Abstract
Three new polyketides myxotritones A-C (2–4), together with a new natural product 7,8-dihydro-7R,8S-dihydroxy-3,7-dimethyl-2-benzopyran-6-one (1) were obtained from the endolichenic fungus Myxotrichum sp. by using OMSAC (One Strain, Many Compounds) method. The planar structures of these new compounds were determined by NMR experiment and HRESIMS data, and the absolute configuration of 1 was established by X-ray diffraction, and the stereochemistry of the new compounds 2-4 were determined by same biosynthesis origin, and similar CD spectra with 1. Allelopathic test showed that compound 4 significantly retarded root elongation of Arabidopsis thaliana seed, indicating that this fungus might contribute to the defense of its host lichen. From the view of biosynthetic pathway, all four compounds 1-4 might be originated from Non-Reduced Polyketide synthase (NR-PKS).
Lichens are combinations of a fungus (the mycobiont) and an algal partner (the photobiont or phycobiont). In addition to fungal mycobionts, some nonobligate fungi, such as endolichenic fungi, are also found to live asymptomatically in the bodies (thalli) of lichens1. Although endolichenic fungi inhabit the lichen thalli similarly to endophytes living in the intercellular spaces of healthy plant tissues, the chemistry of this class of fungi remained largely unexplored2.
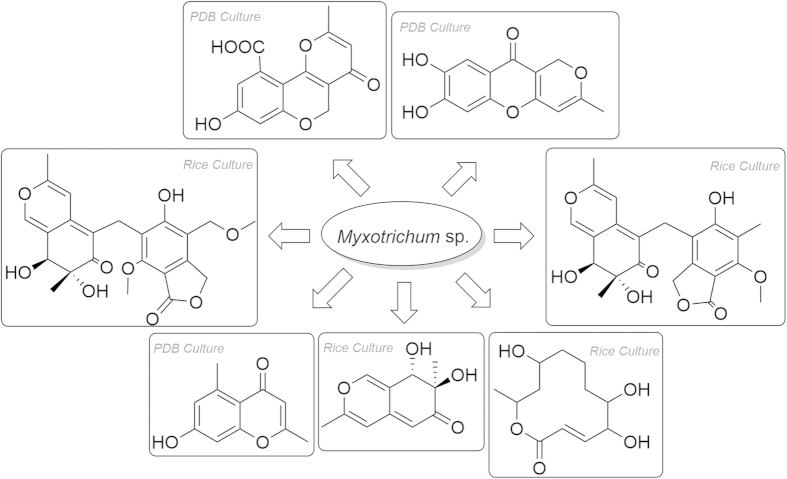
Analysis of a great number genome sequence from different microbes revealed that many secondary metabolite biosynthetic gene clusters are silent under common cultivation conditions, and their metabolic potentials were underestimated. To activate the cryptic gene cluster to express, different methods were innovated3. One easiest way is to vary the culture media to induce different cryptic gene cluster to express and then obtain new/novel secondary metabolites. This approach was termed as “One Strain, Many Compounds” first suggested by Germany natural product chemist Prof. A Zeek4. In our previous report, a series of citromycetin and fulvic acid with unique skeletons have been obtained from the PDB culture of endolichenic fungus Myxotrichum sp5 (Fig. 1). To dig the metabolic potential of endolichenic fungi Myxotrichum sp., rice culture was used to activate the potentially silent gene clusters, from which three new polyketides myxotritones A-C (2–4), together with a new natural product 7, 8-dihydro-7R, 8S-dihydroxy-3, 7-dimethyl-2-benzopyran-6-one (1) were obtained. In this report, the structural elucidation, biological evaluation, and possible biosynthetic pathway were present.
Figure 1. Diverse skeleton polyketides isolated from the endolichenic fungus Myxotrichum sp.
