P-cadherin induces polarization and collective cell migration through an increase in the strength and anisotropy of mechanical forces, which is mediated by the P-cadherin/β-PIX/Cdc42 axis.
Abstract
Collective cell migration (CCM) is essential for organism development, wound healing, and metastatic transition, the primary cause of cancer-related death, and it involves cell–cell adhesion molecules of the cadherin family. Increased P-cadherin expression levels are correlated with tumor aggressiveness in carcinoma and aggressive sarcoma; however, how P-cadherin promotes tumor malignancy remains unknown. Here, using integrated cell biology and biophysical approaches, we determined that P-cadherin specifically induces polarization and CCM through an increase in the strength and anisotropy of mechanical forces. We show that this mechanical regulation is mediated by the P-cadherin/β-PIX/Cdc42 axis; P-cadherin specifically activates Cdc42 through β-PIX, which is specifically recruited at cell–cell contacts upon CCM. This mechanism of cell polarization and migration is absent in cells expressing E- or R-cadherin. Thus, we identify a specific role of P-cadherin through β-PIX–mediated Cdc42 activation in the regulation of cell polarity and force anisotropy that drives CCM.
Introduction
Collective cell migration (CCM), the coordinated movement of cells connected by cell–cell adhesion, is a fundamental process in development, tissue repair, and tumor invasion and metastasis (Friedl and Gilmour, 2009; Rørth, 2009; Friedl et al., 2012). Both epithelial (carcinoma) and mesenchymal (sarcoma) cancer cells undergo CCM (Theveneau and Mayor, 2011). Cells within a moving collective group have two types of interactions: one with the substratum (which is often the ECM, but also other cells) and one with neighboring moving cells through cell–cell interactions. Two protein families typically mediate these interactions and the generation of mechanical forces: integrins (with the underlying ECM) and cadherin trans-dimers (at intercellular adhesion sites). Classic cadherins, a central component of cell–cell and adherens junction formation, are major drivers of CCM (Halbleib and Nelson, 2006). Mechanical coupling between migratory cells may result in the production of force-dependent signals by which the cells can influence their collective behavior (Trepat et al., 2009; Tambe et al., 2011; Mertz et al., 2012, 2013; Hirashima et al., 2013) and also force transmission to the ECM (Jasaitis et al., 2012; Mertz et al., 2013). Besides the physical forces per se, their orientation also influences CCM, because a broad variety of cell types migrate along the direction of maximal intercellular tension (Tambe et al., 2011).
The mechanotransduction pathways whereby cadherin-mediated cell–cell adhesion promotes CCM, and in particular the molecular mechanisms that couple mechanical forces to the correlated cell motion, remain largely to be elucidated. One major process during CCM is the coordination of migration, polarization, and rearrangement of cytoskeletal elements by cells that are moving collectively. Rho GTPases play a crucial role in this coordination (Weber et al., 2012; Das et al., 2015a). RhoA and RhoE activity modulation appears to be involved in decreasing cell contractility at cell–cell contacts, an event that is important for CCM (Hidalgo-Carcedo et al., 2011; Omelchenko and Hall, 2012) and for the formation and maintenance of the migration fingers observed in epithelial MDCK cells (Reffay et al., 2014). In vivo studies using border cells moving in the Drosophila melanogaster ovary—a well-studied model of CCM—or using neural crest cells have demonstrated that Rac1 is activated at the front of migrating cells and participates in CCM (Theveneau et al., 2010; Wang et al., 2010). Recently, positive feedback between E-cadherin and Rac1 signaling was shown to occur in border cell migration (Cai et al., 2014). Moreover, Cdc42 localizes at the astrocyte migrating front and controls polarity during the collective migration of fibroblasts and astrocytes (Cau and Hall, 2005; Osmani et al., 2006). However, it is not known how the cadherin-mediated response regulates Rho GTPase activity during CCM. We decided to tackle this problem by focusing on P-cadherin. Indeed, aberrant P-cadherin expression has been described in many tumor types, including carcinoma and aggressive sarcoma (Paredes et al., 2012; Thuault et al., 2013; van Roy, 2014). In these tumors, P-cadherin is expressed in invasive rather than in in situ lesions, showing that aberrant expression of P-cadherin could be a useful marker for the invasion capacity of tumor cells. Additionally, P-cadherin expression is associated with cell invasiveness (Thuault et al., 2013), and P-cadherin knockdown in MCF10A cells resulted in a reduction of cell migration directionality and persistence during wound healing (Ng et al., 2012).
In this study, to directly investigate the role of P-cadherin during migration, we used mesenchymal C2C12 myoblasts that do not express P-, E-, and R-cadherin and analyzed the impact of P-cadherin expression in a 2D migration assay upon removal of a physical barrier. Because N- and M-cadherin, the two cadherins expressed in C2C12 myoblasts, are downregulated upon P-cadherin expression (Thuault et al., 2013), P-cadherin becomes the main cadherin expressed in these cells. As controls, we generated C2C12 cells that express E- or R-cadherin. By measuring velocity, persistence, directionality, and polarity, we demonstrated that expression of P-cadherin, but not E- or R-cadherin, induces efficient CCM. Using micropatterned cell monolayer and traction forces and monolayer stress microscopy, we dynamically mapped—for the first time in migrating mesenchymal cells—the mechanical forces developed and demonstrated that P-cadherin has a major and specific role in the induction of intercellular and traction force anisotropy in the cell layer. We also found that P-cadherin specifically activates Cdc42 during CCM through the guanine nucleotide exchange factor (GEF) β-PIX and identified the P-cadherin/β-PIX/Cdc42 axis as crucial for P-cadherin–mediated CCM.
Results
P-cadherin induces CCM
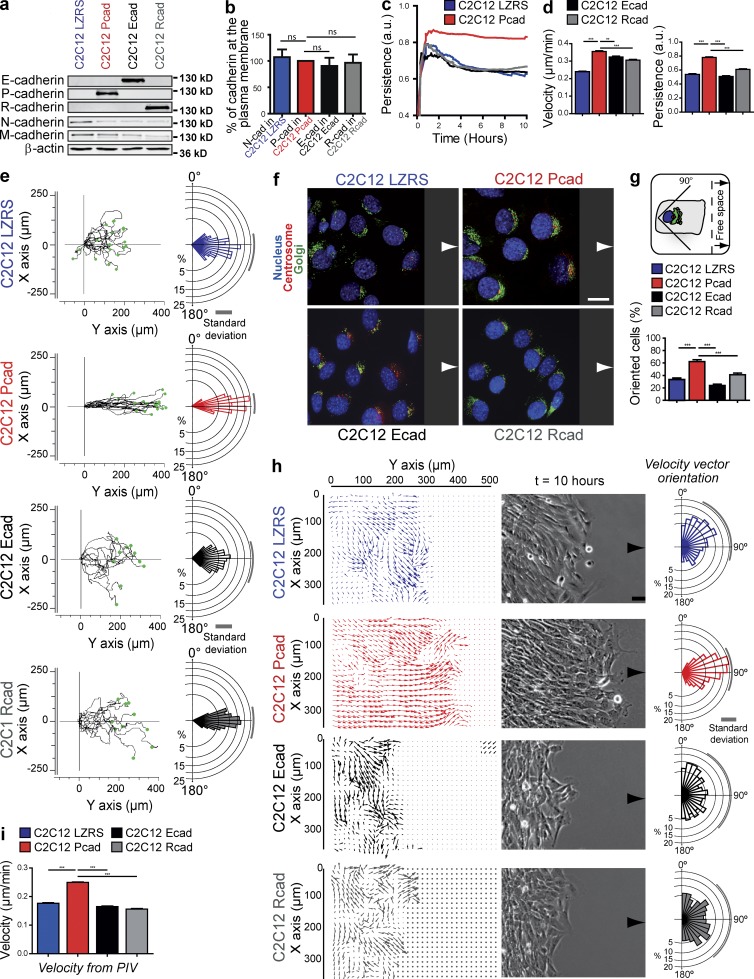
Because P-cadherin is up-regulated in aggressive tumors and associated with cell invasiveness, we investigated its role in cell migration by monitoring mouse C2C12 cells that express P-cadherin (C2C12 Pcad; Fig. 1 a) after removal of a physical barrier to provide free space into which confluent cells could migrate. As a control, C2C12 cells expressing only empty vector (C2C12 LZRS), E-cadherin (C2C12 Ecad), or R-cadherin (C2C12 Rcad) were used (Fig. 1 a). All cadherins were expressed at a similar level and reached the plasma membrane in a similar way (Fig. 1 b and Fig. S1, a and b). By combining time-lapse microscopy, cell tracking, and computational analysis, we characterized cell movements and cell–cell coordination in the sheet from 0 to 15 h after barrier removal (Fig. 1, c–i; and Fig. S1, c and d). Within the first 2 h, cells became progressively motile in the direction perpendicular to the free edges. The mean cell migration velocity and persistence were not significantly different in the four cell lines. Conversely, after 2–3 h, only P-cadherin expression, which induced a reorganization of the cell layer, increased the velocity and persistence of migration (Fig. 1, c and d; Fig. S1, c and d; and Video 1). Tracking the cells in the first three rows indicated directed movement in P-cadherin–expressing cells, whereas control cells (vector alone and E- or R-cadherin–expressing cells) tended to spread in all directions. Measuring the trajectory angle distribution revealed that P-cadherin expression increased the directionality of the movement of individual cells (Fig. 1 e). E- or R-cadherin expression in C2C12 cells also downregulated N- and M-cadherin as in P-cadherin expressing cells (Fig. 1 a and Fig. S1 a), but it did not promote cell layer reorganization (Fig. S1, c and d; and Video 2) and increase in velocity, persistence, and directionality observed upon P-cadherin expression (Fig. 1, c–e). These data demonstrate the specific effect of P-cadherin expression in CCM induction. P-cadherin–mediated CCM requires P-cadherin homotypic interaction because (a) isolated P-cadherin–expressing cells migrated randomly with a persistence similar to that of control cells (Fig. S2, a–e), and (b) calcium chelation by incubation with EGTA perturbed cadherin homophilic cell–cell adhesion and decreased persistence (Fig. S2, f and g). Conversely, mitomycin C treatment did not modify migration speed or persistence, thus excluding the involvement of cell proliferation (unpublished data). We also analyzed cell polarization by monitoring the position of the centrosome and the Golgi apparatus relative to the nucleus (Ridley et al., 2003; Yadav et al., 2009): P-cadherin expression, but not E- or R-cadherin expression, induced a marked orientation of the centrosome and Golgi in front of the nucleus and toward the protruding edges (Fig. 1, f and g). To map cell velocities within entire layers, we used particle image velocimetry (PIV) analysis, a technique that computes correlations between successive images (Petitjean et al., 2010) to visualize individual displacement fields across the cell sheet. P-cadherin expression oriented the velocity fields, and in these cells, the angle distribution was narrower than in control cells (Fig. 1 h). This analysis also confirmed a significant and specific increase in velocity after P-cadherin expression in the entire cell layer (Fig. 1 i).
Figure 1.
P-cadherin expression induces CCM. (a) Protein extracts (20 µg/well) from the indicated cells were immunoblotted to detect E-, P-, R-, N-, and M-cadherin and β-actin. (b) Quantification of the indicated cadherins at the plasma membrane, normalized to the total amount of the corresponding cadherin, calculated from three independent experiments. (c and d) Persistence over 10 h after removal of the insert (c) and mean velocity and persistence measured between 4 and 15 h after removal of the insert (d). n = 242 C2C12 LZRS and 249 C2C12 Pcad cells from 15 independent experiments and 230 C2C12 Ecad and 171 C2C12 Rcad cells from 4 independent experiments. a.u., arbitrary units. (e) Trajectories over 15 h of 17 representative cells. Rose plots of angle trajectories (i.e., directionality). The magnitude of each bar shows the fraction of cells with the indicated angle trajectory. n = 193. (f) Migrating cells (8 h after insert removal) stained for nucleus, centrosome, and Golgi distribution. Arrows indicate the migration direction. (g) Histogram representing the percentage of migrating cells in which the centrosome and Golgi are located in the quadrant facing the free space in front of the nucleus (see cartoon) as an indication of cell polarization. n = 80 cells from five independent experiments. (h) Velocity fields and corresponding phase-contrast images and velocity vector orientation measured using MatPIV in the entire cell layer 10 h after insert removal. n = 983 C2C12 LZRS, 1006 C2C12 Pcad, 677 C2C12 Ecad, and 633 C2C12 Rcad. (i) Mean velocity measured in the entire cell layer from 4 to 15 h. All panels: means ± SEM. *, P < 0.05; **, P < 0.005; ***, P < 0.0005. Bars: (f) 15 µm; (h) 100 µm.
These results indicate that P-cadherin expression specifically promotes efficient CCM by increasing cell velocity, directionality, and persistence toward the free space. Moreover, compared with E- and R-cadherin, P-cadherin expression efficiently and specifically promotes polarization of cells and cell movement.
