Abstract
RNA editing is a process that alters DNA-encoded sequence and is distinct from splicing, 5′ capping and 3′ additions. In thirty years since editing was discovered in mitochondria of trypanosomes, several functionally and evolutionary unrelated mechanisms have been described in eukaryotes, archaea and viruses. Editing events are predominantly post-transcriptional and include nucleoside insertions and deletions, and base substitutions and modifications. Here, we review the mechanism of uridine insertion/deletion mRNA editing in kinetoplastid protists typified by Trypanosoma brucei. This type of editing corrects frameshifts, introduces translation punctuation signals, and often adds hundreds of uridines to create protein coding sequence. We focus on protein complexes responsible for editing reactions and their interactions with other elements of the mitochondrial gene expression pathway.
Keywords: Trypanosoma, mitochondria, RNA editing, guide RNA, polyadenylation, translation
Is there an adaptive advantage in complexity?
In 1986, Rob Benne and co-workers described the insertion of four uridines into cytochrome c oxidase subunit 2 (CO2) mRNA from Trypanosoma brucei as means of correcting the encoded frameshift at the RNA level [1]. Astutely named “RNA editing,” this phenomenon later came to symbolize massive U-insertions [2] and U-deletions [3] that create open reading frames in transcripts of cryptic mitochondrial genes in kinetoplastid protists. This paradigm-shifting discovery stimulated researchers to look closer at discrepancies between DNA and RNA sequences in other organisms and ultimately led to identification of several divergent and largely unrelated editing mechanisms, such as A to I [4] and C to U base deamination [5], 3′-to-5′ polymerization [6], and others. The narrow phylogenetic distribution of editing systems suggests their derived character within lineages in which they currently exist rather than editing being a primordial trait retained from a common evolutionary ancestor in some organisms and lost in others [7]. The sheer mechanistic and component complexity, and the lack of apparent adaptive advantage of having one, positions trypanosomal editing as a fruitful platform for evolutionary debate on the origins of macromolecular assemblies. The constructive neutral evolution (CNE) hypotheses argues that the functional editing machinery may evolve in the absence of positive selection and, importantly, prior to the actual need for the editing process [8]. By virtue of extant proteins forming neutral mutation-driven interactions, e.g., an enzyme with an RNA binding protein, such assembly may acquire a novel capacity. Without pressure from purifying selection, the neutral capacity can persist with no essential cellular function until a mutation arises that can be corrected by such pre-existing activity. Thus, the detrimental impact of a gene mutation may not be compensated unless a functional system to correct the sequence at the RNA level is already in place. It follows that editing is an intrinsically mutagenic process: once evolved, the editing system allows accumulation of mutations that otherwise would be eliminated by the purifying selection [9,10]. Likewise, accumulation of multiple mutations would make reverse changes all but impossible, and render editing an essential pathway. In this context, the composition of the enzymatic core editing complex proved most instructive: catalytic modules implicated in fundamental cellular functions, such as DNA repair and RNA interference, along with proteins likely acquired by horizontal gene transfer, operate as stable protein complex that cleaves mRNA, adds or removes Us, and re-ligates fragments. Finally, when editing becomes an indispensable process, such as generation of a protein coding sequence, it must be incorporated into the overall gene expression pathway. It could be expected that interactions of the editing machinery with RNA processing and translation complexes would be as unique as editing systems themselves. Selection of correctly edited mRNA by the ribosome in a background of partially edited and unedited transcripts is among the most obvious problems that require additional levels of control. These considerations do not rule out possible adaptive advantage of editing once it evolved–indeed two reports indicate that alternative editing may generate protein diversity [11,12]. Editing-dependent protein diversity, both the fact and the function, remain to be firmly established leaving the question wide open to future investigation and hypothesis building. Here, we review the complexity of trypanosomal insertion/deletion editing in terms of underlying biochemistry and potential origins of editing effectors, as well as determinants that direct position-specific insertion and deletion of uridines.
Elemental editing reactions are catalyzed by modular RNA editing core complex (RECC)
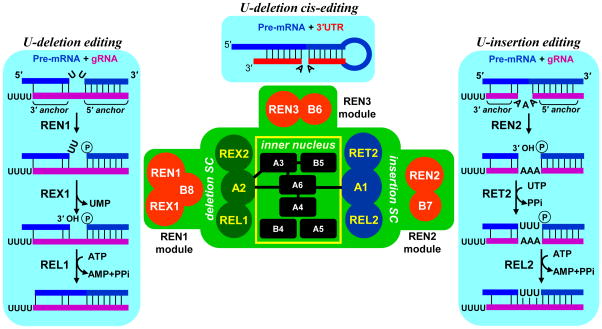
Trypanosoma brucei, the causative agent of African sleeping sickness, and most other representatives of Kinetoplastea, such as Leishmania spp., are characterized by the presence of the kinetoplast (see Glossary). This disc-shaped, high-density nucleoprotein structure is located in the mitochondrial lumen adjacent to the flagellar base. The kinetoplast encloses the mitochondrial genome (kDNA), which is composed of two types of catenated circles. Relatively few maxicircles (~25 kb) encode genes typically found in mitochondrial genomes, such as rRNAs, ribosomal protein RPS12 and subunits of respiratory complexes, while thousands of ~1-kb minicircles constitute the bulk of kDNA. In T. brucei, six of the 18 annotated mRNAs encode predicted polypeptides while the remaining 12 transcripts must undergo editing to acquire open reading frames and translation punctuation signals. The product of trypanosomal mRNA editing is not collinear with DNA as it contains extra nucleotides compared to the gene sequence, and sometime lacks encoded uridines. Historically, the determinants of position-specific U-insertions and deletions have been discovered as short patches of complementarity between edited mRNA and maxicircle DNA in Leishmania tarentolae [13]. By allowing for wobble G-U, in addition to canonical Watson–Crick base-pairing, short (50–60 nt) mitochondrial RNAs transcribed from maxicircles have been recognized as carriers of genetic information and termed guide RNAs (gRNAs). Further work established that most gRNAs are encoded in minicircles [14]. The predicted secondary structure of gRNA-mRNA hybrid instantly suggested a mechanism by which the editing site and the extent of U-insertions/deletions are determined without invoking template-dependent polymerization of nucleic acids [13]. The initial site selection is accomplished via a short (5–10 nt) region of complementarity between gRNA’s 5′ anchor and pre-edited mRNA. The rest of gRNA forms an imperfect duplex (3′ anchor)with mRNA which results in bulging of single-stranded uridines in mRNA (deletion sites) or purine nucleotides in gRNA (insertion sites, Figure 1). At either site, the mRNA is cleaved at the first unpaired nucleotide adjacent to the 5′ anchor duplex. The resultant deletion and insertion intermediates are quite distinct: single-stranded uridines become exposed to a 3′-5′exonucleolytic attack in the former while a gap is created in the latter. Upon trimming uridines to the first paired base in the deletion site, or adding gRNA-specified number of uridines to a 5′ cleavage fragment in the insertion site, the fragments are joined to restore mRNA continuity, which extends the double-stranded anchor region.
Figure 1.
Isoforms of the RNA editing core complex (RECC). Protein-protein interactions within core complex are depicted by overlapping circles or black bars. SC; subcomplex. Trans-guided insertion and deletion, and cis-guided insertion pathways are juxtaposed with corresponding endonuclease modules. Abbreviations: 3′ anchor, base-pairing between gRNA region that directs editing reactions and pre-edited mRNA; 5′ anchor, 5′ part of the gRNA that hybridizes with pre-edited mRNA; A, KREPA; B, KREPB; gRNA, guide RNA; REL, RNA editing ligase; RET, RNA editing TUTase; REX, RNA editing exonuclease; REN, RNA editing endonuclease; UMP, uridine monophosphate.
