Summary
Hydroxyproline O‐arabinosyltransferases (HPATs) are members of a small, deeply conserved family of plant‐specific glycosyltransferases that add arabinose sugars to diverse proteins including cell wall‐associated extensins and small signaling peptides. Recent genetic studies in flowering plants suggest that different HPAT homologs have been co‐opted to function in diverse species‐specific developmental contexts. However, nothing is known about the roles of HPATs in basal plants. We show that complete loss of HPAT function in Arabidopsis thaliana and the moss Physcomitrella patens results in a shared defect in gametophytic tip cell growth. Arabidopsis hpat1/2/3 triple knockout mutants suffer from a strong male sterility defect as a consequence of pollen tubes that fail to fully elongate following pollination. Knocking out the two HPAT genes of Physcomitrella results in larger multicellular filamentous networks due to increased elongation of protonemal tip cells. Physcomitrella hpat mutants lack cell‐wall associated hydroxyproline arabinosides and can be rescued with exogenous cellulose, while global expression profiling shows that cell wall‐associated genes are severely misexpressed, implicating a defect in cell wall formation during tip growth. Our findings point to a major role for HPATs in influencing cell elongation during tip growth in plants.
Keywords: Arabidopsis thaliana, Physcomitrella patens, cell wall, tip growth, glycosylation, arabinosylation, development, extensins, pollination, protonema
Significance Statement
Functionally important genes are evolutionarily conserved, but can be co‐opted for new functions in lineage‐specific contexts. Here we show that the phenotypes of mutants blocked in Hydroyproline O‐arabinosylation in Arabidopsis and in moss are opposite in tip‐growing cells, with shorter pollen tubes but longer protonemal cells, than in their WT counterparts.
Introduction
Glycosylation, or the addition of sugars to proteins, is a common post‐translational modification that serves several functions including regulation of protein folding, stability and structure (Varki et al., 2009). The plant‐specific modification, hydroyxyproline O‐arabinosylation, occurs as linear oligoarabinoside chains primarily on hydroxyproline‐rich glycoproteins (HRGPs), specifically members of the extensin subgroup, as well as secreted signaling peptides including the stem cell regulator CLAVATA3 (CLV3) and related CLE (CLAVATA3/ENDOSPERM SURROUNDING REGION) peptides (Roberts et al., 1985; Ohyama et al., 2009; Hijazi et al., 2014). Though hydroxyproline O‐arabinosylation has been known for decades (Lamport et al., 2011), the enzymes responsible for initiating oligoarabinose chains, the hydroxyproline O‐arabinosyltransferases (HPATs), have only recently been identified (Ogawa‐Ohnishi et al., 2013). A genetic analysis of individual loss‐of‐function mutations in the three Arabidopsis HPAT‐encoding genes initially revealed no phenotypes; however, HPAT1 and HPAT3 were found to be redundantly required for male gamete transmission and hpat1 hpat2 double knockout mutant plants exhibited a wide range of developmental defects, including longer hypocotyls for light‐grown seedlings, early flowering under long‐day conditions and precocious leaf senescence (Ogawa‐Ohnishi et al., 2013). Recently, the single hpat mutants and the hpat1 hpat2 double mutant were further shown to be necessary for full root hair elongation (Velasquez et al., 2015). These pleiotropic phenotypes may be caused by cell wall defects owing to under‐arabinosylation of extensins (Ogawa‐Ohnishi et al., 2013). Following glycosylation of characteristic repeated Ser(Hyp)3–5 motifs (where Hyp is hydroxyproline), extensins are secreted into the cell wall and progressively cross‐linked into a network that may serve as a scaffold for further cell wall assembly (Epstein and Lamport, 1984; Everdeen et al., 1988; Kieliszewski and Lamport, 1994; Held et al., 2004; Cannon et al., 2008; Showalter et al., 2010; Lamport et al., 2011). Extensin3/root‐shoot‐hypocotyl defective (ext3/rsh) is among the few extensins with a reported mutant phenotype; cell plate formation is disrupted in this mutant, leading to severe embryonic growth defects (Hall and Cannon, 2002). In hpat1 hpat2 double and hpat3 single mutants, the level of arabinosylation of EXT3 is reduced, and hypocotyls of hpat1 hpat2 double mutants exhibit thinner cell walls than their wild‐type counterparts (Ogawa‐Ohnishi et al., 2013). Other extensins with described mutant phenotypes include ext6, ‐7, ‐10 and ‐12, which all exhibit shorter root hairs. Similarly, in the root the function of other extensin‐modifying enzymes is also required for full root hair elongation (Velasquez et al., 2011).
Our interest in HPATs and their roles in development arose from our recent finding that mutations in the closest tomato homolog of HPAT3, FASCIATED INFLORESCENCE (FIN), cause dramatically enlarged shoot meristems that result in branched inflorescences and flowers and fruits with more organs, similar to Arabidopsis clavata3 (clv3) mutants (Xu et al., 2015). The mature 12‐amino‐acid CLV3 peptide carries a linear triarabinose chain on a Hyp residue at position 7 (Hyp7), and the presence of these sugars increases the biological activity of exogenously added peptide (Ohyama et al., 2009; Okamoto et al., 2013; Shinohara and Matsubayashi, 2013). Likewise, application of [Ara3]SlCLV3 can rescue fin meristem enlargement, demonstrating a critical role for HPATs in controlling stem cell proliferation (Xu et al., 2015). Notably, members of the CLE family also regulate Rhizobia‐induced nodule formation in legumes (Reid et al., 2011; Okamoto et al., 2013), and a supernodulating mutant from Medicago truncatula, root determined nodulation1 (rdn1), and its pea ortholog (nod3) are defective in the closest homolog of HPAT3 (Schnabel et al., 2011). As HPAT3 has been shown to be largely responsible for arabinosylation of CLE2, which is structurally similar to Rhizobia‐induced CLE peptides, loss of CLE peptide arabinosylation is the likely basis of the rdn1/nod3 phenotype (Ogawa‐Ohnishi et al., 2013). Collectively, these findings indicate that HPATs control many diverse aspects of development in flowering plants.
Given that CLV3 arabinosylation was first discovered in Arabidopsis and that the arabinose chain increases peptide activity in vitro (Ohyama et al., 2009), we found it surprising that none of the single hpat mutants, or the reported double mutant combinations, have defects in shoot meristem size (Ogawa‐Ohnishi et al., 2013). However, the difficulty in recovering hpat1 hpat3 double mutant or hpat1 hpat2 hpat 3 triple mutant plants due to the male transmission defect has precluded a full exploration of possible redundancy among the Arabidopsis HPAT genes. To address this issue, we generated and characterized a complete loss‐of‐function HPAT triple mutant. Simultaneously, we knocked out the two HPAT genes in the model moss Physcomitrella patens (Figures 1a and S1) to investigate HPAT redundancy and specialization in plants more broadly. The simple body plan of Physcomitrella lacks the organs and tissues that are altered in angiosperm hpat mutants, including multicellular meristems (fin), nodules (rdn1) and pollen (hpat1 hpat3). Moreover, despite having diverged from angiosperms about 450 million years ago, the Physcomitrella and Arabidopsis HPAT proteins share 60–65% sequence identity (Figure S1b; Lang et al., 2008). The stark contrast between the strong HPAT sequence conservation and the diverse pleiotropic hpat angiosperm mutant phenotypes raises the question of the ancestral function of hydroxyproline O‐arabinosylation, and how that role was co‐opted into different developmental contexts over evolutionary time and in different plant lineages.
Figure 1.
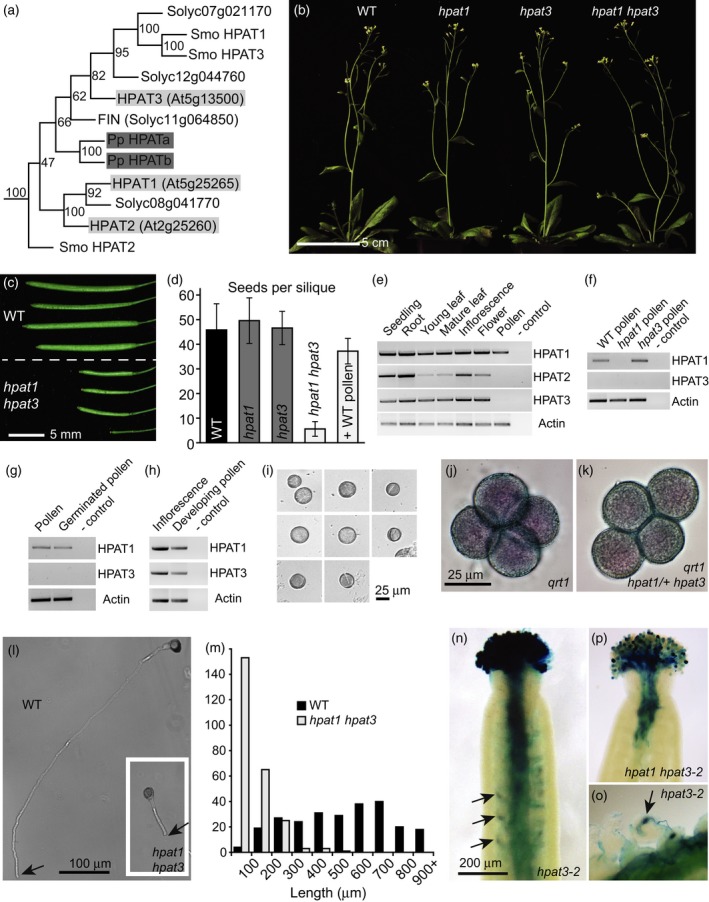
HPAT1 and HPAT3 function redundantly in pollen tube growth.(a) Maximum parsimony phylogenetic tree of the hydroxyproline O‐arabinosyltransferase (HPAT) proteins from Arabidopsis, tomato (Solyc), Physcomitrella patens (Pp) and Selaginella moellendorffii (Smo) with bootstrap support values in nodes. Arabidopsis and Physcomitrella proteins are marked in light and dark grey respectively. (b) Wild type (WT, Columbia‐0), hpat1, hpat3 and hpat1 hpat3 mutant plants all appear morphologically normal. (c) Fully expanded siliques from WT (top) and hpat1 hpat3 double mutants (bottom). (d) Number of seeds per silique (mean ± SD, n = 25) for WT (black bar), single mutants (dark grey bars) and hpat1 hpat3 double mutants (light grey bars), with and without pollination using WT pollen. (e)–(h) HPAT expression. Though generally broadly expressed, HPAT3 transcripts are not detected in mature WT pollen (e), in hpat1 pollen (f), or in WT in vitro germinated pollen (g). However, transcripts are detected in a mixture of developing pollen stages (h), like those shown in (i). (j), (k) qrt1 pollen tetrads stained with simplified Alexander's viability stain. Like qrt1 alone (j), all members of a tetrad from qrt1 hpat1/+ hpat3 plants appear morphologically normal (k). (l) Pollen tubes from WT and hpat1 hpat3 genotypes (inset) grown for 8 h in vitro. Arrows mark the tip of the pollen tube. (m) Distribution of pollen tube lengths after 8 h of in vitro pollen tube growth (n = 250 tubes per genotype). (n)–(p) hpat3‐2 mutants carry a pollen‐specific GUS reporter within the T‐DNA construct allowing mutant pollen to stain blue. Seven hours after pollination of emasculated WT stigmas with hpat3‐2 (n, o) blue pollen tubes can be seen in the ovary and targeting ovules (arrows) and dissection of the style reveals GUS deposition in the ovule upon fertilization (o). After pollination with hpat1 hpat3‐2/+ pollen, the double mutant pollen tubes penetrate poorly and fail to target ovules (p).
