Abstract
Mice deficient in the glucocorticoid‐regenerating enzyme 11β‐HSD1 resist age‐related spatial memory impairment. To investigate the mechanisms and pathways involved, we used microarrays to identify differentially expressed hippocampal genes that associate with cognitive ageing and 11β‐HSD1. Aged wild‐type mice were separated into memory‐impaired and unimpaired relative to young controls according to their performance in the Y‐maze. All individual aged 11β‐HSD1‐deficient mice showed intact spatial memory. The majority of differentially expressed hippocampal genes were increased with ageing (e.g. immune/inflammatory response genes) with no genotype differences. However, the neuronal‐specific transcription factor, Npas4, and immediate early gene, Arc, were reduced (relative to young) in the hippocampus of memory‐impaired but not unimpaired aged wild‐type or aged 11β‐HSD1‐deficient mice. A quantitative reverse transcriptase‐polymerase chain reaction and in situ hybridisation confirmed reduced Npas4 and Arc mRNA expression in memory‐impaired aged wild‐type mice. These findings suggest that 11β‐HSD1 may contribute to the decline in Npas4 and Arc mRNA levels associated with memory impairment during ageing, and that decreased activity of synaptic plasticity pathways involving Npas4 and Arc may, in part, underlie the memory deficits seen in cognitively‐impaired aged wild‐type mice.
Keywords: corticosterone, Y‐maze, spatial memory, microarray, glucocorticoids
Cognitive decline is a prominent feature of normal ageing in humans and rodents. However, large inter‐individual differences exist ranging from little change to mild or severe impairments 1, 2. Glucocorticoids (GCs; largely cortisol in humans, corticosterone in rats and mice), released from the adrenal cortex after stress or diurnal activation of the hypothalamic‐pituitary‐adrenal axis, are implicated in age‐related cognitive impairment. Although short‐term elevated GC levels are generally considered adaptive, prolonged exposure can detrimentally affect brain structure and function, particularly in the hippocampus where they decrease neurogenesis, cause dendritic atrophy and impair memory 3, 4. Hippocampus‐dependent memory impairments are associated with elevated circulating GC levels during ageing in humans and rodents 5, 6, 7.
GCs modulate episodic and working memory processes primarily via activation of two nuclear receptors: the high affinity mineralocorticoid receptors (MR) and low affinity glucocorticoid receptors (GR). Both are ligand‐activated transcription factors and are highly expressed in the hippocampus 8. Before accessing receptors, GCs may be subject to intracellular metabolism. The hippocampus highly expresses 11β‐hydroxysteroid dehydrogenase type 1 (11β‐HSD1), which contributes to intracellular GC levels by catalysing the regeneration of cortisol and corticosterone from inert 11keto forms (cortisone, 11‐dehydrocorticosterone) 9, 10. Our recent studies support a pivotal role for 11β‐HSD1 generated GCs in age‐related cognitive decline 11. Aged 11β‐HSD1 deficient mice show preserved hippocampus‐dependent learning and memory throughout life, resisting age‐related spatial memory deficits observed in wild‐type mice, as shown in both water maze and Y‐maze tasks 12, 13. Conversely, transgenic mice with forebrain specific overexpression of 11β‐HSD1 show accelerated cognitive ageing 14. Short‐term selective inhibition of 11β‐HSD1 reverses spatial memory deficits in aged C57BL/6J mice 15, 16. Improved cognition in aged mice with 11β‐HSD1 deficiency or inhibition associates with reduced intrahippocampal corticosterone (CORT) levels during learning 17, a switch from predominant activation of ‘anti‐cognitive’ GRs to predominant ‘pro‐cognitive’ MRs 18 and enhanced hippocampal long‐term potentiation 13. The downstream genes/pathways beyond receptor activation that underlie age‐related memory deficits associated with 11β‐HSD1 activity are unknown.
Acquisition of long‐term memory requires gene transcription and protein synthesis 19. Gene expression microarrays have identified genes and pathways in rodent hippocampus that associate with cognitive ageing 20, 21, 22, 23. These include down‐regulated immediate early gene (e.g. Arc, Egr‐1, Vgf) and insulin signalling pathways (e.g. Insr, Ide, Stat5b) and up‐regulated general oxidoreductase activity genes (e.g. Acads, Aldh9a1) selectively in aged cognitively impaired animals 22. To dissect the pathways underlying cognitive protection with 11β‐HSD1 deficiency, the Y‐maze was used to define cognitive function in aged wild‐type and 11β‐HSD1‐deficient mice (relative to young controls) prior to microarray analysis of hippocampal gene transcript profiles. Aged wild‐type mice were further separated into memory impaired and unimpaired groups with the aim of identifying differentially expressed hippocampal genes that associate with cognitive ageing.
Materials and methods
Animals
Male mice homozygous for targeted disruption of the Hsd11b1 gene (Hsd11b1 −/−), congenic on the C57BL/6J genetic background 24, 25, and age‐matched C57BL/6J mice as wild‐type (Hsd11b1 +/+) controls were bred and maintained within our biomedical research facility housed under standard conditions under a 12 : 12 h light/dark cycle (lights on 07.00 h), with food and water ad lib. until behavioural testing in the Y‐maze at either 6 months (young) or 24 months (aged). All animal procedures were performed in accordance with the local ethical guidelines of the University of Edinburgh Ethics Committee and those of the UK Animals (Scientific Procedures) Act, 1986.
Y‐maze
Young (6 months) and aged (24 months) wild‐type and Hsd11b1 −/− mice were tested in a two trial Y‐maze task previously validated as hippocampus‐dependent 18, 26 for assessment of their spatial memory. All behavioural testing was carried out in the morning (between 08.00 and 11.00 h) in a dimly lit room. Each mouse was placed at the end of one of the three arms of the maze designated the ‘start arm’ and allowed to explore the maze with one arm blocked (novel arm) for 5 min (trial 1) before returning to their home cage. Fixed spatial cues (various objects such as glass bottle, pipette rack, plastic breaker, etc.) surrounded the maze. After an inter‐trial interval (ITI) (either 1 min or 2 h), the mouse was placed back in the maze start arm and allowed to explore all three arms (trial 2) for 5 min 18. The maze was wiped clean with 70% ethanol in between animals to remove olfactory cues. All mice (young, n = 9/genotype; aged wild‐type, n = 13; aged Hsd11b1 −/− mice, n = 8) were tested first with the 1‐min ITI to ensure they responded to novelty, had no motor deficits and could see the spatial cues around the maze. Spatial memory was tested with the 2‐h ITI, 1 week later. The time spent in each of the arms was calculated with the ANY‐maze software (Stoelting, Dublin, Ireland). Aged mice failing to spend significantly more time in the novel arm compared to the previously visited arms were classed as cognitively impaired, whereas mice showing a preference for the novel arm similar to young controls were cognitively unimpaired. Aged cognitively impaired (AI) and unimpaired (AU) wild‐type mice were randomly selected from the groups for the microarray study (n = 4 per group).
Animals were culled by cervical dislocation between 08.00 and 10.00 h, 3 days after the end of Y‐maze testing. Brains were removed, dissected and the hippocampus was snap frozen in RNase free eppendorf tubes on dry ice and stored at −80 °C. For in situ hybridisation studies, brains were frozen on powdered dry ice and stored at −80 °C.
