Abstract
Cryptic genetic variation is invisible under normal conditions but fuel for evolution when circumstances change. In theory, CGV can represent a massive cache of adaptive potential or a pool of deleterious alleles in need of constant suppression. CGV emerges from both neutral and selective processes and it may inform how human populations respond to change. In experimental settings, CGV facilitates adaptation, but does it play an important role in the real world? We review the empirical support for widespread CGV in natural populations, including its potential role in emerging human diseases and the growing evidence of its contribution to evolution.
Introduction: What is CGV?
Cryptic genetic variation is genetic variation that normally has little or no effect on phenotypic variation but that under atypical conditions, rarely experienced in a population's history, generates heritable phenotypic variation. Though often perceived as mechanistically special and mysterious, CGV is simply a subclass of variation with conditional effects, which has two well-studied forms: gene-by-gene interaction (GxG), including dominance and epistasis, where the effect of an allele is conditional on genetic background, and gene-by-environment interaction (GxE), where the effect of an allele is conditional on the environment (Figure 1). The distinguishing feature of CGV is that the conditions that induce allelic effects are rare or absent in the population's experience, and this rarity limits the opportunities for selection to act on the variation and allows it to accumulate. CGV then provides a pool of standing genetic variation poised to facilitate adaptation when the rare condition becomes common. Cryptic variation hidden from selection may alternatively be maladaptive in the new condition, the premise underlying the hypothesis that modern environments increase the genetic contribution to human disease risk1.
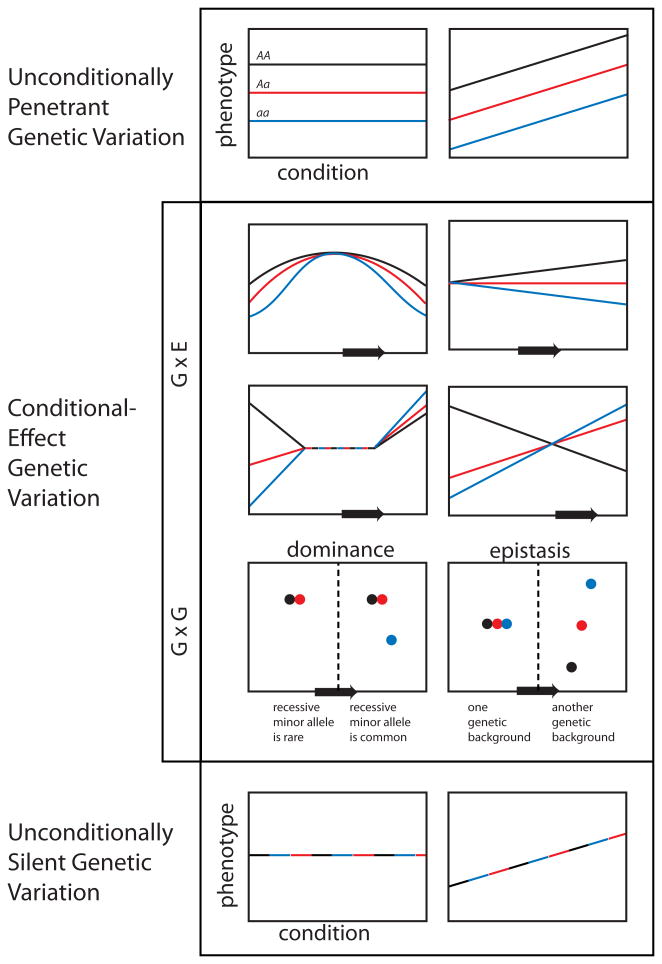
Figure 1.
Cryptic genetic variants are conditional-effect genetic variants. Each plot in the figure shows phenotype (y-axis) as a function of condition (environment or genetic background, x-axis) for three genotypes at a locus (AA homozygote in black, Aa heterozygote in red, and aa homozygote in blue). At the top, unconditionally penetrant genetic variation affects phenotype independent of condition. At the bottom, unconditionally silent variation has no effect under any circumstances (the three lines are superimposed). Between these extremes are variants whose effects depend on circumstances. Under each scenario shown, as conditions change (represented by movement along the arrow), cryptic genetic variation is revealed. In some cases, the genetic variants are completely cryptic in the initial condition, while in others their effect-sizes differ across conditions, hiding cryptic genetic variance in the initial condition. These panels illustrate just a few of the infinite possibilities for conditional-effect genetic variation.
The definition of CGV encompasses both molecular and quantitative genetic perspectives. From the molecular genetics view, cryptic genetic variants are polymorphic loci that have no effect on phenotype until perturbed by unusual conditions2. From the quantitative genetics view, cryptic genetic variance is an increase in heritable phenotypic variation (the additive genetic variance) that arises when a population is exposed to unusual conditions (e.g., 3). This distinction between variants (discrete loci with segregating alleles) and variance (heritable phenotypic variation) parallels the difference between compositional and statistical epistasis4; the former deals with the genotype:phenotype map, the latter with heritability in populations. Both forms are relevant to cryptic genetic variation.
Early investigations into CGV, and their implications, are thoroughly discussed elsewhere (e.g., 2), so here we briefly summarize CGV's historically provocative role in evolutionary theory and the mechanisms by which CGV may accumulate. Most of the research we review derives from work in sexual, outcrossing species, for which CGV is most likely to be important in adaptation 5. We focus our review on the extent of CGV in nature, which appears vast, and the role of CGV in adaptation and disease, which is less clear.
The CGV legacy
The existence of CGV is a longstanding subject of study in evolutionary genetics, motivated by a need to explain the ability of populations to adapt. Why would a population harbor variation that is adaptive in an environment it has never encountered? CGV provided a solution. Dobzhansky, in 1941 (6, page 160), listed JBS Haldane and the Russian geneticist NJ Shapiro, along with himself, as advocates of “a store of concealed genetic variability” containing mutations that were invisible when they arose but which may turn beneficial under new circumstances.
Modern enthusiasm for CGV builds on the iconic work of C. H. Waddington, who provided a clear evolutionary scenario to account for abundant CGV. Waddington noted that, as an empirical matter of fact, wild-type phenotypes develop robustly, with little variation7. The insensitivity of the wild type, he argued, is the result of evolved buffering mechanisms. If departures from the present-day optimum are disadvantageous, stabilizing selection will favor the evolution of mechanisms that dampen the effects of perturbations, yielding a nearly invariant, or canalized phenotype. Critically, Waddington showed that organisms pushed well outside their ordinary conditions, where the dampening mechanisms are overwhelmed, exhibit heritable phenotypic variation that had been invisible though present all along. Waddington's idea that stabilizing selection generates cryptic genetic variation is well supported by population genetic models3.
Waddington argued that given canalization, an alternative path to adaptive evolution, called genetic assimilation, could occur. First using heat shock to induce changes in Drosophila wing veination 8, then ether to induce homeotic transformations of body parts9, he observed variation across heterogeneous lines and artificially selected the most extreme phenotypes. Eventually, the phenotypes were “captured” and the selected lines no longer required the stimulus. Decades later, these early experiments inspired a modern inquiry into CGV. The first gene shown to harbor CGV was Ultrabithorax10, and the first cryptic variants at the nucleotide level were identified in the Epidermal growth factor receptor11, both in Drosophila. Also in Drosophila, disruption of the heat shock chaperone protein Hsp90 was shown to release CGV12, touching off a major research program into the role of this protein as a buffering mechanism (discussed below and in Box 1). These experiments provide a proof of principle for the adaptive potential of CGV (Figure 2).
Box 1. The Hsp90 story.
Rutherford and Lindquist's discovery motivated a renewed experimental effort in exploring genetic assimilation, with Hsp90 as a buffering mechanism in particular. Reduced Hsp90 activity has also been shown to release CGV for phenotypes in Arabidopsis97, cave fish91 and yeast87, and to increase the severity of developmental mutations in zebrafish98. Hsp90 provides a straightforward mechanism for buffering the effects of CGV. As a chaperone, Hsp90 assists in folding other proteins and in refolding misfolded proteins. Mutations in coding sequence can lead to folding error, so reduction in chaperone activity should increase the expressivity and penetrance of protein-coding mutations.
However, two main criticisms have been levied at Hsp90 as a model for releasing CGV and promoting genetic assimilation. One is that reduction of Hsp90 activity affects biogenesis of Piwi-interacting RNA (piRNA), which in turn permits transposable element activity in the germline and can lead to de novo, heritable mutations99. If new mutations account for the variation observed in Hsp90 knockdowns, then the best-studied example of CGV no longer stands. The second criticism is that Hsp90 may be exceptional and hence not a general model for buffering. Hsp90 is an abundant protein and interacts with many molecules in the cell100. Are there other genes that can demonstrate similar buffering of standing genetic variation? And how relevant is synthetic depletion of Hsp90 to the adaptive dynamics of natural populations?