Results
Arabidopsis hpat1 hpat3 double mutants are partially male sterile due to a defect in pollen tube elongation
Individual T‐DNA insertion mutations that eliminated transcription of each of the three Arabidopsis HPAT genes resulted in morphologically normal plants, as reported for both the original null alleles (hpat1‐1, hpat2‐1 and hpat3‐1) and those in use here (hpat1‐2, hpat2‐2, hpat3‐1 and hpat3‐2; Ogawa‐Ohnishi et al., 2013; Figure S2). A redundant requirement for HPAT1 and HPAT3 in male transmission has prevented the recovery of hpat1 hpat3 double mutant plants, which could have sporophytic phenotypes such as a defect in meristem size (Ogawa‐Ohnishi et al., 2013). HPAT1 and HPAT3 are linked on chromosome 5 (approximately 4 Mb apart corresponding to 15 cM), and due to the reported transmission defect (Ogawa‐Ohnishi et al., 2013) homozygous hpat1 hpat3 double mutants would be expected at very low frequencies in F2 populations segregating for mutations in both genes. Therefore, we first isolated recombinants in which one mutation was homozygous and the other heterozygous. From 446 total progeny from self‐pollination of both hpat1‐2/+ hpat3‐1 plants (hereafter these alleles are designated hpat1 and hpat3 unless otherwise noted, n = 240) and hpat1 hpat3/+ (n = 206) plants, we recovered a single hpat1 hpat3 double mutant plant. This double mutant appeared morphologically normal during the vegetative and early reproductive phases (Figure 1b), but developed shorter siliques compared with wild type (WT) plants due to a reduced number of seeds per silique (Figures 1c,d and S3a). Full seed set could be achieved by applying WT pollen to hpat1 hpat3 stigmas (Figure 1d) or by expressing either HPAT1 or HPAT3 under a pollen‐specific promoter in the double mutants (Schneidereit et al., 2003; Figure S3b). When pollen from hpat1/+ or hpat3/+ plants was used to pollinate the WT, we observed no defect in the transmission of the single mutant gametes relative to WT, confirming that these genes act redundantly in pollen function (Table S1).
The redundant requirement for HPAT1 and HPAT3 for efficient transmission of male gametes suggested that both genes control pollen development. We therefore expected to detect expression of both genes in developing pollen grains and/or pollen tubes. HPAT gene transcripts are detected in several tissues by RT‐PCR (Ogawa‐Ohnishi et al., 2013), but we failed to detect expression of HPAT2 or, more surprisingly, HPAT3 in mature pollen (Figure 1e). To test if HPAT3 expression is induced in the absence of HPAT1 through a compensation mechanism, we checked for expression of HPAT3 in hpat1 pollen, but still saw no expression (Figure 1f). Similarly, HPAT3 expression was not induced upon in vitro pollen germination (Figure 1g). However, HPAT3 was expressed in a mixture of developing pollen stages (Figure 1h,i), and microarrays analysis of several precise stages of pollen development showed HPAT3 expression in uninucleate microspores and bicellular pollen but not in immature tricellular pollen or mature pollen grains (Honys and Twell, 2004), in agreement with our observations. A few possible explanations exist for the absence of HPAT3 expression in mature pollen when it is redundantly required for pollen transmission. For example, the HPAT3 protein may be sufficiently stable to maintain activity during later pollen stages in the absence of detectable mRNA, it may be required during earlier developmental stages to modify a target protein that is then stored for later use or HPAT3 could be induced in the pollen only upon interaction with the female tissue. To better understand the function of HPAT in male gamete transmission, we next examined a number of stages of pollen development and function.
Fertilization is a complex process requiring the successful execution of a number of steps in order for the sperm nuclei of the pollen tube to fuse with the egg and central cells of the ovule to ultimately produce a viable seed (Bleckmann et al., 2014). Pollen that fails to properly develop, germinate, elongate or target ovules can lead to unfertilized ovules and siliques with reduced seed set. Therefore, we examined hpat1 hpat3 pollen at several stages. To facilitate these observations, we crossed hpat1/+ hpat3 plants to the quartet1 (qrt1) mutant, whose products of male meiosis fail to separate, resulting in fused pollen tetrads (Preuss et al., 1994). Like qrt1 alone, pollen from hpat1/+ hpat3 qrt1 plants produced four morphologically normal grains per tetrad (Figure 1j,k), suggesting that, at the level of gross morphology, the initial stages of pollen development are unaffected by mutations in HPAT1 and HPAT3.
We next germinated hpat1 hpat3 and WT pollen in vitro and observed that the double mutant pollen tubes were substantially shorter than WT pollen tubes (Figure 1l,m), in agreement with previous observations of pollen from plants segregating for hpat1 hpat3 double mutant pollen (Ogawa‐Ohnishi et al., 2013). To evaluate pollen function in vivo we recovered a second transcriptionally null allele of hpat3 (hpat3‐2) from the Syngenta Arabidopsis Insertion Library (SAIL) collection (Figure S2). A subset of SAIL lines, including hpat3‐2, carries a pollen‐specific promoter driving β‐glucuronidase within the T‐DNA. Pollen and pollen tubes carrying this T‐DNA stain blue in the presence of X‐glucuronide, allowing discrimination between WT and mutant pollen (Sessions et al., 2002). Like the original allele of hpat3, hpat1 hpat3‐2 pollen transmitted poorly, making it difficult to recover double mutants (no double mutants were recovered among 123 progeny from hpat1 hpat3‐2/+ plants). To determine if shorter hpat1 hpat3 pollen tubes were responsible for the male transmission defect, we pollinated WT stigmas with hpat1 hpat3‐2/+ or hpat3‐2 pollen and stained pistils for GUS activity after 7 h. By this time, pollen tubes from hpat3‐2 single mutants had reached ovules and successfully fertilized them, as seen by blue staining in ovules (Figure 1n,o). In contrast, double mutant pollen segregating from hpat1 hpat3‐2/+ flowers germinated and penetrated the top of WT ovaries but no ovules were targeted for fertilization (Figure 1p). At this point, all ovules were probably already fertilized by the hpat1 single mutant pollen. Thus, although at least some double mutant pollen is capable of fertilization, as evidenced from the few seeds produced by homozygous hpat1 hpat3 plants (Figures 1d and S3a), our results suggest that hpat1 hpat3 pollen, due to a defect in pollen tube elongation, is at a strong competitive disadvantage in the presence of WT or single hpat1 or hpat3 mutant pollen.
Arabidopsis hpat triple mutants are not fasciated
Our work in tomato identified the FIN/HPAT3 gene as a major regulator of shoot meristem size acting through the arabinosylation of SlCLV3 (Xu et al., 2015). Meristem enlargement, as seen in tomato fin mutants and the Arabidopsis and tomato clv3 mutants, causes thickened stems, branched inflorescences and the formation of extra flowers and floral organs (Clark et al., 1995; Xu et al., 2015; Figure 2e). None of the single Arabidopsis hpat mutants showed a similar defect (Figure 2f–j). To address potential redundancy between HPAT family members, we used our hpat1 hpat3 double mutant to generate hpat1 hpat2 hpat3 triple mutant plants (Figure S4a–c). Like hpat1 hpat3 double mutant plants, we found that hpat1 hpat2 hpat3 triple mutants exhibited low fertility (Figure S4d,e) and shorter pollen tubes in vitro (Figure S4f). Surprisingly, however, both vegetative and inflorescence shoots were normal (Figure 2a–c), and flowers did not produce extra organs even when grown under short‐day conditions to prolong the activity of the shoot meristem (Figure 2n). Other hpat1 hpat2 phenotypes reported previously (Ogawa‐Ohnishi et al., 2013) were not apparent in our double or triple mutants, possibly due to differences in growth conditions. Thus, the most severe phenotype observed in our complete set of Arabidopsis single and higher‐order hpat mutants was a defect of pollen tube elongation.
Figure 2.
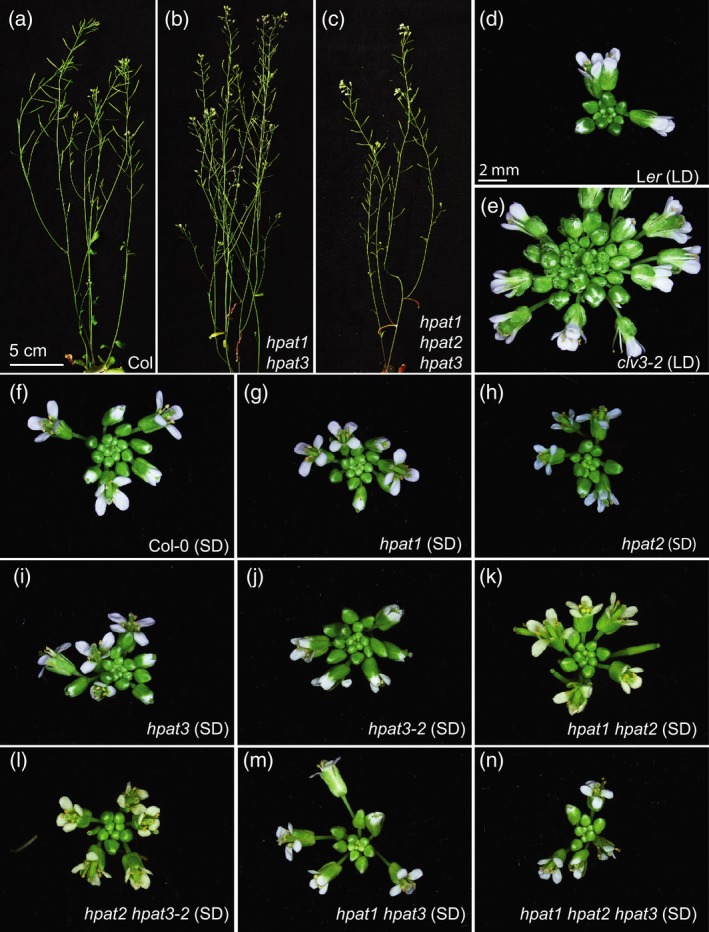
hpat single, double and triple mutants are not fasciated.(a)–(c) Wild type (WT) Columbia‐0 (a), hpat1 hpat3 double mutants (b), and hpat1 hpat2 hpat3 triple mutants (c) show similar growth and morphologies under standard conditions. (d), (e) Compared with the WT background, (Ler, d) clv3‐2 shoots become progressively and severely fasciated due to enlarged meristems, resulting in the initiation of extra floral buds and floral organs in standard long‐day (LD) conditions (e). (f)–(n) Even when flowering and senescence are delayed by growth in short‐day (SD) conditions, hpat single, double and triple mutant inflorescences appear normal and do not initiate extra floral buds or floral organs relative to WT plants (Columbia‐0, f).
