Summary
Plants that form root‐nodule symbioses are within a monophyletic ‘nitrogen‐fixing’ clade and associated signalling processes are shared with the arbuscular mycorrhizal symbiosis. Central to symbiotic signalling are nuclear‐associated oscillations in calcium ions (Ca2+), occurring in the root hairs of several legume species in response to the rhizobial Nod factor signal.
In this study we expanded the species analysed for activation of Ca2+ oscillations, including non‐leguminous species within the nitrogen‐fixing clade.
We showed that Ca2+ oscillations are a common feature of legumes in their association with rhizobia, while Cercis, a non‐nodulating legume, does not show Ca2+ oscillations in response to Nod factors from Sinorhizobium fredii NGR234. Parasponia andersonii, a non‐legume that can associate with rhizobia, showed Nod factor‐induced calcium oscillations to S. fredii NGR234 Nod factors, but its non‐nodulating sister species, Trema tomentosa, did not. Also within the nitrogen‐fixing clade are actinorhizal species that associate with Frankia bacteria and we showed that Alnus glutinosa induces Ca2+ oscillations in root hairs in response to exudates from Frankia alni, but not to S. fredii NGR234 Nod factors.
We conclude that the ability to mount Ca2+ oscillations in response to symbiotic bacteria is a common feature of nodulating species within the nitrogen‐fixing clade.
Keywords: actinorhizal, calcium oscillations, Frankia, legumes, nitrogen‐fixing clade, nodulation, Parasponia, symbiotic signalling
Introduction
Plants have evolved endosymbioses with soil microorganisms that facilitate nutrient acquisition, including the association with arbuscular mycorrhizal (AM) fungi (Harrison, 2005), which evolved some 450 million yr ago and occurs in over 80% of higher plants (Remy et al., 1994; Redecker et al., 2000; Humphreys et al., 2010). Within the last 60 million yr, other symbiotic associations emerged, including the mutually beneficial relationship between plants and different nitrogen‐fixing soil bacteria, which are harboured in nodules on the roots. These root‐nodule symbioses are found in a number of plant families, all of which exist within the ‘nitrogen‐fixing clade’ (Soltis et al., 1995), and recent work reveals that a single innovation drove the emergence of nitrogen‐fixing symbioses resulting in nodulation (Werner et al., 2014). Nodulating plant species within the nitrogen‐fixing clade associate with two different groups of bacteria: filamentous Gram‐positive Frankia bacteria (Stacey et al., 1992) or Gram‐negative rhizobia (Sprent, 2009). Plant species from eight different families (referred to as actinorhizal) can associate with Frankia, but only legumes (Fabaceae) and Parasponia (Cannabaceae) form the rhizobial association (Trinick, 1973, 1979).
The establishment of the rhizobial and Frankia associations involves plant recognition of diffusible signals from the bacteria. Rhizobia produce lipochitooligosaccharides (LCOs), termed Nod factors, with specific modifications to the common LCO backbone, which are important in defining the plant host range of different rhizobial species (Dénarié et al., 1996). Recognition of Nod factors by legume roots involves two distinct LysM receptor kinases, NFR1 and NFR5 in Lotus japonicus and LYK3 and NFP in Medicago truncatula, which function in a heterodimeric complex (Madsen et al., 2003, 2011; Arrighi et al., 2006; Broghammer et al., 2012; Pietraszewska‐Bogiel et al., 2013). Upon perception of Nod factors, these receptors activate a symbiotic signalling pathway that includes an additional LRR‐type transmembrane receptor (SYMRK/DMI2) and nuclear‐localized components among which is a calcium and calmodulin‐dependent protein kinase (CCaMK) (Oldroyd, 2013). In legumes the genes encoding the Nod factor receptors underwent several duplication events, for example the ancestral LjNFR1a/MtLYK3 gene experienced a tandem duplication at the birth of the legume family (De Mita et al., 2014). This event most likely occurred before the evolution of the rhizobial symbiosis, as signatures of the duplication can be found in basal legume species that do not nodulate. Such ancestral duplication in LjNFR1a/MtLYK3 may have provided a degree of freedom to evolve specificity towards rhizobial Nod factors (Op den Camp et al., 2011a; Young et al., 2011).
The structure of the diffusible signal produced by Frankia bacteria has yet to be defined; however, the analysis of exudates and the genome sequences of different Frankia species suggests that these molecules have features that set them apart from canonical rhizobial LCOs (Normand et al., 2007; Kucho et al., 2010). Nevertheless, the Frankia symbiosis relies, at least in part, on the symbiosis signalling pathway, as at least two components are required in actinorhizal species for the Frankia association: SYMRK and CCaMK (Gherbi et al., 2008; Markmann et al., 2008; Svistoonoff et al., 2013). This suggests that even though nodulation evolved independently on multiple occasions, the utilization of symbiosis signalling for recognition of nitrogen‐fixing bacteria is a common feature within the nitrogen‐fixing clade (Markmann & Parniske, 2009).
