Abstract
The objective of the present study was to repurpose L-menthol, which is frequently used in oral health and topical formulations, for cancer therapeutics. In this article, we argue that monoterpenes such as L-menthol might offer veritable potentials in systems medicine, for example, as cheaper anti-cancer compounds. Other monoterpenes such as limonene, perillyl alcohol, and geraniol have been shown to induce apoptosis in various cancer cell lines, but their mechanisms of action are yet to be completely elucidated. Earlier, we showed that L-menthol modulates tubulin polymerization and apoptosis to inhibit cancer cell proliferation. In the present report, we used an apoptosis-related gene microarray in conjunction with proteomics analyses, as well as in silico interpretations, to study gene expression modulation in human adenocarcinoma Caco-2 cell line in response to L-menthol treatment. The microarray analysis identified caspase 10 as the important initiator caspase, instead of caspase 8. The proteomics analyses showed downregulation of HSP90 protein (also corroborated by its low transcript abundance), which in turn indicated inhibition of AKT-mediated survival pathway, release of pro-apoptotic factor BAD from BAD and BCLxL complex, besides regulation of other factors related to apoptosis. Based on the combined microarray, proteomics, and in silico data, a signaling pathway for L-menthol-induced apoptosis is being presented for the first time here. These data and literature analysis have significant implications for “repurposing” L-menthol beyond oral medicine, and in understanding the mode of action of plant-derived monoterpenes towards development of cheaper anticancer drugs in future.
Introduction
Animal studies have suggested the anti-carcinogenic properties of monoterpenes in the past. In this connection, menthol, a cyclic monoterpene, is frequently used in oral health and topical formulations. In this article, we argue that monoterpenes such as L-menthol might offer veritable potentials in systems medicine, for example, as cheaper anti-cancer compounds. Other monoterpenes such as limonene, perillyl alcohol, and geraniol have been shown to induce apoptosis in various cancer cell lines and to hold promise as a novel class of anticancer drugs (Loza-Tavera, 1999).
Limonene and perillyl alcohol have been suggested to play bioactivity against a wide variety of malignancies (Barthelman et al., 1998; Chen et al., 2015; Clark et al., 2003; Haag et al., 1992; Miller et al., 2015; Mills et al., 1995; Reddy et al., 1997; Stark et al., 1995; Stayrook et al., 1997). Perillyl alcohol selectively induces G0/G1 arrest (Sahin et al., 1999) and c-Myc-dependent apoptosis (Clark, 2006) and has a common mechanism of action as compared to geraniol and farnesol (Wiseman et al., 2007). D-Limonene induces apoptosis by inactivation of protein kinase B (AKT) in human colon cancer cells (Jia et al., 2013). In HL-60 cells, D-limonene and D-carvone are reported to induce apoptosis through caspase-8 activation (Yu et al., 2008). The anti-tumor activity and/or mechanism for 37 monoterpenes found in natural essential oils were reviewed by Sobral et al. (2014).
While these monoterpenes are reported to cause apoptosis, their detailed mechanisms of action are not known (Wiseman et al., 2007). Earlier, we reported apoptosis in human colon adenocarcinoma cells by L-menthol (Faridi et al., 2011) with enhanced tubulin polymerization. L-menthol is broadly utilized not only in oral hygiene products but also in confectionary, pharmaceuticals, cosmetics, pesticides, or as a flavoring agent. Positive modulation of tubulin polymerization by L-menthol underscores its destabilizing effect on microtubule assembly similar to that of taxol.
Since the apoptosis signaling pathway involved is yet to be delineated clearly, we conducted a microarray analysis with 368 targets/genes related to apoptosis/cell proliferation and with the help of in silico analysis and proteomics data we were able to propose a putative model. Perillyl alcohol is already in clinical trial for treatment of metastatic breast cancer (Bailey et al., 2008) and malignant gliomas (da Fonseca et al., 2008). As L-menthol is abundantly available from the essential oil of Mentha species, a detailed investigation of the mechanism of apoptotic signaling is very timely both for oral health and systems medicine, not to mention in developing or “repurposing” it towards a potentially cheaper anti-cancer therapy (Kale et al., 2015; Kato et al., 2015; McCabe et al., 2015).
In the present study, the combined use of standard experimental (Burnette, 1981; Faridi et al., 2011; Herrmann et al., 1994;) and in silico (Mi et al., 2013; Sturn et al., 2002) procedures has indicated that L-menthol induces apoptosis through caspase 10 and by suppressing the heat shock protein 90 (HSP90).
Materials and Methods
Cell line treatment
Caco-2 (ATCC HTB37, human colon adenocarcinoma) cells were obtained from National Centre for Cell Science (Pune, India), and treated with L-menthol at IC50 (12 mg/mL) for 48 h (Faridi et al., 2011). Taxol treatment was also carried out as described in our previous study (Faridi et al., 2011).
DNA fragmentation assay
L-menthol-treated cells were trypsinized and pelleted by centrifugation at 1600 g for 5 min. This was followed by washing with 500 μL 1x PBS and addition of 500 μL lysis buffer [1 mL Nonidet P-40, 4 ml EDTA (0.5 M) and 3.33 mL Tris-HCl (1.5 mM) were added and volume was made up to 100 mL] for 10 seconds. Post-treatment, the sample was centrifuged at 1600 g for 5 min and the supernatant was re-extracted with same amount of lysis buffer. To it, added 500 μL of SDS (1%) with 8 μL of RNase (5 μg/μL) and incubated at 56°C for 2 h. Then added 8 μL protinase K (2.5 μg/μL) and incubated for 2 h at 37°C. DNA was precipitated by adding 10% ammonium acetate and 2.5 volume of absolute ethanol followed by centrifugation at 8000 g for 5 min. The precipitated DNA was dissolved in gel loading buffer and observed on 1.5% agarose gel (Herrmann et al., 1994).
Microarray
An oligo-set for 368 apoptosis-related genes (Operon Technologies, USA) was spotted on slides (Genomic Solutions, USA). For each gene, there were 8 technical replicates/spots and the average intensity was calculated.
Labeling and purification for cDNA [synthesized from 10 μg total RNA isolated using TRIzol® (Invitrogen, USA) as per manufacturer's guidelines] were carried out using the ChipShot™ Indirect Labeling and Clean-Up System (Promega, USA). The eluted aminoallyl cDNA was conjugated with CyDye™ NHS Ester as per protocol and the eluted, purified, labeled cDNA was quantified for dye incorporation by measuring absorbance at 260, 550, and 650 nm. The labeled cDNA samples were vacuum dried and dissolved in 120 μL of salt-based hybridization buffer (Ocimum Biosolutions, India), followed by heating at 95°C for 5 min and cooling to room temperature.
Hybridization was carried out for 16 h at 65°C, followed by washing and image acquisition using a Genomic Solutions (USA) platform. The images were scanned with a Gene TAC™ UC 4 Microarray Scanner (Genomic Solutions Inc., a Harward Bioscience Company, MA). Signal intensities for each spot were quantified using Gene TAC Integrator (Version 4; Genomic Solutions). The observed fold change value provides an estimate of the level of differential expression of each gene. Total intensity normalization was performed globally, meaning that for each expression value the same transformation was applied independent of its intensity. Cy5 was used for labeling L-menthol-treated sample and Cy3 was used for labeling untreated sample.
The fold change was calculated as follows: Fold change (FC) value = (Cy5/Cy3) x Normalization factor (N), where N was calculated on the basis of global normalization. For analysis, the fold change value used for upregulated genes was the FC value itself (positive values), whereas for downregulated genes the negative reciprocal of the FC value was used for easier comprehension.
GENETAC Integrator version 4.0 software was used for the analysis (Genomic Solutions). Initially the raw ratios were calculated for both the dyes Cy3 (volume) and Cy 5 (volume) and if the value was less than 1, N fold was calculated by taking negative reciprocal of the raw ratios. The absolute values of all the N- fold changes were calculated. Normalization factor was determined by using reference sample or housekeeping genes within array. The foreground and background intensity values were recorded for every gene in the array using a fixed threshold boundary by Gene TAC Integrator software (Genomic Solutions).
Background intensity values were subtracted from the foreground and normalized using global normalization algorithm used by GeneTAC Integrator software. The resultant comma separated file with the background subtracted, globally normalized intensity ratio between the treated versus control (Cy5/Cy3) values were used for statistical analysis. Dunnett Multiple Comparisons Test (using “Graph Pad InStat Version 3.01”) was performed by comparing the fold change values for all the genes in L-menthol-treated cell line with untreated cell line. Out of 330 genes, only 12 genes in the range of 0 to 2 (SFRP1, DAP13, ING1, HSPA5, MCL1, KIAA0720, MDM4, DFFB, TNFRSF10A, AKT3, TNFRSF14, and IGFALS) and 6 genes in the range of 0 to −2 (TRAF5, PDCD2, CNR2, SPTAN1, DAP3, and P53AIP1) fold expression were having no significant difference with p > 0.05 and the rest of the genes showed significant differences with p < 0.01.
The data from this microarray experiment were submitted to GEO (NCBI) under series accession number GSE53701 [associated sample GSM1299167].
Semi-quantitative RT-PCR for microarray data validation
Total RNA was isolated using TRIzol® (Invitrogen, USA) (as done previously for the microarray analysis) and analyzed through ethidium bromide staining and Nanodrop ND1000 spectrophotometer. Four micrograms of DNaseI-treated total RNA was used for first strand cDNA synthesis using Thermoscript RT-PCR System (Invitrogen). Transcription factor 3C polypeptide 5 (TF3C-epsilon, NM_001122823.1) was amplified as control (Faridi et al., 2011). The primer sequences were designed using Gene Runner version 3.05 (Hastings Software Inc., USA.) (Supplementary Table S1; supplementary material is available online at www.liebertpub.com/omi).
