Abstract
Persistence of latently infected cells in presence of Anti-Retroviral Therapy presents the main obstacle to HIV-1 eradication. Much effort is thus placed on identification of compounds capable of HIV-1 latency reversal in order to render infected cells susceptible to viral cytopathic effects and immune clearance. We identified the BAF chromatin remodeling complex as a key player required for maintenance of HIV-1 latency, highlighting its potential as a molecular target for inhibition in latency reversal. Here, we screened a recently identified panel of small molecule inhibitors of BAF (BAFi's) for potential to activate latent HIV-1. Latency reversal was strongly induced by BAFi's Caffeic Acid Phenethyl Ester and Pyrimethamine, two molecules previously characterized for clinical application. BAFi's reversed HIV-1 latency in cell line based latency models, in two ex vivo infected primary cell models of latency, as well as in HIV-1 infected patient's CD4 + T cells, without inducing T cell proliferation or activation. BAFi-induced HIV-1 latency reversal was synergistically enhanced upon PKC pathway activation and HDAC-inhibition. Therefore BAFi's constitute a promising family of molecules for inclusion in therapeutic combinatorial HIV-1 latency reversal.
Abbreviations: BAF250a, BAF Associated Factor 250 a; BAF, BRG-Brahma Associated Factors; BAFi, BAF inhibitor; bp, base pairs; BRG-1, Brahma Related Gene 1; CAPE, caffeic acid phenetyl esther; cART, combination Antiretroviral Therapy; ChIP, Chromatin Immunoprecipitation; CycA, Cyclophilin A; DHS-1, DNase Hypersensitive Site 1; ES cells, embryonic stem cells; FAIRE, Formaldehyde Assisted Isolation of Regulatory Elements; FBS, Fetal Bovine Serum; GFP, Green Fluorescent Protein; HDAC, histone deacetylase; HIV-1, human immunodeficiency virus type 1; IFNß, Interferon beta; IL10, Interleukin 10; IL12, Interleukin 12; IL4, Interleukin 4; IL6, Interleukin 6; INI-1, Integrase Interactor 1; IRES, Internal Ribosome Entry Site; IκB-α, Inhibitor of NF-κB – alpha; LRA, latency reversal agent; LTR, Long Terminal Repeat; MIP26, Major Intrinsic Protein; MMP9, Matrix Metallopeptidase 9; NF-κB, Nuclear Factor Kappa-light-chain-enhancer of activated B cells; nuc, nucleosome; PBMC, peripheral blood mononuclear cell; PBS, Phosphate Buffered Saline; PKC, Protein Kinase C; PYR, Pyrimethamine; RT-qPCR, Reverse Transcriptase, quantitative Polymerase Chain Reaction; SAHA, Suberoylanilide Hydroxamic Acid; SD, Standard Deviation; siRNA, small interfering RNA; SOCS3, Suppressor Of Cytokine Signaling 3; TGF-ß, Transforming Growth Factor beta; TLR2, Toll-like Receptor 2
Keywords: HIV, Latency, BAF complex, Chromatin remodeling, latency reversal agents
Highlights
-
•
BAF complex inhibitors (BAFi's) activate latent HIV-1 in cell line models of latency.
-
•
BAFi's in combination with HDAC inhibitors and PKC activators synergistically activate latent HIV-1.
-
•
The BAFi's PYR and CAPE reverse HIV-1 latency in primary cell models of latency and in cells obtained from HIV-1 patients.
Access to Combination antiretroviral therapy (cART) has made HIV-1 infection a chronic disease. However, cART is not curative, as a small number of infected cells harboring silent virus with potential to renew the infection persist despite cART. Efforts to cure HIV-1 include activation of these latent cells, making them susceptible to immune clearance. Here we describe the activity of BAF inhibitors in HIV-1 activation in in vitro models of HIV-1 latency as well as in cells obtained from HIV-1 infected patient volunteers. Our data highlight the clinical potential of BAF inhibitors for inclusion in combinatorial therapy to reverse HIV-1 latency.
1. Introduction
The use of fully suppressive combination Anti-Retroviral Therapy (c-ART), aside from efficiently inhibiting viral replication, has provided important insights into the characteristics and dynamics of the persistent latent pool of the HIV-1 virus in infected patients (Pierson et al., 2000, Dahabieh et al., 2015, Geeraert et al., 2008, Chun et al., 1995, Chun et al., 2007). Studies have led to the identification of quiescent latently infected CD4 + memory T cells, which harbor latent HIV-1, as the major barrier to a cure (Finzi et al., 1999, Siliciano et al., 2003). The majority of HIV-1 infected patients are diagnosed during the chronic phase of the infection, when a large reservoir of latently infected cells is already established (Strain et al., 2005, Watanabe et al., 2011, Whitney et al., 2014). Therefore, to achieve a functional cure, recent pharmacological efforts have centered on targeting the reservoir harboring latent HIV-1 for activation in order to induce viral replication. As a second step, following latency reversal, the infected cells would be exposed to viral cytopathic effects and the immune system, thereby eliminating or depleting the infected cells that constitute the latent reservoir (Deeks, 2012).
Latently HIV-1 infected cells harbor replication competent HIV-1 virus, whose gene expression is transcriptionally blocked. Once integrated, the double stranded HIV-1 provirus behaves as a cellular gene and becomes subject to a complex network of molecular mechanisms that determine its transcriptional state (Mbonye and Karn, 2014, Mahmoudi, 2012, Dahabieh et al., 2015). As part of the genome, the HIV-1 chromatin structure at the 5′ LTR, the HIV-1 promoter, is highly organized into specifically deposited nucleosomes: in its latent state, the 5'LTR is organized into nuc-0 and nuc-1, two strictly positioned nucleosomes that are separated by DHS1, a region sensitive to nuclease digestion, which encompasses a loosely positioned nucleosome (Verdin, 1991, Verdin and Van Lint, 1995, Rafati et al., 2011). The positioning of nuc-1, downstream of the core promoter transcription start site, is the hallmark of the repressed 5′ LTR. Upon activation, nuc-1 becomes rapidly and specifically disrupted, allowing for HIV-1 gene expression to occur (Verdin and Van Lint, 1995, Verdin et al., 1993, Van Lint et al., 1996, Rafati et al., 2011).
The chromatin structure of the HIV-1 promoter, as with cellular promoters, is generated through the concerted activity of protein complexes, which remodel chromatin (Mbonye and Karn, 2014, Lusic and Giacca, 2015). This is in part accomplished by loosening or tightening histone-DNA interaction via posttranslational modifications on histone tails, which include acetylation and methylation (Zentner and Henikoff, 2013). Of particular interest for regulation of HIV-1 transcription, histone deacetylases have been shown to play a critical role in repressing gene expression at the HIV-1 LTR (Van Lint et al., 1996, Keedy et al., 2009, Marban et al., 2007, Williams et al., 2006, Barton and Margolis, 2013, Barton et al., 2014, Lu et al., 2014) and represent an important target for therapies aimed at activation of latent virus or latency reversal. Accordingly, a number of small molecule HDAC inhibitors including valproic acid (Edelstein et al., 2009), suberoylanilide hydroxamic acid (SAHA) (Contreras et al., 2009), Panobinostat (Rasmussen et al., 2013), and Romidepsin (Wei et al., 2014) have been or are currently under clinical investigation for their potential in therapeutic HIV-1 latency reversal (Archin et al., 2012, Archin et al., 2014, Elliott et al., 2014, Barton et al., 2015, Rasmussen et al., 2014).
Initial data from preclinical and clinical studies using HDAC inhibitors in therapeutic latency reversal suggest that while promising, HDAC inhibitors alone may not be sufficient in activation of the latent HIV-1 to the extent required to accomplish clinically significant depletion of the latent reservoir in patients. HIV-1 transcriptional latency is a complex molecular phenomenon, driven and controlled by multiple mechanisms, including the position of virus integration in the genome, its orientation and proximity to endogenous genes, and the presence and activity of host transcription factors and cofactors on the HIV-1 promoter (Van Lint et al., 2013, Dahabieh et al., 2015, Mbonye and Karn, 2014, Ruelas and Greene, 2013, Matsuda et al., 2015). Therefore, the specific transcriptional environment of each latently infected cell is likely to impact its responsiveness to activating molecules. Accordingly, the molecular mechanisms at play in transcriptionally silencing the latent reservoir are likely to be diverse and heterogeneous. Indeed, data obtained from in vitro models using latently infected cell lines support the notion that targeting two or more pathways in combination is more effective in activating latent HIV-1 by inducing synergistic activation of the latent promoter (De Crignis and Mahmoudi, 2014, Laird et al., 2015). It is therefore critical to identify and target multiple molecular effectors with distinct mechanisms that drive HIV-1 latency in order to obtain more effective and synergistic activation of the latent reservoir.
ATP dependent chromatin remodeling complexes are another group of factors, which the cell has co-evolved in addition to post-translational histone modifications, in order to alter histone-DNA contacts and change chromatin structure. These multi-subunit protein complexes use energy from ATP hydrolysis to disrupt or move nucleosomes, thereby remodeling chromatin and modulating access to DNA (Narlikar et al., 2013, Petty and Pillus, 2013, Hargreaves and Crabtree, 2011, Mahmoudi and Verrijzer, 2001). We have recently found that the BAF chromatin remodeling complex uses energy from ATP hydrolysis to position the transcriptionally repressive nuc-1 over thermodynamically sub-optimal DNA sequences leading to the establishment and maintenance of HIV-1 latency. Consistent with the pivotal role of BAF complex in HIV-1 latency, we have shown that siRNA mediated depletion of BAF specific subunits significantly decreases the percentage of latent events at the time of infection. Similarly, in cells in which latency is already established, such as in vitro T cell line models of latency, depletion of BAF subunits results in reactivation of latent virus. Our data identified the BAF chromatin remodeling complex as a putative molecular target in therapeutic efforts aimed at HIV-1 latency reversal (Rafati et al., 2011, Mahmoudi, 2012). Recently, a spectrum of potential small molecule inhibitors of the BAF complex were identified in a screen of more than 30,000 compounds, which included synthetic molecules, natural products, drug-like compounds and molecules with known bioactivity (Dykhuizen et al., 2012). 34 BAF inhibitors were initially identified by monitoring mRNA expression of well characterized BAF target genes in mouse embryonic stem cells. A second screen then eliminated molecules with non-specific targets and general repressive activities, leading to the identification of 20 compounds that transcriptionally mimic genetic deletion of BRG1 (the ATPase subunit of the complex), indicating specific activity against the BAF complex.
Here we have tested a panel of BAF inhibitors for their potential to activate latent HIV-1. Following the initial screening, we focused on functional characterization of A01, A11, and C09, the three compounds that displayed most significant activity on the latent LTR with the lowest toxicity. We found that BAF inhibitors (BAFi's) activate latent HIV-1 in both Jurkat cell lines harboring latent full length HIV-1 and HIV-1 derived viruses, in two distinct ex vivo infected primary CD4 + T cell models of HIV-1 latency, as well as in cells obtained from virologically suppressed HIV-1 infected patients. BAFi-mediated activation of latent HIV-1 was accompanied by the displacement of the BAF complex from the HIV-1 LTR, as demonstrated by ChIP assay, and was synergistically enhanced in presence of the HDAC inhibitor SAHA and the PKC agonist Prostratin. Consistently, FAIRE assays demonstrated removal of the repressive positioned nuc-1 in response to treatment with BAFi's, and synergism at the molecular level when cells were co-treated with BAFi's together with Prostratin. While efficiently activating latent HIV-1, treatment with BAFi's did not induce T cell proliferation or general T cell activation of primary CD4 + T cells. Our data identifies BAFi's as a promising family of small molecules for inclusion in therapeutic combinations aiming to reverse HIV-1 latency.
2. Materials and Methods
2.1. Cell Culture and Reagents
Jurkat, J-Lat A2 (LTR-Tat-IRES-GFP), J-Lat 11.1 (integrated full-length HIV-1 genome mutated in env gene and GFP replacing Nef) (Jordan et al., 2001, Jordan et al., 2003) cells were cultured in RPMI-1640 medium (Sigma Aldrich, Zwijndrecht, The Netherlands) supplemented with 10% FBS and 100 μg/ml penicillin-streptomycin at 37 °C in a humidified 95% air-5% CO2 atmosphere. Cells were treated with the following compounds: Phorbol 12-myristate 13-acetate (PMA, Sigma Aldrich); Ionomycin (Sigma Aldrich); SAHA-Vorinostat (Selleck Chemicals); Prostratin (Sigma Aldrich); A01 (N-hydroxy-N′-phenyl-octanediamide 2-hydroxy-N′-[(E)-(5-hydroxy-6-oxocyclohexa-2,4-dien-1-ylidene) methyl]benzohydrazide (BRD-K70161581-001-01-5), Vitas-M Laboratory, Ltd., Apeldoorn, Netherlands.); A11 (5-(4-chlorophenyl)-6-ethyl- 2,4-pyrimidinediamine (BRD-K88429204-001-18-7), Sigma Aldrich); C09 (2-phenylethyl (E)-3-(3,4-dihydroxyphenyl)prop-2-enoate (BRD-K91370081-001-04-6), MP Biomedicals, Eindhoven, The Netherlands).
