Abstract
Glycans play important roles in such cell-cell interactions as signaling and adhesion, including processes involved in pathogenic infections, cancers, and neurological diseases. Glycans are biosynthesized by multiple glycosyltransferases (GTs), which function sequentially. Excluding mucin-type O-glycosylation, the non-reducing terminus of glycans is biosynthesized in the Golgi apparatus after the reducing terminus is biosynthesized in the ER. In the present study, we performed genome-wide analyses of human GTs by investigating the degree of conservation of homologues in other organisms, as well as by elucidating the phylogenetic relationship between cephalochordates and urochordates, which has long been controversial in deuterostome phylogeny. We analyzed 173 human GTs and functionally linked glycan synthesis enzymes by phylogenetic profiling and clustering, compiled orthologous genes from the genomes of other organisms, and converted them into a binary sequence based on the presence (1) or absence (0) of orthologous genes in the genomes. Our results suggest that the non-reducing terminus of glycans is biosynthesized by newly evolved GTs. According to our analysis, the phylogenetic profiles of GTs resemble the phylogenetic tree of life, where deuterostomes, metazoans, and eukaryotes are resolved into separate branches. Lineage-specific GTs appear to play essential roles in the divergence of these particular lineages. We suggest that urochordates lose several genes that are conserved among metazoans, such as those expressing sialyltransferases, and that the Golgi apparatus acquires the ability to synthesize glycans after the ER acquires this function.
Keywords: glycan, phylogenetic profile, evolution, glycosyltransferase, human, ascidian, sialyltransferase
The term glycan encompasses the carbohydrate portion of glycoconjugates but not oligosaccharides or polysaccharides. Glycans are widespread in eukaryotes, bacteria, and archaea [1], and similar structures are present among different taxa, such as yeasts, plants, insects, and vertebrates [2]. The high conservation of glycans across several taxa demonstrates their important biological roles [2]. Glycans are categorized as N-glycans, O-glycans, proteoglycans, glycophosphatidylinositol (GPI) anchors, and glycosphingolipids [3]. More than 50% of all human proteins are glycosylated by processes, such as N-glycosylation and O-glycosylation [4]. Glycosylation has an important role in many cellular functions, including adhesion and signal transduction between cells [5–8], which are also processes associated with pathogenic infection [9], cancer [10], and neurological disorders, such as muscular dystrophy and Alzheimer’s disease [11,12]. Glycosphingolipids comprise a glycan and a ceramide moiety, where the latter is made of an amide-bonded fatty acid and sphingosine. Glycosphingolipids are expressed on the outer surface of the plasma membrane and appear to play roles as constituents of the lipid raft during signal transduction [13]. Glycosphingolipids are separated into two main groups: acidic and neutral glycosphingolipids. The acidic glycosphingolipids include the sialic acid-containing glycosphingolipids referred to as gangliosides, the sulfate-containing glycosphingolipids referred to as sulfatides, and uronic acid-containing glycosphingolipids. Gangliosides are expressed widely in various tissues, where they are involved in cell-cell recognition, myelin-axon interactions, inflammation, and interactions with growth factors on the membrane; they also act as regulatory elements in the immune and nervous systems [14]. Sulfatides are expressed in the nervous system, kidney, immune system, and pancreas, and are involved in many biological processes, including homeostasis, insulin secretion, and bacterial/viral infections [15]. Uronic acid-containing glycosphingolipids have been reported in flies and bivalves [16], but their functions have not yet been elucidated.
Genomic analyses of the ascidian Ciona intestinalis (C. intestinalis) [17–20] and Florida lancelet Branchiostoma floridae (B. floridae) [21] suggest that the lancelet diverged to the mammalian lineage earlier than the ascidian. Previously, we analyzed the structure of glycosphingolipids among ascidians, such as C. intestinalis and Halocynthia roretzi (H. roretzi), where we characterized novel acidic glycosphingolipids, i.e., a sulfatide from C. intestinalis [22] and an uronic acid-containing glycosphingolipid from H. roretzi [23]. However, sialic acid-containing glycosphingolipids, such as gangliosides, were not identified in the ascidians despite the presence of gangliosides in Echinodermata [16] and Vertebrata [24] among Deuterostomia.
Glycosyltransferases (GTs) play central roles in glycan synthesis as membrane proteins, secreted proteins, and soluble-form proteins. GTs have a wide variety of localizations, such as on the membrane or in the lumen of the endoplasmic reticulum (ER) and the Golgi apparatus, as well as on the plasma membrane or in the cytoplasm, nucleus, extracellular regions, and mitochondria. In particular, the GTs considered in the present study are involved in the synthesis of glycans, such as N-glycans, O-glycans, proteoglycans, GPI anchors, and glycosphingolipids, and are generally membrane proteins which are localized to the ER and Golgi apparatus, although several GTs containing C-terminal KDEL-like sequences ([KRHQSA]-[DENQ]-E-L) localize to the lumen rather than the membrane of the ER [25]. During N-glycosylation and other types of O-glycosylation, excluding the mucin-type, the non-reducing terminus is synthesized in the Golgi apparatus, before which the glycan is synthesized at the reducing terminus by GTs in the ER [26]. The Golgi apparatus is divided into cis-, medial-, and trans-cisternae, and the trans-Golgi network. Glycan synthesis proceeds from the cis-cisterna to the trans-Golgi network, where a distinctive distribution of GTs is found in each cisterna [27]. Based on a genome-wide analysis, GTs were separated into two broad groups: enzymes that are widely conserved evolutionarily across different phyla and those that seem to be lineage-specific, appearing within each phylum [28]. For instance, homologous genes of GTs for the synthesis of high-mannose type N-glycan and the GPI anchor are present in all eukaryotic organisms, whereas those of O-glycan and glycosphingolipid are less common among the animal, plant, fungi, and protist kingdoms [28]. All GTs that synthesize N-glycan precursors and GPI anchors are localized in the ER and they comprise several families, including families with common topology and sequence motifs [29]. Therefore, at least some GTs are predicted to be derived from a common ancestral protein [29]. The genealogy of GTs was assessed based on sequence motif [30]. For instance, sialyltransferase sequences were evaluated based on sialyl motifs and analyzed using a phylogenetic tree [31,32]. In addition, a CAZy database was developed, which is dedicated to the display and analysis of genomic, structural, and biochemical information on carbohydrate-active enzymes [33]. In eukaryotic genomes, the GTs were comprehensively analyzed based on structural and functional characterizations of glycans.
Since the whole-genome sequencing of Haemophilus influenzae was completed in 1995 [34], whole-genome sequences of various other organisms have been completed and their phylogenetic relationships have been studied based on comparative genome analysis [35]. For instance, the whole-genome sequence of B. floridae was completed in 2008 and the phylogenetic relationship between Urochordata (ascidian) and Cephalochordata (amphioxus) was elucidated, which has been disputed for many years. In particular, it was shown that Urochordata are related more closely to vertebrates than Cephalochordata [21]. With the increasing availability of whole-genome sequences, techniques for the analysis of these sequences have also been developed [36]. For example, phylogenetic profiling is used to examine the phylogenetic co-occurrence of genes in different genomes [37]. Proteins that are present in the same metabolic pathway tend to be present or absent together in a given organism, thereby exhibiting a similar phylogenic profile. This also applies to proteins that comprise a protein complex. In the present study, we constructed a phylogenetic profile as a binary pattern, which depended on whether the sequences were orthologous (1) or not (0) [37]. This method can be used to identify the partners that interact with each protein, its associated metabolic pathway, subcellular localization, and phylogenetic history [37–39].
