Abstract
Purine nucleosides on position 9 of eukaryal and archaeal tRNAs are frequently modified in vivo by the post-transcriptional addition of a methyl group on their N1 atom. The methyltransferase Trm10 is responsible for this modification in both these domains of life. While certain Trm10 orthologues specifically methylate either guanosine or adenosine at position 9 of tRNA, others have a dual specificity. Until now structural information about this enzyme family was only available for the catalytic SPOUT domain of Trm10 proteins that show specificity toward guanosine. Here, we present the first crystal structure of a full length Trm10 orthologue specific for adenosine, revealing next to the catalytic SPOUT domain also N- and C-terminal domains. This structure hence provides crucial insights in the tRNA binding mechanism of this unique monomeric family of SPOUT methyltransferases. Moreover, structural comparison of this adenosine-specific Trm10 orthologue with guanosine-specific Trm10 orthologues suggests that the N1 methylation of adenosine relies on additional catalytic residues.
INTRODUCTION
Transfer RNA (tRNA) plays a central role in one of the most crucial processes in living organisms: the translation of the genetic code into proteins. Control of the structural integrity and decoding potential of tRNA molecules is therefore of utmost importance. This process is influenced in all domains of life by the presence of post-transcriptional modifications on tRNA. On average 15% of the nucleotides of tRNA are modified and over a 100 chemically distinct tRNA modifications are known to occur in vivo (1). These modifications aid in the correct functioning of tRNA, and, depending on the nature and position of these modifications, they can influence the structure, stability, dynamics or decoding capacity/efficiency of the tRNA molecule (2).
Among the variety of post-transcriptional modifications found in tRNA, the addition of a methyl group is one of the most common. RNA methylation reactions are catalysed by a wide range of structurally distinct methyltransferases, which can add a methyl group on virtually every atom of the base or on the 2′-O-ribose atom (3–6). In particular, N1-methylated purine nucleosides are frequently found in tRNA and these modifications can be essential for the correct functioning of tRNA (1). For example, the N1-methylation of guanosine on position 37 of tRNA serves a dual role: it prevents frame shifts during translation (7) and it helps tRNAAsp in avoiding its mischarging with arginine by arginyl-tRNA synthetases (8). In addition, N1-methylated purines can also be essential for the proper tRNA structure formation as is seen for the human mitochondrial tRNALys, which is unable to fold to its characteristic cloverleaf conformation when it lacks the m1A9 modification (9). This latter modification reaction at position 9 of tRNA is catalysed in Eukarya and Archaea by the methyltransferase Trm10 (10,11). Recently Trm10 has specifically attracted attention due to the discovery that mutations in one of the human cytosolic Trm10 paralogues (TRMT10A) are associated with pathologies involving intellectual disabilities and abnormalities in the glucose homeostasis (12,13).
Among the Trm10 proteins, the representative from Saccharomyces cerevisiae has been characterized most thoroughly both structurally and biochemically. This enzyme belongs to the SpoU-TrmD (SPOUT) family of S-adenosyl-methionine (SAM) dependent methyltransferases (10,14). The SPOUT family proteins are characterised by a Rossmanoidal α/β fold with a C-terminal topological knot, which forms a binding pocket for the methyl donor SAM (15,16). Apart from the SPOUT domain, Trm10 proteins contain additional N- and C-terminal domains, for which the exact function has not yet been unequivocally determined. Surprisingly, the yeast Trm10 proteins were found to be monomeric in solution (14). This makes Trm10 unique within the SPOUT family, where all other known members function as dimers (16). In such ‘classical’ SPOUT dimers the dimeric nature of the protein is believed to aid in tRNA binding, which was also experimentally confirmed for the E. coli TrmL protein where a mutant that forces TrmL in a monomeric form was unable to bind its substrate tRNA (17). Therefore, the monomeric nature of yeast Trm10 raises questions on the mode of tRNA binding by this protein.
In humans, three paralogues of Trm10 are present: the cytosolic TRMT10A and TRMT10B proteins and the mitochondrial TRMT10C. Remarkably, the TRMT10C protein was found to serve as a subunit for the mitochondrial RNAse P (mtRNase P) complex, which is the endonuclease responsible for the removal of the tRNA 5′ extension during tRNA maturation (18). In contrast to cytoplasmic RNase P, which is a ribonucleoprotein, mtRNase P contains only proteins. Human mtRNase P is a complex of the three proteins: PRORP (proteinaceous RNase P), SDR5C1 and TRMT10C. In this complex, the presence of TRMT10C is required for the endonuclease activity of mtRNase P, although this requirement is independent of the methyltransferase activity (19). For its methyltransferase activity, mitochondrial TRMT10C in turn requires the presence of the SDR5C1 protein (19). This contrasts with the S. cerevisiae and human cytosolic Trm10 homologues that are able to independently catalyse the N1-methylation of purines in position 9 of tRNA (19).
The Trm10 orthologues differ with respect to their specificity towards the purine base at position 9 of the tRNA substrate. While the S. cerevisiae Trm10 homologue and the human cytosolic TRMT10A and TRMT10B specifically N1-methylate guanosine in position 9 of tRNA, the mitochondrial TRMT10C protein is able to N1-methylate both guanosine and adenosine (10,19). Such a dual substrate specificity has also been found in the archaeal Trm10 homologue of Thermococcus kodakarensis (TkTrm10), while the Trm10 homologue from the archaeon Sulfolobus acidocaldarius (SaTrm10) is specific for adenosine nucleotides only (11). Thus far the molecular basis of this difference in substrate specificity has remained unknown and it is difficult to imagine how certain enzymes can catalyse the methylation of both adenosine and guanosine considering the different protonation states of their N1 atoms (Supplementary Figure S1).
Currently, structural information is available for three Trm10 proteins. Crystal structures of the isolated SPOUT domains of the G9-specific Trm10 proteins of S. cerevisiae (ScTrm10) and Schizosaccharomyces pombe (SpTrm10) have recently been published (14). Moreover, a structure of the SPOUT domain of the G9-specific human cytosolic Trm10 orthologue TRMT10A, has been deposited in the protein database (PDB: 4fmw). In this paper we present the first crystal structure of a full length Trm10 protein (from the archaeon Sulfolobus acidocaldarius), which shows besides the SPOUT domain also the molecular structure of the N- and C-terminal domain. This is the first experimentally determined structure for an adenosine-specific Trm10 protein. We also present a model for substrate tRNA binding by SaTrm10 and we discuss the implications for the methyltransfer reaction.
MATERIALS AND METHODS
Expression and purification of SaTrm10 constructs
The SaTrm10 open reading frame (ORF) was amplified by polymerase chain reaction from genomic DNA and subsequently cloned in a pET28b vector between the restriction sites NdeI and XhoI as described in Kempenaers et al., 2010 (11). Expression of recombinant protein starting from this vector results in a protein with an N-terminal histidine-tag. The SaTrm10_1–249 construct and point mutants of SaTrm10 were created using the QuickChange site-directed mutagenesis kit (Stratagene). Alternatively, the SaTrm10 ORF (including the stopcodon) was cloned in the pET30 vector between the restriction sites NdeI and XhoI for expression as a tag-less protein.
After transformation of these vectors into E. coli Rosetta (DE3)pLysS, cells were grown at 37°C in Luria Broth media, supplemented with 25 μg/ml chloramphenicol and 50 μg/ml kanamycin, until an optical density at 600 nm (OD600) of 0.8 and protein expression was induced by adding 1mM isopropyl β-D-1-thiogalactopyranoside (IPTG). After overnight induction at 37°C, cells were harvested by centrifugation and resuspended in buffer A (50 mM TRIS pH 8, 1 M NaCl, 1 mM DTT) supplemented with 50 μg/ml DNase I, 5 mM MgCl2, AEBSF (at 0.1 mg/ml) and leupeptin (at 1 μg/ml). After cell disruption using a cell disruptor system (Constant Systems) and clearance of the lysate via centrifugation, all Trm10 constructs containing a histidine-tag were purified by a two-step purification protocol consisting of immobilized metal affinity chromatography followed by size exclusion chromatography. The supernatant was applied to a Ni2+-NTA sepharose column (GE Healthcare), and after extensive washing with buffer A supplemented with 10 mM imidazole, the his-tagged proteins were eluted with buffer A containing 150 mM imidazole. Fractions containing the protein of interest were pooled, concentrated and applied on either a Superdex200 (for full lengthSaTrm10) or Superdex75 (for SaTrm10_1–249) size exclusion chromatography column (GEHealthcare). The running buffer in this step for full length SaTrm10 protein was 20 mM TRIS pH 8, 100 mM NaCl, 50 mM imidazole and 1 mM DTT while for theSaTrm10_1–249 construct 20 mM TRIS pH 8, 100 mM NaCl, 1 mM DTT was used.