Several studies have found that naturally occurring polymorphism at Hsp90 can affect fitness and morphology101,102, and that natural environmental perturbations, such as the low conductivity in aquatic cave habitats, can reduce Hsp90 function91. Other cryptic genetic variants revealed by chemical inhibition of Hsp90 have been confirmed as pre-existing87, and evidence also suggests that the reduction in Hsp90 required to affect transposable element activity is greater than that necessary to reveal CGV103. These results demonstrate that Hsp90 is a legitimate proof of principle for CGV. For now Hsp90 remains the standout example of a mechanism for revealing CGV, but recent experiments in Drosophila suggest that many genes may function to hide CGV at other loci96,104. Future work in this promising area will surely show whether Hsp90 is unique or simply an early herald of an important evolutionary mechanism.
Figure 2.
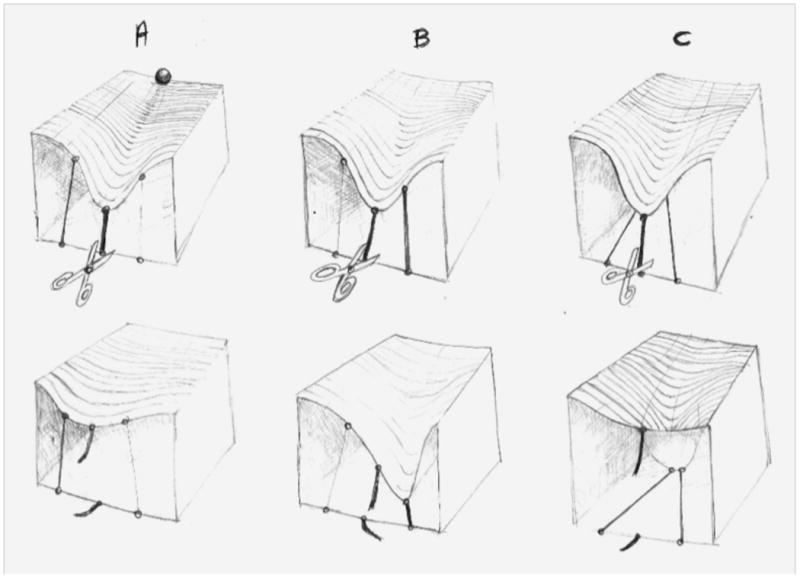
Waddington's epigenetic landscape, repurposed. Waddington's original conception of canalization arose from his observation that as the germ develops, tissues adopt discrete types: the eye or the gut, for example, never an intermediate115. His classic illustration depicts a ball atop a bifurcating landscape, poised to roll down the path of least resistance into valleys, or “canals,” whose endpoints represent terminal differentiation. His brilliantly literal depiction of genetic underpinnings shows guy-ropes pulling down, from the underside of the bifurcating landscape, the undulating topology of the canals and fastening to anchors representing genes7. Here we repurpose Waddington's landscape to illustrate how cryptic genetic underpinnings can induce different phenotypic fates. These genetic underpinnings vary at the molecular level (represented by guy-ropes of different strengths and configurations) but produce a consistent phenotype. After disruption (breakage of the main rope, representing a null mutation in a major gene), variation elsewhere produces deformities to the landscape.
How does CGV accumulate?
Waddington's model of canalization invokes buffering mechanisms that conceal CGV. There are two kinds of evolved buffering mechanisms, each with empirical support13. First, a population may evolve systems that suppress any and all departures from the wild type, under normal conditions. Hsp90, which suppresses the effects of misfolded proteins, remains the standout example of such a generic buffering mechanism (Box 1). Generic buffering systems are called capacitors12, as they have the potential to suppress, and thereby store, an enormous charge of variation that they can release when perturbed. A second type of buffering can arise if stabilizing selection favors mechanisms that suppress perturbations to individual phenotypes. Such targeted suppression could involve phenotype-specific gene networks, through the evolution of duplicate genes, redundant pathways, or shadow enhancers (eg., 14). For example, a cis-regulatory region at the Drosophila melanogaster shavenbaby locus is largely superfluous under ideal conditions but necessary to preserve wild-type expression under thermal stress15. Generic and specific buffering mechanisms thus have different implications for the nature of the stored variation and the perturbations that could expose it.
However, buffering mechanisms are not required for CGV to accumulate. For example, conditional-effect alleles may arise routinely as new mutations, and in the absence of phenotypic effects their frequencies are subject only to genetic drift. A population's pool of CGV is then determined by the product of effective population size, the mutation rate, and the fraction of mutations that have conditional effects. The latter is determined by the biochemical properties of the mutant alleles and the cellular networks in which they reside. For example, biological macromolecules are nonlinearly sensitive to temperature, pH, and ion concentrations; independently, the architecture of pathways and networks can generate automatic conditional neutrality for a large fraction of mutations, in the absence of any evolved buffering mechanism16,17.
In short, under this neutralist scenario, populations may harbor CGV merely because alleles have never been subjected to selection. An extreme example is a rare recessive allele, deleterious in the homozygous state in its present environment, maintained only by mutation-selection-drift balance. Such alleles achieve higher frequencies than additive-effect alleles because their recessive nature makes them cryptic, and when conditions changes they can provide the raw material for adaptation18.
This example of a recessive allele recalls the classic Fisher-Wright debate over the evolution of dominance: why are new mutations usually recessive? Fisher favored evolved suppressors and Wright favored a biochemical explanation. In that debate, which precisely echoes the debate between evolved and neutral CGV, Wright's position was vindicated19,20. In the context of CGV, however, the positions are not mutually exclusive, and both likely contain some truth.
Moreover, it is important to note that the role of CGV in adaptation and disease depends only on its actual realized properties and abundance, not on the mechanisms that create it. CGV is a class of variation, not a process. In other words, we can set aside debate over robustness, canalization, buffering, and capacitance, phenomena that may facilitate the accumulation of CGV but whose relationships to it are contested (Box 2). We don't need to know why CGV exists to ask whether it is important3,21.
Box 2. CGV and robustness.
Robustness describes the relative insensitivity of a system to perturbation, and a robust genotype is one that exhibits little phenotypic variance. Studies of the genetics and evolution of robustness have historically used observations of CGV as evidence that a system is robust. The logic is that the release of CGV demonstrates that those strains or genotypes are phenotypically stable in the face of mutational perturbation—because until they were pushed beyond their tolerance, they hid genetic variation beneath a stable wild-type phenotype.
However, empirical observations of CGV yield little insight into the state of robustness (an argument first made by Hermisson and Wagner3, and most recently, with experimental support, by Richardson et al.21.) Strictly speaking, CGV revealed by perturbation demonstrates that the unperturbed system is robust to that specific suite of CGV. However, it bears no evidence for robustness against other genetic variation, including any new, untested mutations from across the spectrum that may occur in the future. Existing CGV samples the subset of mutations to which the wild-type happens to be robust. Consequently, observations of CGV cannot provide evidence of general robustness.
Studies of robustness also include observations that phenotypic variance can be increased under stressful or extreme environments (reviewed in 66). Nevertheless, some stressful conditions decrease phenotypic variance67, just as perturbations to Hsp90 can both increase and decrease phenotypic variation across genetically distinct lines (87; K. Geiler-Samerotte pers. comm.). A useful way of distinguishing between CGV, which is a class of genetic variation, and robustness, which is a systems-level property, is to recognize that CGV is simply conditionally-neutral genetic variation. Technically, cryptic variation could be revealed, if conditions change and silent mutations become visible, even under increased robustness. This scenario would arise if the new condition—even as it increased additive genetic variation from exisiting alleles—nevertheless sheltered the experession of other mutations, either in the future or actualized21. Specific relationships between CGV and the conditions that reveal it are not necessarily generalizable to other scenarios that affect phenotypic variance.
Just as CGV is not necessarily evidence for robustness, neither is it evidence for canalization, the evolved resistance to perturbation. Canalization can emerge via positive selection on buffering mechanisms, or simply under stabilizing selection wherein the presence of GxG or GxE interactions permit accumulation of CGV105. The process of canalization will promote accumulation of CGV, but its presence indicates neither an evolved resistance to perturbation nor a guaranteed resistance to other perturbations.
What does CGV look like?
If populations harbor genetic variation that is normally invisible and only evident when the population experiences novel conditions, direct experimental manipulation of conditions should reveal it. The foundational work in Drosophila showed this clearly for morphological traits; here, we review more recent experiments in other systems that uncover and characterize CGV.
The nematode vulva: a model for cryptic genetic variation
The vulva is the egg-laying and copulatory organ in C. elegans hermaphrodites and is formed from ventral epidermal precursor cells that undergo highly canalized cell fate specification. Six cells are competent to adopt the vulva fate, but under normal conditions only three do, in response to Ras pathway signaling and morphogen secretion from the gonadal anchor cell. Using mutations and laser ablation of the anchor cell to perturb vulva development, Milloz et al.22 observed CGV in cell fate specification across wild C. elegans isolates: the number of cells achieving the vulva fate and the timing of their induction differed dramatically across strains.
Work in this system elucidates aspects of CGV that may be generalizable. Although the wild C. elegans isolates display morphologically invariant vulval phenotypes under normal conditions, they show twofold differences in Ras pathway activity22. This is a likely example of variation in an “intermediate” phenotype, tolerated either because the differences are too small to disrupt the robust trait, or because they are compensated for elsewhere; searches for variation in intermediate phenotypes may lead to new discovery of CGV23. Surveys for hypervariable traits across closely related taxa may also point to sources of CGV. In the vulva, mechanisms that control cell fate specifications that have evolved most rapidly across species are also the most vulnerable to perturbation within C. elegans24,25. Cell ablations performed on different members of the Caenorhabditis genus revealed different cell induction patterns, indicating divergence in the underlying mechanisms, even though the pathways are conserved and the final vulva phenotype is morphologically invariant26. This last phenomenon is an example of developmental system drift, the interspecific analog to intraspecific CGV and for which the nematode vulva provides an excellent model (Box 3).