2.2. Flow Cytometry
GFP expression in the J-Lat cell lines was analyzed by Flow Cytometry. The live population was defined by forward versus side scatter profiles. Cells were further gated by using forward scatter versus GFP intensity to differentiate between GFP-positive and -negative cells.
In primary models of latency, the phenotypic characteristics of the cell population at the moment of reactivation were determined using flow cytometry: cells were stained with anti-CD4-PB (Becton Dickinson, Breda, The Netherlands), anti-CCR7-PE (eBioscience, Uithoorn, The Netherlands), anti-CD27-APC (Becton Dickinson), and anti CD45RO-FITC (Dako, Heverlee, Belgium). Cells were stained for 30 min at 4 °C, washed with PBS, and re-suspended for flow cytometric analysis.
To determine cell activation following BAF complex inhibition, cells were treated with BAFi's for 72 h. Untreated cells and PMA/Ionomycin treated cells were used as negative and positive control, respectively. At 24 and 72 h post treatment cells were collected and stained with Fixable Viability Dye eFluor® 450 and anti-CD25-PE (Becton Dickinson). Cells were stained for 30 min at 4 °C, washed with PBS, and resuspended for flow cytometric analysis. To determine the expression of KI67, cells were collected and stained with Fixable Viability Dye eFluor® 450 for 15 min. Following 2% PFA fixation, cells were permeabilized with 0.3% saponin for 30 min, incubated with anti-Ki67-PE (eBioscience) antibody for 60 min at 4 °C, washed in PBS and resuspended for flow cytometry. All samples were analyzed with a Becton Dickinson Fortessa instrument.
2.3. Primary CD4 + T Cell Isolation and Infection
Primary CD4 + T cells were isolated from buffy coats from healthy donors by Ficoll gradient followed by magnetic separation with EasyStep Human CD4 + T cell Enrichment kit (StemCells Technologies, Grenoble, France).
Two in vitro models of HIV-1 latency were set up following the method developed by Bosque and Planelles (Bosque and Planelles, 2009) and Lassen and Greene (Lassen et al., 2012). Infections were performed using a pseudotyped virus expressing luciferase. Viral pseudotyped particles were obtained by co-transfecting HXB2 Env together with the HIV-1 backbone plasmid (pNL4.3.Luc.R-E-) into HEK 293 T cells using PEI (Polyethylenimine) transfection reagent. Twenty four, 48 and 72 h post-transfection, the pseudovirus-containing supernatant was collected, filtered through a 0.45 μm filter, aliquoted, and stored at − 80 °C.
For generation of latently infected primary cells according to the Bosque and Planelles method, purified CD4 + T cells were cultured in RPMI 1640 containing 10% FBS in presence of 10 ng/ml) of TGF-β (Sigma-Aldrich), 1 μg/ml α-IL4 (PeproTech, London, UK), 2 μg/ml α-IL12 (PeproTech), and α-CD3-CD28 (Life Technologies, Breda, The Netherlands) coated beads in 1:1 ratio for 3 days. After 3 days α-CD3-CD28 coated beads were removed and cells were cultured in RPMI 1640 containing 10% FBS, 100 μg/ml penicillin-streptomycin and 5 ng/ml of IL2 (Sigma-Aldrich) for 4 days. On day 7 post stimulation cells were infected with HIV-derived virus particles. Primary CD4 + T cells, were infected with the PNL4.3.LUC.R-E- virus by spinoculation (90 min at 1200 g), washed in PBS and cultured in RPMI 1640 containing 10% FBS, 100 μg/ml penicillin-streptomycin, IL2 (5 ng/ml) and Saquinavir Mesylate (5 μM). Six days after infection cells were treated with BAFi's in presence of Raltegravir (30 μM).
Generation of latently infected cells according to Lassen and Greene method was performed as follows: CD4 + T cells were infected with the PNL4.3.LUC.R-E- virus by spinoculation (120 min at 1200 g) right after isolation and cultured for three days in RPMI 10% FBS and 100 μg/ml penicillin-streptomycin in presence of Saquinavir Mesylate (5 μM). Three days after infection cells were treated with BAFi's in presence of Raltegravir (30 μM).
Cells were harvested 24 h after stimulation with BAF inhibitors, washed once in PBS and luciferase activity was measured using Luciferase Assay System (Promega, Leiden, The Netherlands). Relative light units were normalized to protein content determined by Bradford assay (Bio Rad, Veenendaal, The Netherlands).
HIV-1 molecular clone pNL4.3.Luc.R-E-, HIV-1 HXB2-Env expression vector, Saquinavir Mesylate and Raltegravir were kindly provided by the Centre for AIDS Reagents, NIBSC. HIV-1 molecular clone pNL4.3.Luc.R-E- and HIV-1 HXB2-Env expression vector were donated by Dr. Nathaniel Landau and Drs Kathleen Page and Dan Littman, respectively.
2.4. Activation of HIV-1 Transcription in Patient CD4 + T Cells
Total PBMCs were obtained from HIV-infected patients without (overt) clinical symptoms by leukapheresis, and isolated by Ficol gradient density sedimentation. CD4 + T cells were isolated using the CD4 + T-Cell enrichment kit (Stem Cell Technologies) and frozen. Inclusion criteria were as follows: age > 18 years; confirmed HIV-1 infection; plasma HIV-1 RNA viral load below 50 copies/ml for more than 12 months; cART for at least 3 years. 2.5 million CD4 + T cells were plated in 6 wells plates in 2.5 ml of RPMI supplemented with 10% FBS and left untreated or treated with 1 μM C09, 20 μM A11 or 300 nM Prostratin alone or in combination as indicated. As a positive control, cells were treated with CD3-CD28 beads or PMA-Ionomycin. Twelve hours after stimulation cells were collected and lysed in RNA lysis buffer (Promega). RNA was extracted and reverse transcribed using Random primers (Life Technologies) and Superscript II (Life Technologies) following manufacturer's instructions. Levels of cellular associated RNA were quantified using a PCR targeting the HIV-1 POL gene. qPCR was performed in a final volume of 25 μl using 4 μl of cDNA, 2.5 μl of 10 × PCR buffer (Life Technologies), 1.75 μl of 50 mM MgCl2 (Life Technologies), 1 μl of 10 mM dNTPs (Life Technologies), 0.125 μl of 100 μM Pol For (HXB2 genome 4901 → 4924), 0.125 of 100 μM Pol Rev. (HXB2 genome 5060 → 5040), 0.075 μl of 50 μM of Pol Probe, and 0.2 μl Platinum Taq (Life Technologies). The lower limit of detection of this method was of 20 copies of HIV-1 RNA in 1 μg of total RNA. The absolute number of POL copies in PCR was calculated using a standard curves ranging from 4 to 4 × 105 copies of a plasmid containing the full-length HIV-1 genome. The amount of HIV-1 cellular associated RNA was expressed as number of copies/μg of input RNA in reverse transcription. Preparations of cell-associated RNA were tested for potential contamination with HIV-1 DNA and-or host DNA by performing the PCR amplification in the presence and absence of reverse transcriptase.
This study was conducted in accordance with the ethical principles of the Declaration of Helsinki. The patients involved in the study provided signed informed consent and the study protocol was approved by The Netherlands Medical Ethics Committee (MEC-2012-583).
2.5. Total RNA Isolation and Quantitative RT-PCR (RT-qPCR)
Total RNA was isolated from the cells using RealiaPrep RNA Cell Miniprep System (Promega), cDNA synthesis was performed using Superscript II Reverse Transcriptase (Life Technologies) kit following manufactures protocol. RT-qPCR was performed using GoTaq qPCR Master Mix (Promega) following manufacturer protocol. Amplification was performed on the CFX Connect Real-Time PCR Detection System thermocycler (BioRad) using following thermal program starting with 3 min at 95 °C, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. Specificity of the RT-qPCR products was assessed by melting curve analysis. Primers used for real-time PCR are listed in Table 1. Expression data was calculated using 2-ΔΔCt method by Livak Schmittgen (Schmittgen and Livak, 2008). Cyclophyilin A (CycA) and ß-2-microglobulin were used as housekeeping genes for J-Lat cell lines and primary cells, respectively.
Table 1.
List of RT-qPCR primers.
| Primer | Sequence (5′-3′) |
|---|---|
| β – Actin For | CGCAAAGACCTGTACGCCAAC |
| β – Actin Rev | GAGCCGCCGATCCACACG |
| β2Microglobulin For | AGCGTACTCCAAAGATTCAGGTT |
| β2Microglobulin Rev | ATGATGCTGCTTACATGTCTCGAT |
| BAF250a For | CTTCAACCTCAGTCAGCTCCCA (Wang et al., 2012) |
| BAF250a Rev | GGTCACCCACCTCATACTCCTTT (Wang et al., 2012) |
| BRG-1 For | GCAGGCTCGCATCGCACAC |
| BRG-1 Rev | GCTCAATGGTCGCTTTGGTTCG |
| CycA (PPIA) For | TCATCTGCACTGCCAAGACTG |
| CycA (PPIA) Rev | CATGCCTTCTTTCACTTTGCC |
| Gag For | AGTAGTGTGTGCCCGTCTGT |
| Gag Rev | TCGCTTTCAGGTCCCTGTTCG |
| Gfp For | GAAGCAGCACGACTTCTTCAA |
| Gfp Rev | GCTTGTCGGCCATGATATAGA |
| IFNβ For | GGAGGACGCCGCATTGAC |
| IFNβ Rev | TGATAGACATTAGCCAGGAGGTTC |
| IL6 For | CCCTGACCCAACCACAAATGC |
| IL6 Rev | CAACAACAATCTGAGGTGCCCAT |
| IL10 For | ACATCAAGGCGCATGTGAAC |
| IL10 Rev | GCCACCCTGATGTCTCAGTT |
| Luc For | TCTAAGGAAGTCGGGGAAGC (Barakat et al., 2011) |
| Luc Rev | CCCTCGGGTGTAATCAGAAT (Barakat et al., 2011) |
| MIP26 For | AGTGTGTGGCTCTTGATTTCTGAGG |
| MIP26 Rev | CCACCTCCCTTGAGTCCCTTCTC |
| MMP9 For | TGGTCCTGGTGCTCCTGGTG |
| MMP9 Rev | GCTGCCTGTCGGTGAGATTGG |
| SOCS3 For | CCAAGGACGGAGACTTCGAT |
| SOCS3 Rev | GGTACTCGCTCTTGGAGCTG |
| TLR2 For | TGGATGGTGTGGGTCTTGG |
| TLR2 Rev | AGGTCACTGTTGCTAATGTAGG |
| Nuc-0 For | ATCTACCACACACAAGGCTAC |
| Nuc-0 Rev | GTACTAACTTGAAGCACCATCC |
| DHS-1 For | AAGTTTGACAGCCTCCTAGC |
| DHS-1 Rev | CACACCTCCCTGGAAAGTC |
| Nuc-1 For | TCTCTGGCTAACTAGGGAACC |
| Nuc-1 Rev | AAAGGGTCTGAGGGATCTCTAG |
| ChIP CTRL Reg For | GCCAGAGTCAAGCCAGTAGTC |
| ChIP CTRL Reg Rev | TAGCCTAATGTGGAGTGGATGTG |
| Alu For | GCCTCCCAAAGTGCTGGGATTACAG |
| AluGag For | GGTGCGAGAGCGTCAGTAT |
| AluGag Rev | AGCTCCCTGCTTGCCCATA |
| AluGag probe | [6FAM]AAAATTCGGTTAAGGCCAGGGGGAAAGAA[BHQ1] |
| ALB For | TGCATGAGAAAACGCCAGTAA |
| ALB Rev | ATGGTCGCCTGTTCACCAA |
| ALB probe | [FAM]TGACAGAGTCACCAAATGATGCACAGAA[BHQ1] |
| Pol For | GGTTTATTACAGGGACAGCAGAGA |
| Pol Rev | ACCTGCCATCTGTTTTCCATA |
| Pol Probe | [6FAM]ACTACTGCCCCTTCACCTTTCCAGAG[BHQ1] |
2.6. Knock-down Experiments
Pre-designed siRNA pools targeting transcripts of the human ARID1a-BAF250a (L-017,263-00-0005) and non-target control siRNA pool (D-001,810-10-20) were purchased from Dharmacon (Dharmacon, Etten-Leur, The Netherlands). Small-interfering RNAs were delivered by nucleofection using Amaxa Nucleofector (Amaxa AG-Lonza, Cologne, Germany) as previously described (Rafati et al., 2011). Briefly, cells were split to 3 × 105 cells/ml one day before nucleofection. Five million cells were centrifuged at 200 g for 10 min at room temperature, re-suspended in 100 μl of solution R, and nucleofected with 2 μM siRNA using program O28. Nucleofected cells were re-suspended in 500 μl of pre-warmed, serum-free antibiotic-free RPMI at 37 °C for 15 min and then plated in 4 ml of pre-warmed complete media. Seventy-two hours post-nucleofection cells were treated with SAHA [350 nM] or Prostratin [100 nM]. LTR-driven GFP expression was assessed after 24 and 48 h after treatment by FACS. RNA and protein for RT-qPCR and Western blot analysis were isolated 96 h after nucleofection.