Proteomics analysis
Protein samples were isolated from L-menthol-treated and untreated cells and subjected to 2D electrophoresis as described earlier (Faridi et al., 2011). Differentially expressing spots were excised and MS spectra were analyzed through MALDI-TOF-TOF (Faridi et al., 2011). For Western blotting, the protein samples (40 μg each) were loaded on 12% SDS-polyacrylamide gel and electrophoresed for 45 min at 250 V. The proteins were transferred to a nitrocellulose membrane (Burnette, 1981). After complete transfer, the membrane was incubated in blocking buffer (Pierce Biotechnology, Rockford, USA) at room temperature, washed with PBS in 0.1% Tween 20 for 1 h and immune-reacted overnight at 4°C with anti-HSP90 antibody (Cell Signaling Technologies, Danvers, MA).
The membrane was washed four times with PBS with 0.1% Tween 20 (each time for 15 min.) and the HRP-conjugated anti-rabbit IgG (diluted 1:2,000) was applied to the membrane for 2 h at room temperature. Finally the membrane was washed with PBS/0.1% Tween 20 for 1 h and the signal was detected using 4-chloronaphthol as substrate (following the protocol of Thermo Scientific, USA).
In silico analysis
The role of selected genes in cancer-related pathways was visualized using PANTHER (Protein ANalysis THrough Evolutionary Relationship) (Mi et al., 2013). It was used to classify proteins/genes according to family and subfamily, molecular function, biological process, and pathway. The genes involved in apoptosis signaling pathway were clustered according to their expression values. Genesis 1.7.6 clustering tool (Sturn et al., 2002) was used for grouping and heat-map generation. KEGG Mapper (http://www.genome.jp/kegg/mapper.html) was used for visualization of the genes in apoptosis pathway based on their expression values.
Results
DNA fragmentation
DNA in cells undergoing apoptosis shows a typical ladder-like pattern due to fragmentation caused by caspase-activated DNase. The DNA isolated from L-menthol-treated cells was observed to be degraded as compared to the intact DNA from the untreated cells (Fig. 1). Paclitaxel/taxol (positive control) treatment showed complete DNA degradation.
FIG. 1.
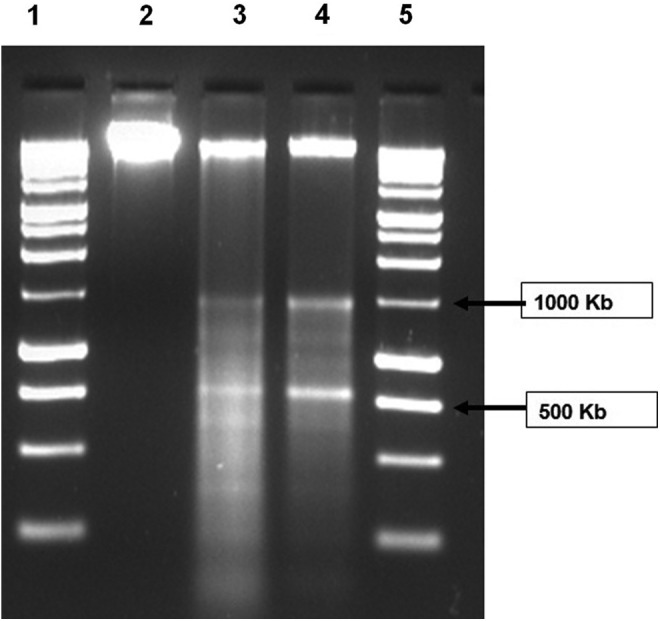
DNA from Caco-2 cell lines. Lanes 1 and 5: molecular weight marker; Lane 2: DNA from untreated cell; Lane 3: DNA from paclitaxel-treated cell; and Lane 4: DNA from L-menthol-treated cell. DNA sample from L-menthol-treated cell shows similar degradation as in case of paclitaxel treatment.
Microarray analysis
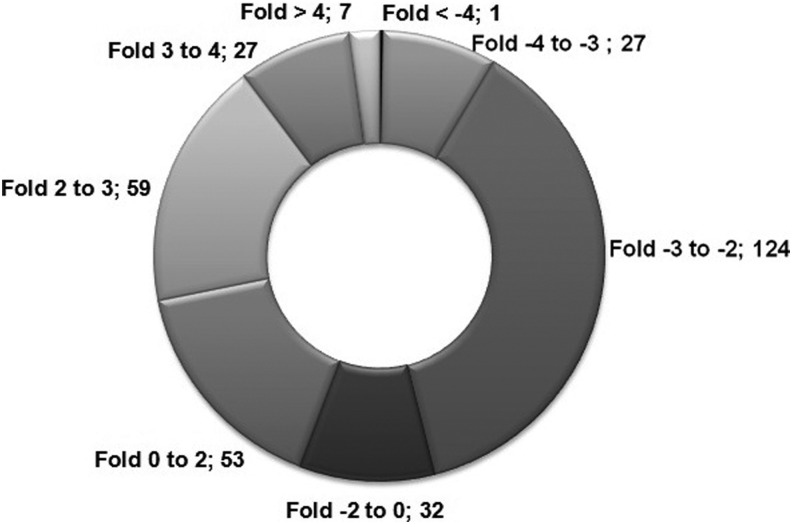
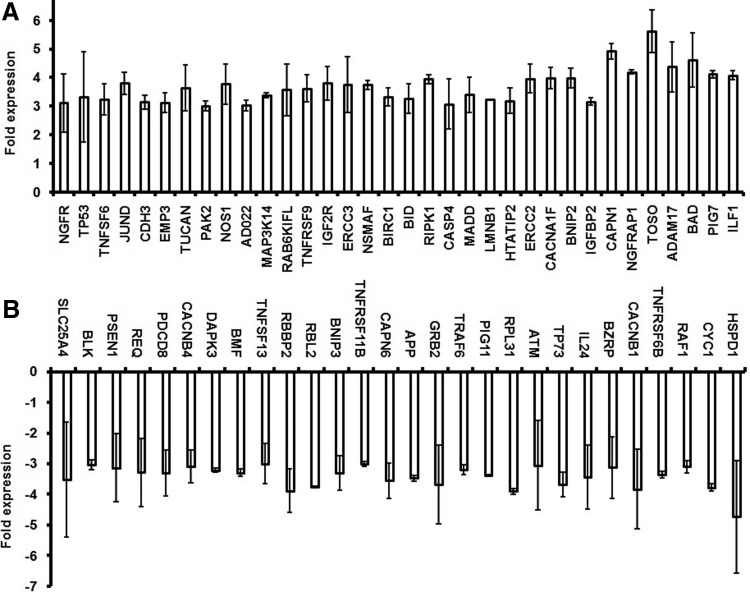
Of the 368 genes, consistent data was obtained for 330 genes (Supplementary Table S2), whereby 146 genes demonstrated upregulation and 184 genes showed downregulation (Fig. 2). The upregulated (above 3-fold) and downregulated (below −3-fold) genes are presented in Figure 3A and 3B, respectively.
FIG. 2.
Doughnut diagram representing the number of differentially expressed genes in L-menthol-treated Caco2 cells as compared to untreated control cells.
FIG. 3.
Differentially expressed genes in L-menthol-treated Caco2 cells. (A) Genes upregulated (more than 3-fold); (B) Genes downregulated (less than −3-fold).
