Abstract
Follicular lymphoma (FL), the most common indolent subtype of non-Hodgkin’s lymphoma, is associated with a relatively long overall survival rate ranging from 6 to 10 years from time of diagnosis. However, in 20–60% of FL patients, transformation to aggressive diffuse large B-cell lymphoma (DLBCL) reduces median survival to only 1.2 years. The specific functional and genetic determinants of FL transformation remain elusive, and genomic alterations underlying disease advancement have only been identified for a subset of cases. Therefore, to identify candidate drivers of FL transformation, we performed systematic analysis of a B-cell-specific regulatory model exhibiting FL transformation signatures using the Master Regulator Inference algorithm (MARINa). This analysis revealed FOXM1, TFDP1, ATF5, HMGA1, and NFYB to be candidate master regulators (MR) contributing to disease progression. Accordingly, validation was achieved through synthetic lethality assays in which RNAi-mediated silencing of MRs individually or in combination reduced the viability of (14;18)-positive DLBCL (t-DLBCL) cells. Furthermore, specific combinations of small molecule compounds targeting synergistic MR pairs induced loss of viability in t-DLBCL cells. Collectively, our findings indicate that MR analysis is a valuable method for identifying bona fide contributors to FL transformation and may therefore guide the selection of compounds to be used in combinatorial treatment strategies.
Keywords: master regulator, follicular lymphoma transformation, small molecule, synergy
Introduction
FL is the second most common non-Hodgkin’s lymphoma (NHL) subtype, followed by DLBCL, comprising about 22% of annually diagnosed cases (1). FL is generally diagnosed in elderly patients (>60 years of age) and is a slow-progressing neoplasm, with a median survival rate of 10 years. Despite an overall indolent course and some improvement in overall survival by rituximab-based therapy (2), FL remains an incurable disease. The canonical t(14;18)(q32;q21) chromosomal translocation represents the most frequent genetic alteration of this disease in the vast majority of these tumors, leading to aberrant expression of the anti-apoptotic protein BCL2 (3). However, this alteration alone is insufficient for tumor initiation, since the translocation is also found in healthy individuals (4). Over time, FL transforms to more aggressive B-cell lymphomas, predominantly DLBCL, an event representing the hallmark of aggressive disease and poor prognosis. The frequency of histological transformation in patients initially diagnosed with FL ranges from 20% to 60% depending on clinical and pathological criteria, with median survival dropping to 1.2 years following transformation (5).
The molecular events leading to FL transformation are poorly characterized. Although, several genomic alterations have been associated with FL transformation, including TP53 mutation, MYC rearrangement, REL amplification and CDKN2A/CDKN2B deletion (6), these represent only ~23% of all transformed FL cases (7). In addition to genetic alterations (8–10), epigenetic mechanisms (11) and microenvironment signals (12) have also been implicated in FL transformation, contributing to a relatively large, heterogeneous, and poorly understood molecular landscape.
Our recent elucidation of MRs of glioma, prostate cancer, and germinal center reaction (13–15) suggests that distinct molecular events may induce aberrant activation of a relatively small number of MR genes, representing the causal, functional drivers of established FL-transformation signature (16). Thus to identify such candidate functional drivers of FL transformation, we interrogated an established human B-cell regulatory network, assembled from a large collection of normal and tumor related gene expression profiles (GEP) using the ARACNe algorithm (17). This approach has been highly successful in discovering novel mechanisms of tumorigenesis and tumor progression, including synergistic gene-gene interactions that could not be elucidated by more conventional analytical approaches (13–15, 18).
The analysis identified novel candidate FL transformation MRs that were experimentally validated, including synthetic-lethal pairs, whose RNAi mediated co-silencing collapsed the FL-transformation signature and induced significant viability reduction. FDA-approved drugs computationally predicted as B-cell specific inhibitors of these MRs were shown to induce t-DLBCL cell death, both individually and in combination.
The proposed drug prioritization methodology is highly general, relying only on the availability of a cell-specific regulatory model and disease-relevant small-molecule signatures. This paves the road to a more efficient precision medicine pipeline for the simultaneous and systematic prioritization of small molecule compounds for either single-agent or combination therapy.
Materials and Methods
Cell lines, Antibodies and Reagents
CB33, SUDHL4 and SUDHL6 cells provided by R. Dalla-Favera (Columbia University, NY) were maintained in IMDM (Life Technology), supplemented with 10% FBS (Gemini) and antibiotics. The HF1 follicular cell line provided by R. Levy (Stanford University, CA) was maintained in DMEM (Life Technology), supplemented with 10% FBS and antibiotics. Cells were tested negative for mycoplasma. Cells were not further authenticated. Antibodies: rabbit anti-MYC (XP) (Cell Signaling Technology); rabbit anti-FOXM1 and mouse anti-GAPDH (SantaCruz); rabbit anti-HMGA1, anti-ATF5, anti-NFYB, mouse anti-TFDP1 (Abcam). Alprostadil, Clemastine, Cytarabine and Troglitazone (Tocris), Econazole nitrate and Promazine hydrochloride (Sigma) were reconstituted in DMSO (Sigma).
Gene silencing, qRT-PCR and Microarray assays
Gene silencing was performed using smart-pool siRNA (Dharmacon) delivered by 96-well Shuttle nucleoporation system (Amaxa) according to the manufacturer (Lonza). Detailed information on nucleoporation, qRT-PCR and Microarray assays in Supplementary Methods. All microarray data have been submitted to Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo - accession number GSE66714).
Cell viability
Cell viability was evaluated by PrestoBlue staining according to the manufacturer (Invitrogen). Fluorescence was measured using VICTOR 3V Plate Reader (Perkin Elmer). Small molecule screening was performed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega) in the Columbia HTS Facility. Cells were plated in 384-well plates, 24h prior to treatment with serial dilutions of the single compounds. Cell viability was analyzed at 48h to assess compound toxicity (Supplementary Fig. S4).
Tissue Microarray Analysis
TMAs construction, diagnostic staining for GCB-origin markers, FISH analysis for t(14;18) and immunohistochemistry staining for MRs were done in the Department of Pathology at Memorial Sloan-Kettering Cancer Center according to (19).
Computational and Statistical Methods
Classification of patient samples and cell lines by MYC activity
GEPs patient samples were obtained from Dataset 1 (16) and Dataset 2 (20). Samples were classified as “low” and “high MYC activity” by clustering methods using MYC targets obtained from (16). An outlier in the cluster analysis was excluded from further analysis. To classify cell lines for experimental validation by MYC activity, we performed clustering analysis using MYC targets on 61 samples from (21). This dataset contained 38 FL samples, 13 transformed DLBCL samples (selected based on BCL2 translocation), 10 normal GCB, 3 DLBCL cell lines (SUDHL4, SUDHL6 and VAL) and LCL-CB33.
MARINa
We performed MR analysis independently for “high activity MYC” and “low activity MYC” for Dataset1 (16) and Dataset 2 (20) samples. Dataset 1 contains 6 paired samples in each group where Dataset 2 have 5 and 7 paired samples in “high activity MYC” and “low activity MYC”, respectively.