In the present study, using this phylogenetic profiling method, we conducted a genome-wide analysis, where we focused on GTs involved in the synthesis of five selected glycan categories (N-glycans, O-glycans, proteoglycans, GPI anchors, and glycosphingolipids), based on the concept that glycans fulfill the biological roles mentioned above by attaching to proteins or lipids, thereby determining the degree of conservation among organisms for each GT. The genome-wide analysis was based on the Gene TO Protein Structures and Functions (GTOP) database [40], which contains information regarding a vast amount of proteins, including homology data for over 1,000 organisms with sequenced genomes. Clustering analysis was performed using the phylogenetic profiles of 173 human GTs and functionally linked glycan synthesis enzymes. Furthermore, we investigated the degree of conservation among sialyltransferases to clarify the evolutionary positions of Urochordata and Cephalochordata.
Material and methods
Dataset
The following procedures were performed to obtain data related to GTs (specifically those involved in the five glycan categories). Data on 250 human GTs and functionally linked glycan synthetic enzymes were obtained from UniProtKB (release 2013_4) [41] using the following keyword search: “glycosyltransferase AND organism: human AND reviewed: yes.” These enzymes were then queried on KEGG pathway maps in the KEGG database (release 66.0) [42] to check their substrates and reaction products. The KEGG pathway maps used for reference were as follows: map00510 (N-glycan biosynthesis), map00512 (mucin type O-glycan biosynthesis), map00514 (other types of O-glycan biosynthesis), map00600 (sphingolipid metabolism), map00601 (glycosphingolipid biosynthesis–lacto and neolacto series), map00603 (glycosphingolipid biosynthesis–globo series), map00604 (glycosphingolipid biosynthesis–ganglio series), map00532 (glycosaminoglycan biosynthesis–chondroitin sulfate), map00534 (glycosaminoglycan biosynthesis–heparan sulfate), map00533 (glycosaminoglycan biosynthesis–keratan sulfate), and map00563 (GPI-anchor biosynthesis). When referring to the KEGG pathway maps, six enzymes that appeared to be involved in glycan syntheses were added to the dataset. These six enzymes were as follows: dolichol phosphate-mannose biosynthesis regulatory protein (UniProt accession: O94777), tumor suppressor candidate 3 (UniProt accession: Q13454), phosphatidylinositol N-acetylglucosaminyltransferase subunit Y (UniProt accession: Q3MUY2), phosphatidylinositol-glycan biosynthesis class X protein (UniProt accession: Q8TBF5), chondroitin sulfate glucuronyltransferase ( UniProt accession: Q9P2E5), and dolichol-phosphate mannosyltransferase subunit 3 (UniProt accession: Q9P2X0). The GENES IDs of human GTs were obtained from the glycan synthesis pathway map and transferred to UniProt accession numbers using the ID mapping tool in UniProtKB. Enzymes that are not related to glycan synthesis, such as glycogen synthase and poly ADP-ribose polymerase, were removed from this dataset. The selection criteria were as follows: the enzyme must catalyze the transfer of a sugar to a protein or lipid, excluding polysaccharide synthases (such as glycogen synthase), the large UGT family (a superfamily of GTs involved in the detoxification pathways of many drugs and pollutants, which is classified as GT1 in CAZy), and poly [ADP-ribose] polymerase. Several enzymes where “function” was not described in the general annotation section were also removed from the dataset. In the present study, 173 human GTs and functionally linked glycan synthetic enzymes listed in the GTOP database (release 2010_10) were present in the final dataset (Table S1).
These GTs were categorized into five categories, i.e., O-glycan, N-glycan, glycosphingolipid, proteoglycan, and GPI anchor, based on the metabolic map in the KEGG database and annotations in the UniProt database. Twenty-one multi-functional GTs were redundantly categorized, e.g., CMP-N-acetylneuraminate-β-galactosamide-α-2,3-sialyltransferase 1 (UniProt accession: Q11201) was categorized into three categories: O-glycan, glycosphingolipid, and proteoglycan.
Phylogenetic profile analysis
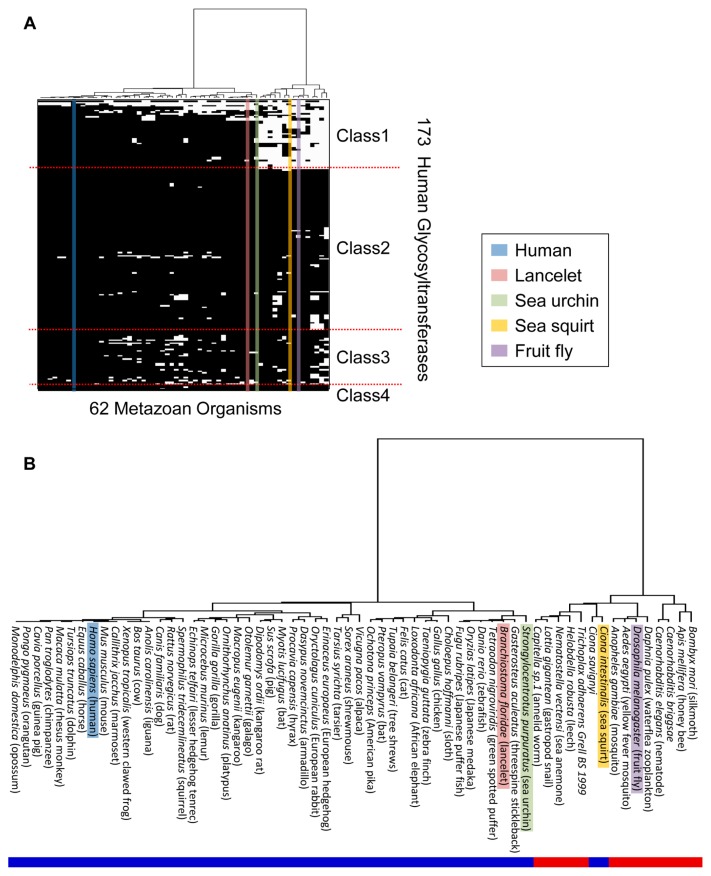
The phylogenetic profiles of 1,341 organisms (eukaryotes: 195; bacteria: 904; archaea: 67; and viruses: 175) were obtained for 173 human GTs and functionally linked glycan synthetic enzymes. The 1,341 organisms comprised 1,340 organisms recorded in the GTOP database and the sea urchin (Strongylocentrotus purpuratus) to represent the phylum Echinodermata, which diverged earlier from the last common ancestor of Chordata but diverged more recently compared with Insecta, thereby allowing a precise comparison among deuterostomes. We analyzed 173 human GTs and functionally linked glycan synthesis enzymes by phylogenetic profiling and clustering. First, proteins homologous to the 173 GTs and functionally linked glycan synthetic enzymes in the genomes of 1,341 organisms were coded as 1 (present) or 0 (absent). Briefly, we searched for each entry name using 173 human GTs and functionally linked glycan synthetic enzymes in UniProt. A genome-wide search for GTs was conducted by entering each UniProt entry name (e.g., GALT1_HUMAN) as a query in the text search field provided by the GTOP database. Among the resulting proteins, we selected a necessary protein derived from Homo sapiens. The following three criteria were adopted for the selection: records starting as “hsap” (abbreviation for Homo sapiens) with the lowest E-value, records having the highest score satisfying the former criterion, and records having the highest coverage satisfying the former criteria. In each of the records retrieved above, homolog information provided as hit count (E-value < 10−10) by the GTOP database was converted to binary data. If the hit count was 0 or ≥1, the homologous protein was defined as 0 (absent) or 1 (present), respectively. These steps were repeated for 173 human GTs and functionally linked glycan synthetic enzymes. Clustering of phylogenetic profiles for 173 GTs and functionally linked glycan synthetic enzymes and 1,341 organisms was performed using Ward’s method [43] based on the Manhattan distance. Subsequently, phylogenetic profiles were obtained for 62 metazoan organisms, which were analyzed by the clustering method. These clustering analyses were performed using gplot (version 2.8.0) in the statistical analysis software R (version 2.13.0; http://cran.r-project.org/).