It has been proposed that within the editing domain overlapping guide RNAs bind sequentially in a 3′–5′ polarity along the mRNA, just as sequence changes directed by the initiating gRNA create binding site for the next one [15]. However, recent deep sequencing studies demonstrated that gRNA coverage of virtually all edited sequences is highly redundant albeit uneven (100–100,000 per nucleotide in mRNA)[16,17]. This observation suggests that multiple overlapping gRNAs may compete for pre-edited and partially-edited sequences and entreats the question whether the full editing potential of each gRNA is indeed realized. Most gRNAs can theoretically direct editing at several closely spaced sites, but as editing progresses within editing block, the tethering interaction between gRNA and mRNA 5′ cleavage fragment (3′ anchor) is supported by fewer base pairs (Figure 1). Stabilizing such interaction in vitro by introducing additional base pairing greatly stimulates the efficiency of U-insertion/deletion and RNA ligation reactions [18–20]. Thus the problem of editing at distal sites may be solved by displacing gRNA with diminishing “3′ anchor” bya molecule capable of directing the same changes while forming a more stable hybrid. The tethering role of the gRNA’s U-tail has been proposed based on purine-rich nature of pre-edited mRNA sequences [21], but the experimental evidence for such function is still lacking.
The asymmetrical U-deletion and U-insertion sites are recognized by RNase III-type endonucleases, KREN1 [22] and KREN2 [23], respectively. The third endonuclease (KREN3) apparently targets the CO2 mRNA that contains a cis-acting guide RNA-like element in its 3′ untranslated region (UTR) [24,25]. In contrast to canonical RNase III enzymes that form functional dimers with two active sites and cut both strands in a double-stranded RNA, editing endonucleases cleave only the mRNA. It seems plausible that a single cleavage is caused by KREN1 and KREN2 forming heterodimers with catalytically-inactive degenerate RNase III-like domains in the T. brucei RECC subunits KREPB4 and KREPB5, respectively (Table 1) [24]. A mutually exclusive binding of KREN1, KREN2 and KREN3 with common set of proteins that contain U-deletion, U-insertion and ligase activities highlights RECC’s modular nature (Figure 1) [24–27]. Apparently, an endonuclease module, e.g., an endonuclease associated with a specific protein(s) such as KREN1/KREX1/KREPB8, KREN2/KREPB7 or KREN3/KREPB6, bind to a common particle conferring a specificity for U-deletion, U-insertion or cis-edited sites, respectively (Figure 1). The interactions responsible for mutually exclusive binding to the core complex are unclear, but may involve dimerization of editing endonucleases with degenerate RNase III motifs in KREPB4 and KREPB5. Alternatively, convergent binding sites for accessory proteins KREPB6, 7 and 8 may enable docking of endonuclease modules to the common core.
Table 1.
Components of the RNA editing core complex (RECC).
| Name | Function | Motifs (Pfam)a | Essential gene | Synonym | T. brucei Gene IDb | Refs. |
|---|---|---|---|---|---|---|
| U-insertion sub-complex | ||||||
| KRET2 | TUTase | NT/PAP/TUTase, PAP/associated | yes | RET2 | Tb927.7.1550 | [31,36, 37,39,83] |
| KREL2 | RNA ligase (U-insertion) | RNA lig/RNL2 | no | REL2 | Tb927.1.3030 | [84–86] |
| KREPA1 | Scaffold for U-insertion sub-complex | C2H2 Zn finger, OB fold | yes | MP81 | Tb927.2.2470 | [29,87, 88] |
| U-deletion sub-complex | ||||||
| KREX2 | 3′-5′ U-specific exonuclease | Exo/endo/phos (EEP) | no | REX2 | Tb927.10.3570 | [32,34] |
| KREL1 | RNA ligase (U-deletion) | RNA lig/RNL2 | yes | REL1 | Tb927.9.4360 | [84–86] |
| KREPA2 | Scaffold for U-deletion sub-complex | C2H2 Zn finger, OB fold | yes | MP63 | Tb927.10.8210 | [30,87, 89] |
| Inner nucleus | ||||||
| KREPA3 | RNA binding | C2H2 Zn finger, OB fold | yes | MP42 | Tb927.8.620 | [90,91] |
| KREPA4 | RNA binding | OB fold | yes | MP24 | Tb927.10.5110 | [92–94] |
| KREPA5 | OB fold | no | MP19 | Tb927.8.680 | [78] | |
| KREPB4 | RNA binding | RNase III, U1-like C2H2 Zn finger | yes | MP46 | Tb927.11.2990 | [26,95] |
| KREPB5 | RNase III, U1-like C2H2 Zn finger | yes | MP44 | Tb927.11.940 | [24,26] | |
| KREPA6 | RNA binding | OB fold | yes | MP18 | Tb927.10.5120 | [96,97] |
| KREN1 module | ||||||
| KREN1 | U-deletion endonuclease | RNase III, dsRBD | yes | REN1 | Tb927.1.1690 | [22,24, 25,34,81] |
| KREPB8 | U1-like C2H2 Zn finger | yes | MP41 | Tb927.8.5690 | [24,98] | |
| KREX1 | 3′-5′ U-specific exonuclease | Exo/endo/phos (EEP) | yes | REX1 | Tb927.7.1070 | [27,32, 34] |
| KREN2 module | ||||||
| KREN2 | U-insertion endonucleasea | RNase III, dsRBD | no | REN2 | Tb927.10.5440 | [23–25,81,98] |
| KREPB7 | U1-like C2H2 Zn finger | yes | MP47 | Tb927.9.5630 | [98] | |
| KREN3 module | ||||||
| KREN3 | U-insertion endonuclease | RNase III, dsRBD | yes | REN3 | Tb927.10.5320 | [24,81, 98] |
| KREPB6 | RNA binding | U1-like C2H2 Zn finger | no | MP49 | Tb927.3.3990 | [24,98] |
| Possible associated factors | ||||||
| MEAT1 | NT/PAP/TUTase, PAP/associated | yes | none | Tb927.1.1330 | [99,100] | |
| KREPB9 | U1-like C2H2 Zn finger | no | none | Tb927.9.4440 | [100] | |
| KREPB10 | U1-like C2H2 Zn finger | no | none | Tb927.8.5700 | [100] | |
Pfam motifs are indicated (http://pfam.xfam.org).
Gene identification numbers are from TriTrypDB Kinetoplastid Genomics Resource (http://tritrypdb.org).
Within the common set of proteins, the U-deletion and U-insertion cascades are spatially separated by virtue of editing enzymes binding to distinct zinc finger proteins, KREPA2 and KREPA1, respectively (Figure 1) [28–31]. KREX1 and KREX2 proteins possess exonuclease-endonuclease-phosphatase (EEP) catalytic domains and display single-stranded uridine-specific 3′–5′ exonuclease activity in vitro [27,32]. However, their interaction networks are remarkably distinct: the essential KREX1 is part of the KREN1 endonuclease module, and likely constitutes the main U-deletion activity; the dispensable KREX2 probably represents a structural component of the U-deletion sub-complex [32,33]. Fittingly, KREX2 lacks a catalytic domain, but remains associated with the U-deletion sub-complex in Leishmania tarentolae [32,34,35]. In the U-insertion sub-complex, KRET2 RNA terminal uridyltransferase (TUTase) binds to KREPA1, which results in a mutual stabilization and stimulation of TUTase activity [31,36–39]. Selectivity of uridine incorporation is determined by KRET2’s intrinsic specificity for UTP [38]; therefore both adenosines and guanines in guiding positions may direct U-insertion editing with equal efficiency. KRET2’s RNA binding properties play a critical role during insertion of multiple Us in the same editing site: a single uridine may be added to the mRNA 5′ cleavage fragment irrespective of potential base pairing with gRNA. However, if a mismatch occurs between the newly added +1U and the gRNA, addition of the +2U would be blocked because of enzyme’s strong preference for the double stranded RNA [36]. The processivity of RET2-catalyzed reaction depends on the chemical nature of the mRNA-gRNA base pair. A distributive +1 addition is predominant when the mRNA 5′ cleavage fragment terminates with a purine base, which is the most common outcome of the endonucleolytic cleavage. Conversely, if the dsRNA substrate terminates with uridine, KRET2 processively fills the gap of up to 12 nucleotides, as specified by the number of guiding nucleotides. U-deletion and U-insertion reactions produce a double-stranded RNA in which nicked mRNA fragments are tethered by gRNA, an optimal substrate for ligation [20]. RNA editing ligases 1 and 2 (KREL1 and KREL2) have been identified as components of U-deletion and U-insertion sub-complexes, respectively [29,31,35]. Although spatial separation [28,30,34,40] is consistent with their specialized roles, only KREL1 is essential for cell viability [41–45]. Remarkable structural similarity between trypanosomal editing ligases and T4 phage RNA ligase 2 (Rnl2) [46–48] indicates that a horizontal gene transfer may be a potential source of editing activities.