P-cadherin expression modifies cellular and focal adhesion (FA) organization and turnover
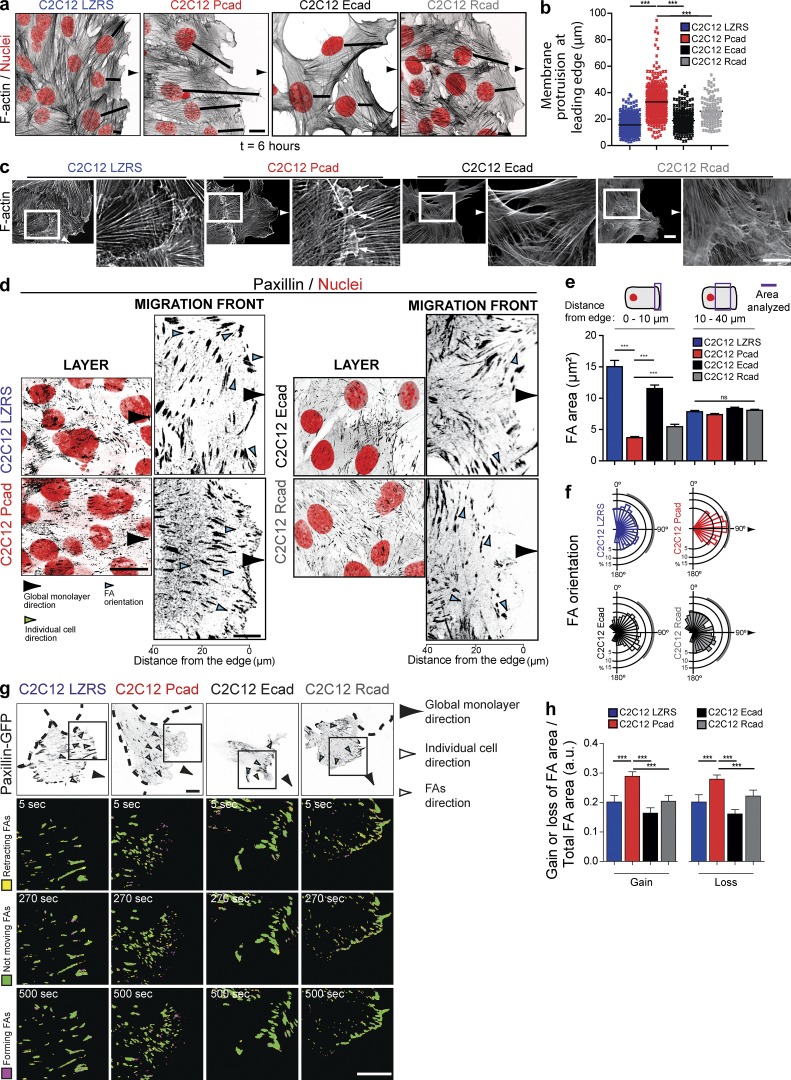
We then analyzed the overall cellular and FA organization in the four cell lines. Immunostaining analysis of the actin cytoskeleton revealed the specific formation of large protrusions in the direction of migration by the first multicellular row of P-cadherin–expressing cells (Fig. 2, a and b). Moreover, cryptic lamellipodia, which are structures characteristic of cells that migrate collectively, were detected at the front edge of cells inside the multicellular layer (Fig. 2 c; Farooqui and Fenteany, 2005). EGTA addition to P-cadherin–expressing cells decreased the orientation of the protrusion in the direction of migration (Fig. S2, h–j). Immunostaining for paxillin, a marker of most adhesion sites (Zaidel-Bar et al., 2003), showed that P-cadherin expression was associated with a marked increase in the number of small nascent FA at the leading edge (within the first 10 µm from the cell edge) and with an important reorientation of mature FAs parallel to the direction of migration across the entire cell layer (Fig. 2, d–f). Moreover, monitoring FA dynamics with paxillin-GFP in migrating cells using time-lapse video microscopy (Fig. 2 g and Video 3) revealed that P-cadherin expression induced a dramatic increase in FA assembly and disassembly rates (Fig. 2 h).
Figure 2.
Specific cellular and FA organization after P-cadherin expression. (a and b) F-actin (black, inverted contrast image) and nuclei (red) visualization indicating the formation of a large protrusion in P-cadherin–expressing cells as illustrated in the box plots. 384 (C2C12 LZRS), 373 (C2C12 Pcad), 115 (C2C12 Ecad), and 263 (C2C12Rcad) cells from four independent experiments were analyzed. Bar, 10 µm. ***, P < 0.0005. (c) F-actin staining of the four cell lines revealed cryptic lamellipodia in C2C12 Pcad cells (arrows). The right panels show higher-magnification images of the outlined region (white rectangle). Bars, 10 µm. (d) Confocal images of migrating cells stained for paxillin (inverted contrast images) and nuclei (red) are shown. Left panels, inside the layer; right panels, at the migration front. Bar, 10 µm. (e) Quantification of the FA area measured 0–10 µm or 10–40 µm from the leading edge. The presented value is the ratio of the area to the total surface. Data represent the means ± SEM of five independent experiments. More than 400 FAs were analyzed from 30 cells. ***, P < 0.0005. (f) Rose plot showing the distribution of the orientation angles of the FAs calculated using the monolayer migration direction as the reference axis (90°). The area of each bin represents the number of FAs in that direction. More than 2000 FAs were analyzed from three independent experiments. (g) Paxillin-GFP dynamics in cells at the first multicellular row were monitored at 8 h after removal of the insert at 5-s intervals for 15 min; inverted contrast images are shown. Dashed lines indicate the nontransfected surrounding cells. The insets indicate a cell area shown at three time points. Inverted contrast images of paxillin-GFP are in gray. Ratio images were generated to illustrate FA dynamics. Bars, 15 µm. (h) FA area was quantified over a period of 15 min, and mean values normalized to the area of the FA as well as the change in mean intensity are given. The means ± SEM of five independent cells are shown. a.u., arbitrary units.
Our results show that P-cadherin expression remodels the actin cytoskeleton with the formation of a large protrusion in the first cell row and of cryptic lamellipodia. It also remodels FAs, with the formation of many nascent FAs at the leading edge of the multicellular migrating row. Moreover, P-cadherin expression specifically promotes the polarization of the membrane protrusion and of FAs toward the direction of migration. These events appeared to be specific of P-cadherin, and not of E- or R-cadherin expression.
P-cadherin expression increases traction forces, intercellular stresses, and plithotaxis
During CCM, mechanical forces play a major role in monolayer expansion. Particularly, traction forces, which are exerted by cells on the underlying substrate, and intercellular stresses allow long-range cell guidance (Trepat et al., 2010; Tambe et al., 2011). Collectively migrating cells also migrate preferentially along the direction of maximum principal stress, a process called plithotaxis (Trepat and Fredberg, 2011).
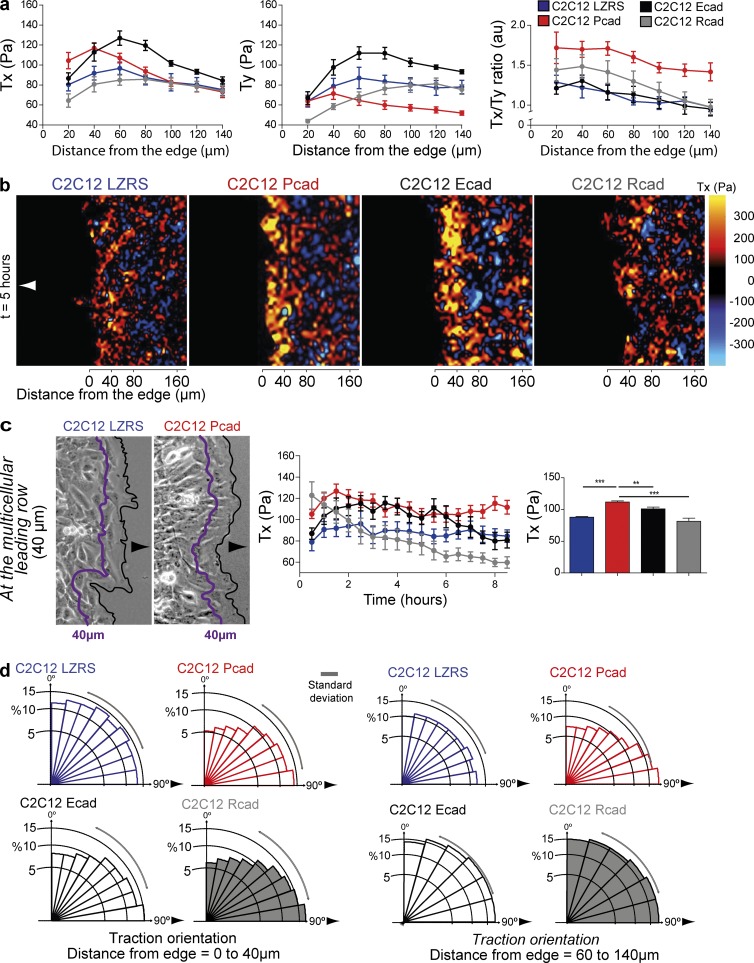
Using a micropatterned cell monolayer and traction-force microscopy (Trepat et al., 2009), we investigated whether P-, E-, or R-cadherin expression influenced the traction forces. C2C12 cells on collagen-coated polyacrylamide gels assumed morphologies comparable to those on a more rigid culture substrate (i.e., glass or plastic; Fig. S3 a and Video 4), and P-cadherin expression similarly increased both the velocity and persistence of migration (Fig. S3, b and c). We measured traction forces along the x direction (Tx; i.e., parallel to the direction of migration) over 8 h within an advancing cell sheet cultured on a collagen-coated polyacrylamide gel. Tx values were higher in P- and E- than in R-cadherin–expressing and control cells in the first 40 µm behind the leading edge (Fig. 3, a and b; and Video 5). The mean Tx value from 0 to 40 µm was highest in P-cadherin–expressing cells (Fig. 3 c). In contrast, Ty values (i.e., traction forces perpendicular to the direction of migration) were significantly decreased after 40 µm in P-cadherin–expressing cells, whereas they were increased in control and E- and R-cadherin–expressing cells (Fig. 3 a). Thus the Tx/Ty ratio was significantly increased in P-cadherin compared with E- or R-cadherin–expressing and control cells, suggesting a polarization of cell movement in P-cadherin–expressing cells. Indeed, the analysis of traction-force orientation from 0 to 40 µm and from 60 to 140 µm from the leading multicellular row toward the center of the cell layer revealed that P-cadherin expression promoted traction-force reorientation in the entire cellular layer toward the direction of migration (Fig. 3 d).
Figure 3.
P-cadherin expression reorganizes and increases traction forces. (a) Traction forces in the x direction (Tx), i.e., parallel to the axis of migration, or in the y direction (Ty), i.e., perpendicular to the axis of migration, measured every 20 µm from the multicellular leading row toward the center of the layer 4–10 h after removal of the PDMS membrane. The Tx/Ty ratio is calculated in the same conditions. Histograms represent the mean ± SEM calculated from n = 6 for each cell line from three independent experiments. au, arbitrary units. (b) Representative image of the traction (Tx) force maps at 6 h after removal of the PDMS membrane. Color bar indicates relative values. Arrowheads indicate the direction of migration. (c) Tx measured from 0 to 40 µm (as indicated in the phase-contrast images; arrowheads indicate the direction of migration) at 0–8 h. Histogram representing the mean ± SEM of Tx over 8 h. n = 6. (d) Overall orientation of traction forces. n = 6 from three independent experiments. All panels: values are means ± SEM. **, P < 0.005; ***, P < 0.0005; ns, nonsignificant.