Statistical analysis
In order to confirm whether the clustered GTs biosynthesize the same category of glycans in each class or not, the GTs were confirmed using Fisher’s exact test (P<0.01) with the statistical analysis software R. Fisher’s exact test was conducted for each of the five glycan categories: O-glycan, N-glycan, glycosphingolipid, proteoglycan, and GPI anchor. Comparisons were performed between classes where a specific class was tested against the other classes, e.g., Class 1 was tested against other groups: a group that comprised Class 1 and a group that comprised classes other than Class 1.
Search for sialyltransferases
All of the protein data of interest (“reference proteome sets”) for the following organisms were retrieved from the UniProt database: Urochordata C. intestinalis (Taxonomy ID: 7719), Cephalochordata B. floridae (Taxonomy ID: 7739), Echinodermata S. purpuratus (Taxonomy ID: 7668), and Arthropoda Drosophila melanogaster (Taxonomy ID: 7719). For each organism searched, the proteins already annotated as sialyltransferases were searched further using a keyword search from the UniProt database and then narrowed down by a BLAST search [44] (version 2.2.28) against 20 human sialyltransferases (E-value <10−10) using protein data for each organism as queries. In this case, query proteins that had the lowest E-values compared with human sialyltransferases were considered to belong to the same family of proteins.
Results
Clustering analysis of the phylogenetic profile of human GTs
UniProt entries for 250 human GTs and functionally linked glycan synthetic enzymes were retrieved by searching UniProtKB. In addition, KEGG gene IDs of 251 human GTs and functionally linked glycan synthetic enzymes were retrieved from the metabolic map of glycan synthesis in the KEGG pathway database. Overall, there were 146 overlapping records and 355 records were present in both sets of retrieved IDs and entries. Among these, 173 human GTs and functionally linked glycan synthetic enzymes were selected from the GTOP database and used as the dataset in the present study, where they belonged to the following five glycan categories: N-glycan, O-glycan, proteoglycan, GPI-anchor, and glycosphingolipid. During the selection process, we excluded the following GTs, which did not appear to be associated with the five glycan categories (N-glycans, O-glycans, proteoglycans, GPI anchors, and glycosphingolipids): GTs related to nucleobase metabolism, ADP-ribosylation, glucuronidation, queuine tRNA-ribosylation, vitamin metabolism, and polysaccharide metabolism, such as glycogen metabolism catalyzed by cytosolic GYG genes or hyaluronan biosynthesis catalyzed by plasma membrane HAS genes.
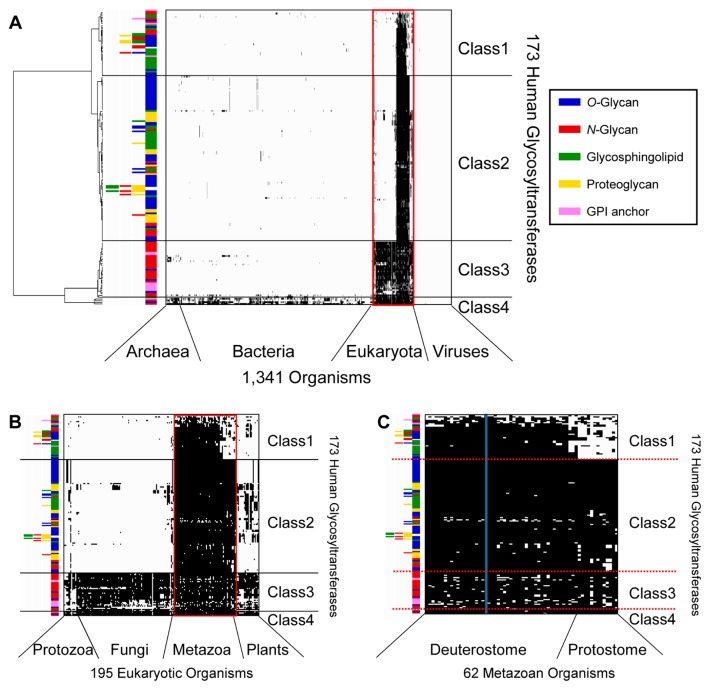
Clustering analysis based on phylogenetic profiling was conducted using the 173 human GTs and functionally linked glycan synthetic enzymes, where they were classified into four classes (Fig. 1 and Supplementary Table S1): 39 GTs mainly conserved in deuterostomes (Class 1), 97 GTs mainly conserved in metazoans (Class 2), 33 GTs mainly conserved in eukaryotes (Class 3), and four GTs conserved widely in eukaryotes, bacteria, and archaea (Class 4). The degree of conservation of GTs in the various biological lineages also reflected their order of appearance during evolution, with deuterostomes (Class 1) as the most recent, followed by metazoans (Class 2), eukaryotes (Class 3), and eukaryotes/bacteria/archaea (Class 4). The 173 GTs and functionally linked glycan synthetic enzymes were grouped into 43 GT families using the CAZy database and were classified in terms of the category of glycan catalyzed: 71 O-glycan-related enzymes, 50 N-glycan-related enzymes, 43 glycosphingolipid-related enzymes, 27 proteoglycan-related enzymes, and 12 GPI anchor-related enzymes. In addition, 21 GTs catalyzed multiple glycans (Supplementary Table S2).
Figure 1.
Clustering analysis of 173 human GTs and functionally linked glycan synthetic enzymes using Ward’s method based on the Manhattan distance. (A) Clustering analysis based on the phylogenetic profiles of 1,341 organisms. The black plot indicates the existence of proteins homologous to human GTs. The colored bars indicate the categories of glycans synthesized by GTs. The branch length on the left shows the distance between GTs. The organisms are arranged in the order in which they are listed in the GTOP database. (B) Magnified view of the 195 eukaryotic organisms enclosed within a red line in (A). (C) Magnified view of the 62 metazoan organisms enclosed within a red line in (B). The blue bar denotes the human profile.