RNA extraction and processing
Total RNA was extracted from hippocampal tissues of young wild‐type (WT_Y), young Hsd11b1 −/−(KO_Y), aged unimpaired wild‐type (WT_AU), aged impaired wild‐type (WT_AI) and aged Hsd11b1 −/− (KO_A) mice using TRIzol reagent (Invitrogen, Paisley, UK) and RNeasy Mini Kit (Qiagen, West Sussex, UK). The concentration and purity of each RNA sample was assessed using a GeneQuant RNA/DNA calculator (GE Healthcare, Little Chalfont, UK). Hippocampal RNA samples were processed through standard Affymetrix protocols, and hybridised to Affymetrix Mouse Genome 430 2.0 GeneChips (n = 4 per group; Affymetrix, Santa Clara, CA, USA). CEL files for all 20 chips were imported into the Affy package of bioconductor, and were processed (background subtraction and normalisation) with the robust multichip average algorithm. Chip data quality control was performed by: (i) visual inspection of the chip images (not shown) that showed no obvious abnormalities and (ii) histogram of raw intensities from all chips that showed no clear outliers and had the usual distribution. Quality control indicated that the data were technically good. Expression levels for each chip and fold changes between genotype for each tissue were calculated. Genes with no or very low expression (i.e. expressed below a normalised expression value of 100 in all, or all but one, of the samples) were excluded. Data were exported in text format and imported into a mysql database (https://www.mysql.com). Annotation data for the genes were obtained from netaffx (Affymetrix). A web accessible front‐end query tool was built that allows query of the data by expression data (normalised expression, fold‐changes, P‐values) and by sequence annotation (gene title and symbol, Entrez gene ID, Affymetrix ID and Gene Ontology data). Microarray data are available in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) with accession number GSE68515. Microarray processing was carried out by ARK‐Genomics (Roslin Institute, Edinburgh UK).
Real‐time quantitative reverse transcriptase‐polymerase chain reaction (RT‐PCR)
Total RNA (1.5 μg) from hippocampal samples from the experimental groups (WT_Y, KO_Y, WT_AU, WT_AI and KO_A) (n = 5–8 per group including overlap with animals used in microarray) was reverse transcribed into cDNA with oligo(dT) primers using the QuantiTect Reverse Transcription Kit (Qiagen) in accordance with the manufacturer's instructions. Gene‐specific mRNA levels were determined in cDNA samples incubated in triplicate with gene‐specific primers and fluorescent probes (using pre‐designed assays from Applied Biosytems, Warrington, UK) in a 1 × Roche LightCyclerR 480 Probes mastermix (Roche, Basel, Switzerland). PCR cycling and detection of fluorescent signal were carried out using a Roche LightCyclerR 480 (Roche). A standard curve was constructed for each primer probe set using a serial dilution of cDNA pooled from all samples. The primers (Invitrogen) were designed for use with intron‐spanning probes from the Roche Universal Probe Library. Software provided with the LightcyclerR 480 was used to analyse the data produced after RT‐PCR. All results were corrected by normalisation to the expression level of the reference (housekeeping) gene Gapdh, which did not differ between groups and expressed arbitrarily as an adjusted ratio.
In situ hybridisation
PCR products corresponding to nucleotides 1859–2349 of the mouse Arc cDNA and 1055–1607 of the mouse Npas4 cDNA were generated from control C57BL/6J mouse hippocampus cDNA and subcloned into the pGEM‐T Easy vector (Promega, Madison, WI, USA). 35S‐UTP (Perkin Elmer, Waltham, MA, USA) labelled sense and antisense cRNA probes were generated using restriction enzyme‐linearised plasmid as template for in vitro transcription using either T7 or SP6 RNA polymerase as appropriate. Cryostat coronal brain sections at the level of the dorsal hippocampus from young and aged WT_AI (WT_AU were omitted as a result of the small sample size) and Hsd11b1 −/− (KO) mice (n = 6–9 per group, including overlap with animals used in microarray) were post‐fixed in 4% paraformaldehyde, acetylated (0.25% acetic anhydride in 0.1m triethanolamine, pH 8.0), washed in phosphate‐buffered saline, dehydrated in graded ethanol and air‐dried. Sections were hybridised with probe overnight at 50 °C, followed by RNaseA treatment and standard saline citrate buffer washes. Slides were dehydrated in graded ethanol and air‐dried before exposure to Biomax MR‐1 film (Amersham, Little Chalfont, UK) for 5–8 days at room temperature. The slides were then dipped in NTB‐2 emulsion (Eastman Kodak Co, Rochester, NY, USA) and exposed for 3 weeks at 4 °C before developing and counterstaining with 1% pyronine. The hybridisation signal was assessed by computer‐assisted grain counting using ks300 image analysis software (Carl Zeiss, Oberkochen, Germany). Silver grains were counted within a fixed circular area under bright‐field illumination using the × 40 magnification objective, over individual cells by an investigator who was blinded to age and genotype. For each animal, 15–18 cells per subregion of hippocampus or cortical layer were assessed and background, counted over areas of white matter, was subtracted. The labelled RNA probes (antisense and sense) were first hybridised onto control brain sections from adult mice to test their specificity at the autoradiographic level. No binding was detected with labelled sense probes of Npas4 and Arc (data not shown).
Statistical analysis
Statistical analysis of the microarray gene expression data was carried using limma r/bioconductor 27 yielding multiple testing corrected P‐values for each comparison of interest. A nonparametric statistical test (rank products; RP) was also used. The RP approach has been shown to be reliable for identifying biologically relevant expression differences, even with highly noisy data 28, 29. Genes were considered differentially expressed between groups when rank product P < 0.05. The Y‐maze (2‐h ITI time spent in novel arm) and in situ gene expression data across the groups were analysed using two‐way anova with age and genotype as the independent variables followed by Tukey's multiple comparison's test as appropriate for between group comparisons. Comparisons of time spent in the arms of the Y‐maze within each group were analysed by Student's paired t‐tests. The quantitative RT‐PCR data with the aged impaired and unimpaired wild‐type mice as separate groups were analysed by one‐way anova with Tukey's multiple comparison's test as appropriate for between group comparisons. Data are reported as the mean ± SEM.
Results
Spatial memory status of aged wild‐type and 11β‐HSD1‐deficient mice in the Y‐maze
All young and aged wild‐type and Hsd11b1 −/− mice spent significantly more time in the novel arm than previously visited arms of the Y‐maze (P < 0.01, paired t‐tests) after a 1‐min ITI (see Supporting information, Fig. S1), confirming the aged mice had no impairment of vision or motor deficits. There were little inter‐individual differences within the aged mice groups (both wild‐type and Hsd11b1 −/−) after the 1‐min ITI, with all mice performing equally well regardless of their later cognitive status after the 2‐h ITI. Analysis of the 2‐h ITI Y‐maze data, as a measure of spatial memory, revealed a significant effect of genotype (F1,33 = 8.4, P < 0.01) and genotype × age interaction (F1,33 = 5.7, P < 0.05) (Fig. 1 a). Aged (24 months) wild‐type mice spent less time in the novel arm during the retention trial after a 2‐h ITI compared to young (6 months) wild‐type controls (P < 0.05) and to aged Hsd11b1 −/− mice (P < 0.01) (Fig. 1 a), recapitulating previous findings 13, 18. Aged wild‐type mice (n = 13) showed an overall impairment in spatial memory (Fig. 1 a), although examination of individual animals revealed four unimpaired mice with intact spatial memory similar to young mice, spending significantly more time in the novel arm (P < 0.05) than previously visited arms of Y‐maze (Fig. 1 b). Aged memory‐unimpaired wild‐type (WT_AU) mice spent a significantly (P < 0.001) longer percentage time in the novel arm than aged memory‐impaired wild‐type mice (WT_AI) (Fig. 1 b). Mice from each age and genotype group were used for microarray analysis of hippocampal gene expression (Fig. 1 b).