Computation of similarity between drug treatment and siRNA signatures
We used Gene Set Enrichment Analysis (GSEA) (22) to assess drug-signature similarity, based on enrichment of the 200 most up-regulated genes and 200 most repressed genes from signature A in the other signature B, and vice versa. Enrichment scores were averaged to obtain a single metric and associated similarity p-value.
Fisher’s exact test (FET)
FET was used to assess MR overlap from each dataset (Table 1), independently for “high” and “low” MYC activity samples and to test if specific MR proteins have positive/negative expression in DLBCL vs FL TMAs (Table 2).
Table 1.
Overlap of MR’s inferred by MARINa from different datasets.
| Subtype | MR mode | Dataset 1 | Dataset 2 | Overlap | P value | Integrated P value |
|---|---|---|---|---|---|---|
| MYC high | Activated MR | 13 | 17 | 10 | 2.00E-07 | 2.00E-03 |
| Repressed MR | 5 | 10 | 2 | 1.30E-01 | ||
| MYC low | Activated MR | 5 | 6 | 5 | 2.00E-06 | 2.00E-03 |
| Repressed MR | 20 | 25 | 12 | 1.00E-03 | ||
| All samples | Activated MR | 9 | 12 | 6 | 3.00E-05 | 6.70E-02 |
| Repressed MR | 23 | 17 | 10 | 2.00E-03 |
Abbreviations: MR, master regulator
Table 2.
MR protein expression in FL and DLBCL patients’ samples by TMA analysis.
| MR | Subtype | Samples (n) | Positive n (%) | Negative n (%) | P value |
|---|---|---|---|---|---|
| HMGA1 | DLBCL-GC | 19 | 13 (68%) | 6 (32%) | 7.00E-10 |
| FL | 58 | 0 (0%) | 58 (100%) | ||
| DLBCL t(14;18) | 5 | 5 (100%) | 0 (0%) | 2.20E-03 | |
| FL t(14;18) | 6 | 0 (0%) | 6 (100%) | ||
| TFDP1 | DLBCL-GC | 20 | 4 (20%) | 16 (80%) | 7.00E-01 |
| FL | 75 | 17 (23%) | 58 (77%) | ||
| DLBCL t(14;18) | 5 | 3 (60%) | 2 (40%) | 4.50E-02 | |
| FL t(14;18) | 7 | 0 (0%) | 7 (100%) | ||
| FOXM1 | DLBCL-GC | 18 | 16 (89%) | 2 (11%) | 5.90E-05 |
| FL | 88 | 33 (38%) | 55 (62%) | ||
| DLBCL t(14;18) | 4 | 3 (75%) | 1 (25%) | 5.30E-01 | |
| FL t(14;18) | 7 | 4 (57%) | 3 (43%) | ||
| NFYB | DLBCL-GC | 18 | 7 (39%) | 11 (61%) | 6.20E-01 |
| FL | 101 | 40 (40%) | 61 (60%) | ||
| DLBCL t(14;18) | 4 | 3 (75%) | 1 (25%) | 9.00E-02 | |
| FL t(14;18) | 7 | 1 (14%) | 6 (85%) | ||
| ATF5 | DLBCL-GC | 18 | 17 (94%) | 1 (6%) | 1.20E-07 |
| FL | 81 | 22 (27%) | 59 (73%) | ||
| DLBCL t(14;18) | 4 | 4 (100%) | 0 (0%) | 3.00E-03 | |
| FL t(14;18) | 7 | 0 (0%) | 7 (100%) |
Abbreviations: TMA, Tissue Microarray; n, number
Results
Inference of Master Regulators of FL transformation
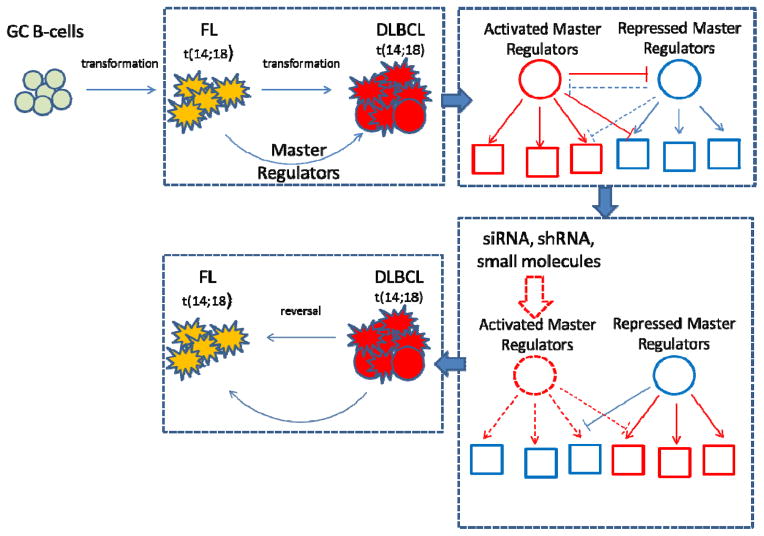
To infer candidate MRs of FL → DLBCL transformation (henceforth FL-transformation), we used the MARINa algorithm (13–15). MARINa assessed the relevance of a transcriptional factor (TF) as a candidate MR of FL transformation by evaluating whether its transcriptional targets are highly enriched in genes differentially expressed in patient samples following transformation. Indeed, if aberrant activity of a TF was responsible for transformation, then its activated and repressed targets should be over- and under-expressed following transformation, respectively (Fig. 1). Prior data from glioma (13) and prostate cancer (14) studies shows that top candidate MRs are highly enriched in genes eliciting essentiality or synthetic lethality. In addition, RNAi mediated silencing of MRs should at least partially revert the FL transformation signature (13, 14).
Figure 1. Model of FL transformation to DLBCL and experimental approach to abrogate transformed gene signature.
GCB derived aberrant FL cells after acquiring new oncogenic events undergo further transformation to DLBCL. These new aberrations cause erroneous downstream signaling of genes called MRs. Targeting of activated MR by siRNA/shRNA silencing or small molecule inhibition should impair their regulatory function and abrogate the FL transformed signature.