Selenomethionine enrichment of the SaTrm10_1–249 construct was accomplished by feedback inhibition of the methionine biosynthesis pathway. Here, minimal media was prepared according to the protocol described in Marley et al. (20) but supplemented with 0.5% glucose and 0.001% thiamine. The cultures were allowed to grow at 37°C until an OD600 of 0.7 was reached. At this point the amino acids lysine, phenylalanine, threonine (at a final concentration of 100 mg/l), isoleucine, leucine, valine (at a concentration of 50 mg/l) and selenomethionine (at a concentration of 60 mg/l) were added. Thirty minutes after addition of these amino acids, over-expression was started by addition of 1 mM IPTG. Expression of selenomethionine enriched protein was continued for 20 h at 37°C prior to harvesting the cells. Hereafter, the selenomethionine enriched proteins were treated and purified similar to the native protein.
The SaTrm10 construct lacking a histidine-tag was recombinantly expressed in a similar way. The purification protocol of this protein included an incubation step at 70°C for 20 min, after which the denatured proteins were removed by centrifugation for 45 min at 39000 rcf. This was followed by an ion exchange chromatography step on a Source 30S cation exchange column (GE Healthcare) run in 20 mM MES pH 6, 50 mM NaCl, 0.2 mM β-mercaptoethanol. Elution of the SaTrm10 protein from the column was accomplished by increasing NaCl concentration. The final purification step consisted of a size exclusion chromatography step identical to the one in the purification protocol of the histidine-tagged protein. The purity of the obtained sample, as determined by SDS-PAGE, was of similar quality as the purified histidine-tagged protein samples, although yields were lower. The histidine-tagged protein was used for all the experiments described in this manuscript, except when explicitly stated otherwise.
Multi-angle light scattering (MALS) and small-angle X-ray scattering (SAXS) analysis
Multi-angle light scatter experiments coupled to size exclusion chromatography (SEC-MALS) were performed with a Dawn Heleos (Wyatt technology) detector (using 9 angles) connected to an Agilent Bio-SEC 3 (size exclusion chromatography) column attached to an HPLC system (Waters). Twenty microlitres of a SaTrm10 protein solution at 10 mg/ml (either with or without histidine-tag) was injected on the column, which was eluted at a flow rate of 0.2 ml/min in 20 mM TRIS pH 8, 100 mM NaCl, 50 mM imidazole, 1 mM β-mercaptoethanol. The molar mass was calculated with the ASTRA 5.3.4.20 software.
All SAXS measurements were performed on the SWING beamline (Soleil, France) with an in line size exclusion chromatography system attached before the SAXS set-up. Here, 50 μl of protein at 10 mg/ml was injected on an Agilent Bio-SEC 3 column (300 Å pore size), which was eluted at 0.2 ml/min in a 20 mM Tris pH 8, 1 M NaCl, 1 mM DTT buffer. Eighteen SAXS frames, each exposed for 1.5 s, were collected at the size exclusion chromatography elution peak. The separate SAXS curves were qualitatively evaluated, scaled and averaged using the ATSAS software package (21). The oligomerisation state of the SaTrm10 protein was determined using the SAXSMoW application over a q-range of 0.01 Å−1 < q < 0.4 Å−1 (22). Fitting of the experimental SAXS curve (between a q-range of 0.01 Å−1 and 0.5 Å−1) with the theoretically calculated SAXS curve of the B-chain of the SaTrm10_SAH crystal structure was performed by CRYSOL and included a constant subtraction to remove systematic errors (23).
Crystallisation and data collection of SaTrm10 constructs
Crystals of selenomethionine enriched SaTrm10_1–249 were obtained by mixing protein (at a concentration of 15 mg/ml) with an equal volume of crystallisation buffer in a hanging drop vapour diffusion set-up. The crystallisation solution contained 8% ethylene glycol, 0.1 M HEPES pH 7.5, 10% PEG8k. Crystals were flash frozen with 25% glycerol as cryoprotectant. Diffraction data of these crystals were collected at 100 K at beamline Proxima I of Soleil (France). The Se-SAD data set of SaTrm10_1–249 (SaTrm10_1–249_SAD) was collected at the selenium absorption peak (0.979 Å) as determined by a fluorescence scan.
Subsequently crystals of selenomethionine enriched SaTrm10_1–249 were obtained using 2% dioxane, 0.05 M Bicine pH 9 and 15% PEG20k as crystallisation condition. Crystals were flash frozen in liquid nitrogen with crystallisation buffer containing 35% PEG200 as cryoprotectant. Diffraction data were collected at 100 K at beamline Proxima I of Soleil (France).
Crystals of full length SaTrm10 were obtained by mixing protein (at a concentration of 8 mg/ml) in the presence of 1 mM SAH with an equal volume of crystallisation buffer in a hanging drop vapour diffusion set-up. The crystallisation solution contained 0.4 M (NH4)2SO4, 0.1 M sodium acetate pH 4.5 and 15–20% PEG2000MME. Plate like crystals appeared several months after setting up the crystallisation plates at 20°C. Crystals were cryoprotected in the crystallisation condition supplemented with 15% PEG200 and 1 mM SAH and flash frozen in liquid nitrogen prior to data-collection at 100 K at beamline I24 of the Diamond light source (UK).
X-ray crystallography data processing, refinement and structure analysis
Data indexing, integration and scaling were done using the XDS suite (24). As criterion for high resolution cut-off an I/σ(I) > 1.5 and CC1/2 value > 50% was used. The selenomethionine substructure of theSaTrm10_1–249_SAD data set was calculated using SHELX C/D (25) in the HKL2MAP graphical user interface. Subsequent phasing, substructure refinement and density modification was performed in the autoSHARP pipeline (26) in ccp4 (27). An atomic model was built within the autoSHARP pipeline using ARP/wARP (28). This model of SaTrm10_1–249_SAD was later used as search model in the molecular replacement using Phaser for the SaTrm10_SAH and SaTrm10_1–249 data sets (29). Automated model building for the SaTrm10_SAH data set was performed by BUCCANEER (30), while ARP/wARP was used for the SaTrm10_1–249 data set (28). Cycles between manual model building using Coot (31) and automated refinement using Refmac5 (32) led to the final molecular models, in which the temperature factors were isotropically refined. The final models were validated with the MolProbity server (33). All data collection, processing and validation statistics are summarized in Supplementary Table S1.
Sequence alignments were performed with the T-coffee server (34) while structural alignments were done using PDBeFOLD and PyMOL (35). The Trm10 secondary structure was assigned by DSSP (36) and the topology map was created by PDBsum (37). The presented SAH omit map shows a Fo-Fc map contoured at 2 sigma created through the FTT program (38) in the CCP4 software package and visualised in pymol. Poisson-Boltzmann electrostatics were calculated with the PARSE force field in PDB2PQR (39) and visualised with APBS (40) in PyMOL. All figures were created in Pymol (www.pymol.org).
Isothermal titration calorimetry (ITC)
Prior to the ITC experiments, WT SaTrm10 and all the protein variants were dialysed extensively against a buffer consisting of 20 mM Tris pH 8, 100 mM NaCl, 50 mM imidazole and 1 mM β-mercaptoethanol. S-adenosyl methionine (SAM) was dissolved in this dialysis buffer and the concentration was determined using the absorbance at 260 nm (ϵ260 nm = 15400 M−1cm−1). ITC experiments were performed with a MicroCal ITC200 Instrument (GE Healthcare, Northampton, MA, USA) using a reference power of 10 microcal/sec. SAM (2 mM stock concentration) was titrated in the cell containing 150 μM SaTrm10 WT, SaTrm10 D220N, SaTrm10 D184N or SaTrm10 G180P protein in a 20 mM Tris pH 8, 100 mM NaCl, 50 mM imidazole and 1 mM β-mercaptoethanol buffer at 25°C. For each experiment a preliminary 0.4 μl injection (not included in data analysis) was followed by 20 injections of each 2 μl with a duration of 4 s at a stirring speed of 400 rpm and intervals of 180 s. The initial delay was 120 s. Data were fitted using MicroCal Origin version 7.0 (GE Healthcare, Northampton, MA, USA). To determine the heat of dilution, 2 mM SAM was titrated against buffer and a buffer solution was titrated against 150 μM SaTrm10.
In silico tRNA-SaTrm10 docking
Since the molecular structure of tRNAMet from Sulfolobus acidocaldarius is not available, we have built its homology model with ModeRNA (41) using the crystal structure of E. coli initiator tRNA (PDB: 3cw5) as a template. The model was refined using the QRNAS optimization tool (J. Stasiewicz and J.M. Bujnicki, unpublished). RNA-binding residues on the SaTrm10 surface were predicted using the PPRINT web server (42) and by identifying regions of positive electrostatic potential.
