Abstract
Astrocytes and neurons form a highly specialized functional unit, and the loss or gain of astrocytic functions can influence the initiation and progression of different neurodegenerative diseases. Neurons depend on the antioxidant protection provided by neighboring astrocytes. Glutathione (γ-l-glutamyl-l-cysteinyl-glycine) is a major component of the antioxidant system that defends cells against the toxic effects of reactive oxygen/nitrogen species. A decline in glutathione levels has been observed in aging and neurodegenerative diseases, and it aggravates the pathology in an amyotrophic lateral sclerosis-mouse model. Using a SILAC-based quantitative proteomic approach, we analyzed changes in global protein expression and lysine acetylation in primary astrocyte cultures obtained from wild-type mice or those deficient in the glutamate-cysteine ligase modifier subunit (GCLM). GCLM knockout astrocytes display an ∼80% reduction in total glutathione levels. We identified potential molecular targets and novel sites of acetylation that are affected by the chronic decrease in glutathione levels and observed a response mediated by Nrf2 activation. In addition, sequence analysis of peptides displaying increased acetylation in GCLM knockout astrocytes revealed an enrichment of cysteine residues in the vicinity of the acetylation site, which suggests potential crosstalk between lysine–acetylation and cysteine modification. Regulation of several metabolic and antioxidant pathways was observed at the level of protein expression and lysine acetylation, revealing a coordinated response involving transcriptional and posttranslational regulation.
Astrocytes represent the largest cell population in the central nervous system (CNS). In addition to providing structural, metabolic, and trophic support to neurons, astrocytes influence neuronal excitability and integrate and process synaptic information defining functional compartments (1–3). In the CNS of higher vertebrates, astrocytes respond to all forms of injury such as trauma, infection, ischemia, and neurodegenerative processes in a typical manner known as reactive astrogliosis (4, 5). Since reactive astrogliosis occurs in all major neurodegenerative diseases, it has long been suggested to be a nonspecific response of glial cells to injury and often is not considered as a primary pathogenic element. However, in the last ten years, it has been shown that altered astrocytic functions can influence the initiation and progression of neurodegenerative diseases, and therefore, astrocytes have emerged as a potential therapeutic target (6, 7).
Oxidative and nitrative stress play a critical role in the pathogenic mechanisms of neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS)1, Alzheimer's disease, Parkinson's disease, and Huntington's disease (8). Astrocytes play a crucial role in the antioxidant defense of the CNS, determining the vulnerability of neurons to noxious stimuli (9). Neurons express antioxidant scavenger/enzymes at low levels, and their survival depends on the antioxidant protection provided by neighboring astrocytes (10). Glutathione (γ-l-glutamyl-l-cysteinyl-glycine) is a key component of the antioxidant system that defends cells against the toxic effects of reactive oxygen/nitrogen species and provides a reducing environment within the cells. Although glutathione is widely distributed in all animal tissues, the concentration of glutathione in neurons is lower than that found in astrocytes (11). Furthermore, glutathione synthesis in neurons depends on their metabolic interaction with astrocytes since it relies on the supply of cysteine or a cysteine-precursor (including glutathione) secreted from astrocytes (12). Glutathione is synthesized by the sequential action of the enzymes glutamate-cysteine ligase (GCL) and glutathione synthetase (13). GCL is a heterodimer composed of a catalytic subunit (GCLC) and a modifier subunit (GCLM), and catalyzes the formation of γ-glutamylcysteine, the rate-limiting reaction in glutathione synthesis. While the lack of GCLC is lethal, GCLM knockout mice [GCLM(−/−)] are viable but display a reduction of 70–80% in total glutathione content when compared with wild-type littermates (14, 15).
A decline in glutathione levels has been documented during aging and in different neurodegenerative diseases (16, 17). We have previously shown that a decrease in glutathione levels aggravates mitochondrial pathology and accelerates the disease in mutant human superoxide dismutase 1 (hSOD1G93A) overexpressing mice, an ALS-mouse model (15). In the present study, we analyzed the effect of a chronic decrease in glutathione content in GLCM(−/−) astrocytes using quantitative proteomics in order to identify molecular targets affected by decreased antioxidant defenses. Moreover, since the reversible acetylation of the ε-amino group of lysine residues regulates diverse cellular functions, including the response to oxidative stress (18–24), we evaluated the effect of decreased glutathione levels on the acetylome profile of GLCM(−/−) astrocytes.
EXPERIMENTAL PROCEDURES
Materials
Culture media, fetal bovine serum, amino acids, and supplements were obtained from Thermo Scientific-Pierce. The following primary antibodies were used: rabbit anti-ACL (Bethyl Laboratories, Inc.); mouse anti-β-actin (clone AC-15; Sigma-Aldrich); rabbit anti-catalase (Thermo Scientific); rabbit anti-calmodulin (clone EP799Y; Abcam); rabbit anti-GCLC (Thermo Scientific); rabbit anti-G6PD (Thermo Scientific); rabbit anti-GPD2 (clone EPR14259; Abcam); rabbit anti-Nrf2 (clone D1Z9C; Cell Signaling); rabbit anti-PGD (Thermo Scientific); and mouse anti-TATA binding protein (Abcam). All other reagents were from Sigma-Aldrich unless otherwise specified.
Animals
Male and female C57BL/6J-GCLM(±) mice (14) were crossed to generate wild-type and GCLM(−/−) littermates. All animal procedures were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The Animal Care and Use Committee of the Medical University of South Carolina (Animal Welfare Assurance number A3428–01) approved the animal protocol pertinent to the experiments reported in this publication.
Primary Astrocyte Cultures
Primary astrocyte cultures were prepared from the cortex of 1 to 2-day-old mice as previously described (25). Astrocyte cultures were metabolically labeled using heavy or light isotopes of lysine from the moment the culture was established. Astrocytes were plated at a density of 2 × 104 cells/cm2 and maintained in SILAC Dulbecco's modified Eagle's media supplemented with NaHCO3 (1.2 g/l), HEPES (3.6 g/l), l-leucine (105 mg/l), l-arginine (84 mg/l), l-lysine 12C6-14N2 (light) or l-lysine-13C6-15N2 (heavy) (100 mg/l), penicillin (100 IU/ml), streptomycin (100 μg/ml), and 10% dialyzed fetal bovine serum. We performed three independent biological replicate experiments. In two of them, GCLM(+/+) (wild-type) astrocytes were cultured in “light” SILAC media supplemented with l-lysine 12C6-14N2, while GCLM(−/−) astrocytes were cultured in “heavy” SILAC media containing l-lysine-13C6-15N2. As a control, in the third experiment, a label swap was used. Astrocyte monolayers were >98% pure as determined by glial fibrillary acidic protein (an astrocytic marker) immunoreactivity and devoid of microglial cells (as reflected by the absence of CD11b-positive cells).
Sample Preparation for Mass Spectrometric Analysis
Confluent astrocyte monolayers were washed twice with PBS and lysed in Tris-HCl buffer, pH 7.6, supplemented with 2 mm EDTA, 150 mm NaCl, 1% Triton X-100, 0.25% Nonidet P-40, complete protease inhibitor mixture EDTA-free (Roche Applied Science), 10 mm nicotinamide, and 1 μm trichostatin A. After sonication, samples were centrifuged at 4 °C for 10 min at 10,000 g. Protein concentration was determined by the bicinchoninic acid method (BCA protein assay; Thermo Scientific).
For differential protein expression analysis, 40 μg aliquots of combined heavy and light labeled protein (1:1 ratio) from each biological replicate were electrophoresed in three lanes of a 4–12% Criterion-XT gel (BioRad). After Coomassie staining, the lanes were cut into 12 bands. Proteins in each gel piece were reduced, alkylated, digested with Lys-C and trypsin, and desalted by C18 ZipTip (EMD Millipore), as previously described (26), prior to LC-MS/MS analysis.
To prepare peptides for differential lysine acetylation analysis, samples were processed as described by Choudhary et al. (27), with minor modifications. Briefly, 10 mg of heavy and 10 mg of light labeled protein were combined and precipitated by adding 4X excess volumes of HPLC-grade acetone following by overnight incubation at −20 °C. Protein pellets were redissolved in 20 mm HEPES buffer, pH 8.0, supplemented with 6 m urea and 2 m thiourea. Samples were then reduced by incubation with 1 mm dithiothreitol for 45 min at room temperature and alkylated with 5.5 mm iodoacetamide for 30 min in the dark at room temperature. After 4.5-fold dilution in water, the protein was digested with Lys-C (1:100 w/w; overnight at room temperature) followed by trypsin digestion (1:100 w/w; overnight at room temperature). After quenching protease activity by acidification, samples were desalted using reversed-phase Sep-Pak C18 peptide purification cartridges (GE Healthcare). Peptides were separated into 12 fractions by strong cation exchange chromatography on a 6.4 mm × 30 mm Resource S methylsulfonate column (15 μm, GE Healthcare) at a flow rate of 1 ml/min using a gradient of 0% B for 10 min; 0–30%B in 30 min; 30–100% B in 5 min, and 100% B for 10 min (Buffer A: 5 mm KH2PO4 (pH 2.7)/30% acetonitrile; Buffer B: 5 mm KH2PO4 (pH 2.7)/350 mm KCl/30% acetonitrile). Acetylated peptides were immunoprecipitated from each fraction.