RNA editing substrate binding complex (RESC): toward definition of the RNA editing holoenzyme
The ‘editosome’ concept has been constantly redressed since detection of RNA ligase-containing heterogeneous macromolecular complexes [49–51]. Studies of the enzymatic RNA editing core complex (RECC) revealed a minimal set of proteins required for the editing cascade, but also highlighted low in vitro efficiency of the purified ~20S RECC [52,53]. Conversely, RECC subunits have been detected in particles with apparent sedimentation coefficient exceeding 40S [54]; sensitivity of these particles to RNase treatment provided initial indication for the ribonucleoprotein nature of RECC-containing supercomplexes. In a parallel line of inquiry, several laboratories pursued guide RNA binding proteins as it is generally held that short unstructured RNAs are maintained in a protein-bound form. Although several candidate proteins have been identified [55–62], only two paralogous proteins initially purified from L. tarentolae [62] fulfilled the expected criteria: 1) gRNA loss upon genetic repression; 2) ensuing inhibition of editing for all but cis-edited CO2 mRNA and 3) RNA binding capacity. Termed gRNA binding complex (GRBC) subunits 1 and 2, these polypeptides lack annotated motifs and similarity to any protein outside Kinetoplastida [63]. GRBC1 and GRBC2, also referred to as GAP2 and GAP1 [64], respectively, form a stable α2β2 heterotetramer which binds gRNA in vitro and participates in a higher order assembly [65], also known as the mitochondrial RNA binding complex 1 (MRB1) [66]. Extensive studies by affinity cross-tagging and yeast two hybrid screens are converging to define a modular RNA editing substrate binding complex (RESC) that is responsible for several critical functions in mitochondrial RNA processing.
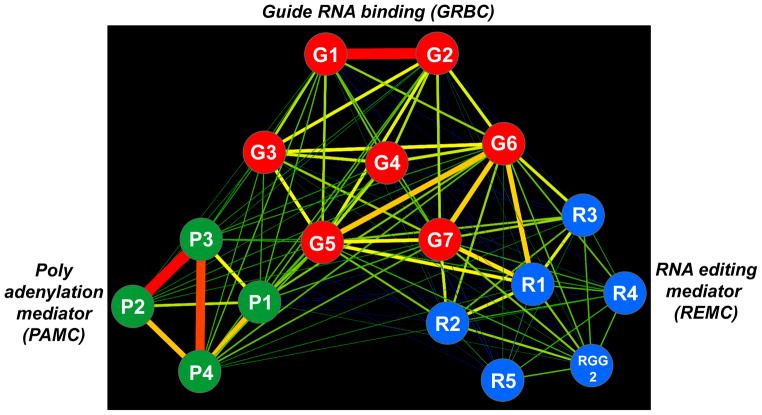
In contrast to RECC, the RESC particle does not contain enzymes and all but one subunits lack discernible motifs or significant similarities to non-kinetoplastid proteins. Since sequence-based predictions are of limited value in asserting functions of specific RESC subunits, a rather uniform RNAi-based approach has been adapted to assess the impact of respective protein depletion on cell viability, gRNA abundance, mRNA editing and the overall complex integrity (Table 2). In conjunction with the proposed tripartite architecture of the RESC particle [17], these studies brought some surprising results. It appears that GRBC1/2 tetramer constitutes a core of the seven-member gRNA binding complex (GRBC), which interacts tightly with a set of proteins clustered around RGG2 RNA binding protein. The RGG2 cluster has been termed RNA editing mediating complex (REMC) to reflect its role in interaction with RECC (Figure 2). Remarkably, GRBC1/2 remain the only subunits essential for gRNA maintenance while knockdowns of most other proteins causes inhibition of RNA editing accompanied by gRNA accumulation [17]. A more detailed analysis of mRNA processing intermediates proved instructive in few cases. For example, gRNA loss in GRBC1/2 knockdown inhibited the editing process and led to accumulation of pre-edited mRNAs [17,63] while depletion of RGG2 from the REMC complex interfered with processivity of editing and exerted particularly strong effect on pan-edited mRNAs [67]. Still another protein module appears to be responsible for RESC’s interaction with the kinetoplast polyadenylation complex [17,68]. Accordingly, RNAi knockdown of polyadenylation mediator complex subunit 1 (PAMC1) within the polyadenylation mediator complex (PAMC) inhibited post-editing polyadenylation/uridylation of edited mRNAs, but had no effect on editing per se [17]. For most other RESC subunits, loss of function studies produced rather generic outcomes (Table 2) and yielded little in terms of exact biological function or mechanism of action.
Table 2.
Components of the RNA editing substrate binding complex (RESC).
| Name | Function | Motifs (Pfam) | Essential gene | Synonym | T. brucei Gene IDc | Refs. |
|---|---|---|---|---|---|---|
| Guide RNA Binding Complex (GRBC) | ||||||
| GRBC1 | gRNA binding | yes | GAP2 | Tb927.7.2570 | [17,63–65] | |
| GRBC2 | gRNA binding | yes | GAP1 | Tb927.2.3800 | [17,63–65] | |
| GRBC3 | nob | MRB8620 | Tb927.11.16860 | [17,101] | ||
| GRBC4 | yes | MRB5390 | Tb11.02.5390 | [17,102] | ||
| GRBC5 | yes | MRB11870 | Tb927.10.11870 | [17,101] | ||
| GRBC6 | yes | MRB3010 | Tb927.5.3010 | [17,103] | ||
| GRBC7 | yes | MRB0880 | Tb927.11.9140 | [17,104] | ||
| RNA Editing Mediator Complex (REMC) | ||||||
| REMC1 | yes | MRB10130 | Tb927.10.10130 | [17,101,104] | ||
| REMC2 | yes | MRB1860 | Tb927.2.1860 | [17,104] | ||
| REMC3 | yes | MRB800 | Tb927.7.800 | [17] | ||
| REMC4 | nob | MRB8180 | Tb927.8.8180 | [17] | ||
| REMC5a | yes | MRB4160 | Tb927.4.4160 | [17,105] | ||
| REMC5Aa | yes | MRB8170 | Tb927.8.8170 | [17,105] | ||
| RGG2 | processivity of editing | RRM | yes | RGG2 | Tb927.10.10830 | [67,106,107] |
| Polyadenylation Mediator Complex (PAMC) | ||||||
| PAMC1 | yes | MRB10130 | Tb927.10.10130 | [17,104] | ||
| PAMC2 | yes | Tb927.6.1200 | [17] | |||
| PAMC3 | no | Tb927.10.1730 | [17] | |||
| PAMC4 | yes | Tb927.1.3010 | [17] | |||
Possible gene duplication.
Inferred from RNAi knockdown.
Gene identification numbers are from TriTrypDB Kinetoplastid Genomics Resource (http://tritrypdb.org).