In the selected 173 GTs, there were six GTs that localized only in the lumen of the ER but not on the membrane, although most of the 173 GTs were transmembrane proteins. Five of the six GTs contained an extreme C-terminal [KRHQSA]-[DENQ]-E-L sequence known as the ER-targeting sequence in the PROSITE database. These five GTs were as follows: UDP-glucose:glycoprotein glucosyl-transferase 1 (UniProt accession: Q9NYU2; CAZy GT24), UDP-glucose:glycoprotein glucosyltransferase 2 (UniProt accession: Q9NYU1; CAZy GT24), Procollagen galactosyl-transferase 1 (UniProt accession: Q8NBJ5; CAZy GT25), Procollagen galactosyltransferase 2 (UniProt accession: Q8IYK4; CAZy GT25), and EGF domain-specific O-linked N-acetylglucosamine transferase (UniProt accession: Q5NDL2; CAZy GT61). The remaining one GT [Protein O-glucosyltransferase 1 (UniProt accession: Q8NBL1; CAZy GT90)] contained an extreme C-terminal KTEL sequence identified as other ER lumen-resident motif [45].
Function and localization of GTs in each class
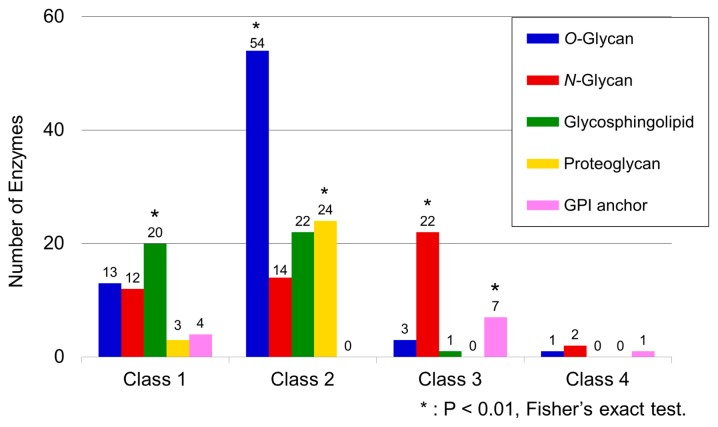
The distribution of GTs of a particular glycan type among the five selected glycan categories was heterogeneous across the different classes. In particular, GTs that catalyzed O-glycans appeared mainly in Class 2, followed by more infrequent appearances in classes 1, 3, and 4 (Fig. 2). GTs catalyzing N-glycan were classified mainly into classes 1, 2, and 3 but less so into Class 4. GTs catalyzing glycosphingolipid were classified mainly into classes 1 and 2, with only a minor appearance in Class 3. GTs involved in proteoglycan biosynthesis were classified mainly into Class 2 but were less abundant in Class 1. GTs catalyzing GPI anchors were classified mainly into Class 3 but were less abundant in Class 1 and only sparsely present in Class 4. The relationship between GT types and phylogenetic classes was not definite or exclusively one-to-one, but a Fisher’s exact test conducted on the 20 possible combinations of associations between four classes and five glycan categories confirmed the existence of the class-specificity expected from the uneven distribution of the GTs in each class. In this test, the class was considered specific if a significantly higher number of GTs of a particular type was present in a class than would have been expected by chance alone. This test detected a statistically significant bias in the presence of GTs for glycosphingolipids in Class 1, O-glycan/proteoglycan in Class 2, and N-glycan/GPI anchor in Class 3 (Fig. 2).
Figure 2.
Frequency distribution of GTs in each glycan type according to class. The numbers of GTs in each category of glycan synthesis are shown for each class. Class-specific glycan categories were assessed using Fisher’s exact test. Asterisks indicate categories that exceeded the numerical expectation value where P<0.01.
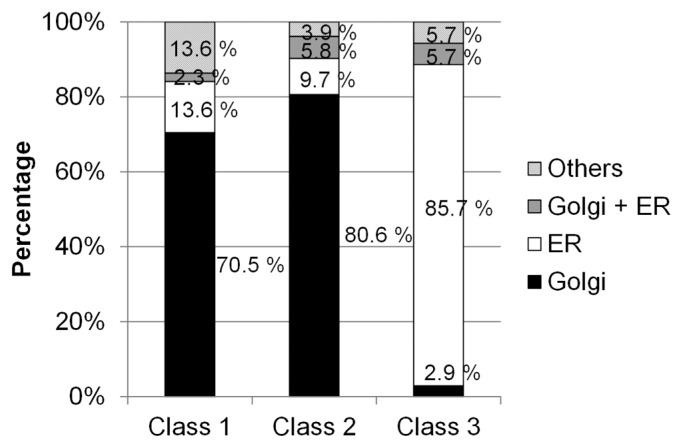
The subcellular localizations of GTs were acquired from UniProt annotation data. The results showed clearly that most of the GTs classified into Class 1 and Class 2 were localized in the Golgi apparatus and that most of the GTs classified into Class 3 were localized in the ER (Fig. 3). This reflects a trend where GTs conserved mainly in multicellular organisms were localized in the Golgi apparatus, whereas GTs that were conserved more widely were localized in the ER. We could not perform an accurate assessment of the class-specific GTs for Class 4 because of the limited number of GTs in this class.
Figure 3.
Subcellular localizations of GTs in each class. Ratios of Golgi-localized and ER-localized GTs. GTs that localized to both the Golgi apparatus and the ER are grouped into “Golgi + ER.” “Others” contain GTs that did not localize to the Golgi apparatus or the ER, i.e., GTs with unknown localizations.
In general, GTs acting in the Golgi apparatus and those acting in the ER were classified into separate classes. Thus, GTs situated upstream or downstream in the glycan synthesis pathway differed in their relative degree of evolution. The early phase of N-glycan processing is common in yeasts and slime molds among eukaryotes (Class 3), insects among metazoans (Class 2), and vertebrates among deuterostomes (Class 1). Therefore, the distribution of N-glycan-related GTs might reflect the evolutionary history of N-glycan processing.