Figure 1.
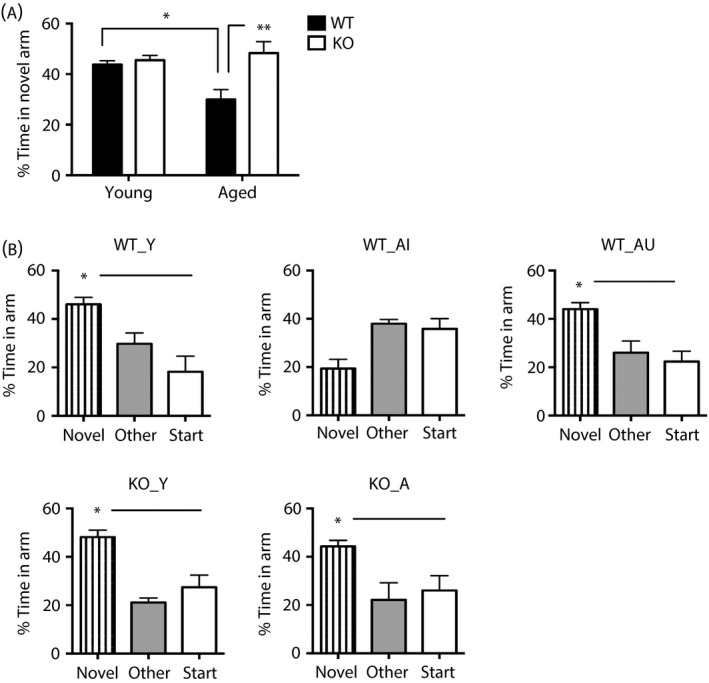
Spatial memory status of aged wild‐type and 11β‐HSD1‐deficient mice in the Y‐maze. (a) Aged (24 months) wild‐type (WT) mice as a group (n = 13) showed impaired spatial memory retention in the Y‐maze after a 2‐h inter‐trial interval (ITI) (less time in novel arm) compared to young (6 months) WT controls (n = 9) and aged (24 months) Hsd11b1 −/− (KO) mice (n = 8). (b) Spatial memory of mice selected for microarray analysis from (a) showing the five groups used: WT_Y (young wild‐type), KO_Y (young Hsd11b1 −/−), KO_A (aged Hsd11b1 −/−), WT_AI (aged wild‐type memory‐impaired) and WT_AU (aged wild‐type memory‐unimpaired) (n = 4 per group). *P < 0.05, **P < 0.01 significant difference between comparisons. Data shown are the mean ± SEM.
Differentially expressed hippocampal genes associated with 11β‐HSD1 deficiency, memory impairment and ageing
Data from aged wild‐type mice were examined as two subgroups [aged memory‐impaired (WT_AI, n = 4) and aged memory unimpaired (WT_AU, n = 4)] and also as a combined group (WT_A, n = 8). These were compared with aged Hsd11b1 −/− mice, which showed no cognitive impairment. Most differentially expressed hippocampal transcripts were increased with age but did not differ between genotypes (an approximate 1.5‐fold increase, < 0.05 RP score compared to corresponding young controls). These include inflammatory/immune response genes (C1qb, C1qbc, B2m, Aif1, Fcgr2b, Fcgr3, Trem2, Lyz2, Mpeg1) and glial/structural genes (Gfap, Vim, Dmp1), as well as those for cholesterol/lipid metabolism (Apod), signal transduction/transport (Anxa3, Cyba, Abca8a) and protein binding (Rtp4, Tyrobp) (see Supporting information, Table S1). Some genes were increased with age (approximately 1.5‐fold, P < 0.05 RP score) in wild‐type mice regardless of cognitive status (WT_A) but not in Hsd11b1 −/− mice, including the rate‐limiting retinoic acid‐synthesising enzyme (Aldh1a2) and the GABA transporter 2 (Slc6a13) (Fig. 2). Only three genes were differentially expressed significantly between WT_AI and WT_AU (Fig. 2; see also Supporting information, Fig. S2) and between WT_AI and KO_A (Fig. 2; see also Supporting information, Fig. S3). Of these, the transcription factor Npas4 and immediate early gene Arc, are of particular interest because of their link with learning and memory 30, 31. Npas4 and Arc mRNA levels were selectively decreased with age in the hippocampus of WT_AI (P < 0.01 and P < 0.05, RP score, respectively) but not WT_AU or KO_A mice (Figs 2 and 3). Npas4 mRNA levels were also lower in KO_Y compared to WT_Y (P < 0.05) (Fig. 2). Interestingly, the level of Agxt2l1 (alanine‐glyoxylate aminotransferase 2‐like 1), a gene whose function is poorly characterised but for which a dysregulation in prefrontal cortex has been associated with mood disorders 32, differed significantly between WT_AU and WT_AI mice (P < 0.01, RP score) and was increased with age in the hippocampus of both WT_AU and KO_A (P < 0.001, RP score) but not WT_AI mice (Figs 2 and 3).
Figure 2.