Disrupted HPAT function in Physcomitrella patens enhances vegetative network growth
The redundant role of Arabidopsis HPAT1 and HPAT3 in pollen tube growth is in stark contrast with the tomato fin meristem and Medicago rdn1 nodulation phenotypes (Schnabel et al., 2011; Xu et al., 2015). These species‐specific mutant phenotypes, which probably reflect loss of arabinosylation on extensins (hpat1 hpat3) and CLV3/CLE proteins (fin and rdn1), suggest that HPATs have been co‐opted to function in distinct developmental contexts in flowering plants. Yet the deep conservation of the HPATs suggests critical functions across the plant kingdom and raises questions about the roles of HPATs in distantly related plants. To address this, we used the model moss P. patens.
Early development in Physcomitrella involves the formation of a branching network of filaments that are collectively known as protonema. Protonema, like pollen tubes, root hairs and rhizoids, elongate by a ‘tip‐growth’ mechanism, in which secretion of new cell wall material is highly polarized and directed to a single growing point, while the remainder of the cell wall does not expand (Rounds and Bezanilla, 2013; Becker et al., 2014). This contrasts with the diffuse growth of other cell types, in which expansion happens more broadly (Braidwood et al., 2013). The protonemal network is composed of two cell types, chloronema and caulonema, that are distinguished by their growth rate, cell length, morphology and further developmental potential (Menand et al., 2007a). Chloronema are produced first following spore germination or subculture, and are distinguished by their larger and more numerous chloroplasts, a slower rate of cell elongation and division, and perpendicular rather than oblique end walls. Interconversion between cell types occurs based on environmental conditions, and because the transition is often gradual, cells of intermediate identity may exist (Jang and Dolan, 2011). Caulonema are further distinguished by their greater ability to initiate the buds that develop into leafy gametophores, which will form anchoring rhizoid filaments and eventually the archegonia and antheridia needed for sexual reproduction.
The Physcomitrella genome contains two members of the HPAT family, HPATa (Pp1s145_88V6.1) and HPATb (Pp1s48_116V6.1), which share 87.0% protein identity (Figures 1a and S1). Both genes are expressed in several tissues, with HPATa expression being consistently higher than that of HPATb (Figure S5a; Hiss et al., 2014). To investigate the role of the HPATa and HPATb genes in moss development, we generated single and double knockout mutants by homologous recombination, which we validated by quantitative PCR (Figure S5; Cove et al., 2009). Since our mutant lines replaced the coding sequences with a selection cassette including a GFP‐GUS reporter, we were able to detect expression from the native promoter by staining for GUS activity. In agreement with the microarray results (Figure S5a), we saw broad GUS expression in the single copy HPATa insertion line #41 (Figure S5c,d).
In multiple hpata mutant lines we observed a significant increase in the diameter of the protonemal network. In contrast, we observed no change in network size in any hpatb mutant lines, and there was no further increase in size in hpata hpatb double mutants relative to hpata (Figures 3a–d and S5e,f). Following a single reference line for each genotype (Figure S5), we observed that the increase in protonemal network size occurred early in development (Figure 3e), and was visible in young networks as an increase in the number and length of filaments leaving the denser central network (Figure 3f,g). This increase in diameter also translated to an increased dry weight biomass at 21 days post‐subculture (Figure 3h). The size and composition of the Physcomitrella protonemal network is controlled by endogenous and environmental cues including hormone and nutrient status (Cove et al., 2006). We found that the larger network phenotype of hpata was robust to variation in media composition (Figure S6a,b), and to treatments with synthetic auxin or cytokinin plant growth regulators (Figure S6c,d), despite major changes in network morphology, suggesting that the larger networks of hpata are not due to disturbances in these pathways.
Figure 3.
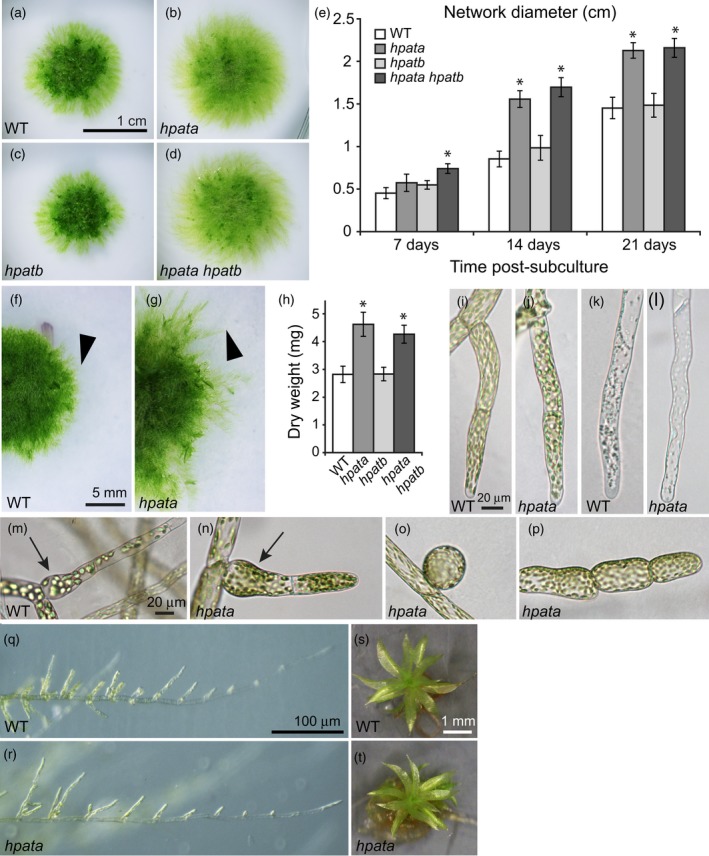
Physcomitrella patens hpata mutants show enhanced protonemal growth.
(a)–(d) Representative plants 21‐days post‐subculture (dps): (a) wild type (WT, Gransden 2004), (b) hpata, (c) hpatb, (d) hpata hpatb. (e) Diameter of the protonemal network over time (mean ± SD, n = 6). Black asterisks mark significant differences from WT at the same time point (P < 0.05) based on Student's t‐tests using the Bonferroni correction for multiple testing. (f), (g) Expanding protonemal networks of WT (f) and hpata mutants (g) showing the extent of filament growth (arrowhead). (h) Dry weight biomass (mg) at 21 dps (mean ± SD, n = 6). Black asterisks mark significant differences compared with WT (P < 0.05) based on Bonferroni corrected Student's t‐tests. (i), (j) Chloronemal tip cells from WT (i) and hpata (j). (k), (l) Caulonemal tip cells from WT (k) and hpata (l). (m)–(p) Cell shape abnormalities. Whereas both WT (m) and hpata (n) plants occasionally produce swollen cells at the initiation of a new branch, hpata mutants occasionally also produce more severe abnormalities, including spherical initial cells (o) and swollen filament cells at filament tips (p). (q), (r) Caulonemal filaments initiate branching in both WT (q) and hpata (r). (s), (t) Gametophores from WT (s) and hpata (t) both appear normal.
hpata mutants produce longer, faster‐growing protonemal cells
To understand the developmental basis for the increased network size in hpata mutants, we looked more closely at the morphology and behavior of individual cells in developing protonemal networks. hpata mutants produced cells with typical chloronemal and caulonemal morphologies (Figure 3i–l); however, they were more prone to rare morphological defects. In all genotypes, including the WT, we observed some side branch initial cells with enlarged basal regions (Figure 3m,n). In hpata and hpata hpatb plants this phenotype was sometimes more severe, occasionally resulting in spherical side branch initials that failed to elongate into tip cells (Figure 3o). We also observed rare swelling of cells at the filament tips in the mutants (Figure 3p). Mutants were largely able to initiate side branches like WT plants, however (Figure 3q,r), and also formed morphologically normal gametophores (Figure 3s,t). These observations suggest that hpata plants, though generally able to maintain normal filament growth, were more prone to cell shape disruption.
We further noted that individual filament cells were longer in hpata networks than in WT networks. Wild‐type chloronemal and caulonemal cells have average lengths of 75–80 μm and 180–200 μm, respectively (Perroud and Quatrano, 2006). Beginning at the branch initial cell of a new filament (position 0 in Figure 4a) we measured cells from the +1 to +7 positions, excluding the elongating tip cells and highly variable branch initial cells. A new filament typically initiates as chloronema, with the tip cell converting to caulonema under appropriate conditions. Average WT cell lengths did not exceed 90 μm at any measured filament positions. However, hpata hpatb cells were significantly longer than WT cells at the +1 position (94.4 ± 16.6 μm), and this size difference was enhanced in later‐formed cells, with +7 position cells in the double mutant reaching 128.6 ± 34.7 μm (Figure 4b). To determine if there was an indiscriminate increase in cell length in hpata mutants, we examined rhizoid cells. Like chloronema and caulonema, rhizoids are tip‐growing filaments, but rhizoids are derived from epidermal cells at the base of gametophores and achieve greater total lengths. We found no significant difference in rhizoid cell length between WT and hpata hpatb double mutants, indicating that only a subset of tip‐growing cells are longer in the mutants (Figure 4c).
Figure 4.
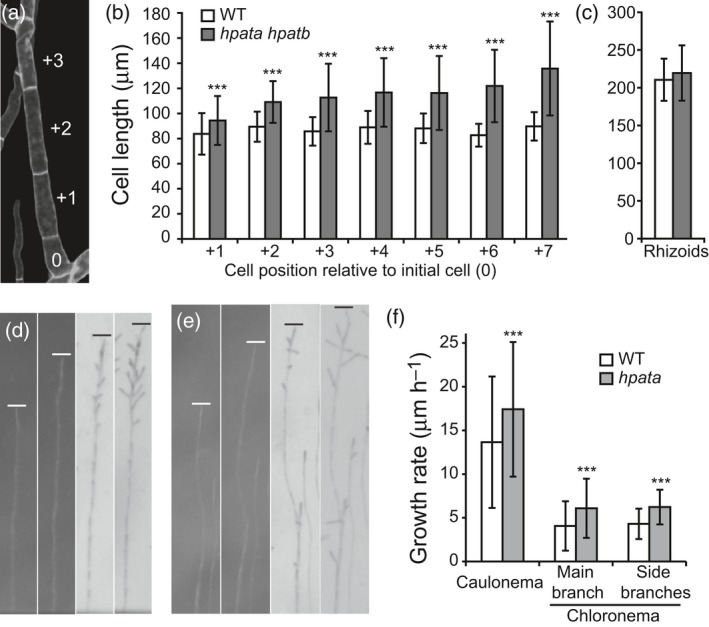
Protonemal cells of hpata mutants grow longer and faster.
(a) Calcoflour white‐stained protonemal filament with the cell positions marked relative to the initial cell (position 0). (b) Cell lengths measured at the indicated position (mean ± SD, n = 11–60). Protonemal cells are initially longer in hpata mutants and this effect enhances as filament growth continues. (c) Rhizoid cells, which are tip‐growing and similar in structure to caulonema, but ontogenetically distinct, are not significantly different in length between mutant and wild type (WT) (mean ± SD, n = 62). (d), (e) Images of single growing filaments from WT (d) and hpata mutants (e) taken at 24‐h intervals. The two left images are during filament growth in unidirectional red light, and the two right images are after movement to overhead white light. Lines mark the position of the filament tip. (f) Filament growth rates for WT and hpata mutants (mean ± SD, n ≥ 100). In (b), (c) and (f): ***P < 0.0005 based on Student's t‐tests.