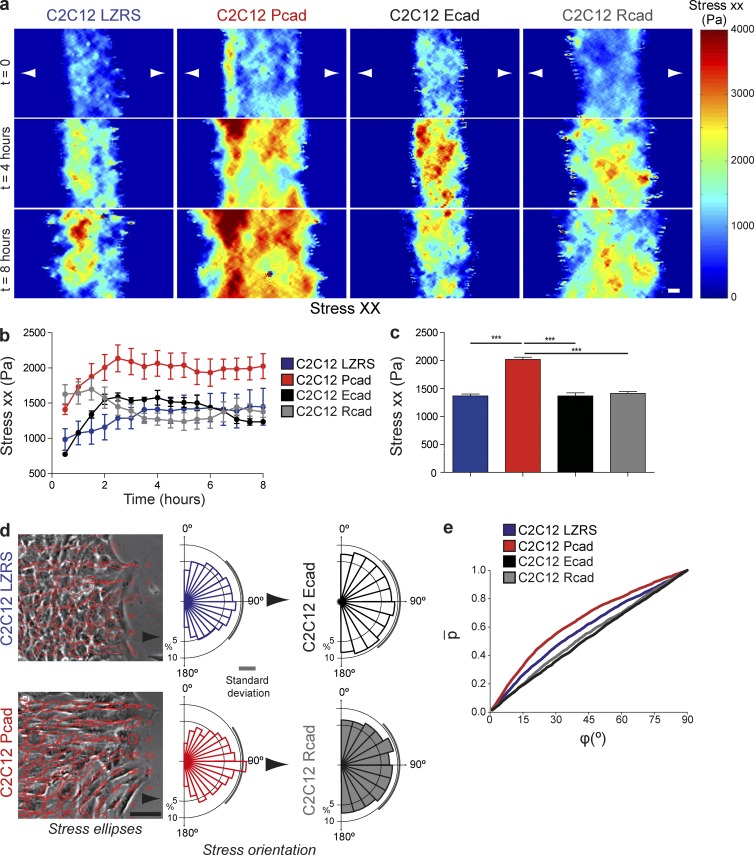
Using micropatterned cell and monolayer stress microscopy, we measured local intercellular stress within the monolayer during cell migration (Tambe et al., 2011). This technique is based on the principle that, according to Newton’s laws, traction forces applied at the cell–gel interface must be balanced by intra- and intercellular forces (Trepat et al., 2009; Maruthamuthu et al., 2011). We measured σxx, the stress component in the direction of expansion of the monolayer. We refer to it interchangeably as monolayer stress or intercellular stress (Tambe et al., 2011). Similarly to persistence, monolayer stress increased monotonically with time and tended to reach a plateau, the magnitude of which was significantly higher for P-cadherin–expressing cells (Fig. 4, a–c; and Video 6). When local monolayer stress was represented as an ellipse with axes corresponding to local maximum and minimum stresses, P-cadherin expression increased stress anisotropy (Fig. 4 d). Moreover, the cell velocity vectors tended to orient with the orientation of the maximum principal stress, and this phenomenon was more pronounced in regions of high stress anisotropy (Fig. 4 e). This mechanism for collective cell guidance is called plithotaxis (Tambe et al., 2011; Trepat and Fredberg, 2011) and was observed exclusively after P-cadherin, but not E- or R-cadherin, expression in C2C12 cells.
Figure 4.
P-cadherin expression increases intercellular stress. (a) Intercellular stress maps parallel to the migration direction (stress xx) measured at the indicated time after removal of the PDMS membrane. n = 6 areas analyzed for each cell line from three independent experiments. (b) Stress xx over 8 h after removal of the insert. (c) Histograms of the maximum principal stress measured over 8 h. n = 6. (d) Principal stress ellipses at 8 h after removal of the PDMS membrane and rose plots of the angle between the principal stress direction and velocity. Arrows indicate the direction of migration. n = 6. (e) Cumulative probability distribution of the angle between cellular velocity and the maximum principal stress for the highest quintile of stress anisotropy (measured using the maximum shear stress). All panels: values are the means ± SEM. ***, P < 0.0005. Bars: (a) 40 µm; (d) 100 µm.
These results show that P-cadherin expression increases the anisotropy and strength of traction forces and of intercellular stresses in the entire layer and also plithotaxis.
P-cadherin expression specifically activates Cdc42 to promote CCM
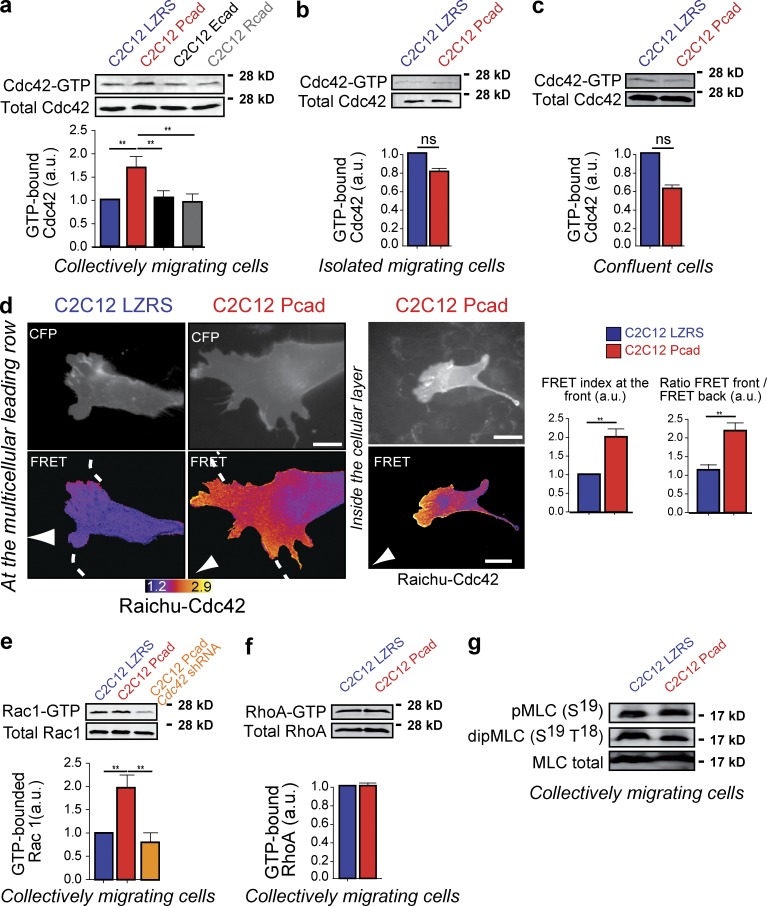
Because P-cadherin expression specifically promoted the polarization of many cell processes and structures (i.e., cell organelles, cell trajectories, intercellular stresses, traction forces, and FAs), we asked whether P-cadherin expression induced the activation of Cdc42, a major regulator of polarity (Etienne-Manneville and Hall, 2001; Zegers and Friedl, 2014). Global Cdc42 activation during migration after massive wounding was assessed using a pull-down assay. Cdc42 was specifically activated in P-cadherin–expressing cells, but not in E- or R-cadherin–expressing and control cells (Fig. 5 a). This P-cadherin–dependent Cdc42 activation required P-cadherin homotypic interactions and CCM, because the level of Cdc42 activation was not increased upon P-cadherin expression in isolated migrating (Fig. 5 b) and nonmigrating (Fig. 5 c) confluent cells. Using the Förster resonance energy transfer (FRET) biosensor Raichu-Cdc42 (Itoh et al., 2002), we mapped Cdc42 activity within migrating cells and detected elevated Cdc42 activity at the leading edge of migrating cells located at the multicellular leading row and inside the multicellular layer in cryptic lamellipodia (Fig. 5 d). Moreover, we measured global Rac1 activity and showed that P-cadherin expression increased Rac1 activity in a Cdc42-dependent manner (Fig. 5 e). In contrast, P-cadherin expression was not correlated with RhoA activation (Fig. 5 f) or with an overall increase in myosin light chain phosphorylation (Fig. 5 g).
Figure 5.
P-cadherin–dependent Cdc42 activation during CCM. (a–c) The level of GTP-bound Cdc42 was measured using GST fused to the CRIB domain of PAK (GST-CRIB) in lysates obtained from cells 5–6 h after wounding (a), in migrating isolated cells (b), and in confluent (c) C2C12 LZRS and C2C12 Pcad cells. Cdc42 was detected by immunoblotting. Histograms represent the GTP-bound Cdc42 normalized to the amount of total protein. The mean ± SEM of five independent experiments is shown. (d) Cdc42 activity was mapped using the FRET reporter Raichu-Cdc42. Examples of increased Cdc42 activity after P-cadherin expression at the leading edge of the migrating cells and at cell–cell contacts inside the cellular layer are shown. Histograms represent the quantification of the FRET index at the front of C2C12 LZRS and C2C12 Pcad migrating cells (top) and the ratio of the FRET index between the front and back in these cells (bottom). n = 42 for C2C12 LZRS and 56 for C2C12 Pcad cells. The mean ± SEM of four independent experiments is shown. (e and f) Levels of GTP-bound Rac1 (e) or RhoA (f) were measured using GST fused to the CRIB domain of PAK (GST-CRIB) or to the RhoA binding domain of Rhotekin, respectively, in lysates obtained from cells 5–6 h after wounding. GTPase was detected by immunoblotting, and histograms represent the GTP-bound GTPase normalized to the amount of total protein. The mean ± SEM of five independent experiments is shown. (g) The level of MLC phosphorylation was analyzed using antibodies that recognize mono- and di-phospho-MLC in cell lysates of C2C12 LZRS and C2C12 Pcad cells 6 h after wounding. Shown are representative Western blot images from three independent experiments. a.u., arbitrary units. **, P < 0.005.
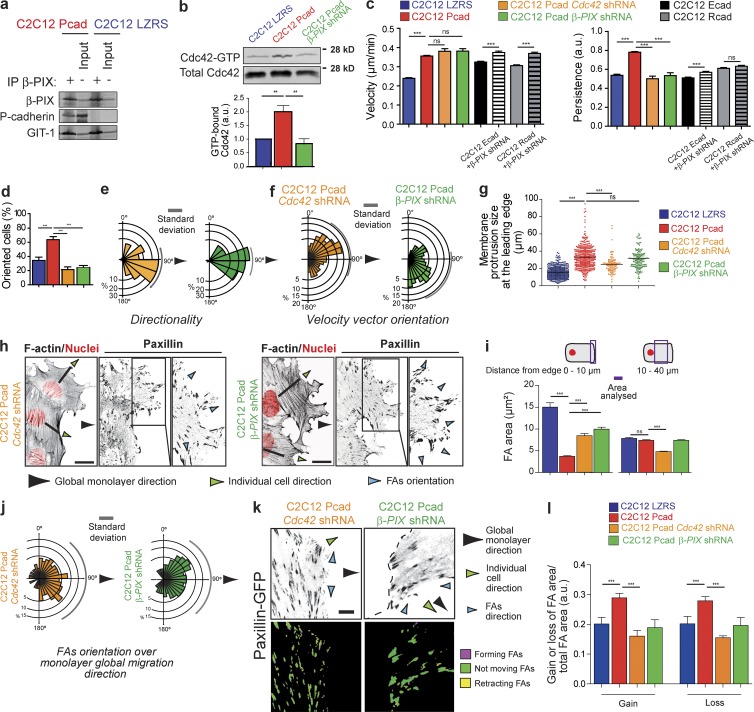
To identify potential regulators of Cdc42 activation downstream of P-cadherin, we performed a proteomic analysis of molecules that were immunoprecipitated with P-cadherin and identified β-PIX (Table S1 and Fig. S4), a GEF involved in Cdc42 and Rac1 activation and CCM in vivo (Osmani et al., 2006; Omelchenko et al., 2014). β-PIX association with P-cadherin was confirmed by coimmunoprecipitation (Fig. 6 a). Knockdown of β-PIX using a specific shRNA (Fig. S5 b) impaired P-cadherin–induced Cdc42 activation during CCM (Fig. 6 b). Cdc42 activation in C2C12 Pcad β-PIX shRNA cells could be rescued by expression of shRNA-resistant wild-type β-PIX (β-PIXWT), but not by a β-PIX GEF dead mutant (β-PIXGD; Fig. S5 g). We then monitored the contribution of Cdc42 to P-cadherin–induced CCM by depleting Cdc42 using specific shRNA or in β-PIX–depleted cells (Fig. S5, a and b). In both cases, P-cadherin accumulation at cell–cell contacts was not affected (Fig. S5, e and f). As a control, we used C2C12 Pcad Luciferase shRNA cells, the migration behavior of which was indistinguishable from that of the parental C2C12 Pcad cells (Fig. S5, c and d). We found that Cdc42 was required for P-cadherin–induced CCM. Cdc42 or β-PIX depletion did not affect migration velocity but did impair the P-cadherin–dependent increase in migration persistence (Fig. 6 c), as did Cdc42 specific inhibition with the chemical inhibitor ML-141 (Fig. S5 c). β-PIX depletion in E- or R-cadherin–expressing cells did not decrease their migration velocity or persistence (Fig. 6 c). Moreover, expression of shRNA-resistant β-PIXWT, but not of β-PIXGD, in C2C12 Pcad β-PIX shRNA cells significantly rescued the decrease in migration persistence (Fig. S5 g). These data show the specific role of β-PIX in P-cadherin–expressing cells.
Figure 6.