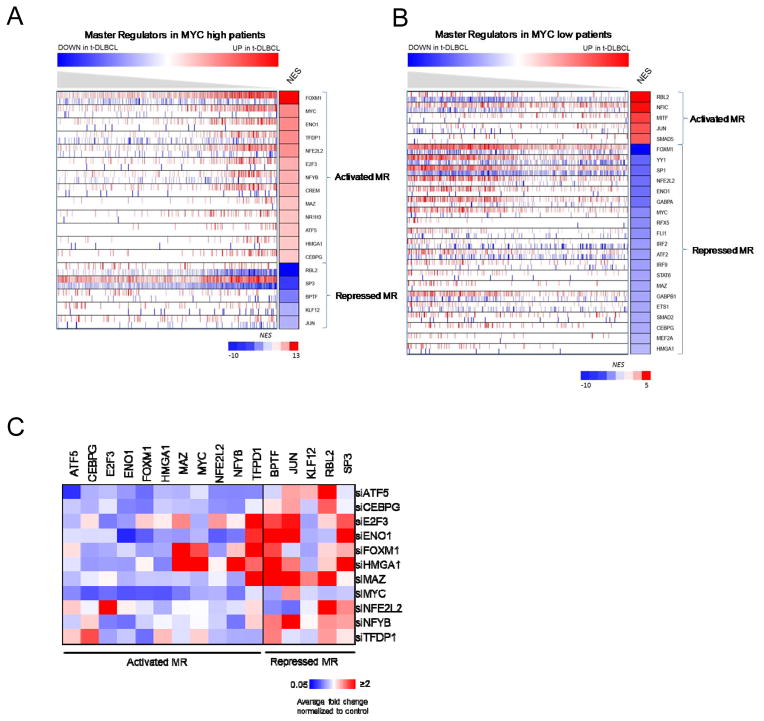
As a regulatory model in this study we used an ARACNe-inferred interactome for human B-cells (21). FL transformation signatures were defined using patient-matched GEPs from 12 patient biopsies at FL diagnosis and following transformation (Dataset 1) (16). As reported, patient signatures stratified into two distinct molecular subtypes, representing high and low MYC activity, respectively (16). Thus, we performed independent MR analysis for each subtype (Fig. 2A, B, Supplementary Table S1A, B). Results were compared with equivalent MARINa analysis of an independent dataset (Dataset 2) (20), comprising 12 additional patient-matched biopsies before and after transformation (Supplementary Table S1C,D). Despite only marginal significance (10% overlap in differentially expressed genes, p ≤ 0.05), which is generally the case when the same phenotype is profiled by different labs using different technologies (13, 23), MRs inferred from these datasets were almost perfectly overlapping (77% identical MRs in the top 13, p = 0.01 by Fisher’s exact test, FET), for both the high and the low MYC activity groups (Table 1). This is consistent with previous reports on the algorithm; see (13, 23).
Figure 2. MR inference and mRNA expression after silencing of activated MRs in SUDHL6 cell line.
(A) MR of high MYC activity patients; (B) MR of low MYC activity patients. Genes are sorted by NES; red, activated-MR; blue, repressed-MR. (C) qRT-PCR analysis of MR mRNA levels at 20h after siRNA silencing in SUDHL6. Relative mRNA expression levels of activated-MR and repressed-MR were normalized to GAPDH. Heatmap represents average fold change normalized to control non-target siRNA. Blue, downregulation; white, no change; red, upregulation (Supplementary Table S2).
Activated Master Regulators are functional drivers of the FL transformation signature
MARINa identified both activated and repressed candidate MRs of FL transformation (Fig. 2, Supplementary Table S1A, B). To experimentally validate these results we concentrated on the high-MYC activity subtype, including 13 activated (FOXM1, MYC, ENO1, TFDP1, NFE2L2, E2F3, NFYB, CREM, MAZ, NR1H3, ATF5, HMGA1, CEBPG), and 5 repressed MRs (SP3, JUN, BPTF, RBL2, KLF12) (Supplementary Table S1A). We selected two t(14;18)-positive DLBCL cell lines, SUDHL6 (24) and SUDHL4 (25) (henceforth t-DLBCL cells), representing Germinal Center B-cell type (GCB) tumors, harboring the canonical BCL2 translocation t(14;18)(q32;q21), the FL-derived cell line HF1 (26), and as a control an immortalized lymphoblastoid cell line LCL-CB33 (27). There are no confirmed cell lines representative of transformed FL. As a result, we used the t(14;18)-positive DLBCL cell lines as the model that best recapitulates the aberrant activity of the master regulators we have identified from the analysis of transformed FL in patients. SUDHL6 and SUDHL4 cells clustered with high-MYC activity FL-transformed patients (Supplementary Fig. S1A, B). mRNA expression levels of activated-MRs were evaluated by qRT-PCR in all B-lymphoma cell lines (Supplementary Fig. S2A). Two genes, NF1H3 and CREM were excluded from the study due to insufficient mRNA expression and availability of Dharmacon library siRNAs, respectively. Activated MRs were systematically silenced by nucleoporation with the targeting siRNA pool in SUDHL6 cells (Supplementary Fig. S2B). These express high levels of MYC protein and overall highest levels of activated MR mRNA, among all tested DLBCL cell lines (Supplementary Fig. S2A). As expected, individual MR silencing, confirmed by qRT-PCR, significantly affected the expression of several other MARINa-inferred MRs, (Fig. 2C and Supplementary Table S2), suggesting cooperative activity, as a regulatory module, and supporting their inference as functional drivers of FL transformation.
Specifically, individual activated-MR silencing inhibited and activated expression of several other activated and repressed MRs, respectively, consistent with their inference as positive regulators of FL-transformation signature. To select dominant activated-MR genes, representing the most upstream regulators in the FL-transformation control module, we ranked them based on their overall effect on other MARINa-inferred MRs, based on qRT-PCR data (Supplementary Table S2B). Following each MR silencing, gene expression of all other MRs was log-transformed and discretized into 3 states: H (fold-change, FC > 0.5), L (FC < −0.5), and M for the others. Silenced MRs were ranked by average effect on all other MRs, using their discretized state. The five highest ranking MRs (FOXM1, TFDP1, ATF5, HMGA1 and NFYB) were selected for further study (henceforth, selected MRs).
Differential MR protein expression is confirmed in patient TMAs
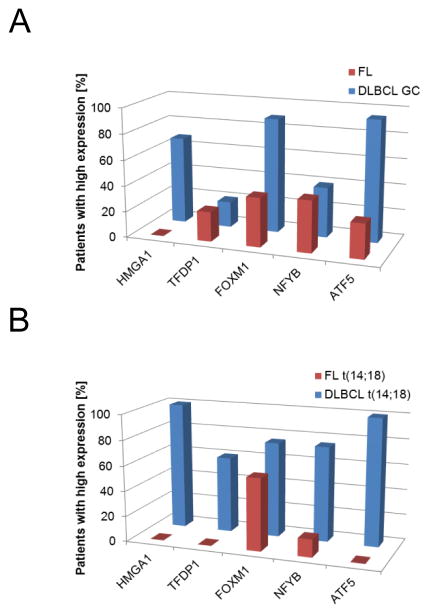
To assess MR protein levels in patients, we compared tissue microarray sections (TMA) from patients diagnosed with DLBCL against those with FL, by immunostaining with specific antibodies (Supplementary Fig. S3A). TMAs were evaluated by a board-certified pathologist and scored using a two tier scale: negative < 5% and positive ≥ 5% positive cells. Staining patterns were analyzed in two complementary ways. First, we compared DLBCL samples identified by common diagnostic markers used to identify GCB-like tumors (BCL2+, CD10+, BCL6+, MUM-) to FL samples (Fig. 3A). The analysis revealed that HMGA1, FOXM1 and ATF5 were statistically significantly overexpressed in GCB-DLBCL vs. FL patients, while TFDP1 and NFYB were not significant (Table 2). Next, we compared only DLBCL patients with the canonical t(14:18) translocation, as detected by FISH, to FL patients (Fig. 3B) to identify those more likely to represent FL transformation. These results confirmed that all selected MRs, including NFYB and TFDP1, had higher protein expression in FL-transformed patients, although the difference for FOXM1 was not statistically significant. These results were consistent with protein expression data from the FL cell line HF1 compared with high MYC t-DLBCL cell lines (Supplementary Fig. S2B), suggesting that overexpression of the selected MRs in FL-transformed patients is associated with the process of transformation and that TFDP1 and NFYB are uniquely overexpressed following FL-transformation.