Figure 2.
Predicted protein interaction network within the RNA editing substrate binding complex (RESC). The network of RNase-resistant interactions was generated from cross-tagging and mass spectroscopy analysis of all subunits (adapted from [17]). The edge thickness and color intensity correlate with a relative strength of bait-prey interactions. Abbreviations: G, guide RNA binding (GRBC); R, RNA editing mediator (REMC); P, polyadenylation mediator (PAMC) complexes are colored red, blue and green, respectively.
Although potential RNA binding motifs are found in many RECC subunits (Table 1), a preferential association of RNA editing substrates and products with the RESC (MRB1) complex has been found in two independent studies. By using rapid affinity pulldowns combined with Northern blotting and deep sequencing analyses of bound RNAs, it has been established that RNA editing substrates (gRNAs and pre-edited mRNAs), intermediates and products are held predominantly by the RESC complex [17]. Enrichment of gRNAs, pre-edited and edited mRNAs in immunopurified material was also detected by RT-PCR analysis [69]. These findings rationalized previous observations of RNA-mediated interactions that draw the well-defined ~ 1 MDa RECC complex into heterogeneous particles exceeding 2 MDa. Indeed, a hypothesis has been put forward that RECC and RESC particles represent catalytic and RNA binding components, respectively, of the RNA editing holoenzyme [17,69,70]. In this setting, gRNAs and pre-edited mRNAs likely remain bound to RESC during the editing process whereas the U-insertion and U-deletion isoforms of the enzymatic core complex engage transiently to target individual editing sites. Elucidating the mRNA binding pathway through the RESC and defining functions of specific subunits remains the work of the future, but there is a high likelihood that contacts with several proteins are responsible for maintaining cleaved mRNA intermediates in a close proximity.
RESC is responsible for orchestrating pre-and post-editing mRNA processing events
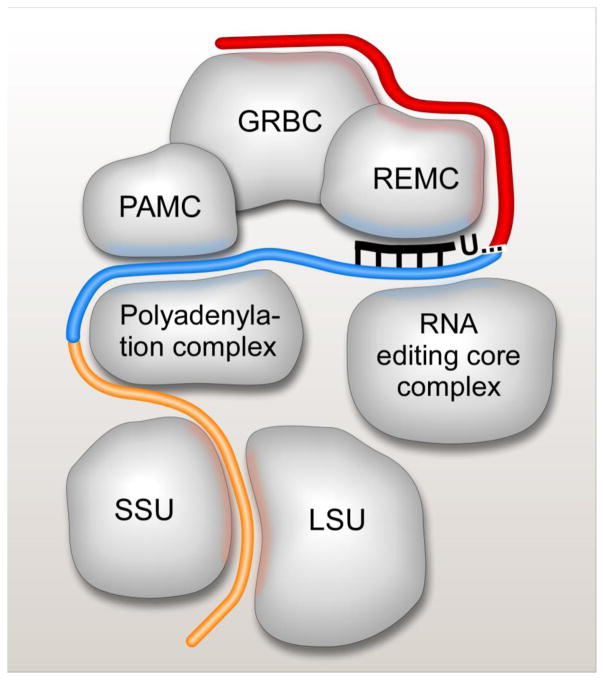
RNA editing is a critical, but one of many mRNA processing steps in mitochondria of trypanosomes. In a striking contrast between the catalytic (RECC) and substrate binding (RESC) components of the editing machinery, it is the latter that appears to be responsible for coordinating pre-and post-editing processing events via RNA-mediated contacts with respective protein complexes.
Mitochondrial pre-mRNAs are believed to originate from multicistronic precursors that span both strands of the maxicircle DNA. Although the nature of mitochondrial promoters and molecular mechanism of precursor partitioning remain a mystery, the pre-edited and unedited mRNAs are characterized by short monophosphorylated 5′ UTRs without apparent ribosome binding sites. Conversely, modifications of the 3′ end are critical for mRNA stability and translation, and are intertwined with the editing process. Kinetoplast poly(A) polymerase KPAP1 adds 20–25 nt A-tail to the 3′ end of pre-edited mRNA and this short A-tail is maintained for the duration of the editing process. Remarkably, presence of a short A-tail does not affect the steady state levels of pre-edited mRNAs, but becomes essential for mRNA stability after very few initial editing events take place adjacent to the polyadenylation site [68,71]. This ‘stability switch’ phenomenon most likely ensures that mRNA editing products are 3′ adenylated and competent for the post-editing 3′ A/U-tailing. The 200–300 nt-long A/U heteropolymers composed of stretches of As interspersed by few Us are added to the pre-existing 3′ A-tails upon completion of the editing process at the 5′ region [68]. This selective reaction requires KPAP1, RET1 TUTase and a heterodimer of pentatricopeptide RNA binding proteins, kinetoplast polyadenylation/uridylation factors (KPAFs) 1 and 2 [72] (Table 3). Remarkably, edited transcripts bearing 200–300 nucleotide-long A/U-tails, but not short A-tails, are found in translating ribosomal complexes [72–74]. Existing evidence points out interactions between key 3′ mRNA modification factors andRESC. Thus, GRBC1 and GRBC2 were detected in the polyadenylation complex [68] while KPAP1, RET1 and KPAF1 were clearly enriched in affinity purified components of the PAMC complex ([17], Figure 2). To that end, edited mRNAs isolated from affinity purified RESC variants possessed mostly short A-tails while the PAMC1 knockdown triggered a loss of long A/U-tails, but not short A-tails [17]. It seems plausible that the A/U-tailing may be coupled with edited mRNA release from the RESC complex which, in turn, implies that the post-editing 3′ modification eventis probably orchestrated by PAMC protein cluster within RESC.
Table 3.
mRNA editing, polyadenylation and stability factors.
| Name | Function | Motifs (Pfam) | Essential gene | Synonym | T. brucei Gene IDb | Refs. |
|---|---|---|---|---|---|---|
| REH1 | gRNA displacement from pan-edited mRNAs | DEADc, HELICc | yes | mHel61 | Tb927.11.8870 | [76,108,109] |
| REH2 | DSRM, DEXDc, HA2, OB-fold | yes | Tb927.4.1500 | [63,64,66, 69,70,77] | ||
| RET1 | gRNA processing, gRNA and rRNA U-tailing | NT/PAP/TUTase, PAP/associated | yes | Tb927.7.3950 | [31,65,72, 83,110,111] | |
| MERS1 | mRNA stability | NUDIX hydrolase | yes | Tb927.11.15640 | [63] | |
| KPAP1 | mRNA polyadenylation | NT/PAP/TUTase, PAP/associated | yes | Tb927.11.7960 | [68] | |
| KPAF1 | mRNA A/U-tailing | Pentatricopeptide repeats | yes | Tb927.2.3180 | [72] | |
| KPAF2 | mRNA A/U-tailing | Pentatricopeptide repeats | noa | Tb927.11.14380 | [72] |
Inferred from RNAi knockdown.
Gene identification numbers are from TriTrypDB Kinetoplastid Genomics Resource (http://tritrypdb.org).
Editing reactions inevitably produce double-stranded mRNA-gRNA hybrids that must be resolved to enable sequential gRNA binding during pan-editing and probably before fully edited mRNAs are translated. This basic premise led to identification of REH1 (Hel61) DEAD/H-box helicase [75], which was found to be essential for editing mediated by two or more overlapping gRNAs [76] (Table 3). Proteomic studies of GRBC1/2-associated proteins in L. tarentolae [63] and MRB1 complex in T. brucei also identified REH2 helicase [66], a large polypeptide representing RHA sub-group within the superfamily 2 of DEAH/RHA RNA helicases [69,70]. A protein essential for cell viability, REH2 is composed of dsRNA binding, DEXDc, HA2 and OB-fold domains and displays an ATP-dependent dsRNA unwinding activity [77]. Although the RNA-mediated nature of a relatively stable REH2-RESC interaction is apparent, initial indications of REH2’s participation in gRNA biogenesis [64,77] have been questioned by more recent study [69]. Nonetheless, the steady-state levels of several edited mRNAs decline in both REH1 and REH2 knockdowns, which justifies further efforts to understand molecular functions of RNA helicases in editing.
