Abstract
Highly transcribed guanine-run containing sequences, in Saccharomyces cerevisiae, become unstable when topoisomerase I (Top1) is disrupted. Topological changes, such as the formation of extended RNA:DNA hybrids or R-loops or non-canonical DNA structures including G-quadruplexes has been proposed as the major underlying cause of the transcription-linked genome instability. Here, we report that R-loop accumulation at a guanine-rich sequence, which is capable of assembling into the four-stranded G4 DNA structure, is dependent on the level and the orientation of transcription. In the absence of Top1 or RNase Hs, R-loops accumulated to substantially higher extent when guanine-runs were located on the non-transcribed strand. This coincides with the orientation where higher genome instability was observed. However, we further report that there are significant differences between the disruption of RNase Hs and Top1 in regards to the orientation-specific elevation in genome instability at the guanine-rich sequence. Additionally, genome instability in Top1-deficient yeasts is not completely suppressed by removal of negative supercoils and further aggravated by expression of mutant Top1. Together, our data provide a strong support for a function of Top1 in suppressing genome instability at the guanine-run containing sequence that goes beyond preventing the transcription-associated RNA:DNA hybrid formation.
INTRODUCTION
Genome instability—recombination and mutagenesis—is elevated by highly active transcription (1–7) (see (8,9) for review). The transcription complex itself can physically obstruct other DNA metabolic processes such as replication, especially when the direction of the transcription is in a collisional orientation with the movement of replication fork (10–12). The unwinding of the duplex DNA, mandatory for transcription process, transiently produces single stranded DNA regions of the non-transcribed strand (NTS) that are highly susceptible to chemical and enzymatic modifications. This effect can be aggravated when the nascent RNA stably anneals to the template DNA strand (transcribed strand – TS) producing extensive stretches of RNA:DNA hybrids or R-loops (13). When unresolved, R-loops lead to genome instability additionally by obstructing DNA replication (14) or by generating DSB via structure specific endonucleotic activity (15). R-loop formation in lieu of duplex DNA restoration ensues when transcription elongation or important co-transcriptional processes such as splicing, ribonucleoprotein (RNP) assembly and resolution of DNA topological stress are disturbed (16–18). Genome instability associated with R-loops is suppressed by RNase H enzymes, which degrades RNA hybridized to DNA and thereby prevents nucleation of RNA:DNA hybrids (19–21). Extensive R-loop accumulation, particularly at the highly transcribed rDNA locus and the tRNA genes, is observed in absence of this family of enzymes (22,23).
Conditions conducive to R-loop formation—open chromatin structure, DNA strand separation, and negative helical torsion—are similarly conducive to the structural transition of the duplex B-form DNA into non-canonical secondary structures when the transcribed regions contain repetitive sequences (24,25). Genome instability associated with these secondary structure-forming sequences becomes exacerbated under high transcription conditions, which promotes the folding of repetitive DNA strands into the non-B structure formation on the NTS as well as the stable formation of RNA:DNA hybrids involving the TS (26,27).
The four-stranded G-quadruplexes or G4 DNA, which contains a stacked array of multiple G-quartets comprised of four guanines interacting in a planar configuration, is another non-B secondary DNA structure capable of inducing genome instability by obstructing normal DNA transactions (28–31). Sequence motifs with potential to form G4 DNA are enriched at telomeres and G-rich immunoglobulin switch regions and frequently associated with unstable genomic loci including proto-oncogenes (32). Recent bioinformatic analyses of genes involved in chromosomal translocations in lymphoid cancers identified potential G4 DNA-forming sequences near many of the mapped translocation breakpoints (33). As for the immunoglobulin switch regions, chromosomal translocations involving this locus have long been observed in various cancer types (34). For several of the translocation-linked G4 motifs Including those at the major breakpoint region of the BCL2 gene involved in t(14;18) translocation in follicular lymphoma (35) and the TCF3 gene involved in t(1;19) in acute lymphoblastic leukemia (36), the transformation into the quadruplex structure have been confirmed in vitro. In the model organism Saccharomyces cerevisiae, the capacity of G4 DNA-forming sequences to stimulate repeat instability and gross chromosomal rearrangements has been demonstrated using reporter assays (37–40).
During transcription, the negative and positive topological stress accruing behind and in front of the RNA polymerase complexes, respectively, are removed by topoisomerase I or Top1 (41). Yeast Top1, a type IB topoisomerase, binds DNA non-specifically and generates a transient nick in DNA by covalently attaching to the 3′ end of cleaved strand through the catalytic tyrosine. After swiveling of the DNA strands, the nicked DNA is re-ligated by the 5′ hydroxyl group on the other side of the strand break attacking the phospho-tyrosyl bond thereby releasing Top1 from the DNA end. Top1 protein mutated at the highly conserved active site tyrosine (i.e. yeast Top1Y727F) cannot cleave DNA but forms a non-covalent complex with DNA (42). Interfering with the re-ligation activity of Top1, either by mutation of another conserved tyrosine (i.e. yeast Top1Y722A) or by the Top1-binding camptothecin-class of drugs, results in accumulation of unresolved covalent complex composed of Top1 and nicked DNA (Top1cc) that is cytotoxic and recombinogenic (43).
We previously reported that, in absence of Top1, a guanine-run (G-run) containing mouse immunoglobulin (Ig) switch Mu region sequence (Sμ) integrated into the yeast genome induces high rates of gene conversions, mitotic crossovers and gross chromosomal rearrangements including loss of chromosome arm and large segmental duplications (44,45). The genome instability was suppressed when the level of transcription was reduced and when the direction of transcription was altered to place the guanine-runs on the TS. In absence of Top1 activity, negative torsional stress increases, especially at highly transcribed regions. The re-annealing of TS to the NTS becomes less efficient in this condition, thereby shifting the equilibrium to hybridizing with the nascent RNA. At the Sμ sequence containing multiple runs of guanines, defective Top1 function also facilitates formation of G4 DNA structures by producing both negatively supercoiled NTS and single-stranded regions on the NTS necessitated by RNA:DNA hybrids. We initially hypothesized that the R-loop accumulation is the major underlying cause of the G4-associated genome instability associated with Top1-deficiency. In this report, we present a clear in vivo demonstration of sequence-specificity in R-loop accumulation; when the G4-forming Sμ is highly transcribed, RNA:DNA hybrids accumulate in an orientation-specific manner that correlates with the orientation-specific increase in genome instability observed in Top1- or RNase H-deficient genetic backgrounds. In absence of Top1, however, RNA:DNA hybrid accumulation did not fully account for the elevated recombination at the G-run containing sequence. Our data further show that the removal of negative helical stress during transcription is necessary but not sufficient for the Top1-mediated suppression of genome instability induced by the G-runs.
MATERIALS AND METHODS
Yeast strains and plasmids
All yeast strains used here were derived from YPH45 (MATa, ura3-52 ade2-101 trp1Δ1; (46)). The construction of pTET-LYS2 and pTET-lys2-GTOP or -GBTM (previously referred to as pTET-lys2-SμF and -SμR) cassettes and the genomic integration on chromosome were previously described (7,44). Gene deletion was carried out through one-step allele replacement by amplification of the loxP-flanked marker cassettes (Euroscarf; (47)). Two-step allele replacement procedure was used to introduce the pol2M644L mutation as previously described (48).
CEN plasmids expressing WT TOP1, top1 Y727F and top1 Y722A contain endogenous TOP1 promoter and URA3 marker. pSTS77 (49) and YEptopA-PGAL1 (50) are 2μ plasmids containing the yeast GAL1 promoter upstream of the fusion protein of E. coli gyrB and gyrA (ecGyrase) or E. coli Top1 (ecTop1), respectively, and were gifts from Dr Joaquim Roca (Molecular Biology Institute of Barcelona, Barcelona, Spain). For vector controls, a CEN plasmid pRS416 or a high copy 2μ plasmid YEP24 was used as indicated.
Determination of recombination rates
Gene conversion rates were determined according to the growth and plating conditions as previously described (44). For complementation assays, yeast strains were transformed with the appropriate plasmid DNA and plated on synthetic complete media with 2% glucose lacking uracil (SCD-Ura) plates. Individual Ura+ transformants were used to inoculate 1 ml cultures in SC-Ura media supplemented either with 2% glucose (for pRS416, WT TOP1, top1 Y727F and top1 Y722A) or with 1% raffinose plus 2% galacotse (YEP24, ecGyrase and ecTop1). After 4 day's growth at 30°C, appropriate dilutions of the cultures were plated on SCD-Ura for determination of total cell numbers or on SCD-Ura-Lys for determination of recombinants. For each strain, 12–36 cultures were used to determine rates and 95% confidence intervals using the Lea-Coulson method of median (51,52).
