Abstract
The ‘Atribacteria' is a candidate phylum in the Bacteria recently proposed to include members of the OP9 and JS1 lineages. OP9 and JS1 are globally distributed, and in some cases abundant, in anaerobic marine sediments, geothermal environments, anaerobic digesters and reactors and petroleum reservoirs. However, the monophyly of OP9 and JS1 has been questioned and their physiology and ecology remain largely enigmatic due to a lack of cultivated representatives. Here cultivation-independent genomic approaches were used to provide a first comprehensive view of the phylogeny, conserved genomic features and metabolic potential of members of this ubiquitous candidate phylum. Previously available and heretofore unpublished OP9 and JS1 single-cell genomic data sets were used as recruitment platforms for the reconstruction of atribacterial metagenome bins from a terephthalate-degrading reactor biofilm and from the monimolimnion of meromictic Sakinaw Lake. The single-cell genomes and metagenome bins together comprise six species- to genus-level groups that represent most major lineages within OP9 and JS1. Phylogenomic analyses of these combined data sets confirmed the monophyly of the ‘Atribacteria' inclusive of OP9 and JS1. Additional conserved features within the ‘Atribacteria' were identified, including a gene cluster encoding putative bacterial microcompartments that may be involved in aldehyde and sugar metabolism, energy conservation and carbon storage. Comparative analysis of the metabolic potential inferred from these data sets revealed that members of the ‘Atribacteria' are likely to be heterotrophic anaerobes that lack respiratory capacity, with some lineages predicted to specialize in either primary fermentation of carbohydrates or secondary fermentation of organic acids, such as propionate.
Introduction
Single-cell amplified genome (SAG) sequencing and metagenomics have proven to be invaluable tools for studying the microbial world by extending the application of genomics to uncultivated microorganisms (Stepanauskas, 2012; Sharon and Banfield, 2013), including phylum-level lineages with no cultivated representatives, that is, candidate phyla. It has been estimated that there may be ⩾100 candidate phyla in the domain Bacteria (Baker and Dick, 2013; Kantor et al., 2013; Yarza et al., 2014), significantly outnumbering phyla with cultivated representatives. Although candidate phyla are typically of low abundance, that is, part of the ‘rare biosphere' (Sogin et al., 2006; Elshahed et al., 2008), they are prominent members of microbial communities in several different environments (Harris et al., 2004; Chouari et al., 2005; Vick et al., 2010; Peura et al., 2012; Cole et al., 2013; Farag et al., 2014; Gies et al., 2014; Parkes et al., 2014) and may have important ecological roles (Sekiguchi, 2006; Yamada et al., 2011). SAG sequencing and metagenomics have yielded partial, nearly-complete or complete genomes for close to 20 candidate bacterial phyla (Glöckner et al., 2010; Siegl et al., 2011; Youssef et al., 2011; Takami et al., 2012; Wrighton et al., 2012; Dodsworth et al., 2013; Kantor et al., 2013; McLean et al., 2013; Rinke et al., 2013; Kamke et al., 2014; Wrighton et al., 2014), as well as several major uncultivated lineages of Archaea (Elkins et al., 2008; Baker et al., 2010; Ghai et al., 2011; Nunoura et al., 2011; Narasingarao et al., 2012; Kozubal et al., 2013; Rinke et al., 2013; Youssef et al., 2015), opening a genomic window to a much better understanding of this so-called ‘microbial dark matter' (Marcy et al., 2007; Rinke et al., 2013). In addition to individual organismal analyses, comparison of genomes from different habitats and from different lineages within a given candidate phylum can yield insight into the phylogeny, conserved features and metabolic diversity within these widespread but poorly understood branches on the tree of life (Kamke et al., 2014).
Candidate phylum OP9 was originally discovered as 1 of the 12 novel lineages (OP1–OP12) in sediments from the hot spring Obsidian Pool in Yellowstone National Park, USA (Hugenholtz et al., 1998). Additional cultivation-independent, 16S rRNA gene-targeted surveys have since recovered sequences related to OP9 in a variety of terrestrial geothermal springs (Costa et al., 2009; Lau et al., 2009; Sayeh et al., 2010; Vick et al., 2010; Wemheuer et al., 2013), wastewater digesters and biogas reactors (Levén et al., 2007; Wrighton et al., 2008; Rivière et al., 2009; Tang et al., 2011), petroleum reservoirs (Gittel et al., 2009, 2012; Kobayashi et al., 2012) and other environments. Similar diversity studies on marine subsurface sediments recovered 16S rRNA gene sequences forming a deeply-branching, monophyletic lineage affiliated with OP9. Based on these results, the marine sediment sister lineage was posited to represent a distinct candidate phylum called JS1 (Webster et al., 2004). A current synthesis of the available data suggests that JS1 is a characteristic denizen of sub-seafloor environments, and is particularly abundant in sediments associated with methane hydrates and hydrocarbon seeps, and on continental margins and shelves (Inagaki et al., 2006; Orcutt et al., 2011; Parkes et al., 2014). Sequences related to JS1 have also been detected in environments such as petroleum reservoirs (Pham et al., 2009; Kobayashi et al., 2012), hypersaline microbial mats (Harris et al., 2013), and landfill leachates (Liu et al., 2011). The phylogenetic relationships between OP9, JS1 and other bacterial phyla have not been fully resolved (McDonald et al., 2012), and to date, no axenic cultures have been reported for either of these lineages, although enrichment cultures containing JS1 have been successfully obtained (Webster et al., 2011).
Several recent studies have reported the first significant genomic data sets corresponding to members of OP9 and JS1. Two genomes of related OP9 species were recovered from terrestrial geothermal springs (Dodsworth et al., 2013), including a co-assembly of 15 SAGs from Little Hot Creek (Vick et al., 2010) representing ‘Candidatus Caldatribacterium californiense' and a metagenome bin recovered from an in situ-enriched, thermophilic cellulosic consortium (77CS) in Great Boiling Spring (Peacock et al., 2013) representing a close relative, ‘Ca. Caldatribacterium saccharofermentans'. Concomitantly, as part of a larger single-cell genome sequencing effort, 13 JS1 SAGs were recovered from different low-oxygen environments, including a terephthalate (TA)-degrading bioreactor, the anaerobic, sulfidic monimolimnion of meromictic Sakinaw Lake, Canada and sediments from Etoliko Lagoon, Greece (Rinke et al., 2013). A single OP9 SAG was also obtained from the TA bioreactor (Rinke et al., 2013). Additional SAGs belonging to JS1 were obtained from marine sediments in Aarhus Bay, Denmark (Lloyd et al., 2013). Other than the ‘Ca. Caldatribacterium' spp., which were predicted to be strictly anaerobic, saccharolytic fermenters, the gene content and metabolic potential of these SAGs have not been described in detail, and a rigorous phylogenomic assessment of the monophyly of these OP9 and JS1 genomic data sets remains to be performed.
In this study, single-cell sequence data were used to further expand the genomic coverage of OP9 and JS1 by identifying sequences corresponding to these lineages in metagenomes from the TA bioreactor and Sakinaw Lake. The SAGs and resulting metagenome bins were then used to assess the monophyly of the ‘Atribacteria' inclusive of OP9 and JS1, to identify metabolic and structural features conserved in ‘Atribacteria' and to predict physiological potential within different ‘Atribacteria' lineages.
Materials and methods
Single-cell genomes and metagenome data sets
JS1 SAGs B17 and I22 were obtained from 10-cm depth in a sediment core taken from Aarhus Bay, Denmark (56° 9' 35.889 N, 10° 28' 7.893 E). Sediment sampling, sample preparation, single-cell sorting and 16S rRNA gene sequencing of these SAGs has previously been described (Lloyd et al., 2013). Secondary genome amplification by multiple strand displacement, sequencing and assembly for SAGs B17 and I22 performed in this study is described in the Supplementary Information. Other SAG data sets used in this study have been published previously (Dodsworth et al., 2013; Rinke et al., 2013). Names and accession numbers of SAGs, metagenomes and metagenome bins used or generated in this study are shown in Supplementary Table S1.