Another enigmatic factor, an essential Nudix hydrolase MERS1 was identified via a relatively stable RNA-mediated interaction with the GRBC complex [63]. Nudix hydrolases catalyze a wide range of reactions on polyphosphates including nucleoside di-and triphosphates, such as those found on gRNA’s 5′ end. However, MERS1 repression causes downregulation of edited mRNAs without affecting gRNA population. It is generally assumed that polycistronic precursors are divided into pre-edited and unedited mRNA by an endonucleolytic cleavage that leaves monophosphorylated 5′ UTRs. Although such nuclease remains unidentified, the processed mRNA 5′ end constitutes an unlikely target for a Nudix hydrolase. Further investigation of MERS1’s in vivo RNA binding sites and possible binding partners is clearly required to resolve the function of this intriguing mRNA stability factor.
Concluding remarks
Current definition of the RNA editing holoenzyme, albeit most likely still incomplete (see Outstanding Questions Box), provides a fruitful ground for understanding molecular details of this process and a plethora of potential targets for developing trypanocides. However, the state-of-the-art techniques adapted by the field have arguably reached their limits. Further progress in elucidating the structure and dynamic interactions of editing complexes requires application of cryoelectron microscopy of purified complexes, assessment of transient protein-protein interactions in vivo, and high resolution mapping of in vivo RNA binding sites for RECC and RESC subunits. The complexity of editing protein assemblies is exacerbated by their interactions with pre- and post-editing mRNA 3′ end modification, and translation complexes. Because of low overall fidelity of the editing process, partially-edited or miss-edited transcripts constitute the bulk of the mRNA population. This raises the problem of how correctly edited mRNAs are selected for ribosome binding and translation. To this end, the signaling mechanism between the completion of the mRNA editing and addition of the long 3′ A/U tail remains to be established. Perhaps the most glaring knowledge gap lies in the origins of RNA editing substrates, which brings into focus the mechanisms of maxicircle and minicircle transcription, and the pathways for processing of primary mitochondrial transcripts. Studies of U-insertion/deletion editing introduced the fundamental concepts, such as guide RNA-directed targeting of protein complexes to mRNA, and discovered the new enzymes, such as TUTases. There remains, however, an intrinsic value in exploring unusual gene expression mechanisms beyond the realm of conventional model organisms.
Outstanding questions.
Significant progress in understanding of mRNA editing and polyadenylation underscores the lack of knowledge about mechanisms of mitochondrial transcription and initial processing events. What is the nature of mitochondrial promoters, the composition of transcription complex, and the structure of primary maxicircle-encoded transcripts? How are they processed into pre-edited and unedited mRNAs, and rRNAs?
Many RESC (MRB1) subunits lack apparent functional motifs, but are essential for editing. Do they bind editing substrates, intermediates or products or facilitate RESC interactions with other RNA processing complexes?
Completion of editing triggers mRNA adenylation/uridylation, a process in which 200–300 nt A/U-tails are added to pre-existing A-tails. What is the signaling event that leads to A/U-tailing? What are the molecular mechanisms involved in A/U-tailing activation or derepression?
What is the mechanism used by mitochondrial ribosomes to distinguish fully edited from partially or missedited mRNAs? Either a sequence-specific mRNA recognition or rapid degradation of abortive translation products can be a potential solution.
Figure 3.
Schematic representation of mRNA interactions with RNA editing substrate binding, RNA editing core and polyadenylation complexes, and the ribosome. The RNA editing substrate binding complex (RESC) consists of three modules: guide RNA binding (GRBC), RNA editing mediator (REMC) and polyadenylation mediator (PAMC). These modules have been implicated in gRNA stabilization, recruitment of RNA editing core and polyadenylation complexes, respectively [17]. Pre-edited mRNA is depicted in red, edited mRNA is shown in blue, and edited A/U-tailed mRNA is colored in orange. The A/U-tail enables mRNA binding to the small ribosomal subunit (SSU) while RNA editing substrates and complexes are predominantly associated with the large ribosomal subunit (LSU).
Box 1. RNA editing core complex (RECC).
The concept of a stable protein complex that displays both U-insertion and U-deletion activities evolved with improvements in purification, detection and reverse genetic approaches [35,52,78]. This particle was referred to as the L-complex to signify the presence of RNA ligases [35]) or 20S editosome to reflect sedimentation properties of a purified complex [79]. The RNA editing core complex (RECC) is a collective term describing three alternative forms distinguished by a mutually exclusive association of endonuclease modules with a common set of structural and enzymatic subunits (Figure 1, [80]). The endonuclease modules contain REN1 and REN2 RNase III-like enzymes that cleave mRNA at deletion and insertions sites, respectively, and one or two accessory proteins. The REN3 module is believed to act on a single CO2 mRNA which contains guiding sequence in the 3′ UTR [24,25,81]. The common set of proteins could be further subdivided into inner nucleus and spatially separated U-insertion and U-deletion sub-complexes. These three-member sub-complexes contain enzymes participating in respective enzymatic cascades. Noteworthy, the KREX2 exonuclease is dispensable for cell viability and lacks catalytic domain in related parasite L. tarentolae.
Box 2. RNA editing substrate binding complex (RESC).
The RESC complex, also referred to as the mitochondrial RNA binding complex 1 (MRB1) [66,82] or the guide RNA binding complex (GRBC) [17,63], emerged recently as the main RNA binding platform, which is also responsible for coupling of mRNA editing, polyadenylation and translation processes. All but one (RGG2) RESC submits lack annotated motifs and few have been assigned specific functions. At least three distinct modules can be distinguished within RESC based on mass spectrometry analysis of affinity purified subunits and yeast two-hybrid mapping of protein interactions: 1) gRNA binding complex (GRBC), of which subunits G1 and G2 are responsible for binding and stabilization of mature gRNAs; 2) RNA editing mediator complex (REMC), of which RGG2 subunit is required for processivity of pan-editing and 3) polyadenylation mediator complex (PAMC), of which subunit P1 is required for post-editing mRNA adenylation/uridylation. Approximately 17 subunits have been verified by cross-tagging experiments.
Trends.
Uridine insertion/deletion editing generates protein coding sequences in most mitochondrial mRNAs of trypanosomes. The emerging architecture of the editing holoenzyme suggests an RNA-mediated assembly of the multi-subunit enzymatic RNA editing core (RECC) and RNA editing substrate binding (RESC) complexes.
Recently characterized RESC complex is composed of approximately 17 polypeptides that can be clustered into GRBC, REMC and PAMC modules. These modules are responsible for guide RNA binding, and mediating interactions with the enzymatic core editing and polyadenylation complexes, respectively.
The majority of RNA editing factors are essential for parasite viability and do not have apparent human homologs. Therefore, RNA editing pathway represents a significant source of therapeutic targets relevant to neglected tropical diseases.
Acknowledgments
We thank members of our laboratories for discussions. This work was supported by NIH grants AI113157 to IA, and AI113157 and AI101057 to RA. We apologize to colleagues whose work was not cited due to space limits.