Cdc42 and β-PIX are required for P-cadherin–induced CCM. (a) Lysates of C2C12 LZRS or C2C12 Pcad cells were immunoprecipitated using an anti–β-PIX antibody and immunoblotted for β-PIX, P-cadherin, and GIT-1, a known β-PIX partner. (b) The level of GTP-bound Cdc42 was measured using GST fused to the CRIB domain of PAK (GST-CRIB) in lysates obtained from cells 5–6 h after wounding in C2C12 LZRS, C2C12 Pcad, and C2C12 Pcad cells in which β-PIX was knocked down. Cdc42 was detected by immunoblotting. Histograms represent the GTP-bound Cdc42 normalized to the amount of total protein. The mean ± SEM of five independent experiments is shown. (c) Velocity and persistence of migration measured 4–15 h after removal of the insert in the indicated cells. n = 45 C2C12 Pcad Cdc42 shRNA cells, 50 C2C12 Pcad β-PIX shRNA cells, 230 C2C12 Ecad, 171 C2C12 Rcad, 97 C2C12 Ecad β-PIX shRNA, and 70 C2C12 Rcad β-PIX shRNA cells. The mean ± SEM of five independent experiments is shown. (d) Histogram quantifying cell polarity shown in Fig. S5 h. n = 120 C2C12 Pcad Cdc42 shRNA cells, 90 C2C12 Pcad β-PIX shRNA cells. (e) Rose plots of angle trajectories (i.e., directionality) shown in Fig. S5 i. (f) Orientation of the velocity vector (in Fig. S5 j) measured using MatPIV software in the entire cell layer at 10 h after insert removal. n = 1006 C2C12 Pcad Cdc42 shRNA; 998 C2C12 Pcad β-PIX shRNA cells. Arrowheads indicate the monolayer global migration direction. Compare with Fig. 1 h. (g) Box plots showing the size of membrane protrusions in the indicated cells. Shown is the mean ± SEM: ***, P < 0.0005; ns: nonsignificant. (h) C2C12 Pcad Cdc42 shRNA or β-PIX shRNA cells 8 h after removal of the insert were stained for nuclei (red), F-actin, and paxillin (inverted contrast images). Bar, 15 µm. (i) Quantification of the FA area measured 0–10 µm or 10–40 µm from the leading edge. More than 700 FAs were analyzed from 50 cells. Data represent the mean ± SEM of five independent experiments. (j) Rose plot showing the distribution of angles of FA orientation calculated using the monolayer migration direction as the reference axis; 90° corresponds to the reference axis. More than 700 FAs were analyzed from 50 cells. Data represent the mean ± SEM of five independent experiments. (k) Cells were transfected with paxillin-GFP, and FA dynamics were analyzed as described in Fig. 2 g. An inverted contrast image of paxillin-GFP is in gray. Ratio images (lower panels) were generated to illustrate FA dynamics, with magenta showing the extension and yellow the FA loss. Green represents the FA area maintained during the analyzed period. Bar, 10 µm. (l) FA area gains and losses were quantified over 15 min. The mean values normalized to the area of the FA as well as the change in mean intensity is given. For all panels, the mean ± SEM is shown: ***, P < 0.0005. a.u., arbitrary units.
We also found that Cdc42 and β-PIX are required for P-cadherin–induced cell polarization (Fig. 6 d and Fig. S5 h; compare with Fig. 1 g). Tracking the cells in the first three rows (Fig. 6 e, and Fig. S5 I, and Video 7) and PIV analysis of the entire cell layer (Fig. 6 f and Fig. S5 j) showed that Cdc42 and β-PIX are required for P-cadherin–induced polarized migration. Finally, we demonstrated that Cdc42 and β-PIX are required for P-cadherin–induced membrane protrusion (Fig. 6 g), FA remodeling and orientation in the direction of the multicellular layer migration (Fig. 6, h–j; compare with Fig. 2, d–f), and FA dynamics (Fig. 6 k and Video 8). These results show that P-cadherin, but not E- or R-cadherin, activates Cdc42 in a β-PIX–dependent manner and that β-PIX and Cdc42 are required for P-cadherin–induced CCM.
β-PIX and Cdc42 are required for P-cadherin–induced mechanical force generation
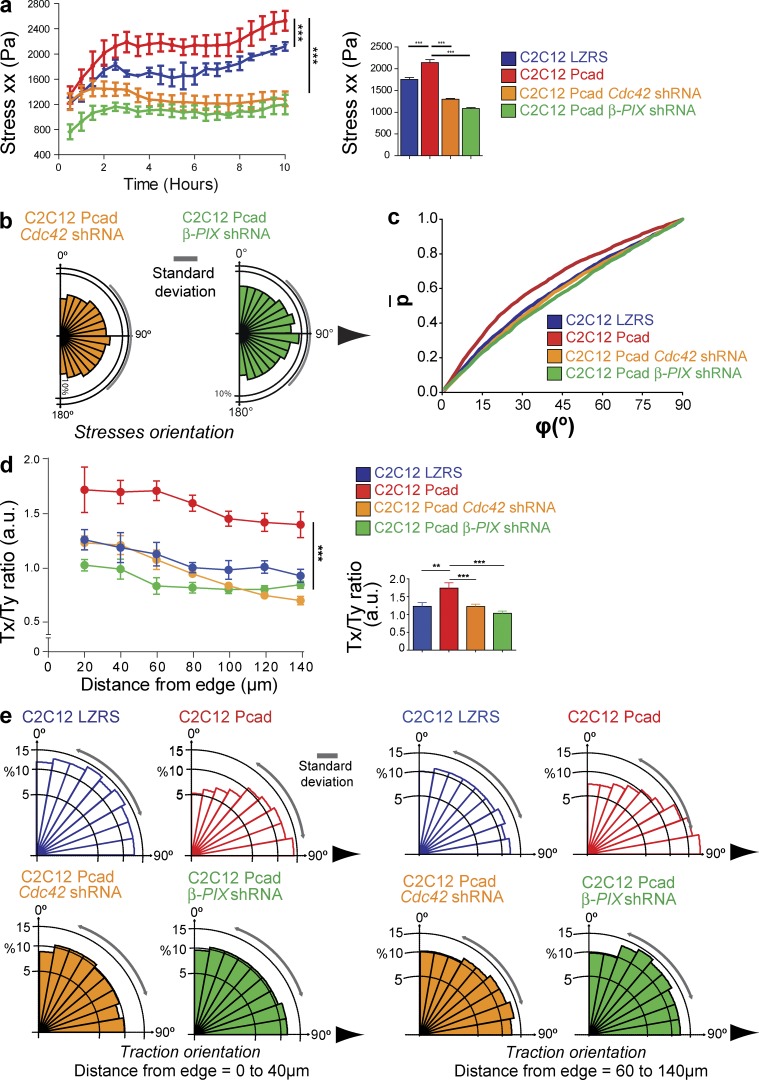
We next investigated whether Cdc42 and β-PIX contribute to P-cadherin–induced intercellular stresses and traction-force generation. Inhibition of Cdc42 (by knockdown) or of β-PIX completely prevented P-cadherin–dependent increases of intercellular stresses (Fig. 7 a and Video 9), stress anisotropy (compare Fig. 7 b with Fig. 4 d), and plithotaxis (Fig. 7 c).
Figure 7.
P-cadherin–induced mechanical force generation requires β-PIX and Cdc42. (a) Intercellular stress maps parallel to the migration direction (stress xx) measured at the indicated migration time and corresponding histograms of the maximum principal stress measured over 10 h. n = 6 for C2C12 Pcad Cdc42 shRNA cells and for C2C12 Pcad β-PIX shRNA cells from three independent experiments. (b) Rose plots of the angle between the principal stress direction and velocity at 8 h after removal of the PDMS membrane. Arrows indicate the migration direction. n = 6. (c) Cumulative probability distribution of the angle between cellular velocity and the maximum principal stress for the highest quintile of stress anisotropy (measured using the maximum shear stress). (d) The Tx/Ty ratio measured every 20 µm from the multicellular leading row toward the center of the layer 4–10 h after removal of the PDMS membrane. Error bars show 95% confidence interval of the mean (95% SEM); all nonoverlapping error bars are statistically significant with P < 0.05. (e) Overall orientation of traction forces 0–40 or 60–140 µm from the multicellular leading row. n = 6 from three independent experiments. For all panels, data are the mean ± SEM: **, P < 0.005; ***, P < 0.0005.
Moreover, Cdc42 inhibition prevented P-cadherin–dependent increase of the traction-force Tx/Ty ratio (Fig. 7 d and Video 10). Analysis of the traction-force orientation from 0 to 40 µm showed that Cdc42 knockdown impaired P-cadherin–induced traction-force reorientation toward the direction of migration (Fig. 7 e). Altogether, these data demonstrate that β-PIX and Cdc42 are required downstream of P-cadherin to increase monolayer stress and traction-force strength and anisotropy.
β-PIX is specifically recruited at P-cadherin–mediated cell–cell junctions (CCJs) during CCM
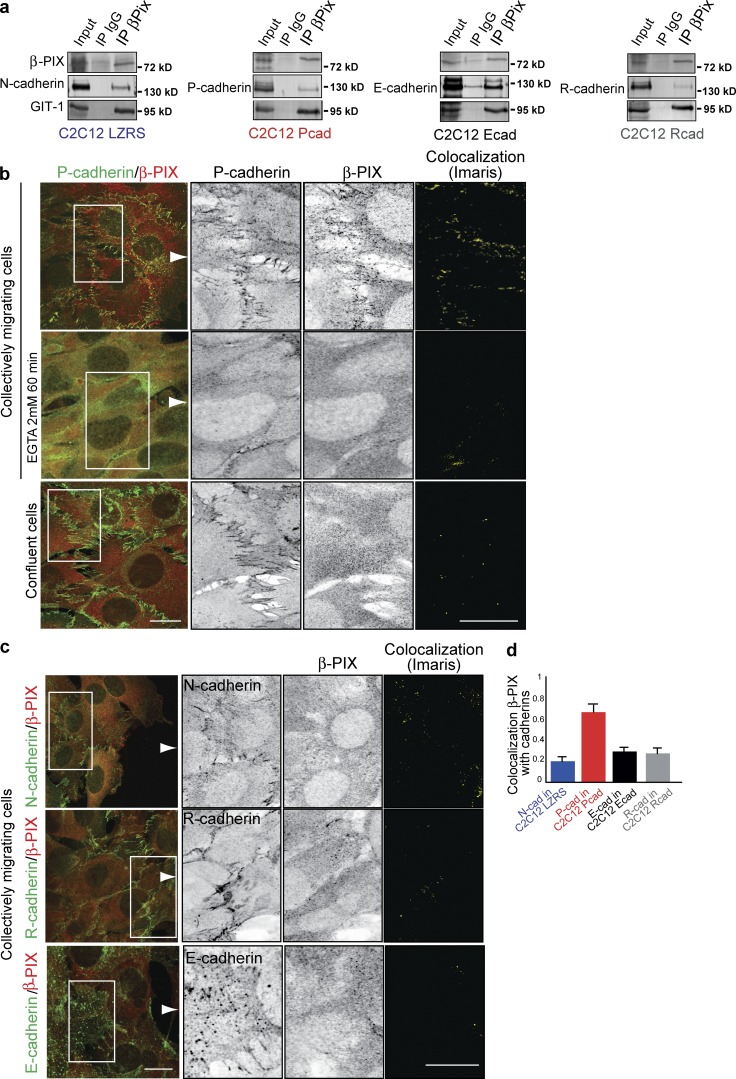
β-PIX could associate with P-cadherin and also with N-, E-, and R-cadherin (Fig. 8 a), but not with α-tubulin or Hsp90, two abundantly expressed proteins (Fig. S5 l). However, detailed analysis of β-PIX colocalization with these cadherins revealed that only P-cadherin recruited β-PIX at CCJs (Fig. 8, b–d). β-PIX recruitment at P-cadherin–mediated CCJs occurred only in collectively migrating cells and not in confluent nonmigrating cells and required P-cadherin homophilic interaction (Fig. 8 b).
Figure 8.
β-PIX is specifically recruited at CCJ by P-cadherin. (a) C2C12 LZRS or C2C12 Pcad, Ecad, or Rcad cell lysates were immunoprecipitated using an anti–β-PIX antibody and immunoblotted to assess the expression of β-PIX, GIT-1, and the indicated cadherins. (b and c) Inverted contrast and merge images of the expression of β-PIX and P-cadherin in C2C12 Pcad cells (b) incubated or not with EGTA at 8 h after removal of the insert or in confluent C2C12 Pcad cells, N-cadherin in C2C12 Pcad cells (c), E-cadherin in C2C12 Ecad cells, and R-cadherin in C2C12 Rcad cells. Colocalization images were generated using the colocalization module of Imaris. Bar, 10 µm. (d) Colocalization of β-PIX and cadherins at the CCJ as Pearson’s correlation coefficient; n = 15 images for each condition.
These results show that although β-PIX can associate with P-, N-, E-, and R-cadherin, it is specifically recruited by P-cadherin at CCJs only during CCM.