Metagenome binning
Binning of metagenomes was performed using Metawatt version 1.7 (Strous et al., 2012), PhylopythiaS (Patil et al., 2011) and an emergent self-organizing maps (ESOM) procedure (Dick et al., 2009). For Metawatt, binning was performed using medium sensitivity with a taxonomic database containing SAG data from Rinke et al. (2013) and Dodsworth et al. (2013), as well as all complete bacterial and archaeal genomes (n=2535) downloaded from http://ftp.ncbi.nlm.nih.gov/genomes/Bacteria on 9 July 2013. The ‘Ca. C. saccharofermentans' bin in the 77CS metagenome (Dodsworth et al., 2013) was refined using Metawatt and manually filtered based on contig coverage profile, retaining contigs with coverage values from 170 to 300. For PhylopythiaS, default settings were used and manual curation was performed based on average contig read depth, BLAST, principle component analysis of tetranucleotide frequency and the presence of single-copy conserved markers (SCMs; Nobu et al., 2015b). For ESOM, metagenome contigs were split into 2500-bp long sequences with a window size of 5000 bp, and tetranucleotide frequencies were calculated for these sequences using tetramer_freqs_esom.pl (https://github.com/tetramerfreqs/binning). This was used as input to esomtrn (http://databionic-esom.sourceforge.net/user.html) to generate an ESOM (Dick et al., 2009). Contig fragments in distinct regions of the ESOM were selected by manual inspection, and corresponding contigs were included in the bin if >50% of their length was represented by these fragments. ClaMS (Patil et al., 2011) was also used for binning of the Etoliko Lagoon metagenome. Other homology-based binning methods such as BLASTN and BLASTP (Altschul et al., 1997) were used on the TA biofilm and Sakinaw Lake metagenomes for comparison. For BLASTN, contigs were binned based on having >95% identity over >1 kb to either the Sakinaw Lake SAGs (for the Sakinaw Lake metagenome) or the TA biofilm SAG 231 (for the TA biofilm metagenome). For BLASTP, metagenome contigs were binned if they contained at least one open reading frame with a top BLASTP hit to the OP9/JS1 SAGs in comparison to the NCBI protein nr database (accessed 1 September 2013), and furthermore if at least half the open reading frames on the contig had BLASTP hits >95% identity over >80% of their length to the OP9/JS1 SAGs. Metawatt was used to screen additional metagenomes identified as containing reads taxonomically assigned to OP9 or JS1 (Rinke et al., 2013).
Phylogenomics and phylogenetics
For phylogenomic analysis, a set of 31 markers conserved in Bacteria were identified in the OP9 and JS1 data sets using Amphora2 (Wu and Scott, 2012), and phylogenetic analysis using RAxML (Stamatakis, 2006) was performed on these and a set of markers in a variety of other bacterial genomes as previously described (Dodsworth et al., 2013). Additionally, SAGs and metagenome bins were scanned for homologues of a set of 83 universally conserved single-copy proteins present in Bacteria (Rinke et al., 2013). Marker genes were detected, aligned with hmmsearch and hmmalign included in the HMMER3 package (Eddy, 2011), and used to build concatenated alignments of up to 83 markers per genome. The phylogenetic inference method used was the maximum likelihood-based FastTree2 (Price et al., 2010) executed using the CAT approximation with 20 rate categories and the Jones–Taylor–Thornton amino-acid evolution model.
For 16S rRNA gene phylogenies, sequences were retrieved from the SAG and metagenomic data sets and aligned with nearly full-length 16S rRNA genes (>1300 bp) from representative ‘Atribacteria' (OP9 and JS1 lineages) and other bacterial phyla from the NCBI database using the SILVA Incremental Aligner (SINA) online tool (Pruesse et al., 2012; http://www.arb-silva.de/). Alignments were then checked and columns containing gaps were removed in BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Phylogenetic trees were constructed using maximum likelihood with the HKY85 substitution model, and 100 bootstrap replicates were implemented with PhyML (Guindon et al., 2010) through the online tool phylogeny.fr (http://phylogeny.lirmm.fr/; Dereeper et al., 2008). Congruent trees were also obtained using other methods, including minimum evolution and LogDet distance, neighbour-joining with Jukes–Cantor algorithm, maximum likelihood with Tamura–Nei model and maximum parsimony in MEGA version 6 (Tamura et al., 2013). Phylogeny was also inferred with a larger data set (744 sequences) as described above using 1344 positions, and the resulting tree was exported into the interactive Tree of Life (http://itol.embl.de/). Sequences were colour-coded by habitat of origin using the interactive Tree of Life online tools (Letunic and Bork, 2011).
Genomic analyses and identification of potentially monophyletic genes
Both the IMG/M-ER (Markowitz et al., 2014) and RAST (Aziz et al., 2008) platforms were used for gene calling, functional prediction and comparison of SAGs and metagenome bins. Because not all of the data sets could be uploaded to IMG/M-ER, RAST was used as a common platform and annotation system for most comparative analyses; however, IMG/M-ER was useful for checking functional annotations and for gene calling near the ends of open reading frames in these fragmented SAG and metagenome bin data sets. SCMs were identified by searching for protein families (PFAMs) using HMMER3 (Eddy, 2011) and marker-specific cutoffs as described (Rinke et al., 2013). Average nucleotide identity (ANI) was calculated by the method of Goris et al. (2007) using the online tool http://enve-omics.ce.gatech.edu/ani/. Phylogenetic analysis of predicted protein sequences from individual genes in putative bacterial microcompartment (BMC) clusters was performed using MUSCLE v3.7 (Edgar, 2004). Alignments were manually adjusted with BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html), and maximum likelihood phylogenies with 100 bootstrap replicates were performed with PhyML (Guindon et al., 2010) using the online tool phylogeny.fr (Dereeper et al., 2008). Prediction of potentially secreted proteins was carried out using PSORTb 3.0 (Yu et al., 2010). To identify genes that are potentially conserved and monophyletic within the ‘Atribacteria', predicted proteins in OP9/JS1 SAGs and metagenome bins were queried by BLASTP against a database composed of ‘Atribacteria' data sets as well as all non-‘Atribactiera' sequences in the NCBI RefSeq protein database (release 65, downloaded 20 July 2014). For each query sequence, results were sorted by bitscore, and top hits were recorded until the first non-'Atribacteria' sequence was encountered. Results for all of the ‘Atribacteria' data sets were cross-referenced. Because of the incomplete nature of the data sets, a predicted protein was considered as being potentially conserved and monophyletic in the ‘Atribacteria' if it had BLASTP hits to predicted proteins in at least one of the OP9 lineages (OP9−1, −2) and at least two of the JS1 lineages (JS1−1, −2, −3, −4) with higher bit scores than hits to any non-‘Atribacteria' proteins.
Results and discussion
Metagenome binning targeting OP9 and JS1
Several tetranucleotide frequency-based binning techniques (PhylopythiaS, Metawatt and ESOM) were compared and used to identify metagenome bins corresponding to OP9 or JS1 based on the available SAG data in the TA biofilm, Sakinaw Lake and other metagenomes (Dodsworth et al., 2013; Rinke et al., 2013). PhylopythiaS, Metawatt and ESOM produced largely overlapping bins of JS1 contigs from the TA biofilm metagenome that had distinct monomodal read depth distribution of 14.8 mean±4.7 s.d. in comparison to a bulk metagenome read depth of 95 mean±273 s.d. Despite the fact that three distinct SAGs (two JS1, one OP9) were obtained from this sample, this was the only clear bin representing a lineage of OP9 or JS1, likely due to its relative abundance and sequencing coverage of the metagenome. These bins also contained a low number of duplicated SCMs (Supplementary Figure S1), consistent with the bins each representing the core genome of a single species or genotype rather than a diverse mixture of related organisms. PhylopythiaS, Metawatt and ESOM produced bins that were significantly larger than the methods based on homology alone (BLASTN and BLASTP), as would be expected for these more generalized, nucleotide frequency-based methods (Supplementary Figure S1). Similar results were obtained through binning of the Sakinaw Lake metagenome, although the bins were somewhat less distinct, possibly because of increased abundance and diversity of JS1 lineages present in this environment (Gies et al., 2014), including at least two prominent, distinct JS1 lineages in the monimolimnion based on previous 16S rRNA gene clone libraries (data not shown). After manual filtering based on contig coverage statistics, the PhylopythiaS bins were selected for further analysis because they were larger and had fewer duplicated SCMs than bins made by Metawatt and ESOM for the TA biofilm (Supplementary Figure S1) and the Sakinaw Lake metagenomes (data not shown). No distinct bins corresponding to OP9 or JS1 were obtained in the Etoliko Lagoon metagenome (source of JS1 SAG 227) or other metagenomes with reads assigned to OP9/JS1 (Rinke et al., 2013), which was probably because of a low abundance of these lineages in the metagenomes that were screened. In addition to generating new metagenome bins, coverage-based curation of the ‘Ca. C. saccharofermentans' bin from the 77CS cellulolytic enrichment metagenome (Dodsworth et al., 2013) significantly enhanced the fidelity of this bin (two duplicated SCMs compared with 12 prior to filtering) without decreasing estimated genomic coverage (Table 1).