Results and Discussion
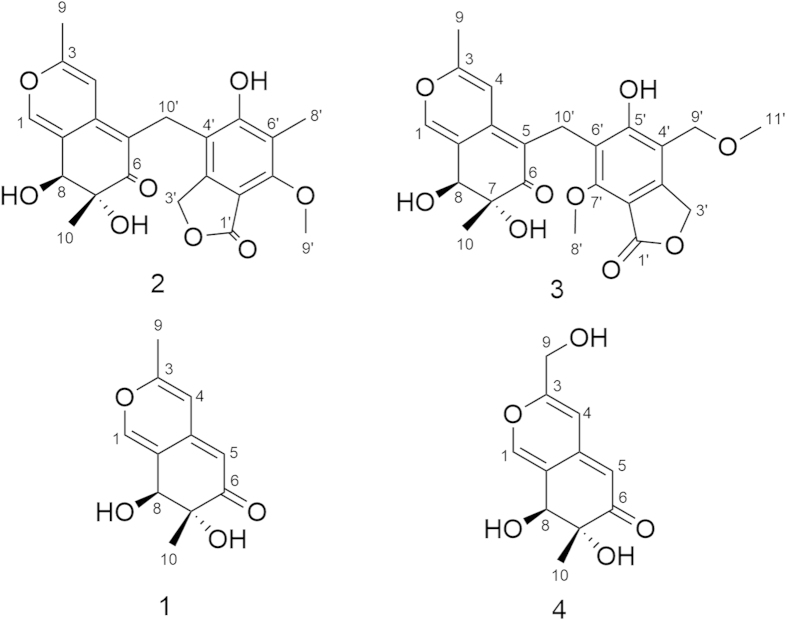
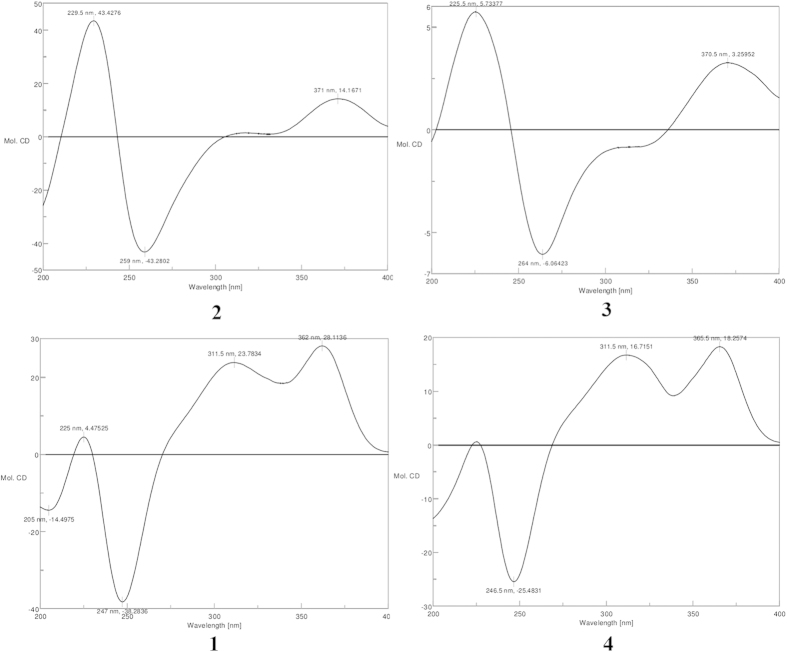
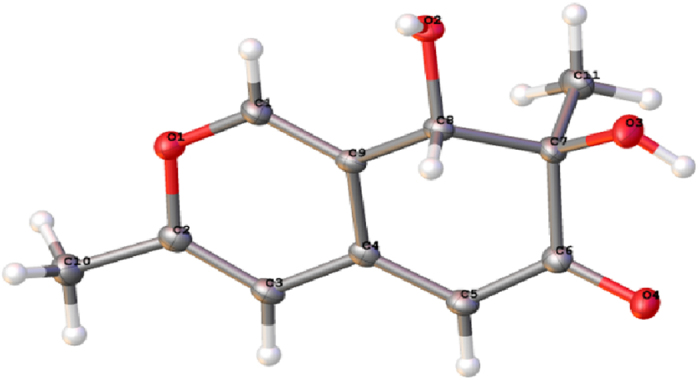
The known compound 1 is identified to be as 7, 8-dihydro-7R, 8S-dihydroxy-3, 7-dimethyl-2- benzopyran-6-one based on the NMR, MS data and optical rotation [α]D22 = +393.0 (c = 0.15, MeOH), which was known as a synthetic compound but never isolated from a natural specimen6,7 (Fig. 2). Fortunately, a suitable crystal was obtained for X-ray diffraction (in MeOH) (Fig. S5). The planar structure and absolute configuration of 1 were confirmed by single-crystal X-ray diffraction analysis with Cu Kα radiation (Fig. 3, CCDC 1419081). The CD spectrum of 1 showed the positive (362 nm, 311 nm and 225 nm) and negative (247 nm) cotton effects (Fig. S4) similar with those of known azaphilones clearly showed (R)-configuration of chiral center at C-78,9,10. Snatzke’ rule was also used to determine the diol of C-7 and C-811,12,13. The positive cotton effect at 327 nm observed in situ dimolybdenum CD spectra permitted the assignment of absolute configuration as 7R, 8S (Fig. S4).