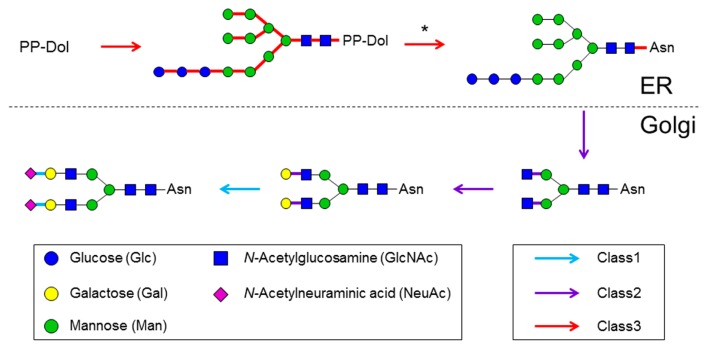
Next, we investigated the relationship between glycan synthesis pathways and the sequential order of GT actions in these pathways. Figure 4 provides a schematic map of the sequence of GTs involved in the N-glycan synthesis pathway. In the case of N-glycan, the GT classes differed between the ER and Golgi apparatus synthesis pathways. All of the high-mannose N-glycan syntheses and protein modifications by oligosaccharide transferases occur in the membrane of the ER [46]. Except for dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit 4 (UniProt accession: P0C6T2), which is a subunit of an oligosaccharide transferase complex, all of the GTs responsible for catalyzing high-mannose N-glycan were classified into Class 3. GTs that catalyze subsequent complex-type N-glycans in the Golgi apparatus were classified into Class 1 and Class 2. The N-glycan synthesis pathway was first catalyzed by Class 2 GTs and catalyzed subsequently by those in Class 1. In particular, all of the sialyltransferases catalyzing non-reducing terminus syntheses were classified into Class 1. Similarly, O-glycan synthases were classified into Class 1 and Class 2. Almost all of the glycosphingolipids are initiated by glucosylceramide synthesis, which involves the transfer of glucose to ceramide in the Golgi apparatus. Glucosylceramide synthase (UniProt accession: Q16739), which is responsible for the reaction, was classified into Class 3. In the glycosphingolipid synthesis pathway, glycan synthesis in the Golgi apparatus following the glucosylceramide synthesis was catalyzed only by GTs classified into Class 1 and Class 2. Most of the glycosphingolipid synthesis pathways were also first catalyzed by Class 2 GTs and then catalyzed subsequently by those in Class 1. Next, the sequential order of function of GTs, excluding those in the N-glycan, mucin type O-glycan, and glycosphingolipid categories, was confirmed. The glycan synthesis pathways were found to be first catalyzed by Class 3 GTs, followed by those in Class 2, and then finally by those in Class 1. Thus, in the glycan synthesis pathway, Class 3 GTs acted upstream of Class 2 GTs, and Class 1 GTs acted downstream of Class 2 GTs. These results show that Class 3 GTs, which are conserved widely in eukaryotes, catalyze reducing terminus syntheses, and that Class 1 GTs, which are conserved only in higher animals, catalyze non-reducing terminus syntheses.
Figure 4.
Color mapping of GT class against the N-glycan synthesis pathway. Pathways were generated based on the “N-glycan biosynthesis” map (KEGG pathway ID map00510) in the KEGG pathway. Arrows and lines between sugar residues are denoted by colors that represent each class. In the pathway of the oligosaccharyltransferase complex, which is denoted by an asterisk, only dolichyl-diphosphooligosaccharide-protein GT subunit 4 was classified into Class 1.
Class 4 comprised four GTs: the UDP-N-acetylglucosamine-peptide N-acetylglucosaminyltransferase 110 kDa subunit (OGT; UniProt accession: O15294), which is related to O-GlcNAc modification; phosphatidylinositol N-acetylglucosaminyltransferase subunit A (UniProt accession: P37287), related to GlcNAc-phosphatidylinositol synthesis; dolichol-phosphate mannose synthase subunit 1 (UniProt accession: O60762), related to mannose-P-dolichol synthesis; and Dol-P-glucosyltransferase (UniProt accession: Q9Y673), related to glucose-P-dolichol synthesis. Two of these GTs catalyze the initial reactions of glycan synthesis, which transfer monosaccharaides to the target protein or lipid molecule. These results demonstrate that Class 4 GTs (conserved widely in eukaryotes, bacteria, and archaea) primarily transfer a monosaccharide to an aglycone to form a glycoconjugate and that GTs other than those classified into Class 4 subsequently transfer a monosaccharide to the resulting glycoconjugate. In summary, we found that glycans were synthesized by GTs in the following order: Class 3/Class 4 GTs, Class 2 GTs, and Class 1 GTs. Thus, the glycan structures in the non-reducing terminus, which are situated downstream in the glycan synthetic pathway, are synthesized by the GTs conserved only in higher animals and those in the reducing terminus, which are situated upstream in the pathway, are synthesized by the widely conserved GTs.
Evolutionary analysis in metazoans
For GTs involved in the five selected glycan categories, we conducted an evolutionary analysis of GTs based on clustering using 62 metazoan organisms, as shown in Figures 1C and 5, which were mainly classified as deuterostomes and protostomes. The results show that conserved GTs differed between these two groups. However, in chordates, ascidians (C. intestinalis and C. savignyi) among deuterostome were arranged as protostomes despite the fact that ascidians are closely related to vertebrates. We found that ascidians were similar to protostomes because they did not contain most of the well-conserved Class 1 GTs (Supplementary Table S3). This result suggests that GT evolution differed in ascidians compared with other deuterostomes.
Figure 5.
Phylogenetic analysis of 62 organisms in metazoa. (A) Clustering analysis using Ward’s method based on the Manhattan distance. The five colors show conserved GTs in representative model organisms. The GTs listed on the vertical axis correspond to those listed on the vertical axis in Figure 1. (B) Magnified view of the phylogenetic tree in (A), as well as the corresponding names of the 62 organisms. The red and blue bars denote deuterostomes and protostomes, respectively.
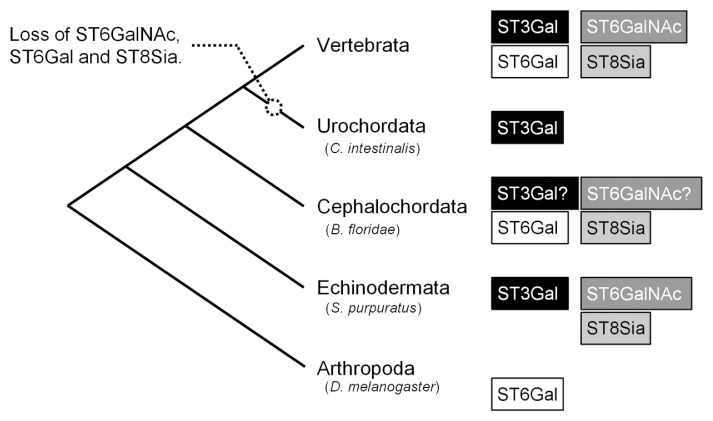
There were 20 sialyltransferases among the 44 human Class 1 GTs. Given their large number, our analysis focused on the evolution of metazoan sialyltransferases. Sialyltransferases, which belong to GT29 in the CAZy database, are grouped into four families (ST3Gal, ST6Gal, ST6GalNAc, and ST8Sia) based on their glycosyl linkages [31,32]. The invertebrate sialyltransferase families recorded in August 2013 in the UniProt database are as follows: ST3Gal family in Urochordata C. intestinalis, ST6Gal/ST8Sia families in Cephalochordata B. floridae, ST6GalNAc/ST8Sia/ST3Gal families in Echinodermata S. purpuratus, and ST6Gal family in Arthropoda D. melanogaster (Fig. 6). Using BLAST search with 20 human sialyltransferases as queries, we confirmed the existence of uncharacterized proteins similar to ST3Gal/ST6GalNAc families in B. floridae (Fig. 6 and Supplementary Figure S1). We found no sialyltransferases other than the previously annotated sialyltransferase family based on BLAST searches in C. intestinalis, S. purpuratus, and D. melanogaster.
Figure 6.
Evolution of sialyltransferases in metazoa. Figure based on four types of sialyltransferase families (ST3Gal, ST6GalNAc, ST6 Gal, and ST8Sia) recorded in the UniProt database. Families denoted by question marks indicate the presence of unknown proteins among Cephalochordata based on BLAST search.