Figure 3. Protein expression of activated MRs in DLBCL patients.
Protein expression by immunohistochemistry analysis in TMAs from FL and DLBCL patients. (A) Expression of MRs in FL and DLBCL patients defined by GCB markers. (B) Expression of MRs in FL and DLBCL patients with t(14;28) translocation. Bars represent number of patients with positive MR expression. See Table 2 for details.
Activated MR are synergistic drivers of DLBCL cell proliferation
A hallmark of FL transformation is an increase in cellular proliferation, which usually correlates with higher MYC expression and activity (20). To evaluate an involvement of the selected MRs in cell proliferation in vitro we assessed viability following their siRNA-pool mediated silencing in SUDHL6, SUDHL4, HF1 and CB33 cell lines. As confirmed by Western blot and qRT-PCR, significant mRNA and protein level reduction was observed at 20h (Supplementary Fig. S2C, D).
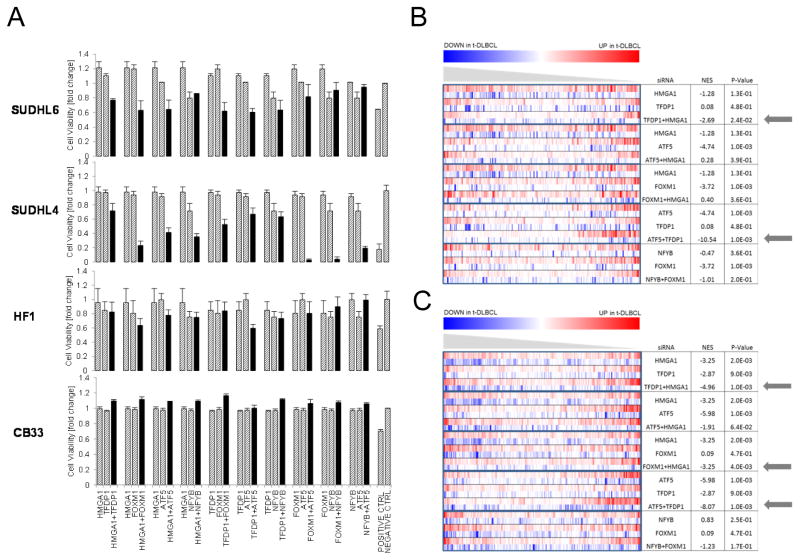
MARINa-inferred MRs frequently participate in synergistic regulation, consistent with their activity within a common regulatory module (13, 14). As a result, we also performed siRNAs mediated co-silencing of all 10 possible selected MR-pairs (Fig. 4A). siRNA targeting Ubiquitin B or AllStars Cell Death Control siRNA were used as positive controls. Results were normalized to control scrambled siRNA (NT). Confirming previous studies (13–15), while individual MR silencing (except for ATF5) did not significantly reduce cell viability at 24h, in either DLBCL cell line, MR co-silencing profoundly affected cell viability for most MR-pairs, (7/10) in SUDHL6 and (10/10) in SUDHL4, supporting their role as candidate synergistic dependencies of t-DLBCL cells. In contrast, there was only minimal effect in control cells, (2/10) in HF1 cells and 0/10 in CB33 cells. This suggests that FL transformation results from and is dependent on the cooperative effect of multiple dysregulated TFs and not from aberrant activity of any individual one in isolation.
Figure 4. Synergistic MR drive t-DLBCL cell proliferation and define transformed FL gene signature.
(A) Cell viability measured by PrestoBlue at 24h after single and paired MRs silencing of in SUDHL6, SUDHL4, HF1 and CB33 cell lines. (B) GSEA of differentially expressed genes after MR silencing in SUDHL6 cell line in patient-derived signatures of FL transformation.
Gene expression analysis of t-DLBCL cells following MR silencing
To further validate the role of inferred MRs in FL-transformation, we analyzed the gene expression signature of t-DLBCL cells following both individual MR and MR-pair silencing. Specifically, if some of the MARINa-inferred MRs are bona fide causal determinants of FL-transformation, their silencing should at least partially abrogate the FL-transformation signature, with a more profound effect when MR-pairs are co-silenced. To test our hypothesis, we selected the five MR-pairs producing the most significant synergistic cell viability reduction in both t-DLBCL cell lines, including HMGA1/TFDP1, HMGA1/ATF5, HMGA1/FOXM1, ATF5/TFDP1, and NFYB/FOXM1, and performed gene expression profiles of SUDHL6 cells at 20h following silencing of each MR and MR-pair (Fig. 4B, C) (Supplementary Methods). Differential gene expression analysis, with respect to NT control SUDHL6 cells, was performed using a SAM test (28). We used GSEA analysis to assess whether genes differentially expressed following MR silencing were negatively enriched in genes differentially expressed in patients following FL-transformation in the high-MYC subtype, using both independent studies (16, 20) (Fig. 4B,C). We also compared normalized enrichment score (NES) for individual MR silencing vs. MR-pair silencing, to further evaluate the synergistic nature of MR regulation. Analysis of patient signatures from both datasets (16, 20) consistently identified two-pairs (HMGA1/TFDP1 and ATF5/TFDP1) as effecting the most striking reversal of t-DLBCL cell gene expression to an FL-like state. For both pairs, co-silencing significantly out-performed individual MR silencing, both by NES and/or p-value assessment. Additionally, the HMGA1/FOXM1 pair was identified and experimentally validated as a candidate synergistic based on signatures from Dataset 2 (20). Taken together, these assays show that most MARINa-inferred MRs indeed regulate genes in the FL-transformation signature, even though, individually, their effect is not sufficient to induce FL-transformation signature collapse. Indeed genes differentially expressed following individual MRs silencing, including HMGA1, TFDP1 and ATF5, were significantly enriched in genes expressed in FL patients before transformation. In sharp contrast, consistent with previous results in other tumors, MR-pairs silencing leads to FL-transformation signature collapse and significant loss of cell viability at 24h.