In order to model the structure of the complex between SaTrm10_SAH and tRNAMet, we used a computational docking approach. A set of 100 alternative models was produced using the hybrid modelling method PyRy3D (in house method available: http://genesilico.pl/pyry3d/) by docking either of the components (tRNA or Trm10) to the other. PyRy3D was configured to minimize violation of the following distance restraints: (i) between the SAM ligand (methyl group donor) and nucleotide 9 of tRNAMet (methyl group acceptor) and (ii) between the protein residue Lys64 (predicted to be involved in RNA binding) and any atom of tRNAMet. Obtained models were clustered according to their mutual similarity with the threshold of 5 Å. As a result we received three clusters consisting of 27, 11 and 8 models, respectively. Representatives (best scored models according to PyRy3D scoring function) of these clusters were selected for further analysis, in particular for experimental validation.
Using additional data from EMSA and activity measurements of Trm10 variants, we filtered models from the three obtained clusters. In particular, we applied FILTREST3D (43) to find a model that fulfils all distance restraints inferred from experimental analyses. This filtering procedure revealed that a highly-scored model from the second cluster (containing 11 models) agreed best with the entire set of experimental data. Therefore, we selected this model as the most probable representation of the structure of the complex. Finally, we optimized local protein-RNA contacts and the binding interface in the selected model by performing an optimisation procedure implemented in the NPDock tool (44). The final model of the complex is available via a publically accessible ftp server at: ftp://ftp.genesilico.pl/iamb/models/trm10.
Preparation of tRNA and in vitro tRNA binding and methyltransferase assays
The two tRNA methyltransferase assays used in this work are described by Droogmans et al. (45). Briefly, the first method consisted in measuring the amount of 14C transferred to total E. coli tRNA using [methyl-14C]SAM as methyl donor. The reaction mixture (400 μl) consisted of 50 mM Tris pH 8, 5 mM MgCl2, 60 μg unfractionated E. coli tRNA, 25 nCi [methyl-14C]SAM (50 mCi/mmol; GE Healthcare) and enzyme (5 to 10 μg). After an incubation of 30 min at 60°C the reaction was stopped by phenol extraction and the tRNAs were TCA (trichloroacetic acid) precipitated. Radioactive tRNA was captured on a filter and washed three times with ethanol prior to the measurement of the radioactivity in a scintillation counter.
The second type of tRNA methyltransferase assay involved in vitro transcribed 32P-labelled tRNAiMet of S. acidocaldarius (11). Radioactive (32P) in vitro transcripts were obtained according to Reyes and Abelson, 1987 (46) using [α32P]ATP. The reaction mixture for the tRNA-methyltransferase assay (400 μl) consisted of 50 mM Tris-HCl pH 8, 5 mM MgCl2, 106 cpm of the radioactive transcript, 500 μM SAM and 0.05–5 μg of purified enzyme. After 30 min of incubation at 60°C the reaction was stopped by phenol extraction and the tRNA was ethanol precipitated. The recovered radioactive tRNA was then completely digested by nuclease P1 (1 μg) in the presence of 5 μg total yeast tRNA as carrier. Modified nucleotides were analysed by 2D thin layer chromatography (TLC) on cellulose plates. Radioactive nucleotides corresponding to 5′ pA and 5′ pm1A (reference map of nucleotide migration described in Grosjean et al. (47)) were visualized by autoradiography and quantified by densitometry.
For the electrophoretic mobility shift assay (EMSA), 2.104 cpm of radioactive [α32P]ATP transcript from tRNAiMet of S. acidocaldarius was incubated in the presence of 1 μg enzyme in 50 mM Tris pH 8, 5 mM MgCl2 at 60°C. The binding reaction (with a total volume of 20 μl) was stopped by the addition of 4 μl of stop solution (0.05% bromophenol blue ogin 30% glycerol) and the mixture was separated by polyacrylamide gel electrophoresis (on a 6% PAGE gel with dimensions 190 mm x 160 mm x 1.5 mm) in TB buffer at room temperature. The electrophoresis was performed at a voltage of 180 V until the samples entered the gel and further at 150 V until the end of the run (1  h).
h).
RESULTS
Both eukaryal and archaeal Trm10 proteins exist as monomers in solution
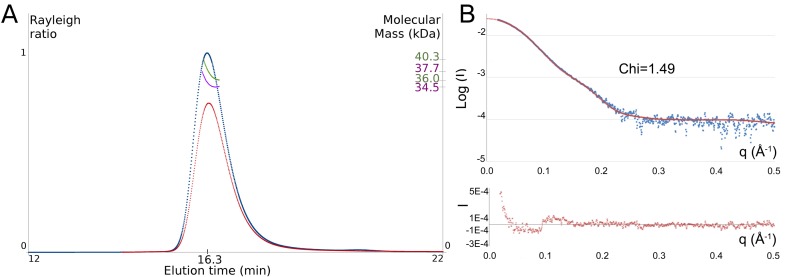
Until recently, it was believed that all SPOUT proteins function as dimers (16). However, in 2013 Shao et al. experimentally showed that yeast Trm10 is monomeric in solution (14). The uncommon monomeric nature of the yeast Trm10 proteins within the dimeric SPOUT family raises questions on the oligomerisation state of the archaeal Trm10 homologues. In order to experimentally determine the oligomerisation state of an archaeal Trm10 protein, we performed multi-angle light scatter (MALS) and small angle X-ray scatter (SAXS) experiments on purified Trm10 of Sulfolobus acidocaldarius (SaTrm10). Our multi-angle light scatter experiments were performed on a 10 mg/ml SaTrm10 protein solution which was run on a size exclusion chromatography column prior to the MALS measurement (SEC-MALS). In order to exclude any influence of the purification tag on the oligomerisation state, the SEC-MALS experiments were performed both on SaTrm10 containing an N-terminal histidine-tag and on the tag-less variant. This analysis yielded a molecular mass for the protein in solution of about 38 kDa for the histidine-tagged protein and 35 kDa for the tag-less protein (Figure 1A), values which corresponds best with the theoretical molecular mass of a SaTrm10 monomer (33.2 kDa for tag-less protein, 35.3 kDa for protein with histidine-tag). This monomeric nature of the SaTrm10 protein in solution was further confirmed by SAXS experiments, in which a 10 mg/ml SaTrm10 solution (with histidine-tag) was run on a size exclusion chromatography column in-line with SAXS data collection. The protein eluted as a single peak from the column and analysis of the scaled and merged scatter curves with the SAXSMoW software yielded an experimental molecular mass of 31.8 kDa, thus confirming the monomeric arrangement of the SaTrm10 protein in solution (Figure 1B). Hence, unlike other SPOUT proteins, both the eukaryal and archaeal Trm10 proteins exist (and likely function) as monomers in solution.
Figure 1.
(A) SEC-MALS analysis of SaTrm10 indicates that the protein is monomeric in solution (theoretical molecular mass of a monomer of SaTrm10 is 33.2 kDa for the tag-less protein and 35.3 kDa for the histidine-tagged variant). Light scatter (on the left axis) is shown as a function of the elution time for the histidine-tagged SaTrm10 (red) and tag-less SaTrm10 (blue). The experimentally calculated molecular mass of the main peak (on the right axis) is shown in green and purple, respectively. (B) Overlay of the experimental SAXS curve (blue) of SaTrm10 (histidine-tagged) with the theoretical SAXS curve calculated from the SaTrm10_SAH crystal structure (red). The residual of the fit is shown below.
Crystallisation and structure determination of SaTrm10
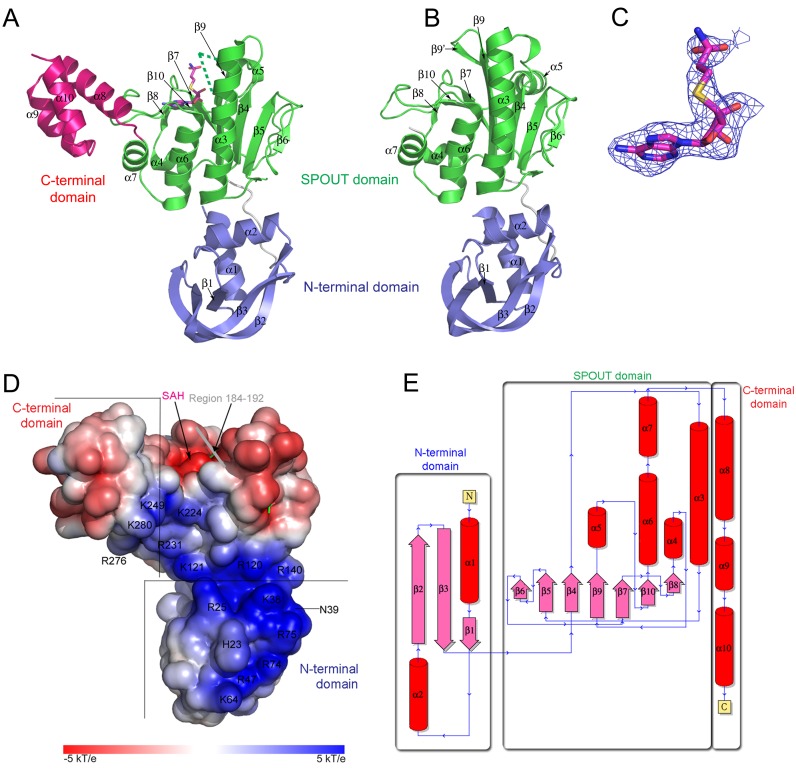
In order to gain insight in the tRNA binding mechanism and the molecular origin for substrate specificity of Trm10 proteins, we set out to solve the crystal structure of the adenosine specific SaTrm10 protein. In first instance a crystal structure of a truncated construct of SaTrm10, lacking the C-terminal domain and enriched in selenomethionine, was solved using SAD phasing at a resolution of 2.4 Å (this structure will be referred upon as SaTrm10_1–249_SAD). This structure was subsequently used to solve the structure of the same protein construct to a resolution of 2.1 Å using molecular replacement (SaTrm10_1–249). Given that theSaTrm10_1–249_SAD and SaTrm10_1–249 crystal structures are virtually identical and that the latter has a higher resolution, the SaTrm10_1–249 structure will be used for further analysis in the remainder of this manuscript. Crystals derived from full length SaTrm10 were obtained in an unrelated crystallisation condition and its crystal structure was determined at 2.5 Å resolution through molecular replacement using the SaTrm10_1–249_SAD structure as search model. In the crystal structure of full length SaTrm10 the reaction product S-adenosyl homocysteine (SAH) is bound in the SAM binding pocket and therefore this structure will be referred to as SaTrm10_SAH in the remainder of the text. The data-collection, processing and refinement statistics for all crystal structures are summarized in Supplementary Table S1.