Box 3. Developmental system drift.
CGV represents hidden polymorphism within populations; developmental system drift (DSD) is hidden divergence among species. The phenomenon describes the divergence of genetic developmental mechanisms even as the phenotypic traits they determine remain static106. Evidence for DSD can be found in observations of species hybrids, in which morphological characters are malformed despite identical trait expression between the parental species (e.g. 107), and in the molecular divergence of conserved processes, including sex-determination in Diptera108.
A well-studied example of DSD is the evolution of the nematode vulva. Within the Caenorhabditis clade, species exhibit conservation of signaling pathways and vulva organogenesis but substantial differences in the relative importance of signals for cell fate specification26. Basic morphology has also been conserved between C. elegans and the distantly related Pristionchus pacificus. In both species, the vulva is derived from the same cells via the same cellular processes109, but its development is induced by different genetic mechanisms, principally EGF signaling in C. elegans and Wnt signaling in P. pacificus (reviewed in 110).
DSD can arise from both selection and neutral processes26,111,112. Natural selection might drive DSD by targeting pleiotropic alleles that are fully penetrant in one tissue but act cryptically in others26. Gene network simulations confirm that selection on pleiotropic targets can lead to rapid evolution of DSD113, and such a process makes sense for DSD of the nematode vulva because signals for vulva induction are known to mediate many other processes114. Indeed, such appears to be the case for a cryptic nucleotide for vulva cell fate specification that also affects egg-laying and sperm production62.
CGV has also been observed in the sex determination pathway of C. elegans, which, unlike most of its relatives, exhibits a male-hermaphrodite mating system. Mutations at two known sex determination genes revealed hidden variation, and QTL mapping identified genomic regions both with and without known genes for sex determination27. The emergence C. elegans as a model for CGV studies, joining Drosophila, suggests that CGV is a general feature of populations, both outcrossers and selfers, easily accessed in genetically tractable organisms but likely abundant in others.
Observations of increased variance
CGV can be inferred from changes in additive genetic variance (VA) across conditions. Estimating VA requires no sophisticated molecular tools and can provide evidence for the existence of cryptic variation without an attempt to identify causal loci. VA represents the transmissible component of phenotypic variation, so an increase in VA under perturbation indicates the presence of conditional, functional genetic variants. Whereas the previous examples demonstrate CGV by the transformation of invariant into variant (and often aberrant) phenotypes, estimating VA allows the possibility of phenotypic variation before perturbation as well. Several recent studies have demonstrated that ecologically relevant changes to environment can increase VA in natural populations, including body size in sticklebacks28, spermathecae number in dung flies29, plasma antioxidant level in gulls30, and traits associated with facultative carnivory in spadefoot toad relatives31.
How much CGV do populations harbor?
The experiments described above involved targeted efforts to identify cryptic genetic variation. But the broader question of the abundance of CGV can be addressed in a more general way, by asking about the prevalence of its proximate genetic mechanisms2,3. In other words, are alleles with GxG and GxE interactions common? We should focus in particular on interactions that have been rarely tested in a population's history.
There are two broad classes of rarely-tested GxG: higher-order epistasis, where alleles at multiple loci may all be at intermediate frequencies but particular genotypes may nevertheless be vanishingly rare, and modifiers of rare mutations, a class well studied in the context of human Mendelian diseases.
Higher-order epistasis
Recent empirical data suggest that higher-order epistasis may be exceptionally abundant, even if its effects are rarely exposed. A key type of evidence comes from near-isogenic lines (NILs or congenics) and chromosome substitution strains (CSSs or consomics). These are inbred lines carrying a fragment of one wild-type genome introgressed into a different wild-type genetic background. Studies in mice have found that these isolated genomic regions have large phenotypic effects in these heterologous backgrounds, effects that can vastly exceed their additive effects averaged across backgrounds32,33. For example, Shao et al.32 found that for 20 of 90 traits of mouse blood, bone, and metabolism, introgressing a chromosome from one strain into another resulted in an effect on phenotype that exceeded the phenotypic difference between the two strains. For two traits, seven different chromosome substitutions each had such large effects.
The genomic regions contained in mouse CSSs often contain multiple separable genetic effects, tightly linked32, suggesting that linkage disequilibrium can store cryptic variation that recombination can release34,35. Similar findings have emerged from analyses of NILs in Arabidopsis and C. elegans (e.g., 36-38). The same conclusions can be drawn from other experimental designs, including the comparison of additive genetic effects between inbred lines and outbred populations in Drosophila melanogaster39 and the genetic complexity of mutational suppression in a cross of yeast strains40. In general, individuals with rare, untested genotypic combinations often exhibit new phenotypes, a principle also demonstrated by transgressive segregation in genetic crosses (e.g., 41).
In these examples, the epistatic effects are probably a byproduct of stabilizing selection, as the phenotypic variance is low across progenitors. Stabilizing selection within isolated lineages can produce eventual incompatibilities between hybrids, even as the polygenic trait in question maintains a shared high-fitness phenotype42. Evolutionary pressure to maintain a stable phenotype can favor compensatory changes, but this pattern does not require compensatory evolution; alleles with no effect when they arise may simply be incompatible with alternative genetic backgrounds. Divergence between lineages can thus draw entirely from substitutions that are neutral within lineages but incompatible across lineages43. From the perspective of an experimental geneticist, this model holds that suppressor alleles fix, by chance, before the alleles they suppress even arise, rendering both cryptic. The idea has been formalized in several models44,45.
Modifiers of rare mutations
The second class of rarely-exposed GxG, modifiers of rare mutations, also appears ubiquitous46. Mendelian genetic disorders in humans are by definition examples of rare mutations inducing phenotype, but even diseases with a simple genetic basis can present as complex physiological disorders, and differences across patients with the same disease-causing allele indicate that genetic background is important. For example, cystic fibrosis is one of the most common monogenic disorders in humans and is caused by expression of recessive mutations in the gene CFTR, affecting the lungs, intestines, pancreas and metabolic homeostasis. Recently, several modifier loci that influence the expression of one or more of these physiological targets have been identified47. Likewise, a mouse model for congenital heart disease caused by mutations in the gene Nkx2-5 mapped multiple modifiers that substantially affect risk48. These genetic modifiers have been explicitly defined using the same language as CGV: as loci that influence the action of a primary locus while remaining silent, or at least “quiet,” with respect to phenotype on their own49.
In experimental systems, mutation modifiers have been explored in the context of genetic background effects. Evidence suggests that genetic background effects are pervasive50; occasionally they have been exposed incidentally in evaluation of the primary genetic defect. In Drosophila, the search for longevity genes has revealed ubiquitous, and occasionally confounding, effects of genetic background on the expression of lifespan-mediating alleles51,52. In C. elegans, microevolution in the signaling network underlying vulva development indicates that genetic screens for vulva determinants will vary depending upon the strain tested22.
Only occasionally have these studies examined genomic background explicitly to test for CGV2,53. However, background effects compete in magnitude with the effect of mutations within the conventional genetic paradigm—that is, the effect of a mutation in a controlled genetic background. Moreover, background effects are themselves genetically complex; modifiers of rare mutations themselves interact epistatically54.
A particularly striking demonstration of the ubiquity of mutation-modifying background effects comes from a study in flies in which mutations affecting startle behavior in the Canton-S strain were introgressed into different wild-type backgrounds55. In each case, genetic background significantly influenced the mutation's effect, and effects were smaller in wild-type backgrounds than in Canton-S. The implication is that the mutations have strong effects in the background where they were identified by phenotypic screens, while their effects in random backgrounds are lower, and in many cases, nonexistent.
Genotype-by-environment interactions
The other basis for CGV, GxE, is undoubtedly prevalent, as the expression of genotype routinely depends on environment in all genetic systems (e.g. 56-60). The type of GxE that underlies CGV is conditional neutrality: when genetic variation is silent (or quieter) with respect to phenotype in one environment, but penetrant (or louder) in another. Studies that examine the genetics of local adaptation—most commonly conducted in plants, which are stationary with respect to their environment—report widespread findings of conditional neutrality61.
The scenario of conditional neutrality raises a possibility that is not often invoked in discussion of CGV: that neutral alleles may be maintained in one environment via positive selection in, and subsequent migration from, another environment. (An analogy to this is the case where pleiotropic alleles are functionally neutral with regard to one trait, but targets of positive selection for another. For example the C. elegans nath-10 allele underlies cryptic variation in vulva development but likely fixed in populations due to increased egg-laying62; whether this phenomenon is rare or common is completely unknown, but in theory it may allow accumulation of alleles that are biased against deleterious fitness effects in the cryptic phenotype.) Theoretical work addressing niche adaptation across heterogeneous environments has shown how, across populations with gene flow, the effect of selection in marginal populations can be negligible63 and that this is explicitly so for the case where alleles are neutral in the predominant environment but not in the marginal environment64. Thus, it may be that CGV accumulates neutrally in populations as cryptic alleles are expressed at low rates, given that environments that reveal CGV are by definition rare. At the same time, if CGV is the result of adaptive canalization, with systemic buffering that breaks down when it would be adaptive to do so, occasional exposure of the CGV in rare environments can create a pool of preadapted alleles in the event that the environment shifts to resemble the previously rare environments65.