Immunoprecipitation of Lysine-Acetylated Peptides
For enrichment of lysine-acetylated peptides, strong cation exchange fractions were dried under vacuum and redissolved in immunoprecipitation buffer (50 mm Tris-HCl, pH 8.0; 100 mm NaCl; 1 mm EDTA; 0.5% Nonidet P-40). Peptides were incubated overnight at 4 °C with antiacetylated lysine antibody conjugated to agarose beads (ImmuneChem). Immune complexes were then washed 3X with immunoprecipitation buffer and 3X with water. Immnoprecipitated peptides were then eluted with 0.1% (v/v) trifluoroacetic acid in water.
LC-MS/MS Analysis
Peptides were loaded on a trap column, eluted, and separated on a 75 μm × 15 cm fused-silica column packed with C18 reversed-phase resin (YMC-ODS-AQ; 5-μm particles; 200-Å pore; Waters). Peptides were separated using an acetonitrile gradient of 5–50% in 120 or 180 min containing 0.2% formic acid on a Dionex Ultimate 3000 nano LC system at flow rate of 200 nl/min. The sample was introduced via nanoelectrospray ionization using an uncoated silica emitter (360 μm outer diameter, 50 μm inner diameter, with a 5 μm tip, New Objective) to a hybrid dual-pressure linear ion trap–orbitrap mass spectrometer (Orbitrap Elite, Thermo Scientific). Mass spectra were acquired in data-dependent mode using a TOP20 method that acquires one FTMS survey MS scan with a mass range of m/z 400–1700 followed by tandem mass spectra (MS/MS) of the 20 most-intense ions in the ion trap. The automatic gain control target value in the Orbitrap was 106 ions, for the survey MS scan the resolution was 60,000 at m/z 400. A lock-mass was used for recalibration during data processing (28). Fragmentation in the ion trap was performed by collision-induced dissociation with a target value of 10,000 ions and a threshold of 500 counts. Three biological replicate experiments were performed. For protein expression analysis, 12 LC-MS/MS analyses per replicate were performed (36 LC-MS/MS analyses). These analyses were performed sequentially under identical conditions to enable peptide matching between chromatographic separations (match between runs). For acetyl peptide analysis, 12 LC-MS/MS analyses were performed for each biological replicate. The first biological replicate was analyzed three times, and the remaining biological replicates were analyzed twice each (84 LC-MS/MS analyses). The LC-MS/MS data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD001834.
Database Searching and Filtering
LC-MS/MS data were searched using the Andromeda algorithm in MaxQuant 1.5.0.30 (MQ) (29). To correct for drift in the calibration during data acquisition, the recalibration option was selected in which a first database search was performed at 20 ppm precursor mass tolerance. Following recalibration of the m/z ratios, the search was repeated with a more stringent mass tolerance of 4.5 ppm for precursor ions. To search for missing SILAC partners after the initial database search, the “requantify” option was employed. Two separate searches were performed to quantitate changes in protein expression and lysine acetylation. LC-MS/MS analyses of acetylated peptides enriched from strong cation exchange fractions were searched simultaneously against a UniProt murine database downloaded on May 16, 2014 containing 82,538 entries and commonly observed potential contaminants. A reversed decoy database (reward) was used to determine the peptide false discovery rate and minimize false assignment of modifications at the peptide N terminus. The false discovery rates for peptide spectral matches, protein identification, and modification site assignment were each set to 1%. The search parameters allowed for two missed cleavages, fragment mass tolerances ± 0.5 Da and a minimum of seven amino acids per peptide. Dynamic modifications of acetyl (protein N-term and Lys), oxidation of Met, cyclization of peptide N-terminal Glu to pyro-Glu, and static cysteine modification with carbamidomethyl were used. A minimum Andromeda score and delta score of 40 and 6, respectively, were required for identification of modified peptides. The site of lysine acetylation was assigned if the acetyl score difference was >5, and the probability of site localization was above 95%. MaxQuant annotated spectra corresponding to the best Andromeda score for each peptide from the acetyl(K) site table were visually inspected. With the exception of unlabeled neutral losses from the precursor ion, acetyl peptide spectra with multiple intense unassigned peaks were omitted. The msms.txt file from MaxQuant was uploaded to enable viewing of the spectra in MS-Viewer function of Protein Prospector (University of California, SanFrancisco).
Similar parameters were used to search the gel-LC-MS/MS spectra for changes in protein expression with the exceptions that identifications within a 1 min window were matched between runs, and a reversed decoy database (revert) was utilized. For protein identification, one unique peptide was required. Protein quantitation was based on lysine containing unique and razor peptides that were unmodified or carbamidomethylated.
The search results were further analyzed and filtered using Perseus v. 1.5.0.15 (Max Planck Institute) with site-specific modification databases downloaded in July 2015 from PhosphoSitePlus (www.phosphosite.org) (30). Protein expression data were extracted from the protein groups, and evidence tables and the acetylation data were extracted from the acetyl(K)sites and evidence tables. Tables were filtered to remove potential contaminants and matches to the reversed database. Normalized ratios from the label swap experiments were inverted to yield experimental/control and all ratios were log2 transformed. Potential contaminants exhibiting ratios equal and opposite between the reversed and forward reactions as revealed by the label swap control were removed.
Statistical Rationale
Log2 transformed ratios were used to calculate a moderated t-statistic in order to determine a p value for each protein or “mono-acetylated” peptide with ratios measured in at least two experiments. To adjust the changes in lysine acetylation by potential changes in protein expression, we corrected each Log2 acetyl peptide ratio by subtracting the mean Log2 ratio of the corresponding protein if it was measured in at least two experiments. For acetylated peptides, this p value was adjusted for multiple hypotheses testing by the Benjamini and Hochberg method (31) with a false discovery rate ≤ 0.1 as previously described by Udeshi et al. (32). Downstream analyses were performed on regulated acetylation sites, including those that could not be normalized by protein expression as these peptides represent “nodes” of regulation.
Pathway Analysis
Ingenuity® Pathway Analysis software (Qiagen) was used to determine the significantly enriched canonical pathways associated with the datasets of regulated protein and lysine-acetylated peptides. The significance of the overrepresented canonical pathways was determined using the Fisher Exact test p value and the enrichment ratio, i.e. number of hits in a pathway divided by the total number of proteins in the pathway.
Peptide Consensus Sequence Analysis
Peptide sequence analysis was performed by iceLogo (33) using the static reference method, with the Mus musculus Swiss-Prot database or the complete list of identified acetylated peptides as reference for background correction. The analysis was visualized using heat maps (p ≤ .05) and iceLogo figures with percentage difference as the scoring system (p ≤ .05).
Immunoprecipitation, Nuclear Extracts, and Western Blot Analysis
To validate the quantification of the SILAC analysis, proteins extracts were prepared as described above from independent astrocyte cultures maintained in Dulbecco's modified Eagle's media supplemented as previously described (25). For immunoprecipitation, 1 mg of protein extracts in lysis buffer (described above) were incubated overnight at 4 °C with rabbit antiacetylated lysine antibody (Cell Signaling). After adding protein A, magnetic particles (Polysciences, Inc.), the incubation was extended for 1 h. After washing three times with lysis buffer, immunoprecipitated proteins were eluted by incubation for 5 min at 95 °C in 2X concentrated reducing Laemmli sample buffer (BioRad). Immnoprecipitated proteins or protein extracts (20 μg) were resolved on sodium dodecyl sulfate-polyacrylamide gels and transferred to nitrocellulose membranes using the Trans-Blot Turbo transfer system (BioRad). Nuclear fractions were prepared as previously described (34), resolved in 4–15% Mini-PROTEAN® TGX™ gels (BioRad) and transferred to nitrocellulose membranes as described above. Membranes were blocked for 1 h in Tris-buffered saline (TBS), 0.1% Tween-20, and 5% bovine serum albumin, followed by an overnight incubation at 4 °C with the primary antibody diluted in the same buffer. After washing with 0.1% Tween-20 in TBS, membranes were incubated with horseradish peroxidase-conjugated secondary antibody (GE Healthcare) for 1 h at room temperature. For immunodetection of immunoprecipitated proteins, membranes were incubated for 1 h at room temperature with clean blot IP detection reagent (HRP) (Thermo Scientific) or peroxidase-conjugated goat anti-mouse IgG light chain specific antibody (Jackson ImmunoResearch Laboratories, Inc.). Membranes were then washed and developed using the ECL Prime chemiluminescent detection system (GE Healthcare-Amersham Biosciences). Membranes were imaged and quantified using the C-DiGit Blot Scanner (LI-COR Biosciences). For immunoblots of cell extracts, protein loading was corrected by β-actin levels. Comparison between groups was performed by Student's t test using GraphPad Prism 6.0 (GraphPad Software). Differences were declared statistically significant if p ≤ .05.
Glutathione Measurement
Total glutathione levels (GSH and GSSG) were determined using the Tietze method as previously described (25). Glutathione content was corrected by protein concentration determined by BCA protein assay (Thermo Scientific).
