Abstract
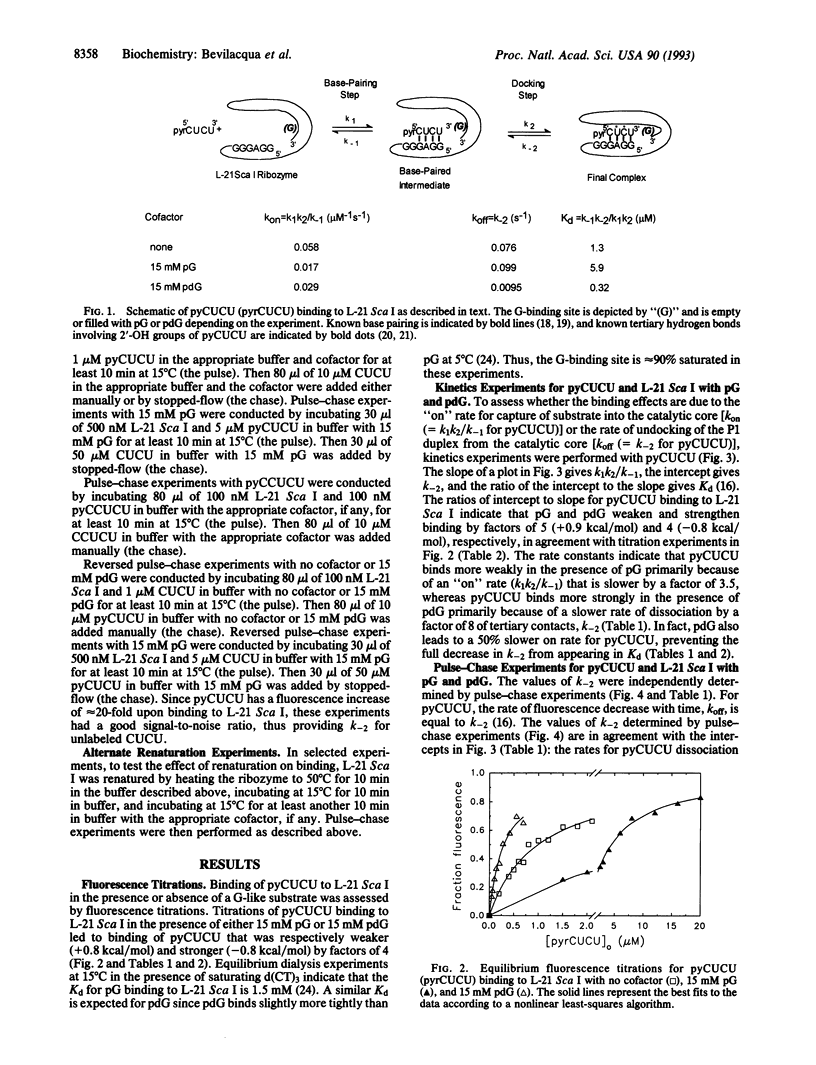
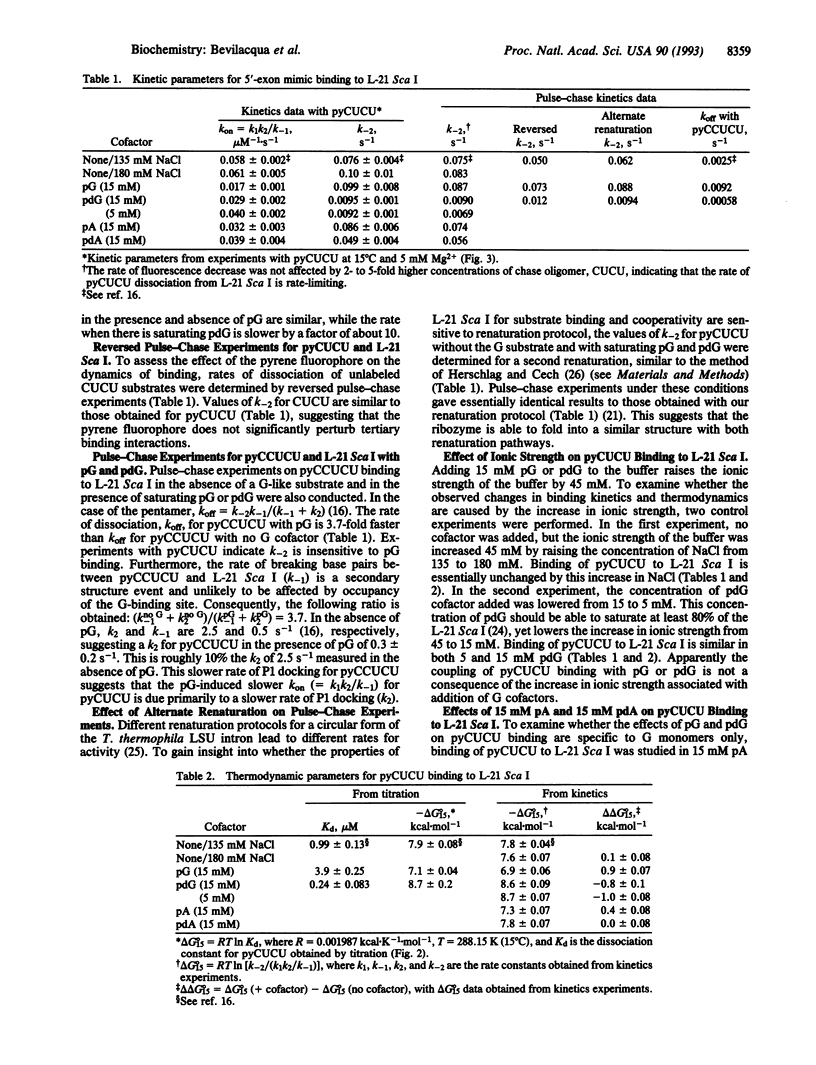
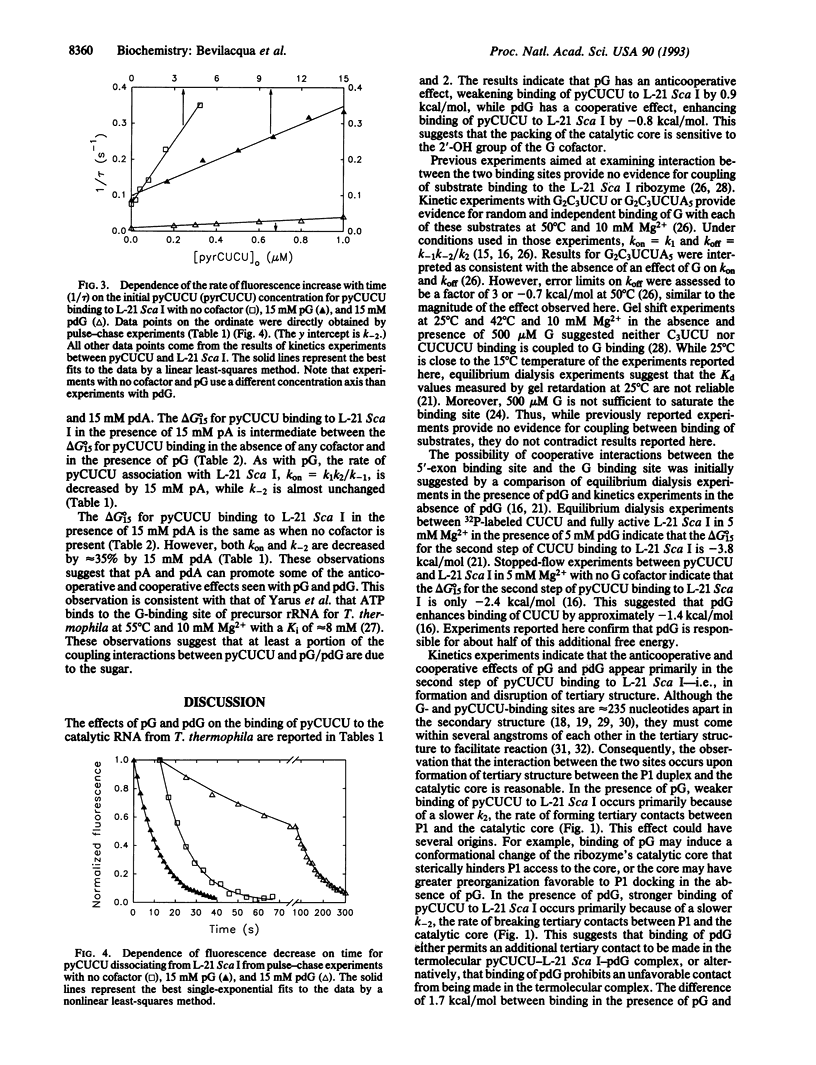
The effects of guanosine 5'-monophosphate and 2'-deoxyguanosine 5'-monophosphate on the thermodynamics and kinetics of pyrene-labeled 5' exon mimic (pyCUCU) binding to the catalytic RNA (ribozyme) from Tetrahymena thermophila have been determined by fluorescence titration and kinetics experiments at 15 degrees C. pyCUCU binding to L-21 Sca I-truncated ribozyme is weaker by a factor of 5 in the presence of saturating guanosine 5'-monophosphate, whereas it is 4-fold stronger in the presence of saturating 2'-deoxyguanosine 5'-monophosphate. Results from kinetics experiments suggest that anticooperative effects in the presence of guanosine 5'-monophosphate arise primarily from slower formation of tertiary contacts between the catalytic core of the ribozyme and the P1 duplex formed by pyCUCU and GGAGGG of the ribozyme. Conversely, cooperative effects in the presence of 2'-deoxyguanosine 5'-monophosphate arise primarily from slower disruption of tertiary contacts between the catalytic core of the ribozyme and the P1 duplex. Additional experiments suggest that these cooperative and anticooperative effects are not a function of the pyrene label, are not caused by a salt effect, and are not specific to one renaturation procedure for the ribozyme.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackers G. K. Linked functions in allosteric proteins: an exact theory for the effect of organic phosphates on oxygen affinity of hemoglobin. Biochemistry. 1979 Jul 24;18(15):3372–3380. doi: 10.1021/bi00582a025. [DOI] [PubMed] [Google Scholar]
- Bass B. L., Cech T. R. Ribozyme inhibitors: deoxyguanosine and dideoxyguanosine are competitive inhibitors of self-splicing of the Tetrahymena ribosomal ribonucleic acid precursor. Biochemistry. 1986 Aug 12;25(16):4473–4477. doi: 10.1021/bi00364a001. [DOI] [PubMed] [Google Scholar]