Survival and recombination frequency in CPT-treated cells
Overnight cultures in YEPD (1% yeast extract, 2% peptone, 2% glucose) were diluted to 2 × 105 cell/ml in fresh YEPD. Camptothecin (CPT, Sigma) to the final concentration of 100 μM or dimethyl sulfoxide (DMSO) was added to each 2 ml cultures and cultured for 24 h at 30°C on a roller drum. The cells were harvested and diluted appropriately before plating on YEPD plates to determine the total cell number and on SD-Lys plates to determine the number of recombinants. 8–24 cultures were used to determine the recombination frequency for each strain.
RNA:DNA hybrid immunoprecipitation
Yeast genomic DNA was prepared from fresh 5 ml cultures grown in YEPD by agarose plug methods by first suspending the cells at the density of 100 mg cells/1ml of 1% low melting agarose. For each immunoprecipitation (IP), DNA in three agarose plugs were dissolved in GenElute Gel extraction solution (Sigma Cat# NA1111) and fragmented to average size of 750 bp using the QSonica sonicator equipped with a microtip. DNA fragments were extracted according to the GenElute Gel extraction kit protocol and resuspended in 700 μl of IP wash solution (0.1% deoxycholic acid, 1 mM EDTA, 50 mM HEPES pH7.5, 140 mM NaCl, 1% Triton X-100). For each IP, 10 μl of protein A-conjugated dynabeads (Life Technologies) were incubated with 2.5 μg of S9.6 antibody (a gift of Dr. Stephen Leppla, NIH/NIAID), washed with IP wash solution and added to the DNA preparation. After incubation at 4°C for 16–20 h, the beads are washed three times with the IP wash solution and once with TE. Both IP and input samples were suspended in 80μl of elution buffer (50 mM Tris–HCl pH7.5, 10 mM EDTA, 1% SDS) and 20 μl of pronase (20 mg/ml, Roche) and incubated for 2 h at 42°C followed by 8 h at 65°C. DNA was purified using the Qiagen MiniElute PCR Purification Kit prior to qPCR using SensiFAST SYBR no-ROX Mastermix (Bioline) and CFX Connect instrument (Biorad). Cycling conditions were 95°C for 3 min followed by 40 cycles of 95°C for 5 s, 60°C for 10 s, and 72°C for 10 s. Primers used in the qPCR analysis are listed in the Supplementary Table S1.
RESULTS
Top1-mediated suppression of genome instability at the highly transcribed G-run sequence requires its catalytic activity
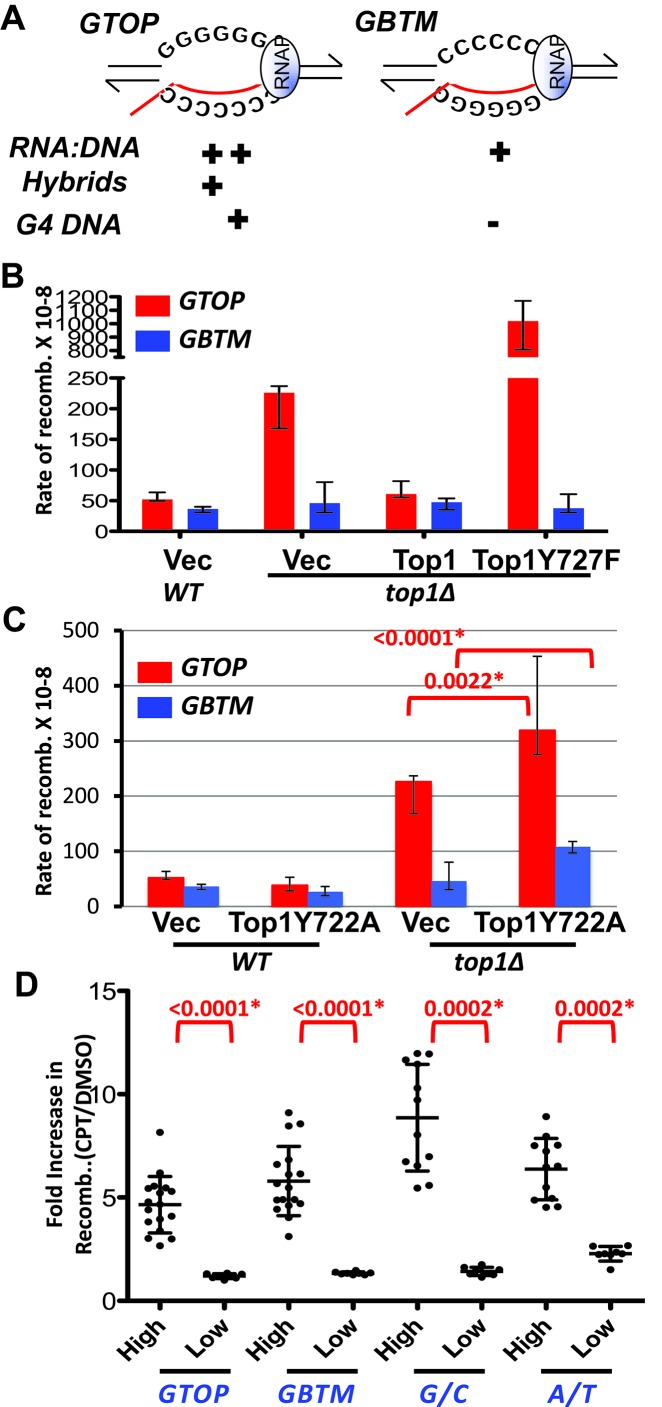
We previously reported on the construction of recombination reporter assay that allowed us to determine the contribution of guanine-run containing sequences to genome instability under high- or low-transcription conditions (44). In this reporter assay, a 770-bp fragment of murine immunoglobulin Switch Mu sequence (Sμ) was inserted into the LYS2 gene in the yeast genome. The Sμ fragment (Pubmed Accession #J00442), comprising 46% guanines overall and 35 runs of three or more guanines, was inserted either in its physiological or ‘GTOP’ orientation with the top or non-transcribed strand (NTS) containing the G-runs or in inverse or ‘GBTM’ orientation with the bottom or the transcribed strand (TS) containing the G-runs (Figure 1A and Supplementary Figure S1). This reporter construct (pTET-lys2-GTOP or -GBTM) replaced the HIS4 gene located on the left arm of Chromosome III and is regulated at the transcription level by the tetracycline-repressible promoter. Based on the demonstrated thermodynamic stability of rG:dC basepairs (53) and the in vitro analysis implicating more stable RNA:DNA hybrid formation when the RNA strand contains high guanine content (54), we postulated that the pTET-lys2-GTOP construct has higher potential for R-loop accumulation than the pTET-lys2-GBTM (+++ versus + in Figure 1A). And G4 DNA assembly is much more likely to occur at the pTET-lys2-GTOP construct, where the guanines in the NTS become unpaired and are free to form the Hoogsten bonding with other guanines during transcription (+ versus – in Figure 1A).
Figure 1.
Effects of WT or mutant Top1 on the rates of recombination at the pTET-lys2-GTOP and -GBTM reporters. (A) Transcription orientations and the guanine-runs in the Sμ-containing cassettes. Guanine-runs are located on the non-transcribed or the transcribed strand in the pTET-lys2-GTOP or pTET-lys2-GBTM cassette, respectively. The red line indicates the nascent RNA. The sequence of Sμ in -GTOP orientation is listed in Supplementary Figure S1A. Relative abundances of RNA:DNA hybrid are inferred from Belotserkovskii et al. (54). For B, C, and D, the rates of recombination at the pTET-lys2-GTOP (red bar) or pTET-lys2-GBTM (blue bar) cassette. The rates were determined by the method of median. Error bars indicate 95% confidence interval. (B) WT or top1Δ cells transformed with the indicated plasmid were grown in synthetic media lacking uracil supplemented with 2% glucose. Vec: pRS416. (C) Recombination induced by Top1Y722A. WT or top1Δ cells transformed with the indicated plasmid were grown in synthetic media lacking uracil supplemented with 2% glucose. Vec: pRS416. The rates of recombination at the pTET-lys2-GTOP (red bar) or pTET-lys2-GBTM (blue bar) cassette. The rates were determined by the method of median. Error bars indicate 95% confidence interval. P values (shown above red brackets) were determined by Mann-Whitney test using the Prism software. *, significantly different; ns, no significant difference. (D) Recombination induced by CPT treatment. WT cells containing the indicated pTET-lys2-derived reporter construct were treated either with DMSO or CPT (see material and methods). Each dot indicates the frequency of Lys+ colonies in CPT treated cells divided by the frequency in DMSO treated cells. Cells were cultured in YEPD (‘High’ transcription condition) or YEPD + 2 μg/ml doxycycline (‘Low’ transcription condition) as indicated in the x-axis label. Error bars indicate standard deviation from 8 to 24 independent measurements. *P values were determined by Mann–Whitney test using the Prism software.