Real-Time PCR
Total RNA was isolated from confluent astrocyte monolayers using TRI Reagent. 2 μg were randomly reverse transcribed using SuperScript II reverse transcriptase (Life Technologies). PCRs were carried out in a StepOnePlus Real-Time PCR System (Life Technologies), in a 20 μl reaction with 1X SYBR® Green PCR Master Mix (Life Technologies) containing 1 μl of cDNA and 20 pmoles of each specific primer. The cycling parameters were as follows: 95 °C, 15 s; 60 °C, 15 s; 72 °C, 30 s. Minus reverse transcriptase controls were included in each assay. The following specific primers were used: G6pdx: 5′-GAAGCTGCCAATGGATACTTAGA-3′, 5′-CCACCGTTCATTCTCCACATAG-3′; Pgd: 5′-AGGACATGGTCTCCAAACTAAAG-3′, 5′-CGTGTCCAACAAGGGTACTAAT-3′; Catalase: 5′-TGAAGACAATGTCACTCAGGTGCG-3′, 5′-ATGTTCTCACACAGGCGTTTCCTC-3′. Gclc and actin primers sequences were previously described (35).
RESULTS
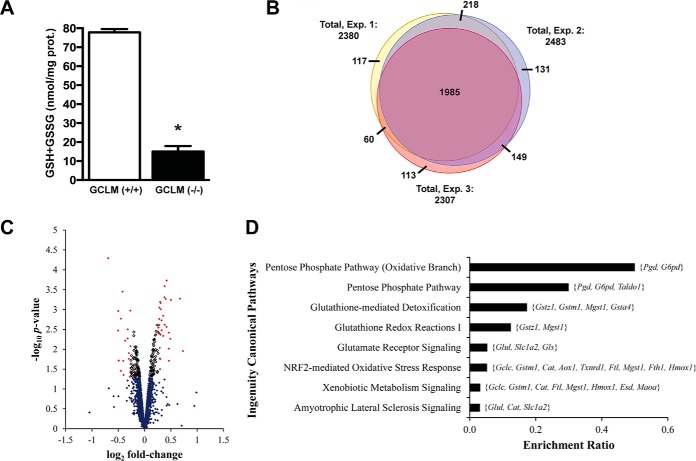
We used SILAC-based quantitative proteomics to identify changes in global protein expression and lysine acetylation induced by chronic decreased glutathione content in astrocytes. The experiments were conducted in primary cortical astrocyte cultures obtained from wild-type or knockout mice lacking the glutamate-cysteine ligase modifier subunit [GCLM(−/−)], which exhibit ∼80% reduction in total glutathione levels (Fig. 1A). We performed three independent biological replicate experiments, including a label swap control. To ensure sufficient labeling, primary astrocytes were cultured in media containing the isotope-encoded lysine from the moment the culture was established. Liquid chromatography-tandem mass spectrometric analysis (LC-MS/MS) confirmed that following 21 days in vitro, protein from primary astrocyte cultures displayed above 97.5% incorporation of isotopically labeled lysine.
Fig. 1.
Changes in protein expression induced by decreased glutathione levels in astrocytes. (A) Total glutathione (GSH+GSSG) content in primary GCLM(+/+) and GCLM(−/−) astrocyte cultures. Each bar represents the mean ± S.D. *Significantly different from GCLM(+/+) (p ≤ .05). (B) Venn diagram showing the overlap of protein groups quantified in the three independent experiments (Exp. 1–3). Exp. 2 corresponds to the experiment in which the SILAC label swap was introduced. (C) Volcano plot of the proteins quantified in at least two independent experiments. eBayes moderated t-statistic was used to determine a p value for each protein. Significantly regulated proteins (p ≤ .05) in GCLM(−/−) astrocytes are shown as empty boxes, while those with an absolute fold-change of 1.2 are shown in red. (D) Major ingenuity canonical pathways overrepresented by proteins differentially expressed in GCLM(−/−) astrocytes (1.2-fold cutoff). Members of each pathway that were present in the dataset of regulated proteins are indicated on the graph (gene symbol). Significance was established at BH adjusted p ≤ .05.
To determine changes in protein expression induced by chronic decreased glutathione levels, SILAC labeled protein from wild-type and GCLM(−/−) astrocytes was combined (1:1 ratio), fractionated by gel electrophoresis, and the in-gel digested peptides were analyzed by LC-MS/MS. A total of 4,526 protein groups were identified, of which 2,773 were quantified using MaxQuant (Supplemental Table I). Approximately 87% were observed in at least two biological replicates (Fig. 1B). Proteins with a 1.2-fold change and a p ≤ .05, as assessed by a moderated t test, were considered significantly regulated. Using these criteria, 52 proteins were differentially expressed in GCLM(−/−) astrocytes (Fig. 1C, Supplemental Table II). Most of the differentially expressed proteins were up-regulated. Among the 19 down-regulated proteins are the glutamate transporter, excitatory amino acid transporter 2 (EAAT2; 0.75-fold change, p = .00036), and the enzyme glutamine synthetase (GS; 0.71-fold change, p = .0011), key components of the glutamate–glutamine shuttle and critical for the regulation of synaptic transmission (36–38).
Pathway analysis of the regulated proteins revealed significant enrichment (p ≤ .05) of the pentose phosphate pathway, glutathione metabolism pathways, glutamate receptor-signaling, and amyotrophic lateral sclerosis (ALS) signaling (Fig. 1D). Another canonical pathway significantly overrepresented is the nuclear factor erythroid-2-related factor 2 (Nrf2)-mediated oxidative stress response. As depicted in Table I, of the 33 proteins that we observed up-regulated, 16 are known targets of Nrf2. The induction of phase II detoxifying and antioxidant enzymes depends almost exclusively on the activation of the transcription factor Nrf2 (9). In addition, Nrf2 also regulates the intermediary metabolism by inhibiting lipogenesis, increasing beta-oxidation of fatty acids and facilitating the flux through the pentose phosphate pathway (PPP) (39). The increased flux rate through the PPP allows the cell to regenerate the pool of NADPH necessary for recycling oxidized glutathione to reduced glutathione and for the catalytic activity of many drug-metabolizing and antioxidant enzymes that are up-regulated by Nrf2 itself (39). Since GCLM(−/−) astrocytes retain about 20% of the glutathione levels, an increase in NADPH could facilitate the maintenance of the remaining small pool of glutathione in a reduced form and available for detoxification processes. It has been shown in other cellular models that Nrf2 regulates the expression of all the NADPH-generating enzymes, i.e. G6PD, 6PGD, malic enzyme 1 (ME1), and isocitrate dehydrogenase 1 (IDH1) (39, 40). In GCLM(−/−) astrocytes, we observed increased expression of G6PD, 6PGD (Fig. 2 and Supplemental Table II), and ME1 (although the expression of ME1 showed a 1.16-fold change (p = .004) and did not pass our 1.2-fold cutoff). While we did not detect a change in IDH1 expression, we observed a 3.03-fold increase in the acetylation status of this enzyme (see below, Lys243; BH adjusted p = .0005).
Table I. Proteins coded by Nrf2-driven genes found differentially expressed in GCLM(−/−) astrocytes.
| Gene symbol | Protein name | Protein ID | Sequence coverage (%) | Mean Log2 ratio GCLM(−/−)/ GCLM(+/+) | p value |
|---|---|---|---|---|---|
| Aox1 | Aldehyde oxidase | Q8R387 | 15.6 | 0.46 | .00384 |
| Cat | Catalase | P24270 | 71.5 | 0.38 | .00049 |
| Fech | Ferrochelatase, mitochondrial | Q91W09 | 23.3 | 0.28 | .00393 |
| Ftl1; Ftl2 | Ferritin; Ferritin light chain 1/2 | Q9CPX4 | 60.7 | 0.33 | .00334 |
| Fth1 | Ferritin heavy chain | P09528 | 76.4 | 0.28 | .00090 |
| Gclc | Glutamate-cysteine ligase catalytic subunit | P97494 | 54.2 | 0.42 | .00019 |
| Gls | Glutaminase kidney isoform, mitochondrial | D3Z7P3 | 24.2 | 0.30 | .01781 |
| Gsta4 | Glutathione S-transferase A4 | P24472 | 41.4 | 0.27 | .00251 |
| Gstm2 | Glutathione S-transferase Mu 2 | P15626 | 83 | 0.26 | .00286 |
| G6pdx | Glucose-6-phosphate 1-dehydrogenase | Q790Y8 | 52.8 | 0.40 | .00055 |
| Hmox1 | Heme oxygenase 1 | Q3U5H8 | 71.6 | 0.67 | .00054 |
| Mgst1 | Microsomal glutathione S-transferase 1 | Q91VS7 | 32.3 | 0.28 | .00173 |
| Pgd | 6-phosphogluconate dehydrogenase, decarboxylating | Q9DCD0 | 71.6 | 0.31 | .00069 |
| Ptgr1 | Prostaglandin reductase 1 | Q91YR9 | 40.1 | 0.29 | .01707 |
| Taldo1 | Transaldolase | Q93092 | 45.4 | 0.28 | .00246 |
| Txnrd1 | Thioredoxin reductase 1, cytoplasmic | Q9JMH6–2 | 65.5 | 0.50 | .00058 |
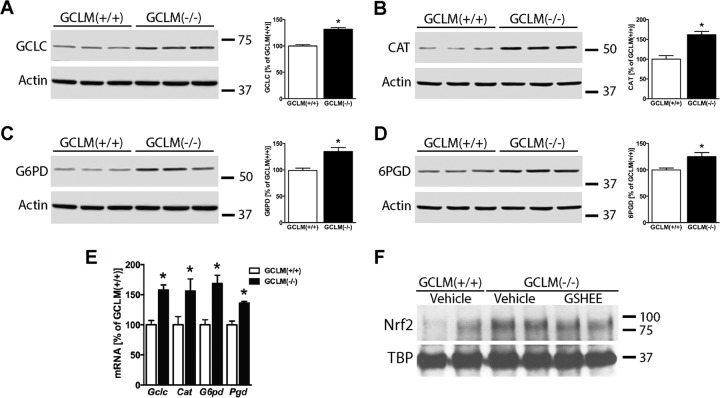
Fig. 2.