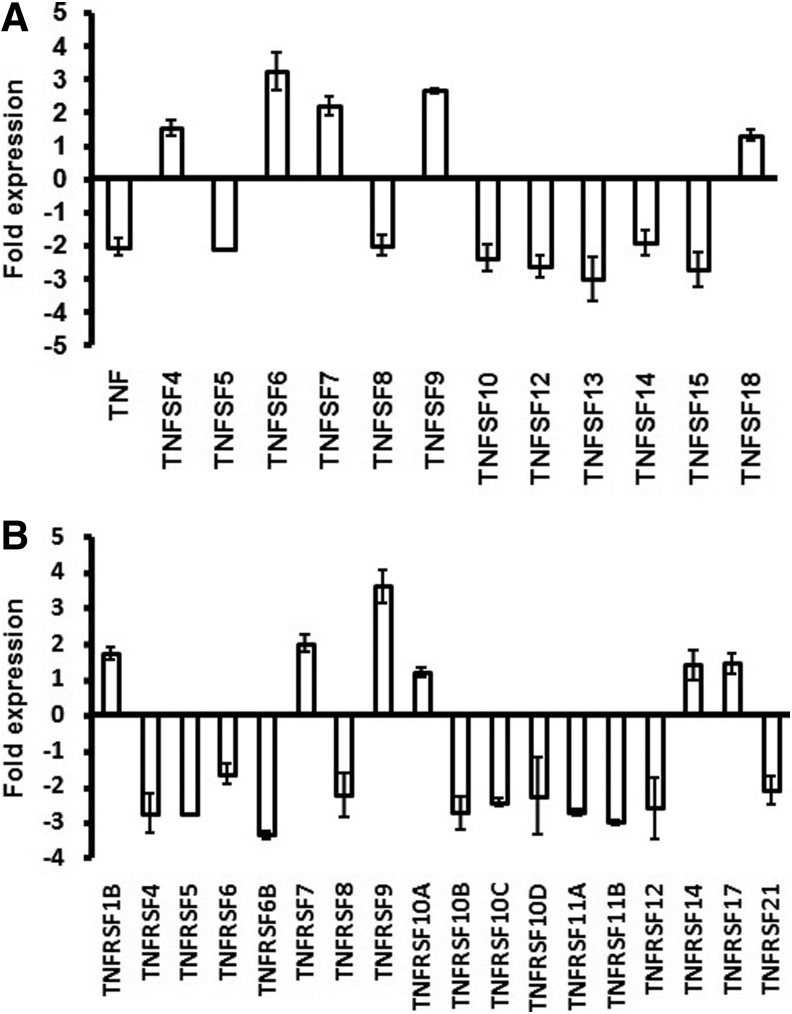
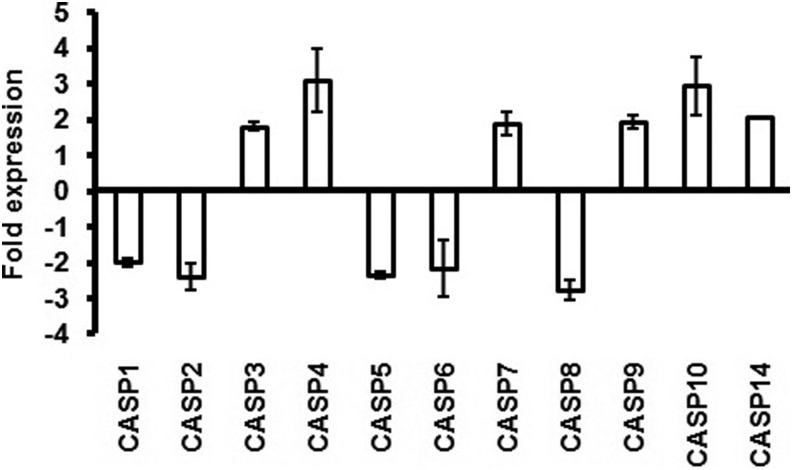
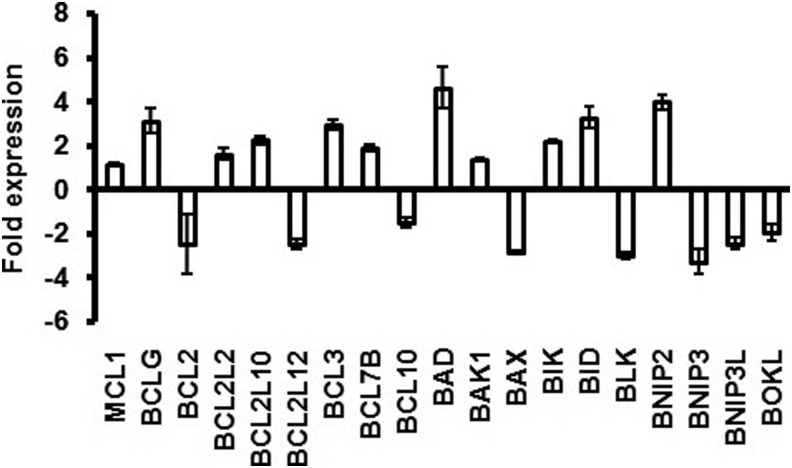
The genes TNFSF 6, 7, and 9 and the receptors TNFRSF 7 and 9 were upregulated (> 2-fold) (Fig. 4A and 4B). Similarly, the genes for caspases CASP 3, 4, 7, 9, 10, and 14 were also upregulated (Fig. 5). Substantially higher expression of BCL-2 such as genes BCLG, BCL2L10, BCL3, BAD, BID, and BNIP2 was also observed (Fig. 6). Semi-quantitative RT-PCR validated the microarray data for some randomly selected genes (TNFSF9, TNFRSF7, CASP3, CASP7, CASP9, BID, and AKT1) (Fig. 7).
FIG. 4.
TNF (A) and TNF receptor (B) genes showing up/down regulation in L-menthol-treated Caco2 cells.
FIG. 5.
Caspase family genes showing up/down regulation in L-menthol-treated Caco2 cells.
FIG. 6.
BCL-2 family genes showing up/down regulation in L-menthol-treated Caco2 cells.
FIG. 7.
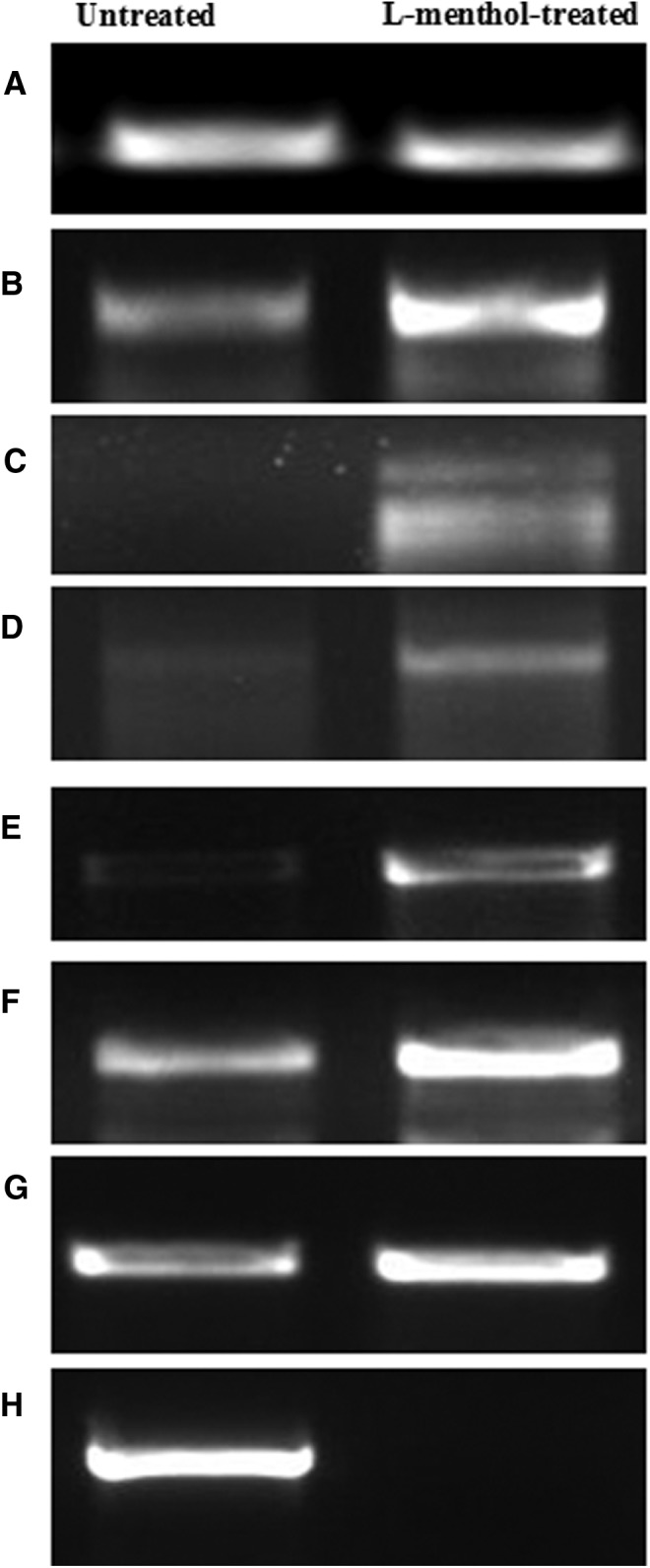
Representative gel image depicting the semi-quantitative RT-PCR-based validation of microarray data for randomly selected differentially expressed genes in L-menthol-treated Caco2 cells. (A) Control (TF3C-epsilon); (B) TNFSF9; (C) TNFRSF7; (D) Caspase 3; (E) Caspase 7; (F) Caspase 9; (G) BID; (H) AKT1.
Proteomics and expression analysis
In 2D electrophoresis and MS-MS analysis (Faridi et al., 2011), 7 proteins were conspicuously absent along with tubulin in case of L-menthol-treated cells (Supplementary Table S3). Absence/decrease in HSP90 expression was confirmed through Western blotting and its transcript abundance was also found to decrease in L-menthol-treated cells (Fig. 8).
FIG. 8.
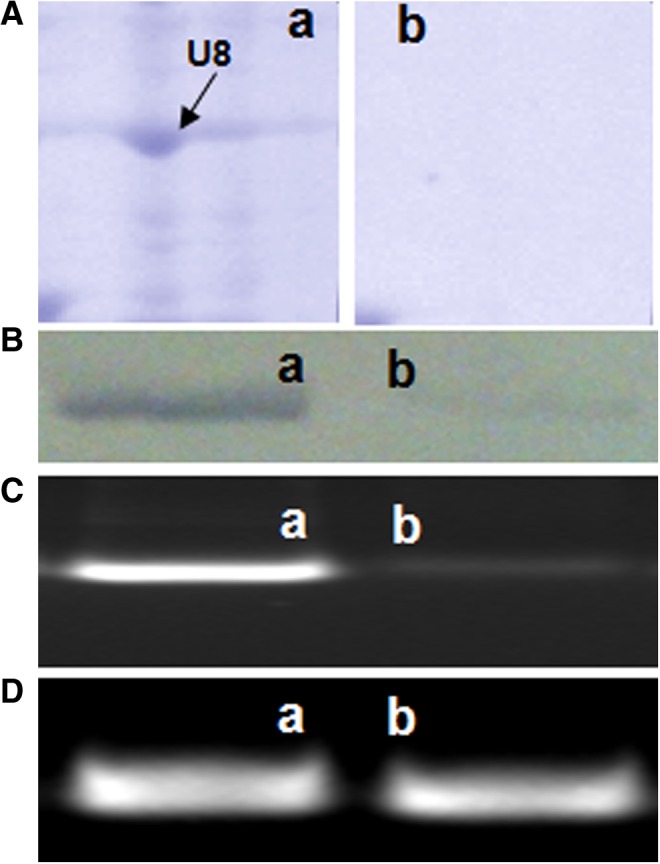
HSP90 protein and gene expression in untreated (a) vs L-menthol-treated (b) Caco-2 cells. (A) Magnified view of the gel portion showing absence of the spot “U8” representing HSP90 protein in L-menthol-treated cells. (B) Western blot showing decrease in concentration of HSP90 in L-menthol-treated cells. (C) Semi-quantitative RT-PCR showing decrease in expression of HSP90 in L-menthol-treated cells. (D) Semi-quantitative RT-PCR of control gene TF3C-epsilon.