Discussion
In the present study, we focused on the five selected glycan categories known as complex glycoconjugate glycans, and investigated the evolutionary origins of glycans in four classes of GTs (Fig. 1). Clustering analyses of GTs and organisms were conducted using phylogenetic profiles, which arranged ascidians, commonly grouped as deuterostomes, as protostomes. We found that ascidians were closely related to invertebrates, where they appeared to have diverged from the common ancestor of invertebrates after the divergence of Cephalochordata and Echinodermata from the common ancestor [21]. However, we found that most of the Class 1 GTs, which are largely conserved in deuterostomes, were not conserved in ascidians. Nearly half of the Class 1 GTs (20 of 44) were sialyltransferases with a conserved and common functional motif, thereby suggesting that sialyltransferases diverged from a common ancestral protein [31,32]. After diverging from Arthropoda, the common ancestor of invertebrates and echinoderms appears to have conserved sialyltransferases in the ST3Gal, ST6GalNAc, ST6Gal, and ST8Sia families. This was inferred based on the minimum evolution principle and presence or absence of sialyltransferase families (Fig. 6). However, three of the four sialyltransferase families were not found in ascidians. Ascidian genomes differ significantly from those of other deuterostomes as follows: gene loss and intron loss, genome shortening by genome reorganization [21], and acquisition of a cellulose synthase enzyme gene via horizontal transfer [47]. The B. floridae genome project showed that B. floridae diverged primarily from the last common ancestor of vertebrates and that C. intestinalis then diverged but whole genome duplication in C. intestinalis was difficult to determine because of high levels of genomic reconstruction/omissions. In particular, our phylogenetic profiling of GTs in ascidians revealed the loss of most sialyltransferases. This finding is consistent with the dramatic loss of genes found in ascidians.
We found that Class 1 GTs were conserved mainly in deuterostomes and all sialyltransferases were classified into Class 1. Sialoglycans exhibit tissue-specific and stage-specific expression and are involved in cellular differentiation, growth control, signaling, and fertilization [48–50]. Sialic acid plays a role in enhancing mucin viscosity on the surface of mucin [51], thereby masking glycans that can be recognized by antigenic glycans exploited by pathogens [52]. Therefore, we suggest that deuterostomes utilized sialic acid during the course of evolution to acquire immunity from pathogens, as well as facilitating complex cellular differentiation and signal transduction systems.
O-glycan synthesis GTs, which are mainly conserved in metazoans, were classified into Class 1 and Class 2. In total, 20 genes for highly O-glycan attached mucin proteins have been genetically cloned, which show tissue-specific expression [53]. Secretory mucins in the digestive tract and trachea help to capture microorganisms and provide protection from digestive enzymes [53]. Therefore, we hypothesize that these O-glycans have been important in the evolution of multi-cellularity by allowing an organism to differentiate its cells and to develop specialized tissues.
GTs classified into Class 1 and Class 2 localized mainly to the Golgi apparatus. Similarly, most GTs used for the synthesis of glycoproteins and glycosphingolipids localized to the Golgi apparatus (Supplementary Table S1). Glycans synthesized in the Golgi apparatus are involved mainly in cell-cell interactions. Transmembrane-type mucins are related to signal transduction and cellular adhesion [53]. Integrin, a cellular adhesion molecule, is attached to complex-type N-glycans, which regulate cellular adhesion by integrin [54]. In addition, glycosphingolipids appear to form lipid rafts with transmembrane-type signaling proteins and are involved in signal transduction [13]. Thus, we suggest that higher animals developed unique mechanisms for cellular recognition and cellular adhesion. GTs for high-mannose type N-glycans were classified into Class 3. High-mannose type N-glycan processing plays an important role in mechanisms related to protein quality control [55]. For example, UDP-glucose: glycoprotein glucosyltransferases 1 (UGGT1; UniProt accession: Q9NYU2) transfers glucose to the N-glycan terminus of proteins and its interaction with the protein ensures normal protein folding; otherwise, it signals the protein for degradation [55]. Similar to the GTs for high-mannose type N-glycan, UGGT1 was classified into Class 3 and is conserved among most eukaryotes. Therefore, we suggest that eukaryotic evolution was stimulated by the acquisition of a quality control mechanism related to protein folding.
The GTs for O-glycans classified into Class 3 were protein O-mannosyltransferases 1 and 2, which transfer mannose to Ser and Thr residues in a protein. O-Man structures by these GTs are regarded as core structures of α-dystroglycans in mammals [56]. GTs in Class 3 are conserved among eukaryotes, most of which are related to the synthesis of high-mannose type N-glycans or GPI anchors. The pathways between N-glycan and GPI-anchor synthesis are similar because glycan is synthesized on the scaffold of a lipid anchor [46,57], where dolichol-phosphate sugars are utilized as donor substrates [29]. Therefore, they are thought to have a common genetic origin [29]. Protein O-mannosyltransferases 1 and 2 may have been derived from a common ancestral protein because they utilize Dol-P-Man as a donor substrate, which is similar to the mannosyltransferases associated with N-glycan or GPI-anchor syntheses.
The UDP-N-acetylglucosamine-peptide N-acetylglucosaminyltransferase 110-kDa subunit (OGT; UniProt accession: O15294), which is localized in the nucleus and cytoplasm, was classified into Class 4, thereby clarifying that OGT has been widely conserved among eukaryotes, bacteria, and archaea. In the present study, we excluded cytosolic GYG genes related to glycogen biosynthesis and plasma membrane HAS genes related to hyaluronan biosynthesis to concentrate on GTs from the five selected glycan categories, i.e., N-glycans, O-glycans, proteoglycans, GPI anchors, and glycosphingolipids. The resulting GTs related to the five glycan categories mainly comprised GTs that localized on the membrane of the Golgi apparatus or the ER. However, OGT was included in the present study because O-GlcNAcylation by OGT may be regarded as a specific type of O-glycan. It is known that O-GlcNAcylation is involved in extensive crosstalk with phosphorylation during the regulation of mRNA transcription and signaling in proximal or identical amino-acid residues [58]. It should be noted that such OGT was acquired in the very early stage of biological evolution.
We found that GTs from the five selected glycan categories tend to function in sequential order according to their class type, starting with Class 3 enzymes and ending with Class 1 enzymes in the glycan synthesis pathway. Among the Animalia, homologous GTs to human GTs were conserved widely in Class 3 GTs among eukaryotes, Class 2 GTs among metazoans, and Class 1 GTs only among deuterostomes. Thus, the order in which GTs have evolved corresponds to the order in which the GTs function. The Golgi apparatus may have acquired the function of glycan synthesis after the acquisition of the ER because Class 3 mainly comprises the ER-localized GTs and because Class 1 and 2 mainly comprise the Golgi apparatus-localized GTs.