Targeting FL-transformation MRs with small molecule perturbations
Identification of MR proteins representing novel functional drivers of tumor-related phenotypes may open relevant therapeutic opportunities (18). As proof of concept, we thus proceeded to assess whether B-cell-specific inhibitors of validated MRs could be systematically identified from small molecule perturbation assays. Following the Connectivity Map rationale (29), we reasoned that the differential expression signature following MR-silencing in human B cells represent an ideal multiplexed gene reporter assays to assess the activity of candidate small molecule inhibitors of the same MR. Since TF-targets are highly conserved across 18 distinct subtypes of human B cells, including FL and DLBCL (the rationale for using the B cell interactome for this analysis) (30), we proceeded to assess 92 compounds for which GEPs were available following perturbation of an ABC (OCI-LY3) and a GCG (OCI-LY7) DLBCL cell lines (31). Specifically, we assessed enrichment of compound-induced signatures in genes differentially expressed following siRNA-mediated silencing of validated FL-transformation MRs in SUDHL6 cells, using a two-tail GSEA (Supplementary Methods) to account for both over and under-expressed genes, to identify compounds that significantly recapitulate relevant MR silencing (Supplementary Table S3A).
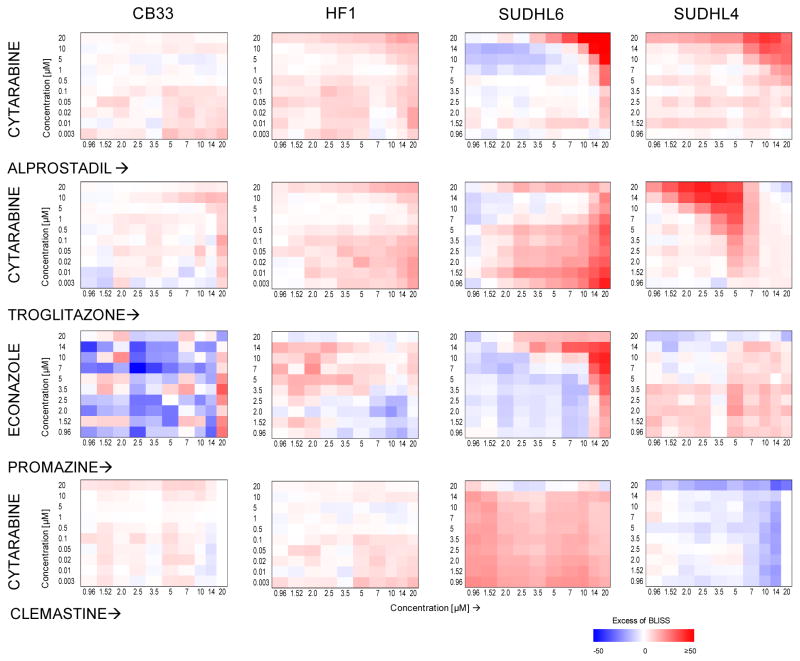
Since individual MR silencing had little effect on tumor viability, we prioritized four compound combinations predicted to target the synergistic MR-pairs inducing greatest viability reduction in SUDHL6 and SUDHL4 cells. These were tested in SUDHL4 and SUDHL6 (t-DLBCL), HF1 (FL), and LCL-CB33 (normal control) cells (Table S3B). For each combination we used a 10×10 dilution matrix, with individual compound concentrations ranging from 0.003 μM to 20 μM (Supplementary Fig. S4). Cell viability was assessed by ATP levels at 48h following compound treatment (see Materials and Methods). To evaluate compound synergy, we used the Excess-over-Bliss (EOB) score (Supplementary Methods), defined as a difference between observed and predicted additive drug combination effect (Fig. 5, Supplementary Table S5). Compound pairs were considered strongly synergistic at EOB 20. As expected, compound-pairs predicted to target synergistic MRs were strongly synergistic in t-DLBCL cells but not in FL and control cells. Among these, alprostadil/cytarabine (targeting the HMGA1/FOXM1 pair) were strongly synergistic in both t-DLBCL cell lines. Troglitazone/cytarabine (targeting the FOXM1/NFYB pair) and Econazole/promazine (targeting the HMGA1/TFDP1 pair) presented stronger synergistic activity in SUDHL4 vs. SUDHL6 cells and vice-versa, respectively, consistent with greater viability reduction following co-silencing of the associated MR-pairs. Finally, clemastine/cytarabine (both targeting FOXM1), showed no synergistic activity in any of the four cell lines, as expected.
Figure 5. Effect of drug combinations on cell viability in B-lymphoma cell lines.
Cell viability of SUDHL6, SUDHL4, HF1 and CB33 cells treated with the combination of compounds for 48hrs was evaluated by ATP assay. Compound synergy is represented by EOB score; we defined EOB 20 as strongly synergistic and EOB −20 as strongly antagonistic. Color coded matrices represent EOS scores; red – positive, blue - negative.
Although the clinical relevance of these results is limited by the relatively small number of profiled compounds and by lack of in vivo validation, all of the four predicted compound combinations induced synergistic t-DLBCL cell death. Thus, these results represent an important proof of concept that MR analysis, combined with straightforward perturbational assays, can help identify compound combinations that are effective in abrogating tumor cell viability in vitro.
Discussion
Precision cancer medicine has been almost universally predicated on the use of targeted inhibitors for oncogenes harboring activating mutations, based on the “oncogene addiction” paradigm (32). While this has been transformational for some tumors, from chronic myelogenous leukemia to lung cancer, it also presents significant limitations. Indeed, oncogene mutations are neither individually sufficient nor necessary for implementing and maintaining molecularly distinct tumor subtypes. Consistently, there are no fully penetrant genomic alterations responsible for inducing FL-transformation of every high- or low-MYC subtype patient, even though the gene expression signatures of patients undergoing FL-transformation to either subtype are virtually identical, suggesting a common, conserved functional regulators set.
As a result, we decided to approach FL-transformation from a different and highly complementary perspective. Rather than looking for recurrent genetic or epigenetic alterations in a transformed patient cohort, we interrogated a B-cell specific regulatory network to identify transcriptional factors that are responsible for the specific regulation of genes that are differentially expressed following patient FL-transformation to either the low-MYC or high-MYC subtype. As previously shown for glioma, breast cancer, and even Alzheimer’s disease, and as confirmed by DIGGIT analysis (18), the MRs of FL-transformation were downstream of most previously reported genetic alterations, including CARD11, CD79A, CD79B, STAT3, CREBBP, TNFRSF14, SOCS1, BCL10, PRKCB and PLCG2 (data not shown) (10). As a result, they represent non-oncogene dependencies, as proposed in (33), whose aberrant regulatory activity is the result of one or more genetic or epigenetic alterations in their upstream pathways.
Even though the MARINa algorithm has already been effectively used to elucidate novel functional drivers in glioblastoma, prostate cancer, leukemia, and breast cancer, our study presents significant novelty in two distinct areas. First, we report a novel tumor checkpoint, comprising 18 MR proteins, whose synergistic activity regulates the genes that are differentially expressed in FL patients following transformation. Co-inhibition of activated MR-pairs induces rapid and specific cell death in t-DLBCL cells, but not in FL related and normal related B cells. Second, we used the MR-silencing signature to elucidate compounds that, in combinations, may induce t-DLBCL specific cell death, opening a new avenue in precision cancer medicine, especially for phenotypes lacking a canonical targetable oncogene dependency.