2.7. Western blot Analysis
Cells were treated with BAF inhibitors for 36 h and then lysed with IP buffer (25 mM HEPES, pH 7.9, 150 mM KCl, 1 mM EDTA, 5 mM MgCl2, 5% glycerol, 1% NP40, 0.5 mM dithiothreitol and a protease inhibitor cocktail (Sigma Aldrich)) for 30 min on ice. Whole-cell protein lysate was used for SDS-PAGE in order to detect BAF250a, BRG-1, INI-1, IκB, and GAPDH. The following antibodies were used in Western blot analysis: anti-SMARCA4-BRG1 (sc-17,796, Santa Cruz Biotechnology; ab4081-100, Abcam), anti-ARID1a-BAF250a (kind gift from C.P. Verrijzer), anti-SMARCAB1-hSNF5 (ab4552, Abcam), anti- IκB (sc-371, Santa Cruz Biotechnology) and anti-GAPDH (ab8245, Abcam). Signal intensity for representative blots was quantified using ImageJ software according to NIH guidelines. Briefly, specific bands, related to protein of interest were marked in order to generate plots of relative density. Area under the plots, representing each band intensity, was normalized to its loading control and then to its untreated sample for BAF250a and INI-1, each band intensity for BRG-1 was normalized to its corresponding loading control and to untreated control, band intensity of treated samples for GAPDH was normalized with untreated controls.
2.8. Latency Establishment Experiment
HIV-derived virus particles were generated as described (Jordan et al., 2001). Briefly, HEK 293 T cells were transfected with VSVG, the R8.9 packaging vector, and the retroviral vector LTR-Tat-IRES-EGFP (pEV731). Virus was harvested every 12 h starting at 24 h after transfection. Jurkat cells were treated with 1 μM BAFi's as indicated 1 h prior to infection. Cells were then infected in presence or absence of BAFi's with the LTR-Tat-IRES-EGFP virus at low multiplicity of infection (MOI), such that approximately 10% of cells were infected. Ninety six hours after infection, the GFP negative cell population was sorted using a FACSAria II cell sorter (Becton Dickinson) and treated with PMA. Percentage of GFP positive (latent) infections was determined by FACS analysis 48 h after PMA stimulation.
For Alu-PCR, 2–5 ∙ 106 cells were subjected to genomic DNA isolation using DNeasy Blood & Tissue Kit (Qiagen, Hilden, The Netherlands). The following reaction mix was used for the first round PCR: 1X PCR buffer (Life Technologies), 1.5 mM MgCl2 (Life Technologies), 0.5 mM dNTPs (Thermo Scientific, Waltham, MA USA) 1 μM Alu forward primer, 6 μM Gag reverse primer, 2.5 U of Platinium Taq polymerase (Life Technologies) and 150 ng of genomic DNA. Amplification was performed in a T100 Thermal Cycler (BioRad) with following thermal program: 2 min at 95 °C, followed by 14 cycles of 95 °C for 30 s, 50 °C for 30 s and 72 °C for 210 s. A nested PCR targeting the Gag gene was performed using the following reaction mix: 1 X PCR buffer, 1.5 mM MgCl2, 0.5 mM dNTPs, 0.5 μM AluGag For and AluGag Rev. primers, 0.15 μM AluGag probe (FAM-AAAATTCGGTTAAGGCCAGGGGGAAAGAA-BHQ1), 1 U Platinium Taq polymerase and 3 ng of genomic DNA or 2 μl of Alu-Gag PCR product. Amplification was performed on the CFX Connect Real-Time PCR Detection System thermocycler (BioRad) using following thermal program starting with 5 min 95 °C, followed by 30 cycles 95 °C for 30 s, 60 °C for 30 s. Albumin gene was used for normalization. Primer sequences used are listed in Table. 1.
2.9. Chromatin Immunoprecipitation (ChIP)
Eighteen hours prior to analysis J-Lat 11.1 cells were untreated or treated with BAFi's as indicated. Twenty million cells were used per sample. Cells were washed with PBS ++ (PBS, 1 mM CaCl2, 1 mM MgCl2) and cross-linked for 30 min at room temperature by adding buffer A (1% HCHO, 10 mM NaCl, 100 μM EDTA, 50 μM EGTA, 2 mM HEPES pH 7.6). The reaction was quenched with 125 mM glycine. Cross-linked cells were washed in ice-cold PBS ++ followed by washes with buffer B (0.25% Triton-X 100, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES, pH 7.6) and buffer C (150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES, pH 7.6) for 10 min at 4 °C. For sonication, cells were re-suspended in ChIP incubation buffer (1% SDS, 1% Triton-X 100, 0.15 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES, pH 7.6, Protease inhibitors cocktail) and chromatin was sheared by sonication to an apparent length of ~ 500 bp using a Bio Ruptor sonicator (Cosmo Bio Co., Ltd., Tokyo, Japan) with 20 times 60-s pulses at maximum setting at 4 °C. At least 150 ul of each sample were collected for input. Chromatin from approximately 20 million cells was diluted 7 times with ChIP incubation buffer lacking SDS, in order to decrease SDS concentration in sonicated samples. Chromatin was immunoprecipitated with 2–5 μg of antibody specific to BAF250a (kind gift of gift from C.P. Verrijzer or sc-32761, SantaCruz Biotechnology) and BSA-blocked protein-A agarose beads (GE Healthcare) at 4 °C overnight. Beads were washed twice with the following buffers buffer 1 (0.1% SDS, 0.1% deoxycholate, 1% Triton-X 100, 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES pH 7.6), buffer 2 (0.1% SDS, 0.1% deoxycholate, 1% Triton-X 100, 500 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES pH 7.6), buffer 3 (250 mM LiCl, 0.5% deoxycho- late, 0.5% NP-40, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES, pH 7.6), and buffer 4 (1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES, pH 7.6). Immunoprecipitated complexes and inputs were eluted in elution buffer (1% SDS, 0.1 M NaHCO3) for 30 min at room temperature, and decrosslinked overnight at 65 °C in presence of 200 mM NaCl2. DNA was extracted with phenol:chloroform:isoamylalcohol 24:24:1 (Sigma) followed by chroloform:isoamylalcohol 24:1 extraction, ethanol precipitated, and resuspended in 100 μl H2O by shaking at 37 °C. Input and immunoprecipitated DNA were subjected to Sybergreen qPCR cycles with specific primers (sequences provided in Table. 1) with an CFX Connect Real-Time PCR Detection System thermocycler (BioRad) and GoTaq qPCR Mastermix (Promega).
2.10. Formaldehyde-Assisted Isolation of Regulatory Elements (FAIRE)
Eighteen hours prior to analysis, J-Lat A2 and 11.1 cells were treated with BAFi's where indicated. J-Lat cells were fixed for 30 min by adding formaldehyde to a final concentration of 1% at room temperature. Twenty million cells were used per FAIRE experiment. The reaction was quenched with 125 mM glycine. Cross-linked cells were washed with PBS followed by washes with buffer B (0.25% Triton-X 100, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES, pH 7.6) and buffer C (150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES, pH 7.6). For sonication, cells were re-suspended in ChIP incubation buffer (1% SDS, 1% Triton-X 100, 0.15 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES, pH 7.6) and chromatin was sheared by sonication to an apparent length of ~ 200–400 bp (corresponding to ~ 100–200 bp of free DNA) using a BioRuptor sonicator (Cosmo Bio Co., Ltd) with 20 times 30-s pulses at maximum setting at 4 °C. Sonicated chromatin was twice phenol:chloroform:isoamyl alcohol (24:24:1) extracted, washed with chloroform:isoamylalcohol (24:1) and ethanol precipitated. Isolated DNA was subjected to Sybergreen qPCR cycles with specific primers (sequences provided in Table. 1) with a CFX Connect Real-Time PCR Detection System (BioRad) and GoTaq qPCR Mastermix (Promega), using following thermal program starting with 3 min at 95 °C, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s.
2.11. Statistical Analysis
Data are shown as the average + S.D. of three or more independent experiments. Statistical significance for multiple comparisons was calculated using Student's t test or, when appropriate, Mann–Whitney non parametric test.
According to the Bliss independence method (Bliss, 1939), threshold for synergism was calculated using the following equation μ(1 + 2)exp. = [1 − (1 – μ1) × (1 − μ2)], where μ(1 + 2)exp. is the expected percentage of cells reactivated after combinatorial treatment in absence of synergism and μ1 and μ2 correspond to the percentage of cells reactivated by the single treatments. The difference between the observed percentage of cells activated by combination treatment and the expected percentage (μ(1 + 2)exp) describes the interaction between two treatments. Combination was considered synergistic if the difference and its 95% normal confidence interval (CI) were > 0. Synergistic effect of BAFi's over SAHA + Prostratin combination was calculated considering Prostratin + SAHA as a single treatment.
3. Results
3.1. Small Molecule Inhibitors of BAF Activate Latent HIV
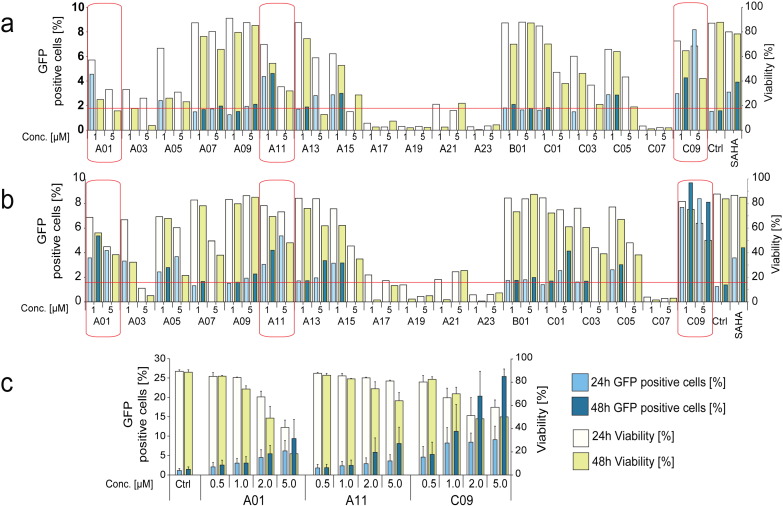
We tested a panel of compounds (Figure S1 and Table S1), identified in a screen for inhibitors of the BAF chromatin remodeling complex (Dykhuizen et al., 2012) for their potential to activate latent HIV-1 in J-Lat A2 (Fig. 1a) and J-Lat 11.1 (Fig. 1b) cells. These Jurkat clonal cell lines contain, respectively, an integrated latent LTR-Tat-IRES-GFP virus or a defective full-length HIV-1 genome expressing GFP in place of Nef (Jordan et al., 2003). Six compounds significantly activated the latent HIV-1 in both cell lines, however 3 of these displayed significant toxicity at the concentrations tested in the screening. Therefore we focused on A01, A11, and C09, the three BAF inhibitors displaying the strongest capacity to activate latent HIV-1 while not significantly affecting cell viability. Titration experiments indicated optimal concentrations for these compounds in HIV-1 activation to be in the 0.5–2 μM range in J-Lat cell lines (Fig. 1c).
Fig. 1.
Small molecule Inhibitors of the BAF complex (BAFi's) re-activate latent HIV.
J-Lat A2 (Panel a) and 11.1 (Panel b) cells were treated with BAFi's at 1 μM and 5 μM concentrations and re-activation quantitated at 24 and 48 h post treatment. Percent of GFP positive cells (left axes, dark colored bars), corresponding to the level of HIV-1 activation, and cell viability (right axes light colored bars) were evaluated by FACS analysis. GFP levels of samples in which cell viability was below 50% are not shown. HIV-1 activation was considered significant when GFP levels exceeded the average + 2SD of untreated controls (red horizontal line). Red squares identify BAFi's which induced activation of HIV-1 without significantly affecting cell viability. These compounds were further investigated for their ability to activate HIV-1 transcription in 11.1 cells (Panel c). Data are presented as mean ± SD.