- Been M. D., Cech T. R. One binding site determines sequence specificity of Tetrahymena pre-rRNA self-splicing, trans-splicing, and RNA enzyme activity. Cell. 1986 Oct 24;47(2):207–216. doi: 10.1016/0092-8674(86)90443-5. [DOI] [PubMed] [Google Scholar]
- Benesch R., Benesch R. E. The effect of organic phosphates from the human erythrocyte on the allosteric properties of hemoglobin. Biochem Biophys Res Commun. 1967 Jan 23;26(2):162–167. doi: 10.1016/0006-291x(67)90228-8. [DOI] [PubMed] [Google Scholar]
- Bevilacqua P. C., Kierzek R., Johnson K. A., Turner D. H. Dynamics of ribozyme binding of substrate revealed by fluorescence-detected stopped-flow methods. Science. 1992 Nov 20;258(5086):1355–1358. doi: 10.1126/science.1455230. [DOI] [PubMed] [Google Scholar]
- Bevilacqua P. C., Turner D. H. Comparison of binding of mixed ribose-deoxyribose analogues of CUCU to a ribozyme and to GGAGAA by equilibrium dialysis: evidence for ribozyme specific interactions with 2' OH groups. Biochemistry. 1991 Nov 5;30(44):10632–10640. doi: 10.1021/bi00108a005. [DOI] [PubMed] [Google Scholar]
- Burke J. M., Esherick J. S., Burfeind W. R., King J. L. A 3' splice site-binding sequence in the catalytic core of a group I intron. Nature. 1990 Mar 1;344(6261):80–82. doi: 10.1038/344080a0. [DOI] [PubMed] [Google Scholar]
- Davies R. W., Waring R. B., Ray J. A., Brown T. A., Scazzocchio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982 Dec 23;300(5894):719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- Gilman A. G. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- HELMREICH E., CORI C. F. THE ROLE OF ADENYLIC ACID IN THE ACTIVATION OF PHOSPHORYLASE. Proc Natl Acad Sci U S A. 1964 Jan;51:131–138. doi: 10.1073/pnas.51.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D., Cech T. R. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990 Nov 6;29(44):10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- Herschlag D. Evidence for processivity and two-step binding of the RNA substrate from studies of J1/2 mutants of the Tetrahymena ribozyme. Biochemistry. 1992 Feb 11;31(5):1386–1399. doi: 10.1021/bi00120a015. [DOI] [PubMed] [Google Scholar]