In absence of Top1, gene conversion, mitotic crossover and gross chromosomal rearrangements at the guanine-run containing Sμ fragment are significantly elevated in transcription- and orientation-dependent manner (44,45). In order to determine whether the catalytic activity of Top1 is required for the suppression of genome instability, we transformed the top1Δ yeast strains containing either pTET-lys2-GTOP or pTET-lys2-GBTM construct with plasmids carrying WT TOP1 gene or the top1Y727F allele and measured the rates of gene conversion occurring at these reporter constructs. With the catalytic tyrosine (Y727) mutated to phenylalanine, the Top1Y727F mutant cannot form the phosphotyrosyl bond necessary to cleave DNA but retains DNA binding activity (55–57). The recombination rate of the pTET-lys2-GBTM reporter was not significantly changed when WT Top1 or Top1Y727F was expressed (Figure 1B). For the pTET-lys2-GTOP, the gene conversion rate was decreased by ∼4-fold with WT Top1 expression and became indistinguishable compared to the rate in WT cells. With Top1Y727F expression, the recombination rate for the pTET-lys2-GTOP was 17-fold higher compared to WT Top1 expression and 4.5-fold higher compared to vector control. When the transcription of the pTET promoter was repressed by the addition of 2μg/ml doxycycline in the growth media, Top1Y727F expression did not significantly elevate recombination rates for either the pTET-lys2-GTOP or pTET-lys2-GBTM construct indicating the hyper-recombinogenic effect of Top1Y727F is transcription-dependent (Supplementary Figure S2A).
We reported previously that the recombination rate in WT strains for the pTET-lys2-GTOP construct is higher than the pTET-lys2-GBTM by about 2-fold (44). This relatively small difference in the rates of recombination was nevertheless statistically significant. To test whether the increased level of Top1 activity can overcome the elevated recombination associated with the pTET-lys2-GTOP construct, we transformed a WT Top1-expressing plasmid into the WT strains containing the pTET-lys2-GTOP or pTET-lys2-GBTM construct. With the control vector, the recombination rate for the pTET-lys2-GTOP is 1.4 fold higher than the rate for the pTET-lys2-GBTM and this difference is statistically significant (P = 0.0009) (Supplementary Figure S2B). Upon WT Top1 expression from the plasmid, the rate of recombination for the pTET-lys2-GTOP was reduced to a level that is statistically indistinguishable from the pTET-lys2–GBTM (P = 0.141) indicating that the limiting factor leading to the relatively higher recombination rate for the pTET-lys2-GTOP construct in WT strains is Top1 activity. In WT backgrounds, Top1 Y727F expression did not significantly change the recombination rate for the pTET-lys2-GTOP or pTET-lys2-GBTM construct.
Recombination induced by the Top1-DNA complex is dependent on the level of transcription
Yeast Top1 with mutation of tyrosine to alanine at the amino acid 722 (Top1Y722A) has intact DNA cleavage activity but significantly reduced re-ligation activity leaving Top1 trapped as a Top1–DNA cleavage complex (Top1cc) (43). Accumulation of Top1cc due to Top1Y722A mutant was shown to elevate recombination between homologous chromosomes in diploid yeast cells and instability at the rDNA and CUP1 tandem arrays (58) and to enhance the 2-nucleotide deletion mutations occurring at short tandem repeat sequences (59). In order to determine whether the genome instability induced by Top1cc is enhanced by G4 DNA structures, we expressed Top1Y722A from a plasmid with the endogenous TOP1 promoter and measured the recombination rate at the highly expressed pTET-lys2-GTOP or -GBTM reporter. In WT strains with functional genomic allele of TOP1, Top1Y722A expression from the plasmid did not have significant effect on the rates of recombination (Figure 1C). When Top1Y722A was expressed in top1Δ backgrounds, the recombination rates for both the pTET-lys2-GTOP and -GBTM were significantly elevated.
Top1 is the target of a widely used anti-cancer chemotherapeutic named campthothecin (CPT) and its derivatives, which bind to Top1-DNA covalent complex and inhibits the re-ligation step (41,60). Similar to Top1Y722A expression, Top1cc accumulate in the genome when treated with CPT. We tested whether CPT treatment can elevate recombination at the Sμ-containing reporter constructs. Recombination frequencies in cells treated with CPT were 5- and 6-fold higher than DMSO treated control cells for the pTET-lys2-GTOP and -GBTM, respectively (Figure 1D). In addition to the 16 GGGG•CCCC runs, Sμ fragment inserted into the LYS2 gene for the construction of the pTET-lys2-GTOP and -GBTM reporter is 60% G/C rich (Supplementary Figure S1A). We tested whether the GC content affects the sensitivity to CPT by measuring CPT-induced recombination at the identical pTET-lys2 reporter with insertion of a 750 bp fragment of chicken β-globin 2 coding sequence. This fragment is 63% GC and contains only four widely spaced G-runs (‘G/C’ in Figure 1D). We also tested the effect of CPT treatment on the recombination occurring at the pTET-lys2-oligo allele with the insertion of 25 nt sequence (GATCTGTCCCTTACTAGCTAGGTAG) (61). LYS2 gene itself is 60% AT and with the short insertion we considered this allele to be AT rich (‘A/T’ in Figure 1D).
CPT treatment led to induction of 9– and 6-folds of recombination at the pTET-lys2-G/C or –A/T construct, respectively. The CPT treatment was repeated for all four constructs, pTET-lys2-GTOP, GBTM, -G/C and -A/T after adding doxycycline to repress transcription from the pTET promoter. The increase in recombination frequency compared to DMSO-treated cells were between 1 and 2 fold and they were all significantly lower than when transcription was fully active (see P-values in Figure 1D). The level of transcription or the G/C content of the pTET-lys2 reporter had no effect on the overall cell survival after CPT treatment (Supplementary Figure S3). Overall, the level of recombination induced by the Top1cc accumulation in CPT treated cells was not dependent on the sequence content but significantly dependent on the level of transcription. The lack of sequence-specificity for the Top1cc-induced hyper-recombination ensuing from CPT treatment (Figure 1D) or Top1Y722A expression (Figure 1C) suggests that Top1cc formation is not relevant to either suppression or aggravation of G4-associated genome instability. Top1 can interact with the C-terminal domain of elongating RNA polymerase (62), which could explain the enhanced level of recombination ensuing from CPT-induced Top1cc formation when transcription is activated.
E. coli Top1 can partially complement the loss of Top1
During transcription, positive and negative supercoils accumulate ahead and behind of the RNA polymerase complexes, respectively (Figure 2A) (63). Because yeast Top1 can remove both negative and positive supercoils associated with transcription, it is not clear which type of supercoils contribute to the orientation-specific elevation in recombination for the pTET-lys2-GTOP construct in absence of Top1. We transformed top1Δ strains with plasmids expressing either E. coli Topoisomerase 1 (ecTop1), which removes only negative supercoils or a fusion protein of E. coli GyrA and GyrB (ecGyrase), which removes only positive supercoils (49,50). ecTop1 and ecGyrase were previously shown to be active in yeast and, furthermore, expression of either bacterial topoisomerase was shown to suppress the slow growth phenotype in strains lacking both yeast Top1 and Top2 activity. The recombination rates at pTET-lys2-GTOP or -GBTM expression did not significantly change upon expression of ecGyrase (Figure 2B). The expression of ecTop1 reduced the recombination rate at the pTET-lys2-GTOP by 3-fold compared to the control vector indicating that the removal of negative supercoils by ecTop1 can partially suppress elevated recombination at the highly transcribed G-run sequence in absence of yeast Top1.
Figure 2.
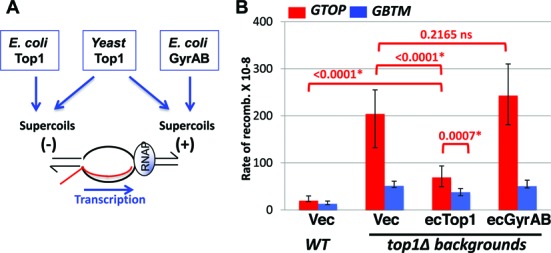
Expression of E. coli Top1 or Gyrases in top1Δ backgrounds. (A) Transcription-associated positive (+) and negative (–) helical stresses are removed by topoisomerases. (B) WT or top1Δ cells transformed with the indicated plasmid were grown in synthetic media lacking uracil supplemented with 2% galactose and 1% raffinose. Vec: YEP24. The rates of recombination at the pTET-lys2-GTOP (red bar) or pTET-lys2-GBTM (blue bar) cassette. The rates were determined by the method of median. Error bars indicate 95% confidence interval. P values (shown above red brackets) were determined by Mann-Whitney test using the Prism software. *, significantly different; ns, no significant difference.