The Golgi apparatus is made of multiple compartments. Initial glycan synthesis reactions occur within the cis-Golgi region, which is close to the ER, whereas late reactions occur within the trans-Golgi region [27]. Among the GTs that localize to the Golgi apparatus, α-1,3-mannosyl-glycoprotein 2-β-N-acetylglucosaminyltransferase (GlcNAc-TI) mainly localizes to the medial-Golgi, β-1,4-galactosyltransferase 1 (β4Gal-T1) localizes to the trans-Golgi, and β-galactoside α-2,6-sialyltransferase 1 (ST6Gal I) localizes to the trans-Golgi and the trans-Golgi network [59]. In the present study, we confirmed that GlcNAc-TI and β4Gal-T1 were classified into Class 2, and that ST6Gal I was classified into Class 1. Therefore, Class 1 GTs may localize closer to the trans-Golgi network side of the pathway.
Most of the CAZy GT families were clearly grouped into a single class except for GT1, GT4, and GT61 (Supplementary Table S2). The members of GT1 and GT4 were distributed unevenly across multiple classes according to their type. GT1 was divided into Class 2 (glycosphingolipid) and Class 3 (N-glycan). GT4 was divided into Class 3 (N-glycan) and Class 4 (GPI anchor). The GT61 proteins, although being involved in O-glycan syntheses, were classified into Class 1 and Class 2.
The present study was conducted based only on a genome level analysis rather than a structure level analysis. If additional molecular level analyses of the GTs referred to in the study are considered, it is likely that the molecular functions of GTs will differ to some extent.
In conclusion, our phylogenetic profiling of GTs clarified the phylogenetic classification of ascidians with respect to deuterostomes, thereby suggesting that organisms have evolved by elongating glycans to the common basal glycan structure.
Supplementary Information
Acknowledgments
We would like to thank Mr. Shigetaka Sakamoto for transferring the genomic sea urchin data to GTOP (National Institute of Genetics, Mishima, Japan). This work was supported partly by a grant to the Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to SF).
Footnotes
Conflict of Interest
All the authors declare that they have no conflict of interest.
Author Contributions
M. I. designed the experiments. T. T. and H. K. performed the experiments. T. T., H. K., S. F., Y. T. and M. I. analyzed the data. H. K., T. T. and M. I. wrote the manuscript.
References
- 1.Esko JD, Doering TL, Raetz CRH. Eubacteria and archaea. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, et al., editors. Essentials of Glycobiology. 2nd edition. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2009. pp. 293–307. [PubMed] [Google Scholar]
- 2.Varki A, Freeze HH.&Gagneux P. Evolution of glycan diversity. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, et al., editors. Essentials of Glycobiology. 2nd edition. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2009. pp. 281–292. [PubMed] [Google Scholar]
- 3.Lowe JB, Marth JD. A genetic approach to Mammalian glycan function. Annu Rev Biochem. 2003;72:643–691. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]
- 4.Apweiler R, Hermjakob H.&Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 5.Fukuda M, Hiraoka N.&Yeh JC. C-type lectins and sialyl Lewis X oligosaccharides Versatile roles in cell-cell interaction. J Cell Biol. 1999;147:467–470. doi: 10.1083/jcb.147.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hakomori S. Structure and function of glycosphingolipids and sphingolipids: Recollections and future trends. Biochim Biophys Acta. 2008;1780:325–346. doi: 10.1016/j.bbagen.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu J, Sato Y, Kariya Y, Isaji T, Taniguchi N.&Fukuda T. A mutual regulation between cell-cell adhesion and N- glycosylation: implication of the bisecting GlcNAc for biological functions. J Proteome Res. 2009;8:431–435. doi: 10.1021/pr800674g. [DOI] [PubMed] [Google Scholar]
- 8.Kitazume S, Imamaki R, Ogawa K, Komi Y, Futakawa S, Kojima S, et al. α2,6-sialic acid on platelet endothelial cell adhesion molecule (PECAM) regulates its homophilic interactions and downstream antiapoptotic signaling. J Biol Chem. 2010;285:6515–6521. doi: 10.1074/jbc.M109.073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlsson KA. Meaning and therapeutic potential of microbial recognition of host glycoconjugates. Mol Microbiol. 1998;29:1–11. doi: 10.1046/j.1365-2958.1998.00854.x. [DOI] [PubMed] [Google Scholar]
- 10.Li M, Song L.&Qin X. Glycan changes: cancer metastasis and anti-cancer vaccines. J Biosci. 2010;35:665–673. doi: 10.1007/s12038-010-0073-8. [DOI] [PubMed] [Google Scholar]
- 11.Wells L. The O-mannosylation pathway: glycosyltransferases and proteins implicated in congenital muscular dystrophy. J Biol Chem. 2013;288:6930–6935. doi: 10.1074/jbc.R112.438978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ariga T, Miyatake T.&Yu RK. Role of proteoglycans and glycosaminoglycans in the pathogenesis of Alzheimer’s disease and related disorders: amyloidogenesis and therapeutic strategies—a review. J Neurosci Res. 2010;88:2303–2315. doi: 10.1002/jnr.22393. [DOI] [PubMed] [Google Scholar]
- 13.Kasahara K.&Sanai Y. Functional roles of glycosphingolipids in signal transduction via lipid rafts. Glycoconj J. 2000;17:153–162. doi: 10.1023/a:1026576804247. [DOI] [PubMed] [Google Scholar]
- 14.Lopez PHH.&Schnaar RL. Gangliosides in cell recognition and membrane protein regulation. Curr Opin Struct Biol. 2009;19:549–557. doi: 10.1016/j.sbi.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi T.&Suzuki T. Role of sulfatide in normal and pathological cells and tissues. J Lipid Res. 2012;53:1437–1450. doi: 10.1194/jlr.R026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itonori S.&Sugita M. Glycophylogenetic aspects of lower animals. In: Boons G, Lee YC, Suzuki A, Taniguchi N, Voragen AGJ, editors. Comprehensive Glycoscience 3: Biochemistry of Glycoconjugate Glycans Carbohydrate-mediated Interactions. Oxford: Elsevier; 2007. pp. 253–284. [Google Scholar]
- 17.Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 18.Satou Y, Yamada L, Mochizuki Y, Takatori N, Kawashima T, Sasaki A, et al. A cDNA resource from the basal chordate Ciona intestinalis. Genesis. 2002;33:153–154. doi: 10.1002/gene.10119. [DOI] [PubMed] [Google Scholar]
- 19.Hubbard TJP, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satou Y, Mineta K, Ogasawara M, Sasakura Y, Shoguchi E, Ueno K, et al. Improved genome assembly and evidence-based global gene model set for the chordate Ciona intestinalis: new insight into intron and operon populations. Genome Biol. 2008;9:R152. doi: 10.1186/gb-2008-9-10-r152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 22.Yamada S, Matsumuro Y, Inoue T, Kitamura T, Itonori S, Sugita M, et al. A Novel sulfatide, GlcCer-I6 sulfate, from the ascidian Ciona intestinalis. J Oleo Sci. 2007;56:129–136. doi: 10.5650/jos.56.129. [DOI] [PubMed] [Google Scholar]