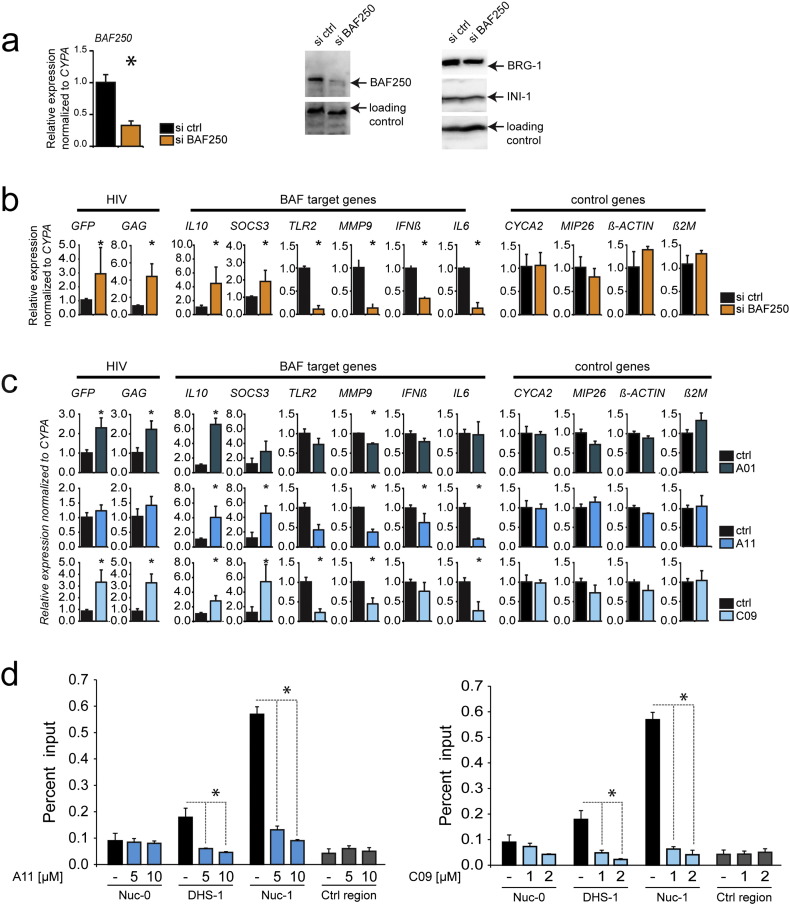
As the BAF inhibitors were identified in a screen in mouse ES cells, we set out to confirm their specificity in inhibition of the BAF complex in human CD4 + T cells. Thus, we analyzed the gene expression profile of human CD4 + T cells after siRNA depletion of the BAF-specific subunit BAF250a and compared it with the gene expression profile obtained after treatment with BAFi's. J-Lat 11.1 cells were transfected with a siRNA targeting the BAF250a subunit and the expression levels of the LTR driven transcripts as well as those of endogenous BAF target genes were analyzed by RT-qPCR 48 h after transfection. Depletion of BAF250a was confirmed by RT-qPCR and Western blot analysis (Fig. 2a). As observed previously (Rafati et al., 2011), depletion of BAF250a resulted in activation of latent HIV-1 as determined by an increase in expression of the HIV-1 LTR-driven P24 and GFP reporter (Fig. 2b). Moreover, qRT-PCR analysis identified a set of endogenous previously described BAF target genes which were positively or negatively modulated after BAF250a knock down. In particular, BAF250a depletion induced the expression of IL10 (Wurster et al., 2012), and SOCS3 (Dykhuizen et al., 2012), and decreased the expression of TLR2, MMP9, IFNß, and IL6 (Barker et al., 2001, Ni and Bremner, 2007) (Fig. 2b). To determine the level of activity and specificity of the BAFi's, we examined the gene expression profile of J-Lat11.1 cells 18 h after treatment with A01, A11, and C09 (1 μM). Mirroring BAF250a depletion, treatment with A11 and C09 upregulated IL-10 and SOCS3 while downregulating the expression of endogenous BAF target genes TLR2, MMP9, INFβ, IL-6, whereas treatment with A01 resulted in significant down-regulation of MMP9 only, together with a significant upregulation of both IL-10 and SOCS3 (Fig. 2c). All BAFi's activated latent HIV-1 as demonstrated by the increase in P24 and GFP expression (Fig. 2c). Thus, inhibition of BAF with BAFi's functionally mimicked the effect of siRNA depletion of BAF250a in modulating the expression of endogenous BAF target genes and resulted in activation of the latent HIV-1 as demonstrated by an increase in GFP and P24 expression.
Fig. 2.
Treatment with BAFi's specifically modulates BAF target genes, and displaces the BAF complex from the latent HIV-1 LTR.
(Panel a) Knockdown of BAF complex was obtained by nucleofecting J-Lat 11.1 cells with siRNA targeting the BAF complex specific subunit BAF250a. RNA levels of BAF250a were quantitated by RT PCR analysis, whereas protein levels of BAF250a as well as of the other subunits of the complex were determined by Western blot. RT-PCR analysis of J-Lat 11.1 cells after BAF250a knockdown (Panel b) or treatment with BAFi's A01 (1 μM), A11 (1 μM) and C09 (1 μM) for 18 h (Panel c). The effect of BAFi's treatment was monitored analyzing the changes in gene expression of HIV-1 LTR driven genes (GFP, GAG) and BAF target genes (TLR2, MMP9, IFNß, IL6, IL10, SOCS3). A set of BAF independent genes (CYCA2, MIP26, ß-ACTIN, ß-2-MICROGLOBULIN) was used as control. RT-qPCR data are presented as mean ± SD. (Panel d) BAFi treatment results in displacement of the BAF complex-specific subunit BAF250a from DHS1 and nuc-1 regions of the HIV-1 LTR. J-Lat 11.1 cells untreated or treated with BAFi's at indicated concentrations were subjected to ChIP using antibodies specific for the BAF250a subunit. Input and immunoprecipitated DNA were analyzed by PCR using primer pairs specific for the HIV-1 LTR nuc-0, DHS1, and nuc-1 regions and for a control region located upstream of the Axin gene promoter. ChIP results are presented as percent of immunoprecipitated material over input, error bars represent the standard deviation of three independent experiments. * indicates the level of significance at p < 0.05.
To determine the direct effects of BAFi treatment on the latent HIV-1 promoter, we examined the BAF complex occupancy of the promoter by ChIP assay. We have shown previously that, while bound to the latent HIV-1 LTR, the BAF complex is displaced upon LTR activation (Rafati et al., 2011). Similarly, as shown in Fig. 2d, the stimulation with the BAFi's A11 and C09 results in the displacement of the BAF specific subunit BAF250a, which is specifically enriched in the DHS1 and nuc-1 regions on the latent HIV-1 promoter. These results provide a direct mechanistic evidence for the function of BAFi's in HIV-1 latency.
To investigate the mechanism mediating the activity of the BAFi's, we analyzed the expression of the BAF subunits at the RNA and at the protein level. To determine whether BAFi's directly affect the BAF complex at the protein level, we quantitated the relative expression of the BAF subunits BAF250a, BRG1, and INI-1 by Western blotting after treatment with increasing concentrations of BAFi's (Figure S2a). Treatment of cells for 36 h with increasing concentrations of A11 and C09 resulted in a decrease in the BAF-specific subunit BAF250a protein levels, while not affecting the BRG1 and INI-1 subunits. We observed a decrease of BAF250a levels of up to 50% after treatment with the highest concentrations of A11, while C09 treatment resulted in a substantial (up to 70%) decrease in BAF250a (Figure S2a). At the RNA level, the expression of BAF250a did not change in response to BAFi's (Figure S2b), excluding an indirect mechanism whereby BAFi's treatment would cause a decrease in transcription of the BAF subunits. Conversely, the negative effect of A11 and C09 on stability of BAF250a is consistent with a mechanism of direct inhibition, in which destabilization and degradation of BAF250a at the protein level results in the specific inhibition of the BAF complex.
3.2. HIV-1 Latency Reversal by BAFi's is Synergistically Enhanced by Co-treatment With Prostratin and SAHA
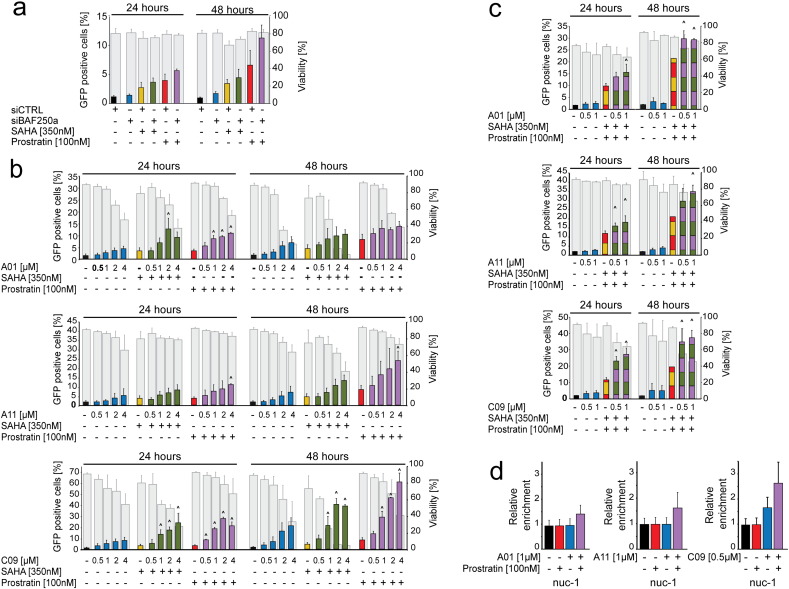
Combination therapy using latency reversal agents (LRAs) targeting different pathways is considered the most effective strategy for reactivation of latent HIV-1 (De Crignis and Mahmoudi, 2014, Laird et al., 2015). HDAC inhibitors (HDACi's) and PKC agonists are two promising classes of compounds currently under clinical investigation for their potential to reverse HIV-1 latency. HDACi's counteract the activity of histone deacetylases which repress HIV-1 transcription by compacting chromatin structure, whereas Prostratin induces translocation of NF-κB to the nucleus where it binds to the HIV-1 LTR and activates its transcription. To examine whether the combination of these mechanisms with BAF inhibition would enhance the activation of latent HIV-1 we evaluated the effect of SAHA and Prostratin after siRNA mediated knock down of BAF complex or treatment with BAFi's. The Bliss independence model for combined drug effects was used to calculate the threshold for synergism (Bliss, 1939, Zhao et al., 2014a).
J-Lat 11.1 cells were treated with SAHA or Prostratin after nucleofection with BAF250a-specific siRNA. siRNA treatment alone induced a limited activation of HIV-1 LTR as shown by the slight increase in the percentage of GFP positive cells at 48 h; when BAF250a-depleted cells were co-treated with Prostratin or SAHA, however, we observed a synergistic or additive, respectively, increase in reactivation of latent HIV-1 (Fig. 3a). Neither treatment significantly affected cell viability. Notably, we observed a similar effect when BAF complex activity was inhibited using BAFi's. J-Lat 11.1 cells were treated with Prostratin or SAHA alone or together with increasing concentrations of A01, A11, and C09 ranging from 0.5 μM to 4 μM (Fig. 3b). C09 demonstrated the most potent activity, synergistically enhancing HIV-1 transcription with both Prostratin and SAHA at all tested concentrations, whereas combination of LRAs with either A01 or A11 induced a synergistic or additive increase in activity depending on the concentration tested as shown (Fig. 3b). Interestingly, all BAFi's showed synergism used in triple treatment, i. e. when cells were treated with a combination of SAHA and Prostratin together with sub-optimal concentrations of BAFi's (Fig. 3c). We performed Formaldehyde Assisted Isolation of regulatory Elements (FAIRE) in order to examine the effect of BAFi's on the positioning of the latent HIV-1 LTR nuc-1 (Fig. 3d). Consistent with the previously observed HIV-1 activation and the displacement of BAF250a from DHS-1 and Nuc-1 demonstrated by ChIP assay, treatment with BAFi's resulted in a dose dependent remodeling of nuc-1 as demonstrated by increased accessibility of the DNA encompassing the positioned nuc-1 (Fig. 3d and Figure S3). Moreover, BAFi-induced remodeling of nuc-1 was also synergistically enhanced at the molecular level after co-treatment with Prostratin (Fig. 3d and Figure S3b).
Fig. 3.
Re-activation of latent HIV-1 by BAF inhibition is synergistically enhanced with co-treatment with HDACi's and PKC agonists.