The SaTrm10_1–249 (and SaTrm10_1–249_SAD) crystal structure contains one protein chain in the asymmetric unit and shows clear electron density for the complete polypeptide except for the three C-terminal amino acids. In contrast, the full length SaTrm10 crystal structure (SaTrm10_SAH) has two molecules in the asymmetric unit and in both molecules one region of the protein, residues 182–202 in the A-chain and residues 182–194 in the B-chain, could not be modelled due to missing electron density. While the SaTrm10_SAH crystal structure has SAH bound in both active sites of the asymmetric unit, the SaTrm10_1–249 crystal structure did not contain a bound ligand in the active site. The SAH found in the SaTrm10_SAH crystal structure was included (at a concentration of 1 mM) in the initial crystallisation condition. It should be noted that the electron density corresponding to the homocysteine moiety of SAH is better defined in the B-chain than in the A-chain.
SaTrm10 consists of a central SPOUT domain linked to an N-terminal domain and a C-terminal helical domain
The crystal structure of monomeric, full length SaTrm10_SAH displays an overall (tRNA resembling) L-shape and can be divided in three regions: a central domain (residues 89–246) which adopts a typical SPOUT fold, an N-terminal domain (residues 1–79) and a C-terminal helical domain (residues 247–292) (Figure 2A and Supplementary Figure S2). The N-terminal domain is formed by a curved β-sheet and two α-helices which together resemble a horse shoe. Two long, curved anti-parallel β-strands (β2 and β3) form the tip of the horse shoe, while the heel is formed by two α-helices (α1 and α2) which are linked to each other by a central β-strand (β1) which runs parallel to β3 (Figure 2). This fold seems to be unique among the known protein structures, since a search using the DALI server did not reveal the presence of a similar conformation in the PDB database (48). The N-terminal domain is connected by a long, flexible linker to the SPOUT domain. Both domains also interact with each other through H-bonds (connecting the backbone of Thr2 with the backbone of Glu234 and the side chain of Gln29 with the backbones of Arg120 and Leu123) and hydrophobic interactions (residues Leu3, Leu36, Leu84 and Trp124). The central SPOUT domain adopts the classical SPOUT fold, consisting of 7 parallel β-strands and 5 α-helices, and is linked to the C-terminal domain of SaTrm10_SAH through its most C-terminal α-helix (α7). The two longest α-helices of the SPOUT domain (α3 and α6) are orientated parallel to each other on one side of the central β-sheet. These helices form the dimerisation surface in classical SPOUT dimers. However, this type of dimerisation is sterically prohibited in monomeric SaTrm10 by the presence of helix α7, which interacts with helix α6, and by the C-terminal domain (Figure 2). Compared to the founding members of the SPOUT family, TrmD and TrmH, SaTrm10 possesses a significantly enlarged helix α3, while helix α6 is shortened (49,50). Finally, the C-terminal domain is composed of three α-helices, from which residues Arg255 and Gln247 of the first helix (α8) interact through H-bonds with loop β10-α6 of the SPOUT domain.
Figure 2.
Crystal structure of SaTrm10_SAH (A) and SaTrm10_1–249 (B), shown in the same orientation (the B molecule of the SaTrm10_SAH crystal structure is shown). In both panels, the domains of SaTrm10 are colour coded as followed: the N-terminal domain in blue, the SPOUT domain in green and the C-terminal domain in pink. In panel A residues 182–194, which are missing in the SaTrm10_SAH model, are indicated by a green dotted line and the reaction product SAH is shown as purple sticks. (C) An Fo-Fc omit map contoured at 2σ of SAH in the SaTrm10_SAH structure. (D) Electrostatic potential mapped on the solvent accessible surface of SaTrm10_SAH. Residues which are part of the positively charged surface are indicated. (E) A topology map of SaTrm10_SAH.
Globally, the SaTrm10 crystal structures (SaTrm10_SAH and SaTrm10_1–249) superimpose well on each other (Supplementary Figure S3). The individual N-terminal and SPOUT domains of both SaTrm10 structures superimpose nearly perfect with root mean square deviations (rmsd) between Cα atoms of 0.7 Å and 0.6 Å, respectively. However, the relative orientation of the N-terminal domain with respect to the SPOUT domain slightly differs in the SaTrm10_SAH and the SaTrm10_1–249 structure (Supplementary Figure S3A). This observation indicates that there is a certain degree of flexibility between the domains. Despite this flexibility, SAXS analysis of the full length SaTrm10 protein shows a very good fit (chi-value of 1.49 for q-values between 0.01 Å−1 and 0.5 Å−1) between the experimentally determined SAXS curve of SaTrm10 and a theoretically calculated SAXS curve from the B-chain of the SaTrm10_SAH crystal structure using the CRYSOL program (Figure 1B)) (23).
Some structural differences between the SaTrm10_SAH and SaTrm10_1–249 crystal structures are also observed within the SPOUT domains. The first difference concerns a small shift in the orientation of the N-terminus of helix α3, which likely also represents flexibility in the SaTrm10 structure (Supplementary Figure S3A). A second, more important, difference concerns the region preceding helix α5 (Supplementary Figure S3B). While this region (residues 182–194) could not be modelled in the SaTrm10_SAH crystal structure, it is clearly visible in the SaTrm10_1–249 crystal structure and forms a small anti-parallel β-sheet (containing strands β9 and β9′). However, superposition on SaTrm10_SAH shows that this region would block the SAM/SAH binding site. In particular the side chains of Ile182 and Arg188 of the SaTrm10_1–249 structure would occupy the same space as the ribose and homocysteine moiety of SAH in SaTrm10_SAH. At present it is unclear whether this conformation is mechanistically relevant (e.g. in facilitating the release of the SAH reaction product) or is an artefact of crystal packing.
The binding mode of the reaction product SAH to SaTrm10, as seen in the SaTrm10_SAH structure, is similar to the SAH binding of other SPOUT family members. Not only is the SAM/SAH binding pocket of SaTrm10_SAH located near the topological knot which characterises this family, also the ‘bended conformation’ of SAH (characterised by a O4′-C4′-C5′-Sδ dihedral angle of 84°) is maintained (Figure 2C) (51). The adenosine ring of SAH binds in a hydrophobic pocket formed by residues Leu158, Pro160, Ile179, Ile209, and Ile225 (Supplementary Figure S4). Additionally, H-bonds are formed between the backbone carbonyl and amide of Ile209 and the N6 and N1 atom of the adenine ring of SAH, and between the backbone of Gly180 (amide) and Leu158 (carbonyl) and the ribose moiety of SAH.