Pathway map
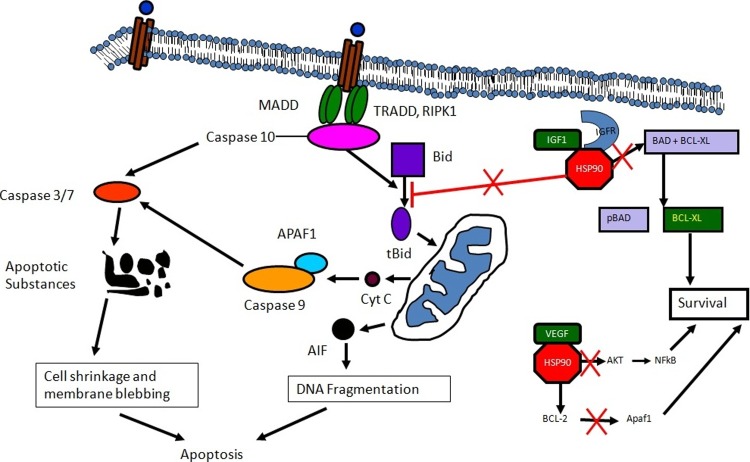
The selected genes with their fold change values were analyzed for their role in different signaling pathways using PANTHER (Supplementary Fig. S1). Fifty-four genes were found associated with apoptosis signaling pathways and were analyzed through a heatmap (and arranged according to their fold change values) (Supplementary Fig. S2). The color range was light blue (downregulated) to orange (upregulated). The genes were visualized in accordance to their expression values in the apoptosis pathway map (Supplementary Fig. S3). Red color denoted upregulation, whereas green color indicated downregulation. Based on microarray, proteomics and in silico data, a signaling pathway for L-menthol-induced apoptosis was proposed (Fig. 9).
FIG. 9.
Model predicting the possible mode of action of L-menthol. As described, the initial response is through Caspase 10 and followed by Caspase 3 and Caspase 7. Caspase 10 may also influence the activation of BID to tBID. This step is inhibited by HSP90, but due to lower expression in L-menthol-treated cell, the availability of HSP90 decreases. After release of cytochrome c from mitochondria, APAF1 and Caspase 9 induce Caspase 3 and Caspase 7 towards apoptosis. HSP90 may not be available in sufficient amount to trigger the AKT- and NFkB-mediated survival pathway. Similarly, BAD (a pro-apoptotic factor) is released from the complex “BAD and BCL-xL” due to low availability of HSP90.
Discussion
Omics biotechnologies such as genomics, proteomics, and metabolomics allow us to repurpose drugs or examine the system scale impacts of molecular perturbations, be they induced by drugs, nutrition, xenobiotics, or changes in the environmental stressors (Dandara et al., 2014; Gong et al., 2014; Kale et al., 2015; Kato et al., 2015; McCabe et al., 2015; Mounayar et al., 2014; Sahu et al., 2014; Sitole et al., 2014; Zheng et al., 2014). Based on the combined microarray, proteomics, and in silico data, a signaling pathway for L-menthol-induced apoptosis is being presented for the first time here. To contextualize the present findings, we provide below the broader rationale and molecular background to consider repurposing L-menthol beyond applications in oral medicine, with a view to anti-cancer therapeutics and systems medicine.
Tumor necrosis factor superfamily (TNFSF) proteins govern many cellular functions including apoptosis (Ware, 2008). They bind to members of the tumor necrosis factor receptor superfamily (TNFRSF) and depending on the proteins involved, it triggers a series of chemical signals that instruct cells to grow/divide, self-destruct, or mature for specialized functions. In this analysis, TNFSF7 and TNFSF9 and their corresponding TNFRSFs (TNFRSF7, TNFRSF9) were upregulated in L-menthol-treated cells (Fig. 4). TNFSF7 expression is epigenetically downregulated during progression in breast cancer cells (Yu et al., 2010) and it is also associated with the tumor necrosis factor (TNF) ligand-receptor death pathway (Hengartner, 2000). There are reports on apoptosis induction by TSNSF7-CD27L (Kashii et al., 1999) and TNFRSF9-TNFSF9 (Seko et al., 2004; Ebmeyer et al., 2011) in tumor cells. Hence, in case of L-menthol-treated cells, the apoptosis signal was predicted to be initiated by TNFSF7-TNFRSF7 and TNFSF9-TNFRSF9.
Induction of apoptosis via death receptors typically activates an initiator caspase (CASP) such as CASP8 or CASP10, which in turn activate other caspases in a cascade leading to the activation of effector caspases, like CASP3 and CASP6, responsible for cleaving key proteins in cells undergoing apoptosis. CASP8 plays an obligatory role in apoptosis initiation, but the role of its structural relative, CASP10, remains to be investigated (Kischkel et al., 2001).
In this investigation, high expression of CASP10 was obtained instead of CASP8 (Fig. 5). CASP10 is closely related in sequence to CASP8 (Fernandes-Alnemri et al., 1996; Hu et al., 1997) and both are encoded on the same region of human chromosome 2, suggesting duplication of an ancestral gene. In vitro experiments have demonstrated the ability of CASP10 to process CASP3 and CASP7 (Fernandes-Alnemri et al., 1996), and CASP10 has also been implicated in apoptosis (Hu et al., 1997; Kischkel et al., 2001; Milhas et al., 2005; Wang et al., 2001). Thus, involvement of CASP10 rather than CASP8 in apoptosis signaling in L-menthol-treated cells seems more likely. Post-activation, CASP10 will activate downstream caspases including CASP3 and CASP7. Here L-menthol was found to induce the expression of genes for pro-apoptotic caspases 3, 7, 9, and 10 (Fig. 5).
Moving on to the next category, BCL-2 family proteins, having either pro- or anti-apoptotic activities, have been studied intensively owing to their importance in regulation of apoptosis, tumor genesis and cellular responses to anticancer therapy. In mammals, there are at least 12 core BCL-2 family proteins, having either three-dimensional structural similarity or a predicted secondary structure that is similar to BCL-2 (Youle and Strasser, 2008).
In this investigation, genes for many BCL-2 family proteins showed higher expression in L-menthol-treated cells (Fig. 6). Their role in induction of apoptosis has been described in detail in many earlier studies (Brocke-Heidrich et al., 2006; Chinnadurai et al., 2008; Guo et al., 2001; Hsu et al., 1997; Inohara et al., 1998; Luo et al., 2009; Valencia et al., 2007; Zhao et al., 2003).
A lot of interplay is also involved among various categories of proteins in effecting apoptosis. APAF1 is a key link in apoptosis (Soengas et al., 2001), which is associated with pro-caspase-9 and cytochrome c, forming the apoptosome complex and activating CASP9 that in turn cleaves and activates effector caspases (Li et al., 1997). BCL2L10 binds to this protein directly and prevents Bcl-xL (apoptosis inhibitor) binding to APAF1 (Inohara et al., 1998). In this analysis, higher expression of APAF1 and BCL2L10 indicated the occurrence of this pathway for apoptosis after L-menthol treatment (Supplementary Table S2).
Although it is well established that the high-affinity NGF receptor plays a pivotal role in cell survival, there are reports about low-affinity NGF receptor inducing apoptosis (Taglialatela et al., 1997). Resveratrol is known to induce apoptosis through a TP53-dependent pathway (Huang et al., 1999). Apoptosis enhancements by JUND (Li et al., 2002), EMP3 (Fumoto et al., 2009), AD022 (Zucchelli et al., 2009), RAB6KIFL (Hill et al., 2000), TNFRSF9 (Ebmeyer et al., 2011), IGFR (Wylie et al., 2003), NSMAF (O'Brien et al., 2003), BID (Zhao and Wang, 2004), RIPK1 (Wang et al., 2008), CASP4 (Hitomi et al., 2004), MADD (Schievella et al., 1997), HTATIP2 (Xiao et al., 2000), BNIP-2 (Valencia et al., 2007), and IGFBP2 (Frommer et al., 2006) are known. TNFSF6 initiates apoptosis by binding to its receptor FAS (TNFRSF6) (Castellano et al., 2006). However, in this investigation TNFRSF6 transcript was downregulated (Fig. 4).
The other important proteins showing more than 4-fold increase in transcript expression in response to L-menthol (Supplementary Table S2) and reported earlier to be regulating apoptosis are CAPN1 (Piñeiro et al., 2007), NGFRAP1 (Naderi et al., 2007), ADAM17 (Wang et al., 2011), BAD (Adachi et al., 2002), PIG7 (Matsumura et al., 2004), and ILF1 (Sadkowski et al., 2008). Taxol induces CASP3-independent apoptosis in NIH3T3 cells by a calpain (CAPN1)-mediated mechanism (Piñeiro et al., 2007). L-menthol has also been found to promote tubulin polymerization (Faridi et al., 2011) such as taxol and increases the expression of CAPN1.
Besides these, several other predominantly anti-apoptotic protein transcripts showing overexpression in response to L-menthol were CDH3 (Wheelock and Johnson, 2003), TUCAN (Pathan et al., 2001), NOS1 (Andoh et al., 2000), MAP3K14 (Liao et al., 2004), BIRC1 (Yin et al., 2008), and CACNA1F (Kotturi et al., 2003) (Supplementary Table S2). PAK2, although anti-apoptotic (Jakobi et al., 2001), can be activated via cleavage by caspases mediating cell death (Rudel and Bokoch, 1997; Rudel et al., 1998). The role of DNASE1l3 (upregulated 2.11-fold) in chromatin cleavage during apoptosis has been proposed earlier (Napirei et al., 2009). Hence, the nucleotide excision repair enzyme excision repair cross-complementing group 2 and 3 genes (ERCC2, ERCC3) (Hsia et al., 2003; Justenhoven et al., 2004) are expected to overexpress both in cancer cells and during apoptosis. But the protein mTOSO (Song and Jacob, 2005) is critical in inhibiting CASP8 activation. Since CASP8 is downregulated in this investigation (Fig. 5), high expression of TOSO in L-menthol-treated cells (Fig. 3) may not have any anti-apoptotic effect, although influence on other proteins cannot be ruled out.
