Abstract
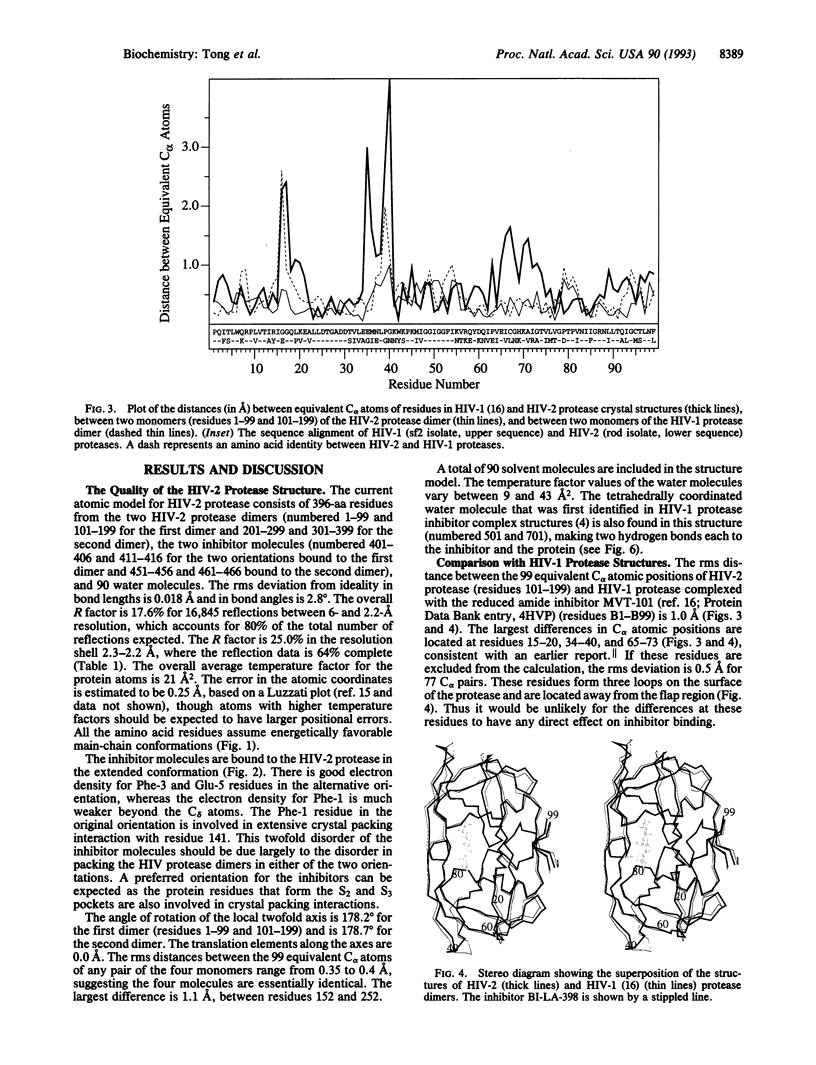
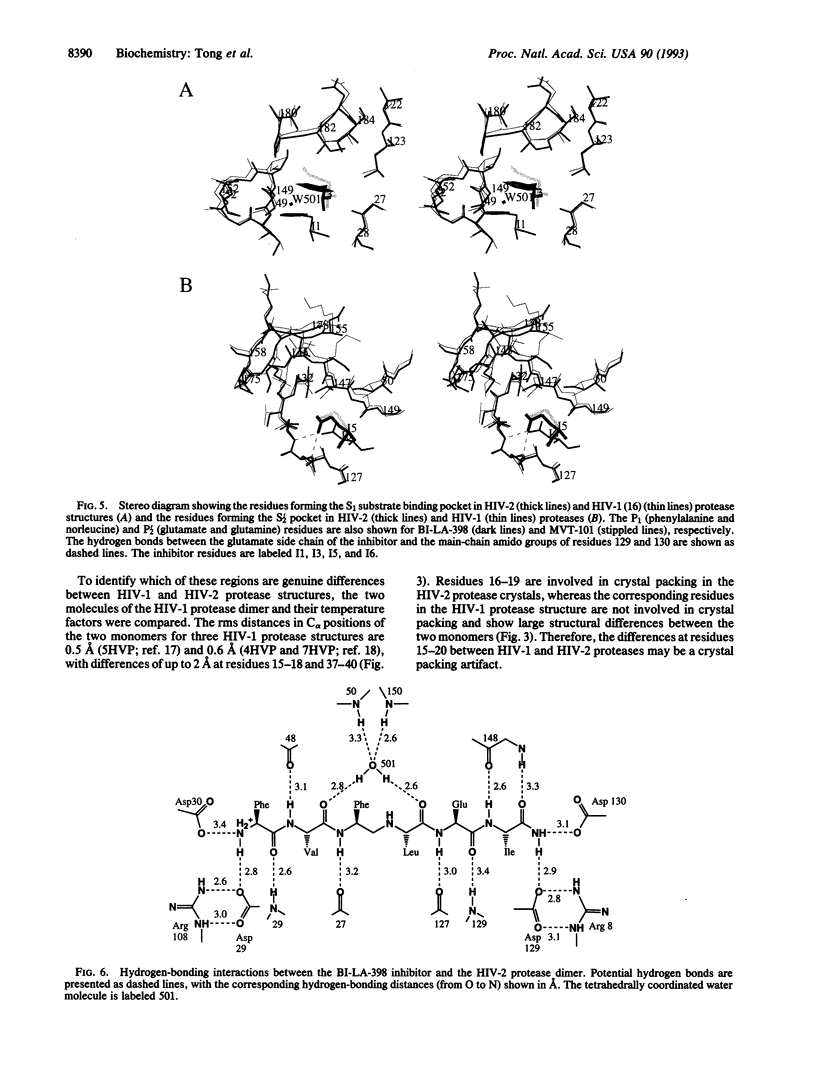
The crystal structure of HIV-2 protease in complex with a reduced amide inhibitor [BI-LA-398; Phe-Val-Phe-psi (CH2NH)-Leu-Glu-Ile-amide] has been determined at 2.2-A resolution and refined to a crystallographic R factor of 17.6%. The rms deviation from ideality in bond lengths is 0.018 A and in bond angles is 2.8 degrees. The largest structural differences between HIV-1 and HIV-2 proteases are located at residues 15-20, 34-40, and 65-73, away from the flap region and the substrate binding sites. The rms distance between equivalent C alpha atoms of HIV-1 and HIV-2 protease structures excluding these residues is 0.5 A. The shapes of the S1 and S2 pockets in the presence of this inhibitor are essentially unperturbed by the amino acid differences between HIV-1 and HIV-2 proteases. The interaction of the inhibitor with HIV-2 protease is similar to that observed in HIV-1 protease structures. The unprotected N terminus of the inhibitor interacts with the side chains of Asp-29 and Asp-30. The glutamate side chain of the inhibitor forms hydrogen bonds with the main-chain amido groups of residues 129 and 130.
Full text
PDF
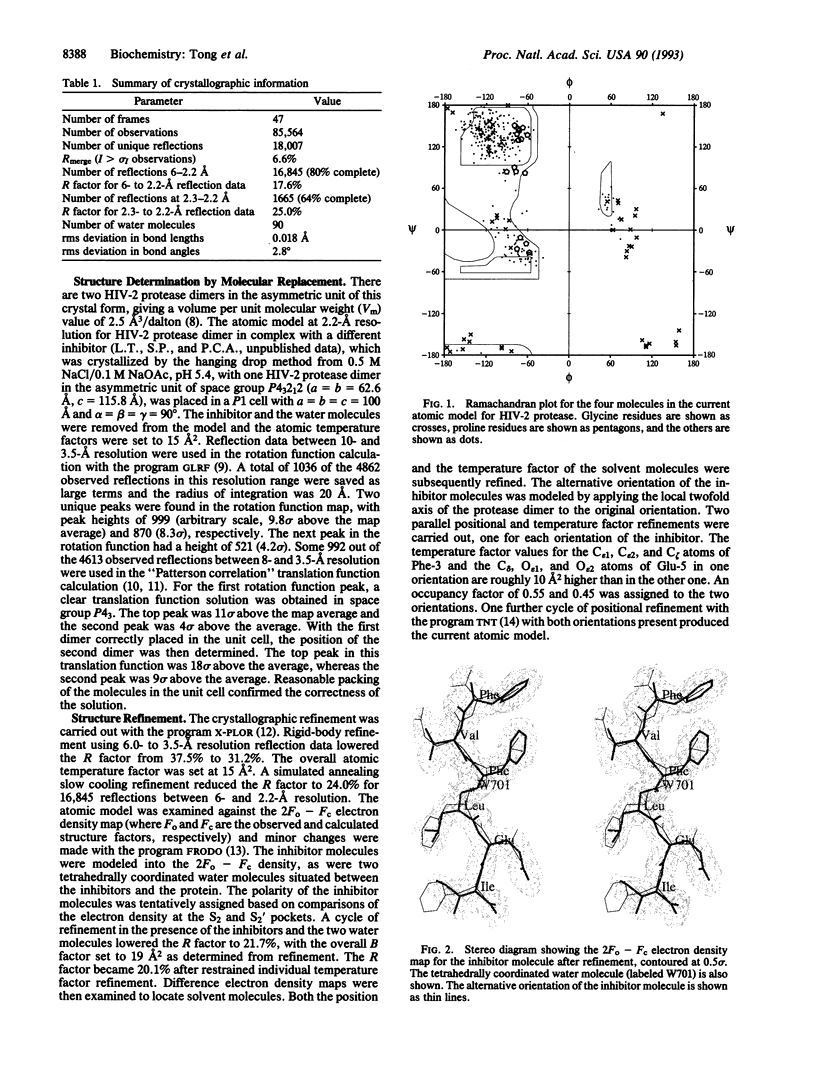

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Fitzgerald P. M., McKeever B. M., VanMiddlesworth J. F., Springer J. P., Heimbach J. C., Leu C. T., Herber W. K., Dixon R. A., Darke P. L. Crystallographic analysis of a complex between human immunodeficiency virus type 1 protease and acetyl-pepstatin at 2.0-A resolution. J Biol Chem. 1990 Aug 25;265(24):14209–14219. [PubMed] [Google Scholar]