Discussion
P-cadherin is expressed in invasive carcinoma and sarcoma and is associated with poor prognosis (Paredes et al., 2012; Thuault et al., 2013). To analyze P-cadherin’s role in cell migration, we used a validated 2D system to investigate cell migration in response to the sudden removal of boundaries (Poujade et al., 2007; Trepat et al., 2009; Petitjean et al., 2010; Tambe et al., 2011; Ng et al., 2012). This in vitro system mimics the collective cell movement of tumor cells occurring as 2D monolayers migrating along tissue clefts (Alexander et al., 2013). It allows investigating questions concerning the complex process of CCM by combining mechanical force measurements and molecular imaging. C2C12 myoblasts are of mesenchymal origin, express only N- and M-cadherins, and form and maintain cell–cell contacts. However, they do not endogenously exhibit CCM as do MCF10A or MDCK epithelial cells. In contrast, upon P-cadherin expression, C2C12 cells adopt a very efficient CCM phenotype with features characteristic of a collective migration model. We show that P-cadherin expression is associated with an increase in cell velocity, persistence, directionality, and polarization. All the cells in a layer expressing P-cadherin exhibit a similar behavior and migrate in a coordinated, directional fashion in a cell–cell interaction–dependent manner. CCM induction in this mesenchymal cell model is specific to P-cadherin expression and is not observed upon expression of E- or R-cadherin. Moreover, N- and M-cadherin were similarly downregulated upon expression of P-, E-, or R-cadherin. This indicates that P-cadherin–induced CCM was not caused by the downregulation of these cadherins, thus further supporting a P-cadherin–specific role in CCM. A previous study demonstrated that P-cadherin knockdown in epithelial MCF10A cells perturbed directionality and persistence during CCM (Ng et al., 2012). Thus, P-cadherin is involved in CCM not only in mesenchymal cells but also in epithelial cells.
Importantly, we demonstrated that P-cadherin specifically increases intercellular forces, both intercellular stress and traction forces. Interestingly, P-cadherin expression was recently shown to be predictive of the level of intercellular tension in epithelial cells (Bazellières et al., 2015). Our study not only confirms this observation in mesenchymal cells by comparing P-, E-, and R-cadherin; it also demonstrates that P-cadherin specifically increases the anisotropy of intercellular stresses across CCJs and the alignment between cell velocities and the maximal principal stress direction. This mode of local cell guidance, called plithotaxis, enables the efficient translocation of the entire cell layer, because cells are aligned and migrate along the direction of the transmitted stress (Tambe et al., 2011). Although described mostly in epithelial cells and tumor cells (Friedl et al., 1995; Friedl and Gilmour, 2009), mesenchymal cells also exhibit CCM (Arboleda-Estudillo et al., 2010; Theveneau and Mayor, 2011). Nevertheless, it was unknown whether and how cadherins expressed in mesenchymal cells could have a mechanical role and how they could be involved in force transmission during CCM. We demonstrate here that P-cadherin specifically induces CCM and polarization through an increase in the strength and anisotropy of mechanical forces. The advancement of the P-cadherin–expressing cell sheet was achieved not in a cell proliferation–dependent process but through the development of traction forces that counterbalance intercellular forces (Maruthamuthu et al., 2011) and drive the movement of the cell layer. Our data showing that P-cadherin increases polarization and intercellular stress nicely confirm the predictive theory of soft active matter on force–velocity relationships in 1D and 2D living systems (Marchetti et al., 2013). This theory proposes that high polarization and high viscosity or stress would lead to rapid and cohesive expansion under high intercellular tension (fast-strong phenotype).
P-cadherin expression not only polarizes mechanical forces but also extensively remodels cells and polarizes them toward the migration direction. At the front, a large and polarized protrusion is generated; inside the multicellular layer, cells display a polarized organization and develop polarized cryptic lamellipodia, which help maintain a coordinated and efficient migration, as recently reported (Das et al., 2015b). P-cadherin–dependent induction of CCM in C2C12 cells is characterized by the formation or development of a multicellular leading row, as reported for other cell types (Khalil and Friedl, 2010). This is in contrast to the pluricellular migration fingers preceded by leader cells in MDCK cell layers (Poujade et al., 2007; Reffay et al., 2014). Interestingly, migration fingers are associated with the activity of RhoA (Reffay et al., 2014), a GTPase not activated in P-cadherin–mediated CCM of C2C12 cells (Fig. 5 f), suggesting that distinct molecular pathways control these two types of collective cell guidance.
In contrast, our data show that Cdc42 is specifically activated by P-cadherin, but not by E- or R-cadherin, during CCM and is required for P-cadherin–mediated polarized cell organization and CCM. Cdc42 activation during P-cadherin–mediated CCM is logical because (a) polarization of cells, trajectories, membrane protrusions, and FA is the obvious P-cadherin–mediated downstream effect, and (b) Cdc42 was previously shown to control polarity during the collective migration of fibroblasts and astrocytes (Cau and Hall, 2005; Osmani et al., 2006). The key finding is that for the first time we demonstrate a Cdc42 role in mechanical force control in general and more specifically in cadherin-dependent force generation. We show that Cdc42 controls both plithotaxis and traction-force polarization and generation to enable the efficient P-cadherin–mediated collective displacement of cells. Our identification of a P-cadherin–mediated/Cdc42-mediated polarity pathway as the major regulator of mechanical-force generation provides insight into the role of cell–cell contacts in directing cell migration. C-cadherin engagement (only when stress is applied) allows Xenopus laevis mesodermal cells to polarize and extend protrusions in the direction of migration (Weber et al., 2012). We confirmed this process here upon P-cadherin expression in C2C12 cells that generates a large and polarized protrusion at the front of the first row of migrating cells and cryptic lamellipodia at the front of migrating cells across the layer. Interestingly, substrate micropatterning was used to demonstrate that traction forces increase with the cell spreading area (Reinhart-King et al., 2003; Tan et al., 2003; Stricker et al., 2011). Moreover, FA polarization in the migration direction may enable force-vector orientation, resulting in an increased Tx/Ty ratio, thus making FA orientation an interesting predictive parameter of traction force across the entire cell. The generation of small FAs, as we observed upon P-cadherin expression, can also participate in increasing traction forces at the leading edge of migrating cells (Beningo et al., 2001). These morphological modifications (i.e., membrane protrusion and small FA formation) were lost in P-cadherin–expressing cells in which Cdc42 was inhibited. These modifications are characteristic of the activation of Rac1, a small G protein downstream of Cdc42 that we found to be activated upon P-cadherin expression.
Proteomic analysis of the molecules that are immunoprecipitated together with P-cadherin revealed the presence of the Cdc42 GEF β-PIX. It was previously reported that β-PIX directs CCM of anterior visceral endoderm cells in early mouse embryos (Omelchenko et al., 2014). Here, we provide data showing that β-PIX is crucial for P-cadherin–mediated plithotaxis, traction force orientation in the direction of migration, and their increase and thus CCM. Although β-PIX can associate with all tested cadherins (P-, E-, R-, and N-cadherin), we show that β-PIX is specifically required for P-cadherin–mediated CCM. β-PIX is activated and recruited at CCJ exclusively by P-cadherin, but not by N-, E-, or R-cadherin, and only during CCM. Future studies should determine whether the higher intercellular tension generated by P-cadherin during monolayer extension, identified in this study by comparing P-, E-, and R-cadherin (Fig. 4), and also in Bazellières et al. (2015) by comparing P- and E-cadherin, could be responsible for β-PIX recruitment to P-cadherin–mediated CCJs. Indeed, tension generated at cadherin adhesion sites is emerging as an important process in protein recruitment at CCJs (Buckley et al., 2014; Yao et al., 2014). Our data also suggest that Cdc42 is activated at the cell front, where P-cadherin is not engaged in CCJs. How β-PIX is activated there and whether an external stimulus is involved, as described during cell migration in fibrillar collagen environments (Kutys and Yamada, 2014), remain to be determined.
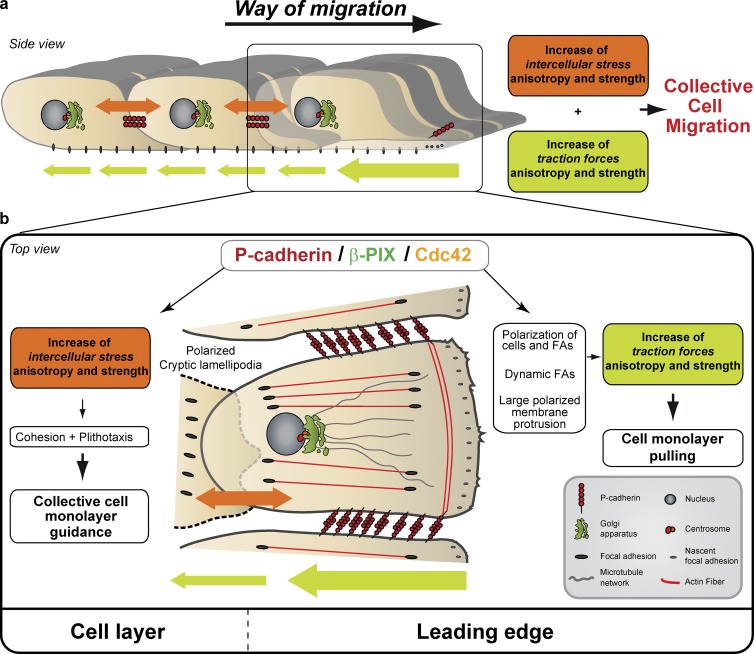
In conclusion, we demonstrated that P-cadherin, a protein upregulated in carcinomas and sarcomas, promotes CCM through an increase in the strength and anisotropy of physical forces (Fig. 9, a and b). Moreover, we identified Cdc42, activated by the GEF β-PIX, as a major regulator of intercellular stresses and traction-force polarization and generation in the induction of P-cadherin–dependent CCM. Our study brings important clues about a biological mechanism that links mechanical forces to the resulting cell movement.
Figure 9.
Model for the role of P-cadherin in CCM. (a) P-cadherin expression promotes CCM through an increase in the strength and anisotropy of physical forces; it is associated with an increase in intercellular stresses anisotropy and strength that promotes collective cell guidance called plithotaxis. P-cadherin expression also increases traction-force anisotropy (through increasing the Tx/Ty ratio) and strength that pull the cell layer. (b) P-cadherin expression induces polarization because it activates Cdc42 through the GEF β-PIX; this generates biological cues, such as polarization of the cell layer, of cryptic lamellipodia and FAs in the migration direction, polarized membrane protrusion generation and FA dynamics, thereby controlling mechanical force anisotropy and strength.
Materials and methods
Cell lines
C2C12 mouse myoblasts expressing either the LZRS empty vector (from K. Johnson, University of Nebraska Medical Center, Omaha, NE; C2C12 LZRS), LZRS-MS-neo-hPcad plasmid (from K. Johnson; C2C12 Pcad), LZRS-MS-neo-Ecad plasmid (from A. Reynolds, Vanderbilt University Medical Center, Nashville, TN; C2C12 Ecad), or LZRS-MS-neo-hRcad (the plasmid pLZRS-Ms-Neo-R-cadherin was generated by cloning the mouse R-cadherin cDNA from K. Johnson between AfeI and SfiI sites in pLZRS-Ms-Neo; C2C12 Rcad) were generated as well as C2C12 Pcad, C2C12 Ecad, and C2C12 Rcad cell lines that stably express β-PIX shRNA using the retroviral vector pSIREN-RetroQ. The pSIREN-RetroQ vector, which targets the firefly luciferase (Luci shRNA), was used as a control. Cell lines were generated by infection with recombinant retroviruses produced by Phoenix cells transfected with the corresponding vectors. Infection was performed on exponentially growing C2C12 myoblasts (5 × 105 cells per 60-mm dish) by using 5 ml viral supernatant. Stably transfected cells were selected with 1 mg/ml G418, and different clones were isolated by limited dilution. Cdc42 knockdown was performed by transient infection of Cdc42 shRNA. Cells were analyzed 48–72 h after infection.
shRNA
shRNA constructs were generated using the retroviral vector pSIREN-RetroQ according to the manufacturer’s protocol (BD Biosciences). To suppress endogenous Cdc42 expression, the annealed double-strand oligonucleotides 5′-GATCCGATTCCCATCGGAATATGTATTCAAGAGATACATATTCCGATGGGAATCTTTTTTACGCGTG-3′ (forward) and 5′-AATTCACGCGTAAAAAAGATTCCCATCGGAATATGTATCTCTTGAATACATATTCCGATGGGAATCG-3′ (reverse; for Cdc42 shRNA1) and 5′-GATCCGCTCACCACTGTCCAAAGACTTCAAGAGAGTCTTTGGACAGTGGTGAGCTTTTTTACGCGTG-3′ (forward) and 5′-AATTCACGCGTAAAAAAGCTCACCACTGTCCAAAGACTCTCTTGAAGTCTTTGGACAGTGGTGAGCG-3′ (reverse; for Cdc42 shRNA2) were inserted into the RNAi-Ready pSIREN-RetroQ vector (Clontech). Bold letters correspond to positions 246–263 or 469–486 of the mouse Cdc42 cDNA sequence (NCBI Reference Sequence accession no. NM_009861.3). To suppress endogenous β-PIX expression, the annealed double-strand oligonucleotides were used: 5′-GATCCGAGGAGTTCGCTGTGCGCAATTCAAGAGATTGCGCACAGCGAACTCCTCTTTTTTACGCGTG-3′ (forward) and 5′-AATTCACGCGTAAAAAAGAGGAGTTCGCTGTGCGCAATCTCTTGAATTGCGCACAGCGAACTCCTCG-3′ (reverse) for β-PIX shRNA1 and 5′-GATCCGGCAAAGACCATGTTCGCCATTCAAGAGATGGCGAACATGGTCTTTGCCTTTTTTACGCGTG-3′ (forward) and 5′-AATTCACGCGTAAAAAAGGCAAAGACCATGTTCGCCATCTCTTGAATGGCGAACATGGTCTTTGCCG-3′ (reverse) for β-PIX shRNA2. Bold letters correspond to position 1,961–1,978 or 3,867–3,884 of the mouse β-PIX (Arhgef7) cDNA sequence (NCBI Reference Sequence accession no. NM_017402.4). The siRNA constructs Luciferase shRNA was made by inserting the oligonucleotide 5′-GATCCGTGCGTTGCTAGTACCAACTTCAAGAGAGTTGGTACTAGCAACGCACTTTTTTGCTAGCGAATTC-3′. Bold letters correspond to oligonucleotide 1,310–1,328 of the Photinus pyralis (firefly) luciferase sequence (GenBank accession no. M15077.1).
