, Opella SJ. Two-dimensional 1H NMR experiments show that the 23-residue magainin antibiotic peptide is an alpha-helix in dodecylphosphocholine micelles, sodium dodecylsulfate micelles, and trifluoroethanol/water solution. J. Biomol. NMR. 1997;9:127–135. doi: 10.1023/a:1018698002314. [DOI] [PubMed] [Google Scholar]
- 163.Shalev DE, Rotem S, Fish A, Mor A. Consequences of N-acylation on structure and membrane binding properties of dermaseptin derivative K4-S4-(1–13) J. Biol. Chem. 2006;281:9432–9438. doi: 10.1074/jbc.M513051200. [DOI] [PubMed] [Google Scholar]
- 164.Conlon JM, Sonnevend A, Davidson C, Smith DD, Nielsen PF. The ascaphins: a family of antimicrobial peptides from the skin secretions of the most primitive extant frog, Ascaphus truei . Biochem. Biophys. Res. Commun. 2004;320:170–175. doi: 10.1016/j.bbrc.2004.05.141. [DOI] [PubMed] [Google Scholar]
- 165.Rozek T, Wegener KL, Bowie JH, Olver IN, Carver JA, Wallace JC, Tyler MJ. The antibiotic and anticancer active aurein peptides from the Australian Bell Frogs Litoria aurea and Litoria raniformis the solution structure of aurein 1.2. Eur. J. Biochem. 2000;267:5330–5341. doi: 10.1046/j.1432-1327.2000.01536.x. [DOI] [PubMed] [Google Scholar]
- 166.Mignogna G, Simmaco M, Kreil G, Barra D. Antibacterial and haemolytic peptides containing D-alloisoleucine from the skin of Bombina variegata. EMBO J. 1993;12:4829–4832. doi: 10.1002/j.1460-2075.1993.tb06172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gibson BW, Tang DZ, Mandrell R, Kelly M, Spindel ER. Bombinin-like peptides with antimicrobial activity from skin secretions of the Asian toad, Bombina orientalis . J. Biol. Chem. 1991;266:23103–23111. [PubMed] [Google Scholar]
- 168.Yi GS, Park CB, Kim SC, Cheong C. Solution structure of an antimicrobial peptide buforin II. FEBS Lett. 1996;398:87–90. doi: 10.1016/s0014-5793(96)01193-3. [DOI] [PubMed] [Google Scholar]
- 169.Wong H, Bowie JH, Carver JA. The solution structure and activity of caerin 1.1, an antimicrobial peptide from the Australian green tree frog, Litoria splendida. Eur. J. Biochem. 1997;247:545–557. doi: 10.1111/j.1432-1033.1997.00545.x. [DOI] [PubMed] [Google Scholar]
- 170.Apponyi MA, Pukala TL, Brinkworth CS, Maselli VM, Bowie JH, Tyler MJ, Booker GW, Wallace JC, Carver JA, Separovic F, Doyle J, Llewellyn LE. Host-defence peptides of Australian anurans: structure, mechanism of action and evolutionary significance. Peptides. 2004;25:1035–1054. doi: 10.1016/j.peptides.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 171.Steinborner ST, Currie GJ, Bowie JH, Wallace JC, Tyler MJ. New antibiotic caerin 1 peptides from the skin secretion of the Australian tree frog Litoria chloris. Comparison of the activities of the caerin 1 peptides from the genus Litoria. J. Pept. Res. 1998;51:121–126. doi: 10.1111/j.1399-3011.1998.tb00629.x. [DOI] [PubMed] [Google Scholar]
- 172.Wegener KL, Wabnitz PA, Carver JA, Bowie JH, Chia BC, Wallace JC, Tyler MJ. Host defence peptides from the skin glands of the Australian blue mountains tree-frog Litoria citropa. Solution structure of the antibacterial peptide citropin 1.1. Eur. J. Biochem. 1999;265:627–637. doi: 10.1046/j.1432-1327.1999.00750.x. [DOI] [PubMed] [Google Scholar]