Figure 2. Structures of compounds (1–4).
Figure 3. X-ray crystallographic structure of (1).
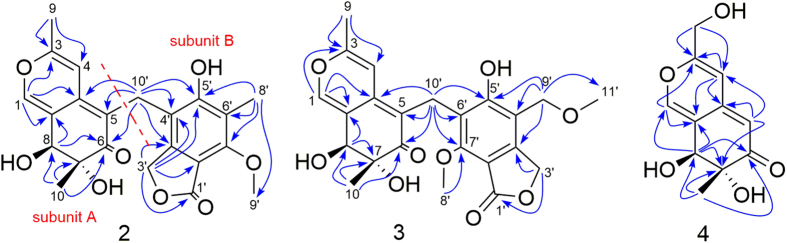
Myxotritone A (2) was isolated as a yellow powder, [α]D22 = +45.6 (c = 0.125, MeOH). Its molecular formula was determined as C22H22O8 (12 degrees of unsaturation) by TOF-ESI-MS spectral data, which showed a pseudomolecular ion at m/z 437.1209 [M + Na]+ (Fig. S10). The UV spectrum of 2 displayed the maximum absorptions at 217 nm (log ε 4.21), 256 nm (log ε 3.86) and 364 nm (log ε 3.97) (Fig. S12), revealing the presence of an extended conjugated system as the characteristic of azaphilones. The 1H, 13C NMR and HMQC spectra revealed that 1 contained four methyls (one methoxyl group), two methylenes with one oxygenated, an oxymethine unit, an oxygenated quaternary carbon, 12 olefinic carbons, an ester carbonyl carbon, and a keto carbonyl group, which explained all carbon signals of 2. Analysis of the 1H and 13C-NMR data of 2 revealed the same structural fragment (subunit A) as 1, except the H-5 in 1 was replaced by other moiety in 2, and this conclusion was supported by HMBC correlations (Fig. 4). The remaining connectivity was solved by detailed analysis of HMBC spectrum. The correlations from 10′-CH2- to C-3′a, C-4′ and C-5′, from 3′-CH2- to C-3′a, C-4′ and C-7′a, 8′-CH3 to C-5′, C-6′ and C-7′ together with correlations of the 9′-methoxyl with C-7′ established a hexa substituted phenyl ring. The key correlation from 3′-CH2- to the ester carbonyl and considering the chemical shift value of C-7′a (δC 107.8) led to construct an isobenzofuran-1(3H)-one fragment (subunit B). The correlations from 10′-CH2- to C-4a, C-5 and C-6 connected the subunit A with subunit B (Fig. 4). Considering the chemical shift values of C-7, C-8, and C-5′ and molecular formula, these three carbons must be anchored a free hydroxyl group, respectively. Thus the planar structure of 2 was determined. Compound 2 showed positive (371 nm, 311 nm and 229 nm) and negative (259 nm) cotton effects in the CD spectrum (Fig. 5). Based on the similar CD data and same biosynthetic pathway with 1, the relative and absolute configurations of 2 were postulated to be 7R, 8S.
Figure 4. Key HMBC correlations of myxotritone (A–C)(2–4).
Figure 5. Experimental ECD of compounds (1–4).