Oscillations in the calcium ion concentration (Ca2+) associated with the nucleus are at the core of the symbiosis signalling pathway (Oldroyd, 2013). Nod factor‐induced Ca2+ oscillations have been observed in a number of different legume species, including M. truncatula (Wais et al., 2000), Pisum sativum (Walker et al., 2000), Phaseolus vulgaris (Cardenas et al., 1999) and L. japonicus (Harris et al., 2003). These species are restricted to Papilionoideae legumes. Non‐nodulating legumes and nodulating non‐legumes have not been analysed. In this study we asked whether Ca2+ oscillations are a common feature of bacterial recognition within the nitrogen‐fixing clade. We analysed calcium responses in a variety of legumes that provided representative species across the diversity of legumes and analysed calcium responses in the nodulating non‐legumes Parasponia andersonii and Alnus glutinosa. Our work reveals that Ca2+ oscillations are a conserved feature of bacterial recognition in all nodulating species analysed.
Materials and Methods
Plant growth
Unless stated otherwise, all plants were grown in a controlled environment at 20°C, under 16 : 8 h, light : dark conditions. The relative humidity was 32% and the light intensity was 300 μmol m−2 s−1. When aminoethoxyvinylglycine (AVG) was added to the medium, it was always to a concentration of 0.1 μM. Seeds were scarified either with sandpaper or with H2SO4 to help germination and subsequently left in bleach for 2–15 min, depending on the species. After washing with dH2O approximately seven times, the seeds were allowed to imbibe while rotating for 4 h at room temperature and then placed on distilled water agar (DWA) media in a Petri dish upside down overnight at room temperature. When the seeds had germinated, they were grown vertically on plates of buffered nodulation medium (BNM) with added AVG. The exception was Cercis siliquastrum, seeds of which were cut open after imbibing, the embryos removed and transferred to woody plant media (WPM) plates. When the plants were large enough to handle, they were moved to EKM (Becking, 1983) plates with added AVG and grown between filter papers. For transformation with NupYC2.1, Parasponia andersonii (clone WU1) and Trema tomentosa (clone WU10) were clonally propagated and transformed using Agrobacterium rhizogenes, as described previously (Op den Camp et al., 2011b; Cao et al., 2012).
NGR234 Nod factor preparation
Strain ANU2811 (Chen et al., 1985), an exopolysaccharide defective derivative of Sinorhizobium sp. NGR234, was grown for 2 d in 2 l of Y medium with added Apigenin (1 μM). The culture was centrifuged at 5°C, 6000 g, for 60 min. The supernatant was then passed through a C18 : 1 reverse‐phase column (Sep Pak, Waters, Elstree, UK) and eluted with 4 ml of MeOH.
The Nod factor activity was tested using root hair deformation in Vicia hirsuta. For this, V. hirsuta seeds were germinated and roots were allowed to grow to c. 1 cm long. Cover slides were attached to sterile microscope glass slides using silicon grease and seedlings placed between the cover slip and the slide, with the root in Fahraeus plant (FP) medium. Dilutions of the NGR234 Nod factor preparation, plus positive and negative controls, were added to the FP medium. These were left at room temperature in the dark overnight and then assessed for root hair deformation.
The NGR234 Nod factors were analysed by high‐pressure liquid chromatography coupled to a Shimadzu IT‐ToF mass spectrometer. Nod factors were separated on a 50 × 2.1 mm 2.7 μ Kinetex XB‐C18 column (Phenomenex, Macclesfield, UK) using the following gradient of acetonitrile (B) vs 0.1% formic acid in water (A), run at 600 μl min−1 and 40°C: 0 min, 5% B; 11.67 min, 95% B; 14.58 min, 95% B; 15.17 min, 5% B; 18.08 min, 5% B. Products were detected by positive and negative mode electrospray, with automatic data‐dependent collection of MS2 data at an isolation width of m/z 3.0, 50% collision energy and 50% collision gas. Spray chamber conditions were 250°C curved desorbation line, 300°C heat block, 1.5 l min−1 nebulizer gas, and drying gas ‘on’. The instrument was calibrated using sodium trifluoroacetate according to the manufacturer's instructions before use. In the negative mode, we observed m/z peaks corresponding to NodNGR‐V (Carb2, NMe, C18 : 1MeFuc, S) and derivatives of that with one or no carbamoyl groups (masses 1594.69, 1551.69 and 1508.68, respectively) in a ratio of c. 11 : 7 : 2 based on peak abundance. There were low numbers (c. 1/20th) of molecules with masses two units higher, presumably corresponding to the C18 : 0 variants. There were also masses of 1568.68, 1525.67 and 1482.66 which would correspond with the C16 : 0 derivatives and these were present at c. 25% of the level of the C18 : 1 compounds. In the positive mode, we observed masses corresponding to the non‐sulphated Nod factors NodNGR‐V (Carb1 or Carb2, NMe, C18 : 1 MeFuc) and NGR‐V (Carb1 or Carb2, NMe, C18 : 1, MeFucAc). All these correspond to the Nod‐factors described previously (Price et al., 1992). Using tandem MS we observed many of the expected fragments as described previously (Price et al., 1992). There were also some ions observed in the negative mode that we were unable to attribute and we did not see the corresponding peaks in the positive mode. We conclude that the mixture of Nod factors that we used is at least as complex as described previously (Price et al., 1992). To quantify the Nod‐factors present, we mixed a known concentration of Sinorhizobium meliloti Nod factor with the NGR234 Nod‐factor preparation and estimated the concentration of NodNGR‐V (Carb2, NMe, C18 : 1, MeFuc S) relative to SmIV (Ac,S, C16 : 2) by estimating relative abundance using negative mode MS; this quantification is based on the assumption that both sulphated Nod‐factors will ionize equally.