Confirmation of differential protein expression by immunoblot analysis. Cell lysates from three independent GCLM(+/+) and GCLM(−/−) astrocyte cultures were analyzed by Western blot to determine the protein expression levels of (A) glutamate-cysteine ligase catalytic subunit (GCLC); (B) catalase (CAT); (C) glucose-6-phosphate dehydrogenase (G6PD); and (D) 6-phosphogluconate dehydrogenase (6PGD). Protein loading was corrected by β-actin levels. The membrane in (A) was stripped and reprobed for G6PD (C) while membrane in (B) was stripped and reprobed for 6PGD (D). Hence, β-actin panels for A and C and B and D, respectively, are the same. Densitometric analysis is expressed as percentage of GCLM(+/+). *Significantly different from GCLM(+/+) (p ≤ .05). (E) Total RNA was extracted from GCLM(+/+) or GCLM(−/−) confluent astrocyte monolayers and GCLC, CAT, G6PD, and PGD mRNA levels were determined by real-time PCR and corrected by actin mRNA levels. mRNA levels are expressed as percentage of GCLM(+/+). *Significantly different from GCLM(+/+) (p ≤ .05). (F) Nuclear fractions were prepared from confluent GCLM(+/+) or GCLM(−/−) astrocyte cultures treated for 48 h with vehicle or glutathione ethyl ester (GSHEE; 5 mm). To determine the level of nuclear Nrf2, samples were analyzed by Western blotting using specific antibodies. As a loading control, membranes were stripped and reprobed for TATA-binding protein (TBP).
To confirm the changes in protein expression detected in the SILAC analysis by an alternative method, the abundance of four representative proteins corresponding to the most overrepresented canonical pathways was determined in three independent biological replicates by immunoblotting using specific antibodies. Figures 2A-2D validate up-regulation of glutamate-cysteine ligase catalytic subunit (GCLC; 1.32-fold change), catalase (1.62-fold change), glucose-6-phosphate dehydrogenase (G6PD; 1.35-fold change), and 6-phosphogluconate dehydrogenase (6PGD; 1.25-fold change) in GCLM(−/−) astrocytes. The fold changes determined by densitometric analysis of the immunoblots were consistent with the corresponding SILAC ratios, which further strengthens the SILAC-based quantification. Increased expression of these proteins correlated with increased mRNA levels of the respective genes in GCLM(−/−) astrocytes (Fig. 2E). Up-regulation at the transcriptional level further supports the activation of the Nrf2-mediated oxidative stress response evidenced by pathway analysis (Fig. 1D). Upon activation, Nrf2 translocates from the cytosol to the nucleus, where it activates the transcription of antioxidant response element-driven genes (39). To confirm the activation of Nrf2, we determined its levels in nuclear fractions obtained from GCLM(+/+) and GCLM(−/−) astrocyte cultures. Compared with GCLM(+/+), GCLM(−/−) astrocytes displayed increased levels of Nrf2 in the nucleus (Fig. 2F). Moreover, treatment of GCLM(−/−) astrocytes for 48 h with glutathione ethyl ester partially reverted Nrf2 nuclear accumulation (Fig. 2F).
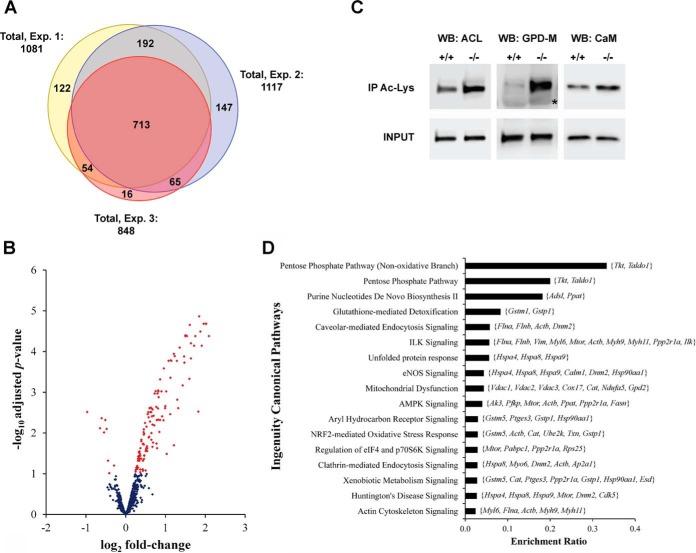
Since lysine acetylation has emerged as a key posttranslational modification affecting diverse cellular functions, including the response to oxidative stress (18–24), we hypothesized that chronic glutathione deficiency could alter the acetylome profile of GCLM(−/−) astrocytes. To identify proteins differentially acetylated, SILAC-labeled proteins from control and GCLM(−/−) astrocytes were combined at 1:1 ratio, digested, and fractionated by strong cation exchange. After enrichment of the acetylated peptides by immunoprecipitation using an anti-acetyl-lysine antibody, a total of 1,317 acetylated peptides were identified by LC-MS/MS (Supplemental Table III). The MaxQuant annotated spectra (41) are provided in Supplemental Fig. 1 and can be accessed in an interactive format in MS-Viewer (42) at http://prospector2.ucsf.edu/prospector/cgi-bin/msform.cgi?form = msviewer using the search key: nhh2vn348o. We identified 367 novel acetylation sites, not previously reported in the mouse proteome in PhosphoSitePlus (July 2015) (30) or observed in databases retrieved with Perseus 1.5.0.15. Of the 1,317 sites of acetylation identified, 381 are known sites of ubiquitinylation.
Approximately 78% of the acetylation sites were observed in at least two biological replicates (Fig. 3A), and the ratios of acetylated peptides observed were reproducible between experiments as evidenced by the Pearson's correlation coefficients (Supplemental Fig. 2). After correcting for changes in protein expression and filtering by a Benjamini–Hochberg corrected p value ≤ .1, we identified 132 regulated sites of lysine acetylation (Supplemental Table IV). Tandem mass spectra of regulated sites of acetylation were manually inspected (43, 44). and the annotated spectra are provided in Supplemental Fig. 3. As depicted in the volcano plot in Fig. 3B, only nine sites displayed decreased acetylation in GLCM(−/−) astrocytes. Two of them correspond to glutathione S-transferase Mu isoforms, which are involved in glutathione metabolism. Thirty-seven novel sites of acetylation were regulated in GCLM(−/−) astrocytes (see Supplemental Table IV). Although not reported in the mouse proteome, 27 of the regulated novel sites have been previously reported in the human or rat protein counterpart.
Fig. 3.
Changes in protein lysine-acetylation induced by chronic decreased glutathione levels in astrocytes. (A) Venn diagram showing the overlap of the acetylated sites quantified in the three independent experiments (Exp. 1–3). Exp. 2 corresponds to the experiment in which the SILAC label swap was introduced. (B) Volcano plot of the acetylation sites quantified in at least two independent experiments. Regulated sites in GCLM(−/−) astrocytes, as determined using moderated t test with a Benjamini–Hochberg corrected p value ≤ .1, are shown in red. (C) Representative images confirming regulated lysine acetylation by immunoprecipitation and Western blot analysis. Lysine-acetylated proteins were immunoprecipitated from protein extracts obtained from three independent GCLM(+/+) and GCLM(−/−) astrocyte cultures and then analyzed by Western blot analysis using antibodies against ATP-citrate synthase (WB: ACL); glycerol-3-phosphate dehydrogenase (mitochondrial) (WB: GPD-M); and calmodulin (WB: CaM). Membranes were stripped for reprobing with the different antibodies. 20 μg of whole protein extracts were analyzed in the same gel as input control (INPUT). The asterisk corresponds to the heavy chain of the IgG used for immunoprecipitation. (D) Major ingenuity canonical pathways overrepresented by proteins with regulated acetylation sites in GCLM(−/−) astrocytes. Members of each pathway that were present in the dataset of regulated sites are indicated on the graph (gene symbol). Significance was established at BH adjusted p ≤ .05.