Myxotritone B (3) was obtained as yellow powder, [α]D22 = +6.0 (c = 0.067, MeOH). The molecular formula of 3 was deduced as C23H24O9 on the basis of its TOF-ESI-MS spectrum, in which a pseudomolecular ion was observed at m/z 467.1313 [M + Na]+ (Fig. S17). The 1H and 13C NMR spectra for 3 were similar with those of 2 except that one more methoxyl signal was observed, and the methyl anchored at the phenyl ring was disappeared in 3, which implied that the methyl group on the phenyl ring in 3 was methoxylation. Yet, careful analysis of the 1H NMR of 2 and 3 revealed that the peak shape of 10′-CH2 in 2 and in 3 was completely different: singlet in 2, whereas two doublets in 3. This phenomenon implied that the substitutes around C-10′ in 3 were different from those in 2, leading to the chemical environment change, which produced anisotropic characteristics of 10′-CH2 in 3. Thus further HMBC spectrum was done to explain the phenomenon. The HMBC correlations clearly revealed that the connection of lactone ring was changed in 3 (Fig. 4), which finally established the planar structure of 3. The relative and absolute configurations of 3 were postulated to be 7R, 8S, due to its similar CD data and same biosynthetic origin with 1.
Myxotritone C (4) was obtained as yellow needle, [α]D22 = +403.3 (c = 0.15, MeOH). Its molecular formula was assigned as C11H12O5 (6 degrees of unsaturation) by TOF-ESI-MS spectral data, which showed a pseudomolecular ion at m/z 225.0760 [M + H]+, 247.0573 [M + Na]+ (Fig. S24). The 1H and 13C NMR spectra of 4 suggested the presence of similar subunit found in 1 except for 9-methyl signal (δC 17.9/δH 2.18) replaced by a methylol signal (δC 59.9/δH 4.29). This hypothesis was further confirmed by HMBC correlations (Fig. S23) (Fig. 4). The HMBC correlations from 9-CH2OH to C-3, C-4 and C-4a confirmed that the additional methylol group was connected with C-3. Similarly, the relative and absolute configurations of 4 were also deduced as 7S and 8S.
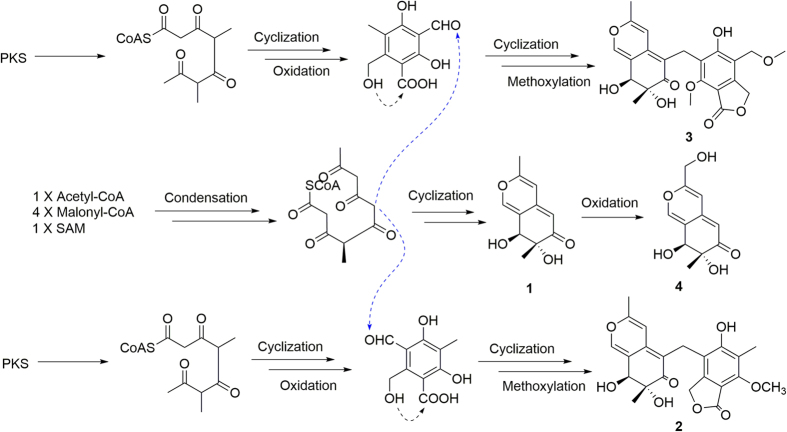
From the structural features of compounds 1–4, all these four compounds might come from same Non-Reduced Polyketide synthase (NR-PKS) origins. The putative biosynthetic pathway was suggested in the Fig. 6.
Figure 6. Possible pathway for the biosynthesis of (1–4).