RNA:DNA hybrids or R-loops accumulate in an orientation-specific manner in top1Δ backgrounds
Annealing of the TS to the nascent RNA strand is one of the major consequences of the negative helical torsion accumulated behind the transcription complex in absence of Top1 activity. Accumulation of such extensive RNA:DNA hybrids or R-loops can lead to elevated genome instability (13,64). In order to determine whether there is a correlation between the observed increase in the recombination at the pTET-lys2-GTOP reporter and RNA:DNA hybrid accumulation, we used the genomic DNA isolated from top1Δ strains containing either the pTET-lys2-GTOP or -GBTM construct in immunoprecipitation (IP) experiment using the monoclonal antibody S9.6 followed by quantitative PCR (qPCR). S9.6 is specific for RNA:DNA hybrids of ≥6 bp in length (65,66) and was recently used in mapping RNA:DNA hybrid accumulation in the yeast genome (22,23). For the negative control, we carried out qPCR with primers targeting an untranscribed intergenic region on Chromosome V (23). At this region, ∼0.3% of DNA was recovered by the IP with S9.6 antibody. Due to the high G/C content and presence of repeated sequences, it was not possible to design efficient qPCR primers within the Sμ fragment in the pTET-lys2-GTOP or -GBTM constructs. Instead, a primer-pair targeting 100 nt upstream of 5′ end of the Sμ fragment was used to quantitate the RNA:DNA hybrids at the Sμ fragment (‘Sμ’ in the Figure 3A and Supplementary Table S1). When normalized to the negative control, there was a 18-fold enrichment of RNA:DNA hybrids proximal to the G-run containing Sμ fragment when it is in the context of the pTET-lys2-GTOP. When the Sμ fragment is in the pTET-lys2-GBTM context, the RNA:DNA hybrid proximal to the Sμ was enriched by ∼5-fold. We also quantitated the level of RNA by qRT-PCR methods using the identical primers used to detect RNA:DNA hybrids using two different housekeeping genes, UBC6 and ALG9, as the reference controls (67) (Supplementary Table S2). No significant difference in the levels of RNA was present in the strains containing pTET-lys2-GTOP or -GBTM construct indicating that the preferential accumulation of RNA:DNA hybrids when the Sμ-fragment is in ‘GTOP’ configuration is not due to the difference in levels of transcription.
Figure 3.
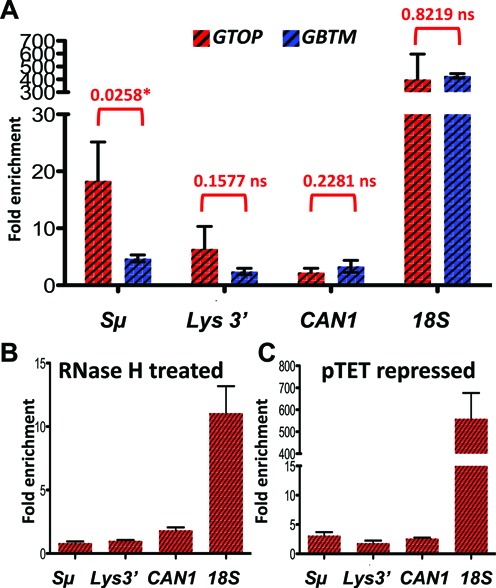
RNA:DNA hybrids in top1Δ backgrounds. RNA:DNA hybrids were quantitated by immunoprecipitation with S9.6 antibody as described in Materials and Methods. Fold enrichment relative to the untranscribed control region was determined by ΔΔCq analysis using the quantitative PCR results. Error bars indicate standard deviation calculated from three independent experiments. (A) Immunoprecipiation with samples from top1Δ cells containing either pTET-lys2-GTOP (red-hashed bars) or pTET-lys2-GBTM (blue-hashed bars). P values were determined by unpaired t-test using the Prism software. *, significantly different; ns, no significant difference. (B) DNA samples were treated with recombinant RNase H after sonication prior to immunoprecipitation. (C) Immunoprecipitation with S9.6 antibody was carried out in samples prepared from cells grown with 2 μg/ml doxycycline.
We additionally measured RNA:DNA hybrid accumulation within the pTET-lys2-GTOP or-GBTM construct but 3.5 kb downstream from the Sμ fragment (‘LYS3′’ in Figure 3A), at a RNA Polymerase II-transcribed gene with a low level of expression (‘CAN1’) and at the highly transcribed rDNA locus (‘18S’). Consistent with previously reported data, very high level of RNA:DNA hybrids were detected at the rDNA locus compared to the CAN1 gene (Figure 3A). And at the three loci tested, there was no significant difference in the level of RNA:DNA hybrid accumulation between the strains containing the pTET-lys2-GTOP or -GBTM construct. R-loops are normally prevented by riboendonucleases called RNase H's that recognizes RNA:DNA hybrids and degrades the RNA strand. When the DNA samples were treated with recombinant bacterial RNase H prior to IP, RNA:DNA hybrids were detected at significantly reduced level at Sμ and at 18S (P = 0.0111 and 0.0272, respectively) (Figure 3B). And when the cells cultured with doxycycline was used for IP with S9.6 antibody, RNA:DNA hybrids were reduced at the Sμ but not at the 18S (P = 0.0164 and 0.9372, respectively) (Figure 3C).
Recombination is elevated at G-run containing sequences in the absence of RNase H activity
Similar to higher eukaryotic species, there are two RNase H class enzymes in yeast; RNase H1 is a single subunit protein encoded by the RNH1 gene whereas RNase H2 is a hetero-trimer comprising gene products of the RNH201, RNH202 and RNH203 (68). RNA:DNA hybrids ≥3 bp-long including co-transcriptionally formed R-loops are substrates for both RNase H1 and H2 whereas single or double ribonucleotide(s) embedded in duplex DNA molecule is recognized by RNase H2 enzyme only (69). Single ribonucleotide-accumulation in DNA in absence of RNase H2 has been previously described as a key source of mutagenesis, recombination and gross chromosomal rearrangements in yeast (70–73) and the lesion responsible for the genome-wide chromosomal anomalies and embryonic lethality in mice (74). We previously showed that simultaneous disruption of RNase H1 and H2 results in significant elevation of recombination for both pTET-lys2-GTOP and -GBTM constructs (44). In order to determine whether the instability observed at these constructs correlates with the failure to remove co-transcriptionally formed R-loops or single ribonucleotides in DNA, we measured the effect of deleting RNH1 or RNH201 singly on the rate of recombination. In the single deletion mutants, rnh1Δ and rnh201Δ, there was no significant elevation in recombination for the pTET-lys2-GTOP or -GBTM construct (Figure 4A). As reported earlier, in strains deficient in both RNase H enzymes, the recombination rate for pTET-lys2-GTOP and -GBTM constructs were elevated by 19- and 11-fold, respectively (44). The functional redundancy of RNase H1 and H2 suggests that the elevated recombination is attributable to R-loops, the substrate common to both enzymes.
Figure 4.
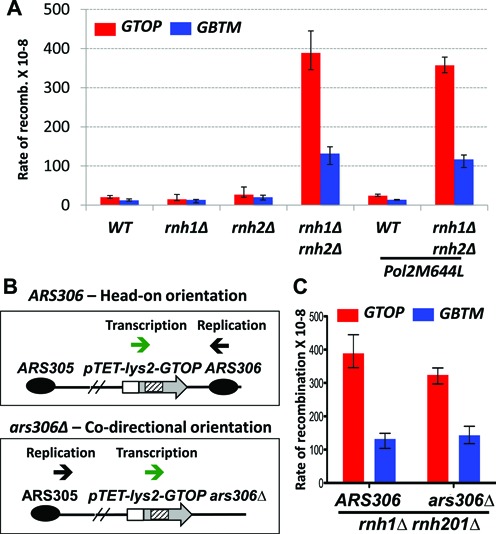
Effect of RNase H-disruption on the recombination rates. (A) The rates of recombination at the pTET-lys2-GTOP (red bar) or -GBTM (blue bar). Cells with indicated genetic backgrounds were grown in YEP supplemented with 2% glycerol and 2% ethanol. The rates were determined by the method of median. Error bars indicate 95% confidence interval. (B) Effect of changing the replication direction. The putative direction of transcription (green arrow) and replication (black arrow) through the pTET-lys2-GTOP or -GBTM construct are indicated for ARS306 or ars306Δ backgrounds. (C) The rates of recombination at the pTET-lys2-GTOP (red bar) or -GBTM (blue bar) in rnh1Δ rnh201Δ ARS306 or rnh1Δ rnh201Δ ars306Δ backgrounds.