Table 1. OP9 and JS1 SAG and metagenome (MG) bin data sets.
| SAGs or MG bin | Group | Size (Mb) | No. of scaffolds | No. of genes | No. of SCMs | No. of SCMs >1 copy | Estimated genomic coverage | Reference |
|---|---|---|---|---|---|---|---|---|
| LHC SAG co-assembly | OP9-1 | 2.09 | 545 | 2477 | 130 | 1 | >99% | Dodsworth et al., 2013 |
| 77CS MG bin | OP9-1 | 1.79 | 153 | 1970 | 134 | 2 | >99% | Dodsworth et al., 2013; this study |
| TA biofilm SAG 232 | OP9-2 | 0.71 | 43 | 687 | 9 | 0 | 7% | Rinke et al., 2013 |
| SL SAG co-assembly | JS1-1 | 2.09 | 153 | 2071 | 102 | 0 | 81% | Rinke et al., 2013 |
| SL MG bin | JS1-1 | 0.34 | 150 | 448 | 39 | 1 | 31% | This study |
| SL SAG 217 | JS1-1 | 0.33 | 27 | 320 | 29 | 0 | 23% | Rinke et al., 2013 |
| Additional SL SAGs (4)a | JS1-1 | 0.15–1.65 | 20–131 | 163–1672 | 7–92 | 0–2 | 6–73% | Rinke et al., 2013 |
| Aarhus Bay SAG I22 | JS1-1 | 1.04 | 291 | 1074 | 28 | 0 | 22% | Lloyd et al., 2013; this study |
| TA biofilm SAG 231 | JS1-2 | 0.95 | 50 | 932 | 32 | 0 | 25% | Rinke et al., 2013 |
| TA biofilm MG bin | JS1-2 | 2.01 | 789 | 2434 | 108 | 1 | 86% | This study |
| Aarhus Bay SAG B17 | JS1-3 | 1.13 | 246 | 1361 | 9 | 1 | 7% | Lloyd et al., 2013; this study |
| Etoliko Lagoon SAG 227 | JS1-3 | 0.32 | 28 | 320 | 11 | 0 | 8% | Rinke et al., 2013 |
| TA biofilm SAG 167 | JS1-4 | 0.49 | 27 | 466 | 42 | 0 | 33% | Rinke et al., 2013 |
Abbreviations: LHC, Little Hot Creek; SAG, single-cell amplified genome; SCM, single-copy conserved marker; SL, Sakinaw Lake; TA, terephthalate.
Sakinaw Lake SAGs 124, 130, 136 and 219.
The bins obtained from the TA biofilm and Sakinaw Lake metagenomes were closely related to existing JS1 SAG data sets and expanded genomic coverage within these lineages. Based on ANI, the Sakinaw Lake bin corresponded to the Sakinaw Lake SAG co-assembly (99.2% ANI). The TA biofilm metagenome bin corresponded to TA biofilm SAG 231 (99.9% ANI) but was not closely related to the other two SAGs recovered from the TA biofilm, JS1 SAG 167 and OP9 SAG 232. Distinct bins corresponding to SAGs 167 and 232 were not detected, possibly due to a low abundance of these lineages in the TA reactor biofilm. Although these ANI values are above the %ANI typically shared by members of the same species (95% Richter and Rosselló-Móra, 2009), co-assembly of the metagenomes with their respective SAG data sets was not performed because of the distinct nature of the data sets (SAG vs metagenome). The TA biofilm metagenome bin was significantly larger and had a much higher estimated genomic coverage (86%) than SAG 231 (25% Table 1). A genomic coverage of 91% was estimated for the species-level lineage represented by pooling the SAG and metagenome bin (designated JS1-2), indicating good coverage within this lineage. The Sakinaw Lake metagenome bin also expanded on genomic coverage of JS1 in Sakinaw Lake, with 110 SCMs suggesting 87% estimated coverage present in both data sets combined in comparison to 81% (101 SCMs) by the SAG co-assembly alone. The modest gains were likely due to the small size of this metagenome bin in relationship to the Sakinaw SAG co-assembly.
Phylogenetic diversity and phylogenomics of OP9 and JS1 data sets
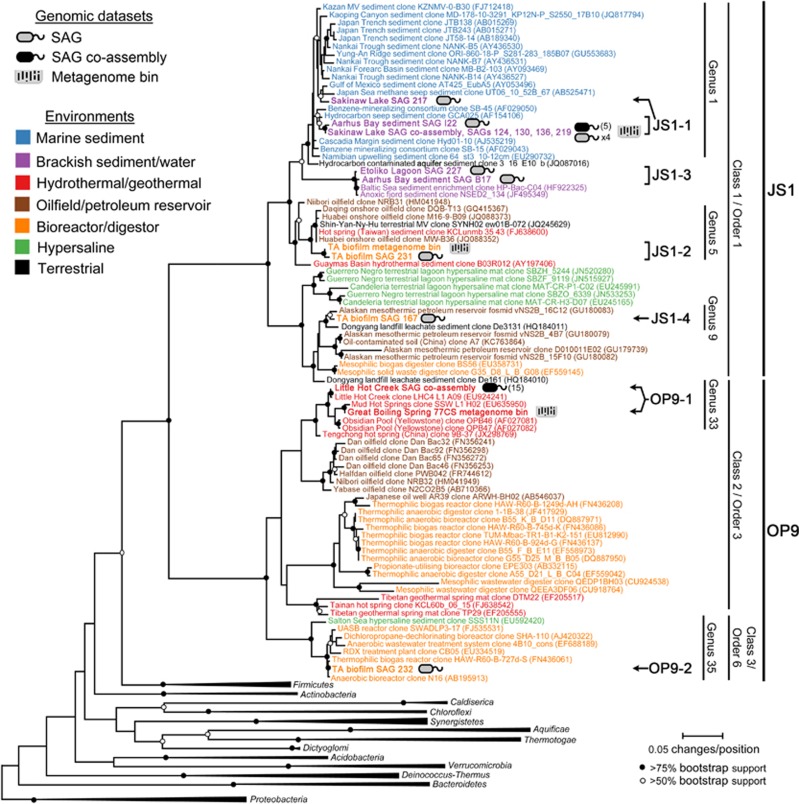
The SAGs and metagenome bins were broadly distributed within OP9 and JS1 and represent a diversity of habitats where these lineages are found. The SAGs and metagenome bins together comprised six groups (Figure 1), including three genus-level groups (OP9-1, JS1-1, JS1-3) at >95% 16S rRNA gene identity and one species-level group (JS1-2) at >95% ANI and >98.7% 16S rRNA gene identity (Richter and Rosselló-Móra, 2009; Yarza et al., 2014). These groups represent most major clades comprising OP9 and JS1, including three of the four order-level groups identified in the Greengenes taxonomy (McDonald et al., 2012), as well as all three classes and major order-level candidate taxonomic units identified by Yarza et al. (2014). Overall, members of the OP9 lineage tend to be found in terrestrial thermal and subsurface environments while JS1 sequences are found mainly in non-thermal systems (Supplementary Figure S2), although there is some overlap. Both lineages, however, appear to be restricted to anaerobic environments, often with considerable amounts of biomass or reduced carbon present. The SAGs and metagenome bins represent most of the major habitat types where OP9 and JS1 sequences are found, including geothermal springs, bioreactors, brackish waters and marine sediments. Of note, the genus-level lineage represented by the group JS1-1 SAGs and metagenome bin also includes sequences that are particularly abundant in some marine sediments (Inagaki et al., 2006; Parkes et al., 2014). In contrast to some previous phylogenetic analyses of OP9 and JS1 (Webster et al., 2004), there is high bootstrap support (>96% of pseudoreplicates) for the monophyly of these two lineages with the broader set of 16S rRNA gene sequences included here (Figure 1, Supplementary Figure S2). This is consistent with some other recent taxonomies (McDonald et al., 2012; Yarza et al., 2014) and likely reflects the increased number and diversity of OP9 and JS1 sequences now available in the Genbank database.
Figure 1.
A 16S rRNA gene phylogeny of SAG and metagenome bin data sets (underlined) within the context of cloned sequences from OP9 and JS1 and of other Bacteria. The number of SAGs used for construction of co-assemblies are indicated in parentheses. Genus, Order and Class candidate taxonomic units proposed by Yarza et al. (2014) that encompass the OP9 and JS1 data sets are indicated. Although the Sakinaw Lake JS1 metagenome bin did not contain a 16S rRNA gene, its affiliation with the Sakinaw Lake co-assembly (based on %ANI) is indicated in this figure.