Several genes downregulated in this investigation were involved with the survival pathway. SLC25A4 functions as a gated pore translocating ADP from the mitochondrial matrix into the cytoplasm (Napoli et al., 2001), whose downregulation leads to mitochondrial dysfunction. BCL-2 proteins are shown to interact and regulated by BH3 domain-containing proteins such as BLK (Zong et al., 2001). Presenilins (PSEN1 and 2) are required for maintenance of neural stem cells in the developing brain (Kim and Shen, 2008). Although the protein requiem (REQ) is described to be a regulator of apoptosis, its role in signaling is unclear (Wong et al., 2006). Apoptosis-inducing factor (AIF) has dual functions, a pro-apoptotic activity in the nucleus and an anti-apoptotic activity through oxidoreductase activity (Klein et al., 2002; Lipton and Bossy-Wetzel, 2002).
TNFSF13 is a NF-kappaB-activating receptor for TALL1 (Shu and Johnson, 2000). The other pro-apoptotic or antiproliferative protein genes downregulated were DAPK3 (Kawai et al., 1998), BMF (Puthalakath et al., 2001), RBL2 (Dick, 2007), RBBP2 (Chicas et al., 2012), BNIP3 (Burton et al., 2013), APP (Takahashi et al., 2009), PIG11 (Wu et al., 2009), RPL31 (Su et al., 2012), ATM (Kim et al., 1999), TP73 (Wang et al., 2013), and IL24 (Nagakawa et al., 2012) (Supplementary Table S2). Similarly, anti-apoptotic/cell proliferation genes downregulated are CACNB4 (Tadmouri et al., 2012), TNFRSF11B (Oliver et al., 2013), CAPN6 (Rho et al., 2008), GRB2 (Kraskouskaya et al., 2013), TRAF6 (Wong et al., 1999), BZRP (Maaser et al., 2002), TNFRSF6B (Chen et al., 2010), RAF1 (McPhillips et al., 2006), CYC1 (Zhu et al., 2012), and HSPD1 (Ghosh et al., 2008) (Supplementary Table S2).
In the proposed model, CASP10 and HSP90 have major roles (Fig. 9). HSP90 modulates tumor cell apoptosis and its inhibition disrupts multiple pathways essential for survival, proliferation, and metastasis of transformed cells, making it a promising target for developing cancer chemotherapeutics. It acts as an anti-apoptotic factor via several mechanisms (Arya et al., 2007) and also forms a cytosolic complex with APAF1, inhibiting cytochrome c-mediated APAF1 oligomerization and pro-caspase-9 activation (Pandey et al., 2000). Besides, HSP90 interacts with BID, a pro-apoptotic member of the Bcl-2 family, and prevents TNF-α-induced BID cleavage, which is involved in cytochrome c release (Zhao and Wang, 2004).
By keeping pro-apoptotic factors inert, HSP90 regulates anti-apoptotic proteins. It also modulates the stability of receptor-interacting protein, which functions as an anti-apoptotic protein by regulating NF-kB activity (Lewis et al., 2000). HSP90 regulates TNF-induced activation of IkB kinase (IKK) and NF-kB by interacting with the kinase domains of IKK and by forming a hetero-complex, containing NF-kB essential modulator and co-chaperone protein CDC37 (Chen et al., 2002). This protein also directly interacts with and maintains the activity of AKT by inhibiting its dephosphorylation, and functions together with AKT to inhibit the activity of pro-apoptotic kinase ASK1 (apoptosis signal-regulating kinase 1) (Basso et al., 2002; Sato et al., 2000; Zhang et al., 2005). Inhibition of CASP8/CASP10 activation inhibits HSP90 cleavage and largely inhibits UV-B-induced apoptosis (Chen et al., 2009).
Conversely, downregulation of HSP90 by expression of its shRNA, which would be expected to relieve its inhibition of pro-apoptotic proteins and the protection of anti-apoptotic proteins, promotes cell death in response to UV-B irradiation (Chen et al., 2009). In this investigation, the effect of L-menthol on downregulation of HSP90 has been shown both at transcript and protein levels (Fig. 8). Further, AKT1 and AKT2 were downregulated and expression of AKT3 was marginally positive (Supplementary Table S2). GRB2 and RAF1, involved in the AKT pathway, were severely downregulated, indicating inhibition of the pathway (Supplementary Table S2). Apoptosis can also be inhibited by the activation of pro-survival signaling pathways, such as insulin-like growth factor (IGF)-I and its HSP90-dependent receptor (Nielsen et al., 2004). Activation of this pathway increases the expression of pro-survival factors, like BCL-2 and BCL-xL, and decreases the expression of pro-apoptotic factors, like BIM (Kooijman, 2006).
Furthermore, IGF-I receptor activation induces the phosphorylation of BAD and phosphorylated BAD dissociates from Bcl-xL, liberating its anti-apoptotic activity (Nielsen et al., 2004; Xu and Neckers, 2007). The mannose-6-phosphate/insulin-like growth factor 2 receptor (M6P/IGF2R) encodes a multifunctional protein involved in lysosomal enzyme trafficking, fetal organogenesis, tumor suppression, and T cell-mediated immunity (Wylie et al., 2003). Perillyl alcohol treatment increases M6P/IGF2R (Belanger, 1998). IGFBP2-induced gene expression is of functional significance for proliferation, cell adhesion, cell migration, and apoptosis, and IGFBP2 can promote apoptosis in tumor cells independent of IGF (Frommer et al., 2006). In this investigation increase in expression of BAD, IGF2R, IGFBP2, and decrease in the expression of BCL2 was observed in L-menthol-treated cells, indicating the inhibition of this pro-survival pathway (Supplementary Table S2).
The expression of another gene VEGF (vascular endothelial growth factor) believed to be inducing HSP90 expression decreased (-2.71-fold) in L-menthol-treated cells (Supplementary Table S2). Upon VEGF stimulation, HSP90 binds to BCL2 and APAF1, an effect mediated through VEGFR2 and involving the activation of the MAP kinase pathway. These actions of VEGF result in increased resistance to apoptosis (Dias et al., 2002). Also, high levels of expression of BID (Fig. 6), the two TNFSFs (TNFSF7, TNFSF9) and the corresponding TNFRSFs (TNFRSF7, TNFRSF9) (Fig. 4) in L-menthol-treated cells indicated their association in apoptosis signaling. FADD demonstrated negative expression, whereas TRADD showed positive expression indicating the involvement of TRADD (Supplementary Table S2).
MADD (3.4-fold) has been shown to be associated with the death domain of the type 1 TNF receptor through its own C-terminal death domain (Schievella et al., 1997). Some of the variants of this gene IG20 are known to activate CASP3 and CASP8 (Al-Zoubi et al., 2001), indicating the possibility of its involvement in apoptosis. In L-menthol-treated cells, receptor-associated protein kinase genes RIPK1, RIPK2, and RIPK3 were upregulated (Supplementary Table S2). RIPK1 is critical for CASP8 activation induced by SMAC mimetic (Wang et al., 2008), but in this experiment CASP8 function is being apparently taken care by CASP10. It may be possible that upregulation of this gene by L-menthol induction may be aiding activation of CASP10.
Whereas the upregulation of different apoptosis-related genes indicates programmed cell death, simultaneous upregulation of anti-apoptotic genes indicates the cell's tendency to maintain a delicate balance between apoptosis and survival. A similar argument can be made to explain the simultaneous downregulation of anti-apoptotic and pro-apoptotic genes. The competition between these genes determines the cellular fate depending on the family member and isoform expressed in the specific cell type. Further future studies need to be carried out for more direct implication of the two target candidates in the apoptotic pathway. Blocking caspase 10 in order to confirm that the apoptotic pathway goes through it, as well as the addition of HSP90 to inhibit apoptosis will be the further experiments needed to be carried out. Another futuristic dimension would be to carry out the experiment on more and varied human cancer cell lines towards establishing the role of L-menthol in inducing apoptosis. However, the data and literature analysis in the present study have significant implications for “repurposing” L-menthol beyond oral medicine, and in understanding the mode of action of plant-derived monoterpenes towards development of cheaper anticancer drugs in future.
Supplementary Material
Acknowledgments
The authors express their sincere gratitude to Director of the CSIR-CIMAP for keen interest and providing facilities for the experiments. The support provided by Dr. Rakesh Kumar Shukla, Scientist, CSIR-CIMAP, during the study is also gratefully acknowledged. Uzma was supported by CSIR SRF. Funding support from the Department of Biotechnology and the Council of Scientific and Industrial Research, India (Twelfth Five Year Plan Project BSC0203) is also acknowledged.
AK Sha conceived the study. SSD, MPD, and AK Sha designed the wet lab experiments. UF, SSD and SP performed the wet lab experiments. SG and AS performed the in silico experiments and analysis. UF, SSD, MPD and AK Sha analyzed the wet lab data. AK Shu and AK Sha compiled the analyzed data and wrote the manuscript. All authors read and approved the final manuscript.
Author Disclosure Statement
The authors declare no conflicting financial interests.