Single ribonucleotides embedded in DNA occur as a byproduct of DNA replication process. We introduced a single missense mutation (M644L) to the one of the two major replicative DNA polymerases Pol ϵ. Pol2M644L mutant was previously shown to have decreased propensity to incorporate ribonucleotides into DNA during replication and to reduce ribonucleotide-initiated mutagenesis (48,59). In WT or rnh1Δ rnh201Δ backgrounds, the recombination rates at the pTET-lys2-GTOP or -GBTM construct was not changed when Pol2M644L mutant was introduced (Figure 4A), further indicating that ribonucleotide incorporated in DNA is not a significant factor leading to the observed hyperrecombination at the highly transcribed Sμ fragment in rnh1Δ rnh201Δ background.
We also tested whether the relative orientation of transcription and replication affects the hyperrecombination observed at the pTET-lys2-GTOP and –GBTM constructs in RNase H-deficient yeast strains. Upon deletion of the replication origin ARS306, the genomic locus containing the pTET-lys2-GTOP or -GBTM construct is replicated from the ARS305, thereby reversing the direction of replication fork to move in co-directional orientation with the transcription initiating at the pTET promoter (Figure 4B). In top1Δ background, reversing the relative orientation of the transcription and replication from head-on to co-directional resulted in 3-fold decrease in the rate of recombination occurring at the pTET-lys2-GTOP reporter (45). In rnh1Δ rnh201Δ backgrounds, however, reversing the direction of the replication fork movement did not significantly change the recombination rate for the pTET-lys2-GTOP or -GBTM construct (Figure 4C).
In absence of RNase H1 and H2, more extensive R-loop accumulation occurs when G-runs are on the NTS
In order to confirm that R-loop accumulation occurs as a consequence of deleting RNH1 and RNH201, we quantitated the RNA:DNA hybrid that can be immunoprecipitated by the specific antibody S9.6. In the rnh1Δ rnh201Δ strains containing the pTET-lys2-GTOP or -GBTM construct, we detected greater than 500-fold enrichment of RNA:DNA hybrids at 18S, compared to the untranscribed control region (Figure 5A). Proximal to the G4-forming Sμ within the pTET-lys2-GTOP and -GBTM constructs (Sμ), RNA:DNA hybrid was enriched by 57- and 11-fold compared to the negative control, respectively. At this site, the level of transcripts for the pTET-lys2-GTOP and -GBTM were indistinguishable to each other and to the level measured in top1Δ backgrounds (Supplementary Table S2). 3.5 kb away from the Sμ (LYS3′), the level of transcription was slightly higher in the strain containing the pTET-lys2-GTOP compared to that containing the pTET-lys2-GBTM construct, which could account for the significant difference in the level of RNA:DNA hybrid enrichment at the LYS3′ location (Figure 5A). As in top1Δ backgrounds, RNA:DNA hybrids at Sμ were significantly reduced by pretreating with recombinant RNase H or by repressing the pTET-driven transcription (P = 0.0075 and 0.0079, respectively) (Figure 5B and C).
Figure 5.
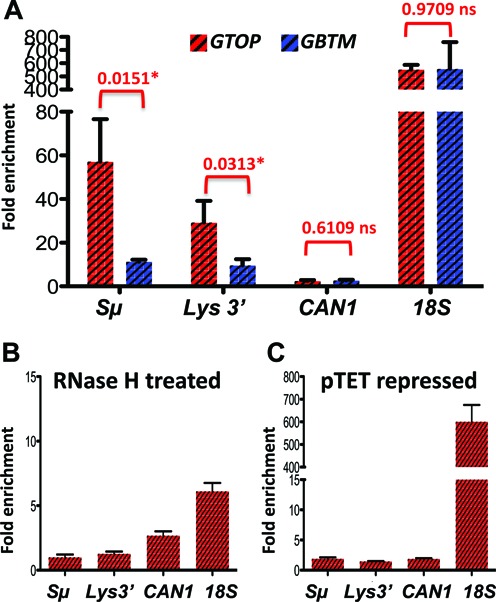
RNA:DNA hybrids in rnh1Δ rnh201Δ backgrounds. RNA:DNA hybrids were quantitated by immunoprecipitation with S9.6 antibody as described in the materials and methods. Fold enrichment relative to the untranscribed control region was determined by ΔΔCq analysis using the quantitative PCR results. Error bars indicate standard deviation calculated from three independent experiments. (A) Immunoprecipiation with samples from rnh1Δ rnh201Δ cells containing either pTET-lys2-GTOP (red hashed bar) or pTET-lys2-GBTM (blue hashed bar). P values were determined by unpaired t-test using the Prism software. *, significantly different; ns, no significant difference. (B) DNA samples were treated with recombinant RNase H after sonication prior to immunoprecipitation. (C) Immunoprecipitation with S9.6 antibody was carried out in samples prepared from cells grown with 2 μg/ml doxycycline.
Removal of negative supercoils partly suppresses the elevated recombination at the G-run containing sequence in rnh1Δ rnh201Δ
As we reported earlier, the hyperrecombination at the pTET-lys2-GTOP and -GBTM constructs in rnh1Δ rnh201Δ backgrounds was suppressed by overexpression of yeast RNH1 (45). In order to test whether reducing helical torsion in the highly transcribed region can counteract the defect in RNA:DNA hybrid processing in absence of RNase H activity, we transformed the plasmids expressing either yeast topoisomerase I or bacterial topoisomerases into rnh1Δ rnh201Δ cells. Expression of yeast Top1 (WT or Top1Y727F) or overexpression of the ecGyrase did not change the rate of recombination for either the pTET-lys2-GTOP or -GBTM construct (Figure 6A and B). Upon overexpression of ecTop1, which removes negative supercoils, the recombination rates for both the pTET-lys2-GTOP and -GBTM were reduced by ∼2-fold but remained significantly higher than the rates in WT backgrounds.
Figure 6.
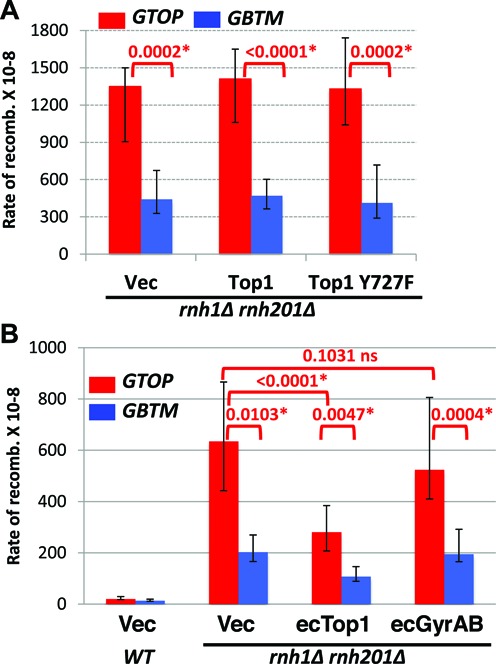
The rates of recombination in rnh1Δ rnh201Δ backgrounds. WT or rnh1Δ rnh201Δ cells transformed with the indicated plasmid were grown in synthetic media lacking uracil supplemented with 2% glucose (A) or 2% galactose and 1% raffinose (B). Vec: pRS416 for A; YEP24 for B. The rates of recombination at the pTET-lys2-GTOP (red bar) or pTET-lys2-GBTM (blue bar) cassette. The rates were determined by the method of median. Error bars indicate 95% confidence interval. P values (shown above red brackets) were determined by Mann–Whitney test using the Prism software. *, significantly different; ns, no significant difference.
DISCUSSION
When a repetitive sequence is actively transcribed, the formation of R-loops and DNA secondary structures could be mutually cooperative. Annealing of the nascent RNA to the TS, which initiates R-loop formation, might be promoted by the unavailability of the NTS when occupied in secondary DNA structure formation. Intra-strand interaction of the NTS, which is required for non-B structure formation, conversely, might be promoted by the unavailability of the TS when stably annealed to the RNA. For the G4 DNA-forming Ig switch region sequence, destabilizing the RNA:DNA hybrids involving the TS led to less robust G4 formation on the NTS (26). We previously found that disruption of RNase Hs or Top1, each of which was shown to lead to R-loop formation at highly transcribed areas such as the rDNA locus (18,22,23), elevates genome instability at a highly transcribed G4 DNA-forming Ig Sμ sequence in orientation-specific manner (44,45). R-loop and G4 DNA, which are likely to cooperatively form during active transcription of guanine-run containing sequence such as Sμ used to construct pTET-lys2-GTOP and -GBTM reporters, are each sufficient to induce genome instability (19,38,39). It is, therefore, difficult to define which of the higher order structures is directly responsible for the elevated recombination in top1Δ and/or rnh1Δ rnh201Δ backgrounds. We compared what factors affect the elevated recombination in top1Δ or rnh1Δ rnh201Δ strains in order to determine whether the intermediates involved in leading to instability at the G-run containing sequence in these two genetic backgrounds are identical or divergent.