Critically, both the B-cell specific regulatory model and the FL transformation signatures were derived from primary patient tissue. Thus, our predictions are independent of potentially idiosyncratic cell line or mouse model dependencies and should have high likelihood of being further recapitulated in vivo. Our model for the transformation process is consistent with the recently proposed linear evolution model (9), suggesting that transformed FL originates from the dominant FL clone as a result of new oncogenic events. However, since MARINa analysis is agnostic to the underlying tumor progression mechanism and only predicts the regulatory proteins that become aberrantly activated as a result of these secondary events our findings would equally support alternative hypotheses, such as divergent evolution.
Confirming results from previous studies (13, 14, 18), MARINa-inferred MRs found to be highly enriched in synthetic lethal pairs. While several of these genes were previously reported as overexpressed in hematologic malignancies, such as MYC (34), FOXM1 (35), TFDP1 (36) or ATF5 (37), the majority of inferred MRs were not previously causally associated with FL transformation nor were they shown to represent individual/synergistic dependencies of transformed DLBCL. Five of these MRs emerged as the strongest causal determinant of FL-transformation signature, including FOXM1, TFDP1, ATF5, HMGA1, MYC and NFYB. Interestingly, with the exception of MYC, these genes had been previously reported among 26 MRs of the germinal center reaction (15), suggesting that dysregulation of proteins presiding over B-cell maturation programs by a complex landscape of genetic and epigenetic alterations may be responsible for transformation of GCB-originated FL to DLBCL. Immunohistochemistry assays in patient-derived TMAs confirmed over-expression of the 5 MR proteins in DLBCL patients harboring the canonical t(14;18)(q32;q21) translocation, compared to FL tumors.
We showed that MRs act synergistically to preserve the transformed FL state. Indeed siRNA-mediated co-silencing of MR-pairs, including HMGA1/TFDP1 and ATF5/TFDP1, and FOXM1/HMGA1, had a profound effect on t-DLBCL cell viability but not on that of FL and lymphoblastoid cell lines. Consistently, analysis of GEPs from SUDHL6 cells following co-silencing of these MR-pairs showed a significant shift toward an FL-like signature. Slight differences in the analyses were likely associated with the differences in Lymphochip cDNA gene sets used in these analyses (10,731 and 4,908 genes respectively).
HMGA1/TFDP1 and ATF5/TFDP1 pairs were consistently identified from both patient signatures. HMGA1 proteins are members of a nonhistone, chromatin binding protein family, detected at high level during the process of embryogenesis in contrast to normal adult tissues and associated with a variety of aggressive human malignancies (reviewed in (38)). A recent study of HMGA1’s role in reprograming somatic cell into pluripotent stem cell (39) suggests that HMGA1 could be an important master regulator of neoplastic transformations, responsible for tumor state plasticity and reprogramming. Yet, our study represents the first where HMGA1 is identified as a mechanistic regulator of FL-transformation and as a potentially synergistic cofactor. TFDP1 is an established heterodimerization partner of E2F family proteins, regulating transcriptional activity of cell cycle progression genes (40). Both TFDP1 and E2F1 can interact and inhibit transcriptional activity of p53 and are expressed in non-Hodgkin lymphomas (36). A member of the E2F family, E2F3, was also inferred as an activated MR by MARINa analysis suggesting that these proteins may represent interacting partners in FL progression. Lastly, ATF5 is a member of the ATF/CREB family of TFs, widely expressed in neoplastic and normal tissues, however only in neoplastic cells was silencing of ATF5 shown to induce cell death (41). ATF5 was also associated with sensitivity to bortezomib-induced apoptosis in SUDHL6, but not SUDHL4 DLBCL cell lines (37), nonetheless it was never reported in the context of FL transformation.
Precise characterization of MR genes representing individual/synergistic oncogene and non-oncogene dependencies of transformed DLBCL opens a range of novel opportunities for targeted pharmacological treatment. Here we demonstrate that relevant MR inhibitors, likely operating indirectly, could be effectively inferred from the analysis of GEPs following small molecule perturbation in representative cell lines.
Specifically, based on our previously ascertained conservation of regulatory interactions across 18 distinct human B cell subtypes, including FL and DLBCL, we used GEPs of DLBCL cell lines following treatment with a library of 92 FDA approved compounds to infer novel candidate inhibitors of FL-transformation MRs. Our analysis identified several compound combinations that were experimentally validated, showing synergistic activity in t-DLBCL cells but not in normal or FL derived cells. Remarkably, even though this study represents only a proof of concept, it prioritized cytarabine, a drug frequently used in combination therapy for the treatment of acute leukemias and lymphomas (42, 43). Indeed, high-dose cytarabine, in combination with cisplatin and dexamethasone (DHAP), etoposide, cisplatin and methylprednisolone (ESHAP), is representative of key chemotherapeutic regimens for NHL and HL treatment (43). Our study identified the novel synergistic interaction of cytarabine and alprostadil (Prostaglandin E1) or troglitazone to induce cell death in DLBCL cells but not in FL and control cells. Prostaglandins are hormone-like lipid metabolites, playing a key role in inflammatory response (44). Although prostaglandins are associated with wide range of cancers they were also shown to induce apoptosis in human leukemia cell lines (45). Troglitazone is an anti-inflammatory drug, initially used for treatment of patients with Type 2 diabetes. It activates peroxisome proliferator-activated receptors (PPARs) and decreases Nf-κB (46). Since PPAR-gamma agonists were shown to induce apoptosis in human B lymphomas (47), troglitazone could prove a very realistic choice in transformed FL. Due to adverse events this drug was withdrawn from the market in 2000. Yet, new troglitazone derivatives with lower toxicity and anti-proliferative activity are now emerging (48). Two other compounds, econazole nitrate and promazine hydrochloride, showed synergy in the SUDHL6 cell line. Econazole is best known as an antifungal medication but there is increasing evidence that it may also have anticancer properties (49). Moreover, it was shown that sensitivity to econazole is specifically mediated by MYC in the HL60 cell line. Indeed MYC negative cells were resistant to this agent (50).
Taken together, these data suggest that the systematic, network-based identification of MR genes may represent an alternative and highly complementary approach to the targeting of classical oncogene dependencies. Once these dependencies are identified, their small molecule inhibitors, including both individual drugs and synergistic drug combinations, can be effectively prioritized. If further validated, such an approach would significantly extend the reach of precision cancer medicine, especially since the analyses performed in this manuscript can be performed in hours to days using high-performance computing platforms. This would allow the efficient prioritization of compound and compound combinations to treat individual tumors.
Supplementary Material
Acknowledgments
Financial support: This work was supported by MAGNeT (Multiscale Analysis of Genomic and Cellular Networks - http://magnet.c2b2.columbia.edu/) grant (5U54CA121852)
We would like to thank Riccardo Dalla-Favera and Laura Pasqualucci for providing cell lines (CB33, SUDHL4 and SUDHL6) and gene expression data for 226 B-cell samples including cell lines and FISH data for BCL2 translocation in DLBCL samples.