Glossary
- 5′ anchor
5′ part of the guide RNA that forms a continuous 5–10 nt duplex with pre-edited mRNA; this region is responsible for initial gRNA-mRNA interaction
- 3′ anchor
refers to scattered base-pairing between gRNA region that directs editing reactions and pre-edited mRNA
- 5′ and 3′ cleavage fragments
mRNA fragments generated by gRNA-directed endonucleolytic cleavage, the first reaction of the editing cascade
- Endo/Exo/phosphatase (EEP)
metal-dependent hydrolase, endonuclease/exonuclease/phosphatase family
- Editing block
mRNA segment covered by a single guide RNA; typically contains both U-insertion and U-deletion sites
- Editing domain
mRNA segment covered by multiple overlapping gRNAs. Sequence changes directed by the initiating gRNA create binding site for a sequential one and so forth; the hierarchical gRNA binding provides for the overall 3′-5′ progression of editing events within the domain
- Editing site
position of the guide RNA-directed mRNA cleavage where uridines are either removed from or added to the 5′ cleavage fragment
- dsRNA
double-stranded RNA
- Fully-edited mRNA
final product of the editing process; contains an open reading frame
- Guide RNA
small non-coding RNA that specifies positions and extent of uridine insertions and deletions. Guide RNAs are typically 40–60 nt in length and possess 5′ triphosphates and 3′ oligo U-tails
- Kinetoplast
a densely packed, disc-like nucleoprotein structure that encloses mitochondrial DNA
- Kinetoplast DNA
kDNA, mitochondrial genome, DNA component of the kinetoplast composed of maxicircles and minicircles. In most cases circular DNA molecules are locked into a catenated network, but in some species circles are not catenated and distributed throughout mitochondrial lumen
- Kinetoplastida (Kinetoplastea)
a group of flagellated protists belonging to the phylum Euglenozoa and characterized by the presence of a kinetoplast
- Maxicircle
An equivalent of a typical mitochondrial genome, maxicircles contain genes for respiratory complex subunits, often ribosomal protein S12, and ribosomal RNAs. Maxicircles are typically longer than 20 kb and are present in few dozen copies per kinetoplast
- Minicircle
minicircles encode guide RNAs (gRNAs) that participate in the editing process and gRNA-like molecules on unknown function. The lengths (0.75–10 kb) and copy numbers (~10,000) vary dramatically among species
- Pan-edited mRNAs
transcripts that undergo massive editing directed by multiple overlapping gRNAs. There can be several editing domains within pan-edited mRNA
- Partially-edited mRNA
intermediate of the editing process
- Pre-edited mRNA
precursor transcript that must undergo editing to acquire an open reading frame and/or translation initiation and termination signals
- RNA helicase
a motor protein capable of harnessing the energy from NTP hydrolysis to unwind double stranded RNAs or to remodel ribonucleoprotein complexes
- RNase III
an enzyme that binds to and cleaves double-stranded RNA leaving 5′ monophosphate and 3′ hydroxyl groups
- TUTase
RNA terminal uridyltransferase, UTP-specific nucleotidyl transferase which adds uridylyl residues to the 3′ end of RNA
- Unedited mRNA
protein coding transcript that contains open reading frame and does not require editing
- UTR
untranslated region of mRNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Benne R, et al. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- 2.Feagin JE, et al. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell. 1988;53:413–422. doi: 10.1016/0092-8674(88)90161-4. [DOI] [PubMed] [Google Scholar]
- 3.Shaw J, et al. Editing of mitochondrial mRNAs by uridine addition and deletion generates conserved amino acid sequences and AUG initiation codons. Cell. 1988;53:401–411. doi: 10.1016/0092-8674(88)90160-2. [DOI] [PubMed] [Google Scholar]
- 4.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanc V, Davidson NO. APOBEC-1-mediated RNA editing. Wiley Interdiscip Rev Syst Biol Med. 2010;2:594–602. doi: 10.1002/wsbm.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackman JE, et al. Doing it in reverse: 3′-to-5′ polymerization by the Thg1 superfamily. RNA. 2012;18:886–899. doi: 10.1261/rna.032300.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray MW. Diversity and evolution of mitochondrial RNA editing systems. IUBMB Life. 2003;55:227–233. doi: 10.1080/1521654031000119425. [DOI] [PubMed] [Google Scholar]
- 8.Stoltzfus A. On the possibility of constructive neutral evolution. J Mol Evol. 1999;49:169–181. doi: 10.1007/pl00006540. [DOI] [PubMed] [Google Scholar]
- 9.Gray MW, et al. Cell biology. Irremediable complexity? Science. 2010;330:920–921. doi: 10.1126/science.1198594. [DOI] [PubMed] [Google Scholar]
- 10.Gray MW. Evolutionary origin of RNA editing. Biochemistry. 2012;51:5235–5242. doi: 10.1021/bi300419r. [DOI] [PubMed] [Google Scholar]
- 11.Ochsenreiter T, Hajduk SL. Alternative editing of cytochrome c oxidase III mRNA in trypanosome mitochondria generates protein diversity. EMBO Rep. 2006;7:1128–1133. doi: 10.1038/sj.embor.7400817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ochsenreiter T, et al. Alternative mRNA editing in trypanosomes is extensive and may contribute to mitochondrial protein diversity. PLoS ONE. 2008;3:e1566. doi: 10.1371/journal.pone.0001566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blum B, et al. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- 14.Sturm NR, Simpson L. Kinetoplast DNA minicircles encode guide RNAs for editing of cytochrome oxidase subunit III mRNA. Cell. 1990;61:879–884. doi: 10.1016/0092-8674(90)90198-n. [DOI] [PubMed] [Google Scholar]
- 15.Maslov DA, Simpson L. The polarity of editing within a multiple gRNA-mediated domain is due to formation of anchors for upstream gRNAs by downstream editing. Cell. 1992;70:459–467. doi: 10.1016/0092-8674(92)90170-h. [DOI] [PubMed] [Google Scholar]
- 16.Koslowsky D, et al. The insect-phase gRNA transcriptome in Trypanosoma brucei. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aphasizheva I, et al. RNA binding and core complexes constitute the U-insertion/deletion editosome. Mol Cell Biol. 2014;34:4329–4342. doi: 10.1128/MCB.01075-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Igo RP, et al. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol Cell Biol. 2000;20:8447–8457. doi: 10.1128/mcb.20.22.8447-8457.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Igo RP, Jr, et al. RNA sequence and base pairing effects on insertion editing in Trypanosoma brucei. Mol Cell Biol. 2002;22:1567–1576. doi: 10.1128/mcb.22.5.1567-1576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanc V, et al. The mitochondrial RNA ligase from Leishmania tarentolae can join RNA molecules bridged by a complementary RNA. J Biol Chem. 1999;274:24289–24296. doi: 10.1074/jbc.274.34.24289. [DOI] [PubMed] [Google Scholar]
- 21.Blum B, Simpson L. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo-(U) tail involved in recognition of the pre-edited region. Cell. 1990;62:391–397. doi: 10.1016/0092-8674(90)90375-o. [DOI] [PubMed] [Google Scholar]
- 22.Trotter JR, et al. A deletion site editing endonuclease in Trypanosoma brucei. Mol Cell. 2005;20:403–412. doi: 10.1016/j.molcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 23.Carnes J, et al. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc Natl Acad Sci U S A. 2005;102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carnes J, et al. Endonuclease associations with three distinct editosomes in Trypanosoma brucei. J Biol Chem. 2011;286:19320–19330. doi: 10.1074/jbc.M111.228965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carnes J, et al. RNA Editing in Trypanosoma brucei requires three different editosomes. Mol Cell Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carnes J, et al. Mutational analysis of Trypanosoma brucei editosome proteins KREPB4 and KREPB5 reveals domains critical for function. RNA. 2012;18:1897–1909. doi: 10.1261/rna.035048.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst NL, et al. Differential functions of two editosome exoUases in Trypanosoma brucei. RNA. 2009;15:947–957. doi: 10.1261/rna.1373009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnaufer A, et al. A protein-protein interaction map of trypanosome ~20S editosomes. J Biol Chem. 2010;285:5282–5295. doi: 10.1074/jbc.M109.059378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnaufer A, et al. Separate Insertion and Deletion Subcomplexes of the Trypanosoma brucei RNA Editing Complex. Mol Cell. 2003;12:307–319. doi: 10.1016/s1097-2765(03)00286-7. [DOI] [PubMed] [Google Scholar]