tRNA is bound via interactions with all three domains of SaTrm10
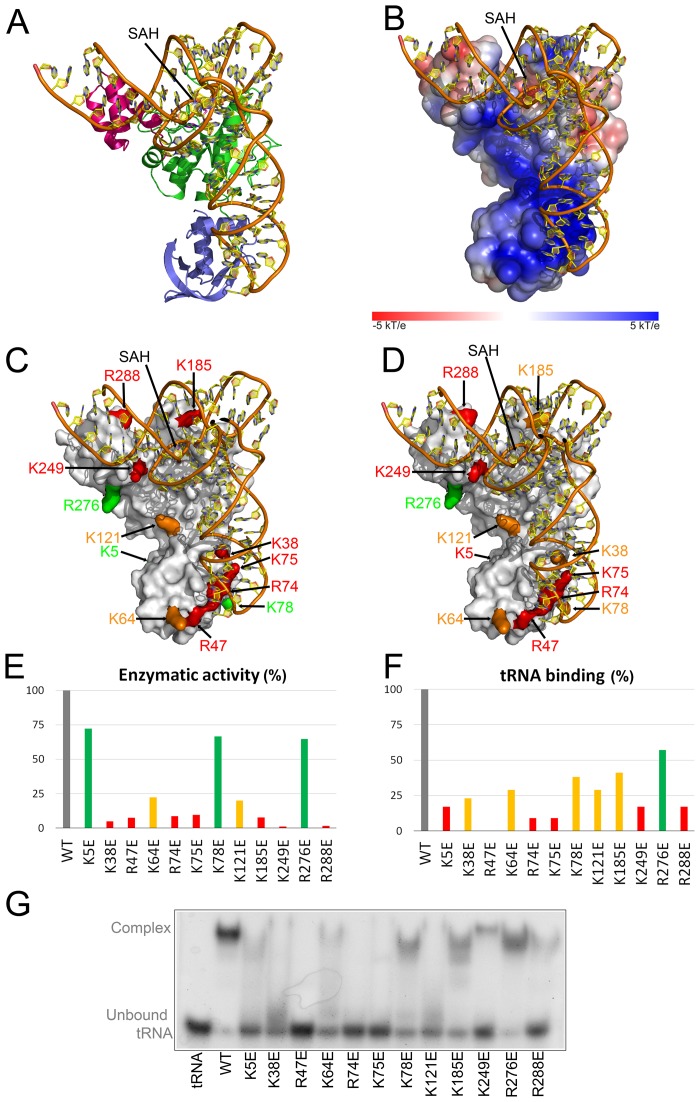
Mapping of the electrostatic potential on the solvent accessible surface of SaTrm10_SAH shows a large surface patch of positively charged residues spanning the N-terminal domain, the interface between the N-terminal domain and the SPOUT domain and finally continuing into the C-terminal domain (Figure 2D). It should also be noted that the unmodelled region 184–192 of SaTrm10-SAH contains three arginine and one lysine residues, which might become ordered and interact with the tRNA upon binding.
To obtain a more detailed image of the binding mode of tRNA to this monomeric SPOUT methyltransferase, we adopted a computational docking strategy as explained in detail in the Materials and Methods section. To this end, a homology model of the substrate, S. acidocaldarius initiator tRNAMet (tRNAiMet) was built using ModeRNA (41), using the crystal structure of E. coli initiator tRNA (PDB: 3cw5) as a template. As a result of the docking procedure (in which the region around the target nucleotide A9 was allowed to flip out from the core of the tRNA molecule to allow entry into the active site pocket of SaTrm10-SAH), three clusters of docking solutions were obtained (called cluster 1–3), of which the best-scored representatives are shown in Supplementary Figure S5 (called models 1–3, respectively). In model 1 and 2, the tRNAs are bound to SaTrm10-SAH in an approximately similar fashion, although they are rotated vis-à-vis each other. Especially in model 2 the L-shape of the tRNA coincides with the L-shape of the protein. In contrast, in model 3 the bound tRNA is rotated about 180° compared to the models 1 and 2.
To further discriminate between our docking models, a number of positively charged residues in the proposed interaction surfaces were substituted by glutamate residues (see Supplementary Figure S6 for an SDS-PAGE of the purified protein variants), and the ability of the resulting protein variants to bind tRNA substrate was determined by electrophoretic mobility shift assays (EMSA) at a single protein concentration (Figure 3G). In order to quantify the effect of the amino acid residue substitutions on tRNA binding the amount of unbound tRNA was measured on gel with densitometry and normalized against the value for the wild-type protein (set as 100% binding, Figure 3F). Additionally the ability of these variants to methylate tRNA was determined and compared to wild-type SaTrm10 (Figure 3E). Considering the well established influence of Mg2+ ions on the correct folding of tRNA and the corresponding effect on modification reactions in the tRNA core (52,53), MgCl2 at a concentration of 5 mM, which is optimal for activity, was added to all assay buffers involving tRNA substrates (i.e. EMSA and tRNA methyltransferase measurements). Three categories of (positively charged) residues were selected for mutagenesis: (i) residues in the ‘tip’ of the N-terminal domain that would interact mainly with tRNA in model 2 and to a lesser extend in model 1 and 3 (K38E, R47E, K64E, R74E and K75E); (ii) residues in the SPOUT domain, with one residue in the long α-helix 3 (K121E) and one residue in the flexible loop 184–192 (K185E); (iii) residues in the C-terminal domain (K249E, R276E and R288E). In addition to the amino acid residue substitutions on the proposed interface, two variants were included as negative controls that affect residues outside of the proposed binding interface (K5E and K78E).
Figure 3.
Docking model of tRNAiMet of S. acidocaldarius onto SaTrm10_SAH (model 2). For S. acidocaldarius tRNAiMet a homology model based on the structure of E. coli tRNAiMet was used. (A) Docking model 2 with both tRNAiMet and SaTrm10_SAH shown in cartoon representation. The N-terminal, SPOUT and C-terminal domains of SaTrm10 are coloured blue, green and pink, respectively. (B) Docking model 2 in the same orientation as in (A) with SaTrm10_SAH shown with the electrostatic potential mapped on its solvent accessible surface. (C and D) Results of the mutagenesis data, as evaluated with a tRNA methylation assay (C) and EMSA measurements (D), mapped on the surface of docking model 2 (an SDS-PAGE of these protein variants is shown in Supplementary Figure S6). The residues that have been substituted to validate the docking model are coloured according to the results of the tRNA methylation assay (C) or EMSA (D) using the following colour code: green: 50–100% activity/binding compared to WT; orange: 50–20% activity/binding compared to WT; red: 20–0% activity/binding compared to WT. (E, F and G) Results of the tRNA methylation assay using [methyl-14C]SAM (E), and of the single concentration EMSA (F and G) of protein variants that were generated in this study to validate the docking model. Panel F displays the fraction of bound tRNA normalized against the wild type protein, as determined from the EMSA shown in panel G. Hereto, the density of the band corresponding to unbound tRNA was measured and compared to the band in absence of protein (labelled ‘tRNA’). The value of the wild type protein was set as 100%. Residues Lys5 and Lys78 were chosen away from the proposed binding surface to serve as negative controls.
Neither of the two protein variants that were included as negative controls (K5E and K78E) significantly affect activity, while, surprisingly, an effect of the K5E variant on tRNA binding is observed. Supplementary Figure S5 shows the results of these mutagenesis data mapped on the accessible surface area in the context of models 1–3. This analysis clearly validates the involvement of the positively charged surface patch of SaTrm10 in tRNA binding. Moreover, the large effects on tRNA binding and methyltransferase activity observed for the mutations affecting residues in the tip of the N-terminal domain (especially true for R47E, R74E and K75E) strongly favour model 2 over model 1 and 3 (Figure 3C and D, and Supplementary Figure S5).
In the preferred docking model 2 the N-terminal domain of SaTrm10_SAH interacts with the anti-codon stem and loop of the tRNA substrate, while the SPOUT domain makes interactions with the variable loop, the T stem and the region around nucleotide A9. The C-terminal domain forms interactions with the acceptor stem and the part of the D-stem that is flipped out to accommodate A9 into the active site pocket (nucleotide 10–12) (Figure 3, panels A and B). Accordingly, two out of three amino acid residue substitutions in the C-terminal domain (K249E and R288A) have large effects on binding and even more pronounced on activity. This large effect on activity agrees with a role of this domain in the flip-out mechanism. Finally, the loop region 184–192, harbouring the catalytic Asp184 residue (see further) would be positioned ideally to close over the active site upon tRNA binding. In agreement with such an induced fit mechanism, a substitution of a positively charged residues in this loop (K185E) affects catalysis more than tRNA binding (Figure 3).