- Griffiths J. T., Phylip L. H., Konvalinka J., Strop P., Gustchina A., Wlodawer A., Davenport R. J., Briggs R., Dunn B. M., Kay J. Different requirements for productive interaction between the active site of HIV-1 proteinase and substrates containing -hydrophobic*hydrophobic- or -aromatic*pro- cleavage sites. Biochemistry. 1992 Jun 9;31(22):5193–5200. doi: 10.1021/bi00137a015. [DOI] [PubMed] [Google Scholar]
- Gustchina A., Weber I. T. Comparative analysis of the sequences and structures of HIV-1 and HIV-2 proteases. Proteins. 1991;10(4):325–339. doi: 10.1002/prot.340100406. [DOI] [PubMed] [Google Scholar]
- Guyader M., Emerman M., Sonigo P., Clavel F., Montagnier L., Alizon M. Genome organization and transactivation of the human immunodeficiency virus type 2. Nature. 1987 Apr 16;326(6114):662–669. doi: 10.1038/326662a0. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kohl N. E., Emini E. A., Schleif W. A., Davis L. J., Heimbach J. C., Dixon R. A., Scolnick E. M., Sigal I. S. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. W. Solvent content of protein crystals. J Mol Biol. 1968 Apr 28;33(2):491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Miller M., Schneider J., Sathyanarayana B. K., Toth M. V., Marshall G. R., Clawson L., Selk L., Kent S. B., Wlodawer A. Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 A resolution. Science. 1989 Dec 1;246(4934):1149–1152. doi: 10.1126/science.2686029. [DOI] [PubMed] [Google Scholar]
- Mitsuya H., Yarchoan R., Broder S. Molecular targets for AIDS therapy. Science. 1990 Sep 28;249(4976):1533–1544. doi: 10.1126/science.1699273. [DOI] [PubMed] [Google Scholar]