We next measured the growth rate of WT and hpata filaments, taking advantage of the response of filaments to different light conditions. When grown under unidirectional red light, filaments adopt a caulonemal identity, grow toward the light and do not initiate side branches (Figure 4d,e, left; Aoyama et al., 2012). Following transfer to white light, tip cells switch to a chloronemal identity and new side branches initiate from young sub‐apical cells (Figure 4d,e, right). We measured individual filament tip positions at 24‐h intervals during growth in red light and after transfer to white light. These measures were done using small fragments of protonema in the absence of gametophores, thus eliminating any potential confounding effects of gametophore development on filament growth. For both red‐light‐grown caulonema and white light‐grown main and side branch chloronema, the hpata filaments elongated at a greater average rate than the WT (Figure 4f), suggesting that the increased network size of hpata is due to a general increase in elongation rates of protonemal filaments and cell lengths.
HPATs are necessary for production of cell‐wall‐associated hydroxyproline arabinosides
Tip‐growing cells, like pollen tubes and protonema, are particularly sensitive to cell wall perturbation. Based on the cell morphology defects and faster growth rates of hpata mutants (Figures 3n–p and 4f), we hypothesized that the mutant phenotype was due to altered cell wall structure. Given that extensins are heavily hydroxyproline O‐arabinosylated and function in the cell wall (Velasquez et al., 2012), these proteins would be the likely targets for HPAT modification in protonema. However, the Physcomitrella genome is unusual among plants in that it does not encode canonical extensins (Lawton and Saidasan, 2011). In the absence of canonical extensins, we searched the Physcomitrella genome for genes encoding the extensin glycosylation motif and found 20 predicted extensin chimeras containing three or more Ser(Pro)3 motifs (Table S2).
Since we hypothesize that these extensin chimeras may be targets of hydroxyproline O‐arabinosylation functioning in the cell wall, we next looked for changes in cell‐wall‐associated Hyp‐arabinoside levels in our mutants. Cell wall fractions (alcohol‐insoluble residue, AIR) were hydrolyzed with Ba(OH)2 to cleave peptide, but not sugar–Hyp bonds. The resulting amino acids were separated by high‐voltage paper electrophoresis (HVPE), a powerful, yet under‐utilized, technique that allows for analysis of small, hydrophilic, ionic compounds, including Hyp and Hyp‐arabinosides (Fry, 2011) that can then be visualized by staining with isatin/ninhydrin reagent (Kolor and Roberts, 1957). Both Arabidopsis WT and Physcomitrella WT samples produced weak but detectable levels of Hyp‐arabinosides (Figure S7), indicating that Physcomitrella harbors stable cell‐wall‐associated proteins carrying this modification, despite the absence of canonical extensins in the Physcomitrella genome. Notably, while Hyp‐arabinosides were maintained in the Arabidopsis hpat1, hpat2 and hpat3 single mutants, this modification was completely lost in Arabidopsis triple mutants and Physcomitrella hpata haptb double mutants, indicating that other, unknown, enzymes are not catalyzing hydroxyproline O‐arabinosylation in the absence of the HPATs, at least for cell‐wall‐associated proteins.
Expression of cell‐wall‐associated genes is altered in hpata mutants
To test the hypothesis that hpata cell walls were disrupted despite the absence of canonical extensins, we compared the transcriptomes of developing networks from hpata mutants and the WT using custom Physcomitrella NimbleGen microarrays. Because tip‐growing cell types exhibit clear transcriptomic signatures at the level of cell wall genes (Becker et al., 2014), we expected to see signs of altered cell wall gene expression if disruption of the cell wall was the basis of the phenotype. We identified 224 genes that were downregulated in hpata [false discovery rate (FDR) <10%; Table S3]. At FDR < 10% we did not identify any significantly upregulated genes, but at the less stringent FDR < 25%, we identified 58 such genes which included several members of the same gene families (Table S4). We further validated differential expression for four of the upregulated and five of the downregulated genes by quantitative (q)RT‐PCR (Figure S8a).
We next used Gene Ontology (GO) analysis to identify overrepresented functional categories among our differentially expressed genes. Though the small number of upregulated genes did not yield any enriched categories, the hpata downregulated genes yielded 15 enriched terms, eight of which were directly related to the cell wall including the biological process term ‘plant‐type cell wall organization’ (23.4‐fold enrichment) and the cellular component term ‘extracellular region’ (4.8‐fold enrichment; Figure 5a, Table S5). We also found weaker enrichment of vesicle‐associated terms which may be due to the higher growth rate of the hpata mutants relative to the WT (Figure 4f), since tip growth is dependent on vesicle trafficking both to deliver new wall material and to recycle excess membrane (Hepler et al., 2001; Cheung and Wu, 2008).
Figure 5.
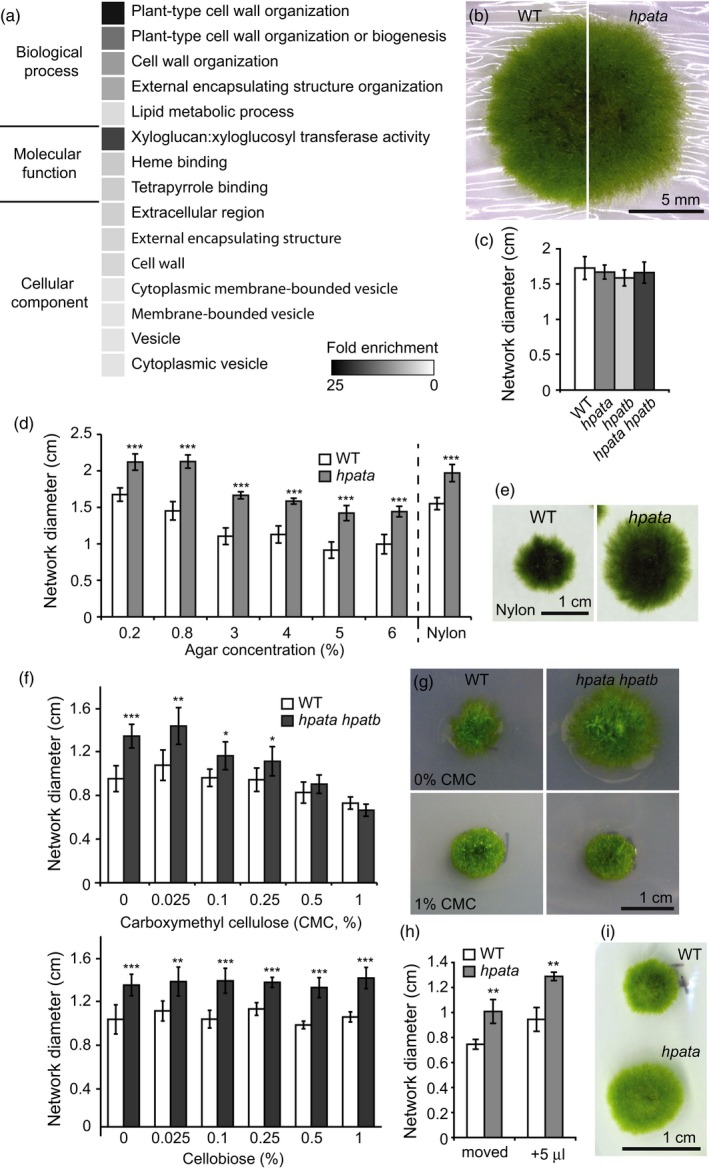
hpata mutants mis‐express cell wall associated genes and can be rescued by exogenous cellulose.
(a) Heat map of the fold enrichment for Gene Ontology terms significantly overrepresented among the genes differentially expressed between the wild type (WT) and hpata mutants (Table S5). (b) The appearance of cellophane‐grown networks of WT (left) and hpata (right) plants. (c) The diameter of the protonemal network after 21 days of growth on standard medium overlaid with cellophane shows no significant differences between genotypes, indicating rescue of the hpata mutant phenotype (mean ± SD, n = 6). (d) Blocking filament invasion by increasing the agar concentration of the medium or by growing on nylon membrane is not sufficient to rescue the hpata phenotype (mean of network diameter ± SD, n = 6). (e) Nylon membrane‐grown WT (left) and hpata (right). (f) Networks grown on medium supplemented with carboxymethyl cellulose (CMC), a soluble cellulose derivative, also show rescue of the hpata at concentrations of 0.5–1% at 21‐days post‐subculture (top). However, plants grown on cellobiose, a disaccharide of β(1→4) linked glucose were not rescued (bottom, mean ± SD, n = 6). (g) The appearance of WT (left) and hpata hpatb (right) networks grown on control medium (top) or 1% CMC (bottom). (h) Plants grown for 17 days on cellophane overlay plates that were either physically moved to a virgin position on the cellophane once a day or received a daily addition of 5 μl of liquid growth medium were not rescued by cellophane, suggesting that a stable interaction is necessary for rescue. (i) Networks that were grown on cellophane for 17 days to which 5 μl of liquid growth medium was added directly over the developing network at 24‐h intervals. In (c), (d), (f) and (h): *P < 0.05; **P < 0.005; ***P < 0.0005 based on Student's t‐tests.
The structure of the Physcomitrella cell wall is similar to that of angiosperms; both are largely composed of cellulose, the major load‐bearing polymer, hemicellulose and pectin (Moller et al., 2007; Roberts et al., 2012). The specific genes differentially expressed fell into categories relating to all of the major cell wall polymers. For example, a cellulose synthase family member was downregulated in hpata, and we identified an up‐ and a downregulated pair of leucine‐rich repeat (LRR) receptor kinases homologous to the Arabidopsis FEI1 protein (Figure S8). Arabidopsis fei1 fei2 double mutants are defective in anisotropic cell expansion and cellulose biosynthesis (Xu et al., 2008). Expansins promote cell expansion by loosening cell walls by non‐hydrolytically disrupting hemicellulose binding (McQueen‐Mason and Cosgrove, 1995). Of the 34 Physcomitrella expansins (Carey and Cosgrove, 2007), six were downregulated in hpata mutants. We also found several downregulated pectin‐digesting pectate lyases (Marin‐Rodriguez, 2002). The variety of polymers affected by the genes misregulated in hpata mutants suggests that many aspects of wall structure are altered and that a complex regulatory change has occurred to compensate for the loss of hydroxyproline O‐arabinosylation.
Given these changes in cell‐wall‐associated gene expression, we next examined cell wall structure by transmission electron microscopy. At this level, however, there were no visible changes in wall structure, consistent with the general maintenance of cell wall function and cell viability in the mutants (Figure S9).