To validate the changes in the level of lysine-acetylation detected by the SILAC analysis, we performed immunoprecipitation in independent biological samples using a pan-specific antibody against acetylated lysine, followed by Western blot analysis of selected candidates. Figure 3C shows representative Western blot images confirming increased acetylation of ATP-citrate synthase (ACL), glycerol-3-phosphate dehydrogenase 2 (mitochondrial) (GPD-M), and calmodulin.
Pathway enrichment analysis of the regulated acetylated proteins also revealed statistically significant overrepresentation (p ≤ .05) of the pentose phosphate pathway, glutathione-mediated detoxification, xenobiotic metabolism signaling, and Nrf2-mediated oxidative stress response (Fig. 3D). Remarkably, the analysis also identified overrepresentation of the purine nucleotides de novo biosynthesis II pathway, in which one of the products of the pentose phosphate pathway, ribose-5-phosphate, through the formation of 5-phospho-α-d-ribosyl-1-pyrophosphate, is converted to AMP or GMP.
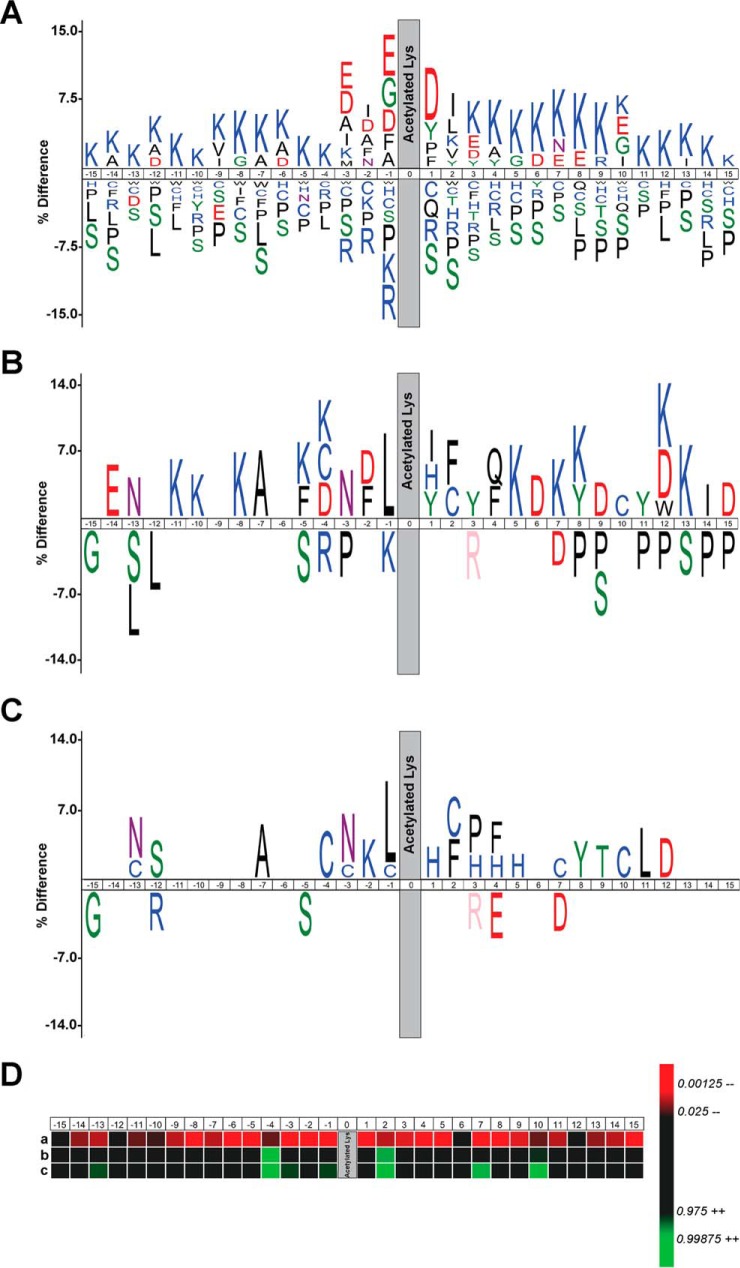
Using iceLogo (33), we analyzed the sequence of the 1,317 acetylated peptides identified and compared it to the average amino acid occurrence for the mouse Swiss-Prot proteome. As previously reported by Lundby et al. (45), we observed that the acetylation occurs in lysine-rich regions with a preference for negatively charged residues around the modified site, glycine residues at position –1, proline/tyrosine/phenylalanine residues at position +1, and isoleucine/leucine/valine residues at position +2 (Fig. 4A). As previously observed (45), we also detected an extensive underrepresentation of cysteine residues surrounding the acetylation site (Figs. 4A and 4D). However, when we analyzed the sequences of the peptides displaying increased acetylation in GCLM(−/−) astrocytes using the mouse Swiss-Prot proteome as background, we observed that the acetylation occurs in lysine-rich regions with an overrepresentation of cysteine residues at positions –4, +2, and +10 (Figs. 4B and 4D). Moreover, when the peptides displaying increased acetylation were analyzed against the total 1,317 acetylated peptides identified in our dataset, again we found a preference for cysteine residues at several positions in the vicinity of the acetylation site, mainly at position –4, +2, and +10 but also in positions +7, –1, –3, and –13 (Figs. 4C and 4D).
Fig. 4.
Consensus sequence analysis of acetylated peptides by iceLogo. (A) iceLogo generated after analyzing aligned sequence windows of the 1,317 lysine-acetylated peptides identified in the SILAC analysis using the mouse Swiss-Prot proteome as reference set for background correction. (B) iceLogo generated after analyzing the sequences of the peptides exhibiting increased lysine acetylation using the mouse Swiss-Prot proteome for background correction. (C) iceLogo generated after analyzing the sequence of the peptides displaying increased lysine acetylation as compared with all 1,317 lysine-acetylated peptides identified in our dataset. Results are expressed using percentage difference as scoring system (p ≤ .05). (D) Heat map showing the frequency for the presence of a cysteine residue in positions –15 to +15 after alignment of the sequences in the acetylated-lysine residue (position 0). Every column represents a position in the sequence (–15 to +15). Row (a) shows the underrepresentation of cysteine residues observed after the iceLogo analysis described in A (i.e. the set of 1,317 acetylated peptides compared with the mouse Swiss-Prot proteome). Row (b) represents the enrichment of cysteine residues observed after the iceLogo analysis described in B (i.e. peptides with increased acetylation in GCLM(−/−) astrocytes compared with the mouse Swiss-Prot proteome). Row (c) indicates the enrichment of cysteine residues observed after the iceLogo analysis described in C (i.e. peptides with increased acetylation in GCLM(−/−) astrocytes as compared with all acetylated peptides identified in our dataset). Significantly (p ≤ .05) regulated positions are colored in a shade of green (increased) and red (decreased), and the correlation between p values and colors is shown on the right.
DISCUSSION
Glutathione is an essential antioxidant that protects cells against oxidative stress (46). In addition to its important function as an antioxidant, glutathione also participates in the detoxification of xenobiotics, provides a reservoir for cysteine, participates in the maintenance of the redox potential of the cell, maintains the essential thiol status of proteins thus regulating redox-dependent cell signaling, and modulates diverse cellular processes such as DNA synthesis, microtubular-related processes, and immune function (47). Here we show that a chronic decrease in glutathione content in GCLM(−/−) astrocytes induces a response that involves changes in protein expression and lysine acetylation.
The reversible acetylation of the ε-amino group of lysine residues is one of the major posttranslational modifications occurring in different subcellular compartments, including nucleus, cytoplasm, mitochondria, and endoplasmic reticulum (22, 45, 48, 49). This posttranslational modification affects protein function in different ways by altering its conformation, subcellular localization, stability, protein–protein or protein–nucleic acid interactions, and/or enzymatic activity (22, 50). Particularly, many metabolic enzymes are targets of lysine acetylation, and indeed, nearly all enzymes involved in glycolysis, gluconeogenesis, glycogen metabolism, tricarboxylic acid cycle, fatty acid oxidation, urea cycle, and nitrogen metabolism are acetylated (50). Lysine acetylation negatively affects the activity of most metabolic enzymes; however, some metabolic enzymes display increased enzymatic activity following acetylation [e.g. sphingosine kinase 1 (SPHK1); phosphoglycerate mutase 1 (PGAM1); malate dehydrogenase (MDH); enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase (EHHADH); and aldehyde dehydrogenase (ALDH2)] (50). These observations preclude a straightforward interpretation of the functional consequences of the changes in lysine acetylation observed in GCLM(−/−) astrocytes. However, our study identified potential molecular targets that are affected by chronic decreased glutathione levels and revealed a coordinated response involving transcriptional and posttranslational regulation.