- Mulichak A. M., Hui J. O., Tomasselli A. G., Heinrikson R. L., Curry K. A., Tomich C. S., Thaisrivongs S., Sawyer T. K., Watenpaugh K. D. The crystallographic structure of the protease from human immunodeficiency virus type 2 with two synthetic peptidic transition state analog inhibitors. J Biol Chem. 1993 Jun 25;268(18):13103–13109. [PubMed] [Google Scholar]
- Peterlin B. M., Luciw P. A. Molecular biology of HIV. AIDS. 1988;2 (Suppl 1):S29–S40. doi: 10.1097/00002030-198800001-00005. [DOI] [PubMed] [Google Scholar]
- Swain A. L., Miller M. M., Green J., Rich D. H., Schneider J., Kent S. B., Wlodawer A. X-ray crystallographic structure of a complex between a synthetic protease of human immunodeficiency virus 1 and a substrate-based hydroxyethylamine inhibitor. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8805–8809. doi: 10.1073/pnas.87.22.8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasselli A. G., Hui J. O., Sawyer T. K., Staples D. J., Bannow C., Reardon I. M., Howe W. J., DeCamp D. L., Craik C. S., Heinrikson R. L. Specificity and inhibition of proteases from human immunodeficiency viruses 1 and 2. J Biol Chem. 1990 Aug 25;265(24):14675–14683. [PubMed] [Google Scholar]
- Tong L. A., Rossmann M. G. The locked rotation function. Acta Crystallogr A. 1990 Oct 1;46(Pt 10):783–792. doi: 10.1107/s0108767390005530. [DOI] [PubMed] [Google Scholar]
- Wlodawer A., Erickson J. W. Structure-based inhibitors of HIV-1 protease. Annu Rev Biochem. 1993;62:543–585. doi: 10.1146/annurev.bi.62.070193.002551. [DOI] [PubMed] [Google Scholar]