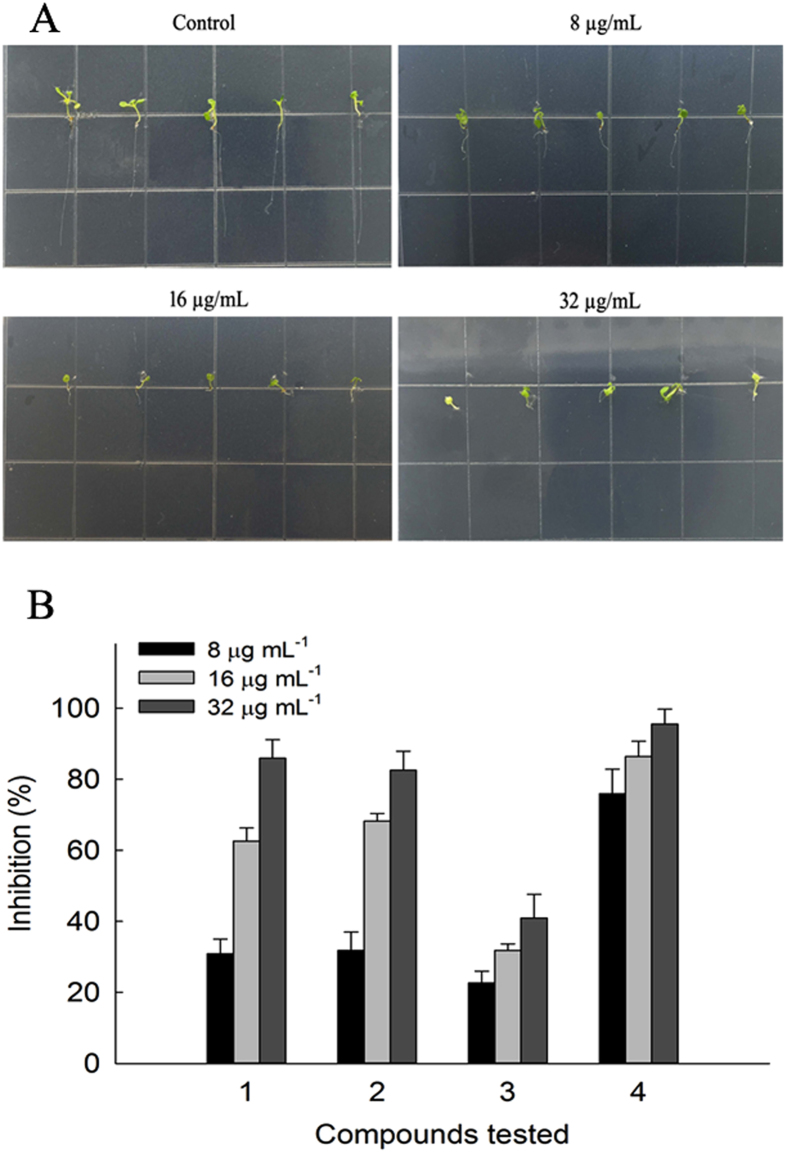
To study the potential effects of these metabolites, allelopathic potential of these compounds was tested with the root elongation of A. thaliana as model. The root growth of A. thaliana was inhibited after treatment with the compounds in a dose dependent manner (Fig. 7). The inhibition of compounds 1–4 at different concentrations was shown in Fig. 7. Compound 4 were found to retard the Arabidopsis seeds root significantly, with the inhibition rate of 75.9% at 8 μg/mL, whereas compounds 1 and 2 showed moderate inhibition activities. The results implied that this fungus might contribute to the defense of its host lichen.
Figure 7. Effect of compounds (1–4) on seedling growth of A. thaliana.
(A) The growth of A.thaliana on Petri dishes with different concentrations of compound (4) at 8, 16 and 32 μg/mL, and DMSO was used as blank control with the same volume. (B) The inhibition on A.thaliana root growth of compounds (1–4) at different concentrations.
Methods
General Experimental Procedures
Optical rotations were measured on a Perkin-Elmer 241 Polarimeter (Perkin-Elmer, Bruker, Billerica, MA, USA) and UV data were obtained on a Shimadzu Biospec-1601 spectrophotometer. CD spectra were obtained on a Chirascan spectropolarimeter. 1H and 13C NMR data were acquired using Bruker 600 and Varian Inova 600 spectrometers using solvent signals (MeOH-d4; δH 4.87, 3.31/δC 49.15) as references. HRESIMS data were acquired using a LTQ Orbitrap XL Mass Spectrometer (Thermo, Waltham, MA, USA).
Fungus and culturing condition
The endolichenic fungus Myxotrichum sp. was isolated from the lichen Cetraria islandica (L.) Ach. collected from Laojun Mountain, Yunnan Province, People’s Republic of China. The isolate was identified on the basis of the internal transcribed spacer region sequences of the rDNA (Genbank Accession No. HQ324780) (Fig. S1) and the fungus assigned the accession no.20081189 was deposited at lichen laboratory’s culture collection in College of Life Sciences, Shandong Normal University, Jinan. The fungal strain was cultured on slants of potato dextrose agar (PDA) at 25 °C for 15 days. Then, the proper fungus strain was inoculated in five Erlenmeyer flasks (500 mL) each containing 200 mL PDB (20% potato and 2% glucose) media. Flask cultures were incubated at 25 °C on a rotary shaker at 110 rpm for seven days as spore seeds. These spore seeds were used to inoculate in Fernbach flasks (500 mL), each containing 60 g of rice, and incubated at 25 °C for 40 days.