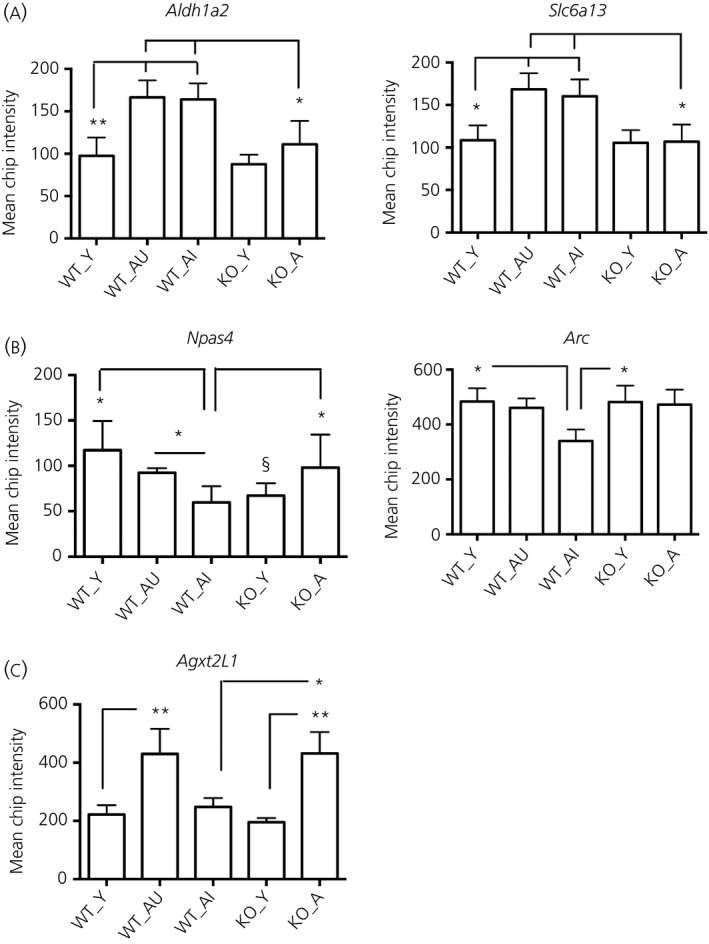
Microarray mean chip intensity of a selection of differentially expressed hippocampal genes in wild‐type and 11β‐HSD1‐deficient mice. (a) Increased with age in WT but not in KO. (b) Decreased with age in WT_AI but not in KO. (c) Increase with age in WT_AU and in KO but not in WT_AI. Comparisons between young and aged wild‐type unimpaired and impaired mice [(WT_Y) and (WT_AU) or (WT_AI), n = 4 per group] and between aged wild‐type and Hsd11b1 −/− mice [WT_AI and KO_A, n = 4 per group] differed by approximately 1.5‐fold (*P < 0.05, **P < 0.01 RP score). §P < 0.05 compared to WT_Y. Data shown are the mean ± SEM.
Figure 3.
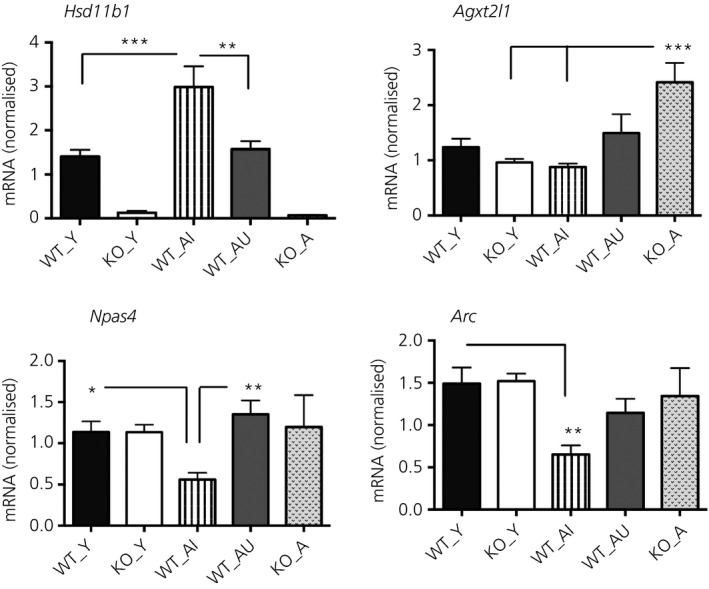
Quantitative real‐time polymerase chain reaction measurement of Hsd11b1, Agxt2l1, Npas4 and Arc mRNA levels in the hippocampus of wild‐type and 11β‐HSD1‐deficient mice. Levels of Hsd11b1, Agxt2l1, Npas4 and Arc mRNA in the hippocampus of young wild‐type (WT_Y), young Hsd11b1 −/− (KO_Y), aged wild‐type impaired (WT_AI), aged wild‐type unimpaired (WT_AU) and aged Hsd11b1 −/− mice (KO_A) were measured relative to Gapdh and expressed as a ratio (n = 5–8 per group). *P < 0.05, **P < 0.01, ***P < 0.001 significance difference between groups. Data shown are the mean ± SEM.
Decreased Npas4 and Arc but increased Hsd11b1 mRNA levels in the hippocampus of aged memory‐impaired wild‐type mice
Differential expression of Npas4 and Arc mRNA levels was validated by quantitative real‐time PCR (qPCR) using total RNA from the hippocampus of WT_AI, WT_AU, KO_A and corresponding young mice of each genotype (n = 5–8 per group). However, the microarray changes in Agxt2l1 mRNA levels were not fully validated by qPCR, with no significant increase in WT_AU compared to WT_AI (P = 0.11) or compared to WT_Y (P = 0.47) (Fig. 3). Although the microarray data analysis did not reveal differences in Hsd11b1 mRNA levels between young and aged wild‐type mice, qPCR showed significantly higher levels of Hsd11b1 mRNA in the WT_AI group compared to WT_Y controls (P < 0.001) (Fig. 3) confirming our previous findings 14. Both hippocampal Npas4 and Arc mRNA levels differed significantly between the groups (F4,27 = 5.1, P < 0.01 and F4,26 = 5.9, P < 0.01, respectively) (Fig. 3). Hippocampal Npas4 and Arc mRNA levels were decreased selectively in WT_AI but not WT_AU mice (WT_Y versus WT_AI, P < 0.05 and P < 0.01, respectively) (Fig. 3). Npas4 mRNA levels were lower in the hippocampus of WT_AI compared to WT_AU mice (P < 0.01) (Fig. 3). By contrast, both hippocampal Npas4 and Arc mRNA levels were not significantly altered with age in Hsd11b1 −/− mice (Fig. 3). However, the lower Npas4 mRNA levels in KO_Y versus WT_Y from the microarray data (n = 4/genotype) were not evident by qPCR in the larger sample size (n = 8 per genotype) (Fig. 3).
Decreased Npas4 and Arc mRNA expression selectively in hippocampal CA1 cells of aged memory‐impaired wild‐type but not 11β‐HSD1‐deficient mice
We performed in situ hybridisation to gain a regional resolution of the reduced hippocampal Npas4 and Arc mRNA expression in aged memory‐impaired wild‐type mice. In young wild‐type mice, Npas4 mRNA expression was greatest in the cortical region (layers 2/3 and 5) and hippocampus, particularly in the CA1 and CA3 cell layers (Fig. 4 a). Within the CA1 subregion of the hippocampus, Npas4 mRNA levels were decreased with age (approximate 40% reduction, F1,26 = 11.8, P < 0.01) in WT_AI but not KO_A mice (Fig. 4 b). Levels of Npas4 mRNA in the CA1 region showed a nonsignificant trend to be lower in WT_AI mice compared to KO_A mice (F1,26 = 3.2, P = 0.08) (Fig. 4 b). CA3 Npas4 mRNA levels were decreased with age regardless of genotype (approximate 40% reduction, F1,24 = 19, P < 0.001) (Fig. 4 b). In the cortex, levels of Npas4 mRNA were affected by age but not genotype (cortical layer 2/3, F1,25 = 12.9, P < 0.01; cortical layer 5, F1,25 = 6.8, P < 0.05) (Fig. 4 b); post‐hoc analysis showed a decrease with age in WT_AI (approximate 37% decrease, P < 0.05) but not KO_A mice (Fig. 4 b).
Figure 4.