Pathway analysis revealed a significant overrepresentation of proteins involved in the Nrf2-mediated oxidative stress response. We have confirmed the activation of Nrf2 in GCLM(−/−) and observed that the translocation of Nrf2 to the nucleus could be partially prevented by increasing glutathione levels, which suggest that Nrf2 activation is a direct consequence of reduced antioxidant defenses. GCLM(−/−) astrocytes displayed increased expression of Nrf2 target genes, including key enzymes of the PPP oxidative-branch, G6PD (the rate-limiting enzyme of the pathway), and 6PGD; and increased expression of transaldolase from the nonoxidative branch, all direct targets of Nrf2 (39, 40). GCLM(−/−) astrocytes also displayed increased acetylation of the nonoxidative branch enzymes, transketolase and transaldolase. These enzymes constitute the reversible link between the PPP and glycolysis, catalyzing the conversion of ribose-5-phosphate into glyceraldehyde 3-phosphate and fructose 6-phosphate. The increased flux through the PPP oxidative branch leads to the production of high levels of ribose-5-phosphate, which can be consequently converted by the activity of 5-phospho-α-d-ribosyl-1-pyrophosphate synthetase to 5-phospho-α-d-ribosyl-1-pyrophosphate, a precursor for both purine and pyrimidine nucleotides biosynthesis. Remarkably, GCLM(−/−) astrocytes also displayed increased acetylation of enzymes from the purine nucleotides de novo biosynthesis II pathway, by which 5-phospho-α-d-ribosyl-1-pyrophosphate is converted to AMP or GMP. We observed increased acetylation of glutamine phosphoribosylpyrophosphate amidotransferase (PPAT; Lys60; 2.83-fold change; BH adjusted p = .00004), the rate-limiting enzyme in purine nucleotide biosynthesis, and increased acetylation of adenylosuccinate lyase (ASL; Lys295; 1.80-fold change; BH adjusted p = .0094), which catalyzes two nonconsecutive steps in the de novo AMP synthesis pathway. Since the levels of AMP can modulate different metabolic processes, the regulation of ASL by acetylation is particularly relevant. Although the functional consequences of increased acetylation of these enzymes is not yet established, posttranslational modulation of these enzymes suggests a fine-tuning of the channeling of metabolites through the PPP, glycolysis and different biosynthetic pathways.
It has been previously shown that in addition to orchestrating glucose entry through the PPP, Nrf2 also plays a critical role in the modulation of glutamine metabolism, redirecting it into anabolic pathways in proliferating cells (40). Glutamine can be partially oxidized, in a pathway known as glutaminolysis, to generate lactate, NADPH and NAD+ in the cytoplasm (51). In this pathway, glutamine is deaminated to generate glutamate, which in turn is converted to α-ketoglutarate, an important intermediate in the tricarboxylic acid cycle, which can be used as an energetic or biosynthetic (anaplerosis) source (51). Glutaminase, the enzyme that catalyzes the hydrolysis of glutamine to glutamate and ammonia is a direct target of Nrf2 (39) and was up-regulated in GCLM(−/−) astrocytes. Moreover, a peptide corresponding to glutamate dehydrogenase (GDH), one of the enzymes able to catalyze the deamination of glutamate to generate α-ketoglutarate, displayed decreased acetylation in GCLM(−/−) astrocytes (Supplemental Table IV). GDH enzymatic activity is negatively regulated by acetylation and it has been shown that deacetylation by sirtuin 3 activates the enzyme (52). Therefore, in GCLM(−/−) astrocytes, glutamine anaplerosis could be further facilitated by the decreased acetylation of GDH. Hence, in culture and in the presence of high levels of exogenous glutamine, glutaminolysis appears as an important bioenergetic and anaplerotic source for GCLM(−/−) astrocytes. This occurs together with a down-regulation of the glutamate transporter EAAT2 and the enzyme glutamine synthetase (Supplemental Table II), key components of the glutamate–glutamine shuttle occurring between astrocytes and neurons. This is particularly relevant due to the critical role of the glutamate–glutamine shuttle in the regulation of synaptic transmission (36–38).
In contrast to the substrate consensus motifs identified for protein phosphorylation, no clear consensus motifs have been described for lysine acetylation. This could be explained by the existence of multiple acetyltransferases with different specificities or the possibility of nonenzymatic acetylation (22, 53–55). However, sequence analysis of different datasets has indicated the prevalence of specific features in the sequence surrounding the acetylated lysine residue (45, 53). Lundby et al. (45) were able to identify, in a subcellular compartment-specific manner, the preference for particular amino acids near the acetylated lysine residues. In all the subcellular compartments analyzed, they observed that the acetylation occurs in lysine-rich regions. However, on nuclear proteins, there was a strong preference for glycine residues in position –1 and proline residues in position +1, whereas on cytoplasmic proteins they observed a clear preference for glutamate residues surrounding the acetylation site. On the other hand, mitochondrial proteins displayed preference for negatively charged residues in the immediate vicinity of the acetylation site and a strong preference for hydrophobic residues (Val/Ile/Leu/Phe) at position +2 (45). We analyzed the sequence of the 1,317 identified acetylated peptides and compared it to the average amino acid occurrence for the mouse Swiss-Prot proteome using iceLogo. We observed that the acetylation sites identified display a similar sequence motif to that described by Lundby et al. (45). However, when we analyzed the sequence of the peptides displaying increased acetylation in GCLM(−/−) astrocytes, we observed that, as previously reported, the acetylated residue occurs in lysine-rich regions, but we also observed a preference for cysteine residues at positions –4, +2, and +10 (Fig. 4B). The preference for cysteine residues was observed at additional positions (+7, –1, –3, and –13) when we used our dataset of identified acetylated peptides for background correction (Fig. 4C). Remarkably, when our entire dataset was analyzed against the mouse Swiss-Prot proteome, an underrepresentation of cysteine residues in almost all the positions surrounding the acetylated lysine residue was observed. These observations further strengthen the relevance of the overrepresentation of cysteine residues in the sequence of differentially acetylated peptides and suggest a potential specific regulation by chronic glutathione deficiency.
The ability of oxidative stress to positively regulate the acetylation status of two transcription factors, FoxO4 and Sp1, has been previously reported (56, 57). Moreover, evidence for both, positive and negative crosstalk between lysine acetylation and other posttranslational modifications, including phosphorylation, methylation, and ubiquitination, has also been previously described (20, 58). Our study revealed a striking overrepresentation of cysteine residues in peptides displaying increased acetylation in astrocytes with chronic glutathione depletion and overall suggests a potential two-way crosstalk between cysteine oxidation and lysine acetylation.
Cysteine is one of the least abundant residues in proteins but it is often present in functionally relevant sites, where it has important catalytic, regulatory (target of posttranslational modification), and/or structure-stabilizing functions (59). The reactivity of the thiol group allows cysteine residues to (i) form covalent interactions with other thiols generating intra- and intermolecular disulfide bonds, (ii) coordinate a variety of metals, and (iii) be target of reversible posttranslational modifications, including nitrosylation, sulfhydration, and glutathionylation, among others (59, 60). Glutathione participates in the regulation of redox-dependent cell signaling by modifying the oxidation state of cysteine residues in target proteins (47). S-glutathionylation occurs not only in response to oxidative or nitrosative stress but also under nonpathological conditions and mediates the redox regulation of protein function (60, 61). Redox-sensitive proteins have specific cysteine residues that are accessible in the three-dimensional structure of the protein and exist as thiolate anions due to a lowering of their pKa at physiological pH by the presence of a basic environment, i.e. the vicinity of basic amino acid residues (Arg, His, or Lys) in the primary or tertiary structure (61). Therefore, the acetylation of a lysine residue, by neutralizing its positive charge, could negatively impact the reactivity of a neighboring cysteine residue. Of the 123 acetylated peptides exhibiting increased lysine acetylation, 55 (i.e. 44.7%) contain a cysteine residue within 15 residues of the acetylation site. As depicted in Table II, 20 of those peptides contain experimentally verified known sites of cysteine S-glutathionylation (dbGSH, database of cysteine S-glutathionylation; http://csb.cse.yzu.edu.tw/dbGSH/). Hence, this set of acetylated peptides containing a cysteine residue proximal to the acetylation site could correspond to a subset of sites in which acetylation is directly regulated to prevent glutathionylation of the neighboring cysteine residue. On the other hand, S-glutathionylation of a cysteine residue might prevent acetylation of a neighboring lysine residue. Thus, a decrease in cysteine glutathionylation due to a reduction in total glutathione levels could facilitate the acetylation of the lysine residue. It is also possible that oxidation of the cysteine residue due to glutathione depletion could facilitate lysine acetylation by inducing conformational changes that facilitate lysine acetylation or by altering the interaction with acetyltransferases or deacetylases. Regardless, the overrepresentation of cysteine residues in the vicinity of the regulated acetylated sites suggests the existence of crosstalk between lysine acetylation and cysteine modification.