References
- Adachi M, and Imai K. (2002). The proapoptotic BH3-only protein BAD transduces cell death signals independently of its interaction with Bcl-2. Cell Death Differ 9, 1240–1247 [DOI] [PubMed] [Google Scholar]
- Al-Zoubi AM, Efimova EV, Kaithamana S, et al. (2001). Contrasting effects of IG20 and its splice isoforms, MADD and DENN-SV, on tumor necrosis factor alpha-induced apoptosis and activation of caspase-8 and −3. J Biol Chem 276, 47202–47211 [DOI] [PubMed] [Google Scholar]
- Andoh T, Lee SY, and Chiueh CC. (2000). Preconditioning regulation of bcl-2 and p66shc by human NOS1 enhances tolerance to oxidative stress. FASEB J 14, 2144–2146 [DOI] [PubMed] [Google Scholar]
- Arya R, Mallik M, and Lakhotia SC. (2007). Heat shock genes—Integrating cell survival and death. J Biosci 32, 595–610 [DOI] [PubMed] [Google Scholar]
- Bailey HH, Attia S, Love RR, et al. (2008). Phase II trial of daily oral perillyl alcohol (NSC 641066) in treatment-refractory metastatic breast cancer. Cancer Chemother Pharmacol 62, 149–157 [DOI] [PubMed] [Google Scholar]
- Barthelman M, Chen W, Gensler HL, Huang C, Dong Z, and Bowden GT. (1998). Inhibitory effects of perillyl alcohol on UV-B-induced murine skin cancer and AP-1 transactivation. Cancer Res 58, 711–716 [PubMed] [Google Scholar]
- Basso AD, Solit DB, Chiosis G, Giri B, Tsichlis P, and Rosen N. (2002). Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. J Biol Chem 277, 39858–39866 [DOI] [PubMed] [Google Scholar]
- Belanger JT. (1998). Perillyl alcohol: Applications in oncology. Altern Med Rev 3, 448–457 [PubMed] [Google Scholar]
- Brocke-Heidrich K, Ge B, Cvijic H, et al. (2006). Bcl-3 is induced by IL-6 via Stat3 binding to intronic enhancer HS4 and represses its own transcription. Oncogene 25, 7297–7304 [DOI] [PubMed] [Google Scholar]
- Burnette WN. (1981). “Western blotting”: Electrophoretic transfer of proteins from sodium dodecyl sulfate–polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem 112, 195–203 [DOI] [PubMed] [Google Scholar]
- Burton TR, Henson ES, Azad MB, Brown M, Eisenstat DD, and Gibson SB. (2013). BNIP3 acts as transcriptional repressor of death receptor-5 expression and prevents TRAIL-induced cell death in gliomas. Cell Death Dis 4,e587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano R, Vire B, Pion M, et al. (2006). Active transcription of the human FASL/CD95L/TNFSF6 promoter region in T lymphocytes involves chromatin remodeling: Role of DNA methylation and protein acetylation suggest distinct mechanisms of transcriptional repression. J Biol Chem 281, 14719–14728 [DOI] [PubMed] [Google Scholar]
- Chen G, Cao P, and Goeddel DV. (2002). TNF-induced recruitment and activation of the IKK complex require Cdc37 and Hsp90. Mol Cell 9, 401–410 [DOI] [PubMed] [Google Scholar]
- Chen G, Rong M, and Luo D. (2010). TNFRSF6B neutralization antibody inhibits proliferation and induces apoptosis in hepatocellular carcinoma cell. Pathol Res Pract 206, 631–641 [DOI] [PubMed] [Google Scholar]
- Chen H, Xia Y, Fang D, Hawke D, and Lu Z. (2009). Caspase-10-mediated heat shock protein 90 beta cleavage promotes UV-B irradiation-induced cell apoptosis. Mol Cell Biol 29, 3657–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TC, Fonseca CO, and Schönthal AH. (2015). Preclinical development and clinical use of perillyl alcohol for chemoprevention and cancer therapy. Am J Cancer Res 5, 1580–1593 [PMC free article] [PubMed] [Google Scholar]
- Chicas A, Kapoor A, Wang X, et al. (2012). H3K4 demethylation by Jarid1a and Jarid1b contributes to retinoblastoma-mediated gene silencing during cellular senescence. Proc Natl Acad Sci USA 109, 8971–8976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnadurai G, Vijayalingam S, and Rashmi R. (2008). BIK, the founding member of the BH3-only family proteins: Mechanisms of cell death and role in cancer and pathogenic processes. Oncogene 27, S20–S29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SS. (2006). Perillyl alcohol induces c-Myc-dependent apoptosis in Bcr/Abl-transformed leukemia cells. Oncology 70, 13–18 [DOI] [PubMed] [Google Scholar]
- Clark SS, Zhong L, Filiault D, et al. (2003). Anti-leukemia effect of perillyl alcohol in Bcr/Abl-transformed cells indirectly inhibits signaling through Mek in a Ras- and Raf-independent fashion. Clin Cancer Res 9, 4494–4504 [PubMed] [Google Scholar]
- Dandara C, Huzair F, Borda-Rodriguez A, et al. (2014). H3Africa and the African life sciences ecosystem: Building sustainable innovation. OMICS 18, 733–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca CO, Schwartsmann G, Fischer J, et al. (2008). Preliminary results from a phase I/II study of perillyl alcohol intranasal administration in adults with recurrent malignant gliomas. Surg Neurol 70, 259–266 [DOI] [PubMed] [Google Scholar]
- Dias S, Shmelkov SV, Lam G, and Rafii S. (2002). VEGF (165) promotes survival of leukemic cells by Hsp90-mediated induction of Bcl-2 expression and apoptosis inhibition. Blood 99, 2532–2540 [DOI] [PubMed] [Google Scholar]
- Dick FA. (2007). Structure-function analysis of the retinoblastoma tumor suppressor protein—Is the whole a sum of its parts? Cell Div 2, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebmeyer J, Leichtle A, Hernandez M, et al. (2011). TNFA deletion alters apoptosis as well as caspase 3 and 4 expression during otitis media. BMC Immunol 12, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faridi U, Sisodia BS, Shukla AK, et al. (2011). Proteomics indicates modulation of tubulin polymerization by L-menthol inhibiting human epithelial colorectal adenocarcinoma cell proliferation. Proteomics 11, 2115–2119 [DOI] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Armstrong RC, Krebs J, et al. (1996). In vitro activation of CPP32 and Mch3 by Mch4, a novel human apoptotic cysteine protease containing two FADD-like domains. Proc Natl Acad Sci USA 93, 7464–7469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer KW, Reichenmiller K, Schutt BS, et al. (2006). IGF-independent effects of IGFBP-2 on the human breast cancer cell line Hs578T. J Mol Endocrinol 37, 13–23 [DOI] [PubMed] [Google Scholar]
- Fumoto S, Tanimoto K, Hiyama E, Noguchi T, Nishiyama M, and Hiyama K. (2009). EMP3 as a candidate tumor suppressor gene for solid tumors. Expert Opin Ther Targets 13, 811–822 [DOI] [PubMed] [Google Scholar]
- Ghosh JC, Dohi T, Kang BH, and Altieri DC. (2008). Hsp60 regulation of tumor cell apoptosis. J Biol Chem 283, 5188–5194 [DOI] [PubMed] [Google Scholar]
- Gong F, Yang L, Tai F, Hu X, and Wang W. (2014). “Omics” of maize stress response for sustainable food production: Opportunities and challenges. OMICS 18, 714–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Godzik A, and Reed JC. (2001). Bcl-G, a novel pro-apoptotic member of the Bcl-2 family. J Biol Chem 276, 2780–2785 [DOI] [PubMed] [Google Scholar]
- Haag JD, Lindstrom MJ, and Gould MN. (1992). Limonene-induced regression of mammary carcinomas. Cancer Res 52, 4021–4026 [PubMed] [Google Scholar]
- Hill E, Clarke M, and Barr FA. (2000). The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. EMBO J 19, 5711–5719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner MO. (2000). The biochemistry of apoptosis. Nature 407, 770–776 [DOI] [PubMed] [Google Scholar]
- Herrmann M, Lorenz HM, Voll R, Grünke M, Woith W, and Kalden JR. (1994). A rapid and simple method for the isolation of apoptotic DNA fragments. Nucleic Acids Res 22, 5506–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi J, Katayama T, Eguchi Y, et al. (2004). Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Aβ-induced cell death. J Cell Biol 165, 347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia KT, Millar MR, King S, et al. (2003). DNA repair gene Ercc1 is essential for normal spermatogenesis and oogenesis and for functional integrity of germ cell DNA in the mouse. Development 130, 369–378 [DOI] [PubMed] [Google Scholar]
- Hsu YT, Wolter KG, and Youle RJ. (1997). Cytosol-to-membrane redistribution of Bax and Bcl-x(L) during apoptosis. Proc Natl Acad Sci USA 94, 3668–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Vincenz C, Ni J, Gentz R, and Dixit VM. (1997). I-FLICE, a novel inhibitor of tumor necrosis factor receptor-1-and CD-95-induced apoptosis. J Biol Chem 272, 17255–17257 [DOI] [PubMed] [Google Scholar]
- Huang C, Ma WY, Goranson A, and Dong Z. (1999). Resveratrol suppresses cell transformation and induces apoptosis through a p53-dependent pathway. Carcinogenesis 20, 237–242 [DOI] [PubMed] [Google Scholar]
- Inohara N, Gourley TS, Carrio R, et al. (1998). Diva, a Bcl-2 homologue that binds directly to Apaf-1 and induces BH3-independent cell death. J Biol Chem 273, 32479–32486 [DOI] [PubMed] [Google Scholar]
- Jakobi R, Moertl E, and Koeppel MA. (2001). p21-activated protein kinase gamma-PAK suppresses programmed cell death of BALB3T3 fibroblasts. J Biol Chem 276, 16624–16634 [DOI] [PubMed] [Google Scholar]
- Jia SS, Xi GP, Zhang M, et al. (2013). Induction of apoptosis by D-limonene is mediated by inactivation of Akt in LS174T human colon cancer cells. Oncol Rep 29, 349–354 [DOI] [PubMed] [Google Scholar]