For rescue experiments, the retroviral vector pMSCV-Hygro-GFP-β-PIXc-“shRNAresist” was generated as follows. Site-directed mutagenesis was performed using peGFP-L11–β-PIXc (from mouse origin; NCBI Reference Sequence accession no. NM_017402) as a template to introduce silent mutations in the sequence targeted by the β-PIXc shRNA: 5′-AGGAGTTCGCTGTGCGCAA-3′ in 5′-AGGAATTCGCTGTACGCAA-3′ (underlined letters are the mutated bases). The obtained GFP–β-PIXc shRNA-resistant sequence was then amplified by PCR and subcloned in pMSCV-Hygro (Clontech) between the XhoI and AgeI restriction sites. The retroviral vector pMSCV-Hygro-GFP-GEF-dead-β-PIXc LL-“shRNAresist” was obtained from the pMSCV-Hygro-GFP-β-PIXc-“shRNAresist” template by site-directed mutagenesis to generate the L238S and L239S mutations.
Antibodies and reagents
Mouse antibodies against P-cadherin (Invitrogen), M-cadherin (Charrasse et al., 2004), E-cadherin, pMLC (S19), and dipMLC (S19T18; Cell Signaling); R-cadherin (MRCD5; gift from Takeichi); Cdc42, Rac1, Paxillin, Hsp90, N-cadherin, and β- and p120 catenin (BD Biosciences); GIT1 (sc-9657; Santa Cruz); and β-actin, Pan-cadherin (from rabbit), FLAG (from rabbit), and myosin light chain (MLC) (Sigma-Aldrich) were used. Rabbit polyclonal antibodies against Pericentrin (Eurogentec) and α-tubulin (Sigma-Aldrich) were also used. Rabbit polyclonal antibodies were raised in-laboratory against the extracellular domain (aa 184–519) of human P-cadherin and against the SH3 domain of β-PIX (aa 1–65). F-actin was labeled with Alexa Fluor 405 or 546 phalloidin conjugates (Molecular Probes and Interchim), nuclei with Hoechst 33342 (Sigma-Aldrich), and Golgi using Alexa Fluor 488 lectin conjugates from Helix pomatia (Molecular Probes).
Primary antibodies were detected with Alexa Fluor 488– or 546–conjugated goat anti–mouse or anti–rabbit IgG (Molecular Probes). The Cdc42 inhibitor (ML-141; Tocris) was used at 20 µM. Cells were pretreated for 4 h before insert removal, and fresh medium with the drug was added before analysis.
Time-lapse imaging
Cells were plated in Ibidi Culture-Inserts (BioValley; 20,000 cells per chamber) until they reached the desired density level. The insert was removed to allow the cells to migrate into the cell-free areas. The cells were imaged using an Axiovert inverted microscope (Carl Zeiss) equipped with a motorized stage, as well as a heated and CO2-regulated incubator. Phase-contrast images were taken every 5–15 min overnight using a 10×/0.3 NA PH1 DIC1 objective and captured with a CCD MicroMax 1300 Y/HS camera (Roper Scientific) controlled with the MetaMorph 7.0 software.
Measurements of cell velocity, persistence, and directionality
Manual cell tracking and x/y cell position recording (of each cell at each time point) were performed using MetaMorph software. Individual cell mean speed, cell persistence, and cell directionality were calculated and illustrated using Ibidi’s Chemotaxis tool. Angles were plotted using Rose.net software.
Particle velocimetry analysis
The velocity field in monolayers was mapped via PIV analysis (available at http://folk.uio.no/jks/matpiv/index2.html; Poujade et al., 2007). Stacks of images were analyzed using the MatPIV toolbox (http://www.mn.uio.no/math/english/people/aca/jks/matpiv/) for MATLAB (MathWorks). We used an in-laboratory loop protocol in MATLAB to analyze sequential time frames for velocity and angle measurements (see dataset in the online supplemental material). Velocities were plotted in GraphPad Prism and angles in Rose.net software. The time between successive analyzed images was 5 or 10 min. The window size was set to 32 or 64 pixels, with no noticeable differences.
Preparation of polyacrylamide gels
In brief, polyacrylamide gels with a Young’s modulus of 12 kPa were prepared as previously described (Yeung et al., 2005; Kandow et al., 2007; Serra-Picamal et al., 2012) using a solution containing 19% acrylamide, 8% bis-acrylamide, 0.5% ammonium persulfate, 0.05% tetramethylethylenediamine, 0.64% of 200-nm-diameter red fluorescent carboxylate-modified beads, and 2 mg⋅ml−1 NH-acrylate. After polymerization, the gels were incubated overnight with 0.1 mg/ml collagen I.
Polydimethylsiloxane (PDMS) membrane fabrication
PDMS membranes were fabricated according to previously described procedures (Ostuni et al., 2000; Poujade et al., 2007; Serra-Picamal et al., 2012). SU8-50 masters containing rectangles of 300 × 2,500 µm were generated using conventional photolithography. Uncured PDMS was spin-coated onto the masters to a thickness lower than the height of the SU8 feature and cured for 2 h at 60°C. A thick border of PDMS was left at the edges of the membranes for handling purposes. The PDMS was then peeled off from the master and stored in ethanol at 4°C until use.
Cell monolayer patterning and time-lapse microscopy
To pattern the cells on top of the polyacrylamide gels, a PDMS membrane with a rectangular opening was deposited on top of the polyacrylamide gel. We then seeded 20,000 cells within the rectangle defined by the PDMS stencils; the cells were allowed to adhere and proliferate on the gel for a few hours. Before the time-lapse analysis, the PDMS membrane was carefully removed, enabling the cells to migrate toward the available substrate. The time-lapse imaging was performed using an automated inverted microscope (10× lens; Eclipse Ti; Nikon) equipped with thermal, CO2, and humidity control with MetaMorph (Universal Imaging) software. The recording started ∼30 min after removal of the PDMS membrane and lasted for 15 h. Images were obtained every 3 min over a period of 1–15 h.
Traction force microscopy
As cells migrate, they exert traction forces on the substrate. Traction forces are computed from the gel deformations induced by the cells at the interface with the gel substrate. These gel deformations are observed by imaging the displacements of fluorescent beads embedded in the gel. Beads displacements between any experimental time point and a reference image obtained after cell trypsinization are computed using custom-made particle imaging velocimetry software. To reduce systematic biases in subpixel resolution and peak-locking effects, we implemented an iterative process (up to four iterations) based on a continuous window shift technique (Serra-Picamal et al., 2012). Traction vectors Ti,j(t) within the field of interest are obtained from displacement vectors ui,j(t) for all time points t = 1,…, n and locations (i,j) of the M × N gel interface matrix. Traction field vectors Ti,j are computed by superimposing the equilibrium solution for a 3D linear-elastic gel substrate in the Fourier space (Trepat et al., 2009).
Monolayer stress microscopy
To compute maps of mechanical inter- and intracellular tension within the monolayer sheets, we used monolayer stress microscopy as previously described (Tambe et al., 2011). According to Newton’s law, traction forces applied at the cell–gel interface must be balanced by intra- and intercellular forces (Tambe et al., 2011, 2013; Serra-Picamal et al., 2012). In a 2D approximation, monolayer stress is fully captured by a tensor possessing two independent normal components (σxx and σyy) and two identical shear components (σxy and σyx; Tambe et al., 2011). At every pixel of the monolayer, these four components of the stress tensor define two particular directions of the plane, one in which the normal stress is maximum and one in which it is minimum. These directions, which are mutually orthogonal, are called principal stress orientations, and the stress values in each principal orientation are called maximum (σ11) and minimum (σ22) stress components. The mean normal stress is defined as σn = (σ11 + σ22)/2, and the maximum shear stress is defined as σs = (σ11 − σ22)/2. The spatial resolution and force precision of MSM are formally set by those in the original traction maps (Tambe et al., 2013).
The alignment angle between the major axis of the principal stress ellipse and the direction of the cellular motion was measured as described (Serra-Picamal et al., 2012). The maximum principal stress direction was obtained at each point by diagonalization of the stress tensor. These principal directions were compared with the directions of cell displacement obtained by PIV to obtain the angle between principal stress and cellular motion.
Gel electrophoresis and immunoblotting
Protein extracts (20–60 µg) prepared as described elsewhere (Thuault et al., 2013) were resolved on 10% or 15% polyacrylamide gels and transferred to Immobilon-FL membranes (Millipore). The membranes were incubated with indicated antibodies as described previously (Thuault et al., 2013). For protein detection and quantification, the Odyssey Infrared Imaging System (LI-COR Biosciences) was used. Immunoblots were quantified via densitometry using Odyssey version 3.0 and ImageJ (National Institutes of Health).
Immunoprecipitation
Cell lysates were obtained as described previously (Thuault et al., 2013). Polyclonal anti–β-PIX or P-cadherin antibodies or IgG (1 µg) were incubated with protein G (Dynabeads; Invitrogen) at RT for 30 min. After washing, 1 mg protein extract was added at RT for 1 h.
Immunohistochemistry
Cells were plated in Ibidi’s culture inserts on 12-mm-diameter coverslips until they reached the desired density level. 5–10 h after insert removal, the cells were fixed in 3.2% paraformaldehyde (in PBS) for 15 min, followed by a 2-min permeabilization with 0.1% Triton X-100 (in PBS) and saturation with 2% BSA (in PBS). The cells were incubated with primary and secondary antibodies in PBS containing 2% BSA.
Images were taken using a confocal SP5-SMD (Leica) with 40×/1.3 or 63×/1.4 oil HCX PL APO CS objectives (Leica) and captured with a hybrid detector (Leica HyD) controlled using C software. Colocalization data originating from multichannel fluorescence stacks were collected using a Leica SP5 microscope. For each stack, a single value of the Pearson’s coefficient (ranging from −1 to 1) was measured at the CCJ, imposing a threshold value (calculated based on the algorithms by Costes et al. [2004]) for green and red channels using the colocalization analysis section of Imaris (Bitplane).
Cell surface biotinylation
Cells grown on 60-mm dishes were incubated in cold PBS for 5 min and then with 1 mg/ml sulfosuccinimidyl-2-(biotinamido)ethyl-dithioproprionate (sulfo-NHS-SS-biotin; Pierce Chemical Co.), followed by washing with sulfo-NHS-SS-biotin blocking reagent (50 mM NH4Cl in PBS containing 1 mM MgCl2 and 0.1 mM CaCl2) to quench free sulfo-NHS-SS-biotin, followed by several further washes in PBS. Cells were then scrapped and lysed as described in Thuault et al. (2013) before centrifugation to obtain a detergent-soluble supernatant. At this stage, an aliquot of 10 µl was kept (= total fraction), and the remainder was incubated with streptavidin beads to collect biotinylated proteins. Samples were then analyzed by SDS-PAGE and immunoblotting to identify cadherins using an anti–pan-cadherin antibody. Quantification of at least three independent experiments was performed using the Odyssey system (LI-COR Biosciences).
GTPase activity assays
For global GTPase activity measurements, we induced CCM by injuring a confluent monolayer using a device designed to perform multiple calibrated and reproducible injuries with a spiral scarificator (Turchi et al., 2002). After 6 h of migration, the cells were processed as described to measure Cdc42 and Rac1 activities (Meriane et al., 2002) and RhoA activity (Charrasse et al., 2002). Migrating isolated and confluent cells were also used.