Frankia factor preparation
Frankia alni strain ACN14a (Normand & Lalonde, 1982) was grown under nitrogen‐replete conditions as described previously (Alloisio et al., 2010). Mycelia from this culture were harvested by centrifugation at 700 g for 5 min. The supernatant was discarded and the bacteria (c. 50 mg) were resuspended in 1 ml of sterile distilled water. After vortexing, the suspension was sonicated for 30 s and centrifugated at 13 000 g for 5 min. The supernatant was collected and filtered using a 0.20 μm syringe filter (Sartorious, Epsom, UK). Supernatant (300 μl) was added to a bath of c. 1 ml of BNM containing the Alnus glutinosa seedling with microinjected cells for analysis of Ca2+.
Calcium imaging
Cameleon NupYC2.1
The nuclear‐targeted cameleon NupYC2.1 was used for analysing Ca2+ responses in P. andersonii and T. tomentosa. The plants were inspected on a florescent stereomicroscope and roots expressing a strong signal were selected. These were removed and transferred to a microscope slide with liquid BNM media. Fluorescence resonance energy transfer (FRET) measurements were taken using an epifluorescence microscope. Cyan fluorescent protein (CFP) was excited by 437 nm with an 11 nm bandpass using an Optoscan Monochromator (Cairn Research, Faversham, Kent, UK). An image splitter (Optosplit, Cairn Research) allowed analysis of the same image through 485 and 535 nm filters to measure CFP and yellow fluorescent protein (YFP) emissions.
Microinjection
This protocol follows Wais et al. (2000). The plants were prepared as described and placed in BNM media on coverslips. The dyes used were Oregon Green, which responds to changes in Ca2+, and Texas Red, which is non‐responsive to Ca2+ and allows pseudoratiometric imaging. Both dyes were fused to a 10 kD Dextran molecule to limit free diffusion. Glass needles were prepared using an electrode puller (model 773; Campden Instruments Ltd, Loughborough, UK) from borosilicate glass capillaries (1B120F‐4; World Precision Instruments Inc., Sarasota, FL, USA). The needle was back‐loaded with 0.5 μl of the dye solution with a Microloader pipette tip. The needle was then back‐filled with c. 10 μl 1 M KCl. Once injected, the dye was forced from the needle into the plant cell using iontophoresis. After injection, only those cells that showed strong cytoplasmic streaming were used in the experiments. Imaging was performed using a Nikon TE2000U inverted microscope (Kingston Upon Thames, UK) and a Hamamatsu Photonics digital CC camera. Images were collected every 5 s with a 1 s exposure.
Results
Nod factor induced calcium oscillations in a range of legume species
In order to address whether Ca2+ oscillations are a common feature of rhizobial entry into legumes, we chose a number of legumes that represent the diversity present in this family (Fig. 1). We used Chamaecrista fasciculata as a representative nodulating species within the Caesalpinoideae (Naisbitt et al., 1992; Singer et al., 2009) and C. siliquastrum was investigated, as a representative basal, non‐nodulating legume. As a representative of the Mimosoideae subfamily, we chose Acacia retinoides. Within the Papilionoideae, we chose species with distinctive bacterial infection modes: Cytisus proliferus, which initiates root hair entry, but this aborts to be replaced by intercellular invasion (Vega‐Hernandez et al., 2001); and Lupinus pilosus, which shows intercellular invasion between epidermal cells (González‐Sama et al., 2004). The selection of these species was also influenced by their rhizobial symbiont: all can be colonized by the broad host range species Sinorhizobium fredii NGR234 (hitherto referred to as NGR234) and this allowed ease of Nod factor production and comparability across the different species (Pueppke & Broughton, 1999).
Figure 1.
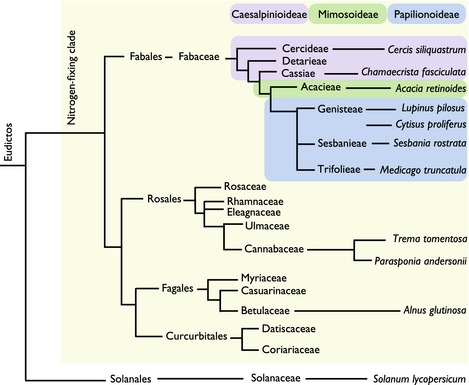
An overview of the phylogenetic relationships of plant species included in this study. The three subfamilies of the Fabaceae are indicated with coloured boxes. The tree is unscaled and Solanum lycopersicum (within the Asterids) is included as an outgroup.
A crude isolation of NGR234 Nod factor was undertaken using published methods (Price et al., 1992) and the activity was validated in a root hair deformation assay on V. hirsuta, which is known to perceive a variety of Nod factors (Schultze et al., 1992). In this assay the NGR234 Nod factor preparation showed strong root hair deformation activity in a 1 : 100 000 dilution (Supporting Information Fig. S1: 10 μl of a 1 : 1000 dilution of the NGR234 NF stock in 1 ml of buffer) and therefore we used this dilution throughout the analyses of Ca2+ oscillations. Analysis of this Nod factor isolation revealed that this is equivalent to 0.7 nM of NodNGR‐V (Carb2, NMe, C18 : 1MeFuc, S), the pentameric Nod factor carrying N‐linked C18 : 1 acyl and methyl groups, two carbamoyl groups on the terminal non‐reducing glucosamine and a sulphated fucose on the reducing glucosamine residue. The other identified Nod factors (lacking one or two carbamoyl groups and with different acyl groups; see the Materials and Methods section) added up to a similar concentration, such that the total concentration of the identified Nod factors applied throughout this work was c. 1.5 nM.