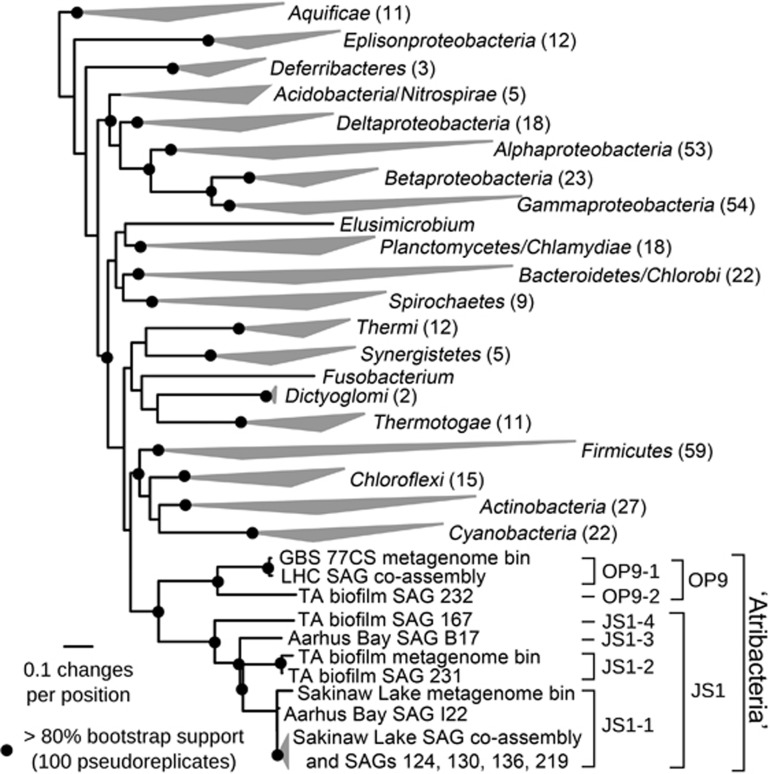
Phylogenies based on conserved protein-coding markers in the SAGs and metagenome bins, as well as several other affiliated bacterial phyla with cultivated representatives, show strong support for the monophyly of the ‘Atribacteria' lineage encompassing both OP9 and JS1 (Figure 2, Supplementary Figure S3). Although support for a node connecting the ‘Atribacteria' and Synergistetes was sometimes observed (Supplementary Figure S3), this affiliation was not supported in a majority of phylogenomic analyses using various outgroups (data not shown) or in previous phylogenomic analyses of larger data sets (Rinke et al., 2013) and is not supported by 16S rRNA gene phylogenies presented here (Figure 1) or elsewhere (Rinke et al., 2013; Yarza et al., 2014). Therefore, inclusion of the ‘Atribacteria' in the phylum Synergistetes is not justified. To address the question of whether OP9 and JS1 represent a single phylum or two distinct sister phyla (Webster et al., 2004), both of which would be consistent with the phylogenomics results, 16S rRNA gene identities were compared among these lineages. The median (80.8%) and minimum (74.2%) 16S rRNA gene identity between members of OP9 and JS1 are within the range of these values suggested for other bacterial and archaeal phyla (Supplementary Table S2; (Yarza et al., 2014)). Furthermore, only two of the 70 784 pairwise comparisons have a sequence identity below the suggested threshold (75%) recently proposed for delineation of a phylum, consistent with the designation of sequences in OP9 and JS1 as a single phylum-level candidate taxonomic unit by Yarza et al. (2014). Thus there is not a compelling argument for designation of OP9 and JS1 as separate phyla, and the most parsimonious analysis of the available data would suggest that the ‘Atribacteria', inclusive of OP9 and JS1, is a single candidate phylum within the Bacteria.
Figure 2.
Phylogenomic analysis of OP9 and JS1 SAGs, metagenome bin data sets and other Bacteria. Maximum likelihood phylogeny inferred with RAxML (Stamatakis, 2006) using a concatenated alignment of 31 conserved markers. The number of organisms represented by each wedge is indicated in parentheses. Etoliko Lagoon SAG 227 is not included because it did not contain any of these markers.
Conserved features and potential roles for bacterial microcompartments in the ‘Atribacteria'
All of the ‘Atribacteria' represented by the genomic data sets appear to be heterotrophic fermenters or syntrophs, because none of the genomes contain genes suggestive of capacity for autotrophy, and no clear evidence of either aerobic or anaerobic respiratory capacity was observed. As noted previously for the Little Hot Creek SAG co-assembly and 77CS metagenome bin (Dodsworth et al., 2013), several key markers associated with bacteria containing an outer membrane (Sutcliffe, 2011) were also present in the other ‘Atribacteria' data sets (Supplementary Table S3), suggesting a diderm cell envelope structure for both OP9 and JS1 lineages. To sift the genomes for additional conserved features that might offer insight into the physiology of the ‘Atribacteria', a set of putatively monophyletic genes was identified, each of which was present in at least one OP9 and two JS1 groups (Table 1,Figure 1) and showed higher BLASTP matches to other OP9 and JS1 putative homologues than to hits outside the ‘Atribacteria'. In total, only 51 genes met these criteria (Supplementary Table S4). The majority of these encode proteins predicted to be involved in cell envelope synthesis, transport/secretion, housekeeping/central metabolism or have no predicted function. Potentially monophyletic genes that may be involved in specific metabolic processes include peptidases of the C11/clostrapain superfamily (Chen et al., 1998; Rawlings et al., 2014) with predicted N-terminal Sec-dependent secretion sequences, which could allow for digestion and utilization of proteins, and the HylB subunit of an electron-bifurcating formate dehydrogenase (Wang et al., 2013) that may be involved in energy conservation, as discussed below.
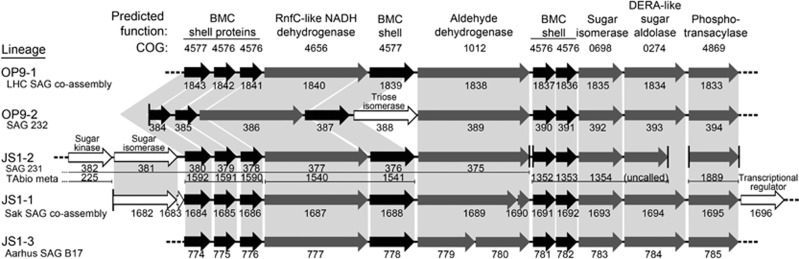
Interestingly, 11 of the 51 potentially monophyletic genes are present in the majority of the ‘Atribacteria' genomic data sets, either at a single locus or on contig fragments consistent with the structure of a single locus (Figure 3). Six of the genes at these loci are predicted to encode homologues of BMC shell proteins (COGs 4576 and 4577, containing PFAMs PF03319 and PF00936, respectively). BMCs are protein-bound bacterial organelles that can function in anabolic (for example, the carboxysome in cyanobacteria) or catabolic (for example, ethanolamine or 1,2-propanediol utilization) processes (Kerfeld et al., 2010). Recent genomic surveys have revealed that BMC gene clusters are broadly distributed within the Bacteria, including the ‘Atribacteria' and several other candidate phyla (Axen et al., 2014; Kamke et al., 2014). The only ‘Atribacteria' lineage for which genomic data are available where these BMC genes were not detected was JS1-4 (represented by SAG 167), quite probably because the estimated genomic coverage for this SAG was relatively low at 33% (Table 1). Three of the genes in these BMC loci encode homologues of PduP (coenzyme A-acylating aldehyde dehydrogenase), PduL (phosphotransacetylase) and PduS (RnfC/quinone oxidoreductase (Nqo)-like NADH dehydrogenase), which are involved in BMC-mediated 1,2-propanediol catabolism in Salmonella spp. (Sampson et al., 2005; Parsons et al., 2010). The presence of PduPL suggests that the putative ‘Atribacteria' BMC may sequester metabolically generated, toxic aldehydes as has been proposed for catabolic BMCs in other organisms (Sampson and Bobik, 2008). However, the lack of several other key Pdu genes required for 1,2-propanediol catabolism suggest that this specific substrate is not utilized by ‘Atribacteria' (Parsons et al., 2010). The remaining two conserved genes in these ‘Atribacteria' BMC loci encode homologues of 2-deoxy-D-ribose 5-phosphate aldolase (DERA) and pentose monophosphate isomerase, suggesting that BMC may link aldehyde and sugar metabolism. Although such aldolases and sugar isomerases are not typically associated with BMC loci in other organisms, comparison of the predicted products of these genes with close homologues revealed an N-terminal 30–40 amino-acid sequence unique to the BMC-associated ‘Atribacteria' proteins (Supplementary Figure S4). This suggests that they may be targeted to the BMC lumen, as several BMC-associated genes from other bacteria have been shown to have N-terminal extensions of similar length for facilitating BMC lumen localization, although prediction of specific residues responsible for targeting is not straightforward (Fan et al., 2010). A putative transcriptional regulator flanking the BMC cluster in the Sakinaw SAG co-assembly (Figure 3) was also identified as a conserved ‘Atribacteria' gene (Supplementary Table S4). Although not apparently closely linked to the BMC cluster in other ‘Atribacteria' lineages, this regulator may be involved in controlling BMC cluster expression.
Figure 3.