Extraction and isolation
The fermented material was extracted with ethyl acetate (6 L for four times). The solution was concentrated to dryness under vacuum to afford a crude extract (22.0 g), which was fractionated by silica gel column chromatography (10 × 100 cm) using CH2Cl2–MeOH gradient elution. The fraction (611 mg) eluted with CH2Cl2–MeOH 50:1 was separated by Sephadex LH-20 (Pharmacia, Uppsala, Sweden) column chromatography eluting with MeOH to afford 4 subfractions. The resulting subfraction 1 was further purified by semipreparative RP HPLC (Lumtech, Berlin, Germany; YMC-Pack ODS-A column; 10 μm; 250 × 10 mm; 2 mL · min−1, 46% MeOH in H2O) to afford 1 (30 mg, tR = 14.3 min), subfraction 4 was purified by RP-HPLC (Lumtech, Berlin, Germany; YMC-Pack ODS-A column; 10 μm; 250 × 10 mm; 2 mL · min−1, 70% MeOH in H2O for 20 min) to afford myxotritone A (2, 4 mg, tR = 10 min) and myxotritone B (3, 1 mg, tR = 13 min). Fraction (863 mg) eluted with CH2Cl2–MeOH 10:1 were fractionated again by Sephadex LH-20 column chromatography using MeOH as eluent. Purification of the subfraction by RP-HPLC (Lumtech; YMC-Pack ODS-A column; 10 μm; 250 × 10 mm; 2 mL · min−1,12% MeOH in H2O) afforded myxotritone C (4, 6 mg, tR = 14 min).
7R, 8S -7, 8-dihydro-7, 8-dihydroxy-3, 7-dimethyl-2- benzopyran-6-one (1): Brown powder, [α]D22 = +393.0° (c 0.15, MeOH); UV(MeOH) λmax (log ε) 349 (4.45) nm; CD (MeOH) 205 (Δε-0.21), 225 (Δε + 0.06), 247 (Δε−0.55), 311 (Δε + 0.34), 362 (Δε + 0.40) nm; IR (KBr) Vmax: 3430, 2981, 2937, 1717, 1645, 1602, 1546, 1457, 1394, 1369, 1326, 1087, 1298, 990 967, 893 cm−1; 1H NMR and 13C NMR, see Table 1; positive HRESIMS m/z 209.0813 [M + H]+ (calcd for C11H13O4, 209.0814).
Table 1. 1H (600MHz) and 13C NMR (150 MHz) spectroscopic data (MeOH-d4) of myxotritone A–C (2–4).
| No. | 2 |
3 |
4 |
|||
|---|---|---|---|---|---|---|
| δC [ppm] | δH [ppm], M(J in Hz) | δC [ppm] | δH [ppm], M(J in Hz) | δC [ppm] | δH [ppm], M(J in Hz) | |
| 1 | 145.3 | 7.48, d (1.8) | 145.9 | 7.57, d (1.8) | 145.3 | 7.47, t (1.2) |
| 2 | ||||||
| 3 | 161.1 | 161.6 | 162.3 | |||
| 4 | 104.2 | 6.69, s | 105.3 | 7.07, s | 104.9 | 6.39, s |
| 5 | 111.6 | 111.9 | 104.7 | 5.37, d (1.2) | ||
| 6 | 199.4 | 201.1 | 200.1 | |||
| 7 | 76.0 | 76.0 | 76.5 | |||
| 8 | 71.5 | 4.44, d (2.4) | 71.4 | 4.47, d (1.8) | 72.1 | 4.54, d (2.4) |
| 9 | 18.4 | 2.28, s | 18.6 | 2.34, s | 59.9 | 4.29, s |
| 10 | 17.5 | 1.11, s | 17.5 | 1.15, s | 17.7 | 1.16, s |
| 4a | 144.3 | 146.2 | 146.7 | |||
| 8a | 121.1 | 121.5 | 121.4 | |||
| 1′ | 170.5 | 170.2 | ||||
| 3′ | 69.1 | 5.28, d (15.6) 5.15, d (15.6) | 69.0 | 5.32, d (15.6) 5.28, d (15.6) | ||
| 4′ | 116.8 | 115.7 | ||||
| 5′ | 161.2 | 160.7 | ||||
| 6′ | 119.4 | 120.5 | ||||
| 7′ | 156.3 | 157.5 | ||||
| 8′ | 7.9 | 2.16, s | 61.7 | 4.05, s | ||