The analysis of calcium responses on these diverse species is rather limited, as no established transformation procedures exist for most of these species and therefore we are restricted to microinjection of calcium responsive dyes. Such an approach is limited to the analysis of young growing root hair cells, as other cell types are intransigent to microinjection. We microinjected growing root hairs on the primary roots of C. fasciculata, A. retinoides, C. proliferus, L. pilosus and C. siliquastrum with a mixture of the Ca2+‐responsive dye Oregon Green and the non‐responsive dye Texas Red to allow pseudo‐ratiometric analysis of Ca2+ oscillations. Chamaecrista fasciculata, A. retinoides, C. proliferus and L. pilosus all responded to NGR234 Nod factor with activation of Ca2+ oscillations (Fig. 2; Table 1). For comparison, Ca2+ traces from M. truncatula and Sesbania rostrata are included in Fig. 2 (Capoen et al., 2009). Similar to Nod factor‐induced Ca2+ oscillations previously reported in M. truncatula, L. japonicus and S. rostrata (Ehrhardt et al., 1996; Wais et al., 2000; Miwa et al., 2006; Capoen et al., 2009), the Ca2+ responses we observed were activated 10–20 min after Nod factor addition, continued for over 1 h (Fig. 2) and were restricted to the nuclear region (Fig. S2). By contrast, root hairs of the non‐nodulating legume C. siliquastrum and the non‐legume rice (Oryza sativa) showed no response to NGR234 Nod factor (Figs 2d, 3d; Table 1). Taken together, this work shows that in legumes Nod factor‐induced Ca2+ oscillations appear to correlate with the capacity to form root nodules.
Figure 2.
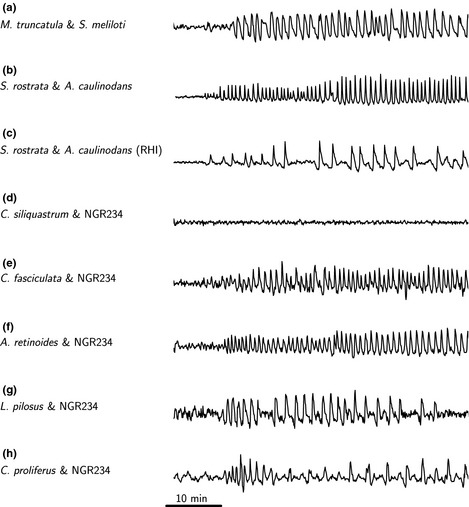
Representative traces showing Nod factor‐induced Ca2+ oscillations in a variety of legume species. Each panel shows a representative root hair Ca2+ trace from the following species: (a) Medicago truncatula treated with Sinorhizobium meliloti Nod factor; (b) Sebania rostrata grown under conditions that promote crack entry and treated with Azorhizobium caulinodans Nod factor (Capoen et al., 2009); (c) S. rostrata grown under conditions that promote root hair invasion and treated with A. caulinodans Nod factor (Capoen et al., 2009); (d) Cercis siliquastrum treated with NGR234 Nod factor; (e) Chamaecrista fasciculata treated with NGR234 Nod factor; (f) Acacia retinoides treated with NGR234 Nod factor; (g) Lupinus pilosus treated with NGR234 Nod factor; (h) Cytisus proliferus treated with NGR234 Nod factor.
Table 1.
Proportion of cells showing Ca2+ oscillations in response to NGR234 Nod factors or Frankia exudates (Alnus glutinosa), measured by either microinjection or cameleon
| Plant species | Ca2+‐responsive cells/total cells analysed |
|---|---|
| Rice (nipponbare) | 0/30 |
| Parasponia andersonii | 17/26 |
| Trema tomentosa | 0/35 |
| Alnus glutinosa | 5/10 |
| Cercis siliquastrum | 0/15 |
| Chamaecrista fasciculata | 13/20 |
| Acacia retinoides | 13/17 |
| Lupinus pilosus | 11/12 |
| Cytisus proliferus | 12/17 |
Figure 3.
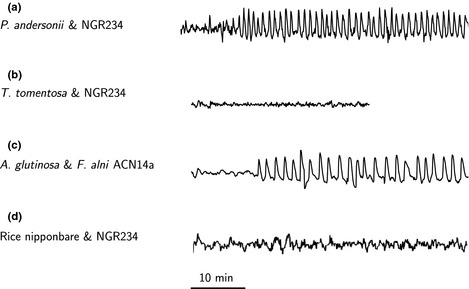
Representative traces showing bacterial‐induced Ca2+ oscillations in non‐legumes. Each panel shows a representative root hair Ca2+ trace from the following species: (a) Parasponia andersonii treated with NGR234 Nod factor; (b) Trema tomentosa treated with NGR234 Nod factor; (c) Alnus glutinosa treated with exudates from Frankia alni ACN14a; (d) rice nipponbare treated with NGR234 Nod factor.