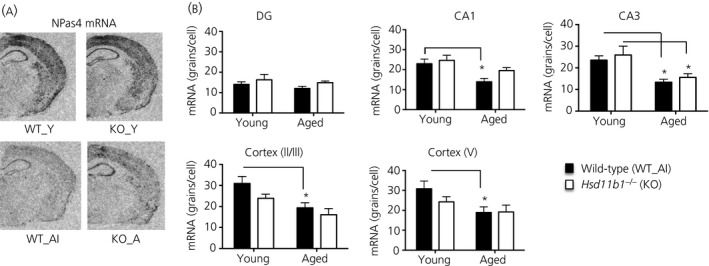
Differentially expressed Npas4 mRNA in hippocampus and cortex of aged wild‐type and 11β‐HSD1‐deficient mice. (a) Representative in situ hydridisation autoradiograms showing Npas4 mRNA expression in coronal mouse brain sections at the level of the dorsal hippocampus from young wild‐type (WT_Y), young Hsd11b1 −/− (KO_Y), aged memory‐impaired wild‐type (WT_AI) and aged Hsd11b1 −/− mice (KO_A) (n = 6–8 per group). Mice were previously tested in the Y‐maze to confirm spatial memory status as in Fig. 1 with only WT_AI mice included for in situ hybridisation analysis. (b) Quantification of Npas4 mRNA levels in hippocampus subregions (dentate gyrus, DG, CA1 and CA3) and cortex (layers 2/3 and V) of wild‐type and Hsd11b1 −/− mice. *P < 0.05 significant difference between groups. Data shown are the mean ± SEM.
Levels of Arc mRNA were highest in the CA1 cell layer of the hippocampus and layers 2/3 and 5 of the cortex (Fig. 5 a). In the hippocampus, there was an age (F1,27 = 14.8, P < 0.001) and age × genotype interaction (F1,27 = 5.0, P < 0.05) selectively in CA1 (Fig. 5 b). Arc mRNA levels were decreased with age in hippocampal CA1 cells of WT_AI mice (approximate 50% reduction, P < 0.001) and CA3 (approximate 33% reduction, P < 0.05) but not KO_A mice (Fig. 5 b). In the cortex, Arc mRNA levels were decreased with age but not genotype (cortical layer 2/3, F1,30 = 32, P < 0.001; cortical layer 5, F1,30 = 40, P < 0.001) (Fig. 5 b). Post‐hoc analysis revealed a decrease with age in both WT_AI (approximate 52–57% decrease, P < 0.001) and KO_A mice (approximate 41–46% decrease, P < 0.05) (Fig. 5 b).
Figure 5.
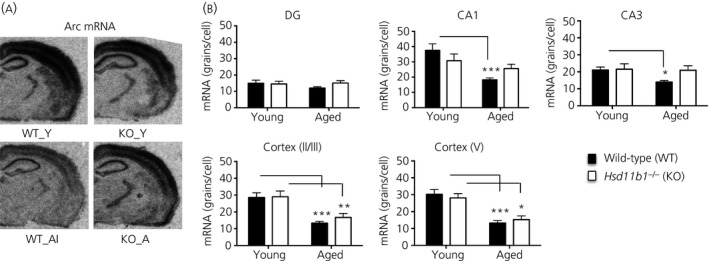
Differentially expressed Arc mRNA in hippocampus and cortex of aged wild‐type and 11β‐HSD1‐deficient mice. (a) Representative in situ hybridisation autoradiograms showing Arc mRNA expression in coronal mouse brain sections at the level of the dorsal hippocampus from young wild‐type (WT_Y), young Hsd11b1 −/− (KO_Y), aged memory‐impaired wild‐type (WT_AI) and aged Hsd11b1 −/− mice (KO_A) (n = 6–9 per group). Mice were previously tested in the Y‐maze to confirm spatial memory status as in Fig. 1 with only WT_AI mice included for in situ hybridisation analysis. (b) Quantification of Arc mRNA levels in hippocampus subregions (dentate gyrus, DG, CA1 and CA3) and cortex (layers 2/3 and V) of wild‐type (WT) and Hsd11b1 −/− mice. *P < 0.05, **P < 0.01, ***P < 0.001 significant difference between groups. Data shown are the mean ± SEM.
Discussion
Lifelong deficiency or short‐term inhibition of 11β‐HSD1 consistently preserves or improves spatial memory in aged mice 12, 13, 15, 18. In the present study, we identified two hippocampal genes, the brain‐specific activity‐dependent transcription factor Npas4 (neuronal Per‐Arnt‐Sim domain protein 4) and the immediate early gene Arc (activity‐regulated cytoskeleton‐associated protein), as being differentially expressed with ageing, cognitive decline and 11β‐HSD1 deficiency. Given the crucial roles of Npas4 and Arc in the regulation of learning and memory 31, 33, 34, 35, 36, 37, and their decreased mRNA levels in the hippocampus of aged memory‐impaired (AI) but not unimpaired (AU) wild‐type or 11β‐HSD1‐deficent mice, it is suggested that these proteins may lie in pathways that are important for the preservation of hippocampus‐associated memory in ageing and are maintained by 11β‐HSD1 deficiency/inhibition.
Several studies have used microarrays to identify hippocampal gene transcripts associated with cognitive ageing under basal (home cage) and memory‐activated (1 h after water maze training) conditions in rats 22, 23, 38, 39, 40 and mice 41, 42. The number of hippocampal genes identified as differentially expressed between aged cognitively impaired and unimpaired animals vary, with some studies revealing more genes altered than others. Importantly, several of the transcripts elevated with ageing regardless of genotype or cognitive status in the present study were also identified as genes regulated by ageing and not memory decline in cognitively tested aged rats, including inflammatory/immunity genes (C1qc, C1qb, B2m, Aif1, Fcgr2b) and structural genes (Gfap, Vim), as well as genes for cholesterol/lipid metabolism (Apod) and signal transduction (Anxa3) 21, 22, 43. This affords some confidence that these reflect ageing per se rather than the processes underlying cognitive variation with age.
By contrast to previous studies in rats that examined memory‐activated hippocampal gene expression profiles 22, 23, only three notable hippocampal genes were differentially expressed between the AI and AU wild‐type mice as characterised in the Y‐maze spatial recognition memory task. This low number of differentially expressed hippocampal genes suggests that the AI and AU characterisation based on a single ‘non‐aversive’ Y‐maze trial may not be as reliable as previous methods that use multiple ‘aversive’ water maze trials as shown in aged mice 42 and rats 6, 44, 45. The AU wild‐type mice would benefit from further characterisation in the water maze to demonstrate a consistent phenotype of preserved memory function. Thus, the implications of the altered gene transcript levels in AU wild‐type mice are less clear and could simply reflect within‐subject variability rather than a consistent distinct subject cognitive phenotype. By contrast, the impaired and preserved cognitive phenotype of the key comparisons between AI wild‐type and aged 11β‐HSD1‐deficient mice, respectively, have been reliably confirmed in previous studies following both Y‐maze and water maze protocols 12, 13, 18. Among the differentially expressed genes, Arc transcript levels were reduced selectively in AI wild‐type mice, a finding consistent with the correlation of hippocampal Arc mRNA levels with spatial memory 46 and previous studies in aged rats with memory deficits 21, 22, 47, 48. Acutely, both a memory‐enhancing dose of CORT and learning have been shown to increase Arc mRNA and protein expression 43, 46, 49 indicating a plausible pathway linking stress and its GC mediators through life, cognition and individual differences in cognitive decline with age. Moreover, hippocampal Arc expression was reduced in CA1 and CA3 but not the dentate gyrus of AI wild‐type mice in line with subregional changes in basal levels of Arc mRNA in aged rats 47. Decreased Arc mRNA levels have been reported during both ‘offline’ periods of rest and following spatial behaviour in the aged hippocampus 47. Indeed, Arc mRNA levels under resting home cage conditions are considered to reflect active information processing in cells that previously transcribed Arc in response to behaviour 50. Thus, reduced levels of Arc mRNA in CA1 and CA3 pyramidal cells of AI wild‐type mice may reflect impaired memory consolidation, as in Arc −/− mice 51, during the rest (home cage) period. Importantly, aged 11β‐HSD1‐deficient mice and AU wild‐type mice do not show reduced Arc mRNA levels in CA1 and CA3 cells and are not impaired in the Y‐maze.