What role does CGV play in evolution?
For CGV to play an important role in evolution, there must be naturally occurring mechanisms that expose it. The influence of newly-exposed CGV will depend strongly on its nature (Figure 4). Does the cryptic variation consist of damaging mutations that are concealed by buffering mechanisms? Is it disproportionately enriched for non-damaging mutations, given that they are, by definition, not unconditionally deleterious? Or are effects of the cryptic mutations completely random, perhaps symmetrically distributed? In the first case, exposure of the globally-buffered variation under new conditions will invariably be deleterious, while release of specific subsets might produce novel beneficial phenotypes. In the second case, CGV will play a disproportionate role in adaptive evolution, providing standing-variation fuel for a response to selection. In the last case, revealed CGV will increase genetic variance with beneficial and deleterious effects, with part of the population preadapted to new conditions and another part saddled with maladaptive phenotypes.
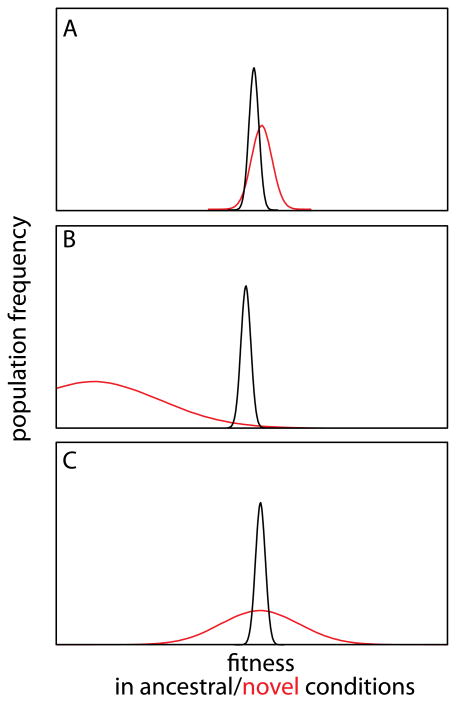
Figure 4. Fitness effect distribution of CGV in new conditions.
Here, three simple scenarios illustrate alternative outcomes for exposure of CGV. In black is the population's heritable variation in fitness under the normal condition; in red, the transformed fitness distribution following a change of environment or genetic background. a | Under a buffering scenario, a large fraction of the cryptic variants will be strongly damaging, and their exposure will primarily generate low-fitness monsters. b | Under an enrichment model, occasional exposure of CGV in a population's history will weed out the strongly deleterious alleles, leaving the CGV pool enriched for variation that improves the population's fit to its environment. c | Under a symmetrical scenario, newly exposed CGV simply increases the heritable phenotypic variance around the same mean.
Environmental exposure of CGV
Stressful conditions are well suited to expose CGV, facilitating adaptation to otherwise hostile environments7. However, there are substantive criticisms of this model, starting with the distinction between stressful conditions and novel conditions66. For CGV to accumulate neutrally, the conditions that expose it to selection must be rare in the population's history. Stress is usefully defined as conditions that reduce fitness relative to that found in a population's optimal realized environment67, and such stress is probably a typical experience for most populations. If particular stresses are routine, populations will adapt to them, evolving generalized buffering mechanisms, such as hsp90. If these mechanisms are particularly effective, they may allow for the fixation of mutations that are strongly deleterious when exposed by rarer or more extreme stresses67. If exposure is sufficiently rare, occurring on the order of once during the expected coalescence time for neutral mutations, fixation of conditional lethals may render CGV useless68.
There are two models that salvage stress-induced loss of buffering as a mechanism for releasing useful variation. One holds that buffering mechanisms are specific, such that particular conditions only expose subsets of the concealed CGV. This variation then is available to selection acting on specific traits, with limited undesirable pleiotropy69. An alternative is that the CGV-releasing conditions are rare but not exceedingly so, such that selection has an opportunity to purge the truly deleterious alleles, leaving a residue of CGV depleted of disadvantageous variation and harboring alleles at mutation-selection balance at frequencies determined by their effects integrated across the historical distribution of exposure68.
Novel conditions need not be stressful. For example, a population introduced to an environment that lacks its competitors and predators and contains new resources is novel but lacks stress, and CGV may cause some individuals to be better fitted to their new circumstances. Specific biotic and physical stresses are well-studied, but how populations respond to novel environments remains poorly understood. A possible line of analysis focuses on CGV due not to evolved buffering mechanisms but to the inherent properties of molecular variants in biological networks. If CGV is due to simple accumulation of conditionally silent variants under stabilizing selection in an ancestral environment, a change of environment might then increase variation symmetrically, yielding an increase in VA without a change in mean phenotype. This new additive variance is fuel for adaptation70. The increase in the fraction of the population far from the ancestral optimum, even without a change in the mean, is also a proposed mechanism for the increase in disease in human populations exposed to the novel circumstances of modernity1.
Genetic exposure of CGV
While environmental change can rapidly expose CGV due to GxE, the mechanisms responsible for release of GxG variation are less obvious. Further, while environmental change can act on many individuals in concert, instantly refiguring a population's pattern of selectable variation, epistasis depends on each individual's genetic constitution, affecting them one at a time.
Models for the release of CGV stored by epistasis have focused on mechanisms that can radically alter genotype frequencies across a population. Bottlenecks and founder events are natural candidates71,72. Although some theoretical work suggests that population contractions do not ordinarily release substantial VA from epistasis (e.g., 73,74), not all epistasis is equivalent. In particular, directional epistasis, whereby interaction effects tend to depart from additive in a consistent direction, can facilitate (or thwart) evolution; allele frequency increases at each locus systematically increase (or decrease) the marginal effects of each other locus (e.g., 75,76). Genetic data suggest that epistasis may be directional more often than not55, and biochemical and gene-network models yield similar implications (reviewed in 76). These results do not render a clear verdict on whether the patterns of directional epistasis are those that would tend to promote or hinder divergence, but empirical data weakly support an increase in VA following bottlenecks77,78.
Selection itself, by changing allele frequencies, is another mechanism that can expose CGV79,80. In an experimental dissection of loci contributing to divergent evolution of chicken body weight under artificial selection, Carlborg and colleagues81 found that divergence required a suite of epistatically interacting loci whose effects reinforced each other, progressively exposing additional VA during the course of the selection.
Although the contribution of epistatic CGV to adaptation remains somewhat opaque, the existence of abundant epistasis indicates that there are large numbers of silent polymorphic loci with the capacity, under specific conditions, to affect phenotypes. If the conditions induced by rare genotype combinations are also accessible to environmental perturbations, this pool of GxG CGV may overlap GxE CGV. Environmental perturbations arising outside the organism will induce physiological responses, such as signaling cascades, that are mediated by the same factors vulnerable to genetic change within the cellular environment66. Further, evolved buffering is expected to conceal both types of CGV by the same mechanisms82.
CGV in adaptation
Early concepts of CGV were explicitly founded on the notion that it might facilitate an alternative path to adaptation (e.g. 9). Under the most dramatic scenario, cryptic genetic variation might underlie the evolution of novelty and major evolutionary transitions. Evolution of complex traits might require multiple changes, each individually deleterious and hence resistant to fixation. However, segregating neutrally, individual alleles may recombine into the same background to reach appreciable frequencies, or prerequisite alleles might even fix, before being revealed in the stimulus environment. This scenario provides a potential mechanism for circumventing low-fitness valleys in an adaptive landscape. At the same time, CGV should prove valuable in ordinary adaptive evolution, when changes in circumstances reposition a population on the flanks of a novel fitness peak. In such situations, CGV provides standing variation for a rapid response to selection (e.g., 83). Whether CGV routinely contributes to either evolutionary scenario is an open empirical question, though data from experimental and natural settings are starting to shed some light.
CGV in adaptation: in vitro experiments
Two of the most definitive demonstrations that CGV can facilitate adaptation come from manipulations of in vitro populations of molecules. Hayden and colleagues84 evolved populations of ribozymes on a novel substrate. Populations that had previously accumulated cryptic variation under stabilizing selection on the ancestral substrate adapted more rapidly than populations that lacked cryptic variation. The populations with cryptic variation harbored genotypes that, while as fit as the wild-type on the ancestral substrate, were also preadapted to an unseen environment. In effect, the exploration of neutral genotype space in one environment left these populations poised to adapt to an environment they had never seen.