We first confirmed whether robust transcription of the pTET-lys2-GTOP or -GBTM reporter results in the enrichment of RNA:DNA hybrids detectable by the monoclonal antibody S9.6. Our data show that, in both RNase H-defective and Top1-defective strain backgrounds, RNA:DNA hybrids accumulate to a significant extent proximal to the G/C rich Sμ insert within the pTET-lys2-GTOP where the transcription is oriented to produce (rG)n:(dC)n base pairing (Figures 3A and 5A). When transcription from the pTET promoter was repressed, no significant enrichment of RNA:DNA hybrids occurred relative to the untranscribed region on Chromosome V used as the negative control (Figures 3C and 5C). While in the pTET-lys2-GBTM context, the RNA:DNA hybrids accumulation proximal to the Sμ insert was observed at a much lower level in top1Δ and rnh1Δ rnh201Δ (Figures 3A and 5A). At the 3′ end of the pTET-lys2-GTOP reporter (LYS3′), where sequence is ∼60% A/T, RNA:DNA hybrids accumulated to much less degree than at Sμ in top1Δ and rnh1Δ rnh201Δ strains. In both strain backgrounds, the levels of steady-state transcripts were apparently about two-fold higher at LYS3′ than at Sμ (Supplementary Table S2) indicating that the level of guanine-content is the overriding factor in formation of R-loops. The orientation-bias or G-C bias in RNA:DNA hybrid accumulation was previously observed during in vitro transcription experiments and is consistent with the biochemical data showing that rG:dC pairing is thermodynamically more stable than rC:dG (26,54,75).
For the pTET-lys2-GBTM reporter in rnh1Δ rnh201Δ background, similar levels of RNA:DNA hybrid enrichment were observed at the Sμ, where the nascent RNA contains runs of Cs, and at the LYS3′, where the nascent RNA and TS strand are both A/T-rich (Figure 5A). In top1Δ cells, the level of RNA:DNA hybrids detected at Sμ and at LYS3′ within the pTET-lys2-GBTM construct was minimal and not significantly different from the level at the CAN1 gene, which is transcribed at 20- to 30-fold lower rate than the pTET-derived reporter constructs (Figure 3A) (76). These results suggest that high number of Cs present on the NTS at Sμ within the pTET-lys2-GBTM is not sufficient to stimulate R-loop formation even when the rate of transcription is very high.
In correlation with the orientation bias in the recombination, R-loop accumulation in top1Δ and rnh1Δ rnh201Δ strains is equally pronounced when the transcription is oriented to place the guanine runs on the NTS (GTOP). However, our data indicate that there are substantial differences in the intermediate steps leading to elevated recombination in these genetic backgrounds. Unlike the disruption of RNase Hs, which elevates recombination of highly transcribed sequences containing biased runs of Gs or Cs or a mix of Gs and Cs (Figure 4 and (44)), the effect of Top1 disruption is highly specific to the Sμ fragment when the guanine-runs are located on the NTS. Several independent reports suggest that, for the most of the genome with the exception of the rDNA and CUP1 loci, which contain arrays of direct repeats, the function of Top1 in maintaining genome stability is nonessential (58,77). The loss of Top1 did not result in significant elevation in recombination between two direct repeats (72), genome wide mitotic crossovers (58) or gross chromosomal rearrangements (GCRs) (73). But, as we have shown previously and in the current report, at the G-run containing sequence cloned from murine Ig Switch Mu region (Sμ fragment) or from the human protooncogene TCF3, Top1-disruption led to significant stimulation of gene conversion recombination and/or GCR (36,45). This indicates that the function of Top1 is specifically required to suppress genome instability when the sequences containing guanine runs are located on the NTS and therefore in conditions conducive to form G4 DNA structure during active transcription. Another important difference from rnh1Δ rnh201Δ is the significance of the R-loop in top1Δ background. According to previous experiments in yeast and mammalian cells, following the disturbance of a co-transcriptional process such as mRNP packaging, RNA-DNA unwinding, splicing, RNA degradation and export, the genome instability associated with R-loop accumulation was suppressed by overexpression of RNase H1 (15,19–21). RNase H1 overexpression was not able to affect recombination when it is elevated due to other factors such as defect in DNA damage response or repair pathways (21). We previously reported that preventing RNA:DNA hybrid accumulation by overexpression of RNH1, which significantly reduced recombination at both the pTET-lys2-GTOP and -GBTM constructs in rnh1Δ rnh201Δ background, failed to complement for the loss of Top1 in suppressing the elevated recombination at the pTET-lys2-GTOP construct (45).
In the absence Top1, R-loop accumulation is thought to occur as a secondary effect of the failure to remove negative supercoils accumulated during transcription. Negatively wound NTS limits the re-annealing with the TS thereby increasing the chance of TS annealing to the nascent RNA to form RNA:DNA hybrid behind the transcription complex. Negative superhelicity also leads to local melting of the duplex DNA and thereby facilitate the transformation of B-DNA into secondary structures including G4 DNA (78,79). We showed that removal of negative supercoils by bacterial topoisomerase I (ecTop1) can functionally compensate for the loss of yeast Top1 and results in reduction in the recombination rate at the pTET-lys2-GTOP reporter (Figure 2). Yeast Top1 expression from a plasmid resulted in complete complementation of TOP1 deletion leading to the recombination rate indistinguishable from that in WT cells at the pTET-lys2-GTOP reporter (Figure 1B). With the ecTop1 expression, however, the recombination rate at the pTET-lys2-GTOP, although significantly less than in top1Δ cells, still remained ∼3.5-fold higher than WT level (Figure 2B). The inability of ecTop1 to fully complement the loss of yeast Top1 indicates that negative supercoil accumulation and the associated R-loop formation is only partially responsible for the genome instability associated with the highly expressed G-run containing sequence and that yeast Top1 has an as-yet-unknown function in addition to the removal of negative helical torsion. Alternatively, the partial complementation by ecTop1 and the complete lack of complementation by ecGyrase could be due to their limited activities in the particular yeast strain used in the current study, although the expression and function of the ecTop1 and ecGyrase in the heterologous yeast cells have been previously verified (49,50).
The effect of catalytically inactive Top1 (Top1Y727F) also suggests that Top1 function at the G-run containing sequence is not limited to removing helical torsion in DNA. When Top1Y727F was expressed in top1Δ cells with the pTET-lys2-GTOP reporter, there was 20- and 4.5-fold increase in recombination rate compared to WT and top1Δ, respectively (Figure 1B). When co-transcriptional formation of G4 DNA is much less likely because the guanine runs were located on the transcribed strand (pTET-lys2-GBTM), there was no significant difference in the rates of recombination in cells expressing no Top1 (top1Δ with the vector plasmid), WT Top1 (WT with the vector plasmid or top1Δ with the WT Top1-expression plasmid) or Top1Y727F (top1Δ with the Top1Y727F-expression plasmid). Since Top1Y727F failed to suppress genome instability at the highly transcribed Sμ sequence, it is clear that the catalytic activity of removing helical torsion is required for this function of Top1. The further increase observed in the rate of recombination at the pTET-lys2-GTOP with Top1Y727F expression in top1Δ backgrounds suggests that Top1Y727F mutant has an additional effect besides the lack of its catalytic properties. We speculate from this result that the role of Top1 could be more complex than its catalytic activity and involve its high-affinity, high-specificity binding of G4 DNA structures as demonstrated by the biochemical analyses with purified human or bovine topoisomerase I (80–82). G4 DNA-binding property of the mammalian topoisomerases I is expected to be shared by the highly conserved yeast Top1. The core and the catalytic domains of the human topoisomerase I, which together form a clamp around the duplex DNA, are 73% and 85% homologous to, respectively, and can functionally replace the corresponding yeast Top1 domains in chimeric protein constructs (Supplementary Figure S4)(83). In addition, out of the 26 residues directly contacting DNA according to the co-crystal structure of human Top1/DNA complex (42), 23 residues are identical in yeast Top1. In support of the direct interaction between Top1 and G4 DNA, chromatin immunoprecipitation (ChIP) analysis showed that both WT Top1 and Top1Y727F are enriched at the G-rich telomeres in yeast cells (84). Specific binding to G4 structure by WT Top1 could be required for recruitment of other factors that facilitate resolution of the secondary structure such as structure-specific helicases. When catalytically active, the dynamic nature of Top1–DNA interaction makes it possible that Top1 deposits and subsequently allows access of G4-resolving factors to the G4 DNA. The catalytically inactive Top1Y727F could exacerbate genome instability at G-run containing sequences by stabilizing the G4 DNA structures through cooperative binding and/or by preventing access of G4 processing factors such as structure-specific helicases. In addition, Top1Y727F, which forms an encircling clamp when bound to the canonical duplex DNA, could assume a different conformation when bound to the four-stranded G4 DNA (41).