Npas4 mRNA levels were also decreased in the hippocampal CA1 region of AI wild‐type but not aged 11β‐HSD1‐deficient mice. Reduced levels of Npas4 mRNA in hippocampus have previously been noted in aged rats 52, although this has not been specifically associated with the subset of animals showing cognitive decline. However, recent evidence indicates a key role for Npas4 in memory formation 33, 34. Moreover, Npas4 influences the survival of neurones, development and the maintenance of synapses, as well as the regulation of synaptic plasticity 30, via downstream target genes, including brain‐derived neurotrophic factor 53. Indeed, in a separate study, we found hippocampal levels of Bdnf mRNA to be reduced in aged wild‐type but not aged 11β‐HSD1‐deficient mice (S. Caughey, A.P. Harris, J.R. Seckl, M.C. Holmes and J.L.W. Yau, unpublished data). This is consistent with the lower Npas4 mRNA levels in AI wild‐type mice, decreased mRNA levels of both Bdnf and Npas4 in the hippocampus of aged rats 52, and reduced transcription of multiple Bdnf isoforms in Npas4 −/− mice 54.
Putative negative glucocorticoid response elements (GREs) found upstream of the Npas4 transcription initiation site suggest regulation by CORT 55. Indeed, in vivo treatment with high CORT doses reduce Npas4 mRNA and protein expression in the mouse hippocampus and frontal cortex 55, 56. This suggests that the maintained NPas4 mRNA levels in AU wild‐type mice and aged 11β‐HSD1‐deficient mice may, at least in part, be a result of lower brain intracellular CORT levels as a consequence of decreased 11β‐HSD1 activity 17. In support of this notion, hippocampal Hsd11b1 mRNA levels were selectively increased in AI but not AU wild‐type mice compared to young wild‐type mice.
It is possible that the greater rise in hippocampal CORT levels induced by learning/training in aged wild‐type mice 17 activates GRs, which in turn reduce Npas4 transcription directly by binding to negative GREs in its promoter 55. A decrease in Npas4 mRNA expression may contribute to reduced transcription of its target gene, Bdnf 52, which regulates neuroplasticity and memory mechanisms 30, 52. Lower Npas4 expression may also affect the expression of Arc indirectly. Indeed, memory‐activated expression of Npas4 mRNA in the dorsal hippocampus of mice appears upstream of several other immediate‐early genes including Arc 33. Moreover, conditional deletion of Npas4 in cultured mouse hippocampal neurones abolished the depolarisation‐induced expression of Arc mRNA 33. Thus, a reduced Npas4 expression could contribute to lower levels of Arc transcripts in the hippocampus of AI wild‐type mice.
These findings implicate both Npas4 and Arc in the pathways that may underlie the impairment and maintenance of spatial memory associated with ageing and 11β‐HSD1 deficiency. However, any causal relationship between 11β‐HSD1 deficiency, resistance to ageing‐associated decline of Npas4 and Arc mRNA levels, and ageing‐associated spatial memory deficits remains to be determined. If 11β‐HSD1 deficiency causes resistance to age‐related decline of hippocampal Npas4 and/or Arc mRNA levels, then short‐term selective inhibition of 11β‐HSD1 in aged C57BL/6J mice, which reverses spatial memory impairments 16, would be anticipated to associate with increased Npas4 and/or Arc mRNA levels. Future studies could examine this in aged mice during 11β‐HSD1 inhibitor treatment when spatial memory is improved, and after stopping treatment when memory reverts back to impaired, to test whether Npas4 and/or Arc mRNA levels are increased and decreased, respectively. Furthermore, intrahippocampal administration of high CORT levels to aged 11β‐HSD1 deficient mice (i.e. to levels equivalent to those found in aged wild‐type mice) could be carried out to establish whether it is the lower hippocampal CORT levels as a consequence of 11β‐HSD1 deficiency 17 that prevent the decreased Npas4 and Arc mRNA levels and/or memory deficits. It is likely that there are other hippocampal synaptic plasticity genes modulated by 11β‐HSD1 activity playing a role in the variable cognitive decline with ageing. Examination of learning‐activated gene transcripts and proteins in the hippocampus and functional characterisation of selected genes in vivo could help provide further insight into the mechanisms whereby 11β‐HSD1 activity contributes to age‐related memory decline.
Supporting information
Figure S1. Initial assessment of young and aged wild‐type and 11β‐HSD1‐deficient mice in the Y‐maze following a 1‐min inter‐trial interval (ITI).
Figure S2. Comparison of scatter plots of log intensity values (four replicates combined) for young and aged wild‐type mice main comparisons.
Figure S3. Comparison of scatter plots of log intensity values (four replicates combined) for young and aged wild‐type and 11β‐HSD1‐deficient mice main comparisons.
Table S1. Hippocampal genes up‐regulated with ageing in wild‐type and 11β‐HSD1‐deficient mice but not affected by genotype.
Acknowledgements
This study was supported by a Medical Research Council (MRC) project grant (G0501596; JLWY, JRS) and Wellcome Trust programme grant (JRS). DRD was funded by the Wellcome Trust Cardiovascular Research Initiative and by a British Heart Foundation Centre of Research Excellence Award. JLWY and JRS are members of The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology (CCACE), part of the cross council Lifelong Health and Wellbeing Initiative (MR/K026992/1). The authors declare that they have no conflicts of interest.