Addition of exogenous cellulose can rescue hpat mutants
Despite the far‐ranging changes in cell‐wall‐associated gene expression in hpata mutants, we fortuitously found that the simple addition of cellulose can rescue the mutant phenotype. Cellophane sheets composed of regenerated cellulose are commonly used in moss culture to provide a diffusible barrier to invasion of the growth media by filaments. When WT and hpata networks were grown on cellophane over standard media, we found the network size difference was abolished (Figure 5b,c). To determine if this effect was simply the result of physical blockage of media invasion, we grew WT and hpata mutants on media in which the concentration of agar varied from the standard 0.8% (w/v) to between 0.2% and 6%. A significant difference in network diameter was maintained under all conditions, even though filament invasion was blocked and overall network size was reduced at higher concentrations (Figure 5d). We also blocked filament invasion by growth on a 0.45‐μm pore nylon membrane. Nylon is not derived from cellulose and did not share the ability of cellophane to rescue the mutant phenotype (Figure 5d,e). However, direct supplementation of the medium with cellulose using carboxymethyl cellulose, a solubilized cellulose derivative, was also able to rescue the mutant phenotype at concentrations of 0.5% (w/v) and greater (Figure 5f, top, g). In contrast, addition of cellobiose, a disaccharide of β(1→4)‐linked glucose did not rescue the size difference (Figure 5f, bottom), indicating that larger polymer fragments are necessary for rescue. Furthermore, we could disrupt the rescue effect of growth on cellophane by either physically moving the developing network to a new position daily (Figure 5h) or by daily addition of just 5 μl of liquid growth medium over the developing network (Figure 5h,i). Therefore, despite its lack of canonical extensins, Physcomitrella relies on HPATs to maintain normal protonemal cell expansion and network morphology, possibly acting through control of cell wall composition.
Though we observed a rescue effect of cellophane on the size of the Physcomitrella mutant network, cellophane was not able to rescue the Arabidopsis pollen tube defect. Our in vitro pollen tube growth method uses cellophane sheets to simulate the dry stigma of Arabidopsis (Figure 1l,m; Rodriguez‐Enriquez et al., 2013), and our results agree with previously reported in vitro data, which did not use cellophane (Ogawa‐Ohnishi et al., 2013), and our in vivo growth observations (Figure 1n,p). The ability to rescue the hpat mutant phenotype with exogenous cellulose in Physcomitrella but not Arabidopsis highlights another difference between these two systems in the context of the loss of HPAT function.
Discussion
Hydroxyproline O‐arabinosylation occurs across the plant kingdom (Lamport and Miller, 1971), and HPATs remain the only identified enzymes capable of initiating an oligoarabinoside chain on hydroxyprolines (Ogawa‐Ohnishi et al., 2013). Results from tomato and Medicago point to roles for HPATs in homeostasis of meristem size and control of nodule numbers, acting through modification of CLE signaling peptides (Schnabel et al., 2011; Okamoto et al., 2013; Xu et al., 2015). Although CLV3 is a major regulator of meristem size in Arabidopsis (Clark et al., 1995), disruption of arabinosylation by the triple hpat mutants did not alter meristem size in this species (Figure 2). This striking phenotypic difference between tomato and Arabidopsis hpat mutants could be due to either an unknown enzyme functioning in the absence of the HPATs to modify CLV3, or to the non‐arabinosylated peptide maintaining sufficient signaling activity to limit meristem size. While our observation that the triple hpat mutants failed to produce detectable levels of Hyp‐arabinosides (Figure S7) suggests that, at least for cell‐wall‐associated targets, there are no other enzymes acting in the absence of the HPATs, we cannot formally exclude the possibility that other enzymes are acting to modify the CLE peptides. However, previously reported evidence also supports a non‐essential role for this modification in CLV3 function. Though exogenously applied [Ara3]CLV3 reduces meristem size more strongly than unmodified peptide (Ohyama et al., 2009), the proline‐7 that carries this modification can be changed to an alanine and is still capable of rescuing clv3 mutants transgenically (Song et al., 2012). Thus, although arabinosylation enhances CLV3 activity, the significance of this modification for controlling stem cell proliferation and meristem size seems to vary between species.
While the CLE peptides have been employed in a number of developmental processes (Miyawaki et al., 2013), the extensins, as critical cell wall structural proteins, have a deeply conserved function and are probably ancient in origin (Bollig et al., 2007). Due to their structure, tip‐growing cell types are particularly sensitive to cell wall disruption. In Arabidopsis and Physcomitrella hpat mutants, we observe d alteration in cell length of tip‐growing pollen and protonema. Interestingly, the phenotypic effects of loss of HPAT function were opposite in these species, with hpat1 hpat3 pollen tubes being shorter and hpata protonemal cells being longer than their WT counterparts. Why these two systems exhibit opposite responses to loss of HPAT function is not clear, but may be related to differences in (i) growth rate, (ii) cell wall composition, or (iii) potential target proteins. First, Arabidopsis pollen tubes grow more quickly than either chloronema or caulonema, by about an order of magnitude (Rounds and Bezanilla, 2013). Given the competitive nature of fertilization, with multiple pollen tubes racing to reach a limited number of ovules, there is a clear selective advantage to achieving the maximum possible growth rate. In such a system any genetic perturbation of the cell wall would either reduce elongation or cause premature tube rupture. Meanwhile, protonema do not suffer the same selective pressure to maximize growth rate, and in these cells the relevant HPAT targets may help stabilize the cell wall and therefore limit elongation. Disrupted glycosylation of these targets would reduce wall stability, causing growth abnormalities (Figure 3n–p) and an increased growth rate, as the checks on elongation are removed (Figure 4f).
Second, cell wall composition differs between pollen tubes and protonema. Pollen tube cell walls are unusual in that they contain little cellulose (β‐1,4‐glucan) but an abundance of callose (β‐1,3‐glucan) (Galway, 2006; Chebli et al., 2012). Physcomitrella cell walls contain similar wall glycans to those found in higher plants (Moller et al., 2007), and though Physcomitrella does produce callose it is not reported as a typical wall constituent in expanding protonema (Roberts et al., 2012). Apart from callose, other wall components are likely to vary between these systems as well, and how the different milieus of the cell wall will respond to disrupting hydroxyproline O‐arabinosylation is difficult to predict, since much remains unknown about how wall components interact in vivo. These differences in cell wall composition may also account for the ability of cellulose to rescue the hpat mutant phenotype in Physcomitrella protonema, but not Arabidopsis pollen (Figure 5b–g).
Third, the HPATs, as members of glycosyltransferase superfamily, will exert their effects through the proteins they target for modification. Despite the lack of canonical extensins (Lawton and Saidasan, 2011), the Physcomitrella genome encodes not only the HPATs, members of the GT8 glycosyltransferase family (Nikolovski et al., 2012, Carbohydrate Active enZYmes Database; http://www.cazy.org/), but also all known glycosyltransferases associated with extensin modification including the later‐acting arabinosyltransferases of the GT77 family, RRA1‐3 and XEG113 (Egelund et al., 2007; Gille et al., 2009; Velasquez et al., 2011; Harholt et al., 2012). Serine in the Ser(Hyp)4 context is also modified with a single galactose by SGT1 (Lamport et al., 1973; Saito et al., 2014), two homologs of which are encoded in the Physcomitrella genome. In the absence of canonical extensins, extensin chimeras carrying regions with several extensin‐like Ser(Pro)3–5 motifs may be targeted by the above enzymes and account for the Hyp‐arabinosides detected in the Physcomitrella cell wall fraction (Figure S7).
Among the extensin chimeras we identified was a small group similar to LRR‐extensin (LRX) proteins (Figure S10, Table S5). The LRXs are frequently associated with tip‐growing cell types; Arabidopsis LRX1 and LRX2 have a demonstrated role in maintaining root hair structure (Baumberger et al., 2001, 2003a), and several LRX family members are expressed specifically in pollen (Rubinstein et al., 1995; Stratford et al., 2001; Baumberger et al., 2003b). However, there are important differences between the angiosperm and Physcomitrella LRXs. The Physcomitrella proteins have shorter extensin‐like regions that lack the tyrosines needed for covalent cross‐linking (Stratford et al., 2001; Held et al., 2004) and four of the five Physcomitrella proteins also contain a c‐type lectin domain at the C‐terminus (Figure S8b,c). Though common in mammals, where they often function in carbohydrate recognition and immune response (Cambi et al., 2005; Dambuza and Brown, 2015), the calcium‐dependent c‐type lectin domain is rare in plants, with only a single example of unknown function in Arabidopsis and no other examples in Physcomitrella (Bouwmeester and Govers, 2009). Though changes in gene expression and the ability of exogenous cellulose to rescue the phenotype suggest a critical role for the cell wall and, presumably, cell‐wall‐associated HPAT targets, in the mutant phenotype, we cannot rule out the possibility that other targets, particularly the CLE signaling peptides, could be responsible for the Physcomitrella hpata mutant phenotype. Future molecular analyses of modifications on these and/or other potential target proteins should help reveal precisely how cell walls are disrupted in Physcomitrella hpat mutants and provide a foundation for future comparative evolutionary studies between the roles of HPAT genes and glycosylation in controlling tip growth in different developmental contexts in flowering and basal plants.
Experimental procedures
Phylogenetic analysis
Protein sequences were aligned using Clustal Omega, and maximum parsimony phylogenetic trees were generated with phylip with 1000 bootstrap replicates with 10 global rearrangements per replicate. A consensus tree was calculated by extended majority rule. Gene identification numbers for the Selaginella moellendorffii HPATs are, in order of numbering in Figure 1(a): 235499, 150302 and 231983. For LRX‐like proteins, only the conserved N‐terminal 450 amino acids were used since the C‐terminal extensin‐like region aligns poorly (Baumberger et al., 2003b).
Arabidopsis material and growth conditions
The following lines were obtained from the Arabidopsis Biological Resource Center: hpat1‐2 (Salk_120066C), hpat2‐2 (SM_3_38225), hpat3‐1 (Salk_047668), hpat3‐2 (SAIL_301_C09), qrt1‐1 and clv3‐2. Genotypes were confirmed by PCR using the primers in Table S6. We noted that seed recovered from hpat1 hpat3 plants is prone to contamination due to fertilization by ambient non‐mutant pollen. Therefore, all double or triple mutant individuals were genotyped before subsequent use in assays or seed collection.
RT‐PCR
For RT‐PCR of hpat alleles (Figure S2) total RNA was extracted from 7 day‐old seedlings by an RNeasy Mini Kit (Qiagen, http://www.qiagen.com/). One microgram was converted to cDNA using oligo (dT) primers and a Superscript III First Strand Synthesis Kit (Invitrogen, http://www.invitrogen.com/). One microliter of cDNA (corresponding to 50 ng input RNA) was used per 20 μl Phusion polymerase (New England BioLabs, https://www.neb.com/) reaction for the following cycle numbers: HPAT1 = 23, HPAT2 = 32, HPAT3 = 23, Actin = 26. The protocol was the same for RT‐PCR for different tissues (Figure 1e) except that, due to limiting amounts of starting material, 500 ng (Figure 1f) or 100 ng (Figure 1g,h) of total RNA was used, and reactions proceeded for 27 cycles. Developing pollen of various stages was extracted from unopened flowers by coarse mechanical disruption followed by filtering through a 70‐μm nylon membrane to remove floral debris and centrifugation at 106g for 30 sec to pellet the pollen. Isolated pollen was then washed three times in water before RNA extraction as above.