Table II. Regulated lysine-acetylated peptides containing cysteine residues experimentally verified as targets of glutathionylation (see Supplemental Table IV for full list of regulated lysine-acetylated sites).
| Gene symbol | Protein name | Protein ID | Peptide sequence | Position K-ac | Position C-GSH a |
|---|---|---|---|---|---|
| Anxa1 | Annexin A1 | P10107 | IRIMVSRSEIDMNEIKVFYQKKYGISLCQAI | 312 | 324 |
| Anxa2 | Annexin A2 | P07356 | KWISIMTERSVCHLQKVFERYKSYSPYDMLE | 227 | 223 |
| Ddx39b | Spliceosome RNA helicase Ddx39b | Q9Z1N5 | PGRILALARNKSLNLKHIKHFILDECDKMLE | 188 | 198 b |
| Echs1 | Enoyl-CoA hydratase, mitochondrial | Q8BH95 | VLTGGDKAFAAGADIKEMQNRTFQDCYSSKF | 101 | 111 |
| Esd | S-formylglutathione hydrolase | Q9R0P3 | KKAFSGYLGPDESKWKAYDATCLVKAYSGSQ | 200 | 206 |
| Hspa8 | Heat shock cognate 71 kDa protein | P63017 | DKNQTAEKEEFEHQQKELEKVCNPIITKLYQ | 597 | 603 |
| Hspa9 | Stress-70 protein, mitochondrial | P38647 | EEFKDQLPADECNKLKEEISKMRALLAGKDS | 612 | 608 |
| Hsp90aa1 | Heat shock protein HSP 90-alpha | P07901 | APFDLFENRKKKNNIKLYVRRVFIMDNCEEL | 363 | 375 |
| Lgals1 | Galectin-1 | P16045 | PGECLKVRGEVASDAKSFVLNLGKDSNNLCL | 29 | 43 |
| Prdx6 | Peroxiredoxin-6 | O08709 | SEEEAKQCFPKGVFTKELPSGKKYLRYTPQP | 209 | 201 |
| Ptges3 | Prostaglandin E synthase 3 | Q9R0Q7 | EFCVEDSKDVNVNFEKSKLTFSCLGGSDNFK | 33 | 20, 40 |
| Pura | Transcriptional activator protein Pur-alpha | P42669 | TVPYKVWAKFGHTFCKYSEEMKKIQEKQREK | 272 | 271 |
| S100a11 | Protein S100-A11 | P50543 | RCIESLIAVFQKYSGKDGNNTQLSKTEFLSF | 22 | 8 |
| Tcp1 | T-complex protein 1 subunit alpha | P11983 | CDEMERSLHDALCVVKRVLELKSVVPGGGAV | 400 | 385, 397 |
| Tkt; Tktl2 | Transketolase; Transketolase-like protein 2 | P40142 | LGAACGMAYTGKYFDKASYRVYCMLGDGEVS | 144 | 133 |
| Ube2n | Ubiquitin-conjugating enzyme E2 N | P61089 | HPNVDKLGRICLDILKDKWSPALQIRTVLLS | 92 | 87 |
| Vdac1 | Voltage-dependent anion-selective channel protein 1 | Q60932 | AWTAGNSNTRFGIAAKYQVDPDACFSAKVNN | 237 | 245 |
| Vdac2 | Voltage-dependent anion-selective channel protein 2 | Q60930 | SSNTDTGKVSGTLETKYKWCEYGLTFTEKWN | 73 | 77 |
| Vdac3 | Voltage-dependent anion-selective channel protein 3 | Q60931 | HAYTDTGKASGNLETKYKVCNYGLTFTQKWN | 61 | 65 |
| Vdac3 | Voltage-dependent anion-selective channel protein 3 | Q60931 | YTDTGKASGNLETKYKVCNYGLTFTQKWNTD | 63 | 65 |
Supplementary Material
Acknowledgments
We thank Jennifer Rutherford Bethard for her support in data analysis and instrumentation maintenance. We also thank Dr. Peter R. Baker and Dr. Robert J. Chalkley (University of California, San Francisco) for uploading the MaxQuant results of the acetyl peptide dataset into MS-Viewer to facilitate viewing of the tandem mass spectra.
Footnotes
Author contributions: M.P., L.E.B., and M.R.V. designed the research; M.P., L.E.B., D.R.S., B.A.H., S.C., and M.R.V. performed the research; M.P., L.E.B., S.C., B.A.N., and M.R.V. analyzed the data; and M.P., L.E.B., and M.R.V. wrote the paper.
* This work was supported by National Institutes of Health Grants GM103542 and ES019186 (M.R.V) and S10 D010731 (Orbitrap Elite, L.E.B.). Mice were housed in a facility constructed with support from the NIH, Grant Number C06 RR015455 from the Extramural Research Facilities Program of the National Center for Research Resources. The MUSC Mass Spectrometry Facility is supported by the Office of the Provost and the South Carolina COBRE in Oxidants, Redox Balance and Stress Signaling (GM103542). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material Supplement Tables I-IV and Supplemental Figs. 1–3.
This article contains supplemental material Supplement Tables I-IV and Supplemental Figs. 1–3.
1 The abbreviations used are:
- ALS
- amyotrophic lateral sclerosis
- CNS
- central nervous system
- GCL
- glutamate-cysteine ligase
- GCLC
- glutamate-cysteine ligase catalytic subunit
- GCLM
- glutamate-cysteine ligase modifier subunit
- hSOD1
- human superoxide dismutase 1
- MS/MS
- tandem mass spectrometry
- Nrf2
- nuclear factor erythroid-2-related factor 2
- PPP
- pentose phosphate pathway.
REFERENCES
- 1.Nedergaard M., Ransom B., and Goldman S. A. (2003) New roles for astrocytes: Redefining the functional architecture of the brain. Trends Neurosci. 26, 523–530 [DOI] [PubMed] [Google Scholar]
- 2.Perea G., Navarrete M., and Araque A. (2009) Tripartite synapses: Astrocytes process and control synaptic information. Trends Neurosci. 32, 421–431 [DOI] [PubMed] [Google Scholar]
- 3.Perea G., and Araque A. (2010) GLIA modulates synaptic transmission. Brain Res. Rev. 63, 93–102 [DOI] [PubMed] [Google Scholar]
- 4.Sofroniew M. V., and Vinters H. V. (2010) Astrocytes: Biology and pathology. Acta Neuropathol. 119, 7–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ridet J. L., Malhotra S. K., Privat A., and Gage F. H. (1997) Reactive astrocytes: Cellular and molecular cues to biological function. Trends Neurosci. 20, 570–577 [DOI] [PubMed] [Google Scholar]
- 6.Verkhratsky A., Sofroniew M. V., Messing A., deLanerolle N. C., Rempe D., Rodriguez J. J., and Nedergaard M. (2012) Neurological diseases as primary gliopathies: A reassessment of neurocentrism. ASN Neuro Online publication 4, e0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vargas M. R., and Johnson J. A. (2010) Astrogliosis in amyotrophic lateral sclerosis: Role and therapeutic potential of astrocytes. Neurotherapeutics 7, 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ischiropoulos H., and Beckman J. S. (2003) Oxidative stress and nitration in neurodegeneration: cause, effect, or association? J. Clinical Invest. 111, 163–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vargas M. R., and Johnson J. A. (2009) The Nrf2-ARE cytoprotective pathway in astrocytes. Expert Rev. Mol. Med. 11, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez-Fernandez S., Almeida A., and Bolaños J. P. (2012) Antioxidant and bioenergetic coupling between neurons and astrocytes. Biochem. J. 443, 3–11 [DOI] [PubMed] [Google Scholar]
- 11.Dringen R., Pawlowski P. G., and Hirrlinger J. (2005) Peroxide detoxification by brain cells. J. Neurosci. Res. 79, 157–165 [DOI] [PubMed] [Google Scholar]
- 12.Dringen R., Gutterer J. M., and Hirrlinger J. (2000) Glutathione metabolism in brain metabolic interaction between astrocytes and neurons in the defense against reactive oxygen species. Eur. J. Biochem. FEBS 267, 4912–4916 [DOI] [PubMed] [Google Scholar]
- 13.Meister A. (1988) Glutathione metabolism and its selective modification. J. Biol. Chem. 263, 17205–17208 [PubMed] [Google Scholar]
- 14.Yang Y., Dieter M. Z., Chen Y., Shertzer H. G., Nebert D. W., and Dalton T. P. (2002) Initial characterization of the glutamate-cysteine ligase modifier subunit GCLM(-/-) knockout mouse. Novel model system for a severely compromised oxidative stress response. J. Biol. Chem. 277, 49446–49452 [DOI] [PubMed] [Google Scholar]
- 15.Vargas M. R., Johnson D. A., and Johnson J. A. (2011) Decreased glutathione accelerates neurological deficit and mitochondrial pathology in familial ALS-linked hSOD1(G93A) mice model. Neurobiol. Disease 43, 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulz J. B., Lindenau J., Seyfried J., and Dichgans J. (2000) Glutathione, oxidative stress and neurodegeneration. Eur. J. Biochem. FEBS 267, 4904–4911 [DOI] [PubMed] [Google Scholar]
- 17.Dringen R. (2000) Metabolism and functions of glutathione in brain. Progress Neurobiol. 62, 649–671 [DOI] [PubMed] [Google Scholar]
- 18.Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., and Guan K. L. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albaugh B. N., Arnold K. M., and Denu J. M. (2011) KAT(ching) metabolism by the tail: Insight into the links between lysine acetyltransferases and metabolism. Chembiochem. Eur. J. Chem. Biol. 12, 290–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Z., Cheng Z., Zhao Y., and Volchenboum S. L. (2011) Bioinformatic analysis and post-translational modification crosstalk prediction of lysine acetylation. PloS One 6, e28228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merksamer P. I., Liu Y., He W., Hirschey M. D., Chen D., and Verdin E. (2013) The sirtuins, oxidative stress and aging: An emerging link. Aging 5, 144–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choudhary C., Weinert B. T., Nishida Y., Verdin E., and Mann M. (2014) The growing landscape of lysine acetylation links metabolism and cell signalling. Nature Rev. Mol. Cell Biol. 15, 536–550 [DOI] [PubMed] [Google Scholar]