BMC gene loci in representatives of different ‘Atribacteria' lineages. Genes predicted to encode BMC shell proteins (black) and enzymes (grey) conserved in the BMC loci, as well as intervening or peripheral genes that may be involved in BMC function (white), are indicated by arrows, with corresponding RAST gene numbers below each arrow. Predicted function and COGs are indicated at the top. Homologous genes are indicated by light grey shading, and truncated contigs are indicated by vertical lines. For JS1-2, individual gene numbers and contig fragments (thin lines below gene numbers) are indicated separately in SAG 231 and the TA biofilm metagenome bin that together contain a complete set of the conserved genes in ‘Atribacteria' BMC loci on multiple, truncated contigs.
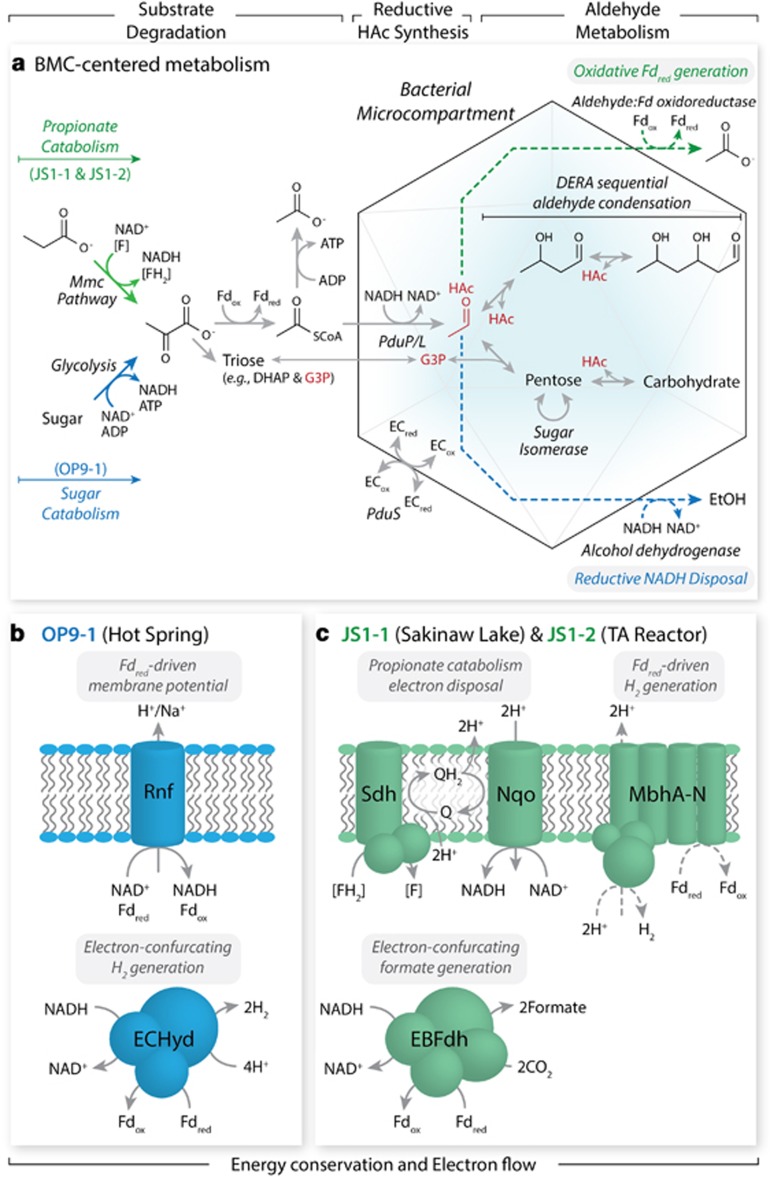
DERA is known to perform aldol condensation of acetaldehyde and glyceraldehyde-3-phosphate, forming deoxyribose-5-phosphate (Jennewein et al., 2006), and it has been suggested that the BMC in ‘Atribacteria' may be involved in catabolism of deoxyribose-5-phosphate or other sugar phosphates (Axen et al., 2014). Compared with the Escherichia coli DERA, the ‘Atribacteria' DERA consistently encode residue substitutions indicative of high resistance to aldehyde inhibition (K172F and V206I), increased capacity to perform sequential aldol reactions (F200I) and substrate specificity modifications (F200I and M185I; Jennewein et al., 2006; Sakuraba et al., 2007). Tolerance to high aldehyde concentrations, implied by these substitutions, may enable the ‘Atribacteria' DERA to condense aldehydes within an aldehyde-accumulating BMC. Although the implications of the substrate specificity modifications are unclear, we speculate that the ‘Atribacteria' DERA performs sequential condensation of aldehydes involving a sugar intermediate that is not necessarily deoxyribose-5-phosphate. To generate aldehydes for such metabolism, PduPL are thought to reduce acetyl-CoA to acetaldehyde utilizing the BMC lumen NAD+/NADH pool, which can be isolated from (Huseby and Roth, 2013) or exchanged with the cytoplasmic NAD+/NADH pool (Cheng et al., 2012). If BMC and cytoplasmic NADH are exchangeable in ‘Atribacteria', aldehyde generation within the BMC could effectively serve as a cytoplasmic NADH sink (Figure 4a), which could have important implications in energy conservation in ‘Atribacteria' lineages as discussed below. Although PduS may also facilitate reducing power transfer between the BMC lumen and cytoplasm (Figure 4a), its exact role remains unclear. In this alternative scenario, the sugar isomerase and aldolase in the compartment interior may allow the BMC to function as a potential sugar storage compartment. Phylogenetic analyses support the monophyly of each of the 11 conserved genes in the ‘Atribacteria' BMC cluster in comparison to representative homologues (from appropriate COGs, PFAMs or conserved domains) and top BLASTP hits (Supplementary Figures S5−S12), and the overall order of the genes within the loci is highly conserved. The broad distribution, monophyly and conserved synteny of these genes in the ‘Atribacteria' suggest that this putative BMC is an ancestral trait within the phylum, and it is reasonable to deduce that BMC-mediated aldehyde conversion to sugars is central to ‘Atribacteria' metabolism.
Figure 4.
Predicted catabolism, BMC function and energy conservation in ‘Atribacteria' JS1-1, JS1-2 and OP9-1 lineages. (a) Catabolic degradation of propionate via the Mmc pathway in JS1-1 and JS1-2 (green arrows) and fermentation of sugars in OP9-1 (blue) converge on pyruvate, which can be further processed to acetyl-CoA, acetate and acetaldehyde (HAc) in all these lineages (grey). NADH-dependent acetyl-CoA reduction in the BMC by PduPL produces HAc, which can further serve either as an electron sink via alcohol dehydrogenase for OP9-1 (blue dotted line) or as a high energy electron source via aldehyde:Fd oxidoreductase (producing reduced Fd and acetate) for JS1-1 and JS1-2 (green dotted line). HAc and pyruvate-derived glyceraldehyde-3-phosphate (G3P) may also undergo sequential aldehyde condensation through DERA and sugar isomerase, facilitating carbon storage and later use as an electron source or sink. See text and Supplementary Table S5 for additional details. (b) Energy conservation in OP9-1 via NADH:Fd oxidoreductase (Rnf complex) and electron-confurcating hydrogenase (ECHyd). (c) Energy-conserving formate/H2 generation in JS1-1 and JS1-2 through EBFdh, membrane-bound hydrogenase (MbhA-N), succinate dehydrogenase (Sdh) and NADH:Nqo using nicotinamide adenine dinucleotide (NAD+/NADH), ferredoxin (Fd), flavin (F/FH2) and quinones (Q/QH2) as electron carriers.
Predicted catabolic substrates and energy conservation in OP9-1, JS1-1 and JS1-2 lineages
The high genomic coverage in lineages OP9-1, JS1-1 and JS1-2 allow a more detailed discussion of their physiological potential, including predicted substrates, energy conservation mechanisms and the possible role of BMC in catabolic processes. ‘Atribacteria' members associated with the hot spring environment (‘Ca. Caldatribacterium' spp. in the OP9-1 lineage) have been predicted to perform saccharolytic fermentation from cellulosic substrates, including xyloglucan (Dodsworth et al., 2013). Sugar oxidation generates NADH and reduced ferredoxin (Fdred) as reduced electron carriers and thus requires complementary pathways to dispose this reducing power (Figure 4a). These genomes encode pathways for reoxidation of NADH via acetyl-CoA reduction to ethanol (aldehyde and alcohol dehydrogenases) and H2 production (NiFe hydrogenase, not shown in Figure 4) and concomitant re-oxidation of both NADH and Fdred by an electron-confurcating hydrogenase (Schut and Adams, 2009; Sieber et al., 2012; Figure 4b). In addition, they possess an NADH:Fd oxidoreductase (Rnf complex; Biegel et al., 2011) that may allow ‘Ca. Caldatribacterium' to balance oxidation of NADH and Fdred (Figure 4b) and generate a proton- (or sodium-) motive force when Fdred is in excess. Consumption of NADH via reduction of acetyl-CoA to acetaldehyde within the BMC (Figure 4a), as well as reduction of acetaldehyde to ethanol in the cytoplasm, are potential mechanisms for generating excess Fdred and driving Rnf-mediated energy conservation.