| 9′ | 61.0 | 3.90, s | 66.8 | 4.60, s | ||
| 10′ | 21.1 | 3.59, s | 19.0 | 3.79, d (14.4) 3.53, d (14.4) | ||
| 11′ | 57.3 | 3.40, s | ||||
| 3′a | 147.2 | 148.2 | ||||
| 7′a | 107.8 | 107.9 | ||||
Myxotritone A (2): Yellow needle, [α]D22 = +45.6 (c 0.125, MeOH), UV (MeOH) λmax (log ε) 217 (4.21) nm; 256 (3.86) nm; 364 (3.97) nm; CD (MeOH) 230 (Δε + 1.47), 259 (Δε −1.47), 371 (Δε + 0.48) nm; IR (KBr) Vmax: 3436, 2951, 2843, 1732, 1674, 1661, 1599, 1485, 1329, 1279, 1187, 1146, 1098, 1020, 962, 909, 879, 847, 825 cm−1; 1H NMR and 13C NMR, see Table 1; positive HRESIMS m/z 415.1394 [M + H]+ (calcd for C22H23O8, 415.1393), 437.1209 [M + Na] + (calcd for C22H23O8Na, 437.1212).
Myxotritone B (3): Yellow powder, [α]D22 = +6.0° (c 0.067, MeOH); UV (MeOH) λmax(log ε) 221 (3.91), 256 (3.56) and 370 (3.61) nm; CD (MeOH) 226 (Δε + 0.77), 264 (Δε -0.82), 371 (Δε + 0.44) nm; IR (KBr) Vmax: 3436, 2954, 2844, 1636, 1517, 1457, 1448, 1387, 1350, 1339, 1277, 1187, 1101, 1064, 1032, 1017, 966 cm−1; 1H NMR and 13C NMR, see Table 1; positive HRESIMS m/z 467.1313 [M + H]+ (calcd for C23H25O9Na, 467.1318).
Myxotritone C (4): Brown powder, [α]D22 = +403.3° (c 0.15, MeOH); UV (MeOH) λmax(log ε) 233 (3.86), 243 (3.83) and 348 (4.46) nm; CD (MeOH) 247 (Δε -1.94), 312 (Δε + 1.28), 366 (Δε + 1.39) nm; IR (KBr) Vmax: 3430, 2913, 2864, 1674, 1653, 1623, 1616, 1545, 1512, 1444, 1398, 1373, 1326, 1270, 1235, 1160, 1123, 1061, 1015, 970, 930, 883, 830 cm−1; 1H NMR and 13C NMR, see Table 1; positive HRESIMS m/z 225.0760 [M + H]+ (calcd for C11H13O5, 225.0762).
Seedling growth test
Arabidopsis thaliana seeds were surface sterilized by 5% sodium hypochlorite for 5 min, followed by washing with sterile distilled water for five times. Compounds were dissolved with DMSO to final concentration of 40 mg/mL. Then 20 μL of them were added to 25 mL 1/2 MS medium supplemented with 0.8% (w/v) agar to get plates with different concentrations of compounds (8, 16, 32 μg/mL). To eliminate the effect of DMSO on the growth of A. thaliana, plates with 20 μL DMSO were used as blank control. Fifteen seeds were distributed on each Petri dishes described before. Each concentration was conducted in triplicate. The Petri dishes were placed in a growth chamber at 23 ± 1 °C under light for 8 h and darkness for 6 h. The lengths of roots were measured after 9 days. The percentage of growth inhibition of root lengths was calculated as the following equation:
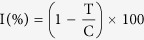 |
where T stands for the average length of treatment (cm) and C stands for the average length of control (cm)14,15.
Additional Information
How to cite this article: Yuan, C. et al. Allelopathic Polyketides from an Endolichenic Fungus Myxotrichum SP. by Using OSMAC Strategy. Sci. Rep. 6, 19350; doi: 10.1038/srep19350 (2016).