- 173.Jin LL, Song SS, Li Q, Chen YH, Wang QY, Hou ST. Identification and characterisation of a novel antimicrobial polypeptide from the skin secretion of a Chinese frog (Rana chensinensis) Int. J. Antimicrob. Agents. 2009;33:538–542. doi: 10.1016/j.ijantimicag.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 174.Charpentier S, Amiche M, Mester J, Vouille V, Le Caer JP, Nicolas P, Delfour A. Structure, synthesis, and molecular cloning of dermaseptins B, a family of skin peptide antibiotics. J. Biol. Chem. 1998;273:14690–14697. doi: 10.1074/jbc.273.24.14690. [DOI] [PubMed] [Google Scholar]
- 175.Mor A, Nicolas P. Isolation and structure of novel defensive peptides from frog skin. Eur. J. Biochem. 1994;219:145–154. doi: 10.1111/j.1432-1033.1994.tb19924.x. [DOI] [PubMed] [Google Scholar]
- 176.Lequin O, Ladram A, Chabbert L, Bruston F, Convert O, Vanhoye D, Chassaing G, Nicolas P, Amiche M. Dermaseptin S9, an alpha-helical antimicrobial peptide with a hydrophobic core and cationic termini. Biochemistry. 2006;45:468–480. doi: 10.1021/bi051711i. [DOI] [PubMed] [Google Scholar]
- 177.Batista CV, Scaloni A, Rigden DJ, Silva LR, Rodrigues Romero A, Dukor R, Sebben A, Talamo F, Bloch C. A novel heterodimeric antimicrobial peptide from the tree-frog Phyllomedusa distincta. FEBS Lett. 2001;494:85–89. doi: 10.1016/s0014-5793(01)02324-9. [DOI] [PubMed] [Google Scholar]
- 178.Jackway RJ, Bowie JH, Bilusich D, Musgrave IF, Surinya-Johnson KH, Tyler MJ, Eichinger PC. The fallaxidin peptides from the skin secretion of the Eastern Dwarf Tree Frog Litoria fallax.Sequence determination by positive and negative ion electrospray mass spectrometry: antimicrobial activity and cDNA cloning of the fallaxidins. Rapid Commun. Mass Spectrom. 2008;22:3207–3216. doi: 10.1002/rcm.3723. [DOI] [PubMed] [Google Scholar]
- 179.Raftery MJ, Waugh RJ, Bowie JH, Wallace JC, Tyler MJ. The structures of the frenatin peptides from the skin secretion of the giant tree frog Litoria infrafrenata. J. Pept. Sci. 1996;2:117–124. doi: 10.1002/psc.60. [DOI] [PubMed] [Google Scholar]
- 180.Prates MV, Sforca ML, Regis WC, Leite JR, Silva LP, Pertinhez TA, Araujo AL, Azevedo RB, Spisni A, Bloch C., Jr The NMR-derived solution structure of a new cationic antimicrobial peptide from the skin secretion of the anuran Hyla punctata. J. Biol. Chem. 2004;279:13018–13026. doi: 10.1074/jbc.M310838200. [DOI] [PubMed] [Google Scholar]
- 181.Castro MS, Ferreira TC, Cilli EM, Crusca E, Jr, Mendes-Giannini MJ, Sebben A, Ricart CA, Sousa MV, Fontes W. Hylin a1, the first cytolytic peptide isolated from the arboreal South American frog Hypsiboas albopunctatus (“spotted treefrog”) Peptides. 2009;30:291–296. doi: 10.1016/j.peptides.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 182.Mattute B, Knoop FC, Conlon JM. Kassinatuerin-1: a peptide with broad-spectrum antimicrobial activity isolated from the skin of the hyperoliid frog, Kassina senegalensis. Biochem. Biophys. Res. Commun. 2000;268:433–436. doi: 10.1006/bbrc.2000.2136. [DOI] [PubMed] [Google Scholar]
- 183.Rozek T, Waugh RJ, Steinborner ST, Bowie JH, Tyler MJ, Wallace JC. The maculatin peptides from the skin glands of the tree frog Litoria genimaculata: a comparison of the structures and antibacterial activities of maculatin 1.1 and caerin 1.1. J. Pept. Sci. 1998;4:111–115. doi: 10.1002/(sici)1099-1387(199804)4:2<111::aid-psc134>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 184.Conlon JM, Sonnevend A, Patel M, Camasamudram V, Nowotny N, Zilahi E, Iwamuro S, Nielsen PF, Pal T. A melittin-related peptide from the skin of the Japanese frog, Rana tagoi, with antimicrobial and cytolytic properties. Biochem. Biophys. Res. Commun. 2003;306:496–500. doi: 10.1016/s0006-291x(03)00999-9. [DOI] [PubMed] [Google Scholar]