N1-adenosine methylation by SaTrm10 requires two catalytic aspartate residues
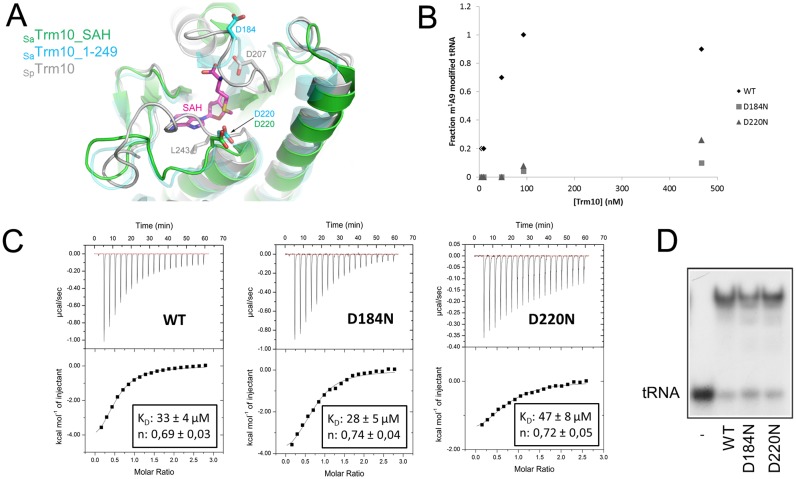
Based on the crystal structures of the SPOUT domain of the guanosine specific yeast Trm10 proteins and on mutagenesis data, Shao et al. suggested that an aspartate residue (Asp207 in SpTrm10 and Asp210 in ScTrm10) might function as a catalytic base, which is needed to deprotonate the N1 of the target guanosine during the methyl transfer reaction (14). Deprotonation of this nitrogen will significantly increase its nucleophilicity hence facilitating a nucleophilic attack on the methyl group of SAM. Although adenosine is not protonated at N1 at physiological pH, the proposed catalytic aspartate residue is strictly conserved in all eukaryal and archaeal Trm10 homologues and corresponds to Asp184 in the A9-specific SaTrm10 and Asp206 in A9/G9-specific TkTrm10 (Supplementary Figure S2). In the SaTrm10_SAH structure Asp184 is located on the unmodelled flexible loop (residues 184–192) in the vicinity of the SAM binding pocket.
Close examination of the SaTrm10_SAH crystal structure revealed the presence of another aspartate residue (Asp220) pointing into the active site and located close to the sulphur atom of the bound SAH (Figure 4A). Interestingly, this aspartate residue is conserved in the other archaeal (A9/G9 specific) Trm10 protein from T. kodakarensis (Asp245), but not in the eukaryal guanosine-specific Trm10 homologues (Supplementary Figure S2). To evaluate the importance of both aspartate residues in the tRNA methyltransferase mechanism of SaTrm10, we mutated them to asparagine residues (D184N and D220N). These conservative substitutions would preserve the hydrogen-bonding capabilities while disrupting any charge-charge interactions or proton transfer reactions at these positions. Subsequently, the methyltransferase activity of both protein variants was measured at different enzyme concentrations and compared to the corresponding wild-type SaTrm10 activity (Figure 4B). This analysis revealed that the activity of both variants is reduced to less than 10% of the wild-type activity, pointing to an important role of both residues. To test whether any of the above substitutions has an effect on the binding of the methyl donor SAM, the binding affinity for this substrate was determined using isothermal titration calorimetry (ITC) measurements (Figure 4C). Moreover, as an extra control also the heat of dilution was measured by titrating SAM into buffer or by titrating buffer into a SaTrm10 solution (Supplementary Figure S7). As a positive control, SAM binding to a protein variant, G180P, affecting an amino acid residue directly interacting with SAM was also measured (Supplementary Figures S4 and S7). Wild-type SaTrm10 binds SAM with an equilibrium dissociation constant (KD) of 33 ± 4 μM. In some cases very mild precipitation of the protein in the measure cell was observed after the experiment which might lead to the observed stoichiometry slightly below 1. The equilibrium dissociation constant of SaTrm10 for SAM is higher than the corresponding values reported for S. cerevisiae and S. pombe Trm10 (7.63 μM and 4.46 μM, respectively), but is in the same range as found for other SPOUT methyltransferases (14,54). The D184N and D220N variants have binding affinities for SAM which are comparable to the one of the wild-type protein (KD = 28 ± 5 μM and KD = 47 ± 8 μM, respectively), indicating that their observed effect on activity is not caused by an impaired SAM binding. This contrasts with what has been found in S. cerevisiae and S. pombe Trm10, where mutation of the residue corresponding to D184 (D210 and D207, respectively) abrogates SAM binding (14). Reassuringly, no binding under the used conditions is observed for the G180P variant, in agreement with the interaction of the latter residue with both hydroxyl groups of the ribose moiety of SAM. Next, the tRNA binding of both protein variants (D184N and D220N) was compared to the wild-type protein using an EMSA at one single protein concentration (using the same conditions as for the interface mutants described above). This analysis shows that tRNA binding is nearly unaltered by both protein variations (Figure 4D). Together these data show that SaTrm10 needs two aspartate residues to catalyse the methyl transfer reaction to the N1 atom of A9. Thus, although the reaction mechanism is anticipated to be significantly different for the transfer of a methyl group to a protonated N1 of guanosine compared to an unprotonated N1 of adenosine, the crucial role of Asp184 seems to be conserved in both G9- and A9-specific methyltransferases. On the other hand SaTrm10 requires a second aspartate residue for catalysis (Asp220). Since this residue is present in both the archaeal Trm10 homologues that can methylate adenosine (SaTrm10 and TkTrm10), but is absent in the guanosine specific eukaryal proteins, this hints toward a specific role of this residue in the N1-methylation of adenosine.
Figure 4.
Role of Asp184 and Asp220 in the catalytic mechanism of SaTrm10. (A) Overlay of the crystal structures of SaTrm10_SAH (green), SaTrm10_1–249 (blue) and SpTrm10 (grey) showing the position of the catalytic residues (Asp184/Asp220 in SaTrm10 and Asp207 in SpTrm10). The bound reaction product SAH is shown as purple sticks. Note that residue Asp184 could not be modelled in the SaTrm10_SAH crystal structure. (B) Methyltransferase activity assay (with in vitro transcribed 32P-labelled SatRNAiMet) comparing the catalytic activity of wild-type SaTrm10 with the one of the D184N and D220N variants at different protein concentrations (and a fixed tRNA concentration). (C) Isothermal titration calorimetry (ITC) assay comparing the binding of SAM to either wild-type SaTrm10 or the D184N or D220N variant. Extra control experiments to determine the heat of dilution and using a protein variant affected directly in the SAM binding pocket can be found in Supplementary Figure S7. (D) Electrophoretic mobility shift assay (EMSA) showing the binding of SatRNAiMet on the wild-type, D184N or D220N SaTrm10 variant.
DISCUSSION
SPOUT enzymes form a single superfamily of RNA methyltransferases that are characterized by an unusual α/β fold with a deep topological knot (15,16). This knot forms the binding site for the SAM methyl donor, and, in the common case where the SPOUT methyltransferases functions as dimers, the active site is located at the dimer interface. Besides a few exceptions like TrmL and RlmH, most SPOUT proteins have N- and/or C- terminal extensions fused to the SPOUT domain, which are assumed to participate in substrate (RNA) binding (16). Trm10 is a SPOUT superfamily member, conserved in eukaryotes and archaea, which is involved in the methylation of the N1 atom of purine nucleosides at position 9 of tRNA. Previously crystal structures of the SPOUT domain of the m1G9-forming Trm10 proteins from S. cerevisiae and S. pombe have been reported (14). Surprisingly, it was shown that these proteins function as monomers in solution, unlike all other so far studied SPOUT methyltransferases. In this paper we present the first experimental structure of a full length Trm10 homologue from the archaeon S. acidocaldarius (SaTrm10_SAH), that catalyses the N1-methylation of adenosine residues at position 9 of tRNA (m1A9). The crystal structure as well as our biochemical data (SAXS and SEC-MALS) clearly show that this protein also exists as a monomer in solution, indicating that this is a general feature of both eukaryal and archaeal Trm10 enzymes. The structure of full length SaTrm10 shows that the protein adopts an overall L-shaped form which seems to mimic the overall shape of a tRNA molecule. This fold is constructed from three distinguishable domains, consisting next to the SPOUT domain of an N-terminal domain with a unique fold and an α-helical C-terminal domain. Sequence analysis reveals that also the ScTrm10, SpTrm10, human TRMT10A and TkTrm10 homologues have such N- and C-terminal extensions with regard to the SPOUT domain, although the sequence conservation in these regions is low between eukaryal and archaeal Trm10 proteins (Supplementary Figure S2). Therefore, it remains to be determined whether the N- and C-terminal domains of all Trm10 proteins are structurally identical to the here described domains of SaTrm10.
So far, very little information has been available on the RNA binding mode of SPOUT methyltransferases. Only one crystal structure of a SPOUT methyltransferase/RNA complex was previously reported: that of the rRNA methyltransferase Nep1 in complex with a 14-base RNA fragment (55). In addition, the crystal structure of a TrmD/tRNA complex was released during the revision process of this manuscript (56). In both these structures the RNA binds in a cleft on the interface of the subunits of the dimer. A similar RNA binding mode is clearly impossible for the monomeric Trm10 proteins. The SPOUT domain of the ScTrm10 and SpTrm10 homologues was shown to be able to bind to substrate tRNA on its own, although the N-terminal domain also significantly contributes to binding (14). However, we find that in SaTrm10 amino acid substitutions in both the N-terminal and C-terminal domain completely disrupt tRNA binding, indicating differences in the binding mechanism between eukaryal and archaeal Trm10 homologues. This difference is also reflected in the electrostatics of the SPOUT domain of SaTrm10_SAH, which is mostly negatively charged, while the SPOUT domains of the eukaryal Trm10 proteins contain a clear positively charged surface (Supplementary Figure S8, panels C–F). In this regard, a particular structural difference between the SPOUT domains of archaeal and eukaryal Trm10 homologues concerns the region between β5 and β6 (residues 133–142 in SaTrm10) (Supplementary Figure S8A and B). In all eukaryal proteins an α-helix is present in this region, which is absent in the SaTrm10 structure. Two residues in this α-helix have been shown to be involved in tRNA binding in SpTrm10 (14). Interestingly, this α-helix also seems to be important for the correct in vivo functioning of ScTrm10 since a serine to proline variant in this helix influences the translational termination efficiency in S. cerevisiae (57). The exact molecular mechanism of how a mutation in this helix of ScTrm10 is influencing the translational termination efficiency is still unclear at the moment, but a possibility would be that this mutation in ScTrm10 prevents the binding and subsequent methylation of certain substrate tRNAs.