(Panel a) Percent of GFP positive cells was evaluated in BAF complex depleted J-Lat 11.1 cells 24 and 48 h after treatment with suboptimal concentrations of Prostratin and SAHA. (Panel b and c) J-Lat 11.1 cells were treated with BAFi's A01, A11 and C09 alone or together with SAHA and-or Prostratin. HIV-1 re-activation was monitored by measuring the percentage of cells expressing GFP: colored bars show the percent of GFP positive cells after treatment, whereas gray bars show cell viability. (Panel d) Levels of nucleosome occupancy of the HIV-1 5'LTR Nuc-1 region following treatment with BAFi's alone and in combination with Prostratin were analyzed using FAIRE assay. Data are presented as mean ± SD. The (^) symbol indicates a synergistic interaction between treatments.
The striking synergism observed when cells were co-treated with the BAFi C09 and the PKC agonist Prostratin was unexpected, as C09 or Caffeic Acid Phenethyl Ester (CAPE) has been previously described to be an inhibitor of NF-κB, albeit at significantly higher concentrations (Marquez et al., 2004, Natarajan et al., 1996, Wang et al., 2010). We therefore examined the activity of C09 in a range of concentrations on Prostratin-induced IκBα degradation and HIV-1 activation. At concentrations effective in latency reversal (0.5–2 μM), C09 did not inhibit IκBα degradation induced by Prostratin (Figure S4a). Moreover, while synergistic with Prostratin in activation of the latent HIV-1 promoter at low concentrations, C09 interfered with the ability of Prostratin to re-activate latent HIV-1 when used at higher concentrations (30–100 μM) (Figure S4b), consistent with its previously described activity as an NF-κB inhibitor.
3.3. BAFi's Reverse HIV-1 Latency in Primary Infected CD4 + T Cells Without Inducing Cell Activation
The main reservoir of latent HIV-1 in infected patients resides in resting CD4 + T cells. Due to their extended life span and the capacity of self-renewal these latently infected cells constitute the major barrier to HIV-1 eradication and are the most relevant target to determine the activity and efficiency of potential LRAs. Therefore, while T cell line models of HIV-1 latency represent useful models for compound screens and the initial molecular characterization of putative LRAs that requires large scale techniques, it is critical that potential LRAs are characterized also in primary T cell models of HIV-1 latency.
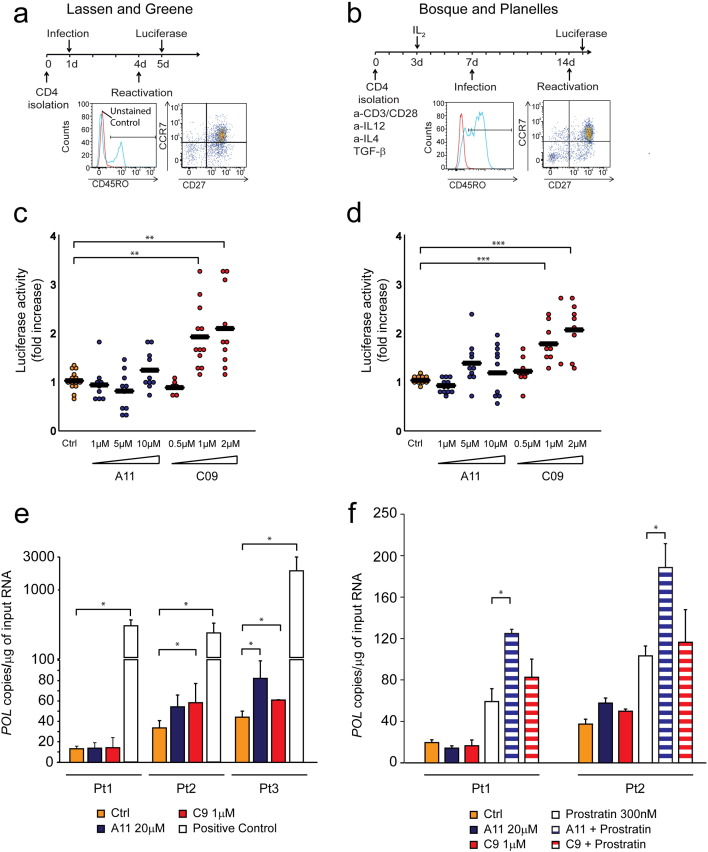
Several primary cell models of HIV-1 latency have been developed and validated in past years. In this study we first tested the activity of BAFi's using two in vitro primary models that recapitulate two different mechanisms that are thought to mediate the establishment of HIV-1 latency in vivo (Fig. 4a and b). In the first model, initially developed by O′ Doherty (Swiggard et al., 2005) and further modified by Lassen and Greene (Lassen et al., 2012), primary cells are infected without activation; the lack of specific transcription factors and the reduced metabolic rate in resting cells prevents the establishment of a productive infection and drives the virus into the latent state (Fig. 4a). In this model, infected cells mainly belong to the Transitional (CD4 + CD45RO + CCR7- CD27 +) and Central Memory (CD4 + CD45RO + CCR7 + CD27 +) populations. In the second method, developed by Bosque and Planelles, primary CD4 + T cells are stimulated for three days and infection is performed after removing the activating stimuli, i.e. during reversion to the resting state (Bosque and Planelles, 2009) (Fig. 4b). Cells generated via this model acquire a phenotype that closely resembles the CD4 + Central Memory population (CD4 + CD45RO + CCR7 + CD27 +), which is thought to harbor the majority of latent proviruses in HIV-1 infected individuals under suppressive cART (Chomont et al., 2009). In both protocols, cells were infected with a defective virus harboring luciferase in place of the envelope gene and latency reversal activity was monitored determining the levels of luciferase activity 24 h after treatment with BAFi's.
Fig. 4.
BAFi treatment re-activates latent HIV-1 in two distinct primary model systems of HIV-1 latency.
Latency reversal activity of the BAFi's A11 and C09 was tested on two models of HIV-1 latency. Panel a (Lassen and Greene) and b (Bosque and Planelles) depict the protocols for latency establishment in primary human CD4 + T cells. FACS plots show the characteristic of the cells population at the moment of reactivation for each method. Dotplots (Panels c and d) show the fold increase in luciferase activity after treatment with different concentrations of A11 and C09. Each dot represents a single measurement, black horizontal lines show the average fold increase for each treatment. Experiments were performed in duplicate using cells obtained from 6 healthy blood donors. Panel e shows the levels of cell associated HIV-1 RNA in CD4 + T cells isolated from three virologically suppressed HIV-infected patients after treatment with the BAFi's C09 and A11. Cells obtained from Patient 1 and Patient 2 were further analyzed and treated with BAFi's in combination with Prostratin (Panel f). Cell associated HIV-1 RNA was measured as the number of copies of HIV-1 POL per μg of input RNA; for each condition, bars represent the average of experiments performed at least in triplicate. Statistical significance was calculated within patient, based on at least three independent replicates performed for each condition. Asterisks indicate the level of significance (* p < 0.05, ** p < 0.01, *** p < 0.001).
To verify the activity of BAFi's on latent HIV-1, primary cells were treated with increasing concentrations of A11 and C09, the two compounds showing the maximum activity in cell line models of latency. The range of concentrations was selected based on their toxicity profile on primary cells. We observed a dose dependent increase in luciferase activity after treatment with C09 (Fig. 4c and d) with significantly higher levels of luciferase activity compared to untreated control at 1 μM and 2 μM concentrations (p < 0.05). Treatment with A11 did not induce significant activation of latent HIV-1, however we observed a tendency towards activation at the highest concentration (10 μM).
CD4 + T cells obtained from virologically suppressed HIV-1 infected patients are considered the most relevant model system for assessing the efficacy of potential latency reversal compounds. In light of our results showing the activity of A11 and C09 in latency reversal in primary ex-vivo infected cells, we sought to confirm these observations in CD4 + T cells obtained from HIV-1 infected patients. Three patients who maintained HIV-1 viremia below 50 copies/ml for at least two years (Pt1 3 years; Pt2 2 years; Pt 3 3 years) were enrolled in the study. Untouched total CD4 + T cells were purified from PBMCs and used to assess the effect of BAFi's on the latent HIV-1 reservoir. Treatment with BAFi's alone increased the levels of cellular associated HIV-1 RNA in CD4 + T cells obtained from two out of three patients. In this model, increase in HIV-1 transcription after BAFi treatment mirrored the results obtained in the ex vivo infected primary cells: C09 elicited HIV-1 transcription at 1 μM concentration, and A11 showed reversal activity at 20 μM, confirming the trend observed in the ex vivo infected model (Fig. 4e). A variable response to treatment with de-repressors in primary cells obtained from HIV-1 infected patients has been observed previously (Archin et al., 2012, Wei et al., 2014, Laird et al., 2015); however, even in absence of a detectable effect, the removal of a repressive mechanism has been suggested to facilitate triggering of transcription by enhancing noise and increasing the likelihood of stochastic promoter activation (Dar et al., 2014). We therefore examined the effect of the co-treatment of BAFi's together with the PKC agonist Prostratin on cells obtained from Patients 1 and 2. CD4 + T cells obtained from Patient 1, which did not respond to BAFi's alone, and CD4 + T cells obtained from Patient 2, which showed a BAFi-induced increase in HIV-1 cellular associated RNA, were treated with BAFi's alone or in combination with a sub-optimal concentration of Prostratin. As shown in Fig. 4f, treatment with BAFi's enhanced Prostratin-mediated HIV-1 transcription, independently of the response of cells to BAFi treatment alone. In particular, combination treatment with A11 together with Prostratin resulted in a significant enhancement of Prostratin-induced increase in cellular associated HIV-1 RNA in both patients. Interestingly, C09, although capable of significantly inducing latency reversal in patient cells when used alone, did not significantly enhance Prostratin-mediated latency reversal, consistent with its role in NF-κB pathway inhibition. Given this limitation, C09 will likely be more effective in latency reversal combinations lacking PKC activators. Taken together, these results showed that treatment with BAFi's either alone or in combination with Prostratin resulted in latency reversal in all three patients included in the study.
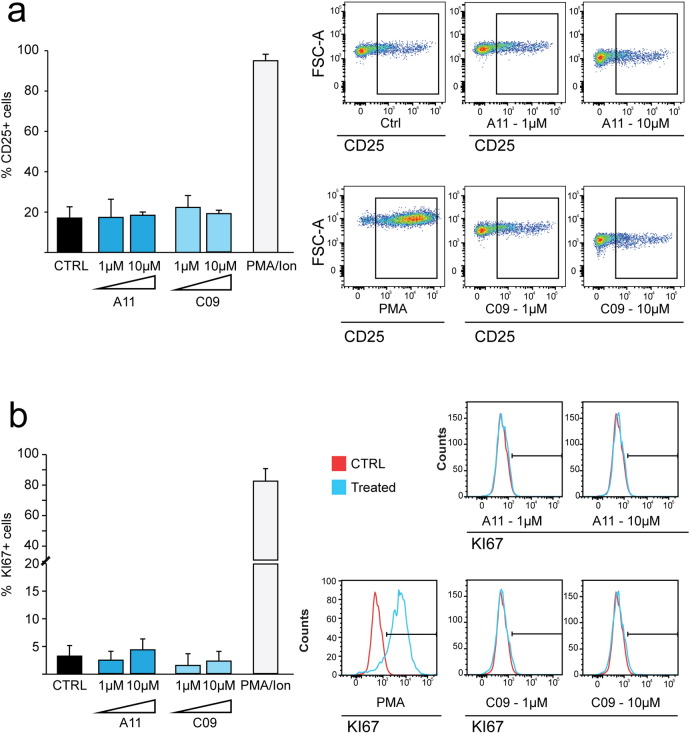
Activation of bystander uninfected cells is a side effect which can be observed after treatment with LRAs (Dechristopher et al., 2012, Korin et al., 2002). Massive stimulation of T cells can result in general immune activation and thus will decrease the potential usefulness of latency reversal molecules in clinical settings. To investigate the effect of BAFi's on primary cells we treated human CD4 + T cells isolated from healthy donors with A11 and C09 for 72 h, and determined the levels of T cell activation and proliferation by flow cytometric analysis of CD25 and Ki67, respectively. Treatment with PMA-Ionomycin was used as a positive control. As shown in Fig. 5, in contrast to PMA, treatment with BAFi's did not induce CD25 (Fig. 5a) and Ki67 (Fig. 5b) expression compared to untreated control during 3 days of treatment.
Fig. 5.
BAF inhibitors do not induce T cell activation or proliferation.
Percentage of cells expressing the markers of cell activation (CD25, panel a) and proliferation (KI67, panel b) in primary CD4 + T cells treated with A11 and C09. Treatment with PMA/Ionomycin was used as a positive control. Bars represent the average ± SD of experiments performed on samples deriving from three healthy blood donors. Images on the figure shows representative FACS plots for each marker.