- 23.Ito M, Matsumuro Y, Yamada S, Kitamura T, Itonori S.&Sugita M. Isolation and characterization of a novel uronic acid-containing acidic glycosphingolipid from the ascidian Halocynthia roretzi. J Lipid Res. 2007;48:96–103. doi: 10.1194/jlr.M600296-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Yu RK, Yanagisawa M.&Ariga T. Glycosphingolipid structures. In: Boons G, Lee YC, Suzuki A, Taniguchi N, Voragen AGJ, editors. Comprehensive Glycoscience 1: Introduction to Glycoscience Synthesis of Carbohydrates. Elsevier; Oxford: 2007. pp. 73–122. [Google Scholar]
- 25.The KDEL-like sequences are described in the PROSITE entry PDOC00014 (PS00014/ER_TARGET)
- 26.Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA.&Tabak LA. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 2012;22:736–756. doi: 10.1093/glycob/cwr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moremen KW, Tiemeyer M.&Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashimoto K, Tokimatsu T, Kawano S, Yoshizawa AC, Okuda S, Goto S, et al. Comprehensive analysis of glycosyltransferases in eukaryotic genomes for structural and functional characterization of glycans. Carbohydr Res. 2009;344:881–887. doi: 10.1016/j.carres.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Oriol R, Martinez-Duncker I, Chantret I, Mollicone R.&Codogno P. Common origin and evolution of glycosyltransferases using Dol-P-monosaccharides as donor substrate. Mol Biol Evol. 2002;19:1451–1463. doi: 10.1093/oxfordjournals.molbev.a004208. [DOI] [PubMed] [Google Scholar]
- 30.Togayachi A, Sato T.&Narimatsu H. Comprehensive enzymatic characterization of glycosyltransferases with a β3GT or β4GT motif. Methods Enzymol. 2006;416:91–102. doi: 10.1016/S0076-6879(06)16006-1. [DOI] [PubMed] [Google Scholar]
- 31.Harduin-Lepers A, Mollicone R, Delannoy P.&Oriol R. The animal sialyltransferases and sialyltransferase-related genes: a phylogenetic approach. Glycobiology. 2005;15:805–817. doi: 10.1093/glycob/cwi063. [DOI] [PubMed] [Google Scholar]
- 32.Harduin-Lepers A. Comprehensive analysis of sialyltransferases in vertebrate genomes. Glycobiol Insights. 2010;2:29–61. [Google Scholar]
- 33.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzyme database (CAZy) in 2013. Nucleic Acids Res. 2013;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 35.Kawashima T, Shoguchi E, Satou Y.&Satoh N. Comparative genomics of invertebrates. In: Brown JR, editor. Comparative genomics: Basic and Applied Research. CRC Press; Boca Raton: 2008. pp. 87–104. [Google Scholar]
- 36.Hanson AD, Pribat A, Waller JC.&de Crécy-Lagard V. ‘Unknown’ proteins and ‘orphan’ enzymes: the missing half of the engineering parts list–and how to find it. Biochem J. 2010;425:1–11. doi: 10.1042/BJ20091328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D.&Yeates TO. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcotte EM, Xenarios I, van Der Bliek AM, Eisenberg D. Localizing proteins in the cell from their phylogenetic profiles. Proc Natl Acad Sci USA. 2000;97:12115–12120. doi: 10.1073/pnas.220399497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamanishi Y, Itoh M.&Kanehisa M. Extraction of organism groups from phylogenetic profiles using independent component analysis. Genome Inform. 2002;13:61–70. [PubMed] [Google Scholar]
- 40.Fukuchi S, Homma K, Sakamoto S, Sugawara H, Tateno Y, Gojobori T, et al. The GTOP database in 2009: updated content and novel features to expand and deepen insights into protein structures and functions. Nucleic Acids Res. 2009;37:D333–D337. doi: 10.1093/nar/gkn855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The UniProt Consortium. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanehisa M, Goto S, Sato Y, Furumichi M.&Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward HJ. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58:236–244. [Google Scholar]
- 44.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raykhel I, Alanen H, Salo K, Jurvansuu J, Nguyen VD, Latva-Ranta M, et al. A molecular specificity code for the three mammalian KDEL receptors. J Cell Biol. 2007;179:1193–1204. doi: 10.1083/jcb.200705180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones J, Krag SS.&Betenbaugh MJ. Controlling N-linked glycan site occupancy. Biochim Biophys Acta. 2005;1726:121–137. doi: 10.1016/j.bbagen.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 47.Nakashima K, Yamada L, Satou Y, Azuma J.&Satoh N. The evolutionary origin of animal cellulose synthase. Dev Genes Evol. 2004;214:81–88. doi: 10.1007/s00427-003-0379-8. [DOI] [PubMed] [Google Scholar]
- 48.Saito M.&Sugiyama K. Tissue-specific expression of c-series gangliosides in the extraneural system. Biochim Biophys Acta. 2000;1474:88–92. doi: 10.1016/s0304-4165(99)00222-6. [DOI] [PubMed] [Google Scholar]
- 49.Varki A.&Schauer R. Sialic Acids. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, et al., editors. Essentials of Glycobiology. 2nd edition. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2009. pp. 199–217. [PubMed] [Google Scholar]
- 50.Yu RK, Tsai YT.&Ariga T. Functional roles of gangliosides in neurodevelopment: an overview of recent advances. Neurochem Res. 2012;37:1230–1244. doi: 10.1007/s11064-012-0744-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishibashi Y, Takayama G, Inouye Y.&Taniguchi A. Carbocisteine normalizes the viscous property of mucus through regulation of fucosylated and sialylated sugar chain on airway mucins. Eur J Pharmacol. 2010;641:226–228. doi: 10.1016/j.ejphar.2010.05.045. [DOI] [PubMed] [Google Scholar]
- 52.Varki A.&Gagneux P. Multifarious roles of sialic acids in immunity. Ann NY Acad Sci. 2012;1253:16–36. doi: 10.1111/j.1749-6632.2012.06517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brockhausen I, Schachter H.&Stanley P. O-GalNAc glycans. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, et al., editors. Essentials of Glycobiology. 2nd edition. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2009. pp. 115–127. [PubMed] [Google Scholar]
- 54.Gu J, Isaji T, Xu Q, Kariya Y, Gu W, Fukuda T, et al. Potential roles of N-glycosylation in cell adhesion. Glycoconj J. 2012;29:599–607. doi: 10.1007/s10719-012-9386-1. [DOI] [PubMed] [Google Scholar]
- 55.Moremen KW.&Molinari M. N-linked glycan recognition and processing: the molecular basis of endoplasmic reticulum quality control. Curr Opin Struct Biol. 2006;16:592–599. doi: 10.1016/j.sbi.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobson CM, Hempel SJ, Stalnaker SH, Stuart R.&Wells L. O-Mannosylation and human disease. Cell Mol Life Sci. 2013;70:2849–2857. doi: 10.1007/s00018-012-1193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Orlean P.&Menon AK. Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J Lipid Res. 2007;48:993–1011. doi: 10.1194/jlr.R700002-JLR200. [DOI] [PubMed] [Google Scholar]
- 58.Butkinaree C, Park K, Hart GW. O-Linked β-N-Acetylglucosamine (O-GlcNAc): Extensive Crosstalk with Phosphorylation to Regulate Signaling and Transcription in Response to Nutrients and Stress. Biochim Biophys Acta. 2010;1800:96–106. doi: 10.1016/j.bbagen.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Opat AS, van Vliet C.&Gleeson PA. Trafficking and localization of resident Golgi glycosylation enzymes. Biochimie. 2001;83:763–773. doi: 10.1016/s0300-9084(01)01312-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.