Similar work demonstrated the same principle in a cytochrome protein. Bloom and colleagues85 mutagenized two cytochrome P450 molecules and then tested their activities on novel substrates. The two starting molecules shared function on their initial substrate, but one was an evolved variant of the other, differing by 8 amino acid residues fixed by selection for thermostability. Following mutagenesis, the highly thermostable molecule better retained its ability to fold, which in turn permitted activity on the novel substrates85. That is, newly beneficial mutations that were accessible to the thermostable P450 were inaccessible to the ancestor due to epistasis with the stability-confering mutations. Ancestral cryptic variation in thermostability would therefore facilitate adaptation to novel environments. These experiments show compelling evidence for the adaptive potential of CGV, though their applicability to in vivo systems, specifically those with recombination, is as yet unclear86.
CGV in adaptation: in vivo experiments
Few demonstrations of adaptation fed by cryptic genetic variation beat Waddington's original genetic assimilation experiment, selection on heat- or ether-induced phenotypes. More recently, a number of studies in Drosophila and Arabidopsis have shown that selection on phenotypic variation revealed by Hsp90 depletion also yield responses. (Box 1). In yeast, the release of CGV by inhibiting Hsp90 activity demonstrated substantial variation in growth rates across many environments, in some cases to increase fitness. A similar result was achieved under high temperature stress, which may deplete the Hsp90 folding reservoir and offers a potential mechanism by which natural populations both maintain robust phenotypes and facilitate rapid adaptation under new environments87.
Evolved plasticity can also draw on cryptic variation88,89. In an experimental test of this scenario, Suzuki and Nijhout90 selected for a temperature-dependent larval color polyphenism in the hormworm Manduca sexta, which produces larvae whose color is insensitive to the ordinary range of developmental temperatures. They used acute heat-shock to expose cryptic variation for larval color, and after 13 generations of selection on heat-shock-exposed CGV, they had evolved a polyphenic line whose larval color exhibited a switch-like dependence on temperature within its ordinary range. This polyphenism matches a naturally occurring (and putatively adaptive) one in the related species M. quinquemaculata.
CGV in adaptation: evidence in nature
Going beyond the lab and into the field, the gaps in evidence of CGV's role in adaptation become apparent67. However, several studies provide compelling demonstrations of how ecologically relevant conditions might have facilitated adaptive change. Oceanic stickebacks reared in low salinity exhibited dramatic increases in VA for body size, indicating that CGV in ancestral oceanic populations may have facilitated the adaptive evolution of smaller size in freshwater habitats28. Similarly, surface fish reared in low conductivity water, mimicking cave conditions, showed increased variation for eye size that may underlie the adaptive morphology of blind cavefish91. Spadefoot toads exhibit a novel feeding strategy, facultative carnivory, accompanied by a derived body morphology. When a related species, standing as a proxy for the spadefoot ancestor, was fed a carnivorous diet, it exhibited increased heritability for body size, developmental stage and gut length, indicating that diet may have released adaptive morphological variation31.
A striking suggestion of CGV's direct role in phenotypic evolution comes from a study of the genetic origins of domesticated maize, the product of centuries of artificial selection. CGV for seven traits in teosinte, the ancestor to domesticated maize, was observed in crosses between heterogeneous teosinte and a single inbred strain of maize92. The contribution of the maize parental acted as a genomic perturbation to the teosinte genotypes, which were the only source of genetic variation in the experiment. Although the pure teosinte strains were phenotypically invariant, substantial variation in traits relating to branch and inflorescence morphology was released in the test cross. QTL mapping identified multiple causal regions, and provided some evidence that loci already identified to account for phenotypic differences between teosinte and maize harbor CGV for the same traits in teosinte.
Given the number of high-confidence findings about CGV — its abundance in populations, it potential, in theory, to fuel evolution, and its demonstrated ability to do so in experimental settings — the paucity of evidence for its role in adaptation in natural populations is striking. On the other hand, well-understood examples of the genetic basis of adaptive evolution are scarce generally, and the higher evidential threshold required by CGV — the demonstration that phenotypic effects of alleles are conditional on ancestral and derived circumstances — makes the task daunting.
CGV and complex human disease
The flip side to CGV's role in adaptation is its role in disease. Alleles that accumulate while hidden may play an important role in the emergence of complex human diseases, though as yet empirical evidence for this hypothesis is scant. The recent move of human populations into novel conditions, including changes to hygiene, diet, and exposure to environmental insults (e.g., smoking and industrial pollutants) and to new pathogens (e.g., HIV), are hypothesized to have revealed preexisting allelic variation for modern disease susceptibility93. These alleles may have accumulated via phenotypic canalization on fundamental aspects of mammalian physiology, only to be exposed by previously unseen conditions1. Body mass index (BMI), for example, is associated with many modern diseases94 and demonstrates a substantial global increase in the last century95. Alleles that influence BMI, and alleles whose effects depend on BMI, are likely to have changed in their contributions to phenotypes in contemporary human populations, potentially increasing the genetic variance. Further tentative support for this scenario is the observation that ancestral (as opposed to novel) alleles underlie disease susceptibility for multiple common diseases1. The question that demands testing, then, is whether the environmental and cultural conditions associated with modern complex diseases actually increase heritability by exposing cryptic genetic variance.
Conclusions
More than seventy years after the recognition that populations harbor a cryptic store of standing variation, the nature and importance of this CGV is better understood in theory3,68 than in nature. Nevertheless, some facts are clearly established. CGV, concealed by GxG and GxE, is abundant in natural populations and can be released under novel conditions. This cryptic variation has the potential to fuel a selective response; it represents, if not standing variation, at least crouching variation96. Further, although CGV is connected to hotly debated topics including capacitance, robustness, and canalization, its study is separable from those issues and its occurrence is not dependent on them.
One of the key questions for the future is about the extent to which CGV in natural populations is shaped by selection. Our classical definition of cryptic genetic variants supposes that these alleles accumulate in populations under strict neutrality, never tested by selection. Quantitative and population genetic arguments suggest that this definition may be too strict. Alleles whose contribution to additive genetic variance increases under novel conditions may comprise a more useful class, as these variants will have been filtered of those that are strictly deleterious when exposed. The challenge then turns toward a more continuous account of conditional variation, integrating the both the degree of condition-dependence of effect sizes and the frequency distribution of conditions. The latter is a question that demands ecological investigations outside the normal ken of molecular geneticists.
Figure 3.
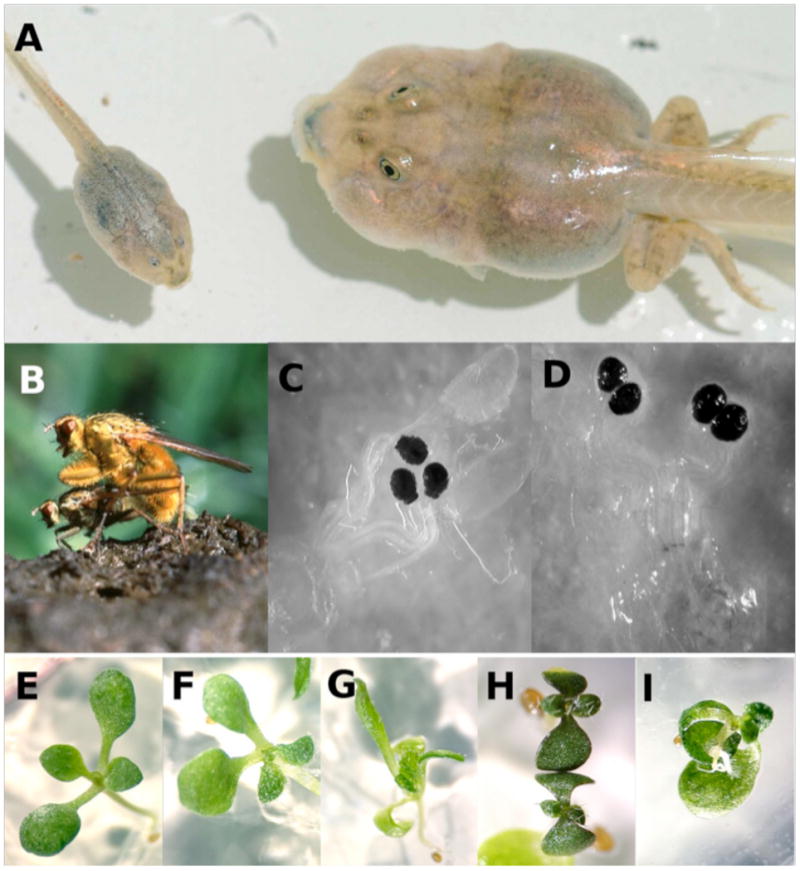
A sampling of experimental systems. a | Spadefoot toad (Spea) tadpoles are facultatively carnivorous, and meat-eating tadpoles are larger and have shorter guts than their conspecifics that consume a plant-based diet. These two siblings are the same age, but the tadpole on the left developed on a diet of plants and detritus. Ledón-Rettig et al.31 fed a related species, the non-carnivorous Scaphiopus couchii, a shrimp diet and observed increased heritability for body size, developmental stage and gut length, indicating that the dietary transition to the novel carnivorous feeding strategy in the Spea ancestor may have released cryptic genetic variation for these resource-use traits. (Image courtesy of David Pfennig.) b-d | Female yellow dung flies almost always have three sperm storage compartments, or spermathecae; Berger et al.29 perturbed spermathecae development by increasing rearing temperature to reveal cryptic genetic variation for four spermathecae. (Mating pair image courtesy of Peter Jann; spermathecae images courtesy of David Berger and reproduced with permission from John Wiley and Sons.) e-f | Queitsch et al.97 demonstrated that Arabidopsis plants exposed to the drug geldanamycin (GDA), an Hsp90 inhibitor, exhibit a variety of morphological abnormalities. Untreated, different accessions consistently develop into the wild-type phenotype (e). On GDA, different accessions exhibited abnormalities at different frequencies. For example, the Shadara accession was most likely to exhibit juxtaposed cotyledons (f) and deformed and radially symmetrical true leaves (g). Col more frequently exhibited dwarf plants with dark pigmentatio (h) and Ler more frequently produced curled hypocotyls (i). (Images courtesy of Christine Queitsch and reproduced with permission from Nature Publishing Group.)