In summary, using reporter constructs containing the identical G/C rich sequence from mouse Ig Sμ placed in two different orientations, we demonstrated the preferential accumulation of RNA:DNA hybrids when guanine-runs are located on the NTS of highly transcribed region. This specificity of RNA:DNA hybrid accumulation dependent on the transcriptional orientation correlates with the orientation-specific elevation in genome instability in absence of Top1 or RNase Hs. In top1Δ backgrounds, however, removal of negative helical torsion by overexpression of E. coli Top1 and prevention of RNA:DNA hybrids by overexpression of RNase H1 had partial to no effect on the recombination occurring at the highly transcribed G4-forming sequence. This data suggest that accumulation of R-loops due to negative supercoils does not constitute the sufficient basis for the G4-specific genome instability in absence of Top1. Our current model is that yeast Top1 suppresses genome instability associated with guanine-runs first by preventing the G-runs present on the NTS from folding into G4 DNA through removal of the transcription-associated negative supercoils and also by recruiting proteins that can resolve G4 structures through physical interaction with G4 DNA. In the future, the technically challenging demonstration of the presence of G4 DNA structure and its physical interaction with Top1 and Top1Y727F in vivo will be necessary to provide concrete evidence for this model.
Supplementary Material
Acknowledgments
Authors would like to thank Dr Stephen Leppla of National Institutes of Health at Bethesda, Maryland for the gift of S9.6 antibody and Dr Joaquim Roca of Molecular Biology Institute of Barcelona at Barcelona, Spain for the gift of ecTop1 and ecGyrase plasmids. We also thank the members of Kim lab for discussion and critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of Texas Health Science Center at Houston (to N.K.). Funding for open access charge: University of Texas Health Science Center at Houston.
Conflict of interest statement. None declared.
REFERENCES
- 1.Voelkel-Meiman K., Keil R.L., Roeder G.S. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987;48:1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- 2.Nickoloff J., Reynolds R. Transcription stimulates homologous recombination in mammalian cells. Mol. Cell. Biol. 1990;10:4837–4882. doi: 10.1128/mcb.10.9.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nickoloff J.A. Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol. Cell. Biol. 1992;12:5311–5318. doi: 10.1128/mcb.12.12.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prado F., Piruat J.I., Aguilera A. Recombination between DNA repeats in yeast hpr1delta cells is linked to transcription elongation. EMBO J. 1997;16:2826–2835. doi: 10.1093/emboj/16.10.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saxe D., Datta A., Jinks-Robertson S. Stimulation of mitotic recombination events by high levels of RNA polymerase II transcription in yeast. Mol. Cell. Biol. 2000;20:5404–5414. doi: 10.1128/mcb.20.15.5404-5414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datta A., Jinks-Robertson S. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]
- 7.Kim N., Abdulovic A.L., Gealy R., Lippert M.J., Jinks-Robertson S. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair (Amst.) 2007;6:1285–1296. doi: 10.1016/j.dnarep.2007.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim N., Jinks-Robertson S. Transcription as a source of genome instability. Nat. Rev. Genet. 2012;13:204–214. doi: 10.1038/nrg3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguilera A., Garcia-Muse T. Causes of genome instability. Annu. Rev. Genet. 2013;47:1–32. doi: 10.1146/annurev-genet-111212-133232. [DOI] [PubMed] [Google Scholar]
- 10.Wang J.D., Berkmen M.B., Grossman A.D. Genome-wide coorientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5608–5613. doi: 10.1073/pnas.0608999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srivatsan A., Tehranchi A., MacAlpine D.M., Wang J.D. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet. 2010;6:e1000810. doi: 10.1371/journal.pgen.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mirkin E.V., Mirkin S.M. Mechanisms of transcription-replication collisions in bacteria. Mol. Cell. Biol. 2005;25:888–895. doi: 10.1128/MCB.25.3.888-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamperl S., Cimprich K.A. The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair (Amst.) 2014;19:84–94. doi: 10.1016/j.dnarep.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wellinger R.E., Prado F., Aguilera A. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol. Cell. Biol. 2006;26:3327–3334. doi: 10.1128/MCB.26.8.3327-3334.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sollier J., Stork C.T., Garcia-Rubio M.L., Paulsen R.D., Aguilera A., Cimprich K.A. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell. 2014;56:777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chavez S., Beilharz T., Rondon A.G., Erdjument-Bromage H., Tempst P., Svejstrup J.Q., Lithgow T., Aguilera A. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 2000;19:5824–5834. doi: 10.1093/emboj/19.21.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X., Manley J.L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 18.El Hage A., French S.L., Beyer A.L., Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huertas P., Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 20.Mischo H.E., Gomez-Gonzalez B., Grzechnik P., Rondon A.G., Wei W., Steinmetz L., Aguilera A., Proudfoot N.J. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol. Cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wahba L., Amon J.D., Koshland D., Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol. Cell. 2011;44:978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El Hage A., Webb S., Kerr A., Tollervey D. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet. 2014;10:e1004716. doi: 10.1371/journal.pgen.1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan Y.A., Aristizabal M.J., Lu P.Y., Luo Z., Hamza A., Kobor M.S., Stirling P.C., Hieter P. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 2014;10:e1004288. doi: 10.1371/journal.pgen.1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mirkin S.M. Discovery of alternative DNA structures: a heroic decade (1979–1989) Front. Biosci. 2008;13:1064–1071. doi: 10.2741/2744. [DOI] [PubMed] [Google Scholar]
- 25.Wang G., Vasquez K.M. Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability. DNA Repair (Amst.) 2014;19:143–151. doi: 10.1016/j.dnarep.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duquette M.L., Handa P., Vincent J.A., Taylor A.F., Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grabczyk E., Mancuso M., Sammarco M.C. A persistent RNA.DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res. 2007;35:5351–5359. doi: 10.1093/nar/gkm589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bochman M.L., Paeschke K., Zakian V.A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis L., Maizels N. G4 DNA: at risk in the genome. EMBO J. 2011;30:3878–3879. doi: 10.1038/emboj.2011.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 31.Maizels N., Gray L.T. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duquette M.L., Huber M.D., Maizels N. G-rich proto-oncogenes are targeted for genomic instability in B-cell lymphomas. Cancer Res. 2007;67:2586–2594. doi: 10.1158/0008-5472.CAN-06-2419. [DOI] [PubMed] [Google Scholar]
- 33.Katapadi V.K., Nambiar M., Raghavan S.C. Potential G-quadruplex formation at breakpoint regions of chromosomal translocations in cancer may explain their fragility. Genomics. 2012;100:72–80. doi: 10.1016/j.ygeno.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 34.Willis T.G., Dyer M.J. The role of immunoglobulin translocations in the pathogenesis of B-cell malignancies. Blood. 2000;96:808–822. [PubMed] [Google Scholar]
- 35.Nambiar M., Goldsmith G., Moorthy B.T., Lieber M.R., Joshi M.V., Choudhary B., Hosur R.V., Raghavan S.C. Formation of a G-quadruplex at the BCL2 major breakpoint region of the t(14;18) translocation in follicular lymphoma. Nucleic Acids Res. 2011;39:936–948. doi: 10.1093/nar/gkq824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams J.D., Fleetwood S., Berroyer A., Kim N., Larson E.D. Sites of instability in the human TCF3 (E2A) gene adopt G-quadruplex DNA structures in vitro. Front. Genet. 2015;6:177. doi: 10.3389/fgene.2015.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piazza A., Boule J.B., Lopes J., Mingo K., Largy E., Teulade-Fichou M.P., Nicolas A. Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res. 2010;38:4337–4348. doi: 10.1093/nar/gkq136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piazza A., Serero A., Boule J.B., Legoix-Ne P., Lopes J., Nicolas A. Stimulation of gross chromosomal rearrangements by the human CEB1 and CEB25 minisatellites in Saccharomyces cerevisiae depends on G-quadruplexes or Cdc13. PLoS Genet. 2012;8:e1003033. doi: 10.1371/journal.pgen.1003033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ribeyre C., Lopes J., Boule J.B., Piazza A., Guedin A., Zakian V.A., Mergny J.L., Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paeschke K., Bochman M.L., Garcia P.D., Cejka P., Friedman K.L., Kowalczykowski S.C., Zakian V.A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pommier Y., Leo E., Zhang H., Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Redinbo M.R., Stewart L., Kuhn P., Champoux J.J., Hol W.G. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 43.Megonigal M.D., Fertala J., Bjornsti M.A. Alterations in the catalytic activity of yeast DNA topoisomerase I result in cell cycle arrest and cell death. J. Biol. Chem. 1997;272:12801–12808. doi: 10.1074/jbc.272.19.12801. [DOI] [PubMed] [Google Scholar]