3.4. Treatment with BAF Inhibitors Prevents Latency Establishment in Jurkat Cells
We have previously demonstrated that, in addition to its role in maintenance of HIV-1 latency, the BAF complex also contributes to the establishment of latency, as siRNA depletion of BAF subunits significantly decreased the incidence of latent infections in T cell lines (Rafati et al., 2011). We therefore examined whether inhibition of the BAF complex with A01, A11, and C09 interfered with the establishment of HIV-1 latency.
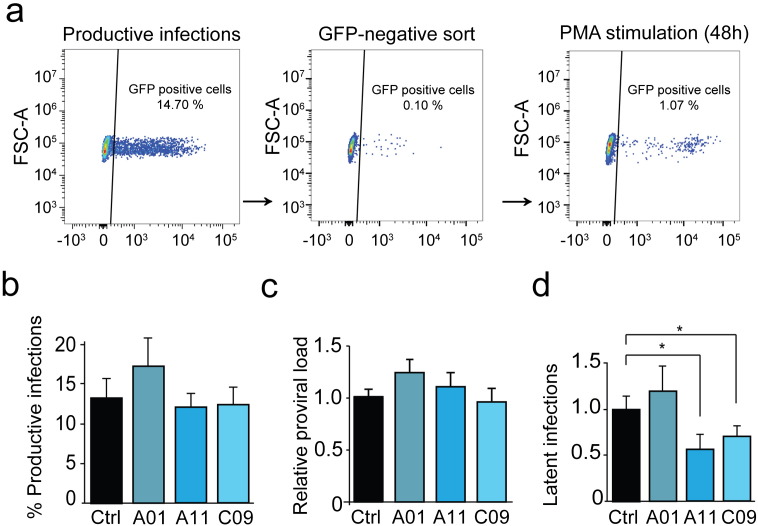
Jurkat cells were pre-treated with 1 μM A01, A11, or C09 for 30 min prior to infection with an HIV-1 derived virus containing a GFP reporter, LTR-Tat-IRES-GFP in presence of BAFi's. Percentage of GFP positive cells and relative amount of integrated DNA were measured five days after infection to evaluate infection efficiency. GFP-negative cell population consisting of uninfected and potentially latently infected cells were sorted by flow cytometry and stimulated with PMA. The percentage of GFP positive, latently infected cells was then determined and quantitate by FACS analysis (Fig. 6a). While treatment with A01 caused a slight increase in productive integrations, treatment of cells with A11 and C09 did not significantly affect the percentage of cells productively infected with the HIV-1 derived virus (Fig. 6b and c). However, when cells were treated with either A11 or C09, we observed a significant decrease in the percentage of GFP positive cells after PMA treatment, corresponding to a lower incidence of latent infections (Fig. 6d). Taken together these data show that inhibition of BAF activity using A11 and C09 interferes with the establishment of latency.
Fig. 6.
Inhibition of BAF prevents the establishment of latent HIV + infections.
(Panel a) FACS plots show the protocol for quantification of latently infected cells. GFP-negative cells were sorted (left panel) five days after infections. Following sorting, the GFP-negative population (middle panel) was stimulated with PMA and the percentage of reactivated cells was measured 48 h after reactivation (right panel). (Panel b) Infection efficiency in untreated vs BAFi's treated samples was determined by FACS. Bars represent the percent of GFP positive cells (i.e. productive infections) 5 days after infection. (Panel c) Proviral load from the infected population was measured by Alu-PCR. Bars represent the relative amount of HIV-1 DNA compared to untreated control. (Panel d) GFP-negative population (containing uninfected as well as latently infected cells) was treated with PMA for 48 h. Bars represent the percentage of GFP positive cells (i.e. latently infected cells) for each treatment. Data are presented as mean ± SD, * indicates the level of significance at p < 0.05.
4. Discussion
Recent pharmacological efforts aimed at HIV-1 eradication have focused on the development of “shock and kill” strategies, where HIV-1 is induced from latency in order to render infected cell recognizable to the immune system for clearance. We showed previously that the BAF chromatin remodeling complex plays an important role in the establishment and maintenance of HIV-1 latency, highlighting BAF as an attractive molecular target in HIV-1 eradication efforts. We have now tested a panel of small molecules, recently discovered in a screen for inhibition of BAF activity, for their potential to reverse HIV-1 latency. Treatment with BAFi's results in activation of latent virus in cell line as well as primary cell models harboring latent HIV-1. BAFi's constitute a group of molecules including compounds with different chemical structures targeting BAF activity with different degrees of specificity and potency. In the present screening we found the highest levels of HIV-1 activation for two compounds, A11 and C09.
A11, or Pyrimethamine (PYR), is an FDA approved licensed anti-protozoan drug which has been used to control opportunistic infections in HIV-1 infected patients (Rosenblatt, 1999, Klinker et al., 1996, Mathanga et al., 2011). Consistent with our results describing PYR as an activator of HIV-1 transcription, a previous report has shown that PYR, in the 10–100 μM range, can enhance HIV-1 replication in vitro in human PBMCs (Oguariri et al., 2010). C09, caffeic acid phenetyl esther (CAPE), is a bioactive compound found in many plants and isolated from honey bee propolis (Wu et al., 2011) shown to possess anti-inflammatory and immunomodulatory capacities (Tolba et al., 2013). CAPE, currently under clinical investigation (Clinical trial.gov identifier: NCT02050334 and NCT02351622), has been proposed in the treatment of several diseases including thrombocytopenia and cancer. CAPE has also been shown to function as a non-competitive inhibitor of the enzyme HIV-1 integrase and thus inhibits HIV-1 replication (Fesen et al., 1994). In the 0.5–2 μM concentration range in which CAPE efficiently reactivates latent HIV-1, we did not observe an inhibitory effect on HIV-1 integration. We did however observe a decreased propensity for latency establishment in Jurkat cells in presence of low concentrations of both PYR and CAPE, mimicking decreased latency establishment upon siRNA mediated depletion of BAF shown previously (Rafati et al., 2011). The ability of BAFi's to prevent establishment of latency is of great interest in light of their potential use as LRAs. Indeed, despite the high efficiency of cART, the concentrations of antiretroviral drugs might not reach the optimal levels in some anatomical compartments. If reactivation of latent HIV-1 takes place in these compartments, de novo infections may occur and possibly lead to the establishment of a new reservoir. The use of LRAs that also inhibit infection and latency establishment, such as BAFi's and Romidepsin (Jonsson et al., 2015), could therefore increase the overall safety of latency reactivation strategies.
One important clinical consideration to be made in evaluating the potential of candidate molecules for latency reversal is whether they are able to generate sufficient levels of HIV-1 activation without inducing immune activation. Using two primary cell models of latency, we found that BAFi's induce reversal of HIV-1 latency in primary CD4 + T cells, which represent the main target of HIV-1 infection and harbor the majority of latent virus. Importantly, treatment of primary cells with BAFi's, at concentrations which efficiently induce latency reversal, did not result in cell proliferation nor caused general activation of primary CD4 + T cells. We also examined the activity of BAFi's in CD4 + T cells obtained from three long-term virologically suppressed HIV-1 infected patients, as this model is currently considered the most relevant model of latency. The patients included in our study were treated for at least 3 years and showed no sign of viral replication for at least 2 years. In fully suppressed c-ART treated patients, the presence of HIV-1 in activated T cells has been shown to be negligible (Finzi et al., 1997). To assess BAFi-mediated latency reversal, we therefore used total CD4 + patient T cells that include both activated and resting T cells as a surrogate for resting CD4 + T cell response, an approach also used in other recent studies to examine latency reversal ex vivo (Wei et al., 2014, Jiang et al., 2014, Jiang et al., 2015).
In Patient 2 and 3, treatment with BAFi's alone increased the levels of cellular associated HIV-1 RNA. Although limited, the two fold increase in HIV-1 cellular associated RNA levels observed is similar to the modest effects of BAFi treatment alone observed in both cell line models and in vitro infected primary cells, and is consistent with de-repression, which results from removal of an LTR-bound repressive complex. Similar levels of activity have been observed for other candidate LRAs such as Vorinostat, Disulfiram, JQ1, and Romidepsin (Jiang et al., 2014, Laird et al., 2015, Wei et al., 2014). Importantly, many of the putative LRAs which showed limited latency reversal activity when used alone, have been shown to effectively induce HIV-1 transcription when used in combination (Laird et al., 2015, Spivak et al., 2015, Barton et al., 2014). Similarly, in cells obtained from both Patient 1, in which treatment with BAFi's alone did not upregulate the levels of HIV-1 RNA, and Patient 2, which responded to BAFi treatment, co-treatment using PYR together with the bona fide activator Prostratin resulted in significant enhancement of Prostratin mediated HIV-1 activation.
Interestingly, although CAPE when used alone induced significant latency reversal in cells from two of the three patients examined, it did not result in significant enhancement of Prostratin-mediated latency reversal. This lack of synergy is likely due to the described effect of CAPE in inhibition of NF-κB. At high concentrations (10–85 μM) CAPE has been shown to block phosphorylation and degradation of IκB (Zhao et al., 2014b, Wang et al., 2010, Shvarzbeyn and Huleihel, 2011), and to inhibit translocation and DNA binding of p65 (Natarajan et al., 1996, Marquez et al., 2004). It is noteworthy that, in our experimental setting, CAPE was effective in activating latent HIV-1 at concentrations ranging from 0.5 to 2 μM. At these relatively low concentrations, CAPE showed significant synergy in latency reversal and did not show an inhibitory effect towards either Prostratin-induced activation of latent HIV-1 or Prostratin-mediated degradation of IκB in cell lines. The difference between cell line and patient cell response to CAPE in latency reversal highlights the importance of validation of potential LRA activity in primary cells.
Another clinical consideration in appraising the potential for candidate LRAs, is the question of target specificity; most LRAs currently under investigation target chromatin modifying factors or transcriptional pathways, which are essential for normal regulation of host gene expression. Targeting these complexes will therefore have pleiotropic consequences and result in mis-regulation of host gene expression as it was recently shown for SAHA (Elliott et al., 2014). Similarly to SAHA, treatment with BAFi's, particularly at high concentrations, may result in adverse pleiotropic effects, including induction of genes counteracting HIV-1 activation (White et al., 2015). Further studies, such as transcriptome and proteome analysis, are needed to determine the array of cellular modifications induced by these compounds. In this context, inclusion of distinct LRAs, which function via different molecular targets in a combination therapy, would decrease the adverse pleiotropic effects of each compound, decrease toxicity issues, and provide a higher degree of specificity for the HIV-1 LTR.
Transcription of integrated HIV-1 is stochastic, and reversal of the latent state using a single agent can be limited by specific factors, including restrictions posed by the site of integration of the virus and its molecular environment, subtypes of the latent virus and their susceptibility to a single agent, and the cell type in which latency is established. Data obtained from in vitro cell line HIV-1 latency models support the effectiveness of combinatorial approaches for the synergistic activation of HIV-1 transcription. These synergistic effects are likely the result of the combination of two mechanisms: the removal or decrease in a repressive chromatin environment together with the triggering and recruitment of bona fide activators. Combination of Prostratin with HDACi's results in synergistic activation of HIV-1 in vitro (Reuse et al., 2009, Ying et al., 2012, Qu et al., 2013, Burnett et al., 2010, Pandeló et al., 2014, Micheva-Viteva et al., 2011) as well as ex vivo (Laird et al., 2015). Similarly, BAFi-mediated reversal of transcriptional latency was synergistically enhanced when latent cells were co-treated with the HDAC inhibitor SAHA or the PKC agonist Prostratin. FAIRE assays indicate remodeling of the repressive LTR nuc-1 in response to BAFi treatment at the molecular level, and demonstrate the molecular synergism between Prostratin and BAFi's leading to eviction of nuc-1. These findings point to the importance of combining molecules that target multiple pathways to disrupt and reverse the latent transcriptional state of the virus in shock and kill strategies.
Our data highlights the potential of BAFi's as a source of molecules for inclusion in latency reversal therapies. BAFi's effectively reverse latency either alone or in combination with Prostratin in all three patients enrolled in this study. A caveat of the current study is the limited number of patients enrolled, which does not allow for a strong statistical validation of the effect of BAFi's across patients. Nevertheless, although not sufficient to fully recapitulate the variability in response to LRAs across patients, the data from the three included patients support our observations in cell lines and other primary models of latency. As with other molecules currently under clinical characterization in latency reversal, it is conceivable that with BAF inhibitors too, there will be some variability in patient response. Thus ex vivo studies performed on a larger number of HIV-1 infected patients are required in order to statistically validate the present observations. These studies, together with proof-of-concept clinical studies are the necessary next steps in order to further characterize the potency and applicability of BAFi's to reverse the latent HIV-1 reservoir in vivo. Given their demonstrated promising activity and the available information concerning their toxicity and pharmokinetics profiles, CAPE and PYR, represent attractive candidates for future clinical development. Further screenings will likely identify additional molecules targeting HIV-1 latency via counteracting the activity of the repressive BAF complex. Importantly, since targeting a previously unexplored pathway, this promising class of drugs has high potential for inclusion in combination therapy.