- Justenhoven C, Hamann U, Pesch B, et al. (2004). ERCC2 genotypes and a corresponding haplotype are linked with breast cancer risk in a German population. Cancer Epidemiol Biomarkers Prev 13, 2059–2064 [PubMed] [Google Scholar]
- Kale VP, Amin SG, and Pandey MK. (2015) Targeting ion channels for cancer therapy by repurposing the approved drugs. Biochim Biophys Acta 1848, 2747–2755 [DOI] [PubMed] [Google Scholar]
- Kato S, Moulder SL, Ueno NT, et al. (2015) Challenges and perspective of drug repurposing strategies in early phase clinical trials. Oncoscience 2, 576–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashii Y, Giorda R, Herberman RB, Whiteside TL, and Vujanovic NL. (1999). Constitutive expression and role of the TNF family ligands in apoptotic killing of tumor cells by human NK cells. J Immunol 163, 5358–5366 [PubMed] [Google Scholar]
- Kawai T, Matsumoto M, Takeda K, Sanjo H, and Akira S. (1998). ZIP kinase, a novel serine/threonine kinase which mediates apoptosis. Mol Cell Biol 18, 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, and Kastan MB. (1999). Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem 274, 37538–37543 [DOI] [PubMed] [Google Scholar]
- Kim WY, and Shen J. (2008). Presenilins are required for maintenance of neural stem cells in the developing brain. Mol Neurodegener 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kischkel FC, Lawrence DA, Tinel A, et al. (2001). Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. J Biol Chem 276, 46639–46646 [DOI] [PubMed] [Google Scholar]
- Klein JA, Longo-Guess CM, Rossmann MP, et al. (2002). The harlequin mouse mutation downregulates apoptosis-inducing factor. Nature 419, 367–374 [DOI] [PubMed] [Google Scholar]
- Kooijman R. (2006). Regulation of apoptosis by insulin-like growth factor (IGF)-I. Cytokine Growth Factor Rev 17, 305–323 [DOI] [PubMed] [Google Scholar]
- Kotturi MF, Carlow DA, Lee JC, Ziltener HJ, and Jefferies WA. (2003). Identification and functional characterization of voltage-dependent calcium channels in T lymphocytes. J Biol Chem 278, 46949–46960 [DOI] [PubMed] [Google Scholar]
- Kraskouskaya D, Duodu E, Arpin CC, and Gunning PT. (2013). Progress towards the development of SH2 domain inhibitors. Chem Soc Rev 42, 3337–3370 [DOI] [PubMed] [Google Scholar]
- Lewis J, Devin A, Miller A, et al. (2000). Disruption of hsp90 function results in degradation of the death domain kinase, receptor interacting protein (RIP), and blockage of tumor necrosis factor-induced nuclear factor-kappaB activation. J Biol Chem 275, 10519–10526 [DOI] [PubMed] [Google Scholar]
- Li T, Dai W, and Lu L. (2002). Ultraviolet-induced junD activation and apoptosis in myeloblastic leukemia ML-1 cells. J Biol Chem 277, 32668–32676 [DOI] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, et al. (1997). Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91, 479–489 [DOI] [PubMed] [Google Scholar]
- Liao G, Zhang M, Harhaj EW, and Sun SC. (2004). Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J Biol Chem 279, 26243–26250 [DOI] [PubMed] [Google Scholar]
- Lipton SA, and Bossy-Wetzel E. (2002). Dueling activities of AIF in cell death versus survival: DNA binding and redox activity. Cell 111, 147–150 [DOI] [PubMed] [Google Scholar]
- Loza-Tavera H. (1999). Monoterpenes in essential oils: Biosynthesis and properties. Adv Exp Med Biol 464, 49–62 [DOI] [PubMed] [Google Scholar]
- Luo N, Wu Y, Chen Y, et al. (2009). Upregulated BclG(L) expression enhances apoptosis of peripheral blood CD4+ T lymphocytes in patients with systemic lupus erythematosus. Clin Immunol 132, 349–361 [DOI] [PubMed] [Google Scholar]
- Maaser K, Grabowski P, Sutter AP, et al. (2002). Overexpression of the peripheral benzodiazepine receptor is a relevant prognostic factor in stage III colorectal cancer. Clin Cancer Res 8, 3205–3209 [PubMed] [Google Scholar]
- Matsumura Y, Matsumura Y, Nishigori C, Horio T, and Miyachi Y. (2004). PIG7/LITAF gene mutation and overexpression of its gene product in extramammary Paget's disease. Int J Cancer 111, 218–223 [DOI] [PubMed] [Google Scholar]
- McCabe B, Liberante F, and Mills KI. (2015) Repurposing medicinal compounds for blood cancer treatment. Ann Hematol 94, 1267–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhillips F, Mullen P, MacLeod KG, et al. (2006). Raf-1 is the predominant Raf isoform that mediates growth factor-stimulated growth in ovarian cancer cells. Carcinogenesis 27, 729–739 [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, and Thomas PD. (2013). Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8, 1551–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milhas D, Cuvillier O, Therville N, et al. (2005). Caspase-10 triggers bid cleavage and caspase cascade activation in FasL-induced apoptosis. J Biol Chem 280, 19836–19842 [DOI] [PubMed] [Google Scholar]
- Miller JA, Pappan K, Thompson PA, et al. (2015). Plasma metabolomic profiles of breast cancer patients after short-term limonene intervention. Cancer Prev Res (Phila) 8, 86–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JJ, Chari RS, Boyer IJ, Gould MN, and Jirtle RL. (1995). Induction of apoptosis in liver tumors by the monoterpene perillyl alcohol. Cancer Res 55, 979–983 [PubMed] [Google Scholar]
- Mounayar R, Morzel M, Brignot H, et al. (2014) Nutri-metabolomics applied to taste perception phenotype: Human subjects with high and low sensitivity to taste of fat differ in salivary response to oleic acid. OMICS 18, 666–672 [DOI] [PubMed] [Google Scholar]
- Naderi A, Teschendorff AE, Beigel J, et al. (2007). BEX2 is overexpressed in a subset of primary breast cancers and mediates nerve growth factor/nuclear factor-kappaB inhibition of apoptosis in breast cancer cell lines. Cancer Res 67, 6725–6736 [DOI] [PubMed] [Google Scholar]
- Nagakawa H, Shimozato O, Yu L, et al. (2012). Expression of a murine homolog of apoptosis-inducing human IL-24/MDA-7 in murine tumors fails to induce apoptosis or produce anti-tumor effects. Cell Immunol 275: 90–97 [DOI] [PubMed] [Google Scholar]
- Napirei M, Ludwig S, Mezrhab J, Klockl T, and Mannherz HG. (2009). Murine serum nucleases—Contrasting effects of plasmin and heparin on the activities of DNase1 and DNase1-like 3 (DNase1l3). FEBS J 276, 1059–1073 [DOI] [PubMed] [Google Scholar]
- Napoli L, Bordoni A, Zeviani M, et al. (2001). A novel missense adenine nucleotide translocator-1 gene mutation in a Greek adPEO family. Neurology 57, 2295–2298 [DOI] [PubMed] [Google Scholar]
- Nielsen TO, Andrews HN, Cheang M, et al. (2004). Expression of the insulin-like growth factor I receptor and urokinase plasminogen activator in breast cancer is associated with poor survival: potential for intervention with 17-allylamino geldanamycin. Cancer Res 64, 286–291 [DOI] [PubMed] [Google Scholar]
- O'Brien NW, Gellings NM, Guo M, Barlow SB, Glembotski CC, and Sabbadini RA. (2003). Factor associated with neutral sphingomyelinase activation and its role in cardiac cell death. Circ Res 92, 589–591 [DOI] [PubMed] [Google Scholar]
- Oliver JL, Alexander MP, Norrod AG, Mullins IM, and Mullins DW. (2013). Differential expression and tumor necrosis factor-mediated regulation of TNFRSF11b/osteoprotegerin production by human melanomas. Pigment Cell Melanoma Res 26, 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P, Saleh A, Nakazawa A, et al. (2000). Negative regulation of cytochrome c-mediated oligomerization of Apaf-1 and activation of procaspase-9 by heat shock protein 90. EMBO J 19, 4310–4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan N, Marusawa H, Krajewska M, et al. , (2001). TUCAN, an antiapoptotic caspase-associated recruitment domain family protein overexpressed in cancer. J Biol Chem 276, 32220–32229 [DOI] [PubMed] [Google Scholar]
- Piñeiro D, Martín ME, Guerra N, Salinas M, and González VM. (2007). Calpain inhibition stimulates caspase-dependent apoptosis induced by taxol in NIH3T3 cells. Exp Cell Res 313, 369–379 [DOI] [PubMed] [Google Scholar]
- Puthalakath H, Villunger A, O'Reilly LA, et al. (2001). Bmf: A proapoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science 293, 1829–1832 [DOI] [PubMed] [Google Scholar]
- Reddy BS, Wang CX, Samaha H, et al. (1997). Chemoprevention of colon carcinogenesis by dietary perillyl alcohol. Cancer Res 57, 420–425 [PubMed] [Google Scholar]
- Rho SB, Byun HJ, Park SY, and Chun T. (2008). Calpain 6 supports tumorigenesis by inhibiting apoptosis and facilitating angiogenesis. Cancer Lett 271, 306–313 [DOI] [PubMed] [Google Scholar]
- Rudel T, and Bokoch GM. (1997). Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science 276, 1571–1574 [DOI] [PubMed] [Google Scholar]
- Rudel T, Zenke FT, Chuang TH, and Bokoch GM. (1998). p21-activated kinase (PAK) is required for Fas-induced JNK activation in Jurkat cells. J Immunol 160, 7–11 [PubMed] [Google Scholar]
- Sadkowski T, Jank M, Zwierzchowski L, Siadkowska E, Oprzadek J, and Motyl T. (2008). Gene expression profiling in skeletal muscle of Holstein-Friesian bulls with single-nucleotide polymorphism in the myostatin gene 5′-flanking region. J Appl Genet 49, 237–250 [DOI] [PubMed] [Google Scholar]