- Johnson K. A. Rapid kinetic analysis of mechanochemical adenosinetriphosphatases. Methods Enzymol. 1986;134:677–705. doi: 10.1016/0076-6879(86)34129-6. [DOI] [PubMed] [Google Scholar]
- Kay P. S., Inoue T. Catalysis of splicing-related reactions between dinucleotides by a ribozyme. 1987 May 28-Jun 3Nature. 327(6120):343–346. doi: 10.1038/327343a0. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Cech T. R. Three-dimensional model of the active site of the self-splicing rRNA precursor of Tetrahymena. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8788–8792. doi: 10.1073/pnas.84.24.8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D. E., Jr, Némethy G., Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966 Jan;5(1):365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- MONOD J., WYMAN J., CHANGEUX J. P. ON THE NATURE OF ALLOSTERIC TRANSITIONS: A PLAUSIBLE MODEL. J Mol Biol. 1965 May;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Michel F., Hanna M., Green R., Bartel D. P., Szostak J. W. The guanosine binding site of the Tetrahymena ribozyme. Nature. 1989 Nov 23;342(6248):391–395. doi: 10.1038/342391a0. [DOI] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Moran S., Kierzek R., Turner D. H. Binding of guanosine and 3' splice site analogues to a group I ribozyme: interactions with functional groups of guanosine and with additional nucleotides. Biochemistry. 1993 May 18;32(19):5247–5256. doi: 10.1021/bi00070a037. [DOI] [PubMed] [Google Scholar]
- Nowak T., Mildvan A. S. Nuclear magnetic resonance studies of the function of potassium in the mechanism of pyruvate kinase. Biochemistry. 1972 Jul 18;11(15):2819–2828. doi: 10.1021/bi00765a014. [DOI] [PubMed] [Google Scholar]