Footnotes
Disclosure of Potential Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.A clinical evaluation of the International Lymphoma Study Group classification of non-Hodgkin’s lymphoma. The Non-Hodgkin’s Lymphoma Classification Project. Blood. 1997;89(11):3909–18. Epub 1997/06/01. [PubMed] [Google Scholar]
- 2.Guirguis HR, Cheung MC, Piliotis E, Spaner D, Berinstein NL, Imrie K, et al. Survival of patients with transformed lymphoma in the rituximab era. Annals of hematology. 2014;93(6):1007–14. doi: 10.1007/s00277-013-1991-y. [DOI] [PubMed] [Google Scholar]
- 3.Vaandrager JW, Schuuring E, Raap T, Philippo K, Kleiverda K, Kluin P. Interphase FISH detection of BCL2 rearrangement in follicular lymphoma using breakpoint-flanking probes. Genes Chromosomes Cancer. 2000;27(1):85–94. Epub 1999/11/24. [PubMed] [Google Scholar]
- 4.Limpens J, Stad R, Vos C, de Vlaam C, de Jong D, van Ommen GJ, et al. Lymphoma-associated translocation t(14;18) in blood B cells of normal individuals. Blood. 1995;85(9):2528–36. Epub 1995/05/01. [PubMed] [Google Scholar]
- 5.Montoto S, Davies AJ, Matthews J, Calaminici M, Norton AJ, Amess J, et al. Risk and clinical implications of transformation of follicular lymphoma to diffuse large B-cell lymphoma. J Clin Oncol. 2007;25(17):2426–33. doi: 10.1200/JCO.2006.09.3260. Epub 2007/05/09. [DOI] [PubMed] [Google Scholar]
- 6.Freedman AS. Biology and management of histologic transformation of indolent lymphoma. Hematology/the Education Program of the American Society of Hematology American Society of Hematology Education Program. 2005:314–20. doi: 10.1182/asheducation-2005.1.314. Epub 2005/11/24. [DOI] [PubMed] [Google Scholar]
- 7.Goff LK, Neat MJ, Crawley CR, Jones L, Jones E, Lister TA, et al. The use of real-time quantitative polymerase chain reaction and comparative genomic hybridization to identify amplification of the REL gene in follicular lymphoma. Br J Haematol. 2000;111(2):618–25. doi: 10.1046/j.1365-2141.2000.02352.x. Epub 2000/12/21. [DOI] [PubMed] [Google Scholar]
- 8.Berglund M, Enblad G, Thunberg U, Amini RM, Sundstrom C, Roos G, et al. Genomic imbalances during transformation from follicular lymphoma to diffuse large B-cell lymphoma. Mod Pathol. 2007;20(1):63–75. doi: 10.1038/modpathol.3800708. Epub 2006/12/16. [DOI] [PubMed] [Google Scholar]
- 9.Pasqualucci L, Khiabanian H, Fangazio M, Vasishtha M, Messina M, Holmes AB, et al. Genetics of follicular lymphoma transformation. Cell reports. 2014;6(1):130–40. doi: 10.1016/j.celrep.2013.12.027. Epub 2014/01/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okosun J, Bodor C, Wang J, Araf S, Yang CY, Pan C, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nature genetics. 2014;46(2):176–81. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayslip J, Montero A. Tumor suppressor gene methylation in follicular lymphoma: a comprehensive review. Mol Cancer. 2006;5:44. doi: 10.1186/1476-4598-5-44. Epub 2006/10/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rawal S, Chu F, Zhang M, Park HJ, Nattamai D, Kannan S, et al. Cross talk between follicular Th cells and tumor cells in human follicular lymphoma promotes immune evasion in the tumor microenvironment. J Immunol. 2013;190(12):6681–93. doi: 10.4049/jimmunol.1201363. Epub 2013/05/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–25. doi: 10.1038/nature08712. Epub 2009/12/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aytes A, Mitrofanova A, Lefebvre C, Alvarez MJ, Castillo-Martin M, Zheng T, et al. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer cell. 2014;25(5):638–51. doi: 10.1016/j.ccr.2014.03.017. Epub 2014/05/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefebvre C, Rajbhandari P, Alvarez MJ, Bandaru P, Lim WK, Sato M, et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Molecular systems biology. 2010;6:377. doi: 10.1038/msb.2010.31. Epub 2010/06/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lossos IS, Alizadeh AA, Diehn M, Warnke R, Thorstenson Y, Oefner PJ, et al. Transformation of follicular lymphoma to diffuse large-cell lymphoma: alternative patterns with increased or decreased expression of c-myc and its regulated genes. Proc Natl Acad Sci U S A. 2002;99(13):8886–91. doi: 10.1073/pnas.132253599. Epub 2002/06/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nature genetics. 2005;37(4):382–90. doi: 10.1038/ng1532. Epub 2005/03/22. [DOI] [PubMed] [Google Scholar]
- 18.Chen JC, Alvarez MJ, Talos F, Dhruv H, Rieckhof GE, Iyer A, et al. Identification of Causal Genetic Drivers of Human Disease through Systems-Level Analysis of Regulatory Networks. Cell. 2014;159(2):402–14. doi: 10.1016/j.cell.2014.09.021. Epub 2014/10/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hedvat CV, Hegde A, Chaganti RS, Chen B, Qin J, Filippa DA, et al. Application of tissue microarray technology to the study of non-Hodgkin’s and Hodgkin’s lymphoma. Hum Pathol. 2002;33(10):968–74. doi: 10.1053/hupa.2002.127438. Epub 2002/10/24. [DOI] [PubMed] [Google Scholar]
- 20.Davies AJ, Rosenwald A, Wright G, Lee A, Last KW, Weisenburger DD, et al. Transformation of follicular lymphoma to diffuse large B-cell lymphoma proceeds by distinct oncogenic mechanisms. Br J Haematol. 2007;136(2):286–93. doi: 10.1111/j.1365-2141.2006.06439.x. Epub 2007/02/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basso K, Saito M, Sumazin P, Margolin AA, Wang K, Lim WK, et al. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood. 2010;115(5):975–84. doi: 10.1182/blood-2009-06-227017. Epub 2009/12/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim WK, Lyashenko E, Califano A. Master regulators used as breast cancer metastasis classifiers. Pac Symp Biocomput. 2009;14:504–15. [PMC free article] [PubMed] [Google Scholar]
- 24.Siminovitch KA, Jensen JP, Epstein AL, Korsmeyer SJ. Immunoglobulin gene rearrangements and expression in diffuse histiocytic lymphomas reveal cellular lineage, molecular defects, and sites of chromosomal translocation. Blood. 1986;67(2):391–7. Epub 1986/02/01. [PubMed] [Google Scholar]
- 25.Bakhshi A, Jensen JP, Goldman P, Wright JJ, McBride OW, Epstein AL, et al. Cloning the chromosomal breakpoint of t(14;18) human lymphomas: clustering around JH on chromosome 14 and near a transcriptional unit on 18. Cell. 1985;41(3):899–906. doi: 10.1016/s0092-8674(85)80070-2. Epub 1985/07/01. [DOI] [PubMed] [Google Scholar]