Pollen methods
qrt1 pollen tetrads were stained with simplified Alexander's viability stain (Peterson et al., 2010) before imaging (Figure 1j,k). For in vitro pollen germination we used the protocol described by Rodriguez‐Enriquez et al. (2013). Pollen from five flowers that had opened less than 24 h previously was applied to solid growth media overlaid with cellophane and allowed to grow for 8 h at which point the pollen was fixed in 10% sucrose, 100 mm piperazine‐N,N′‐bis(2‐ethanesulfonic acid) (PIPES) buffer pH 6.9, 4 mm MgSO4 and 5% formaldehyde. Tubes were stained with calcofluor white stain (Fluka Analytical, Sigma), then imaged under UV light and measured in imageJ. For in vivo staining of hpat3‐2 (Figure 1n–p), unopened flowers were emasculated, allowed to mature for 24 h then pollinated with either hpat3‐2 or hpat1 hpat3‐2/+ pollen. Seven hours after pollination, flowers were stained for GUS activity overnight (16 h) and then cleared in 70% ethanol.
Generation of transgenic Arabidopsis
The DNA cloning was done using the Gateway system (Invitrogen). Fragments were amplified using the primers in Table S6 and recombined into the appropriate pDONR vector. For HPAT expression under the pollen‐specific AtSTP9 promoter, the promoter (first fragment) and cDNA (second fragment) were recombined into pMDC99 (Curtis and Grossniklaus, 2003). Transgenic Arabidopsis plants were generated using the standard floral dip method.
Generation of Physcomitrella mutants
Physcomitrella knockout cassettes were generated using the primers in Table S6 with approximately 1 kb of sequence from the left and right sides of the targeted coding region. Selection cassettes were amplified from BNRF or BHSNR plasmids (Menand et al., 2007b), using G418 resistance for HPATa and hygromycin B resistance for HPATb transformation. A GFP–GUS coding region was added 5′ to the resistance cassette by overlap PCR. The flanking regions and selection cassettes were cloned into the appropriate pDONR vector and recombined into pMDC99 (Curtis and Grossniklaus, 2003). The full knockout cassettes were PCR amplified from the resulting plasmids using Phusion DNA polymerase (New England BioLabs) and the primers in Table S6. The selection cassette fragments were purified by phenol extraction and alcohol precipitation and transformed into Gransden 2004 protoplasts by polyethylene glycol (PEG)‐mediated transformation (Cove et al., 2009). Proper insertion (Figure S5b) was confirmed by PCR using the primers in Table S6, and disrupted gene expression (Figure S5g) was confirmed by RT‐PCR using the same method as above for the Arabidopsis alleles. Insert copy number was estimated by qPCR using the ∆∆Ct method (Livak and Schmittgen, 2001) with the primers in Table S6. The amplification of a fragment of the left border homology region was compared between mutant strains and WT with normalization to a single locus control gene.
Physcomitrella growth conditions and assays
The standard medium used in this study was BCDAT (1 mm MgSO, 1 mm CaCl2, 10 mm KNO3, 45 μm FeSO4, 1.8 mm KH2PO4 [pH 6.5 adjusted with KOH], 5 mm di‐ ammonium (+)‐tartrate, 0.22 μm CuSO4, 0.19 μm ZnSO4, 10 μm H3BO3, 0.10 μm Na2MoO4, 2 μm MnCl2, 0.23 μm CoCl2, and 0.17 μm KI solidified with 0.8% agar) (Nishiyama et al., 2000). Low‐phosphate and low‐nitrate media contained 5% of the standard amount of KH2PO4 or KNO3. Media with carboxymethyl cellulose were supplemented with the indicated concentration (w/v) of low‐viscosity carboxymethyl cellulose sodium salt, as was d‐(+)‐cellobiose (Sigma‐Aldrich, http://www.sigmaaldrich.com/). Plants were grown in a Percival Scientific (http://www.percival-scientific.com/) chamber at 24°C in 12‐h light, 12‐h dark cycles. For network diameter measurements, plates were inoculated with protonema fragments of approximately 2 mm, and network diameter (extending from the filament tips and passing through the middle of the network) was measured for six plants per genotype at 21 days post‐inoculation using imageJ software. Values for the WT and mutants were compared by Student's t‐test, using the Bonferroni correction for multiple testing. To determine dry weight biomass, 21‐day post‐subculture networks were extracted from the agar by room‐temperature (25°C) incubation with QC buffer (Qiagen), washed four times with distilled water and then dried under heat in vacuo. To determine filament growth rate, small fragments of isolated filaments were grown on BCDAT medium with 0.5% glucose in unidirectional red‐light chambers in the absence of gametophores. Filaments were imaged at 24‐h intervals over several days followed by several more days of imaging in overhead white light. The positions of individual filament tips were compared between subsequent images to determine the growth rate.
Hyp‐oligoarabinoside detection by high‐voltage paper electrophoresis
Tissue samples of Arabidopsis inflorescences and leaves and developing protonemal networks grown on solid BCDAT medium overlaid with a 0.45‐μm pore nylon membrane (Amersham Hybond −N+; GE Healthcare, http://www.gehealthcare.com/) for 5 days were harvested into 70% ethanol. Further sample preparation and HVPE were carried out at EDIPOS (http://fry.bio.ed.ac.uk//edipos.html) by Professor Stephen C. Fry (Fry, 2011). Briefly, AIR corresponding to the cell wall fraction was prepared by tissue incubation in 70% ethanol at 70°C for 2 × 2 h then 1 × 16 h. The resulting residue was rinsed with acetone and dried. The AIR (9–44 mg) was then subjected to alkaline hydrolysis in 3.0 ml of 233 mm Ba(OH)2 for 17 h at 105°C, then 5 h at 110°C. A sample of cultured tomato cell AIR, an abundant source of Hyp‐oligoarabinosides, was processed in parallel. The hydrolyzed components were isolated by an anion exchange column following the addition of 0.1 kBq [14C]proline to monitor recovery, along with sufficient H2SO4 to reduce the pH to 2.5. After centrifugation, the supernatant and 2.5 ml of water wash were brought to 6.0 ml with water. The sample was passed through a 1.2‐ml Dowex 50 column (H+ form), and non‐cations were washed out with 15 ml of 10 mm formic acid followed by 10 ml of water. Cations were then eluted with 3.5 ml of 1 m NH4OH (recovery of [14C]proline = 61.6 ± 2.7%, mean ± SD). The remainder of the eluate was dried in vacuo and re‐dissolved in 0.2 m NH4OH, using 10 μl per mg of starting AIR. Next, Asp, Glu, Lys, Arg and His were removed by preparative electrophoresis at pH 6.5. A portion of the remaining ‘no‐net‐charge’ fraction (equivalent to 2.5 mg of input AIR) along with an added internal marker of 1.5 μg N ε‐(2,4‐dinitrophenyl)‐L‐lysine (DNP‐Lys) and an external marker mixture (Hyp, Asp, Pro, Ser, 2 μg each) were then electrophoresed in pH 2.0 buffer at 4.5 kV for 40 min. The yellow DNP‐Lys spots were circled in pencil, then the paper was treated with mild acid to hydrolyze Hyp‐Arans to free Hyp in situ (S. C. Fry, University of Edingurgh, Scotland, UK, pers. comm.) and stained with isatin/ninhydrin reagent (Kolor and Roberts, 1957). The paper was then washed in running tap‐water for 1 h, dried and scanned.
Physcomitrella transcriptome profiling
Three biological replicates of hpata mutants and WT plants were grown as for HVPE and RNA was extracted with a Qiagen RNeasy plant RNA extraction kit. Samples were treated with TURBO DNase at 37°C for 30 min (Ambion, Life Technologies, http://www.thermofisher.com/us/en/home/brands/invitrogen/ambion.html). The integrity and quantity of RNA were assessed on an Agilent 2100 Bioanalyzer using a 6000 Pico Assay (Agilent Technologies, http://www.agilent.com/). On average 7.5 ng of RNA was used to synthesize cDNA with an Ovation Pico WTA System V2® amplification kit (NuGen Technologies, Inc., http://www.nugen.com/). The concentration of cDNA obtained was in the same range (220–260 ng μl−1) for all samples, and the quality of the cDNA was confirmed using a Bioanalyzer. Seven hundred and fifty nanograms of cDNA was used for labeling and hybridization on custom Nimblegen 12 × 135K arrays (Roche NimbleGen, Inc., http://sequencing.roche.com/) following the manufacturer's instructions in the IRB Barcelona Functional Genomics Core Facility (FGC). Raw data were obtained using the deva software, applying robust multichip average (RMA) normalization to all arrays (Roche NimbleGen, Inc.). Differential expression analysis was conducted using dchip software (Li and Wong, 2001).
To determine differentially expressed genes, pair‐wise comparisons of normalized data were conducted. A lower‐confidence bound fold‐change (LCB FC) cutoff was estimated for each comparison, ranging from 2 to 2.6, according the lowest false discovery rate (FDR) possible: below 10% for downregulated genes and 25% for upregulated genes. The FDR was estimated by applying LCB FC to sample‐wise comparison permutations as described by Tusher et al. (2001). At least 100 permutations were done for each comparison. The P. patens genome annotation v.1.6 release 2012.3 was used (http://cosmoss.org/), combined with annotations obtained through several alignments against non‐plant organisms and STRING information (http://string-db.org/) (Franceschini et al., 2013). Functional enrichment analysis was performed using the FatiGO functions (Al‐Shahrour et al., 2004) integrated into Blast2GO (Conesa et al., 2005). Physcomitrella GO annotations were downloaded from http://cosmoss.org/ as a reference set (nightly build as of 11 February 2015). Enrichment of GO annotation functional categories between hpata versus WT differentially expressed gene lists was analyzed by Fisher's exact test with a significance cut‐off of a FDR corrected P‐value <0.05. We confirmed differential expression of a subset of identified genes using triplicate independent RNA samples by quantitative RT‐PCR using the KAPA SYBR FAST qPCR kit and the primers in Table S6 and adenine phosphoribosyltransferase as a normalization control (Le Bail et al., 2013).
Transmission electron microscopy
Filaments were grown in the same manner as for transcriptome profiling and were fixed in 2.5% glutaraldehyde, 4% paraformaldehyde, 0.18 m sucrose, 100 mm sodium phosphate buffer pH 7.0 for 24 h at 4°C followed by post‐fixation in 1% osmium tetroxide at 25°C for 4 h. After washing in 100 mm sodium phosphate buffer, samples were dehydrated in an acetone series for 20–30 min (for each step) before exchange to propylene oxide. Dehydrated samples were embedded in Spurr's resin (Electron Microscopy Sciences, https://www.emsdiasum.com/microscopy/) before sectioning and mounting to grids. Grids were stained with uranyl acetate replacement stain (Electron Microscopy Sciences) and lead citrate before imaging on an FEI/Philips CM100 Biotwin transmission electron microscope (http://www.fei.com/) with a Kodak 4.2i, bottom‐mount digital camera (http://www.kodak.com).
Supporting information
Figure S1. Comparison of Arabidopsis and Physcomitrella hydroxyproline O‐arabinosyltransferase protein sequences.