In contrast with ‘Ca. Caldatribacterium', ‘Atribacteria' members from the JS1-1 and JS1-2 lineages found in Sakinaw Lake and the TA-degrading bioreactor (‘Ca. Atricorium thermopropionicum' Nobu et al., 2015b) lack such sugar fermentation pathways but appear to have the capacity to catabolize organic acids such as acetate (Gies et al., 2014) or propionate (Figure 4a). Previous studies have proposed that this lineage may oxidize acetate through syntrophy or sulfate reduction based on observation of Wood–Ljungdahl pathway genes and acetate uptake in marine enrichment cultures, respectively (Webster et al., 2011; Gies et al., 2014). In network analysis of Sakinaw Lake microbial communities, ‘Atribacteria' co-occurred with H2- and formate-scavenging methanogens and putative propionate-metabolizing Cloacimonetes, also suggesting involvement in syntrophic degradation of propionate (Gies et al., 2014). Further supporting a role in propionate catabolism, metatranscriptomics of the TA-degrading community revealed the expression of ‘Ca. Atricorium' genes potentially involved in propionate degradation via the methylmalonyl-CoA (Mmc) pathway (Nobu et al., 2015b). Members of both JS1-1 and JS1-2 lineages encode Mmc mutase, epimerase and decarboxylase (alpha subunit) genes with high similarity (>70, 70 and 60%, respectively) to those from a representative thermophilic propionate-degrading syntroph, Pelotomaculum thermopropionicum strain SI (Imachi et al., 2002), along with other genes that could enable conversion of propionate to pyruvate (Supplementary Figure S13). Moreover, they possess complementary energy conservation genes for facilitating thermodynamically limited disposal of reducing power derived from syntrophic propionate catabolism (Figure 4c). Specifically, an electron-bifurcating formate dehydrogenase (EBFdh, containing Fdh and HylABC, where Fdh and HylA are fused; Supplementary Table S5) and membrane-bound hydrogenase could serve as potential mechanisms for energy-conserving formate (Wang et al., 2013) and H2 production (Vignais and Colbeau, 2004) possibly involved in syntrophic metabolism, based on identification of homologues in Pelobacter carbinolicus (Nobu et al., 2015b) and Moorella thermoacetica (Pierce et al., 2008). Succinate dehydrogenase and NADH:Nqo may also be involved in energy conservation, allowing for reduction of NAD+ by reduced flavin produced in propionate catabolism by the Mmc pathway (Figure 4c). Syntrophic propionate oxidation by these ‘Atribacteria' lineages may therefore depend on formate and H2 transfer as observed with Syntrophobacter (De Bok et al., 2002).
Predicted catabolism of both saccharides and propionate by the ‘Atribacteria' takes advantage of electron confurcation to accomplish endergonic NADH oxidation (Figures 4b and c). As this requires exergonic Fdred oxidation as a driving force, the organism must either have sufficient Fdred or Fdred-independent NADH sinks. In addition to the BMC, OP9-1 lineages appear to have other Fd-independent NADH sinks, such as NiFe hydrogenase and alcohol/aldehyde dehydrogenase (Dodsworth et al., 2013). However, the putative propionate-oxidizing ‘Atribacteria' lineages lack these enzymes as well as NADH:Fd oxidoreductases such as Rnf (Sieber et al., 2012; Nobu et al., 2015a) that syntrophic propionate oxidizers typically rely on to circumvent this issue. The only obvious alternative NADH sink encoded by the putative propionate-oxidizing JS1-1 and JS1-2 lineages involves acetyl-CoA reduction to acetaldehyde by BMC-associated PduPL (Figure 4a), suggesting that the BMC has a critical role in propionate catabolism. As BMC are thought to allow aldehyde concentration and storage (Sampson and Bobik, 2008), this could serve as both a NADH sink and carbon storage mechanism analogous to polyhydroxyalkanoate synthesis (Nobu et al., 2014c). The association of aldehyde-condensing DERA and sugar isomerase with the BMC locus suggests potential conversion of aldehydes to sugar. To reoxidize the stored acetaldehyde, JS1-1 and JS1-2 genomes encode homologues of aldehyde:Fd oxidoreductase; coupling this oxidoreductase with Mbh could allow acetaldehyde oxidation, Fdred generation and ultimately H2 generation even under limitation of exogenous substrates, such as propionate. Although it is not likely required in ‘Ca. Caldatribacterium', this BMC-mediated electron sink and aldehyde storage could nonetheless enhance sugar fermentation by providing flexibility in using an aldehyde reservoir as an electron donor (that is, acetaldehyde oxidation) or acceptor (that is, reduction to ethanol). Therefore, we speculate that the BMC-associated aldehyde metabolism may interact with syntrophic metabolism (propionate catabolism, EBFdh and Mbh) in JS1-1 and JS1-2 lineages and sugar fermentation in OP9-1 lineages to facilitate phylum-wide energy-conserving catabolism and carbon storage.
Conclusions
The ‘Atribacteria', inclusive of the OP9 and JS1 lineages, is a globally distributed candidate phylum that appears restricted to anaerobic environments. Notably, many of these environments, such as the TA reactor (Nobu et al., 2015b), Etoliko Lagoon sediment (Chamalaki et al., 2014) and Sakinaw Lake monimolimnion (Gies et al., 2014), contain considerable amounts of organic carbon but have relatively low availability of inorganic compounds suitable for use in anaerobic respiration and thus represent the so-called ‘low-energy' ecosystems where fermentation and syntrophy are likely important metabolic strategies. These characteristics coincide with the potential catabolisms and lack of respiratory capacity predicted for the ‘Atribacteria' lineages represented by the genomic data sets analysed in this study. BMC-mediated metabolism of sugar phosphates such as deoxyribose 5-phosphate by the ‘Atribacteria', as suggested by Axen et al., (2014), could also have an important role in nutrient recycling in these environments. The capacity for syntrophic propionate catabolism predicted for the JS1-1 lineage points to an ecological role for the ‘Atribacteria' in sediments in the ‘dark ocean biosphere', especially those associated with methane hydrates, hydrocarbon seeps and on continental margins and shelves where this candidate phylum is often abundant (Orcutt et al., 2011; Parkes et al., 2014). Although the existing genomic coverage only scratches the surface of the diversity encompassed by this candidate phylum, we posit that primary and secondary fermentation and syntrophy may be a common catabolic theme among members of the ‘Atribacteria'. The results presented here can inform enrichment or cultivation efforts targeting specific members of the ‘Atribacteria' and provide a platform for probing cooperative interactions, physiological capacities and the role of the BMC in members of this lineage.