Calcium oscillations in the non‐legume P. andersonii
Plant species in the genus Parasponia within the Cannabaceae are the only non‐legumes known to form nodules with rhizobia, and P. andersonii does so with NGR234 (Pueppke & Broughton, 1999). Parasponia andersonii is a tropical tree that can be propagated vegetatively (Op den Camp et al., 2011b) and plantlets from vegetative propagation were used in this study. P. andersonii showed Ca2+ oscillations in root hair cells in response to NGR234 Nod factor following microinjection (Fig. 3a; Table 1). However, microinjection of P. andersonii root hairs was extremely challenging and this made it difficult to gather data from a sufficient number of cells. We therefore used A. rhizogenes to introduce the Ca2+ reporter cameleon (NupYC2.1; Sieberer et al., 2009) into P. andersonii roots. When challenged with NGR234 Nod factor, these cameleon‐transformed roots revealed Ca2+ oscillations (Table 1). Plant species of the sister genus of Parasponia, Trema, do not nodulate and to assess Nod factor‐induced Ca2+ oscillations in this close relative, we transformed T. tomentosa roots with NupYC2.1. We observed no Ca2+ oscillations in T. tomentosa treated with NGR234 Nod factors (Fig. 3b; Table 1). Taken together, these results show that also outside the legume family, rhizobium root nodule formation is causally associated with Nod factor‐induced Ca2+ oscillations.
Frankia factor(s) induce Ca2+ oscillations in the actinorhizal tree A. glutinosa
Alnus glutinosa belongs to the Betulaceae in the order Fagales and is an important colonizer in many ecosystems because of its capacity to form nitrogen‐fixing symbioses with Frankia bacteria (Wall, 2000). While Frankia produce diffusible signals, their structure(s) have yet to be defined (Cérémonie et al., 1999; Pawlowski et al., 2011). However, exudates from Frankia bacteria can induce root hair deformation (Pawlowski et al., 2011) and, therefore, we asked whether such exudates could also activate Ca2+ oscillations. We cultured the slow‐growing F. alni ACN14a for a number of months and used sonication to break up the hyphal mats. The bacteria were centrifuged and the exudates were used to treat Oregon Green/Texas Red microinjected root hairs of A. glutinosa. This exudate could activate Ca2+ oscillations in A. glutinosa root hair cells (Fig. 3c; Table 1) and also induced root hair deformation (Fig. S3). By contrast, A. glutinosa did not respond to NGR234 Nod factor, and M. truncatula was unable to mount calcium oscillations in response to the exudates from F. alni (Fig. S4). We observed a comparable delay before induction of Ca2+ oscillations in A. glutinosa and similar nuclear‐localization as previously observed in legumes (Ehrhardt et al., 1996).
Discussion
All plant species that form root‐nodule symbioses are confined to the monophyletic nitrogen‐fixing clade, but most species within this clade do not nodulate (Soltis et al., 1995). This suggests that a genetic event within a common ancestor predisposed this group to evolve root‐nodule symbioses, followed by multiple gains and losses of the symbiosis (Werner et al., 2014). Here we show that bacterial activation of Ca2+ oscillations in root hair cells is a common feature of diverse legumes, the actinorhizal species A. glutinosa and the rhizobial‐associating non‐legume P. andersonii. This is consistent with genetic work indicating the requirement for components of the symbiosis signalling pathway in the actinorhizal species Casuarina (Gherbi et al., 2008; Markmann et al., 2008; Svistoonoff et al., 2013).
Symbiosis signalling initially evolved to support the AM association and this signalling pathway was recruited during the evolution of nodulation (Markmann & Parniske, 2009; Oldroyd, 2013). AM fungi and their exudates can activate calcium oscillations in both legumes and non‐legumes (Kosuta et al., 2008; Chabaud et al., 2011; Genre et al., 2013) and this is probably the result of LCOs and chitooligosaccharides (COs) present in the mycorrhizal exudates (Maillet et al., 2011; Genre et al., 2013). One possible scenario for the predisposition event is a modification to the symbiosis signalling pathway that allowed recognition of nitrogen‐fixing bacteria. We analysed two non‐nodulating species within the nitrogen‐fixing clade, T. tomentosa and C. siliquastrum, and neither were able to activate Ca2+ oscillations in response to NGR234 Nod factor. If these species are representative of non‐nodulating species containing the predisposition event, then we must conclude that bacterial recognition is not associated with the evolution of the predisposition to nodulate. However, caution is neccessary when concluding this based on a negative result using only NGR234 Nod factor.
Parasponia species are the only non‐legumes reported to form root‐nodule symbioses with rhizobia (Geurts et al., 2012). A very closely related sister species, T. tomentosa, does not nodulate and does not respond to NGR234 Nod factors. This suggests that the ability to respond to NGR234 Nod factor may have evolved very recently and could have faciliated the emergence of nodulation in P. andersonii. Molecular studies have revealed that the same receptor in P. andersonii, PaNFP, is involved in the recognition of symbiotic signalling molecules from both AM fungi and rhizobial bacteria (Op den Camp et al., 2011b). Possibly a minor difference within the receptor between Trema and Parasponia explains this shift in Nod factor sensitivity, or, alternatively, evolution of other LysM receptor‐like kinases could be the cause.