Funding Source
This work was supported by a Dutch ‘Aids Fonds’ grant (2014021), an Erasmus MC mRACE Research Grant and an ERC Starting Grant (337116 - Trxn-PURGE).
Conflicts of Interest
The authors declare no conflict of interest.
Author Contributions
MS, EDC and RJP carried out the experiments and performed data analysis. MMK participated in gene expression analysis experiments. TWK and CL participated in FACS experiments. RJP performed FAIRE experiments. CR and AV recruited patients. CB provided expertise and contributed to the writing of the manuscript. ECD provided compounds, expertize and contributed to the writing of the manuscript. MS, EDC and TM conceived the study and wrote the manuscript. All authors read and approved the final manuscript. MS and EDC contributed equally to this work.
Acknowledgements
We would like to thank Dr. Michelle Palmer and Dr. Christina Scherer (Broad Institute of MIT and Harvard) for kindly providing the compounds used in the screening, and Professor C.P. Verrijzer for providing BAF250a antibodies. We would like to thank the Centre for AIDS Reagents (NIBSC, UK) for providing the PL4.3.Luc.R-E- and the HIV-1 HXB2-Env expression vectors, Saquinavir Mesylate, and Raltegravir.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2015.11.047.
Appendix A. Supplementary Material
Supplementary material.
References
- Archin N.M., Liberty A.L., Kashuba A.D., Choudhary S.K., Kuruc J.D., Crooks A.M., Parker D.C., Anderson E.M., Kearney M.F., Strain M.C., Richman D.D., Hudgens M.G., Bosch R.J., Coffin J.M., Eron J.J., Hazuda D.J., Margolis D.M. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin N.M., Bateson R., Tripathy M.K., Crooks A.M., Yang K.H., Dahl N.P., Kearney M.F., Anderson E.M., Coffin J.M., Strain M.C., Richman D.D., Robertson K.R., Kashuba A.D., Bosch R.J., Hazuda D.J., Kuruc J.D., Eron J.J., Margolis D.M. HIV-1 expression within resting CD4 + T cells after multiple doses of vorinostat. J. Infect. Dis. 2014;210:728–735. doi: 10.1093/infdis/jiu155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat T.S., Gunhanlar N., Pardo C.G., Achame E.M., Ghazvini M., Boers R., Kenter A., Rentmeester E., Grootegoed J.A., Gribnau J. RNF12 activates Xist and is essential for X chromosome. PLoS Genet. 2011;7(1):2011. doi: 10.1371/journal.pgen.1002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N., Hurlstone A., Musisi H., Miles A., Bienz M., Clevers H. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 2001;20:4935–4943. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton K., Margolis D. Selective targeting of the repressive transcription factors YY1 and cMyc to disrupt quiescent human immunodeficiency viruses. AIDS Res. Hum. Retrovir. 2013;29:289–298. doi: 10.1089/aid.2012.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton K.M., Archin N.M., Keedy K.S., Espeseth A.S., Zhang Y.L., Gale J., Wagner F.F., Holson E.B., Margolis D.M. Selective HDAC inhibition for the disruption of latent HIV-1 infection. PLoS One. 2014;9 doi: 10.1371/journal.pone.0102684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton K., Hiener B., Palmern S., Rasmussen T.A., Tolstrup M., Østergaard L., Søgaard O., Solomon A., Lewin S., Shao W. Year. Panobinostat Broadly Activates Latent HIV-1 Proviruses in Patients; 2015. CROI, February 23–26, 2015. (Seattle, Washington) [Google Scholar]
- Bliss C.I. The toxicity of poisons applied jointly. Ann. Appl. Biol. 1939;26:585–615. [Google Scholar]
- Bosque A., Planelles V. Induction of HIV-1 latency and reactivation in primary memory CD4 + T cells. Blood. 2009;113:58–65. doi: 10.1182/blood-2008-07-168393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett J.C., Lim K.I., Calafi A., Rossi J.J., Schaffer D.V., Arkin A.P. Combinatorial latency reactivation for HIV-1 subtypes and variants. J. Virol. 2010;84:5958–5974. doi: 10.1128/JVI.00161-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomont N., El-Far M., Ancuta P., Trautmann L., Procopio F.A., Yassine-Diab B., Boucher G., Boulassel M.R., Ghattas G., Brenchley J.M., Schacker T.W., Hill B.J., Douek D.C., Routy J.P., Haddad E.K., Sekaly R.P. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T.W., Finzi D., Margolick J., Chadwick K., Schwartz D., Siliciano R.F. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat. Med. 1995;1:1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- Chun T.W., Justement J.S., Moir S., Hallahan C.W., Maenza J., Mullins J.I., Collier A.C., Corey L., Fauci A.S. Decay of the HIV reservoir in patients receiving antiretroviral therapy for extended periods: implications for eradication of virus. J. Infect. Dis. 2007;195:1762–1764. doi: 10.1086/518250. [DOI] [PubMed] [Google Scholar]
- Contreras X., Schweneker M., Chen C.S., Mccune J.M., Deeks S.G., Martin J., Peterlin B.M. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J. Biol. Chem. 2009;284:6782–6789. doi: 10.1074/jbc.M807898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahabieh M.S., Battivelli E., Verdin E. Understanding HIV latency: the road to an HIV cure. Annu. Rev. Med. 2015;66:407–421. doi: 10.1146/annurev-med-092112-152941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar R.D., Hosmane N.N., Arkin M.R., Siliciano R.F., Weinberger L.S. Screening for noise in gene expression identifies drug synergies. Science. 2014;344:1392–1396. doi: 10.1126/science.1250220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Crignis E., Mahmoudi T. HIV eradication: combinatorial approaches to activate latent viruses. Virus. 2014;6:4581–4608. doi: 10.3390/v6114581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechristopher B.A., Loy B.A., Marsden M.D., Schrier A.J., Zack J.A., Wender P.A. Designed, synthetically accessible bryostatin analogues potently induce activation of latent HIV reservoirs in vitro. Nat. Chem. 2012;4:705–710. doi: 10.1038/nchem.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks S.G. HIV: shock and kill. Nature. 2012;487:439–440. doi: 10.1038/487439a. [DOI] [PubMed] [Google Scholar]
- Dykhuizen E.C., Carmody L.C., Tolliday N., Crabtree G.R., Palmer M.A. Screening for inhibitors of an essential chromatin remodeler in mouse embryonic stem cells by monitoring transcriptional regulation. J. Biomol. Screen. 2012;17:1221–1230. doi: 10.1177/1087057112455060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein L.C., Micheva-Viteva S., Phelan B.D., Dougherty J.P. Short communication: activation of latent HIV type 1 gene expression by suberoylanilide hydroxamic acid (SAHA), an HDAC inhibitor approved for use to treat cutaneous T cell lymphoma. AIDS Res. Hum. Retrovir. 2009;25:883–887. doi: 10.1089/aid.2008.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott J.H., Wightman F., Solomon A., Ghneim K., Ahlers J., Cameron M.J., Smith M.Z., Spelman T., Mcmahon J., Velayudham P., Brown G., Roney J., Watson J., Prince M.H., Hoy J.F., Chomont N., Fromentin R., Procopio F.A., Zeidan J., Palmer S., Odevall L., Johnstone R.W., Martin B.P., Sinclair E., Deeks S.G., Hazuda D.J., Cameron P.U., Sekaly R.P., Lewin S.R. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesen M.R., Pommier Y., Leteurtre F., Hiroguchi S., Yung J., Kohn K.W. Inhibition of HIV-1 integrase by flavones, caffeic acid phenethyl ester (CAPE) and related compounds. Biochem. Pharmacol. 1994;48:595–608. doi: 10.1016/0006-2952(94)90291-7. [DOI] [PubMed] [Google Scholar]
- Finzi D., Hermankova M., Pierson T., Carruth L.M., Buck C., Chaisson R.E., Quinn T.C., Chadwick K., Margolick J., Brookmeyer R., Gallant J., Markowitz M., Ho D.D., Richman D.D., Siliciano R.F. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- Finzi D., Blankson J., Siliciano J.D., Margolick J.B., Chadwick K., Pierson T., Smith K., Lisziewicz J., Lori F., Flexner C., Quinn T.C., Chaisson R.E., Rosenberg E., Walker B., Gange S., Gallant J., Siliciano R.F. Latent infection of CD4 + T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- Geeraert L., Kraus G., Pomerantz R.J. Hide-and-seek: the challenge of viral persistence in HIV-1 infection. Annu. Rev. Med. 2008;59:487–501. doi: 10.1146/annurev.med.59.062806.123001. [DOI] [PubMed] [Google Scholar]
- Hargreaves D.C., Crabtree G.R. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G., Mendes E.A., Kaiser P., Sankaran-Walters S., Tang Y., Weber M.G., Melcher G.P., Thompson G.R., Tanuri A., III, Pianowski L.F., Wong J.K., Dandekar S. Reactivation of HIV latency by a newly modified Ingenol derivative via protein kinase Cdelta-NF-kappaB signaling. AIDS. 2014;28:1555–1566. doi: 10.1097/QAD.0000000000000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G., Mendes E.A., Kaiser P., Wong D.P., Tang Y., Cai I., Fenton A., Melcher G.P., Hildreth J.E., Thompson G.R., Wong J.K., Dandekar S. Synergistic reactivation of latent HIV Expression by ingenol-3-angelate, PEP005, targeted NF-kB signaling in combination with JQ1 induced p-TEFb activation. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson K.L., Tolstrup M., Vad-Nielsen J., Kjaer K., Laustsen A., Andersen M.N., Rasmussen T.A., Sogaard O.S., Ostergaard L., Denton P.W., Jakobsen M.R. Histone deacetylase inhibitor romidepsin inhibits de novo HIV-1 Infections. Antimicrob. Agents Chemother. 2015 doi: 10.1128/AAC.00574-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A., Defechereux P., Verdin E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 2001;20:1726–1738. doi: 10.1093/emboj/20.7.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A., Bisgrove D., Verdin E. HIV reproducibly establishes a latent infection after Acute infection of T cells in vitro. EMBO J. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keedy K.S., Archin N.M., Gates A.T., Espeseth A., Hazuda D.J., Margolis D.M. A limited group of class I histone deacetylases acts to repress human immunodeficiency virus type 1 expression. J. Virol. 2009;83:4749–4756. doi: 10.1128/JVI.02585-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinker H., Langmann P., Richter E. Plasma pyrimethamine concentrations during long-term treatment for cerebral toxoplasmosis in patients with AIDS. Antimicrob. Agents Chemother. 1996;40:1623–1627. doi: 10.1128/aac.40.7.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korin Y.D., Brooks D.G., Brown S., Korotzer A., Zack J.A. Effects of prostratin on T-cell activation and human immunodeficiency virus latency. J. Virol. 2002;76:8118–8123. doi: 10.1128/JVI.76.16.8118-8123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird G.M., Bullen C.K., Rosenbloom D.I., Martin A.R., Hill A.L., Durand C.M., Siliciano J.D., Siliciano R.F. Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J. Clin. Invest. 2015 doi: 10.1172/JCI80142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen K.G., Hebbeler A.M., Bhattacharyya D., Lobritz M.A., Greene W.C. A flexible model of HIV-1 latency permitting evaluation of many primary CD4 T-cell reservoirs. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H.K., Gray L.R., Wightman F., Ellenberg P., Khoury G., Cheng W.J., Mota T.M., Wesselingh S., Gorry P.R., Cameron P.U., Churchill M.J., Lewin S.R. Ex vivo response to histone deacetylase (HDAC) inhibitors of the HIV long terminal repeat (LTR) derived from HIV-infected patients on antiretroviral therapy. PLoS One. 2014;9 doi: 10.1371/journal.pone.0113341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusic M., Giacca M. Regulation of HIV-1 latency by chromatin structure and nuclear architecture. J. Mol. Biol. 2015;427:688–694. doi: 10.1016/j.jmb.2014.07.022. [DOI] [PubMed] [Google Scholar]
- Mahmoudi T. The BAF complex and HIV latency. Transcription. 2012;3:171–176. doi: 10.4161/trns.20541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi T., Verrijzer C.P. Chromatin silencing and activation by polycomb and trithorax group proteins. Oncogene. 2001;20:3055–3066. doi: 10.1038/sj.onc.1204330. [DOI] [PubMed] [Google Scholar]