References
- 1. Nyberg L, Lovden M, Riklund K, Lindenberger U, Backman L. Memory aging and brain maintenance. Trends Cogn Sci 2012; 16: 292–305. [DOI] [PubMed] [Google Scholar]
- 2. Gallagher M, Rapp PR. The use of animal models to study the effects of aging on cognition. Annu Rev Psychol 1997; 48: 339–370. [DOI] [PubMed] [Google Scholar]
- 3. McEwen BS, Seeman T. Protective and damaging effects of mediators of stress. Elaborating and testing the concepts of allostasis and allostatic load. Ann N Y Acad Sci 1999; 896: 30–47. [DOI] [PubMed] [Google Scholar]
- 4. Herbert J, Goodyer IM, Grossman AB, Hastings MH, de Kloet ER, Lightman SL, Lupien SJ, Roozendaal B, Seckl JR. Do corticosteroids damage the brain? J Neuroendocrinol 2006; 18: 393–411. [DOI] [PubMed] [Google Scholar]
- 5. Lupien SJ, Schwartz G, Ng YK, Fiocco A, Wan N, Pruessner JC, Meaney MJ, Nair NP. The Douglas Hospital Longitudinal Study of Normal and Pathological Aging: summary of findings. J Psychiatry Neurosci 2005; 30: 328–334. [PMC free article] [PubMed] [Google Scholar]
- 6. Issa AM, Rowe W, Gauthier S, Meaney MJ. Hypothalamic‐pituitary‐adrenal activity in aged, cognitively impaired and cognitively unimpaired rats. J Neurosci 1990; 10: 3247–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yau JL, Olsson T, Morris RG, Meaney MJ, Seckl JR. Glucocorticoids, hippocampal corticosteroid receptor gene expression and antidepressant treatment: relationship with spatial learning in young and aged rats. Neuroscience 1995; 66: 571–581. [DOI] [PubMed] [Google Scholar]
- 8. Herman J, Patel P, Akil H, Watson S. Localization and regulation of glucocorticoid and mineralocorticoid receptor messenger RNAs in the hippocampal formation of the rat. Mol Endocrinol 1989; 3: 1886–1894. [DOI] [PubMed] [Google Scholar]
- 9. Moisan M‐P, Seckl JR, Edwards CRW. 11ß‐hydroxysteroid dehydrogenase bioactivity and messenger RNA expression in rat forebrain: localization in hypothalamus, hippocampus and cortex. Endocrinology 1990; 127: 1450–1455. [DOI] [PubMed] [Google Scholar]
- 10. Wyrwoll CS, Holmes MC, Seckl JR. 11beta‐hydroxysteroid dehydrogenases and the brain: from zero to hero, a decade of progress. Front Neuroendocrinol 2011; 32: 265–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yau JL, Seckl JR. Local amplification of glucocorticoids in the aging brain and impaired spatial memory. Front Aging Neurosci 2012; 4: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yau JL, Noble J, Kenyon CJ, Hibberd C, Kotelevtsev Y, Mullins JJ, Seckl JR. Lack of tissue glucocorticoid reactivation in 11beta ‐hydroxysteroid dehydrogenase type 1 knockout mice ameliorates age‐related learning impairments. Proc Natl Acad Sci USA 2001; 98: 4716–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yau JL, McNair KM, Noble J, Brownstein D, Hibberd C, Morton N, Mullins JJ, Morris RG, Cobb S, Seckl JR. Enhanced hippocampal long‐term potentiation and spatial learning in aged 11beta‐hydroxysteroid dehydrogenase type 1 knock‐out mice. J Neurosci 2007; 27: 10487–10496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holmes MC, Carter RN, Noble J, Chitnis S, Dutia A, Paterson JM, Mullins JJ, Seckl JR, Yau JL. 11beta‐hydroxysteroid dehydrogenase type 1 expression is increased in the aged mouse hippocampus and parietal cortex and causes memory impairments. J Neurosci 2010; 30: 6916–6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sooy K, Webster SP, Noble J, Binnie M, Walker BR, Seckl JR, Yau JLW. Partial deficiency or short‐term inhibition of 11β‐hydroxysteroid dehydrogenase type 1 improves cognitive function in ageing mice. J Neurosci 2010; 30: 13867–13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wheelan N, Webster SP, Kenyon CJ, Caughey S, Walker BR, Holmes MC, Seckl JR, Yau JL. Short‐term inhibition of 11beta‐hydroxysteroid dehydrogenase type 1 reversibly improves spatial memory but persistently impairs contextual fear memory in aged mice. Neuropharmacology 2015; 91C: 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yau JL, Wheelan N, Noble J, Walker BR, Webster SP, Kenyon CJ, Ludwig M, Seckl JR. Intrahippocampal glucocorticoids generated by 11beta‐HSD1 affect memory in aged mice. Neurobiol Aging 2015; 36: 334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yau JL, Noble J, Seckl JR. 11beta‐hydroxysteroid dehydrogenase type 1 deficiency prevents memory deficits with aging by switching from glucocorticoid receptor to mineralocorticoid receptor‐mediated cognitive control. J Neurosci 2011; 31: 4188–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Athos J, Impey S, Pineda VV, Chen X, Storm DR. Hippocampal CRE‐mediated gene expression is required for contextual memory formation. Nat Neurosci 2002; 5: 1119–1120. [DOI] [PubMed] [Google Scholar]
- 20. Blalock EM, Chen KC, Stromberg AJ, Norris CM, Kadish I, Kraner SD, Porter NM, Landfield PW. Harnessing the power of gene microarrays for the study of brain aging and Alzheimer's disease: statistical reliability and functional correlation. Ageing Res Rev 2005; 4: 481–512. [DOI] [PubMed] [Google Scholar]
- 21. Blalock EM, Chen KC, Sharrow K, Herman JP, Porter NM, Foster TC, Landfield PW. Gene microarrays in hippocampal aging: statistical profiling identifies novel processes correlated with cognitive impairment. J Neurosci 2003; 23: 3807–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rowe WB, Blalock EM, Chen KC, Kadish I, Wang D, Barrett JE, Thibault O, Porter NM, Rose GM, Landfield PW. Hippocampal expression analyses reveal selective association of immediate‐early, neuroenergetic, and myelinogenic pathways with cognitive impairment in aged rats. J Neurosci 2007; 27: 3098–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haberman RP, Colantuoni C, Koh MT, Gallagher M. Behaviorally activated mRNA expression profiles produce signatures of learning and enhanced inhibition in aged rats with preserved memory. PLoS ONE 2013; 8: e83674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kotelevtsev Y, Holmes MC, Burchell A, Houston PM, Schmoll D, Jamieson P, Best R, Brown R, Edwards CRW, Seckl JR, Mullins JJ. 11ß‐Hydroxysteroid dehydrogenase type 1 knockout mice show attenuated glucocorticoid‐inducible responses and resist hyperglycemia on obesity or stress. Proc Natl Acad Sci USA 1997; 94: 14924–14929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carter RN, Paterson JM, Tworowska U, Stenvers DJ, Mullins JJ, Seckl JR, Holmes MC. Hypothalamic‐pituitary‐adrenal axis abnormalities in response to deletion of 11beta‐HSD1 is strain‐dependent. J Neuroendocrinol 2009; 21: 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Conrad CD, Galea LA, Kuroda Y, McEwen BS. Chronic stress impairs rat spatial memory on the Y maze, and this effect is blocked by tianeptine pretreatment. Behav Neurosci 1996; 110: 1321–1334. [DOI] [PubMed] [Google Scholar]
- 27. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Res 2015; 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 2004; 573: 83–92. [DOI] [PubMed] [Google Scholar]
- 29. Hong F, Breitling R. A comparison of meta‐analysis methods for detecting differentially expressed genes in microarray experiments. Bioinformatics 2008; 24: 374–382. [DOI] [PubMed] [Google Scholar]