We have shown that a diffusible signal produced by F. alni can activate symbiosis signalling in A. glutinosa with resultant Ca2+ oscillations. This has analogies with LCO signalling in rhizobia; however, research in Frankia has implied that the diffusible signal is not a canonical LCO (Cérémonie et al., 1999; Normand et al., 2007). The nature of this signal and the receptors that are required for its recognition remain to be resolved. Our work reveals commonalities in the activation of Ca2+ oscillations by diffusible bacterial signalling molecules across all nodulating species tested, both legumes and non‐legumes. This highlights the importance of symbiosis signalling and related Ca2+ oscillations for the recognition of nitrogen‐fixing bacteria within the nitrogen‐fixing clade. The fact that mycorrhizal fungi also produce LCO and CO signalling molecules (Maillet et al., 2011; Genre et al., 2013) suggests that the capacity to perceive nitrogen‐fixing bacteria may have evolved from the perception of mycorrhizal‐associated LCO and CO signalling molecules. A full understanding of the evolutionary path that led to the perception of nitrogen‐fixing bacteria requires a detailed analysis of mycorrhizal recognition processes of plants within the nitrogen‐fixing clade. This is complicated by the fact that Myc‐LCO and CO responses appear restricted to atrichoblasts (Sun et al., 2015), which are intransigent to microinjection. While such an analysis of mycorrhizal‐induced calcium responses is necessary for a full understanding of the evolution of nodulation, it will require the long‐term development of transformation procedures in non‐model species.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Root hair deformation responses of Vicia hirsuta to the NGR234 NF purifications.
Fig. S2 The nuclear localization of the calcium oscillation response in a variety of legume species.
Fig. S3 Root hair deformation in Alnus glutinosa following treatment with Frankia exudates.
Fig. S4 Calcium traces in Alnus glutinosa treated with NGR234 NF and in Medicago truncatula treated with Frankia exudates.
Acknowledgements
Giulia Morieri helped with the purification of the NGR234 Nod factors. We thank Kew Gardens Millenium Seed Bank and Milagros León Barrios for providing seeds. Thanks also to Janet Sprent and Euan James for advice early in the project. This work was supported by the BBSRC as BB/J004553/1 and by the European Research Council as SYMBIOSIS. E.G. was funded by the John Innes Foundation and R.G. by NWO VICI 865.13.001. Thanks are also expressed to the French ANR (Sesam, ANR‐10‐BLAN‐1708).
References
- Alloisio N, Queiroux C, Fournier P, Pujic P, Normand P, Vallenet D, Médigue C, Yamaura M, Kakoi K, Kucho K. 2010. The Frankia alni symbiotic transcriptome. Molecular Plant‐Microbe Interactions 23: 593–607. [DOI] [PubMed] [Google Scholar]
- Arrighi J‐F, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho‐Niebel F, Journet E‐P, Ghérardi M, Huguet T et al 2006. The Medicago truncatula lysin motif‐receptor‐like kinase gene family includes NFP and new nodule‐expressed genes. Plant Physiology 142: 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becking J. 1983. The Parasponia parviflora–Rhizobium symbiosis. Host specificity, growth and nitrogen fixation under various conditions. Plant and Soil 75: 309–342. [Google Scholar]
- Broghammer A, Krusell L, Blaise M, Sauer J, Sullivan JT, Maolanon N, Vinther M, Lorentzen A, Madsen EB, Jensen KJ et al 2012. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proceedings of the National Academy of Sciences, USA 109: 13859–13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Op den Camp R, Seifi Kalhor M, Bisseling T, Geurts R. 2012. Efficiency of Agrobacterium rhizogenes‐mediated root transformation of Parasponia and Trema is temperature dependent. Plant Growth Regulation 68: 459–465. [Google Scholar]
- Capoen W, Den Herder J, Sun J, Verplancke C, De Keyser A, De Rycke R, Goormachtig S, Oldroyd G, Holsters M. 2009. Calcium spiking patterns and the role of the calcium/calmodulin‐dependent kinase CCaMK in lateral root base nodulation of Sesbania rostrata . Plant Cell 21: 1526–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas L, Feijo JA, Kunkel JG, Sanchez F, Holdaway‐Clarke T, Hepler PK, Quinto C. 1999. Rhizobium nod factors induce increases in intracellular free calcium and extracellular calcium influxes in bean root hairs. Plant Journal 19: 347–352. [DOI] [PubMed] [Google Scholar]
- Cérémonie H, Debellé F, Fernandez MP. 1999. Structural and functional comparison of Frankia root hair deforming factor and rhizobia Nod factor. Canadian Journal of Botany 77: 1293–1301. [Google Scholar]
- Chabaud M, Genre A, Sieberer BJ, Faccio A, Fournier J, Novero M, Barker DG, Bonfante P. 2011. Arbuscular mycorrhizal hyphopodia and germinated spore exudates trigger Ca2+ spiking in the legume and nonlegume root epidermis. New Phytologist 189: 347–355. [DOI] [PubMed] [Google Scholar]
- Chen H, Batley M, Redmond J, Rolfe BG. 1985. Alteration of the effective nodulation properties of a fast growing broad host range Rhizobium due to changes in exoploysaccharide synthesis. Journal of Plant Physiology 120: 331–349. [Google Scholar]