- 26.Knuutila S, Klefstrom J, Szymanska J, Lakkala T, Peltomaki P, Eray M, et al. Two novel human B-cell lymphoma lines of lymphatic follicle origin: cytogenetic, molecular genetic and histopathological characterisation. European journal of haematology. 1994;52(2):65–72. doi: 10.1111/j.1600-0609.1994.tb01288.x. [DOI] [PubMed] [Google Scholar]
- 27.Lombardi L, Newcomb EW, Dalla-Favera R. Pathogenesis of Burkitt lymphoma: expression of an activated c-myc oncogene causes the tumorigenic conversion of EBV-infected human B lymphoblasts. Cell. 1987;49(2):161–70. doi: 10.1016/0092-8674(87)90556-3. [DOI] [PubMed] [Google Scholar]
- 28.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–35. doi: 10.1126/science.1132939. Epub 2006/09/30. [DOI] [PubMed] [Google Scholar]
- 30.Mani KM, Lefebvre C, Wang K, Lim WK, Basso K, Dalla-Favera R, et al. A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas. Molecular systems biology. 2008;4:169. doi: 10.1038/msb.2008.2. Epub 2008/02/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woo HJ, Shimoni Y, Yang SW, Subramaniam P, Iyer A, Nicoletti P. Elucidating Compound Mechanism of Action by Network Dysregulation Analysis in Perturbed Cells. Cell. 2015 doi: 10.1016/j.cell.2015.05.056. In submission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297(5578):63–4. doi: 10.1126/science.1073096. Epub 2002/07/06. [DOI] [PubMed] [Google Scholar]
- 33.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136(5):823–37. doi: 10.1016/j.cell.2009.02.024. Epub 2009/03/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yano T, Jaffe ES, Longo DL, Raffeld M. MYC rearrangements in histologically progressed follicular lymphomas. Blood. 1992;80(3):758–67. Epub 1992/08/01. [PubMed] [Google Scholar]
- 35.Uddin S, Hussain AR, Ahmed M, Siddiqui K, Al-Dayel F, Bavi P, et al. Overexpression of FoxM1 offers a promising therapeutic target in diffuse large B-cell lymphoma. Haematologica. 2012;97(7):1092–100. doi: 10.3324/haematol.2011.053421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan JA, Olvera M, Lai R, Naing W, Rezk SA, Brynes RK. Immunohistochemical expression of the transcription factor DP-1 and its heterodimeric partner E2F-1 in non-Hodgkin lymphoma. Applied immunohistochemistry & molecular morphology : AIMM/official publication of the Society for Applied Immunohistochemistry. 2002;10(4):322–6. doi: 10.1097/00129039-200212000-00006. [DOI] [PubMed] [Google Scholar]
- 37.Shringarpure R, Catley L, Bhole D, Burger R, Podar K, Tai YT, et al. Gene expression analysis of B-lymphoma cells resistant and sensitive to bortezomib. Br J Haematol. 2006;134(2):145–56. doi: 10.1111/j.1365-2141.2006.06132.x. [DOI] [PubMed] [Google Scholar]
- 38.Shah SN, Resar LM. High mobility group A1 and cancer: potential biomarker and therapeutic target. Histology and histopathology. 2012;27(5):567–79. doi: 10.14670/HH-27.567. [DOI] [PubMed] [Google Scholar]
- 39.Shah SN, Kerr C, Cope L, Zambidis E, Liu C, Hillion J, et al. HMGA1 reprograms somatic cells into pluripotent stem cells by inducing stem cell transcriptional networks. PloS one. 2012;7(11):e48533. doi: 10.1371/journal.pone.0048533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Connor DJ, Lam EW, Griffin S, Zhong S, Leighton LC, Burbidge SA, et al. Physical and functional interactions between p53 and cell cycle co-operating transcription factors, E2F1 and DP1. The EMBO journal. 1995;14(24):6184–92. doi: 10.1002/j.1460-2075.1995.tb00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monaco SE, Angelastro JM, Szabolcs M, Greene LA. The transcription factor ATF5 is widely expressed in carcinomas, and interference with its function selectively kills neoplastic, but not nontransformed, breast cell lines. International journal of cancer Journal international du cancer. 2007;120(9):1883–90. doi: 10.1002/ijc.22469. [DOI] [PubMed] [Google Scholar]
- 42.Bishop JF, Matthews JP, Young GA, Bradstock K, Lowenthal RM. Intensified induction chemotherapy with high dose cytarabine and etoposide for acute myeloid leukemia: a review and updated results of the Australian Leukemia Study Group. Leukemia & lymphoma. 1998;28(3–4):315–27. doi: 10.3109/10428199809092687. [DOI] [PubMed] [Google Scholar]
- 43.McCarthy J, Gopal AK. Successful use of full-dose dexamethasone, high-dose cytarabine, and cisplatin as part of initial therapy in non-hodgkin and hodgkin lymphoma with severe hepatic dysfunction. Clinical lymphoma & myeloma. 2009;9(2):167–70. doi: 10.3816/CLM.2009.n.039. [DOI] [PubMed] [Google Scholar]
- 44.Ricciotti E, FitzGerald GA. Prostaglandins and inflammation. Arteriosclerosis, thrombosis, and vascular biology. 2011;31(5):986–1000. doi: 10.1161/ATVBAHA.110.207449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soleymani Fard S, Jeddi Tehrani M, Ardekani AM. Prostaglandin E2 induces growth inhibition, apoptosis and differentiation in T and B cell-derived acute lymphoblastic leukemia cell lines (CCRF-CEM and Nalm-6) Prostaglandins, leukotrienes, and essential fatty acids. 2012;87(1):17–24. doi: 10.1016/j.plefa.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 46.Wolffenbuttel BH, Graal MB. New treatments for patients with type 2 diabetes mellitus. Postgraduate medical journal. 1996;72(853):657–62. doi: 10.1136/pgmj.72.853.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Padilla J, Leung E, Phipps RP. Human B lymphocytes and B lymphomas express PPAR-gamma and are killed by PPAR-gamma agonists. Clinical immunology. 2002;103(1):22–33. doi: 10.1006/clim.2001.5181. [DOI] [PubMed] [Google Scholar]
- 48.Bordessa A, Colin-Cassin C, Grillier-Vuissoz I, Kuntz S, Mazerbourg S, Husson G, et al. Optimization of troglitazone derivatives as potent anti-proliferative agents: Towards more active and less toxic compounds. European journal of medicinal chemistry. 2014;83:129–40. doi: 10.1016/j.ejmech.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 49.Ho YS, Wu CH, Chou HM, Wang YJ, Tseng H, Chen CH, et al. Molecular mechanisms of econazole-induced toxicity on human colon cancer cells: G0/G1 cell cycle arrest and caspase 8-independent apoptotic signaling pathways. Food and chemical toxicology : an international journal published for the British Industrial Biological Research Association. 2005;43(10):1483–95. doi: 10.1016/j.fct.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 50.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133(6):1106–17. doi: 10.1016/j.cell.2008.04.043. Epub 2008/06/17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.