Figure S2. Transfer DNA insertion mutations disrupting Arabidopsis HPAT expression.
Figure S3. Reduced seed set in hpat1 hpat3 was exclusively due to a pollen defect.
Figure S4. Generation and phenotypic analysis of hpat1 hpat2 hpat3 triple mutant plants.
Figure S5. Generation of Physcomitrella hpat mutants.
Figure S6. hpata phenotypes on various medium compositions and hormone treatments.
Figure S7. Cell‐wall‐associated hydroxyproline arabinosides are undetectable in full hpat mutants.
Figure S8. FEI1 related genes are up‐ and downregulated in hpata.
Figure S9. Transmission electron micrographs of wild‐type and hpata hpatb filaments.
Figure S10. The Physcomitrella genome encodes leucine‐rich repeat extensin (LRX)‐like extensin chimeras.
Table S1. Transmission rate of hpat1 hpat3 mutations.
Table S2. Physcomitrella genes containing Ser(Pro)3–5 motifs.
Table S3. List of genes downregulated in Physcomitrella hpata mutants.
Table S4. List of genes upregulated in Physcomitrella hpata mutants.
Table S5. Enriched categories of differentially expressed genes.
Table S6. Primer sequences used in this study.
Acknowledgements
We thank Dr Yevgeniy Plavskin for assistance in establishing Physcomitrella culture and transformation, Tim Mulligan for plant care, Dr Benjamin Roche for assistance identifying Ser(Pro)2–5‐containing Physcomitrella protein models and Sophie Thomain for assistance in generating the Arabidopsis triple hpat mutant. We thank Professor Stephen C. Fry (University of Edinburgh) for performing the HVPE and Dr Gregg Sobocinski (University of Michigan) for assistance with TEM. We thank Stefan Rensing (University of Marburg, Germany) for allowing us to use the Nimblegen_Ppat_SR_exp_HX12 array design and the IRB Functional Genomics Core (Barcelona, Spain) for microarray processing. CO‐R, JAF and JDB gratefully acknowledge funding by EU‐FP7‐PEOPLE‐ITN‐2008 ‘PLANT developmental biology: discovering the ORIGINS of form (PLANTORIGINS)’. JAF acknowledges additional funding by grants PTDC/BEX‐BCM/0376/2012 and PTDC/BIA‐PLA/4018/2012 from Fundacão para a Ciência e a Tecnologia‐FCT, Portugal. CAM was supported by the Gordon and Betty Moore Foundation through grant GBMF 2550.01 to the Life Sciences Research Foundation. This research was supported by an Agriculture and Food Research Initiative competitive grant (2015‐67013‐22823) of the US Department of Agriculture National Institute of Food and Agriculture to ZBL. The authors declare no conflict of interest.
Accession numbers: Microarray data can be found in the Array‐Express data libraries under accession number E‐MTAB‐3638.
References
- Al‐Shahrour, F. , Díaz‐Uriarte, R. and Dopazo, J. (2004) FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics, 20, 578–580. [DOI] [PubMed] [Google Scholar]
- Aoyama, T. , Hiwatashi, Y. , Shigyo, M. , Kofuji, R. , Kubo, M. , Ito, M. and Hasebe, M. (2012) AP2‐type transcription factors determine stem cell identity in the moss Physcomitrella patens . Development, 139, 3120–3129. [DOI] [PubMed] [Google Scholar]
- Baumberger, N. , Ringli, C. and Keller, B. (2001) The chimeric leucine‐rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev. 15, 1128–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger, N. , Steiner, M. , Ryser, U. , Keller, B. and Ringli, C. (2003a) Synergistic interaction of the two paralogous Arabidopsis genes LRX1 and LRX2 in cell wall formation during root hair development. Plant J. 35, 71–81. [DOI] [PubMed] [Google Scholar]
- Baumberger, N. , Doesseger, B. , Guyot, R. et al (2003b) Whole‐genome comparison of leucine‐rich repeat extensins in Arabidopsis and rice. A conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol. 131, 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, J.D. , Takeda, S. , Borges, F. , Dolan, L. and Feijó, J.A. (2014) Transcriptional profiling of Arabidopsis root hairs and pollen defines an apical cell growth signature. BMC Plant Biol. 14, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckmann, A. , Alter, S. and Dresselhaus, T. (2014) The beginning of a seed: regulatory mechanisms of double fertilization. Front. Plant Sci. 5, 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollig, K. , Lamshöft, M. , Schweimer, K. , Marner, F.‐J.J. , Budzikiewicz, H. and Waffenschmidt, S. (2007) Structural analysis of linear hydroxyproline‐bound O‐glycans of Chlamydomonas reinhardtii‐conservation of the inner core in Chlamydomonas and land plants. Carbohydr. Res. 342, 2557–2566. [DOI] [PubMed] [Google Scholar]
- Bouwmeester, K. and Govers, F. (2009) Arabidopsis L‐type lectin receptor kinases: phylogeny, classification, and expression profiles. J. Exp. Bot. 60, 4383–4396. [DOI] [PubMed] [Google Scholar]
- Braidwood, L. , Breuer, C. and Sugimoto, K. (2013) My body is a cage: mechanisms and modulation of plant cell growth. New Phytol. 201, 388–402. [DOI] [PubMed] [Google Scholar]
- Cambi, A. , Koopman, M. and Figdor, C.G. (2005) How C‐type lectins detect pathogens. Cell. Microbiol. 7, 481–488. [DOI] [PubMed] [Google Scholar]
- Cannon, M.C. , Terneus, K. , Hall, Q. , Tan, L. , Wang, Y. , Wegenhart, B.L. , Chen, L. , Lamport, D.T.A. , Chen, Y. and Kieliszewski, M.J. (2008) Self‐assembly of the plant cell wall requires an extensin scaffold. Proc. Natl Acad. Sci. USA, 105, 2226–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey, R.E. and Cosgrove, D.J. (2007) Portrait of the expansin superfamily in Physcomitrella patens: comparisons with angiosperm expansins. Ann. Bot. 99, 1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebli, Y. , Kaneda, M. , Zerzour, R. and Geitmann, A. (2012) The cell wall of the Arabidopsis pollen tube–spatial distribution, recycling, and network formation of polysaccharides. Plant Physiol. 160, 1940–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A.Y. and Wu, H.‐M. (2008) Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annu. Rev. Plant Biol. 59, 547–572. [DOI] [PubMed] [Google Scholar]
- Clark, S.E. , Running, M.P. and Meyerowitz, E.M. (1995) CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development, 121, 2057–2067. [Google Scholar]
- Conesa, A. , Götz, S. , García‐Gómez, J.M. , Terol, J. , Talón, M. and Robles, M. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21, 3674–3676. [DOI] [PubMed] [Google Scholar]
- Cove, D. , Bezanilla, M. , Harries, P. and Quatrano, R. (2006) Mosses as model systems for the study of metabolism and development. Annu. Rev. Plant Biol. 57, 497–520. [DOI] [PubMed] [Google Scholar]
- Cove, D.J. , Perroud, P.‐F. , Charron, A.J. , McDaniel, S.F. , Khandelwal, A. and Quatrano, R.S. (2009). Transformation of the moss Physcomitrella patens using direct DNA uptake by protoplasts. Cold Spring Harb. Protoc. 2009, pdb.prot5143. [DOI] [PubMed] [Google Scholar]
- Curtis, M.D. and Grossniklaus, U. (2003) A gateway cloning vector set for high‐throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambuza, I.M. and Brown, G.D. (2015) C‐type lectins in immunity: recent developments. Curr. Opin. Immunol. 32, 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelund, J. , Obel, N. , Ulvskov, P. , Geshi, N. , Pauly, M. , Bacic, A. and Petersen, B.L. (2007) Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol. Biol. 64, 439–451. [DOI] [PubMed] [Google Scholar]
- Epstein, L. and Lamport, D.T.A. (1984) An intramolecular linkage involving isodityrosine in extensin. Phytochemistry, 23, 1241–1246. [Google Scholar]
- Everdeen, D.S. , Kiefer, S. , Willard, J.J. , Muldoon, E.P. , Dey, P.M. , Li, X.‐B. and Lamport, D.T.A. (1988) Enzymic cross‐linkage of monomeric extensin precursors in vitro. Plant Physiol. 87, 616–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini, A. , Szklarczyk, D. , Frankild, S. et al (2013) STRING v9.1: protein‐protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, S.C. (2011). High‐voltage paper electrophoresis (HVPE) of cell‐wall building blocks and their metabolic precursors In The Plant Cell Wall: Methods and Protocols (Popper Z.A., ed). New York: Humana Press, pp. 55–80. [DOI] [PubMed] [Google Scholar]
- Galway, M.E. (2006) Root hair cell walls: filling in the framework. Can. J. Bot. 84, 613–621. [Google Scholar]
- Gille, S. , Hänsel, U. , Ziemann, M. and Pauly, M. (2009) Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc. Natl Acad. Sci. USA, 106, 14699–14704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, Q.Q. and Cannon, M.C. (2002) The cell wall hydroxyproline‐rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell, 14, 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt, J. , Sørensen, I. , Fangel, J. , Roberts, A. , Willats, W.G.T. , Scheller, H.V. , Petersen, B.L. , Banks, J.A. and Ulvskov, P. (2012) The glycosyltransferase repertoire of the spikemoss Selaginella moellendorffii and a comparative study of its cell wall. PLoS ONE, 7, e35846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held, M.A. , Tan, L. , Kamyab, A. , Hare, M. , Shpak, E. and Kieliszewski, M.J. (2004) Di‐isodityrosine is the intermolecular cross‐link of isodityrosine‐rich extensin analogs cross‐linked in vitro. J. Biol. Chem. 279, 55474–55482. [DOI] [PubMed] [Google Scholar]
- Hepler, P.K. , Vidali, L. and Cheung, A.Y. (2001) Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17, 159–187. [DOI] [PubMed] [Google Scholar]
- Hijazi, M. , Velasquez, S.M. , Jamet, E. , Estevez, J.M. and Albenne, C. (2014) An update on post‐translational modifications of hydroxyproline‐rich glycoproteins: toward a model highlighting their contribution to plant cell wall architecture. Front. Plant Sci. 5, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiss, M. , Laule, O. , Meskauskiene, R.M. et al (2014) Large‐scale gene expression profiling data for the model moss Physcomitrella patens aid understanding of developmental progression, culture and stress conditions. Plant J. 79, 530–539. [DOI] [PubMed] [Google Scholar]
- Honys, D. and Twell, D. (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5, R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, G. and Dolan, L. (2011) Auxin promotes the transition from chloronema to caulonema in moss protonema by positively regulating PpRSL1and PpRSL2 in Physcomitrella patens . New Phytol. 192, 319–327. [DOI] [PubMed] [Google Scholar]
- Kieliszewski, M.J. and Lamport, D.T.A. (1994) Extensin: repetitive motifs, functional sites, post‐translational codes, and phylogeny. Plant J. 5, 157–172. [DOI] [PubMed] [Google Scholar]