- De Mita S, Streng A, Bisseling T, Geurts R. 2014. Evolution of a symbiotic receptor through gene duplications in the legume–rhizobium mutualism. New Phytologist 201: 961–972. [DOI] [PubMed] [Google Scholar]
- Dénarié J, Debellé F, Promé JC. 1996. Rhizobium lipo‐chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annual Review of Biochemistry 65: 503–535. [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Wais R, Long SR. 1996. Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681. [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Balzergue C, Puech‐Pagès V, Novero M, Rey T, Fournier J, Rochange S, Bécard G, Bonfante P et al 2013. Short‐chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytologist 198: 190–202. [DOI] [PubMed] [Google Scholar]
- Geurts R, Lillo A, Bisseling T. 2012. Exploiting an ancient signalling machinery to enjoy a nitrogen fixing symbiosis. Current Opinion in Plant Biology 15: 438–443. [DOI] [PubMed] [Google Scholar]
- Gherbi H, Markmann K, Svistoonoff S, Estevan J, Autran D, Giczey G, Auguy F, Péret B, Laplaze L, Franche C et al 2008. SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankia bacteria. Proceedings of the National Academy of Sciences, USA 105: 4928–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Sama A, Lucas MM, de Felipe MR, Pueyo JJ. 2004. An unusual infection mechanism and nodule morphogenesis in white lupin (Lupinus albus). New Phytologist 163: 371–380. [DOI] [PubMed] [Google Scholar]
- Harris JM, Wais R, Long SR. 2003. Rhizobium‐lnduced calcium spiking in Lotus japonicus . Molecular Plant‐Microbe Interactions 16: 335–341. [DOI] [PubMed] [Google Scholar]
- Harrison MJ. 2005. Signaling in the arbuscular mycorrhizal symbiosis. Annual Review of Microbiology 59: 19–42. [DOI] [PubMed] [Google Scholar]
- Humphreys CP, Franks PJ, Rees M, Bidartondo MI, Leake JR, Beerling DJ. 2010. Mutualistic mycorrhiza‐like symbiosis in the most ancient group of land plants. Nature Communications 1: 103. [DOI] [PubMed] [Google Scholar]
- Kosuta S, Hazledine S, Sun J, Miwa H, Morris RJ, Downie JA, Oldroyd GED. 2008. Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proceedings of the National Academy of Sciences, USA 105: 9823–9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucho K‐I, Hay A‐E, Normand P. 2010. The determinants of the actinorhizal symbiosis. Microbes and Environments 25: 241–252. [DOI] [PubMed] [Google Scholar]
- Madsen EB, Antolín‐Llovera M, Grossmann C, Ye J, Vieweg S, Broghammer A, Krusell L, Radutoiu S, Jensen ON, Stougaard J et al 2011. Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor‐mediated signalling in cooperation with Nod factor receptor 5. Plant Journal 65: 404–417. [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N et al 2003. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640. [DOI] [PubMed] [Google Scholar]
- Maillet F, Poinsot V, André O, Puech‐Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A et al 2011. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63. [DOI] [PubMed] [Google Scholar]
- Markmann K, Giczey G, Parniske M. 2008. Functional adaptation of a plant receptor‐kinase paved the way for the evolution of intracellular root symbioses with bacteria. PLoS Biology 6: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmann K, Parniske M. 2009. Evolution of root endosymbiosis with bacteria: how novel are nodules? Trends in Plant Science 14: 77–86. [DOI] [PubMed] [Google Scholar]
- Miwa H, Sun J, Oldroyd GED, Downie JA. 2006. Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant Journal 48: 883–894. [DOI] [PubMed] [Google Scholar]
- Naisbitt T, James EK, Sprent JI. 1992. The evolutionary significance of the legume genus Chamaecrista, as determined by nodule structure. New Phytologist 122: 487–492. [DOI] [PubMed] [Google Scholar]
- Normand P, Lalonde M. 1982. Evaluation of Frankia strains isolated from provenances of two Alnus species. Canadian Journal of Microbiology 28: 1133–1142. [Google Scholar]
- Normand P, Lapierre P, Tisa LS, Gogarten JP, Alloisio N, Bagnarol E, Bassi CA, Berry AM, Bickhart DM, Choisne N et al 2007. Genome characteristics of facultatively symbiotic Frankia sp. strains reflect host range and host plant biogeography. Genome Research 17: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED. 2013. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nature Reviews Microbiology 11: 252–263. [DOI] [PubMed] [Google Scholar]
- Op den Camp RHM, De Mita S, Lillo A, Cao Q, Limpens E, Bisseling T, Geurts R. 2011a. A phylogenetic strategy based on a legume‐specific whole genome duplication yields symbiotic cytokinin type‐A response regulators. Plant Physiology 157: 2013–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Op den Camp R, Streng A, De Mita S, Cao Q, Polone E, Liu W, Ammiraju JSS, Kudrna D, Wing R, Untergasser A et al 2011b. LysM‐type mycorrhizal receptor recruited for rhizobium symbiosis in the nonlegume Parasponia . Science 331: 909–912. [DOI] [PubMed] [Google Scholar]
- Pawlowski K, Bogusz D, Ribeiro A, Berry A. 2011. Progress on research on actinorhizal plants. Functional Plant Biology 38: 633–638. [DOI] [PubMed] [Google Scholar]