Supplementary Material
Acknowledgments
This work was financially supported by National Natural Science Foundation of China (no. 31400110), PUMC Youth Fund and the Fundamental Research Funds for the Central Universities (no. 3332015146), Program for Innovative Research Team in IMPLAD (IT1305), and Special Fund for Basic Scientific Research from Yunnan Branch of IMPLAD at CAMS & PUMC (YZYN-15-03).
Footnotes
Author Contributions Z.-M.Z. and G.D. designed the study; C.Y. and Y.-H.G. performed the experiments with the help of T.J. and J.-L.Z.; H.-Y.W. provided and identified the fungus; X.-J.M. helped to revise the manuscript. C.Y. wrote the paper with the help of Z.-M.Z. and G.D. All authors discussed the results and their interpretation and commented on the manuscript at all stages.
References
- Paranagama P. et al. Heptaketides from corynespora sp. inhabiting the Cavern Beard lichen, Usnea cavernosa: first report of metabolites of an endolichenic fungus (1). J Nat Prod 70, 1700−1705 (2007). [DOI] [PubMed] [Google Scholar]
- Fan Zhang. et al. A thiopyranchromenone and other chromone derivatives from an endolichenic fungus, Preussia Africana. J Nat Prod 75, 230–237 (2012). [DOI] [PubMed] [Google Scholar]
- Harvey A. L., Edrada-Ebel R. & Quinn R. J. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov 14, 111–129 (2015). [DOI] [PubMed] [Google Scholar]
- Grond S., Papastavrou I. & Zeeck A. Novel α-L-Rhamnopyranosides from a single strain of streptomyces by supplement-induced biosynthetic steps. Eur J Org Chem 19, 3237–3242 (2002). [Google Scholar]
- Yuan C. et al. Austdiol, fulvic acid and citromycetin derivatives from an endolichenic fungus, Myxotrichum sp. Phyto Lett 6, 662–666 (2013). [Google Scholar]
- Vleggaar R., Steyn P. S. & Nagel D. W. Constitution and absolute configuration of austdiol, the main toxic metabolite from Aspergillus ustus. J Chem Soc, Perkin Trans. I, 45–49 (1974). [DOI] [PubMed] [Google Scholar]
- Pieter S Steyn & Robert Vleggaar. The Structure of dihydrodeoxy-8-epi-austdiol and the absolute configuration of the azaphilones. J Chem Soc, Perkin Trans. I, 204–206 (1976). [PubMed] [Google Scholar]
- Quang D. N. et al. Sassafrins A–D, new antimicrobial azaphilones from the fungus Creosphaeria sassafras. Tetrahedron 61, 1743–1748 (2005). [Google Scholar]
- Takahashi M., Koyama K. & Natori S. Four new azophilones from Chaetomium globosum var. flavor-viridae. Chem Pharm Bull 38, 625–628 (1990). [Google Scholar]
- Yoshida E., Fujimoto H. & Yamazaki M. Isolation of three new azaphilones, Luteusins C, D, and E, from an Ascomycete, Talaromyces luteus. Chem Pharm Bull 44, 284–287 (1996). [DOI] [PubMed] [Google Scholar]
- Di Bari L., Pescitelli G., Pratelli C., Pini D. & Salvadori P. Determination of absolute configuration of acyclic 1,2-Diols with Mo2(OAc)4. 1. Snatzke’s Method Revisited. J Org Chem 66, 4819–4825 (2001). [DOI] [PubMed] [Google Scholar]
- Gorecki M. et al. Practical method for the absolute configuration assignment of tert/tert 1, 2-diols using their complexes with Mo2 (OAc)4. J Org Chem 72, 2906–2916 (2007). [DOI] [PubMed] [Google Scholar]
- Willian J Andrioli. et al. Mycoleptones A−C and Polyketides from the Endophyte Mycoleptodiscus indicus. J Nat Prod 77, 70−78 (2014). [DOI] [PubMed] [Google Scholar]
- Fan P., Hostettmann K. & Lou H. Allelochemicals of the invasive neophyte Polygonum cuspidatum Sieb. & Zucc.(Polygonaceae). Chemoecology 20, 223–227 (2010). [Google Scholar]
- Jiao Y. et al. Metabolites from Penicillium sp., an endophytic fungus from the liverwort Riccardia ultifida (L.) S. Gray. Phyto Lett 6, 14–17 (2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.