- 185.Park S, Park SH, Ahn HC, Kim S, Kim SS, Lee BJ. Structural study of novel antimicrobial peptides, nigrocins, isolated from Rana nigromaculata. FEBS Lett. 2001;507:95–100. doi: 10.1016/s0014-5793(01)02956-8. [DOI] [PubMed] [Google Scholar]
- 186.Rollins-Smith LA, King JD, Nielsen PF, Sonnevend A, Conlon JM. An antimicrobial peptide from the skin secretions of the mountain chicken frog Leptodactylus fallax (Anura: Leptodactylidae) Regul. Pept. 2005;124:173–178. doi: 10.1016/j.regpep.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 187.Li J, Xu X, Xu C, Zhou W, Zhang K, Yu H, Zhang Y, Zheng Y, Rees HH, Lai R, Yang D, Wu J. Anti-infection peptidomics of amphibian skin. Mol. Cell. Proteom. 2007;6:882–894. doi: 10.1074/mcp.M600334-MCP200. [DOI] [PubMed] [Google Scholar]
- 188.Andreu D, Aschauer H, Kreil G, Merrifield RB. Solid-phase synthesis of PYLa and isolation of its natural counterpart, PGLa [PYLa-(4–24)] from skin secretion of Xenopus laevis . Eur. J. Biochem. 1985;149:531–535. doi: 10.1111/j.1432-1033.1985.tb08957.x. [DOI] [PubMed] [Google Scholar]
- 189.Leite JR, Silva LP, Rodrigues MI, Prates MV, Brand GD, Lacava BM, Azevedo RB, Bocca AL, Albuquerque S, Bloch C., Jr Phylloseptins: a novel class of anti-bacterial and anti-protozoan peptides from the Phyllomedusa genus . Peptides. 2005;26:565–573. doi: 10.1016/j.peptides.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 190.Pierre TN, Seon AA, Amiche M, Nicolas P. Phylloxin, a novel peptide antibiotic of the dermaseptin family of antimicrobial/opioid peptide precursors. Eur. J. Biochem. 2000;267:370–378. doi: 10.1046/j.1432-1327.2000.01012.x. [DOI] [PubMed] [Google Scholar]
- 191.Olson L, 3rd, Soto AM, Knoop FC, Conlon JM. Pseudin-2: an antimicrobial peptide with low hemolytic activity from the skin of the paradoxical frog. Biochem. Biophys. Res. Commun. 2001;288:1001–1005. doi: 10.1006/bbrc.2001.5884. [DOI] [PubMed] [Google Scholar]
- 192.Clark DP, Durell S, Maloy WL, Zasloff M. Ranalexin. A novel antimicrobial peptide from bullfrog (Rana catesbeiana) skin, structurally related to the bacterial antibiotic, polymyxin. J. Biol. Chem. 1994;269:10849–10855. [PubMed] [Google Scholar]
- 193.Simmaco M, Mignogna G, Canofeni S, Miele R, Mangoni ML, Barra D. Temporins, antimicrobial peptides from the European red frog Rana temporaria . Eur. J. Biochem. 1996;242:788–792. doi: 10.1111/j.1432-1033.1996.0788r.x. [DOI] [PubMed] [Google Scholar]
- 194.Abbassi F, Oury B, Blasco T, Sereno D, Bolbach G, Nicolas P, Hani K, Amiche M, Ladram A. Isolation, characterization and molecular cloning of new temporins from the skin of the North African ranid Pelophylax saharica. Peptides. 2008;29:1526–1533. doi: 10.1016/j.peptides.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 195.Chia BC, Carver JA, Mulhern TD, Bowie JH. The solution structure of uperin 3.6, an antibiotic peptide from the granular dorsal glands of the Australian toadlet, Uperoleia mjobergii . J. Pept. Res. 1999;54:137–145. doi: 10.1034/j.1399-3011.1999.00095.x. [DOI] [PubMed] [Google Scholar]
- 196.Ali MF, Soto A, Knoop FC, Conlon JM. Antimicrobial peptides isolated from skin secretions of the diploid frog, Xenopus tropicalis (Pipidae) Biochim. Biophys. Acta. 2001;1550:81–89. doi: 10.1016/s0167-4838(01)00272-2. [DOI] [PubMed] [Google Scholar]