Acknowledgments
We thank Lars Schreiber, Karen Lloyd, Ramunas Stepanauskas and the Single Cell Genomics Center at the Bigelow Laboratory for Ocean Sciences for single-cell sorting and providing access to the Aarhus SAGs. This research is supported by NASA Exobiology grant EXO-NNX11AR78G to BPH and JAD; US National Science Foundation grants MCB 0546865 and OISE 0968421 to BPH; US Department of Energy (DOE) grants DE-EE-0000716 and DE-SC0006771 to BPH; the Nevada Renewable Energy Consortium, funded by the DOE, to BPH; an Amazon Web Services Education Research Grant award to BPH and SKM; Natural Environment Research Council, UK grant NE/J011177/1 to AJW, PK, RJP and HS; Cardiff University Research Leave Fellowship to AJW; Tula Foundation, Natural Sciences and Engineering Research Council (NSERC) of Canada, Canada Foundation for Innovation (CFI) and the Canadian Institute for Advanced Research (CIFAR) through grants awarded to SJH; a 4-year fellowship from the University of British Columbia awarded to EAG. Sampling and sorting of the Aarhus Bay SAGs was funded by the Danish National Research Foundation given to the Center for Geomicrobiology, Aarhus University and their sequencing funded by NERC NBAF awards 628 and 744 to AJW. The work conducted by the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported under Contract No. DE-AC02-05CH11231. BPH acknowledges the generous support of Greg Fullmer through the UNLV Foundation.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axen SD, Erbilgin O, Kerfeld CA. (2014). A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput Biol 10: e1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BJ, Comolli LR, Dick GJ, Hauser LJ, Hyatt D, Dill BD et al. (2010). Enigmatic, ultrasmall, uncultivated Archaea. Proc Natl Acad Sci USA 107: 8806–8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BJ, Dick GJ. (2013). Omic approaches in microbial ecology: charting the unknown. Microbe 8: 353–360. [Google Scholar]
- Biegel E, Schmidt S, González JM, Müller V. (2011). Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell Mol Life Sci 68: 613–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bok FAM, Luijten MLGC, Stams AJM. (2002). Biochemical evidence for formate transfer in syntrophic propionate-oxidizing cocultures of Syntrophobacter fumaroxidans and Methanospirillum hungatei. Appl Environ Microbiol 68: 4247–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamalaki A, Gianni A, Kehayias G, Zacharias I, Tsiamis G, Bourtzis K. (2014). Bacterial diversity and hydrography of Etoliko, an anoxic semi-enclosed coastal basin in Western Greece. Ann Microbiol 64: 661–670. [Google Scholar]
- Cheng S, Fan C, Sinha S, Bobik TA. (2012). The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the Pdu microcompartment of Salmonella enterica. PLoS One 7: e47144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, Rawlings ND, Stevens RA, Barrett AJ. (1998). Identification of the active site of legumain links it to caspases, clostripain and gingipains in a new clan of cysteine endopeptidases. FEBS Lett 441: 361–365. [DOI] [PubMed] [Google Scholar]
- Chouari R, Le Paslier D, Dauga C, Daegelen P, Weissenbach J, Sghir A. (2005). Novel major bacterial candidate division within a municipal anaerobic sludge digester. Appl Environ Microbiol 71: 2145–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JK, Peacock JP, Dodsworth JA, Williams AJ, Thompson DB, Dong H et al. (2013). Sediment microbial communities in Great Boiling Spring are controlled by temperature and distinct from water communities. ISME J 7: 718–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa KC, Navarro JB, Shock EL, Zhang CL, Soukup D, Hedlund BP. (2009). Microbiology and geochemistry of great boiling and mud hot springs in the United States Great Basin. Extremophiles 13: 447–459. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, Yelton AP et al. (2009). Community-wide analysis of microbial genome sequence signatures. Genome Biol 10: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodsworth JA, Blainey PC, Murugapiran SK, Swingley WD, Ross CA, Tringe SG et al. (2013). Single-cell and metagenomic analyses indicate a fermentative and saccharolytic lifestyle for members of the OP9 lineage. Nat Commun 4: 1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. (2011). Accelerated profile HMM searches. PLoS Comput Biol 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins JG, Podar M, Graham DE, Makarova KS, Wolf Y, Randau L et al. (2008). A korarchaeal genome reveals insights into the evolution of the Archaea. Proc Natl Acad Sci USA 105: 8102–8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshahed MS, Youssef NH, Spain AM, Sheik C, Najar FZ, Sukharnikov LO et al. (2008). Novelty and uniqueness patterns of rare members of the soil biosphere. Appl Environ Microbiol 74: 5422–5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Cheng S, Liu Y, Escobar CM, Crowley CS, Jefferson RE et al. (2010). Short N-terminal sequences package proteins into bacterial microcompartments. Proc Natl Acad Sci USA 107: 7509–7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag IF, Davis JP, Youssef NH, Elshahed MS. (2014). Global patterns of abundance, diversity and community structure of the Aminicenantes (candidate phylum OP8). PloS One 9: e92139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghai R, Pašić L, Fernández AB, Martin-Cuadrado A-B, Mizuno CM, McMahon KD et al. (2011). New abundant microbial groups in aquatic hypersaline environments. Sci Rep 1: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gies EA, Konwar KM, Beatty JT, Hallam SJ. (2014). Illuminating microbial dark matter in meromictic sakinaw lake. Appl Environ Microbiol 80: 6807–6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittel A, Kofoed MVW, Sørensen KB, Ingvorsen K, Schramm A. (2012). Succession of Deferribacteres and Epsilonproteobacteria through a nitrate-treated high-temperature oil production facility. Syst Appl Microbiol 35: 165–174. [DOI] [PubMed] [Google Scholar]
- Gittel A, Sørensen KB, Skovhus TL, Ingvorsen K, Schramm A. (2009). Prokaryotic community structure and sulfate reducer activity in water from high-temperature oil reservoirs with and without nitrate treatment. Appl Environ Microbiol 75: 7086–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glöckner J, Kube M, Shrestha PM, Weber M, Glöckner FO, Reinhardt R et al. (2010). Phylogenetic diversity and metagenomics of candidate division OP3. Environ Microbiol 12: 1218–1229. [DOI] [PubMed] [Google Scholar]
- Goris J, Konstantinos KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57: 81–91. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Harris JK, Caporaso JG, Walker JJ, Spear JR, Gold NJ, Robertson CE et al. (2013). Phylogenetic stratigraphy in the Guerrero Negro hypersaline microbial mat. ISME J 7: 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JK, Kelley ST, Pace NR. (2004). New perspective on uncultured bacterial phylogenetic division OP11. Appl Environ Microbiol 70: 845–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. (1998). Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol 180: 366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huseby DL, Roth JR. (2013). Evidence that a metabolic microcompartment contains and recycles private cofactor pools. J Bacteriol 195: 2864–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imachi H, Sekiguchi Y, Kamagata Y, Hanada S, Ohashi A, Harada H. (2002). Pelotomaculum thermopropionicum gen. nov., sp. nov., an anaerobic, thermophilic, syntrophic propionate-oxidizing bacterium. Int J Syst Evol Microbiol 52: 1729–1735. [DOI] [PubMed] [Google Scholar]
- Inagaki F, Nunoura T, Nakagawa S, Teske A, Lever M, Lauer A et al. (2006). Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc Natl Acad Sci USA 103: 2815–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennewein S, Schürmann M, Wolberg M, Hilker I, Luiten R, Wubbolts M et al. (2006). Directed evolution of an industrial biocatalyst: 2-deoxy-D-ribose 5-phosphate aldolase. Biotechnol J 1: 537–548. [DOI] [PubMed] [Google Scholar]
- Kamke J, Rinke C, Schwientek P, Mavromatis K, Ivanova N, Sczyrba A et al. (2014). The candidate phylum Poribacteria by single-cell genomics: new insights into phylogeny, cell-compartmentation, eukaryote-like repeat proteins, and other genomic features. PLoS One 9: e87353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor RS, Wrighton KC, Handley KM, Sharon I, Hug LA, Castelle CJ et al. (2013). Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. mBio 4: e00708–e00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld CA, Heinhorst S, Cannon GC. (2010). Bacterial microcompartments. Annu Rev Microbiol 64: 391–408. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Endo K, Sakata S, Mayumi D, Kawaguchi H, Ikarashi M et al. (2012). Phylogenetic diversity of microbial communities associated with the crude-oil, large-insoluble-particle and formation-water components of the reservoir fluid from a non-flooded high-temperature petroleum reservoir. J Biosci Bioeng 113: 204–210. [DOI] [PubMed] [Google Scholar]
- Kozubal MA, Romine M, Jennings R deM, Jay ZJ, Tringe SG, Rusch DB et al. (2013). Geoarchaeota: a new candidate phylum in the Archaea from high-temperature acidic iron mats in Yellowstone National Park. ISME J 7: 622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau MCY, Aitchison JC, Pointing SB. (2009). Bacterial community composition in thermophilic microbial mats from five hot springs in central Tibet. Extrem Life Extreme Cond 13: 139–149. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. (2011). Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39: W475–W478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levén L, Eriksson ARB, Schnürer A. (2007). Effect of process temperature on bacterial and archaeal communities in two methanogenic bioreactors treating organic household waste. FEMS Microbiol Ecol 59: 683–693. [DOI] [PubMed] [Google Scholar]
- Liu J, Wu W, Chen C, Sun F, Chen Y. (2011). Prokaryotic diversity, composition structure, and phylogenetic analysis of microbial communities in leachate sediment ecosystems. Appl Microbiol Biotechnol 91: 1659–1675. [DOI] [PubMed] [Google Scholar]