- 30.Kang X, et al. Disruption of the zinc finger motifs in the Leishmania tarentolae LC-4 (=TbMP63) L-complex editing protein affects the stability of the L-complex 6. J Biol Chem. 2004;279:3893–3899. doi: 10.1074/jbc.M310185200. [DOI] [PubMed] [Google Scholar]
- 31.Aphasizhev R, et al. A tale of two TUTases. Proc Natl Acad Sci U S A. 2003;100:10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogers K, et al. Uridylate-specific 3′ 5′-exoribonucleases involved in uridylate-deletion RNA editing in trypanosomatid mitochondria. J Biol Chem. 2007;282:29073–29080. doi: 10.1074/jbc.M704551200. [DOI] [PubMed] [Google Scholar]
- 33.Carnes J, et al. KREX2 is not essential for either procyclic or bloodstream form Trypanosoma brucei. PLoS One. 2012;7:e33405. doi: 10.1371/journal.pone.0033405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang X, et al. Reconstitution of full-round uridine-deletion RNA editing with three recombinant proteins. Proc Natl Acad Sci U S A. 2006;103:13944–13949. doi: 10.1073/pnas.0604476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aphasizhev R, et al. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ringpis GE, et al. Mechanism of U Insertion RNA Editing in Trypanosome Mitochondria: The Bimodal TUTase Activity of the Core Complex. J Mol Biol. 2010;399:680–695. doi: 10.1016/j.jmb.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ringpis GE, et al. Mechanism of U-insertion RNA Editing in Trypanosome Mitochondria: Characterization of RET2 Functional Domains by Mutational Analysis. J Mol Biol. 2010;399:696–706. doi: 10.1016/j.jmb.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng J, et al. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J. 2005;24:4007–4017. doi: 10.1038/sj.emboj.7600861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ernst NL, et al. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol Cell. 2003;11:1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 40.Gao G, et al. Functional complementation of Trypanosoma brucei RNA in vitro editing with recombinant RNA ligase. Proc Natl Acad Sci U S A. 2005;102:4712–4717. doi: 10.1073/pnas.0500553102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnaufer A, et al. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–2161. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- 42.Gao G, Simpson L. Is the Trypanosoma brucei REL1 RNA ligase specific for U-deletion RNA editing, and is the REL2 RNA ligase specific for U-insertion editing? J Biol Chem. 2003;278:27570–27574. doi: 10.1074/jbc.M303317200. [DOI] [PubMed] [Google Scholar]
- 43.Huang CE, et al. Roles for ligases in the RNA editing complex of Trypanosoma brucei: band IV is needed for U-deletion and RNA repair. EMBO J. 2001;20:4694–4703. doi: 10.1093/emboj/20.17.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cruz-Reyes J, et al. Distinct functions of two RNA ligases in active Trypanosoma brucei RNA editing complexes. Mol Cell Biol. 2002;22:4652–4660. doi: 10.1128/MCB.22.13.4652-4660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Hearn SF, et al. Trypanosoma brucei RNA editing complex: band II is structurally critical and maintains band V ligase, which is nonessential. Mol Cell Biol. 2003;23:7909–7919. doi: 10.1128/MCB.23.21.7909-7919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho CK, et al. Structure and mechanism of RNA ligase. Structure (Camb ) 2004;12:327–339. doi: 10.1016/j.str.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 47.Deng J, et al. High resolution crystal structure of a key editosome enzyme from Trypanosoma brucei: RNA editing ligase 1. J Mol Biol. 2004;343:601–613. doi: 10.1016/j.jmb.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 48.Ho CK, Shuman S. Bacteriophage T4 RNA ligase 2 (gp24.1) exemplifies a family of RNA ligases found in all phylogenetic domains. Proc Natl Acad Sci U S A. 2002;99:12709–12714. doi: 10.1073/pnas.192184699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pollard VW, et al. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 1992;11:4429–4438. doi: 10.1002/j.1460-2075.1992.tb05543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peris M, et al. Characterization of two classes of ribonucleoprotein complexes possibly involved in RNA editing from Leishmania tarentolae mitochondria. EMBO J. 1994;13:1664–1672. doi: 10.1002/j.1460-2075.1994.tb06430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corell RA, et al. Complexes from Trypanosoma brucei that exhibit deletion editing and other editing-associated properties. Mol Cell Biol. 1996;16:1410–1418. doi: 10.1128/mcb.16.4.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rusche LN, et al. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li F, et al. Structure of the core editing complex (L-complex) involved in uridine insertion/deletion RNA editing in trypanosomatid mitochondria. Proc Natl Acad Sci U S A. 2009;106:12306–12310. doi: 10.1073/pnas.0901754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Osato D, et al. Uridine insertion/deletion RNA editing in trypanosomatid mitochondria: In search of the editosome. RNA. 2009;15:1338–1344. doi: 10.1261/rna.1642809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muller UF, et al. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J. 2001;20:1394–1404. doi: 10.1093/emboj/20.6.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allen TE, et al. Association of guide RNA binding protein gBP21 with active RNA editing complexes in Trypanosoma brucei. Mol Cell Biol. 1998;18:6014–6022. doi: 10.1128/mcb.18.10.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koller J, et al. Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J Biol Chem. 1997;272:3749–3757. doi: 10.1074/jbc.272.6.3749. [DOI] [PubMed] [Google Scholar]
- 58.Fisk JC, et al. Distinct and overlapping functions of MRP1/2 and RBP16 in mitochondrial RNA metabolism. Mol Cell Biol. 2009;29:5214–5225. doi: 10.1128/MCB.00520-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller MM, et al. RBP16 stimulates trypanosome RNA editing in vitro at an early step in the editing reaction. RNA. 2006;12:1292–1303. doi: 10.1261/rna.2331506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayman ML, Read LK. Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J Biol Chem. 1999;274:12067–12074. doi: 10.1074/jbc.274.17.12067. [DOI] [PubMed] [Google Scholar]
- 61.Bringaud F, et al. Mitochondrial glutamate dehydrogenase from Leishmania tarentolae is a guide RNA-binding protein. Mol Cell Biol. 1997;17:3915–3923. doi: 10.1128/mcb.17.7.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aphasizhev R, et al. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA. 2003;9:62–76. doi: 10.1261/rna.2134303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weng J, et al. Guide RNA-Binding Complex from Mitochondria of Trypanosomatids. Mol Cell. 2008;32:198–209. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hashimi H, et al. Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA. 2009;15:588–599. doi: 10.1261/rna.1411809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aphasizheva I, Aphasizhev R. RET1-catalyzed Uridylylation Shapes the Mitochondrial Transcriptome in Trypanosoma brucei. Mol Cell Biol. 2010;30:1555–1567. doi: 10.1128/MCB.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Panigrahi AK, et al. Mitochondrial complexes in Trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol Cell Proteomics. 2007;7:534–545. doi: 10.1074/mcp.M700430-MCP200. [DOI] [PubMed] [Google Scholar]