The availability of the full length structure of SaTrm10 allowed us to calculate plausible docking models, which illustrate how tRNA can bind to a monomeric SPOUT protein. Using this strategy, three alternative docking solutions were obtained. Subsequent experimental analysis clearly favoured model 2 over the other two models (Figure 3). This docking model shows how the L-shape of the protein nearly perfectly complements the L-shape of the tRNA molecule. Interestingly, the SPOUT domain forms interactions with the tRNA using its helices α3 and α6. In other SPOUT proteins, these helices form the dimerization surface between protomers (16), indicating that the Trm10 proteins have retuned their dimerization surface to serve in tRNA binding. Apart from the SPOUT domain, major interactions are formed between the substrate tRNA and the N-terminal and C-terminal domains, where the N-terminal domain interacts with the anti-codon stem and loop while the C-terminal domain forms interactions with the acceptor stem and the part of the D-stem that is flipped out to accommodate A9 into the active site pocket. A flexible loop that could not be modelled in the crystal structure (amino acids 184–192) would be ideally positioned to close over the active site upon tRNA binding. This loop harbours residue Asp184, which our data shows to be indispensable for catalysis of the m1A9 modification reaction.
A remarkable aspect of the Trm10 proteins is the difference in substrate specificity between different homologues of this protein (10,11). While the SaTrm10 protein, investigated in this study, is specific for the N1-methylation of adenosine residues at position 9 of tRNA, the yeast Trm10 and human TRMT10A and TRMT10B orthologues are specific for the N1-methylation of guanosine. Moreover, the human mitochondrial TRMT10C orthologue and the orthologue from the archaeon T. kodakarensis show a dual adenosine/guanosine specificity (11,19). This latter promiscuity in substrate specificity is surprising, considering that the N1 atoms of guanosine and adenosine adopt different protonation states under physiological conditions: whereas the N1 atom of guanosine is mainly protonated (pKa ≈ 9.2), the N1 atom of adenosine is not (pKa ≈ 3.5) (58). Therefore, the currently described reaction mechanisms for the N1-methylation of adenosine or guanosine are fundamentally different. The N1-methylation of guanosine in tRNA has been most thoroughly studied in the structurally unrelated m1G37 methyltransferases Trm5 and TrmD (59,60). Both these enzymes rely on a catalytic base (aspartate or glutamate) to deprotonate the N1 atom of guanosine in order to facilitate the nucleophilic attack of the N1 atom on the methyl group of SAM. In contrast, N1-methylation of adenosine does not require deprotonation of the N1 atom, although the nucleophilicity of the N1 atom is expected to be intrinsically low considering its low pKa. In this context it has been observed that the m1A58 methyltransferase TrmI from Thermus thermophilus also relies on a catalytic aspartate residue (61). Here, two possible roles were suggested for this aspartate: (i) It could minimise the Gibbs free energy of the transition state by positioning the methyl donor SAM and target adenosine in an optimal orientation and distance for the N1-methylation reaction or (ii) it could deprotonate the N6 atom of adenosine to generate the exocyclic imino tautomer. This would subsequently activate the lone electron pair on the N1 atom of adenosine, which would in the same elementary step perform a nucleophilic attack on the methyl group of SAM (61).
A catalytically important aspartate residue has been identified in the yeast Trm10 proteins (Asp210 in ScTrm10 and Asp207 in SpTrm10) which is also conserved in the guanosine-specific human TRMT10A (Asp210) (14). In accordance with the mechanism of other m1G methyltransferases, a role as catalytic base in the deprotonation of the N1 atom of the target guanosine has been proposed. Remarkably, this aspartate residue is also conserved in the m1A-specific SaTrm10 (Asp184) and the m1A/m1G-specific TkTrm10 (Asp206). Although the N1 position is a priori expected to be unprotonated in adenosine, we found that this Asp184 residue is required for catalysis of the m1A formation in SaTrm10. Moreover, structural comparison and sequence alignments revealed the presence of a second aspartate residue in enzymes able to catalyse the m1A formation (Asp220 in SaTrm10 and Asp245 in TkTrm10), but absent in m1G-specific methyltransferases. Our mutagenesis and kinetic/binding analysis shows that also this residue is required for efficient catalysis of m1A formation in SaTrm10. Currently, the exact role of the two aspartate residues required for activity of SaTrm10 is still unclear. Further detailed kinetic analyses will be required to shed more light on the reaction mechanism and on the molecular basis of the observed differences in substrate specificity among different Trm10 orthologues.
Supplementary Material
Acknowledgments
We would like to thank the staff at the beamlines Proxima 1 and Swing of Soleil in France and at the beamline I24 of Diamond in the UK for assistance during data collection. We further acknowledge M. Kempenaers and J. Somme for their contribution to the early stages of this work, and Dr. Guy Vandenbussche and Prof. Geert Baggerman for help with setting up experiments and discussions.
Footnotes
Present address: Bart Van Laer, ESRF - European Synchrotron Radiation Facility, Structural Biology Group, 71 Avenue des Martyrs, 38000 Grenoble, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fonds voor Wetenschappelijk Onderzoek (FWO); Agentschap voor Innovatie door Wetenschap en Technologie (IWT); Hercules foundation and a Strategic Research Program Financing of the VUB [to B.V.L., L.W., E.D., W.V.]; Fonds Brachet and the Fonds D. et A. Van Buren [to M.R., L.D.]; European Research Council [grant 261351 to J.M.B.]; European Community's Seventh Framework Programme (FP7/2007–2013) [grant 316125 to Jacek Kuznicki and under BioStruct-X, 283570]. Funding for open access charge: VUB Strategic Research Program (SRP34).
Conflict of interest statement. None declared.