- Lloyd KG, Schreiber L, Petersen DG, Kjeldsen KU, Lever MA, Steen AD et al. (2013). Predominant archaea in marine sediments degrade detrital proteins. Nature 496: 215–218. [DOI] [PubMed] [Google Scholar]
- Marcy Y, Ouverney C, Bik EM, Lösekann T, Ivanova N, Martin HG et al. (2007). Dissecting biological ‘dark matter' with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci USA 104: 11889–11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen I-MA, Chu K, Szeto E, Palaniappan K, Pillay M et al. (2014). IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res 42: D568–D573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean JS, Lombardo M-J, Badger JH, Edlund A, Novotny M, Yee-Greenbaum J et al. (2013). Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc Natl Acad Sci USA 110: E2390–E2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasingarao P, Podell S, Ugalde JA, Brochier-Armanet C, Emerson JB, Brocks JJ et al. (2012). De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J 6: 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobu MK, Narihiro T, Hideyuki T, Qiu Y-L, Sekiguchi Y, Woyke T et al. (2015. a). The genome of Syntrophorhabdus aromaticivorans strain UI provides new insights for syntrophic aromatic compound metabolism and electron flow. Environ Microbiol e-pub ahead print 3 March 2014; doi:10.1111/1462-2920.12444. [DOI] [PubMed]
- Nobu MK, Narihiro T, Rinke C, Kamagata Y, Tringe SG, Woyke T et al. (2015. b). Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. ISME J e-pub ahead print 23 January 2015; doi:10.1038/ismej.2014.256. [DOI] [PMC free article] [PubMed]
- Nobu MK, Tamaki H, Kubota K, Liu W-T. (2014. c). Metagenomic characterization of ‘Candidatus Defluviicoccus tetraformis' strain TFO71, a tetrad-forming organism, predominant in an anaerobic-aerobic membrane bioreactor with deteriorated biological phosphorus removal. Environ Microbiol 16: 2739–2751. [DOI] [PubMed] [Google Scholar]
- Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H et al. (2011). Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res 39: 3204–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orcutt BN, Sylvan JB, Knab NJ, Edwards KJ. (2011). Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol Mol Biol Rev 75: 361–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes RJ, Cragg B, Roussel E, Webster G, Weightman A, Sass H. (2014). A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere:geosphere interactions. Mar Geol 352: 409–425. [Google Scholar]
- Parsons JB, Lawrence AD, McLean KJ, Munro AW, Rigby SEJ, Warren MJ. (2010). Characterisation of PduS, the pdu metabolosome corrin reductase, and evidence of substructural organisation within the bacterial microcompartment. PLoS One 5: e14009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil KR, Haider P, Pope PB, Turnbaugh PJ, Morrison M, Scheffer T et al. (2011). Taxonomic metagenome sequence assignment with structured output models. Nat Methods 8: 191–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock JP, Cole JK, Murugapiran SK, Dodsworth JA, Fisher JC, Moser DP et al. (2013). Pyrosequencing reveals high-temperature cellulolytic microbial consortia in Great Boiling Spring after in situ lignocellulose enrichment. PLoS One 8: e59927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peura S, Eiler A, Bertilsson S, Nykänen H, Tiirola M, Jones RI. (2012). Distinct and diverse anaerobic bacterial communities in boreal lakes dominated by candidate division OD1. ISME J 6: 1640–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VD, Hnatow LL, Zhang S, Fallon RD, Jackson SC, Tomb J-F et al. (2009). Characterizing microbial diversity in production water from an Alaskan mesothermic petroleum reservoir with two independent molecular methods. Environ Microbiol 11: 176–187. [DOI] [PubMed] [Google Scholar]
- Pierce E, Xie G, Barabote RD, Saunders E, Han CS, Detter JC et al. (2008). The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum. Environ Microbiol 10: 2550–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Waller M, Barrett AJ, Bateman A. (2014). MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 42: D503–D509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106: 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437. [DOI] [PubMed] [Google Scholar]
- Rivière D, Desvignes V, Pelletier E, Chaussonnerie S, Guermazi S, Weissenbach J et al. (2009). Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J 3: 700–714. [DOI] [PubMed] [Google Scholar]
- Sakuraba H, Yoneda K, Yoshihara K, Satoh K, Kawakami R, Uto Y et al. (2007). Sequential aldol condensation catalyzed by hyperthermophilic 2-deoxy-d-ribose-5-phosphate aldolase. Appl Environ Microbiol 73: 7427–7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson EM, Bobik TA. (2008). Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol 190: 2966–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson EM, Johnson CLV, Bobik TA. (2005). Biochemical evidence that the pduS gene encodes a bifunctional cobalamin reductase. Microbiology 151: 1169–1177. [DOI] [PubMed] [Google Scholar]
- Sayeh R, Birrien JL, Alain K, Barbier G, Hamdi M, Prieur D. (2010). Microbial diversity in Tunisian geothermal springs as detected by molecular and culture-based approaches. Extrem Life Extreme Cond 14: 501–514. [DOI] [PubMed] [Google Scholar]
- Schut GJ, Adams MWW. (2009). The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J Bacteriol 191: 4451–4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi Y. (2006). Yet-to-be cultured microorganisms relevant to methane fermentation processes. Microbes Environ 21: 1–15. [Google Scholar]
- Sharon I, Banfield JF. (2013). Genomes from metagenomics. Science 342: 1057–1058. [DOI] [PubMed] [Google Scholar]
- Sieber JR, McInerney MJ, Gunsalus RP. (2012). Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu Rev Microbiol 66: 429–452. [DOI] [PubMed] [Google Scholar]
- Siegl A, Kamke J, Hochmuth T, Piel J, Richter M, Liang C et al. (2011). Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J 5: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored ‘rare biosphere'. Proc Natl Acad Sci USA 103: 12115–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Stepanauskas R. (2012). Single cell genomics: an individual look at microbes. Curr Opin Microbiol 15: 613–620. [DOI] [PubMed] [Google Scholar]
- Strous M, Kraft B, Bisdorf R, Tegetmeyer HE. (2012). The binning of metagenomic contigs for microbial physiology of mixed cultures. Front Microbiol 3: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe IC. (2011). Cell envelope architecture in the Chloroflexi: a shifting frontline in a phylogenetic turf war. Environ Microbiol 13: 279–282. [DOI] [PubMed] [Google Scholar]
- Takami H, Noguchi H, Takaki Y, Uchiyama I, Toyoda A, Nishi S et al. (2012). A deeply branching thermophilic bacterium with an ancient acetyl-CoA pathway dominates a subsurface ecosystem. PLoS One 7: e30559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y-Q, Ji P, Hayashi J, Koike Y, Wu X-L, Kida K. (2011). Characteristic microbial community of a dry thermophilic methanogenic digester: its long-term stability and change with feeding. Appl Microbiol Biotechnol 91: 1447–1461. [DOI] [PubMed] [Google Scholar]
- Vick TJ, Dodsworth JA, Costa KC, Shock EL, Hedlund BP. (2010). Microbiology and geochemistry of Little Hot Creek, a hot spring environment in the Long Valley Caldera. Geobiology 8: 140–154. [DOI] [PubMed] [Google Scholar]
- Vignais PM, Colbeau A. (2004). Molecular biology of microbial hydrogenases. Curr Issues Mol Biol 6: 159–188. [PubMed] [Google Scholar]
- Wang S, Huang H, Kahnt J, Thauer RK. (2013). Clostridium acidurici electron-bifurcating formate dehydrogenase. Appl Environ Microbiol 79: 6176–6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster G, Parkes RJ, Fry JC, Weightman AJ. (2004). Widespread occurrence of a novel division of bacteria identified by 16 S rRNA gene sequences originally found in deep marine sediments. Appl Environ Microbiol 70: 5708–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster G, Sass H, Cragg BA, Gorra R, Knab NJ, Green CJ et al. (2011). Enrichment and cultivation of prokaryotes associated with the sulphate–methane transition zone of diffusion-controlled sediments of Aarhus Bay, Denmark, under heterotrophic conditions. FEMS Microbiol Ecol 77: 248–263. [DOI] [PubMed] [Google Scholar]
- Wemheuer B, Taube R, Akyol P, Wemheuer F, Daniel R. (2013). Microbial diversity and biochemical potential encoded by thermal spring metagenomes derived from the Kamchatka Peninsula. Archaea 2013: 136714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KC, Agbo P, Warnecke F, Weber KA, Brodie EL, DeSantis TZ et al. (2008). A novel ecological role of the Firmicutes identified in thermophilic microbial fuel cells. ISME J 2: 1146–1156. [DOI] [PubMed] [Google Scholar]
- Wrighton KC, Castelle CJ, Wilkins MJ, Hug LA, Sharon I, Thomas BC et al. (2014). Metabolic interdependencies between phylogenetically novel fermenters and respiratory organisms in an unconfined aquifer. ISME J 8: 1452–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC et al. (2012). Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337: 1661–1665. [DOI] [PubMed] [Google Scholar]
- Wu M, Scott AJ. (2012). Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28: 1033–1034. [DOI] [PubMed] [Google Scholar]
- Yamada T, Kikuchi K, Yamauchi T, Shiraishi K, Ito T, Okabe S et al. (2011). Ecophysiology of uncultured filamentous anaerobes belonging to the phylum KSB3 that cause bulking in methanogenic granular sludge. Appl Environ Microbiol 77: 2081–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer K-H et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16 S rRNA gene sequences. Nat Rev Microbiol 12: 635–645. [DOI] [PubMed] [Google Scholar]
- Youssef NH, Blainey PC, Quake SR, Elshahed MS. (2011). Partial genome assembly for a candidate division OP11 single cell from an anoxic spring (Zodletone Spring, Oklahoma). Appl Environ Microbiol 77: 7804–7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef NH, Rinke C, Stepanauskas R, Farag I, Woyke T, Elshahed MS. (2015). Insights into the metabolism, lifestyle and putative evolutionary history of the novel archaeal phylum 'Diapherotrites'. ISME J 9: 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26: 1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.