- Kolor, M.G. and Roberts, H.R. (1957) A new reagent for the detection of hydroxyproline on paper chromatograms. Arch. Biochem. Biophys. 70, 620–622. [DOI] [PubMed] [Google Scholar]
- Lamport, D.T.A. and Miller, D.H. (1971) Hydroxyproline arabinosides in the plant kingdom. Plant Physiol. 48, 454–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport, D.T. , Katona, L. and Roerig, S. (1973) Galactosylserine in extensin. Biochem. J. 133, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport, D.T.A. , Kieliszewski, M.J. , Chen, Y. and Cannon, M.C. (2011) Role of the extensin superfamily in primary cell wall architecture. Plant Physiol. 156, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, D. , Zimmer, A.D. , Rensing, S.A. and Reski, R. (2008) Exploring plant biodiversity: the Physcomitrella genome and beyond. Trends Plant Sci. 13, 542–549. [DOI] [PubMed] [Google Scholar]
- Lawton, M.A. and Saidasan, H. (2011) Cell wall genomics in the recombinogenic moss Physcomitrella patens In Routes to Cellulosic Ethanol (Buckeridge M.S. and Goldman G.H., eds). Berlin: Springer, pp. 241–261. [Google Scholar]
- Le Bail, A. , Scholz, S. and Kost, B. (2013) Evaluation of reference genes for RT qPCR analyses of structure‐specific and hormone regulated gene expression in Physcomitrella patens gametophytes. PLoS ONE, 8, e70998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. and Wong, W.H. (2001) Model‐based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl Acad. Sci. USA, 98, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Marin‐Rodriguez, M.C. (2002) Pectate lyases, cell wall degradation and fruit softening. J. Exp. Bot. 53, 2115–2119. [DOI] [PubMed] [Google Scholar]
- McQueen‐Mason, S.J. and Cosgrove, D.J. (1995) Expansin mode of action on cell walls. Analysis of wall hydrolysis, stress relaxation, and binding. Plant Physiol. 107, 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand, B. , Calder, G. and Dolan, L. (2007a) Both chloronemal and caulonemal cells expand by tip growth in the moss Physcomitrella patens . J. Exp. Bot. 58, 1843–1849. [DOI] [PubMed] [Google Scholar]
- Menand, B. , Yi, K. , Jouannic, S. , Hoffmann, L. , Ryan, E. , Linstead, P. , Schaefer, D.G. and Dolan, L. (2007b) An ancient mechanism controls the development of cells with a rooting function in land plants. Science, 316, 1477–1480. [DOI] [PubMed] [Google Scholar]
- Miyawaki, K. , Tabata, R. and Sawa, S. (2013) Evolutionarily conserved CLE peptide signaling in plant development, symbiosis, and parasitism. Curr. Opin. Plant Biol. 16, 598–606. [DOI] [PubMed] [Google Scholar]
- Moller, I. , Sørensen, I. , Bernal, A.J. et al (2007) High‐throughput mapping of cell‐wall polymers within and between plants using novel microarrays. Plant J. 50, 1118–1128. [DOI] [PubMed] [Google Scholar]
- Nikolovski, N. , Rubtsov, D. , Segura, M.P. , Miles, G.P. , Stevens, T.J. , Dunkley, T.P.J. , Munro, S. , Lilley, K.S. and Dupree, P. (2012) Putative glycosyltransferases and other plant Golgi apparatus proteins are revealed by LOPIT proteomics. Plant Physiol. 160, 1037–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, T. , Hiwatashi, Y. , Sakakibara, I. , Kato, M. and Hasebe, M. (2000) Tagged mutagenesis and gene‐trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res. 7, 9–17. [DOI] [PubMed] [Google Scholar]
- Ogawa‐Ohnishi, M. , Matsushita, W. and Matsubayashi, Y. (2013) Identification of three hydroxyproline O‐arabinosyltransferases in Arabidopsis thaliana. Nat. Chem. Biol. 9, 726–730. [DOI] [PubMed] [Google Scholar]
- Ohyama, K. , Shinohara, H. , Ogawa‐Ohnishi, M. and Matsubayashi, Y. (2009) A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5, 578–580. [DOI] [PubMed] [Google Scholar]
- Okamoto, S. , Shinohara, H. , Mori, T. , Matsubayashi, Y. and Kawaguchi, M. (2013) Root‐derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat. Commun. 4, 2191. [DOI] [PubMed] [Google Scholar]
- Perroud, P.‐F. and Quatrano, R.S. (2006) The role of ARPC4 in tip growth and alignment of the polar axis in filaments of Physcomitrella patens . Cell Motil. Cytoskeleton, 63, 162–171. [DOI] [PubMed] [Google Scholar]
- Peterson, R. , Slovin, J.P. and Chen, C. (2010) A simplified method for differential staining of aborted and non‐aborted pollen grains. Int. J. Plant Biol. 1, e13. [Google Scholar]
- Preuss, D. , Rhee, S. and Davis, R. (1994) Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science, 264, 1458–1460. [DOI] [PubMed] [Google Scholar]
- Reid, D.E. , Ferguson, B.J. , Hayashi, S. , Lin, Y.‐H. and Gresshoff, P.M. (2011) Molecular mechanisms controlling legume autoregulation of nodulation. Ann. Bot. 108, 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, K. , Grief, C. , Hills, G.J. and Shaw, P.J. (1985) Cell wall glycoproteins: structure and function. J. Cell Sci. 1985, 105–127. [DOI] [PubMed] [Google Scholar]
- Roberts, A.W. , Roberts, E.M. and Haigler, C.H. (2012) Moss cell walls: structure and biosynthesis. Front. Plant Sci. 3, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Enriquez, M.J. , Mehdi, S. , Dickinson, H.G. and Grant‐Downton, R.T. (2013) A novel method for efficient in vitro germination and tube growth of Arabidopsis thaliana pollen. New Phytol. 197, 668–679. [DOI] [PubMed] [Google Scholar]
- Rounds, C.M. and Bezanilla, M. (2013) Growth mechanisms in tip‐growing plant cells. Annu. Rev. Plant Biol. 64, 243–265. [DOI] [PubMed] [Google Scholar]
- Rubinstein, A.L. , Broadwater, A.H. , Lowrey, K.B. and Bedinger, P.A. (1995) Pex1, a pollen‐specific gene with an extensin‐like domain. Proc. Natl Acad. Sci. USA, 92, 3086–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, F. , Suyama, A. , Oka, T. , Yoko‐O, T. , Matsuoka, K. , Jigami, Y. and Shimma, Y.‐I. (2014) Identification of novel peptidyl serine α‐galactosyltransferase gene family in plants. J. Biol. Chem. 289, 20405–20420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel, E.L. , Kassaw, T.K. , Smith, L.S. , Marsh, J.F. , Oldroyd, G.E. , Long, S.R. and Frugoli, J.A. (2011) The ROOT DETERMINED NODULATION1 gene regulates nodule number in roots of Medicago truncatula and defines a highly conserved, uncharacterized plant gene family. Plant Physiol. 157, 328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidereit, A. , Scholz‐Starke, J. and Büttner, M. (2003) Functional characterization and expression analyses of the glucose‐specific AtSTP9 monosaccharide transporter in pollen of Arabidopsis. Plant Physiol. 133, 182–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A. , Burke, E. , Presting, G. et al (2002) A high‐throughput Arabidopsis reverse genetics system. Plant Cell, 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara, H. and Matsubayashi, Y. (2013) Chemical synthesis of Arabidopsis CLV3 glycopeptide reveals the impact of hydroxyproline arabinosylation on peptide conformation and activity. Plant Cell Physiol. 54, 369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter, A.M. , Keppler, B. , Lichtenberg, J. , Gu, D. and Welch, L.R. (2010) A bioinformatics approach to the identification, classification, and analysis of hydroxyproline‐rich glycoproteins. Plant Physiol. 153, 485–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X.‐F. , Yu, D.‐L. , Xu, T.‐T. , Ren, S.‐C. , Guo, P. and Liu, C.‐M. (2012) Contributions of individual amino acid residues to the endogenous CLV3 function in shoot apical meristem maintenance in Arabidopsis. Mol. Plant, 5, 515–523. [DOI] [PubMed] [Google Scholar]
- Stratford, S. , Barne, W. , Hohorst, D.L. , Sagert, J.G. , Cotter, R. , Golubiewski, A. , Showalter, A.M. , McCormick, S. and Bedinger, P. (2001) A leucine‐rich repeat region is conserved in pollen extensin‐like (Pex) proteins in monocots and dicots. Plant Mol. Biol. 46, 43–56. [DOI] [PubMed] [Google Scholar]
- Tusher, V.G. , Tibshirani, R. and Chu, G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA, 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki, A. , Freeze, H.H. and Gagneux, P. (2009). Evolution of glycan diversity In Essentials of Glycobiology (Varki A., Cummings R.D., Esko J.D., Freeze H.H., Stanley H.H., Bertozzi C.R., Hart G.W. and Etzler M.E., eds). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, pp. 281–292. [PubMed] [Google Scholar]
- Velasquez, S.M. , Ricardi, M.M. , Dorosz, J.G. et al (2011) O‐glycosylated cell wall proteins are essential in root hair growth. Science, 332, 1401–1403. [DOI] [PubMed] [Google Scholar]
- Velasquez, M. , Salter, J.S. , Dorosz, J.G. , Petersen, B.L. and Estevez, J.M. (2012) Recent advances on the posttranslational modifications of EXTs and their roles in plant cell walls. Front. Plant Sci. 3, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez, S.M. , Ricardi, M.M. , Poulsen, C.P. et al (2015) Low sugar is not always good: impact of specific O‐glycan defects on tip growth in Arabidopsis. Plant Physiol. 168, 808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S.‐L. , Rahman, A. , Baskin, T.I. and Kieber, J.J. (2008) Two leucine‐rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell, 20, 3065–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C. , Liberatore, K.L. , MacAlister, C.A. et al (2015) A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 47, 784–792. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Comparison of Arabidopsis and Physcomitrella hydroxyproline O‐arabinosyltransferase protein sequences.
Figure S2. Transfer DNA insertion mutations disrupting Arabidopsis HPAT expression.
Figure S3. Reduced seed set in hpat1 hpat3 was exclusively due to a pollen defect.
Figure S4. Generation and phenotypic analysis of hpat1 hpat2 hpat3 triple mutant plants.
Figure S5. Generation of Physcomitrella hpat mutants.
Figure S6. hpata phenotypes on various medium compositions and hormone treatments.
Figure S7. Cell‐wall‐associated hydroxyproline arabinosides are undetectable in full hpat mutants.
Figure S8. FEI1 related genes are up‐ and downregulated in hpata.
Figure S9. Transmission electron micrographs of wild‐type and hpata hpatb filaments.
Figure S10. The Physcomitrella genome encodes leucine‐rich repeat extensin (LRX)‐like extensin chimeras.
Table S1. Transmission rate of hpat1 hpat3 mutations.
Table S2. Physcomitrella genes containing Ser(Pro)3–5 motifs.
Table S3. List of genes downregulated in Physcomitrella hpata mutants.
Table S4. List of genes upregulated in Physcomitrella hpata mutants.
Table S5. Enriched categories of differentially expressed genes.
Table S6. Primer sequences used in this study.