- 67.Ammerman ML, et al. TbRGG2 facilitates kinetoplastid RNA editing initiation and progression past intrinsic pause sites. RNA. 2010;16:2239–2251. doi: 10.1261/rna.2285510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Etheridge RD, et al. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008;27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Madina BR, et al. Native Variants of the MRB1 Complex Exhibit Specialized Functions in Kinetoplastid RNA Editing. PLoS ONE. 2015;10:e0123441. doi: 10.1371/journal.pone.0123441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Madina BR, et al. Native mitochondrial RNA-binding complexes in kinetoplastid RNA editing differ in guide RNA composition. RNA. 2014;20:1142–1152. doi: 10.1261/rna.044495.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kao CY, Read LK. Opposing effects of polyadenylation on the stability of edited and unedited mitochondrial RNAs in Trypanosoma brucei. Mol Cell Biol. 2005;25:1634–1644. doi: 10.1128/MCB.25.5.1634-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aphasizheva I, et al. Pentatricopeptide Repeat Proteins Stimulate mRNA Adenylation/Uridylation to Activate Mitochondrial Translation in Trypanosomes. Mol Cell. 2011;42:106–117. doi: 10.1016/j.molcel.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ridlon L, et al. The Importance of the 45S Ribosomal Small Subunit-related Complex for Mitochondrial Translation in Trypanosoma brucei. J Biol Chem. 2013;288:32963–32978. doi: 10.1074/jbc.M113.501874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aphasizheva I, et al. Kinetoplast DNA-encoded ribosomal protein S12: a possible functional link between mitochondrial RNA editing and translation in Trypanosoma brucei. RNA Biol. 2013;10:1679–1688. doi: 10.4161/rna.26733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Missel A, et al. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol Cell Biol. 1997;17:4895–4903. doi: 10.1128/mcb.17.9.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li F, et al. Trypanosome REH1 is an RNA helicase involved with the 3′-5′ polarity of multiple gRNA-guided uridine insertion/deletion RNA editing. Proc Natl Acad Sci U S A. 2011;108:3542–3547. doi: 10.1073/pnas.1014152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hernandez A, et al. REH2 RNA helicase in kinetoplastid mitochondria: ribonucleoprotein complexes and essential motifs for unwinding and guide RNA (gRNA) binding. J Biol Chem. 2010;285:1220–1228. doi: 10.1074/jbc.M109.051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Panigrahi AK, et al. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9:484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stuart KD, et al. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 80.Simpson L, et al. Guide to the nomenclature of kinetoplastid RNA editing: a proposal. Protist. 2010;161:2–6. doi: 10.1016/j.protis.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Panigrahi AK, et al. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA. 2006;12:1038–1049. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hashimi H, et al. Dual core processing: MRB1 is an emerging kinetoplast RNA editing complex. Trends Parasitol. 2013;29:91–99. doi: 10.1016/j.pt.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aphasizhev R, Aphasizheva I. RNA Editing Uridylyltransferases of Trypanosomatids. Methods Enzymol. 2007;424:51–67. doi: 10.1016/S0076-6879(07)24003-0. [DOI] [PubMed] [Google Scholar]
- 84.Panigrahi AK, et al. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol Cell Biol. 2001;21:380–389. doi: 10.1128/MCB.21.2.380-389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rusché LN, et al. The two RNA ligases of the Trypanosoma brucei RNA editing complex: Cloning the essential band IV gene and identifying the band V gene. Mol Cell Biol. 2001;21:979–989. doi: 10.1128/MCB.21.4.979-989.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McManus MT, et al. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA. 2001;7:167–175. doi: 10.1017/s1355838201002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Panigrahi AK, et al. Four Related Proteins of the Trypanosoma brucei RNA Editing Complex. Mol Cell Biol. 2001;21:6833–6840. doi: 10.1128/MCB.21.20.6833-6840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Drozdz M, et al. TbMP81 is required for RNA editing in Trypanosoma brucei. EMBO J. 2002;21:1791–1799. doi: 10.1093/emboj/21.7.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang CE, et al. Assembly and function of the RNA editing complex in Trypanosoma brucei requires band III protein. Mol Cell Biol. 2002;22:3194–3203. doi: 10.1128/MCB.22.9.3194-3203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guo X, et al. The KREPA3 zinc finger motifs and OB-fold domain are essential for RNA editing and survival of Trypanosoma brucei. Mol Cell Biol. 2008;28:6939–6953. doi: 10.1128/MCB.01115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guo X, et al. The zinc-fingers of KREPA3 are essential for the complete editing of mitochondrial mRNAs in Trypanosoma brucei. PLoS ONE. 2010;5:e8913. doi: 10.1371/journal.pone.0008913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Salavati R, et al. KREPA4, an RNA binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA. 2006;12:819–831. doi: 10.1261/rna.2244106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kala S, Salavati R. OB-fold domain of KREPA4 mediates high-affinity interaction with guide RNA and possesses annealing activity. RNA. 2010;16:1951–1967. doi: 10.1261/rna.2124610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kala S, et al. The oligonucleotide binding (OB)-fold domain of KREPA4 is essential for stable incorporation into editosomes. PLoS ONE. 2012;7:e46864. doi: 10.1371/journal.pone.0046864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Babbarwal VK, et al. An essential role of KREPB4 in RNA editing and structural integrity of the editosome in Trypanosoma brucei. RNA. 2007;13:737–744. doi: 10.1261/rna.327707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tarun Z, Jr, et al. KREPA6 is an RNA-binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA. 2008;14:347–358. doi: 10.1261/rna.763308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu M, et al. Structures of a key interaction protein from the Trypanosoma brucei editosome in complex with single domain antibodies. J Struct Biol. 2010 doi: 10.1016/j.jsb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guo X, et al. KREPB6, KREPB7, and KREPB8 are important for editing endonuclease function in Trypanosoma brucei. RNA. 2012;18:308–320. doi: 10.1261/rna.029314.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aphasizheva I, et al. Novel TUTase associates with an editosome-like complex in mitochondria of Trypanosoma brucei. RNA. 2009;15:1322–1337. doi: 10.1261/rna.1538809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lerch M, et al. Editosome accessory factors KREPB9 and KREPB10 in Trypanosoma brucei. Eukaryot Cell. 2012;11:832–843. doi: 10.1128/EC.00046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ammerman ML, et al. A core MRB1 complex component is indispensable for RNA editing in insect and human infective stages of Trypanosoma brucei. PLoS ONE. 2013;8:e78015. doi: 10.1371/journal.pone.0078015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Acestor N, et al. The MRB1 complex functions in kinetoplastid RNA processing. RNA. 2009;15:277–286. doi: 10.1261/rna.1353209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ammerman ML, et al. MRB3010 is a core component of the MRB1 complex that facilitates an early step of the kinetoplastid RNA editing process. RNA. 2011;17:865–877. doi: 10.1261/rna.2446311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ammerman ML, et al. Architecture of the trypanosome RNA editing accessory complex, MRB1. Nucleic Acids Res. 2012;40:5637–5650. doi: 10.1093/nar/gks211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kafkova L, et al. Functional characterization of two paralogs that are novel RNA binding proteins influencing mitochondrial transcripts of Trypanosoma brucei. RNA. 2012;18:1846–1861. doi: 10.1261/rna.033852.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fisk JC, et al. TbRGG2, an essential RNA editing accessory factor in two Trypanosoma brucei life cycle stages. J Biol Chem. 2008;283:23016–23025. doi: 10.1074/jbc.M801021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Foda BM, et al. Multifunctional G-rich and RRM-containing domains of TbRGG2 perform separate yet essential functions in trypanosome RNA editing. Eukaryot Cell. 2012;11:1119–1131. doi: 10.1128/EC.00175-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Missel A, et al. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol Cell Biol. 1997;17:4895–4903. doi: 10.1128/mcb.17.9.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Missel A, Goringer HU. Trypanosoma brucei mitochondria contain RNA helicase activity. Nucleic Acids Res. 1994;22:4050–4056. doi: 10.1093/nar/22.20.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aphasizheva I, et al. RNA-editing terminal uridylyl transferase 1: identification of functional domains by mutational analysis. J Biol Chem. 2004;279:24123–24130. doi: 10.1074/jbc.M401234200. [DOI] [PubMed] [Google Scholar]
- 111.Aphasizhev R, et al. Trypanosome Mitochondrial 3′ Terminal Uridylyl Transferase (TUTase): The Key Enzyme in U-insertion/deletion RNA Editing. Cell. 2002;108:637–648. doi: 10.1016/s0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]