Acknowledgments
Our work is supported by the Charles H. Revson Foundation (ABP) and the NIH (R01GM089972). We thank the reviewers for valuable advice and guidance.
Glossary
- Capacitor
A gene whose effect is to conceal the phenotypic effects of mutations at other loci, allowing the population to build up a store of cryptic genetic variation available for evolutionary response when the gene's effects are overcome by environmental challenge or mutation
- Mutation-selection-drift balance
An equilibrium arising from the balance between the introduction of alleles by mutation and their elimination by genetic drift and natural selection
- Canalization
Evolved resistance to perturbations, such that an invariant phenotype is produced across a range of genotypes and environments
- Robustness
A state of reduced phenotypic variance, not necessarily evolved, and which can be defined relative to specific perturbations (such as standing genetic variation) or to perturbations in general (such as the full mutational spectrum)
- Additive genetic variance (VA)
The transmissible component of a population's phenotypic variation. This is the variation due to the additive effects of segregating alleles
- Near-isogenic line
An inbred strain genetically identical to a progenitor strain except for a small region of genome that derives from a second strain
- Transgressive segregation
The appearance in the progeny of a cross of phenotypes outside the range of phenotypes present in the parental generation
- Stabilizing selection
natural selection favoring an intermediate phenotype, disfavoring phenotypes that depart from it in any direction
- Antagonistic pleiotropy
Allelic effects that are beneficial with respect to one aspect of fitness but detrimental with respect to another
- Standing variation
Genetic variation present within a population, as opposed to new mutations
- Genetic assimilation
The process by which selection can convert phenotypes revealed by environmental stimuli into phenotypes reliably produced in the absence of the stimulus. Genetic assimilation relies on genetic variation revealed by the stimulus
- Polyphenism
Multiple discrete phenotypic states produced under different conditions by a single genotype
References
- 1.Gibson G. Decanalization and the origin of complex disease. Nat Rev Genet. 2009;10:134–140. doi: 10.1038/nrg2502. This opinion article introduces the hypothesis that the genetic basis for diseases like diabetes and asthma may include cryptic alleles that increase susceptibility in modern environments. [DOI] [PubMed] [Google Scholar]
- 2.Gibson G, Dworkin I. Uncovering cryptic genetic variation. Nat Rev Genet. 2004;5:681–690. doi: 10.1038/nrg1426. [DOI] [PubMed] [Google Scholar]
- 3.Hermisson J, Wagner GP. The population genetic theory of hidden variation and genetic robustness. Genetics. 2004;168:2271–2284. doi: 10.1534/genetics.104.029173. This paper builds the theory showing that genetic or environmental perturbations will release hidden variation under general conditions of GxG or GxE interactions, indicating that cryptic genetic variation is not necessarily evidence of canalization or robustness. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips PC. Epistasis - the essential role of gene interactions in the structure and evolution of genetic systems. Nature Reviews Genetics. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masel J, Trotter MV. Robustness and evolvability. Trends Genet. 2010;26:406–414. doi: 10.1016/j.tig.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobzhansky T. Genetics and the Origin of Species. 2nd. Columbia University Press; 1941. [Google Scholar]
- 7.Waddington CH. The strategy of the genes. George Allen & Unwin Ltd; 1957. [Google Scholar]
- 8.Waddington CH. Genetic assimilation of an acquired character. Evolution. 1953;7:118–126. [Google Scholar]
- 9.Waddington CH. Genetic assimilation of the bithorax phenotype. Evolution. 1956;10:1–13. In this classic paper, exposure to ether revealed cryptic variation in D. melanogaster haltere development, which was captured over generations of selection via genetic assimilation. [Google Scholar]
- 10.Gibson G, Hogness DS. Effect of polymorphism in the Drosophila regulatory gene Ultrabithorax on homeotic stability. Science. 1996;271:200–203. doi: 10.1126/science.271.5246.200. [DOI] [PubMed] [Google Scholar]
- 11.Dworkin I, Palsson A, Birdsall K, Gibson G. Evidence that Egfr contributes to cryptic genetic variation for photoreceptor determination in natural populations of Drosophila melanogaster. Curr Biol. 2003;13:1888–1893. doi: 10.1016/j.cub.2003.10.001. This study identified the first cryptic nucleotides and presents an overview of the scope and nature of cryptic genetic variation at a single locus. [DOI] [PubMed] [Google Scholar]
- 12.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. This landmark experimental study demonstrated that reducing Hsp90 activity in D. melanogaster reveals extensive morphological variation, which can be selected upon and genetically assimilated. [DOI] [PubMed] [Google Scholar]
- 13.Burga A, Casanueva MO, Lehner B. Predicting mutation outcome from early stochastic variation in genetic interaction partners. Nature. 2011;480:250–253. doi: 10.1038/nature10665. [DOI] [PubMed] [Google Scholar]
- 14.Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frankel N, et al. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergman A, Siegal ML. Evolutionary capacitance as a general feature of complex gene networks. Nature. 2003;424:549–552. doi: 10.1038/nature01765. A simulation study shows that cryptic variation is an inherent feature of gene regulatory network architecture, arising without selection for capacitance, and perturbations to many genes will reveal it. [DOI] [PubMed] [Google Scholar]
- 17.Gjuvsland AB, Hayes BJ, Omholt SW, Carlborg O. Statistical epistasis is a generic feature of gene regulatory networks. Genetics. 2007;175:411–420. doi: 10.1534/genetics.106.058859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orr HA, Betancourt AJ. Haldane's sieve and adaptation from the standing genetic variation. Genetics. 2001;157:875–884. doi: 10.1093/genetics/157.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kacser H, Burns JA. The molecular basis of dominance. Genetics. 1981;97:639–666. doi: 10.1093/genetics/97.3-4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orr HA. A test of Fisher's theory of dominance. Proc Natl Acad Sci U S A. 1991;88:11413–11415. doi: 10.1073/pnas.88.24.11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson JB, Uppendahl LD, Traficante MK, Levy SF, Siegal ML. Histone variant HTZ1 shows extensive epistasis with, but does not increase robustness to, new mutations. PLoS Genet. 2013;9:e1003733. doi: 10.1371/journal.pgen.1003733. This paper provides experimental validation of the claim that cryptic genetic variation is not evidence of robustness, by demonstrating that mutation-accumulation yeast lines are phenotypically different, but equally diverse, with and without perturbation to a chromatin regulator. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milloz J, Duveau F, Nuez I, Felix MA. Intraspecific evolution of the intercellular signaling network underlying a robust developmental system. Genes Dev. 2008;22:3064–3075. doi: 10.1101/gad.495308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felix MA, Wagner A. Robustness and evolution: concepts, insights and challenges from a developmental model system. Heredity (Edinb) 2008;100:132–140. doi: 10.1038/sj.hdy.6800915. [DOI] [PubMed] [Google Scholar]
- 24.Braendle C, Baer CF, Felix MA. Bias and evolution of the mutationally accessible phenotypic space in a developmental system. PLoS Genet. 2010;6:e1000877. doi: 10.1371/journal.pgen.1000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penigault JB, Felix MA. Evolution of a system sensitive to stochastic noise: P3.p cell fate in Caenorhabditis. Dev Biol. 2011;357:419–427. doi: 10.1016/j.ydbio.2011.05.675. [DOI] [PubMed] [Google Scholar]
- 26.Felix MA. Cryptic quantitative evolution of the vulva intercellular signaling network in Caenorhabditis. Curr Biol. 2007;17:103–114. doi: 10.1016/j.cub.2006.12.024. [DOI] [PubMed] [Google Scholar]
- 27.Chandler CH. Cryptic intraspecific variation in sex determination in Caenorhabditis elegans revealed by mutations. Heredity (Edinb) 2010;105:473–482. doi: 10.1038/hdy.2010.62. [DOI] [PubMed] [Google Scholar]
- 28.McGuigan K, Nishimura N, Currey M, Hurwit D, Cresko WA. Cryptic genetic variation and body size evolution in threespine stickleback. Evolution. 2011;65:1203–1211. doi: 10.1111/j.1558-5646.2010.01195.x. This paper showed that oceanic sticklebacks reared in low-saline conditions revealed substantial cryptic genetic variation for body size, describing a compelling case of putative release of cryptic variation and subsequent adaptive evolution in the wild. [DOI] [PubMed] [Google Scholar]
- 29.Berger D, Bauerfeind SS, Blanckenhorn WU, Schafer MA. High temperatures reveal cryptic genetic variation in a polymorphic female sperm storage organ. Evolution. 2011;65:2830–2842. doi: 10.1111/j.1558-5646.2011.01392.x. [DOI] [PubMed] [Google Scholar]