- Marban C., Suzanne S., Dequiedt F., De Walque S., Redel L., Van Lint C., Aunis D., Rohr O. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional Silencing. EMBO J. 2007;26:412–423. doi: 10.1038/sj.emboj.7601516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez N., Sancho R., Macho A., Calzado M.A., Fiebich B.L., Munoz E. Caffeic acid phenethyl ester inhibits T-cell activation by targeting both nuclear factor of activated T-cells and NF-kappaB transcription factors. J. Pharmacol. Exp. Ther. 2004;308:993–1001. doi: 10.1124/jpet.103.060673. [DOI] [PubMed] [Google Scholar]
- Mathanga D.P., Uthman O.A., Chinkhumba J. Intermittent preventive treatment regimens for malaria in HIV-positive pregnant women. Cochrane Database Syst. Rev. 2011 doi: 10.1002/14651858.CD006689.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y., Kobayashi-Ishihara M., Fujikawa D., Ishida T., Watanabe T., Yamagishi M. Epigenetic heterogeneity in HIV-1 latency establishment. Sci. Rep. 2015;5:7701. doi: 10.1038/srep07701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbonye U., Karn J. Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology. 2014;454-455:328–339. doi: 10.1016/j.virol.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheva-Viteva S., Kobayashi Y., Edelstein L.C., Pacchia A.L., Lee H.L., Graci J.D., Breslin J., Phelan B.D., Miller L.K., Colacino J.M., Gu Z., Ron Y., Peltz S.W., Dougherty J.P. High-throughput screening uncovers a compound that activates latent HIV-1 and acts cooperatively with a histone deacetylase (HDAC) inhibitor. J. Biol. Chem. 2011;286:21083–21091. doi: 10.1074/jbc.M110.195537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar G.J., Sundaramoorthy R., Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K., Singh S., Burke T.R., Jr., Grunberger D., Aggarwal B.B. Caffeic acid phenethyl ester is a potent and specific inhibitor of Activation of nuclear transcription factor NF-kappa B. Proc. Natl. Acad. Sci. U. S. A. 1996;93:9090–9095. doi: 10.1073/pnas.93.17.9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z., Bremner R. Brahma-related gene 1-dependent STAT3 recruitment at IL-6-inducible genes. J. Immunol. 2007;178:345–351. doi: 10.4049/jimmunol.178.1.345. [DOI] [PubMed] [Google Scholar]
- Oguariri R.M., Adelsberger J.W., Baseler M.W., Imamichi T. Evaluation of the effect of pyrimethamine, an anti-malarial drug, on HIV-1 replication. Virus Res. 2010;153:269–276. doi: 10.1016/j.virusres.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandeló J.D., Bartholomeeusen K., Da Cunha R.D., Abreu C.M., Glinski J., Da Costa T.B., Bacchi Rabay A.F., Pianowski Filho L.F., Dudycz L.W., Ranga U., Peterlin B.M., Pianowski L.F., Tanuri A., Aguiar R.S. Reactivation of latent HIV-1 by new semi-synthetic ingenol esters. Virology. 2014;462-463:328–339. doi: 10.1016/j.virol.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty E., Pillus L. Balancing chromatin remodeling and histone modifications in transcription. Trends Genet. 2013;29:621–629. doi: 10.1016/j.tig.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson T., Mcarthur J., Siliciano R.F. Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu. Rev. Immunol. 2000;18:665–708. doi: 10.1146/annurev.immunol.18.1.665. [DOI] [PubMed] [Google Scholar]
- Qu X., Ying H., Wang X., Kong C., Zhou X., Wang P., Zhu H. Histone deacetylase inhibitor MC1293 induces latent HIV-1 reactivation by histone modification in vitro latency cell lines. Curr. HIV Res. 2013;11:24–29. [PubMed] [Google Scholar]
- Rafati H., Parra M., Hakre S., Moshkin Y., Verdin E., Mahmoudi T. Repressive LTR nucleosome positioning by the BAF complex is required for HIV latency. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen T.A., Schmeltz Sogaard O., Brinkmann C., Wightman F., Lewin S.R., Melchjorsen J., Dinarello C., Ostergaard L., Tolstrup M. Comparison of HDAC inhibitors in clinical development: effect on HIV production in latently infected cells and T-cell activation. Hum. Vaccin Immunother. 2013;9:993–1001. doi: 10.4161/hv.23800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen T.A., Tolstrup M., Brinkmann C., Olesen R., Erikstrup C., Solomon A., Winckelmann A., Palmer S., Dinarello C., Buzon M., Lichterfeld M., Lewin S., Østergaard L., Søgaard O. CROI; Boston: 2014. Year. Panobinostat Induces HIV Transcription and Plasma Viremia in HIV Patients on Suppressive cART. [DOI] [PubMed] [Google Scholar]
- Reuse S., Calao M., Kabeya K., Guiguen A., Gatot J.S., Quivy V., Vanhulle C., Lamine A., Vaira D., Demonte D., Martinelli V., Veithen E., Cherrier T., Avettand V., Poutrel S., Piette J., DE Launoit Y., Moutschen M., Burny A., Rouzioux C., De Wit S., Herbein G., Rohr O., Collette Y., Lambotte O., Clumeck N., Van Lint C. Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: implications for treatment of latent infection. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt J.E. Antiparasitic agents. Mayo Clin. Proc. 1999;74:1161–1175. doi: 10.4065/74.11.1161. [DOI] [PubMed] [Google Scholar]
- Ruelas D.S., Greene W.C. An integrated overview of HIV-1 latency. Cell. 2013;155:519–529. doi: 10.1016/j.cell.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Shvarzbeyn J., Huleihel M. Effect of propolis and caffeic acid phenethyl ester (CAPE) on NFkappaB activation by HTLV-1 Tax. Antivir. Res. 2011;90:108–115. doi: 10.1016/j.antiviral.2011.03.177. [DOI] [PubMed] [Google Scholar]
- Siliciano J.D., Kajdas J., Finzi D., Quinn T.C., Chadwick K., Margolick J.B., Kovacs C., Gange S.J., Siliciano R.F. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4 + T cells. Nat. Med. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- Spivak A.M., Bosque A., Balch A.H., Smyth D., Martins L., Planelles V. Ex vivo bioactivity and HIV-1 latency reversal by ingenol Dibenzoate and panobinostat in resting CD4 + T cells from aviremic patients. Antimicrob. Agents Chemother. 2015;59:5984–5991. doi: 10.1128/AAC.01077-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strain M.C., Little S.J., Daar E.S., Havlir D.V., Gunthard H.F., Lam R.Y., Daly O.A., Nguyen J., Ignacio C.C., Spina C.A., Richman D.D., Wong J.K. Effect of treatment, during primary infection, on establishment and clearance of cellular reservoirs of HIV-1. J. Infect. Dis. 2005;191:1410–1418. doi: 10.1086/428777. [DOI] [PubMed] [Google Scholar]
- Swiggard W.J., Baytop C., Yu J.J., Dai J., Li C., Schretzenmair R., Theodosopoulos T., O'doherty U. Human immunodeficiency virus type 1 can establish latent infection in resting CD4 + T cells in the absence of activating stimuli. J. Virol. 2005;79:14179–14188. doi: 10.1128/JVI.79.22.14179-14188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolba M.F., Azab S.S., Khalifa A.E., Abdel-Rahman S.Z., Abdel-Naim A.B. Caffeic acid phenethyl ester, a promising component of propolis with a plethora of biological activities: a review on its anti-inflammatory, neuroprotective, hepatoprotective, and cardioprotective effects. IUBMB Life. 2013;65:699–709. doi: 10.1002/iub.1189. [DOI] [PubMed] [Google Scholar]
- Van Lint C., Emiliani S., Ott M., Verdin E. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 1996;15:1112–1120. [PMC free article] [PubMed] [Google Scholar]
- Van Lint C., Bouchat S., Marcello A. HIV-1 transcription and latency: an update. Retrovirology. 2013;10:67. doi: 10.1186/1742-4690-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E. DNase I-hypersensitive sites are associated with both long terminal repeats and with the intragenic enhancer of integrated human immunodeficiency virus type 1. J. Virol. 1991;65:6790–6799. doi: 10.1128/jvi.65.12.6790-6799.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E., Van Lint C. Internal transcriptional regulatory elements in HIV-1 and other retroviruses. Cell. Mol. Biol. 1995;41:365–369. [PubMed] [Google Scholar]
- Verdin E., Paras P., Jr., Van Lint C. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 1993;12:3249–3259. doi: 10.1002/j.1460-2075.1993.tb05994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.D., Chen Y.B., Pan K., Wang W., Chen S.P., Chen J.G., Zhao J.J., Lv L., Pan Q.Z., Li Y.Q. Decreased expression of the ARID1A gene is associated with poor prognosis in primary gastric cancer. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.C., Chu K.H., Liang Y.C., Lin Y.L., Chiang B.L. Caffeic acid phenethyl ester inhibits nuclear factor-kappaB and protein kinase B signalling pathways and induces caspase-3 expression in primary human CD4 + T cells. Clin. Exp. Immunol. 2010;160:223–232. doi: 10.1111/j.1365-2249.2009.04067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe D., Ibe S., Uehira T., Minami R., Sasakawa A., Yajima K., Yonemoto H., Bando H., Ogawa Y., Taniguchi T., Kasai D., Nishida Y., Yamamoto M., Kaneda T., Shirasaka T. Cellular HIV-1 DNA levels in patients receiving antiretroviral therapy strongly correlate with therapy initiation timing but not with therapy duration. BMC Infect. Dis. 2011;11:146. doi: 10.1186/1471-2334-11-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D.G., Chiang V., Fyne E., Balakrishnan M., Barnes T., Graupe M., Hesselgesser J., Irrinki A., Murry J.P., Stepan G., Stray K.M., Tsai A., Yu H., Spindler J., Kearney M., Spina C.A., Mcmahon D., Lalezari J., Sloan D., Mellors J., Geleziunas R., Cihlar T. Histone deacetylase inhibitor romidepsin induces HIV expression in CD4 T cells from patients on suppressive antiretroviral therapy at concentrations achieved by clinical dosing. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C.H., Johnston H.E., Moesker B., Manousopoulou A., Margolis D.M., Richman D.D., Spina C.A., Garbis S.D., Woelk C.H., Beliakova-Bethell N. Mixed effects of suberoylanilide hydroxamic acid (SAHA) on the host transcriptome and proteome and their implications for HIV reactivation from latency. Antivir. Res. 2015;123:78–85. doi: 10.1016/j.antiviral.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney J.B., Hill A.L., Sanisetty S., Penaloza-Macmaster P., Liu J., Shetty M., Parenteau L., Cabral C., Shields J., Blackmore S., Smith J.Y., Brinkman A.L., Peter L.E., Mathew S.I., Smith K.M., Borducchi E.N., Rosenbloom D.I., Lewis M.G., Hattersley J., Li B., Hesselgesser J., Geleziunas R., Robb M.L., Kim J.H., Michael N.L., Barouch D.H. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature. 2014;512:74–77. doi: 10.1038/nature13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.A., Chen L.F., Kwon H., Ruiz-Jarabo C.M., Verdin E., Greene W.C. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 2006;25:139–149. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Omene C., Karkoszka J., Bosland M., Eckard J., Klein C.B., Frenkel K. Caffeic acid phenethyl ester (CAPE), derived from a honeybee product propolis, exhibits a diversity of anti-tumor effects in pre-clinical models of human breast cancer. Cancer Lett. 2011;308:43–53. doi: 10.1016/j.canlet.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurster A.L., Precht P., Becker K.G., Wood W.H., Zhang Y., III, Wang Z., Pazin M.J. IL-10 transcription is negatively regulated by BAF180, a component of the SWI/SNF chromatin remodeling enzyme. BMC Immunol. 2012;13:9. doi: 10.1186/1471-2172-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H., Zhang Y., Zhou X., Qu X., Wang P., Liu S., Lu D., Zhu H. Selective histonedeacetylase inhibitor M344 intervenes in HIV-1 latency through increasing histone acetylation and activation of NF-kappaB. PLoS One. 2012;7 doi: 10.1371/journal.pone.0048832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner G.E., Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 2013;20:259–266. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- Zhao W., Sachsenmeier K., Zhang L., Sult E., Hollingsworth R.E., Yang H. A new bliss independence model to analyze drug combination data. j. biomol. screen. 2014;19:817–821. doi: 10.1177/1087057114521867. [DOI] [PubMed] [Google Scholar]
- Zhao W.X., Wang L., Yang J.L., Li L.Z., Xu W.M., Li T. Caffeic acid phenethyl ester attenuates pro-inflammatory and fibrogenic phenotypes of LPS-stimulated hepatic stellate cells through the inhibition of NF-kappaB signaling. Int. J. Mol. Med. 2014;33:687–694. doi: 10.3892/ijmm.2013.1613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.