Cdc42 activity measurement
To analyze local Cdc42 activity, we used FRET measurement with the Raïchu-Cdc42 biosensor (1,054×). More details about the probes are available at http://www.fret.lif.kyoto-u.ac.jp/e-phogemon/vector.htm. Cells transfected with the biosensor were plated in Ibidi inserts. After 24 h, the inserts were removed, and after an additional 5 h the cells were imaged using an inverted microscope (TE Eclipse; Nikon) equipped for a 37°C and 5% CO2 humidified atmosphere. The CFP and FRET mages were taken using a Nikon 100× PL APO VC 1.4 oil objective, an excitation filter and dichroic mirror for CFP, and an emission filter wheel containing the CFP emission wavelength filter and another filter for the YFP emission wavelength. Fluorescence was captured with an electron-multiplying CCD camera (C9100-13; Hamamatsu ImagEM) controlled using MetaMorph 7.0 software. A ratio image of FRET/CFP was created to represent FRET efficiency, which correlated with the activity of Cdc42. Pseudocolored ratio images were generated from images from CFP and FRET channels as described previously (Hodgson et al., 2006). Background values were calculated from a mean intensity value within a region of interest outside the cell and subtracted from the entire image. We create a binary mask based on the intensity threshold performed on the FRET image. We applied this mask to CFP and FRET images. Image ratios were calculated by pixel-wise divisions of the masked CFP and masked FRET images. ImageJ Fire LUT was applied to a fixed dynamic range of the 32-bit ratio image.
Cell polarity measurement
5–10 h after Ibidi insert removal, the cells were fixed, and the nuclei, centrosomes, and Golgi apparatus were stained. The cells in the first four rows were analyzed. Measurements of cell polarity toward a global monolayer direction (centrosomes located in front of the nucleus and behind the Golgi apparatus within the quadrant facing the wound) were scored as correctly oriented (Osmani et al., 2006).
FA quantification and orientation
Confocal images of paxillin staining were taken 6 h after removal of the Ibidi insert. The images were binarized to create a mask based on a manually selected threshold level that enabled the visualization of all FAs containing paxillin using an in-laboratory protocol with Cell Profiler software (dataset in supplemental material). The subareas 0–10 µm or 10–40 µm from the leading edge were scored manually using masks, and the FA surface area was measured from binarized images using ImageJ.
FA orientation was measured using ImageJ (dataset in supplemental material), and angle values were plotted using Rose.Net software. FA orientation was determined according to the monolayer migration direction as a reference axis.
FA dynamics
Cells were transfected with the pBabe-paxillin-GFP vector (a gift from C. Albiges-Rizo, Université Joseph Fourier, Grenoble, France); after 24 h, the FAs were imaged during cell migration at 37°C using a confocal microscope (SP5-SMD) equipped with thermal, CO2, and humidity control. Images were taken using a 63×/1.4 oil HCX PL APO CS objective (Leica) and captured with a hybrid detector (Leica HyD) controlled using Leica LAS AF software. Images were taken every 5 s at 5 h after Ibidi insert removal and lasted for 10 to 15 min.
Confocal images of paxillin-GFP–expressing cells were binarized and thresholded using ImageJ. The rate of FA area gains and losses was quantified over 15 min, and mean values were normalized to the FA area in addition to the change in mean intensity. Ratio images were generated using the same procedure.
Statistical analysis
For experiments with n > 30, a Student’s t test was used to assess significant differences between two experimental conditions. For experiments with n < 30, the nonparametric Mann–Whitney U test was used. At least three independent experiments were performed.
Protein separation and identification by LC–MS/MS
C2C12 Pcad cells grown in light and heavy stable isotope labeling with amino acids in cell culture (SILAC) media for 1 wk were used to compare changes in specific P-cadherin interaction partners between control immunoprecipitation using an anti-FLAG antibody (heavy) and immunoprecipitation with a rabbit anti–P-cadherin antibody (light) of cell extracts obtained from confluent cells (IP1) or cells after 6 h of collective migration (IP2) using a device designed to perform multiple calibrated and reproducible injuries with a spiral scarificator (Turchi et al., 2002). 5 mg cell lysates obtained as described in Thuault et al. (2013) were incubated overnight at 4°C with antibody/cross-linked protein A beads, as indicated by the manufacturer (Abcam). After three washes with lysis buffer, beads were recovered by centrifugation and boiled in 15 µl DTT at 95°C for 5 min. Samples from heavy and light conditions were then mixed to obtain a final volume of 30 μl.
Proteins were separated on SDS-PAGE gels (12% polyacrylamide; Mini-Protean TGX Precast Gels; Bio-Rad) and stained with Page Blue Stain (Fermentas). Gel lanes were cut into several gel pieces and destained by three washes in 50% acetonitrile and 50 mM triethylammonium bicarbonate (TEABC). After protein reduction (with 10 mM dithiothreitol in 50 mM TEABC at 56°C for 45 min) and alkylation (55 mM iodoacetamide TEABC at RT for 30 min), proteins were in-gel digested using trypsin (1 µg/band; Gold; Promega). Digested products were dehydrated in a vacuum centrifuge and reduced to 3 µl. The generated peptides were analyzed online by nano-flow HPLC–nanoelectrospray ionization using an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) coupled to an Ultimate 3000 HPLC (Thermo Fisher Scientific). Sample desalting and preconcentration were performed on-line on a Pepmap precolumn (0.3 × 10 mm; Dionex). A gradient consisting of 0–40% B for 60 min and 80% B for 15 min (A = 0.1% formic acid, 2% acetonitrile in water; B = 0.1% formic acid in acetonitrile) at 300 nL/min was used to elute peptides from the capillary reverse-phase column (0.075 × 150 mm, Acclaim Pepmap 100 C18; Thermo Fisher Scientific). Eluted peptides were electrosprayed online at a voltage of 2.2 kV into an LTQ Orbitrap XL mass spectrometer. A cycle of one full-scan mass spectrum (400–2,000 m/z) at a resolution of 60,000 (at 400 m/z), followed by five data-dependent MS/MS spectra was repeated continuously throughout the nanoLC separation. All MS/MS spectra were recorded using normalized collision energy (35%, activation Q 0.25, and activation time 30 ms) with an isolation window of 3 m/z. Data were acquired using Xcalibur software (version 2.0.7). For all full-scan measurements with the Orbitrap detector, a lock-mass ion from ambient air (m/ 445.120024) was used as an internal calibrator. Raw data analysis was performed using MaxQuant software (version 1.5.0.0). Peak lists from IP1 and IP2 were searched against the UniProt mouse database, 250 frequently observed contaminants, as well as the reversed sequences of all entries. Search parameters were the default parameters when using MaxQuant: the search precursor mass tolerance was set at 20 ppm and the main search (after recalibration) at 7 ppm. Enzyme specificity was set to trypsin, and a maximum of two missed cleavages were allowed. Carbamidomethylation was set as fixed cysteine modification and oxidation was set as variable methionine modification for searches. The false discovery rate was set at 0.01 for peptides and proteins and the minimal peptide length at 7. Quantification was also performed using standard parameters; we considered only proteins with at least two peptides identified/quantified, after elimination of reverse and contaminant entries. The statistical validity of the results and the determination of significant proteins were assessed using significance A, as defined using Perseus (version 1.5.2.6, standard parameters), on the logarithmized ratio (base 2). Identified proteins and peptides are included in the supplementary table. The graphic representation (Fig. S4) was generated using Perseus software.
Online supplemental material
Fig. S1 and Videos 1 and 2 show the migration of the four used cell lines and the quantification of cadherin expression levels. Fig. S2 shows that P-cadherin engagement is required for CCM and membrane protrusion polarization. Fig. S3 and Video 4 show C2C12 LZRS and C2C12 Pcad cell morphology and behavior on acrylamide gels. Videos 3 and 8 illustrate FA organization and dynamics. Fig. S4 shows the proteomic analysis. Videos 5, 6, 9, and 10 show the mechanical forces generated by parental and Cdc42 or β-PIX knockdown C2C12 LZRS and C2C12 Pcad cell lines. Video 7 shows the migration of C2C12 Pcad cells in which Cdc42 or β-PIX was knocked down. Fig. S5 shows Cdc42 and β-PIX shRNA efficiency and their effects on velocity, persistence, cadherin accumulation at CCJ, polarity, directionality, and FA number. Rescue experiments show the effect on Cdc42 activity and persistence of expressing β-PIXWT or β-PIX GD in silenced cells. Three datasets are provided for the MatPIV toolbox used to analyze sequential time frames for velocity and angle measurements, the method for FA detection using Cell Profiler software, and the ImageJ macro to measure FA orientation. Table S1 lists all the proteomic data. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201505105/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201505105.dv.
Supplementary Material
Acknowledgments
We thank Virginie Georget, Sylvain de Rossi, and Vicky Dialou from Montpellier Imaging, Halima Amnai and Edith Demettre from Montpellier Proteomic Facilities, Dimitris Liakopoulos for critical reading of the manuscript, and Corinne Albiges-Rizo, Albert Reynolds, and Keith Johnson for plasmids.
This work was supported by the Ligue Nationale contre le Cancer (“Equipe labellisée”) and the Institut National du Cancer. C. Gauthier-Rouvière was supported by Institut National de la Santé et de la Recherche Médicale, C. Plutoni by the Association pour la Recherche contre le Cancer, and M. Le Borgne-Rochet by the Ligue Nationale contre le Cancer, Spanish Ministry of Economy and Competitiveness (grant BFU2012-38146), the Generalitat de Catalunya (grant 2014-SGR-927), and the European Research Council (grant StG-242993 and CoG-616480).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CCJ
- cell–cell junction
- CCM
- collective cell migration
- FA
- focal adhesion
- FRET
- Förster resonance energy transfer
- GEF
- guanine nucleotide exchange factor
- PDMS
- polydimethylsiloxane
- PIV
- particle image velocimetry
- SILAC
- stable isotope labeling with amino acids in cell culture
- TEABC
- triethylammonium bicarbonate
References
- Alexander S., Weigelin B., Winkler F., and Friedl P.. 2013. Preclinical intravital microscopy of the tumour-stroma interface: Invasion, metastasis, and therapy response. Curr. Opin. Cell Biol. 25:659–671. 10.1016/j.ceb.2013.07.001 [DOI] [PubMed] [Google Scholar]
- Arboleda-Estudillo Y., Krieg M., Stühmer J., Licata N.A., Muller D.J., and Heisenberg C.P.. 2010. Movement directionality in collective migration of germ layer progenitors. Curr. Biol. 20:161–169. 10.1016/j.cub.2009.11.036 [DOI] [PubMed] [Google Scholar]
- Bazellières E., Conte V., Elosegui-Artola A., Serra-Picamal X., Bintanel-Morcillo M., Roca-Cusachs P., Muñoz J.J., Sales-Pardo M., Guimerà R., and Trepat X.. 2015. Control of cell-cell forces and collective cell dynamics by the intercellular adhesome. Nat. Cell Biol. 17:409–420. 10.1038/ncb3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beningo K.A., Dembo M., Kaverina I., Small J.V., and Wang Y.L.. 2001. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J. Cell Biol. 153:881–888. 10.1083/jcb.153.4.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley C.D., Tan J., Anderson K.L., Hanein D., Volkmann N., Weis W.I., Nelson W.J., and Dunn A.R.. 2014. Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science. 346:1254211 10.1126/science.1254211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D., Chen S.C., Prasad M., He L., Wang X., Choesmel-Cadamuro V., Sawyer J.K., Danuser G., and Montell D.J.. 2014. Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell. 157:1146–1159. 10.1016/j.cell.2014.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau J., and Hall A.. 2005. Cdc42 controls the polarity of the actin and microtubule cytoskeletons through two distinct signal transduction pathways. J. Cell Sci. 118:2579–2587. 10.1242/jcs.02385 [DOI] [PubMed] [Google Scholar]
- Charrasse S., Meriane M., Comunale F., Blangy A., and Gauthier-Rouvière C.. 2002. N-cadherin-dependent cell-cell contact regulates Rho GTPases and β-catenin localization in mouse C2C12 myoblasts. J. Cell Biol. 158:953–965. 10.1083/jcb.200202034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrasse S., Comunale F., Gilbert E., Delattre O., and Gauthier-Rouvière C.. 2004. Variation in cadherins and catenins expression is linked to both proliferation and transformation of rhabdomyosarcoma. Oncogene. 23:2420–2430. 10.1038/sj.onc.1207382 [DOI] [PubMed] [Google Scholar]