- Sahin MB, Perman SM, Jenkins G, and Clark SS. (1999). Perillyl alcohol selectively induces G0/G1 arrest and apoptosis in Bcr/Abl-transformed myeloid cell lines. Leukemia 13, 1581–1591 [DOI] [PubMed] [Google Scholar]
- Sahu J, Sen P, Choudhury MD, et al. (2014). Rediscovering medicinal plants' potential with OMICS: microsatellite survey in expressed sequence tags of eleven traditional plants with potent antidiabetic properties. OMICS 18, 298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitole L, Steffens F, Krüger TP, and Meyer D. (2014). Mid-ATR-FTIR spectroscopic profiling of HIV/AIDS sera for novel systems diagnostics in global health. OMICS 18, 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Fujita N, and Tsuruo T. (2000). Modulation of Akt kinase activity by binding to Hsp90. Proc Natl Acad Sci USA 97, 10832–10837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schievella AR, Chen JH, Graham JR, and Lin LL. (1997). MADD, a novel death domain protein that interacts with the type 1 tumor necrosis factor receptor and activates mitogen-activated protein kinase. J Biol Chem 272, 12069–12075 [DOI] [PubMed] [Google Scholar]
- Seko Y, Sugishita K, Sato O, et al. (2004). Expression of costimulatory molecules (4-1BBL and Fas) and major histocompatibility class I chain-related A (MICA) in aortic tissue with Takayasu's arteritis. J Vasc Res 41, 84–90 [DOI] [PubMed] [Google Scholar]
- Shu HB, and Johnson H. (2000). B cell maturation protein is a receptor for the tumor necrosis factor family member TALL-1. Proc Natl Acad Sci USA 97, 9156–9161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobral MV, Xavier AL, Lima TC, and de Sousa DP. (2014). Antitumor activity of monoterpenes found in essential oils. Sci World J 2014, 953451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soengas MS, Capodieci P, Polsky D, et al. (2001). Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature 409, 207–211 [DOI] [PubMed] [Google Scholar]
- Song Y, and Jacob CO. (2005). The mouse cell surface protein TOSO regulates Fas/Fas ligand-induced apoptosis through its binding to Fas-associated death domain. J Biol Chem 280, 9618–9626 [DOI] [PubMed] [Google Scholar]
- Stark MJ, Burke YD, McKinzie JH, Ayoubi AS, and Crowell PL. (1995). Chemotherapy of pancreatic cancer with the monoterpene perillyl alcohol. Cancer Lett 96, 15–21 [DOI] [PubMed] [Google Scholar]
- Stayrook KR, McKinzie JH, Burke YD, Burke YA, and Crowell PL. (1997). Induction of the apoptosis-promoting protein Bak by perillyl alcohol in pancreatic ductal adenocarcinoma relative to untransformed ductal epithelial cells. Carcinogenesis 18, 1655–1658 [DOI] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, and Trajanoski Z. (2002). Genesis: Cluster analysis of microarray data. Bioinformatics 18, 207–208 [DOI] [PubMed] [Google Scholar]
- Su XL, Hou YL, Yan XH, et al. (2012). Expression, purification, and evaluation for anticancer activity of ribosomal protein L31 gene (RPL31) from the giant panda (Ailuropoda melanoleuca). Mol Biol Rep 39, 8945–8954 [DOI] [PubMed] [Google Scholar]
- Tadmouri A, Kiyonaka S, Barbado M, et al. (2012). Cacnb4 directly couples electrical activity to gene expression, a process defective in juvenile epilepsy. EMBO J 31, 3730–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglialatela G, Robinson R, and Perez-Polo JR. (1997). Inhibition of nuclear factor kappa B (NFκB) activity induces nerve growth factor-resistant apoptosis in PC12 cells. J Neurosci Res 47, 155–162 [PubMed] [Google Scholar]
- Takahashi K, Niidome T, Akaike A, Kihara T, and Sugimoto H. (2009). Amyloid precursor protein promotes endoplasmic reticulum stress-induced cell death via C/EBP homologous protein-mediated pathway. J Neurochem 109, 1324–1337 [DOI] [PubMed] [Google Scholar]
- Valencia CA, Cotten SW, and Liu R. (2007). Cleavage of BNIP-2 and BNIP-XL by caspases. Biochem Biophys Res Commun 364, 495–501 [DOI] [PubMed] [Google Scholar]
- Wang J, Chun HJ, Wong W, Spencer DM, and Lenardo MJ. (2001). Caspase-10 is an initiator caspase in death receptor signaling. Proc Natl Acad Sci USA 98, 13884–13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Du F, and Wang X. (2008). TNF-α induces two distinct caspase-8 activation pathways. Cell 133, 693–703 [DOI] [PubMed] [Google Scholar]
- Wang Y, Radhakrishnan D, He X, Peehl DM, and Eng C. (2013). Transcription factor KLLN inhibits tumor growth by AR suppression, induces apoptosis by TP53/TP73 stimulation in prostate carcinomas, and correlates with cellular differentiation. J Clin Endocrinol Metab 98, E586–E594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Robertson JD, and Walcheck B. (2011). Different signaling pathways stimulate a disintegrin and metalloprotease-17 (ADAM17) in neutrophils during apoptosis and activation. J Biol Chem 286, 38980–38988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CF. (2008). The TNF superfamily-2008. Cytokine Growth Factor Rev 19, 183–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelock MJ, and Johnson KR. (2003). Cadherins as modulators of cellular phenotype. Annu Rev Cell Dev Biol 19, 207–235 [DOI] [PubMed] [Google Scholar]
- Wiseman DA, Werner SR, and Crowell PL. (2007). Cell cycle arrest by the isoprenoids perillyl alcohol, geraniol, and farnesol is mediated by p21Cip1 and p27Kip1 in human pancreatic adenocarcinoma cells. J Pharmacol Exp Ther 320, 1163–1170 [DOI] [PubMed] [Google Scholar]
- Wong BR, Besser D, Kim N, et al. (1999). TRANCE, a TNF family member, activates Akt/PKB through a signaling complex involving TRAF6 and c-Src. Mol Cell 4, 1041–1049 [DOI] [PubMed] [Google Scholar]
- Wong DCF, Wong KTK, Lee YY, Morin PN, Heng CK, and Yap MGS. (2006). Transcriptional profiling of apoptotic pathways in batch and fed-batch CHO cell cultures. Biotechnol Bioeng 94, 373–382 [DOI] [PubMed] [Google Scholar]
- Wu Y, Liu XM, Wang XJ, Zhang Y, Liang XQ, and Cao EH. (2009). PIG11 is involved in hepatocellular carcinogenesis and its over-expression promotes Hepg2 cell apoptosis. Pathol Oncol Res 15, 411–416 [DOI] [PubMed] [Google Scholar]
- Wylie AA, Pulford DJ, McVie-Wylie AJ, et al. (2003). Tissue-specific inactivation of murine M6P/IGF2R. Am J Pathol 162, 321–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Palhan V, Yang Y, and Roeder RG. (2000). TIP30 has an intrinsic kinase activity required for up-regulation of a subset of apoptotic genes. EMBO J 19, 956–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, and Neckers L. (2007). Targeting the molecular chaperone heat shock protein 90 provides a multifaceted effect on diverse cell signaling pathways of cancer cells. Clin Cancer Res 13, 1625–1629 [DOI] [PubMed] [Google Scholar]
- Yin Y, Huang WW, Lin C, Chen H, MacKenzie A, and Ma L. (2008). Estrogen suppresses uterine epithelial apoptosis by inducing birc1 expression. Mol Endocrinol 22, 113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, and Strasser A. (2008). The BCL-2 protein family: Opposing activities that mediate cell death. Nat Rev Mol Cell Biol 9, 47–59 [DOI] [PubMed] [Google Scholar]
- Yu SE, Park SH, and Jang YK. (2010). Epigenetic silencing of TNFSF7 (CD70) by DNA methylation during progression to breast cancer. Mol Cells 29, 217–221 [DOI] [PubMed] [Google Scholar]
- Yu Z, Wang W, Xu L, Dong J, and Jing Y. (2008). d-Limonene and d-carvone induce apoptosis in HL-60 cells through activation of caspase-8. Asian J Trad Med 3, 134–143 [Google Scholar]
- Zhang R, Luo D, Miao R, et al. (2005). Hsp90-Akt phosphorylates ASK1 and inhibits ASK1-mediated apoptosis. Oncogene 24, 3954–3963 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Yu B, Alexander D, Couper DJ, and Boerwinkle E. (2014) Medium-term variability of the human serum metabolome in the Atherosclerosis Risk in Communities (ARIC) study. OMICS 18, 364–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ding WX, Qian T, Watkins S, Lemasters JJ, and Yin XM. (2003). Bid activates multiple mitochondrial apoptotic mechanisms in primary hepatocytes after death receptor engagement. Gastroenterology 125, 854–867 [DOI] [PubMed] [Google Scholar]
- Zhao C, and Wang E. (2004). Heat shock protein 90 suppresses tumor necrosis factor alpha induced apoptosis by preventing the cleavage of Bid in NIH3T3 fibroblasts. Cell Signal 16, 313–321 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Li M, Wang X, et al. (2012). Caspase cleavage of cytochrome c1 disrupts mitochondrial function and enhances cytochrome c release. Cell Res 22, 127–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong WX, Lindsten T, Ross AJ, MacGregor GR, and Thompson CB. (2001). BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes Dev 15, 1481–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchelli S, Vilotti S, Calligaris R, et al. (2009). Aggresome-forming TTRAP mediates pro-apoptotic properties of Parkinson's disease-associated DJ-1 missense mutations. Cell Death Differ 16, 428–438 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.