- Pietraszewska‐Bogiel A, Lefebvre B, Koini MA, Klaus‐Heisen D, Takken FLW, Geurts R, Cullimore JV, Gadella TWJ. 2013. Interaction of Medicago truncatula lysin motif receptor‐like kinases, NFP and LYK3, produced in Nicotiana benthamiana induces defence‐like responses. PLoS ONE 8: e65055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NP, Relić B, Talmont F, Lewin A, Promé D, Pueppke SG, Maillet F, Dénarié J, Promé JC, Broughton WJ. 1992. Broad‐host‐range Rhizobium species strain NGR234 secretes a family of carbamoylated, and fucosylated, nodulation signals that are O‐acetylated or sulphated. Molecular Microbiology 6: 3575–3584. [DOI] [PubMed] [Google Scholar]
- Pueppke SG, Broughton WJ. 1999. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Molecular Plant‐Microbe Interactions 12: 293–318. [DOI] [PubMed] [Google Scholar]
- Redecker D, Kodner R, Graham LE. 2000. Glomalean fungi from the Ordovician. Science 289: 1920–1921. [DOI] [PubMed] [Google Scholar]
- Remy W, Taylor TN, Hass H, Kerp H. 1994. Four hundred‐million‐year‐old vesicular arbuscular mycorrhizae. Proceedings of the National Academy of Sciences, USA 91: 11841–11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze M, Quiclet‐Sire B, Kondorosi E, Virelizer H, Glushka JN, Endre G, Géro SD, Kondorosi A. 1992. Rhizobium meliloti produces a family of sulfated lipooligosaccharides exhibiting different degrees of plant host specificity. Proceedings of the National Academy of Sciences, USA 89: 192–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer BJ, Chabaud M, Timmers AC, Monin A, Fournier J, Barker DG. 2009. A nuclear‐targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiology 151: 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer SR, Maki SL, Farmer AD, Ilut D, May GD, Cannon SB, Doyle JJ. 2009. Venturing beyond beans and peas: what can we learn from Chamaecrista? Plant Physiology 151: 1041–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Morgan DR, Swensen SM, Mullin BC, Dowd JM, Martin PG. 1995. Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proceedings of the National Academy of Sciences, USA 92: 2647–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent JI. 2009. Legume nodulation: a global perspective. Chichester, UK: Wiley‐Blackwell. [Google Scholar]
- Stacey GS, Burris RH, Evans HJ. 1992. Biological nitrogen fixation. New York, NY, USA: Chapman and Hall. [Google Scholar]
- Sun J, Miller JB, Granqvist E, Wiley‐Kalil A, Gobbato E, Maillet F, Cottaz S, Samain E, Venkateshwaran M, Fort S et al 2015. Activation of symbiosis signaling by arbuscular mycorrhizal fungi in legumes and rice. Plant Cell 27: 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S, Benabdoun FM, Nambiar‐Veetil M, Imanishi L, Vaissayre V, Cesari S, Diagne N, Hocher V, de Billy F, Bonneau J et al 2013. The independent acquisition of plant root nitrogen‐fixing symbiosis in Fabids recruited the same genetic pathway for nodule organogenesis. PLoS ONE 8: e64515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinick MJ. 1973. Symbiosis between Rhizobium and the non‐legume, Trema aspera . Nature 244: 459–460. [Google Scholar]
- Trinick MJ. 1979. Structure of nitrogen‐fixing nodules formed by Rhizobium on roots of Parasponia andersonii Planch. Canadian Journal of Microbiology 25: 565–578. [DOI] [PubMed] [Google Scholar]
- Vega‐Hernandez MC, Perez‐Galdona R, Dazzo FB, Jarabo‐Lorenzo A, Alfayate MC, Leon‐Barrios M. 2001. Novel infection process in the indeterminate root nodule symbiosis between Chamaecytisus proliferus (tagasaste) and Bradyrhizobium sp. New Phytologist 150: 707–721. [Google Scholar]
- Wais RJ, Galera C, Oldroyd G, Catoira R, Penmetsa RV, Cook D, Gough C, Denarie J, Long SR. 2000. Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula . Proceedings of the National Academy of Sciences, USA 97: 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SA, Viprey V, Downie AJ. 2000. Dissection of nodulation signaling using pea mutants defective for calcium spiking induced by Nod factors and chitin oligomers. Proceedings of the National Academy of Sciences, USA 97: 13413–13418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall LG. 2000. The actinorhizal symbiosis. Journal of Plant Growth Regulation 19: 167–182. [DOI] [PubMed] [Google Scholar]
- Werner GD, Cornwell WK, Sprent JI, Kattge J, Kiers ET. 2014. A single evolutionary innovation drives the deep evolution of symbiotic N2‐fixation in angiosperms. Nature Communications 5: 4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GED, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KFX, Gouzy J, Schoof H et al 2011. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Root hair deformation responses of Vicia hirsuta to the NGR234 NF purifications.
Fig. S2 The nuclear localization of the calcium oscillation response in a variety of legume species.
Fig. S3 Root hair deformation in Alnus glutinosa following treatment with Frankia exudates.
Fig. S4 Calcium traces in Alnus glutinosa treated with NGR234 NF and in Medicago truncatula treated with Frankia exudates.