- 23.Tao R., Vassilopoulos A., Parisiadou L., Yan Y., and Gius D. (2014) Regulation of MnSOD enzymatic activity by SIRT3 connects the mitochondrial acetylome signaling networks to aging and carcinogenesis. Antioxidants Redox Signal. 20, 1646–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dittenhafer-Reed K. E., Richards A. L., Fan J., Smallegan M. J., Fotuhi Siahpirani A., Kemmerer Z. A., Prolla T. A., Roy S., Coon J. J., and Denu J. M. (2015) SIRT3 mediates multi-tissue coupling for metabolic fuel switching. Cell Metabolism 21, 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vargas M. R., Pehar M., Cassina P., Beckman J. S., and Barbeito L. (2006) Increased glutathione biosynthesis by Nrf2 activation in astrocytes prevents p75NTR-dependent motor neuron apoptosis. J. Neurochem. 97, 687–696 [DOI] [PubMed] [Google Scholar]
- 26.Klein A. L., Berkaw M. N., Buse M. G., and Ball L. E. (2009) O-linked N-acetylglucosamine modification of insulin receptor substrate-1 occurs in close proximity to multiple SH2 domain binding motifs. Mol. Cell. Proteomics 8, 2733–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., and Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 28.Cox J., Michalski A., and Mann M. (2011) Software lock mass by two-dimensional minimization of peptide mass errors. J. Am. Soc. Mass Spectrom. 22, 1373–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 30.Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., and Sullivan M. (2012) PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smyth G. K. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genetics Mol. Biol. 3, Article3 [DOI] [PubMed] [Google Scholar]
- 32.Udeshi N. D., Mani D. R., Eisenhaure T., Mertins P., Jaffe J. D., Clauser K. R., Hacohen N., and Carr S. A. (2012) Methods for quantification of in vivo changes in protein ubiquitination following proteasome and deubiquitinase inhibition. Mol. Cell. Proteomics 11, 148–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colaert N., Helsens K., Martens L., Vandekerckhove J., and Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nature Meth. 6, 786–787 [DOI] [PubMed] [Google Scholar]
- 34.Lahiri D. K., and Ge Y. (2000) Electrophoretic mobility shift assay for the detection of specific DNA-protein complex in nuclear extracts from the cultured cells and frozen autopsy human brain tissue. Brain Res. Protocols 5, 257–265 [DOI] [PubMed] [Google Scholar]
- 35.Vargas M. R., Johnson D. A., Sirkis D. W., Messing A., and Johnson J. A. (2008) Nrf2 activation in astrocytes protects against neurodegeneration in mouse models of familial amyotrophic lateral sclerosis. J. Neurosci. 28, 13574–13581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ortinski P. I., Dong J., Mungenast A., Yue C., Takano H., Watson D. J., Haydon P. G., and Coulter D. A. (2010) Selective induction of astrocytic gliosis generates deficits in neuronal inhibition. Nature Neurosci. 13, 584–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danbolt N. C. (2001) Glutamate uptake. Progress Neurobiol. 65, 1–105 [DOI] [PubMed] [Google Scholar]
- 38.Zhou Y., and Danbolt N. C. (2014) Glutamate as a neurotransmitter in the healthy brain. J. Neural Transmission 121, 799–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayes J. D., and Dinkova-Kostova A. T. (2014) The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 39, 199–218 [DOI] [PubMed] [Google Scholar]
- 40.Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H., Yamamoto M., and Motohashi H. (2012) Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22, 66–79 [DOI] [PubMed] [Google Scholar]
- 41.Neuhauser N., Michalski A., Cox J., and Mann M. (2012) Expert system for computer-assisted annotation of MS/MS spectra. Mol. Cell. Proteomics 11, 1500–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker P. R., and Chalkley R. J. (2014) MS-viewer: A web-based spectral viewer for proteomics results. Mol. Cell. Proteomics 13, 1392–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Medzihradszky K. F. (2005) Peptide sequence analysis. Meth. Enzymol. 402, 209–244 [DOI] [PubMed] [Google Scholar]
- 44.Medzihradszky K. F., and Chalkley R. J. (2015) Lessons in de novo peptide sequencing by tandem mass spectrometry. Mass Spectrom. Rev. 34, 43–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lundby A., Lage K., Weinert B. T., Bekker-Jensen D. B., Secher A., Skovgaard T., Kelstrup C. D., Dmytriyev A., Choudhary C., Lundby C., and Olsen J. V. (2012) Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Reports 2, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dringen R., and Hirrlinger J. (2003) Glutathione pathways in the brain. Biol. Chem. 384, 505–516 [DOI] [PubMed] [Google Scholar]
- 47.Lu S. C. (2013) Glutathione synthesis. Biochim. Biophys. Acta 1830, 3143–3153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pehar M., Lehnus M., Karst A., and Puglielli L. (2012) Proteomic assessment shows that many endoplasmic reticulum (ER)-resident proteins are targeted by N(epsilon)-lysine acetylation in the lumen of the organelle and predicts broad biological impact. J. Biol. Chem. 287, 22436–22440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pehar M., and Puglielli L. (2013) Lysine acetylation in the lumen of the ER: A novel and essential function under the control of the UPR. Biochim. Biophys. Acta 1833, 686–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiong Y., and Guan K. L. (2012) Mechanistic insights into the regulation of metabolic enzymes by acetylation. J. Cell Biol. 198, 155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeBerardinis R. J., Lum J. J., Hatzivassiliou G., and Thompson C. B. (2008) The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metabolism 7, 11–20 [DOI] [PubMed] [Google Scholar]
- 52.Schlicker C., Gertz M., Papatheodorou P., Kachholz B., Becker C. F., and Steegborn C. (2008) Substrates and regulation mechanisms for the human mitochondrial sirtuins SIRT3 and SIRT5. J. Mol. Biol. 382, 790–801 [DOI] [PubMed] [Google Scholar]
- 53.Basu A., Rose K. L., Zhang J., Beavis R. C., Ueberheide B., Garcia B. A., Chait B., Zhao Y., Hunt D. F., Segal E., Allis C. D., and Hake S. B. (2009) Proteome-wide prediction of acetylation substrates. Proc. Natl. Acad. Sci. U.S.A. 106, 13785–13790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagner G. R., and Hirschey M. D. (2014) Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Molecular Cell 54, 5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baeza J., Smallegan M. J., and Denu J. M. (2015) Site-specific reactivity of nonenzymatic lysine acetylation. ACS Chem. Biol. 10, 122–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dansen T. B., Smits L. M., van Triest M. H., de Keizer P. L., van Leenen D., Koerkamp M. G., Szypowska A., Meppelink A., Brenkman A. B., Yodoi J., Holstege F. C., and Burgering B. M. (2009) Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nature Chem. Biol. 5, 664–672 [DOI] [PubMed] [Google Scholar]
- 57.Ryu H., Lee J., Olofsson B. A., Mwidau A., Dedeoglu A., Escudero M., Flemington E., Azizkhan-Clifford J., Ferrante R. J., and Ratan R. R. (2003) Histone deacetylase inhibitors prevent oxidative neuronal death independent of expanded polyglutamine repeats via an Sp1-dependent pathway. Proc. Natl. Acad. Sci. U.S.A. 100, 4281–4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang X. J., and Seto E. (2008) Lysine acetylation: Codified crosstalk with other posttranslational modifications. Mol. Cell 31, 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marino S. M., and Gladyshev V. N. (2012) Analysis and functional prediction of reactive cysteine residues. J. Biol. Chem. 287, 4419–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grek C. L., Zhang J., Manevich Y., Townsend D. M., and Tew K. D. (2013) Causes and consequences of cysteine S-glutathionylation. J. Biol. Chem. 288, 26497–26504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dalle-Donne I., Rossi R., Colombo G., Giustarini D., and Milzani A. (2009) Protein S-glutathionylation: A regulatory device from bacteria to humans. Trends Biochem. Sci. 34, 85–96 [DOI] [PubMed] [Google Scholar]
- 62.Su D., Gaffrey M. J., Guo J., Hatchell K. E., Chu R. K., Clauss T. R., Aldrich J. T., Wu S., Purvine S., Camp D. G., Smith R. D., Thrall B. D., and Qian W. J. (2014) Proteomic identification and quantification of S-glutathionylation in mouse macrophages using resin-assisted enrichment and isobaric labeling. Free Radical Biol. Med. 67, 460–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chiang B. Y., Chou C. C., Hsieh F. T., Gao S., Lin J. C., Lin S. H., Chen T. C., Khoo K. H., and Lin C. H. (2012) In vivo tagging and characterization of S-glutathionylated proteins by a chemoenzymatic method. Angew. Chem. 51, 5871–5875 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.