- Costes S.V., Daelemans D., Cho E.H., Dobbin Z., Pavlakis G., and Lockett S.. 2004. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 86:3993–4003. 10.1529/biophysj.103.038422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A.M., Eggermont A.M., and ten Hagen T.L.. 2015a A ring barrier-based migration assay to assess cell migration in vitro. Nat. Protoc. 10:904–915. 10.1038/nprot.2015.056 [DOI] [PubMed] [Google Scholar]
- Das T., Safferling K., Rausch S., Grabe N., Boehm H., and Spatz J.P.. 2015b A molecular mechanotransduction pathway regulates collective migration of epithelial cells. Nat. Cell Biol. 17:276–287. 10.1038/ncb3115 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., and Hall A.. 2001. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 106:489–498. 10.1016/S0092-8674(01)00471-8 [DOI] [PubMed] [Google Scholar]
- Farooqui R., and Fenteany G.. 2005. Multiple rows of cells behind an epithelial wound edge extend cryptic lamellipodia to collectively drive cell-sheet movement. J. Cell Sci. 118:51–63. 10.1242/jcs.01577 [DOI] [PubMed] [Google Scholar]
- Friedl P., and Gilmour D.. 2009. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10:445–457. 10.1038/nrm2720 [DOI] [PubMed] [Google Scholar]
- Friedl P., Noble P.B., Walton P.A., Laird D.W., Chauvin P.J., Tabah R.J., Black M., and Zänker K.S.. 1995. Migration of coordinated cell clusters in mesenchymal and epithelial cancer explants in vitro. Cancer Res. 55:4557–4560. [PubMed] [Google Scholar]
- Friedl P., Locker J., Sahai E., and Segall J.E.. 2012. Classifying collective cancer cell invasion. Nat. Cell Biol. 14:777–783. 10.1038/ncb2548 [DOI] [PubMed] [Google Scholar]
- Halbleib J.M., and Nelson W.J.. 2006. Cadherins in development: Cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 20:3199–3214. 10.1101/gad.1486806 [DOI] [PubMed] [Google Scholar]
- Hidalgo-Carcedo C., Hooper S., Chaudhry S.I., Williamson P., Harrington K., Leitinger B., and Sahai E.. 2011. Collective cell migration requires suppression of actomyosin at cell-cell contacts mediated by DDR1 and the cell polarity regulators Par3 and Par6. Nat. Cell Biol. 13:49–58. 10.1038/ncb2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirashima T., Hosokawa Y., Iino T., and Nagayama M.. 2013. On fundamental cellular processes for emergence of collective epithelial movement. Biol. Open. 2:660–666. 10.1242/bio.20134523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson L., Nalbant P., Shen F., and Hahn K.. 2006. Imaging and photobleach correction of Mero-CBD, sensor of endogenous Cdc42 activation. Methods Enzymol. 406:140–156. 10.1016/S0076-6879(06)06012-5 [DOI] [PubMed] [Google Scholar]
- Itoh R.E., Kurokawa K., Ohba Y., Yoshizaki H., Mochizuki N., and Matsuda M.. 2002. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol. Cell. Biol. 22:6582–6591. 10.1128/MCB.22.18.6582-6591.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasaitis A., Estevez M., Heysch J., Ladoux B., and Dufour S.. 2012. E-cadherin-dependent stimulation of traction force at focal adhesions via the Src and PI3K signaling pathways. Biophys. J. 103:175–184. 10.1016/j.bpj.2012.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandow C.E., Georges P.C., Janmey P.A., and Beningo K.A.. 2007. Polyacrylamide hydrogels for cell mechanics: steps toward optimization and alternative uses. Methods Cell Biol. 83:29–46. 10.1016/S0091-679X(07)83002-0 [DOI] [PubMed] [Google Scholar]
- Khalil A.A., and Friedl P.. 2010. Determinants of leader cells in collective cell migration. Integr. Biol. (Camb.). 2:568–574. 10.1039/c0ib00052c [DOI] [PubMed] [Google Scholar]
- Kutys M.L., and Yamada K.M.. 2014. An extracellular-matrix-specific GEF-GAP interaction regulates Rho GTPase crosstalk for 3D collagen migration. Nat. Cell Biol. 16:909–917. 10.1038/ncb3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti M.C., Joanny S., Rmamswamy T.B., Prost J., Rao M., and Aditi Simba R.. 2013. Hydrodynamics of soft active matter. Rev. Mod. Phys. 85:1143–1189. 10.1103/RevModPhys.85.1143 [DOI] [Google Scholar]
- Maruthamuthu V., Sabass B., Schwarz U.S., and Gardel M.L.. 2011. Cell-ECM traction force modulates endogenous tension at cell-cell contacts. Proc. Natl. Acad. Sci. USA. 108:4708–4713. 10.1073/pnas.1011123108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meriane M., Charrasse S., Comunale F., Méry A., Fort P., Roux P., and Gauthier-Rouvière C.. 2002. Participation of small GTPases Rac1 and Cdc42Hs in myoblast transformation. Oncogene. 21:2901–2907. 10.1038/sj.onc.1205396 [DOI] [PubMed] [Google Scholar]
- Mertz A.F., Banerjee S., Che Y., German G.K., Xu Y., Hyland C., Marchetti M.C., Horsley V., and Dufresne E.R.. 2012. Scaling of traction forces with the size of cohesive cell colonies. Phys. Rev. Lett. 108:198101 10.1103/PhysRevLett.108.198101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz A.F., Che Y., Banerjee S., Goldstein J.M., Rosowski K.A., Revilla S.F., Niessen C.M., Marchetti M.C., Dufresne E.R., and Horsley V.. 2013. Cadherin-based intercellular adhesions organize epithelial cell-matrix traction forces. Proc. Natl. Acad. Sci. USA. 110:842–847. 10.1073/pnas.1217279110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M.R., Besser A., Danuser G., and Brugge J.S.. 2012. Substrate stiffness regulates cadherin-dependent collective migration through myosin-II contractility. J. Cell Biol. 199:545–563. 10.1083/jcb.201207148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omelchenko T., and Hall A.. 2012. Myosin-IXA regulates collective epithelial cell migration by targeting RhoGAP activity to cell-cell junctions. Curr. Biol. 22:278–288. 10.1016/j.cub.2012.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omelchenko T., Rabadan M.A., Hernández-Martínez R., Grego-Bessa J., Anderson K.V., and Hall A.. 2014. β-Pix directs collective migration of anterior visceral endoderm cells in the early mouse embryo. Genes Dev. 28:2764–2777. 10.1101/gad.251371.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani N., Vitale N., Borg J.P., and Etienne-Manneville S.. 2006. Scrib controls Cdc42 localization and activity to promote cell polarization during astrocyte migration. Curr. Biol. 16:2395–2405. 10.1016/j.cub.2006.10.026 [DOI] [PubMed] [Google Scholar]
- Ostuni E., Kane R.S., Chen C.S., Ingber D.E., and Whitesides G.M.. 2000. Patterning mammalian cells using elastomeric membranes. Langmuir. 16:7811–7819. 10.1021/la000382m [DOI] [Google Scholar]
- Paredes J., Figueiredo J., Albergaria A., Oliveira P., Carvalho J., Ribeiro A.S., Caldeira J., Costa A.M., Simões-Correia J., Oliveira M.J., et al. . 2012. Epithelial E- and P-cadherins: Role and clinical significance in cancer. Biochim. Biophys. Acta. 1826:297–311. 10.1016/j.bbcan.2012.05.002 [DOI] [PubMed] [Google Scholar]
- Petitjean L., Reffay M., Grasland-Mongrain E., Poujade M., Ladoux B., Buguin A., and Silberzan P.. 2010. Velocity fields in a collectively migrating epithelium. Biophys. J. 98:1790–1800. 10.1016/j.bpj.2010.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poujade M., Grasland-Mongrain E., Hertzog A., Jouanneau J., Chavrier P., Ladoux B., Buguin A., and Silberzan P.. 2007. Collective migration of an epithelial monolayer in response to a model wound. Proc. Natl. Acad. Sci. USA. 104:15988–15993. 10.1073/pnas.0705062104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reffay M., Parrini M.C., Cochet-Escartin O., Ladoux B., Buguin A., Coscoy S., Amblard F., Camonis J., and Silberzan P.. 2014. Interplay of RhoA and mechanical forces in collective cell migration driven by leader cells. Nat. Cell Biol. 16:217–223. 10.1038/ncb2917 [DOI] [PubMed] [Google Scholar]
- Reinhart-King C.A., Dembo M., and Hammer D.A.. 2003. Endothelial cell traction forces on RGD-derivatized polyacrylamide substrata. Langmuir. 19:1573–1579. 10.1021/la026142j [DOI] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., and Horwitz A.R.. 2003. Cell migration: integrating signals from front to back. Science. 302:1704–1709. 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Rørth P. 2009. Collective cell migration. Annu. Rev. Cell Dev. Biol. 25:407–429. 10.1146/annurev.cellbio.042308.113231 [DOI] [PubMed] [Google Scholar]
- Serra-Picamal X., Conte V., Vincent R., Anon E., Tambe D.T., Bazellieres E., Butler J.P., Fredberg J.J., and Trepat X.. 2012. Mechanical waves during tissue expansion. Nat. Phys. 8:628–634. 10.1038/nphys2355 [DOI] [Google Scholar]
- Stricker J., Aratyn-Schaus Y., Oakes P.W., and Gardel M.L.. 2011. Spatiotemporal constraints on the force-dependent growth of focal adhesions. Biophys. J. 100:2883–2893. 10.1016/j.bpj.2011.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambe D.T., Hardin C.C., Angelini T.E., Rajendran K., Park C.Y., Serra-Picamal X., Zhou E.H., Zaman M.H., Butler J.P., Weitz D.A., et al. . 2011. Collective cell guidance by cooperative intercellular forces. Nat. Mater. 10:469–475. 10.1038/nmat3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambe D.T., Croutelle U., Trepat X., Park C.Y., Kim J.H., Millet E., Butler J.P., and Fredberg J.J.. 2013. Monolayer stress microscopy: Limitations, artifacts, and accuracy of recovered intercellular stresses. PLoS One. 8:e55172 10.1371/journal.pone.0055172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J.L., Tien J., Pirone D.M., Gray D.S., Bhadriraju K., and Chen C.S.. 2003. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc. Natl. Acad. Sci. USA. 100:1484–1489. 10.1073/pnas.0235407100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E., and Mayor R.. 2011. Can mesenchymal cells undergo collective cell migration? The case of the neural crest. Cell Adhes. Migr. 5:490–498. 10.4161/cam.5.6.18623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E., Marchant L., Kuriyama S., Gull M., Moepps B., Parsons M., and Mayor R.. 2010. Collective chemotaxis requires contact-dependent cell polarity. Dev. Cell. 19:39–53. 10.1016/j.devcel.2010.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuault S., Hayashi S., Lagirand-Cantaloube J., Plutoni C., Comunale F., Delattre O., Relaix F., and Gauthier-Rouvière C.. 2013. P-cadherin is a direct PAX3-FOXO1A target involved in alveolar rhabdomyosarcoma aggressiveness. Oncogene. 32:1876–1887. 10.1038/onc.2012.217 [DOI] [PubMed] [Google Scholar]
- Trepat X., and Fredberg J.J.. 2011. Plithotaxis and emergent dynamics in collective cellular migration. Trends Cell Biol. 21:638–646. 10.1016/j.tcb.2011.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepat X., Wasserman M.R., Angelini T.E., Millet E., Weitz D.A., Butler J.P., and Fredberg J.J.. 2009. Physical forces during collective cell migration. Nat. Phys. 5:426–430. 10.1038/nphys1269 [DOI] [Google Scholar]
- Trepat X., Fabry B., and Fredberg J.J.. 2010. Pulling it together in three dimensions. Nat. Methods. 7:963–965. 10.1038/nmeth1210-963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchi L., Chassot A.A., Rezzonico R., Yeow K., Loubat A., Ferrua B., Lenegrate G., Ortonne J.P., and Ponzio G.. 2002. Dynamic characterization of the molecular events during in vitro epidermal wound healing. J. Invest. Dermatol. 119:56–63. 10.1046/j.1523-1747.2002.01805.x [DOI] [PubMed] [Google Scholar]
- van Roy F. 2014. Beyond E-cadherin: Roles of other cadherin superfamily members in cancer. Nat. Rev. Cancer. 14:121–134. 10.1038/nrc3647 [DOI] [PubMed] [Google Scholar]
- Wang X., He L., Wu Y.I., Hahn K.M., and Montell D.J.. 2010. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell Biol. 12:591–597. 10.1038/ncb2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G.F., Bjerke M.A., and DeSimone D.W.. 2012. A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration. Dev. Cell. 22:104–115. 10.1016/j.devcel.2011.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S., Puri S., and Linstedt A.D.. 2009. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol. Biol. Cell. 20:1728–1736. 10.1091/mbc.E08-10-1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M., Qiu W., Liu R., Efremov A.K., Cong P., Seddiki R., Payre M., Lim C.T., Ladoux B., Mège R.M., and Yan J.. 2014. Force-dependent conformational switch of α-catenin controls vinculin binding. Nat. Commun. 5:4525 10.1038/ncomms5525 [DOI] [PubMed] [Google Scholar]
- Yeung T., Georges P.C., Flanagan L.A., Marg B., Ortiz M., Funaki M., Zahir N., Ming W., Weaver V., and Janmey P.A.. 2005. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton. 60:24–34. 10.1002/cm.20041 [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R., Ballestrem C., Kam Z., and Geiger B.. 2003. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 116:4605–4613. 10.1242/jcs.00792 [DOI] [PubMed] [Google Scholar]
- Zegers M.M., and Friedl P.. 2014. Rho GTPases in collective cell migration. Small GTPases. 5:e28997 10.4161/sgtp.28997 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.