- 30. Maya‐Vetencourt JF. Activity‐dependent NPAS4 expression and the regulation of gene programs underlying plasticity in the central nervous system. Neural Plast 2013; 2013: 683909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bramham CR, Alme MN, Bittins M, Kuipers SD, Nair RR, Pai B, Panja D, Schubert M, Soule J, Tiron A, Wibrand K. The Arc of synaptic memory. Exp Brain Res 2010; 200: 125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shao L, Vawter MP. Shared gene expression alterations in schizophrenia and bipolar disorder. Biol Psychiatry 2008; 64: 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ramamoorthi K, Fropf R, Belfort GM, Fitzmaurice HL, McKinney RM, Neve RL, Otto T, Lin Y. Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science 2011; 334: 1669–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ploski JE, Monsey MS, Nguyen T, DiLeone RJ, Schafe GE. The neuronal PAS domain protein 4 (Npas4) is required for new and reactivated fear memories. PLoS ONE 2011; 6: e23760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramirez‐Amaya V, Angulo‐Perkins A, Chawla MK, Barnes CA, Rosi S. Sustained transcription of the immediate early gene Arc in the dentate gyrus after spatial exploration. J Neurosci 2013; 33: 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nonaka M, Kim R, Sharry S, Matsushima A, Takemoto‐Kimura S, Bito H. Towards a better understanding of cognitive behaviors regulated by gene expression downstream of activity‐dependent transcription factors. Neurobiol Learn Mem 2014; 115: 21–29. [DOI] [PubMed] [Google Scholar]
- 37. Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA. Inhibition of activity‐dependent arc protein expression in the rat hippocampus impairs the maintenance of long‐term potentiation and the consolidation of long‐term memory. J Neurosci 2000; 20: 3993–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haberman RP, Colantuoni C, Stocker AM, Schmidt AC, Pedersen JT, Gallagher M. Prominent hippocampal CA3 gene expression profile in neurocognitive aging. Neurobiol Aging 2011; 32: 1678–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burger C, Lopez MC, Feller JA, Baker HV, Muzyczka N, Mandel RJ. Changes in transcription within the CA1 field of the hippocampus are associated with age‐related spatial learning impairments. Neurobiol Learn Mem 2007; 87: 21–41. [DOI] [PubMed] [Google Scholar]
- 40. Burger C, Lopez MC, Baker HV, Mandel RJ, Muzyczka N. Genome‐wide analysis of aging and learning‐related genes in the hippocampal dentate gyrus. Neurobiol Learn Mem 2008; 89: 379–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Verbitsky M, Yonan AL, Malleret G, Kandel ER, Gilliam TC, Pavlidis P. Altered hippocampal transcript profile accompanies an age‐related spatial memory deficit in mice. Learn Mem 2004; 11: 253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pawlowski TL, Bellush LL, Wright AW, Walker JP, Colvin RA, Huentelman MJ. Hippocampal gene expression changes during age‐related cognitive decline. Brain Res 2009; 1256: 101–110. [DOI] [PubMed] [Google Scholar]
- 43. Chen KC, Blalock EM, Curran‐Rauhut MA, Kadish I, Blalock SJ, Brewer L, Porter NM, Landfield PW. Glucocorticoid‐dependent hippocampal transcriptome in male rats: pathway‐specific alterations with aging. Endocrinology 2013; 154: 2807–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gallagher M, Burwell R, Burchinal M. Severity of spatial learning impairment in aging: development of a learning index for performance in the Morris water maze. Behav Neurosci 1993; 107: 618–626. [DOI] [PubMed] [Google Scholar]
- 45. Rowe WB, Spreekmeester E, Meaney MJ, Quirion R, Rochford J. Reactivity to novelty in cognitively‐impaired and cognitively‐ unimpaired aged rats and young rats. Neuroscience 1998; 83: 669–680. [DOI] [PubMed] [Google Scholar]
- 46. Guzowski JF, Setlow B, Wagner EK, McGaugh JL. Experience‐dependent gene expression in the rat hippocampus after spatial learning: a comparison of the immediate‐early genes Arc, c‐fos, and zif268. J Neurosci 2001; 21: 5089–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Penner MR, Roth TL, Chawla MK, Hoang LT, Roth ED, Lubin FD, Sweatt JD, Worley PF, Barnes CA. Age‐related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol Aging 2011; 32: 2198–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fletcher BR, Hill GS, Long JM, Gallagher M, Shapiro ML, Rapp PR. A fine balance: regulation of hippocampal Arc/Arg3.1 transcription, translation and degradation in a rat model of normal cognitive aging. Neurobiol Learn Mem 2014; 115: 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McReynolds JR, Donowho K, Abdi A, McGaugh JL, Roozendaal B, McIntyre CK. Memory‐enhancing corticosterone treatment increases amygdala norepinephrine and Arc protein expression in hippocampal synaptic fractions. Neurobiol Learn Mem 2010; 93: 312–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Marrone DF, Schaner MJ, McNaughton BL, Worley PF, Barnes CA. Immediate‐early gene expression at rest recapitulates recent experience. J Neurosci 2008; 28: 1030–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, Kobalz U, Stawrakakis A, Fernandez E, Waltereit R, Bick‐Sander A, Therstappen E, Cooke SF, Blanquet V, Wurst W, Salmen B, Bosl MR, Lipp HP, Grant SG, Bliss TV, Wolfer DP, Kuhl D. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron 2006; 52: 437–444. [DOI] [PubMed] [Google Scholar]
- 52. Calabrese F, Guidotti G, Racagni G, Riva MA. Reduced neuroplasticity in aged rats: a role for the neurotrophin brain‐derived neurotrophic factor. Neurobiol Aging 2013; 34: 2768–2776. [DOI] [PubMed] [Google Scholar]
- 53. Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME. Activity‐dependent regulation of inhibitory synapse development by Npas4. Nature 2008; 455: 1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bloodgood BL, Sharma N, Browne HA, Trepman AZ, Greenberg ME. The activity‐dependent transcription factor NPAS4 regulates domain‐specific inhibition. Nature 2013; 503: 121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Furukawa‐Hibi Y, Yun J, Nagai T, Yamada K. Transcriptional suppression of the neuronal PAS domain 4 (Npas4) gene by stress via the binding of agonist‐bound glucocorticoid receptor to its promoter. J Neurochem 2012; 123: 866–875. [DOI] [PubMed] [Google Scholar]
- 56. Yun J, Koike H, Ibi D, Toth E, Mizoguchi H, Nitta A, Yoneyama M, Ogita K, Yoneda Y, Nabeshima T, Nagai T, Yamada K. Chronic restraint stress impairs neurogenesis and hippocampus‐dependent fear memory in mice: possible involvement of a brain‐specific transcription factor Npas4. J Neurochem 2010; 114: 1840–1851. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Initial assessment of young and aged wild‐type and 11β‐HSD1‐deficient mice in the Y‐maze following a 1‐min inter‐trial interval (ITI).
Figure S2. Comparison of scatter plots of log intensity values (four replicates combined) for young and aged wild‐type mice main comparisons.
Figure S3. Comparison of scatter plots of log intensity values (four replicates combined) for young and aged wild‐type and 11β‐HSD1‐deficient mice main comparisons.
Table S1. Hippocampal genes up‐regulated with ageing in wild‐type and 11β‐HSD1‐deficient mice but not affected by genotype.