- 30.Kim SY, Noguera JC, Tato A, Velando A. Vitamins, stress and growth: the availability of antioxidants in early life influences the expression of cryptic genetic variation. Journal of Evolutionary Biology. 2013;26:1341–1352. doi: 10.1111/jeb.12136. [DOI] [PubMed] [Google Scholar]
- 31.Ledon-Rettig CC, Pfennig DW, Crespi EJ. Diet and hormonal manipulation reveal cryptic genetic variation: implications for the evolution of novel feeding strategies. Proc Biol Sci. 2010;277:3569–3578. doi: 10.1098/rspb.2010.0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao H, et al. Genetic architecture of complex traits: large phenotypic effects and pervasive epistasis. Proc Natl Acad Sci U S A. 2008;105:19910–19914. doi: 10.1073/pnas.0810388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spiezio SH, Takada T, Shiroishi T, Nadeau JH. Genetic divergence and the genetic architecture of complex traits in chromosome substitution strains of mice. BMC Genet. 2012;13:38. doi: 10.1186/1471-2156-13-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen TF. The evolution of genetic architecture. Annu Rev Ecol Evol Syst. 2006;37:123–157. [Google Scholar]
- 35.Mather K. Variation and selection of polygenic characters. Journal of Genetics. 1941;41:159–193. [Google Scholar]
- 36.Gaertner BE, Parmenter MD, Rockman MV, Kruglyak L, Phillips PC. More than the sum of its parts: a complex epistatic network underlies natural variation in thermal preference behavior in Caenorhabditis elegans. Genetics. 2012;192:1533–1542. doi: 10.1534/genetics.112.142877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kroymann J, Mitchell-Olds T. Epistasis and balanced polymorphism influencing complex trait variation. Nature. 2005;435:95–98. doi: 10.1038/nature03480. [DOI] [PubMed] [Google Scholar]
- 38.Glater EE, Rockman MV, Bargmann CI. Multigenic Natural Variation Underlies Caenorhabditis elegans Olfactory Preference for the Bacterial Pathogen Serratia marcescens. G3 (Bethesda) 2013 doi: 10.1534/g3.113.008649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang W, et al. Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1213423109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dowell RD, et al. Genotype to phenotype: a complex problem. Science. 2010;328:469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brem RB, Kruglyak L. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc Natl Acad Sci U S A. 2005;102:1572–1577. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fierst JL, Hansen TF. Genetic architecture and postzygotic reproductive isolation: evolution of Bateson-Dobzhansky-Muller incompatibilities in a polygenic model. Evolution. 2010;64:675–693. doi: 10.1111/j.1558-5646.2009.00861.x. [DOI] [PubMed] [Google Scholar]
- 43.Haag ES. Compensatory vs. pseudocompensatory evolution in molecular and developmental interactions. Genetica. 2007;129:45–55. doi: 10.1007/s10709-006-0032-3. [DOI] [PubMed] [Google Scholar]
- 44.Wagner A. Neutralism and selectionism: a network-based reconciliation. Nature Reviews Genetics. 2008;9:965–974. doi: 10.1038/nrg2473. [DOI] [PubMed] [Google Scholar]
- 45.Gavrilets S. Evolution and speciation on holey adaptive landscapes. Trends Ecol Evol. 1997;12:307–312. doi: 10.1016/S0169-5347(97)01098-7. [DOI] [PubMed] [Google Scholar]
- 46.Badano JL, Katsanis N. Beyond Mendel: an evolving view of human genetic disease transmission. Nature Reviews Genetics. 2002;3:779–789. doi: 10.1038/nrg910. [DOI] [PubMed] [Google Scholar]
- 47.Cutting GR. Modifier genes in Mendelian disorders: the example of cystic fibrosis. Ann N Y Acad Sci. 2010;1214:57–69. doi: 10.1111/j.1749-6632.2010.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winston JB, et al. Complex trait analysis of ventricular septal defects caused by Nkx2-5 mutation. Circ Cardiovasc Genet. 2012;5:293–300. doi: 10.1161/CIRCGENETICS.111.961136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamilton BA, Yu BD. Modifier genes and the plasticity of genetic networks in mice. PLoS Genet. 2012;8:e1002644. doi: 10.1371/journal.pgen.1002644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chandler CH, Chari S, Dworkin I. Does your gene need a background check? How genetic background impacts the analysis of mutations, genes, and evolution. Trends Genet. 2013;29:358–366. doi: 10.1016/j.tig.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spencer CC, Howell CE, Wright AR, Promislow DE. Testing an ‘aging gene’ in long-lived Drosophila strains: increased longevity depends on sex and genetic background. Aging Cell. 2003;2:123–130. doi: 10.1046/j.1474-9728.2003.00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torjek O, et al. Segregation distortion in Arabidopsis C24/Col-0 and Col-0/C24 recombinant inbred line populations is due to reduced fertility caused by epistatic interaction of two loci. Theor Appl Genet. 2006;113:1551–1561. doi: 10.1007/s00122-006-0402-3. [DOI] [PubMed] [Google Scholar]
- 53.Dworkin I, et al. Genomic consequences of background effects on scalloped mutant expressivity in the wing of Drosophila melanogaster. Genetics. 2009;181:1065–1076. doi: 10.1534/genetics.108.096453. This study places cryptic genetic variation in the context of genetic background effects, by characterizing extensive phenotypic and gene-expression consequences of a specific mutation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chari S, Dworkin I. The conditional nature of genetic interactions: the consequences of wild-type backgrounds on mutational interactions in a genome-wide modifier screen. PLoS Genet. 2013;9:e1003661. doi: 10.1371/journal.pgen.1003661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamamoto A, Anholt RR, MacKay TF. Epistatic interactions attenuate mutations affecting startle behaviour in Drosophila melanogaster. Genet Res (Camb) 2009;91:373–382. doi: 10.1017/S0016672309990279. [DOI] [PubMed] [Google Scholar]
- 56.Clausen J, Keck DD, Hiesey W. Experimental studies on the nature of species I Effects of varied environments on western North American plants. Carnegie Institute: 1940. [Google Scholar]
- 57.Hodgins-Davis A, Adomas AB, Warringer J, Townsend JP. Abundant gene-by-environment interactions in gene expression reaction norms to copper within Saccharomyces cerevisiae. Genome Biology and Evolution. 2012;4:1061–1079. doi: 10.1093/gbe/evs084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas D. Methods for investigating gene-environment interactions in candidate pathway and genome-wide association studies. Annu Rev Public Health. 2010;31:21–36. doi: 10.1146/annurev.publhealth.012809.103619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ungerer MC, Halldorsdottir SS, Purugganan MD, Mackay TF. Genotype-environment interactions at quantitative trait loci affecting inflorescence development in Arabidopsis thaliana. Genetics. 2003;165:353–365. doi: 10.1093/genetics/165.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vieira C, et al. Genotype-environment interaction for quantitative trait loci affecting life span in Drosophila melanogaster. Genetics. 2000;154:213–227. doi: 10.1093/genetics/154.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anderson JT, Lee CR, Rushworth CA, Colautti RI, Mitchell-Olds T. Genetic trade-offs and conditional neutrality contribute to local adaptation. Mol Ecol. 2013;22:699–708. doi: 10.1111/j.1365-294X.2012.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duveau F, Felix MA. Role of pleiotropy in the evolution of a cryptic developmental variation in Caenorhabditis elegans. PLoS Biol. 2012;10:e1001230. doi: 10.1371/journal.pbio.1001230. This study identifies a cryptic nucleotide variant affecting vulva development in C. elegans, which likely experienced positive selection via pleiotropy on non-cryptic, fitness-related traits. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holt RD, Gaines MS. Analysis of Adaptation in Heterogeneous Landscapes - Implications for the Evolution of Fundamental Niches. Evolutionary Ecology. 1992;6:433–447. [Google Scholar]
- 64.Kawecki TJ, Barton NH, Fry JD. Mutational collapse of fitness in marginal habitats and the evolution of ecological specialisation. Journal of Evolutionary Biology. 1997;10:407–429. [Google Scholar]
- 65.Eshel I, Matessi C. Canalization, genetic assimilation and preadaptation. A quantitative genetic model. Genetics. 1998;149:2119–2133. doi: 10.1093/genetics/149.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schlichting CD. Hidden reaction norms, cryptic genetic variation, and evolvability. Ann N Y Acad Sci. 2008;1133:187–203. doi: 10.1196/annals.1438.010. [DOI] [PubMed] [Google Scholar]
- 67.McGuigan K, Sgro CM. Evolutionary consequences of cryptic genetic variation. Trends Ecol Evol. 2009;24:305–311. doi: 10.1016/j.tree.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 68.Masel J. Cryptic genetic variation is enriched for potential adaptations. Genetics. 2006;172:1985–1991. doi: 10.1534/genetics.105.051649. This theoretical paper addresses the fundamental issue of the fitness distribution of cryptic genetic variation, and finds that cryptic alleles can improve fitness under a wide range of realistic parameter values. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rutherford SL. From genotype to phenotype: buffering mechanisms and the storage of genetic information. Bioessays. 2000;22:1095–1105. doi: 10.1002/1521-1878(200012)22:12<1095::AID-BIES7>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 70.Fisher RA. The genetical theory of natural selection. Oxford University Press; 1930. [Google Scholar]