- Pyle A. M., Cech T. R. Ribozyme recognition of RNA by tertiary interactions with specific ribose 2'-OH groups. Nature. 1991 Apr 18;350(6319):628–631. doi: 10.1038/350628a0. [DOI] [PubMed] [Google Scholar]
- Pyle A. M., McSwiggen J. A., Cech T. R. Direct measurement of oligonucleotide substrate binding to wild-type and mutant ribozymes from Tetrahymena. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8187–8191. doi: 10.1073/pnas.87.21.8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelander L., Reichard P. Reduction of ribonucleotides. Annu Rev Biochem. 1979;48:133–158. doi: 10.1146/annurev.bi.48.070179.001025. [DOI] [PubMed] [Google Scholar]
- Van Schaftingen E., Hue L., Hers H. G. Fructose 2,6-bisphosphate, the probably structure of the glucose- and glucagon-sensitive stimulator of phosphofructokinase. Biochem J. 1980 Dec 15;192(3):897–901. doi: 10.1042/bj1920897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WYMAN J., Jr LINKED FUNCTIONS AND RECIPROCAL EFFECTS IN HEMOGLOBIN: A SECOND LOOK. Adv Protein Chem. 1964;19:223–286. doi: 10.1016/s0065-3233(08)60190-4. [DOI] [PubMed] [Google Scholar]
- Walstrum S. A., Uhlenbeck O. C. The self-splicing RNA of Tetrahymena is trapped in a less active conformation by gel purification. Biochemistry. 1990 Nov 20;29(46):10573–10576. doi: 10.1021/bi00498a022. [DOI] [PubMed] [Google Scholar]
- Weber G. Energetics of ligand binding to proteins. Adv Protein Chem. 1975;29:1–83. doi: 10.1016/s0065-3233(08)60410-6. [DOI] [PubMed] [Google Scholar]
- Yarus M., Illangesekare M., Christian E. Selection of small molecules by the Tetrahymena catalytic center. Nucleic Acids Res. 1991 Mar 25;19(6):1297–1304. doi: 10.1093/nar/19.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug A. J., Grosshans C. A., Cech T. R. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988 Dec 13;27(25):8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]