REFERENCES
- 1.Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M., et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackman J.E., Alfonzo J.D. Transfer RNA modifications: nature's combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA. 2013;4:35–48. doi: 10.1002/wrna.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Somme J., Van Laer B., Roovers M., Steyaert J., Versees W., Droogmans L. Characterization of two homologous 2′-O-methyltransferases showing different specificities for their tRNA substrates. RNA. 2014;20:1257–1271. doi: 10.1261/rna.044503.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fislage M., Roovers M., Tuszynska I., Bujnicki J.M., Droogmans L., Versees W. Crystal structures of the tRNA:m2G6 methyltransferase Trm14/TrmN from two domains of life. Nucleic Acids Res. 2012;40:5149–5161. doi: 10.1093/nar/gks163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamdane D., Argentini M., Cornu D., Myllykallio H., Skouloubris S., Hui-Bon-Hoa G., Golinelli-Pimpaneau B. Insights into folate/FAD-dependent tRNA methyltransferase mechanism: role of two highly conserved cysteines in catalysis. J. Biol. Chem. 2011;286:36268–36280. doi: 10.1074/jbc.M111.256966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boal A.K., Grove T.L., McLaughlin M.I., Yennawar N.H., Booker S.J., Rosenzweig A.C. Structural basis for methyl transfer by a radical SAM enzyme. Science. 2011;332:1089–1092. doi: 10.1126/science.1205358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hagervall T.G., Tuohy T.M., Atkins J.F., Björk G.R. Deficiency of 1-methylguanosine in tRNA from Salmonella typhimurium induces frameshifting by quadruplet translocation. J. Mol. Biol. 1993;232:756–765. doi: 10.1006/jmbi.1993.1429. [DOI] [PubMed] [Google Scholar]
- 8.Pütz J., Florentz C., Benseler F., Giegé R. A single methyl group prevents the mischarging of a tRNA. Nat. Struct. Biol. 1994;1:580–582. doi: 10.1038/nsb0994-580. [DOI] [PubMed] [Google Scholar]
- 9.Helm M., Brulé H., Degoul F., Cepanec C., Leroux J.P., Giegé R., Florentz C. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res. 1998;26:1636–1643. doi: 10.1093/nar/26.7.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackman J.E., Montange R.K., Malik H.S., Phizicky E.M. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA N. Y. N. 2003;9:574–585. doi: 10.1261/rna.5070303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kempenaers M., Roovers M., Oudjama Y., Tkaczuk K.L., Bujnicki J.M., Droogmans L. New archaeal methyltransferases forming 1-methyladenosine or 1-methyladenosine and 1-methylguanosine at position 9 of tRNA. Nucleic Acids Res. 2010;38:6533–6543. doi: 10.1093/nar/gkq451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillis D., Krishnamohan A., Yaacov B., Shaag A., Jackman J.E., Elpeleg O. TRMT10A dysfunction is associated with abnormalities in glucose homeostasis, short stature and microcephaly. J. Med. Genet. 2014;51:581–586. doi: 10.1136/jmedgenet-2014-102282. [DOI] [PubMed] [Google Scholar]
- 13.Igoillo-Esteve M., Genin A., Lambert N., Désir J., Pirson I., Abdulkarim B., Simonis N., Drielsma A., Marselli L., Marchetti P., et al. tRNA methyltransferase homolog gene TRMT10A mutation in young onset diabetes and primary microcephaly in humans. PLoS Genet. 2013;9:e1003888. doi: 10.1371/journal.pgen.1003888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao Z., Yan W., Peng J., Zuo X., Zou Y., Li F., Gong D., Ma R., Wu J., Shi Y., et al. Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate. Nucleic Acids Res. 2014;42:509–525. doi: 10.1093/nar/gkt869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anantharaman V., Koonin E.V., Aravind L. SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 2002;4:71–75. [PubMed] [Google Scholar]
- 16.Tkaczuk K.L., Dunin-Horkawicz S., Purta E., Bujnicki J.M. Structural and evolutionary bioinformatics of the SPOUT superfamily of methyltransferases. BMC Bioinformatics. 2007;8:73. doi: 10.1186/1471-2105-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu R.-J., Zhou M., Fang Z.-P., Wang M., Zhou X.-L., Wang E.-D. The tRNA recognition mechanism of the minimalist SPOUT methyltransferase, TrmL. Nucleic Acids Res. 2013;41:7828–7842. doi: 10.1093/nar/gkt568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holzmann J., Frank P., Löffler E., Bennett K.L., Gerner C., Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Vilardo E., Nachbagauer C., Buzet A., Taschner A., Holzmann J., Rossmanith W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase–extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 2012;40:11583–11593. doi: 10.1093/nar/gks910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marley J., Lu M., Bracken C. A method for efficient isotopic labeling of recombinant proteins. J. Biomol. NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 21.Petoukhov M.V., Franke D., Shkumatov A.V., Tria G., Kikhney A.G., Gajda M., Gorba C., Mertens H.D.T., Konarev P.V., Svergun D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer H., de Oliveira Neto M., Napolitano H.B., Polikarpov I., Craievich A.F. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J. Appl. Crystallogr. 2009;43:101–109. [Google Scholar]
- 23.Koch M.H.J., Barberato C., Svergun D.I. CRYSOL - a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- 24.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheldrick G.M. Experimental phasing with SHELXC / D / E: combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vonrhein C., Blanc E., Roversi P., Bricogne G. Macromolecular Crystallography Protocols, Volume 2. Vol. 364. New Jersey: Humana Press; 2006. Automated structure solution With autoSHARP; pp. 215–230. [DOI] [PubMed] [Google Scholar]
- 27.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A., et al. Overview of the CCP 4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langer G., Cohen S.X., Lamzin V.S., Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B., Snoeyink J., Richardson J.S., et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Notredame C., Higgins D.G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 35.Krissinel E., Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 36.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 37.Laskowski R.A. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 2001;29:221–222. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Read R.J., Schierbeek A.J. A phased translation function. J. Appl. Crystallogr. 1988;21:490–495. [Google Scholar]
- 39.Dolinsky T.J., Czodrowski P., Li H., Nielsen J.E., Jensen J.H., Klebe G., Baker N.A. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rother M., Rother K., Puton T., Bujnicki J.M. ModeRNA: a tool for comparative modeling of RNA 3D structure. Nucleic Acids Res. 2011;39:4007–4022. doi: 10.1093/nar/gkq1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z.-P., Wu L.-Y., Wang Y., Zhang X.-S., Chen L. Prediction of protein-RNA binding sites by a random forest method with combined features. Bioinforma. Oxf. Engl. 2010;26:1616–1622. doi: 10.1093/bioinformatics/btq253. [DOI] [PubMed] [Google Scholar]
- 43.Gajda M.J., Tuszynska I., Kaczor M., Bakulina A.Y., Bujnicki J.M. FILTREST3D: discrimination of structural models using restraints from experimental data. Bioinforma. Oxf. Engl. 2010;26:2986–2987. doi: 10.1093/bioinformatics/btq582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuszynska I., Magnus M., Jonak K., Dawson W., Bujnicki J.M. NPDock: a web server for protein-nucleic acid docking. Nucleic Acids Res. 2015;43:W425–W430. doi: 10.1093/nar/gkv493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Droogmans L., Roovers M., Bujnicki J.M., Tricot C., Hartsch T., Stalon V., Grosjean H. Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res. 2003;31:2148–2156. doi: 10.1093/nar/gkg314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reyes V.M., Abelson J. A synthetic substrate for tRNA splicing. Anal. Biochem. 1987;166:90–106. doi: 10.1016/0003-2697(87)90551-3. [DOI] [PubMed] [Google Scholar]
- 47.Grosjean H., Droogmans L., Roovers M., Keith G. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 2007;425:55–101. doi: 10.1016/S0076-6879(07)25003-7. [DOI] [PubMed] [Google Scholar]
- 48.Holm L., Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nureki O., Watanabe K., Fukai S., Ishii R., Endo Y., Hori H., Yokoyama S. Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme. Structure. 2004;12:593–602. doi: 10.1016/j.str.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 50.Elkins P.A., Watts J.M., Zalacain M., van Thiel A., Vitazka P.R., Redlak M., Andraos-Selim C., Rastinejad F., Holmes W.M. Insights into catalysis by a knotted TrmD tRNA methyltransferase. J. Mol. Biol. 2003;333:931–949. doi: 10.1016/j.jmb.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 51.Kurowski M.A., Sasin J.M., Feder M., Debski J., Bujnicki J.M. Characterization of the cofactor-binding site in the SPOUT-fold methyltransferases by computational docking of S-adenosylmethionine to three crystal structures. BMC Bioinformatics. 2003;4:9. doi: 10.1186/1471-2105-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Draper D.E. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Serebrov V., Vassilenko K., Kholod N., Gross H.J., Kisselev L. Mg2+ binding and structural stability of mature and in vitro synthesized unmodified Escherichia coli tRNAPhe. Nucleic Acids Res. 1998;26:2723–2728. doi: 10.1093/nar/26.11.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boundy S., Safo M.K., Wang L., Musayev F.N., O'Farrell H.C., Rife J.P., Archer G.L. Characterization of the Staphylococcus aureus rRNA methyltransferase encoded by orfx, the gene containing the staphylococcal chromosome Cassette mec (SCCmec) insertion site. J. Biol. Chem. 2013;288:132–140. doi: 10.1074/jbc.M112.385138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas S.R., Keller C.A., Szyk A., Cannon J.R., Laronde-Leblanc N.A. Structural insight into the functional mechanism of Nep1/Emg1 N1-specific pseudouridine methyltransferase in ribosome biogenesis. Nucleic Acids Res. 2011;39:2445–2457. doi: 10.1093/nar/gkq1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ito T., Masuda I., Yoshida K., Goto-Ito S., Sekine S., Suh S.W., Hou Y.-M., Yokoyama S. Structural basis for methyl-donor–dependent and sequence-specific binding to tRNA substrates by knotted methyltransferase TrmD. Proc. Natl. Acad. Sci. 2015;112:E4197–E4205. doi: 10.1073/pnas.1422981112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Torabi N., Kruglyak L. Variants in SUP45 and TRM10 underlie natural variation in translation termination efficiency in Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1002211. doi: 10.1371/journal.pgen.1002211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sober H. A. Handbook of biochemistry: Selected data for molecular biology. 2nd edn. Cleveland: The Chemical Rubber Company; 1970. [Google Scholar]
- 59.Ahn H.J., Kim H.-W., Yoon H.-J., Lee B.I., Suh S.W., Yang J.K. Crystal structure of tRNA(m1G37)methyltransferase: insights into tRNA recognition. EMBO J. 2003;22:2593–2603. doi: 10.1093/emboj/cdg269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Christian T., Lahoud G., Liu C., Hoffmann K., Perona J.J., Hou Y.-M. Mechanism of N-methylation by the tRNA m1G37 methyltransferase Trm5. RNA N. Y. N. 2010;16:2484–2492. doi: 10.1261/rna.2376210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barraud P., Golinelli-Pimpaneau B., Atmanene C., Sanglier S., Van Dorsselaer A., Droogmans L., Dardel F., Tisné C. Crystal structure of Thermus thermophilus tRNA m1A58 methyltransferase and biophysical characterization of its interaction with tRNA. J. Mol. Biol. 2008;377:535–550. doi: 10.1016/j.jmb.2008.01.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.