- 44.Kim N., Jinks-Robertson S. Guanine repeat-containing sequences confer transcription-dependent instability in an orientation-specific manner in yeast. 2011;10:953–960. doi: 10.1016/j.dnarep.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yadav P., Harcy V., Argueso J.L., Dominska M., Jinks-Robertson S., Kim N. Topoisomerase I plays a critical role in suppressing genome instability at a highly transcribed g-quadruplex-forming sequence. PLoS Genet. 2014;10:e1004839. doi: 10.1371/journal.pgen.1004839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gueldener U., Heinisch J., Koehler G.J., Voss D., Hegemann J.H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nick McElhinny S.A., Kumar D., Clark A.B., Watt D.L., Watts B.E., Lundstrom E.B., Johansson E., Chabes A., Kunkel T.A. Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trigueros S., Roca J. A GyrB-GyrA fusion protein expressed in yeast cells is able to remove DNA supercoils but cannot substitute eukaryotic topoisomerase II. Genes Cells. 2002;7:249–257. doi: 10.1046/j.1365-2443.2002.00516.x. [DOI] [PubMed] [Google Scholar]
- 50.Trigueros S., Roca J. Failure to relax negative supercoiling of DNA is a primary cause of mitotic hyper-recombination in topoisomerase-deficient yeast cells. J. Biol. Chem. 2002;277:37207–37211. doi: 10.1074/jbc.M206663200. [DOI] [PubMed] [Google Scholar]
- 51.Lea D.E., Coulson C.A. The distribution of the numbers of mutants in bacterial populations. J. Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 52.Spell R.M., Jinks-Robertson S. Determination of mitotic recombination rates by fluctuation analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 2004;262:3–12. doi: 10.1385/1-59259-761-0:003. [DOI] [PubMed] [Google Scholar]
- 53.Sugimoto N., Nakano S., Katoh M., Matsumura A., Nakamuta H., Ohmichi T., Yoneyama M., Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 54.Belotserkovskii B.P., Liu R., Tornaletti S., Krasilnikova M.M., Mirkin S.M., Hanawalt P.C. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eng W.K., Pandit S.D., Sternglanz R. Mapping of the active site tyrosine of eukaryotic DNA topoisomerase I. J. Biol. Chem. 1989;264:13373–13376. [PubMed] [Google Scholar]
- 56.Lynn R.M., Bjornsti M.A., Caron P.R., Wang J.C. Peptide sequencing and site-directed mutagenesis identify tyrosine-727 as the active site tyrosine of Saccharomyces cerevisiae DNA topoisomerase I. Proc. Natl. Acad. Sci. U.S.A. 1989;86:3559–3563. doi: 10.1073/pnas.86.10.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pourquier P., Bjornsti M.A., Pommier Y. Induction of topoisomerase I cleavage complexes by the vinyl chloride adduct 1,N6-ethenoadenine. J. Biol. Chem. 1998;273:27245–27249. doi: 10.1074/jbc.273.42.27245. [DOI] [PubMed] [Google Scholar]
- 58.Andersen S.L., Sloan R.S., Petes T.D., Jinks-Robertson S. Genome-destabilizing effects associated with top1 loss or accumulation of top1 cleavage complexes in yeast. PLoS Genet. 2015;11:e1005098. doi: 10.1371/journal.pgen.1005098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cho J.E., Kim N., Li Y.C., Jinks-Robertson S. Two distinct mechanisms of Topoisomerase 1-dependent mutagenesis in yeast. 2013. [DOI] [PMC free article] [PubMed]
- 60.Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem. Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Freedman J.A., Jinks-Robertson S. Genetic requirements for spontaneous and transcription-stimulated mitotic recombination in Saccharomyces cerevisiae. Genetics. 2002;162:15–27. doi: 10.1093/genetics/162.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu J., Phatnani H.P., Hsieh T.S., Greenleaf A.L. The phosphoCTD-interacting domain of Topoisomerase I. Biochem. Biophys. Res. Commun. 2010;397:117–119. doi: 10.1016/j.bbrc.2010.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu H.Y., Shyy S.H., Wang J.C., Liu L.F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 64.Aguilera A., Gaillard H. Transcription and recombination: when RNA meets DNA. Cold Spring Harbor Perspect. Biol. 2014;6 doi: 10.1101/cshperspect.a016543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boguslawski S.J., Smith D.E., Michalak M.A., Mickelson K.E., Yehle C.O., Patterson W.L., Carrico R.J. Characterization of monoclonal antibody to DNA:RNA and its application to immunodetection of hybrids. J. Immunol. Methods. 1986;89:123–130. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- 66.Phillips D.D., Garboczi D.N., Singh K., Hu Z., Leppla S.H., Leysath C.E. The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J. Mol. Recognit.: JMR. 2013;26:376–381. doi: 10.1002/jmr.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Teste M.A., Duquenne M., Francois J.M., Parrou J.L. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol. Biol. 2009;10:99. doi: 10.1186/1471-2199-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cerritelli S.M., Crouch R.J. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chon H., Sparks J.L., Rychlik M., Nowotny M., Burgers P.M., Crouch R.J., Cerritelli S.M. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res. 2013;41:3130–3143. doi: 10.1093/nar/gkt027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim N., Huang S.Y., Williams J.S., Li Y.C., Clark A.B., Cho J.E., Kunkel T.A., Pommier Y., Jinks-Robertson S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim N., Cho J.E., Li Y.C., Jinks-Robertson S. RNAratioDNA hybrids initiate quasi-palindrome-associated mutations in highly transcribed yeast DNA. PLoS Genet. 2013;9:e1003924. doi: 10.1371/journal.pgen.1003924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Potenski C.J., Niu H., Sung P., Klein H.L. Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms. Nature. 2014;511:251–254. doi: 10.1038/nature13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allen-Soltero S., Martinez S.L., Putnam C.D., Kolodner R.D. A Saccharomyces cerevisiae RNase H2 interaction network functions to suppress genome instability. Mol. Cell. Biol. 2014;34:1521–1534. doi: 10.1128/MCB.00960-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reijns M.A., Rabe B., Rigby R.E., Mill P., Astell K.R., Lettice L.A., Boyle S., Leitch A., Keighren M., Kilanowski F., et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu K., Chedin F., Hsieh C.L., Wilson T.E., Lieber M.R. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 76.Takahashi T., Burguiere-Slezak G., Van der Kemp P.A., Boiteux S. Topoisomerase 1 provokes the formation of short deletions in repeated sequences upon high transcription in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2011;108:692–697. doi: 10.1073/pnas.1012582108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim R.A., Wang J.C. A subthreshold level of DNA topoisomerases leads to the excision of yeast rDNA as extrachromosomal rings. Cell. 1989;57:975–985. doi: 10.1016/0092-8674(89)90336-x. [DOI] [PubMed] [Google Scholar]
- 78.Napierala M., Bacolla A., Wells R.D. Increased negative superhelical density in vivo enhances the genetic instability of triplet repeat sequences. J. Biol. Chem. 2005;280:37366–37376. doi: 10.1074/jbc.M508065200. [DOI] [PubMed] [Google Scholar]
- 79.Sun D., Hurley L.H. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J. Med. Chem. 2009;52:2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marchand C., Pourquier P., Laco G.S., Jing N., Pommier Y. Interaction of human nuclear topoisomerase I with guanosine quartet-forming and guanosine-rich single-stranded DNA and RNA oligonucleotides. J. Biol. Chem. 2002;277:8906–8911. doi: 10.1074/jbc.M106372200. [DOI] [PubMed] [Google Scholar]
- 81.Arimondo P.B., Riou J.F., Mergny J.L., Tazi J., Sun J.S., Garestier T., Helene C. Interaction of human DNA topoisomerase I with G-quartet structures. Nucleic Acids Res. 2000;28:4832–4838. doi: 10.1093/nar/28.24.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shuai L., Deng M., Zhang D., Zhou Y., Zhou X. Quadruplex-duplex motifs as new topoisomerase I inhibitors. Nucleosides Nucleotides Nucleic Acids. 2010;29:841–853. doi: 10.1080/15257770.2010.530635. [DOI] [PubMed] [Google Scholar]
- 83.Wright C.M., van der Merwe M., DeBrot A.H., Bjornsti M.A. DNA topoisomerase I domain interactions impact enzyme activity and sensitivity to camptothecin. J. Biol. Chem. 2015;290:12068–12078. doi: 10.1074/jbc.M114.635078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lotito L., Russo A., Chillemi G., Bueno S., Cavalieri D., Capranico G. Global transcription regulation by DNA topoisomerase I in exponentially growing Saccharomyces cerevisiae cells: activation of telomere-proximal genes by TOP1 deletion. J. Mol. Biol. 2008;377:311–322. doi: 10.1016/j.jmb.2008.01.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.