- 71.Cheverud JM, Routman EJ. Epistasis as a source of increased additive genetic variance at population bottlenecks. Evolution. 1996;50:1042–1051. doi: 10.1111/j.1558-5646.1996.tb02345.x. [DOI] [PubMed] [Google Scholar]
- 72.Goodnight CJ. Epistasis and the Effect of Founder Events on the Additive Genetic Variance. Evolution. 1988;42:441–454. doi: 10.1111/j.1558-5646.1988.tb04151.x. [DOI] [PubMed] [Google Scholar]
- 73.Barton NH, Turelli M. Effects of genetic drift on variance components under a general model of epistasis. Evolution. 2004;58:2111–2132. doi: 10.1111/j.0014-3820.2004.tb01591.x. [DOI] [PubMed] [Google Scholar]
- 74.Turelli M, Barton NH. Will population bottlenecks and multilocus epistasis increase additive genetic variance? Evolution. 2006;60:1763–1776. [PubMed] [Google Scholar]
- 75.Carter AJ, Hermisson J, Hansen TF. The role of epistatic gene interactions in the response to selection and the evolution of evolvability. Theor Popul Biol. 2005;68:179–196. doi: 10.1016/j.tpb.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 76.Hansen TF. Why epistasis is important for selection and evolution. Evolution. 2013;67:3501–3511. doi: 10.1111/evo.12214. [DOI] [PubMed] [Google Scholar]
- 77.Taft HR, Roff DA. Do bottlenecks increase additive genetic variance? Conservation Genetics. 2012;13:333–342. [Google Scholar]
- 78.van Heerwaarden B, Willi Y, Kristensen TN, Hoffmann AA. Population bottlenecks increase additive genetic variance but do not break a selection limit in rain forest Drosophila. Genetics. 2008;179:2135–2146. doi: 10.1534/genetics.107.082768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hallander J, Waldmann P. The effect of non-additive genetic interactions on selection in multi-locus genetic models. Heredity. 2007;98:349–359. doi: 10.1038/sj.hdy.6800946. [DOI] [PubMed] [Google Scholar]
- 80.Fuerst C, James JW, Solkner J, Essl A. Impact of dominance and epistasis on the genetic make-up of simulated populations under selection: A model development. Journal of Animal Breeding and Genetics-Zeitschrift Fur Tierzuchtung Und Zuchtungsbiologie. 1997;114:163–175. doi: 10.1111/j.1439-0388.1997.tb00502.x. [DOI] [PubMed] [Google Scholar]
- 81.Carlborg O, Jacobsson L, Ahgren P, Siegel P, Andersson L. Epistasis and the release of genetic variation during long-term selection. Nat Genet. 2006;38:418–420. doi: 10.1038/ng1761. [DOI] [PubMed] [Google Scholar]
- 82.Meiklejohn CD, Hartl DL. A single mode of canalization. Trends Ecol Evol. 2002;17:468–473. [Google Scholar]
- 83.Hermisson J, Pennings PS. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169:2335–2352. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hayden EJ, Ferrada E, Wagner A. Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature. 2011;474:92–95. doi: 10.1038/nature10083. This study employed in vitro populations of RNA molecules to demonstrate the adaptive potential of cryptic variation: populations with accumulated, cryptic mutations adapted faster to a novel substrate than did the wild-type population. [DOI] [PubMed] [Google Scholar]
- 85.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc Natl Acad Sci U S A. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rajon E, Masel J. Compensatory evolution and the origins of innovations. Genetics. 2013;193:1209–1220. doi: 10.1534/genetics.112.148627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jarosz DF, Lindquist S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science. 2010;330:1820–1824. doi: 10.1126/science.1195487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Braendle C, Flatt T. A role for genetic accommodation in evolution? Bioessays. 2006;28:868–873. doi: 10.1002/bies.20456. [DOI] [PubMed] [Google Scholar]
- 89.West-Eberhard MJ. Developmental plasticity and evolution. Oxford University Press; 2003. [Google Scholar]
- 90.Suzuki Y, Nijhout HF. Evolution of a polyphenism by genetic accommodation. Science. 2006;311:650–652. doi: 10.1126/science.1118888. [DOI] [PubMed] [Google Scholar]
- 91.Rohner N, et al. Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science. 2013;342:1372–1375. doi: 10.1126/science.1240276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lauter N, Doebley J. Genetic variation for phenotypically invariant traits detected in teosinte: implications for the evolution of novel forms. Genetics. 2002;160:333–342. doi: 10.1093/genetics/160.1.333. A clever experimental design revealed abundant cryptic genetic variation in the teosinte population. Crosses between teosinte isolates and a common tester strain of maize exposed phenotypic diversity that was concealed by the teosinte genetic background. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gibson G. It takes a genome: How a clash between our genes and modern life is making us sick. FT Press; 2009. [Google Scholar]
- 94.Ezzati M, Lopez AD, Rodgers A, Vander Hoorn S, Murray CJ. Selected major risk factors and global and regional burden of disease. Lancet. 2002;360:1347–1360. doi: 10.1016/S0140-6736(02)11403-6. [DOI] [PubMed] [Google Scholar]
- 95.Finucane MM, et al. National, regional, and global trends in body-mass index since 1980. Lancet. 2011;377:557–567. doi: 10.1016/S0140-6736(10)62037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Siegal ML. Crouching variation revealed. Mol Ecol. 2013;22:1187–1189. doi: 10.1111/mec.12195. [DOI] [PubMed] [Google Scholar]
- 97.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 98.Yeyati PL, Bancewicz RM, Maule J, van Heyningen V. Hsp90 selectively modulates phenotype in vertebrate development. PLoS Genet. 2007;3:e43. doi: 10.1371/journal.pgen.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Specchia V, et al. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature. 2010;463:662–665. doi: 10.1038/nature08739. [DOI] [PubMed] [Google Scholar]
- 100.Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen B, Wagner A. Hsp90 is important for fecundity, longevity, and buffering of cryptic deleterious variation in wild fly populations. BMC Evol Biol. 2012;12:25. doi: 10.1186/1471-2148-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sgro CM, Wegener B, Hoffmann AA. A naturally occurring variant of Hsp90 that is associated with decanalization. Proc Biol Sci. 2010;277:2049–2057. doi: 10.1098/rspb.2010.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Siegal ML, Masel J. Hsp90 depletion goes wild. BMC Biol. 2012;10:14. doi: 10.1186/1741-7007-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Takahashi KH. Multiple capacitors for natural genetic variation in Drosophila melanogaster. Mol Ecol. 2013;22:1356–1365. doi: 10.1111/mec.12091. [DOI] [PubMed] [Google Scholar]
- 105.Siegal ML, Bergman A. Waddington's canalization revisited: developmental stability and evolution. Proc Natl Acad Sci U S A. 2002;99:10528–10532. doi: 10.1073/pnas.102303999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.True JR, Haag ES. Developmental system drift and flexibility in evolutionary trajectories. Evol Dev. 2001;3:109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- 107.Takano TS. Loss of notum macrochaetae as an interspecific hybrid anomaly between Drosophila melanogaster and D. simulans. Genetics. 1998;149:1435–1450. doi: 10.1093/genetics/149.3.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schutt C, Nothiger R. Structure, function and evolution of sex-determining systems in Dipteran insects. Development. 2000;127:667–677. doi: 10.1242/dev.127.4.667. [DOI] [PubMed] [Google Scholar]
- 109.Kolotuev I, Podbilewicz B. Pristionchus pacificus vulva formation: polarized division, cell migration, cell fusion, and evolution of invagination. Dev Biol. 2004;266:322–333. doi: 10.1016/j.ydbio.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 110.Sommer RJ. Evolution of regulatory networks: nematode vulva induction as an example of developmental systems drift. Adv Exp Med Biol. 2012;751:79–91. doi: 10.1007/978-1-4614-3567-9_4. [DOI] [PubMed] [Google Scholar]
- 111.Kiontke K, et al. Trends, stasis, and drift in the evolution of nematode vulva development. Curr Biol. 2007;17:1925–1937. doi: 10.1016/j.cub.2007.10.061. [DOI] [PubMed] [Google Scholar]
- 112.Nahmad M, Glass L, Abouheif E. The dynamics of developmental system drift in the gene network underlying wing polyphenism in ants: a mathematical model. Evol Dev. 2008;10:360–374. doi: 10.1111/j.1525-142X.2008.00244.x. [DOI] [PubMed] [Google Scholar]
- 113.Johnson NA, Porter AH. Evolution of branched regulatory genetic pathways: directional selection on pleiotropic loci accelerates developmental system drift. Genetica. 2007;129:57–70. doi: 10.1007/s10709-006-0033-2. [DOI] [PubMed] [Google Scholar]
- 114.Haag ES, True JR. Evolution and development: anchors away! Curr Biol. 2007;17:R172–174. doi: 10.1016/j.cub.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 115.Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]